Significance
Therapy-loaded nanoparticles may be directed to the brain by functionalization with ligands targeting blood–brain barrier (BBB)-associated proteins. However, such targeting strategies have inherent brain-specificity limitations, as the target proteins are also significantly expressed in peripheral organs. Here, we present a counterintuitive targeting strategy that exploits the high impermeability of the BBB to selectively retain ligand labels on the surface of brain endothelium. Nanoparticles capable of binding the displayed ligands are consequently directed specifically to the brain microvasculature. This two-step targeting strategy paves the way to overcome the peripheral “off-target” nanoparticle accumulation, increasing the clinical translation of nanoparticle-based therapies. In addition, it shifts the identification of brain-target proteins from focusing on differential expression to instead focus on differential endocytic rates.
Keywords: blood–brain barrier, microvasculature labeling, differential endocytic rate, brain-specific target generation, nanoparticle brain delivery
Abstract
Current strategies to direct therapy-loaded nanoparticles to the brain rely on functionalizing nanoparticles with ligands which bind target proteins associated with the blood–brain barrier (BBB). However, such strategies have significant brain-specificity limitations, as target proteins are not exclusively expressed at the brain microvasculature. Therefore, novel strategies which exploit alternative characteristics of the BBB are required to overcome nonspecific nanoparticle targeting to the periphery, thereby increasing drug efficacy and reducing detrimental peripheral side effects. Here, we present a simple, yet counterintuitive, brain-targeting strategy which exploits the higher impermeability of the BBB to selectively label the brain endothelium. This is achieved by harnessing the lower endocytic rate of brain endothelial cells (a key feature of the high BBB impermeability) to promote selective retention of free, unconjugated protein-binding ligands on the surface of brain endothelial cells compared to peripheral endothelial cells. Nanoparticles capable of efficiently binding to the displayed ligands (i.e., labeled endothelium) are consequently targeted specifically to the brain microvasculature with minimal “off-target” accumulation in peripheral organs. This approach therefore revolutionizes brain-targeting strategies by implementing a two-step targeting method which exploits the physiology of the BBB to generate the required brain specificity for nanoparticle delivery, paving the way to overcome targeting limitations and achieve clinical translation of neurological therapies. In addition, this work demonstrates that protein targets for brain delivery may be identified based not on differential tissue expression, but on differential endocytic rates between the brain and periphery.
Treatment of neurological disorders, such as Alzheimer’s disease and Parkinson’s disease, is severely hindered by the poor penetration of the vast majority of potential therapies into the brain due to the presence of the blood–brain barrier (BBB), a highly impermeable cellular barrier composed primarily by the specialized endothelial cells (ECs) lining the brain microvasculature (1, 2). Therefore, strategies are needed to actively and specifically transport therapies across the BBB to successfully combat these conditions. The application of nanotechnology in the medical field has achieved modest success in delivering therapies into the brain by designing nanoparticles conjugated with ligands targeting proteins associated with the BBB (e.g., LRP1, TfR1, Glut1, and SLC7A5) (3–6). However, such strategies have significant brain-specificity limitations, as protein expression is not limited to the BBB. For instance, LRP1/TfR1, Glut1, and SLC7A5 are also highly expressed on lung, kidney, and intestine epithelium/endothelium, respectively (7–10), leading to high “off-target” accumulation of functionalized nanoparticles into peripheral organs (11). Exemplifying such limitation, functionalization of nanocarriers with peptide ligands targeting TfR1/RAGE has recently been shown to significantly increase nanocarrier accumulation in the brain (circa [c.] threefold). However, peptide functionalization resulted in stronger targeting (c. 10-fold) of nanocarriers into the lung (12). Such peripheral accumulation limits the clinical translation of therapies by reducing the therapeutic dose reaching the brain and increasing detrimental peripheral side effects (13, 14). Therefore, novel strategies which harness alternative BBB characteristics other than expression levels of BBB-associated proteins are required to achieve truly specific brain targeting of nanoparticles with minimal increase in peripheral uptake.
As such, the high impermeability of the BBB may, counterintuitively, be exploited to specifically deliver nanoparticles to the brain by selectively labeling the brain endothelium. A crucial requirement for the high impermeability of the BBB is a markedly reduced level of constitutive endocytosis into brain ECs compared to peripheral ECs (15–17). This differential endocytic rate may therefore be harnessed to endocytose, and hence remove, free, unconjugated protein-binding ligands at a faster rate from the surface of peripheral endothelium than from brain endothelium, resulting in ligand retention specifically at the surface of brain ECs. Nanoparticles capable of efficiently binding to the displayed ligands (i.e., labeled endothelium) by functionalization with a ligand-recognizing linker are consequently targeted specifically to the brain microvasculature with minimal “off-target” accumulation in peripheral organs.
Here, we present in vitro and in vivo data demonstrating the feasibility of such brain-targeting strategy by selectively labeling brain ECs with biotinylated ligands to promote targeting of avidin-functionalized polymeric nanomicelles specifically to the brain microvasculature with minimal targeting into peripheral organs.
Results
Nanomicelle Synthesis, Functionalization, and Characterization.
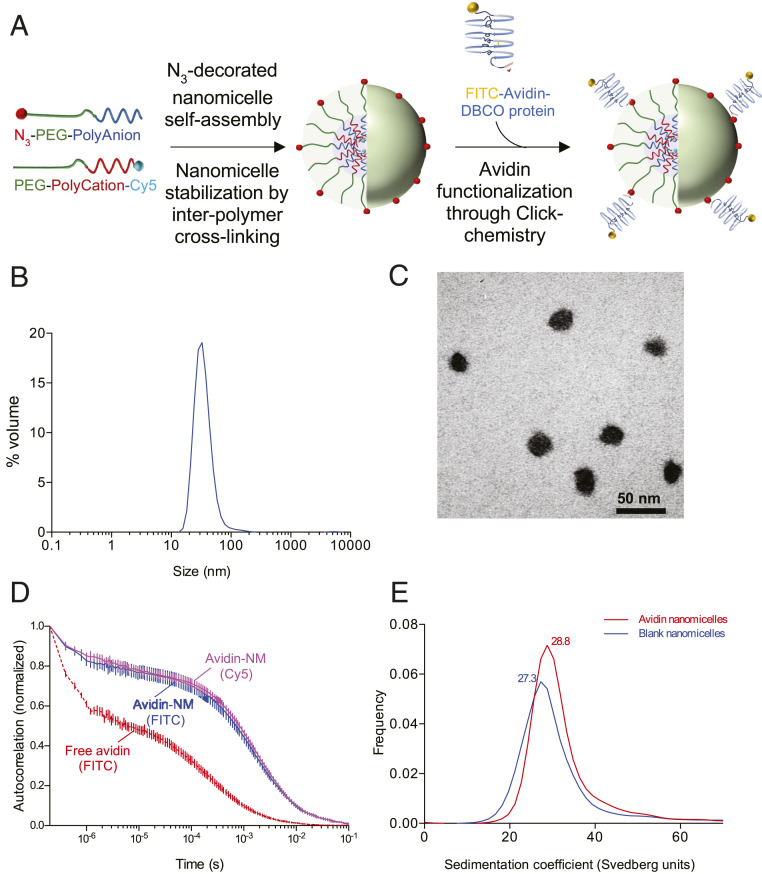
In order to create nanoparticles capable of recognizing biotinylated targets on ECs, avidin-functionalized nanomicelles (avidin-NMs) were synthesized by reacting N3-decorated PEG-ylated polyion-complexed nanomicelles with dibenzocyclooctyne (DBCO)-avidin (Fig. 1A). The resulting avidin-NMs had a hydrodynamic diameter (Z-average) of 44.6 ± 4.3 nm with a monodispersed size distribution, as determined by single-peak dynamic light scattering (DLS) histogram distributions (as percent volume) and a polydispersity index (PDI) value of 0.149 ± 0.02 (Fig. 1B and Table 1). The spherical morphology of the nanomicelles was further confirmed through transmission electron microscopy (TEM), demonstrating well-dispersed individual nanomicelles with a core diameter (calculated as number average) of 22.4 ± 0.3 nm (Fig. 1C and Table 1). Z-potential measurements indicated that the avidin-NMs had a near neutral surface charge (Table 1).
Fig. 1.
Synthesis and characterization of avidin-NMs. (A) Polymeric nanomicelles were assembled from oppositely charged ionic block-copolymers consisting of a PEG block attached to either an anionic (PEG-polyanion) or cationic (PEG-polycation) poly-amino acid block. The nanomicelle structure was stabilized by cross-linking adjacent amino acid polymers through EDC coupling. PEG-polyanions were capped with an azide (N3) group to allow binding of DBCO-linked proteins onto the nanomicelle surface through Click chemistry. PEG-polycations were tagged with a Cy5 fluorophore for fluorescence labeling of the nanomicelle core. (B and C) The size and monodispersity of the protein-functionalized nanomicelles was characterized through DLS (B) and TEM (C). (D and E) Protein functionalization was verified and quantified through FCS (through detection of FITC signal from free or nanomicelle-bound avidin, as well as Cy5 signal from nanomicelles) (D) and analytical ultracentrifugation (E). Results represent averages of six (B and D), three (Cy5 detection in D), or one (E) independent nanomicelle syntheses.
Table 1.
Characterization of avidin-NM size, distribution (n = 6 independent syntheses), and Z-potential (n = 2) through DLS and core-size quantification through TEM (quantified from 369 individual nanomicelles)
| Sample | Hydrodynamic size (Z-average), nm | PDI | Z-potential, mV | Core size, nm |
| Avidin-NMs | 44.6 ± 4.3 | 0.149 ± 0.02 | 0.11 ± 0.91 | 22.4 ± 0.27 |
Appropriate functionalization of avidin proteins onto the nanomicelle surface was confirmed by tracking the movement of individual avidin proteins (fluorescein isothiocyanate [FITC]-tagged) through fluorescence correlation spectroscopy (FCS). Reaction of avidin-DBCO with N3-nanomicelles resulted in a marked right shift in the autocorrelation curve of avidin (Fig. 1D, blue plot) compared to free avidin proteins (Fig. 1D, red plot), which translated to an increase in diffusion time from 228 ± 25 μs to 1,577 ± 78 μs (Table 2). Furthermore, the autocorrelation curve of avidin-NM obtained through FITC detection (i.e., detecting avidin movement) closely followed the autocorrelation curve obtained through detection of Cy5 (i.e., detecting nanomicelle movement) (Fig. 1D, magenta plot), demonstrating proper functionalization of avidin proteins onto the nanomicelles. FCS was further employed to calculate the functionalization number of avidin proteins per nanomicelle by dividing the FITC fluorescence counts per molecule (cpm) obtained from avidin-NMs by the cpm of free avidin proteins, yielding a functionalization number of 3.8 ± 0.7 avidin proteins per nanomicelle (Table 2). Avidin functionalization number was further confirmed through analytical ultracentrifugation (AUC), which demonstrated an increase of 1.5 Svedberg units in the sedimentation coefficient between blank and avidin-NMs (Fig. 2E), translating to a molecular-mass difference of 191 kDa (Table 3). Taking the molecular mass of individual (deglycosylated) avidin proteins as 60 kDa, AUC indicated a functionalization number of 3.2 avidins per nanomicelle, closely matching the estimation obtained through FCS.
Table 2.
Quantification of avidin functionalization by FCS (n = 6 independent syntheses)
| Sample | Diffusion time, μs | Cpm | Avidin per nanomicelle |
| Free avidin | 228 ± 24.9 | 4.7 ± 0.3 | — |
| Avidin-NMs | 1,577 ± 77.9 | 17.8 ± 3.8 | 3.8 ± 0.7 |
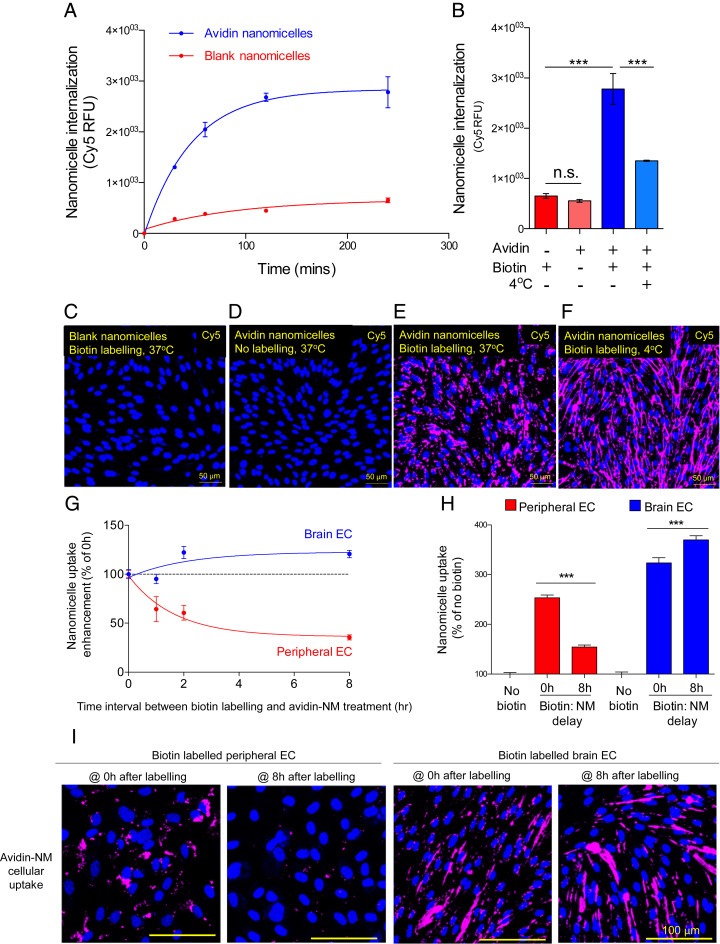
Fig. 2.
Specific internalization of avidin-NMs into brain ECs through selective retention of biotin targets on the cell surface. (A) Avidin-NM or blank-NM uptake into brain ECs displaying biotin targets was assessed through fluorescence (Cy5) quantification. (B) In addition, nanomicelle uptake (4-h treatment) was quantified in brain ECs with or without biotin targets at 37 °C or 4 °C. (C–F) Cellular internalization was confirmed through CLSM visualization of nanomicelle localization. (G and H) Avidin-NM uptake was also assessed in brain ECs and peripheral ECs (HUVECs) at increasing time intervals between biotin target generation and avidin-NM treatment. (I) Cellular internalization was confirmed through CLSM visualization of nanomicelle localization. (Scale bars: 100 μm.) Results are displayed as mean ± SEM of three (A and B) or six (G and H) independent wells. ***P < 0.001 (vs. respective column as determined by a one-way ANOVA with Tukey’s post hoc test). n.s., not significant.
Table 3.
Quantification of avidin functionalization by analytical ultracentrifugation (single nanomicelle synthesis)
| Sample | Sedimentation coefficient | Molecular mass, kDa | Molecular mass difference, kDa | Avidin per nanomicelle |
| Blank-nanomicelles | 27.2727 | 1,950 | — | — |
| Avidin-NMs | 28.7879 | 2,141 | 191 | 3.2 |
Specific Avidin-NM Internalization into Brain ECs through Selective Retention of Cell-Surface Biotin Targets.
Following confirmation of avidin functionalization onto N3-decorated nanomicelles, the ability of avidin-NMs to be internalized into brain ECs displaying biotin targets was examined in vitro. To this end, biotin targets were generated on the cell surface of primary brain ECs through conjugation of the cell-impermeable reagent sulfosuccinimidyl-6-[biotin-amido]hexanoate (sulfo-NHS-LC-biotin) to cell-membrane proteins. Cell-surface biotin targets induced a significant increase in avidin-NM internalization compared to blank (i.e., nonfunctionalized) nanomicelles (blank-NMs), as demonstrated by a marked time-dependent increase in cell-associated Cy5 fluorescence over a 4-h period (Fig. 2A). Avidin-NM internalization was specifically mediated by the avidin–biotin interaction, as no increase in avidin-NM internalization was seen in ECs lacking biotin targets (Fig. 2B). Furthermore, the avidin-NM uptake was significantly inhibited by incubation at 4 °C (Fig. 2B), demonstrating the requirement of an energy-dependent endocytic process for avidin-NM internalization. The active internalization of avidin-NMs was further confirmed through confocal laser-scanning microscopy (CLSM) of ECs following 4-h nanomicelle treatment. Both biotin-displaying ECs treated with blank-NMs (Fig. 2C) and control ECs treated with avidin-NMs (Fig. 2D) had negligible Cy5 fluorescence compared to biotin-displaying ECs treated with avidin-NMs (Fig. 2E). Furthermore, Cy5 fluorescence (i.e., nanomicelle signal) of biotin-displaying ECs treated with avidin-NMs strongly colocalized with FITC fluorescence (i.e., avidin signal) within the cells (SI Appendix, Fig. S1 A–C), indicating that the avidin-NM complex entered the cells as a unit. Importantly, while there was still Cy5 fluorescence in biotin-displaying ECs treated with avidin-NMs at 4 °C, fluorescence was limited to the EC wall with no localization in the cytosol (Fig. 2F; expanded image, colocalization with FITC signal, and cell-body visualization through Cell Mask staining and phase-contrast microscopy in SI Appendix, Fig. S1 D–H), demonstrating that avidin-NMs are able to bind to the cell-surface biotin targets, but are not internalized under energy-depleting conditions. While we have previously demonstrated that isolated primary brain ECs retain a strong BBB phenotype (6), we cannot discard the possibility that a certain amount of dedifferentiation might occur during isolation (18), which would increase the endocytic capacity of cultured brain ECs.
Similarly to brain ECs, cell-surface biotin-target generation on peripheral ECs induced a significant increase in avidin-NM internalization in an energy-dependent manner (SI Appendix, Fig. S2).
Following confirmation of avidin-NM internalization through biotin-target generation on both brain and peripheral ECs, the ability of the lower endocytic rate of brain ECs to promote retention of the biotin target on the cell surface of brain ECs compared to peripheral ECs as a function of time was examined. To this end, biotin targets on the cell surface of brain and peripheral ECs were generated as before. However, the cells were then incubated for increasing time periods to allow for biotin-target removal (endocytosis) before treatment with avidin-NMs. Quantification of Cy5 fluorescence (above ECs without biotin targets) indicated that the internalization of avidin-NMs by biotin targets was lost at a significantly faster rate in peripheral ECs compared to brain ECs (Fig. 2G), so that at 8 h postbiotinylation, peripheral ECs had only 35.5% of the initial avidin-NM targeting (Fig. 2H). Conversely, the avidin-NM internalization response in brain ECs did not decrease as a function of time (Fig. 2G). Interestingly, there was actually a small, yet significant, enhancement in avidin-NM uptake in brain ECs compared to the initial internalization response (Fig. 2H). CLSM imaging confirmed that avidin-NM uptake into peripheral ECs reverted close to control levels when treated at 8 h following biotin-target generation, while brain ECs had comparably high avidin-NM uptake at both 0 and 8 h following biotin-target generation (Fig. 2I).
In Vivo Nanoparticle Brain Targeting through Selective Target Display on the Brain Microvasculature of Mice.
Next, whether nanoparticles could be targeted to the brain through generation of brain-specific targets was examined in vivo. To this end, biotin targets on microvascular ECs of both the brain and peripheral organs were generated by employing a biotinylated anti-PECAM-1 antibody (biotin–α-PECAM1) to promote uptake of avidin-NMs.
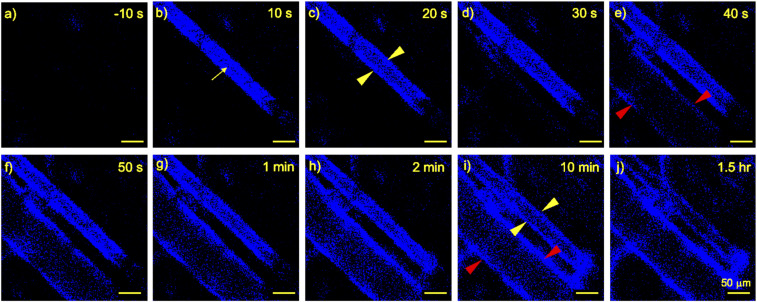
Firstly, the temporal dynamics of α-PECAM1 antibody binding to the microvasculature were examined by visualizing the earlobe microvasculature of mice. Fluorescently labeled α-PECAM1 was detected by in vivo CLSM circulating within the vascular lumen (artery) within 10 s following intravenous (i.v.) injection (Fig. 3 A and B, yellow arrow). Antibodies began to accumulate on the arterial vascular wall within 20 s (Fig. 3C, yellow arrowheads) and on the venous vascular wall within 40 s (Fig. 3E, red arrowheads). By 10 min, the antibodies were detected lining both arterial and venous vascular walls (Fig. 3I, yellow and red arrowheads, respectively). By 1.5 h, the vascular wall localization of the antibodies was further pronounced, with minor antibody detection within the vascular lumen (Fig. 3J), indicating that the majority of the antibodies present were associated with the vascular walls (i.e., ECs) and a comparably negligible number of antibodies remained circulating in the bloodstream. Because of its relatively low resolution, in vivo CLSM does not allow for accurate discrimination between antibodies bound to the EC wall and antibodies internalized into the ECs. Therefore, the degree of label display with respect to time was not able to be measured.
Fig. 3.
In vivo binding of α-PECAM1 antibody to microvascular walls. BALB/c mice were i.v. (tail-vein) injected with fluorescently labeled α-PECAM1 antibodies (eFluor450 fluorophore, 25-μg antibody injection). Antibody localization was detected in the ear vasculature through in vivo CLSM. Yellow arrow, arterial lumen; yellow and red arrowheads, arterial and venous vascular wall, respectively. A–J depict a single imaged localization at progressive time points before and after antibody injection. (Scale bars: 50 μm.) Images are from a single mouse representative of two mice.
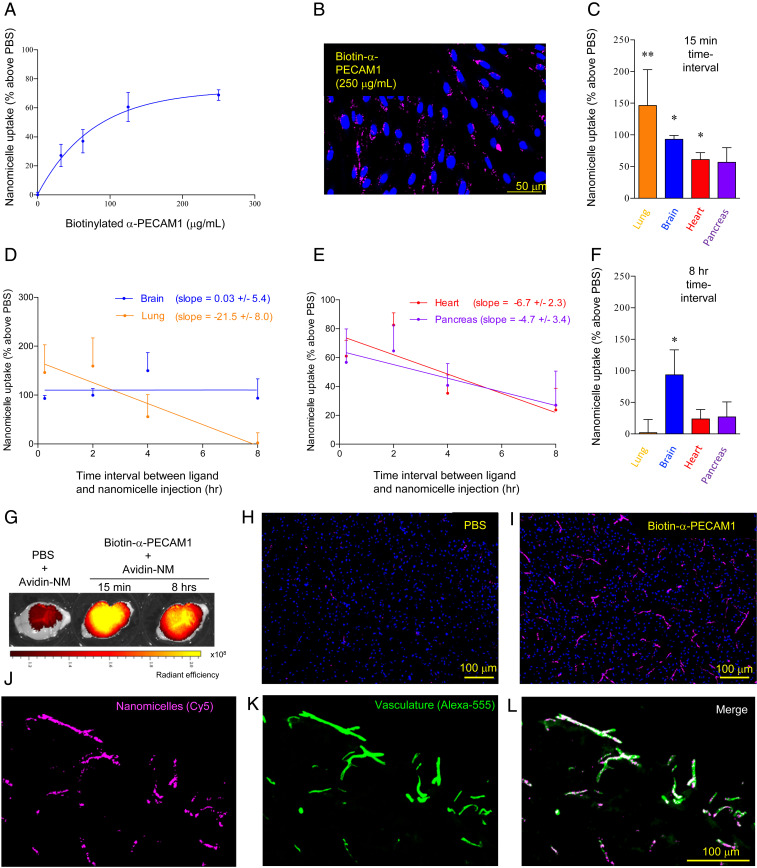
Secondly, the ability of cell-surface-bound biotin–α-PECAM1 to induce cellular internalization of avidin-NMs was examined in vitro. Display of biotin–α-PECAM1 on the cell surface of brain ECs led to a dose-dependent increase in avidin-NM uptake (Fig. 4A). Furthermore, CLSM visualization revealed that avidin-NMs were not limited to the cell surface, but were properly internalized into brain ECs (Fig. 4B), similar to internalization induced by indiscriminately biotinylating surface proteins to generate biotin targets (Fig. 2).
Fig. 4.
Specific in vivo brain targeting of avidin-NMs by retention of a biotin–α-PECAM1 ligand on the brain microvasculature surface. (A and B) Brain ECs were labeled (in vitro) with biotin–α-PECAM1, followed by incubation with avidin-NMs. Nanomicelle uptake was assessed through fluorescence (Cy5) quantification (A) and CLSM imaging (B). (C–L) BALB/c mice were i.v. injected with biotin–α-PECAM1 (25 μg) (or PBS) followed by i.v. avidin-NM injection after indicated time intervals. Nanomicelle accumulation in perfused animals was assessed through quantification of nanomicelle (Cy5) fluorescence in homogenized organs (above PBS control) (C–E), ex vivo fluorescence imaging (G), and immunohistochemical CLSM imaging of fixed tissue (H and I). (J–L) Colocalization of nanomicelles (Cy5 fluorescence) (J) with the microvasculature (visualized by staining with an independent nonbiotinylated α-PECAM1 antibody) (K) was confirmed through merging of the two fluorescence channels (L). Results are displayed as mean ± SEM of triplicate independent wells (A) or seven mice (five for 15-min time interval) (C–F). Slopes were calculated through linear regression analysis. *P < 0.05; **P < 0.01 (vs. respective PBS condition, as determined by an ANOVA with Dunnett’s post hoc test comparing PBS, 15-min and 8-h conditions).
Targeting of avidin-NMs to the brain through increased retention of biotin–α-PECAM1 on the brain microvasculature was then examined by i.v. injecting BALB/c mice with free, unconjugated biotin–α-PECAM1, followed by i.v. injection of avidin-NMs at increasing time periods (SI Appendix, Fig. S3). Quantification of Cy5 fluorescence in harvested organs revealed that avidin-NMs injected a short time period (15 min) following biotin–α-PECAM1 injection had increased targeting into the lung, heart, pancreas, and brain, with no targeting seen for the liver, kidney, spleen, and muscle (Fig. 4C and SI Appendix, Fig. S4) (pancreatic targeting reached statistical significance through a Student’s t test, though marginally failed to reach statistical significance through an ANOVA). Importantly, this biodistribution pattern was also seen in independent experiments employing biotin–α-PECAM1 preconjugated to avidin-NMs before injection into mice (SI Appendix, Fig. S5), indicating that the short-time-period biodistribution pattern was due to differences in microvascular expression of PECAM1 and not due to artifactual effects associated with circulating biotin–α-PECAM1 antibodies. Furthermore, preliminary experiments demonstrated biotin–α-PECAM1 administration dose-dependently increased avidin-NM accumulation into the brain and lung (SI Appendix, Fig. S6)
While injection of avidin-NMs a short time interval following unconjugated biotin–α-PECAM1 injection resulted in significant nanomicelle targeting into peripheral organs in addition to the brain, increasing the time interval between injections induced a marked decrease in nanomicelles targeting to the lung (−21.5 ± 8.0% per h), heart (−6.7 ± 2.3% per h), and pancreas (−4.7 ± 3.4% per h), while targeting to the brain remained nearly constant (+0.03 ± 5.4% per h) (Fig. 4 D and E). The differential targeting-decrease rate between peripheral organs and the brain resulted in significant avidin-NM targeting only to the brain when nanomicelles were injected 8 h after the initial biotin–α-PECAM1 injection (Fig. 4F). The shift in the avidin-NM targeting profile between short (15 min) and long (8 h) injection delay translated into a brain-to-organ targeting ratio increase of ×71.4 against the lung, ×2.6 against the heart, and ×2.1 against the pancreas.
Targeting of avidin-NMs into the brain was further confirmed through ex vivo fluorescence imaging of freshly excised whole brains and immunohistochemical staining of cryosectioned fixed brain tissue. Visualization of nanomicelle fluorescence (Cy5) revealed that treatment with biotinylated α-PECAM1 ligand led to a robust increase in the accumulation of avidin-NMs throughout the brain (Fig. 4G). Furthermore, avidin-NM accumulation was comparable whether nanomicelles were injected 15 min or 8 h following biotin–α-PECAM1 ligand injection.
CLSM imaging of fixed tissue sections revealed a similarly marked increase in avidin-NM localization in the brain tissue of biotin–α-PECAM1 treated (8-h interval) vs. phosphate-buffered saline (PBS)-treated mice (Fig. 4 H and I). Nanomicelle content appeared restricted to the microvasculature, with a long, microvessel-like fluorescence pattern seen throughout the tissue. Nanomicelle accumulation in the brain microvasculature was confirmed through microvessel immunostaining. Strong colocalization between the nanomicelle signal and microvessel signal was seen throughout (Fig. 4 J–L). Interestingly, high-magnification CLSM imaging revealed that the nanomicelle signal distribution had a discrete, punctate pattern (SI Appendix, Fig. S7), similar to the distribution of peptides targeting cell-junction proteins in the brain (19), indicating that avidin-NMs successfully bound to the biotin-labeled interendothelial PECAM-1 proteins.
Discussion
In the present study, we show that brain-specific targets for nanoparticle delivery may be generated by exploiting the reduced endocytosis of brain ECs to promote retention of free, unconjugated ligands on the brain microvasculature. We have established the in vivo feasibility of this approach by demonstrating that binding of a biotinylated anti–PECAM-1 antibody ligand to both the peripheral and the brain microvasculature leads to faster antibody removal from peripheral ECs, resulting in preferential retention of biotin targets on brain ECs. Avidin-NM targeting to the brain is consequently increased as a function of the time interval between ligand and nanomicelle injection.
PECAM-1 was selected as a target protein as it is present in the vasculature of the majority of tissues, yet its expression is higher in the lung, heart, and pancreas compared to the brain (20, 21). In line with this, functionalization with α-PECAM-1 antibodies has been shown to promote higher targeting of nanoparticles into the lung and heart over the brain (22, 23). In addition, PECAM-1 has been shown to be endocytically removed from the surface of peripheral ECs within hours (24, 25), as well as being highly associated with caveolae-mediated endocytosis (26–28). Therefore, PECAM-1 was an ideal target protein to examine whether the higher impermeability of the BBB (which is a direct consequence of reduced constitutive caveolar endocytosis) could be harnessed to achieve specific nanoparticle brain targeting employing ligands lacking intrinsic brain-targeting capacity.
One apparent limitation of targeting a nontransporter protein such as PECAM-1 to generate brain-specific targets would be its inability to induce nanoparticle transcytosis across brain endothelia to reach the brain parenchyma. However, increasing evidence indicates that brain penetration of nanoparticles is not strictly reliant on the transporter function of target proteins. For instance, nanoparticles targeting the membrane channel transporters Glut-1, nAchR, SLC7A5, or ChT (5, 29–32) have been shown to be transcytosed into the brain, despite the fact that these target proteins, due to their size, are unable to transport nanoparticles through their “normal” biological functions. Similarly, functionalization with α-PECAM-1 antibodies has been shown to successfully transcytose iron oxide nanoparticles across the BBB into the brain parenchyma (23). Instead, nanoparticle brain entry may rely on increasing nanoparticle interaction with the cell surface, as is the case for the well-described brain-entry mechanism of adsorptive-mediated transcytosis (33). Indeed, both the nanometrical curvature (34) and the protein-clustering ability of multivalent nanoparticles (35, 36) have been shown to promote endocytic cellular internalization, which, under appropriate conditions, would promote release into the brain parenchyma. Therefore, the retention of avidin-NMs in the brain microvasculature seen in the current study is more readily explained by the high affinity of avidin to biotin, resulting in the inability of the nanomicelle to dissociate from the internalized endocytic membrane. Indeed, nanoparticle avidity is well known to modulate the passage of nanoparticles into the brain parenchyma (37, 38). Therefore, future studies will optimize nanoparticle design by employing linkers with tuned affinity (e.g., aptamer–aptamer interaction) or pH-sensitive nanomicelle functionalization to promote detachment from target proteins within the endocytic vesicle and promote release into the brain parenchyma. In addition, the use of aptamers instead of antibodies as display ligands may promote more efficient transcytosis due to their smaller size and, hence, steric hinderance. In addition, aptamers may provide a better safety profile due to reduced immunogenicity.
Having said this, the current strategy’s ability to achieve specific targeting of drug-loaded nanoparticles to brain ECs, even without actual penetration into the brain parenchyma, would have significant therapeutic advantages for strategies aiming to induce neuroprotection through modulation of endothelial function (39–41) or by employing the brain microvasculature as a cellular reservoir for therapeutic proteins (42). Regarding Alzheimer’s disease in particular, the current targeting strategy would have important advantages for selectively modulating the impaired vascular-mediated clearance of amyloid-β from the brain (43, 44), or selective opening of the BBB to allow brain entry of drug compounds (45), without affecting endothelial function in the periphery. While organs with higher endocytic vasculature might more efficiently transport nanoparticles to the parenchyma, a point which we have not been able to assess with the use of whole-organ homogenates, our strategy would still result in a significant decrease in nanoparticle entry into the parenchyma of peripheral organs due to reduced vascular labeling, resulting in a significant increase in brain-to-periphery nanoparticle delivery.
A more significant limitation of employing PECAM-1 to generate brain-specific targets would be its interendothelial localization. While PECAM-1 is known to be expressed by venous, capillary, and arterial ECs in the brain microvasculature (20), such relatively limited interendothelial distribution would allow for only a fraction of the cell surface to display targets, thereby not achieving maximal nanoparticle uptake. Importantly, however, the current in vitro studies, given that they achieved preferential uptake of nanoparticles into brain ECs by indiscriminately biotinylating all cell-surface proteins, indicate that brain-specific target generation would be achievable by employing other proteins besides PECAM-1. Therefore, it will be of interest in future studies to identify proteins with more general cell-surface distribution which have significantly different endocytic rates between the brain and peripheral endothelium, thereby allowing maximal brain-specific target generation for increased nanoparticle uptake. To maximize clinical translation of this targeting strategy, it will be important to ensure that endocytic rates of identified proteins are comparable between animal models and patients, as well as fully characterizing the processing of the labeling ligands to properly understand their toxicity profile.
Conclusions
The present work demonstrates a simple, yet counterintuitive, strategy to specifically target nanoparticles to the brain through generation of brain-specific targets by exploiting the high impermeability of the BBB. Such an approach has the potential to revolutionize brain-targeting strategies by 1) moving away from directly functionalizing nanoparticles with targeting ligands, to instead adopt a two-step delivery strategy capable of displaying free targeting ligands on the brain microvasculature to achieve genuine nanoparticle brain targeting over peripheral organs; and 2) shift the target-protein identification process from having a focus on proteins with overexpression at the brain microvasculature to instead identify proteins with significantly different endocytic rates between the peripheral and brain microvasculature, thereby expanding the target-protein repertoire to achieve genuine brain targeting capable of increasing therapeutic delivery to the brain, as well as decreasing clinically restrictive peripheral side effects.
Materials and Methods
Nanomicelle Synthesis and Avidin Functionalization.
Nanomicelles were assembled from oppositely charged block copolymers consisting of a poly(ethylene glycol) (PEG) (2.2 k) segment tandemly coupled to either an anionic polypeptide [poly(α,β-aspartic acid)] or a cationic polypeptide [poly(5-aminopentyl-α,β-aspartamide)] (average degree of polymerization = 75) segment. A reactive azide (N3) group was attached to the α end of the PEG segment of anionic block copolymers (hereafter, N3-anions) to allow functionalization of DBCO-linked proteins through Click chemistry. A fraction of cationic block copolymers were tagged with a Cy5 fluorophore onto the ω end of the polypeptide chain (hereafter, Cy5-cation) to allow for nanomicelle quantification and imaging. Copolymers were blended in 10 mM phosphate buffer (PB; pH 7.4) (N3-anion:Cy5-cation:untagged-cation volume ratio = 10:4:16 at 1 mg/mL) to promote self-assembly of nanomicelles with a hydrophilic, noncharged PEG shell with azide reactive group-surface decoration, and Cy5 fluorescence at the core of the nanomicelle. The nanomicelle structure was then stabilized by cross-linking cationic and anionic polypeptide segments with 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC; 15 h, room temperature), followed by washing through Vivaspin filters (100 kDa molecular mass cutoff [MMCO], polyethersulfone [PES] membrane) (Sartorius) with 10 mM PB (pH 7.4) to remove EDC. The functionality of the decorating N3 groups was confirmed by synthesizing nanomicelles with increasing percentage of N3 anions (by varying the ratio of N3-terminated to MeO-terminated anions) and reacting them with fluorescently labeled DBCO. As expected, increasing the percentage of N3 anions composing the nanomicelle resulted in increased DBCO incorporation, with no incorporation seen for 0% N3 nanomicelles (SI Appendix, Fig. S8), thereby demonstrating the capacity of the N3 decoration to mediate Click chemistry on the nanomicelle surface.
Deglycosylated tetrameric avidin (Neutravidin, 60 kDa molecular mass, FITC-tagged) (ThermoFisher) was reacted with maleimide–PEG4–DBCO (Mal-PEG4-DBCO) (×10 molar excess Mal-PEG4-DBCO) in 10 mM PB at pH 8.5 (15 h, room temperature) to promote binding of maleimide onto primary amines on the protein (46). Unreacted Mal-PEG4-DBCO was removed by dialysis (3.6 kDa MMCO, PES membrane) against 10 mM PB (pH 7.4), with 200 mM arginine (10 mM PB/Arg) to prevent protein aggregation.
DBCO-linked avidin was then attached to N3-decorated nanomicelles by mixing at a 5:1 (protein:nanomicelle) molar ratio in 10 mM PB/Arg (15 h, room temperature). Unreacted avidin proteins were removed by washing through Vivaspin filters (100 kDa MMCO) with 10 mM PB/Arg. To remove contaminating avidin aggregates, avidin-NMs were filtered through a 0.2-μm PES filter (Millipore) with Dulbecco’s PBS (D-PBS) without calcium or magnesium. Avidin-NMs were diluted to a concentration of 1 mg/mL (polymer weight) and kept in D-PBS to allow for direct in vivo injection.
DLS.
Avidin-NM population size (hydrodynamic diameter) and dispersity were measured through DLS at 25 °C in D-PBS (1 mg/mL) with a Zetasizer Nano ZS Zen3500 (173° backscatter measurement angle) (Malvern Instruments Ltd.) in a Malvern QS quartz cuvette (Zen2112).
TEM.
TEM micrographs of the core structure of polymeric micelle were acquired with a JEM-1400 electron microscope (JOEL). Briefly, a copper TEM grid with a carbon-coated collodion membrane (Nisshin EM) was glow-discharged in an Eiko IB-3 ion coater (Eiko). The grid was captured with forceps at its carbon side up by gripping the outermost edge. Two microliters of the polymeric micelle solution was applied onto the grid, followed by mixing of 2 μL of uranyl acetate (2% [wt/vol]) solution. The solution was placed on the grid for 30 s to allow deposition of polymeric micelle onto the collodion membrane. The excess solution was blotted with a piece of filter paper, and the sample was air-dried for 15 min at room temperature. The grid was mounted into the TEM machine for imaging, conducted at an acceleration voltage of 120 kV. The nanomicelle core diameter was calculated from 369 individual nanomicelles by using ImageJ 1.48v software (NIH).
FCS.
Diffusion times were calculated from the autocorrelation functions of free DBCO-avidin proteins and avidin-NMs (measured in separate samples, both in D-PBS, at 100 nM avidin concentration) through FCS employing an LSM880 microscope equipped with a C-Apochromat 40× (water immersion) objective (Carl Zeiss). Autocorrelation functions were obtained from either the fluorescence signals of FITC or Cy5 separately. In-built software was employed to convert autocorrelation functions to diffusion times. The number of avidin proteins per nanomicelle was calculated by dividing the FITC fluorescence cpm of avidin-NMs by the cpm of free avidin-DBCO.
AUC.
AUC was performed by using an analytical ultracentrifuge (Optima XL-1, Beckman Coulter) equipped with an AnTi60 rotor cell that was able to house three sets of 1.2-mm two-channel Epon-filled centerpieces. The measurement was carried out by setting the detection of absorbance at 650 nm and the rotation speed to 18,000 rpm (26,081 × g), at 20 °C. Radial absorbance data were collected in continuous-scanning mode at 0.002-cm increments and 3-min intervals for a total of 120 scans. Acquired sedimentation boundaries were fitted based on the Lamm equation using the continuous-sedimentation coefficient model in the SEDFIT software to calculate the distribution of sedimentation coefficient (in Svedberg units) with the following parameters: resolution, 100 S; smin, 0 S; smax, 150 S; friction, 1.2 μ; buffer density, 1.02 g/cm3; viscosity, 0.01002 poise; and partial specific volume, 0.73 cm3/g. Sedimentation coefficient was converted to molecular mass (M) by the Svedberg equation:
where s is the sedimentation coefficient, R is the gas constant, T is the absolute temperature, v is the partial specific volume, ρ is the solution density, and D is the diffusion coefficient (calculated by using the Einstein–Stokes equation with the DLS diameter).
Primary Brain EC Isolation and Culturing.
Primary brain ECs were isolated as described (6). Rats (Sprague–Dawley, female, 8 wk old) were killed, and the brain cortex was isolated over dissection buffer (Hanks’ Balanced Salt Solution [HBSS]/1% bovine serum albumin [BSA]/2% penicillin/streptomycin [Pen/Strep, 4 °C). Following removal of the meninges and visible blood vessels, the cortex was homogenized and incubated in digestion buffer (Dulbecco’s modified Eagle medium/Ham’s F-12 with collagenase, dispase, DNase type I, and trypsin) at 37 °C. The digest was centrifuged in separation gradient buffer (25% BSA in HBSS), and the microvessel pellet was further incubated in digestion buffer (1 h, 37 °C). The microvessel fragments were collected by centrifugation and plated on Cell-BIND T75 cell-culture flasks (Corning) coated with calf-skin collagen type I and bovine plasma fibronectin in EC growth medium MV2 (PromoCell), supplemented with vascular endothelial growth factor (VEGF; 0.5 ng/mL) (unless otherwise stated), insulin-like growth factor 1 (20 ng/mL), epidermal growth factor (5 ng/mL), basic fibroblast growth factor (10 ng/mL), hydrocortisone (200 ng/mL), ascorbic acid (1,000 ng/mL), 5% (vol/vol) fetal calf serum, and 5% Pen/Strep (hereafter termed full MV2 medium) with 4 μg/mL puromycin and 100 μg/mL EC growth supplement. After 5 d culturing (37 °C, 5% CO2), the culture medium was changed to low puromycin (1 μg/mL) until reaching confluency. For experimentation, cells were detached with trypsin/ethylenediaminetetraacetic acid solution and plated in appropriate cell-culture plates (coated with collagen/fibronectin) in full MV2 medium. Once cells reached 90% confluency (after c. 4 d), the medium was changed to full MV2 medium with no VEGF (hereafter termed NV-MV2) for 3 d to allow full adoption of a BBB phenotype before experimentation began.
Peripheral EC Culturing.
Human umbilical vein ECs (HUVECs), employed as an in vitro model of peripheral vasculature ECs (47–49) were obtained from the Japanese Collection of Research Bioresources cell bank and employed between passages 2 and 7. HUVEC culturing, collection, and plating conditions were identical to primary brain ECs (see above), with the exception of culturing in NV-MV2 medium.
In Vitro Nanomicelle Uptake into Biotinylated ECs.
Biotin targets were generated on EC surfaces by employing the cell-impermeable biotinylating agent sulfo-NHS-LC-biotin (EZ-Link, ThermoScientific), capable of binding to primary amines of cell-surface proteins through its N-hydroxysuccinimide moiety. Cells were washed with ice-cold HBSS and incubated with sulfo-NHS-LC-biotin (500 μg/mL in HBSS, 20 min, 4 °C). Cells were then washed with ice-cold HBSS and either directly treated with nanomicelles (either blank nanomicelles or avidin-NMs at a concentration of 200 μg/mL in culturing medium) (at 37 °C or 4 °C for specified time points) or incubated (37 °C) in culturing medium for specified time points before nanomicelle treatment (as above). Cells were then thoroughly washed with culturing medium, stained with Hoechst dye (in culturing medium), and fixed (4% paraformaldehyde [PFA]). Nanomicelle uptake was assessed by quantifying Cy5 fluorescence in an Infinite M1000 Pro fluorescence microplate reader (Tecan) and by visualizing nanomicelle localization through CLSM imaging of Cy5 or FITC fluorescence signal (LSM880 Carl Zeiss microscope).
In experiments employing biotin–α-PECAM1 antibody to generate biotin targets on the EC surface, the same above protocol was applied, except that cells were incubated with biotin–α-PECAM1 (in culturing medium) at 37 °C for 1 h, followed by incubation with avidin-NMs (200 μg/mL in culturing medium) for 90 min after appropriate washes (all HBSS washes were done with room-temperature HBSS).
In Vivo CLSM.
Mice were anesthetized with 2.5% isolfuorane and directly placed on a thermoplate (Tokai Hit). The ear was placed flat against a viewing glass coverslip, and a canulae was inserted into the tail vein. An A1R CLSM system attached to an upright ECLIPSE FN1 microscope (Nikon) was employed for image capture. Ear-vasculature imaging region was set by using autofluorescence (488 laser excitation) of the tissue. Intravascular antibody (eFluor450 conjugated) circulation was imaged with a 405 laser excitation. Fluorescence signal was detected within a c. 10-μm-thick plane (pinhole size = 109.8 μm; magnification ×20) at 10-s (first 5 min) or 1-min (subsequent captures) intervals. Antibody injection (25 μg) was administered through the tail-vein canulae during the first 10-s interval following the first image capture.
In Vivo Nanomicelle Uptake following Microvascular Biotin-Target Generation.
Balb-c mice (female, 5 wk old) were systemically (tail-vein) injected with D-PBS or biotin–α-PECAM1 (monoclonal rat anti-mouse, 25 μg; ThermoFisher). Following appropriate time intervals, the mice were systemically (tail-vein) injected with avidin-NM (200 μg in D-PBS). After 16 h, the mice were anesthetized (isofluorane) and perfused with D-PBS (transhepatic perfusion followed by transcardial perfusion) before organ collection into D-PBS and weighing. Organs were then homogenized in a multibead shocker homogenizer (Yasui Kikai) in passive lysis buffer (Promega), followed by nanomicelle fluorescence quantification in the homogenates with an Infinite M1000 Pro fluorescence microplate reader (Tecan). A subset of brains from selected groups was dissected into two hemispheres before homogenization in order to process one hemisphere for immunohistochemistry. The nonhomogenized hemisphere was submerged in 4% PFA (4 °C, 24 h), followed by cryoprotection in 20% sucrose (4 °C, 24 h). The hemisphere was then cryosectioned into 20-μm slices and mounted onto glass slides for immunostaining.
Biotin–α-PECAM1 Antibody Ligand Conjugation onto Avidin-NMs.
To examine the biodistribution of avidin-NMs preconjugated with biotin–α-PECAM1, avidin-NMs (200 μL, 1 mg/mL) were incubated with biotin–α-PECAM1 (50 μL, 500 μg/mL) at 4 °C for 72 h. Mice were then injected with the total 250 μL mixture solution to maintain nanomicelle and antibody ligand amount equal to those in endothelial-labeling experiments (see above). The mice were perfused 16 h following nanomicelle injection, and nanomicelle accumulation in organs was quantified as before.
Ex Vivo Whole-Brain Fluorescence Imaging.
Whole brains were excised following perfusion, as described above. The brains were placed inside an In Vivo Imaging System (Perkin-Elmer), and single Cy5 fluorescence images were captured for all comparison brains.
Immunohistochemistry.
Tissue slices were blocked with 100% Blocking One solution (Nacalai Tesque) prior to staining with α-PECAM-1 (rat anti-mouse, SantaCruz) (700 ng/mL diluted in PBS/0.1% Tween) (16 h, 4 °C). Following washing with PBS/0.1% Tween, tissue was stained with Alexa 555 goat anti-rat (1:300 dilution, 2 h, room temperature). Cell nuclei were visualized by saturating the tissue with Prolong Gold DAPI mounting medium (Invitrogen) before covering with a glass coverslip for confocal microscopy visualization (LSM880 Carl Zeiss microscope).
Minimal staining was seen when the primary antibody was omitted (SI Appendix, Fig. S9), indicating that Alexa 555 goat anti-rat secondary antibody recognized the α-PECAM-1 antibody employed for immunostaining and not the biotin–α-PECAM-1 antibody previously administered as a targeting ligand.
Statistics.
Statistical analysis was carried out through linear and nonlinear (one-phase decay) regression analysis, one-way ANOVA (with Tukey’s post hoc tests), and t tests (unpaired, two-way) for indicated experiments with the use of GraphPad Prism software.
Supplementary Material
Acknowledgments
The TEM work was conducted at Advanced Characterization Nanotechnology Platform of the University of Tokyo, supported by “Nanotechnology Platform” of the Ministry of Education, Culture, Sports, Science and Technology. We thank Dr. Saed Abbasi (Innovation Center of NanoMedicine, Japan) for his comments on how to improve the quality of the manuscript. This work was supported by a Japan Society for the Promotion of Science “Early Career Scientist” Grant 18K14100 (to D.G.-C.) and the Japan Science and Technology Agency Center of Innovation program.
Footnotes
Competing interest statement: D.G.-C. and K.K. hold a pending patent on the developed brain-targeting strategy.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002016117/-/DCSupplemental.
Data Availability.
Full experimental data sets may be found in the public repository Open Science Framework through https://osf.io/N39XK/.
References
- 1.Palmer A. M., The role of the blood brain barrier in neurodegenerative disorders and their treatment. J. Alzheimers Dis. 24, 643–656 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Abbott N. J., Patabendige A. A., Dolman D. E., Yusof S. R., Begley D. J., Structure and function of the blood-brain barrier. Neurobiol. Dis. 37, 13–25 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Tian X. et al., LRP-1-mediated intracellular antibody delivery to the central nervous system. Sci. Rep. 5, 11990 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnsen K. B. et al., Targeting transferrin receptors at the blood-brain barrier improves the uptake of immunoliposomes and subsequent cargo transport into the brain parenchyma. Sci. Rep. 7, 10396 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anraku Y. et al., Glycaemic control boosts glucosylated nanocarrier crossing the BBB into the brain. Nat. Commun. 8, 1001 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gonzalez-Carter D. A. et al., L-DOPA functionalized, multi-branched gold nanoparticles as brain-targeted nano-vehicles. Nanomedicine 15, 1–11 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Kim J., Wessling-Resnick M., The role of iron metabolism in lung inflammation and injury. J. Allergy Ther. 3 (suppl. 4), 4 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin S. et al., A role for low density lipoprotein receptor-related protein 1 in the cellular uptake of tissue plasminogen activator in the lungs. Pharm. Res. 33, 72–82 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farrell C. L., Yang J., Pardridge W. M., GLUT-1 glucose transporter is present within apical and basolateral membranes of brain epithelial interfaces and in microvascular endothelia with and without tight junctions. J. Histochem. Cytochem. 40, 193–199 (1992). [DOI] [PubMed] [Google Scholar]
- 10.Fraga S., Pinho M. J., Soares-da-Silva P., Expression of LAT1 and LAT2 amino acid transporters in human and rat intestinal epithelial cells. Amino Acids 29, 229–233 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Lam F. C. et al., Enhanced efficacy of combined temozolomide and bromodomain inhibitor therapy for gliomas using targeted nanoparticles. Nat. Commun. 9, 1991 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu L. P. et al., Crossing the blood-brain-barrier with nanoligand drug carriers self-assembled from a phage display peptide. Nat. Commun. 10, 4635 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sevigny J. et al., The antibody aducanumab reduces Aβ plaques in Alzheimer’s disease. Nature 537, 50–56 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Nutt J. G. et al.; ICV GDNF Study Group. Implanted intracerebroventricular. Glial cell line-derived neurotrophic factor , Randomized, double-blind trial of glial cell line-derived neurotrophic factor (GDNF) in PD. Neurology 60, 69–73 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Andreone B. J. et al., Blood-brain barrier permeability is regulated by lipid transport-dependent suppression of caveolae-mediated transcytosis. Neuron 94, 581–594.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ben-Zvi A. et al., Mfsd2a is critical for the formation and function of the blood-brain barrier. Nature 509, 507–511 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hawkins B. T., Davis T. P., The blood-brain barrier/neurovascular unit in health and disease. Pharmacol. Rev. 57, 173–185 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Sabbagh M. F., Nathans J., A genome-wide view of the de-differentiation of central nervous system endothelial cells in culture. eLife 9, e51276 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang F. H. F. et al., A ligand motif enables differential vascular targeting of endothelial junctions between brain and retina. Proc. Natl. Acad. Sci. U.S.A. 116, 2300–2305 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vanlandewijck M. et al., A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475–480 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Eppihimer M. J. et al., Differential expression of platelet-endothelial cell adhesion molecule-1 (PECAM-1) in murine tissues. Microcirculation 5, 179–188 (1998). [PubMed] [Google Scholar]
- 22.Parhiz H. et al., PECAM-1 directed re-targeting of exogenous mRNA providing two orders of magnitude enhancement of vascular delivery and expression in lungs independent of apolipoprotein E-mediated uptake. J. Control. Release 291, 106–115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dan M., Cochran D. B., Yokel R. A., Dziubla T. D., Binding, transcytosis and biodistribution of anti-PECAM-1 iron oxide nanoparticles for brain-targeted delivery. PLoS One 8, e81051 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mamdouh Z., Chen X., Pierini L. M., Maxfield F. R., Muller W. A., Targeted recycling of PECAM from endothelial surface-connected compartments during diapedesis. Nature 421, 748–753 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Goldberger A. et al., Biosynthesis and processing of the cell adhesion molecule PECAM-1 includes production of a soluble form. J. Biol. Chem. 269, 17183–17191 (1994). [PubMed] [Google Scholar]
- 26.Noel J. et al., PECAM-1 and caveolae form the mechanosensing complex necessary for NOX2 activation and angiogenic signaling with stopped flow in pulmonary endothelium. Am. J. Physiol. Lung Cell. Mol. Physiol. 305, L805–L818 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bakhshi F. R. et al., Nitrosation-dependent caveolin 1 phosphorylation, ubiquitination, and degradation and its association with idiopathic pulmonary arterial hypertension. Pulm. Circ. 3, 816–830 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kronstein R. et al., Caveolin-1 opens endothelial cell junctions by targeting catenins. Cardiovasc. Res. 93, 130–140 (2012). [DOI] [PubMed] [Google Scholar]
- 29.You L. et al., Targeted brain delivery of rabies virus glycoprotein 29-modified deferoxamine-loaded nanoparticles reverses functional deficits in Parkinsonian mice. ACS Nano 12, 4123–4139 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Li L. et al., Large amino acid transporter 1 mediated glutamate modified docetaxel-loaded liposomes for glioma targeting. Colloids Surf. B Biointerfaces 141, 260–267 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Li J. et al., Choline-derivate-modified nanoparticles for brain-targeting gene delivery. Adv. Mater. 23, 4516–4520 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Li J. et al., Choline transporter-targeting and co-delivery system for glioma therapy. Biomaterials 34, 9142–9148 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Goulatis L. I., Shusta E. V., Protein engineering approaches for regulating blood-brain barrier transcytosis. Curr. Opin. Struct. Biol. 45, 109–115 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Zhao W. et al., Nanoscale manipulation of membrane curvature for probing endocytosis in live cells. Nat. Nanotechnol. 12, 750–756 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu A. P., Aguet F., Danuser G., Schmid S. L., Local clustering of transferrin receptors promotes clathrin-coated pit initiation. J. Cell Biol. 191, 1381–1393 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muro S. et al., A novel endocytic pathway induced by clustering endothelial ICAM-1 or PECAM-1. J. Cell Sci. 116, 1599–1609 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Clark A. J., Davis M. E., Increased brain uptake of targeted nanoparticles by adding an acid-cleavable linkage between transferrin and the nanoparticle core. Proc. Natl. Acad. Sci. U.S.A. 112, 12486–12491 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiley D. T., Webster P., Gale A., Davis M. E., Transcytosis and brain uptake of transferrin-containing nanoparticles by tuning avidity to transferrin receptor. Proc. Natl. Acad. Sci. U.S.A. 110, 8662–8667 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lutton E. M. et al., Endothelial targeted strategies to combat oxidative stress: Improving outcomes in traumatic brain injury. Front. Neurol. 10, 582 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu Q. J., Tao H., Wang X., Li M. C., Targeting brain microvascular endothelial cells: A therapeutic approach to neuroprotection against stroke. Neural Regen. Res. 10, 1882–1891 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuwahara H. et al., Efficient in vivo delivery of siRNA into brain capillary endothelial cells along with endogenous lipoprotein. Mol. Ther. 19, 2213–2221 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y. H., Chang M., Davidson B. L., Molecular signatures of disease brain endothelia provide new sites for CNS-directed enzyme therapy. Nat. Med. 15, 1215–1218 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deane R. et al., RAGE mediates amyloid-beta peptide transport across the blood-brain barrier and accumulation in brain. Nat. Med. 9, 907–913 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Ramanathan A., Nelson A. R., Sagare A. P., Zlokovic B. V., Impaired vascular-mediated clearance of brain amyloid beta in Alzheimer’s disease: The role, regulation and restoration of LRP1. Front. Aging Neurosci. 7, 136 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yanagida K. et al., Size-selective opening of the blood-brain barrier by targeting endothelial sphingosine 1-phosphate receptor 1. Proc. Natl. Acad. Sci. U.S.A. 114, 4531–4536 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brewer C., Riehm J., Evidence for possible nonspecific reactions between N-ethylmaleimide and proteins. Anal. Biochem. 18, 248–255 (1967). [Google Scholar]
- 47.Azcutia V. et al., Inflammation determines the pro-adhesive properties of high extracellular d-glucose in human endothelial cells in vitro and rat microvessels in vivo. PLoS One 5, e10091 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Altannavch T., Roubalova K., Kucera P., Andel M., Effect of high glucose concentrations on expression of ELAM-1, VCAM-1 and ICAM-1 in HUVEC with and without cytokine activation. Physiol. Res. 53, 77–82 (2004). [PubMed] [Google Scholar]
- 49.Wautier J. L. et al., Advanced glycation end products (AGEs) on the surface of diabetic erythrocytes bind to the vessel wall via a specific receptor inducing oxidant stress in the vasculature: A link between surface-associated AGEs and diabetic complications. Proc. Natl. Acad. Sci. U.S.A. 91, 7742–7746 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Full experimental data sets may be found in the public repository Open Science Framework through https://osf.io/N39XK/.