Abstract
In the last two decades there have been dramatic advances in development of rapid diagnostic tests. Turnaround time of the assays have significantly been shortened which led to reductions in time to appropriate antimicrobial therapy and improvement of patient clinical outcomes. Molecular-based assays generally have better sensitivity than conventional methods, but the cost is higher. The results need to be interpreted cautiously as detection of colonized organisms, pathogen detection in asymptomatic patients, and false negative/positive can occur. Indications and cost-effectiveness need to be considered for appropriate utilization of rapid diagnostic tests.
Introduction
Detection of pathogens plays a major role in the clinical care of the patients with infectious diseases. Traditional diagnostic testing for infectious diseases such as microscopic examination, antigen detection, serology, cultures, and biochemical reactions are still being used, and sometimes essential for determination of infectious disease etiologies. However, those traditional methods tend to suffer from long turn-around time. For example, bacteria generally require 1–2 days to have growth on culture media, and conventional biochemical identification and susceptibility tests require additional one to two days to result. In the meantime, the patients tend to remain on empiric broad spectrum antibiotics which could lead to selection of antimicrobial resistance.
New technologies such as nucleic-acid amplification, mass spectrometry, and genomic sequencing have revolutionized diagnostic testing for infectious diseases by providing rapid and robust results. Introduction of rapid diagnostic testing in collaboration with antimicrobial stewardship has led to appropriate antimicrobial use and improvement of clinical outcomes.1
This article is a concise summary of rapid diagnostic tests for infectious diseases currently available in the United States.
Rapid Molecular Assays Molecular Multiplex Syndromic Panel Testing
There are currently 4 molecular multiplex syndromic panels available in the U.S.: Blood stream infections, respiratory tract infections, gastrointestinal infections and meningitis/encephalitis. The common advantages of those panels are simultaneous detection of multiple targets and rapid turnaround time that could shorten time to appropriate antimicrobial treatment and improve associated clinical outcomes. The common disadvantages of those panels are the limited numbers of detectable pathogens and higher cost than conventional methods.
Blood Stream Infections
Blood stream infections are often associated with significant morbidity and mortality.2 Multiple studies demonstrated associations between delay in initiation of appropriate antimicrobial therapy and increased mortality for patients with blood stream infections.3–6 Identification of the pathogens in blood cultures is essential for directing appropriate antimicrobial therapy and improving patient outcomes. There are currently four FDA-cleared molecular multiplex assays for blood culture pathogen identification: FilmArray BCID panel (bioMérieux, Marcy l’Etoile, France), Verigene Gram Positive Blood Culture ID assay (BCID-GP) and Gram Negative Blood Culture ID assay (BCID-GN) (Luminex Molecular Diagnostics, Toronto, Canada), Accelerate PhenoTest BC kit (Accelerate Diagnostics, Tucson, AZ), and ePlex Blood Culture Identification Gram-Positive Panel (BCID-GP) and Gram-Negative Panel (BCID-GN) (GenMark Diagnostics, Carlsbad, CA).
The assays are performed directly on positive blood culture specimens detected by a continuous monitoring blood culture system. The turnaround time and pathogen targets in each panel are shown in Table 1. While FilmArray BCID and PhenoTest BC include targets for both Gram positive and Gram negative organisms in one test kit, Verigene and ePlex have Gram-positive and Gram-negative panels separately and Gram staining results determines selection of the appropriate test panels. Of note the PhenoTest BC is currently the only assay that enables rapid antimicrobial susceptibility testing in addition to pathogen identification. The antimicrobial susceptibility panel of the PhenoTest BC includes amikacin, ampicillin, ampicillin/sulbactam, aztreonam, ceftazidime, ceftaroline, cefepime, ceftriaxone, ciprofloxacin, daptomycin, erythromycin, ertapenem, gentamicin, linezolid, meropenem, piperacillin/tazobactam, tobramycin, and vancomycin. There have been no major differences in the performance of those multiplex blood culture panels to support superiority one over another.7–22 In general, their performance is lower with polymicrobial blood culture specimens than monomicrobial ones.7,8,13,14,19,20,22 Improvement of clinical outcomes such as length of stay, time to optimal antibiotic therapy, and 30-day mortality rate with antimicrobial stewardship interventions based on the results of rapid blood culture identification have been demonstrated.23–34
Table 1.
Turnaround time and pathogen targets of FDA-cleared multiplex blood culture identification panels
| FilmArray | Verigene | PhenoTest | ePlex | |
|---|---|---|---|---|
| Turnaround time | 1 hour | 2–2.5 hours | 7 hours a | 1.5 hours |
| Pathogen targets | ||||
| Gram positive bacteria | ||||
| Staphylococcus species | X | X | X | |
| S. aureus | X | X | X | X |
| Coagulase-negative Staphylococcus spp. | X | |||
| S. epidermidis | X | X | ||
| S. lugdunensis | X | X | X | |
| Streptococcus species | X | X | X | X |
| S. pneumoniae | X | X | X | |
| S. pyogenes | X | X | X | |
| S. agalactiae | X | X | X | |
| S. anginosus group | X | X | ||
| Enterococcus species | X | X | ||
| E. faecalis | X | X | X | |
| E. faecium | X | X | X | |
| Listeria species | X | X | ||
| L. monocytogenes | X | X | ||
| Lactobacillus species | X | |||
| Micrococcus species | X | |||
| Bacillus cereus group | X | |||
| Bacillus subtilis group | X | |||
| Corynebacterium species | X | |||
| Cutibacterium acnes | X | |||
| Gram negative bacteria | ||||
| Enterobacteriaceae | X | |||
| Escherichia coli | X | X | X | X |
| Klebsiella species | X | |||
| Klebsiella pneumoniae | X | X | X | |
| Klebsiella oxytoca | X | X | X | |
| Proteus species | X | X | X | X |
| Proteus mirabilis | X | |||
| Enterobacter species | X | X | ||
| Enterobacter (non-cloacae complex) | X | |||
| Enterobacter cloacae complex | X | X | ||
| Citrobacter species | X | X | X | |
| Serratia species | X | |||
| Serratia marcescens | X | X | X | |
| Pseudomonas aeruginosa | X | X | X | X |
| Acinetobacter species | X | |||
| Acinetobacter baumanii | X | X | X | |
| Haemophilus influenzae | X | X | ||
| Neisseria meningitidis | X | X | ||
| Bacteroides fragilis | X | |||
| Fusobacterium necropholum | X | |||
| Fusobacterium nucleatum | X | |||
| Morganella morganii | X | |||
| Coronobacter sakazakii | X | |||
| Salmonella species | X | |||
| Stenotrophomonas maltophilia | X | |||
| Yeast | ||||
| Candida species | X | |||
| Candida albicans | X | X | ||
| Candida glabrata | X | X | ||
| Candida krusei | X | |||
| Candida parapsilosis | X | |||
| Candida tropicalis | X | |||
| Antimicrobial resistance markers | ||||
| mecA | X | X | X | |
| mecC | X | |||
| VanA/B | X | |||
| VanA | X | X | ||
| VanB | X | X | ||
| KPC (blaKPC) | X | X | X | |
| OXA (blaoxa) | X | X | ||
| VIM (blaVIM) | X | X | ||
| IMP (blaIMP) | X | X | ||
| CTX-M (blaCTX-M) | X | X | ||
| NDM (blaNDM) | X | X |
Including antimicrobial susceptibility testing for amikacin, ampicillin, ampicillin/sulbactam, aztreonam, ceftazidime, ceftaroline, cefepime, ceftriaxone, ciprofloxacin, daptomycin, erythromycin, ertapenem, gentamicin, linezolid, meropenem, piperacillin/tazobactam, tobramycin, and vancomycin.
Meningitis and Encephalitis
Meningitis and encephalitis are potentially life-threatening infections and could leave severe neurological sequela. Rapid pathogen detection to guide appropriate treatment is critically important to improve clinical outcomes. Currently FilmArray Meningitis/Encephalitis (ME) panel (bioMérieux, Marcy l’Etoile, France) is the only FDA-cleared assay available in the U.S. The pathogen targets included in the FilmArray ME panel are Escherichia coli K1, Haemophilus influenzae, Listeria monocytogenes, Neisseria meningitidis (encapsulated), Streptococcus agalactiae, Streptococcus pneumoniae, Cytomegalovirus, Enterovirus, Herpes simplex virus 1, Herpes simplex virus 2, Human herpesvirus 6, Human parechovirus, Varicella zoster virus, and Cryptococcus neoformans/gattii. The advantages of the FilmArray ME panel compared to conventional cultures and individual molecular-based tests are rapid turnaround time (one hour) and a small amount of CSF (0.2mL) required for detection of 14 targets. In the meta-analysis of eight studies accounting for 3,059 patients met the inclusion criteria of the diagnostic accuracy test review of the FilmArray ME panel, mean sensitivity was 90% (95% CI 86 – 93%) and mean specificity was 97% (95% CI 94 – 99%).35 The studies that specifically evaluated false positive and negative results were also analyzed, and it was found that 4% and 1.5% of specimens were determined as false positive and false negative respectively by the FilmArray ME panel compared with reference standard methods after implementing adjudication for discrepant results.35 The highest proportion of false positive was observed for Streptococcus pneumoniae followed by Streptococcus agalactiae, and the highest proportion of false negative was observed for Herpes simplex virus 1 and 2, enterovirus, and Cryptococcus neoformans/gattii.35 Most of the false negative Cryptococcus neoformans/gattii cases were those who were on antifungal treatment.35
Respiratory Tract Infections
There are currently five FDA-cleared multiplex panels for respiratory pathogens: NxTAG Respiratory Pathogen Panel (Luminex Molecular Diagnostics, Toronto, Canada), FilmArray Respiratory Panel (bioMérieux, Marcy l’Etoile, France), Verigene Respiratory Pathogens Flex Nucleic Acid Test (RP Flex) (Luminex Molecular Diagnostics, Toronto, Canada), eSensor Respiratory Viral Panel (RVP) (GenMark Diagnostics, Carlsbad, CA), and ePlex Respiratory Pathogen (RP) Panel (GenMark Diagnostics, Carlsbad, CA). The assays are performed on nasopharyngeal swab specimens collected from individuals who are suspected to have respiratory tract infections. NxTAG Respiratory Pathogen Panel and Verigene RP Flex allow panel customization to avoid detection of unnecessary targets and minimize the cost. The turnaround time and pathogen targets included in each panel are shown in Table 2. The assay performance generally demonstrated high concordance rates with positive percent agreement 84.5 – 98.8% and negative percent agreement 99.2 – 100% when compared between different multiplex panels or with a laboratory-developed polymerase chain reaction (PCR) assay as a reference method.36–40 However, more frequent discrepancies have been reported for adenovirus,36,40 influenza B virus,36 human metapneumovirus,39 parainfluenza 3,39 coronavirus,40 and rhinovirus/enterovirus.40 It is important to keep in mind that detection of a certain virus does not necessarily mean that the virus is a causative pathogen of respiratory symptoms that a patient has because viruses can be colonized in the respiratory tract asymptomatically. In addition, detection of multiple targets is not uncommon.36,37 Although immunocompromised patients can develop severe viral respiratory infections and are more likely to benefit from the multiplex panel testing than immunocompetent individuals, they can shed viruses for a prolonged period of time without clinical symptoms. The results of the multiplex respiratory panel testing need to be interpreted carefully in a clinical context. Further work up may be indicated as there are many respiratory pathogens that are not included in multiplex respiratory panels.
Table 2.
Turnaround time and pathogen targets of FDA-cleared multiplex respiratory panels
| FilmArray | Verigene | NxTAG | eSensor | ePlex | |
|---|---|---|---|---|---|
| Turnaround time | 1 hour | 2 hours | 5 hoursa | 6 hours | 1.5 hours |
| Pathogen targets | |||||
| Viruses | |||||
| Influenza A | X | X | X | X | X |
| Influenza A H1 | X | X | X | X | X |
| Influenza A H3 | X | X | X | X | X |
| Influenza 2009 H1N1 | X | X | X | ||
| Influenza B | X | X | X | X | X |
| Respiratory Syncytial Virus | X | ||||
| Respiratory Syncytial Virus A | X | X | X | X | |
| Respiratory Syncytial Virus B | X | X | X | X | |
| Parainfluenza virus 1 | X | X | X | X | X |
| Parainfluenza virus 2 | X | X | X | X | X |
| Parainfluenza virus 3 | X | X | X | X | X |
| Parainfluenza virus 4 | X | X | X | X | |
| Coronavirus (229E/HKU1/NL63/OC43) | X | ||||
| Coronavirus HKU1 | X | X | X | ||
| Coronavirus NL63 | X | X | |||
| Coronavirus 229E | X | X | |||
| Coronavirus OC43 | X | X | |||
| Adenovirus | X | X | X | ||
| Adenovirus B/E | X | ||||
| Adenovirus C | X | ||||
| Rhinovirus | X | X | |||
| Rhinovirus/Enterovirus | X | X | X | ||
| Human Metapneumovirus | X | X | X | ||
| Human Bocavirus | X | ||||
| Bacteria | |||||
| Mycoplasma pneumoniae | X | X | X | ||
| Chlamydophila pneumoniae | X | X | X | ||
| Bordetella pertussis | X | X | |||
| Bordetella parapertussis/Bordetella bronchiseptica | X | ||||
| Bordetella holmesii | X |
For 96 samples
Gastrointestinal Infections
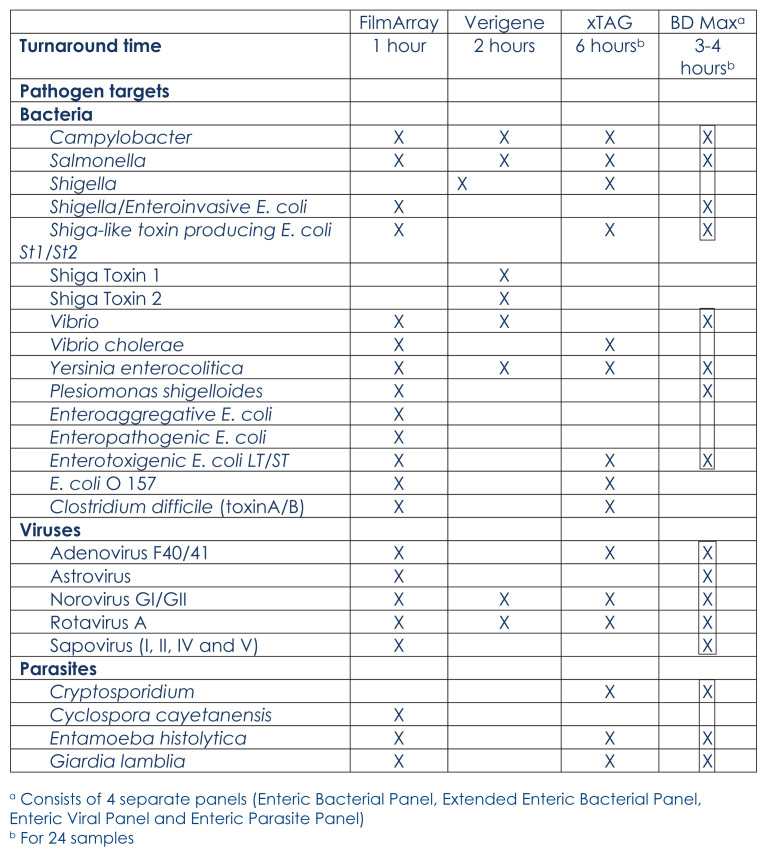
Infectious diarrheal illness is very common worldwide. Since 1990, diarrhea has been ranked among the top ten causes of death and disability-adjusted life-years (DALYs) among all ages, and one of the top five causes of death and DALYs for children younger than five years.41 Conventional methods for diarrheal pathogen detections include microscopic examination, culture, and enzyme-linked immunosorbent assays (ELISA). Microscopic examination for parasite detection requires specific expertise and the results are operator dependent which causes variability in sensitivity. Stool culture is labor intensive and takes two to three days to result. ELISA assays are generally less sensitive than PCR.42–44 There are currently three FDA-cleared multiplex gastrointestinal pathogen panels in one kit available in the U.S. FilmArray Gastrointestinal (GI) Panel (bioMérieux, Marcy l’Etoile, France), Verigene Enteric Pathogen Test (Luminex Molecular Diagnostics, Toronto, Canada), and xTAG Gastrointestinal Pathogen Panel (Luminex Molecular Diagnostics, Toronto, Canada). BD MAX (BD Diagnostics, Sparks, Maryland, USA) is also a FDA-cleared multiplex assay, but has 4 separate panels for gastrointestinal pathogens: Enteric Bacterial Panel, Extended Enteric Bacterial Panel, Enteric Parasite Panel, and Enteric Viral Panel. The turnaround time and pathogen targets included in those assays are shown in Table 3. Overall multiplex panel tests had higher positivity rates compared to conventional methods in performance evaluation studies.45–52 Simultaneous detection of multiple pathogens is not uncommon as well as detection of pathogens in asymptomatic patients which can make interpretation of the test results and management challenging for clinicians.53,54
Table 3.
Turnaround time and pathogen targets of FDA-cleared multiplex
Point-of-care Molecular Based tests
There are a variety of FDA-cleared nucleic acid amplification tests available for one to several targets for surveillance as well as diagnosis of specific infections. Examples include methicillin-resistant Staphylococcus aureus, vancomycin-resistant Enterococcus, Carbapenem resistance genes (blaKPC, blaNDM, blaVIM, blaOXA-48, and blaIMP), Group A Streptococcus, Group B Streptococcus, Influenza virus A & B/RSV, Clostridium difficile with or without NAP 027, Norovirus, Human simplex virus 1 & 2, Mycobacterium tuberculosis with rifampin resistance, Trichomonas vaginalis, Chlamydia trachomatis/Neisseria gonorrhea, and Bordetella pertussis/Bordetella parapertussis.
T2 system
Candidemia is associated with high mortality rates. Although the rates are variable depending on the clinical setting, mortality rates range from 20% to 60%.55,56 Blood culture is the gold standard for diagnosis of Candidemia. However, sensitivity of blood culture is as low as 50%.57,58 T2Candida Panel (T2 Biosystems, Lexington, Massachusetts, USA) is an FDA-cleared qualitative T2 Magnetic Resonance (T2MR) assay for detection of Candida species directly from whole blood specimens. In T2 system blood-compatible polymerase chain reaction is followed by hybridization of the amplified pathogen DNA to capture probe–decorated nanoparticles. Hybridization yields nanoparticle micro-clusters that cause large changes in the sample’s T2MR signal.59 T2Candida Panel identifies five species of Candida by categorizing them in three groups: Candida albicans/Candida tropicalis, Candida parapsilosis, and Candida glabrata /Candida krusei. Time to result is three to four hours. In a multicenter clinical trial including 14 centers, the assay sensitivity was 89%.60 In patients receiving antifungal therapy, T2Candida Panel identified bloodstream infections that were missed by conventional blood cultures.60 In another multicenter clinical trial including 12 centers, the overall sensitivity was 91.1% and specificity was 99.4%. 61 The pooled analysis including eight studies conducted to evaluate accuracy of T2Candida Panel demonstrated the pooled sensitivity of 91% and specificity of 94%.62 T2 system also has FDA-cleared Bacterial Panel which can detect Enterococcus faecium, Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Staphylococcus aureus directly from whole blood samples. In a diagnostic accuracy study T2Bacterial Panel sensitivity and specificity for proven blood stream infections were 90% and 90% respectively.63 Mean time to identification was 3.61 hours (SD, 0.2) for one sample and 7.70 hours (SD, 1.38) for seven samples.63
MALDI-TOF MS
Matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry (MS) is a rapid, accurate, and cost-effective identification method for bacterial and fungal culture isolates with direct smear. In mass spectrometry analysis, sample protein molecules are converted into ions in the gas phase by laser ablation. The ionized molecules are subsequently accelerated by a potential difference and fly through the flight tube towards the detector, and the system measures the analytes’ time of flight to the detector which produces a characteristic spectrum. The advantages of MALDI-TOF MS are ability to identify broad range of pathogens and low cost. Turnaround time for the MALDI-TOF MS assay itself is short, but it takes one to two days until the culture isolate for direct smear becomes available. In an effort to shorten time to pathogen identification in blood culture, various sample preparation procedures including Sepsityper kit (Bruker Daltonics, Bremen, Germany) and inhouse methods were developed to perform MALDI-TOF MS directly from positive blood culture bottles without subculture. Overall correct identification rates to species level and genus level range 39.9 – 89.7% and 76.4 –100% respectively, and the rates were higher in Gram negative organisms compared to Gram positive organisms.64–79 Performance was poor in polymicrobial cultures65,67–69,71–73,75–78 and yeast identifications.66,70,71,79 Recently, an alternative method with subculture has been developed. The positive blood culture specimens are plated on solid media. After short incubation (four to six hours) 1 – 2 mm of the bacterial lawn is transferred with a 1 μL inoculation loop for direct smear. Using short incubation overall correct identification rates to species level range 69.7 – 99.5%.80–83
Conclusions
Rapid diagnostic tests are powerful tools for the timely optimization of antimicrobial use. However, interpretations of the results such as potential false positive or negative and detection of colonized microorganisms require careful evaluation of the clinical settings and background of the patients. Costeffectiveness also needs to be considered to determine indications for expensive rapid diagnostic tests to prevent overutilization of those tests.
Footnotes
Masako Mizusawa, MD, PhD, MS, is in the Section of Infectious Diseases, Department of Internal Medicine, University of Missouri - Kansas City, Kansas City, Missouri.
Disclosure
None reported.
SARS-CoV-2 Serology Testing: An Epilogue
There are currently more than 200 SARS-CoV-2 commercial serology tests available in the U.S. Serology assays for SARS-CoV-2 initially did not require FDA emergency use authorization (EUA), and submission of assay validation data to Food and Drug Administration (FDA) was a voluntary process until May 4, 2020. Therefore, many laboratories notified FDA that they had validated their assays and started patient testing, but their assay performances were not reviewed by FDA prior to market release. A revised guidance from FDA has provided specific assay performance thresholds, and validation data submission for EUA has become manufacturers’ requirements. A list of SARS-Co-V-2 serology tests that have been removed from the notification list due to manufacturers’ voluntary withdrawal and lack of pending EUA request or issued EUA is available on the FDA website (https://www.fda.gov/medical-devices/emergency-situations-medicaldevices/faqs-testing-sars-cov-2#nolonger). There is also a list of EUA authorized serology tests (https://www.fda.gov/medical-devices/emergency-situations-medicaldevices/eua-authorized-serology-test-performance). It is advised to select an EUA authorized SARS-Co-V-2 serology test to assure the appropriate assay performance.
Commercial serology assays for SARS-CoV-2 are variable in different formats (lateral flow assays, enzyme immunosorbent assays, and chemiluminescent immunoassays) and antibody classes (IgM, IgA, IgG, and IgM/IgG total antibody) with using different antigens (nucleocapsid, S1 and/or S2 spike glycoproteins, and spike glycoprotein receptor binding protein).1 The majority of patients seem to develop antibody response between seven and eleven days following exposure to SARS-CoV-22 although available data regarding timing of antibody appearance following disease onset are variable.2–6 Wu et al. collected plasma samples from 175 COVID-19 recovered patients with mild symptoms and found that SARS-CoV-2 specific neutralizing antibodies were detected in patients from day 10–15 after the onset of disease.7 They also found that elderly and middle-age patients had significantly higher plasma neutralizing antibody titers than young patients, and approximately 30% of recovered patients generated a very low level of neutralizing antibody titers (10 patients had undetectable levels).7
Antibody testing can be used for surveillance to identify how many people have been exposed to SARS-CoV- 2 in the community as well as contact tracing. Another use of antibody testing is donor screening for convalescent plasma which is used to treat patients with SARS-CoV-2 infection. In the future, when a vaccine for SARS-CoV-2 becomes available, antibody testing would play a role in screening vaccine candidates and monitoring immune responses of vaccinated individuals.
Not all the antibodies produced are neutralizing antibodies that block viral entry to host cells.8 Commercial serology assays do not distinguish between neutralizing antibodies and other antibodies. Therefore, detection of IgG antibodies by those assays does not mean that detectable levels of neutralizing antibodies are present, and antibody testing should not be used as a surrogate marker for protective immunity against SARS-CoV-2 infection. Even if a serology test that specifically detects neutralizing antibodies is developed, detection of neutralizing antibodies still does not equal to protective immunity because what levels of antibody titers would protect patients from SARS-CoV-2 infection is still unknown.
Antibody testing should not be used for diagnosing acute/recent SARS-Co-V-2 infection by itself because antibody may not be detected in the early days of the infection when the risk of transmission is the highest, and negative results do not rule out acute/recent SARS-CoV- 2 infection. In general IgM suffers from false positivity more than other classes of antibodies,9 and positive IgM results do not rule in acute/recent SARS-CoV- 2 infection either. Grifoni et al. found SARS-CoV-2-reactive CD4+ T cells in ~40%–60% of unexposed individuals, suggesting cross-reactive T cell recognition between circulating “common cold” coronaviruses (HCoV-OC43, HCoV-HKU1, HCoV-NL63, and HCoV-229E) and SARS-CoV- 2,10 and false positive results due to cross-reactivity with other coronaviruses are also possible. It is advised to check whether cross-reactivity with common cold coronaviruses has been validated or not prior to selection of antibody assays. It is also important to keep in mind that positive and negative predictive values of antibody assays depend on disease prevalence. When disease prevalence is low, false positivity rate is increased even with the excellent specificity.
References
- 1.Theel ES, Slev P, Wheeler S, Couturier MR, Wong SJ, Kadkhoda K. The Role of Antibody Testing for SARS-CoV-2: Is There One? J Clin Microbiol. doi: 10.1128/JCM.00797-20. Published online April 29, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel R, Babady E, Theel ES, et al. Report from the American Society for Microbiology COVID-19 International Summit 23 March 2020: Value of Diagnostic Testing for SARS–CoV-2/COVID-19. mBio. 2020;11(2) doi: 10.1128/mBio.00722-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo L, Ren L, Yang S, et al. Profiling Early Humoral Response to Diagnose Novel Coronavirus Disease (COVID-19) Clin Infect Dis Off Publ Infect Dis Soc Am. doi: 10.1093/cid/ciaa310. Published online March 21, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lou B, Li T-D, Zheng S-F, et al. Serology characteristics of SARS-CoV- 2 infection since exposure and post symptom onset. Eur Respir J. doi: 10.1183/13993003.00763-2020. Published online January 1, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okba NMA, Müller MA, Li W, et al. Early Release - Severe Acute Respiratory Syndrome Coronavirus 2–Specific Antibody Responses in Coronavirus Disease 2019 Patients. Emerging Infectious Diseases journal - CDC. 2020 Jul;26(7) doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suhandynata RT, Hoffman MA, Kelner MJ, McLawhon RW, Reed SL, Fitzgerald RL. Longitudinal Monitoring of SARS-CoV-2 IgM and IgG Seropositivity to Detect COVID-19. J Appl Lab Med. doi: 10.1093/jalm/jfaa079. Published online May 19, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu F, Wang A, Liu M, et al. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv. doi: 10.1101/2020.03.30.20047365. Published online April 20, 2020:2020.03.30.20047365. [DOI] [Google Scholar]
- 8.Lv H, Wu NC, Tsang OT-Y, et al. Cross-reactive Antibody Response between SARS-CoV-2 and SARS-CoV Infections. Cell Rep. 2020;31(9):107725. doi: 10.1016/j.celrep.2020.107725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landry ML. Immunoglobulin M for Acute Infection: True or False? Clin Vaccine Immunol. 2016;23(7):540–545. doi: 10.1128/CVI.00211-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grifoni A, Weiskopf D, Ramirez SI, et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. doi: 10.1016/j.cell.2020.05.015. Published online May 20, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 1.Beganovic M, McCreary EK, Mahoney MV, Dionne B, Green DA, Timbrook TT. Interplay between Rapid Diagnostic Tests and Antimicrobial Stewardship Programs among Patients with Bloodstream and Other Severe Infections. The Journal of Applied Laboratory Medicine. 2019;3(4):601–616. doi: 10.1373/jalm.2018.026450. [DOI] [PubMed] [Google Scholar]
- 2.Laupland KB, Church DL. Population-Based Epidemiology and Microbiology of Community-Onset Bloodstream Infections. Clinical Microbiology Reviews. 2014;27(4):647–664. doi: 10.1128/CMR.00002-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen H-C, Lin W-L, Lin C-C, et al. Outcome of inadequate empirical antibiotic therapy in emergency department patients with community-onset bloodstream infections. J Antimicrob Chemother. 2013;68(4):947–953. doi: 10.1093/jac/dks475. [DOI] [PubMed] [Google Scholar]
- 4.Corl KA, Zeba F, Caffrey AR, et al. Delay in Antibiotic Administration Is Associated With Mortality Among Septic Shock Patients With Staphylococcus aureus Bacteremia*. Read Online: Critical Care Medicine | Society of Critical Care Medicine. 2020;48(4):525–532. doi: 10.1097/CCM.0000000000004212. [DOI] [PubMed] [Google Scholar]
- 5.Falcone M, Bassetti M, Tiseo G, et al. Time to appropriate antibiotic therapy is a predictor of outcome in patients with bloodstream infection caused by KPC-producing Klebsiella pneumoniae. Crit Care. 2020;24(1):1–12. doi: 10.1186/s13054-020-2742-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robineau O, Robert J, Rabaud C, et al. Management and outcome of bloodstream infections: a prospective survey in 121 French hospitals (SPA-BACT survey) Infect Drug Resist. 2018;11:1359–1368. doi: 10.2147/IDR.S165877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altun O, Almuhayawi M, Ullberg M, Ozenci V. Clinical evaluation of the FilmArray blood culture identification panel in identification of bacteria and yeasts from positive blood culture bottles. J Clin Microbiol. 2013;51(12):4130–4136. doi: 10.1128/JCM.01835-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blaschke AJ, Heyrend C, Byington CL, et al. Rapid identification of pathogens from positive blood cultures by multiplex polymerase chain reaction using the FilmArray system. Diagnostic Microbiology and Infectious Disease. 2012;74(4):349–355. doi: 10.1016/j.diagmicrobio.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhatti MM, Boonlayangoor S, Beavis KG, Tesic V. Evaluation of FilmArray and Verigene Systems for Rapid Identification of Positive Blood Cultures. Journal of Clinical Microbiology. 2014;52(9):3433–3436. doi: 10.1128/JCM.01417-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng X, Polanco W, Carter D, Shulman S. Rapid Identification of Pathogens from Pediatric Blood Cultures by Use of the FilmArray Blood Culture Identification Panel. Journal of Clinical Microbiology. 2014;52(12):4368–4371. doi: 10.1128/JCM.02133-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Southern TR, VanSchooneveld TC, Bannister DL, et al. Implementation and performance of the BioFire FilmArray® Blood Culture Identification panel with antimicrobial treatment recommendations for bloodstream infections at a midwestern academic tertiary hospital. Diagnostic Microbiology and Infectious Disease. 2015;81(2):96–101. doi: 10.1016/j.diagmicrobio.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 12.Buchan BW, Ginocchio CC, Manii R, et al. Multiplex Identification of Gram-Positive Bacteria and Resistance Determinants Directly from Positive Blood Culture Broths: Evaluation of an Automated Microarray-Based Nucleic Acid Test. PLOS Medicine. 2013;10(7):e1001478. doi: 10.1371/journal.pmed.1001478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dodémont M, De Mendon¸a R, Nonhoff C, Roisin S, Denis O. Evaluation of Verigene Gram-positive blood culture assay performance for bacteremic patients. Eur J Clin Microbiol Infect Dis. 2015;34(3):473–477. doi: 10.1007/s10096-014-2250-4. [DOI] [PubMed] [Google Scholar]
- 14.Samuel LP, Tibbetts RJ, Agotesku A, Fey M, Hensley R, Meier FA. Evaluation of a Microarray-Based Assay for Rapid Identification of Gram-Positive Organisms and Resistance Markers in Positive Blood Cultures. Journal of Clinical Microbiology. 2013;51(4):1188–1192. doi: 10.1128/JCM.02982-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ward C, Stocker K, Begum J, Wade P, Ebrahimsa U, Goldenberg SD. Performance evaluation of the Verigene® (Nanosphere) and FilmArray® (BioFire®) molecular assays for identification of causative organisms in bacterial bloodstream infections. Eur J Clin Microbiol Infect Dis. 2015;34(3):487–496. doi: 10.1007/s10096-014-2252-2. [DOI] [PubMed] [Google Scholar]
- 16.Wojewoda CM, Sercia L, Navas M, et al. Evaluation of the Verigene Gram-positive blood culture nucleic acid test for rapid detection of bacteria and resistance determinants. J Clin Microbiol. 2013;51(7):2072–2076. doi: 10.1128/JCM.00831-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arroyo MA, Denys GA. Parallel Evaluation of the MALDI Sepsityper and Verigene BC-GN Assays for Rapid Identification of Gram-Negative Bacilli from Positive Blood Cultures. Journal of Clinical Microbiology. 2017;55(9):2708–2718. doi: 10.1128/JCM.00692-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chantell C. Multiplexed Automated Digital Microscopy for Rapid Identification and Antimicrobial Susceptibility Testing of Bacteria and Yeast Directly from Clinical Samples. Clinical Microbiology Newsletter. 2015;37(20):161–167. doi: 10.1016/j.clinmicnews.2015.10.001. [DOI] [Google Scholar]
- 19.Charnot-Katsikas A, Tesic V, Love N, et al. Use of the Accelerate Pheno System for Identification and Antimicrobial Susceptibility Testing of Pathogens in Positive Blood Cultures and Impact on Time to Results and Workflow. J Clin Microbiol. 2018;56(1) doi: 10.1128/JCM.01166-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lutgring JD, Bittencourt C, McElvania TeKippe E, Cavuoti D, Hollaway R, Burd EM. Evaluation of the Accelerate Pheno System: Results from Two Academic Medical Centers. J Clin Microbiol. 2018;56(4) doi: 10.1128/JCM.01672-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pancholi P, Carroll KC, Buchan BW, et al. Multicenter Evaluation of the Accelerate PhenoTest BC Kit for Rapid Identification and Phenotypic Antimicrobial Susceptibility Testing Using Morphokinetic Cellular Analysis. J Clin Microbiol. 2018;56(4) doi: 10.1128/JCM.01329-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang T-D, Melnik E, Bogaerts P, Evrard S, Glupczynski Y. Evaluation of the ePlex Blood Culture Identification Panels for Detection of Pathogens in Bloodstream Infections. J Clin Microbiol. 2019;57(2) doi: 10.1128/JCM.01597-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banerjee R, Teng CB, Cunningham SA, et al. Randomized Trial of Rapid Multiplex Polymerase Chain Reaction–Based Blood Culture Identification and Susceptibility Testing. Clin Infect Dis. 2015;61(7):1071–1080. doi: 10.1093/cid/civ447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Messacar K, Hurst AL, Child J, et al. Clinical Impact and Provider Acceptability of Real-Time Antimicrobial Stewardship Decision Support for Rapid Diagnostics in Children With Positive Blood Culture Results. J Pediatric Infect Dis Soc. 2017;6(3):267–274. doi: 10.1093/jpids/piw047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pardo J, Klinker KP, Borgert SJ, Butler BM, Giglio PG, Rand KH. Clinical and economic impact of antimicrobial stewardship interventions with the FilmArray blood culture identification panel. Diagnostic Microbiology and Infectious Disease. 2016;84(2):159–164. doi: 10.1016/j.diagmicrobio.2015.10.023. [DOI] [PubMed] [Google Scholar]
- 26.Carreno JJ, Lomaestro BM, Jacobs AL, Meyer RE, Evans A, Montero CI. Assessment of Time to Clinical Response in Patients with Sepsis Treated Before and After Implementation of a Matrix-Assisted Laser Desorption Ionization Time-of-Flight Blood Culture Identification Algorithm. Infection Control & Hospital Epidemiology. 2016;37(8):916–923. doi: 10.1017/ice.2016.105. [DOI] [PubMed] [Google Scholar]
- 27.Ray STJ, Drew RJ, Hardiman F, Pizer B, Riordan A. Rapid Identification of Microorganisms by FilmArray Blood Culture Identification Panel Improves Clinical Management in Children. Pediatr Infect Dis J. 2016;35(5):e134–138. doi: 10.1097/INF.0000000000001065. [DOI] [PubMed] [Google Scholar]
- 28.Verroken A, Despas N, Rodriguez-Villalobos H, Laterre PF. The impact of a rapid molecular identification test on positive blood cultures from critically ill with bacteremia: A pre-post intervention study. PLoS ONE. 2019;14(9):e0223122. doi: 10.1371/journal.pone.0223122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Box MJ, Sullivan EL, Ortwine KN, et al. Outcomes of Rapid Identification for Gram-Positive Bacteremia in Combination with Antibiotic Stewardship at a Community-Based Hospital System. Pharmacotherapy: The Journal of Human Pharmacology and Drug Therapy. 2015;35(3):269–276. doi: 10.1002/phar.1557. [DOI] [PubMed] [Google Scholar]
- 30.Sango A, McCarter YS, Johnson D, Ferreira J, Guzman N, Jankowski CA. Stewardship Approach for Optimizing Antimicrobial Therapy through Use of a Rapid Microarray Assay on Blood Cultures Positive for Enterococcus Species. Journal of Clinical Microbiology. 2013;51(12):4008–4011. doi: 10.1128/JCM.01951-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neuner EA, Pallotta AM, Lam SW, et al. Experience With Rapid Microarray-Based Diagnostic Technology and Antimicrobial Stewardship for Patients With Gram-Positive Bacteremia. Infection Control & Hospital Epidemiology. 2016;37(11):1361–1366. doi: 10.1017/ice.2016.175. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki H, Hitomi S, Yaguchi Y, et al. Prospective intervention study with a microarray-based, multiplexed, automated molecular diagnosis instrument (Verigene system) for the rapid diagnosis of bloodstream infections, and its impact on the clinical outcomes. Journal of Infection and Chemotherapy. 2015;21(12):849–856. doi: 10.1016/j.jiac.2015.08.019. [DOI] [PubMed] [Google Scholar]
- 33.Beal SG, Ciurca J, Smith G, et al. Evaluation of the Nanosphere Verigene Gram-Positive Blood Culture Assay with the VersaTREK Blood Culture System and Assessment of Possible Impact on Selected Patients. Journal of Clinical Microbiology. 2013;51(12):3988–3992. doi: 10.1128/JCM.01889-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker T, Dumadag S, Lee CJ, et al. Clinical Impact of Laboratory Implementation of Verigene BC-GN Microarray-Based Assay for Detection of Gram-Negative Bacteria in Positive Blood Cultures. Journal of Clinical Microbiology. 2016;54(7):1789–1796. doi: 10.1128/JCM.00376-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tansarli GS, Chapin KC. Diagnostic test accuracy of the BioFire® FilmArray® meningitis/encephalitis panel: a systematic review and meta-analysis. Clin Microbiol Infect. 2020;26(3):281–290. doi: 10.1016/j.cmi.2019.11.016. [DOI] [PubMed] [Google Scholar]
- 36.Popowitch EB, O’Neill SS, Miller MB. Comparison of the Biofire FilmArray RP, Genmark eSensor RVP, Luminex xTAG RVPv1, and Luminex xTAG RVP Fast Multiplex Assays for Detection of Respiratory Viruses. J Clin Microbiol. 2013;51(5):1528–1533. doi: 10.1128/JCM.03368-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rand KH, Rampersaud H, Houck HJ. Comparison of two multiplex methods for detection of respiratory viruses: FilmArray RP and xTAG RVP. J Clin Microbiol. 2011;49(7):2449–2453. doi: 10.1128/JCM.02582-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pierce VM, Hodinka RL. Comparison of the GenMark Diagnostics eSensor respiratory viral panel to real-time PCR for detection of respiratory viruses in children. J Clin Microbiol. 2012;50(11):3458–3465. doi: 10.1128/JCM.01384-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen JHK, Lam H-Y, Yip CCY, et al. Clinical Evaluation of the New High-Throughput Luminex NxTAG Respiratory Pathogen Panel Assay for Multiplex Respiratory Pathogen Detection. J Clin Microbiol. 2016;54(7):1820–1825. doi: 10.1128/JCM.00517-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Babady NE, England MR, Jurcic Smith KL, et al. Multicenter Evaluation of the ePlex Respiratory Pathogen Panel for the Detection of Viral and Bacterial Respiratory Tract Pathogens in Nasopharyngeal Swabs. J Clin Microbiol. 2018;56(2) doi: 10.1128/JCM.01658-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Troeger C, Blacker BF, Khalil IA, et al. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of diarrhoea in 195 countries: a systematic analysis for the Global Burden of Disease Study 2016. The Lancet Infectious Diseases. 2018;18(11)(18):1211–1228. 30362–1. doi: 10.1016/S1473-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goñi P, Martín B, Villacampa M, et al. Evaluation of an immunochromatographic dip strip test for simultaneous detection of Cryptosporidium spp, Giardia duodenalis, and Entamoeba histolytica antigens in human faecal samples. Eur J Clin Microbiol Infect Dis. 2012;31(8):2077–2082. doi: 10.1007/s10096-012-1544-7. [DOI] [PubMed] [Google Scholar]
- 43.Elsafi SH, Al-Maqati TN, Hussein MI, Adam AA, Hassan MMA, Al Zahrani EM. Comparison of microscopy, rapid immunoassay, and molecular techniques for the detection of Giardia lamblia and Cryptosporidium parvum. Parasitol Res. 2013;112(4):1641–1646. doi: 10.1007/s00436-013-3319-1. [DOI] [PubMed] [Google Scholar]
- 44.Gonin P, Trudel L. Detection and Differentiation of Entamoeba histolytica and Entamoeba dispar Isolates in Clinical Samples by PCR and Enzyme-Linked Immunosorbent Assay. Journal of Clinical Microbiology. 2003;41(1):237–241. doi: 10.1128/JCM.41.1.237-241.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buss SN, Leber A, Chapin K, et al. Multicenter Evaluation of the BioFire FilmArray Gastrointestinal Panel for Etiologic Diagnosis of Infectious Gastroenteritis. J Clin Microbiol. 2015;53(3):915–925. doi: 10.1128/JCM.02674-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khare R, Espy MJ, Cebelinski E, et al. Comparative Evaluation of Two Commercial Multiplex Panels for Detection of Gastrointestinal Pathogens by Use of Clinical Stool Specimens. J Clin Microbiol. 2014;52(10):3667–3673. doi: 10.1128/JCM.01637-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spina A, Kerr KG, Cormican M, et al. Spectrum of enteropathogens detected by the FilmArray GI Panel in a multicentre study of community-acquired gastroenteritis. Clinical Microbiology and Infection. 2015;21(8):719–728. doi: 10.1016/j.cmi.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 48.Wessels E, Rusman LG, van Bussel MJAWM, Claas ECJ. Added value of multiplex Luminex Gastrointestinal Pathogen Panel (xTAG® GPP) testing in the diagnosis of infectious gastroenteritis. Clinical Microbiology and Infection. 2014;20(3):O182–O187. doi: 10.1111/1469-0691.12364. [DOI] [PubMed] [Google Scholar]
- 49.Huang RSP, Johnson CL, Pritchard L, Hepler R, Ton TT, Dunn JJ. Performance of the Verigene® enteric pathogens test, Biofire FilmArrayTM gastrointestinal panel and Luminex xTAG® gastrointestinal pathogen panel for detection of common enteric pathogens. Diagnostic Microbiology and Infectious Disease. 2016;86(4):336–339. doi: 10.1016/j.diagmicrobio.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 50.Anderson NW, Buchan BW, Ledeboer NA. Comparison of the BD MAX Enteric Bacterial Panel to Routine Culture Methods for Detection of Campylobacter, Enterohemorrhagic Escherichia coli (O157), Salmonella, and Shigella Isolates in Preserved Stool Specimens. J Clin Microbiol. 2014;52(4):1222–1224. doi: 10.1128/JCM.03099-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simner PJ, Oethinger M, Stellrecht KA, et al. Multisite Evaluation of the BD Max Extended Enteric Bacterial Panel for Detection of Yersinia enterocolitica, Enterotoxigenic Escherichia coli, Vibrio, and Plesiomonas shigelloides from Stool Specimens. Journal of Clinical Microbiology. 2017;55(11):3258–3266. doi: 10.1128/JCM.00911-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stokes W, Simner PJ, Mortensen J, et al. Multicenter Clinical Validation of the Molecular BD Max Enteric Viral Panel for Detection of Enteric Pathogens. Journal of Clinical Microbiology. 2019;57(9) doi: 10.1128/JCM.00306-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mengelle C, Mansuy JM, Prere MF, et al. Simultaneous detection of gastrointestinal pathogens with a multiplex Luminex-based molecular assay in stool samples from diarrhoeic patients. Clinical Microbiology and Infection. 2013;19(10):E458–E465. doi: 10.1111/1469-0691.12255. [DOI] [PubMed] [Google Scholar]
- 54.Enserink R, Scholts R, Bruijning-Verhagen P, et al. High Detection Rates of Enteropathogens in Asymptomatic Children Attending Day Care. PLoS One. 2014;9(2) doi: 10.1371/journal.pone.0089496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bassetti M, Righi E, Montravers P, Cornely OA. What has changed in the treatment of invasive candidiasis? A look at the past 10 years and ahead. J Antimicrob Chemother. 2018;73(suppl_1):i14–i25. doi: 10.1093/jac/dkx445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Horn DL, Neofytos D, Anaissie EJ, et al. Epidemiology and Outcomes of Candidemia in 2019 Patients: Data from the Prospective Antifungal Therapy Alliance Registry. Clin Infect Dis. 2009;48(12):1695–1703. doi: 10.1086/599039. [DOI] [PubMed] [Google Scholar]
- 57.Clancy CJ, Nguyen MH. Finding the “Missing 50%” of Invasive Candidiasis: How Nonculture Diagnostics Will Improve Understanding of Disease Spectrum and Transform Patient Care. Clin Infect Dis. 2013;56(9):1284–1292. doi: 10.1093/cid/cit006. [DOI] [PubMed] [Google Scholar]
- 58.Avni T, Leibovici L, Paul M. PCR Diagnosis of Invasive Candidiasis: Systematic Review and Meta-Analysis. Journal of Clinical Microbiology. 2011;49(2):665–670. doi: 10.1128/JCM.01602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neely LA, Audeh M, Phung NA, et al. T2 Magnetic Resonance Enables Nanoparticle-Mediated Rapid Detection of Candidemia in Whole Blood. Science Translational Medicine. 2013;5(182):182ra54–182ra54. doi: 10.1126/scitranslmed.3005377. [DOI] [PubMed] [Google Scholar]
- 60.Clancy CJ, Pappas PG, Vazquez J, et al. Detecting Infections Rapidly and Easily for Candidemia Trial, Part 2 (DIRECT2): A Prospective, Multicenter Study of the T2Candida Panel. Clin Infect Dis. 2018;66(11):1678–1686. doi: 10.1093/cid/cix1095. [DOI] [PubMed] [Google Scholar]
- 61.Mylonakis E, Clancy CJ, Ostrosky-Zeichner L, et al. T2 Magnetic Resonance Assay for the Rapid Diagnosis of Candidemia in Whole Blood: A Clinical Trial. Clin Infect Dis. 2015;60(6):892–899. doi: 10.1093/cid/ciu959. [DOI] [PubMed] [Google Scholar]
- 62.Tang D-L, Chen X, Zhu C-G, Li Z-W, Xia Y, Guo XG. Pooled analysis of T2 Candida for rapid diagnosis of candidiasis. BMC Infect Dis. 2019;19(1):798. doi: 10.1186/s12879-019-4419-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nguyen MH, Clancy CJ, Pasculle AW, et al. Performance of the T2Bacteria Panel for Diagnosing Bloodstream Infections: A Diagnostic Accuracy Study. Ann Intern Med. 2019;170(12):845–852. doi: 10.7326/M18-2772. [DOI] [PubMed] [Google Scholar]
- 64.Buchan BW, Riebe KM, Ledeboer NA. Comparison of the MALDI Biotyper System Using Sepsityper Specimen Processing to Routine Microbiological Methods for Identification of Bacteria from Positive Blood Culture Bottles. Journal of Clinical Microbiology. 2012;50(2):346–352. doi: 10.1128/JCM.05021-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen JHK, Ho P-L, Kwan GSW, et al. Direct Bacterial Identification in Positive Blood Cultures by Use of Two Commercial Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry Systems. Journal of Clinical Microbiology. 2013;51(6):1733–1739. doi: 10.1128/JCM.03259-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ferreira L, Sánchez-Juanes F, Porras-Guerra I, et al. Microorganisms direct identification from blood culture by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Clinical Microbiology and Infection. 2011;17(4):546–551. doi: 10.1111/j.1469-0691.2010.03257.x. [DOI] [PubMed] [Google Scholar]
- 67.Haigh JD, Green IM, Ball D, Eydmann M, Millar M, Wilks M. Rapid identification of bacteria from bioMerieux BacT/ALERT blood culture bottles by MALDI-TOF MS. British Journal of Biomedical Science. 2013;70(4):149–155. doi: 10.1080/09674845.2013.11669949. [DOI] [PubMed] [Google Scholar]
- 68.Kok J, Thomas LC, Olma T, Chen SCA, Iredell JR. Identification of Bacteria in Blood Culture Broths Using Matrix-Assisted Laser Desorption- Ionization SepsityperTM and Time of Flight Mass Spectrometry. PLoS One. 2011;6(8) doi: 10.1371/journal.pone.0023285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lagacé-Wiens PRS, Adam HJ, Karlowsky JA, et al. Identification of Blood Culture Isolates Directly from Positive Blood Cultures by Use of Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry and a Commercial Extraction System: Analysis of Performance, Cost, and Turnaround Time. Journal of Clinical Microbiology. 2012;50(10):3324–3328. doi: 10.1128/JCM.01479-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Leli C, Cenci E, Cardaccia A, et al. Rapid identification of bacterial and fungal pathogens from positive blood cultures by MALDI-TOF MS. International Journal of Medical Microbiology. 2013;303(4):205–209. doi: 10.1016/j.ijmm.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 71.Martinez RM, Bauerle ER, Fang FC, Butler-Wu SM. Evaluation of Three Rapid Diagnostic Methods for Direct Identification of Microorganisms in Positive Blood Cultures. Journal of Clinical Microbiology. 2014;52(7):2521–2529. doi: 10.1128/JCM.00529-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martiny D, Dediste A, Vandenberg O. Comparison of an in-house method and the commercial SepsityperTM kit for bacterial identification directly from positive blood culture broths by matrix-assisted laser desorption-ionisation time-of-flight mass spectrometry. Eur J Clin Microbiol Infect Dis. 2012;31(9):2269–2281. doi: 10.1007/s10096-012-1566-1. [DOI] [PubMed] [Google Scholar]
- 73.Meex C, Neuville F, Descy J, et al. Direct identification of bacteria from BacT/ALERT anaerobic positive blood cultures by MALDI-TOF MS: MALDI Sepsityper kit versus an in-house saponin method for bacterial extraction. Journal of Medical Microbiology. 2012;61 doi: 10.1099/jmm.0.044750-0. [DOI] [PubMed] [Google Scholar]
- 74.Prod’hom G, Bizzini A, Durussel C, Bille J, Greub G. Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry for Direct Bacterial Identification from Positive Blood Culture Pellets. Journal of Clinical Microbiology. 2010;48(4):1481–1483. doi: 10.1128/JCM.01780-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saffert RT, Cunningham SA, Mandrekar J, Patel R. Comparison of three preparatory methods for detection of bacteremia by MALDI-TOF mass spectrometry. Diagnostic Microbiology and Infectious Disease. 2012;73(1):21–26. doi: 10.1016/j.diagmicrobio.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 76.Schubert S, Weinert K, Wagner C, et al. Novel, Improved Sample Preparation for Rapid, Direct Identification from Positive Blood Cultures Using Matrix-Assisted Laser Desorption/Ionization Time-of-Flight (MALDI-TOF) Mass Spectrometry. The Journal of Molecular Diagnostics. 2011;13(6):701–706. doi: 10.1016/j.jmoldx.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stevenson LG, Drake SK, Murray PR. Rapid Identification of Bacteria in Positive Blood Culture Broths by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry. Journal of Clinical Microbiology. 2010;48(2):444–447. doi: 10.1128/JCM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moussaoui W, Jaulhac B, Hoffmann A-M, et al. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry identifies 90% of bacteria directly from blood culture vials. Clinical Microbiology and Infection. 2010;16(11):1631–1638. doi: 10.1111/j.1469-0691.2010.03356.x. [DOI] [PubMed] [Google Scholar]
- 79.Azrad M, Keness Y, Nitzan O, et al. Cheap and rapid in-house method for direct identification of positive blood cultures by MALDI-TOF MS technology. BMC Infect Dis. 2019;19(1):1–7. doi: 10.1186/s12879-019-3709-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bhatti MM, Boonlayangoor S, Beavis KG, Tesic V. Rapid Identification of Positive Blood Cultures by Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry Using Prewarmed Agar Plates. J Clin Microbiol. 2014;52(12):4334–4338. doi: 10.1128/JCM.01788-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Florio W, Cappellini S, Giordano C, Vecchione A, Ghelardi E, Lupetti A. A new culture-based method for rapid identification of microorganisms in polymicrobial blood cultures by MALDI- TOF MS BMC Microbiology. 2019;19(1):267. doi: 10.1186/s12866-019-1641-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kohlmann R, Hoffmann A, Geis G, Gatermann S. MALDI-TOF mass spectrometry following short incubation on a solid medium is a valuable tool for rapid pathogen identification from positive blood cultures. International Journal of Medical Microbiology. 2015;305(4):469–479. doi: 10.1016/j.ijmm.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 83.Köck R, Wüllenweber J, Horn D, Lanckohr C, Becker K, Idelevich EA. Implementation of short incubation MALDI-TOF MS identification from positive blood cultures in routine diagnostics and effects on empiric antimicrobial therapy. Antimicrob Resist Infect Control. 2017;6:12. doi: 10.1186/s13756-017-0173-4. [DOI] [PMC free article] [PubMed] [Google Scholar]