Significance
How does enamel achieve its remarkable microstructure? This study reveals that the cleaved enamel protein amelogenin adopts a ribbon-like supramolecular structure that controls the growth of mineral into nanofibers. Protein nanoribbons of 15 to 20 nm in width were identified in dental enamel and replicated from a recombinant protein mimicking an enzymatic cleavage product of human amelogenin. In mineralization experiments, these protein structures templated the growth of highly oriented apatite nanofibers along their backbone from an amorphous precursor. In accordance with the natural process of enamel development, this in vitro model suggests that the ability of amelogenin nanoribbons to guide fibrous apatite growth is regulated by enzymatic processing and their interaction with acidic nonamelogenin proteins in the developing enamel matrix.
Keywords: biomineralization, protein assembly, nanoribbon structure, hydroxyapatite, enamel
Abstract
As the hardest tissue formed by vertebrates, enamel represents nature’s engineering masterpiece with complex organizations of fibrous apatite crystals at the nanometer scale. Supramolecular assemblies of enamel matrix proteins (EMPs) play a key role as the structural scaffolds for regulating mineral morphology during enamel development. However, to achieve maximum tissue hardness, most organic content in enamel is digested and removed at the maturation stage, and thus knowledge of a structural protein template that could guide enamel mineralization is limited at this date. Herein, by examining a gene-modified mouse that lacked enzymatic degradation of EMPs, we demonstrate the presence of protein nanoribbons as the structural scaffolds in developing enamel matrix. Using in vitro mineralization assays we showed that both recombinant and enamel-tissue–based amelogenin nanoribbons are capable of guiding fibrous apatite nanocrystal formation. In accordance with our understanding of the natural process of enamel formation, templated crystal growth was achieved by interaction of amelogenin scaffolds with acidic macromolecules that facilitate the formation of an amorphous calcium phosphate precursor which gradually transforms into oriented apatite fibers along the protein nanoribbons. Furthermore, this study elucidated that matrix metalloproteinase-20 is a critical regulator of the enamel mineralization as only a recombinant analog of a MMP20-cleavage product of amelogenin was capable of guiding apatite mineralization. This study highlights that supramolecular assembly of the scaffold protein, its enzymatic processing, and its ability to interact with acidic carrier proteins are critical steps for proper enamel development.
Amelogenin is a highly conserved protein during the evolution of vertebrates (1). It makes up at least 90% of the developing enamel matrix (DEM) secreted by ameloblast during amelogenesis (2). Transgenic mouse models have shown that amelogenin, along with other enamel matrix proteins, are essential for proper enamel formation (3–5). Especially, the self-assembly of amelogenin has widely been recognized as a crucial factor in controlling enamel structural development (6). However, the study of the exact process of how amelogenin regulates enamel structure formation has been challenging because the majority of the organic content is enzymatically processed in the secretory stage and almost fully removed during the maturation stage (2, 7). To circumvent this obstacle and to provide possible models, in vitro recombinant amelogenin self-assembly systems have been developed (8). A widely studied model argues that amelogenin proteins assemble into nanospheres as basic structural units to regulate enamel mineralization (9–11). Recently, our group discovered that nanospheres from recombinant full-length amelogenin can further assemble into uniform nanoribbons in the presence of calcium and phosphate (7, 12, 13). The constructed amelogenin nanoribbons feature signature amyloid characteristics (7, 14) and share corresponding X-ray diffraction (XRD) parameters that previously were characterized in the DEM of biological samples (15–17). Moreover, numerous early tissue-based transmission electron microscopy (TEM) studies also show the presence of filamentous protein assemblies (18–20) that share great structural similarity with the recombinant amelogenin nanoribbons (7, 12). Even though previous XRD and TEM characterizations suggest that nanoribbons may represent the structural nature of the in vivo observed filamentous proteins, they do not provide sufficient resolution to make detailed comparison. In addition, the filamentous proteins in the DEM match the size and alignment of apatite crystal ribbons that initially formed during secretory stage (20), suggesting that the protein superstructure may act as a template for oriented apatite mineralization. Previous attempts, however, to induce templated growth of fibrous apatite crystals along recombinant full-length amelogenin (rH174) nanoribbons failed (7), presumably due to two reasons: 1) full-length amelogenin is quickly processed by matrix metalloproteinase-20 (MMP20) after secretion, and, thus, the major component that accumulates in the DEM is a MMP20 cleavage product and not the full-length amelogenin (21, 22) and 2) studies have shown that nonamelogenin proteins, like enamelin, are critical to induce mineralization in the DEM and possibly act through interaction with amelogenin assemblies (23, 24).
In this study, we aim to obtain direct evidence that connects the in vitro-constructed recombinant amelogenin nanoribbons with the biological matrix structures in enamel tissue and to prove their ability to template apatite crystal formation.
Results
Construction of In Vitro Amelogenin Nanoribbons.
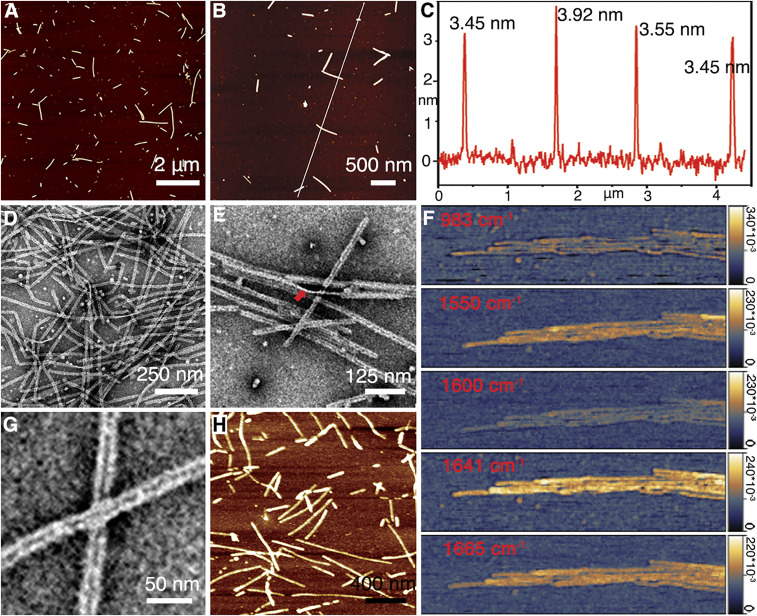
Inspired by the natural proteolytic process during early amelogenesis (21, 25), the MMP20 cleavage product of amelogenin (rH146) was selected as the relevant building block for in vitro assembly studies. rH146 lacks the hydrophilic C terminus (28 residues) compared to the full-length amelogenin (detailed sequence provided in Materials and Methods). Large numbers of rH146 amelogenin nanoribbons were successfully assembled in calcium phosphate solution (Fig. 1). The sample solutions at various assembly times were vortexed evenly and deposited on a mica surface for atomic force microscope (AFM) characterization in ambient conditions: the formation of amelogenin nanoribbons was first observed around 19 h and increased in numbers over time (SI Appendix, Fig. S1). The nanoribbons have a homogeneous thickness at 3.76 ± 0.28 nm (Fig. 1 A–C and SI Appendix, Fig. S2). TEM images provided additional characterization for the assembly process (SI Appendix, Fig. S3): at early time points, protein-building blocks aggregate as spherical or irregular precursors, which can further associate together and adopt nanoribbon structures (SI Appendix, Fig. S3 A–C). As the assembly system matures, amelogenin nanoribbons prevail to be the dominant structure (Fig. 1D and SI Appendix, Fig. S3D). The in vitro-constructed nanoribbon features a homogeneous width of 16.41 ± 1.24 nm (SI Appendix, Fig. S4). The thin edge as seen at the location of twisting further confirms a ribbon-like morphology (Fig. 1E). Atomic force microscope-infrared spectroscopy (AFM-IR) characterizations with high Amide I absorption at 1,641 cm−1 and Amide II absorption at 1,550 cm−1 (Fig. 1F) suggest that rH146 nanoribbons retain the beta-sheet–rich, amyloid-like structure (26) as previously characterized for the rH174 ribbons (14). Significant absorption at 1,665 cm−1 indicates that an alpha helix/beta turn secondary structure (27) is also present. Low absorption at 1,600 and 983 cm−1 was characterized as negative controls. As previously identified for the full-length rH174 nanoribbons (12), rH146 nanoribbons also feature a central dark line under negative staining TEM as a structural signature (Fig. 1G and SI Appendix, Fig. S4). Real-time AFM in aqueous protein solutions (Fig. 1H) and cryogenic electron microscopy studies (SI Appendix, Fig. S5) demonstrate that the ribbon morphology exists in situ in solution and is not a drying artifact.
Fig. 1.
Structural characterization of rH146 nanoribbons. (A and B) AFM images at 8 d show the presence of nanoribbons. (C) Height profile along the white line in B shows the representative measurements of nanoribbon thickness. (D) TEM image of rH146 assembly system at 4 d: The nanoribbon structure has become the dominant assembly product. (E) At a twist of the nanoribbon (indicated with the red arrow) the thin edge of the structure is observed. (F) AFM-IR image of rH146 nanoribbons at various wavelengths between 983 and 1,665 cm−1. (G) Negative stained TEM image of rH146 nanoribbons show unique central dark lines. (H) Real-time AFM image of protein solution with 1 mg/mL rH146 on mica surface reveals nanoribbons exist in solution.
Characterization of Tissue-Based Amelogenin Nanoribbons.
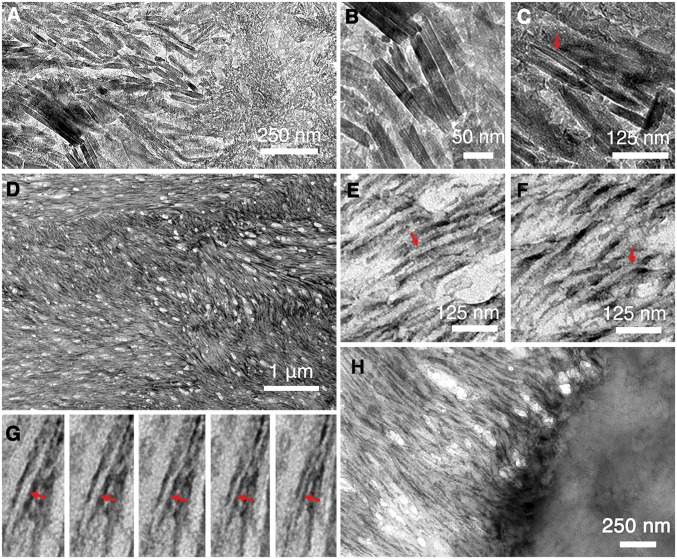
Kallikrein-4 (KLK4) is the enzyme that hydrolyzes the organic content of the DEM during enamel maturation (28). Thus, enamel from mice that lack KLK4 (KLK4−/−) largely comprises a late secretory stage composition and structure (29), where matrix protein is prevalent and its structure possibly preserved. To investigate and obtain direct tissue-based evidence of the protein template structure in the DEM, electron microscopy studies on microtomed KLK4−/− incisor sections were performed. TEM images show that both KLK4−/− enamel and dentin are well mineralized with hydroxyapatite crystals (SI Appendix, Fig. S6). The enamel from KLK4−/− mice is prismatic and maintains the signature interwoven texture (SI Appendix, Fig. S6A). Alignment of uniaxial apatite crystals within enamel rods results in the arcing selected area electron diffraction (SAED) signal of the (002) and (004) planes (SI Appendix, Fig. S6B). In contrast, the mineral phase in the dentin region presents random ordering (SI Appendix, Fig. S6C) with the SAED signal showing as a complete ring (SI Appendix, Fig. S6D). A crystalline phase transition (Fig. 2A) can be observed near the dentin-enamel junction (DEJ) region. Individual apatite crystals can be resolved (Fig. 2B), and some of them present a typical, flexible ribbon-like morphology (Fig. 2C). Even though the amelogenin nanoribbons (Fig. 1) appear as an ideal template to guide the formation of such ribbon-like apatite crystals, a direct observation of an underlying protein matrix by TEM is not feasible without removal of the mineral coating.
Fig. 2.
TEM characterization of KLK4−/− mice incisor sections before and after demineralization. (A) TEM image near the DEJ region before demineralization. (B) Close-up image shows individual apatite crystals. (C) Some apatite crystals present with a typical flexible ribbon-like morphology (indicated with red arrow). (D) TEM image of demineralized enamel region reveals an extensive protein matrix network. (E) Close-up images reveal that the protein matrix network is composed of individual protein nanoribbon structures. Some of these share the unique central dark line (indicated with red arrow) as observed with rH146 nanoribbons in Fig. 1G. (F) Twisting of the nanoribbon structures (indicated with red arrow) is frequently observed. (G) From left to right, a series of TEM images of the same protein nanoribbon (indicated with red arrow) with increasing tilting angles counterclockwise shows the transition from the ribbon top view to its side view. (H) TEM image near the DEJ region after demineralization. Collagen structures can be observed in the demineralized dentin region, and amelogenin nanoribbons can be seen to grow out from the dentin surface directly.
To this end, we performed demineralization studies. Notably, to maintain the structure and organization of the organic matrix, the tissue was chemically crosslinked immediately after mouse scarification (detailed procedures are described in Materials and Methods). In addition, we chose a low-concentration acid (1 mM HCl) instead of ethylenediaminetetraacetic acid (EDTA) to demineralize the sections because the strong chelating agent EDTA has been shown to disassemble in vitro-constructed amelogenin nanoribbons (12) and may also disintegrate in vivo protein matrix structures. Ultra-thin KLK4−/− incisor sections were first picked up on a TEM grid and subsequently demineralized by floating upside down on top of a 1-mM HCl droplet for 5 min. Demineralized sections revealed the presence of extensive filamentous protein networks featuring the signature enamel texture (Fig. 2D and SI Appendix, Fig. S7), which suggests that the unique organization of enamel rods and interrods architecture are directly associated and templated by the protein matrix structures. Close-up images (Fig. 2 E and F) revealed that the protein matrix networks are composed of individual protein nanoribbons. Some of the nanoribbons share the unique central dark line (Fig. 2E and SI Appendix, Fig. S8) as observed for rH146 nanoribbons (Fig. 1G). Twisting of the nanoribbon structures was also commonly seen (Fig. 2F); in contrast, most in vitro-constructed rH146 nanoribbons lay flat on top of the electron microscopy (EM) grid with random orientations (Fig. 1). Tomographic TEM was utilized to further demonstrate the occurrence of a ribbon-like morphology in the tissue: by continuously recording the same nanoribbon during specimen tilting, the thin edge of the nanoribbon was revealed at high angles (Fig. 2G). The width of the tissue-based ribbons was measured to be 16.90 ± 1.81 nm (SI Appendix, Fig. S8), which closely matches that of the rH146 ribbons (SI Appendix, Fig. S4). Immunostaining experiments were carried out to investigate the components of the nanoribbon structures via incubating demineralized sections in primary amelogenin antibody from rabbit and secondary antibody from goat with 5-nm nanogold particle labels subsequently. Even though the antibodies act like spacers, which makes the nanogold labels flexible and difficult to delineate the targeted structures, in this experiment the nanogolds can still be observed to bind with the nanoribbon networks and form a trend line following the local growing direction of the ribbon structures (SI Appendix, Fig. S9 A–C). The binding specificity was confirmed with the control sample in which the primary antibody was diluted 10 times and the secondary antibody concentration was kept constant, and very few gold nanoparticles were observed (SI Appendix, Fig. S9D). Considering that amelogenin comprises more than 90% of the DEM (2), the immunostaining study demonstrates that amelogenin protein is the predominant building block of the nanoribbons in the enamel matrix of KLK4−/− mice.
The interfacial DEJ region is known for its function of providing a crack-arresting barrier (30), and it features unique a microstructure in which type-I collagen fibrils emanate from the dentin and project perpendicularly to the DEJ interface (31). The demineralized DEJ region showed that the organic phases of the dentin and enamel are strongly connected (Fig. 2H) and retain a rugged interface (SI Appendix, Fig. S10A). Amelogenin ribbons are less organized near the DEJ and seem to grow directly from the collagenous dentin surface (Fig. 2H), which may relate to previous observations of enamel crystallites growing from mineral in dentin collagen fibrils (32). Farther out into the enamel region, amelogenin nanoribbons become more organized (SI Appendix, Fig. S10) and reach into tens of micrometers in length that often extend out of the TEM view (SI Appendix, Fig. S10B).
Amorphous Mineralization of Amelogenin Nanoribbon Scaffolds.
To demonstrate the ability of amelogenin nanoribbons to template enamel apatite crystal growth, we designed mineralization studies via the polymer-induced liquid precursor (PILP) process (33) for controlled delivery of calcium and phosphate ions onto the protein scaffolds. Polyaspartic acid (pAsp) has been exploited in the PILP mineralization process to mimic the activity of acidic noncollagen proteins and has proven effective in remineralizing the collagen fibril scaffolds in bone (34) and dentin (35). Here we endeavored to extend the application scope of pAsp as a substitute of acidic nonamelogenin proteins in the DEM (36) to interact with the amelogenin nanoribbon scaffolds.
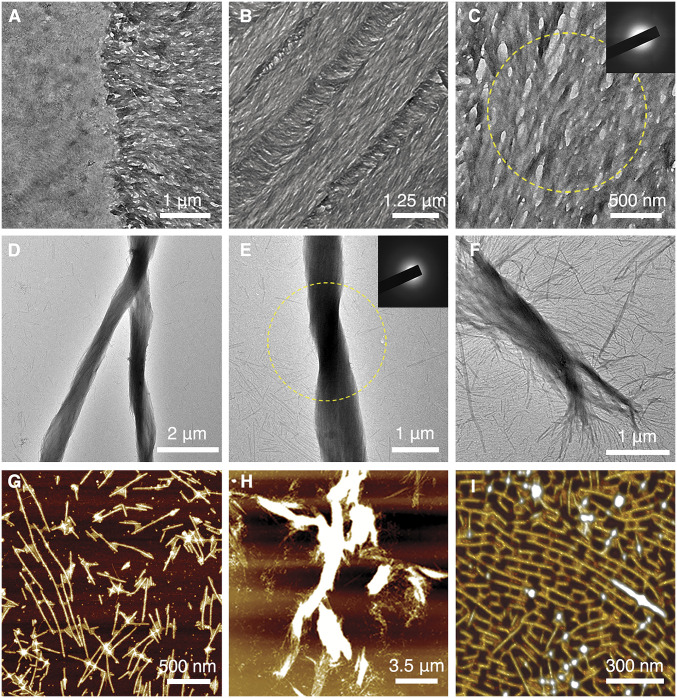
Demineralized KLK4−/− incisor sections were incubated in PILP solution with 0.1 mg/mL pAsp at 37 °C, and the mineral formation was monitored over time. Amorphous calcium phosphate (ACP) readily formed as early as 1 h (SI Appendix, Fig. S11 A and B). At the DEJ region (represented in Fig. 2H), the binding of the mineral phase to the organic matrix is selective and mineral can be observed to preferably form on the enamel region over the dentin region (Fig. 3A). The formed mineral phase follows the protein matrix structure (represented in Fig. 2D and SI Appendix, Fig. S7) and develops into the signature enamel texture (Fig. 3B). The mineral was amorphous at 4.5 h of incubation time at 37 °C (Fig. 3C) and remained amorphous at 2 d (SI Appendix, Fig. S11 C and D) without transforming to crystalline mineral.
Fig. 3.
PILP-induced mineral formation on both tissue-based amelogenin nanoribbons and rH146 amelogenin nanoribbons. (A) TEM image near the DEJ region after incubating demineralized KLK4−/− mouse incisor section with PILP solution at 37 °C for 1 h. (B) TEM image of the demineralized KLK4−/− mouse incisor section after incubating in PILP solution for 4.5 h. (C) Close-up TEM image of amorphous mineral coating over amelogenin ribbons; yellow circle corresponds to area of SAED analysis (Inset), which indicates absence of crystalline phase. (D) TEM image of the rH146 nanoribbon and PILP-solution mixing sample after incubating for 15 d shows the formation of large bundled structures. (E) TEM image of mineralized rH146 ribbon bundle; yellow circle corresponds to area of SAED analysis, SAED (Inset) shows that mineral within the bundled structures remained amorphous at 15 d. (F) Close-up image reveals the process where individual nanoribbons associate together and align to form larger bundles. (G) AFM image of the rH146 nanoribbon and PILP-solution mixing sample. PILP droplets can be seen to bind with rH146 nanoribbons. (H) AFM image of the rH146 nanoribbon and PILP-solution mixing sample shows the formation of large bundles. (I) Three-dimensional rending of AFM image reveals formation of bridges between nanoribbons when exposed to PILP solution, which generates a unique pattern on the mica substrate.
We further investigated if the same binding mechanisms and mineral formation apply when using in vitro-constructed rH146 nanoribbons as scaffolds in a solution environment. The rH146 nanoribbon assembly solution was mixed with PILP solution with 0.1 mg/mL pAsp at a 1:1 volume ratio and incubated at 37 °C. PILP droplets (SI Appendix, Fig. S12 A and B) can be observed to associate with rH146 nanoribbons (SI Appendix, Fig. S12 C and D) and aggregate into elongated structures at 2 h (SI Appendix, Fig. S12 E and F). At 5 h, PILP droplets were absorbed, and bundled organic-inorganic complexes containing amorphous mineral were formed (SI Appendix, Fig. S12 G and H). Similar to the mineral formed on demineralized tissue sections, the mineral phase within the bundled structures remained amorphous for extended periods of time (Fig. 3 D and E and SI Appendix, Fig. S13). A close-up image reveals the process where rH146 nanoribbons self-organize into bundled structures and initial ACP formation occurs (Fig. 3F). The nature of the bundled structures as a protein-apatite mineral complex was further confirmed by scanning transmission electron microscopy-energy dispersive X-ray spectroscopy (STEM-EDS) studies (SI Appendix, Fig. S14). The PILP-rH146 nanoribbon interaction (Fig. 3G) and the formation of the large-scale bundled complex (Fig. 3H) can also be characterized by AFM. Unique patterns (Fig. 3I and SI Appendix, Fig. S15) can be observed on the two-dimensional AFM substrate as a result of induced association of rH146 nanoribbons.
Amorphous Phase Transforms through Uniaxial Crystallization along Protein Nanoribbons.
The amorphous phase has been recognized as a key precursor stage in biomineralization (33), and the conversion of amorphous calcium phosphate to oriented hydroxyapatite has been proven to be biologically relevant in the formation of enamel (37). To facilitate phase transformation, increased incubation temperature was utilized to provide the thermal mobilities for ion reorganization and the exclusion of water molecules from the amorphous phase (33).
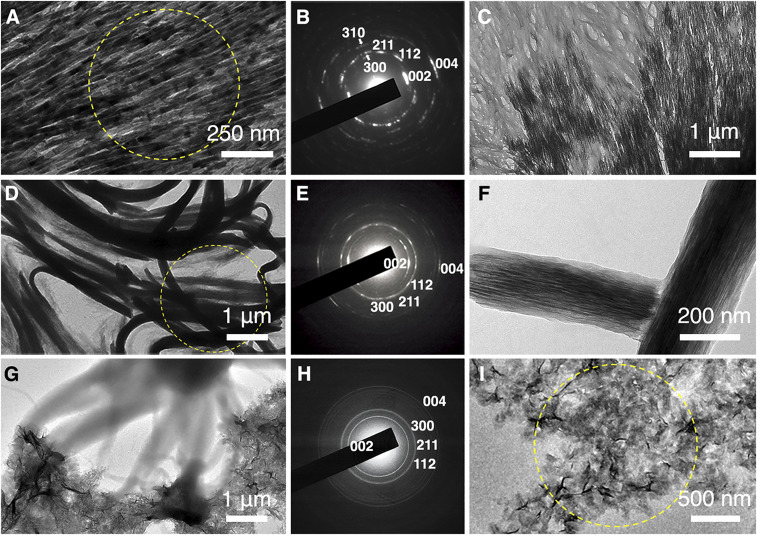
Demineralized KLK4−/− incisor sections were incubated in PILP solution at 50 °C. TEM-SAED studies showed the formation of fibrous hydroxyapatite crystals following the signature enamel texture (Fig. 4A and SI Appendix, Fig. S16A). The formed crystals feature a high degree of order as indicated by the defined diffraction pattern (Fig. 4B). Especially, the (002) and (004) reflections orient in parallel to the ribbon long axis. In fact, the reformed crystalline phase is so perfect that its morphology and texture resemble the original hydroxyapatite fibers before demineralization (SI Appendix, Fig. S16). Notably, we were able to capture the phase transformation event: Fig. 4C shows the front end where the translucent unstructured amorphous phase transformed into well-defined fibrous crystals following the pattern of the underlying protein network. Such observation confirms that the observed hydroxyapatite crystal formation is a result of the induced remineralization experiment and not due to incomplete etching of the tissue sections.
Fig. 4.
Protein-guided transformation from amorphous phase into crystalline phase. (A) TEM image of remineralized KLK4−/− enamel section after incubating in PILP solution at 50 °C for 17 h. (B) SAED data (corresponding area indicated with yellow ring in A shows the crystalline phase is highly ordered hydroxyapatite). The (002) and (004) reflections are in parallel with the ribbon long axis. (C) TEM image of the remineralized KLK4−/− enamel section shows the front end where the amorphous phase transforms into the fibrous crystalline apatite phase. (D) TEM image of the bundled structures after incubating at 85 °C for 1.5 h and subsequently at 37 °C for another 3 d. (E) SAED data (corresponding area indicated with yellow ring in D) suggest that the formed crystals are hydroxyapatite with their c-axes parallel to the bundles’ long axes. (F) Close-up TEM image reveals the formed crystal fibers are oriented along the long axis of the bundled structures. (G) TEM image of the rH174 nanoribbon and PILP-solution mixing sample incubated at 37 °C for 15 d. (H) SAED data (corresponding selected area indicated with yellow circle in I) suggest that the formed crystal is hydroxyapatite with random orientations. (I) Close-up TEM image reveals that formed hydroxyapatite crystals adopt an irregular appearance.
Phase-transformation experiments were also applied to the bundled structures of aligned rH146 nanoribbons formed by interaction with PILP solution. For the large-scale bundled structures (SI Appendix, Fig. S17), phase transformation was achieved at an elevated temperature of 85 °C. TEM-SAED experiments suggest that the crystalline phase is formed at as early as 1.5 h (SI Appendix, Fig. S18), and the sample was further incubated at 37 °C for another 3 d to allow for crystal growth. A TEM image (Fig. 4D) then showed that the bundled superstructures were highly crystallized with hydroxyapatite (Fig. 4E). The (002) and the (004) reflections are in parallel with the major orientation of the bundled structures in the selected area (Fig. 4E). High-resolution analysis shows that long, thin crystal fibers formed within the bundled superstructures (Fig. 4F). The crystals can be observed to follow the long axis of the bundles (Fig. 4F), which is essentially determined by the rH146 amelogenin nanoribbon subunits.
In addition, we tested the influence of amelogenin C terminus on the assembly structures and the induced mineral formation process using the rH174 assembly system as a control. As shown in Fig. 1, rH146 ribbons are generally evenly distributed on substrate surfaces and some aggregation is only due to physical entanglement of the lengthy structures. In contrast, rH174 ribbons (12) tend to aggregate on the TEM grid and AFM substrates (SI Appendix, Fig. S19), which is possibly due to the electrostatic interactions between the charged C termini. When the rH174 ribbon solution was mixed with PILP solution, irregular hydroxyapatite crystals readily formed without the need to increase the temperature (Fig. 4G). Even though the formed crystals are also hydroxyapatite, they feature random orientation (Fig. 4 H and I) and their morphologies are independent of the protein scaffolds (Fig. 4G).
Discussion
A major pillar of enamel mineralization is built on the premise that proteins in the DEM create a supermolecular structure that can guide the growth of oriented, ribbon-like apatite nanocrystals (6, 8, 38, 39). Assembly of nanoribbons from recombinant amelogenin proteins was described about a decade ago (12, 40), and a series of papers has shown the presence of filamentous proteins with cross-beta structure in TEM and X-ray diffraction analysis over the past 60 years (15–20). However, none of these studies succeeded in demonstrating a convincing relationship between the filamentous protein structure and the observed nanofibrillar apatite crystals that comprise mammalian enamel.
This study, on the contrary, was first able to provide high-resolution evidence of nanoribbons (Fig. 2) in enamel by investigating mice that lack the ability to fully degrade enamel proteins. The KLK4 knockout mouse model (29, 41) proved to be a powerful tool for providing a stable enamel matrix that allowed for a detailed analysis of supramolecular structures in the DEM and facilitated the identification of nanoribbons as the organic template that underlies fibrous apatite crystals in developing enamel.
Second, this study demonstrated that amelogenin nanoribbons, both native or recombinant, are able to template uniaxial growth of apatite nanocrystals with high-aspect ratios in vitro (Fig. 4 A and D), further substantiating the evidence that these protein nanostructures are indeed the relevant assemblies in the DEM to control biomineralization of enamel in vivo.
Third, it appears that the in vitro mineralization approach chosen in this study emulated the natural process of enamel mineralization. Currently, it is well understood from ameloblast biology, tissue histology and various characterizations of a series of knockout mice models that enzymatic processing (25, 29, 41) and protein–protein interactions (36, 38) are critical for proper enamel formation. Here, it was demonstrated that the recombinant full-length amelogenin assembly was not capable of controlling fibrous apatite formation along its backbone: When exposed to mineralizing PILP solution, random mineralization was observed without association with the underlying protein structure and without control over crystal morphology (Fig. 4G). In contrast, the study found that assembly of the major MMP-20 cleavage product of amelogenin, which lacked the hydrophilic C terminus of the full-length protein, was able to guide the formation of an amorphous layer of calcium phosphate that further transformed into crystalline ribbon-like apatite structures following the protein supramolecular backbone (Fig. 4 A and D). Transient amorphous phases that transform into fibrous apatite crystals have been described during the early secretory stage of amelogenesis (37). Here, this process appears to have been replicated in vitro and recorded in Fig. 4C, which exactly pinpoints the moment of amorphous to crystalline transformation.
Fourth, in enamel as well as in other mineralized tissues including dentin, bone, and shell, recent evidence suggests that biomineralization requires not only an organic scaffold protein that can template the size and morphology of the mineral but also nonscaffolding proteins which appear to be critical for transport of mineral ions and site-specific mineral deposition on the protein scaffold (24, 34, 42–44). As an example, during dentin formation, the mineralization of collagen fibrils is orchestrated by acidic proteins such as dentin matrix protein-1 and dentin sialophosphoprotein (45). In the case of enamel, there is substantial evidence that enamelin may act as a carrier and nucleating protein in the DEM by interacting with amelogenin proteins (46–48). The absence of a substantial amount of any mineral on teeth from enamelin knockout mice suggests a critical role of this molecule in mineral induction (24). Due to the lack of access to a sufficient amount of enamelin (49), which makes up only about 2% of the DEM and is degraded during enamel maturation, we attempted the use of polyaspartic acid as a mineralizing protein analog and applied pAsp through the PILP method to amelogenin nanoribbon scaffolds. Interestingly, this approach was able to reproduce the formation of oriented fibrous apatite crystals, suggesting that the anionic pAsp 27-kD molecule mimics the activity of other DEM components, possibly the 32-kD fraction of enamelin, as described by others (50, 51). Therefore, this mineralization approach not only appears to emulate the process of early mineral formation in secretory stage amelogenesis (Fig. 4 A and C) but also provides a method to grow nanofibrillar apatite in vitro. The organic nanoribbons developed here are organic scaffolds that can guide uniaxial growth of inorganic minerals over several tens of micrometers while maintaining control over morphology at the nanometer range (Fig. 4 D and F), thus illustrating their potential for use in the synthesis of nanostructured materials.
Materials and Methods
Expression and Purification of Recombinant Amelogenin Protein.
Matrix metalloproteinase-20 proteolytic product (rH146) and recombinant full-length amelogenin (rH174) were expressed in Escherichia coli BL21(DE3) and purified as previously described (52). The purity of the protein batches was >95% as assessed by high-performance liquid chromatography. rH146 and rH174 lacks the first amino acid residue (Met) compared to the native human amelogenin sequences. rH146 lacks the hydrophilic C terminus (28 residues) compared to the rH174 amelogenin. Sequences for rH174 (the C terminus 28 residues are highlighted in boldface) are the following: PLPPHPGHPGYINFSYEVLTPLKWYQSIRPPYPSYGYEPMGGWLHHQIIPVLSQQHPPTHTLQPHHHIPVVPAQQPVIPQQPMMPVPGQHSMTPIQHHQPNLPPPAQQPYQPQPVQPQPHQPMQPQPPVHPMQPLPPQPPLPPMFPMQPLPPMLPDLTLEAWPSTDKTKREEVD.
In Vitro Self-Assembly of Amelogenin Protein.
Utilizing a similar protocol as previously described (7), rH146 protein powder was measured and dissolved in 1 mM HCl at 2 mg/mL, and the resulting solution was vortexed thoroughly and allowed to incubate at 4 °C overnight to ensure dissolution. Calcium and phosphate stock solutions were prepared using reagent grade CaCl2 (132 mM) and KH2PO4 (84 mM) and filtered (0.22 μm) before use. An assembly sample solution of 1 mL total volume containing 1 mg/mL of amelogenin were prepared by adding 250 μL KH2PO4 stock solution to one sample vial containing 250 μL rH146 stock solution and 250 μL CaCl2 stock solution to a second vial containing the same amount of rH146 stock solution. The two vials were combined, resulting in a final concentration of 33 mM CaCl2 and 21 mM KH2PO4. The final pH of the resulting solution was adjusted to 5.3 ± 0.2 with 1 M KOH. The final sample solution was then incubated in a closed vial at 37 °C. The assembly of rH174 amelogenin follows the same procedure.
AFM in Ambient Conditions.
All solutions were vortexed gently 5 to 10 times to get an even distribution of the samples before applying onto AFM substrate. Sample solution (20µL) was deposited on freshly cleaved mica substrate (Electron Microscopy Science) and further incubated for 30 min in a wet cell for immobilization. Excess sample solution was removed with pipette, and the sample surface was immediately washed with 100µL deionized (DI) water followed by air drying. AFM imaging was performed on a Bruker Multimode 8 with Nanoscope V controller (Bruker) under ScanAsyst in air mode using a silicon probe (Bruker ScanAsyst Air-HR). Micrographs were flattened and analyzed using Nanoscope Analysis v1.8 software.
AFM in Liquid Conditions.
A total of 20 μL of rH146 assembly solution was deposited on freshly cleaved mica substrate (Electron Microscopy Science) and further incubated for 30 min in a wet cell for immobilization. Excessive sample solution was removed, but a thin layer of liquid was left on the sample surface to prevent drying, and 20 μL of DI water was added to the sample surface immediately. A silicon probe with nitride coating (Bruker ScanAsyst-Fluid) was loaded on a fluid cell holder and immersed under the liquid droplet on the mica surface. AFM imaging was performed on a Bruker Multimode 8 with Nanoscope V controller (Bruker) under ScanAsyst in fluid mode. Micrographs were flattened and analyzed using Nanoscope Analysis v1.8 software.
AFM-IR.
A total of 20 μL of rH146 assembly solution was deposited on silicon wafer substrate (Ted Pella) and further incubated for 30 min in a wet cell for immobilization. Excessive sample solution was removed with a pipette, and the sample surface was immediately washed with 100 μL DI water followed by air drying. We chose a silicon wafer as substrate for this experiment to obtain a clean infrared signal background. Infrared nano-imaging was performed at neaspec GmbH, Germany, with a commercial neaSNOM microscope. Monochromatic infrared imaging at selected laser excitation frequencies was achieved by detecting the backscattered light interferometrically while scanning the rH146 ribbon topography via tapping AFM.
Cryo-EM Imaging of rH146 Ribbons.
A total of 2.5 µL of in vitro rH146 assembly sample was applied to holey carbon grids (Quantifoil 400 mesh Cu R1.2/1.3). The grids were blotted by Whatman No. 1 filter paper and plunge-frozen in liquid ethane using a Mark IV Vitrobot (Thermo Fisher Scientific) with blotting times of 3 to 6 s at room temperature and >90% humidity. Cryo-EM datasets were collected using SerialEM Talos Arctica microscopes (operated at 300 kV) equipped with Field Emission source and K3 camera (Gatan) in superresolution mode.
Mice Teeth Samples Fixation and Embedding.
Heads from kallikrein-4 knockout (KLK4−/−) mice (provided by the Simmer laboratory, University of Michigan) were immediately fixed in 10% neutral buffered formalin after mouse scarification. After dissection of the mandibles and maxilla, additional fixation with 4% paraformaldehyde in 0.1 M cacodylate buffer was applied for 2 d, and then mandibles and maxilla were rinsed with 0.1 M cacodylate buffer. The fixation and rinsing cycle were repeated one more time. Samples were then dehydrated with increasing alcohol content in steps and eventually stored in 100% ethanol before being embedded in Spurr’s resin.
The described animal experiments were done according to the basic protocols reviewed and approved by the Institutional Animal Care and Use Committees at the University of Michigan and in an agreement with the US Public Health Service Policy on Human Care and Use of Laboratory Animals.
KLK4−/− Teeth Samples Ultra-microtome Sectioning.
Embedded KLK4−/− mice incisor samples (n = 3) were cut and ground into a cylinder shape that could fit in the sample holder for the microtome (Ultratome Nova, LKB Bromma). Maturation stage of the KLK4−/− mice incisors (SI Appendix, Fig. S20A) were located with the first molar and cut with a diamond knife (Diamond knife-9342 MT, microstar) at about 80 nm thickness. For each sectioning session, 10 to 20 sections were collected in the boat of the diamond knife (SI Appendix, Fig. S20B), and 2 to 5 intact sections were carefully picked up with the TEM grid for further characterizations (SI Appendix, Fig. S20C). Fresh specimens were prepared immediately before each tissue-section related experiment including direct TEM characterization, demineralization studies (SI Appendix, Fig. S20D), immunostaining studies, and interaction with PILP solution.
TEM.
TEM samples were prepared on Formvar-coated 200-mesh copper grids (Ted Pella). TEM imaging was performed on a JEOL JEM-1400 (JEOL USA) operating at 120 kV. Data were recorded with a 4k Gatan Ultra Scan CCD camera (Gatan). All solution samples were vortexed gently 5 to 10 times to get an even distribution before depositing on EM grids.
For the in vitro rH146 and rH174 nanoribbon assembly solutions.
A droplet of 10 μL of sample solution was incubated for 5 min on an EM grid. Excessive sample solution was removed with a pipette. The grid was further washed with filtered DI water. Excessive moisture was wicked away with filter paper, and the grid was subsequently stained with a droplet of 2% methylamine tungstate (Nano-W, Nanoprobes) for 45 s.
For KLK4−/− mice incisor sections.
TEM grids were used to pick up the sections, and moisture was wicked away with filter paper. The resulting sample was imaged directly.
For demineralized KLK4−/− mice incisor sections.
We used the EM grid to pick up the original KLK4−/− incisor sections and position the EM grid upside down on top of a 1-mM HCl droplet for 5 min to demineralize the tissue. The EM grid was subsequently washed with DI water. Excessive moisture was wicked away with filter paper, and the grid was stained with a droplet of 2% methylamine tungstate (Nano-W, Nanoprobes) for 45 s.
For the amelogenin nanoribbon and PILP-solution mixing systems.
A droplet of 10 μL of mixture sample solution was incubated on the EM grid for 5 min. Excessive sample solution was removed with a pipette. The grid was further washed with filtered DI water. Excessive moisture was wicked away, and no staining was performed in order to characterize the formed minerals.
Selected Area Electron Diffraction.
Selected area electron diffraction was performed on JEOL JEM-1400 (JEOL USA) operating at 120 kV. Data were recorded with a 4K Gatan Ultra Scan CCD camera (Gatan) with a camera length at 25 cm. A standard evaporated aluminum sample (Ted Pella) was measured as a reference for d-spacing calculation.
Tomography Study on Demineralized KLK4−/− Enamel Section.
Stained demineralized KLK4−/− mice incisor sections were imaged on a Tecnai T20 microscope (Thermo Fisher Scientific) operated at 200 kV with a TemCaM F816 scintillator-based CMOS camera (TVIPS). Images were recorded by SerialEM Tilt Series collection mode at a magnification of 50,000 × g. Defocus was set to −2 to −2.5 µm. For data processing, the tilt series was processed by the workflow of Etomo (53). All of the results were visualized by IMOD.
Immunostaining of Demineralized KLK4−/− Mice Incisor Sections.
Primary amelogenin antibody (0.9 mg/mL) from rabbit (provided by the Li laboratory, School of Dentistry, University of California, San Francisco) was used at 1:100 dilution in phosphate-buffered saline buffer. An EM grid with demineralized KLK4−/− mice incisor sections on top was incubated in the diluted primary amelogenin antibody solution for 6 h at 37 °C. Excessive primary antibody was washed off with DI water, the sections were further incubated in secondary anti-rabbit antibody from goat with 5-nm colloidal gold labeling (MilliporeSigma, used as purchased) for 1 h under room temperature. Excessive antibody was washed away with DI water, and the sections were stained with a droplet of 2% methylamine tungstate (Nano-W, Nanoprobes) for 45 s, wicked dry, and stored in a grid box for TEM imaging.
Incubation of Demineralized KLK4−/− Mice Incisor Sections with PILP Solution.
pAsp powder (27 kDa) was dissolved in 25 mM, pH 7.4, Hepes buffer to make the pAsp stock solution at a concentration of 0.8 mg/mL. Notably, we chose Hepes buffer over the traditional Tris buffer because Tris has been proved to be able to bind with metal ions (54), thus competing with Ca2+ binding with pAsp, and may also disrupt the amelogenin nanoribbon structures (12). Stock solutions were prepared by dissolving reagent grade CaCl2, NaCl, and KH2 PO4 in Hepes buffer, respectively. The final PILP solution was prepared freshly per use with concentration of 0.1 mg/mL pAsp, 4.5 mM Ca2+, 2.1 mM PO43-, and 150 mM NaCl following established protocols (44). Vortex was applied at each mixing step.
TEM grids with demineralized KLK4−/− mice incisor sections were floated on 2 mL of PILP solution in a sealed tube. Incubation temperatures were set at 37 °C to induce amorphous apatite formation. The transformation to crystalline apatite was achieved with an increased incubation temperature at 50 °C.
Incubation of In Vitro rH146 Nanoribbons with PILP Solution.
Fresh PILP solution as described above was mixed with mature rH146 assembly solution at a 1:1 volume ratio. The mixture solution was incubated at 37 °C in a sealed vial and characterized by TEM/AFM methods at various time points to investigate the formation of big bundled structures. The transformation to crystalline apatite within the bundled structures was achieved with an incubation temperature at 85 °C.
STEM-EDS.
STEM-EDS analysis was performed at the Eurofin Nanolab with an aberration-corrected FEI Titan microscope performing at 200 kV. Images were collected in STEM mode with a HAADF detector, and EDS elemental maps were collected by a ChemiSTEM system which has a X-FEG module and four Bruker SDD detectors.
Data Availability Statement.
All study data are included in the article and SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. James Simmer (University of Michigan) for providing the KLK4−/− mice incisors and Dr. James De Yoreo from Pacific Northwest National Laboratory for helpful discussions. We also thank Stefan Mastel from neaspec GmbH, Germany, for performing the AFM-IR experiment. This work was supported by NIH/National Institute of Dental and Craniofacial Research Grant R01-DE025709.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2007838117/-/DCSupplemental.
References
- 1.Lyngstadaas S. P., Risnes S., Nordbø H., Flønes A. G., Amelogenin gene similarity in vertebrates: DNA sequences encoding amelogenin seem to be conserved during evolution. J. Comp. Physiol. B 160, 469–472 (1990). [DOI] [PubMed] [Google Scholar]
- 2.Gibson C. W., The amelogenin proteins and enamel development in humans and mice. J. Oral Biosci. 53, 248–256 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen E. et al., The small bovine amelogenin LRAP fails to rescue the amelogenin null phenotype. Calcif. Tissue Int. 73, 487–495 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Pugach M. K., Gibson C. W., Analysis of enamel development using murine model systems: Approaches and limitations. Front. Physiol. 5, 313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibson C. W. et al., Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J. Biol. Chem. 276, 31871–31875 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Moradian-Oldak J., Protein-mediated enamel mineralization. Front. Biosci. 17, 1996–2023 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carneiro K. M. M. et al., Amyloid-like ribbons of amelogenins in enamel mineralization. Sci. Rep. 6, 23105 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Margolis H. C., Beniash E., Fowler C. E., Role of macromolecular assembly of enamel matrix proteins in enamel formation. J. Dent. Res. 85, 775–793 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Fang P.-A., Conway J. F., Margolis H. C., Simmer J. P., Beniash E., Hierarchical self-assembly of amelogenin and the regulation of biomineralization at the nanoscale. Proc. Natl. Acad. Sci. U.S.A. 108, 14097–14102 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fincham A. G. et al., Evidence for amelogenin “nanospheres” as functional components of secretory-stage enamel matrix. J. Struct. Biol. 115, 50–59 (1995). [DOI] [PubMed] [Google Scholar]
- 11.Du C., Falini G., Fermani S., Abbott C., Moradian-Oldak J., Supramolecular assembly of amelogenin nanospheres into birefringent microribbons. Science 307, 1450–1454 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Martinez-Avila O. et al., Self-assembly of filamentous amelogenin requires calcium and phosphate: From dimers via nanoribbons to fibrils. Biomacromolecules 13, 3494–3502 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engelberth S. A. et al., Progression of self-assembly of amelogenin protein supramolecular structures in simulated enamel fluid. Biomacromolecules 19, 3917–3924 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanii B., Martinez-Avila O., Simpliciano C., Zuckermann R. N., Habelitz S., Matching 4.7-Å XRD spacing in amelogenin nanoribbons and enamel matrix. J. Dent. Res. 93, 918–922 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glimcher M. J., Daniel E. J., Travis D. F., Kamhi S., Electron optical and X-ray diffraction studies of the organization of the inorganic crystals in embryonic bovine enamel. J. Ultrastruct. Res. 50 (suppl. 7), 1–77 (1965). [PubMed] [Google Scholar]
- 16.Jodaikin A., Traub W., Weiner S., Protein conformation in rat tooth enamel. Arch. Oral Biol. 31, 685–689 (1986). [DOI] [PubMed] [Google Scholar]
- 17.Pautard F. G. E., An X-ray diffraction pattern from human enamel matrix. Arch. Oral Biol. 3, 217–220 (1961). [DOI] [PubMed] [Google Scholar]
- 18.Kallenbach E., Crystal-associated matrix components in rat incisor enamel. An electron-microscopic study. Cell Tissue Res. 246, 455–461 (1986). [DOI] [PubMed] [Google Scholar]
- 19.Meyer J. M., Bodier-Houllé P., Cuisinier F. J. G., Lesot H., Ruch J. V., Initial aspects of mineralization at the dentino-enamel junction in embryonic mouse incisor in vivo and in vitro: A tem comparative study. In Vitro Cell. Dev. Biol. Anim. 35, 159–168 (1999). [DOI] [PubMed] [Google Scholar]
- 20.Travis D. F., Glimcher M. J., The structure and organization of, and the relationship between the organic matrix and the inorganic crystals of embryonic bovine enamel. J. Cell Biol. 23, 447–497 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagano T. et al., Mmp-20 and Klk4 cleavage site preferences for amelogenin sequences. J. Dent. Res. 88, 823–828 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fukae M. et al., Enamelysin (matrix metalloproteinase-20): Localization in the developing tooth and effects of pH and calcium on amelogenin hydrolysis. J. Dent. Res. 77, 1580–1588 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Iijima M., Fan D., Bromley K. M., Sun Z., Moradian-Oldak J., Tooth enamel proteins enamelin and amelogenin cooperate to regulate the growth morphology of octacalcium phosphate crystals. Cryst. Growth Des. 10, 4815–4822 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu J. C. C. et al., Enamel defects and ameloblast-specific expression in Enam knock-out/lacz knock-in mice. J. Biol. Chem. 283, 10858–10871 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu Y. et al., Functions of KLK4 and MMP-20 in dental enamel formation. Biol. Chem. 389, 695–700 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyazawa T., Blout E. R., The infrared spectra of polypeptides in various conformations: Amide I and II Bands1. J. Am. Chem. Soc. 83, 712–719 (1961). [Google Scholar]
- 27.Kong J., Yu S., Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta Biochim. Biophys. Sin. (Shanghai) 39, 549–559 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Núñez S. M. et al., Maturation stage enamel malformations in Amtn and Klk4 null mice. Matrix Biol. 52–54, 219–233 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith C. E. et al., Relationships between protein and mineral during enamel development in normal and genetically altered mice. Eur. J. Oral Sci. 119 (suppl. 1), 125–135 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imbeni V., Kruzic J. J., Marshall G. W., Marshall S. J., Ritchie R. O., The dentin-enamel junction and the fracture of human teeth. Nat. Mater. 4, 229–232 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Habelitz S., Balooch M., Marshall S. J., Balooch G., Marshall G. W. Jr., In situ atomic force microscopy of partially demineralized human dentin collagen fibrils. J. Struct. Biol. 138, 227–236 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Smith C. E., Hu Y., Hu J. C. C., Simmer J. P., Ultrastructure of early amelogenesis in wild-type, Amelx−/−, and Enam−/− mice: Enamel ribbon initiation on dentin mineral and ribbon orientation by ameloblasts. Mol. Genet. Genomic Med. 4, 662–683 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gower L. B., Biomimetic model systems for investigating the amorphous precursor pathway and its role in biomineralization. Chem. Rev. 108, 4551–4627 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olszta M. J. et al., Bone structure and formation: A new perspective. Mater. Sci. Eng. Rep. 58, 77–116 (2007). [Google Scholar]
- 35.Bacino M. et al., Integrating the PILP-mineralization process into a restorative dental treatment. Dent. Mater. 35, 53–63 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bartlett J. D., et al. , “Protein–protein interactions of the developing enamel matrix” in Current Topics in Developmental Biology, Schatten G. P., Ed. (Academic Press, 2006), vol. 74, pp. 57–115. [DOI] [PubMed] [Google Scholar]
- 37.Beniash E., Metzler R. A., Lam R. S. K., Gilbert P. U. P. A., Transient amorphous calcium phosphate in forming enamel. J. Struct. Biol. 166, 133–143 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lacruz R. S., Habelitz S., Wright J. T., Paine M. L., Dental enamel formation and implications for oral health and disease. Physiol. Rev. 97, 939–993 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Habelitz S., Materials engineering by ameloblasts. J. Dent. Res. 94, 759–767 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinez-Avila O. M. et al., Self-assembly of amelogenin proteins at the water-oil interface. Eur. J. Oral Sci. 119 (suppl. 1), 75–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamakoshi Y. et al., Enamel proteins and proteases in Mmp20 and Klk4 null and double-null mice. Eur. J. Oral Sci. 119 (suppl. 1), 206–216 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nudelman F., Nacre biomineralisation: A review on the mechanisms of crystal nucleation. Semin. Cell Dev. Biol. 46, 2–10 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Nurrohman H. et al., Repair of dentin defects from DSPP knockout mice by PILP mineralization. J. Mater. Res. 31, 321–327 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nudelman F., Lausch A. J., Sommerdijk N. A. J. M., Sone E. D., In vitro models of collagen biomineralization. J. Struct. Biol. 183, 258–269 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Deshpande A. S., Beniash E., Bio-inspired synthesis of mineralized collagen fibrils. Cryst. Growth Des. 8, 3084–3090 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan W.-J. et al., The importance of a potential phosphorylation site in enamelin on enamel formation. Int. J. Oral Sci. 9, e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deutsch D. et al., Sequencing of bovine enamelin (“tuftelin”) a novel acidic enamel protein. J. Biol. Chem. 266, 16021–16028 (1991). [PubMed] [Google Scholar]
- 48.Hu J. C. C., Yamakoshi Y., Enamelin and autosomal-dominant amelogenesis imperfecta. Crit. Rev. Oral Biol. Med. 14, 387–398 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Limeback H., Isolation and characterization of pig enamelins. Biochem. J. 243, 385–390 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fan D., Lakshminarayanan R., Moradian-Oldak J., The 32kDa enamelin undergoes conformational transitions upon calcium binding. J. Struct. Biol. 163, 109–115 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brookes S. J., Kingswell N. J., Barron M. J., Dixon M. J., Kirkham J., Is the 32-kDa fragment the functional enamelin unit in all species? Eur. J. Oral Sci. 119 (suppl. 1), 345–350 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Svensson Bonde J., Bulow L., One-step purification of recombinant human amelogenin and use of amelogenin as a fusion partner. PLoS One 7, e33269 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mastronarde D. N., Held S. R., Automated tilt series alignment and tomographic reconstruction in IMOD. J. Struct. Biol. 197, 102–113 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Radford R. J., Tezcan F. A., A superprotein triangle driven by nickel(II) coordination: Exploiting non-natural metal ligands in protein self-assembly. J. Am. Chem. Soc. 131, 9136–9137 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.