Significance
It is the dream of many scientists to develop antimicrobial peptides (AMPs) into potent antibiotics to combat resistant pathogens and their biofilms. However, the success is low due to limited knowledge on peptide design. Natural AMPs are diverse in sequence, activity, and source. We found a linear relationship between the averaged contents of arginine (R) and hydrophobic amino acids (Pho) of over 3,000 AMPs in the current Antimicrobial Peptide Database (http://aps.unmc.edu/AP). Based on this R–Pho relationship, we identified a peptide template and designed two representative amphipathic peptides with distinct structures and activities. Remarkably, horine and verine showed systemic efficacy in mice, but no nephrotoxicity to rodents. Our study advances peptide design and provides two antibiotics to combat drug-resistant pathogens.
Keywords: antibiotic resistance, nephrotoxicity, NMR, peptide antibiotics, systemic efficacy
Abstract
Antimicrobial peptides are important candidates for developing new classes of antibiotics because of their potency against antibiotic-resistant pathogens. Current research focuses on topical applications and it is unclear how to design peptides with systemic efficacy. To address this problem, we designed two potent peptides by combining database-guided discovery with structure-based design. When bound to membranes, these two short peptides with an identical amino acid composition can adopt two distinct amphipathic structures: A classic horizontal helix (horine) and a novel vertical spiral structure (verine). Their horizontal and vertical orientations on membranes were determined by solid-state 15N NMR data. While horine was potent primarily against gram-positive pathogens, verine showed broad-spectrum antimicrobial activity. Both peptides protected greater than 80% mice from infection-caused deaths. Moreover, horine and verine also displayed significant systemic efficacy in different murine models comparable to conventional antibiotics. In addition, they could eliminate resistant pathogens and preformed biofilms. Significantly, the peptides showed no nephrotoxicity to mice after intraperitoneal or intravenous administration for 1 wk. Our study underscores the significance of horine and verine in fighting drug-resistant pathogens.
The growing bacterial resistance to conventional antibiotics has led to a health crisis in our era. Approximately 90% of hospital-associated antibiotic-resistant infections are related to the ESKAPE pathogens, including gram-positive Enterococcus faecium, Staphylococcus aureus, gram-negative Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species (1). Consequently, it is critical to discover a new generation of potent antibiotics to curb such pathogens. Naturally occurring antimicrobial peptides (AMPs) are important sources for new antibiotics (2–5). These peptides are key components of the host innate immune system, which play an essential role in protecting the host from pathogenic infection. According to the Antimicrobial Peptide Database (APD), AMPs have been identified in all of the life kingdoms, ranging from bacteria to animals (6). Recently, Lewis and colleagues (7) extended the search to uncultivable soil bacteria and identified teixobactin by utilizing the i-chip technology. New peptides could also be identified in silico (8–11). However, there is a great knowledge gap from in vitro activity to in vivo efficacy. In addition, there is little knowledge on designing AMPs with systemic efficacy. This study succeeded in identifying two short peptides with systemic efficacy. Importantly, the peptide did not display toxicity to the kidney of both mice and rats after daily administration for a week. Our study has raised new hope for developing a new generation of antibiotics based on AMPs. Here we report the results of the peptides and discuss their potential as novel antibiotics.
Results
Database-Guided Peptide Design and Structure-Based Refinement.
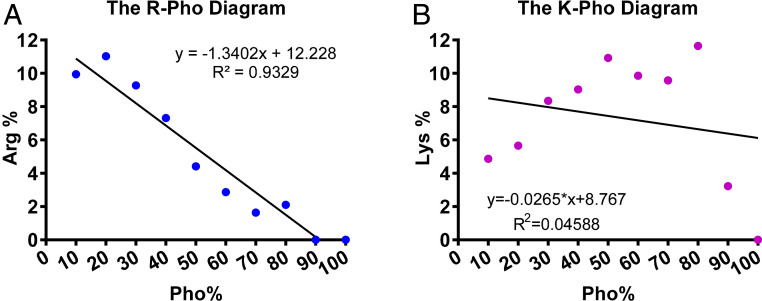
After decades of AMP research, there is no theory to guide the design of peptides with systemic efficacy. To facilitate our peptide discovery, we created the APD (http://aps.unmc.edu/AP) (6). To ensure high quality, our database registered the peptides based on a set of criteria. The APD laid the foundation for peptide screening (10) and developing the database-filtering technology (11). Here we designed two peptides by combining database with 3D structure. The primary steps are as follows. First, we found a linear relationship between arginine (R) and hydrophobic contents (Pho) (correlation coefficient 0.93) of the 3,177 peptides in the current APD (Fig. 1A). Such a relationship defines the ratio between R and Pho and may be useful for peptide design. Note that there is no correlation between lysine (K) and Pho (correlation coefficient 0.05) (Fig. 1B), although lysine is a frequently occurring amino acid in the APD (6). Second, we decided on the peptide design parameters: Low R and high Pho based on our recent discovery (12). Third, we then filtered the thousands of peptides in the database to identify candidates with such properties. To narrow down the possibilities, we focused on short peptides (<10 amino acids) for cost-effective production. Because helical peptides in the APD tend to have an increased content of phenylalanine with a decrease in peptide length (13), we selected temporin-SHf (14) with four Phe residues and only one arginine as a starting template. We found this peptide template showed a moderate activity against methicillin-resistant S. aureus (MRSA) with a minimal inhibitory concentration (MIC) at 25 µM (15). Fourth, we enhanced the peptide potency by replacing four Phe with Trp residues because the Trp fused aromatic rings prefer membrane interfaces (16). Our practice led to a unique Trp-rich peptide WW291 (Fig. 2) with anti-MRSA activity increased eightfold (15).
Fig. 1.
Relationships between averaged net charge and hydrophobic residue content. (A) A linear correlation between hydrophobic residue content (Pho) and arginine content (R) of AMPs based on 3,177 peptides in the current APD (6). (B) There is no such correlation between lysine contents and Pho. The peptides in the APD (6) were separated into 10 bins based on hydrophobic contents (0–10, 11–20, 21–30, 31–40, 41–50, 51–60, 61–70, 71–80, 81–90, and 91–100%), which are represented as 10, 20, 30, 40, 50, 60, 70, 80, 90, and 100 on the x axis of this plot. These relationships remained the same from 2017 to 2020. Data were obtained from the APD in March 2020.
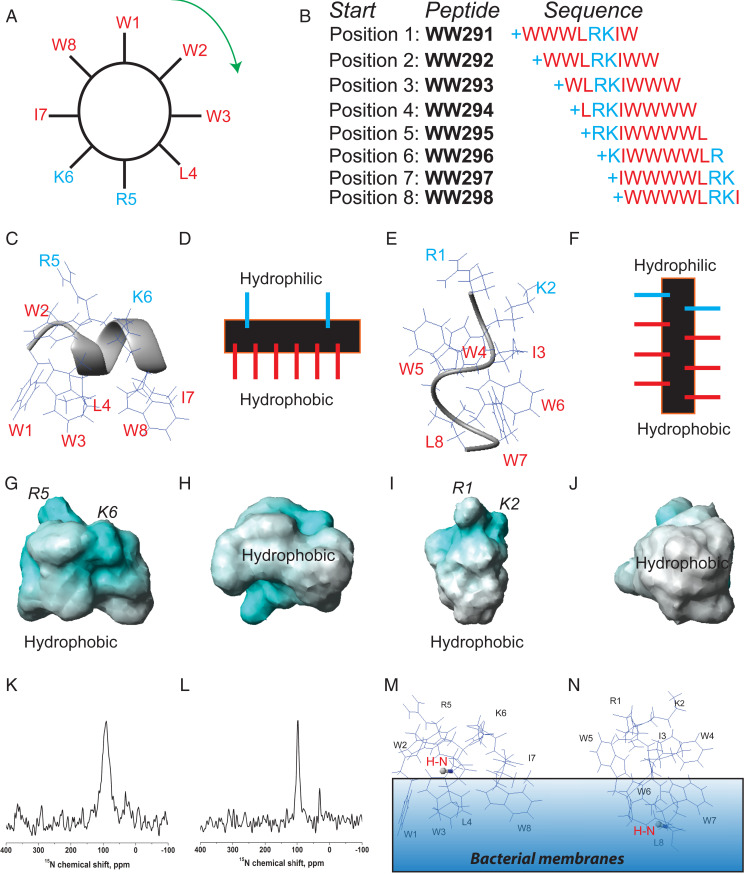
Fig. 2.
Design and NMR characterization of the two distinct amphipathic peptides. (A) The amino acid sequence of WW291 is distributed evenly on the wheel. Reading from position 1 (W1) clockwise generated WW291. Reading from position 2 (W2) in the same manner produced WW292. Repeating this reading from position 1 to position 8 (W8) led to eight peptides (B). From this sequence permutation, we identified WW295 with increased activity against Klebsiella (SI Appendix, Table S1). NMR structural determination revealed two distinct amphipathic models: Classic helix of WW291 with a horizontal axis (C and D) and nonclassic spiral structure of WW295 with a vertical axis (E and F), all relative to the hydrophobic surface. As depicted in the potential surfaces (G–J), both structures possess positive charges (cyan) for recognition of the negative bacterial surfaces and hydrophobic surfaces (white) for membrane anchoring: G and H for WW291 and I and J for WW295. NMR sample conditions are given in the legend to SI Appendix, Table S2. (K and L) Peptide membrane orientation determination by solid-state NMR spectroscopy. The solid-state NMR 15N chemical shift (<100 ppm) of a single-site 15N-labeled leucine in horine (K) and verine-L (L) indicates an H-N vector (red) parallel to membrane surface. This enables us to position the 3D structure of horine (M) and verine-L (N) on the lipid bilayer so that the H-N vector (in ball-and-stick) is approximately parallel to the bacterial membranes.
Fifth, we also attempted to identify peptides against gram-negative bacteria as sequence shuffling alters peptide activity (10). A clockwise sequence permutation of WW291 (read the sequence from positions W1 to W8, respectively, in Fig. 2A) generated seven new peptides (WW292 to -298 in Fig. 2B). While these sequence-permutated peptides all inhibited MRSA, WW295 gained up to fourfold activity against gram-negative K. pneumoniae compared to WW291 (SI Appendix, Table S1). WW295 became an interesting template for us to design new antibiotics against K. pneumoniae because clinically there is no effective drug to treat the recent Klebsiella infections in the United States (17).
Sixth, we determined the 3D structures of WW291 and WW295 bound to membrane-mimetic micelles (18), since these two peptides permeated bacterial membranes. For high-quality signal assignments, we used the improved 2D NMR method (19), which also records heteronuclear 15N- and 13C-involved correlated spectra at natural abundance. The 13C chemical shifts were found to be critical here to distinguish the multiple aromatic protons from the four Trp residues (50%) of these short peptides assigned based on the standard 2D 1H-correlated NMR method (18). A combined use of distance (18) and dihedral angle restraints (20) generated well-defined structures (SI Appendix, Fig. S1). On the hydrophobic surfaces, the aromatic rings of both peptides superimposed well (SI Appendix, Fig. S1 B and D). The dihedral angles of both peptide structures are in the allowed region (SI Appendix, Table S2). In the 3D structure of WW291 we determined previously (21), a short C-terminal helical turn is stabilized by the N-terminal WWW motif (Fig. 2C), whereas WW295 determined here has a novel, spiral structure with hydrophilic amino acids R1 and K2 at the top and hydrophobic residues clustered at the bottom (Fig. 2E). Hence, the 3D structure of WW295 constitutes an alternative amphipathic model where the peptide chain folds along a vertical axis relative to the hydrophobic surface (Fig. 2F and SI Appendix, Fig. S1C). Such a vertical amphipathic structure differs drastically from the classic amphipathic helix of WW291 where the peptide chain folded horizontally relative to the hydrophobic surface (Fig. 2D and SI Appendix, Fig. S1A). The amphipathic nature of these structures is also evident on their potential surfaces (Fig. 2 G and I, white), where the hydrophobic surfaces face the readers after a 90° rotation (Fig. 2 H and J).
Finally, we aimed at improving peptide cell selectivity. Based on the 3D structure of WW291 (Fig. 2C), we changed both isoleucine and lysine to arginines (WW304 in SI Appendix, Table S3) because of the preferred association between tryptophan and arginine (15). This allowed us to retain peptide potency against MRSA and decrease toxicity to erythrocytes. We also improved WW295 by changing the terminal L8 to V8, which doubled the 50% hemolytic concentration (HL50) (WW307 in SI Appendix, Table S4). Because the peptide chains of WW291 and WW295 have horizontal and vertical folding axes relative to the hydrophobic surface (Fig. 2), respectively, we refer to the improved versions of these two peptides as horine and verine.
In Vitro Antimicrobial Robustness.
We found that the peptides did not reduce the antibacterial activity in the presence of 150 mM NaCl, indicating the designed AMPs retain activity under physiological conditions. When the pH of the media was reduced from 8.0 to 6.8, there was a twofold increase in the MIC of both peptides against S. aureus USA300. Human serum (5 to 10%) compromised the activity of horine (four- to eightfold) more than verine (twofold). However, peptide activity in the presence of 5% of different mouse tissue homogenates showed essentially no change in peptide MIC values (SI Appendix, Fig. S2A), indicating that these peptides are likely to work in mice. We also evaluated peptide stability. In a liver microsomal stability assay, horine and verine showed 50% degradation in 1 h (SI Appendix, Fig. S2B). Notably, they were stable after incubation with murine peritoneal fluids or plasma for 3 h (SI Appendix, Fig. S2C). In addition, the peptide remained stable after incubation with 5% human serum for 24 h at 37 °C (SI Appendix, Fig. S2D).
We then tested the potency of horine and verine using a set of resistant bacteria that cover most of the ESKAPE pathogens (Table 1). Both peptides were active against gram-positive pathogens, such as vancomycin-resistant Enterococci (VRE) and MRSA (MIC 2 to 4 µM). At a twofold MIC, they rapidly killed S. aureus USA300 LAC (Fig. 3A), including persisters (22) (Fig. 3B). In addition, verine killed both VRE and K. pneumoniae, while tobramycin at 8 µM failed to inhibit such pathogens (Table 1). Similarly, this peptide, but not doripenem (8 µM), was potent against A. baumannii. We also tested the antibacterial susceptibility of the peptides using a panel of 31 clinical S. aureus strains (SI Appendix, Table S5). Both peptides showed an MIC50 value of 4 µM. While nafcillin did not inhibit S. aureus Mu50 and USA200 strains at 8 µM, both horine and verine were able to. For verine, we also evaluated 22 Klebsiella clinical strains and found an MIC50 of 8 to 16 µM (SI Appendix, Table S6). In contrast, seven strains already developed resistance to tobramycin (MIC >32 to 64 µM). All of these results illustrate the potency of our designer peptides against clinically resistant pathogens and persisters.
Table 1.
Minimal inhibitory concentrations (μM) of horine and verine
| Compound | E. faecium V286-17 (VRE) | S. aureus USA300 (MRSA) | K. pneumoniae E406-17 | A. baumannii B28-16 | P. aeruginosa E411-17 | E. coli E423-17 |
| Horine | 2 | 4 | >32 | 32 | >32 | 16–32 |
| D-horine | 4 | 4 | >32 | >32 | 32 | 16 |
| Verine | 2 | 2–4 | 8–16 | 8 | 4–8 | 2 |
| D-verine | 2 | 4 | 32 | 4 | 4 | 4 |
| Vancomycin | >8 | 1 | >8 | >8 | >8 | >8 |
| Rifamycin | >8 | 0.25 | >8 | >8 | >8 | >8 |
| Daptomycin | 4 | 4 | >8 | >8 | >8 | >8 |
| Colistin | >8 | >8 | >8 | 1 | 2 | 1 |
| Tobramycin | >8 | 4 | >8 | 4 | 2 | 8 |
| Doripenem | >8 | 0.25 | 0.5 | >8 | 2 | 1 |
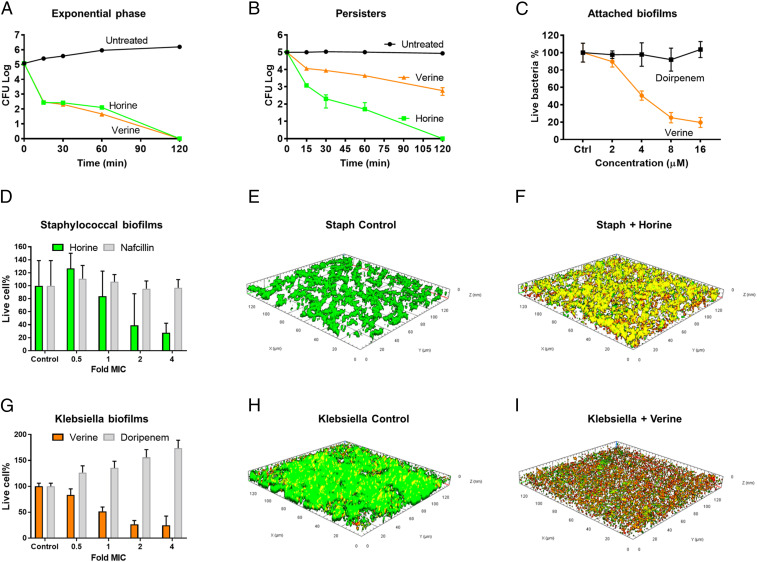
Fig. 3.
In vitro potential of horine (green) and verine (gold). (A) Horine and verine (2 × MIC) killed the exponential phase S. aureus USA300 LAC in 120 min. (B) The two peptides (2 × MIC) also killed nafcillin-induced persisters of S. aureus. (C) Verine, but not doripenem, inhibited the attachment of K. pneumoniae E406-17 in a dose-dependent manner. (D) Horine disrupted the 48-h established biofilms of S. aureus. In the confocal images, live bacteria in the untreated control are in green (E) and dead bacteria (F) treated at 16 µM of horine are in red. (G) Verine was effective in disrupting 48-h established Klebsiella biofilms. In the confocal images, live cells are green in untreated K. pneumoniae (H), and dead cells are red in verine-treated K. pneumoniae (I).
To further demonstrate the merits of the new peptides, we also evaluated their efficacy against biofilms, which are notoriously difficult to treat with conventional antibiotics. Pathogens attach to surfaces for biofilm formation. We found that verine inhibited the attachment of K. pneumoniae (Fig. 3C). In addition, these peptides also had an effect on preformed biofilms. While horine demonstrated a dose-dependent activity against the 48-h preformed biofilms of S. aureus USA300 (Fig. 3D, green), nafcillin did not (Fig. 3D, gray). Similarly, verine disrupted the 48-h preformed biofilms of K. pneumoniae (Fig. 3G, gold). In contrast, doripenem showed no antibiofilm effect in the same experiment (Fig. 3G, gray). Both peptides were able to kill the pathogens in biofilms as observed by confocal microscopy. Untreated bacteria were live (Fig. 3 E and H, green) while treated bacteria were dead (Fig. 3 F and I, red).
Mechanisms of Action of Peptides and Bacterial Resistance Development.
The similar MIC values of the L- and D-forms of horine and verine in Table 1 imply membrane targeting. To further substantiate membrane targeting, we made use of a Staphylococcal mutant from the Nebraska Transposon Mutant Library (23). When the mprF gene was inactivated, S. aureus became more susceptible to cationic peptide killing since this mutant was unable to transfer a basic lysine to acidic phosphatidylglycerol (PG) to make the membrane surface more positive (24). Indeed, the mprF-disrupted mutant became more susceptible to killing by either horine or verine compared to the wild-type Staphylococcal strains JE2 or USA300 (SI Appendix, Fig. S2E).
To provide evidence for membrane targeting, we conducted membrane permeation and depolarization experiments. Both horine and verine rapidly permeated the membranes of S. aureus USA300 in a propidium iodide-based fluorescent experiment (SI Appendix, Fig. S3A). As a positive control, daptomycin permeated bacterial membranes in tryptic soy broth, but rifamycin, which inhibits the RNA polymerase, did not. Moreover, these two peptides could depolarize the membranes of S. aureus USA300 (SI Appendix, Fig. S3B), implying a smaller breach in membranes due to proton flux (25). To view bacterial damage, we used the scanning electron microscopy. Without treatment, we observed the smooth S. aureus surface (SI Appendix, Fig. S3D), but membrane blebs after horine treatment (SI Appendix, Fig. S3E). In the case of K. pneumoniae, the bacterial surface (SI Appendix, Fig. S3F) was entirely damaged after verine treatment (SI Appendix, Fig. S3G).
To provide additional insight into the mechanism of action of horine and verine, we conducted 15N, 2H, and 31P solid-state NMR studies in a well-defined Escherichia coli membrane-mimetic lipid bilayer model. Verine is more potent than horine in killing E. coli in terms of MIC (Table 1) and membrane permeation at 2 µM (SI Appendix, Fig. S3C). Because glass-plate–supported lipid bilayers have a uniaxial orientation relative to the magnetic field of the NMR spectrometer, the resulting solid-state NMR spectra provide information for the peptide alignment (26). To this end, we introduced a single 15N-labeled leucine by replacing normal leucine 4 in horine and the C-terminal valine 8 in verine (verine-L). In a well-established lipid system (PG/phosphatidylethanolamine [PE] = 1:3) mimicking the E. coli membranes, the 15N chemical shift in horine at 91 ± 13 ppm (Fig. 2K) indicates this backbone 15N-1H vector of Leu4 is parallel to the membrane surface. For verine-L, a similar chemical shift at 97 ± 5 ppm (Fig. 2L) suggests a similar orientation for the 15N-1H vector of Leu8 in verine-L. Based on this peptide orientational information, we then placed the 3D structures of horine and verine on the membrane surface. The helical peptide horine is located on the membrane surface (Fig. 2M), exactly as proposed in Fig. 2D. The chemical shift of the C-terminal leucine in verine-L enabled us to place this peptide in a vertical position (Fig. 2N), consistent with the proposed orientation in Fig. 2F as well.
To shine light on peptide–membrane interactions, we measured the 2H order parameters (27) by deuterating one of the lipids in the mixture at a time (SI Appendix, Fig. S3 H and K). Both horine and verine-L resulted in a decreased 2H order of palmitoyloleoyl PG (POPG)-d31 (SI Appendix, Fig. S3I), indicative of increased dynamics and additional gauche isomerization of the palmitoyl chain. In contrast, the spectra of palmitoyloleoyl PE (POPE)-d31 were hardly affected by the peptides (SI Appendix, Fig. S3L), indicative of a preferred interaction of these two cationic peptides with the anionic lipid in the mixture, which could lead to lipid clustering. It appeared that relative deuterium order parameters (SCD) for acyl chains of deuterated POPG or POPE (SI Appendix, Fig. S3 J and M) were slightly higher in the presence of verine-L than horine. In addition, 31P NMR spectra provide evidence for the liquid crystalline state of the lipid bilayer and indicate that horine and verine-L exert considerable curvature strain on the bacterial membrane (SI Appendix, Fig. S4). A positive curvature would suggest that the peptides kill bacteria via forming a pore on membranes, consistent with membrane permeation and depolarization experiments (SI Appendix, Fig. S3 A–C).
Membrane targeting of horine and verine poses difficulty to bacterial resistance development. To illustrate this, we conducted an in vitro multiple-passage experiment (28) in the presence of a sub-MIC level of peptides or antibiotics. While the MIC of nafcillin increased 1,000-fold against S. aureus USA300 after 16 passages, there was no change in the MIC for horine and verine (SI Appendix, Fig. S2F). Collectively, these results support the robustness of horine and verine in eliminating drug-resistant pathogens without developing resistance.
Peptide In Vitro and In Vivo Toxicity.
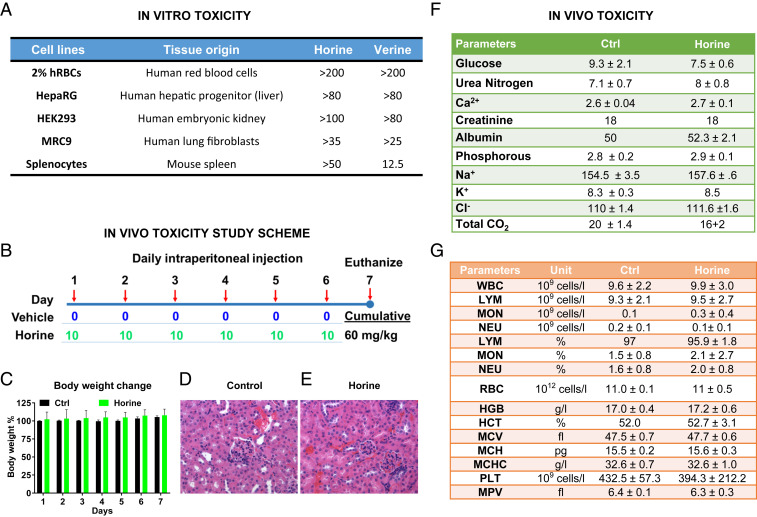
Toxicity constitutes a major hurdle in developing novel antibiotics. In particular, some antibiotics, such as vancomycin and colistin, are known to have nephrotoxicity (29, 30). To further evaluate the potential toxicity of the peptides in vitro, we also tested the effects of horine and verine on other human and murine cells. Based on their LD50 concentrations, they are less toxic to human liver and kidney cells (LD50 > 80 µM) than human lung fibroblasts (LD50 > 25 µM). It appeared that verine was slightly more toxic to murine splenocytes than horine. These results indicate that both peptides were not toxic to human cells (Fig. 4A) at the MIC (2 to 4 µM) required to inhibit VRE and MRSA (Table 1).
Fig. 4.
Peptide toxicity in vitro and in vivo. Toxicity of horine and verine to mammalian cells in vitro (A) or mice (B–G). (A) Peptide toxicity to liver, kidney, lung, and spleen cells determined by the XTT method. (B) Scheme of the peptide toxicity study in mice. C57BL/6 mice (n = 3) were intraperitoneally administered with vehicle or horine (10 mg/kg) daily for a week. Shown are (C) body weight change percentages, kidney histology images (H&E stained) for (D) vehicle control- and (E) horine-treated, (F) kidney chemistry profile, and (G) whole-blood cell analysis. The abbreviations for blood cells can be found in SI Appendix, Additional Methods. The data represent the mean ± SD. This figure indicates no significant kidney histologic differences between the vehicle control (D) and treatment group (E). There is no significant acute tubular injury or interstitial inflammation. D and E amplified by 400-fold.
Because these two peptides showed similar in vitro toxicity (Fig. 4A), we used horine as an example to evaluate toxicity in both mice and rats. After a daily intraperitoneal injection of the peptide for 1 wk at the therapeutic dose (Fig. 4B), we did not observe any of the following: Murine behavior changes, body weight loss (Fig. 4C), differences in kidney histological morphology (Fig. 4 D and E), the kidney chemistry profiles (Fig. 4F), and whole-blood cell profiles of C57BL/6 mice (Fig. 4G). The similar levels of blood urea nitrogen and creatine indicate a lack of renal toxicity. Similarly, we also evaluated the toxicity of horine after 1 wk of daily intravenous injection and again found no sign of renal toxicity (SI Appendix, Fig. S5). These data indicate no murine nephrotoxicity of horine after multiple doses in 1 wk administered via different routes.
To evaluate the cumulative toxicity, we used Sprague–Dawley rats (6-wk-old) to conduct a multiple ascending dose study for 1 wk. Three groups of rats were daily treated with vehicle, peptide dose I and peptide dose II, respectively, at an increasing dose, as depicted in SI Appendix, Fig. S6A, based on animal body weight. All of the animals behaved similarly until the peptide dose reached 20 mg/kg per rat, where we observed a transient effect on rat mobility for ∼30 min. However, we observed no change in other behaviors nor body weight loss (SI Appendix, Fig. S6B). A liver function marker (SI Appendix, Fig. S6C) and kidney profile analysis (SI Appendix, Fig. S6D) revealed no statistical differences between the vehicle- and horine-treated groups. No significant increase in blood urea nitrogen and creatine indicates no acute toxicity to kidney. Moreover, most of blood cell counts were comparable for the three groups. There appeared a slight increase in white blood cells and lymphocytes proportional to the peptide dose (SI Appendix, Fig. S6E). In agreement with a lack of toxicity, no significant kidney histologic differences were identified between the control group (SI Appendix, Fig. S6F) and either treatment group (SI Appendix, Fig. S6 G and H). There was no significant acute tubular injury or interstitial inflammation. Taking these data together, we find that horine was not toxic to rat kidney at a cumulative dose of 82.5 or 120 mg/kg for a period of 1 wk.
Peptide In Vivo Efficacy.
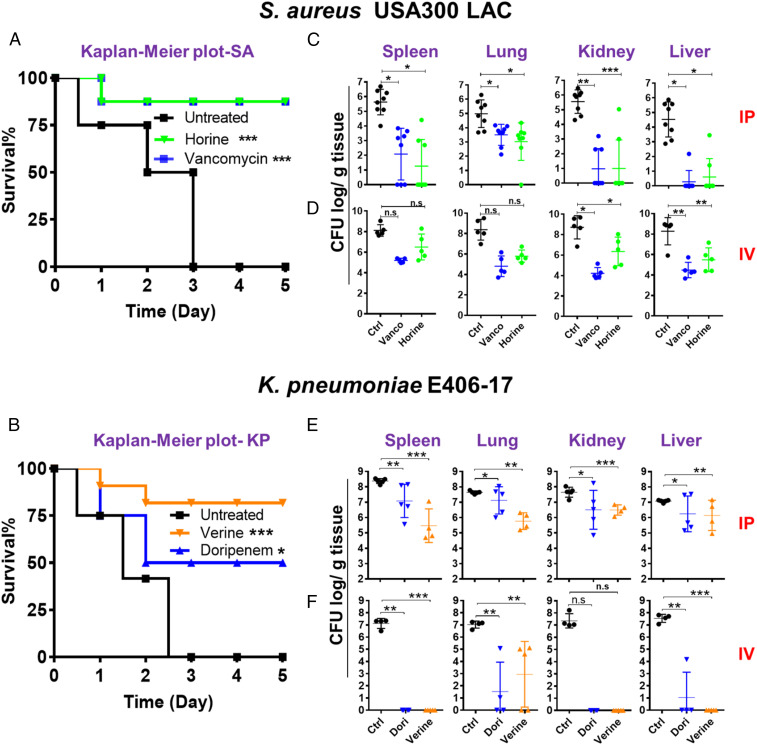
Neutropenic mice are widely accepted for systemic efficacy evaluation of antibiotics and peptide mimics (12, 31, 32). This model enables better evaluation of compound potency by minimizing the effect of the immune system. C57BL/6 mice were made neutropenic by two injections of cyclophosphamide intraperitoneally. Using this established laboratory model (12), we tested the efficacy of horine against S. aureus USA300 LAC and verine against K. pneumonia E406-17. In the survival experiment where mice were intraperitoneally infected with 1 × 107 colony forming units (CFU) of S. aureus, all of the mice in the untreated group died within 3 d. However, 87.5% of the mice survived in the horine-treated group during the 5-d observation, indicating a protective effect of the peptide (Fig. 5A). Such a protective effect of horine is identical to vancomycin treated at the same dose. Similarly, untreated mice died in 2 to 3 d when intraperitoneally infected with 9 × 106 CFU of K. pneumoniae. After horine treatment, however, we observed 81.8% protection, whereas doripenem showed 50% protection at day 5 (Fig. 5B).
Fig. 5.
Systemic efficacy of horine and verine in neutropenic mice administered intraperitoneally (IP) or intravenously (IV). (A) Mouse survival after S. aureus USA300 LAC infection (∼2 × 107 CFU) without (black) and with a single dose treatment (intraperitoneally, 2 h postinfection) of horine (green) or vancomycin (vanco, blue) at 10 mg/kg. (B) Mouse survival after K. pneumoniae E406-17 infection (∼1 × 107 CFU) without (black) and with a single dose of verine (orange) and doripenem (dori, blue) at 15 mg/kg. Survival of the C57BL/6 mice (n = 8 in each group) was observed for 5 d postinfection. (C–F) The CFU burden in spleen, lung, kidney, and liver 24 h after infection of neutropenic mice treated with the peptide via either the intraperitoneally (C and E) or intravenously (D and F) route. (C and D) Mice were intraperitoneally infected with S. aureus USA300 LAC at 2 × 106 CFU per animal without (black) and with horine (green) or vancomycin (blue) treatment at 10 mg/kg 2 h postinfection (n = 8, C57BL/6 mice intraperitoneally, and n = 5, BALB/c mice intravenously). (E and F) Mice were intraperitoneally infected with K. pneumoniae E406-17 at 5 × 105 CFU per animal without (black) and with verine (orange) or doripenem (blue) treatment at 15 mg/kg 2 h postinfection, n = 5 (C57BL/6 mice intraperitoneally and BALB/c mice intravenously). The bacterial loads from each mouse were plotted as individual points and error bars represent the deviation within the experimental group. *P < 0.05; **P < 0.01; and ***P < 0.001 (determined by t test); n.s., not significant.
To confirm a systemic infection, we infected each mouse with a sublethal S. aureus inoculum at 2 × 106 CFU and S. aureus was detected in various tissues 2 h postinfection (12). We then initiated peptide treatment via either the intraperitoneal or intravenous route. The mice were euthanized 24 h postinfection. As a positive control, we observed a significant CFU decrease (1 to 4 logs) in spleen, lung, kidney, and liver when the infected mice were treated with vancomycin at 10 mg/kg per mouse. Remarkably, we also observed a similar CFU drop in these mouse tissues up to 4 logs when treated with horine intraperitoneally at the same dose (Fig. 5C). Moreover, although less effective than vancomycin, intravenous injection of horine at the same 10 mg/kg into the infected BALB/c mice also caused a decrease of the S. aureus burden in various organs. These data established a systemic efficacy for horine (Fig. 5D).
We also tested the in vivo efficacy of verine in a neutropenic mouse model infected with K. pneumoniae (∼5 × 105 CFU per mouse), which was able to establish a systemic infection in 2 h (SI Appendix, Fig. S7). Following a single-dose intraperitoneal injection of verine, we observed a dose-dependent antimicrobial effect in different tissues of C57BL/6 mice, indicating that the effect originates from the peptide. Then we treated the infected mice with verine once at 15 mg/kg and found that bacterial CFU was reduced by 1 to 3 logs in spleen, lung, kidney, and liver (Fig. 5E). In addition, verine also displayed outstanding efficacy when intravenously administered to BALB/c mice (Fig. 5F). It is notable that such efficacies were comparable to those of doripenem injected at the same dose in the same experiment. These results documented the systemic efficacy for verine against K. pneumoniae.
We also investigated the efficacy of horine and verine in normal mice against S. aureus USA300 LAC. A higher bacterial inoculum at 2 × 108 CFU was used. The infected mice were treated either intraperitoneally or intravenously. Again, we observed CFU reduction up to 2.5 logs in spleen, lung, kidney, and liver (SI Appendix, Fig. S8). These experiments established a similar in vivo efficacy of the two peptides against S. aureus and the peptide efficacy is not due to the use of neutropenic mice.
There are cases where the compounds work effectively in mice, but failed during clinical trials. To substantiate the efficacy of these peptides, we also utilized a humanized mouse model (33). The deployment of human immune cells in NSG mice provides a more relevant immunological setting, leading to severe S. aureus infection (34). We found it necessary to increase the peptide dose at 2 and 3 h postinfection to achieve a similar efficacy (SI Appendix, Fig. S9), as observed above using normal mice after a one-time single injection (SI Appendix, Fig. S8). Since primarily male mice were used in this study, our peptides can be utilized to treat the infections of both female and male animals. Our results not only underscore the efficacy of horine but also imply a translational value of the humanized mouse model that may allow a better definition of the relationship between bacterial infection and treatment regimens.
Peptide Pharmacokinetics and Tissue Distribution.
To evaluate the peptide pharmacokinetics, we followed the plasma clearance rate of horine after intravenous injection of 10 mg/kg into BALB/c mice. Horine was rapidly cleared in mouse plasma (266 ng/mL at 5 min) and became undetectable within 1 h (half-life < 15 min) (SI Appendix, Fig. S10A). In contrast, we were able to detect D-horine, made of all D-amino acids, at 192 µg/mL at 5 min and 0.17 µg/mL at 24 h (half-life ∼1 h), indicating that D-horine is more stable in mice (SI Appendix, Fig. S10B). Since the D-form has a longer half-life, we also determined its distribution in spleen, lung, kidney, and liver of BALB/c mice at various time points from 0.5 to 24 h (SI Appendix, Fig. S10C). D-horine was detected in all tissues up to 24 h. The decrease of the peptide in the blood plasma was accompanied by an increase in other tissues. Because the peptide could reach different tissues, these data provide a basis for the systemic efficacy we observed in mice. Interestingly, we observed an in vivo efficacy for D-horine comparable to L-form after treating for 24, 48, or 72 h (SI Appendix, Fig. S11).
Discussion
Novel antibiotics are in urgent need since the ESKAPE pathogens can escape the killing of conventional antibiotics. Current antimicrobial peptide development mainly focuses on topical applications (2, 5, 35), leading to little knowledge on designing peptides with systemic efficacy. This study revealed a linear relationship between averaged contents of arginine and hydrophobic amino acids (Fig. 1), despite the diversity of AMPs in terms of sequence, activity, and scaffolds. We selected a template with a low arginine content based on our recent discovery (12). Sequence permutation led to two representative amphipathic peptides with different 3D structures and antimicrobial activities (Fig. 2). While horine has a classic amphipathic structure, resembling hundreds of natural AMPs in the APD (6), there are no sequences in the database similar to verine. We used the well-established neutropenic mouse model to observe systemic efficacy (12, 31, 32). Different pathogens were utilized to demonstrate the establishment of systemic infections in neutropenic mice 2 h postinfection, including E. coli (31), S. aureus USA300 (12), and K. pneumoniae (present study). Mor and colleagues (31) reported systemic efficacy for peptide mimetics but not typical cationic AMPs. Similarly, we observed no systemic efficacy for an LL-37–derived peptide in this model (12). Therefore, our discovery of the two peptides, horine and verine, in this study advanced our design of AMPs with systemic efficacy. We may attribute the systemic efficacy of these two peptides to their small size with merely eight amino acids and highly packed 3D structures (Fig. 2), minimizing the likelihood of their association with host factors. The compact and regular structures of horine and verine are different from the extended and irregular structures of currently known Trp-rich peptides, none of which has been illustrated to have such an in vivo efficacy (36, 37).
It is remarkable that the in vivo efficacies of horine and verine are comparable to conventional antibiotics tested at the same dose although they differ in MIC. It appears that MIC values may not be the sole indicator for in vivo efficacy of designed peptides, as also noticed by Hoffmann and colleagues (38). Superior to conventional antibiotics, however, horine and verine are also capable of eliminating resistant pathogens, persisters, and biofilms, including dozens of clinical strains. Also, the membrane-targeting of horine and verine makes it difficult for pathogens to develop resistance (3–7), in line with multiple passage experiments we conducted (SI Appendix, Fig. S2F). Unlike vancomycin and colistin, which show nephrotoxicity (29, 30), the new peptides did not display toxicity to murine kidney after daily injection for 1 wk either intraperitoneally or intravenously. There were no noticeable adverse effects of the peptide on murine behavior, kidney chemistry profiles, and histological images at the treatment level (Fig. 4 and SI Appendix, Fig. S5). To validate peptide safety, we also conducted a multiple ascending dose study in rats. Again, we did not observe nephrotoxicity and hepatotoxicity after 1-wk treatment at a high cumulative dose (SI Appendix, Fig. S6). Taken together, our results imply that these peptides may be used to treat resistant pathogens without causing adverse effects on the host.
Our pharmacokinetic studies confirm that the L- and D-forms of horine have an in vivo half-life of up to 1 h (SI Appendix, Fig. S10), indicating the importance of rapid killing. Indeed, we observed similar in vivo efficacies for these two forms of horine (SI Appendix, Fig. S11). It may be Nature’s wisdom that a rapid clearance of the defense peptide, as generally designed in innate immune systems, minimizes the chance of bacterial resistance development as well as potential toxicity to the host due to compound accumulation. This is clearly a desired feature for the antibiotics reported here and represents a strategy for designing precise future medicine. Our consideration of the natural forms may accelerate the development of AMPs into a new generation of antibiotics.
Conclusions
Although AMPs differ in sequence, scaffold, and activity, they can be unified into four general classes (39). This study discovered a linear equation between the average arginine and hydrophobic contents of all of the AMPs in the APD (40). Such a relationship may be useful to design a peptide library for screening new antimicrobials (41). Based on this relationship, we designed here two unique amphipathic peptides that target bacterial membranes. In particular, verine has a novel 3D structure, leading to a unique model for peptide design. Both horine and verine showed systemic efficacies in different mouse models. Our peptide efficacy appears to be robust because it could be observed in C57BL/6 and BALB/c mice infected with different pathogens and treated via different routes. The peptides show advantages over conventional antibiotics used here in several aspects: 1) High potency against drug-resistant pathogens; 2) lack of toxicity to rodents; and 3) no bacterial resistance development. Their highly packed 3D structures may be important for their in vivo efficacy and small size makes them more cost-effective for chemical synthesis. Horine and verine may reignite the hope to develop AMPs into new antibiotics to combat drug-resistant pathogens.
Materials and Methods
Peptides, Bacteria, Antibacterial Assays, and Killing Kinetics.
All peptides were chemically synthesized and purified to >95% (Genemed Synthesis). The quality of each peptide was determined based on mass spectrometry (MS) and high-performance liquid chromatography. A list of bacteria and the procedures for antibacterial and killing kinetics assays are detailed in SI Appendix.
Structural and Mechanistic Studies by Liquid and Solid-State NMR Spectroscopy.
The 3D structures of horine and verine were determined by using an improved 2D NMR method as we described previously (12, 42). Peptide membrane orientations and interactions with lipid bilayers were determined by solid-state 15N NMR spectroscopy. To provide complementary information from the membrane side, both 2H and 31P nuclei were used in this study. Additional details can be found in SI Appendix.
Toxicity to Rodents.
All animal studies were performed by following the protocols approved by the Institutional Animal Care and Use Committee (IACUC #16-076-08-FC, 17-104-12-FC, and 19-097-10-FC) at the University of Nebraska Medical Center. Female C57BL/6, BALB/c mice, and CD Sprague–Dawley rats (6-wk-old) were purchased from Charles River and humanized mice were transferred from our Core Facility. Animal housing and management conditions are the same as described previously (12).
Peptide in vivo toxicity was evaluated by injecting a single dose of horine to C57BL/6 mice intraperitoneally or BALB/c mice intravenously via the tail vein (n = 3 to 5). Animals were also injected daily for a week and monitored for adverse effects and mortality at least twice a day. Body weights and animal behaviors were recorded and scored. Prior to being killed by CO2, blood was collected for blood chemistry and cell count evaluation. Organs such as kidney and liver were harvested for histological analysis via the standard H&E staining.
Animal Survival, Systemic Efficacy of Peptides in Neutropenic, Normal, and Humanized Mice.
In these studies, a sublethal bacterial CFU was inoculated. Mice were infected, treated, and processed as described in SI Appendix.
Statistical Analysis.
Experiments were replicated and repeated. The results were represented as mean ± SD. However, MIC values were reported as a range, indicating the minimal concentrations needed to fully inhibit the growth of pathogens. For in vivo studies, the bacterial loads from mouse tissues were plotted as individual points and error bars represent the deviations from the average within the experimental group. *P < 0.05, **P < 0.01, ***P < 0.001, and n.s. represents no significance (determined by t test). Statistical analysis was performed with GraphPad Prism 7 or Microsoft excel software.
Supplementary Material
Acknowledgments
We thank Dr. Paul D. Fey for bacterial strains, helpful suggestions, and manuscript editing; Drs. Manju George and David Oupicky for help with our proposed rat study protocol; the University of Nebraska Medical Center Core Facilities (nanomaterial characterization, tissue, confocal microscopy, NMR, and electron microscopy) for excellent service; and Ms. Yan Cheng and Dr. Lili Guo for humanized mouse generation. This work was supported by Grants AI128230 and AI105147 from the National Institutes of Health and, in part, by a Nebraska Research Initiative Proof of Concept (to G.W.). Humanized mice were supported by R24 OD018546-01 (to S.G.). For the solid-state NMR study, B.B. is gratefully supported by the Agence Nationale de la Recherche (projects Biosupramol 17-CE18-0033-3 and the LabEx Chemistry of Complex Systems 10-LABX-0026_CSC) and the Réseau Thématique de Recherche Avancée International Center of Frontier Research in Chemistry. Visiting scholars were supported by the India Raman Postdoctoral Fellowship (D.Z. from Jawaharlal Nehru Technological University Anantapur), Henan Visiting Scholarship (F.W. from Henan Academy of Agricultural Sciences), China Scholarship Council (Y.Z. from Hainan University and X.D. from Anhui Agricultural University), and Yangzhou University Scholarship (Q.W. from Yangzhou University).
Footnotes
Competing interest statement: The authors have submitted worldwide patent applications for the reported new peptides.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2005540117/-/DCSupplemental.
Data Availability.
Additional methods, figures, and tables can be found in SI Appendix. The atomic coordinates have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6NM2 for WW291 and PDB ID code 6NM3 for WW295).
References
- 1.Mulani M. S., Kamble E. E., Kumkar S. N., Tawre M. S., Pardesi K. R., Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: A review. Front. Microbiol. 10, 539 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mangoni M. L., McDermott A. M., Zasloff M., Antimicrobial peptides and wound healing: Biological and therapeutic considerations. Exp. Dermatol. 25, 167–173 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salzman N. H. et al., Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol. 11, 76–83 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Hoek M. L., Antimicrobial peptides in reptiles. Pharmaceuticals (Basel) 7, 723–753 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mishra B., Reiling S., Zarena D., Wang G., Host defense antimicrobial peptides as antibiotics: Design and application strategies. Curr. Opin. Chem. Biol. 38, 87–96 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang G., Li X., Wang Z., APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 44, D1087–D1093 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ling L. L. et al., A new antibiotic kills pathogens without detectable resistance. Nature 517, 455–459 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hilpert K., Volkmer-Engert R., Walter T., Hancock R. E., High-throughput generation of small antibacterial peptides with improved activity. Nat. Biotechnol. 23, 1008–1012 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Loose C., Jensen K., Rigoutsos I., Stephanopoulos G., A linguistic model for the rational design of antimicrobial peptides. Nature 443, 867–869 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Wang G., Watson K. M., Peterkofsky A., Buckheit R. W. Jr., Identification of novel human immunodeficiency virus type 1-inhibitory peptides based on the antimicrobial peptide database. Antimicrob. Agents Chemother. 54, 1343–1346 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mishra B., Wang G., Ab initio design of potent anti-MRSA peptides based on database filtering technology. J. Am. Chem. Soc. 134, 12426–12429 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mishra B., Lakshmaiah Narayana J., Lushnikova T., Wang X., Wang G., Low cationicity is important for systemic in vivo efficacy of database-derived peptides against drug-resistant Gram-positive pathogens. Proc. Natl. Acad. Sci. U.S.A. 116, 13517–13522 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang G., The antimicrobial peptide database provides a platform for decoding the design principles of naturally occurring antimicrobial peptides. Protein Sci. 29, 8–18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abbassi F. et al., Temporin-SHf, a new type of phe-rich and hydrophobic ultrashort antimicrobial peptide. J. Biol. Chem. 285, 16880–16892 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mishra B., Lushnikova T., Golla R. M., Wang X., Wang G., Design and surface immobilization of short anti-biofilm peptides. Acta Biomater. 49, 316–328 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wimley W. C., White S. H., Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 3, 842–848 (1996). [DOI] [PubMed] [Google Scholar]
- 17.Szabo L., “Nightmare” bacteria, resistant to almost every drug, stalks U.S. hospitals. USA Today. https://www.usatoday.com/story/news/nation/2018/04/03/nightmare-bacteria-antibiotic-resistant-stalk-hospitals/482162002/. Accessed 1 March 2020.
- 18.Wüthrich K., NMR of Proteins and Nucleic Acids, (John Wiley & Sons, New York, 1986). [Google Scholar]
- 19.Wang G., Li Y., Li X., Correlation of three-dimensional structures with the antibacterial activity of a group of peptides designed based on a nontoxic bacterial membrane anchor. J. Biol. Chem. 280, 5803–5811 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Cornilescu G., Delaglio F., Bax A., Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13, 289–302 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Zarena D., Mishra B., Lushnikova T., Wang F., Wang G., The π configuration of the WWW motif of a short Trp-rich peptide is critical for targeting bacterial membranes, disrupting preformed biofilms, and killing methicillin-resistant Staphylococcus aureus. Biochemistry 56, 4039–4043 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis K., Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5, 48–56 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Fey P. D. et al., A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. MBio 4, e00537-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peschel A. et al., Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J. Exp. Med. 193, 1067–1076 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Epand R. F., Pollard J. E., Wright J. O., Savage P. B., Epand R. M., Depolarization, bacterial membrane composition, and the antimicrobial action of ceragenins. Antimicrob. Agents Chemother. 54, 3708–3713 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bechinger B., Zasloff M., Opella S. J., Structure and dynamics of the antibiotic peptide PGLa in membranes by solution and solid-state nuclear magnetic resonance spectroscopy. Biophys. J. 74, 981–987 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salnikov E. S., Mason A. J., Bechinger B., Membrane order perturbation in the presence of antimicrobial peptides by (2)H solid-state NMR spectroscopy. Biochimie 91, 734–743 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Ge Y. et al., In vitro antibacterial properties of pexiganan, an analog of magainin. Antimicrob. Agents Chemother. 43, 782–788 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ciarambino T. et al., Acute kidney injury and vancomycin/piperacillin/tazobactam in adult patients: A systematic review. Intern. Emerg. Med. 15, 327–331 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Ordooei Javan A., Shokouhi S., Sahraei Z., A review on colistin nephrotoxicity. Eur. J. Clin. Pharmacol. 71, 801–810 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Radzishevsky I. S. et al., Improved antimicrobial peptides based on acyl-lysine oligomers. Nat. Biotechnol. 25, 657–659 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Craig W. A., Redington J., Ebert S. C., Pharmacodynamics of amikacin in vitro and in mouse thigh and lung infections. J. Antimicrob. Chemother. 27 (suppl. C), 29–40 (1991). [DOI] [PubMed] [Google Scholar]
- 33.Parker D., Humanized mouse models of Staphylococcus aureus infection. Front. Immunol. 8, 512 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prince A., Wang H., Kitur K., Parker D., Humanized mice exhibit increased susceptibility to Staphylococcus aureus pneumonia. J. Infect. Dis. 215, 1386–1395 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hancock R. E., Sahl H. G., Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 24, 1551–1557 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Chan D. I., Prenner E. J., Vogel H. J., Tryptophan- and arginine-rich antimicrobial peptides: Structures and mechanisms of action. Biochim. Biophys. Acta 1758, 1184–1202 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Wang G., “Structural insight into the mechanisms of action of antimicrobial peptides and structure-based design” in Antimicrobial Peptides: Discovery, Design and Novel Therapeutic Strategies, Wang G., Ed. (CABI, 2017), pp. 169–187. [Google Scholar]
- 38.Knappe D., Adermann K., Hoffmann R., Oncocin Onc72 is efficacious against antibiotic-susceptible Klebsiella pneumoniae ATCC 43816 in a murine thigh infection model. Biopolymers 104, 707–711 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Wang G., Improved methods for classification, prediction, and design of antimicrobial peptides. Methods Mol. Biol. 1268, 43–66 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z., Wang G., APD: The antimicrobial peptide database. Nucleic Acids Res. 32, D590–D592 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wimley W. C., Application of synthetic molecular evolution to the discovery of antimicrobial peptides. Adv. Exp. Med. Biol. 1117, 241–255 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang G., Structures of human host defense cathelicidin LL-37 and its smallest antimicrobial peptide KR-12 in lipid micelles. J. Biol. Chem. 283, 32637–32643 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Additional methods, figures, and tables can be found in SI Appendix. The atomic coordinates have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6NM2 for WW291 and PDB ID code 6NM3 for WW295).