Significance
Whether HIV-1 infection triggers cGAS-mediated immune responses in CD4+ T cells remains debated. It is important to investigate to which extent HIV-1–infected T cells contribute to IFN production and expression of antiviral interferon-stimulated genes. By analyzing cellular responses upon productive HIV-1 infection or transduction, we demonstrate that lentiviruses and gammaretroviruses can infect and spread in primary CD4+ T cells and T cell lines without alarming the cGAS-mediated DNA sensing machinery, probably due to their replication strategy that minimizes the abundance of cGAS-sensitive DNA PAMPs.
Keywords: HIV-1, HSV-1, cGAS, innate sensing, T cells
Abstract
The DNA sensor cGAS catalyzes the production of the cyclic dinucleotide cGAMP, resulting in type I interferon responses. We addressed the functionality of cGAS-mediated DNA sensing in human and murine T cells. Activated primary CD4+ T cells expressed cGAS and responded to plasmid DNA by upregulation of ISGs and release of bioactive interferon. In mouse T cells, cGAS KO ablated sensing of plasmid DNA, and TREX1 KO enabled cells to sense short immunostimulatory DNA. Expression of IFIT1 and MX2 was downregulated and upregulated in cGAS KO and TREX1 KO T cell lines, respectively, compared to parental cells. Despite their intact cGAS sensing pathway, human CD4+ T cells failed to mount a reverse transcriptase (RT) inhibitor-sensitive immune response following HIV-1 infection. In contrast, infection of human T cells with HSV-1 that is functionally deficient for the cGAS antagonist pUL41 (HSV-1ΔUL41N) resulted in a cGAS-dependent type I interferon response. In accordance with our results in primary CD4+ T cells, plasmid challenge or HSV-1ΔUL41N inoculation of T cell lines provoked an entirely cGAS-dependent type I interferon response, including IRF3 phosphorylation and expression of ISGs. In contrast, no RT-dependent interferon response was detected following transduction of T cell lines with VSV-G-pseudotyped lentiviral or gammaretroviral particles. Together, T cells are capable to raise a cGAS-dependent cell-intrinsic response to both plasmid DNA challenge or inoculation with HSV-1ΔUL41N. However, HIV-1 infection does not appear to trigger cGAS-mediated sensing of viral DNA in T cells, possibly by revealing viral DNA of insufficient quantity, length, and/or accessibility to cGAS.
The ability of mammalian cells to sense invading pathogens by cellular pattern recognition receptors is crucial for mounting of an effective cellular defense response and for initiating adequate adaptive immunity. cGMP-AMP synthetase (cGAS) senses DNA of aberrant subcellular localization in a sequence-independent, but length-dependent manner. Binding of DNA by cGAS is implicated in antiviral and antimicrobial defense, and autoimmunity. Activated cGAS catalyzes the cyclization of ATP and GTP, resulting in the small molecule and cyclic dinucleotide cGAMP (1, 2). cGAMP induces a STING/TBK1/IRF3-dependent signaling cascade that culminates in the expression of IRF-3–induced genes and type I interferons (IFNs), which, in turn, elicit an antiviral state by transactivating multiple IFN-stimulated genes (ISGs). The exonuclease TREX1 constitutes an important counterplayer of cGAS and prevents immune hyperactivation by cleaving cytoplasmic DNA that would otherwise be sensed (3, 4).
It is widely accepted that herpesviral infections trigger cGAS/STING-dependent cellular responses that are essential for the host to overcome the infection in vivo. Herpesviruses have evolved numerous strategies to dampen this cellular surveillance machinery (reviewed in refs. 5 and 6). It is debated to which extent retroviruses, including HIV-1, are prone to cGAS-mediated sensing, especially in primary HIV-1 target cells. Many studies on HIV-1 and cGAS have been conducted in immortalized adherent cell lines of limited physiological relevance, including A549 lung epithelial cells, monocytic THP-1 cells, mouse L929 fibroblast cells (7–10), and primary human dendritic cells. The latter produce reverse transcripts efficiently only if transduced with HIV-1 particles that copackage SIV or HIV-2 Vpx (11, 12), a scenario that does not occur during natural HIV-1 infection. HIV-1 infection fails to trigger a detectable type I IFN expression in primary monocyte-derived macrophages unless capsids are destabilized genetically or pharmacologically (13), interferences which may induce leakage of reverse transcripts into the cytoplasm. The limited data available on the role of cGAS in HIV-1–infected T cells is controversial. One study in HIV-1–infected T cells proposed cGAS-dependent responses, which appeared to be triggered postintegration, potentially through release of mitochondrial DNA in the course of productive infection (14). In that study, the accessory protein Vpr was reported to potentiate, while Vpu dampened these responses (14). In contrast, another study suggested that DNA sensing is generally blunted in T cells (15). Our work revealed that productive HIV-1 infection does not induce type I IFN expression in IL-2/PHA-activated PBMCs (16). Here, we readdressed the integrity of cGAS-mediated DNA sensing in primary and immortalized human and mouse CD4+ T cells. We find that the integral components of the cGAS-mediated DNA sensing pathway are expressed and functional in CD4+ T cells from both species. Challenge of T cells with a HSV-1 mutant with reduced ability to counteract cGAS elicited dramatic cGAS-dependent responses. HIV-1 infection, on the contrary, failed to induce detectable cGAS-dependent or reverse transcriptase (RT) inhibitor-sensitive type I IFN responses.
Results
Plasmid DNA Challenge Elicits a Type I IFN Response in Activated Human and Murine Primary CD4+ T Cells.
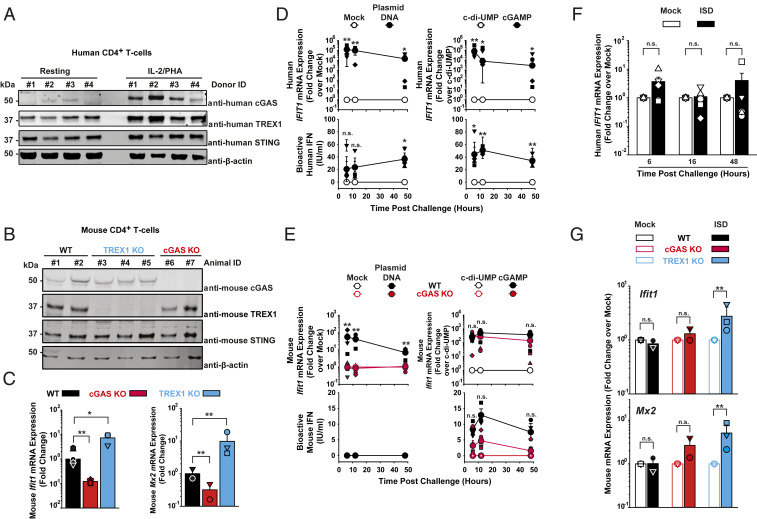
We first assessed cGAS expression in purified, resting, and activated primary human and mouse CD4+ T cells. Immunoblotting revealed detectable levels of cGAS expression in IL-2/PHA–stimulated human CD4+ T cells (Fig. 1A and SI Appendix, Fig. S1A) and anti-CD3/28–stimulated mouse CD4+ T cells (Fig. 1B and SI Appendix, Fig. S1B). In human CD4+ T cells, cGAS messenger RNA (mRNA) and protein levels were drastically lower in resting than in activated cultures, consistent with published data (17) (Fig. 1A and SI Appendix, Fig. S1 A and C–D), while cGAS expression was clearly detectable in both unstimulated and anti-CD3/28–activated mouse CD4+ T cells (SI Appendix, Fig. S1B). STING and the exonuclease TREX1 were expressed in activated CD4+ T cells of humans (Fig. 1A) and WT mice (Fig. 1B). In murine-activated CD4+ T cells, basal expression levels of Ifit1 and Mx2 mRNAs were reduced and elevated in the context of cGAS KO and TREX1 KO, respectively (Fig. 1C), suggesting that basal activities of cGAS and TREX1 modulate steady-state expression levels of cellular ISGs in T cells.
Fig. 1.
Plasmid DNA challenge elicits a type I IFN response in activated human and murine primary CD4+ T cells. (A) Immunoblotting of lysates of primary human CD4+ T cells using indicated antibodies. (B) Immunoblotting of lysates of primary anti-CD3/28–activated and IL-2–activated CD4+ T cells from WT, TREX1 KO, and cGAS KO mice using indicated antibodies. (C) Relative steady-state Ifit1 and Mx2 mRNA levels were determined by qRT-PCR in activated CD4+ T cells of mice of indicated genotype (n = 2–3). Error bars indicate SEM from values obtained from cell cultures of two to three individual animals of each genotype. Small symbols represent levels obtained in individual animals; bars represent the arithmetic mean of values of all cell cultures of a given condition. (D) Activated human CD4+ T cells were either mock-electroporated or electroporated with plasmid DNA (Left), and electroporated either with c-di-UMP or cGAMP (Right). Cultures were monitored at indicated time points postchallenge for relative IFIT1 mRNA expression by qRT-PCR (Upper) and release of bioactive type I IFN using a luminometric HL116-based assay (Lower). Error bars indicate SEM from values obtained from cells from four individual donors whose values are depicted as small symbols. Large symbols represent the arithmetic mean of values of all cell cultures of a given condition. (E) Activated mouse CD4+ T cells of WT and cGAS KO animals were either mock-electroporated or electroporated with plasmid DNA (Left), and electroporated either with c-di-UMP or cGAMP (Right). Cultures were monitored at indicated time points postchallenge for Ifit1 mRNA expression by qRT-PCR (Upper) and release of bioactive type I IFN using a luminometric MEF-based assay (Lower). Error bars indicate SEM from values obtained from cells from three individual animals of each genotype whose values are depicted as small symbols. Large symbols represent the arithmetic mean of values of all cell cultures of a given condition. Statistical significance was calculated for T cells from cGAS KO versus WT animals. (F) Activated human CD4+ T cells were mock-electroporated or electroporated with short ISD and monitored at indicated hours postchallenge for IFIT1 mRNA expression by qRT-PCR. Error bars indicates SEM from values obtained from cells from four to five individual donors whose values are depicted as small symbols; bars represent the arithmetic mean of values of all cell cultures of a given condition. (G) Activated mouse CD4+ T cells of indicated animals were mock-electroporated or electroporated with short ISD and monitored at 6 h postchallenge for Ifit1 mRNA expression by qRT-PCR. Error bars indicates SEM from cells of three individual animals of each genotype whose values are depicted as small symbols; bars represent the arithmetic mean of values of all cell cultures of a given condition. P values <0.05 were considered significant (*) and <0.01 very significant (**); n.s. = not significant (≥0.05).
Next, we tested the DNA sensing abilities of activated CD4+ T cells of both species. Electroporation of IL-2/PHA–activated human CD4+ lymphocytes with endotoxin-free plasmid DNA gave rise to a more than 100,000-fold induction of IFIT1 mRNA expression compared to mock electroporation (Fig. 1 D, Upper Left). Importantly, mock electroporation of primary CD4+ T cells did not induce detectable cellular responses (SI Appendix, Fig. S2A). Of note, IFIT1 is transactivated directly by IRF3 and independently of type I IFNs (18). Up to 60 IU bioactive type I IFN/mL were secreted in the culture supernatant following plasmid DNA challenge (Fig. 1 D, Lower Left), while mock electroporation failed to induce significant changes of both readouts. Direct challenge with cGAMP, the catalytic product of cGAS, but not c-di-UMP, which does not activate STING, up-regulated IFIT1 mRNA expression up to 100,000-fold (Fig. 1 D, Upper Right) and triggered the release of up to 80 IU bioactive IFN/mL (Fig. 1 D, Lower Right).
Analogous experiments in mouse CD4+ T cells (Fig. 1E) revealed that Ifit1 mRNA induction upon plasmid DNA electroporation is strictly cGAS-dependent, suggesting that cGAS is the unique functional cytosolic DNA sensor expressed in this cell type, at least in the mouse species (Fig. 1 E, Upper Left). In contrast to human CD4+ T cells, intriguingly, no secretion of bioactive type I IFN into the culture supernatant was detectable upon DNA challenge (Fig. 1 E, Lower Left). Irrespective of their cGAS expression status, mouse CD4+ T cells scored positive for Ifit1 mRNA induction (Fig. 1 E, Upper Right) and type I IFN release (Fig. 1 E, Lower Right) upon delivery of cGAMP, indicating that IFN release is not inherently blunted in this cell line. Sensing of short immunostimulatory DNA (ISD) was effective and cGAS-dependent in THP-1 cells (SI Appendix, Fig. S3, but occurred neither in human CD4+ T cells (Fig. 1F) nor in CD4+ T cells from WT and cGAS KO mice (Fig. 1G), but was detectable in cells from TREX1 KO mice (Fig. 1G), suggesting that TREX1 reduces the abundance of ISD molecules that are otherwise susceptible to cGAS sensing.
De Novo HIV-1 Infection, as Opposed to HSV-1 Infection, Fails to Trigger a Type I IFN Response in Human CD4+ T Cells.
We next interrogated to which extent HIV-1 infection triggers cell-intrinsic innate immunity in human CD4+ T cells. Primary IL-2/PHA-activated CD4+ T cells were inoculated with HIV-1Ba-L and monitored for different parameters over a period of up to 13 d postinfection. We used R5-tropic HIV-1Ba-L because the extent of HIV-1Ba-L–induced CD4+ T cell death was moderate and allowed us to conduct the experiment over a prolonged time period. In contrast, extensive cell death in cultures infected with the prototypic X4-tropic HIV-1 NL4.3 strain precluded any long-term kinetic experiments. Furthermore, HIV-1Ba-L stock was produced by serial passaging on susceptible T cell lines and did not rely on transfection of producer cells, a procedure that is inherently linked to contamination of viral stocks with proviral plasmid DNA. This potential contamination is problematic especially in the context of studies addressing DNA sensing and can only be removed by excessive DNase treatment of particles (19, 20).
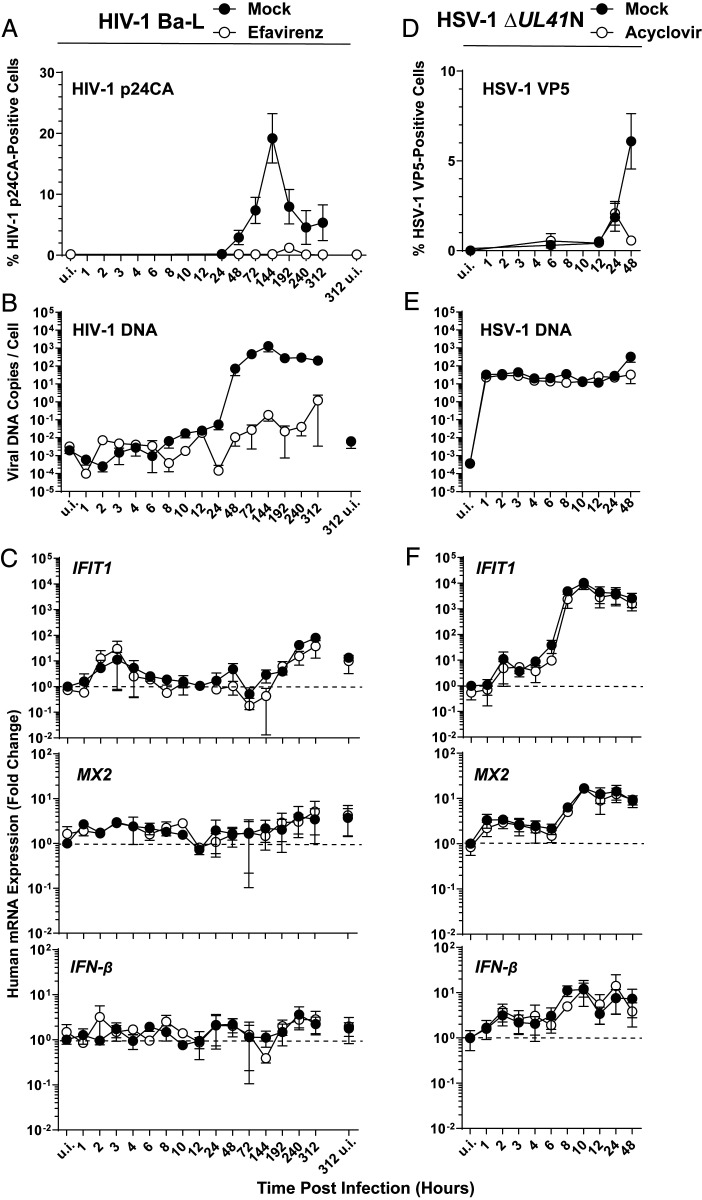
RT inhibitor efavirenz (EFV)-sensitive and, thus, de novo production of HIV-1 p24 capsid, was detectable from 2 d postinfection on and typically peaked at day 6 postinfection, followed by a steady decrease until the end of the experiment at day 13 postinfection (Fig. 2A). Detectable de novo synthesis of HIV-1 late RT products started at 8 h postinfection (Fig. 2B). HIV-1 DNA synthesis peaked at day 6 postinfection, reaching 1,000 copies per cell and subsequently plateaued at around 200–300 copies per cell (Fig. 2B), suggesting multiple infection events per cell under these experimental conditions that favor viral spread from cell-to-cell. Strikingly, in these very same cultures, IFIT1 mRNA expression was only 10-fold increased at 2 to 4 h. At 10 and 13 d postinfection, a 40- and 80-fold induction of IFIT1 mRNA expression was detected. However, these increases were neither sensitive to EFV treatment nor grossly exceeded the level of IFIT1 mRNA expressed in uninfected cells at the end of the experiment (day 13 postinfection) (Fig. 2 C, Top). This result suggests that the increases in IFIT1 mRNA expression very early (3 and 4 h) and late (10 and 13 d) after viral inoculation indicate sensing of virus-associated PAMPs or DAMPs that are independent of reverse transcription. Alternatively, they may simply reflect fluctuations of the steady-state IFIT1 mRNA levels in these activated primary CD4+ T cell cultures. Similarly, levels of MX2 (Fig. 2 C, Middle) and IFN-β mRNA (Fig. 2 C, Bottom) expression fluctuated between unchanged and 4-fold increased and almost superimposed the levels detected in cells that had been treated with EFV prior to HIV-1 inoculation.
Fig. 2.
De novo HIV-1 infection, as opposed to HSV-1 infection, fails to trigger a type I IFN response in human CD4+ T cells. (A–C) Primary human CD4+ T cells were infected with HIV-1Ba-L in the absence and presence of EFV and monitored, at indicated time points, for: (A) HIV-1 p24 capsid expression by intracellular immunostaining followed by FACS analysis. (B) De novo synthesis of HIV-1 late RT products by absolute qPCR. (C) Relative expression of IFIT1, MX2, and IFN-β by qRT-PCR. (D–F) Primary human CD4+ T cells were inoculated with HSV-1 ΔUL41N in the absence and presence of ACV and monitored, at indicated time points, for: (D) HSV-1 VP5 capsid protein expression by intracellular immunostaining followed by FACS analysis. (E) Genomic HSV-1 DNA copy numbers by absolute qPCR. (F) Relative expression of IFIT1, MX2, and IFN-β by qRT-PCR. Error bars show SEM from values obtained from CD4+ T cells from three to five individual donors.
The small molecule inhibitor PF74, which binds a groove in the N-terminal domain of HIV-1 capsid, has been suggested to provoke premature disassembly of the viral core post entry (21–23). Interestingly, PF74-induced premature HIV-1 uncoating has been recently linked to increased cGAS-dependent immune responses (10, 24). We thus explored if PF74 treatment of CD4+ T cells during HIV-1 infection may force viral DNA leakage and accumulation in the cytoplasm, potentially followed by sensing. As expected, PF74 treatment potently inhibited productive infection of CD4+ T cells in the absence of detectable cell death (SI Appendix, Fig. S4A). However, no statistically significant induction of mRNA expression of IFIT1, MX2, and IFN-β was detectable (SI Appendix, Fig. S4B). Although values in PF74-treated, HIV-1–infected T cell cultures tended to be elevated compared to mock-treated, HIV-1–infected counterparts, no statistical significant difference between those two conditions could be established. In addition, treatment of cells with PF74 per se slightly induced IFN-β mRNA expression, although again, no statistically significant difference to levels of HIV-1–infected samples was obtained.
In order to explore whether primary human CD4+ T cells are competent for sensing of a DNA-viral infection at all, we assessed their response to HSV-1 inoculation. In vivo, HSV-2–infected T cells have been detected in lesions (25). Ex vivo, T cell lines and activated primary T cells appear to be susceptible to HSV-1 infection, as judged by detection of intracellular HSV-1 antigens upon exposure to virus (25–27). Owing to its genomic DNA of 152-kbp length, which is directly introduced into the infected cell upon entry of the capsid associated to tegument proteins and which has been reported to be released from a subset of capsids into the cytosol (28–30), we reasoned that HSV-1 might represent a useful control with a potentially high susceptibility to cGAS-mediated DNA sensing in T cells. We made use of an HSV-1 mutant (HSV-1 ΔUL41N), which lacks a large part of the 5′ part of the immunoevasion gene UL41, whose gene product pUL41 targets cGAS mRNA to degradation (31). Inoculation with HSV-1 ΔUL41N yielded 6% VP5 capsid protein-positive CD4+ T cells at day 2 postinfection, and sensitivity of the VP5 positivity to acyclovir (ACV) provides first evidence for a productive HSV-1 infection in this cell type, at least ex vivo (Fig. 2D). We detected an average of 32 viral DNA copies per cell directly upon HSV-1 inoculation, a quantity that was maintained over 24 h irrespective of absence or presence of ACV. This DNA most likely represents incoming genomic DNA prior to HSV-1 DNA replication. At 48 h, the level of HSV-1 ΔUL41N DNA copies per cell increased 10-fold in untreated cells in an ACV-sensitive fashion, suggesting active viral DNA replication in CD4+ T cells (Fig. 2E). In these very same infected T cell cultures, IFIT1 mRNA expression clearly increased over time in an ACV-insensitive manner and reached up to 14,000-fold higher levels than uninfected cells at 8 h postinfection, suggesting that the incoming viral DNA or a PAMP/DAMP independent of viral DNA replication triggered IRF3-mediated IFIT1 mRNA expression (Fig. 2 F, Top). mRNA expression of MX2 (Fig. 2 F, Middle) and IFN-β (Fig. 2 F, Bottom) was increased 29-fold and 25-fold, respectively.
Together, these results point toward the absence of a robust cell-intrinsic immunity response to HIV-1 infection in primary human CD4+ T cells despite productive infection and abundant synthesis of de novo HIV-1 RT products. In contrast, a control HSV-1 ΔUL41N infection, although accompanied by the intracellular delivery of viral DNA copies 10-fold less in numbers or at best approaching those measured at the peak of spreading HIV-1 infection, clearly elicited cell-intrinsic innate immunity.
Global Analysis of the Cellular Transcriptome of HIV-1Ba-L–Infected Primary CD4+ T Cells.
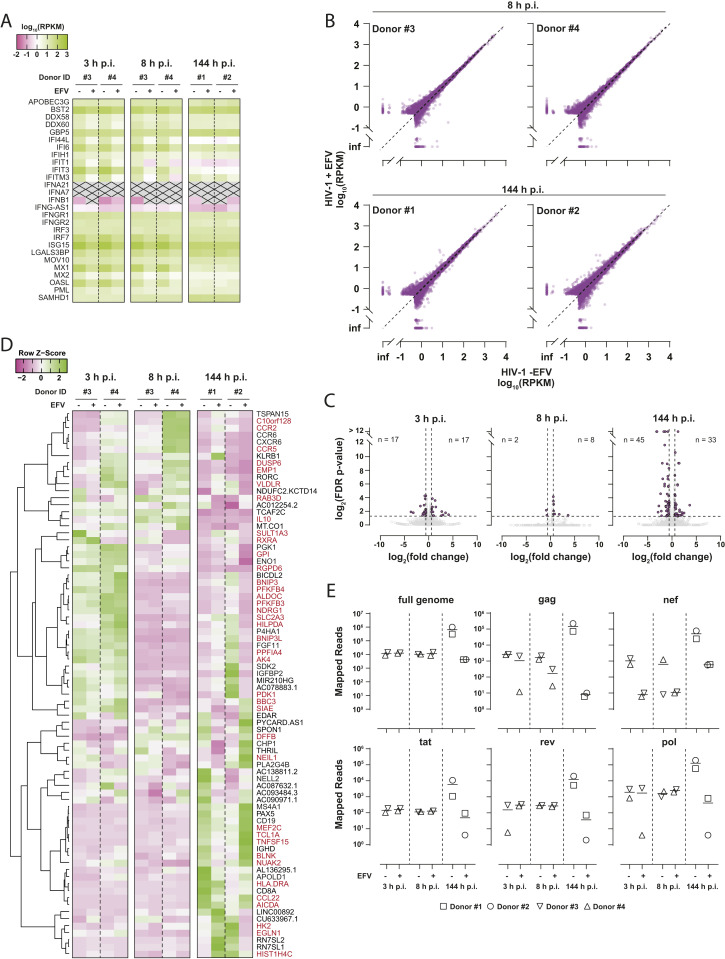
To assess the cellular response of primary CD4+ T cells to HIV-1 infection on a global scale, we sequenced RNA isolated from infected cultures. Transcript abundance for 28 individual genes implicated in or induced by the IFN signaling pathway were calculated and mean RPKM values were compared (Fig. 3A). None of them were notably deregulated with respect to EFV treatment, indicating that productive infection did not provoke a detectable type I IFN response, in line with our qRT-PCR–based measurements of IFIT1, MX2, and IFN-β mRNA expression (Fig. 2C). Plotting of raw RPKM values greater than 0.5 of all RNAs revealed only very minor differences in the overall transcriptome of cells productively infected compared to cells treated with EFV prior to infection (Fig. 3B). Those genes that were statistically significantly deregulated (34 at 3 h, 10 at 8 h, and 78 at 144 h) were only modulated at very mild levels (Fig. 3C) and are predominantly associated with cell cycle-associated pathways. Complete linkage clustering did not reveal patterns of ISGs differentially regulated in cells with productive HIV-1 replication over time (Fig. 3D). Finally, HIV-1 RNA reads retrieved from the identical samples confirmed de novo production of HIV-1 mRNAs at 144 h postinfection (Fig. 3E).
Fig. 3.
Global analysis of the cellular transcriptome of HIV-1Ba-L–infected primary CD4+ T cells. (A) RPKM values of 28 individual genes implicated in or induced by the IFN signaling pathway are depicted as heatmap. Shown is the temporal expression in ex vivo HIV-1Ba-L–infected CD4+ T cells at 3, 8, and 144 h postinfection (p.i.), in the presence or absence of EFV treatment. Transcripts with RPKM < 0.01 are crossed out. (B) Plot of raw RPKM values greater than 0.5 of RNAs of all donors obtained from productively infected cells (HIV-1 -EFV) versus cells treated with EFV (HIV-1 +EFV) prior to and during infection. (C) Plot of fold change (untreated versus EFV-treated cells) versus FDR P value of all mapped human genes at indicated time points p.i. with HIV-1Ba-L (gray circles). The numbers of transcripts deregulated more than twofold with FDR P values <0.05 (purple circles) are shown in the plots. (D) Heatmap displaying row z-scores of the 78 genes that were statistically significantly deregulated in untreated versus EFV-treated infected cells at 144 h p.i. Complete linkage clustering of genes using the Pearson distance measurement method was performed with heatmapper 2 (www2.heatmapper.ca). Genes labeled in red are known ISGs according to interferome v2.01 (www.interferome.org; search parameters were set to “any”, except, Species: “Homo sapiens” and System: “Haemopoietic/Immune”). (E) Number of RNA-seq reads mapping to HIV-1Ba-L reference sequence at indicated time points postinfection.
The Human T Cell Line PM1 Expresses Functional cGAS.
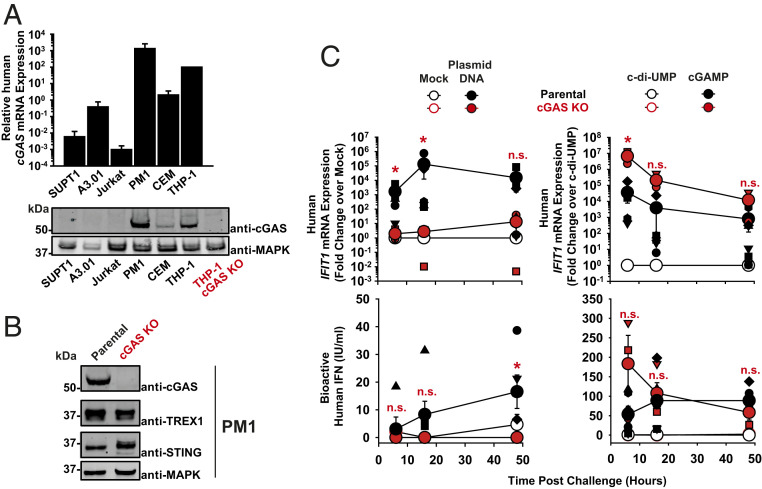
We next studied the role of cGAS during HIV-1 and HSV-1 ΔUL41N infection of T cells. To this end, we first screened a panel of immortalized human T cell lines frequently used in HIV-1 research for endogenous cGAS expression. Interestingly, the collection displayed drastic cell line-specific differences in cGAS mRNA and protein expression. PM1 and CEM T cell lines displayed high and moderate cGAS levels, respectively, whereas SUPT1, A3.01, and Jurkat T cell lines expressed low levels of cGAS mRNA and scored negative for cGAS protein (Fig. 4A). Because PM1 T cells shared high levels of endogenous cGAS expression with primary human activated CD4+ T cells, they were chosen for mechanistic studies and subjected to CRISPR/Cas9-mediated knock-out of cGAS (Fig. 4B). Delivery of plasmid DNA into the cytoplasm of parental PM1 T cells by electroporation increased the abundance of IFIT1 mRNAs up to 133,000-fold (Fig. 4 C, Upper Left) and induced release of up to 17 IU/mL bioactive IFN (Fig. 4 C, Lower Left). Mock electroporation per se did not induce cellular responses (SI Appendix, Fig. S2B). These responses were almost entirely abrogated in cGAS KO PM1 T cells. The efficiency of plasmid DNA sensing in the other T cell lines largely correlated with their individual cGAS expression status, with the exception of A3.01 T cells, which reacted to plasmid DNA in the absence of detectable endogenous cGAS protein expression (SI Appendix, Fig. S5A), pointing toward the potential contribution of other DNA sensors than cGAS in this cell line to DNA sensing. Reconstitution of normally cGAS-negative Jurkat T cells with WT, but not with catalytically inactive, cGAS (G212A/S213A) mutant enabled IFIT1 up-regulation as an early response to plasmid DNA challenge (SI Appendix, Fig. S5A). cGAMP-triggered enhancement of IFIT1 mRNA expression and IFN release was generally detectable irrespective of the cGAS expression status (SI Appendix, Fig. S5B), although appearing more pronounced in cGAS KO PM1 T cells as compared to parental PM1 T cells (Fig. 4 C, Right). Conclusively, PM1 T cells recapitulate the expression status of endogenous cGAS and the plasmid DNA sensing ability of primary activated human CD4+ T cells, suggesting that they may serve as a valuable model for investigating cGAS-mediated sensing of DNA in T cells upon viral infections. Most importantly, cGAS appears to be the only and essential sensor of plasmid DNA in this model T cell line.
Fig. 4.
The human T cell line PM1 expresses functional cGAS. (A) Relative levels of cGAS mRNA expression in indicated cell lines (bar diagram). The level detected in THP-1 cells is set to 100. Immunoblotting of indicated cell lysates using anti-cGAS and anti-MAPK antibodies. Lysates of parental and cGAS KO THP-1 cells serve as specificity control for the anti-human cGAS antibody. (B) Immunoblotting of lysates of parental and cGAS KO PM1 T cell lines using indicated antibodies. (C) Parental and cGAS KO PM1 T cells were either mock-electroporated or electroporated with plasmid DNA (Left), or electroporated either with c-di-UMP or cGAMP (Right). Cultures were monitored at indicated time points postchallenge for relative IFIT1 mRNA expression by qRT-PCR (Upper) and release of bioactive type I IFN using a luminometric HL116-based assay (Lower). Error bars show SEM from three to four independent experiments whose values are shown as small symbols. The arithmetic means of values of all cell cultures of a given condition are shown as large symbols. Statistical significance was calculated for cGAS KO versus parental T cells. P values <0.05 were considered significant (*) and <0.01 very significant (**); n.s. = not significant (≥0.05).
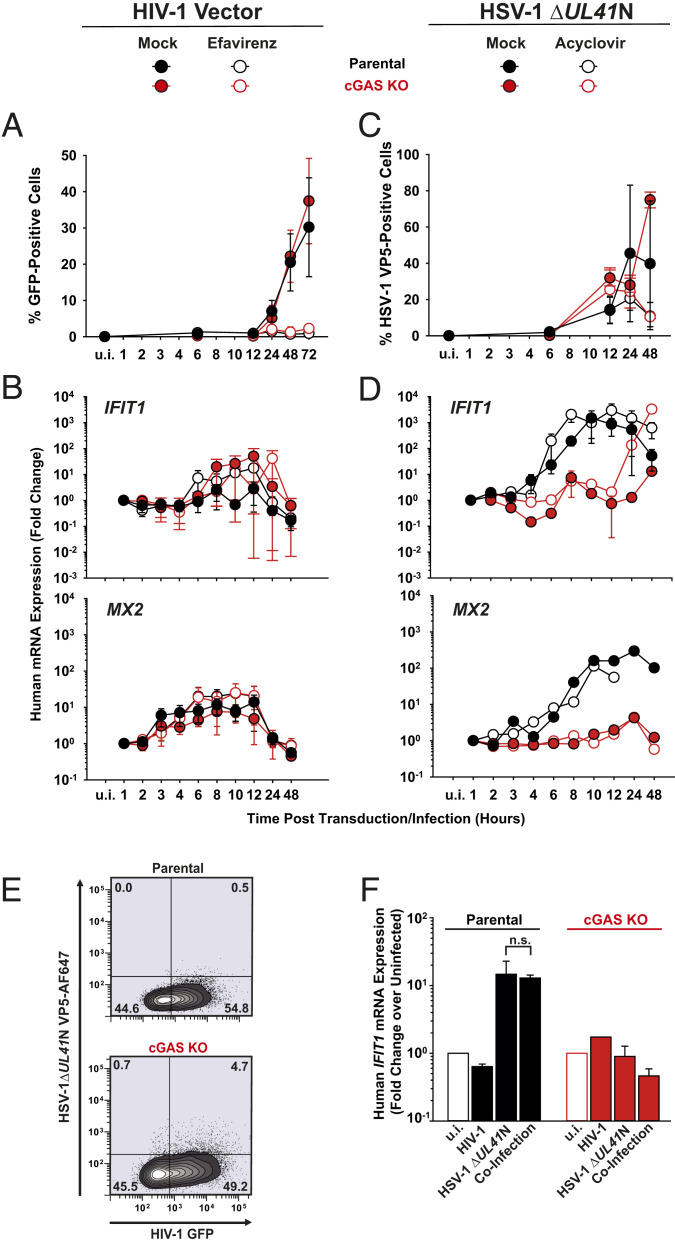
Absence of cGAS-Mediated Innate Immune Responses in PM1 T Cells upon Lentiviral Vector Transduction, as Opposed to HSV-1 Infection.
To unravel the contribution of cGAS in PM1 T cells during sensing of HIV-1 infection, we transduced parental and cGAS KO cells with VSV-G–pseudotyped lentiviral vectors containing a CMV-driven GFP-encoding transfer vector and monitored the cellular response over time. VSV-G–pseudotyped vectors do not express HIV-1 accessory genes, some of which have been suggested to counteract cell-intrinsic immune sensing (14, 32) and enable a robust transduction efficiency during a single round of replication. We thus hypothesized that any potential cellular sensing of HIV-1 should have the highest chance of being detected in this experimental system. Inoculation of PM1 T cell lines resulted in 30–35% GFP-positive cells at 3 d postinfection, irrespective of the cGAS expression status, and EFV-mediated inhibition of RT almost entirely abolished transduction (Fig. 5A). During the entire experiment, transduced parental PM1 T cells displayed virtually unchanged IFIT1 and slightly (10-fold) elevated MX2 mRNA expression, which however was neither cGAS-dependent nor EFV-sensitive (Fig. 5B). Genetic modification of the HIV-1 capsid at specific positions (N74D and P90A) has been reported to prevent the interaction with capsid-stabilizing cellular cofactors and, thereby, reduce the stability of the viral core (33–35), inducing a premature capsid disassembly resulting in a type I IFN response in THP-1 cell lines and primary macrophages (13, 36). As expected (36), individual inoculation of parental PM1 T cells with equal p24 capsid equivalents of HIV-1 (CA N74D) and HIV-1 (CA P90A) (SI Appendix, Fig. S6A) resulted in a 10-fold reduced transduction efficiency, as compared to cells infected with WT counterpart (SI Appendix, Fig. S6B). Our data do not exclude the possibility that the capsid mutations resulted in less-efficient reverse transcription and nuclear import. However, both mutations are well-described to destabilize the capsid (33–35) and, even if reduced in quantity, leaky and/or abortive RT products were shown to become accessible to cellular sensors at least in other cell types (13, 36). Strikingly, HIV-1 (CA N74D) and HIV-1 (CA P90A) failed to trigger a detectable elevation of IFIT1 and MX2 mRNA expression in PM1 T cells (SI Appendix, Fig. S6C).
Fig. 5.
Absence of cGAS-mediated innate immune responses in PM1 T cells upon lentiviral vector transduction as opposed to HSV-1 infection. (A and B) Parental and cGAS KO PM1 T cells were transduced with VSV-G–pseudotyped HIV-1 GFP vectors in the absence and presence of EFV and monitored, at indicated time points, for: (A) Reporter GFP expression by FACS analysis. (B) Relative expression of IFIT1 and MX2 by qRT-PCR. (C and D) Parental and cGAS KO PM1 T cells were inoculated with HSV-1 ΔUL41N in the absence and presence of ACV and monitored, at indicated time points, for: (C) HSV-1 VP5 capsid protein expression by intracellular immunostaining followed by FACS analysis. (D) Relative expression of IFIT1 and MX2 by qRT-PCR. (E) Parental and cGAS KO T cells were transduced with HIV-1 vectors, followed by inoculation with HSV-1 ΔUL41N 24 h later. Shown is a representative dot plot of dually infected cells at 48 h post-HSV-1 inoculation. (F) Relative IFIT1 mRNA expression in indicated cells 48 h post-HSV-1 inoculation. P values <0.05 were considered significant (*) and <0.01 very significant (**); n.s. = not significant (≥0.05).
In stark contrast, inoculation of parental PM1 T cell lines with HSV-1 ΔUL41N (Fig. 5C) was followed by an up to 1,500-fold up-regulated IFIT1 mRNA expression, which was strictly cGAS-dependent until 12 h post infection (Fig. 5 D, Upper), and a constantly cGAS-dependent, up to 300-fold up-regulated MX2 mRNA expression (Fig. 5 D, Lower). ACV treatment did not prevent IFIT1 and MX2 mRNA expression, consistent with the idea that the incoming viral genome is sensed by cGAS. In line with our observations in primary human CD4+ T cells infected with HIV-1Ba-L, parental PM1 T cells fail to induce robust immune responses upon lentiviral transduction, despite a high transduction efficiency and absence of HIV-1 accessory gene expression. In strong contrast, PM1 T cells mount drastic cell-intrinsic responses to HSV-1 ΔUL41N inoculation in a strictly cGAS-dependent fashion early after infection.
Our data did not exclude the possibility of an active suppression of innate sensing by an incoming component of the lentiviral particle, e.g., capsid-mediated inhibition of IRF3 phosphorylation. However, pulsing cells with the GFP-expressing lentivirus (Fig. 5E) did not interfere with cGAS-mediated innate sensing of a subsequent HSV-1 ΔUL41N infection (Fig. 5F), arguing against the idea of a lentivirus-induced suppression of the innate sensing machinery.
cGAS and TREX1 Modulate Sensing of Plasmid DNA and of ISD, Respectively.
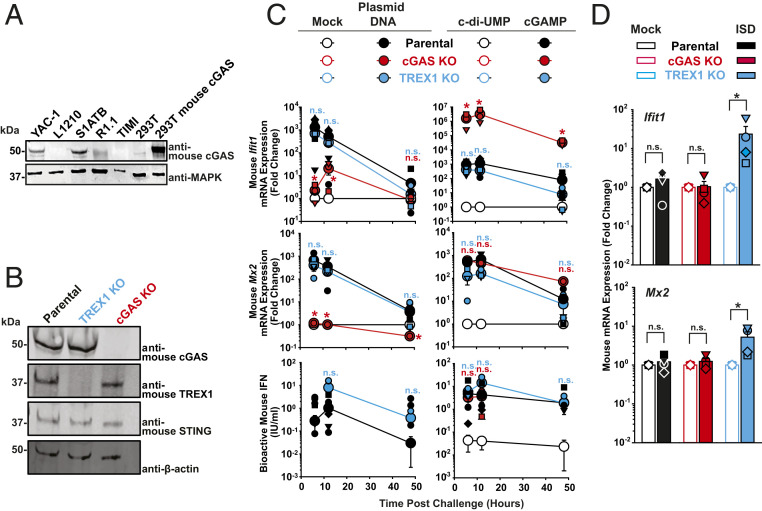
Mouse T cell lines are refractory to productive HIV-1 infection, even upon circumvention of the mouse-specific entry block by usage of VSV-G–pseudotyped virus (37–40). We initially hypothesized that this postentry block may be related to a mouse T cell-specific sensing property. Since mouse CD4+ T cells expressed functional cGAS (Fig. 1 B and E and SI Appendix, Fig. S1B), we tested its role during lentiviral and gammaretroviral transduction. In analogy to the human T cell line screen, we identified a remarkable diversity in cGAS expression levels among a panel of mouse T cell lines. While YAC-1 and S1A.TB T cells expressed cGAS, L1210, R1.1, and TIMI.4 cell lines scored negative (Fig. 6A). We thus introduced cGAS and TREX1 knock-outs in YAC-1 T cell lines via CRISPR/Cas9 gene editing (Fig. 6B).
Fig. 6.
cGAS and TREX1 modulate sensing of plasmid DNA and of ISD, respectively. (A) Immunoblotting of indicated cell lysates using anti-mouse cGAS and anti-MAPK antibodies. Lysates of parental HEK293T and mouse cGAS-expressing HEK293T serve as specificity controls for the anti-mouse cGAS antibody. (B) Immunoblotting of lysates of parental and cGAS KO YAC-1 T cell lines using indicated antibodies. (C) Parental, cGAS KO and TREX1 KO YAC-1 T cells were either mock-electroporated or electroporated with plasmid DNA (Left), or electroporated either with c-di-UMP or cGAMP (Right). Cultures were monitored at indicated time points postchallenge for relative Ifit1 and Mx2 mRNA expression by qRT-PCR (Top and Middle) and release of bioactive type I IFN using a luminometric MEF-based assay (Bottom). (D) Indicated YAC-1 T cell lines were mock-electroporated or electroporated with short ISD and monitored at 6 h postchallenge for Ifit1 and Mx2 mRNA expression by qRT-PCR. P values <0.05 were considered significant (*) and <0.01 very significant (**); n.s. = not significant (≥0.05).
In mouse YAC-1 T cells, the induction of Ifit1 and Mx2 mRNA expression upon plasmid DNA challenge was almost entirely dependent on endogenous cGAS expression (Fig. 6 C, Top and Middle). The remaining four cell lines failed to induce significant Ifit1 mRNA expression upon plasmid DNA electroporation (SI Appendix, Fig. S7A). Among the parental T cell line panel, only YAC-1 T cells secreted detectable amounts of bioactive type I IFN in the culture supernatant upon plasmid DNA electroporation, and this process occurred in an entirely cGAS-dependent manner (Fig. 6 C, Bottom and SI Appendix, Fig. S7B). TREX1 KO and parental YAC-1 T cells shared a common Ifit1 and Mx2 mRNA induction pattern upon DNA challenge, and differed only by a slightly elevated, but very transient (only at 6 h) secretion of bioactive type I IFN in the culture supernatant of TREX1 KO cells. These results probably reflect the overall resistance of the electroporated circular plasmid DNA to the exonuclease activity of TREX1 (41). All tested mouse T cell lines reacted to cGAMP by up-regulation of Ifit1 mRNA expression (SI Appendix, Fig. S7C) and most of them by release of bioactive type I IFN. Paralleling our results obtained in cGAMP-transfected human PM1 T cells (Fig. 4 C, Right), cGAMP challenge of cGAS KO YAC-1 T cells was followed by an enhanced Ifit1 mRNA expression as compared to parental YAC-1 T cells (Fig. 6 C, Top Right). In contrast, expression of Mx2 mRNA and release of bioactive IFN were of similar quantities in cGAS KO and parental YAC-1 T cells (Fig. 6 C, Middle and Bottom Right), suggesting that cGAS expression does not grossly modulate the magnitude of cGAMP-induced IFN production, but may potentially enforce IRF3-mediated responses. TREX1 KO did not display overt changes in any readout monitored after cGAMP challenge compared to parental cells.
While eletroporation of parental and cGAS KO YAC-1 T cells with short ISD did not induce detectable changes of Ifit1 and Mx2 mRNA expression as compared to mock treatment, TREX1 KO clearly augmented cell-intrinsic immune responses (Fig. 6D) in accordance with our findings in primary mouse T cells (Fig. 1G) and in line with the predominant exonuclease activity of TREX1 (41).
Conclusively, in analogy to human PM1 T cells, YAC-1 T cells express endogenous cGAS and sense plasmid DNA in a cGAS-dependent manner. In addition, TREX1 prevents sensing of short ISD, while it does not prevent sensing of circular and/or long DNA species.
Lack of cGAS-Mediated Innate Immune Sensing of HIV-1 and MLV Transduction in Mouse YAC-1 T Cells.
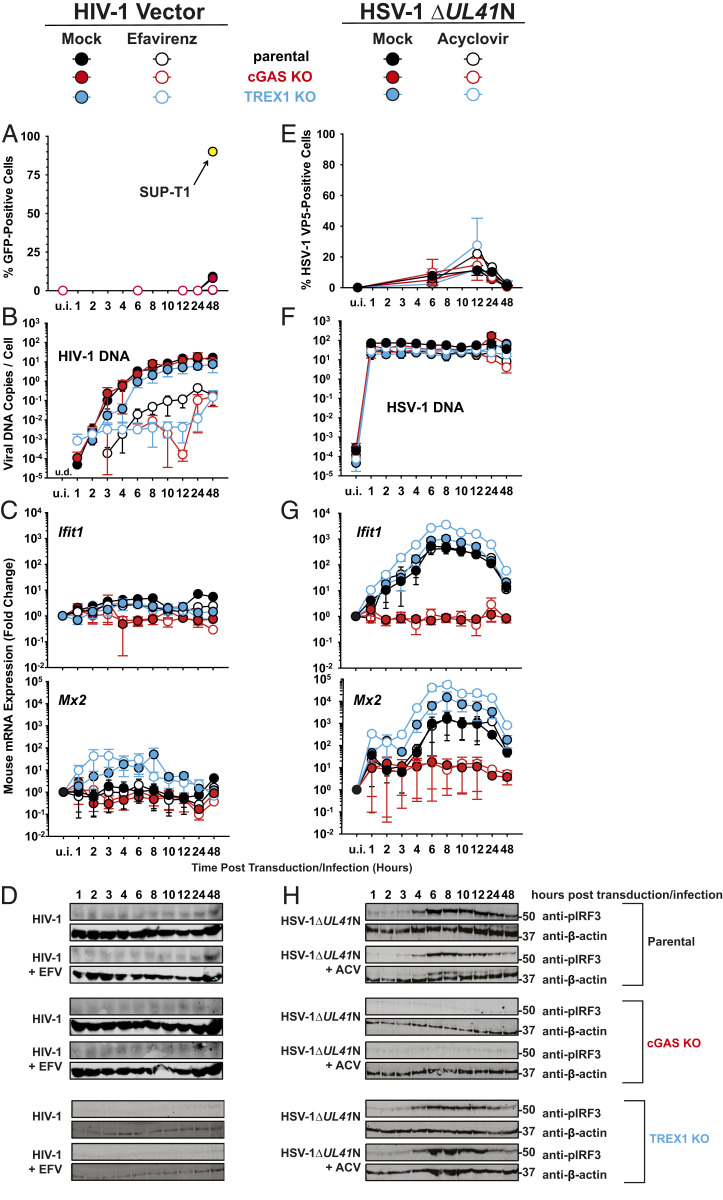
Lentiviral transduction of YAC-1 T cells yielded very low GFP positivity (Fig. 7A), in agreement with their documented block at the level of nuclear import/integration in mouse T cells (37–40). A parallel transduction of human SUP-T1 T cells confirmed the infectivity of the used lentiviral stock. Despite the occurrence of EFV-sensitive reverse transcription, reaching up to 20 HIV-1 DNA copies per cell (Fig. 7B), Ifit1 and Mx2 mRNA expression hardly increased upon transduction (Fig. 7C). In addition, phosphorylation of IRF3 was undetectable in transduced YAC-1 T cells (Fig. 7D). All values obtained from cGAS KO T cells equaled those obtained for parental T cells, supporting the idea that HIV-1 transduction does not induce cGAS-dependent type I IFN responses in T cells. TREX1 KO did not result in any phenotypic changes, except a mild, but EFV-insensitive, trend toward higher Mx2, but not Ifit1, mRNA induction.
Fig. 7.
Lack of cGAS-mediated innate immune sensing of HIV-1 and MLV transduction in mouse YAC-1 T cells. (A–D) Parental, cGAS KO, and TREX1 KO T cell lines were inoculated with VSV-G HIV-1 GFP vectors in the absence and presence of EFV and monitored, at indicated time points, for: Reporter GFP expression by FACS analysis (A), de novo synthesis of HIV-1 late RT products by absolute qPCR (B), relative expression of Ifit1 and Mx2 by qRT-PCR (C), and phosphorylated IRF3 by immunoblotting using a phospho-IRF3 antibody (D). (E–H) Parental, cGAS KO, and TREX1 KO T cell lines were inoculated with HSV-1 ΔUL41N in the absence and presence of ACV and monitored, at indicated time points, for: HSV-1 VP5 capsid protein expression by intracellular immunostaining followed by FACS analysis (E), genomic HSV-1 DNA copy numbers by absolute qPCR (F), relative expression of Ifit1 and Mx2 by qRT-PCR (G), phosphorylated IRF3 by immunoblotting using a phospho-IRF3 antibody (H).
HSV-1 ΔUL41N inoculation of parental YAC-1 T cells resulted in 10% VP5-positive cells (Fig. 7E) and the delivery of up to 100 copies of HSV-1 genomes per cell (Fig. 7F), which was accompanied by an up to 500-fold and 1,500-fold up-regulation of Ifit1 and Mx2 mRNA expression, respectively (Fig. 7G), and a clear induction of phosphorylation of IRF3 (Fig. 7H). Again, those responses were insensitive to ACV, but strictly cGAS-dependent (Fig. 7 G and H), suggesting that the incoming HSV-1 genome or a DNA replication-independent PAMP or DAMP is sensed by cGAS. HSV ΔUL41N-inoculated TREX1 KO T cells displayed a slightly enhanced induction of Ifit1 and Mx2 mRNA expression. Finally, transduction of YAC-1 T cells with a cognate retrovirus, murine leukemia virus (MLV) pseudotyped with VSV-G, failed to trigger detectable innate immune responses, despite robust transduction efficiency (SI Appendix, Fig. S8).
Discussion
The cGAS/STING-mediated DNA sensing pathway is implicated in surprisingly diverse biological processes in different cell types, ranging from antiviral and antimicrobial defense, autoimmunity, sensing of endogenous retroviruses, senescence, DNA repair, and even to inflammation after myocardial infarction (reviewed in ref. 42). Viruses which replicate via DNA or DNA intermediates are generally considered as likely candidates prone to cGAS sensing. In addition, viral infection-caused stress responses may trigger release of mitochondrial DNA, which may result in cGAS-dependent responses to both DNA- and RNA-viral infections (43). Consequently, many viruses have evolved to prevent recognition by cGAS (5, 6).
The initial goal of our study was to define whether T cells contribute to sensing of DNA PAMPs or DAMPs in the context of exogenous DNA challenge and of viral infections. Activated CD4+ T cells are the predominant target cell type of productive HIV-1 infection and support the de novo synthesis of reverse transcription products more efficiently than resting CD4+ T cells, macrophages, and dendritic cells, probably due to reasons including optimal intracellular dNTP levels. Furthermore, human cGAS is expressed in activated T cells (14, 17), as opposed to resting T cells (14, 17). Although not belonging to classical antigen-presenting cells, we wondered whether T cells contribute to the immune system’s defense against viral infection via cGAS/STING.
A first interesting observation was the markedly reduced and enhanced level of Ifit1 and Mx2 mRNA expression in mouse CD4+ T cells devoid of cGAS and TREX1 expression, respectively. These results indicate a basal catalytic activity of cGAS, even in uninfected T cells, which has been also proposed for other cell types, including bone marrow-derived mouse macrophages (44). Basal activity of cGAS has been suggested to be triggered when cGAS is expressed at high levels (1), by binding to DNA originating from endogenous retroviruses and by the release of mitochondrial DNA (45, 46). cGAS activity is counteracted by TREX1 that degrades cytosolic DNA and, thus, precludes interaction of DNA with cGAS (3). The steady-state cGAS activity might therefore represent an important innate barrier to any invading pathogen that is sensitive to proteins expressed in the context of basal cGAS stimulation, since it may ensure maintenance of a basal antiviral state (44).
The ability of activated CD4+ T cells to respond to electroporated plasmid DNA and cGAMP indicated that this cell type has a functional DNA sensing machinery, which, as shown in mouse T cells, is cGAS-dependent. On the contrary, transfection of short ISD of 45 base pairs length failed to induce detectable responses. This is in line with cGAS′s reported ability to sense DNA in a length-dependent manner and to form intracellular cGAS protein-DNA ladder-like structures, thus requiring longer DNA molecules, and potentially cellular cofactors (47). On the contrary, in vitro stimulation of purified cGAS was reported to be effective with DNA fragments as small as 10–20 base pairs, at least when high concentration of both cGAS and DNA are present (47). Furthermore, our experiments in mouse CD4+ T cells suggest that transfected ISDs might be prone to TREX1-mediated nucleic acid cleavage, since a detectable, but moderate, induction of Ifit1 and Mx2 mRNA expression was detected in TREX1 KO, but not in WT and cGAS KO cells. Together, this suggests that the discrepancy in the reported results regarding the response to ISD challenge in vitro and in living cells might be partially explained by the activity of TREX1.
In untreated CD4+ T cells infected with HIV-1Ba-L, intracellular p24 capsid positivity and quantities of reverse transcription products peaked 6 d postinfection, confirming productive infection at a level that is typically reached in primary CD4+ T cells infected ex vivo with authentic HIV-1 (48). The biphasic IFIT1 mRNA expression up-regulation observed in this context at early (2–3 h) and late (10–13 d) time points postinoculation could neither be attributed to sensing of de novo-synthesized viral DNA nor to productive infection, since EFV treatment did not alter the expression profile of IFIT1. These results were corroborated in a global transcriptomic approach, which indicated that IRF3-transactivated and IFN-related genes are generally not induced in productively infected cells at multiple time points, as compared to EFV-treated cultures. The lack of impact of both pharmacological and genetic destabilization of the viral capsid in T cells contrasts findings in macrophages and THP-1 cells (13, 24, 36), suggesting that viral DNA escaping the protective capsid is still unaccesssible to or quantitatively insufficient for cGAS-mediated sensing. An interesting hypothesis for this apparent cell type-specific phenomenon may be differential levels of TREX1 expression. Future studies are required to define the contribution of TREX1 to avoidance of HIV-1 sensing in T cells and macrophages. Short abortive RT products most likely resemble ISD, an experimental stimulus that triggered an immune response only in TREX1 KO T cells. Along this line, siRNA-mediated threefold reduction of TREX1 mRNA expression resulted in a more potent induction of IFN genes in macrophages than in T cells (4). Nuclear export of unspliced viral RNA has been proposed to induce sensing of viral RNAs (49, 50). While our lentiviral virus particle approach is not an appropriate system to test this aspect in T cells based on a simple transfer vector, we believe that the absence of RT inhibitor-sensitive immune responses in HIV-1-Ba-L–infected T cells argues against this possibility regarding induction of the tested ISGs. We conclude that HIV-1 can efficiently spread in activated CD4+ T cells, the most susceptible target cell type, without provoking a notable type I IFN-mediated antiviral response. In stark contrast, efficient and ACV-insensitive induction of expression of IFIT1, MX2, and IFN-β mRNA in HSV-1 ΔUL41N-infected cells suggest that the incoming HSV-1 genomic DNA is prone to sensing in CD4+ T cells. Interestingly, while the majority of HSV-1 DNA genomes are believed to be enclosed within a protective capsid, their leakage has already been suggested to occur in myeloid cells (28–30), and our data support the idea that this phenomenon occurs also in T cells.
The study by Vermeire et al. reporting cGAS-dependent sensing of HIV-1 infection postintegration in human CD4+ T cells (14) is reminiscent of a reported sensing of HIV-1/Vpx in dendritic cells (11) and appears to contradict our findings. The reasons for these discrepancies may include the use of different virus production schemes and different virus strains. We chose HIV-1Ba-L because its production through serial passaging excludes the possibility of contaminating HIV-1 plasmid DNA in virion preparations and because it allowed us to monitor infection for up to 13 d in the absence of extensive cell death. In contrast, Vermeire et al. generated virus stocks by calcium phosphate transfection and used, for the majority of experiments, the X4-tropic NL4.3 that, in our hands, induces massive death of CD4+ T cells 2–3 d postinfection, a well-documented property of many X4-tropic HIV-1 strains. Whether the differential ability of NL4.3 and Ba-L to induce cell stress and death upon productive infection explain, at least partially, the different observations remains to be investigated.
Screening human and mouse cell lines revealed a surprising heterogeneity in cGAS expression among tested T cells. We therefore advise that DNA sensing studies in T cell lines should carefully validate the cGAS expression profile in the chosen T cell system. The absence of cGAS-dependent cellular responses upon productive HIV-1 and MLV vector transduction of PM1 and YAC-1 T cells, despite their full capability to sense DNA via cGAS, is consistent with the idea that HIV-1 infection either evades or antagonizes this DNA sensing machinery. Since no accessory HIV-1 proteins are expressed by the lentiviral vectors employed, absence of cGAS-dependent responses in transduced cells let us hypothesize that cGAS is not activated, neither in the context of spreading infection nor of lentiviral and retroviral transduction. Along this line, superinfection of lentivirally transduced cells with HSV-1 did not alleviate the induction of HSV-triggered IFIT1 mRNA expression, arguing against a putative ability of lentiviruses to directly interfere with the cGAS-dependent signaling pathway. During lentiviral and gammaretroviral infection, a single full-length DNA genome is produced progressively during capsid migration to the nucleus and is then integrated into the cellular chromosome; no DNA replication during the rest of the replication cycle is required. In contrast, herpesviral genomes are 20-fold larger than HIV-1 genomes, and, in addition to being inevitably present as integral part of incoming virions, are also generated anew in large quantities in the context of progeny virus production. Thus, retroviruses including HIV-1 may have evolved a replication strategy that reduces the abundance of cytoplasmic DNA intermediates to a minimum, thus avoiding susceptibility to cGAS-mediated sensing in infected T cells.
Methods
Animals.
cGAS KO mice on the C57BL/6 background (Mb21d1tm1d(EUCOMM)Hmgu/J) were kindly obtained from Charles Rice. TREX1 KO mice were kindly obtained from Tomas Lindahl (Francis Crick Institute, London). C57BL/6 (WT), cGAS KO and TREX1 KO mice were bred under specific pathogen-free conditions. Mouse experimental work was carried out using 8- to 14-wk-old mice in compliance with regulations of the German animal welfare law.
Cell Lines and Primary Cells.
A3.01, SUPT1, Jurkat, CEM T cells were purchased from American Type Culture Collection. PM1 T cells were obtained from the NIH AIDS Reagent Program. S1A.TB, R1.1, TIMI.4 were kindly obtained from Oliver Keppler. Parental YAC-1 and L1210 cells were kindly obtained from Roland Jacobs, Hanover Medical School. CRISPR/Cas9-mediated knockouts were generated by transient electroporation of an EF1alpha-Cas9-2A-EGFP/U6-sgRNA expression plasmid, followed by FACS-sorting and single-cell cloning by limiting dilution. Parental HEK293T cells and HEK293T cells expressing cGAS (clone 17) are described in ref. 51. Parental THP-1, cGAS KO THP-1, and Jurkat T cells stably expressing WT cGAS and cGAS (G212A/S213A) are described in ref. 16. Withdrawal of blood samples from healthy humans and cell isolation were conducted with approval of the full study of the local ethics committees (ethical review committee of Hanover Medical School, vote ID 3025-2016; Ethical review committee of Charité–Universitätsmedizin Berlin, vote ID EA4/167/19). Purification of human CD4+ T cells was performed by negative selection.
Single-cell suspensions of mouse splenocytes were prepared by pushing spleen tissue pieces through a 70-μm pore size nylon mesh screen (Fisher Scientific). Washed splenocytes were subjected to CD4+ T cell isolation using the EasySep Mouse CD4+ T-Cell Isolation kit (Stemcell Technologies).
Viruses.
HIV-1Ba-L was obtained from the NIH AIDS Reagent Program and propagated on PM1 T cells. HSV-1 ΔUL41N (HSV-1(KOS) UL41NHB) was kindly provided by David A. Leib (52). It encodes a truncated version of pUL41, which fails to induce the degradation of cellular mRNAs and is unable to counteract cGAS. To prepare concentrated stocks, extracellular virions were pelleted from the medium of cells infected with a multiplicity of infection of 0.01 pfu per cell for 3 d (53, 54). Virus stocks were plaque-titrated on Vero cells (53, 55). To determine the genome/pfu ratio of HSV-1 stocks, we measured the number of HSV-1 genomes by qPCR as described previously (54, 56).
Lentiviral and Gammaretroviral Particles.
VSV-G–pseudotyped HIV-1 GFP particles were generated by calcium phosphate-based transfection of HEK293T cells with the packaging plasmid pCMV ΔR8.91 (57) expressing WT capsid or CA(P90A) or pCMV ΔR8.2 (CA N74D), the GFP-encoding transfer plasmid pHR.GFP (58), and the pCMV-VSV-G plasmid (59). VSV-G–pseudotyped MLV GFP particles were generated by calcium phosphate-based transfection of HEK293T cells with the packaging plasmid pCMVi gag-pol (60), the GFP-encoding transfer plasmid pSER S11 SF GFP (61), and the pCMV-VSV-G plasmid (59).
Intracellular HIV-1 p24CA and HSV-1 VP5 Immunostaining.
PBS-washed cells were paraformaldehyde (PFA)-fixed and immunostained for intracellular HIV-1 p24CA using fluorescein isothiocyanate (FITC)-conjugated mAb KC57 (Beckman Coulter) in 0.1% Triton in phosphate-buffered saline (PBS). VP5 immunostaining was performed with rabbit anti-HSV-1 VP5 (SY4563; ref. 62) and an appropriate fluorochrome-conjugated secondary antibody in 0.1% Triton in PBS.
qPCR of Viral DNA.
DNA extraction from cells were performed with Maxwell 16 Blood DNA purification kit (Promega). Quantification of absolute copy numbers of HIV-1 late RT products was performed with the 7500 Fast Real-Time PCR System (Applied Biosystems) using a published Taq-Man–based PCR (63). The number of HSV-1 genomes was quantified as described previously (54, 56) using the LightCycler FastStart DNA Master HybProbe kit (Roche Diagnostics).
Human and Mouse Type I IFN Bioactivity Assays.
Secretion of human type I IFN bioactivity was quantified using the human reporter cell line HL116 that carries the luciferase gene under the control of the IFN-inducible 6–16 promoter (ref. 64, a kind gift from Sandra Pellegrini, Institut Pasteur, France). Secretion of murine, bioactive type I IFN was quantified using the mouse reporter cell line MEF that expresses the luciferase gene under the control of the mouse Mx2 promoter (ref. 65, a kind gift of Mario Köster, Helmholtz Center for Infection Research, Brunswick, Germany).
Supplementary Material
Acknowledgments
We thank Charles Rice (The Rockefeller University, New York) and Tomas Lindahl for providing the cGAS KO and TREX1 KO mouse lines, respectively. We thank Sandra Pelligrini (Institut Pasteur, Paris), Frank Pessler (TWINCORE, Hanover, Germany), and Mario Köster for the kind gift of the HL116 cell line, THP-1 cells, and MEF-luciferase cells, respectively. We thank Oya Cingöz (Robert Koch Institute, Berlin) for kindly providing the ΔR8.92 CA (N74D) plasmid. We thank Oliver Dittrich-Breiholz (Hanover Medical School, Hanover, Germany) and Victor Tarabykin (Charité - Universitätsmedizin Berlin, Berlin) for granting access to the Step One Plus Real-Time PCR System and the ABI7500 Real-Time PCR System, respectively. We thank the NIH AIDS Research & Reference Reagent Program for providing essential reagents. We thank Thomas Pietschmann (TWINCORE, Hanover, Germany) and Christian Drosten (Charité - Universitätsmedizin Berlin, Berlin) for constant support. This work was supported by a postdoctoral fellowship from the Foundation Ernst & Margarete Wagemann (to A.D.); by a DAAD thesis completion fellowship (to A.P.); funding of Joint French-German Project cGAS-VAC Project 406922110 (to U.K.); funding from Deutsche Forschungsgemeinschaft (DFG) for Germany’s Excellence Strategy, EXC 2155, Project 390874280 (to B.S.); DFG Collaborative Research Centre CRC900 “Microbial Persistence and its Control” Project 158989968, project C2 (to B.S.) and project C8 (to C.G.); and DFG SPP Priority Programm 1923 “Innate Sensing and Restriction of Retroviruses” GO2153/4 Grant (to C.G.); funding from Boehringer Ingelheim Foundation (Exploration Grant) (to C.G.); and funding of the Helmholtz Center for Infection Research and Berlin Institute of Health (to C.G.).
Footnotes
Competing interest statement: W.B. is an employee of IFM Therapeutics.
This article is a PNAS Direct Submission.
Data deposition: The RNA-sequencing (seq) data discussed in this publication has been deposited in National Center for Biotechnology Information’s Gene Expression Omnibus (GEO), https://www.ncbi.nlm.nih.gov/geo (accession no. GSE150753).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002481117/-/DCSupplemental.
Data Availability.
The RNA-sequencing data discussed in this publication have been deposited in National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) (66), https://www.ncbi.nlm.nih.gov/geo (accession no. GSE150753).
All relevant data and protocols are included in the paper. Requests for reagents should be directed to C.G.
Additional Materials and Methods are posted in SI Appendix.
References
- 1.Ablasser A., et al. , cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 498, 380–384 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao P., et al. , Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell 153, 1094–1107 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ablasser A., et al. , TREX1 deficiency triggers cell-autonomous immunity in a cGAS-dependent manner. J. Immunol. 192, 5993–5997 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Yan N., Regalado-Magdos A. D., Stiggelbout B., Lee-Kirsch M. A., Lieberman J., The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat. Immunol. 11, 1005–1013 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma Z., et al. , Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses. Proc. Natl. Acad. Sci. U.S.A. 112, E4306–E4315 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stempel M., Chan B., Brinkmann M. M., Coevolution pays off: Herpesviruses have the license to escape the DNA sensing pathway. Med. Microbiol. Immunol. (Berl.) 208, 495–512 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Cingöz O., Goff S. P., HIV-1 is a poor inducer of innate immune responses. MBio 10, e02834-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao D., et al. , Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 341, 903–906 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar S., Morrison J. H., Dingli D., Poeschla E., HIV-1 activation of innate immunity depends strongly on the intracellular level of TREX1 and sensing of incomplete reverse transcription products. J. Virol. 92, e00001-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siddiqui M. A., et al. , A novel phenotype links HIV-1 capsid stability to cGAS-mediated DNA sensing. J. Virol. 93, e00706-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lahaye X., et al. , The capsids of HIV-1 and HIV-2 determine immune detection of the viral cDNA by the innate sensor cGAS in dendritic cells. Immunity 39, 1132–1142 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Yoh S. M., et al. , PQBP1 is a proximal sensor of the cGAS-dependent innate response to HIV-1. Cell 161, 1293–1305 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rasaiyaah J., et al. , HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature 503, 402–405 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vermeire J., et al. , HIV triggers a cGAS-dependent, Vpu- and Vpr-regulated type I interferon response in CD4+ T cells. Cell Rep. 17, 413–424 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Berg R. K., et al. , T cells detect intracellular DNA but fail to induce type I IFN responses: Implications for restriction of HIV replication. PLoS One 9, e84513 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu S., et al. , cGAS-mediated innate immunity spreads intercellularly through HIV-1 env-induced membrane fusion sites. Cell Host Microbe 20, 443–457 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Cerboni S., et al. , Intrinsic antiproliferative activity of the innate sensor STING in T lymphocytes. J. Exp. Med. 214, 1769–1785 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grandvaux N., et al. , Transcriptional profiling of interferon regulatory factor 3 target genes: Direct involvement in the regulation of interferon-stimulated genes. J. Virol. 76, 5532–5539 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goffinet C., et al. , Primary T-cells from human CD4/CCR5-transgenic rats support all early steps of HIV-1 replication including integration, but display impaired viral gene expression. Retrovirology 4, 53 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koh K. B., Fujita M., Adachi A., Elimination of HIV-1 plasmid DNA from virus samples obtained from transfection by calcium-phosphate co-precipitation. J. Virol. Methods 90, 99–102 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Matreyek K. A., Yücel S. S., Li X., Engelman A., Nucleoporin NUP153 phenylalanine-glycine motifs engage a common binding pocket within the HIV-1 capsid protein to mediate lentiviral infectivity. PLoS Pathog. 9, e1003693 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi J., Zhou J., Shah V. B., Aiken C., Whitby K., Small-molecule inhibition of human immunodeficiency virus type 1 infection by virus capsid destabilization. J. Virol. 85, 542–549 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Márquez C. L., et al. , Kinetics of HIV-1 capsid uncoating revealed by single-molecule analysis. eLife 7, e34772 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sumner R. P., et al. , Disrupting HIV-1 capsid formation causes cGAS sensing of viral. bioRxiv:10.1101/838011 (11 November 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aubert M., Yoon M., Sloan D. D., Spear P. G., Jerome K. R., The virological synapse facilitates herpes simplex virus entry into T cells. J. Virol. 83, 6171–6183 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jerome K. R., et al. , Herpes simplex virus inhibits apoptosis through the action of two genes, Us5 and Us3. J. Virol. 73, 8950–8957 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lahmidi S., Strunk U., Smiley J. R., Pearson A., Duplay P., Herpes simplex virus 1 infection of T cells causes VP11/12-dependent phosphorylation and degradation of the cellular protein Dok-2. Virology 511, 66–73 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Horan K. A., et al. , Proteasomal degradation of herpes simplex virus capsids in macrophages releases DNA to the cytosol for recognition by DNA sensors. J. Immunol. 190, 2311–2319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rasmussen S. B., et al. , Activation of autophagy by α-herpesviruses in myeloid cells is mediated by cytoplasmic viral DNA through a mechanism dependent on stimulator of IFN genes. J. Immunol. 187, 5268–5276 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun C., et al. , Cellular requirements for sensing and elimination of incoming HSV-1 DNA and capsids. J. Interferon Cytokine Res. 39, 191–204 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Su C., Zheng C., Herpes simplex virus 1 abrogates the cGAS/STING-mediated cytosolic DNA-sensing pathway via its virion host shutoff protein, UL41. J. Virol. 91, e02414-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trotard M., et al. , Sensing of HIV-1 infection in tzm-bl cells with reconstituted expression of STING. J. Virol. 90, 2064–2076 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee K., et al. , Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe 7, 221–233 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price A. J., et al. , CPSF6 defines a conserved capsid interface that modulates HIV-1 replication. PLoS Pathog. 8, e1002896 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaller T., et al. , HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog. 7, e1002439 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bulli L., et al. , Complex interplay between HIV-1 capsid and MX2-independent alpha interferon-induced antiviral factors. J. Virol. 90, 7469–7480 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baumann J. G., et al. , Murine T cells potently restrict human immunodeficiency virus infection. J. Virol. 78, 12537–12547 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tervo H. M., Goffinet C., Keppler O. T., Mouse T-cells restrict replication of human immunodeficiency virus at the level of integration. Retrovirology 5, 58 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsurutani N., et al. , Nuclear import of the preintegration complex is blocked upon infection by human immunodeficiency virus type 1 in mouse cells. J. Virol. 81, 677–688 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang J. X., Diehl G. E., Littman D. R., Relief of preintegration inhibition and characterization of additional blocks for HIV replication in primary mouse T cells. PLoS One 3, e2035 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stetson D. B., Ko J. S., Heidmann T., Medzhitov R., Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134, 587–598 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Motwani M., Pesiridis S., Fitzgerald K. A., DNA sensing by the cGAS-STING pathway in health and disease. Nat. Rev. Genet. 20, 657–674 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Sun B., et al. , Dengue virus activates cGAS through the release of mitochondrial DNA. Sci. Rep. 7, 3594 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schoggins J. W., et al. , Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505, 691–695 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maekawa H., et al. , Mitochondrial damage causes inflammation via cGAS-STING signaling in acute kidney injury. Cell Reports 29, 1261–1273.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 46.West A. P., et al. , Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520, 553–557 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andreeva L., et al. , cGAS senses long and HMGB/TFAM-bound U-turn DNA by forming protein-DNA ladders. Nature 549, 394–398 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Cooper A., et al. , HIV-1 causes CD4 cell death through DNA-dependent protein kinase during viral integration. Nature 498, 376–379 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Akiyama H., et al. , HIV-1 intron-containing RNA expression induces innate immune activation and T cell dysfunction. Nat. Commun. 9, 3450 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCauley S. M., et al. , Intron-containing RNA from the HIV-1 provirus activates type I interferon and inflammatory cytokines. Nat. Commun. 9, 5305 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ablasser A., et al. , Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature 503, 530–534 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strelow L. I., Leib D. A., Role of the virion host shutoff (vhs) of herpes simplex virus type 1 in latency and pathogenesis. J. Virol. 69, 6779–6786 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sodeik B., Ebersold M. W., Helenius A., Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 136, 1007–1021 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Döhner K., Radtke K., Schmidt S., Sodeik B., Eclipse phase of herpes simplex virus type 1 infection: Efficient dynein-mediated capsid transport without the small capsid protein VP26. J. Virol. 80, 8211–8224 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Döhner K., et al. , Function of dynein and dynactin in herpes simplex virus capsid transport. Mol. Biol. Cell 13, 2795–2809 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Engelmann I., et al. , Rapid quantitative PCR assays for the simultaneous detection of herpes simplex virus, varicella zoster virus, cytomegalovirus, Epstein-Barr virus, and human herpesvirus 6 DNA in blood and other clinical specimens. J. Med. Virol. 80, 467–477 (2008). [DOI] [PubMed] [Google Scholar]
- 57.Zufferey R., Nagy D., Mandel R. J., Naldini L., Trono D., Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 15, 871–875 (1997). [DOI] [PubMed] [Google Scholar]
- 58.Miyoshi H., Takahashi M., Gage F. H., Verma I. M., Stable and efficient gene transfer into the retina using an HIV-based lentiviral vector. Proc. Natl. Acad. Sci. U.S.A. 94, 10319–10323 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stewart S. A., et al. , Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9, 493–501 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fletcher A. J., et al. , Trivalent RING assembly on retroviral capsids activates TRIM5 ubiquitination and innate immune signaling. Cell Host Microbe 24, 761–775.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schambach A., et al. , Overcoming promoter competition in packaging cells improves production of self-inactivating retroviral vectors. Gene Ther. 13, 1524–1533 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Döhner K., et al. , Importin α1 is required for nuclear import of herpes simplex virus proteins and capsid assembly in fibroblasts and neurons. PLoS Pathog. 14, e1006823 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goffinet C., Allespach I., Keppler O. T., HIV-susceptible transgenic rats allow rapid preclinical testing of antiviral compounds targeting virus entry or reverse transcription. Proc. Natl. Acad. Sci. U.S.A. 104, 1015–1020 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Uzé G., et al. , Domains of interaction between alpha interferon and its receptor components. J. Mol. Biol. 243, 245–257 (1994). [DOI] [PubMed] [Google Scholar]
- 65.Kugel D., Pulverer J. E., Köster M., Hauser H., Staeheli P., Novel nonviral bioassays for mouse type I and type III interferon. J. Interferon Cytokine Res. 31, 345–349 (2011). [DOI] [PubMed] [Google Scholar]
- 66.Edgar R., Domrachev M., Lash A. E., Gene expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-sequencing data discussed in this publication have been deposited in National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) (66), https://www.ncbi.nlm.nih.gov/geo (accession no. GSE150753).
All relevant data and protocols are included in the paper. Requests for reagents should be directed to C.G.
Additional Materials and Methods are posted in SI Appendix.