Significance
Improving cytotoxicity, proliferation, and infiltration ability of CD8+ T cells is critical in T cell immunotherapies against tumors. This study has identified LRCH1 as a negative regulator of LAT-mediated TCR signal transduction, as LRCH1 inhibits LAT signalosome formation and facilitates the endocytosis of LAT on the plasma membrane. LRCH1-deficient CD8+ T cells are more proliferative and effective at pathogen control and tumor elimination. Importantly, CRISPR-Cas9–mediated knockout of LRCH1 in human T cells also increases IFN-γ production, cell proliferation, and migration ability in vitro. These data suggest LRCH1 as a potential target to improve CD8+ T cell responses against tumors and pathogens.
Keywords: LRCH1, LAT, CD8+ T cell, cytotoxicity, migration
Abstract
CD8+ T cells play pivotal roles in eradicating pathogens and tumor cells. T cell receptor (TCR) signaling is vital for the optimal activation of CD8+ T cells. Upon TCR engagement, the transmembrane adapter protein LAT (linker for activation of T cells) recruits other key signaling molecules and forms the “LAT signalosome” for downstream signal transduction. However, little is known about which functional partners could restrain the formation of the LAT signalosome and inhibit CD8+ cytotoxic T lymphocyte (CTL)-mediated cytotoxicity. Here we have demonstrated that LRCH1 (leucine-rich repeats and calponin homology domain containing 1) directly binds LAT, reduces LAT phosphorylation and interaction with GRB2, and also promotes the endocytosis of LAT. Lrch1−/− mice display better protection against influenza virus and Listeria infection, with enhanced CD8+ T cell proliferation and cytotoxicity. Adoptive transfer of Lrch1−/− CD8+ CTLs leads to increased B16-MO5 tumor clearance in vivo. Furthermore, knockout of LRCH1 in human chimeric antigen receptor (CAR) T cells that recognize the liver tumor-associated antigen glypican-3 could improve CAR T cell migration and proliferation in vitro. These findings suggest LRCH1 as a potential translational target to improve T cell immunotherapy against infection and tumors.
CD8+ T cells are key cytotoxic immune cells responsible for the elimination of pathogen-infected cells and cancer cells. Our understanding of T cell receptor (TCR) signaling for T cell activation, migration, proliferation, and differentiation into effector or memory subsets has contributed to therapeutic applications against tumors and pathogens (1). T cells expressing chimeric antigen receptors (CARs; CAR T cells), which combine the antigen-binding property of monoclonal antibodies with the lytic capacity and self-renewal of T cells, have been developed to kill tumor cells independent of the major histocompatibility complex (MHC) and overcome the lack of costimulation by tumor cells. CAR T cell therapy has demonstrated impressive clinical results in eradicating hematologic malignancies, such as CD19 CARs in leukemias. Despite this, CAR T cell infiltration, persistent ability of proliferation, and cytotoxicity in hostile tumor microenvironments are still challenges in the treatment of solid tumors (2). Thus, targeting inhibitory signaling proteins to improve CAR T cell therapy has been recently implicated, such as depleting diacylglycerol kinase (3) and all three NR4A transcription factors NR4A1, NR4A2, and NR4A3 (4, 5).
Upon TCR engagement, CD3 is phosphorylated by the Src family kinase LCK, enabling the recruiting and activation of the tyrosine kinase ZAP70 that in turn phosphorylates LAT (linker for activation of T cells). LAT has no enzymatic or kinase activity but serves as a transmembrane scaffold protein via the multiple tyrosine residues in its cytoplasmic tail. Phosphorylated LAT directly binds to PLC-γ1, GRB2, and GADs (GRB2-related adapter protein), and each of them further recruits other signaling proteins, such as SLP-76, ADAP, and VAV1, to generate a multiprotein complex known as the “LAT signalosome.” The LAT signalosome is indispensable for TCR-induced activation of transcription factors regulating cell proliferation and effector functions (6–9). LAT-deficient cytotoxic T lymphocytes (CTLs) fail to up-regulate FasL and produce interferon γ (IFN-γ) after engagement with target cells and have impaired granule-mediated killing (10). Targeted disruption of the Lat gene in mice causes early arrest of thymocyte development and the absence of mature αβT cells in peripheral lymphoid organs (11). Importantly, patients with defective LAT signaling present from early childhood suffer from combined immunodeficiency and severe autoimmune disease (12). Although the LAT signalosome is critical to favor T cell activation and proliferation, excessive T cell activation can also lead to autoimmune diseases. Therefore, precise control of T cell signaling by both positive and negative regulators is essential to maintain T cell homeostasis. However, only a few indirect negative regulators of the LAT signalosome have been found, such as SHIP-1 (8). A previous study suggests that LAT endocytosis and subsequent degradation provide an efficient way of terminating TCR signaling (13). K52 and K204 in LAT could be ubiquitinated by c-Cbl, followed by rapid internalization of LAT-nucleated signaling clusters (14, 15). Intriguingly, direct negative regulators of the LAT signalosome remain to be discovered.
Our laboratory has recently identified LRCH1 (leucine-rich repeats and calponin homology domain containing 1) as a new binding partner of the guanine nucleotide exchange factor protein DOCK8 in T cells, which interferes with Cdc42 activation and restrains CD4+ T cell migration into the central nervous system to ameliorate the development of experimental autoimmune encephalomyelitis (16). LRCH1 was first reported in a large-scale association analysis of single-nucleotide polymorphisms (SNPs) in knee osteoarthritis (OA) patients, depicting a C/T polymorphism in intron 1 of LRCH1 (rs912428) that may associate with the risk of knee OA (17). However, it remains controversial since other reports suggest no association between the LRCH1 SNP and OA (18, 19). Nevertheless, the functions of LRCH1 and the underlying mechanisms in CD8+ T cells in antiinfection and antitumor responses are still unknown. In this study, we have demonstrated that LRCH1 directly binds LAT to disturb LAT signalosome formation and promote the endocytosis of LAT. LRCH1 deficiency potentiates CD8+ T cell proliferation, migration, and killing ability, which result in an improved protection against bacterial or viral infections, and the clearance of tumors in vivo. This indicates that LRCH1 might be a potential target to improve T cell immunotherapy.
Results
LRCH1 Inhibits CD8+ T Effector Function and Cytotoxicity.
To study the function of LRCH1, Lrch1−/− (knockout; KO) mice were generated by transcription activator-like effector nuclease technology. Lrch1−/− mice were viable and fertile and displayed normal T cell development in the thymus when compared with Lrch1+/+ (wild-type; WT) littermates (SI Appendix, Fig. S1A). Total cell numbers and the percentages of CD11b+ F4/80+ macrophages, B220+ B cells, and CD11c+ dendritic cells (DCs) in the bone marrow or spleen were normal in Lrch1−/− mice (SI Appendix, Fig. S1 B and C). The percentages and numbers of CD8+ T cells in the spleen of Lrch1−/− mice were slightly lower (SI Appendix, Fig. S1D). We next examined the phenotype of Lrch1−/− CD8+ T cells. In response to anti-CD3/CD28 stimulation, Lrch1−/− CD8+ T cells significantly enhanced the production of IFN-γ, granzyme B (GzmB), and perforin at messenger RNA (mRNA) (Fig. 1A) and protein levels (Fig. 1B and SI Appendix, Fig. S1E). Lrch1−/− CD8+ T cells displayed up-regulated expression of Tbx21 and eomesodermin (Eomes) compared with WT CD8+ T cells upon anti-CD3/CD28 stimulation (Fig. 1C). This was consistent with previous findings that the transcription factors T-bet and Eomes are critical for CD8+ T effector function by inducing the expression of IFN-γ, granzyme B, and perforin (20–22).
Fig. 1.
LRCH1 inhibits CD8+ T cell effector function and cytotoxicity. (A and C) qRT-PCR analysis of effector genes Ifng, Gzmb, and Prf1 (A) or transcriptional genes Tbx21 and Eomes (C) in anti-CD3/CD28–stimulated WT and KO CD8+ T cells. (B) Flow cytometry (FACS) analysis of intracellular IFN-γ and GzmB in anti-CD3/CD28–stimulated WT and KO CD8+ T cells; n = 4. (D and E) RNA-sequencing analysis of the differentially expressed genes between anti-CD3/CD28–stimulated WT and KO CD8+ T cells; n = 3. (F and H) The percentages of CD69+ (F; n = 6) and PD-1+ (H; n = 4) CD8+ T cells after coculture with WT splenocytes, which were depleted of CD8+ T cells and pulsed with OVA257–264 peptide. (G) FACS analysis of intracellular IFN-γ in OVA257–264–stimulated OT1-Tg WT and KO CD8+ T cells; n = 4. MFI, median fluorescence intensity. (I) The percentages of apoptotic (Annexin V+) CD8+ T cells after OVA257–264 (OT1+; n = 4; Left) or anti-CD3/CD28 (OT1−; n = 8; Right) stimulation. (J) The in vitro killing assay of OT1-Tg WT and KO CD8+ CTLs. CTLs were incubated with 10 nM OVA257–264–pulsed EL-4 cells at different effector:target (E:T) ratios. Lactate dehydrogenase release was measured to assess the cytotoxic efficiency. (K) The in vivo killing assay of OT1-Tg WT and KO CD8+ CTLs. CTLs (3 × 106) were i.v. injected into wild-type C57BL/6 mice, followed by injection of nonpulsed (CFSEhi) or 10 nM OVA257–264–pulsed (CFSElow) splenocytes (5 × 106). The frequencies of CFSElow cells in the spleen were analyzed by FACS; n = 4. (L) The in vitro killing assay of WT and Lrch1 Tg CD8+ CTLs. (M) The transcription levels of Ifng, Prf1, and Tbx21 in the vector and LRCH1-transduced T8.1 cells after anti-CD3/CD28 stimulation. ns, not significant (P > 0.05); *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (unpaired Student’s t test). Data are representative of three independent experiments (A, C, J, L, and M, mean ± SD; B, F, G–I, and K, mean ± SEM).
In order to obtain a comprehensive understanding of LRCH1 function in CD8+ T cells, RNA sequencing was performed in anti-CD3/CD28–stimulated WT and Lrch1−/− CD8+ T cells. Compared with WT CD8+ T cells, Lrch1−/− CD8+ T cells displayed 214 up-regulated genes and 94 down-regulated genes (twofold change). Besides Ifng, Gzmb, Prf1, and Il2, many other key regulators of chemotaxis, the cell cycle, survival, or metabolism were up-regulated, such as Cxcr3, Cdca7, Bcl2, Tnfrsf9, Myc, Hk2, and Shmt1 (Fig. 1 D and E). Interestingly, Tnfrsf9, which encodes 4-1BB, is an important activation-induced costimulator and is often used as the signaling domain of CAR T cells. In addition, functional pathway enrichment analysis confirmed that multiple signaling pathways were regulated by LRCH1 (SI Appendix, Fig. S2A). qRT-PCR was performed to verify the RNA-sequencing data (SI Appendix, Fig. S2B). Additionally, LRCH1 deficiency enhanced the expression levels of Il17a and Il17f (Fig. 1E and SI Appendix, Fig. S2B). Also, it has been reported that interleukin-17 (IL-17)/IFN-γ double-producing CD8+ T cells exhibit strong antitumor activity (23).
To further investigate how LRCH1 deficiency affects CD8+ T cell cytotoxicity, WT and Lrch1−/− mice were crossed with OT-1 TCR transgenic mice (termed OT1-Tg WT/KO). The percentages and numbers of CD8+ T cells in the spleen of OT1-Tg KO mice were also fewer than those of WT littermates (SI Appendix, Fig. S1F). Splenocytes (deprived of T cells) were used as antigen-presenting cells and pulsed with OVA257–264 peptide, followed by incubation with OT1-Tg WT or KO CD8+ T cells. OT1-Tg KO CD8+ T cells expressed higher levels of the early activation marker CD69 (Fig. 1F) and produced more IFN-γ (Fig. 1G). However, LRCH1 deficiency did not affect surface PD-1 expression (Fig. 1H) or activation-induced cell death (Fig. 1I; Annexin V+ CD8+ T cells). OVA257–264–specific WT/KO CD8+ CTLs were then incubated at different ratios with the targets, namely OVA257–264–pulsed EL-4 tumor cells, and LRCH1 deficiency enhanced the CD8+ T cell-mediated cytotoxicity in vitro (Fig. 1J).
We also measured the in vivo killing ability of CD8+ CTLs. OVA257–264–pulsed splenocytes (i.e., carboxyfluorescein succinimidyl ester [CFSE]low) and nonpulsed splenocytes (i.e., CFSEhigh) were used as target cells, which were intravenously (i.v.) injected at a 1:1 ratio with OT1-Tg WT/KO CD8+ CTLs into the recipient mice. Compared with WT controls, OT1-Tg KO CTL cells killed more OVA257–264–pulsed splenocytes in vivo (Fig. 1K). To further validate our findings, Lrch1 transgenic mice were bred with OT1-Tg mice, and then WT and Lrch1 Tg CD8+ CTLs were generated by OVA257–264–pulsed splenocytes. Lrch1 Tg CD8+ CTLs reduced the in vitro killing ability against OVA257–264–pulsed EL-4 cells (Fig. 1L). Additionally, the transcription levels of Ifng, Prf1, and Tbx21 were markedly decreased in LRCH1-overexpressing T8.1 cells (a murine T cell line) upon anti-CD3/CD28 stimulation (Fig. 1M). The inhibitory effect of LRCH1 in T8.1 cells was stronger than that of Lrch1 Tg CD8+ CTLs (Fig. 1 M and L), and this might be due to the much higher expression levels of LRCH1 in LRCH1-transduced T8.1 cells compared with Lrch1 Tg CD8+ CTLs (SI Appendix, Fig. S1G). Taken together, these results suggest that LRCH1 negatively regulates CD8+ T effector function and cytotoxicity upon TCR stimulation.
To examine whether LRCH1 also suppresses CD4+ T cell function, CD4+ T cells were isolated from WT and KO mice and induced into Th1 and Th2 subsets in vitro. LRCH1 deficiency enhanced the transcription levels of Ifng and Tbx21 in Th0 and Th1 cells, along with increased IFN-γ production (SI Appendix, Fig. S1 H and I). However, LRCH1 deficiency did not influence the differentiation of Th2 cells (SI Appendix, Fig. S1 J and K). These data indicate that LRCH1 can suppress IFN-γ production in both CD4+ and CD8+ T cells.
Lrch1−/− Mice Clear Pathogens More Effectively.
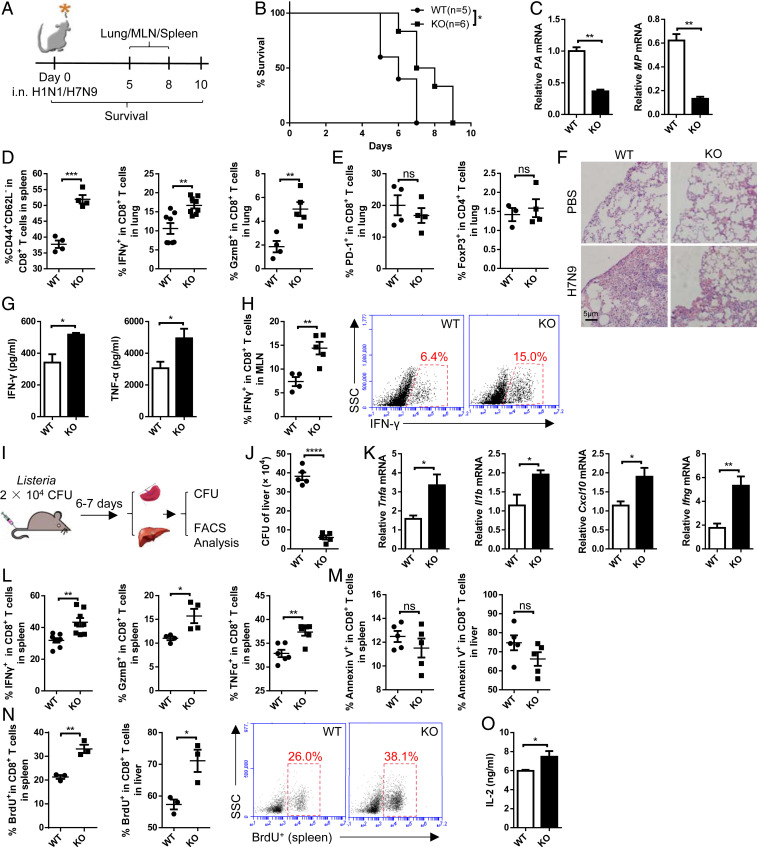
The influenza viral genome has high mutation rates which result in antigen drift and limited vaccine efficacy. CD8+ CTLs that target the conserved proteins of the influenza virus could overcome viral resistance and provide long-term protection for the host (24, 25). DCs in the airways and alveoli express MHC and the costimulation molecules CD80/CD86 for CD8+ T cell priming and activation (26). We found that LRCH1 deficiency did not influence CD80/CD86 expression in resting or lipopolysaccharide (LPS)-stimulated DCs (SI Appendix, Fig. S3A). WT and Lrch1−/− mice were infected intranasally with the PR8 strain of H1N1 influenza A virus or the highly pathogenic avian influenza H7N9 virus (Fig. 2A) and monitored daily for mortality. Lrch1−/− mice increased survival rates upon H1N1 infection (Fig. 2B). The amount of H1N1 viral RNA in the lung, including PA (polymerase acidic protein) (27) and MP (matrix protein) (28), was lower in Lrch1−/− mice (Fig. 2C). Furthermore, higher percentages of CD44+ CD62L− effector CD8+ T cells were observed in Lrch1−/− mice, and Lrch1−/− CD8+ T cells in the lung produced more IFN-γ and GzmB compared with the WT controls (Fig. 2D). LRCH1 deficiency did not affect PD-1 expression on CD8+ T cells or the proportion of CD4+ FoxP3+ regulatory T cells (Treg) upon PR8 infection (Fig. 2E). However, except for Il1b, the transcription levels of Il6, Ifnb, and IFN-stimulated genes including Cxcl10 and Ccl5 (SI Appendix, Fig. S3B) and the frequencies of other subsets of immune cells (SI Appendix, Fig. S3C) in the lung were comparable between WT and KO mice.
Fig. 2.
Lrch1−/− mice clear pathogens more effectively. (A) Schematic of the experimental design of influenza virus infection. MLN, mediastinal lymph node. (B–E) WT and KO mice were intranasally (i.n.) infected with H1N1 influenza strain PR8. (B) Survival rates of WT (n = 5) and KO (n = 6) mice. (C) The relative mRNA levels of the PA and MP genes of PR8 in the lung. (D) The percentage of effector (CD44+ CD62L−) CD8+ T cells in the spleen, and the frequencies of IFN-γ– and GzmB-producing CD8+ T cells in the lung; n = 4 to 8. (E) The percentages of PD-1+ CD8+ T cells and Treg (FoxP3+) cells in the lung (n = 3 or 4). (F–H) WT (n = 4) and KO (n = 5) mice were i.n. infected with the H7N9 influenza virus. (F) Hematoxylin and eosin staining of the lung. (Scale bar, 5 μm.) (G) Enzyme-linked immunosorbent assay of IFN-γ and TNF-α in lung homogenates. (H) The percentage of IFN-γ–producing CD8+ T cells in the MLN. (I–O) WT and KO mice were i.v. infected with L. monocytogenes and killed at day 6 or 7 post infection. (I) Schematic of the experimental design. CFU, colony-forming unit. (J) Liver bacteria titer; n = 5. (K) The mRNA expression levels of Tnfa, Il1b, Cxcl10, and Ifng in the liver; n = 5. (L) FACS analysis of the frequencies of IFN-γ–, GzmB-, and TNF-α–producing CD8+ T cells in the spleen; n = 4 to 8. (M) Apoptotic CD8+ T cells in the spleen and liver; n = 5. (N) BrdU incorporation rates of CD8+ T cells in the spleen and liver; n = 3. (O) IL-2 concentration after HKLM stimulation; n = 7. ns, not significant (P > 0.05); *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (B, log-rank test; C–E, G, H, and J–O, unpaired Student’s t test). Data are representative of three or more independent experiments (mean ± SEM).
Similarly, H7N9-infected Lrch1−/− mice exhibited decreased morbidity (SI Appendix, Fig. S3D) and less severe lung damage (Fig. 2F). Higher levels of IFN-γ and tumor necrosis factor α (TNF-α) were detected in the lung of KO mice (Fig. 2G). Lrch1−/− mice increased the percentages of activated CD8+ T cells (SI Appendix, Fig. S3E), and mediastinal lymph node CD8+ T cells produced more IFN-γ (Fig. 2H) without affecting PD-1 expression (SI Appendix, Fig. S3F). The percentages of Treg cells and DCs in the lung were comparable between WT and KO mice (SI Appendix, Fig. S3 G and H). Although the percentages of macrophages in the lung of KO mice were slightly enhanced (SI Appendix, Fig. S3H), the transcription levels of inflammatory cytokines or chemokines were comparable in the lungs of WT and KO mice (SI Appendix, Fig. S3I).
Apart from viral infection, CD8+ T cells also play vital roles during bacterial infection. We generated systemic Listeria monocytogenes infection in mice (Fig. 2I) (29, 30). Since macrophages ingest Listeria through phagocytosis (31), we first examined whether LRCH1 affected the phagocytic ability of macrophages. WT and KO macrophages engulfed similar numbers of serum-coated latex beads at different incubation times (SI Appendix, Fig. S3J). In addition, the costimulation molecules CD80/CD86 were expressed at similar levels on WT and KO macrophages with or without LPS stimulation (SI Appendix, Fig. S3K). These findings suggest that LRCH1 may not affect macrophage activation and phagocytosis. Seven days post Listeria infection, Lrch1−/− mice displayed markedly enhanced immune responses, as demonstrated by reduced bacterial load in the liver (Fig. 2J) and spleen (SI Appendix, Fig. S3L). Besides, higher levels of Tnfa, Il1b, Cxcl10, and Ifng were detected in the liver of KO mice (Fig. 2K). Additionally, there were more antigen-specific effector CD8+ T cells, with enhanced IFN-γ, GzmB, and TNF-α production, in the spleen and liver of Lrch1−/− mice (Fig. 2L and SI Appendix, Fig. S3M). Similar to H1N1 or H7N9 infection, PD-1 expression on CD8+ T cells and percentages of Treg cells were not affected in Lrch1−/− mice upon Listeria infection (SI Appendix, Fig. S3 N and O). The percentages of Annexin V+ apoptotic CD8+ T cells in the spleen and liver were comparable between WT and KO mice (Fig. 2M). Intriguingly, after Listeria infection, comparable numbers of CD8+ T cells were detected in the spleen and liver of WT and KO mice (SI Appendix, Fig. S3P). Since Lrch1−/− mice displayed lower numbers of CD8+ T cells at resting stage (SI Appendix, Fig. S1D), this prompted us to analyze whether LRCH1 could inhibit CD8+ T cell proliferation upon infection. Indeed, Lrch1−/− mice increased the percentages of bromodeoxyuridine-positive (BrdU+) CD8+ T cells in the spleen and liver upon Listeria infection (Fig. 2N). Lrch1−/− CD8+ T cells also showed increased IL-2 production in response to heat-killed L. monocytogenes (HKLM) restimulation (Fig. 2O). Collectively, Lrch1−/− CD8+ T cells are hypersensitive, showing enhanced activation, proliferation, and IFN-γ production abilities, which lead to better protection against influenza and Listeria infection in vivo. Consistent with the in vitro data (SI Appendix, Fig. S1 H and I), the percentages of activated CD4+ T cells were increased in Lrch1−/− mice, with more IFN-γ production (SI Appendix, Fig. S3 Q and R). Despite the levels of CD80 and CD86 on DCs or macrophages not being affected by LRCH1 deficiency (SI Appendix, Fig. S3T), we detected higher percentages of B cells, natural killer cells, and DCs in the liver of KO mice (SI Appendix, Fig. S3S). These increased immune cells might contribute to the greater impact on Listeria clearance in Lrch1−/− mice.
LRCH1 Directly Binds LAT to Inhibit TCR Signaling and IFN-γ Production.
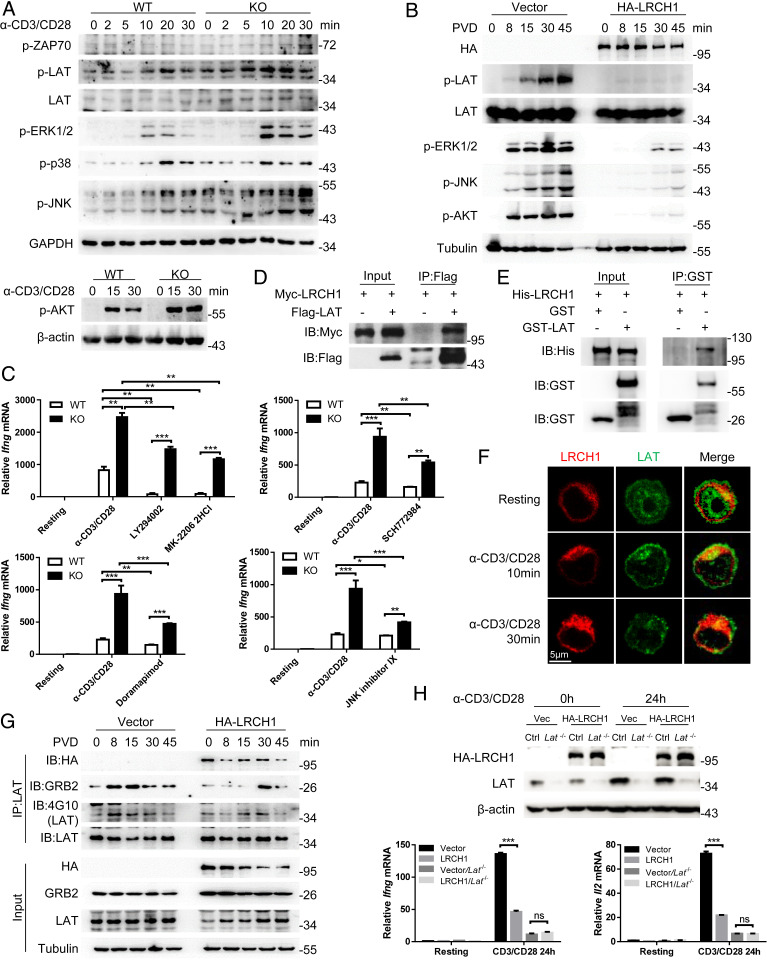
TCR signaling is crucial for the generation of effector CD8+ T cells, and therefore we next stimulated WT and KO CD8+ T cells with anti-CD3/CD28 to analyze the phosphorylation of various signaling molecules in the TCR signaling pathway. LRCH1 deficiency enhanced the phosphorylation levels of LAT, ERK1/2, and JNK (Fig. 3A), while LRCH1 overexpression substantially suppressed ERK1/2 and JNK phosphorylation (Fig. 3B) upon TCR activation. WT and KO CD8+ T cells were then pretreated with inhibitors against PI3K (LY294001), AKT (MK-2206 2HCl), ERK (SCH772984), p38 (doramapimod), or JNK (JNK inhibitor IX) to detect anti-CD3/CD28–induced Ifng transcription. The inhibitors could significantly decrease Ifng transcription in both WT and KO CD8+ T cells, but LRCH1-deficient CD8+ T cells still showed higher Ifng levels (Fig. 3C). This indicates that LRCH1 might inhibit multiple pathways downstream of TCR signaling.
Fig. 3.
LRCH1 directly binds LAT to inhibit TCR signaling and IFN-γ production. (A) Immunoblot analysis of the indicated proteins in anti-CD3/CD28–stimulated CD8+ T cells from WT and KO mice. (B) Immunoblot analysis of the indicated proteins in PVD–stimulated T8.1 cells transduced with the vector control or HA-LRCH1. (C) The transcription level of Ifng after stimulation with anti-CD3/CD28. CD8+ T cells were pretreated with the indicated inhibitors. (D) Coimmunoprecipitation analysis of Myc-tagged LRCH1 and Flag-tagged LAT in 293T cells. IB, immunoblot; IP, immunoprecipitation. (E) In vitro pull down of GST, GST-LAT, and His-LRCH1. (F) Confocal images of the relative localization of LRCH1 and LAT in T8.1 cells transduced with HA-LRCH1. (Scale bar, 5 μm.) (G) Coimmunoprecipitation analysis of endogenous LAT interaction with GRB2 or LRCH1 in PVD-stimulated T8.1 cells used in B. (H) CRISPR-Cas9–mediated knockout efficiency of Lat was detected by immunoblot. The relative mRNA levels of Ifng and Il2 in anti-CD3/CD28–stimulated T8.1 cells used in B and T8.1 cells without LAT. ns, not significant (P > 0.05); *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired Student’s t test). Data are representative of two or more independent experiments (C and H, mean ± SD).
To further investigate which signaling partners bind to LRCH1, we performed antihemagglutinin (anti-HA) immunoprecipitation in T8.1 cells expressing HA-tagged LRCH1 and the vector control, followed by mass spectrometry (MS) analysis. Possible binding partners of LRCH1 are listed in SI Appendix, Table S1 (spectral counts > 8). Consistent with our and others’ reports (16, 32), DOCK8 was detected; interestingly, the transmembrane adapter LAT was identified in our MS data. An immunoprecipitation assay was next performed in 293T cells, and confirmed the binding between Flag-tagged LAT and Myc-tagged LRCH1 (Fig. 3D). To further examine their direct binding, the recombinant proteins His-LRCH1 and glutathione S-transferase (GST)-LAT were purified from Escherichia coli (SI Appendix, Fig. S4A). His-LRCH1 was pulled down by GST-LAT (Fig. 3E). In addition, immunofluorescence staining data showed that anti-CD3/CD28 stimulation enhanced LRCH1 colocalization with LAT (Fig. 3F). The interaction between Myc-LRCH1 and Flag-LAT was also enhanced when tyrosine phosphorylation was maintained by treatment with the phosphatase inhibitor pervanadate (PVD) (SI Appendix, Fig. S4B) (33). The phosphorylated tyrosine residues in LAT, especially Tyr132, 171, 191, and 226, are important docking sites for SH2 domain-containing proteins such as PLC-γ1, GRB2, and GADs (9). We thus generated single or a combination of tyrosine-to-phenylalanine mutations in LAT, which, however, did not affect their interaction with LRCH1 (SI Appendix, Fig. S4C).
Instead, we observed that LRCH1 overexpression reduced LAT interaction with GRB2, and decreased LAT tyrosine phosphorylation levels at the early time points (Fig. 3G). GRB2 is reported to bind to the three tyrosine residues Tyr171, 191, and 226 in LAT to promote LAT oligomerization and optimal T cell activation (9). Consistent with this, we found that LRCH1-overexpressing T cells impaired TCR signaling (Fig. 3B). In addition, LRCH1 itself could form a dimer, which was increased after PVD treatment (SI Appendix, Fig. S4D). This in part explains why LRCH1 was concentrated upon TCR stimulation (Fig. 3F) and increased its interaction with LAT (SI Appendix, Fig. S4B).
To further analyze whether LAT functions together with LRCH1, we used the CRISPR-Cas9 system to knock out Lat in HA-LRCH1–transduced T8.1 cells (Fig. 3H) and analyzed mRNA levels of Ifng and Il2. In LAT-deficient T cells, LRCH1 overexpression no longer inhibited Ifng and Il2 production (Fig. 3H), suggesting that LRCH1 exerts its inhibitory function through LAT.
LRCH1 Facilitates the Endocytosis of Membrane LAT into Lysosomes.
LRCH1 consists of three domains: the N-terminal leucine-rich repeats (LRRs), the intermediate region (IR), and the C-terminal calponin homology (CH) domain. We constructed various LRCH1 truncation mutants (Fig. 4A) and performed coimmunoprecipitation to identify that both the LRR and CH domains could bind LAT, while the IR domain was dispensable (Fig. 4B and SI Appendix, Fig. S5A). Besides, tyrosine mutations in LAT did not influence its interaction with the LRR or the CH domain (SI Appendix, Fig. S4 E and F). Interestingly, the IR interacted with LAT upon PVD treatment in 293T cells (SI Appendix, Fig. S5B) or upon anti-CD3/CD28 stimulation in T8.1 cells (SI Appendix, Fig. S5C). We next investigated how these domains in LRCH1 affected Ifng transcription in anti-CD3/CD28–stimulated T8.1 cells. The ∆LRR mutant and the ∆CH mutant still inhibited Ifng transcription, but deletion of the IR totally relieved the inhibitory effect (Fig. 4C). In addition, the LRR alone inhibited Ifng transcription (Fig. 4C). These findings suggest that the LRR and the inducible interaction between the IR and LAT are indispensable for the inhibitory function of LRCH1.
Fig. 4.
LRCH1 facilitates the endocytosis of membrane LAT into lysosomes. (A) Schematic diagram of human LRCH1 and its truncations. (B) Coimmunoprecipitation of Flag-tagged LAT and the indicated HA-tagged LRCH1 mutants. (C) The transcription level of Ifng in T8.1 cells transduced with the indicated LRCH1 mutants or the vector control after anti-CD3/CD28 stimulation. (D and E) Immunoblot analysis of the indicated proteins in the plasma membrane (PM) and whole-cell lysates after stimulation for the indicated time in primary WT/KO CD8+ T cells (D) or HA-LRCH1–overexpressed T8.1 cells (E). ATP1B3 is a membrane protein used as a loading control. (F) Schematic diagram of membrane compartment isolation by density gradient centrifugation of T8.1 cells transduced with HA-LRCH1 (Left). Immunoblot analysis of LRCH1 and LAT relative locations in different layers at different times (Right). (G) Immunofluorescence analysis of LRCH1, LAT, and LAMP1 colocalization in HA-LRCH1–transduced T8.1 cells. (Scale bar, 1 μm.) ns, not significant (P > 0.05); *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired Student’s t test). Data are representative of two or more independent experiments (C, mean ± SD).
Opposite LAT signalosome formation, LAT can also undergo endocytosis driven by its long cytoplasmic tail that contains several potential endocytic sorting motifs (13). Then, ubiquitination at the lysine residues of LAT by c-Cbl promotes its degradation. Functionally, T cells reconstituted with the LAT mutants resistant to ubiquitylation can elevate TCR signaling (15). Interestingly, tertiary structure analysis by Phyre2 predicts a transmembrane sequence in LRCH1. Besides, LRCH1 could form a dimer (SI Appendix, Figs. S4D and S5 D and E), and self-association of proteins to form dimers is a mechanism commonly employed to regulate protein stability and endocytosis (34). We therefore wondered whether LRCH1 is involved in LAT endocytosis. Anti-CD3/CD28–stimulated WT CD8+ T cells gradually decreased the amount of LAT at the plasma membrane; in contrast, Lrch1−/− CD8+ T cells retained higher levels of LAT at the cell membrane (Fig. 4D). In agreement with this, LRCH1-overexpressing T8.1 cells reduced total and phosphorylated LAT on the plasma membrane (Fig. 4E).
To better understand how LRCH1 affects LAT endocytosis, we purified different components of the membranes by density gradient centrifugation (Fig. 4F) (35, 36). LRCH1 and LAT were mainly found in fractions 3 to 6, along with LAMP1 (lysosome-associated membrane protein 1), a lysosome marker (Fig. 4F). In anti-CD3/CD28–stimulated T8.1 cells, an enhanced amount of LRCH1 and LAT colocalized with LAMP1 (Fig. 4G and Movies S1 and S2). Besides, the ubiquitin levels of LAT were reduced in Lrch1−/− CD8+ T cells (SI Appendix, Fig. S5F).
STAT1 is reported to be regulated by c-Cbl (37), and the JAK-STAT signaling pathway was enriched according to the RNA-sequencing data (SI Appendix, Fig. S2A). However, the total and phosphorylated levels of various STATs were not affected by LRCH1 deficiency in anti-CD3/CD28–stimulated CD8+ T cells (SI Appendix, Fig. S6 A and B). Stat2 was not detected in CD8+ T cells, and the transcription levels of Stat1, Stat3, Stat5, and Stat6 were comparable between WT and Lrch1−/− CD8+ T cells (SI Appendix, Fig. S2B). Besides, we did not detect the interaction between STAT1 and LRCH1 or LAT (SI Appendix, Fig. S6C). These findings suggest that LRCH1 might not affect TCR-induced STAT expression. Collectively, we have demonstrated that LRCH1 binds to LAT with its LRR and CH domains to promote LAT endocytosis and degradation, resulting in the termination of TCR signaling.
LRCH1 Deficiency Potentiates the Antitumor Activity of CD8+ T Cells.
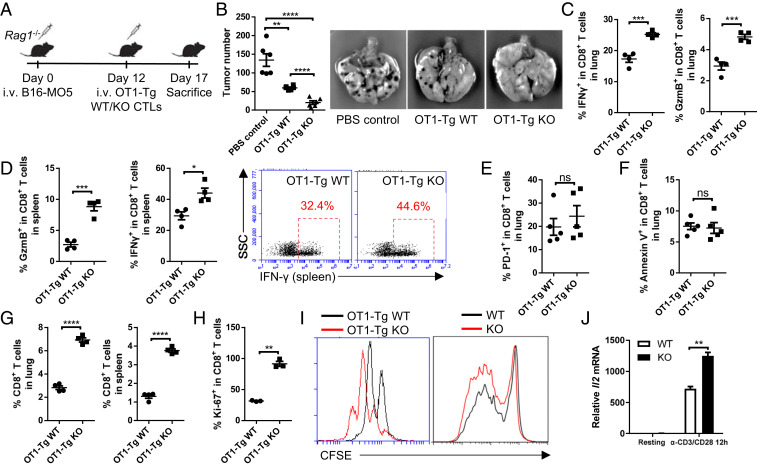
As LRCH1 is an inhibitory regulator of TCR signaling, we next tested whether LRCH1-deficient CD8+ T cells could enhance the in vivo antitumor efficacy. We used a T cell adoptive transfer model to exclusively examine the therapeutic effect of Lrch1−/− CD8+ T cells (Fig. 5A). B16-MO5 melanoma cells were i.v. injected into Rag1−/− mice at day 0, and equal numbers of OT1-Tg WT or KO CD8+ CTLs were transferred into the tumor-bearing Rag1−/− mice at day 12. Compared with the phosphate-buffered saline (PBS)-treated group, adoptively transferred WT CTLs effectively eradicated lung tumors; Lrch1−/− CD8+ CTL-treated mice cleared even more tumor nodules (Fig. 5B). Lung tumor-infiltrating Lrch1−/− CTLs had enhanced production of IFN-γ and GzmB (Fig. 5C), with little effect on PD-1 expression (Fig. 5E). In addition, Lrch1−/− CD8+ T cells in the peripheral lymphoid organs, namely spleen, also produced higher levels of IFN-γ and GzmB (Fig. 5D), which might help to clear tumor cells in the circulatory system.
Fig. 5.
LRCH1 deficiency potentiates the antitumor activity of CD8+ T cells. (A) Schematic diagram of the tumor treatment mouse model. (B) The number of tumor nodes in the lung; n = 6. (C and D) The percentages of IFN-γ– and GzmB-producing CD8+ T cells in the lung (C) and spleen (D); n = 4. (E and F) The percentages of PD-1+ CD8+ T cells (E) and Annexin V+ CD8+ T cells (F) in the lung; n = 5. (G) CD8+ T cell frequencies in the spleen and lung of the recipient mice; n = 4. (H) The proportion of Ki-67+ CD8+ T cells in the spleen; n = 3. (I) Cell proliferation assays (based on CFSE dilution) of WT and KO CD8+ T cells post OVA257–264 or anti-CD3/CD28 stimulation. (J) The relative mRNA level of Il2 in anti-CD3/CD28–stimulated WT/KO CD8+ T cells. ns, not significant (P > 0.05); *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (unpaired Student’s t test). Data are representative of two or more independent experiments (B–H, mean ± SEM; J, mean ± SD).
We next examined the apoptosis and proliferation of CD8+ T cells in the lung. Annexin V staining showed that LRCH1 deficiency did not affect apoptosis (Fig. 5F), consistent with the in vitro results (Fig. 1I). However, more Lrch1−/− CD8+ T cells were detected in the lung and spleen of the recipient mice (Fig. 5G). This might be due to the superior proliferative capacity of Lrch1−/− CD8+ T cells in the tumor microenvironment, indicated by Ki-67 staining (Fig. 2H). This was consistent with the BrdU incorporation data of the Listeria infection model (Fig. 2N). To further validate this, CFSE-labeled WT and KO CD8+ T cells from either OT1 transgenic mice or nontransgenic mice were stimulated in vitro with OVA257–264–pulsed splenocytes or anti-CD3/CD28 antibodies, respectively. Lrch1−/− CD8+ T cells were more proliferative (Fig. 5I). IL-2 is an important cytokine for T cell proliferation, and Lrch1−/− CD8+ T cells had an increased Il2 transcription level (Fig. 5J). These data together suggest that LRCH1 deficiency enhances CD8+ T cell proliferation and antitumor activity both in vivo and in vitro.
LRCH1 Deficiency Promotes the Effector Function of Human T Cells.
Tremendous efforts have been made to improve or maintain cytotoxic T cell function for immunotherapy, including CAR T or TCR-T cell therapy. One of the current main challenges is to improve T cell entry into solid tumors and maintain CTL proliferation and killing ability (2, 38). We previously reported that LRCH1 inhibits the migration of CD4+ T cells (16). In this study, we observed that more Lrch1−/− CD8+ T cells migrated toward CXCL10 in the transwell assay (SI Appendix, Fig. S7A). Since the proliferation and killing ability of Lrch1−/− CD8+ T cells against tumors were enhanced (Fig. 5), we proposed LRCH1 as a potential target to improve the therapeutic effect of CAR T cells.
LRCH1 could also bind LAT in primary human T cells (Fig. 6A and SI Appendix, Fig. S7 B, Right and D). Single-guide RNAs (sgRNAs) targeting LRCH1 were electroporated into primary human T cells. sgRNA7 and sgRNA10 showed better knockout efficiency, and promoted IFN-γ production upon phorbol 12-myristate 13-acetate (PMA) and ionomycin stimulation (Fig. 6B).
Fig. 6.
LRCH1 deficiency promotes the effector function of human T cells. (A) Coimmunoprecipitation analysis of endogenous LAT and LRCH1 in primary human T cells. (B) Knockout efficiency of sgRNAs of LRCH1 (Left) and FACS analysis of intracellular IFN-γ (Right) in WT and LRCH1−/− human T cells. (C) Migration ability of GPC3-CAR–transfected WT and LRCH1−/− human T cells toward human CXCL10 (Upper). Knockout efficiency of sgRNA7 and sgRNA10 (Lower). (D) Cell proliferation assay of GPC3-CAR–transfected WT and LRCH1−/− human T cells after a 48-h coculture with PLC or Huh7. (E and F) Coimmunoprecipitation analysis of LRCH3 binding with LRCH1 (E) or LAT (F) in 293T cells. (G) Knockout efficiency of different sgRNAs of LRCH3 (Upper) and FACS analysis of intracellular IFN-γ (Lower) in WT and LRCH3-deficient human T cells. ns, not significant (P > 0.05); *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired Student’s t test). Data are representative of two or more independent experiments (C, mean ± SD).
Hepatocellular carcinoma (HCC) is a solid tumor with high mortality worldwide. GPC3 (glypican-3) is highly expressed in HCC as a liver tumor-associated antigen, and GPC3-CAR T cells have been generated to defend HCC (39). LRCH1 was targeted in GPC3-CAR T cells using sgRNA7 or sgRNA10. The migration ability of LRCH1-deficient GPC3-CAR cells was enhanced (Fig. 6C and SI Appendix, Fig. S7C). LRCH1-deficient GPC3-CAR cells were more proliferative when cocultured with two GPC3-expressing cell lines (PLC/PRF/5 and Huh-7) (Fig. 6D).
Since LRCH1 deficiency did not substantially enhance the migration and proliferation of GPC3-CAR T cells, we wondered whether this was caused by the functional redundancy of LRCH3, a homolog of LRCH1. Moreover, LRCH3 interacted with LRCH1 and LAT (Fig. 6 E and F and SI Appendix, Fig. S7D). Several sgRNAs targeting human LRCH3 were designed, and sgRNA3, sgRNA4, and sgRNA5 showed high knockout efficiency (Fig. 6G). LRCH3-deficient T cells produced more IFN-γ after PMA and ionomycin stimulation (Fig. 6G). LRCH3 deficiency also increased human T cell migration, and the migration of LRCH1/3 double-knockout T cells was further enhanced slightly in the transwell assay (SI Appendix, Fig. S7C). Collectively, LRCH1 and LRCH3 in human T cells inhibit IFN-γ production, cell proliferation, and migration in vitro, which might be targeted to improve T cell immunotherapy.
Discussion
In this study, we identify LRCH1 as a negative regulator of TCR signaling. LRCH1-deficient CD8+ T cells proliferate faster with enhanced migration ability, and produce higher levels of multiple effector molecules to potentiate the killing ability. Moreover, Lrch1−/− mice display improved protection against influenza and Listeria infection. Furthermore, adoptively transferred Lrch1−/− CTLs clear B16-MO5 metastatic lung tumors more effectively than WT CTLs.
Studies have shown that LRCH family membranes are evolutionarily conserved across animal species, and may be functionally redundant. LRCH gene polymorphisms are frequently reported to be associated with various diseases. Despite the uncertainty of the association between knee OA and rs912428 in LRCH1 (17), the most significant association is observed at rs754106 in intron 1 of the LRCH1 gene (40), in the genome-wide association study meta-analysis from 2,118 radiographically defined hip OA cases with superior maximal joint space narrowing. Besides, LRCH1 is up-regulated in rapid Parkinson disease (PD) progression, and is a potential progression rate-associated biomarker of PD (41). In addition, LRCH3 has previously been implicated in nuclear factor κB activation (42), and is associated with the humoral response to Mycobacterium avium paratuberculosis (43). LRCH4 deficiency attenuates cytokine induction by LPS and multiple other Toll-like receptor ligands, and reduces both cell-surface gangliosides and surface display of CD14 (44). Importantly, both LRCH1 and LRCH4 are involved in the progression of colorectal cancer (CRC). LRCH1 overexpression contributes to the 13q-driven adenoma-to-carcinoma progression (45), and LRCH4 expression level is increased in more advanced CRC stages (46).
In this study, we have demonstrated that LRCH3 could also interact with LAT and form a heterodimer with LRCH1. LRCH3 is highly homogeneous and functionally redundant with LRCH1. LRCH3−/− human T cells migrate faster and produce higher levels of IFN-γ. Double knockout of LRCH1 and LRCH3 could further promote the migration of human CAR T cells in vitro. Infiltration, proliferation, and persistence of the cytotoxicity of T cells are critical in tumor eradication, especially in T cell immunotherapy for solid tumors (38). Since proteins that regulate these functions of T cells are potential targets to enhance CAR T cells’ therapeutic effects, LRCH1 and LRCH3 may be potential targets in T cell immunotherapy. It is important to investigate in the future whether LRCH1 and LRCH3 could be targeted to improve human CAR T cells against solid tumors in vivo.
Mechanistically, we have elucidated that LRCH1 binds directly to LAT via the LRR and CH domain of LRCH1, independent of the tyrosine residues in LAT. The dimerization of the LRR and CH domain may further enhance the interaction between LRCH1 and LAT. Functionally, LRCH1 overexpression impedes LAT phosphorylation and disrupts GRB2 recruitment, resulting in the impaired activation of ERK and AKT. This is in agreement with the previous findings that phosphorylated Tyr171, 191, and 226 residues of LAT are critical docking sites for GRB2 and the recruitment of GRB2 is essential to activate the Ras-MAPK cascade (9). Deficiency of LAT impairs both cytotoxicity and expansion during the priming phase and upon secondary infection of CD8+ T cells (10, 47). GRB2 has a structural role in driving the clustering of LAT, and GRB2-deficient T cells had substantially reduced production of IL-2 and IFN-γ (48, 49). Besides, ERK and AKT play pivotal roles in regulating the proliferation, survival, and cytotoxic functions of CD8+ T cells (50–52). Considering our and others’ findings, we propose that the disruption of the GRB2–LAT interaction by LRCH1 interferes with LAT signalosome formation, resulting in defective CD8+ T cell responses.
Apart from this mechanism, we have found that LRCH1 promotes LAT endocytosis, as evidenced by the decreased level of LAT on the plasma membrane of LRCH1-overexpressing T8.1 cells. LRCH1 displays features of cytoskeletal scaffolding proteins, and some other cytoskeleton-related proteins, including Cdc42, Rac1, Rac3, and Arp2/3, are detected by mass spectrometry in anti-LRCH1 immunoprecipitation samples. Cytoskeletal remodeling plays crucial roles in regulating T cell activation, migration, and killing of synapse formation (53), and the Rho family GTPases are critical regulators of T cell polarity, migration, and vesicle trafficking processes (54). Endocytic mechanisms control protein composition of the plasma membrane, and Cdc42 has been reported to regulate endocytosis and vesicular trafficking through interaction with Wiskott–Aldrich syndrome protein and the Arp2/3 complex, leading to changes in actin dynamics (54). Rac1 is also reported to mediate endocytic and exocytic vesicle trafficking of the low-density lipoprotein–SR-B1 complex (55). As we and others have reported that LRCH1 directly binds DOCK8 and forms a complex with Cdc42 or Rac1 (16, 32), it is possible that LRCH1 is involved in vesicular trafficking. In particular, previous studies have elucidated that the LAT cytoplasmic domain contains two lysine residues for ubiquitylation, and LAT could be ubiquitinated by c-Cbl for degradation (14). This study has provided evidence that LRCH1 directly binds LAT and promotes LAT endocytosis for degradation. Collectively, we propose that LRCH1 inhibits TCR signaling through promoting LAT endocytosis as well as interfering with LAT signalosome formation.
Materials and Methods
Detailed materials and methods used in this article are in SI Appendix. All materials, data, and associated protocols, including data and methods included in SI Appendix, will be made available to readers upon request. Contact the corresponding author (H.W.) for related data or materials.
Supplementary Material
Acknowledgments
This work was supported by grants from the Ministry of Science and Technology of China (2016YFD0500207, 2016YFD0500407, 2016YFC0905902, and 2018YFA0800702), Strategic Priority Research Program of the Chinese Academy of Sciences (CAS) (XDB19000000), and National Natural Science Foundation of China (81930038, 81825011, 81630043, 81671572, 81571552, 81701569, 31700781, and 81801575). We thank the Genome Tagging Project Center, Shanghai Institute of Biochemistry and Cell Biology (SIBCB), CAS, and National Center for Protein Science Shanghai for technical support. H.W. is supported by the NSF for Distinguished Young Scholars. We thank the Animal Core Facility, Core Facility of Molecular Biology, and Core Facility of Cell Biology of the SIBCB for their technical support and help.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2000970117/-/DCSupplemental.
Data Availability.
The RNA-sequencing data reported in this paper have been deposited in the Gene Expression Omnibus under accession no. GSE150634.
References
- 1.Golstein P., Griffiths G. M., An early history of T cell-mediated cytotoxicity. Nat. Rev. Immunol. 18, 527–535 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Newick K., O’Brien S., Moon E., Albelda S. M., CAR T cell therapy for solid tumors. Annu. Rev. Med. 68, 139–152 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Jung I. Y. et al., CRISPR/Cas9-mediated knockout of DGK improves antitumor activities of human T cells. Cancer Res. 78, 4692–4703 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Chen J. et al., NR4A transcription factors limit CAR T cell function in solid tumours. Nature 567, 530–534 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu X. et al., Genome-wide analysis identifies NR4A1 as a key mediator of T cell dysfunction. Nature 567, 525–529 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaud G., Lesourne R., Love P. E., Regulatory mechanisms in T cell receptor signalling. Nat. Rev. Immunol. 18, 485–497 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Zhang W. et al., Association of Grb2, Gads, and phospholipase C-gamma 1 with phosphorylated LAT tyrosine residues. Effect of LAT tyrosine mutations on T cell antigen receptor-mediated signaling. J. Biol. Chem. 275, 23355–23361 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Balagopalan L., Coussens N. P., Sherman E., Samelson L. E., Sommers C. L., The LAT story: A tale of cooperativity, coordination, and choreography. Cold Spring Harb. Perspect. Biol. 2, a005512 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balagopalan L., Kortum R. L., Coussens N. P., Barr V. A., Samelson L. E., The linker for activation of T cells (LAT) signaling hub: From signaling complexes to microclusters. J. Biol. Chem. 290, 26422–26429 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ou-Yang C. W. et al., Role of LAT in the granule-mediated cytotoxicity of CD8 T cells. Mol. Cell. Biol. 32, 2674–2684 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang W. et al., Essential role of LAT in T cell development. Immunity 10, 323–332 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Keller B. et al., Early onset combined immunodeficiency and autoimmunity in patients with loss-of-function mutation in LAT. J. Exp. Med. 213, 1185–1199 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brignatz C., Restouin A., Bonello G., Olive D., Collette Y., Evidences for ubiquitination and intracellular trafficking of LAT, the linker of activated T cells. Biochim. Biophys. Acta 1746, 108–115 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Balagopalan L. et al., c-Cbl-mediated regulation of LAT-nucleated signaling complexes. Mol. Cell. Biol. 27, 8622–8636 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balagopalan L. et al., Enhanced T-cell signaling in cells bearing linker for activation of T-cell (LAT) molecules resistant to ubiquitylation. Proc. Natl. Acad. Sci. U.S.A. 108, 2885–2890 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu X. et al., LRCH1 interferes with DOCK8-Cdc42-induced T cell migration and ameliorates experimental autoimmune encephalomyelitis. J. Exp. Med. 214, 209–226 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spector T. D. et al., Association between a variation in LRCH1 and knee osteoarthritis: A genome-wide single-nucleotide polymorphism association study using DNA pooling. Arthritis Rheum. 54, 524–532 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Snelling S., Sinsheimer J. S., Carr A., Loughlin J., Genetic association analysis of LRCH1 as an osteoarthritis susceptibility locus. Rheumatology (Oxford) 46, 250–252 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Jiang Q. et al., Lack of association of single nucleotide polymorphism in LRCH1 with knee osteoarthritis susceptibility. J. Hum. Genet. 53, 42–47 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Kaech S. M., Cui W., Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 12, 749–761 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pearce E. L. et al., Control of effector CD8+ T cell function by the transcription factor eomesodermin. Science 302, 1041–1043 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Sullivan B. M., Juedes A., Szabo S. J., von Herrath M., Glimcher L. H., Antigen-driven effector CD8 T cell function regulated by T-bet. Proc. Natl. Acad. Sci. U.S.A. 100, 15818–15823 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tajima M., Wakita D., Satoh T., Kitamura H., Nishimura T., IL-17/IFN-γ double producing CD8+ T (Tc17/IFN-γ) cells: A novel cytotoxic T-cell subset converted from Tc17 cells by IL-12. Int. Immunol. 23, 751–759 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Duan S., Thomas P. G., Balancing immune protection and immune pathology by CD8(+) T-cell responses to influenza infection. Front. Immunol. 7, 25 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.La Gruta N. L., Turner S. J., T cell mediated immunity to influenza: Mechanisms of viral control. Trends Immunol. 35, 396–402 (2014). [DOI] [PubMed] [Google Scholar]
- 26.McGill J., Van Rooijen N., Legge K. L., Protective influenza-specific CD8 T cell responses require interactions with dendritic cells in the lungs. J. Exp. Med. 205, 1635–1646 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jelley-Gibbs D. M. et al., Persistent depots of influenza antigen fail to induce a cytotoxic CD8 T cell response. J. Immunol. 178, 7563–7570 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Song Y. et al., Repeated low-dose influenza virus infection causes severe disease in mice: A model for vaccine evaluation. J. Virol. 89, 7841–7851 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mannering S. I., Zhong J., Cheers C., T-cell activation, proliferation and apoptosis in primary Listeria monocytogenes infection. Immunology 106, 87–95 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khan S. H., Badovinac V. P., Listeria monocytogenes: A model pathogen to study antigen-specific memory CD8 T cell responses. Semin. Immunopathol. 37, 301–310 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zenewicz L. A., Shen H., Innate and adaptive immune responses to Listeria monocytogenes: A short overview. Microbes Infect. 9, 1208–1215 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi H. et al., Hippo kinases Mst1 and Mst2 sense and amplify IL-2R-STAT5 signaling in regulatory T cells to establish stable regulatory activity. Immunity 49, 899–914.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Shea J. J., McVicar D. W., Bailey T. L., Burns C., Smyth M. J., Activation of human peripheral blood T lymphocytes by pharmacological induction of protein-tyrosine phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 89, 10306–10310 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marianayagam N. J., Sunde M., Matthews J. M., The power of two: Protein dimerization in biology. Trends Biochem. Sci. 29, 618–625 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Carpier J. M. et al., Rab6-dependent retrograde traffic of LAT controls immune synapse formation and T cell activation. J. Exp. Med. 215, 1245–1265 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hivroz C., Larghi P., Jouve M., Ardouin L., Purification of LAT-containing membranes from resting and activated T lymphocytes. Methods Mol. Biol. 1584, 355–368 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Blesofsky W. A. et al., Regulation of STAT protein synthesis by c-Cbl. Oncogene 20, 7326–7333 (2001). [DOI] [PubMed] [Google Scholar]
- 38.Martinez M., Moon E. K., CAR T cells for solid tumors: New strategies for finding, infiltrating, and surviving in the tumor microenvironment. Front. Immunol. 10, 128 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao H. et al., Development of T cells redirected to glypican-3 for the treatment of hepatocellular carcinoma. Clin. Cancer Res. 20, 6418–6428 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Panoutsopoulou K. et al.; arcOGEN Consortium , Radiographic endophenotyping in hip osteoarthritis improves the precision of genetic association analysis. Ann. Rheum. Dis. 76, 1199–1206 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan Y., Xiao S., Progression rate associated peripheral blood biomarkers of Parkinson’s disease. J. Mol. Neurosci. 65, 312–318 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Gewurz B. E. et al., Genome-wide siRNA screen for mediators of NF-κB activation. Proc. Natl. Acad. Sci. U.S.A. 109, 2467–2472 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGovern S. P. et al., Candidate genes associated with the heritable humoral response to Mycobacterium avium ssp. paratuberculosis in dairy cows have factors in common with gastrointestinal diseases in humans. J. Dairy Sci. 102, 4249–4263 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Aloor J. J. et al., Leucine-rich repeats and calponin homology containing 4 (Lrch4) regulates the innate immune response. J. Biol. Chem. 294, 1997–2008 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Groen F. L. et al., Gene-dosage dependent overexpression at the 13q amplicon identifies DIS3 as candidate oncogene in colorectal cancer progression. Genes Chromosomes Cancer 53, 339–348 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Huo T., Canepa R., Sura A., Modave F., Gong Y., Colorectal cancer stages transcriptome analysis. PLoS One 12, e0188697 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ou-Yang C. W., Zhu M., Sullivan S. A., Fuller D. M., Zhang W., The requirement of linker for activation of T cells in the primary and memory responses of CD8 T cells. J. Immunol. 190, 2938–2947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bilal M. Y., Houtman J. C., GRB2 nucleates T cell receptor-mediated LAT clusters that control PLC-γ1 activation and cytokine production. Front. Immunol. 6, 141 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jang I. K. et al., Grb2 functions at the top of the T-cell antigen receptor-induced tyrosine kinase cascade to control thymic selection. Proc. Natl. Acad. Sci. U.S.A. 107, 10620–10625 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dong C., Davis R. J., Flavell R. A., MAP kinases in the immune response. Annu. Rev. Immunol. 20, 55–72 (2002). [DOI] [PubMed] [Google Scholar]
- 51.D’Souza W. N., Chang C. F., Fischer A. M., Li M., Hedrick S. M., The Erk2 MAPK regulates CD8 T cell proliferation and survival. J. Immunol. 181, 7617–7629 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Phu T., Haeryfar S. M., Musgrave B. L., Hoskin D. W., Phosphatidylinositol 3-kinase inhibitors prevent mouse cytotoxic T-cell development in vitro. J. Leukoc. Biol. 69, 803–814 (2001). [PubMed] [Google Scholar]
- 53.Billadeau D. D., Nolz J. C., Gomez T. S., Regulation of T-cell activation by the cytoskeleton. Nat. Rev. Immunol. 7, 131–143 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Chi X., Wang S., Huang Y., Stamnes M., Chen J. L., Roles of rho GTPases in intracellular transport and cellular transformation. Int. J. Mol. Sci. 14, 7089–7108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang L. et al., SR-B1 drives endothelial cell LDL transcytosis via DOCK4 to promote atherosclerosis. Nature 569, 565–569 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-sequencing data reported in this paper have been deposited in the Gene Expression Omnibus under accession no. GSE150634.