Significance
Schistosomes are parasitic flatworms infecting hundreds of millions of people. As they alternate between mammalian and molluscan hosts, their survival and propagation depend upon stem cell proliferation and differentiation. Tracking the fate of these stem cells during early intramammalian development, we find that the esophageal gland, an accessory digestive organ, develops before the rest of the digestive system is formed and feeding begins, suggesting a role for this organ beyond nutrient uptake. We show that schistosomes lacking the esophageal gland die in naive mice but survive in immunodeficient mice lacking B cells; they are unable to lyse ingested immune cells before passing them into the gut. These results unveil an immune-evasion mechanism, which is essential for schistosome survival and pathogenesis.
Keywords: stem cells, host–parasite interaction, foxA, intramammalian parasite development, parasitic helminths
Abstract
Schistosomes are parasitic flatworms that cause schistosomiasis, a neglected tropical disease affecting over 200 million people. Schistosomes develop multiple body plans while navigating their complex life cycle, which involves two different hosts: a mammalian definitive host and a molluscan intermediate host. Their survival and propagation depend upon proliferation and differentiation of stem cells necessary for parasite homeostasis and reproduction. Infective larvae released from snails carry a handful of stem cells that serve as the likely source of new tissues as the parasite adapts to life inside the mammalian host; however, the role of these stem cells during this critical life cycle stage remains unclear. Here, we characterize stem cell fates during early intramammalian development. Surprisingly, we find that the esophageal gland, an accessory organ of the digestive tract, develops before the rest of the digestive system is formed and blood feeding is initiated, suggesting a role in processes beyond nutrient uptake. To explore such a role, we examine schistosomes that lack the esophageal gland due to knockdown of a forkhead-box transcription factor, Sm-foxA, which blocks development and maintenance of the esophageal gland, without affecting the development of other somatic tissues. Intriguingly, schistosomes lacking the esophageal gland die after transplantation into naive mice, but survive in immunodeficient mice lacking B cells. We show that parasites lacking the esophageal gland are unable to lyse ingested immune cells within the esophagus before passing them into the gut. These results unveil an immune-evasion mechanism mediated by the esophageal gland, which is essential for schistosome survival and pathogenesis.
Schistosomes are parasitic flatworms that cause schistosomiasis, a neglected tropical disease affecting over 200 million individuals (1). Schistosomes reside in the vasculature of a wide range of vertebrate hosts; there, they lay eggs that cause inflammation and damage to host tissues, leading to schistosomiasis (2). Their complex life cycle requires passage through molluscan and mammalian hosts (Fig. 1A): aquatic snails release cercariae, free-living, infective larvae that swim to find and penetrate mammalian host skin. Upon infection, the cercarial tail is lost at the infection site, and the parasite body transforms into a schistosomulum. This early intramammalian-stage parasite immediately faces major challenges as it adapts to this new host environment: it must migrate from the skin into the bloodstream, then through the lungs, to reach its niche in the hepatic vasculature (3). There, it will begin feeding on blood, which fuels growth, sexual maturation, and reproduction. Failure at any step would disrupt the life cycle, yet it manages these challenges, even in the face of the host’s immune system (4, 5). Recent work has identified heterogeneous populations of schistosome stem cells (6–10) and described their roles in asexual reproduction (7, 11), tegument (the parasite’s outer surface) production during homeostasis (12, 13), and germ cell development (7, 8, 10, 14). Thus, stem cells help drive schistosome viability and transmission.
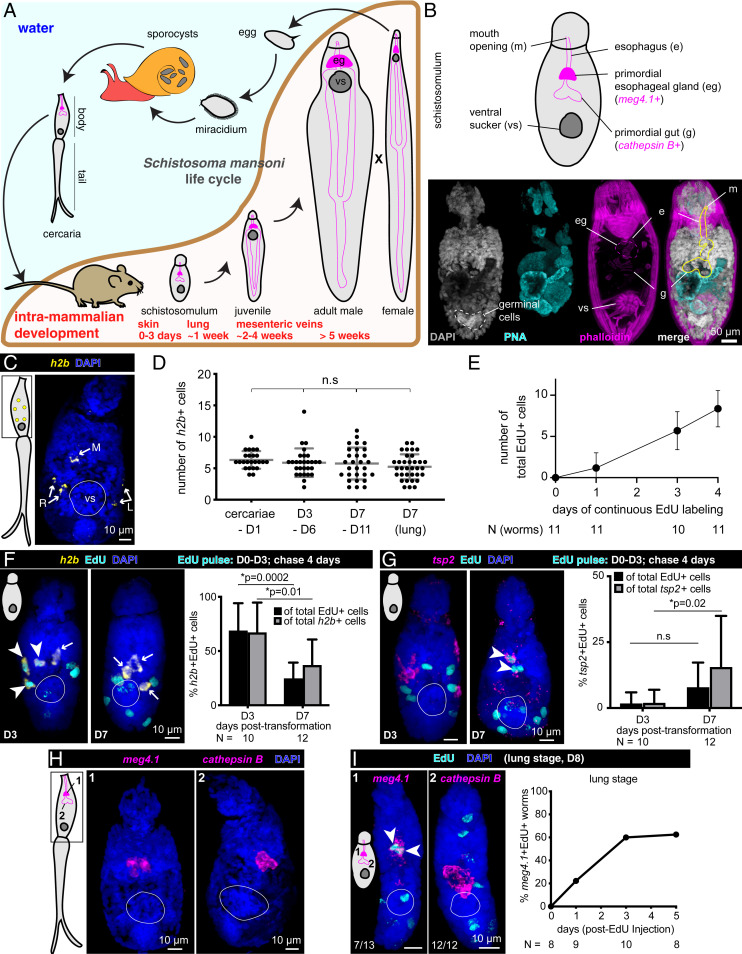
Fig. 1.
Stem cells drive early development of the schistosome esophageal gland. (A) Life cycle of Schistosoma mansoni. eg, esophageal gland; vs, ventral sucker. Each worm schematic depicts the esophageal gland (magenta-filled bean shapes), gut branches that stretch posteriorly (magenta outlines), and the ventral sucker (dark gray disk). The intramammalian developmental curve was adapted from ref. 40. (B) Anatomical features of schistosomula. (Top) Schematic view of a schistosomulum with the primordial digestive tract outlined. Genes expressed in distinct parts of the digestive tract are indicated. (Bottom) Day 1 posttransformation schistosomulum stained with PNA lectin and phalloidin, maximum-intensity projection (MIP). PNA (cyan) labels cercarial penetration gland used for mammalian skin penetration; this gland is largely unused after mechanical removal of tails. Phalloidin staining (magenta) labels muscular features surrounding the primordial digestive tract. (C) h2b fluorescence in situ hybridization (FISH) in cercaria, MIP. R and L, right and left, each showing pairs of h2b+ stem cells; M, medially located stem cell. vs, ventral sucker, indicated by white circles throughout (C–I). (D) Quantification of h2b+ cells in cercariae and schistosomula at various time points in vitro and in vivo (lung) (SI Appendix, Fig. S1A). Gray lines: Mean ± SD. Statistical analysis: one-way ANOVA. (E) Quantification of EdU+ cells at indicated days posttransformation after continuous EdU labeling in vitro. Mean ± SD. (F, Left) h2b FISH and EdU labeling at day 3 or 7 posttransformation in vitro, MIP. Arrowheads: double-positive cells; Arrows: h2b+ EdU− cells. (Right) Percent of h2b+ EdU+ cells over total EdU+ cells (black) or h2b+ cells (gray); mean ± SD. (G, Left) tsp2 FISH and EdU labeling as in F. Arrowheads: double-positive cells; Arrows: tsp2+ EdU− cells. (Right) Percentage of tsp2+ EdU+ cells over total EdU+ cells (black) or total tsp2+ cells (gray); mean ± SD. (H) cathepsin B (gut primordia) and meg4.1 (esophageal gland) FISH in cercariae. (I, Left) cathepsin B and meg4.1 FISH in lung-stage schistosomula (day 8 postinfection) 5 d after a single EdU injection into host on day 3, MIP. (Right) Percentage of worms with meg4.1+ EdU+ cells.
Within the snail host, cercariae develop organs necessary for infection (e.g., penetration glands) and primordia of organs (e.g., the digestive tract) required during the next parasitic stage (Fig. 1B). Cercariae also contain approximately five to six stem cells that are localized stereotypically: one pair of cells on each side, and one cell medially (7). Previously, we showed that upon infection of the mammalian host and transformation into schistosomula, these cells begin to proliferate (7), but the fates of their progeny were unexplored. In this work, we examine the contributions of these stem cells to early intramammalian development. Based on the unexpected finding that stem cells contribute to development of an accessory digestive organ, the esophageal gland, before the rest of the digestive system is formed, we characterize the gland’s function in vivo, revealing a mechanism for evasion of the host immune system.
Results
Stem Cells in Early Schistosomula Generate Parasite–Host Interfaces.
In flatworms, differentiated cells do not proliferate; thus, genes regulated in a cell cycle-dependent manner, such as histone 2b (h2b), can be used as generic stem cell markers (6, 15–17). Using h2b as a marker of these cells in cercariae (Fig. 1C) and schistosomula (Fig. 1D), we tracked their behavior by examining incorporation of 5-ethynyl-2′-deoxyuridine (EdU) in pulse-chase experiments either during in vitro transformation or in vivo (SI Appendix, Fig. S1A and Materials and Methods). The number of (h2b+) stem cells remained unchanged during 1 wk of in vitro culture, analogous to lung-stage in vivo schistosomula (Fig. 1D), whereas the number of EdU+ (progeny) cells increased after continuous labeling posttransformation (Fig. 1E), suggesting that these early stem cells divide to produce differentiating daughter cells. Indeed, the majority (∼70%) of h2b+ or EdU+ cells were doubly positive after an EdU pulse (Fig. 1F and SI Appendix, Fig. S1 A and B). These proportions decreased significantly after a chase, suggesting that most stem cell progeny (EdU-labeled cells) lost h2b expression. Moreover, in parallel in vivo experiments, we observed a similar proportional decrease in h2b+ EdU+ cells in lung-stage schistosomula (SI Appendix, Fig. S1 A and C). These results suggest that, at this early stage, stem cells divide to produce cell types required for the transition from cercariae to schistosomula rather than to expand the stem cell pool.
To follow the fates of stem cell progeny, we focused on the tegument and the digestive tract, the two major parasite–host interfaces. Previous work suggested that the tegument’s unique double-lipid bilayer and the turnover of its surface molecules protect schistosomes from host immunity (13, 18–20). Less is known about the role of the digestive tract in host–parasite interactions, but feeding and digestion of blood provide additional exposure to host immune cells (21–28). Considering the remodeling of the tegument in early intramammalian development (18–20) and the tegumental bias of adult stem cells (12, 13), we first examined when stem cells give rise to tegument in schistosomula. In cercariae, we detected ∼30 cells expressing tetraspanin-2 (tsp2), a marker of tegument progenitors (12), throughout the body (SI Appendix, Fig. S1 D and E). Following in vivo or in vitro transformation of cercariae into schistosomula (SI Appendix, Fig. S1A), the number of tsp2+ cells decreased dramatically during the first week (SI Appendix, Fig. S1E), while the number of tsp2+ EdU+ cells increased following EdU pulse chases (Fig. 1G and SI Appendix, Fig. S1 B and C). Thus, similar to adults, a substantial fraction of stem cells is dedicated to tegument production (12, 13).
Because cercariae contain a nonfunctional gut primordium (29) that must be remodeled after infection to enable ingestion of host blood, we next examined digestive system development. The digestive tract is composed of an anterior mouth opening, followed by an esophagus with associated glands (the anterior cell mass, and more posteriorly, the esophageal gland), and syncytial gut branches (30) (Fig. 1B). To distinguish between these regions, we used markers of the esophageal gland, microexon gene 4.1 (meg4.1) (21, 31–33), and gut, cathepsin B (6, 12, 13, 34). We observed a handful of cells expressing either meg4.1 or cathepsin B in cercarial digestive system primordia (29) (Fig. 1H). A functional digestive system begins to develop in schistosomula, although the parasites do not yet feed on blood at this stage. Interestingly, we found a differential contribution of stem cells to distinct portions of the digestive system: the esophageal gland forms before the gut proper (Fig. 1I). In vivo EdU pulse-chase experiments revealed that >50% of lung-stage schistosomula contained meg4.1+ EdU+ cells (Fig. 1I). Thus, stem cell progeny begin contributing to the esophageal gland at this early stage. By contrast, cathepsin B+ cell numbers in gut primordia remained constant during in vitro culture or in lung-stage schistosomula (SI Appendix, Fig. S1F). Moreover, we were unable to detect any cathepsin B+ EdU+ cells at this stage after various EdU pulse-chase paradigms (SI Appendix, Fig. S1G), suggesting that stem cells do not produce additional gut cells in lung-stage schistosomula. We first detected gut differentiation in 3-wk-old juveniles, when stem cells produce tegument, esophageal gland, and gut; notably, the kinetics of differentiation was faster than observed in adults (SI Appendix, Fig. S2 A–I), consistent with the rapid growth and development of juvenile parasites.
foxA Is Expressed in the Esophageal Gland and Neighboring Stem Cells.
Given that esophageal gland cells are produced before the gut in schistosomula, we suspected that the gland might play an additional role beyond blood digestion for nutrient uptake. In adult male parasites, the esophageal gland consists of ∼1,000 cell bodies with cytoplasmic extensions that reach into the esophagus lumen (25). Esophageal gland proteins are highly glycosylated, seen from the preferential binding of a lectin, peanut agglutinin (PNA) (Fig. 2A). Based on cytological and gene expression studies, the esophageal gland has been proposed to serve as the initial processing site of ingested blood cells (22, 25, 27, 30). However, to date no functional evidence shows its requirement in blood processing.
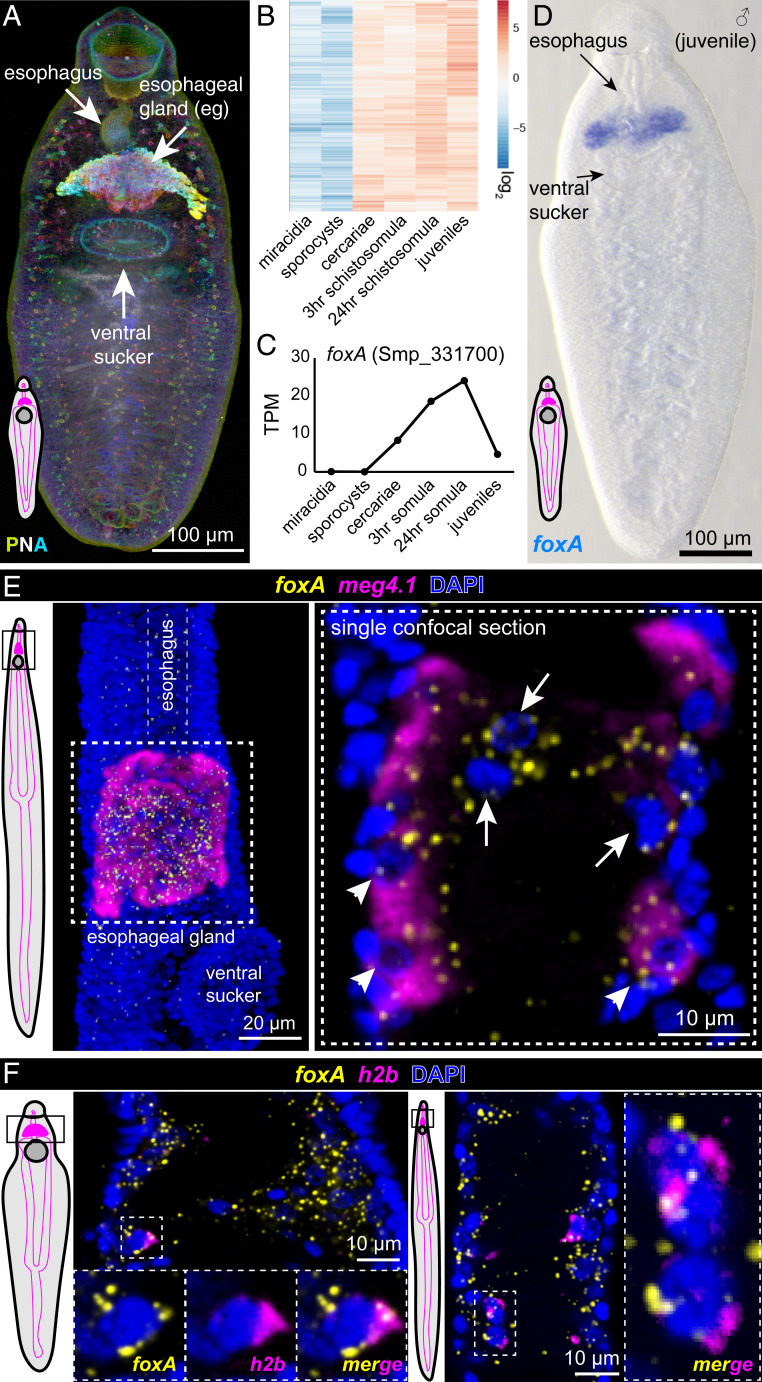
Fig. 2.
foxA expression is enriched in the esophageal gland and neighboring stem/progenitor cells. (A) Peanut agglutinin (PNA) staining of a juvenile male, MIP, depth color coded. Each worm schematic depicts the esophageal gland (magenta-filled bean shape), gut branches that stretch posteriorly (magenta outlines), and the ventral sucker (dark gray disk). (B) Heatmap showing genes up-regulated greater than twofold in cercariae and mammalian stages relative to miracidia and sporocysts (n = 681 genes). (C) foxA TPM (transcripts per million mapped reads) values from whole-worm transcriptomes of different parasite stages. (D) foxA WISH in juvenile worm. (E, Left) Adult female schematic showing the imaged region. (Middle) Double FISH of foxA with meg4.1, MIP. (Right) Magnified view of the boxed area, single confocal section. Arrowheads: foxA+ meg4.1+; arrows: foxA+. (F) Single confocal sections of double FISH of h2b and foxA in adult male (Left) and female (Right) worms. Outlined boxes on worm schematics in E and F indicate imaged regions.
To characterize esophageal gland development and function, we queried previously published whole-worm transcriptomes from different stages of the life cycle (6, 11, 35) for genes up-regulated in the mammalian stage relative to the molluscan stage (Fig. 2B). We mined these genes for transcriptional regulators with known roles in intestinal differentiation, and identified a forkhead-box transcription factor with conserved functions in endodermal differentiation in many animal species (36, 37), Sm-foxA (foxA) (Smp_331700) (38) (Fig. 2C). Phylogenetic analysis showed that the forkhead domain of FoxA clustered with those of FoxA proteins from many organisms (SI Appendix, Fig. S3A), including the planarian Schmidtea mediterranea, in which foxA plays a role in pharynx regeneration (39). Colorimetric whole-mount in situ hybridization (WISH) revealed that foxA, like meg4.1, was highly enriched in the esophageal gland in juvenile and adult parasites (Fig. 2D and SI Appendix, Fig. S3 B and C). Double fluorescence in situ hybridization (FISH) with meg4.1 showed that the majority of foxA-expressing cells also expressed meg4.1 (Fig. 2E). Closer examination revealed that some foxA+ cells closer to the esophagus showed weak or no detectable meg4.1 expression, and instead coexpressed markers of either stem cells (h2b+) (Fig. 2F) or esophageal tegument (tsp2+) (SI Appendix, Fig. S3D). In addition, a recent single-cell transcriptomic study found that foxA was also enriched in a subset of stem cells in adult schistosomes, as with hepatocyte nuclear factor 4 (hnf4), a transcription factor required for gut cell differentiation and maintenance (8). Strong expression of foxA in the esophageal gland and neighboring stem/progenitor cells suggested a potential role for this transcription factor in committing stem cells to the esophageal-gland lineage.
foxA Is Required for Development and Maintenance of the Esophageal Gland.
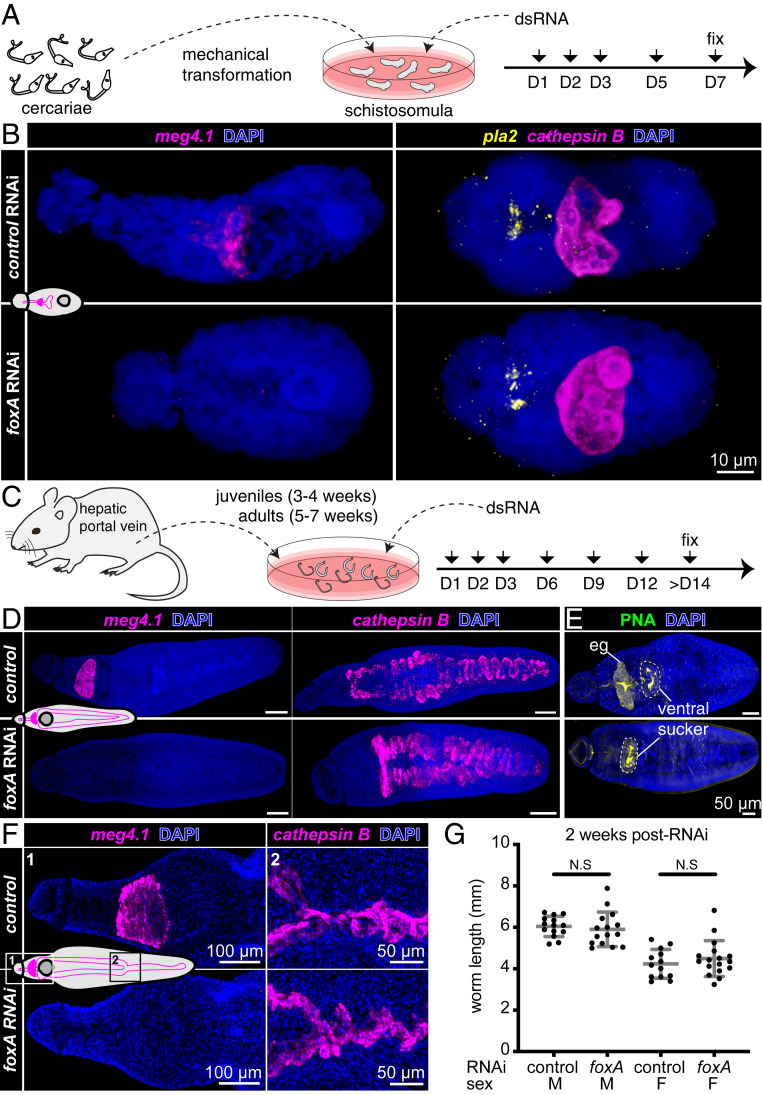
To assess its function in digestive system development, we knocked down foxA using RNA interference (RNAi) in schistosomula (Fig. 3A). meg4.1 expression was undetectable in foxA RNAi schistosomula, whereas markers of the gut (cathepsin B) and anterior cell mass (phospholipase A2 [pla2]) were unaffected (Fig. 3B). Reduced expression of foxA and other esophageal gland genes was confirmed by quantitative real-time PCR (qPCR) (27) (SI Appendix, Fig. S4A). meg4.1 expression was not detected in foxA RNAi juvenile parasites, whereas cathepsin B expression was unaffected (Fig. 3 C and D). Furthermore, PNA staining was eliminated from the esophageal gland region (Fig. 3E), revealing loss of differentiated gland cells. Instead, stem and progenitor cells accumulated in place of the gland (SI Appendix, Fig. S4B). These results suggest that foxA is required for esophageal gland cell differentiation and, thus, for development of the esophageal gland.
Fig. 3.
foxA knockdown parasites fail to develop or maintain the esophageal gland. (A) Experimental scheme for RNAi in schistosomula. (B) meg4.1, cathepsin B, and pla2 FISH in foxA RNAi schistosomula, MIP. n = 18 control RNAi; n = 17 foxA RNAi. (C) Experimental scheme for RNAi in juveniles and adults. (D) meg4.1 and cathepsin B FISH in RNAi juveniles, MIP. n = 17 control RNAi; n = 18 foxA RNAi. (E) PNA labeling in RNAi juveniles, MIP. n = 9 control RNAi; n = 8 foxA RNAi. (F) meg4.1 and cathepsin B FISH in RNAi adults, MIP. n = 9 control RNAi; n = 12 foxA RNAi. (G) Adult worm length after foxA knockdown. Mean ± SD. Statistical analysis: Welch’s t test.
Next, we analyzed the effects of foxA RNAi during adult homeostasis (Fig. 3F). FISH and qPCR confirmed knockdown of foxA and esophageal gland-enriched genes (Fig. 3F and SI Appendix, Fig. S4C), similar to knockdown at earlier stages. Knocking down foxA in adults in vitro did not lead to significant changes in worm length (Fig. 3G), which typically correlates with parasite growth and development (40). Neither male nor female worms showed any behavioral defects, and both were able to attach to the dish and remain paired. Finally, FISH using other known cell-type markers did not reveal any gross morphological differences between control and foxA RNAi parasites (SI Appendix, Fig. S4D). Together, these data demonstrate that foxA is essential for the development and maintenance of the esophageal gland, and that the gland can be ablated without affecting parasite viability, pairing, or behavior in vitro.
The Esophageal Gland Is Essential for Survival inside the Mammalian Host.
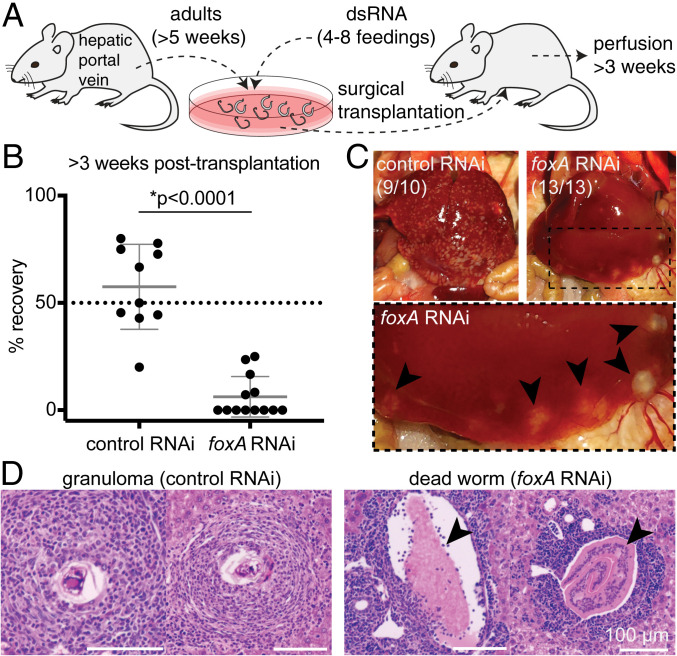
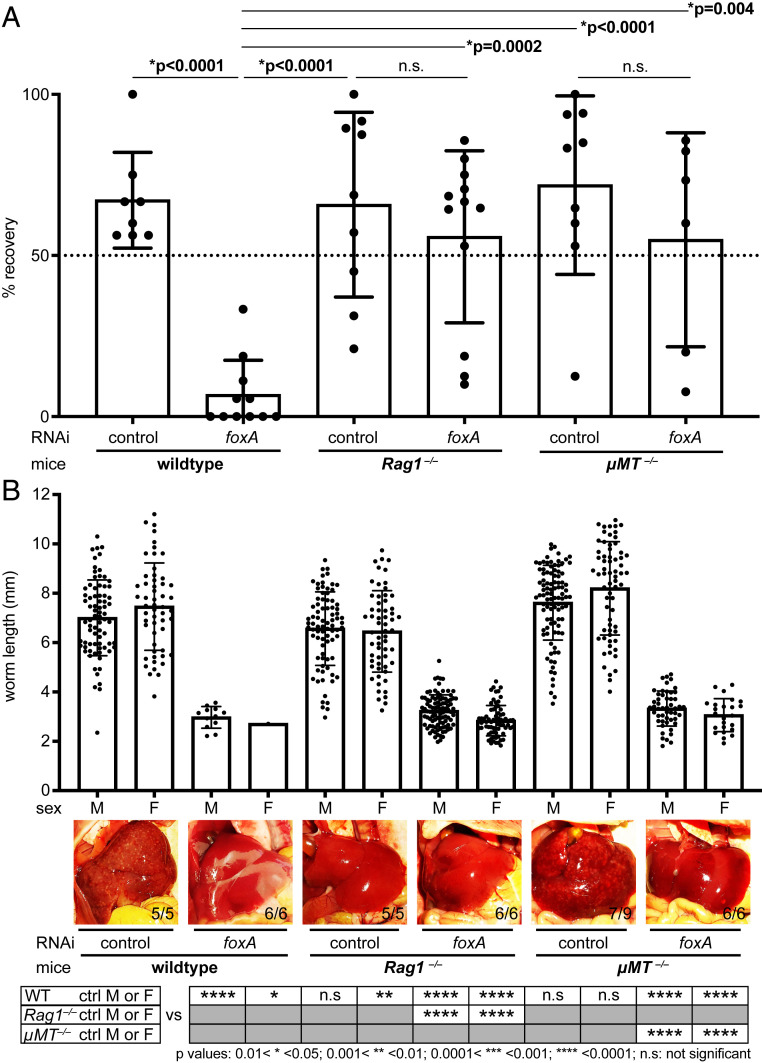
Although ablating the esophageal gland did not overtly affect the health of worms in vitro, current culture conditions do not fully recapitulate the in vivo environment of the host vasculature. To examine the role of the esophageal gland in vivo, we ablated the gland by foxA knockdown in adult parasites in vitro, surgically transplanted them into the cecal vein of wild-type (WT) naive mice, and then counted the number of male parasites recovered after ∼3 wk (Fig. 4A and Materials and Methods) (31, 41). Consistent with previous work (31), we recovered on average ∼60% of control RNAi parasites from mice ∼3 wk after transplantation (Fig. 4B). In contrast, we recovered only ∼6% of the injected foxA RNAi parasites (Fig. 4B). The livers of mice harboring control RNAi parasites contained numerous granulomas, whereas the livers of mice harboring foxA RNAi parasites showed few or no granulomas (Fig. 4C). Instead, these livers had large deposits containing dead worms that had been flushed into the hepatic sinusoids (Fig. 4D), as described in previous work in which schistosomes were transplanted after knocking down a gene required for viability in the mammalian host (31). The few foxA RNAi parasites that survived inside the host were markedly smaller than recovered control RNAi worms, or foxA RNAi worms before transplantation (SI Appendix, Fig. S5A and Movie S1). Despite the stunting of foxA RNAi parasites, these worms nevertheless contained black hemozoin pigment from hemoglobin breakdown (28, 42); thus, these parasites were still able to digest red blood cells in the absence of the esophageal gland. In addition, these parasites showed active movements of their oral sucker, and were able to expel regurgitant containing hemozoin pigment (Movie S1). We conclude that foxA RNAi parasites exhibit typical wild-type feeding behaviors (28, 30), suggesting that the mechanics of ingestion and expulsion were not affected by loss of the esophageal gland.
Fig. 4.
The esophageal gland is required for parasite survival inside the mammalian host. (A) Experimental scheme for the transplantation of adult schistosomes after RNAi. (B) Worm recovery >3 wk after the surgical transplantation. Each dot represents one host mouse. Mean ± SD. Statistical analysis: Welch’s t test. Raw numbers are shown in SI Appendix, Table S1. (C) Representative images of whole livers harvested from mice transplanted with control (Left) or foxA RNAi (Right) parasites after >3 wk. Magnified region inside the dotted box is shown below. Black arrowheads: dead worms flushed into the liver. The numbers indicate how many livers show the respective phenotype. (D, Left) Representative H&E staining of granulomas in liver sections of mouse transplanted with control RNAi parasites. (Right) Representative H&E staining showing a cross section of dead worms (arrowheads) in liver sections of mouse transplanted with foxA RNAi parasites. See SI Appendix, Fig. S6B for additional H&E staining.
B Cells Mediate the Killing of Schistosomes Lacking an Esophageal Gland.
Parasites lacking an esophageal gland appear otherwise normal in in vitro culture and retain basic blood-digestive functions, implying that their increased mortality in mammalian hosts is not merely due to defects in nutrient uptake. Previous work reported various leukocytes clustered in the schistosome esophagus; such cells were not observed in the gut downstream of the esophageal gland (25). Furthermore, leukocytes were surrounded by secreted esophageal gland proteins, suggesting that the gland could be coating and/or disrupting immune cells (25). To test this idea directly, we surgically transplanted foxA RNAi parasites into immunodeficient mouse strains. Rag1−/− mice are deficient in recombination activation gene 1, which is required for proper development of mature B and T cells (43); these mice lack functional adaptive immunity. Unexpectedly, foxA RNAi parasites transplanted into Rag1−/− mice were viable, at levels comparable to control RNAi parasites (Fig. 5A). Thus, lack of host adaptive immunity rescued worms from the mortality associated with loss of the esophageal gland. This observation indicates that the esophageal gland protects the parasite against the host’s adaptive immune system. To further dissect which adaptive immune cell types might mediate parasite death, we next transplanted foxA RNAi parasites into μMT−/− mice, which lack mature B cells but produce functional T cells (44). We recovered a similar proportion of parasites from such mice as from transplants into Rag1−/− mice (Fig. 5A), suggesting that the increased mortality of gland-lacking schistosomes in wild-type mice is attributable to B cells and/or antibody production, but not T cells.
Fig. 5.
Schistosomes without an esophageal gland survive in hosts lacking an adaptive immune response. (A) Recovery of worms from control and immunodeficient mice >3 wk after transplantation. Each dot represents one host mouse (SI Appendix, Table S1). Mean ± SD. Statistical analysis: one-way ANOVA followed by Tukey’s multiple comparisons test. (B, Top) Length of parasites recovered >3 wk posttransplantation. Each dot represents an individual worm (SI Appendix, Table S2). Worms were collected from five or more mice for each condition. Mean ± SD. (Middle) Representative livers of infected mice from each group. The numbers indicate how many livers display the respective phenotype. (Bottom) Table showing the statistical significance of comparisons to control RNAi male or female counterparts. Statistical analysis: one-way ANOVA followed by Tukey’s multiple-comparison test.
Although transplantation into immunocompromised hosts rescued the viability of foxA RNAi parasites, the recovered foxA RNAi worms were significantly smaller than control RNAi worms (Fig. 5B). FISH to detect various cell type markers revealed that different tissue/cell types in these stunted worms were similar to those in control RNAi worms, except for their enlarged cathepsin B+ gut branches (SI Appendix, Fig. S5B) and their reproductive systems, which exhibited reduced gonads in both sexes (SI Appendix, Fig. S6A). The regressed gonads were consistent with the absence of disease pathology in the hosts: no eggs/granulomas were found in the livers of mice that received foxA RNAi parasites (Fig. 5B and SI Appendix, Fig. S6B). In contrast to the differences in recovery of control vs. foxA RNAi parasites 3 wk posttransplantation, at 6 d posttransplantation (prior to full activation of the adaptive immune response) control and foxA RNAi parasites were recovered at similar frequencies (SI Appendix, Fig. S6C), with a mild but statistically significant reduction in worm length (SI Appendix, Fig. S6D). Thus, host innate immunity is insufficient to kill parasites lacking the esophageal gland; instead, killing requires a longer-term process in the host.
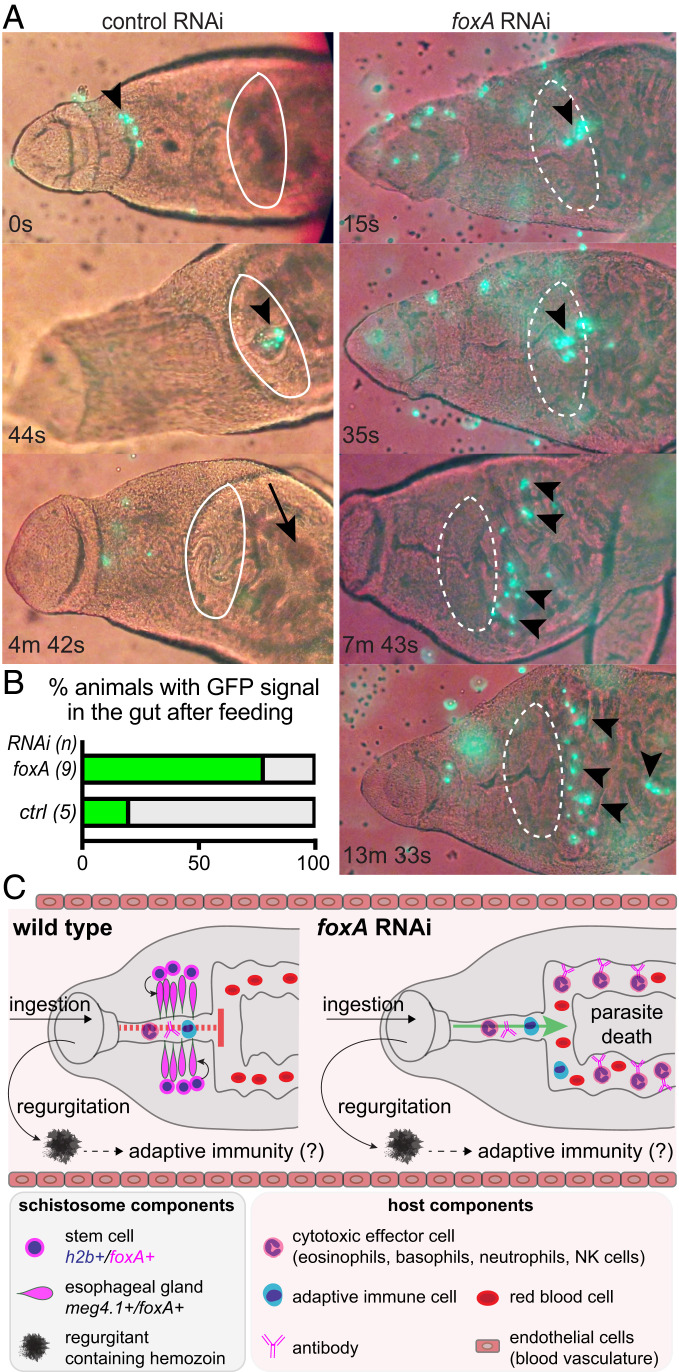
Finally, to characterize the interaction between immune cells and the esophageal gland, we isolated total splenocytes and whole-blood mononuclear cells (leukocytes) from UBC-GFP mice (45) (Materials and Methods), in which all cells of the hematopoietic lineage express GFP. We then introduced GFP+ splenocytes or leukocytes into the parasites’ in vitro culture media (Fig. 6A). Consistent with the blood-feeding behaviors of schistosomes described previously (25), control RNAi worms rapidly and periodically contracted the muscular rim of their oral sucker to grab and ingest GFP+ cells, which entered the esophagus and accumulated posteriorly over the course of a few minutes. Most of the immune-cell GFP signal within the esophagus was lost before “gulping” movements passed the ingested cells into the gut via opening of a muscular sphincter (Fig. 6 A and B and Movie S2). These observations suggest that ingested immune cells are degraded within the esophagus before they are delivered to the gut. By contrast, although GFP+ cells were also retained in the esophagus of foxA RNAi parasites, gulping movements brought these cells into the gut, where they persisted without losing GFP signal (Fig. 6 A and B). This signal could still be detected inside the gut after 1 h (Movie S2), suggesting inefficient immune-cell lysis inside the gut lumen. We conclude that the esophageal gland acts as a physical barrier, blocking entry of ingested immune cells into the gut and contributing to their degradation, thereby protecting the parasite from the host immune system.
Fig. 6.
Schistosomes lacking the esophageal gland fail to lyse ingested immune cells. (A) In vitro feeding of foxA RNAi parasites with GFP-expressing immune cells (Materials and Methods and Movie S2). Time stamps indicate time elapsed after initiation of a grabbing motion. White ellipse indicates presence (solid) or absence (dotted) of the esophageal gland. Arrowheads: GFP+ cells; arrow: gut with no GFP signal. (B) Percentage of animals with GFP signal in the gut after feeding. n: total number of worms. (C) Model for esophageal gland-mediated immune evasion. Regurgitant from the parasite, which contains schistosome gut proteins, can trigger host adaptive immunity and production of antischistosome antibodies. In the presence of an esophageal gland, cytotoxic effector cells (and perhaps antibodies) are blocked from entering into the schistosome gut. In the absence of an esophageal gland, however, antischistosomal antibodies and cytotoxic effector cells have access to the parasite’s gut, where they can trigger death from within the worm.
Discussion
Blood flukes are known for their long-term survival inside the host: previous studies have found schistosomes in infected patients over a decade after their initial exposure, with no possible route of reinfection in the interim (4, 46–48). Such longevity within the hostile environment of the host vasculature has been attributed to the ability of these parasites to evade the host immune system (19). Previous ultrastructural, biochemical, and cell biological studies have revealed the importance of the schistosome tegument in evading the host immune system (12, 13, 18, 19, 49, 50), but less attention has been paid to the function of the digestive tract in immune evasion. Since schistosomes process large amounts of blood throughout their lives (24, 25, 30), there must be a mechanism in place for the parasites to filter out host immune components. Here, we report the functional role of the schistosome esophageal gland in defending against host immunity. We find schistosome FoxA as a key regulator of the development and maintenance of the esophageal gland. Schistosomes lacking an esophageal gland die in immunocompetent hosts, but survive in hosts lacking B cells, and therefore antibodies. Our data suggest that the lack of an esophageal gland does not alter either digestion of red blood cells (evident from the black hemozoin pigment observed in the gut) or regurgitation of waste products (evident from the expulsion of regurgitant containing hemozoin). Importantly, the lack of an esophageal gland grants leukocytes access to the parasite’s gut, where they can persist for up to an hour.
Since neither B cells nor antibodies directly exert cytotoxicity, how might parasites lacking an esophageal gland be killed? Schistosomes, which possess a blind gut, consume and digest blood cells and regurgitate waste products through the same mouth opening (30). In addition to hemozoin, these regurgitated gut contents have been shown to contain schistosomal proteins (28, 42). Therefore, gut antigens and other parasite-derived molecules expelled from the gut may activate host immunity, leading to the production of antibodies recognizing antigens internal to the worm; such antibodies would be generated whether or not there is an esophageal gland. However, in the absence of an esophageal gland, cytotoxic immune effector cells (e.g., eosinophils, macrophages) would have access to and could persist in the worm’s gut, enabling their recognition of antibody-bound tissues and enhancing the killing of parasites from the inside out (Fig. 6C). Such antibody-mediated responses seem to be essential for killing schistosomes lacking an esophageal gland; we find that these parasites can survive in immunocompetent hosts for a short period (∼1 wk), during which the adaptive immune response has not been fully activated. In addition, parasites that survive in hosts lacking B cells and antibodies (but have a basal level of innate immunity) are stunted, which may be due to cytotoxic effector cells that enter the gut and damage parasite tissues.
This model is consistent with previous observations that show the importance of antibodies and exposing inner parasite tissues for a successful immune response for eliminating schistosomes (51–55). In vitro studies have shown that the addition of antibodies is necessary for effector cells (e.g., eosinophils) to kill schistosomula (51–53). Similarly, in vivo, praziquantel-treated adult schistosomes were only killed when immune cells and antibodies gained access to inner parasite tissues through the damaged tegument (54, 55). However, our data do not exclude alternative or complementary models. For instance, the esophageal gland may secrete factors that destroy antibodies (or complement proteins), block their binding to worm tissues, or inhibit their interactions with Fc receptors on host leukocytes. Any of these activities would impair antibody-dependent cellular cytotoxicity and/or activation of the complement pathway within the worm and have a protective effect for the parasite.
Future efforts will seek to decipher the cellular and molecular mechanisms underlying the interactions between the esophageal gland and the host immune system. This work has opened several potential directions for such efforts, including transcriptomic and proteomic analyses of foxA RNAi parasites. FoxA plays a conserved role in endodermal development throughout the animal kingdom (36, 37), acting as a pioneer factor that binds DNA and opens the chromatin of downstream target genes (56). Thus, schistosome FoxA likely regulates transcription of genes underlying differentiation and/or proper function of the esophageal gland. Identifying genes downstream of FoxA may reveal new targets to selectively disrupt esophageal gland function, facilitating schistosome killing by host immunity (57). Because the esophageal gland develops during larval stages, such targets also have the potential to act on immature worms, in contrast to praziquantel, which is only effective against adults (1, 58). Furthermore, comparative studies on the role of FoxA in other flukes could elucidate immune-evasion mechanisms and define potential therapeutic targets in a wide range of parasites.
Materials and Methods
Parasites.
Various stages of Schistosoma mansoni parasites were used in this study. To collect cercariae, infected snails obtained from Biomedical Research Institute (Rockville, MD) were placed under light at 26 °C for 1 h. To obtain schistosomula, cercariae were passed between syringes 12 to 14 times to mechanically remove the tail (59–61). After mechanical transformation, bodies (schistosomula) and tails were separated using a 70% Percoll gradient at 500 × g for 15 min at 4 °C. Schistosomula were collected and cultured in vitro in Basch Media 169 (62). Lung-stage schistosomula, juveniles, and adult parasites were harvested from in-house–infected mice by perfusion 1, 3, and 7 wk postinfection, respectively.
FISH/WISH and Lectin Staining.
FISH and WISH were performed as described previously (6, 7, 11, 63, 64). Parasites were fixed with 4% formaldehyde for 4 h (adults), or 4% formaldehyde containing 1% Nonidet P-40 and 0.2% Triton X-100 for 0.5 to 1 h (cercariae and schistosomula) or 1 to 2 h (juveniles), and then dehydrated in 100% methanol for storage. Five to 10 adult worms (male and female combined) and 5 to 15 juveniles and schistosomula were used for each gene analyzed in each biological replicate. Labeled FISH and WISH probes were generated using either DIG (digoxigenin)-12-UTP (Roche), or fluorescein-12-UTP (Roche). Probes were synthesized by in vitro transcription of partial gene sequences cloned in the plasmid vector pJC53.2, as previously described (63). Primers used for gene isolation by RT-PCR are listed in SI Appendix, Table S3. Following probe hybridization, specimens were incubated with anti–DIG-AP (MilliporeSigma; 11093274910) for WISH, and anti-DIG-POD (MilliporeSigma; 11207733910) or anti–FITC-POD (MilliporeSigma; 11426346910) for FISH, between 1:1,000 and 1:2,000 dilution, and enzymatic labeling reactions carried out as previously described (64). Fluorescein-labeled PNA (Vector Labs) was used at 1:500 dilution in a FISH blocking solution overnight at 4 °C (65).
EdU Detection.
For schistosomula, 10 µM EdU (Invitrogen) dissolved in DMSO was added to Basch media at 1:1,000 to 1:2,000 dilution for pulse labeling. To chase EdU label, pulsed schistosomula were washed with fresh Basch media. For lung and juvenile parasites, EdU (Invitrogen) was dissolved in PBS to 10 mg/mL. Infected mice (1 to 3 wk postinfection) were injected with 500 µL of 10 mg/mL EdU stock intraperitoneally and euthanized after 0 to 7 d. The click reaction was performed as previously described (6, 7, 11) using 20 to 50 μM Alexa Fluor 488 (A10266)-, 555 (A20012)-, 647 (A10277)-, or Oregon Green 488 (O10180)-conjugated azide (Invitrogen).
qPCR.
Total RNA was extracted from schistosomula using TRIzol (Life Technologies; 15596-026) and purified using phenol-chloroform. cDNA was synthesized using iScript Reverse Transcription Supermix (Bio-Rad). qPCRs were run on a StepOnePlus Real-Time PCR System (Applied Biosystems) using GoTaq qPCR Master Mix (Promega). ΔΔCt comparison was used to analyze the relative fold changes in expression between control and foxA RNAi specimens. The nucleotide sequences of primers used for qPCR are listed in SI Appendix, Table S3.
RNAi.
dsRNA against foxA was made via in vitro transcription reaction using previously published methods (66). Oligonucleotide sequences used to generate clones for transcription are listed in SI Appendix, Table S3. Approximately 15 adult pairs, ∼30 juvenile parasites, or >1,000 freshly transformed schistosomula were incubated at 37 °C with dsRNA (∼10 to 20 ng/mL final concentration) in Basch 169 medium or in ABC medium (67). Media with dsRNA were refreshed every day for 3 d, and every 3 to 4 d thereafter for 2 to 4 wk. Schistosomula were collected for qPCR and FISH. Juvenile parasites harvested from mice 3-wk postinfection, were incubated in vitro in Basch 169 media containing dsRNA, and were pulsed with 0.1% (vol/vol) mouse red blood cells during RNAi treatment. For adults, we included lipid mix and ascorbic acid, similar to recently reported improved culture conditions (67). In some cases, blood cells were omitted for adult parasites.
Animals and Surgical Transplantation of Adult Parasites.
All mice were handled in accordance with the Institutional Animal Care Use Committee protocol at the University of Wisconsin–Madison (M005569). For cercariae infection, female Swiss-Webster mice (Taconic) were used. For harvesting of GFP-expressing immune cells, UBC-GFP (Jax; #004353) or CAG-EGFP (Jax; #006567) were used. For surgical transplantation of adult parasites, different mouse genotypes were used: WT (Swiss Webster, Taconic; BL6, Jax), Rag1−/− (Jax; #002216), and μMT−/− (Jax; #002288). Surgical transplantation of schistosomes was based on a procedure performed in hamsters (41) as previously described (31). Briefly, schistosomes were recovered from mice 5 or 7 wk postinfection and treated with nonspecific control (bacterial ccdB gene from pJC53.2) (66) or foxA (Smp_331700) dsRNA for five to eight feedings over 2 to 4 wk in Basch media (62, 67). For each mouse, 15 to 20 male and female paired parasites were loaded into a 1-mL syringe before attaching a custom 23G extrathin-wall hypodermic needle (Cadence). Air and media were expelled, leaving ∼100 µL of media in the syringe, and worms were allowed to settle into the needle hub. The syringe was then placed horizontally until use. WT female Swiss-Webster mice were used for initial transplantation comparing the recovery of control and foxA RNAi parasites. For comparing parasite survival in immunocompetent and immunodeficient mice, male BL6 mice were used as a control strain for male Rag1−/−, μMT −/− mice (BL6 background), each ∼25 to 35 g in weight. Mice were anesthetized with isoflurane (Patterson Veterinary) using a vaporizer system with both an induction chamber and nose cone. For analgesia, 200 μL of 1 mg/mL carprofen (Putney) was administered subcutaneously to each mouse preincision. Eyes were kept moist with eye lubricant (Akorn Animal Health). Mice abdomens were shaved and sterilized with povidone-iodine 7.5% (Betadine scrub) and ethanol. The abdomen was opened with a ∼1.5-cm longitudinal incision centered on the navel and draped with sterile gauze with a slit, moistened with warm sterile saline. The intestines were gently pulled out to expose the large vein running along the cecum and kept moist with warm sterile saline. The tip of the needle, bevel facing down, was inserted beside the vein, sliding parallel to the vein before puncturing the vein wall and injecting the schistosomes. To stop bleeding, a small piece of hemostatic gauze (Blood Stop; LifeScience PLUS) was applied with gentle pressure over the injection site as the needle was removed. Once bleeding stopped (∼1 to 2 min), the intestines were gently returned into the abdominal cavity. The cavity was filled with warm sterile saline, and the mouse was gently shaken to return the intestine to its natural position. The abdominal wall was sutured (Maxon; absorbable sutures; taper point; size 4-0; needle V20; 1/2 circle) and the skin closed with 9-mm surgical staples (AutoClip wound closing system; MikRon Precision). Mice were allowed to recover in a warming chamber. After injection, needles were flushed with media to determine the number of parasites delivered into each mouse. Mice were housed in groups (maximum, five per cage) and individual mice were tracked by ear hole punches. Additional doses of analgesia, carprofen MediGel CFP, 1 mg/2-oz cup (ClearH2O) were administered orally, 1 cup per cage as the sole source of food for 2 d. For quantification of recovery, we only counted male parasites as a readout, because female worm development is dependent on pairing status (4, 67), which could affect female parasite viability in vivo.
Isolation of Total Immune Cells from Spleen and Whole Blood.
UBC-GFP or CAG-EGFP mice were infected with 200 to 800 cercariae by tail immersion and euthanized 3 wk later. A previously published splenocyte isolation protocol (68) was used with a few modifications. Briefly, spleens were collected in splenocyte wash medium (SWM) (RPMI [Thermo Fisher Scientific; A1049101] plus 5% heat-inactivated FBS [GE Healthcare; SH30071.03HI] plus 1× antibiotic/antimycotic solution [Sigma-Aldrich; A5955]) at 4 °C. Spleens were either processed immediately or kept at 4 °C for 1 d before cell isolation. Spleens were mashed through a 40-µm cell strainer (Fisher Scientific; 352340) using a plunger from a 3-mL syringe. The collected suspension was flowed through again with a 40-µm strainer. Collected cell suspension was centrifuged at 500 × g for 5 min (4 °C) to pellet the cells. The supernatant was discarded and 2 to 5 mL of ACK lysis buffer (Thermo Fisher Scientific; A1049201) was added to the pellet (to lyse erythrocytes) for 3 to 5 min at room temperature. SWM (∼25 mL) was added to the cells and centrifuged (500 × g, 5 min, 4 °C) to pellet the cells. This SWM wash was repeated one to two times more. The final pellet was resuspended in 3 to 10 mL of SWM depending on the size of the pellet. In parallel to spleens, whole blood (anticoagulant heparin treated) obtained by perfusion was collected in 50-mL tubes. Cells were spun down (500 × g, 5 min, 4 °C), and supernatant was removed carefully. Two to 3 mL of ACK lysis buffer was added to the cell pellet for 2 to 3 min. The wash step was repeated twice, and cells were resuspended in 2 to 3 mL of SWM. Viable cell density was measured by counting trypan blue-excluding cells with a hemocytometer. GFP expression was confirmed using epifluorescence microscopy.
In Vitro Feeding of GFP+ Immune Cells.
Control and foxA RNAi parasites were placed in µ-channels (Ibidi; µ-Slide VI 0.4) and incubated with 100,000 to 200,000 GFP+ total immune cells harvested from spleen or whole blood. Videos were recorded using an Axio Observer (Carl Zeiss) with a built-in incubation chamber to maintain the temperature at 37 °C.
Imaging and Image Processing.
All FISH and immunolabeled images were taken using a Zeiss LSM 710, Zeiss LSM 880 with Airyscan (Carl Zeiss), or Andor WDb spinning disk confocal microscope (Andor Technology). WISH images were taken using an AxioZoom.V16 stereomicroscope (Carl Zeiss). Imaris (Bitplane) and Photoshop (Adobe Systems) were used to process acquired images of maximum-intensity projections (of z stacks) and single confocal sections for linear adjustment of brightness and contrast. Parasite length was measured using ImageJ.
Protein Sequence and Phylogenetic Analysis.
All S. mansoni forkhead domain-containing genes were identified from GeneDB and WormBase ParaSite (35, 69, 70) and aligned to other forkhead domain-containing proteins (71) across multiple animals using CLC Genomics Workbench 9. Default settings were used for both construction of alignments (gap open cost, 10; gap extension cost, 1; end gap cost, as any other; alignment, very accurate), and estimation of the maximum-likelihood phylogeny (construction method, neighbor joining; protein substitution model, WAG).
Data Availability Statement.
All data are available in the main text and SI Appendix.
Supplementary Material
Acknowledgments
B. glabrata snails were provided by the National Institute of Allergy and Infectious Diseases (NIAID) Schistosomiasis Resource Center of Biomedical Research Institute (Rockville, MD) through NIH–NIAID Contract HHSN272201700014I for distribution through BEI Resources. We thank Research Animal Resources Center and Biomedical Research Model Services at the University of Wisconsin–Madison for animal breeding and husbandry. We thank Jim Collins (University of Texas Southwestern Medical Center) for training on the schistosome transplantation experiments; John Brubacher, Melanie Issigonis, Tania Rozario, and Wan-Lin Lo (University of California, San Francisco) for comments on the manuscript; Jiarong Gao, Janmesh Patel, and Srishti Gupta for help with parasite harvesting and maintenance of the life cycle; and all of the members of the P.A.N. laboratory for discussions and feedback. J.L. is the recipient of a Morgridge Postdoctoral Fellowship, and P.A.N. is an investigator of the Howards Hughes Medical Institute.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2006553117/-/DCSupplemental.
References
- 1.Hoffmann K. F., Brindley P. J., Berriman M., Medicine. Halting harmful helminths. Science 346, 168–169 (2014). [DOI] [PubMed] [Google Scholar]
- 2.McManus D. P. et al., Schistosomiasis. Nat. Rev. Dis. Primers 4, 13 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Wilson R. A., The saga of schistosome migration and attrition. Parasitology 136, 1581–1592 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Basch P. F., Schistosomes: Development, Reproduction, and Host Relations, (Oxford University Press, 1991). [Google Scholar]
- 5.Pearce E. J., MacDonald A. S., The immunobiology of schistosomiasis. Nat. Rev. Immunol. 2, 499–511 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Collins J. J. 3rd et al., Adult somatic stem cells in the human parasite Schistosoma mansoni. Nature 494, 476–479 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang B. et al., Stem cell heterogeneity drives the parasitic life cycle of Schistosoma mansoni. eLife 7, e35449 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wendt G., et al. , A single-cell RNAseq atlas of the pathogenic stage of Schistosoma mansoni identifies a key regulator of blood feeding. bioRxiv:10.1101/2020.02.03.932004 (3 February 2020).
- 9.Diaz Soria C. L., et al. , Single-cell atlas of the first intra-mammalian developmental stage of the human parasite Schistosoma mansoni. bioRxiv: 10.1101/2019.09.11.754713 (11 September 2019). [DOI] [PMC free article] [PubMed]
- 10.Li P., et al. , Single-cell analysis of Schistosoma mansoni reveals a conserved genetic program controlling germline stem cell fate. bioRxiv: 10.1101/2020.07.06.190033 (6 July 2020). [DOI] [PMC free article] [PubMed]
- 11.Wang B., Collins J. J. 3rd, Newmark P. A., Functional genomic characterization of neoblast-like stem cells in larval Schistosoma mansoni. eLife 2, e00768 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins J. J. 3rd, Wendt G. R., Iyer H., Newmark P. A., Stem cell progeny contribute to the schistosome host-parasite interface. eLife 5, e12473 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wendt G. R. et al., Flatworm-specific transcriptional regulators promote the specification of tegumental progenitors in Schistosoma mansoni. eLife 7, e33221 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J., Collins J. J. 3rd, Identification of new markers for the Schistosoma mansoni vitelline lineage. Int. J. Parasitol. 46, 405–410 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo T., Peters A. H., Newmark P. A., A Bruno-like gene is required for stem cell maintenance in planarians. Dev. Cell 11, 159–169 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Rozario T., Quinn E. B., Wang J., Davis R. E., Newmark P. A., Region-specific regulation of stem cell-driven regeneration in tapeworms. eLife 8, e48958 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solana J. et al., Defining the molecular profile of planarian pluripotent stem cells using a combinatorial RNAseq, RNA interference and irradiation approach. Genome Biol. 13, R19 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hockley D. J., McLaren D. J., Schistosoma mansoni: Changes in the outer membrane of the tegument during development from cercaria to adult worm. Int. J. Parasitol. 3, 13–25 (1973). [DOI] [PubMed] [Google Scholar]
- 19.Skelly P. J., Wilson R. A., Making sense of the schistosome surface. Adv. Parasitol. 63, 185–284 (2006). [DOI] [PubMed] [Google Scholar]
- 20.McLaren D. J., Schistosoma mansoni: The Parasite Surface in Relation to Host Immunity, (Tropical Medicine Research Studies Series, Research Studies Press, Chichester, UK, 1980). [Google Scholar]
- 21.Dillon G. P., Illes J. C., Isaacs H. V., Wilson R. A., Patterns of gene expression in schistosomes: Localization by whole mount in situ hybridization. Parasitology 134, 1589–1597 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Figueiredo B. C. et al., Kicking in the guts: Schistosoma mansoni digestive tract proteins are potential candidates for vaccine development. Front. Immunol. 6, 22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martins V. P. et al., Sm10.3, a member of the micro-exon gene 4 (MEG-4) family, induces erythrocyte agglutination in vitro and partially protects vaccinated mice against Schistosoma mansoni infection. PLoS Negl. Trop. Dis. 8, e2750 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawrence J. D., The ingestion of red blood cells by Schistosoma mansoni. J. Parasitol. 59, 60–63 (1973). [PubMed] [Google Scholar]
- 25.Li X. H. et al., The schistosome oesophageal gland: Initiator of blood processing. PLoS Negl. Trop. Dis. 7, e2337 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orcia D. et al., Interaction of an esophageal MEG protein from schistosomes with a human S100 protein involved in inflammatory response. Biochim. Biophys. Acta Gen. Subj. 1861, 3490–3497 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Wilson R. A. et al., The schistosome esophagus is a “hotspot” for microexon and lysosomal hydrolase gene expression: Implications for blood processing. PLoS Negl. Trop. Dis. 9, e0004272 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall S. L. et al., Insights into blood feeding by schistosomes from a proteomic analysis of worm vomitus. Mol. Biochem. Parasitol. 179, 18–29 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Dorsey C. H., Cousin C. E., Lewis F. A., Stirewalt M. A., Ultrastructure of the Schistosoma mansoni cercaria. Micron 33, 279–323 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Skelly P. J., Da’dara A. A., Li X. H., Castro-Borges W., Wilson R. A., Schistosome feeding and regurgitation. PLoS Pathog. 10, e1004246 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collins J. N., Collins J. J. 3rd, Tissue degeneration following loss of Schistosoma mansoni cbp1 is associated with increased stem cell proliferation and parasite death in vivo. PLoS Pathog. 12, e1005963 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeMarco R. et al., Protein variation in blood-dwelling schistosome worms generated by differential splicing of micro-exon gene transcripts. Genome Res. 20, 1112–1121 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X. H. et al., Microexon gene transcriptional profiles and evolution provide insights into blood processing by the Schistosoma japonicum esophagus. PLoS Negl. Trop. Dis. 12, e0006235 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caffrey C. R., McKerrow J. H., Salter J. P., Sajid M., Blood “n” guts: An update on schistosome digestive peptidases. Trends Parasitol. 20, 241–248 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Protasio A. V. et al., A systematically improved high quality genome and transcriptome of the human blood fluke Schistosoma mansoni. PLoS Negl. Trop. Dis. 6, e1455 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaestner K. H., The FoxA factors in organogenesis and differentiation. Curr. Opin. Genet. Dev. 20, 527–532 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mango S. E., The C. elegans pharynx: A model for organogenesis. WormBook, 1–26 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Howe K. L., Bolt B. J., Shafie M., Kersey P., Berriman M., WormBase ParaSite—a comprehensive resource for helminth genomics. Mol. Biochem. Parasitol. 215, 2–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adler C. E., Seidel C. W., McKinney S. A., Sánchez Alvarado A., Selective amputation of the pharynx identifies a FoxA-dependent regeneration program in planaria. eLife 3, e02238 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clegg J. A., In vitro cultivation of Schistosoma mansoni. Exp. Parasitol. 16, 133–147 (1965). [DOI] [PubMed] [Google Scholar]
- 41.Cioli D., Transfer of Schistosoma mansoni into the mesenteric veins of hamsters. Int. J. Parasitol. 6, 349–354 (1976). [DOI] [PubMed] [Google Scholar]
- 42.Xiao S. H., Sun J., Schistosoma hemozoin and its possible roles. Int. J. Parasitol. 47, 171–183 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Mombaerts P. et al., RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68, 869–877 (1992). [DOI] [PubMed] [Google Scholar]
- 44.Kitamura D., Roes J., Kühn R., Rajewsky K., A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature 350, 423–426 (1991). [DOI] [PubMed] [Google Scholar]
- 45.Schaefer B. C., Schaefer M. L., Kappler J. W., Marrack P., Kedl R. M., Observation of antigen-dependent CD8+ T-cell/dendritic cell interactions in vivo. Cell. Immunol. 214, 110–122 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Harris A. R., Russell R. J., Charters A. D., A review of schistosomiasis in immigrants in Western Australia, demonstrating the unusual longevity of Schistosoma mansoni. Trans. R. Soc. Trop. Med. Hyg. 78, 385–388 (1984). [DOI] [PubMed] [Google Scholar]
- 47.Hornstein L. et al., Persistent Schistosoma mansoni infection in Yemeni immigrants to Israel. Isr. J. Med. Sci. 26, 386–389 (1990). [PubMed] [Google Scholar]
- 48.Payet B. et al., Prolonged latent schistosomiasis diagnosed 38 years after infestation in a HIV patient. Scand. J. Infect. Dis. 38, 572–575 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Mclaren D. J., Hockley D. J., Blood flukes have a double outer membrane. Nature 269, 147–149 (1977). [DOI] [PubMed] [Google Scholar]
- 50.Wilson R. A., Barnes P. E., The tegument of Schistosoma mansoni: Observations on the formation, structure and composition of cytoplasmic inclusions in relation to tegument function. Parasitology 68, 239–258 (1974). [PubMed] [Google Scholar]
- 51.Capron A. et al., IgE and cells in schistosomiasis. Am. J. Trop. Med. Hyg. 26, 39–47 (1977). [DOI] [PubMed] [Google Scholar]
- 52.Capron M., Rousseaux J., Mazingue C., Bazin H., Capron A., Rat mast cell-eosinophil interaction in antibody-dependent eosinophil cytotoxicity to Schistosoma mansoni schistosomula. J. Immunol. 121, 2518–2525 (1978). [PubMed] [Google Scholar]
- 53.Capron M., Camus D., Carlier Y., Figueiredo J. F., Capron A., Immunological studies in human schistosomiasis. II. Antibodies cytotoxic for Schistosoma mansoni schistosomules. Am. J. Trop. Med. Hyg. 26, 248–253 (1977). [DOI] [PubMed] [Google Scholar]
- 54.Brindley P. J., Sher A., The chemotherapeutic effect of praziquantel against Schistosoma mansoni is dependent on host antibody response. J. Immunol. 139, 215–220 (1987). [PubMed] [Google Scholar]
- 55.Brindley P. J., Strand M., Norden A. P., Sher A., Role of host antibody in the chemotherapeutic action of praziquantel against Schistosoma mansoni: Identification of target antigens. Mol. Biochem. Parasitol. 34, 99–108 (1989). [DOI] [PubMed] [Google Scholar]
- 56.Zaret K. S., Carroll J. S., Pioneer transcription factors: Establishing competence for gene expression. Genes Dev. 25, 2227–2241 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li X. H., Vance G. M., Cartwright J., Cao J. P., Wilson R. A., Castro-Borges W., Mapping the epitopes of Schistosoma japonicum esophageal gland proteins for incorporation into vaccine constructs. PLoS One. 15, e0229542 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matthews K. R., Controlling and coordinating development in vector-transmitted parasites. Science 331, 1149–1153 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lewis F., Schistosomiasis. Curr. Protoc. Immunol. Chapter 19, Unit 19.1 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Protasio A. V., Dunne D. W., Berriman M., Comparative study of transcriptome profiles of mechanical- and skin-transformed Schistosoma mansoni schistosomula. PLoS Negl. Trop. Dis. 7, e2091 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tucker M. S., Karunaratne L. B., Lewis F. A., Freitas T. C., Liang Y. S., Schistosomiasis. Curr. Protoc. Immunol. 103, 19.1.1-19.1.58 (2013). [DOI] [PubMed] [Google Scholar]
- 62.Basch P. F., Cultivation of Schistosoma mansoni in vitro. I. Establishment of cultures from cercariae and development until pairing. J. Parasitol. 67, 179–185 (1981). [PubMed] [Google Scholar]
- 63.Cogswell A. A., Collins J. J. 3rd, Newmark P. A., Williams D. L., Whole mount in situ hybridization methodology for Schistosoma mansoni. Mol. Biochem. Parasitol. 178, 46–50 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.King R. S., Newmark P. A., In situ hybridization protocol for enhanced detection of gene expression in the planarian Schmidtea mediterranea. BMC Dev. Biol. 13, 8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Collins J. J. 3rd, King R. S., Cogswell A., Williams D. L., Newmark P. A., An atlas for Schistosoma mansoni organs and life-cycle stages using cell type-specific markers and confocal microscopy. PLoS Negl. Trop. Dis. 5, e1009 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Collins J. J. 3rd et al., Genome-wide analyses reveal a role for peptide hormones in planarian germline development. PLoS Biol. 8, e1000509 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang J., Chen R., Collins J. J. 3rd, Systematically improved in vitro culture conditions reveal new insights into the reproductive biology of the human parasite Schistosoma mansoni. PLoS Biol. 17, e3000254 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Davies S. J. et al., Modulation of blood fluke development in the liver by hepatic CD4+ lymphocytes. Science 294, 1358–1361 (2001). [DOI] [PubMed] [Google Scholar]
- 69.Berriman M. et al., The genome of the blood fluke Schistosoma mansoni. Nature 460, 352–358 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Howe K. L. et al., WormBase 2016: Expanding to enable helminth genomic research. Nucleic Acids Res. 44, D774–D780 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hannenhalli S., Kaestner K. H., The evolution of Fox genes and their role in development and disease. Nat. Rev. Genet. 10, 233–240 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text and SI Appendix.