SUMMARY
The novel coronavirus SARS-CoV-2, the causative agent of COVID-19 respiratory disease, has infected over 2.3 million people, killed over 160,000, and caused worldwide social and economic disruption1,2. There are currently no antiviral drugs with proven clinical efficacy, nor are there vaccines for its prevention, and these efforts are hampered by limited knowledge of the molecular details of SARS-CoV-2 infection. To address this, we cloned, tagged and expressed 26 of the 29 SARS-CoV-2 proteins in human cells and identified the human proteins physically associated with each using affinity-purification mass spectrometry (AP-MS), identifying 332 high-confidence SARS-CoV-2-human protein-protein interactions (PPIs). Among these, we identify 66 druggable human proteins or host factors targeted by 69 compounds (29 FDA-approved drugs, 12 drugs in clinical trials, and 28 preclinical compounds). Screening a subset of these in multiple viral assays identified two sets of pharmacological agents that displayed antiviral activity: inhibitors of mRNA translation and predicted regulators of the Sigma1 and Sigma2 receptors. Further studies of these host factor targeting agents, including their combination with drugs that directly target viral enzymes, could lead to a therapeutic regimen to treat COVID-19.
MAIN
SARS-CoV-2 is an enveloped, positive-sense, single-stranded RNA betacoronavirus of the family Coronaviridae3,4. Coronaviruses infecting humans historically included several mild common cold viruses, including hCoV-OC43, HKU, and 229E5. However, over the past two decades, highly pathogenic human coronaviruses have emerged, including SARS-CoV in 2002 with 8,000 cases worldwide and a death rate of ~10%, and MERS-CoV in 2012, with 2,500 confirmed cases and a death rate of 36%. Infection with these highly pathogenic coronaviruses can result in Acute Respiratory Distress Syndrome (ARDS), which may lead to long-term reduction in lung function, arrhythmia, or death. Compared to MERS-CoV or SARS-CoV, SARS-CoV-2 has a lower case-fatality rate but spreads more efficiently6, making it difficult to contain. To devise therapeutic strategies to counteract SARS-CoV-2 infection and the associated COVID-19 (Coronavirus Disease 2019) pathology, it is crucial to understand how this coronavirus hijacks the host during infection, and to apply this knowledge towards developing new drugs and repurposing existing ones.
So far, no clinically available antiviral drugs have been developed for SARS-CoV, SARS-CoV-2 or MERS-CoV. Clinical trials are ongoing for treatment of COVID-19 with the nucleoside analog RNA-dependent RNA Polymerase (RdRP) inhibitor Remdesivir7, and recent data suggests a new nucleoside analog may be effective against SARS-CoV-2 infection in laboratory animals8. Clinical trials on several vaccine candidates are also underway9, as are trials of repurposed compounds inhibiting the human protease TMPRSS210. We believe there is great potential in systematically exploring the host dependencies of the SARS-CoV-2 virus to identify other host proteins already targeted by existing drugs. Therapies targeting the host-virus interface, where the emergence of mutational resistance is arguably less likely, could potentially present durable, broad-spectrum treatment modalities11. Unfortunately, limited knowledge of the molecular details of SARS-CoV-2 infection precludes a comprehensive evaluation of small molecule candidates for host-directed therapies. We sought to address this gap by systematically mapping the interaction landscape between SARS-CoV-2 proteins and human proteins.
Cloning and expression of predicted SARS-CoV-2 proteins
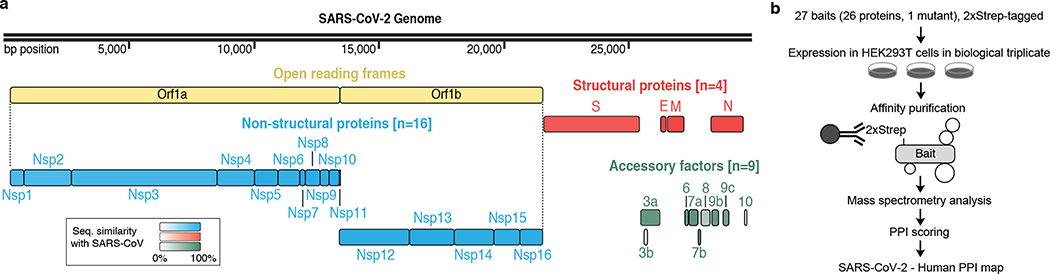
Sequence analysis of SARS-CoV-2 isolates suggests that the 30kb genome encodes as many as 14 open reading frames (Orfs). The Orf1a / Orf1ab encodes a polyprotein, which is auto-proteolytically processed into 16 non-structural proteins (Nsp1–16) that form the replicase / transcriptase complex (RTC) (Fig. 1a). The RTC consists of multiple enzymes, including the papain-like protease (Nsp3), the main protease (Nsp5), the Nsp7-Nsp8 primase complex, the primary RNA-dependent RNA polymerase (Nsp12), a helicase / triphosphatase (Nsp13), an exoribonuclease (Nsp14), an endonuclease (Nsp15), and N7- and 2’O-methyltransferases (Nsp10/Nsp16)1,12,13. At the 3’ end of the viral genome, as many as 13 Orfs are expressed from nine predicted sub-genomic RNAs. These include four structural proteins: Spike (S), Envelope (E), Membrane (M) and Nucleocapsid (N)13, and nine putative accessory factors (Fig. 1a)1,12. The SARS-CoV-2 genome is very similar to SARS-CoV. While both have an Orf1ab encoding 16 predicted Nsps as well as the four typical coronavirus structural proteins, they differ in their complement of 3’ Orfs: SARS-CoV-2 possesses an Orf3b and Orf10 with limited detectable homology to SARS-CoV1,12 (Extended Data Fig. 1a).
Figure 1: AP-MS workflow for identification of SARS-CoV-2 host protein-protein interactions.
(a) SARS-CoV-2 genome annotation, color intensity is proportional to protein sequence similarity with SARS-CoV homologs (when homologs exist). (b) Experimental workflow for AP-MS studies.
Mature Nsps, with the exception of Nsp3 and Nsp16, and all predicted proteins expressed from other SARS-CoV-2 Orfs (27 proteins plus one mutant) were codon optimized and cloned into a mammalian expression vector with a 2xStrep affinity tag fused for affinity purification mass spectrometry (AP-MS) based proteomics upon expression in HEK293T/17 cells. High-confidence interactors were identified using SAINTexpress and MiST scoring algorithms14,15.
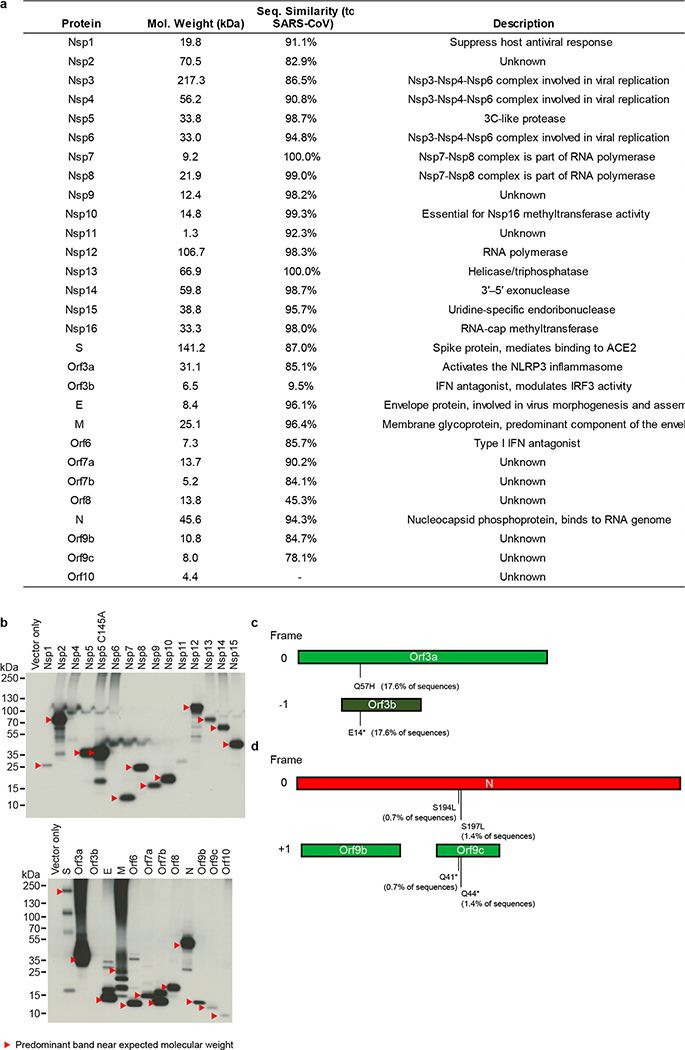
To verify viral protein expression, we performed an anti-Strep western blot on input cell lysate, and with the exception of Nsp4, Nsp6, Nsp11, and Orf3b, we observed bands consistent with predicted protein sizes (24 of 28 constructs; Extended Data Fig. 1b). Despite the lack of detection via western blot, we detected expression of viral peptides Nsp4, Nsp6, and Orf3b in the proteomic analysis. The fourth construct not confirmed by western blot, the small peptide Nsp11, had a predicted molecular mass of 4.8 kDa (including tag) but an apparent mass of approximately 30 kDa (Extended Data Fig. 1b).
Alignment of 2,784 SARS-CoV-2 sequences revealed a premature stop codon at position 14 of Orf3b in 17.6% of isolates (Extended Data Fig. 1c), and two mutations were also observed resulting in premature stop codons in Orf9c (Extended Data Fig. 1d). These data suggest that Orf3b and Orf9c might not be bonafide SARS-CoV-2 reading frames, or are dispensable for replication. Pending a comprehensive evaluation of viral protein expression, we nevertheless proceeded with analysis for all possible viral proteins. Out of the 27 bait proteins (Fig. 1b), the affinity purification of Orf7b showed an unusually high number of background proteins and was therefore excluded from protein interaction analysis. We have thus far sent these plasmids to almost 300 laboratories in 35 countries.
Global analysis of SARS-CoV-2 host interacting proteins
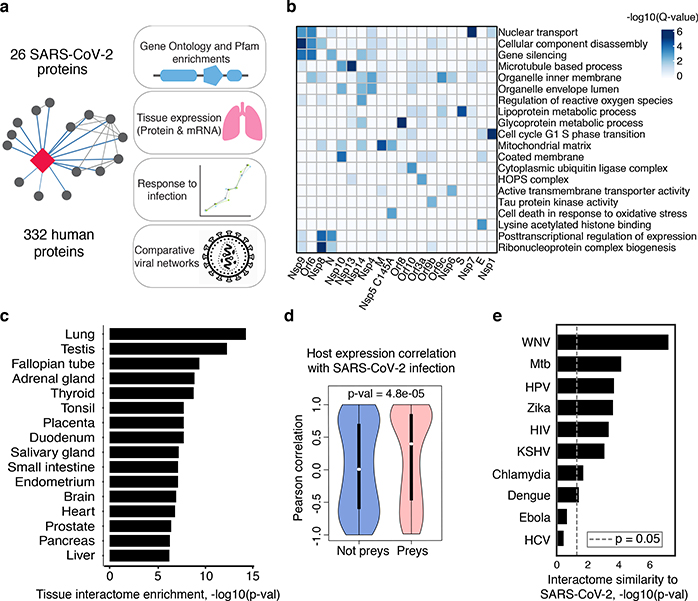
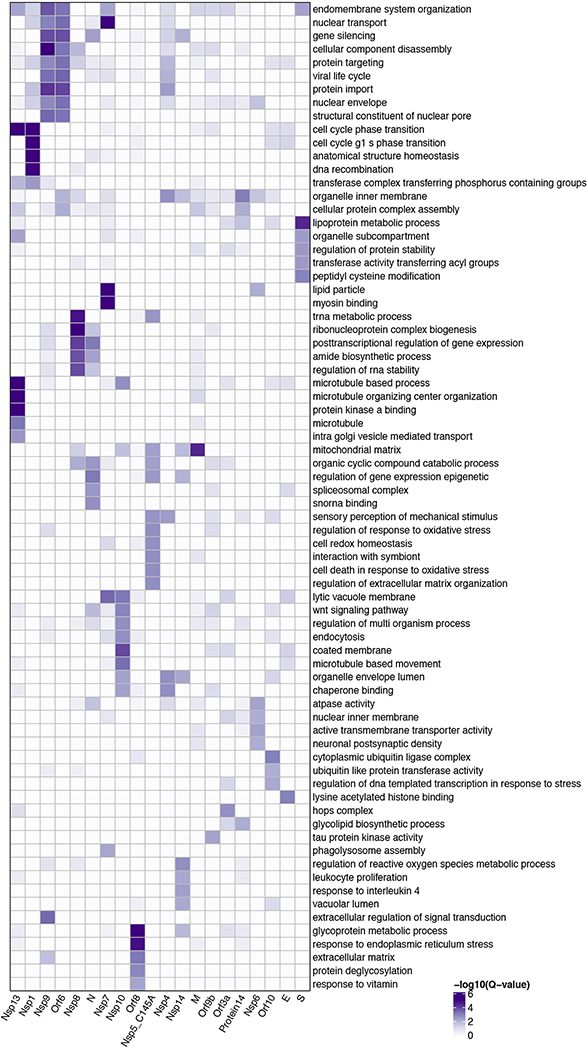
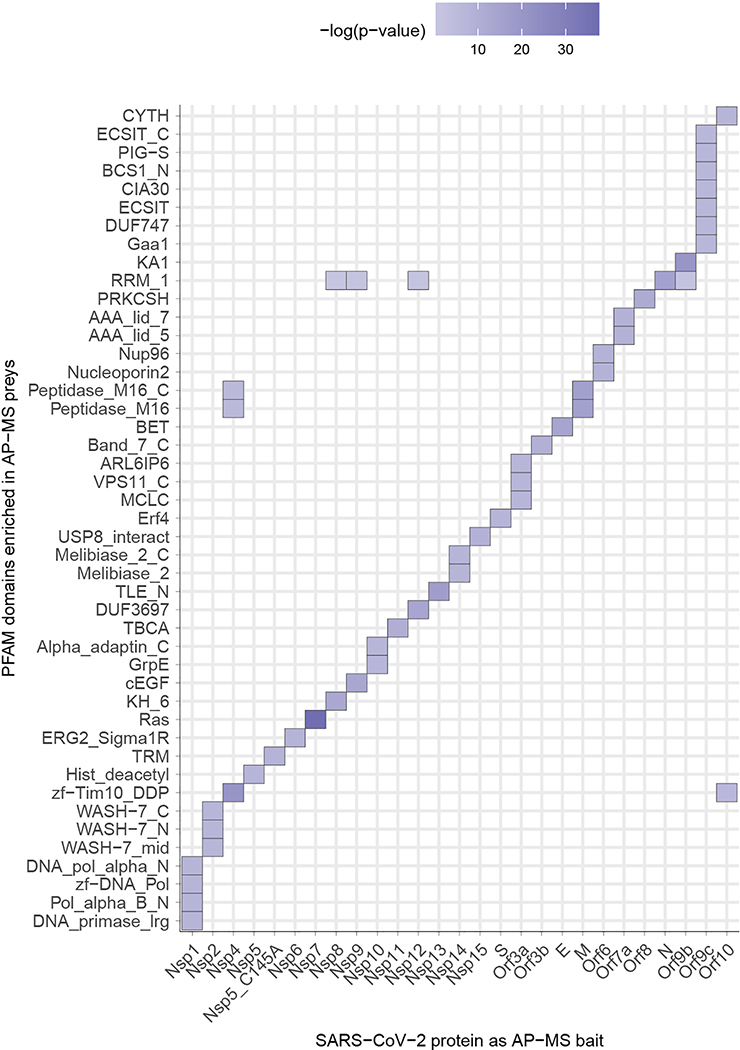
Our AP-MS analysis identified 332 high-confidence protein interactions between SARS-CoV-2 proteins and human proteins, observing correlation between replicate experiments of each viral bait (R = 0.46 – 0.72) (Extended Data Fig. 2, Supplementary Tables 1 and 2). We studied the interacting human proteins in regards to their cell biology, anatomical expression patterns, expression changes during SARS-CoV-2 infection16 and in relation to other maps of host-pathogen interacting proteins15,17 (Fig. 2a). We analyzed each viral protein for Gene Ontology enrichment (Fig. 2b, Extended Data Fig. 3), identifying the major cell processes of the interacting proteins, including lipoprotein metabolism (S), nuclear transport (Nsp7), and ribonucleoprotein complex biogenesis (Nsp8). To discover potential binding interfaces, we enriched for domain families within the interacting proteins of each viral bait (Extended Data Fig. 4). For instance, DNA polymerase domains are enriched among Nsp1 interactors, while bromodomains and extra-terminal domain (BET) family domains are enriched among E interactors (see Supplementary Discussion and Methods).
Figure 2: Global analysis of SARS-CoV-2 protein interactions.
(a) Overview of global analyses performed. (b) Gene Ontology (GO) enrichment analysis was performed on the human interacting proteins of each viral protein, p-values calculated by hypergeometric test and a false discovery rate was used to account for multiple hypothesis testing (Methods). The top GO term of each viral protein was selected for visualization. (c) Degree of differential protein expression for the human interacting proteins (n=332) across human tissues. We obtained protein abundance values for the proteome in 29 human tissues and calculated the median level of abundance for the human interacting proteins (top 16 tissues shown). This was then compared with the abundance values for the full proteome in each tissue and summarized as a Z-score from which a p-value was calculated and false discovery rate was used to account for multiple hypothesis testing. (d) The distribution of correlation of protein level changes during SARS-CoV-2 infection for pairs of viral-human proteins (median is shown) is higher than non-interacting pairs of viral-human proteins (p-value=4.8e-05, Kolmogorov–Smirnov test) The violin plots show each viral to human protein correlation for preys (n=210, min=−0.986, max=0.999, Q1=−0.468, Q2=0.396, Q3=0.850) and non-preys (n=54765, min= −0.999, max=0.999, Q1=−0.599, Q2=0.006, Q3=0.700). (e) Significance of the overlap of human interacting proteins between SARS-CoV-2 and other pathogens using a hypergeometric test (unadjusted for multiple testing). The background gene set for the test consisted of all unique proteins detected by mass spectrometry across all pathogens (n=10,181 proteins).
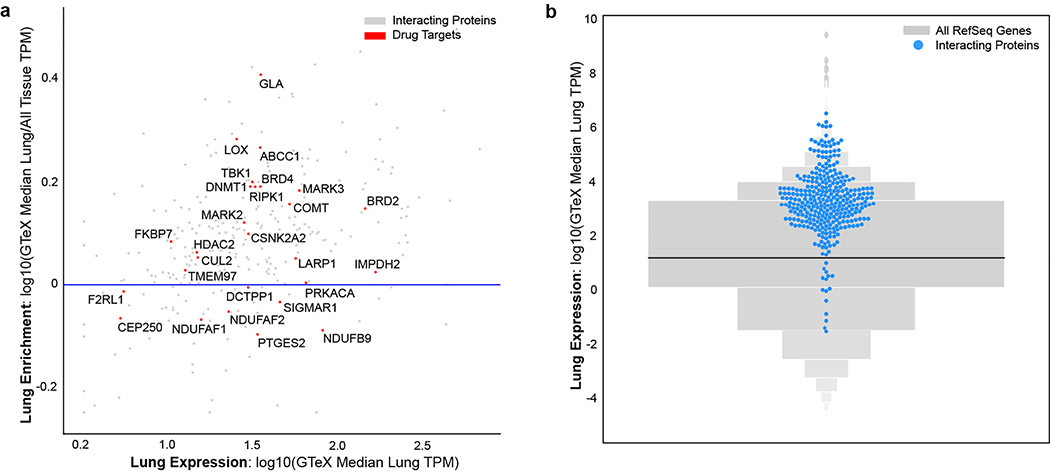
While the cell line used for these AP-MS experiments, HEK-293T/17, is permissive to SARS-CoV-2 infection18, it does not represent the primary physiological site of infection, lung tissue. From 29 human tissues19, we identified the lung as the tissue with the highest expression of the prey proteins relative to the average proteome (Fig. 2c). Consistent with this, the interacting proteins were enriched in the lung relative to other tissues (Extended Data Fig. 5a), and compared to overall RefSeq gene expression in the lung (median=3.198 TPM) interactors were expressed at a higher level (median=25.52 TPM, p=0.0007; t-test) (Extended Data Fig. 5b), supporting the hypothesis that SARS-CoV-2 preferentially hijacks proteins expressed in lung tissue.
We also studied the evolutionary properties of the host proteins (Supplementary Table 3 and Supplementary Discussion and Methods) and analyzed protein abundance changes during SARS-CoV-2 infection16. We calculated, when possible, the correlation between changes in abundance of viral proteins and their human interaction partners across four time points. Interacting pairs typically have stronger correlated changes than other pairs of viral-human proteins (Fig. 2d, KS test p-value=4.8e−05), arguing that the AP-MS derived interactions are relevant for the target tissue and the infection context. Comparing our SARS-CoV-2 interaction map with those for ten other pathogens (Fig. 2e), West Nile Virus (WNV)20 and Mycobacterium tuberculosis (Mtb)21 had the most similar host protein interaction partners. The association with Mtb is particularly interesting considering it also infects lung tissue.
The SARS-CoV-2 interactome reveals novel aspects of SARS-CoV-2 biology
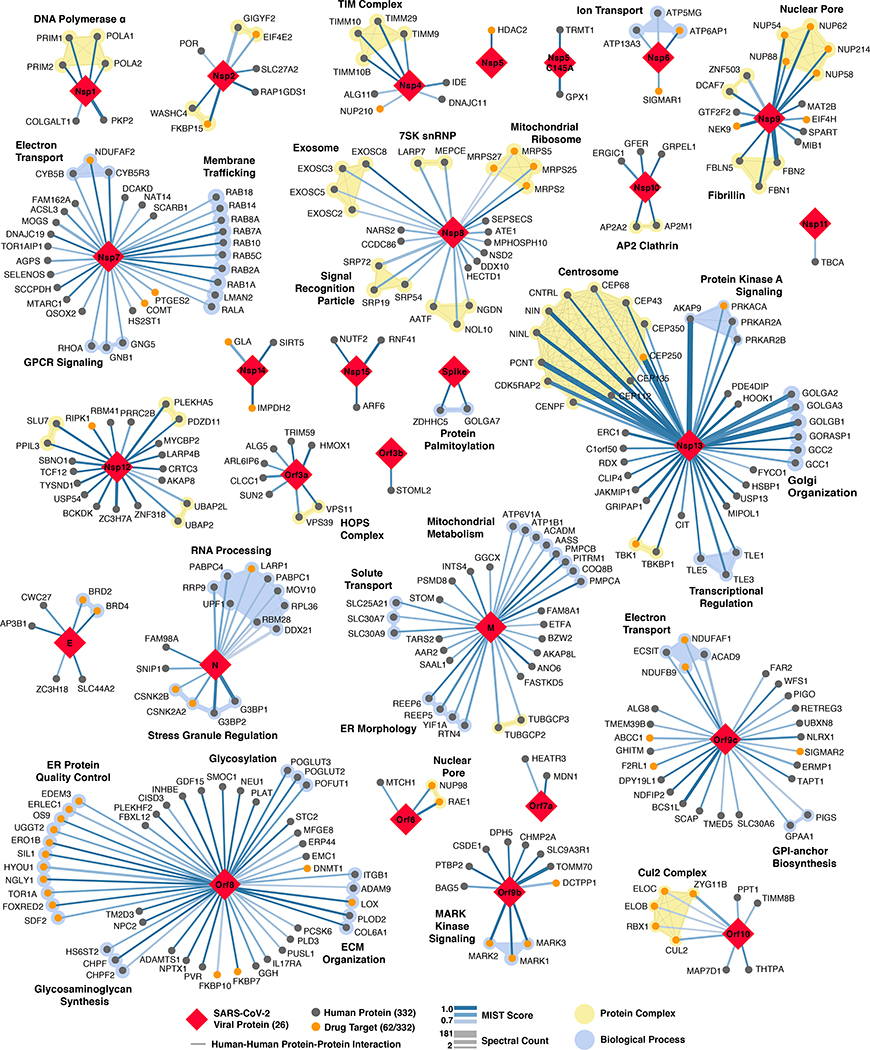
Our study highlighted interactions between SARS-CoV-2 proteins and human proteins involved in several complexes and biological processes (Fig.3). These included DNA replication (Nsp1), epigenetic and gene expression regulators (Nsp5, Nsp8, Nsp13, E), vesicle trafficking (Nsp2, Nsp6, Nsp7, Nsp10, Nsp13, Nsp15, Orf3a, E, M, Orf8), lipid modification (Spike), RNA processing and regulation (Nsp8, N), ubiquitin ligases (Orf10), signaling (Nsp8, Nsp13, N, Orf9b), nuclear transport machinery (Nsp9, Nsp15, Orf6), cytoskeleton (Nsp1, Nsp13), mitochondria (Nsp4, Nsp8, Orf9c), and extracellular matrix (Nsp9).
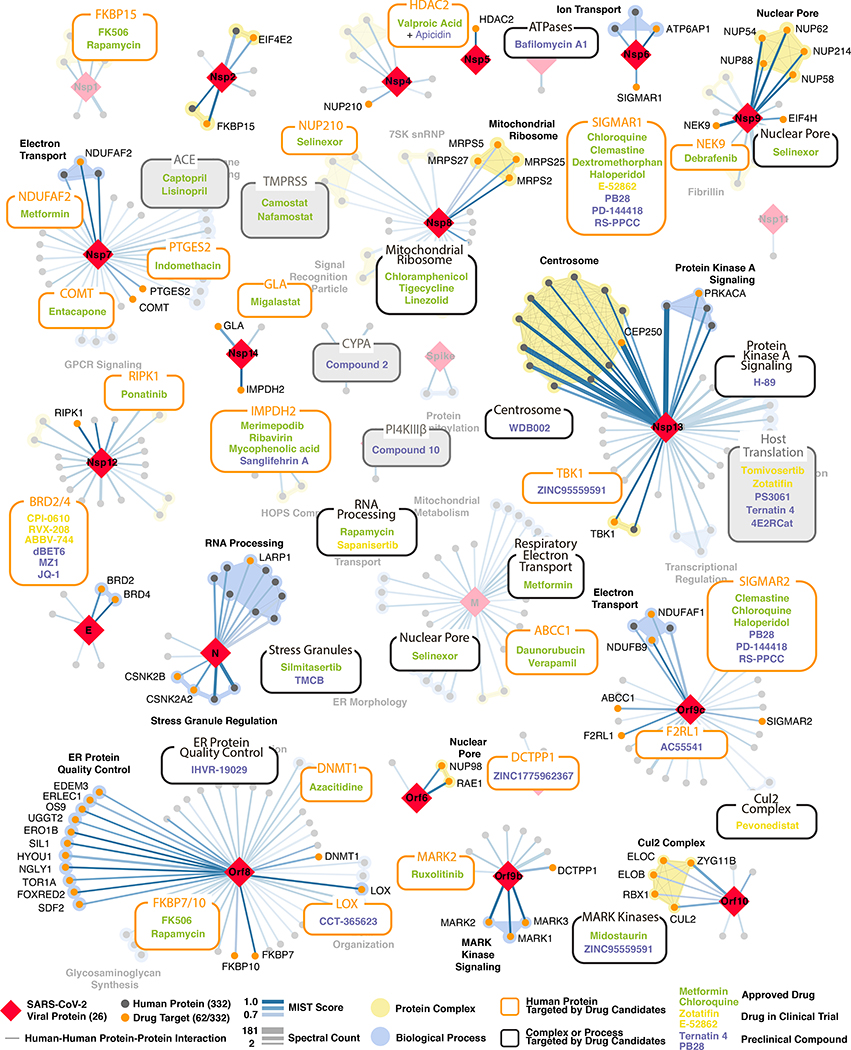
Figure 3: SARS-CoV-2 protein-protein interaction network.
332 high-confidence interactions between 26 SARS-CoV-2 proteins (red diamonds) and human proteins (circles; drug targets: orange; protein complexes: yellow; proteins in the same biological process: blue). Edge color proportional to MiST score; edge thickness proportional to spectral counts. Physical interactions among host proteins (thin black lines) were curated from CORUM, IntAct, and Reactome. An interactive PPI map can be found at kroganlab.ucsf.edu/network-maps. n=3 biologically independent samples.
Approximately 40% of SARS-CoV-2 interacting proteins were associated with endomembrane compartments or vesicle trafficking pathways. Host interactions of Nsp8 (signal recognition particle; SRP), Orf8 (endoplasmic reticulum protein quality control), M (ER morphology), and Nsp13 (centrosome and golgi organization) may facilitate the dramatic reconfiguration of ER/Golgi trafficking during coronavirus infection, and interactions in peripheral compartments by Nsp2 (WASH), Nsp6 and M (vacuolar ATPase), Nsp7 (Rabs), Nsp10 (AP2), E (AP3), and Orf3a (HOPS) may also modify endomembrane compartments to favor coronavirus replication. Nsp6 and Orf9c interact with Sigma receptors that are implicated in lipid remodeling and ER stress response; these proteins interact with many human drugs (see below).
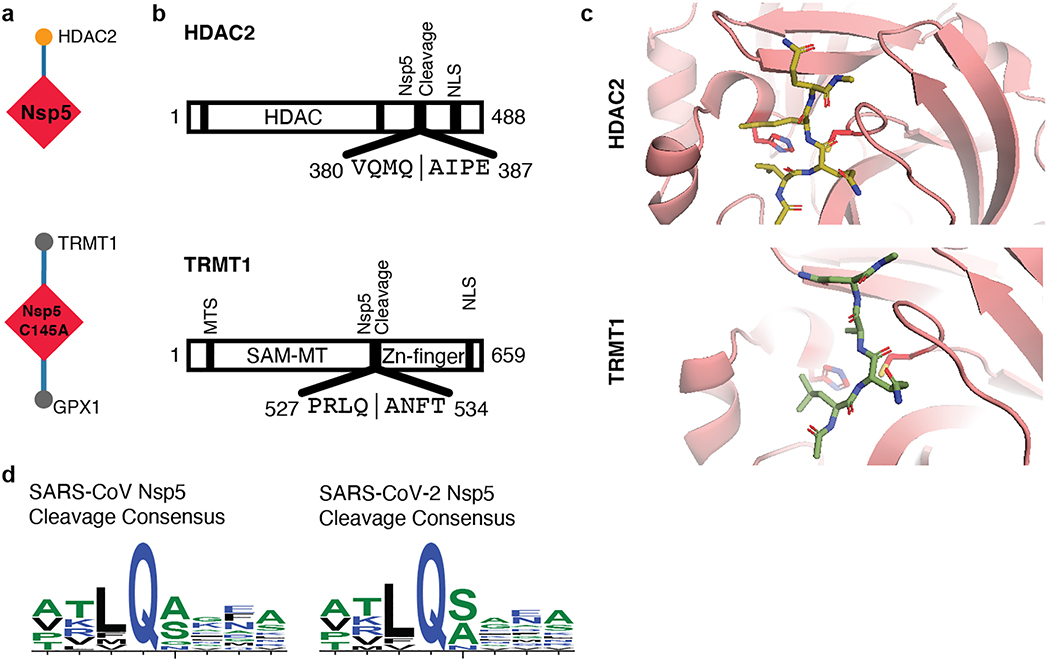
Trafficking into the ER and mitochondria may also be impacted by the SARS-CoV-2 main protease, Nsp5. We identified one high-confidence interaction of wild-type Nsp5 with the epigenetic regulator histone deacetylase 2 (HDAC2), and predicted a cleavage site between the HDAC domain and the nuclear localization sequence (Extended Data Fig. 6a-d), suggesting that Nsp5 may inhibit HDAC2 transport into the nucleus, potentially impacting HDAC2’s mediation of inflammation and interferon response22,23. We also identified an interaction of catalytically dead Nsp5 (C145A) with tRNA methyltransferase 1 (TRMT1), which is responsible for the dimethylation of guanosine (m2,2G) on both nuclear and mitochondrial tRNAs24. We predict TRMT1 is also cleaved by Nsp5 (Extended Data Fig. 6a-d), removing its zinc finger and nuclear localization signal, likely resulting in an exclusively mitochondrial localization.
SARS-CoV-2 interacts with multiple innate immune pathways
Several innate immune signaling proteins are targeted by SARS-CoV-2 viral proteins. The IFN pathway is targeted by Nsp13 (TBK1 and TBKBP1), Nsp15 (RNF41 / Nrdp1) and Orf9b (TOMM70); while the NF-κB pathway was targeted by Nsp13 (TLE1, 3, and 5) Orf9c (NLRX1, F2RL1, NDFIP2). Also, two other E3 ubiquitin ligases that regulate antiviral innate immune signaling, TRIM59 and MIB1, are bound by Orf3a and Nsp9, respectively25,26
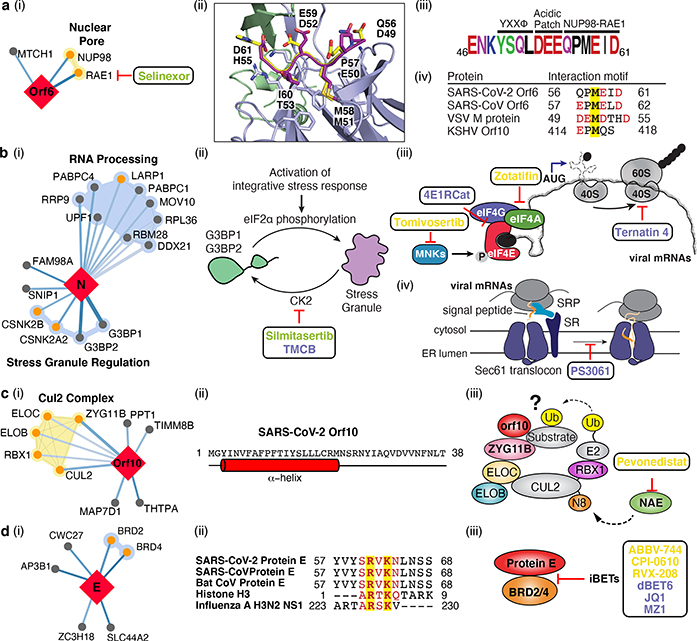
We also identified interactions between SARS-CoV-2 Orf6 and NUP98-RAE1 (Fig. 4a i), an interferon-inducible mRNA nuclear export complex27 that is hijacked or degraded by multiple viruses including VSV, Influenza-A, KSHV, and Polio, and is a restriction factor for Influenza-A infection28–31. The X-ray structure of VSV M protein complexed with NUP98-RAE132 reveals key binding interactions, including a buried methionine residue on the M protein packing into a hydrophobic pocket in RAE1, and neighboring acidic residues interacting with a basic patch on the NUP98-RAE1 complex32. These features are also present in a conserved motif in the C-terminal region of SARS-CoV-2 Orf6 (Fig. 4a ii-iv, Extended Data Fig. 7a,b), providing a structural hypothesis for the observed interaction. Orf6 of SARS-CoV antagonizes host interferon signaling by perturbing nuclear transport33, and the NUP98-RAE1 interaction with Orf6 may perform the same function for SARS-CoV-2.
Figure 4: The SARS-CoV-2 interactome reveals novel aspects of SARS-CoV-2 biology and pharmacological targets.
(a) Orf6 interacts with an mRNA nuclear export complex that (i) can be targeted by Selinexor. (ii) Carboxy-terminal peptide of SARS-CoV-2 Orf6 (dark purple) modeled into the binding site of the VSV M protein (yellow)-NUP98 (green)-RAE1 (light purple) complex (PDB ID: 4OWR). Orf6 and M protein residues labeled. (iii) C-terminal sequence of SARS-CoV-2 Orf6, highlighting described trafficking motifs and putative NUP98-RAE1 binding sequence. Chemical properties of amino acids: polar (green), neutral (purple), basic (blue), acidic (red), and hydrophobic (black). (iv) Putative NUP98-RAE1 interaction motifs (negatively charged residues (red) surrounding a conserved methionine (yellow)) from several viral species. (b) Protein N targets stress granule (SG) proteins (i). (ii) Inhibition of Casein kinase II (silmitasertib or TMCB) disrupting SGs. (iii) Translation initiation inhibition: MNK inhibitor (tomivosertib) prevents phosphorylation of eIF4E; 4ER1Cat blocks the interaction of eIF4E with eIF4G. Inhibition of eIF4A (zotatifin) may prevent unwinding of the viral 5’ UTR to thwart its translation. Targeting translation elongation factor-1A ternary complex (ternatin-4) or (iv) Sec61 translocon (PS3061) can prevent viral protein production and membrane insertion, respectively. (c) Orf10 interacts with the CUL2ZYG11B complex (i). (ii) Orf10 predicted secondary structure. (iii) Orf10 might hijack CUL2ZYG11B for ubiquitination of host proteins which can be inhibited by pevonedistat. (d) Envelope (E) protein interacts with bromodomain proteins (i). (ii) Alignment of proteins E of SARS-CoV-2, SARS-CoV and bat CoV with histone H3 and NS1 protein of Influenza A H3N2. Identical and similar amino acids are highlighted. (iii) Bromodomain inhibitors (iBETs) might disrupt the interaction between protein E and BRDs. Figure shows FDA approved drugs (green), clinical candidates (yellow), and preclinical candidates (purple).
SARS-CoV-2 interacts with the host translation machinery
SARS-CoV-2 nucleocapsid (N) binds the stress granule (SG) proteins G3BP1/2, and to other host mRNA binding proteins including the mTOR-regulated translational repressor LARP1, the protein kinases CK2, and mRNA decay factors UPF1 and MOV10 (Fig. 4b i). Manipulation of SG and related RNA biology is common among coronaviridae34–36 and stress granule formation is thought to be a primarily antiviral response. The promotion of G3BP aggregation by eIF4A inhibitors28,37 may partially explain their antiviral activity (see below).
All coronavirus mRNAs rely on cap-dependent translation to produce their proteins, a process enhanced in trans by the SARS-CoV N protein38. Key eIF4F-cap binding complex constituents – the cap binding protein eIF4E, scaffold protein eIF4G, and the DEAD-box helicase eIF4A – are candidates for therapeutic targeting of coronaviruses39,40. Therapeutic targeting (Fig. 4b ii-iii) of viral translation by interfering with the eIF4F complex formation or the interactions between viral proteins N, Nsp2, Nsp8, and the translational machinery may have therapeutic benefits (see below and Fig. 6).
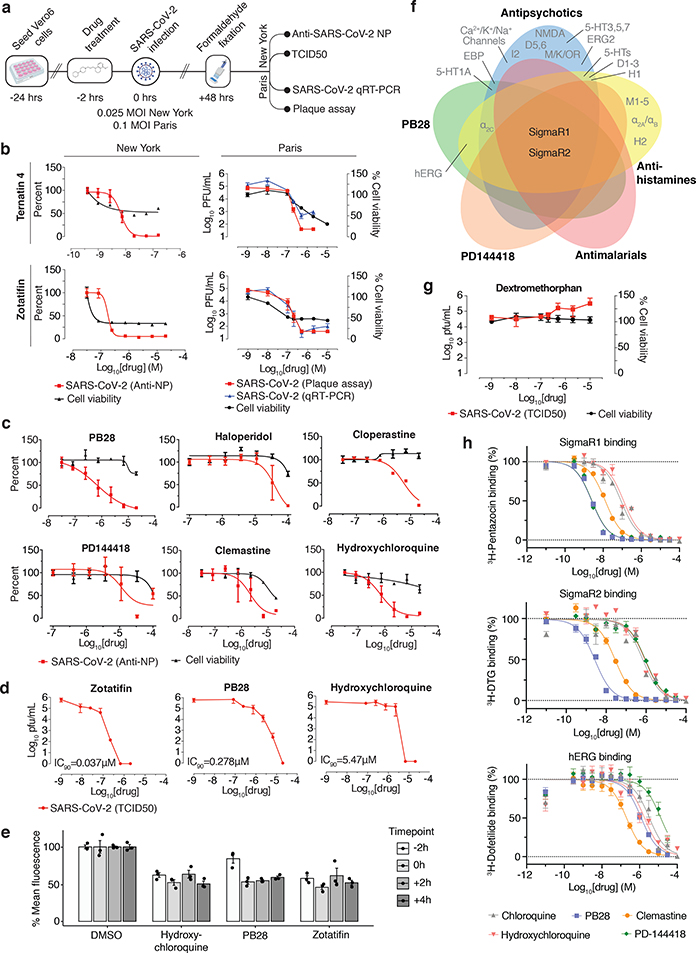
Figure 6: The anti-viral activity of the translation inhibitors and Sigma receptor ligands.
(a) Schema of viral infectivity assays. (b) The mRNA translation inhibitors (zotatifin, ternatin 4) reduce viral infectivity in a concentration-dependent matter (viral infectivity: red, Anti-NP or Plaque assay, blue, qRT-PCR; cell viability: black, with the initial decline likely reflecting cytostatic and not cytotoxic effects). Data=mean±SD; n=6 biologically independent samples for cell viability data and DMSO controls from Paris; all others n=3. (c) Sigma drugs and preclinical molecules inhibit viral infectivity (colored as in b). Data=mean±SD; n=3 biologically independent samples. (d) TCID50 assays using zotatifin, PB28 and hydroxychloroquine (e) Drugs added before or after high titer virus (MOI=2) had similar antiviral effects (viral infectivity: Anti-NP). Data=mean±SD; n=3 biologically independent samples. (f) SigmaR1 and SigmaR2 are the common targets of the Sigma ligands at the 1 μM activity threshold56. (g) Dextromethorphan increases viral titers (viral titer TCID50: red; cell viability: black). Data=mean±SD; n=3 biologically independent samples. (h) SigmaR1/R2 on-target Kd values vs. those for the hERG ion channel. PB28 and PD-144418 show 500 to 5000-fold, while chloroquine and hydroxychloroquine ~30-fold selectivity between these targets. pKi values for hERG vs. SigmaR1 vs. SigmaR2 are: chloroquine (5.5±0.1; 7.1±0.1; 6.3±0.1); hydroxychloroquine (5.6±0.2; 6.9±0.2; 6.0±0.1); PB28 (6.0±0.1; 8.7±0.1; 8.6±0.1); PD-144418 (5.0±0.2; 8.7±0.1; 6.1±0.1); clemastine (6.8±0.2; 8.0±0.1; 7.6±0.1). All data are shown as mean±SD; PB28, clemastine, PD-144418 n=9 biologically independent samples for SigmaR1/R2 and hERG; chloroquine, hydroxychloroquine n=6 for SigmaR1/R2 and n=4 for hERG.
Cotranslational entry into the secretory pathway is a potential target for SARS-CoV-2 inhibition. Up to ten SARS-CoV-2 proteins are predicted to undergo ER membrane insertion mediated by the Sec61 translocon, which localizes to SARS-CoV replication complexes41. Furthermore, high-confidence interactions between Nsp8 and three SRP components suggest viral hijacking of Sec61-mediated protein translocation into the ER. Sec61 inhibitors of protein biogenesis such as PS3061 (Fig. 4b iv), previously shown to inhibit other enveloped RNA viruses42,43, may also block SARS-CoV-2 replication and assembly.
The novel Orf10 of SARS-CoV-2 interacts with a Cullin ubiquitin ligase complex
Viruses commonly hijack ubiquitination pathways for replication and pathogenesis44. The novel Orf10 of SARS-CoV-2 interacts with members of a Cullin 2 (CUL2) RING E3 ligase complex (Fig. 4c i), specifically the CUL2ZYG11B complex. ZYG11B is the highest scoring protein in the Orf10 interactome, suggesting a direct interaction. Despite its small size (38aa), Orf10 appears to contain an alpha helical region (Fig 4c ii) that could be adopted in complex with CUL2ZYG11B. The ubiquitin transfer to a substrate requires neddylation of CUL2 via NEDD8-activating enzyme (NAE), a druggable target 45 (Fig. 4c iii). Orf10 may bind to the CUL2ZYG11B complex and hijack it for ubiquitination and degradation of restriction factors, or alternatively, ZYG11B may bind the N-terminal glycine in Orf10 to target it for degradation31.
SARS-CoV-2 envelope interacts with bromodomain proteins
We found that the transmembrane E protein, likely resident on ERGIC and golgi membranes, binds to BRD2 and BRD4 (Fig. 4d i), members of the bromodomain and extra-terminal (BET) domain family of epigenetic readers that bind acetylated histones to regulate gene transcription46. The C-terminal region of E mimics the N-terminal segment of histone H3, a known interacting partner of bromodomains47. Importantly, this region of E is highly conserved in SARS and bat coronaviruses, suggesting conserved function (Fig. 4d ii). A similar short peptide motif has also been identified in the NS1 protein of influenza A H3N2 strain, where it interferes with transcriptional processes that support antiviral response47,48. Bromodomain inhibitors (iBETs) might disrupt the interaction between protein E and BRDs (Fig. 4d iii).
For a more comprehensive overview of virus-host interactions, see Supplementary Discussion and Methods.
Identification of existing drugs targeting SARS-CoV-2 human host factors
To disrupt the SARS-CoV-2 interactome, we sought ligands of the human interacting proteins (Methods). Molecules were prioritized by the statistical significance of the interaction between the human and viral proteins; by their status as approved drugs, investigational new drugs (INDs, “clinical”), or as preclinical candidates; by their selectivity; and by their availability (Supplementary Tables 4 and 5). Chemoinformatics searches from the IUPHAR/BPS Guide to Pharmacology (2020–3-12) and the ChEMBL25 database on the human interactors yielded 16 approved drugs, 3 investigational new drugs (clinical), and 18 pre-clinical candidates (Supplementary Table 4); while target- and pathway-specific literature search revealed 13 approved drugs, 9 investigational new drugs (clinical), and 10 preclinical candidates (Supplementary Table 5). Of the 332 human targets that interact with the viral bait proteins with high significance (Fig. 3), 63 have 69 drugs/INDs/preclinical molecules that modulate them and can be overlaid on our protein interaction network (Fig. 5).
Figure 5: Drug-human target network.
PPIs of SARS-CoV-2 baits with approved drugs (green), clinical candidates (yellow), and preclinical candidates (purple) with experimental activities against the host proteins (white background) or previously known host factors (grey background) are shown.
Antiviral activity of host-directed drugs and compounds
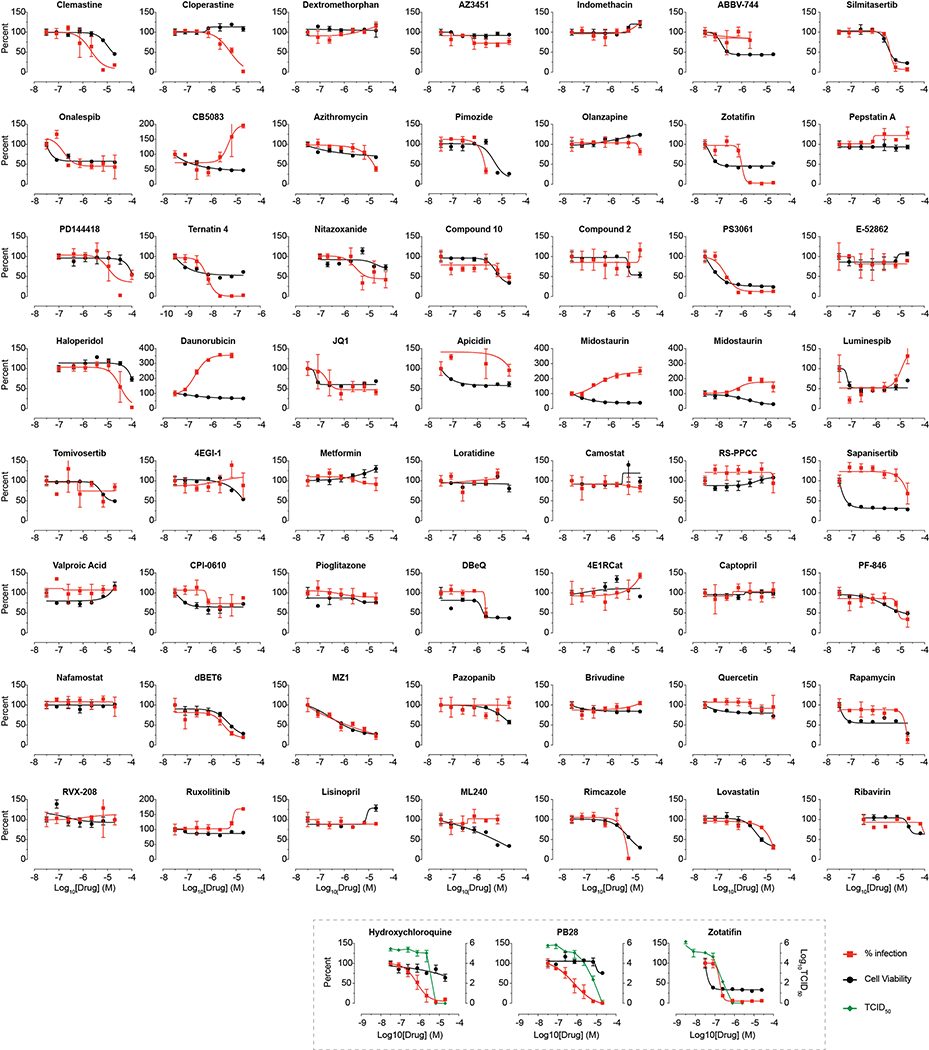
We next investigated the antiviral activity of these drugs and compounds, employing two viral assays (Fig. 6a). First, at Mt Sinai Hospital in New York, we developed a medium-throughput immunofluorescence-based assay (detecting the viral NP protein) to screen 37 compounds for inhibition of SARS-CoV-2 infection in the Vero E6 cell line. Second, at the Institut Pasteur in Paris, viral RNA was monitored using qRT-PCR upon treatment with 44 drugs and compounds. Together, both locations tested 47 of the 69 compounds we identified, plus 13 to expand testing at the SigmaR1/R2 receptors and mRNA translation targets, and 15 additional molecules prioritized by other methods (see Methods and Supplementary Table 6). Viral growth and cytotoxicity were monitored at both institutions (Extended Data Figs. 8 & 9; Supplementary Table 6). Two classes of molecules emerged as effectively reducing viral infectivity: protein biogenesis inhibitors (zotatifin, ternatin-4, and PS3061; Fig. 6b, Extended Data Fig. 9) and ligands of the Sigma1 and Sigma2 receptors: haloperidol, PB28, PD-144418 and hydroxychloroquine, which is undergoing clinical trials in COVID-19 patients49; we also subsequently found the SigmaR1/R2 active drugs clemastine, cloperastine, and progesterone (Fig. 6c, Extended Data Fig. 9) and the clinical molecule siramesine (Extended Data Figure 9) to be antiviral. TCID50 assays on supernatants from infected cells treated with PB28 (IC90 0.278 μM) and zotatifin (IC90 0.037 μM) revealed a more potent inhibition than was observed in the NP-staining assay (Fig. 6d). Interestingly, in this assay, PB28 was ~20 times more potent than hydroxychloroquine (IC90 5.78 μM).
To better understand the mechanism by which these inhibitors exert their antiviral effects, we performed a time course assay where the drugs were added at different times relative to infection (Fig. 6e). This was a single cycle infection at high MOI (2) over the course of 8 hours, where the drugs were either added 2 hours prior to infection or at 0, 2 or 4 hours post infection. PB28, zotatifin, and hydroxychloroquine all decreased the detection of the viral NP protein even in this single cycle assay, indicating the antiviral effect occurs before viral egress from the cell (Fig. 6e). Furthermore, all three molecules inhibited NP expression when added up to 4 hours post-infection, after viral entry has occurred. Thus, these molecules seem to exert their antiviral effect during viral replication.
Coronaviruses rely on cap-dependent mRNA translation through the host translation machinery. eIF4H, an interactor of Nsp9, is a partner of eIF4A, and we observed a strong antiviral effect by the eIF4A inhibitor zotatifin (Fig. 6b), which is currently in a phase I clinical trial for cancer therapy. We also observed potent antiviral effects of the elongation factor-1A (eEF1A) inhibitor ternatin-450 (Fig. 6b), which may suggest that the rate of translation elongation is critical for obtaining optimal levels of viral proteins. Of note, the eEF1A inhibitor Plitidepsin is used clinically in multiple myeloma patients51. Multiple SARS-CoV-2 proteins are predicted to undergo SRP- and Sec61-mediated co-translational insertion into the endoplasmic reticulum, and SRP19/54/72 were identified as Nsp8 interacting proteins (Fig. 3). Consistent with previous studies of flaviviruses42, the Sec61 inhibitor PS3061 also blocked SARS-CoV-2 replication (Extended Data Fig. 9). The two translation inhibitors had cytostatic effect in uninfected Vero cells, which are immortalized cell lines with indefinite proliferative capacity, harboring mutations in key cell cycle inhibitors. Unsurprisingly, these cells are more sensitive to anti-cancer compounds, which affect the cell cycle state of immortalized cells more strongly than normal cells. A critical question going forward is whether these or related inhibitors of viral protein biogenesis will show therapeutic benefit in COVID-19 patients. Plitidepsin is currently under consideration by the Spanish Medicines Agency for a Phase II trial in hospitalized COVID-19 patients.
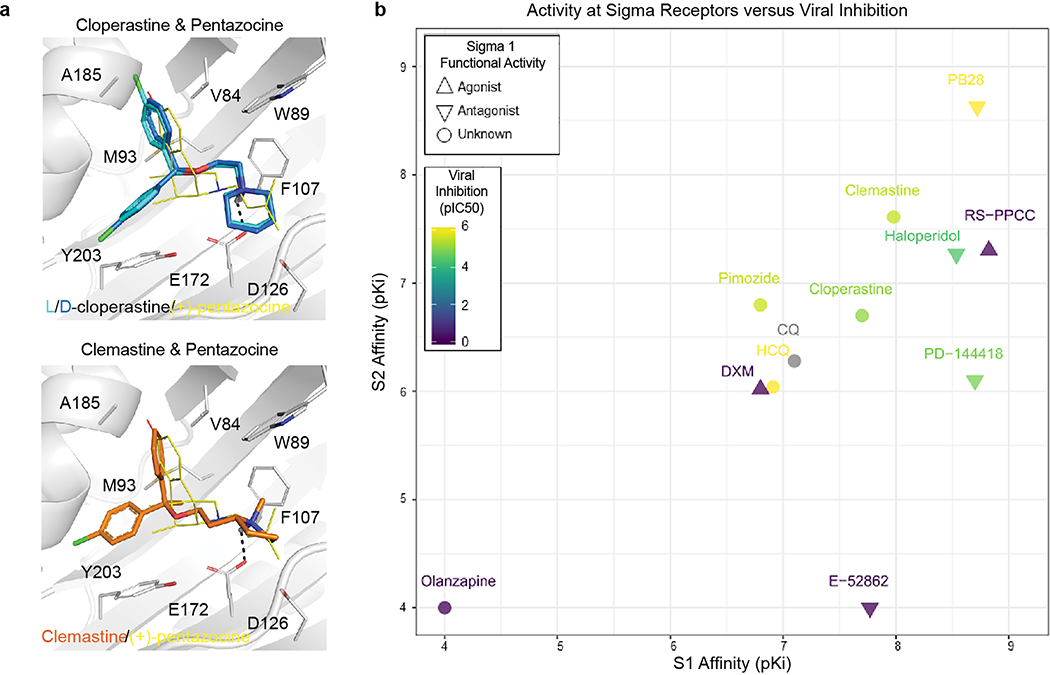
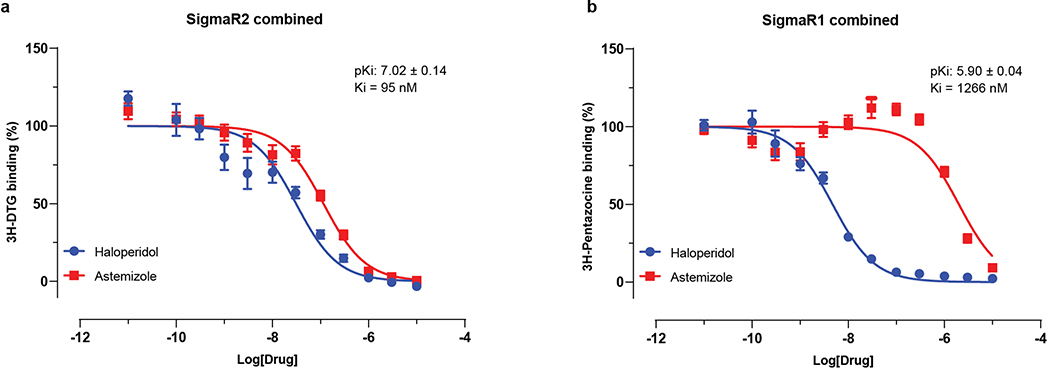
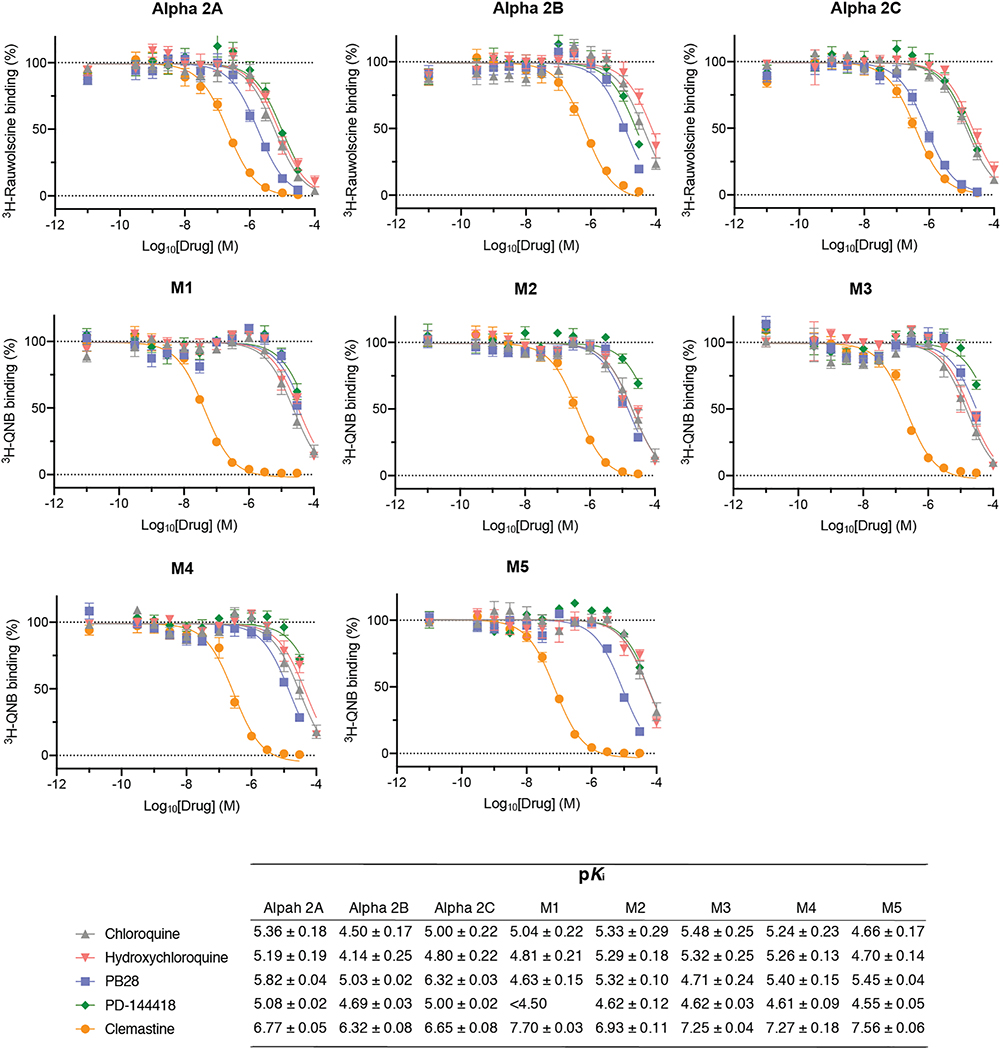
Molecules that target the Sigma1 and Sigma2 receptors perturb the virus through different mechanisms than the translation inhibitors, potentially including cell stress response52. These molecules are also active against other aminergic receptors, but the only ones shared among all of them are the Sigma1 and Sigma2 receptors (Fig. 6f), into which these drugs can be readily modeled (Extended Data Fig. 10a). For instance, the antipsychotic haloperidol inhibits the dopamine D2 and histamine H1 receptors, while clemastine and cloperastine are themselves antihistamines; each also are Sigma receptor ligands with antiviral activity (Fig. 6c). Conversely, the antipsychotic olanzapine, which also inhibits H1 and D2 receptors, has little Sigma receptor activity and is not antiviral (Extended Data Fig. 10b). Which of the Sigma receptors might be most responsible for activity remains uncertain, as does the role of pharmacologically-related targets, such as EBP and related sterol isomerases, whose ligand recognition resembles those of the Sigma receptors. Intriguingly, the Sigma1 benzomorphan agonist, dextromethorphan, actually has pro-viral activity (Fig. 6g), further supporting the role of these receptors in viral infection. Overall, two features merit emphasis. First, several of the Sigma active molecules, like clemastine, cloperastine, and progesterone, are approved drugs with a long history in human therapy. Many other widely-used drugs, active on Sigma receptors, remain to be tested; and indeed, several such as astemizole, which we find to be a 95 nM Sigma2 receptor ligand (Extended Data Fig. 11), verapamil, and amiodarone, have been reported by others to be active in viral replication assays, though this has not been linked to their Sigma receptor activity53,54. Second, the Sigma ligands have a clear separation between antiviral and cytotoxic effects (Fig. 6b and c), and ligands like PB28 have substantial selectivity for the Sigma receptors versus side-effect targets, like the hERG ion channel. Indeed, the lack of selectivity of chloroquine and hydroxychloroquine versus hERG (Fig. 6h) and other off-targets (Extended Data Fig. 12) may be related to the cardiac adverse drug reactions55 that have limited their use.
Discussion
In this study, we have identified 332 high-confidence SARS-CoV-2-human PPIs connected to multiple biological processes, including protein trafficking, translation, transcription and ubiquitination regulation. Against these targets we found 69 ligands, including FDA approved drugs, compounds in clinical trials, and preclinical compounds. Antiviral tests in two different laboratories reveal two broad sets of active drugs and compounds; those impinging on translation, and those modulating Sigma1 and Sigma2 receptors. Within these sets are at least five targets and over ten different chemotypes, suggesting a rich landscape for optimization.
The chemo-proteomic analysis that emerges from this study not only highlights clinically actionable drugs that target human proteins in the interactome, it provides a context for interpreting their mechanism of action. The potent efficacy of the translation inhibitors on viral infectivity—in the 10 to 100 nM range—makes these molecules attractive as candidate antivirals, and also highlights this pathway as a point of intervention. While the mechanism of action of the drugs targeting the Sigma1 and Sigma2 receptors remains less defined, their activity as both anti- and pro-viral agents is mechanistically suggestive. The relatively strong efficacy of PB28, at 280 nM IC90 in the viral titer assay, and its high selectivity against off-targets, suggests that molecules of this class may be optimized towards therapeutics. Whereas it is unclear that approved drugs like clemastine and cloperastine, which are used as antihistamines and antitussives, have pharmacokinetics suitable for antiviral therapy, nor are they free of binding to side-effect targets (Fig. 6f and Extended Data Fig. 12), they have been used for decades. We do caution against their use outside of controlled studies, due to their side-effect liabilities. By the same standard, we find that the widely used antitussive dextromethorphan harbors proviral activity and therefore its use should merit caution and further study in the context of COVID-19. More positively, there are dozens of approved drugs that are active against Sigma receptors that remain untested, some of which, intriguingly, have begun to appear in other studies, although not recognized as Sigma ligands53,54. Therefore, this area of pharmacology has great promise for repurposing and for the optimization of new agents in the fight against COVID-19.
Our approach of host-directed intervention as an antiviral strategy overcomes problems associated with drug resistance and may also provide pan-viral therapies as we prepare for the next pandemic. Furthermore, the possibilities for co-therapies are expanded, for example with drugs directly targeting the virus, including remdesivir, and, as we demonstrate in this study, a rich set of repurposing opportunities are illuminated. More broadly, the pipeline described here represents a new approach for drug discovery not only for pan-viral strategies, but for many diseases, and illustrates the speed in which science can be moved forward using a multi-disciplinary and collaborative approach.
METHODS
Genome annotation
The genbank sequence for SARS-CoV-2 isolate 2019-nCoV/USA-WA1/2020, accession MN985325, was downloaded on January 24, 2020. In total, we annotated 29 possible open reading frames and proteolytically mature proteins encoded by SARS-CoV-21,12. Proteolytic products resulting from Nsp3 and Nsp5-mediated cleavage of the Orf1a / Orf1ab polyprotein were predicted based on the protease specificity of SARS-CoV proteases57, and 16 predicted nonstructural proteins (Nsps) were subsequently cloned (Nsp1-Nsp16). For the Nsp5 protease (3Clike / 3CLpro), we also designed the catalytic mutant Nsp5 C145A 58,59. Open reading frames at the 3’ end of the viral genome annotated in the original genbank file included 4 structural proteins: S, E, M, N, and the additional open reading frames Orf3a, Orf6, Orf7a, Orf8, and Orf10. Based on analysis of open reading frames in the genome and comparisons with other annotated SARS-CoV open reading frames, we annotated a further four open reading frames: Orf3b, Orf7b, Orf9b, and Orf9c.
Cloning
Open reading frames and proteolytically mature Nsps annotated in the SARS-CoV-2 genome were human codon optimized using the IDT codon optimization tool (https://www.idtdna.com/codonopt) and internal EcoRI and BamHI sites eliminated. Start and stop codons were added as necessary to Nsps 1–16, a Kozak sequence was added before each start codon, and a 2×-Strep tag with linker was added to either the N- or C-terminus. To guide our tagging strategy we utilized GPS-Lipid to predict protein lipid modification on termini (http://lipid.biocuckoo.org/webserver.php)60,61, TMHMM Server v. 2.0 to predict transmembrane / hydrophobic regions (http://www.cbs.dtu.dk/services/TMHMM/)62, and SignalP v. 5.0 to predict signal peptides (http://www.cbs.dtu.dk/services/SignalP/)63. IDT gBlocks were ordered for all reading frames with 15-bp overlaps corresponding to flanking sequences of the EcoRI and BamHI restriction sites in the lentiviral constitutive expression vector pLVX-EF1alpha-IRES-Puro (Takara). Vectors were digested and gel purified, and gene fragments were cloned using InFusion (Takara). The Spike protein was synthesized and cloned into pTwist-EF1alpha-IRES-Puro (Twist Biosciences). Nsp16 displayed multiple mutations which could not be repaired prior to the time-sensitive preparation of this manuscript, and Nsp3 was too large to be synthesized in time to be included in this study. Strep-tagged constructs encoding Nsp3, Nsp3 C857A (catalytic mutant), and Nsp16 will be used in future AP-MS experiments.
Cell culture
HEK293T/17 cells were cultured in Dulbecco’s Modified Eagle’s Medium (Corning) supplemented with 10% Fetal Bovine Serum (Gibco, Life Technologies) and 1% Penicillin-Streptomycin (Corning) and maintained at 37°C in a humidified atmosphere of 5% CO2. HEK-293T/17 cells were procured from the UCSF Cell Culture Facility, now available through UCSF’s Cell and Genome Engineering Core ((https://cgec.ucsf.edu/cell-culture-and-banking-services); cell line collection listed here: https://ucsf.app.box.com/s/6xkydeqhr8a2xes0mbo2333i3k1lndqv (CCLZR076)). STR analysis by the Berkeley Cell Culture Facility on August 8, 2017 authenticates HEK-293T/17 cells with 94% probability. Cells were tested on July 3, 2019 using the MycoAlertTM Mycoplasma Detection Kit (Lonza LT07–318) and were negative: B/A ratio < 1 (no detected mycoplasma).
Transfection
For each affinity purification (26 wild-type and one catalytically dead SARS-CoV-2 baits, one GFP control, one empty vector control), ten million HEK293T/17 cells were plated per 15-cm dish and transfected with up to 15 μg of individual Strep-tagged expression constructs after 20–24 hours. Total plasmid was normalized to 15 μg with empty vector and complexed with PolyJet Transfection Reagent (SignaGen Laboratories) at a 1:3 μg:μl ratio of plasmid to transfection reagent based on manufacturer’s recommendations. After more than 38 hours, cells were dissociated at room temperature using 10 ml Dulbecco’s Phosphate Buffered Saline without calcium and magnesium (D-PBS) supplemented with 10 mM EDTA for at least 5 minutes and subsequently washed with 10 ml D-PBS. Each step was followed by centrifugation at 200 × g, 4°C for 5 minutes. Cell pellets were frozen on dry ice and stored at −80°C. For each bait, n=3 independent biological replicates were prepared for affinity purification.
Affinity purification
Frozen cell pellets were thawed on ice for 15–20 minutes and suspended in 1 ml Lysis Buffer [IP Buffer (50 mM Tris-HCl, pH 7.4 at 4°C, 150 mM NaCl, 1 mM EDTA) supplemented with 0.5% Nonidet P 40 Substitute (NP40; Fluka Analytical) and cOmplete mini EDTA-free protease and PhosSTOP phosphatase inhibitor cocktails (Roche)]. Samples were then frozen on dry ice for 10–20 minutes and partially thawed at 37°C before incubation on a tube rotator for 30 minutes at 4°C and centrifugation at 13,000 × g, 4°C for 15 minutes to pellet debris. After reserving 50 μl lysate, up to 48 samples were arrayed into a 96-well Deepwell plate for affinity purification on the KingFisher Flex Purification System (Thermo Scientific) as follows: MagStrep “type3” beads (30 μl; IBA Lifesciences) were equilibrated twice with 1 ml Wash Buffer (IP Buffer supplemented with 0.05% NP40) and incubated with 0.95 ml lysate for 2 hours. Beads were washed three times with 1 ml Wash Buffer and then once with 1 ml IP Buffer. To directly digest bead-bound proteins as well as elute proteins with biotin, beads were manually suspended in IP Buffer and divided in half before transferring to 50 μl Denaturation-Reduction Buffer (2 M urea, 50 mM Tris-HCl pH 8.0, 1 mM DTT) and 50 μl 1× Buffer BXT (IBA Lifesciences) dispensed into a single 96-well KF microtiter plate, respectively. Purified proteins were first eluted at room temperature for 30 minutes with constant shaking at 1,100 rpm on a ThermoMixer C incubator. After removing eluates, on-bead digestion proceeded (below). Strep-tagged protein expression in lysates and enrichment in eluates were assessed by western blot and silver stain, respectively. The KingFisher Flex Purification System was placed in the cold room and allowed to equilibrate to 4°C overnight before use. All automated protocol steps were performed using the slow mix speed and the following mix times: 30 seconds for equilibration/wash steps, 2 hours for binding, and 1 minute for final bead release. Three 10 second bead collection times were used between all steps.
On-bead digestion
Bead-bound proteins were denatured and reduced at 37°C for 30 minutes and after bringing to room temperature, alkylated in the dark with 3 mM iodoacetamide for 45 minutes and quenched with 3 mM DTT for 10 minutes. Proteins were then incubated at 37°C, initially for 4 hours with 1.5 μl trypsin (0.5 μg/μl; Promega) and then another 1–2 hours with 0.5 μl additional trypsin. To offset evaporation, 15 μl 50 mM Tris-HCl, pH 8.0 were added before trypsin digestion. All steps were performed with constant shaking at 1,100 rpm on a ThermoMixer C incubator. Resulting peptides were combined with 50 μl 50 mM Tris-HCl, pH 8.0 used to rinse beads and acidified with trifluoroacetic acid (0.5% final, pH < 2.0). Acidified peptides were desalted for MS analysis using a BioPureSPE Mini 96-Well Plate (20mg PROTO 300 C18; The Nest Group, Inc.) according to standard protocols.
Mass spectrometry data acquisition and analysis
Samples were re-suspended in 4% formic acid, 2% acetonitrile solution, and separated by a reversed-phase gradient over a nanoflow C18 column (Dr. Maisch). Each sample was directly injected via a Easy-nLC 1200 (Thermo Fisher Scientific) into a Q-Exactive Plus mass spectrometer (Thermo Fisher Scientific) and analyzed with a 75 min acquisition, with all MS1 and MS2 spectra collected in the orbitrap; data were acquired using the Thermo software Xcalibur (4.2.47) and Tune (2.11 QF1 Build 3006). For all acquisitions, QCloud was used to control instrument longitudinal performance during the project64. All proteomic data was searched against the human proteome (uniprot reviewed sequences downloaded February 28th, 2020), EGFP sequence, and the SARS-CoV-2 protein sequences using the default settings for MaxQuant (version 1.6.11.0)65,66. Detected peptides and proteins were filtered to 1% false discovery rate in MaxQuant, and identified proteins were then subjected to protein-protein interaction scoring with both SAINTexpress (version 3.6.3)14 and MiST (https://github.com/kroganlab/mist)15,67. We applied a two-step filtering strategy to determine the final list of reported interactors which relied on two different scoring stringency cutoffs. In the first step, we chose all protein interactions that possess a MiST score ≥ 0.7, a SAINTexpress BFDR ≤ 0.05 and an average spectral count ≥ 2. For all proteins that fulfilled these criteria we extracted information about stable protein complexes they participate in from the CORUM68 database of known protein complexes. In the second step we then relaxed the stringency and recovered additional interactors that (1) form complexes with interactors determined in filtering step 1 and (2) fulfill the following criteria: MiST score ≥ 0.6, SAINTexpress BFDR ≤ 0.05 and average spectral counts ≥ 2. Proteins that fulfilled filtering criteria in either step 1 or step 2 were considered to be HC-PPIs and visualized with Cytoscape (version 3.7.1)69. Using this filtering criteria, nearly all of our baits recovered a number of HC-PPIs in close alignment with previous datasets reporting an average of ~6 PPIs per bait70. However, for a subset of baits (Orf8, Nsp8, Nsp13, and Orf9c) we observed a much higher number of PPIs passing these filtering criteria. For these four baits, the MiST scoring was instead performed using a larger in-house database of 87 baits that were prepared and processed in an analogous manner to this SARS-CoV-2 dataset. This was done to provide a more comprehensive collection of baits for comparison, to minimize the classification of non-specifically binding background proteins as HC-PPIs. All mass spectrometry raw data and search results files have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD01811771,72. PPI networks have also been uploaded to NDEx.
Gene ontology over-representation analysis
The targets of each bait were tested for enrichment of Gene Ontology (GO Biological Process) terms. The over-representation analysis (ORA) was based on the hypergeometric distribution and performed using the enricher function of clusterProfiler package in R with default parameters. The gene ontology terms were obtained from the c5 category of Molecular Signature Database (MSigDBv6.1). Significant GO terms (1% FDR) were identified and further refined to select non-redundant terms. In order to select non-redundant gene sets, we first constructed a GO term tree based on distances (1-Jaccard Similarity Coefficients of shared genes) between the significant terms. The GO term tree was cut at a specific level (h=0.99) to identify clusters of non-redundant gene sets. For a bait with multiple significant terms belonging to the same cluster, we selected the broadest term i.e. largest gene set size.
Virus interactome similarity analysis
Interactome similarity was assessed by comparing the number of shared human interacting proteins between pathogen pairs, using a hypergeometric test to calculate significance. The background gene set for the test consisted of all unique proteins detected by mass spectrometry across all pathogens (N=10,181 genes).
Orf6 peptide modeling
The proposed interaction between Orf6 and the NUP98-RAE1 complex was modeled in PyRosetta 473 (release v2020.02-dev61090) using the crystal structure of Vesicular stomatitis virus matrix (M) protein bound to NUP98-RAE1 as a template32 (PDB 4OWR downloaded from the PDB-REDO server74). The M protein chain (C) was truncated after residue 54 to restrict the model to the putative interaction motif in Orf6 (M protein residues 49–54, sequence DEMDTH). These residues were mutated to the Orf6 sequence, QPMEID, using the mutate_residue function in the module pyrosetta.toolbox, without repacking at this initial step. After all six residues were mutated, the full model was relaxed to a low energy conformation using the FastRelax protocol in the module pyrosetta.rosetta.protocols.relax. FastRelax was run with constraints to starting coordinates and scored with the ref2015 score function. The resulting model was inspected for any large energetic penalties associated with the modeled peptide residues or those NUP98 and RAE1 residues interacting with the peptide, and was found to have none. The model was visualized in PyMOL (The PyMOL Molecular Graphics System, Version 2.3.4 Schrödinger, LLC.).
Orf10 secondary structure prediction
The secondary structure of Orf10 was predicted using JPRED (https://www.compbio.dundee.ac.uk/jpred/index.html)75.
Protein E alignment
Protein E sequences from SARS-CoV-2 (YP_009724392.1), SARS-CoV (NP_828854.1), and bat SARS-like CoV (AGZ48809.1) were aligned using Clustal Omega76, and then manually aligned to the sequences of histone H3 (P68431) and Influenza A H3N2 NS1 (YP_308845.1).
Chemoinformatic analysis of SARS-CoV-2 interacting partners
To identify drugs and reagents that modulate the 332 host factors interacting with SARS-CoV-2-HEK293T/17 (MiST >= 0.70), we used two approaches: 1) a chemoinformatic analysis of open-source chemical databases and 2) a target- and pathway-specific literature search, drawing on specialist knowledge within our group. Chemoinformatically, we retrieved 2,472 molecules from the IUPHAR/BPS Guide to Pharmacology (2020–3-12)56 (Supplementary Table 7) that interacted with 30 human “prey” proteins (38 approved, 71 in clinical trials), and found 10,883 molecules (95 approved, 369 in clinical trials) from the ChEMBL25 database77 (Supplementary Table 8). For both approaches, molecules were prioritized on their FDA approval status, activity at the target of interest better than 1 μM, and commercial availability, drawing on the ZINC database78. FDA approved molecules were prioritized except when clinical candidates or preclinical research molecules had substantially better selectivity or potency on-target. In some cases, we considered molecules with indirect mechanisms of action on the general pathway of interest based solely on literature evidence (e.g., captopril modulates ACE2 indirectly via its direct interaction with Angiotensin Converting Enzyme, ACE). Finally, we predicted 6 additional molecules (2 approved, 1 in clinical trials) for proteins with MIST scores between 0.7–0.6 to viral baits (Supplementary Tables 4 and 5). Complete methods can be found here (https://github.com/momeara/BioChemPantry/tree/master/vignette/COVID19).
Molecular docking
After their chemoinformatic assignment to the Sigma1 receptor, cloperastine and clemastine were docked into the agonist-bound state structure of the receptor (6DK1)79 using DOCK3.780. The best scoring configurations that ion-pair with Glu172 are shown; both l-cloperastine and clemastine receive solvation-corrected docking scores between −42 and −43 kcal/mol, indicating high complementarity.
Viral growth and cytotoxicity assays in the presence of inhibitors (Mt. Sinai)
2,000 Vero E6 cells were seeded into 96-well plates in DMEM (10% FBS) and incubated for 24 h at 37C, 5% CO2. Vero E6 cells used were purchased from ATCC and thus authenticated (VERO C1008 [Vero 76, clone E6, Vero E6] (ATCC® CRL-1586™); tested negative for mycoplasma contamination prior to commencement). Two hours before infection, the medium was replaced with 100ul of DMEM (2% FBS) containing the compound of interest at concentrations 50% greater than those indicated, including a DMSO control. The Vero E6 cell line used in this study is a kidney cell line; therefore, we cannot exclude that lung cells yield different results for some inhibitors (also see Methods Institut Pasteur). Plates were then transferred into the BSL3 facility and 100 PFU (MOI 0.025) was added in 50ul of DMEM (2% FBS), bringing the final compound concentration to those indicated. Plates were then incubated for 48 h at 37C. After infection, supernatants were removed and cells were fixed with 4% formaldehyde for 24 hours prior to being removed from the BSL3 facility. The cells were then immunostained for the viral NP protein (anti-sera produced in the Garcia-Sastre lab; 1:10,000) with a DAPI counterstain. Infected cells (488nM) and total cells (DAPI) were quantified using the Celigo (Nexcelcom) imaging cytometer. Infectivity is measured by the accumulation of viral NP protein in the nucleus of the Vero E6 cells (fluorescence accumulation). Percent infection was quantified as ((Infected cells/Total cells) - Background) *100 and the DMSO control was then set to 100% infection for analysis. The IC50 and IC90 for each experiment were determined using the Prism (GraphPad Software) software. For select inhibitors, infected supernatants were assayed for infectious viral titer using the Median Tissue Culture Infectious Dose (TCID)50 method. For this, infectious supernatants were collected at 48h post infection and frozen at −80°C until later use. Infectious titers were quantified by limiting dilution titration on Vero E6 cells. Briefly, Vero E6 cells were seeded in 96-well plates at 20,000 cells/well. The next day, SARS-CoV-2-containing supernatant was applied at serial 10-fold dilutions ranging from 10−1 to 10−6 and, after 5 days, viral CPE was detected by staining cell monolayers with crystal violet. TCID50/mL were calculated using the method of Reed and Muench. Cytotoxicity was also performed using the MTT assay (Roche), according to the manufacturer’s instructions. Cytotoxicity was performed in uninfected VeroE6 cells with same compound dilutions and concurrent with viral replication assay. All assays were performed in biologically independent triplicates.
Cells and viruses (Institut Pasteur)
African green monkey kidney epithelial Vero E6 (ATCC, CRL-1586, authenticated by ATCC and tested negative for mycoplasma contamination prior to commencement [Vero 76, clone E6, Vero E6] (ATCC® CRL-1586™)) were maintained in a humidified atmosphere at 37°C with 5% CO2, in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% (v/v) fetal bovine serum (FBS, Invitrogen) and 5 units/mL penicillin and 5 μg/mL streptomycin (Life Technologies). The Vero E6 cell line used in this study is a kidney cell line; therefore, we cannot exclude that lung cells yield different results for some inhibitors (also see Methods Mt. Sinai). SARS-CoV-2, isolate France/IDF0372/2020, was supplied by the National Reference Centre for Respiratory Viruses hosted by Institut Pasteur (Paris, France) and headed by Pr. Sylvie van der Werf. The human sample from which strain BetaCoV/France/IDF0372/2020 was isolated has been provided by Dr. X. Lescure and Pr. Y. Yazdanpanah from the Bichat Hospital, Paris, France. he BetaCoV/France/IDF0372/2020 strain was supplied through the European Virus Archive goes Global (Evag) platform, a project that has received funding from the European Union’s Horizon 2020 research and innovation programme under the grant agreement No 653316. Viral stocks were prepared by propagation in Vero E6 cells in DMEM supplemented with 2% FBS and 1 μg/ml TPCK-trypsin (Sigma-Aldrich). Viral titers were determined by plaque assay in Minimum Essential Media supplemented with 2% (v/v) FBS (Invitrogen) and 0.05% agarose. All experiments involving live SARS-CoV-2 were performed at Institut Pasteur Paris (IPP) in compliance with IPP’s guidelines following Biosafety Level 3 (BSL-3) containment procedures in approved laboratories. All experiments were performed in at least three biologically independent samples.
Antiviral activity assays (Institut Pasteur)
Vero E6 cells were seeded at 1.5×104 cells per well in 96-well plates 18h prior to the experiment. Two hours prior to infection, the cell culture supernatant of triplicate wells was replaced with media containing 10 μM, 2 μM, 500 nM, 200 nM, 100 nM or 10 nM of each compound or the equivalent volume of maximum DMSO vehicle used as a control. At the time of infection, the drug-containing media was removed, and replaced with virus inoculum (MOI of 0.1 PFU/cell) containing TPCK-trypsin (Sigma-Aldrich). Following a one-hour adsorption at 37 °C, the virus inoculum was removed and 200μL of drug- (or vehicle-) containing media added. 48h post infection (p.i.), the cell culture supernatant was used to extract RNA using the Direct-zol-96 RNA extraction kit (Zymo) following the manufacturer’s instructions. Detection of viral genomes in the extracted RNA was performed by RT-qPCR, using previously published SARS-CoV-2 specific primers81. Specifically, the primers target the N gene region: 5′-TAATCAGACAAGGAACTGATTA-3′ (Forward) and 5′-CGAAGGTGTGACTTCCATG-3′ (Reverse). RT-qPCR was performed using the Luna Universal One-Step RT-qPCR Kit (NEB) in an Applied Biosystems QuantStudio 6 thermocycler, using the following cycling conditions: 55°C for 10 min, 95°C for 1 min, and 40 cycles of 95°C for 10 sec, followed by 60°C for 1 min. The quantity of viral genomes is expressed as PFU equivalents, and was calculated by performing a standard curve with RNA derived from a viral stock with a known viral titer. In addition to measuring viral RNA in the supernatant derived from drug-treated cells, infectious virus was quantified by plaque assay.
Cell viability assays (Institut Pasteur)
Cell viability in drug-treated cells was measured using AlamarBlue reagent (ThermoFisher). Briefly, 48 h post treatment, the drug-containing media was removed and replaced with AlamarBlue and incubated for 1h at 37°C and fluorescence measured in a Tecan Infinity 2000 plate reader. Percentage viability was calculated relative to untreated cells (100% viability) and cells lysed with 20% ethanol (0% viability), included in each plate.
Plaque assays (Institut Pasteur)
Viruses were quantified by plaque assays. For this, Vero E6 cells were seeded in 24-well plates at a concentration of 7.5× 104 cells per well. The following day, 10-fold serial dilutions of individual virus samples in serum-free MEM media were added to infect the cells at 37°C for 1 hour. After the adsorption time the overlay media was added at final concentration of 2% FBS / MEM media and 0.05% Agarose to achieve a semi-solid overlay. Plaque assays were incubated at 37° C for 3 days before fixation with 4% formalin and visualization using crystal violet solution.
Off-target assays for Sigma receptor drugs and ligands
hERG binding assays were carried out as previously described82. Briefly, compounds were incubated with hERG membranes, prepared from HEK293 cells stably expressing hERG channels, and [3H]-Dofetilide (5 nM final) in a total of 150 ul for 90 min at room temperature in the dark. Reactions were stopped by filtering the mixture onto a glass fiber, quickly washed three times to remove unbound [3H]-Dofetilide. The filter was dried in a microwave, melted with a scintillant cocktail, and wrapped in a plastic film. Radioactivity was counted on a MicroBeta counter and results were analyzed in Prism by fitting to the built-in one binding function to obtain affinity Ki. Radioligand binding assays for the muscarinic and alpha-adrenergic receptors were performed as previously described83. Detailed protocols are available on the NIMH PDSP website at https://pdspdb.unc.edu/html/tutorials/UNC-CH%20Protocol%20Book.pdf
Extended Data
Extended Data Figure 1. Mutations in overlapping coding regions result in premature termination of Orf3a and Orf9c.
(a) Table of the SARS-CoV-2 proteins, including molecular weight, sequence similarity with the SARS-CoV homolog, and inferred function based on the SARS-CoV homolog. (b) Immunoblot detection of 2xStrep tag demonstrates expression of each bait in input samples, as indicated by red arrowhead. For each bait, input from one of the three replicates prepared and affinity purified for mass spectrometry was used for western blot (n=1). For gel source data, see Supplementary Figure 1. (c) Schematic representation of Orf3a (light green) and Orf3b (dark green) overlapping regions. A premature stop codon in Orf3b at position 14 (E14*) corresponds to a Q57H mutation in Orf3a. (d) Schematic of the N gene (red), Orf9b (green) and Orf9c (green) overlapping regions. Two mutations in the N protein (S194L and S197L) correspond to premature stop codons at positions 41 and 44 in Orf9c. The analysis is based on 2,784 sequences obtained from GISAID on April 4, 2020.
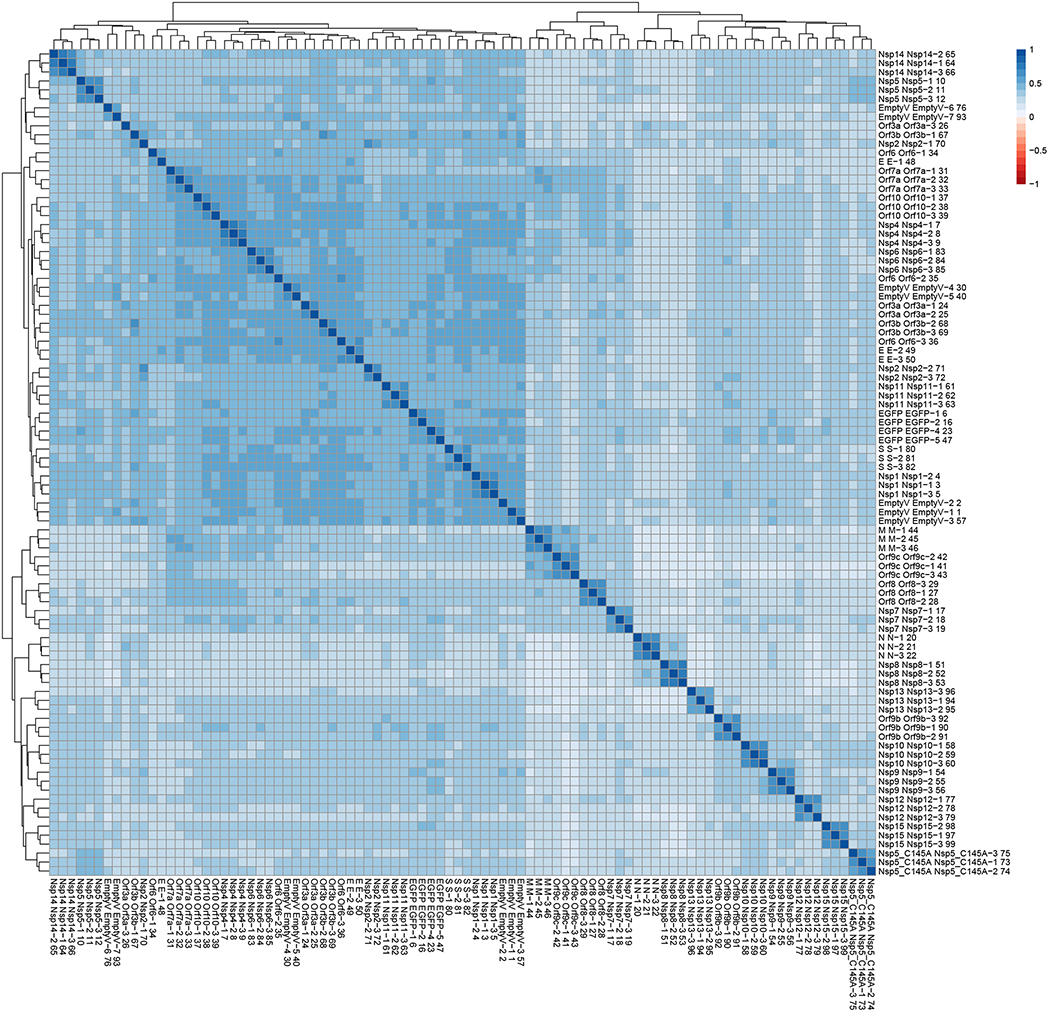
Extended Data Figure 2. Clustering analysis of AP-MS dataset reveals biological replicates of individual baits are well correlated.
All MS runs (n=3 biologically independent samples, run in replicates) were compared and clustered using artMS (David Jimenez-Morales, Alexandre Rosa Campos, and John Von Dollen, and Nevan Krogan. (2019). artMS: Analytical R tools for Mass Spectrometry. R package version 1.3.9. https://github.com/biodavidjm/artMShttps://github.com/bio-davidjm/artMS). This figure depicts all Pearson’s pairwise correlations between MS runs, and is clustered according to similar correlation patterns. Correlation between replicates for individual baits ranges from 0.46–0.72, and in most cases the experiments corresponding to each bait cluster together, with the exception of a couple of baits with lower numbers of specific host interactions (e.g. E, Nsp2, Orf6, Orf3a, and Orf3b).
Extended Data Figure 3. Gene ontology biological process enrichments for SARS-CoV-2 host factors.
We performed GO biological process enrichments (see Methods) for the host factors identified as binding to each SARS-CoV-2 viral protein and represent here the top 5 most significant terms for each viral protein. The p-values were calculated by a hypergeometric test and a false discovery rate was used to account for multiple hypothesis testing.
Extended Data Figure 4. Pfam protein families enrichments for SARS-CoV-2 host factors.
The enrichment of individual PFAM domains was calculated using a hypergeometric test where success is defined as the number of domains, and the number of trials is the number of individual preys affinity purified with each viral bait. The population values were the numbers of individual PFAM domains in the human proteome. The p-values were not adjusted for multiple testing. To make sure that the p-values that signify enrichment were meaningful, we only considered PFAM domains that have been affinity purified at least three times with any SARS-CoV-2 protein, and which occur in the human proteome at least five times. Here, we show PFAM domains with the lowest p-value for a given viral bait protein.
Extended Data Figure 5. Lung mRNA expression and specificity of SARS-CoV-2-interacting human proteins relative to other proteins.
(a) Scatterplot of the lung mRNA expression (TPM) versus enrichment of lung mRNA expression (lung TPM/median all tissue TPM) for human interacting proteins. Red points denote drug targets that are labeled with their gene names. Points above the horizontal blue line represent interacting proteins that are enriched in lung expression and show how most SARS-CoV-2 interacting proteins tend to be enriched in the lung. (b) Gene expression in the lung of the high-confidence human interacting proteins was observed to be higher when compared to all other proteins (blue=interacting proteins; n=332; median=25.52 TPM; grey=all other proteins; n=13583; median=3.198 TPM, p=.0007 using a t-test).
Extended Data Figure 6. Candidate targets for the viral Nsp5 protease.
(a) Nsp5 WT and Nsp5 C145A (catalytic dead mutant) interactome. (b) Domain maps of HDAC2 and TRMT1 illustrating predicted cleavage sites (using NetCorona 1.0). HDAC: Histone Deacetylase Domain, NLS: Nuclear Localization Sequence, MTS: Mitochondrial Targeting Sequence, SAM-MT: S-adenosylmethionine-Dependent Methyltransferase Domain. (c) Peptide docking of predicted cleavage peptides into the crystal structure of SARS-CoV Nsp5. (d) Nsp5 cleavage consensus site for SARS-CoV (left) and SARS-CoV-2 (right).
Extended Data Figure 7. Consensus analysis of SARS-CoV-2 Orf6 homologs.
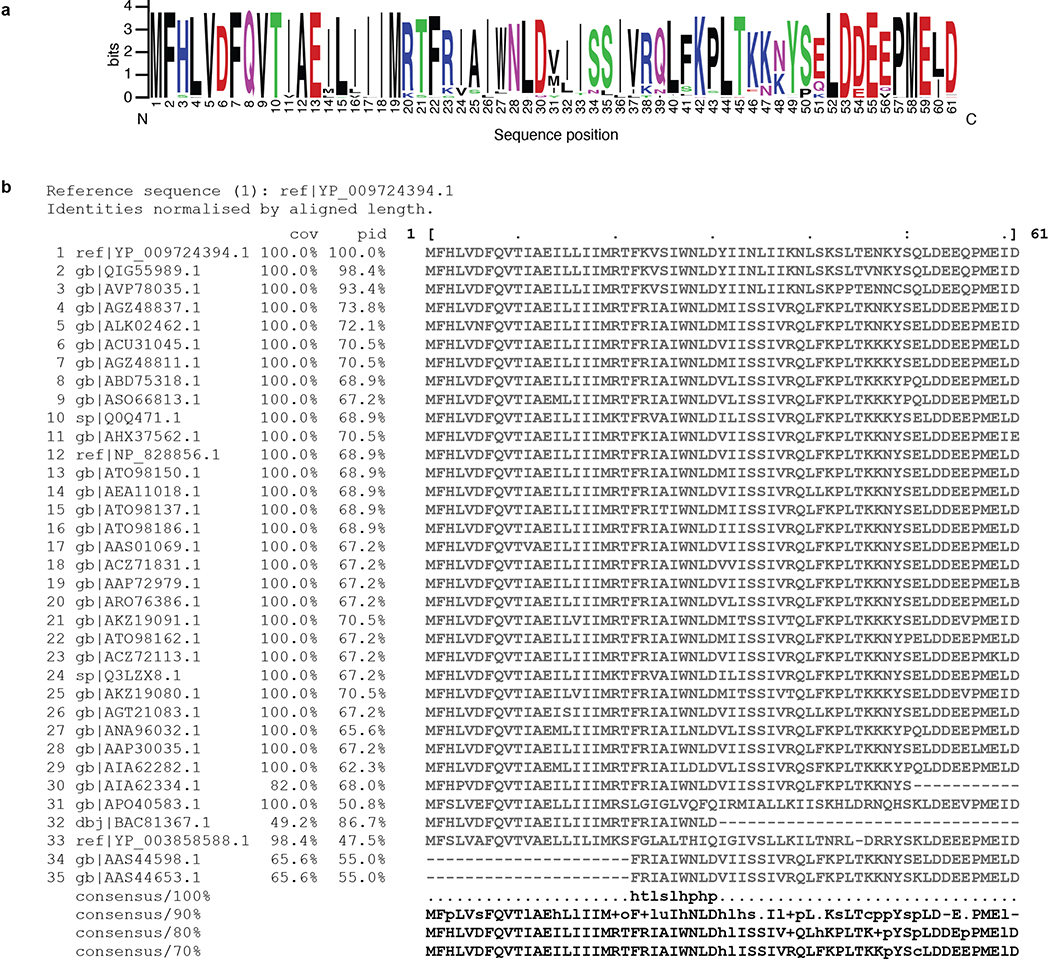
(a) Sequence logo of SARS-CoV-2 Orf6 homologs, showing sequence conservation at each position computed from a multiple sequence alignment of 35 sequences. The key methionine M58, and the acidic residues E55, E59, and D61 of the putative NUP98-RAE1 binding motif are shown to be highly conserved. Homology determined from alignments to full length sequences. Colors indicated chemical properties of amino acids: polar (G, S, T, Y, C, green), neutral (Q, N, purple), basic (K, R, H, blue), acidic (D, E, red), and hydrophobic (A, V, L, I, P, W, F, M, black). (b) Multiple sequence alignment of SARS-CoV-2 Orf6 homologs. Query sequence shown at top (sequence 1 ref|YP_009724394.1). Sequence coverage (cov) and percent identity (pid) shown for each homologous sequence.
Extended Data Figure 8. Viral growth and cytotoxicity for compounds tested in New York.
Viral growth (percent infection; red) and cytotoxicity (black) results for compounds tested at Mount Sinai in New York. Zotatifin, hydroxychloroquine, and PB28 were also tested in Median Tissue Culture Infectious Dose assay (TCID50; green). Zotafitin and Midostaurin were tested in two independent experiments and data are shown in two individual panels. Data=mean±SD; all n=3 biologically independent samples.
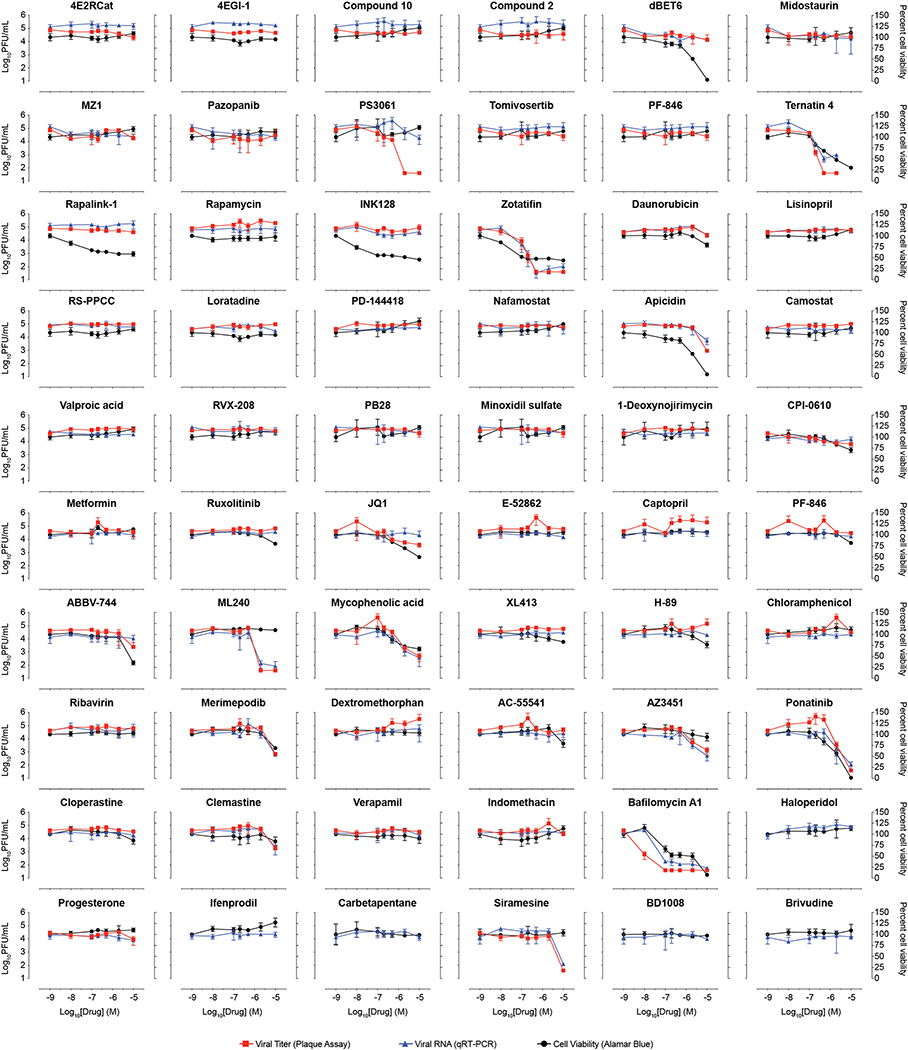
Extended Data Figure 9. Virus plaque assays, qRT-PCR, and cell viability for compounds tested in Paris.
Plaque assay (viral titer; red), qRT-PCR (viral RNA; blue) and cell viability (Alamar Blue; black) results for compounds tested at the Pasteur Institute in Paris. PF-846 was tested in two independent experiments and data are shown in two individual panels. Data=mean±SD; n=3 biologically independent samples for drug-treated cells and n=5 for PS3061, n=6 for DMSO controls.
Extended Data Figure 10. Activity of Sigma ligands.
(a) The drugs cloperastine and clemastine can be readily fit into the agonist-bound structure of the Sigma1 receptor. (b) Compounds tested for antiviral activity with annotated Sigma 1 Receptor (SIGMAR1) and/or Sigma 2 Receptor (SIGMAR2/TMEM97) activity are scatter-plotted. Inhibition pIC50 values of SARS-CoV-2 infection is shown from blue to yellow, mode of functional activity at SIGMA1R is shown by mark shape (upwards triangle: agonist, downwards triangle: antagonist, circle: binding), and pKi values for SIGMA1R and SIGMAR2 are shown along the x- and y-axes. We have not yet tested chloroquine for antiviral activity. E-52862 binding at SIGMAR2/TMEM97 is only reported to be greater than 1 μM. Activities of pimozide and olanzapine at SIGMAR2/TMEM97 have not been reported. Activity of olanzapine at SIGMAR1 is reported to be greater than 5 μM.
Extended Data Figure 11. Astemizole is a potent Sigma2 Receptor Ligand.
Concentration response curves of astemizole from radio-ligand displacement assays for (a) the Sigma2 (95 nM IC50) and (b) the Sigma1 (1.3 uM IC50) receptors are shown. Data=mean±SEM; n= 4 independent assays on each receptor.
Extended Data Figure 12. Off-target activities for characteristic Sigma receptor ligands.
Dose response curves against a panel of eight targets that can confer adverse cardiac, respiratory, and dry-mouth effects for chloroquine, hydroxychloroquine, PB28, PD-144418, and clemastine. These results are not meant to represent or replace a comprehensive test against off-target panels, as might commonly be assayed in drug progression for clinical use. The 8 targets include the Alpha-2A adrenergic receptors: Alpha 2A (ADRA2A), Alpha 2B (ADRA2B), and Alpha 2C (ADRA2C); as well as the Muscarinic acetylcholine receptors: M1 (CHRM1), M2, (CHRM2), M3 (CHRM3), M4 (CHRM4) and M5 (CHRM5). Data=mean±SD; all n= 3 biologically independent samples.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Joshua Sarlo for his artistic contributions to the manuscript, Andy Kruse for his advice on Sigma receptor functional activity and pharmacologically related enzymes, and Steve Worland and eFFECTOR Therapeutics for providing zotatifin.
This research was funded by grants from the National Institutes of Health (P50AI150476, U19AI135990, U19AI135972, R01AI143292, R01AI120694, P01A1063302, and R01AI122747 to N.J.K.; R35GM122481 to B.K.S.; 1R01CA221969 and 1R01CA244550 to K.M.S.; K08HL124068 to J.S.C.; 1F32CA236347–01 to J.E.M., NIMH Psychoactive Drug Screening Contract to B.L.R.); from the Defense Advance Research Projects Agency HR0011–19-2–0020 (to B.K.S., N.J.K., A. G-S., K.M.S., D.A.A., & K.M.V.); by CRIP (Center for Research for Influenza Pathogenesis), a NIAID supported Center of Excellence for Influenza Research and Surveillance (CEIRS, contract # HHSN272201400008C) and by supplements to NIAID grant U19AI135972 and DoD grant W81XWH-19-PRMRP-FPA to A.G.-S.; by the Laboratoire d’Excellence “Integrative Biology of Emerging Infectious Diseases” (grant ANR-10-LABX-62-IBEID) to M.V., O.S. and C.d’E.; funding from F. Hoffmann-La Roche and Vir Biotechnology and gifts from The Ron Conway Family. K.M.S is an investigator of the Howard Hughes Medical Institute. The views, opinions, and findings contained in this study are those of the authors and do not represent the official views, policies, or endorsement of the Department of Defense or the U.S. Government. The authors would like to thank Todd from FedEx for his heroic effort helping us to ship out drugs and SARS-CoV-2 expression constructs across the globe. The authors acknowledge their partners and families for support in child care and other matters during this time.
Abbreviations
- HC-PPIs
High-confidence protein-protein interactions
- PPIs
protein-protein interaction
- AP-MS
affinity purification-mass spectrometry
- COVID-19
Coronavirus Disease-2019
- ACE2
angiotensin converting enzyme 2
- Orf
open reading frame
- Nsp3
papain-like protease
- Nsp5
main protease
- Nsp
nonstructural protein
- TPM
transcripts per million
Footnotes
COMPETING INTERESTS
The Krogan Laboratory has received research support from Vir Biotechnology and F. Hoffmann-La Roche. Kevan Shokat has consulting agreements for the following companies involving cash and/or stock compensation: Black Diamond Therapeutics, BridGene Biosciences, Denali Therapeutics, Dice Molecules, eFFECTOR Therapeutics (zotatifin & tomivosertib), Erasca, Genentech/Roche, Janssen Pharmaceuticals, Kumquat Biosciences, Kura Oncology, Merck, Mitokinin, Petra Pharma, Qulab Inc. Revolution Medicines (WDB002), Type6 Therapeutics, Venthera, Wellspring Biosciences (Araxes Pharma). Jack Taunton is a cofounder and shareholder of Global Blood Therapeutics, Principia Biopharma, Kezar Life Sciences, and Cedilla Therapeutics. Jack Taunton and Phillip P. Sharp are listed as inventors on a patent application describing the cyclic depsipeptide, PS3061 (PCT/US2019/024731 by the Regents of the University of California and Kezar Life Sciences). Davide Ruggero is a shareholder of eFFECTOR Therapeutics, Inc., and a member of its scientific advisory board (zotatifin & tomivosertib).
Data availability: The mass spectrometry raw data and search results files generated during the current study are available in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD018117 (https://www.ebi.ac.uk/pride/archive/projects/PXD018117) and PPI networks have also been uploaded to NDEx (https://public.ndexbio.org/#/network/43803262-6d69-11ea-bfdc-0ac135e8bacf). An interactive version of these networks, including relevant drug and functional information, can be found at the following website: http://kroganlab.ucsf.edu/network-maps. All data generated or analysed during this study are included in this published article (and its Supplementary Information files and as Source Data).
Code availability: Complete methods for chemoinformatic analysis can be found here (https://github.com/momeara/BioChemPantry/tree/master/vignette/COVID19); details on MIST scoring can be found here (https://github.com/kroganlab/mist).
Expression vectors used in this study are readily available from the authors to biomedical researchers and educators in the non-profit sector.
Supplementary Information is available for this paper.
Reprints and permissions information is available at www.nature.com/reprints
The authors have not filed for patent protection on the SARS-CoV-2 host interactions or the use of predicted drugs for treating COVID-19 to ensure all the information is freely available to accelerate the discovery of a treatment.
REFERENCES
- 1.Wu F et al. A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Novel Coronavirus (2019-nCoV) situation reports. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports.
- 3.Wang C, Horby PW, Hayden FG & Gao GF A novel coronavirus outbreak of global health concern. The Lancet vol. 395 470–473 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu N et al. China Novel Coronavirus Investigating and Research Team. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su S et al. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 24, 490–502 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gates B Responding to Covid-19 — A Once-in-a-Century Pandemic? New England Journal of Medicine (2020) doi: 10.1056/nejmp2003762. [DOI] [PubMed] [Google Scholar]
- 7.Sheahan TP et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 11, 222 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheahan TP et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 and multiple endemic, epidemic and bat coronavirus. bioRxiv 2020.03.19.997890 (2020) doi: 10.1101/2020.03.19.997890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paton J Moderna’s Coronavirus Vaccine Trial Set to Begin This Month. Bloomberg News (2020). [Google Scholar]
- 10.Hoffmann M et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell (2020) doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prussia A, Thepchatri P, Snyder JP & Plemper RK Systematic approaches towards the development of host-directed antiviral therapeutics. Int. J. Mol. Sci. 12, 4027–4052 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan JF-W et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 9, 221–236 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fehr AR & Perlman S Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 1282, 1–23 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teo G et al. SAINTexpress: improvements and additional features in Significance Analysis of INTeractome software. J. Proteomics 100, 37–43 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jäger S et al. Global landscape of HIV-human protein complexes. Nature 481, 365–370 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bojkova D et al. SARS-CoV-2 infected host cell proteomics reveal potential therapy targets. In Review (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckhardt M, Hultquist JF, Kaake RM, Hüttenhain R & Krogan NJ A systems approach to infectious disease. Nat. Rev. Genet. (2020) doi: 10.1038/s41576-020-0212-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harcourt J et al. Severe Acute Respiratory Syndrome Coronavirus 2 from Patient with 2019 Novel Coronavirus Disease, United States. Emerg. Infect. Dis. 26, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang D et al. A deep proteome and transcriptome abundance atlas of 29 healthy human tissues. Mol. Syst. Biol. 15, e8503 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li M et al. Identification of antiviral roles for the exon-junction complex and nonsense-mediated decay in flaviviral infection. Nat Microbiol (2019) doi: 10.1038/s41564-019-0375-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Penn BH et al. An Mtb-Human Protein-Protein Interaction Map Identifies a Switch between Host Antiviral and Antibacterial Responses. Mol. Cell 71, 637–648.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barnes PJ Role of HDAC2 in the pathophysiology of COPD. Annu. Rev. Physiol. 71, 451–464 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Xu P et al. NOS1 inhibits the interferon response of cancer cells by S-nitrosylation of HDAC2. J. Exp. Clin. Cancer Res. 38, 483 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dewe JM, Fuller BL, Lentini JM, Kellner SM & Fu D TRMT1-Catalyzed tRNA Modifications Are Required for Redox Homeostasis To Ensure Proper Cellular Proliferation and Oxidative Stress Survival. Mol. Cell. Biol. 37, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kondo T, Watanabe M & Hatakeyama S TRIM59 interacts with ECSIT and negatively regulates NF-κB and IRF-3/7-mediated signal pathways. Biochem. Biophys. Res. Commun. 422, 501–507 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Li S, Wang L, Berman M, Kong Y-Y & Dorf ME Mapping a dynamic innate immunity protein interaction network regulating type I interferon production. Immunity 35, 426–440 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faria PA et al. VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Mol. Cell 17, 93–102 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Slaine PD, Kleer M, Smith NK, Khaperskyy DA & McCormick C Stress Granule-Inducing Eukaryotic Translation Initiation Factor 4A Inhibitors Block Influenza A Virus Replication. Viruses 9, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reineke LC et al. Casein Kinase 2 Is Linked to Stress Granule Dynamics through Phosphorylation of the Stress Granule Nucleating Protein G3BP1. Mol. Cell. Biol. 37, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kindrachuk J et al. Antiviral potential of ERK/MAPK and PI3K/AKT/mTOR signaling modulation for Middle East respiratory syndrome coronavirus infection as identified by temporal kinome analysis. Antimicrob. Agents Chemother. 59, 1088–1099 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Timms RT et al. A glycine-specific N-degron pathway mediates the quality control of protein N-myristoylation. Science 365, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quan B, Seo H-S, Blobel G & Ren Y Vesiculoviral matrix (M) protein occupies nucleic acid binding site at nucleoporin pair (Rae1 • Nup98). Proc. Natl. Acad. Sci. U. S. A. 111, 9127–9132 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frieman M et al. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 81, 9812–9824 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakagawa K, Narayanan K, Wada M & Makino S Inhibition of Stress Granule Formation by Middle East Respiratory Syndrome Coronavirus 4a Accessory Protein Facilitates Viral Translation, Leading to Efficient Virus Replication. J. Virol. 92, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raaben M, Groot Koerkamp MJA, Rottier PJM & de Haan CAM Mouse hepatitis coronavirus replication induces host translational shutoff and mRNA decay, with concomitant formation of stress granules and processing bodies. Cell. Microbiol. 9, 2218–2229 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivanov P, Kedersha N & Anderson P Stress Granules and Processing Bodies in Translational Control. Cold Spring Harb. Perspect. Biol. 11, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson PA et al. Abstract 2698: eFT226, a potent and selective inhibitor of eIF4A, is efficacious in preclinical models of lymphoma. Experimental and Molecular Therapeutics (2019) doi: 10.1158/1538-7445.am2019-2698. [DOI] [Google Scholar]
- 38.Nakagawa K, Lokugamage KG & Makino S Viral and Cellular mRNA Translation in Coronavirus-Infected Cells. Adv. Virus Res. 96, 165–192 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Müller C et al. Broad-spectrum antiviral activity of the eIF4A inhibitor silvestrol against corona- and picornaviruses. Antiviral Res. 150, 123–129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cencic R et al. Blocking eIF4E-eIF4G interaction as a strategy to impair coronavirus replication. J. Virol. 85, 6381–6389 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knoops K et al. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6, e226 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shah PS et al. Comparative Flavivirus-Host Protein Interaction Mapping Reveals Mechanisms of Dengue and Zika Virus Pathogenesis. Cell 175, 1931–1945.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heaton NS et al. Targeting Viral Proteostasis Limits Influenza Virus, HIV, and Dengue Virus Infection. Immunity 44, 46–58 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mahon C, Krogan NJ, Craik CS & Pick E Cullin E3 ligases and their rewiring by viral factors. Biomolecules 4, 897–930 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soucy TA et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Faivre EJ et al. Selective inhibition of the BD2 bromodomain of BET proteins in prostate cancer. Nature 578, 306–310 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Filippakopoulos P et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149, 214–231 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marazzi I et al. Suppression of the antiviral response by an influenza histone mimic. Nature 483, 428–433 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Outcomes Related to COVID-19 Treated With Hydroxychloroquine Among In-patients With Symptomatic Disease - Full Text View - ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT04332991.
- 50.Carelli JD et al. Ternatin and improved synthetic variants kill cancer cells by targeting the elongation factor-1A ternary complex. Elife 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spicka I et al. Randomized phase III study (ADMYRE) of plitidepsin in combination with dexamethasone vs. dexamethasone alone in patients with relapsed/refractory multiple myeloma. Ann. Hematol. 98, 2139–2150 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mitsuda T et al. Sigma-1Rs are upregulated via PERK/eIF2α/ATF4 pathway and execute protective function in ER stress. Biochem. Biophys. Res. Commun. 415, 519–525 (2011). [DOI] [PubMed] [Google Scholar]
- 53.Si L et al. Human organs-on-chips as tools for repurposing approved drugs as potential influenza and COVID19 therapeutics in viral pandemics. bioRxiv 2020.04.13.039917 (2020) doi: 10.1101/2020.04.13.039917. [DOI] [Google Scholar]
- 54.Riva L et al. A Large-scale Drug Repositioning Survey for SARS-CoV-2 Antivirals. bioRxiv 2020.04.16.044016 (2020) doi: 10.1101/2020.04.16.044016. [DOI] [Google Scholar]
- 55.White NJ Cardiotoxicity of antimalarial drugs. Lancet Infect. Dis. 7, 549–558 (2007). [DOI] [PubMed] [Google Scholar]
- 56.Armstrong JF et al. The IUPHAR/BPS Guide to PHARMACOLOGY in 2020: extending immunopharmacology content and introducing the IUPHAR/MMV Guide to MALARIA PHARMACOLOGY. Nucleic Acids Res. 48, D1006–D1021 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang D & Leibowitz JL The structure and functions of coronavirus genomic 3’ and 5’ ends. Virus Res. 206, 120–133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang H et al. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. U. S. A. 100, 13190–13195 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thiel V et al. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84, 2305–2315 (2003). [DOI] [PubMed] [Google Scholar]
- 60.Xie Y et al. GPS-Lipid: a robust tool for the prediction of multiple lipid modification sites. Sci. Rep. 6, 28249 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ren J et al. CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng. Des. Sel. 21, 639–644 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krogh A, Larsson B, von Heijne G & Sonnhammer EL Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 (2001). [DOI] [PubMed] [Google Scholar]
- 63.Almagro Armenteros JJ et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423 (2019). [DOI] [PubMed] [Google Scholar]
- 64.Chiva C et al. QCloud: A cloud-based quality control system for mass spectrometry-based proteomics laboratories. PLoS One 13, e0189209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cox J & Mann M MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 66.Cox J et al. Accurate Proteome-wide Label-free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Verschueren E et al. Scoring Large-Scale Affinity Purification Mass Spectrometry Datasets with MiST. Curr. Protoc. Bioinformatics 49, 8.19.1–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Giurgiu M et al. CORUM: the comprehensive resource of mammalian protein complexes-2019. Nucleic Acids Res. 47, D559–D563 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shannon P et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huttlin EL et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 162, 425–440 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vizcaíno JA et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deutsch EW et al. The ProteomeXchange consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 45, D1100–D1106 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chaudhury S, Lyskov S & Gray JJ PyRosetta: a script-based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics 26, 689–691 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Joosten RP, Long F, Murshudov GN & Perrakis A The PDB_REDO server for macromolecular structure model optimization. IUCrJ 1, 213–220 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Drozdetskiy A, Cole C, Procter J & Barton GJ JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 43, W389–94 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sievers F et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gaulton A et al. The ChEMBL database in 2017. Nucleic Acids Res. 45, D945–D954 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sterling T & Irwin JJ ZINC 15--Ligand Discovery for Everyone. J. Chem. Inf. Model. 55, 2324–2337 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schmidt HR, Betz RM, Dror RO & Kruse AC Structural basis for σ receptor ligand recognition. Nat. Struct. Mol. Biol. 25, 981–987 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mysinger MM & Shoichet BK Rapid context-dependent ligand desolvation in molecular docking. J. Chem. Inf. Model. 50, 1561–1573 (2010). [DOI] [PubMed] [Google Scholar]
- 81.Chu DKW et al. Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia. Clin. Chem. 66, 549–555 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang X-P, Mangano T, Hufeisen S, Setola V & Roth BL Identification of human Ether-à-go-go related gene modulators by three screening platforms in an academic drug-discovery setting. Assay Drug Dev. Technol. 8, 727–742 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Besnard J et al. Automated design of ligands to polypharmacological profiles. Nature 492, 215–220 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.