Significance
To date, most industrially relevant bioproduction processes rely on sugar-based feedstocks, whereas utilization of ever-increasing protein wastes or hydrolysates, e.g., amino acids (AAs), for chemical production has been less explored. Currently, AAs that generated from cheap protein-rich wastes or overproduced from industrial fermentation mainly serve as animal feed additives and flavor ingredients. Considering the natural abundance and structural diversity of AAs, they may serve as new platform chemicals for chemical expansion in microbial systems. Through combination of a common catabolism of charged AAs and a diol production pathway, we devise a general metabolic platform that converts multiple AAs and glucose into C3-C5 diols. Our work presents a platform that enriches the pathway repertoire for nonnatural diols with expanded feedstock flexibility.
Keywords: metabolic engineering, amino acid catabolism, C3-C5 diols
Abstract
Amino acids are naturally occurring and structurally diverse metabolites in biological system, whose potentials for chemical expansion, however, have not been fully explored. Here, we devise a metabolic platform capable of producing industrially important C3-C5 diols from amino acids. The presented platform combines the natural catabolism of charged amino acids with a catalytically efficient and thermodynamically favorable diol formation pathway, created by expanding the substrate scope of the carboxylic acid reductase toward noncognate ω-hydroxylic acids. Using the established platform as gateways, seven different diol-convertible amino acids are converted to diols including 1,3-propanediol, 1,4-butanediol, and 1,5-pentanediol. Particularly, we afford to optimize the production of 1,4-butanediol and demonstrate the de novo production of 1,5-pentanediol from glucose, with titers reaching 1.41 and 0.97 g l−1, respectively. Our work presents a metabolic platform that enriches the pathway repertoire for nonnatural diols with feedstock flexibility to both sugar and protein hydrolysates.
Microbial hosts, like the most widely used Escherichia coli and Saccharomyces cerevisiae, can natively generate an exquisite precursor chemical pool and cofactors from renewable feedstocks, offering a microbial-based platform for production of a variety of natural or new-to-nature chemicals, including biofuels, biopolymers, commodity chemicals, and phytochemicals (1–4). Extending from existing microbial biochemical infrastructures, carbon flux can be streamed into products of interest via purpose-driven biosynthetic pathways. However, most industrial applications are currently focused on utilization of only a limited subset of native metabolites or pathways, for instance, to produce short-chain alcohols or isoprenoids from glycolysis intermediates like pyruvate and acetyl-CoA, to produce long-chain fatty acids, alcohols, or alkanes from fatty acid pathway, and to produce short-chain dicarboxylic acids from tricarboxylic acid (TCA) cycle (5–7). Although substantial developments have been made in expanding the microbial capacity (8, 9), the chemical space accessible from microbial-based system has been significantly restrained by the narrow scope of the precursor pools, as well as the scarcity of natural or synthetic pathways for new products.
Amino acids (AAs) are constitutional units for proteins and also one of the major endogenous metabolites in almost all microbes, whose utilities as building blocks for chemical production, however, have been overlooked. Currently, most of the 20 proteinogenic AAs are microbially produced as end and bulk biochemicals with around 5 million tons per year, and are mainly used as animal feed additives and flavor ingredients (10, 11). Protein hydrolysates from protein-rich biomass (e.g., fast-growing photosynthetic microalgae), or protein wastes from food-processing industry and industrial fermentation residues, serve as inexpensive and readily available external sources for AAs (12–14). Therefore, the structural diversity, ease of achieving high titers in microorganisms, and ever-increasing accessibility from protein feedstocks, have substantiated AAs with potentials as versatile starting scaffolds for various industrially relevant natural or synthetic products (15, 16). In recent decades, intensive efforts have been taken toward converting branched-chain AAs (alanine, valine, leucine, isoleucine, etc.) to advanced alcohols or acids via the Ehrlich pathway (17–19), and aromatic AAs (phenylalanine, tyrosine, and tryptophan) to medically or nutritionally important phytochemicals via phenylpropanoic or alkaloid pathways (2, 20). However, the development of efficient biocatalytic routes to produce functionalized chemicals from charged AAs (aspartate, glutamate, lysine, etc.), which account for >40% of all AAs in protein hydrolysates and >80% of all of the AA market, are comparatively less tapped (10, 21). Charged AAs harbor bifunctional terminal groups, making them recalcitrant to be directly metabolized through central carbon metabolism. Additionally, the paucity of natural or synthetic pathways to utilize charged AAs significantly limited their valorization.
In the present study, we explored the potential of migrating the AA industry to chemical industry by establishing nonnatural pathways capable of converting charged AAs to C3-C5 diols. C3-C5 diols, including 1,3-propanediol (1,3-PDO), 1,4-butanediol (1,4-BDO), and 1,5-pentanediol (1,5-PDO), have wide use in commodity and fine chemical industries such as fuels, solvents, polymer monomers, and pharmaceutical precursors (22–24). These short-chain diols represent an annual commercial market over $7 billion with a nearly 7% annual growth rate (25). The C3 diol 1,3-PDO can be naturally produced from anaerobic reduction of glycerol, which however involves supplementation of expensive vitamin B12 for glycerol dehydratase (22). Although a new glycerol-independent 1,3-PDO route has been developed from homoserine degradation, it suffered from limited pathway performance (26). As for 1,4-BDO and 1,5-PDO, there is no natural pathway for their direct production. Until recently, two major artificial biocatalytic routes to 1,4-BDO were developed: one that involves reduction of succinyl-CoA or decarboxylation of α-ketoglutarate to 4-hydroxybutyrate (4HB) with subsequent CoA-dependent reduction of 4HB to 1,4-BDO, and a second that involves nonphosphorylative xylose metabolism (27, 28). Notably, the former CoA-dependent pathway suffers from drawbacks including low enzyme kinetics and lactonization or hydrolysis of CoA intermediates that lead to incomplete conversion of 4HB to 1,4-BDO, while the latter is specialized for C5 sugar feedstocks (e.g., xylose and arabinose) and requires energy input from additional carbon sources (27–29). With respect to 1,5-PDO, its petroleum-based synthesis has been restricted due to limited access to C5 petroleum feedstocks, while its biobased production from renewable feedstocks has not been developed to date (30). Due to their linear infrastructures and the existence of natural catabolism, charged AAs can potentially serve as direct precursors for C3-C5 diols.
Here we describe the development of a metabolic platform that engenders direct conversion of charged AAs into C3-C5 diols. The centerpiece of the platform is to release the carbon skeletons from charged AAs into “C-1” ω-hydroxylic acids (ω-HAs) via native or heterologous AA catabolism, followed by exploiting the catalytic capabilities of carboxylic acid reductase (Car) to reduce noncognate ω-HA substrates to diols. The synthetic diol pathways bypass either expensive coenzyme supplementation or CoA-dependent chemistry, and is featured by broad host applicability and feedstock flexibility. With the presented platform, a panel of seven AAs were converted into C3-C5 diols, based upon which we were able to achieve and optimize a synthetic pathway of 1,4-BDO and demonstrate de novo production of 1,5-PDO from glucose. Our platform highlighted the feasibility of expanding the chemical space of microbial systems by extending AA catabolism, which could potentially serve as a gateway for diol production from renewable biomass or protein-derived feedstocks.
Results
Design and Validation of a Synthetic Platform for C3-C5 Diols.
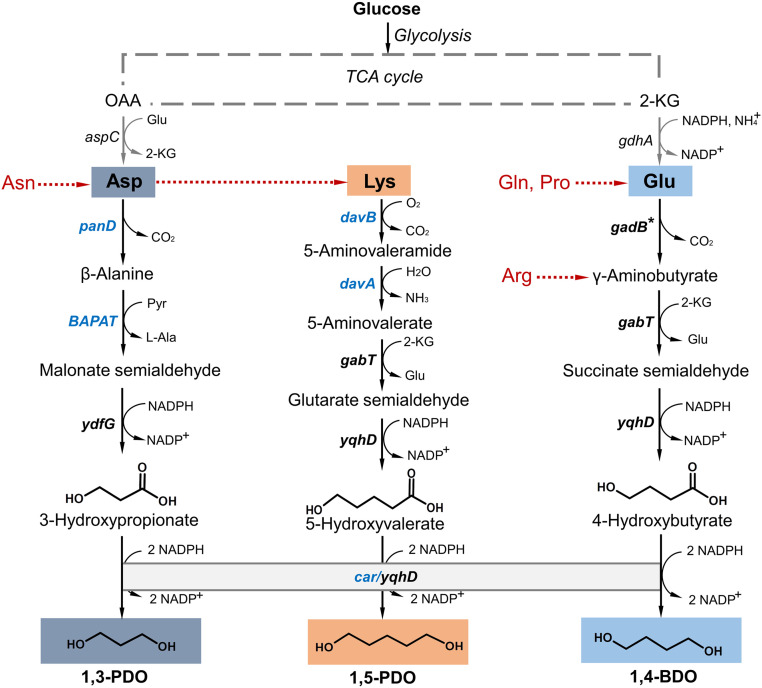
The proposed AA-to-diol platform proceeds through two parts: the upstream degradation of charged AAs into corresponding “C-1” ω-HAs, followed by the downstream reduction of ω-HAs into counterpart diols (Fig. 1). Typical charged AAs, including aspartate (Asp), glutamate (Glu), and lysine (Lys), share a common degradation process of sequential decarboxylation and transamination to counterpart C-1 semialdehydes, which are further reduced to ω-HAs via aldehyde reductases (ALRs)/alcohol dehydrogenases (ADHs). With the existence of well-elucidated native or heterologous AA catabolism supporting our hypothesized platform, the ω-HA-to-diol conversion becomes the most pivotal step for diol production.
Fig. 1.
Establishing a diol production platform via extending AA catabolism in E. coli. The scheme represents native anabolism and designed catabolism of charged AAs for diol production. Asp, Glu, and Lys are biosynthesized from TCA cycle intermediates. Via the synthetic platform, Asp can be converted to 1,3-PDO, Glu converted to 1,4-BDO and Lys converted to 1,5-PDO. Asn, Gln, Pro, and Arg can be directed to the diol production platform via corresponding degradation pathways. Endogenous genes are indicated in black and heterologous genes are indicated in blue. panD, l-aspartate-α-decarboxylase; BAPAT, β-alanine pyruvate transaminase; ydfG, malonate semialdehyde reductase; gadB*, mutant glutamate decarboxylase (GadB E89QΔ452–466); gabT, γ-aminobutyrate aminotransferase; yqhD, alcohol dehydrogenase; davB, lysine 2‐monooxygenase; davA, 5‐aminovaleramidase; davT, 5-aminovalerate transaminase; car, carboxylic acid reductase.
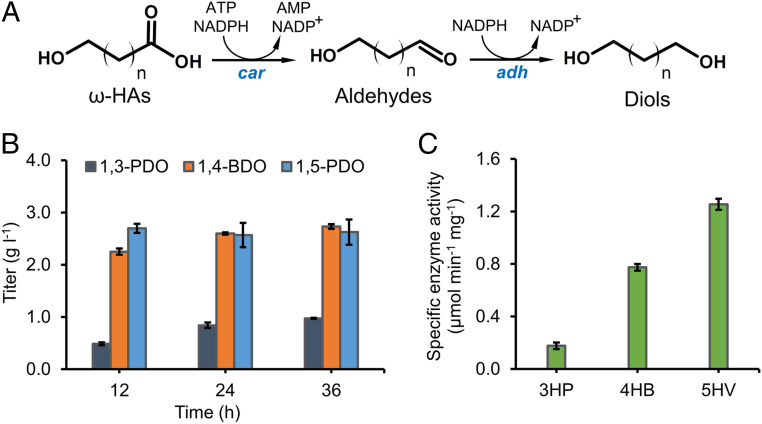
The classical pathway of ω-HA-to-diol conversion harnesses a CoA-dependent reduction route that involves activation of carboxylate group by a CoA-transferase, followed by reduction of the CoA intermediate to aldehyde and then diol by a bifunctional alcohol dehydrogenase or a CoA-dependent aldehyde dehydrogenase and an alcohol dehydrogenase with input of 2 NAD(P)H (27, 31). We here proposed a CoA-independent reduction route that utilizes a promiscuous carboxylic acid reductase (Car) and endogenous ALRs/ADHs to reduce carboxylate group to alcohol group at the costs of 1 ATP and 2 NADPH (Fig. 2A). Car was first characterized to catalyze the reduction of aromatic carboxylic acids, and was later revealed to accept a wide range of aliphatic fatty acids (C6-C18) (32). We attempted to explore its catalytic promiscuity toward noncognate substrates C3-C5 ω-HAs to produce counterpart short-chain diols. To compare the thermodynamics of the two reduction routes of 4HB to 1,4-BDO, we performed the max-min Driving Force (MDF) analysis via eQuilibrator (33). The Car-based reduction pathway operates at a higher MDF (38.0 kJ mol−1) than the CoA-dependent pathway (28.8 kJ mol−1), indicating a higher pathway flux and/or a lower enzyme requirement under physiological conditions in E. coli (SI Appendix, Table S3).
Fig. 2.
Validation of Car-based diol production. (A) Car-based biosynthetic route of ω-HAs to diols. (B) Time-course conversion of 3 g l−1 3HP, 4HB, and 5HV to 1,3-PDO, 1,4-BDO, and 1,5-PDO, respectively, by HA-1 expressing car and sfp. (C) Enzymatic activity assay of Car toward 3HP, 4HB, and 5HV. Error bars represent SD (n = 3).
To experimentally assess the pathway performance, the Car-based reduction route was expressed in E. coli and tested by feeding C3-C5 ω-HAs. The Car-based pathway, comprising car from Mycobacterium marinum and phosphopantetheine transferase (sfp) from Bacillus subtilis, were expressed in the wildtype E. coli BW25113 (F′) on the high-copy-number plasmid (pCar), yielding strain HA-1. Sfp is expressed as a phosphopantetheinyl transferase responsible for activation of the apoenzyme Car by phosphopantetheinylation (34). For the sake of simplicity, we excluded the expression of ALRs/ADHs as it has been reported that the endogenous ones could efficiently reduce various aldehydes to alcohols (35). Since 4HB and 5-hydroxyvalerate (5HV) are not commercially available, they were produced, respectively, from γ-butyrolactone and σ-valerolactone by saponification under alkaline conditions. When fed with 3-hydroxypropionate (3HP), the recombinant E. coli strain HA-1 produced 0.97 g l−1 1,3-PDO from 3 g l−1 3HP in 36 h, representing a conversion efficiency of 38.6% (Fig. 2B). HA-1 showed high conversion capability toward 4HB and 5HV, which produced 2.60 g l−1 1,4-BDO in 24 h and 2.70 g l−1 1,5-PDO in 12 h, accounting for complete conversion of both substrates (Fig. 2B). The bioconversion results underlined the high performance of the Car-based pathway in ω-HA-to-diol reduction. Noteworthy, Car exhibited a catalytic preference toward longer-chain ω-HAs (C5 > C4 > C3), which is consistent with its preference toward long-chain fatty acids (32, 36). This was further confirmed by the increase of the specificity enzyme activity of Car toward substrates with increased chain lengths (0.18 μmol min−1⋅mg−1 for 3HP versus 1.25 μmol min−1⋅mg−1 for 5HV) during in vitro enzyme assay using crude enzyme extracts of HA-2 (Fig. 2C).
Establishing the AA Degradation Pathways in E. coli.
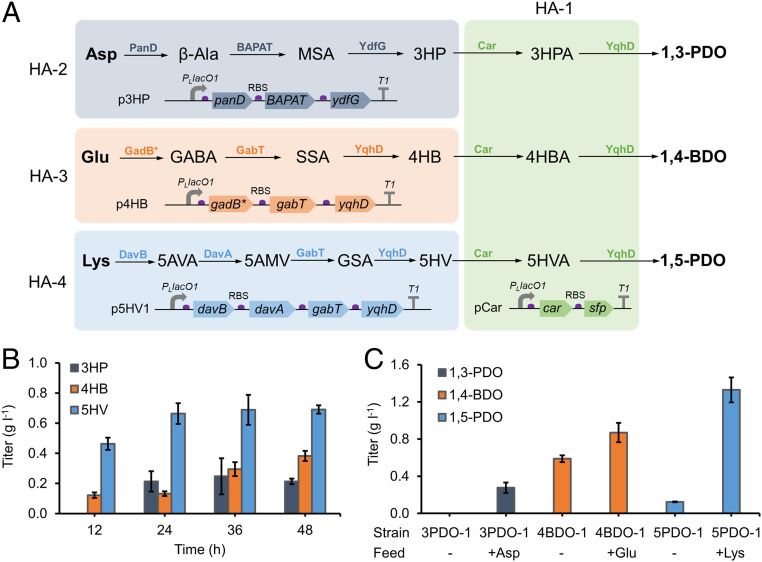
With validation of an efficient ω-HA-to-diol conversion pathway, our second goal is to assemble the upstream AA degradation pathways capable of converting charged AAs into C-1 ω-HAs. E. coli harbors native degradation pathways for Asp, Glu, and Lys, which however, are committed to either CoA biosynthesis or acid resistance and do not naturally yield counterpart ω-HAs (37–39). For Asp degradation, a synthetic 3HP pathway was constructed consisting of l-aspartate-α-decarboxylase (PanD), β-alanine pyruvate transaminase (BAPAT), and malonate semialdehyde (MSA) reductase (40). The native Glu metabolism, also known as γ-aminobutyrate (GABA) pathway, generates GABA via glutamate decarboxylase (GadB). GABA can be transaminated to succinate semialdehyde (SSA) by γ-aminobutyrate aminotransferase (transaminase) (GabT), followed by reduction to 4HB via endogenous NAD(P)H-dependent ADHs like YqhD (41, 42). It was very recently elucidated that the native Lys degradation pathway converts Lys to glutarate semialdehyde (GSA) via a four-step pathway comprising lysine decarboxylase (CadA), putrescine transaminase (PatA), 5-aminopentanal dehydrogenase (PatD), and GabT (43). Alternatively, a heterologous three-step Lys degradation pathway from Pseudomonas putida, known as AMV pathway, converts Lys to GSA with lysine 2‐monooxygenase (DavB), 5‐aminovaleramidase (DavA) and 5-aminovalerate transaminase (DavT) (44). Further reduction of GSA by endogenous ADHs will release 5HV.
To render conversion of AAs to C3-C5 ω-HAs, we constructed and expressed each ω-HA pathway in E. coli. The synthetic 3HP pathway, consisting of panD from Corynebacterium glutamicum, BAPAT from P. putida, and the MSA reductase (ydfG) from E. coli (40), was integrated into the medium-copy plasmid pCS27, yielding p3HP. Similarly, the native GABA pathway for Glu degradation consisting of gadB* (encoding GadB mutant E89Q Δ452–466) (45), gabT, and yqhD from E. coli, was integrated into p4HB, while the synthetic AMV pathway for Lys degradation consisting of davBA from P. putida, gabT, and yqhD from E. coli, was integrated into p5HV1. These three plasmids were transferred into E. coli BW25113 (F′), yielding strains HA-2 to HA-4 (Fig. 3A). When fed with 3 g l−1 corresponding AAs, HA-2 produced 247.3 mg l−1 3HP from Asp, HA-3 produced 382.5 mg l−1 4HB from Glu, and HA-4 produced 690.8 mg l−1 5HV from Lys in 48 h of cultivation (Fig. 3B). These productions demonstrated the functionality of all AA degradation pathways.
Fig. 3.
Direct conversion of AAs to diols. (A) Scheme of the bioconversion experiments via recombinant E. coli strains HA-2, -3, and -4. HA-2 (harboring p3HP), -3 (harboring p4HB), and -4 (harboring p5HV1) converted charged AAs to C3-C5 ω-HAs. MSA, malonate semialdehyde; 3HPA, 3-hydroxypropionaldehyde; SSA, succinate semialdehyde; 4HBA, 4-hydroxybutyraldehyde; 5AMV, 5-aminovalerate; GSA, glutarate semialdehyde; 5HVA, 5-hydroxyvaleraldehyde. (B) Time-course conversion of 3 g l−1 Asp, Glu, and Lys to 3HP, 4HB, and 5HV by HA-2, -3, and -4, respectively. (C) Bioconversion of 3 g l−1 Asp, Glu, and Lys to 1,3-PDO, 1,4-BDO, and 1,5-PDO by strain 3PDO-1, 4BDO-1, and 5PDO-1, respectively. Error bars represent SD (n = 3).
Direct Conversion of Charged AAs to C3-C5 Diols.
Validation of both upstream and downstream parts made it possible to achieve the direct conversion of charged AAs including Asp, Glu, and Lys to diols. Considering the low performance of 3HP and 4HB pathway on medium-copy plasmids, the 3HP and 4HB pathway were reassembled into the high-copy plasmid pCar, yielding p3PDO and p4BDO, respectively. E. coli BW25113 (F′) harboring p3PDO (strain 3PDO-1) did not show any 1,3-PDO production with no exogenous Asp feeding, while it produced 276.6 mg l−1 1,3-PDO in 48 h when fed with 3 g l−1 Asp, accounting for a conversion efficiency of 11.8% (Fig. 3C). This validated the functional expression but low activity of the complete 1,3-PDO pathway, likely because of the cumulative inefficiency of the upper 3HP pathway and the low activity of Car toward 3HP. Upon expressing the 1,4-BDO and 1,5-PDO pathway in E. coli BW25113 (F′), strain 4BDO-1 containing p4BDO produced 588.3 mg l−1 1,4-BDO and strain 5PDO-1 containing pHV1 and pCar produced 123.9 mg l−1 1,5-PDO without Glu or Lys feeding (Fig. 3C). This substantiated the function of both 1,4-BDO and 1,5-PDO pathways and their capabilities of converting intracellular Glu and Lys to counterpart diols. High titer of 1,4-BDO is consistent with the higher abundance of intracellular Glu in E. coli cells (46). Feeding 3 g l−1 Glu to strain 4BDO-1 further increased 1,4-BDO titer by 47.8% to 869.7 mg l−1, accounting for a conversion efficiency of 15.3% (Fig. 3C). Considering low Glu-to-4HB but high 4HB-to-1,4-BDO conversion, Glu degradation was suggested to be a potential bottleneck for 1,4-BDO production. This also clearly indicated that 1,4-BDO titer could not be significantly improved by simply imposing glutamate supply. Strikingly, feeding 3 g l−1 Lys to strain 5PDO-1 drastically increased 1,5-PDO titer by 10.7-fold to 1.33 g l−1 (Fig. 3C), accounting for a conversion efficiency of 56.4%. This indicated that 1,5-PDO pathway is rather efficient as high Lys-to-1,5PDO conversion efficiency was achieved even without pathway or cultivation optimization. As such, unlike the 1,4-BDO pathway, lysine supply might be a major bottleneck for de novo 1,5-PDO production.
Bioconversion of Diol-Convertible AAs.
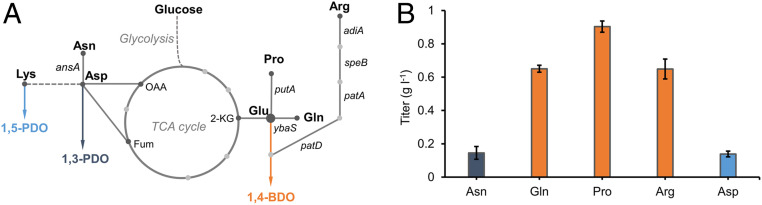
On construction and validation of the C3-C5 diol production platform, we sought to broaden its substrate scope by rerouting all potential AAs into counterpart diols. E. coli natively harbors AA degradation network that enables convergence of four additional AAs into our platform (Fig. 4A). Asparagine (Asn) is directly converted to Asp via asparaginases (AnsA and AnsB). Glutamine (Gln) can be converted to Glu via glutaminase (YbaS) and proline (Pro) can be converted to Glu via bifunctional proline utilization A (PutA). Arginine (Arg) can be degraded to GABA via a four-step pathway comprising arginine decarboxylase (AdiA), agmatinase (SpeB), PatA, and PatD. All these AAs, hereafter referred to as diol-convertible AAs, have previously remained unexplored for deriving new chemical entities due to their structural complexities.
Fig. 4.
Bioconversion of diol-convertible AAs. (A) Engineering endogenous AA degradation pathways of different AAs to diols. (B) Conversion of diol-convertible AAs including Asn, Gln, Pro, Arg, and Asp (3 g l−1) to diols. Error bars represent SD (n = 3).
We then adopted these native AA degradation pathways to enable diol release from diol-convertible AAs. To this end, we cotransferred each diol pathway plasmid and AA degradation pathway plasmid into E. coli BW25113 (F′), and the resulting strains (DO-1 to DO-4) were cultured aerobically in M9Y media supplemented with 3 g l−1 AA substrates for 48 h. When fed with Asn, strain DO-1 harboring p3PDO and pCS-ansA produced 145.3 mg l−1 1,3-PDO (Fig. 4B). Regarding 1,4-BDO conversion, strain DO-2 expressing YbaS enabled a titer of 650.1 mg l−1 from Gln, strain DO-3 expressing PutA enabled a titer of 903.1 mg l−1 from Pro, and strain DO-4 expressing the four-step Arg degradation pathway enabled a titer of 648.8 mg l−1 from Arg (Fig. 4B). The bioconversion experiments demonstrated that, with overexpression of AA degradation pathways, more AAs including Asn, Gln, Pro, and Arg could be converted to counterpart diols. Since Asp is the direct precursor for Lys biosynthetic pathway, we also test if feeding Asp could lead to 1,5-PDO production. Unexpectedly, feeding Asp to strain 5PDO-1 could just slightly increase 1,5-PDO titer (138.7 mg l−1), further suggesting that the Asp-to-Lys pathway is a limiting step for the synthesis of 1,5-PDO.
Production of 1,4-BDO from Glucose by Harnessing the Glu-To-Diol Pathway.
The establishment of AA-to-diol platform expands the feedstocks to AAs, and also implies the feasibility of direct diol production from sugar by connecting it to the endogenous AA pathway in microbial chassis. To test that, we first chose to demonstrate and optimize the 1,4-BDO production from glucose in E. coli. The C4 diol 1,4-BDO is a large-volume commodity chemical with extensive applications in polymer industry, and its biobased production at the commercial scale has been achieved via the CoA-dependent pathway with extensive enzyme screening, host engineering, and fermentative process optimization (47). Given that glutamate is the most abundant intracellular AA serving as the major nitrogen donor in glucose-grown E. coli cells (46), and thus, it can be readily channeled into 1,4-BDO pathway. This is supported by the observation of 1,4-BDO production (588.3 mg l−1) upon expressing the complete 1,4-BDO pathway in E. coli (Fig. 5A), even without further optimization.
Fig. 5.
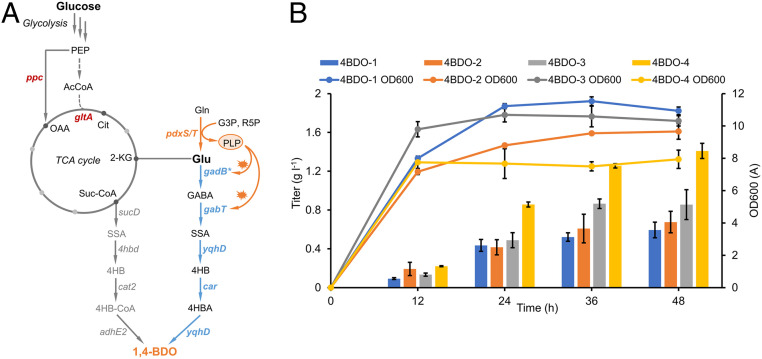
Production of 1,4-BDO from glucose via the Glu-to-1,4-BDO pathway in engineered E. coli. (A) De novo biosynthesis pathway of 1,4-BDO. Enzymes in gray indicate the CoA-dependent reduction pathway, and enzymes in blue indicate the Car-based pathway. PLP is a cofactor for GadB* and GabT. (B) 1,4-BDO production and growth profiles of strain 4BDO-1–4BDO-4 when cultivated in M9Y media. Samples were taken every 12 h for 48 h. All data points are reported as mean from three independent experiments. Error bars represent SD (n = 3).
To increase 1,4-BDO production, we sought to improve Glu decarboxylation efficiency. The wildtype glutamate decarboxylase (GadB) of E. coli exerts catalytic activity in acidic conditions (pH < 6), and its mutant GadB* (E89Q Δ452–466) showed improved activity at neutral pH (45). Considering that both GadB* and GABA aminotransferase GabT are pyridoxal 5′-phosphate (PLP)-dependent enzymes, we therefore resorted to improving glutamate decarboxylation efficiency by increasing the cofactor PLP supply. The native de novo PLP pathway in E. coli starts from erythrose-4-phosphate (E4P) via a seven-step deoxyxylulose-5-phosphate (DXP)-dependent pathway (48). To gain insights into the contribution of increasing PLP supply to 1,4-BDO production and reduce metabolic burden, we instead overexpressed an alternative and short ribose 5-phosphate (R5P)-dependent PLP pathway from B. subtilis that involves only two enzymes, PdxS (PLP synthase subunit) and PdxT (glutamine hydrolase subunit) (49). An additional benefit from PdxST pathway is the concomitant release of glutamate from glutamine, which can be taken up by 1,4-BDO pathway (Fig. 5A). When cultivating strain 4BDO-2 harboring p4BDO and pCS-pdxST in M9Y medium, 675.4 mg l−1 1,4-BDO was produced in 48 h, accounting for 13.7% increase than that produced by control strain 4BDO-1 (Fig. 5B). When further fed with 3 g l−1 glutamate, the 1,4-BDO titer was almost doubled (1.21 g l−1), reaching a conversion efficiency of 34.0% (SI Appendix, Fig. S1). These results implied that PLP surplus would significantly promote 1,4-BDO production, especially when glutamate flux was not limiting.
Hence, we next sought to increase carbon flux to Glu. To redirect carbon flux from glycolysis to TCA cycle, we overexpressed an NADH-insensitive citrate synthase mutant (gltA*, R163L) (27, 50), and phosphoenolpyruvate carboxylase (ppc) (Fig. 5A). The resultant strain 4BDO-3 increased 1,4-BDO titer by 45.6% (0.87 g l−1) than that of control strain 4BDO-1 (Fig. 5B), confirming that redirecting carbon flux to glutamate could augment Glu availability and thus 1,4-BDO production. When further introducing pdxST into 4BDO-3, the resultant strain BDO-4 afforded 1.41 g l−1 1,4-BDO in 48 h with a yield of 0.14 mol mol−1 glucose (14.0% of the theoretical maximum), representing a 2.40-fold increase of titer in relative to its parental strain 4BDO-1 (Fig. 5B). Noteworthy, only trace amounts of acetate were accumulated in the strain BDO-4 culture (SI Appendix, Fig. S2). As a proof-of-concept demonstration, our results highlight the potential of producing 1,4-BDO from glucose via the glutamate degradation machinery in microbial hosts.
De novo 1,5-PDO Production by Harnessing the Lys-To-Diol Pathway.
As a safer replacement for 1,4-BDO, 1,5-PDO is also a high-value and nonnatural short-chain diol with wide applications as a monomer for fibers and polyurethanes. However, its relatively high price and low production capacity limit its commercial use (25, 51). Currently, 1,5-PDO is chemically produced from biomass-derived furfural and tetrahydrofurfuryl alcohol via expensive metal catalysts (52, 53), while direct biocatalytic routes from renewable carbon source to 1,5-PDO have not been reported. With the lysine conversion platform, it is possible to fill the gap by achieving de novo production of 1,5-PDO from glucose.
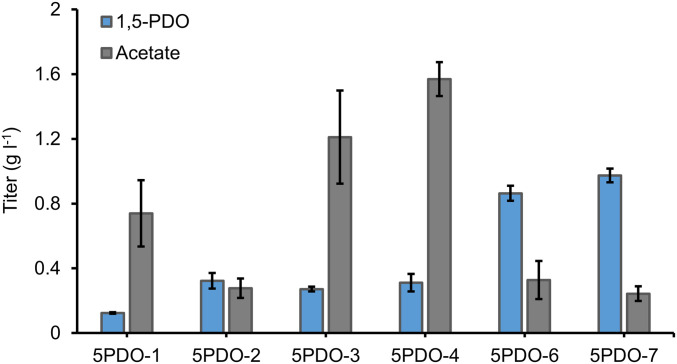
Insufficient lysine supply is a major rate-limiting step for de novo 1,5-PDO production. As observed earlier, lysine supplementation increased 1,5-PDO titer by 10.7-fold, suggesting the need to increase carbon flux to lysine (Fig. 3C). The native lysine pathway involves 10 enzymatic steps starting from oxaloacetate (OAA), within which its biosynthesis-related genes are transcriptionally repressed by lysine via ArgP while aspartate kinase (LysC) and dihydrodipicolinate synthetase (DapA) are subject to feedback inhibition by lysine (54, 55). To enhance the downstream lysine pathway, we introduced lysC and dapA on p5HV1 and the resultant plasmid p5HV2 were cotransferred into E. coli BW25113 (F′) with pCar, yielding strain 5PDO-2. Meanwhile, to desensitize lysine-mediated feedback inhibition (56, 57), LysC mutant T253R and DapA mutant H56K or E84T were also recruited, yielding strain 5PDO-3 and 5PDO-4. Strain 5PDO-2 expressing the wildtype LysC and DapA produced 322.7 mg l−1 1,5-PDO, representing 2.6-fold increase to that of the parental strain 5PDO-1 (Fig. 6), whereas strain 5PDO-3 and 5PDO-4 could not further increase 1,5-PDO production in comparison with strain 5PDO-2, with titers of 271.5 and 311.3 mg l−1, respectively. This suggested that lysine degradation via the 1,5-PDO pathway might alleviate lysine-mediated feedback inhibition, which in turn led to higher 1,5-PDO titer with the wildtype pathway enzymes. Therefore, to reinforce lysine degradation, we incorporated the davB-davA-gabT-yqhD operon into a high-copy-number plasmid pSC74 that was derived from pSA74 with CloDF13 ori (20–40 copies per cell), resulting in plasmid p5HV5. Notably, feeding 3 g l−1 Lys to strain 5PDO-5 containing pCar and p5HV5 enabled 1.67 g l−1 1,5-PDO, reaching a conversion efficiency of 72.4% (SI Appendix, Fig. S3). When cotransferring pCar, p5HV5, and pCS-lysC-dapA into E. coli BW25113 (F′), the resultant strain 5PDO-6 significantly increased 1,5-PDO titer to 0.86 g l−1, representing a 6.94-fold increase than that of the starter strain 5PDO-1 (Fig. 6). Finally, to increase the supply of lysine pathway precursor OAA, we knocked out iclR, which encodes a repressor regulator of the glyoxylate bypass that serves as an alternative mechanism of replenishing oxaloacetate (21, 58). The final 1,5-PDO titer produced by strain 5PDO-7 with iclR deletion reached 0.97 g l−1 at 48 h with a yield of 0.08 mol mol−1 glucose (11.2% of the theoretical maximum). Interestingly, 5PDO-7 also accumulated the least acetate in the culture (0.24 g l−1) (Fig. 6). Taken together, our results established and optimized a 1,5-PDO pathway in E. coli, and demonstrated the de novo production of 1,5-PDO in microbial hosts.
Fig. 6.
Engineering and optimization of de novo 1,5-PDO production from glucose via Lys-to-1,5-PDO pathway. Production performance of different 1,5-PDO producers in M9Y media. All data points are reported as mean from three independent experiments and the peak titers at 48 h were applied for comparison. Error bars represent SD (n = 3).
Discussion
To expand the accessible chemical space for microbial production systems, exploiting new catalytic capabilities by extending existing metabolic networks can expedite the creation of nonnatural pathways for new target products. As such, extending the existing microbial pathways took advantage of native metabolic highways and starting materials, which is superior over assembling long synthetic pathways from scratch, to produce new products of desire (19). AAs are one of such primary and versatile native metabolites naturally produced both in E. coli and in all other microorganisms, and are also easily accessible from renewable protein wastes. Their structural diversity and the existence of AA anabolism and natural degradation machinery make them ideal precursors to derive new functionalized chemicals (2, 17–20). Here, we uncovered a previously unexplored path to extend AA catabolism to industrially important C3-C5 diols, based upon which we enabled diol production from expanded substrates including both AAs and sugars. The established platform serves as an important step toward the utilization of protein feedstocks and offers opportunities to expand the chemical space from charged AAs in microbial systems.
Charged AAs have been used to produce bifunctional chemicals, including short-chain ω-HAs, dicarboxylates, diamines, lactams, etc. (5, 59). However, production of highly reduced short-chain diols from AAs has remained unexplored. In this work, we first devised an enzymatically efficient and thermodynamically favorable Car-based reduction route for ω-HA conversion to diol. Car exhibited rather high activity toward short-chain ω-HAs and achieved complete conversion of 4HB or 5HV to counterpart diols (Fig. 2C). This offered a strategy to circumvent the 4HB accumulation or 4-HB-CoA lactonization issues that occurred in the commercial CoA-dependent 1,4-BDO pathway (27, 47). By adopting three native or heterologous catabolic pathways of major charged AAs (Asp, Glu, and Lys), C3-C5 ω-HAs were released. Final combination of the two segments successfully established a platform capable of biorefining C3-C5 diols from AAs (Fig. 1). Considering the cross-linking of AA degradation network, four additional AAs were converted into counterpart diols using the established platform as entry routes. The upstream degradation of AAs was a major limiting step for diol production. This has been indicated by our results and previous research on Glu or Lys degradation (60, 61). Herein, to improve the AA degradation, increasing cofactor PLP supply increased the Glu-to-1,4-BDO conversion efficiency to 34.0%, while enhancing the expression of Lys degradation pathway enabled a Lys-to-1,5-PDO conversion efficiency to 72.4%. Our results demonstrated the feasibility of diol production from protein hydrolysates via a diol platform.
Connecting the AA-to-diol platform with endogenous AA pathway in microbial systems could readily lead to sustainable production of diols from cheap sugar feedstocks, which is more economically appealing. We chose to demonstrate and optimize 1,4-BDO and 1,5-PDO production from glucose in E. coli. Glutamate is the most abundant metabolite in E. coli and synthesized from TCA α-ketoglutarate (46). Even without host engineering and pathway optimization, 588.3 mg l−1 1,4-BDO could be produced. We endeavored to increase the cofactor PLP supply for glutamate degradation pathway enzymes and increase carbon flux to α-ketoglutarate, which collectively enhanced 1,4-BDO titer by 2.40-fold (1.41 g l−1) (Fig. 5). Lysine, however, could not accumulate in high concentrations without pathway optimization. This was underlined by the low 1,5-PDO titer without lysine feeding or with aspartate feeding in strain 5PDO-1. By elevating lysine degradation, transcriptional or allosteric inhibition of the lysine pathway could be significantly relieved, thus driving 1,5-PDO production. With additional host engineering by deleting iclR to increase OAA supply, 0.97 g l−1 1,5-PDO was finally produced from glucose (Fig. 6). These results demonstrated that, by way of AA-to-diol platform, any carbon feedstock can be directed to diols via existing metabolic pathways in microbial hosts. Noteworthy, these diol pathways offer several potential benefits including oxygen insensitivity and reduced acetate accumulation, in comparison with the CoA-dependent pathways.
In summary, the work outlined here describes the development of a bacterial platform for diol production via AA-based biorefining that has not been described previously. The diol platform involves a CoA-independent chemistry and affords the synthesis of C3-C5 diols from seven different AAs, which lays a basis for diol bioprocesses using both protein hydrolysates and sugars as raw materials. To the best of our knowledge, this work also demonstrated de novo production of the nonnatural diol 1,5-PDO from glucose. Due to the fact of limited degradation efficiency and low productivity of AAs in wildtype E. coli host, the titers and yields of diols achieved in the current study are still low to satisfy the commercialization benchmark. To further improve the platform performance, debottlenecking the upstream AA-to-ω-HA conversion by screening superior enzymes would be most pivotal, considering the high performance of the Car-catalyzed downstream ω-HA-to-diol conversion. Second, increasing the AA import or reducing the intermediate export via transporter systems would maximize AA utilization and thus the final product yield, as extensively exemplified in previous work (58, 62, 63). Finally, it is expected that additional genetic manipulations to enhancing AA flux in microbial host, or switching to the other well-developed AA hyperproducers like C. glutamicum, will further improve diol production. Thus, we established and demonstrated an AA-based platform for diol production that can be further exploited in expanding chemical space and feedstock flexibility in microbes.
Materials and Methods
Bacterial Strains and Chemicals.
All strains and plasmids used in this study are listed in SI Appendix, Table S1. E. coli strain XL1-Blue (Stratagene) was used for plasmids construction and BW25113 (F′) was used as production host. pSC74 that was derived from pSA74 by replacing its pSC101 ori with CloDF13 ori from pCDFDuet-1. Plasmids pZE12-luc (high-copy), pCS27 (medium-copy), and pSC74 (high-copy) were used for pathway construction and pCP20 was used for elimination of kanamycin resistance marker during gene disruption. Phusion High Fidelity DNA polymerase, restriction endonucleases and Quick Ligation kit were purchased from New England Biolabs. Standard chemicals were purchased from Sigma-Aldrich unless otherwise specified. Owing to the absence of commercially available standards, 4HB and 5HV standards used in this study were prepared, respectively, from γ-butyrolactone (GBL) and σ-valerolactone through saponification. Briefly, 664.5 μL GBL and 706.9 μL σ-valerolactone were added into 15-mL tubes and the pH was adjusted by 10 N NaOH to pH 12.0. The tubes were incubated at 37 °C for over 12 h and then the pH was adjusted to pH 7.0 by H2SO4. Pure water was added to a final volume of 3 mL, yielding a 300 g l−1 solution for 4HB and 5HV.
Plasmid Construction.
All DNA manipulations were performed following the standard molecular cloning protocols (64). All primers used in this study are listed in SI Appendix, Table S2. Gene products amplified by PCR were digested with appropriate restriction enzymes and ligated into similarly digested vectors. The amplified panD from C. glutamicum, Pp0596 from P. putida KT2440, and ydfG from E. coli were digested and then integrated into pCS27 to yield 3HP pathway plasmid p3HP. The mutant gadB*(gadB from E. coli harboring E89Q with truncation of C-terminal 452–466) is obtained by overlap extension PCR. The gadB*, along with gabT and yqhD from E. coli, were inserted into pCS27 in one operon, yielding 4HB pathway plasmid p4HB. The davB and davA from P. putida KT2440, gabT, and yqhD from E. coli, were amplified and inserted into pCS27 in one operon, yielding 5HV pathway plasmid p5HV1. The lysC and dapA from E. coli were inserted into pCS27 in one operon, yielding pCS-lysC-dapA. Replacing lysC or dapA with corresponding mutant genes created two plasmids, pCS-lysC (T253R)-dapA (H56K) and pCS-lysC (T253R)-dapA (E84T). p5HV2 was created by inserting the PLlacO1-lysC-dapA cassette into p5HV1. Similarly, inserting PLlacO1-lysC (T253R)-dapA (H56K) or PLlacO1-lysC (T253R)-dapA (E84T) cassette into pHV1 created p5HV3 or p5HV4. p5HV5 was created by inserting the davB-davA-gabT-yqhD cassette into pSC74. To construct 1,3-PDO and 1,4-BDO pathways, the PLlacO1-panD-Pp0596-ydfG and PLlacO1-gadB*-gabT-yqhD cassettes were amplified from p3HP and p4HB, and cloned into previously constructed pCar (see Car sequence in SI Appendix, Fig. S4) (65), yielding plasmids p3PDO and p4BDO. The gltA* (gltA harboring R163L), obtained from overlap extension PCR, and ppc from E. coli were constructed into pCS27 in one operon, resulting in pCS-gltA*-ppc. The pdxS and pdxT were amplified from genomic DNA of Bacillus subtilis 168 and cloned to pCS27 in one operon to generate pCS-pdxST. Insertion of the PLlacO1-pdxST cassette into pCS-gltA*-ppc yielded plasmid pCS-gltA*-ppc-pdxST. The ansA and ybaS from E. coli were, respectively, constructed into pCS27, creating pCS-ansA and pCS-ybaS. The putA from E. coli was cloned into pCS27 to obtain pCS-putA. The adiA and speB from E. coli were cloned into pCS27 to form pCS-adiA-speB. Similarly, the gene patA and patD from E. coli were cloned into pCS27 to yield pCS-patAD. The cassette PLlacO1-patAD from pCS-patAD was then integrated into pCS-adiA-speB to obtain pCS-adiA-speB-patAD.
Culture Media.
Luria–Bertani (LB) medium containing 10 g l−1 NaCl, 10 g l−1 tryptone, and 5 g l−1 yeast extract was used for plasmid propagation and cell inoculation. The M9Y medium, prepared from M9 minimal medium (6.78 g l−1 Na2HPO4, 0.5 g l−1 NaCl, 3 g l−1 KH2PO4, 1 g l−1 NH4Cl, 246.5 mg l−1 MgSO4·7H2O and 14.7 mg l−1 CaCl2·2H2O) containing 20 g l−1 glucose and 5 g l−1 BD Bacto yeast extract, Technical (BD), was used for feeding experiments and de novo production of 1,3-PDO, 1,4-BDO and 1,5-PDO. The antibiotics ampicillin (100 μg ml−1), kanamycin (50 μg ml−1), and chloramphenicol (34 μg ml−1) were added into the medium when needed.
Shake-Flask Experiments.
Seed cultures of recombinant E. coli strains were first inoculated in 3 mL LB medium at 37 °C for 8–12 h, and then transferred to 125-mL shake flasks containing 20 mL fresh M9Y medium at an inoculation volume of 2%. After inoculation at 37 °C until the optical density (OD600) of cultures reached 0.6–0.8, isopropyl β-d-1-thiogalactopyranoside (IPTG) was added at a final concentration of 0.5 mM for protein induction and the shake-flask cultures were then inoculated at 30 °C on a rotary shaker at 270 rpm. For bioconversion experiments, substrates (3HP, 4HB, 5HV, or AAs) were added at a final concentration of 3 g l−1 into the cultures at the same time of IPTG addition. The cultures were sampled every 12 or 48 h postinoculation. The samples were subjected to centrifugation for 15 min at 12,000 rpm and filtration by a 0.22-μm filter membrane to remove any cell pellets. The processed samples were assayed by high-performance liquid chromatography (HPLC) analysis. Shake-flask experiments were performed in triplicates, and data are presented as the averages and SD (n = 3). For bioconversion experiment, the conversion efficiency was calculated as the ratio of product to substrate fed. For AA to diol conversion experiment, the diol produced from control group was subtracted.
In Vitro Enzyme Assays.
The seed culture of E. coli BW25113 (F′) containing plasmid pCar was prepared in 3 mL LB medium at 37 °C for 8–12 h, and then transferred to 250-mL shake flasks containing 50 mL LB medium at an inoculation volume of 2%. After inoculation at 37 °C until the optical density (OD600) of cultures reached 0.6–0.8, 0.5 mM IPTG was added for protein induction and the shake-flask cultures were then inoculated at 30 °C on a rotary shaker at 270 rpm for 9–12 h. The E. coli BW25113 (F′) with pZE12-luc was used as the control. The induced cells were harvested by centrifugation at 5,000 rpm at 4 °C for 10 min, and then resuspended by 1 mL distilled H2O. The cells were lysed using mini bead beater (Biospec Products), and the crude enzyme extracts were obtained by collecting the supernatants after centrifugation at 10,000 rpm for 10 min. The in vitro assay system contains 1 mM MgCl2, 25 mM Tris⋅HCl (pH 7.5), 4 mM NADPH, and 4 mM ATP, and varied concentration of substrates (3HP, 4HB, and 5HV). The crude enzyme extracts were added to a final concentration of 0.14 mg ml−1. The absorbance at 340 nm of the reaction was measured using a Genesys 10S UV-Vis Spectrophotometer (Thermo Scientific). Each assay was performed in triplicates. The changes of NADPH concentration (extinction coefficient 6220 m−1cm−1 at 340 nm) was used to determine the specific reaction rates of Car toward hydroxylic acid substrates.
HPLC Analysis.
Glucose, acetate, 3HP, 4HB, 5HV, 1,3-PDO, 1,4-BDO and 1,5-PDO, were quantified using corresponding standard by Dionex Ultimate 3000 (Ultimate 3000 Photodiode Array Detector) with a Coregel-64H column (Transgenomic). For HPLC analysis, 4 mM H2SO4 was used as mobile phase at a flow rate of 0.40 mL min−1. The oven temperature was set to 45 °C. This HPLC method was modified from our previous research (66).
Supplementary Material
Acknowledgments
This work was supported by the College of Engineering, The University of Georgia, Athens.
Footnotes
Competing interest statement: An invention disclosure has been filed for this work.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2003032117/-/DCSupplemental.
Data Availability.
All data supporting the findings of this study are included in this manuscript and SI Appendix.
References
- 1.Peralta-Yahya P. P., Zhang F., del Cardayre S. B., Keasling J. D., Microbial engineering for the production of advanced biofuels. Nature 488, 320–328 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Wang J., Guleria S., Koffas M. A., Yan Y., Microbial production of value-added nutraceuticals. Curr. Opin. Biotechnol. 37, 97–104 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Lee S. Y. et al., A comprehensive metabolic map for production of bio-based chemicals. Nat. Catal. 2, 18–33 (2019). [Google Scholar]
- 4.Lee S. Y., Kim H. M., Cheon S., Metabolic engineering for the production of hydrocarbon fuels. Curr. Opin. Biotechnol. 33, 15–22 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Chae T. U. et al., Metabolic engineering for the production of dicarboxylic acids and diamines. Metab. Eng. 58, 2–16 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Choi Y. J., Lee S. Y., Microbial production of short-chain alkanes. Nature 502, 571–574 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Vuoristo K. S., Mars A. E., Sanders J. P. M., Eggink G., Weusthuis R. A., Metabolic engineering of TCA cycle for production of chemicals. Trends Biotechnol. 34, 191–197 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Curran K. A., Alper H. S., Expanding the chemical palate of cells by combining systems biology and metabolic engineering. Metab. Eng. 14, 289–297 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Smanski M. J. et al., Synthetic biology to access and expand nature’s chemical diversity. Nat. Rev. Microbiol. 14, 135–149 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirasawa T., Shimizu H., Recent advances in amino acid production by microbial cells. Curr. Opin. Biotechnol. 42, 133–146 (2016). [DOI] [PubMed] [Google Scholar]
- 11.D’Este M., Alvarado-Morales M., Angelidaki I., Amino acids production focusing on fermentation technologies–A review. Biotechnol. Adv. 36, 14–25 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Li S.-Y., Ng I.-S., Chen P. T., Chiang C.-J., Chao Y.-P., Biorefining of protein waste for production of sustainable fuels and chemicals. Biotechnol. Biofuels 11, 256 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z. et al., Biofuels from food processing wastes. Curr. Opin. Biotechnol. 38, 97–105 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Kumar M. B., Gao Y., Shen W., He L., Valorisation of protein waste: An enzymatic approach to make commodity chemicals. Front. Chem. Sci. Eng. 9, 295–307 (2015). [Google Scholar]
- 15.Lammens T., Franssen M., Scott E., Sanders J., Availability of protein-derived amino acids as feedstock for the production of bio-based chemicals. Biomass Bioenergy 44, 168–181 (2012). [Google Scholar]
- 16.Huo Y.-X. et al., Conversion of proteins into biofuels by engineering nitrogen flux. Nat. Biotechnol. 29, 346–351 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Hazelwood L. A., Daran J.-M., van Maris A. J., Pronk J. T., Dickinson J. R., The Ehrlich pathway for fusel alcohol production: A century of research on Saccharomyces cerevisiae metabolism. Appl. Environ. Microbiol. 74, 2259–2266 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang K., Sawaya M. R., Eisenberg D. S., Liao J. C., Expanding metabolism for biosynthesis of nonnatural alcohols. Proc. Natl. Acad. Sci. U.S.A. 105, 20653–20658 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atsumi S., Hanai T., Liao J. C., Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451, 86–89 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Ehrenworth A. M., Peralta-Yahya P., Accelerating the semisynthesis of alkaloid-based drugs through metabolic engineering. Nat. Chem. Biol. 13, 249–258 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Becker J., Wittmann C., Systems and synthetic metabolic engineering for amino acid production–The heartbeat of industrial strain development. Curr. Opin. Biotechnol. 23, 718–726 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y., Liu D., Chen Z., Production of C2-C4 diols from renewable bioresources: New metabolic pathways and metabolic engineering strategies. Biotechnol. Biofuels 10, 299 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeng A.-P., Sabra W., Microbial production of diols as platform chemicals: Recent progresses. Curr. Opin. Biotechnol. 22, 749–757 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Firlotte N., Cooper D. G., Marić M., Nicell J. A., Characterization of 1, 5‐pentanediol dibenzoate as a potential “green” plasticizer for poly (vinyl chloride). J. Vinyl Addit. Technol. 15, 99–107 (2009). [Google Scholar]
- 25.Huang K. et al., Improving economics of lignocellulosic biofuels: An integrated strategy for coproducing 1, 5-pentanediol and ethanol. Appl. Energy 213, 585–594 (2018). [Google Scholar]
- 26.Zhong W., Zhang Y., Wu W., Liu D., Chen Z., Metabolic engineering of a homoserine-derived non-natural pathway for the de novo production of 1, 3-propanediol from glucose. ACS Synth. Biol. 8, 587–595 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Yim H. et al., Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat. Chem. Biol. 7, 445–452 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Tai Y.-S. et al., Engineering nonphosphorylative metabolism to generate lignocellulose-derived products. Nat. Chem. Biol. 12, 247–253 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Martin C. H. et al., A platform pathway for production of 3-hydroxyacids provides a biosynthetic route to 3-hydroxy-γ-butyrolactone. Nat. Commun. 4, 1414 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Huang K. et al., Conversion of furfural to 1, 5-pentanediol: Process synthesis and analysis. ACS Sustain. Chem.& Eng. 5, 4699–4706 (2017). [Google Scholar]
- 31.Farmer W. R., et al. , “Green process and compositions for producing poly (5HV) and 5 carbon chemicals.” US Patent 9090898B2 (2015).
- 32.Akhtar M. K., Turner N. J., Jones P. R., Carboxylic acid reductase is a versatile enzyme for the conversion of fatty acids into fuels and chemical commodities. Proc. Natl. Acad. Sci. U.S.A. 110, 87–92 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hädicke O., von Kamp A., Aydogan T., Klamt S., OptMDFpathway: Identification of metabolic pathways with maximal thermodynamic driving force and its application for analyzing the endogenous CO2 fixation potential of Escherichia coli. PLOS Comput. Biol. 14, e1006492 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venkitasubramanian P., Daniels L., Rosazza J. P., Reduction of carboxylic acids by Nocardia aldehyde oxidoreductase requires a phosphopantetheinylated enzyme. J. Biol. Chem. 282, 478–485 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez G. M., Tashiro Y., Atsumi S., Expanding ester biosynthesis in Escherichia coli. Nat. Chem. Biol. 10, 259–265 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kramer L. et al., Characterization of carboxylic acid reductases for biocatalytic synthesis of industrial chemicals. ChemBioChem 19, 1452–1460 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Richard H., Foster J. W., Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J. Bacteriol. 186, 6032–6041 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moreau P. L., The lysine decarboxylase CadA protects Escherichia coli starved of phosphate against fermentation acids. J. Bacteriol. 189, 2249–2261 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nozaki S., Webb M. E., Niki H., An activator for pyruvoyl-dependent l-aspartate α-decarboxylase is conserved in a small group of the γ-proteobacteria including Escherichia coli. MicrobiologyOpen 1, 298–310 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song C. W., Kim J. W., Cho I. J., Lee S. Y., Metabolic engineering of Escherichia coli for the production of 3-hydroxypropionic acid and malonic acid through β-alanine route. ACS Synth. Biol. 5, 1256–1263 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Atsumi S. et al., Engineering the isobutanol biosynthetic pathway in Escherichia coli by comparison of three aldehyde reductase/alcohol dehydrogenase genes. Appl. Microbiol. Biotechnol. 85, 651–657 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi S., Kim H. U., Kim T. Y., Lee S. Y., Systematic engineering of TCA cycle for optimal production of a four-carbon platform chemical 4-hydroxybutyric acid in Escherichia coli. Metab. Eng. 38, 264–273 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Knorr S. et al., Widespread bacterial lysine degradation proceeding via glutarate and L-2-hydroxyglutarate. Nat. Commun. 9, 5071 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Revelles O., Espinosa-Urgel M., Fuhrer T., Sauer U., Ramos J. L., Multiple and interconnected pathways for L-lysine catabolism in Pseudomonas putida KT2440. J. Bacteriol. 187, 7500–7510 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thu Ho N. A., Hou C. Y., Kim W. H., Kang T. J., Expanding the active pH range of Escherichia coli glutamate decarboxylase by breaking the cooperativeness. J. Biosci. Bioeng. 115, 154–158 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Bennett B. D. et al., Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5, 593–599 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burgard A., Burk M. J., Osterhout R., Van Dien S., Yim H., Development of a commercial scale process for production of 1,4-butanediol from sugar. Curr. Opin. Biotechnol. 42, 118–125 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Rosenberg J., Ischebeck T., Commichau F. M., Vitamin B6 metabolism in microbes and approaches for fermentative production. Biotechnol. Adv. 35, 31–40 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Ma W. et al., Engineering a pyridoxal 5′-phosphate supply for cadaverine production by using Escherichia coli whole-cell biocatalysis. Sci. Rep. 5, 15630 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stokell D. J. et al., Probing the roles of key residues in the unique regulatory NADH binding site of type II citrate synthase of Escherichia coli. J. Biol. Chem. 278, 35435–35443 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Brentzel Z. J. et al., Chemicals from biomass: Combining ring‐opening tautomerization and hydrogenation reactions to produce 1, 5‐pentanediol from furfural. ChemSusChem 10, 1351–1355 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Nakagawa Y., Tomishige K., Production of 1, 5-pentanediol from biomass via furfural and tetrahydrofurfuryl alcohol. Catal. Today 195, 136–143 (2012). [Google Scholar]
- 53.Xu W. et al., Direct catalytic conversion of furfural to 1,5-pentanediol by hydrogenolysis of the furan ring under mild conditions over Pt/Co2AlO4 catalyst. Chem. Commun. (Camb.) 47, 3924–3926 (2011). [DOI] [PubMed] [Google Scholar]
- 54.Contador C. A., Rizk M. L., Asenjo J. A., Liao J. C., Ensemble modeling for strain development of L-lysine-producing Escherichia coli. Metab. Eng. 11, 221–233 (2009). [DOI] [PubMed] [Google Scholar]
- 55.Marbaniang C. N., Gowrishankar J., Role of ArgP (IciA) in lysine-mediated repression in Escherichia coli. J. Bacteriol. 193, 5985–5996 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Z., Rappert S., Sun J., Zeng A.-P., Integrating molecular dynamics and co-evolutionary analysis for reliable target prediction and deregulation of the allosteric inhibition of aspartokinase for amino acid production. J. Biotechnol. 154, 248–254 (2011). [DOI] [PubMed] [Google Scholar]
- 57.Geng F., Chen Z., Zheng P., Sun J., Zeng A.-P., Exploring the allosteric mechanism of dihydrodipicolinate synthase by reverse engineering of the allosteric inhibitor binding sites and its application for lysine production. Appl. Microbiol. Biotechnol. 97, 1963–1971 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Li W. et al., Targeting metabolic driving and intermediate influx in lysine catabolism for high-level glutarate production. Nat. Commun. 10, 3337 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chae T. U., Ko Y.-S., Hwang K.-S., Lee S. Y., Metabolic engineering of Escherichia coli for the production of four-, five- and six-carbon lactams. Metab. Eng. 41, 82–91 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Park S. J. et al., Metabolic engineering of Escherichia coli for the production of 5-aminovalerate and glutarate as C5 platform chemicals. Metab. Eng. 16, 42–47 (2013). [DOI] [PubMed] [Google Scholar]
- 61.Somasundaram S., Lee S. H., Park S. J., Hong S. H., Gamma-aminobutyric acid production through GABA shunt by synthetic scaffolds introduction in recombinant Escherichia coli. Biotechnol. Bioprocess Eng. 21, 261–267 (2016). [Google Scholar]
- 62.Le Vo T. D., Kim T. W., Hong S. H., Effects of glutamate decarboxylase and gamma-aminobutyric acid (GABA) transporter on the bioconversion of GABA in engineered Escherichia coli. Bioprocess Biosyst. Eng. 35, 645–650 (2012). [DOI] [PubMed] [Google Scholar]
- 63.Li Z. et al., Overexpression of transport proteins improves the production of 5-aminovalerate from l-lysine in Escherichia coli. Sci. Rep. 6, 30884 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sambrook J., Fritsch E. F., Maniatis T., Molecular Cloning: A Laboratory Manual, (Cold Spring Harbor Laboratory Press, 1989). [Google Scholar]
- 65.Chen Z., Sun X., Li Y., Yan Y., Yuan Q., Metabolic engineering of Escherichia coli for microbial synthesis of monolignols. Metab. Eng. 39, 102–109 (2017). [DOI] [PubMed] [Google Scholar]
- 66.Wang J. et al., Microbial production of branched-chain dicarboxylate 2-methylsuccinic acid via enoate reductase-mediated bioreduction. Metab. Eng. 45, 1–10 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are included in this manuscript and SI Appendix.