Significance
Since the discovery of penicillin, humans have widely developed and used antibiotics to protect themselves from microbial infections. However, the intensive use of these compounds has led to the emergence of pathogens resistant to all classes of antibiotics. This major public health threat has led scientists to find new molecules with different structures and modes of action to overcome resistance phenomena. A promising alternative concerns bacteriocins secreted by certain bacteria. Of a peptidic nature, their ribosomal synthesis differentiates them from conventional antibiotics. The recently identified RumC1 antimicrobial peptide presents a remarkable bactericidal activity for multidrug-resistant strains. Added to this efficacy, RumC1 is not toxic against a number of human cell lines and is safe for human gut tissues.
Keywords: RiPPs, Ruminococcin C, sactipeptide, gut microbiome, antibiotic
Abstract
The emergence of superbugs developing resistance to antibiotics and the resurgence of microbial infections have led scientists to start an antimicrobial arms race. In this context, we have previously identified an active RiPP, the Ruminococcin C1, naturally produced by Ruminococcus gnavus E1, a symbiont of the healthy human intestinal microbiota. This RiPP, subclassified as a sactipeptide, requires the host digestive system to become active against pathogenic Clostridia and multidrug-resistant strains. Here we report its unique compact structure on the basis of four intramolecular thioether bridges with reversed stereochemistry introduced posttranslationally by a specific radical-SAM sactisynthase. This structure confers to the Ruminococcin C1 important clinical properties including stability to digestive conditions and physicochemical treatments, a higher affinity for bacteria than simulated intestinal epithelium, a valuable activity at therapeutic doses on a range of clinical pathogens, mediated by energy resources disruption, and finally safety for human gut tissues.
Rbosomally synthesized and posttranslationally modified peptides (RiPPs) are an important group of compounds that have stimulated research interest, notably as natural antimicrobial agents with bacteriocins (1). During RiPPs biogenesis, a precursor peptide composed of at least a leader and a core sequence is synthesized by the ribosome. The core peptide is modified by tailoring enzymes, and then the leader sequence is cleaved by one or two peptidases to produce the final active product (1–4). Among the RiPPs, sulfur-to-alpha carbon thioether cross-linked peptides (sactipeptides) are a subgroup that has emerged in recent years (5–7). Despite spectacular soaring made with genomic tools, the sactipeptide subclass is currently limited to only seven members. They include subtilosin A (SboA) (8, 9); thurincin H (10); the sporulation killing factor (SkfA) (11); thuricin CD that consists of two peptides, Trn-α and Trn-β (12); thuricin Z or huazacin (13, 14); and the recently characterized Ruminococcin C1 (RumC1) (15, 16).
The precise mechanistic details of sactipeptide cross-link formation are not fully understood. However, from a chemical point of view, the thioether bonds in these natural products are distinct from those of the well-studied lanthipeptides, such as nisin, in which they are formed between a Cys residue and a β-carbon of a dehydrated Thr/Ser residue (17). In contrast to the two-step redox neutral mechanism used for the maturation of lanthipeptides, radical–S-adenosylmethionine (SAM) sactisynthase enzymes introduce chemically equivalent thioether bonds by a one-step radical-based mechanism (5–7). The enzymes within this superfamily contain the canonical CysX3CysX2Cys motif, which binds the radical-SAM [4Fe-4S]2+/1+ cluster (referred to as the RS cluster), in which the fourth, unique iron is used to bind SAM cofactor (18). In its reduced state, the [4Fe-4S]1+ cluster catalyzes the reductive cleavage of SAM to generate a 5′-deoxyadenosyl radical (5′-Ado•). This radical abstracts an H- atom from the cognate substrate to initiate catalysis (18). In the case of the characterized radical-SAM enzymes involved in the sactipeptide biosynthesis, H-atom abstraction was shown to occur from the α carbon of the acceptor residue (5–7). A survey of available structural and functional data indicates that all radical-SAM enzymes involved in the sactipeptides biosynthesis contain a C-terminal extension appended to the radical-SAM domain called SPASM/Twitch domain that houses additional [4Fe-4S] clusters (19, 20). In contrast to the Twitch domains, which bind only one additional cluster, SPASM domains present a conserved cysteine-rich motif that coordinates two additional iron-sulfur clusters. The role of these clusters remains to be clarified, but it was suggested that they possibly interact with the substrate during catalysis or may also be implicated in electron transfer (5–7, 21).
Among the seven sactipeptides reported to date, four are structurally characterized (SboA, thurincin H, Trn-α, and Trn-β), and all of them have been purified from the genus Bacillus. SboA is a cyclic peptide with three thioether bridges involving two phenylalanines and one threonine. Thuricin CD (i.e., Trn-α and Trn-β) and thurincin H are not cyclic, they present three and four thioether bridges, respectively. One common feature of SboA, Trn-α, Trn-β, and thurincin H is that they all present Cys residues in the N-terminal half and the corresponding partners at the C-terminal part, a property that folds the peptide in a single hairpin-shaped form with the hydrophobic residues facing outward as shown by the available structures (8–10, 12, 22). Sequence analysis of the last identified sactipeptide, namely, RumC1, suggests a new fold. Indeed, one pair of cysteines is located at the N-terminal part of the sequence, while the other one is in the C-terminal end (Fig. 1A). Recent data on the characterization of mature RumC1 by mass spectrometry strongly suggest that the thioether network folds the peptide in a double hairpin-like structure that differs from the currently reported ones (16). Furthermore, sactipeptides and by extension antimicrobial peptides (including RiPPs but not only) have emerged as a potential trove of new weapons and alternatives to conventional antibiotics to fight multidrug-resistant (MDR) bacteria. However, their clinical use remains a challenge due to high cost of production and sensitivity to physiological or manufacturing conditions, as well as toxicity for human tissues (23, 24). In this context, we previously showed that RumC1 has a potent activity against gram-positive bacteria and is harmless for human cells (16). Obviously, the next step was to study RumC1 in a clinical context and to address the above-mentioned reasons that prevent antimicrobial peptides from being considered for pharmaceutical development.
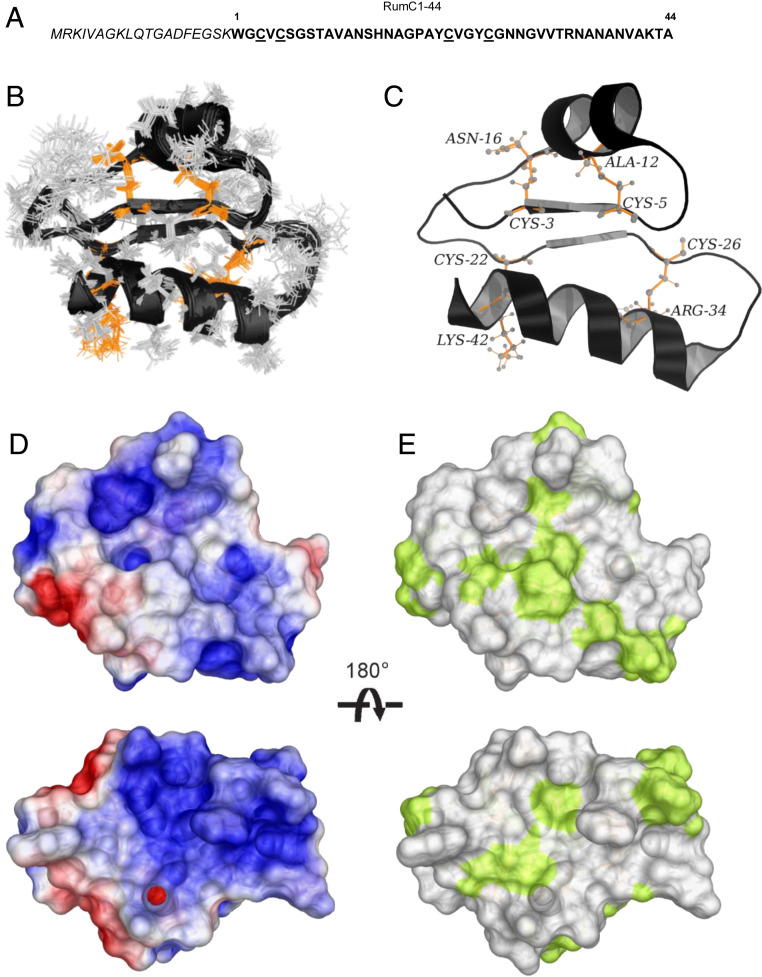
Fig. 1.
Sequence and 3D structure of Ruminococcin C1. (A) Sequence of RumC1 containing leader peptide (italics) and core peptide (RumC1-44). Cysteine residues are underlined. (B) Backbone overlay of the 20 lowest target function value conformers for the DDDD stereoisomer of RumC1. (C) Cartoon backbone representation of the 3D solution structure of RumC1 with the D stereochemistry at Ala12 (α-S), Asn16 (α-S), Arg34 (α-S), and Lys42 (α-S). Cysteine sulfur to α-carbon thioether cross-links are colored in orange and the position are indicated. (D) Electrostatic surface potential of RumC1, where blue indicates positive charge and red indicates negative charge. (E) Surface hydrophobicity of RumC1, where yellow represents hydrophobic residues and white represents hydrophilic residues.
In this work, we sought to determine the three-dimensional (3D) structure of RumC1. The rapid and large-scale production of 13C- and 15N-labeled mature RumC1, by heterologous expression in Escherichia coli (16, 25), allowed extensive NMR analyses to solve the structure of RumC1 and to propose the thioether network stereochemistry. In the meantime, combined electronic paramagnetic resonance (EPR) and Mössbauer spectroscopies enabled us to characterize the Fe-S clusters in RumMc1 sactisynthase. Finally, we point out that the fold of RumC1 confers resistance to physical, chemical, and digestive constraints, features essential for consideration in pharmaceuticals. The clinical properties of RumC1 also covers activity against clinical pathogens, including resistant strains, maintained in a mammalian environment and mediated through energy resources depletion, without any impact to human tissues.
Results
RumC1 Sactipeptide Displays a Double Hairpin-Like Structure.
The 2D [1H, 15N] HSQC spectrum of RumC1 is well resolved with 39 peptide amide peaks out of 42 expected, thus attesting for the folding of the protein (SI Appendix, Fig. S1). No NH peaks were observed for residues C26, G27, and N28, probably due to fast amide exchange with the solvent and/or high flexibility of the region. We assigned the backbone carbon, nitrogen, and proton resonances using a combined strategy of sequential residue correlations based on HNCACB, CBCA(CO)NH, HNCA, HN(CO)CA, HNCO, and HN(CA)CO triple resonance experiments and through-space nOe connectivities using 2D [1H, 1H] nuclear overhauser effect spectroscopy (NOESY), 3D [1H, 15N, 1H], and 3D [1H, 13C, 1H] NOESY experiments. NMR studies of Subtilosin A, Thuricin CD, and Thurincin H, by Vederas and coworkers have shown that cysteine sulfur to α-carbon thioether linkages induce a 10- to 15-ppm downfield shift for the α-carbon atoms of bridged residues (8–10, 12, 22). Moreover, through-space nOe interactions were observed between the β protons of cysteines and the amide proton (NH) of modified residues (8–10, 12, 22). In the same manner, we have demonstrated that RumC1 contains four sulfur to α-carbon thioether cross-links between Cys3 and Asn16, Cys5 and Ala12, Cys22 and Lys42, and Cys26 and Arg34 (16). Since there are four thioether bridges, each can adopt one of the two possible configurations at the α-carbon atom (L or D). Consequently, 16 stereoisomers must be considered to establish the 3D structure of RumC1. Calculations for all 16 stereoisomers were carried out using the combined assignment and dynamics algorithm for NMR applications (CYANA) software, and seven rounds performed on each stereoisomer using the same NMR restraints file (26). The structural statistics and constraint violations (SI Appendix, Table S1) allowed us to identify the stereoisomer with the D stereochemistry at Ala12 (α-S), Asn16 (α-S), Arg34 (α-S), and Lys42 (α-S) as a representative structure given 1) the absence of thioether bridge constraint violations, 2) the great number of nOe connectivities used in the structure calculation, and 3) the lowest average target function value and a low root mean square deviation (rmsd). To improve the structure of the DDDD stereoisomer, an additional refinement step by returning to the NOESY spectra to eliminate the ambiguities found during the structure calculation by the CYANA software was added. The resulting structural statistics of the 20 conformers for the DDDD isomer of RumC1 are summarized in SI Appendix, Table S2. The backbones of the 20 lowest target function value conformers for the DDDD isomer of RumC1 superimpose quite well with an rmsd value of 0.81 Å for the backbone (Fig. 1B). The 3D structure of RumC1 is thus composed on both sides by two α helices and in the middle by a two-stranded parallel β-sheet fragment and the whole stiffened by four cysteine sulfur to α-carbon thioether cross-links (Fig. 1C).
The electrostatic surface potentials present an overall positive charge (Fig. 1D), and the surface hydrophobicity of the DDDD stereoisomer shows a majority of hydrophilic residues (Fig. 1E). The compact 3D solution structure of RumC1 reveals a new sactipeptide fold and by extension a new antimicrobial peptide fold (SI Appendix, Fig. S2A). Four sulfur to α-carbon thioether bridges with a DDDD stereochemistry have been already reported for thurincin H (10). However, the presence of an additional two-stranded parallel β-sheet fragment in the middle of RumC1 induces a new fold, thus resulting in a different location of the thioether cross-links (SI Appendix, Fig. S2 B–E) (9, 10, 22).
Expression, Purification, and Characterization of the RumMc1 Sactisynthase.
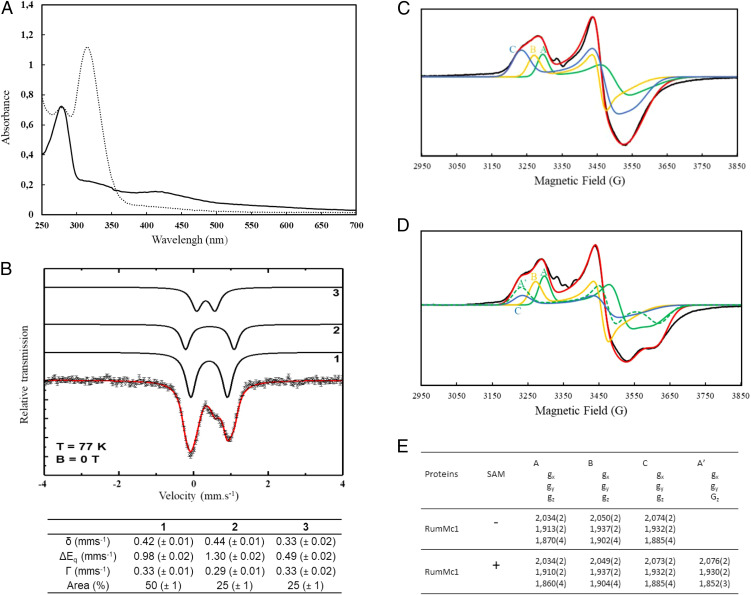
RumMc1 is predicted to contain accessory Fe-S clusters, in addition to the RS cluster; thus, we decided to coexpress the rumMc1 gene with the pDB1282 plasmid, which encodes for a set of proteins involved in iron-sulfur cluster biogenesis (IscS, IscU, IscA, HscB, HscA, and Fdx) (27). After the final step of anaerobic purification, the purity was evaluated by sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS/PAGE) to be over 95% (SI Appendix, Fig. S3). As expected, the holo-RumMc1 protein was dark brown in color, and the iron titration revealed, on average, 10 metal centers per monomer. In good agreement, the UV-vis spectrum of holo-RumMc1 suggests the presence of [4Fe-4S] clusters (Fig. 2A, solid line). Mössbauer spectroscopy on 57Fe-enriched oxidized holo-RumMc1 was then used to deeply investigate the nature of the Fe-S clusters. The experimental spectrum recorded at T = 77 K (dashed line) could be simulated (red line) with the three components 1, 2, and 3 in a 2:1:1 ratio (Fig. 2B). Components 1 and 2 are characterized by similar isomer shifts (δ) with δ = 0.42 mms−1 for component 1 and δ = 0.44 mms−1 for component 2, while the quadrupole splitting (ΔEq) varies with ΔEq = 0.98 mms−1 for component 1 and ΔEq = 1.30 mms−1 for component 2. These values are in a typical range for Fe2,5+ ions in a diamagnetic [4Fe-4S]2+ cluster (28–30). This is supported by simulations of the spectra recorded at T = 4.2 K in external fields of B = 0.1 T and B = 5 T (SI Appendix, Fig. S4 A and B) based on the spin Hamilton formalism assuming a total spin of S = 0. The 2:1 ratio suggests that holo-RumMc1 contains three [4Fe-4S]2+ clusters. This is supported by the sequence alignment of RumMc1 with other related proteins that contain conserved cysteine residues localized at the C-terminal half of the protein (SI Appendix, Fig. S5). Hence, we can conclude that component 1 represents eight Fe2,5+ ions in the same tetrahedral sulfur environment belonging to two [4Fe-4S]2+ clusters while component 2 corresponds to four indistinguishable Fe2,5+ ions present in the single [4Fe-4S]2+ cluster supposed to contain the SAM-binding iron site (RS cluster). The difference in ΔEq between components 1 and 2 points to a slightly different distribution of electron density. Component 3 is characterized by δ = 0.33 mms−1 and ΔEq = 0.49 mms−1 typical for Fe3+ high spin (S = 5/2) in a tetrahedral sulfur environment (28). This component was adequately simulated (SI Appendix, Fig. S4A, with an external magnetic field) by combining contributions from three distinct but antiferromagnetically coupled Fe3+ centers, thus resulting in a global S = 1/2 spin state as described for a paramagnetic [3Fe-4S]1+ cluster (31, 32). The respective hyperfine parameters are characteristic for [3Fe-4S]1+ clusters (SI Appendix, Fig. S4B) (31, 32). Upon addition of dithionite on the holo-RumMc1, the absorption decreased over the entire 310 to 420 nm range as expected for the conversion of the S = 0 [4Fe-4S]2+ chromophore to the S = 1/2 [4Fe-4S]1+ level (Fig. 2A, dashed line). This reduced form has been investigated by X-band EPR spectroscopy in order to gain insights into the individual features of the three [4Fe-4S] clusters involved. The experimental EPR spectrum of holo-RumMc1 is extremely rich, suggesting the presence of multiple S = 1/2 [4Fe-4S]1+ clusters (Fig. 2C, black line). It was satisfactorily simulated considering the three following sets of g tensors: [2.034, 1.913, 1.870] (set A), [2.050, 1.937, 1.902] (set B), and [2.074, 1.932, 1.885] (set C) (Fig. 2 C and E). Addition of the natural SAM cofactor then induced a substantial change in the spectrum that was well simulated with four sets of g tensors: [2.034, 1.910, 1.860] (set A), [2.049, 1.937, 1.904] (set B), [2.073, 1.932, 1.885] (set C), and [2.076, 1.930, 1.852] (set A′) instead of three (Fig. 2 D and E). As the contribution of set A has to be lowered to achieve a reliable simulation, these experiments support that set A originates from the RS cluster, while sets B and C rely on two additional clusters. The new component A′ was assigned to the amount of the RS cluster, which is bound to the SAM as previously described in ref. 33.
Fig. 2.
Spectroscopic characterizations of holo-RumMc1. (A) UV-vis spectrum of holo-RumMc1 in the absence (solid trace) or the presence of dithionite (dashed trace). (B) Mössbauer spectrum of holo-RumMc1 taken at T = 77 K with the simulation (red solid line) representing the sum of the subcomponents 1, 2, and 3 (black lines). Components 1 and 2 are simulated in a ratio of 2:1 and represent two diamagnetic [4Fe4S]2+ clusters. One [4Fe-4S]2+ cluster is supposed to bind SAM. Component 3 was assigned to Fe3+ ions present in a [3Fe-4S]1+ cluster in a S = 1/2 state. The respective Mössbauer parameters obtained from the simulation of the spectra at 77 K are summarized below the spectrum. (C) X-band continuous wave (CW) EPR spectra of dithionite-reduced holo-RumMc1. The black line represents the experimental spectrum, while the red trace is the simulated spectrum by using three components, A, B and C. (D) X-band CW EPR spectra of dithionite-reduced holo-RumMc1 in the presence of SAM. The black trace represents the experimental spectrum, while the red trace is the simulated spectrum by using four components, A, B, C, and A′. (E) g values of components A, B, C, and A′ used to simulate the experimental EPR spectra.
Efficient Maturation of RumC1: Both the Leader and the Core Sequences Are Crucial.
Here we sought to gain insights into the features of RumC1 that are required for its efficient maturation by RumMc1. It is established that the posttranslational modification of RiPPs is an event that is dependent on the presence of the leader peptide (2). For example, in the case of the sactipeptide subclass, Marahiel and coworkers have shown that AlbA failed to transform a variant of the SboA peptide without its leader sequence in a mature sactipeptide (34). We investigated the maturation of RumC1 by the way of an in vitro assay in the presence of RumMc1, the SAM cofactor, and dithionite as an external electron source. These experiments were performed with a full-length RumC1 precursor peptide, a leaderless variant of 44 amino acids (RumC1-44), and a third condition in which the leader sequence and the core peptide were dissociated but both present in the solution (RumC1-44-LS). The reaction mixtures were analyzed by liquid chromatography-mass spectrometry (LC-MS) to detect the formation of thioether bonds within the three substrates (SI Appendix, Fig. S6). Since the full maturation of RumC1 leads to the formation of four thioether bonds, the percentages of maturation (SI Appendix, Fig. S6A) account for the species that present a loss of 8 Da in mass compared to the unmodified substrate. The data clearly indicate that only the RumC1 peptide is fully modified, both RumC1-44 and RumC1-44-LS led to less than one and ten percent of species with four thioether linkages, respectively (SI Appendix, Fig. S6 A and B). It has to be noted that attempts to yield mature peptides form the latter two substrates lead mainly to a mixture of peptides harboring partial maturation with 2 and 3 thioether bonds (SI Appendix, Fig. S6 C and D). In good agreement with the lack of maturation of RumC1-44 during the in vitro enzymatic assay, we also observed that no mature peptide is obtained when this leader less form of RumC1 is produced in vivo using the heterologous expression and maturation protocol in E. coli (SI Appendix, Fig. S7A).
In contrast to the sactipeptides identified so far, the thioether connectivity in RumC1 folds the peptide in a double hairpin-like structure. Sequence alignments of the five RumC isoforms shows a strictly conserved Gly18/Pro19 motif in the loop region between the two hairpins (16). We therefore sought to determine whether this motif is necessary for inducing a turn in the sequence and allowing the peptide maturation by RumMc1. For this, we replaced the Gly18/Pro19 motif by Ala18/Ala19. LC-MS analysis of the heterologously expressed Ala18/Ala19 RumC1 variant clearly demonstrates the lack of maturation, thus suggesting that the loop alteration in RumC1 may disturb substrate–enzyme interactions that are crucial for the formation of the thioether network (SI Appendix, Fig. S7B). As reported by Grove et al. for CteA (21), these results suggest that the binding of RumC1 by RumMc1 involves determinants from both the leader and the core sequences.
The Compact Structure Confers a High Stability to RumC1.
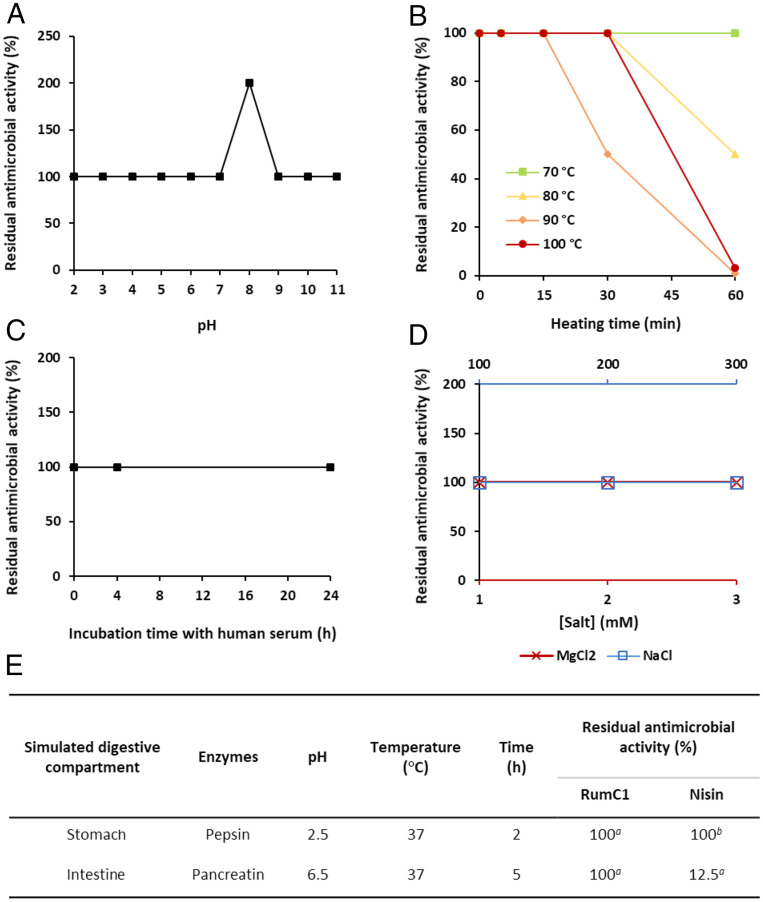
Studies have proven the potent activity of sactipeptides (12, 35); however, there is a lack of evidence showing that they possess the physiochemical properties necessary for in vivo administration and, from an applied point of view, for pharmaceutical development (35). Consequently, we assayed the tolerance of RumC1 to such properties. RumC1 showed no loss of activity when exposed to acidic or basic pH from 2 to 11 (Fig. 3A). Furthermore, RumC1 was resistant to 70 and 100 °C for up to 1 h and more than 15 min, respectively, meaning that RumC1 possesses the intrinsic thermal resistance that is required for drug formulation processes (Fig. 3B). Because of their sensitivities to enzymatic digestion or to physiological salts concentrations and blood enzymes, the administration of antimicrobial peptides by oral route and/or systemic injection is limited. Interestingly, the activity of RumC1 was not impaired by salt concentrations higher than 150 mM for NaCl and 2 mM for MgCl2 (Fig. 3C), which are considered as physiological saline conditions. Likewise, incubation in human serum at 37 °C up to 24 h did not affect the activity of RumC1 (Fig. 3D). RumC1 was finally treated with pepsin at pH 2.5 for 2 h at 37 °C and with pancreatin at pH 6.5 for 5 h at 37 °C in order to mimic the human gastric and intestinal compartments, respectively. RumC1 showed no loss of activity, and its integrity was revealed by MS analysis after these treatments, whereas the lanthipeptide nisin used as a positive control for pancreatin activity showed reduced antimicrobial potency (Fig. 3E and SI Appendix, Fig. S8 A and B). Pepsin activity was confirmed as well by hydrolysis of Bovine Serum Albumine (BSA) (SI Appendix, Fig. S8A). Therefore, the thioether network leading to a compact structure of RumC1 confers to the sactipeptide high resistance to physicochemical treatments and to the physiological but harsh conditions encountered in blood or in the digestive tract after systemic or oral administration.
Fig. 3.
Stability of RumC1. RumC1 was exposed (A) to a range of pH for 1 h, (B) to high temperatures up to 1 h, and (C) to human serum up to 24 h, before measuring its MIC. (D) MIC of RumC1 was determined in MH medium supplemented with NaCl or MgCl2. In A–D, all MICs were determined against C. perfringens. Residual antimicrobial activity was calculated based on the MIC of untreated RumC1. (E) Stability of RumC1 in conditions mimicking the GI tract. MIC of RumC1 against C. perfringens was determined after exposure to digestive enzymes. Nisin was used as a positive control of pancreatin activity. Residual antimicrobial activity was calculated based on the MIC of untreated bacteriocins. Superscript a indicates that residual antimicrobial activity was measured in this study. Superscript b indicates data published by Jarvis and Mahoney (36).
RumC1 Is Able to Act on a Simulated Infected Intestinal Epithelium.
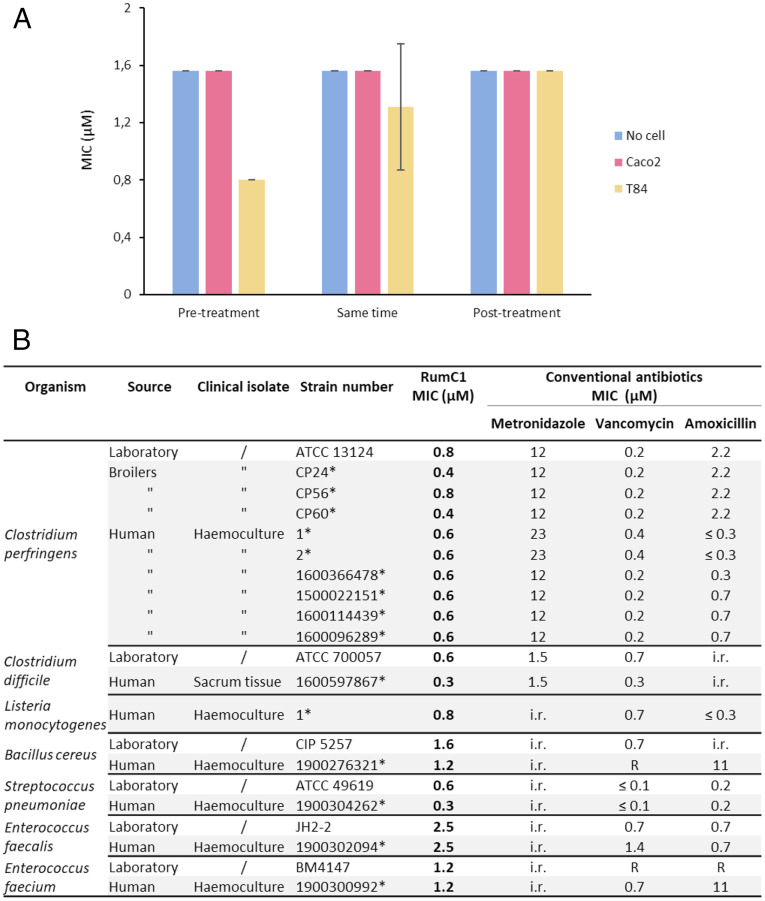
Many studies on antimicrobial peptides show their direct activity on bacterial cultures, without considering their efficiency in a mammalian environment, such as an infected epithelium. Indeed, antimicrobial peptides can act on bacteria but can also insert into eukaryotic cells or merely bind to their surface (24, 37), which could cause a partial loss of activity against extracellular pathogens. On the contrary, some peptides have higher affinity for bacterial cells than host cells (38). Therefore, we assayed the potency of RumC1 against Bacillus cereus, an aerotolerant human intestinal opportunistic pathogen, on simulated gut epitheliums using Caco2 and T84 cells as models of human small intestinal and colonic epithelium, respectively. B. cereus was able to grow on untreated monolayers of Caco2 and T84, whereas treatment with RumC1 was effective to clear the infection (SI Appendix, Fig. S9). The minimum inhibitory concentration (MIC) of RumC1 against B. cereus was measured in the eukaryotic cell line culture medium in the presence or the absence of human intestinal cells. The MIC value did not increase in the presence of intestinal cells when RumC1 was added 30 min before, concomitantly with, or 30 min after the bacterial cells (Fig. 4A). Indeed, no loss of activity was detected as well when B. cereus was allowed to colonize the epithelium for 30 min before adding RumC1 (Fig. 4A). Therefore, it appears that RumC1 has a preferential affinity for bacterial cells rather than host cells. Finally, these results suggest that the physiological environment of the two cell lines did not affect significantly the activity of RumC1.
Fig. 4.
Antimicrobial activity of RumC1. (A) Activity assays of RumC1 on B. cereus grown in eukaryotic cell culture medium in the absence or the presence of a simulated intestinal epithelium. Small and colic intestine compartments were simulated by Caco2 and T84 culture cells monolayer respectively, whereas RumC1 was added before (30 min), concomitantly with, and after infection (30 min) with 5 × 105 CFU/mL B. cereus culture. (B) Activity spectrum of RumC1 against laboratory and clinical gram-positive pathogens. Collection strains and clinical isolates are indicated. The table includes MIC of conventional antibiotics commonly used for clinical treatment and considered here as references (i.e., metronidazole, vancomycin, and amoxicillin). Acquired resistance to antibiotics were determined following the European Committee on Antimicrobial Susceptibility Testing 2019 clinical breakpoint tables and are indicated by R, whereas i.r. refers to intrinsic resistance, and asterisk refers to the laboratory isolate references.
RumC1 Displays a Potent Activity Against Gram-Positive Pathogenic Clinical Isolates.
We previously showed that RumC1 is active against a broad range of gram-positive bacteria including resistant and multiresistant strains (16) but focusing only on collection strains. In order to evaluate the potentiality of RumC1 being considered for pharmaceutical development, we investigated its activity against gram-positive strains isolated in a clinical context from humans or animals (i.e., broiler chickens). As RumC1 was first identified for its anti-Clostridium perfringens activity (16, 39, 40), we measured the MIC of the sactipeptide against a large panel of C. perfringens clinical isolates (Fig. 4B). RumC1 was active under the micromolar range (between 0.4 and 0.8 µM) on all of the 10 strains tested. In comparison with the reference molecules, usually used to eradicate C. perfringens, RumC1 showed a higher activity than metronidazole (12 to 23 µM) and an activity similar to vancomycin (0.2 to 0.4 µM). Interestingly, in a livestock context, RumC1 was active not only against the CP24 C. perfringens strain isolated from a healthy broiler chicken but also against strains isolated from animals suffering from dysbiosis or necrotic enteritis, CP56 and CP60, respectively (41). In addition, RumC was also active against C. perfringens strains isolated from humans suffering from bacteremia (n = 6) (Fig. 4B).
Then, we studied the effect of RumC1 on another main intestinal pathogen from the Clostridium genus, Clostridium difficile. RumC1 showed activity against collection and clinical strains of C. difficile with lower MIC (0.3 to 0.6 µM) than two of the most common antibiotics used for C. difficile intestinal infections, i.e., metronidazole (1.5 µM) and vancomycin (0.3 to 0.7 µM) (Fig. 4B). Moreover, RumC1 is also efficient at the micromolar range against other intestinal pathogens (clinical and collection strains) of main importance such as Listeria monocytogenes, B. cereus, Enterococcus faecalis, and Enterococcus faecium including strains resistant to amoxicillin and/or vancomycin (Fig. 4B). Beside its activity against pathogens causing gut infections, RumC1 is also active against a clinical Streptococcus pneumoniae strain at a low MIC (0.3 µM), a pathogen responsible of multiple types of infection (including respiratory tract infection, meningitis, and septicemia) (Fig. 4B).
The Killing Mechanism of RumC1 Involves Energy Resource Shortage.
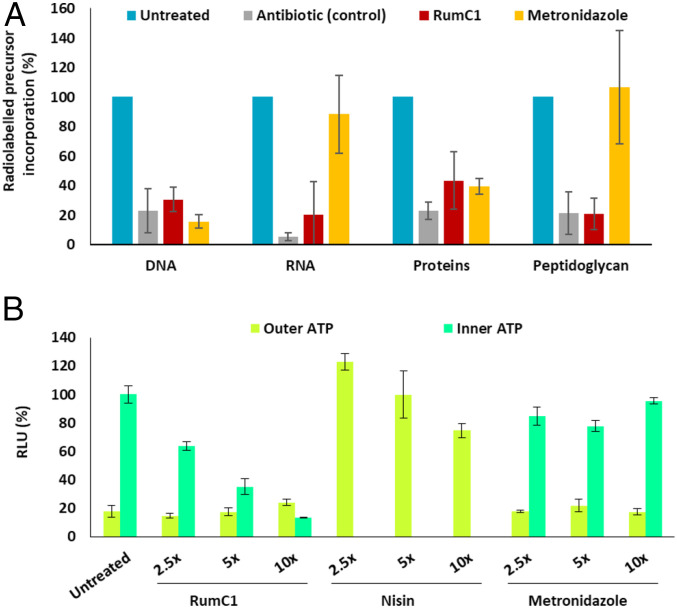
Unlike many bacteriocins, RumC1 does not have a pore forming action (16); we therefore investigated its effects on the main macromolecules synthesis pathways usually targeted by antibiotics. C. perfringens cells were exposed to RumC1, and the synthesis of DNA, RNA, proteins and peptidoglycan was followed by measuring the incorporation of radiolabeled precursors. Remarkably, RumC1 was able to inhibit the synthesis pathways of the four macromolecules with efficacies similar to conventional antibiotics targeting each of these pathways (Fig. 5A). As these synthesis pathways are all energy-dependent, we assayed the potency of RumC1 to break such energy resources. Thus, we measured by bioluminescent assay the ATP released by C. perfringens cells in the extracellular medium and then lysed cells to derive the intracellular ATP content (Fig. 5B). Upon treatment with RumC1 for 15 min, outer ATP was not increased compared to cells untreated, whereas inner ATP decreased in a dose-dependent manner. Treatment with nisin, a pore-forming bacteriocin led to drastic increase of outer ATP, whereas metronidazole does not impact ATP synthesis pathway. Since the synthesis of DNA, RNA, proteins, and peptidoglycan accounts for the consumption of ∼70% of the total ATP content of bacteria, their observed inhibition is likely linked to ATP depletion, and therefore, the specificity of RumC1 could be related to an inhibition of the ATP synthesis pathway (42).
Fig. 5.
Antibacterial mode of action of RumC1. (A and B) Cells of C. perfringens in early log phase were either treated with RumC1, control antibiotics, metronidazole, or nisin or left untreated. Each experiment was done in independent triplicates. (A) After 15 min of treatment at 5× MIC, C. perfringens cells were incubated with radiolabeled precursors of DNA, RNA, proteins, and peptidoglycan for 45 min. The synthesis of each pathway was measured by radioactivity counts. Gemifloxacin, rifampicin, tetracycline, and vancomycin were used as control antibiotics for the inhibition of DNA, RNA, proteins, and peptidoglycan, respectively. Radiolabeled precursor incorporation is expressed as a percentage of maximum incorporation determined with the untreated condition. (B) After 15 min of treatment, the ATP present in the extracellular media (outer ATP) was measured by bioluminescence, then cells were lysed and the ATP content in the extracellular media was measured again. The inner ATP content was deduced from the difference between ATP content in the extracellular media before and after cell lysis. Cells were treated with 2.5×, 5×, and 10× MIC for each condition. Relative light units (RLU) are expressed as percentages normalized by the value of the inner ATP content of untreated cells.
Integrity of Human Tissues Treated with RumC1.
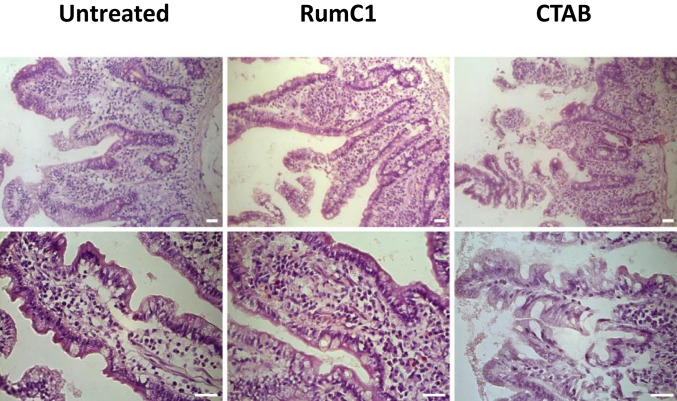
We previously reported that RumC1 was a safe new antimicrobial peptide because of its lack of in vitro toxicity on gastric and intestinal cell lines culture (16). To further characterize the safety of RumC1, we assayed the toxicity of the sactipeptide ex vivo, directly on human ileocecal tissues. Intestinal explants were isolated from unaffected area of ileocecal resection from two patients diagnosed with intestinal carcinoma and were incubated with RumC1 at 100 µM or the detergent CTAB at 300 µM for 4 h. Control and RumC1 exposed human intestinal explants displayed normal tissue organization with normal crypt and villosity lengths and no sign of epithelial desquamation or erosion, in accordance with data we obtained previously in vitro on human intestinal cell lines. Conversely, exposure to CTAB, used as positive control of tissue damages, caused important lesions to the human intestinal epithelium with cell desquamation and shortening of the crypts and villosities (Fig. 6).
Fig. 6.
Untreated (Left) and RumC1 (Middle, 100 µM) exposed human intestinal explants display normal tissue organization with normal crypt and villosity lengths and no sign of epithelial desquamation or erosion. CTAB (Right, 300 µM) exposure caused important lesions to the human intestinal epithelium with cell desquamation and shortening of the crypts and villosities. Human intestinal explants were treated with RumC1 or CTAB for 4 h before H&E staining and microscopic observations as described in Materials and Methods. Images are representative of overall observed effects. (Scale bar, 50 µm.)
Discussion
In this study, we elucidated the 3D structure of mature Ruminococcin C1 from Ruminococcus gnavus E1. NMR experiments clearly showed that the thioether network of RumC1, involving four sulfur to α-carbon bridges, folds the peptide into a double hairpin-like motif, which differs from the currently reported structures within the sactipeptide family. By extension, the fold not described so far includes two-stranded parallel β sheets into the core enclosed between two parallel α helixes. In contrast to a recent report by Berteau and coworkers, where the residues involved in the thioether bridges were identified to be l-configurated, after hydrolysis and derivatization (15), here we found that the stereoisomer that fits best with the NMR data featured D-configurations at Ala12, Asn16, Arg34, and Lys42. RumC1 has a global positive charge at physiological pH and displays a constrained and rigid backbone structure that presents a mostly hydrophilic surface unlike sactipeptides with known structures presenting mostly hydrophobic surfaces. These sactipeptides, like most of the bacteriocins that target gram-positive strains, have a pore-forming mode of action. Despite an overall cationic charge, RumC1 is unable to insert into lipids extracts most likely due to its hydrophilic surface and exerts probably an intracellular mode of action reaching one or more specific targets by active transport.
The double hairpin-like structure of RumC1 provides stability to pH and high temperatures, which could facilitate pharmaceutical manufacturing processes. We also showed that RumC1 is resistant to digestive proteolytic conditions, most likely because the sites of cleavage are protected by the four thioether bonds and the tridimensional folding of the peptide. Many RiPPs including sactipeptides, such as Subtilosin A and Thuricin CD, are not resistant to both pepsin and pancreatic proteases (43, 44), implying that the original folding of RumC1 offers additional protection. Moreover, RumC1 is also resistant to physiological salts conditions as well as to human serum, which can both be detrimental for an exogenous compound. Based on these considerations, we followed our investigations of RumC1 in a preclinical context. The first step was to ensure that RumC1 could cure infections in the mammalian environment. Therefore, we treated with RumC1 a B. cereus-infected simulated epithelium. RumC1 remained active at very low doses, exactly at the same level as the MIC obtained in the absence of eukaryotic cells. This observation confirms the stronger affinity of RumC1 for bacterial versus host cells and the specific primary function of RumC1 as antibiotic compound. Then we checked that RumC1 has potent activity on gram-positive clinical pathogens and not only on collections strains. RumC1 showed strong activity under the micromolar range toward Clostridia pathogens from both human and animal origins. The measured MICs are equivalent to vancomycin, one of the reference antibiotics used in therapy against these pathogens, and lower than metronidazole, another reference antibiotic known to generate resistance (16). Therefore, the use of RumC1 to treat infections caused by C. perfringens or C. difficile should be considered. Indeed, from a livestock point of view, C. perfringens can cause necrotic enteritis, which is one of the most common infections in broiler chickens with mortality rates as high as 50% and resulting in dehydration and lesions on intestinal mucosa or other organs like the liver, spleen, heart, or kidneys (45). On the other hand, C. difficile is a major human pathogen causing nosocomial infections and community-associated diarrhea and is responsible for 250,000 intestinal infections per year in the United States, associated with a mortality rate of 15 to 20%. In 2019, C. difficile has been classified by the Centers for Disease Control and Prevention (CDC) as an urgent threat for which new antimicrobials are needed (46, 47). In the same line, RumC1 is active against amoxicillin and vancomycin resistant E. faecium, registered on all priority lists of human pathogens for which new antimicrobials are urgently needed. It is registered by the CDC as a serious concern that requires prompt action and by the World Health Organization (WHO) on the “antibiotic-resistant priority pathogens” as high concern. E. faecium is also pointed to by the Infectious Diseases Society of America across the “ESKAPE pathogens” list of bacteria, which includes E. faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp. (48), for its ability to escape the effects of the commonly used antibiotics through evolutionarily developed mechanisms of resistance (49). E. faecium, characterized by drug resistance mechanisms, commonly causes life-threatening nosocomial infections among critically ill and immunocompromised individuals. Consequently, it is important to note that RumC1 is particularly active against this pathogen with a MIC of 1,2 µM; thus, it could be considered as a potential therapeutic solution. In the case of S. pneumoniae, with a prevalence of 1.2 million of infections per year in the United States, leading to its inclusion into the CDC and the WHO lists of priority pathogens, the use of RumC1 with a MIC of 300 nM may also be relevant. It should be noted that RumC1 is a broad-spectrum anti–gram-positive molecule directed against resistant anaerobic and aerobic clinical pathogens, which is rare in the antibiotics market with a few exceptions such as vancomycin, thus enhancing its clinical potential. Moreover, we showed that RumC1 inhibits the synthesis of macromolecules including DNA, RNA, proteins, or peptidoglycan and the production of ATP. At this stage, since the main biosynthetic pathways are inhibited, it is conceivable that RumC1 applies its antimicrobial activity intracellularly through a nonspecific mechanism or, conversely, targeting specifically ATP synthases and thus disrupting all necessary energy resources, such as bedaquiline, the only antibiotic currently on the market targeting ATP synthases but used only for the treatment of infections caused by Mycobacterium (50). Otherwise, we have previously identified that the phenotype induced by RumC1 treatment resembles the one induced by metronidazole in C. perfringens (16). However, we have shown here that metronidazole only inhibits DNA synthesis and, to a lesser extent, protein synthesis. Metronidazole is known to impact DNA synthesis and repair systems and most likely inhibits the activity of the strictly ATP-dependent ribonucleotide reductase by modulating the redox state of cells, but its precise mechanism remains elusive (51). Here we suggest that the phenotypic similarities induced by RumC1 and metronidazole might arise from a DNA synthesis inhibition and possibly other common events, although they certainly do not share the same mechanism. Therefore, further in-depth investigations are needed to resolve the potentially unique intracellular mode of action of RumC1. Finally, we followed our preclinical studies by assessing the safety of RumC1 on gut tissues directly sourced from patients. RumC1 did not induce epithelial lesions at a dose about a hundred times higher than the effective antimicrobial dose. This observation could be expected as humans have been exposed to RumC1 through evolution as this peptide is produced by a symbiotic bacterium present in the gut microbiota of healthy adults. Overall, RumC1 encompasses properties essential for a drug candidate to cure intestinal infections, especially since 1) it can be administrated by oral route, 2) it is active in the intestinal epithelium environment, 3) RumC1 shows activity at therapeutic doses against clinical intestinal resistant pathogens, and finally, 4) it is safe for gut tissues. Only a few AMPs meet all of the conditions that are necessary for reaching the marketing step, i.e., respecting the conditions of stability, presenting activity under physiological conditions (including in the presence of eukaryotic cells), owning antimicrobial effect against clinical pathogens at very low doses including resistant or MDR, and finally, retaining safety for human cells and tissues. It is interesting to note that RumC1 fulfills all these conditions, except one remaining bottleneck for the industrial-scale use, which concerns the cost of production. RumC-like synthetic molecule development could be addressed to solve this concern and highlight the great potential of this original sactipeptide.
Materials and Methods
Detailed descriptions of materials and methods, including expression, purification of (13C, 15N)-labeled mature RumC1, NMR studies, structure calculations, EPR and Mössbauer analyses of RumMc1, stability assays of RumC1, and MIC determinations are given in SI Appendix.
Supporting Information.
SI Appendix, Figs. S1–S9, as well as SI Appendix, Tables S1 and S2, are provided.
Supplementary Material
Acknowledgments
We thank Prof. Richard Ducatelle and Prof. Filip Van Immerseel from UGent for providing us with clinical isolates of C. perfringens from broiler chickens. We also thank Prof. Lhousseine Touqui for scientific advising. Finally, we thank the people from the AVB platform (iSm2 CNRS UMR 7313, Marseille). This study was supported by grants from the French National Agency for Research (Agence Nationale de la Recherche) through the Projet de Recherche Collaboratif (RUMBA project, ANR-15-CE21-0020), the Investissement d’Avenir Infrastructures Nationales en Biologie et Santé programme (ProFI project, ANR-10-INBS-08), and partial financial support from the Labex ARCANE and CBH-EUR-GS (ANR-17-EURE-0003). We are grateful to ADISSEO France company and the Association Nationale Recherche Technologie for funding the doctoral fellowship of C.R. titled “Bacteriocins RumC, a novel antimicrobial peptide family as alternative to conventional antibiotics.” This grant, Convention Industrielle de Formation par la Recherche no. 2016/0657, runs from 1 March 2017 to 1 March 2020. V.S. and C.S.M. acknowledge the support of the German Science Foundation within SPP 1927 (SCHU 1251/17-1, 2).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. I.B.S. is a guest editor invited by the Editorial Board.
Data deposition: All of the peak lists and the complete 1H, 13C, and 15N backbone and side chain chemical shift assignments of RumC1 have been deposited into the Biological Magnetic Resonance Databank (www.bmrb.wisc.edu) under accession code 50027. Coordinates of the 20 conformations of DDDD stereoisomer of RumC1 have been deposited into the Protein Data Bank under accession code 6T33.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2004045117/-/DCSupplemental.
Data Availability.
All data needed to evaluate the conclusions in the paper are present in the paper and the SI Appendix. All of the peak lists and the complete 1H, 13C, and 15N backbone and side chain chemical shift assignments of RumC1 have been deposited into the Biological Magnetic Resonance Databank (www.bmrb.wisc.edu) under accession code 50027. Coordinates of the 20 conformations of DDDD stereoisomer of RumC1 have been deposited into the Protein Data Bank under accession code 6T33.
References
- 1.Arnison P. G. et al., Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oman T. J., van der Donk W. A., Follow the leader: The use of leader peptides to guide natural product biosynthesis. Nat. Chem. Biol. 6, 9–18 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hudson G. A., Mitchell D. A., RiPP antibiotics: Biosynthesis and engineering potential. Curr. Opin. Microbiol. 45, 61–69 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chekan J. R., Ongpipattanakul C., Nair S. K., Steric complementarity directs sequence promiscuous leader binding in RiPP biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 116, 24049–24055 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahanta N., Hudson G. A., Mitchell D. A., Radical S-adenosylmethionine enzymes involved in RiPP biosynthesis. Biochemistry 56, 5229–5244 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benjdia A., Balty C., Berteau O., Radical SAM enzymes in the biosynthesis of ribosomally synthesized and post-translationally modified peptides (RiPPs). Front Chem. 5, 87 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flühe L., Marahiel M. A., Radical S-adenosylmethionine enzyme catalyzed thioether bond formation in sactipeptide biosynthesis. Curr. Opin. Chem. Biol. 17, 605–612 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Kawulka K. et al., Structure of subtilosin A, an antimicrobial peptide from Bacillus subtilis with unusual posttranslational modifications linking cysteine sulfurs to α-carbons of phenylalanine and threonine. J. Am. Chem. Soc. 125, 4726–4727 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Kawulka K. E. et al., Structure of subtilosin A, a cyclic antimicrobial peptide from Bacillus subtilis with unusual sulfur to alpha-carbon cross-links: Formation and reduction of alpha-thio-alpha-amino acid derivatives. Biochemistry 43, 3385–3395 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Sit C. S., van Belkum M. J., McKay R. T., Worobo R. W., Vederas J. C., The 3D solution structure of thurincin H, a bacteriocin with four sulfur to α-carbon crosslinks. Angew. Chem. Int. Ed. Engl. 50, 8718–8721 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Liu W.-T. et al., Imaging mass spectrometry of intraspecies metabolic exchange revealed the cannibalistic factors of Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 107, 16286–16290 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rea M. C. et al., Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc. Natl. Acad. Sci. U.S.A. 107, 9352–9357 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mo T. et al., Thuricin Z: A narrow-spectrum sactibiotic that targets the cell membrane. Angew. Chem. Int. Ed. Engl. 58, 18793–18797 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Hudson G. A. et al., Bioinformatic mapping of radical S-adenosylmethionine-dependent ribosomally synthesized and post-translationally modified peptides identifies new Cα, Cβ, and Cγ-linked thioether-containing peptides. J. Am. Chem. Soc. 141, 8228–8238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balty C. et al., Ruminococcin C, an anti-clostridial sactipeptide produced by a prominent member of the human microbiota Ruminococcus gnavus. J. Biol. Chem. 294, 14512–14525 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiumento S. et al., Ruminococcin C, a promising antibiotic produced by a human gut symbiont. Sci. Adv. 5, eaaw9969 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knerr P. J., van der Donk W. A., Discovery, biosynthesis, and engineering of lantipeptides. Annu. Rev. Biochem. 81, 479–505 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Broderick J. B., Duffus B. R., Duschene K. S., Shepard E. M., Radical S-adenosylmethionine enzymes. Chem. Rev. 114, 4229–4317 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grell T. A. J., Goldman P. J., Drennan C. L., SPASM and twitch domains in S-adenosylmethionine (SAM) radical enzymes. J. Biol. Chem. 290, 3964–3971 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Latham J. A., Barr I., Klinman J. P., At the confluence of ribosomally synthesized peptide modification and radical S-adenosylmethionine (SAM) enzymology. J. Biol. Chem. 292, 16397–16405 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grove T. L. et al., Structural insights into thioether bond formation in the biosynthesis of sactipeptides. J. Am. Chem. Soc. 139, 11734–11744 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sit C. S., McKay R. T., Hill C., Ross R. P., Vederas J. C., The 3D structure of thuricin CD, a two-component bacteriocin with cysteine sulfur to α-carbon cross-links. J. Am. Chem. Soc. 133, 7680–7683 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Mahlapuu M., Håkansson J., Ringstad L., Björn C., Antimicrobial peptides: An emerging category of therapeutic agents. Front. Cell. Infect. Microbiol. 6, 194 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aoki W., Ueda M., Characterization of antimicrobial peptides toward the development of novel antibiotics. Pharmaceuticals 6, 1055–1081 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Himes P. M., Allen S. E., Hwang S., Bowers A. A., Production of sactipeptides in Escherichia coli: Probing the substrate promiscuity of subtilosin A biosynthesis. ACS Chem. Biol. 11, 1737–1744 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Güntert P., Mumenthaler C., Wüthrich K., Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273, 283–298 (1997). [DOI] [PubMed] [Google Scholar]
- 27.Cicchillo R. M. et al., Lipoyl synthase requires two equivalents of S-adenosyl-L-methionine to synthesize one equivalent of lipoic acid. Biochemistry 43, 6378–6386 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Pandelia M.-E., Lanz N. D., Booker S. J., Krebs C., Mössbauer spectroscopy of Fe/S proteins. Biochim. Biophys. Acta 1853, 1395–1405 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Middleton P., Dickson D. P. E., Johnson C. E., Rush J. D., Interpretation of the Mössbauer spectra of the four-iron ferredoxin from Bacillus stearothermophilus. Eur. J. Biochem. 88, 135–141 (1978). [DOI] [PubMed] [Google Scholar]
- 30.Layer G. et al., Radical S-adenosylmethionine enzyme coproporphyrinogen III oxidase HemN: Functional features of the [4Fe-4S] cluster and the two bound S-adenosyl-L-methionines. J. Biol. Chem. 280, 29038–29046 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Huynh B. H. et al., On the active sites of the [NiFe] hydrogenase from Desulfovibrio gigas. Mössbauer and redox-titration studies. J. Biol. Chem. 262, 795–800 (1987). [PubMed] [Google Scholar]
- 32.Teixeira M. et al., Redox intermediates of Desulfovibrio gigas [NiFe] hydrogenase generated under hydrogen. Mössbauer and EPR characterization of the metal centers. J. Biol. Chem. 264, 16435–16450 (1989). [PubMed] [Google Scholar]
- 33.Bruender N. A., Wilcoxen J., Britt R. D., Bandarian V., Biochemical and spectroscopic characterization of a radical S-adenosyl-L-methionine enzyme involved in the formation of a peptide thioether cross-link. Biochemistry 55, 2122–2134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flühe L. et al., The radical SAM enzyme AlbA catalyzes thioether bond formation in subtilosin A. Nat. Chem. Biol. 8, 350–357 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Mathur H., Rea M. C., Cotter P. D., Hill C., Ross R. P., The sactibiotic subclass of bacteriocins: An update. Curr. Protein Pept. Sci. 16, 549–558 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Jarvis B., Mahoney R. R., Inactivation of nisin by alpha-chymotrypsin. J. Dairy Sci. 52, 1448–1450 (1969). [DOI] [PubMed] [Google Scholar]
- 37.Takeshima K., Chikushi A., Lee K.-K., Yonehara S., Matsuzaki K., Translocation of analogues of the antimicrobial peptides magainin and buforin across human cell membranes. J. Biol. Chem. 278, 1310–1315 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Welling M. M., Paulusma-Annema A., Balter H. S., Pauwels E. K. J., Nibbering P. H., Technetium-99m labelled antimicrobial peptides discriminate between bacterial infections and sterile inflammations. Eur. J. Nucl. Med. 27, 292–301 (2000). [DOI] [PubMed] [Google Scholar]
- 39.Ramare F. et al., Trypsin-dependent production of an antibacterial substance by a human Peptostreptococcus strain in gnotobiotic rats and in vitro. Appl. Environ. Microbiol. 59, 2876–2883 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crost E. H. et al., Ruminococcin C, a new anti-Clostridium perfringens bacteriocin produced in the gut by the commensal bacterium Ruminococcus gnavus E1. Biochimie 93, 1487–1494 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Gholamiandekhordi A. R., Ducatelle R., Heyndrickx M., Haesebrouck F., Van Immerseel F., Molecular and phenotypical characterization of Clostridium perfringens isolates from poultry flocks with different disease status. Vet. Microbiol. 113, 143–152 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Stokes J. M., Lopatkin A. J., Lobritz M. A., Collins J. J., Bacterial metabolism and antibiotic efficacy. Cell Metab. 30, 251–259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sutyak K. E., Wirawan R. E., Aroutcheva A. A., Chikindas M. L., Isolation of the Bacillus subtilis antimicrobial peptide subtilosin from the dairy product-derived Bacillus amyloliquefaciens. J. Appl. Microbiol. 104, 1067–1074 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rea M. C. et al., Bioavailability of the anti-clostridial bacteriocin thuricin CD in gastrointestinal tract. Microbiology 160, 439–445 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Van Immerseel F. et al., Clostridium perfringens in poultry: An emerging threat for animal and public health. Avian Pathol. 33, 537–549 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Centers for Disease Control and Prevention , Antibiotic Resistance Threats in the United States, (US Department of Health and Human Services, CDC, 2019), p. 148. [Google Scholar]
- 47.Smits W. K., Lyras D., Lacy D. B., Wilcox M. H., Kuijper E. J., Clostridium difficile infection. Nat. Rev. Dis. Primers 2, 16020 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rice L. B., Progress and challenges in implementing the research on ESKAPE pathogens. Infect. Control Hosp. Epidemiol. 31 (suppl. 1), S7–S10 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Cattoir V., Giard J.-C., Antibiotic resistance in Enterococcus faecium clinical isolates. Expert Rev. Anti Infect. Ther. 12, 239–248 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Andries K. et al., A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307, 223–227 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Dingsdag S. A., Hunter N., Metronidazole: An update on metabolism, structure-cytotoxicity and resistance mechanisms. J. Antimicrob. Chemother. 73, 265–279 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper and the SI Appendix. All of the peak lists and the complete 1H, 13C, and 15N backbone and side chain chemical shift assignments of RumC1 have been deposited into the Biological Magnetic Resonance Databank (www.bmrb.wisc.edu) under accession code 50027. Coordinates of the 20 conformations of DDDD stereoisomer of RumC1 have been deposited into the Protein Data Bank under accession code 6T33.