Significance
Analysis of synthetic lethal relationships identified in Saccharomyces cerevisiae was used to identify candidate therapeutic targets for cancers with BRCA1 or BRCA2 defects. Inhibition of one of these targets, flap endonuclease 1 (FEN1), with small-molecule inhibitors or siRNA knockdown of FEN1 expression preferentially killed BRCA1 and BRCA2 mutant cancer cell lines as well as some additional cancer cell lines without BRCA1 or BRCA2 defects. FEN1 inhibitors also preferentially reduced the growth of mouse xenografts established from FEN1 inhibitor-sensitive cancer cell lines. These studies support the use of S. cerevisiae genetics in identifying human therapeutic targets and establish FEN1 as a potential therapeutic target for further investigation and possible development.
Keywords: synthetic lethality, homologous recombination, cancer therapy, DNA repair, DNA replication
Abstract
Synthetic lethality strategies for cancer therapy exploit cancer-specific genetic defects to identify targets that are uniquely essential to the survival of tumor cells. Here we show RAD27/FEN1, which encodes flap endonuclease 1 (FEN1), a structure-specific nuclease with roles in DNA replication and repair, and has the greatest number of synthetic lethal interactions with Saccharomyces cerevisiae genome instability genes, is a druggable target for an inhibitor-based approach to kill cancers with defects in homologous recombination (HR). The vulnerability of cancers with HR defects to FEN1 loss was validated by studies showing that small-molecule FEN1 inhibitors and FEN1 small interfering RNAs (siRNAs) selectively killed BRCA1- and BRCA2-defective human cell lines. Furthermore, the differential sensitivity to FEN1 inhibition was recapitulated in mice, where a small-molecule FEN1 inhibitor reduced the growth of tumors established from drug-sensitive but not drug-resistant cancer cell lines. FEN1 inhibition induced a DNA damage response in both sensitive and resistant cell lines; however, sensitive cell lines were unable to recover and replicate DNA even when the inhibitor was removed. Although FEN1 inhibition activated caspase to higher levels in sensitive cells, this apoptotic response occurred in p53-defective cells and cell killing was not blocked by a pan-caspase inhibitor. These results suggest that FEN1 inhibitors have the potential for therapeutically targeting HR-defective cancers such as those resulting from BRCA1 and BRCA2 mutations, and other genetic defects.
Synthetic lethality (SL) results when nonlethal mutations in different genes cause lethality when they are combined in cells (1, 2). SL is often suggested to result from the inactivation of redundant pathways; however, other mechanisms can underlie SL interactions (2, 3). Examples include combining mutations that result in increased levels of DNA damage and reduced DNA repair capacity, and combining several partial loss-of-function mutations targeting an essential multiprotein complex. The success in developing poly(adenosine diphosphate-ribose) polymerase (PARP) inhibitors for maintenance therapy of BRCA1- and BRCA2-defective breast and ovarian cancers that have homologous recombination (HR) defects has caused growing interest in exploiting SL to identify new therapeutic targets (4–6).
Due to the interest in exploiting SL as a therapeutic tool (7, 8), a number of methods have been developed for evaluating SL in mammalian cells (9, 10). Examples include screens using arrayed small interfering RNAs (siRNAs) and small hairpin RNAs (shRNAs) and, more recently, different types of CRISPR dropout screens (9, 10). SL has also been extensively studied and used as a robust genetic tool in many model organisms. In Saccharomyces cerevisiae, early studies focused on dissecting specific genetic interactions or individual pathways, such as 1) the redundancy between histone H2B subtypes (11), 2) cell-cycle regulation pathways (12, 13), and 3) DNA repair genes (14, 15). SL interactions identified in S. cerevisiae between DNA repair genes include 1) mismatch repair gene defects and mutations affecting the editing exonuclease activities of DNA polymerases (15–17); 2) RAD27 defects, which affect Okazaki fragment processing and base-excision repair (18), and RAD51 or RAD52 defects, which affect HR (14, 19, 20); and 3) SGS1 defects, which affect the S. cerevisiae homolog of the BLM helicase, and multiple DNA repair and DNA damage response genes (21, 22). Interestingly, genes encoding HR proteins constitute a striking hub for SL interactions (20). As a result of the extension of S. cerevisiae SL/SGD (synthetic growth defect) screen methodology to genome-wide approaches, extremely robust databases of S. cerevisiae SL–SGD interactions are available for use as genetic tools (21).
In a previous study, we used SL relationships in S. cerevisiae and other functional genomics datasets to construct a network of genes that were predicted to act in the suppression of genome rearrangements (23, 24). Genetic screens based on these network predictions and validation studies identified 266 genome instability-suppressing (GIS) genes and an additional 38 candidate GIS genes, which then implicated their corresponding human homologs and pathway genes as candidate human GIS genes (24, 25). Analysis of The Cancer Genome Atlas data has suggested that the human GIS genes are frequently defective in cancers that exhibit genome instability (24, 25). In the present study, we have performed experiments to determine if S. cerevisiae SL networks can predict possible therapeutic targets for cancers with defects in GIS genes, initially focusing on cancers with HR defects caused by BRCA1 and BRCA2 defects, thereby identifying the nuclease Rad27/FEN1 as an attractive candidate therapeutic target.
Results
Identification of RAD27/FEN1 as a GIS Gene Synthetic Lethal Target.
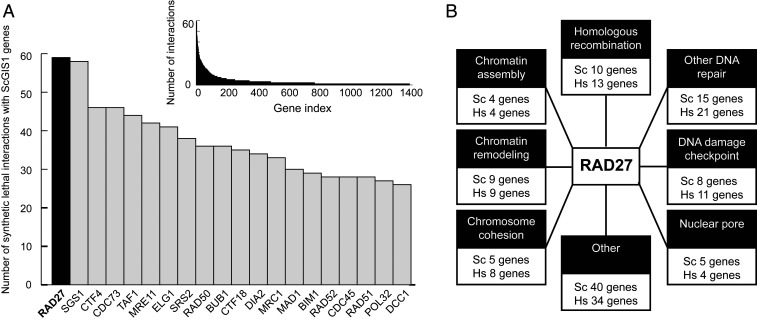
Evaluation of known S. cerevisiae SL interactions (21) demonstrated that RAD27 had the greatest number of SL relationships with the GIS genes identified in our studies (23–25) (59 SL partners; Fig. 1A and Dataset S1). RAD27 and FEN1 encode an evolutionarily conserved endonuclease that cleaves DNA flaps for Okazaki fragment maturation during lagging-strand DNA synthesis and during long-patch base-excision repair (18). The RAD27 SL partners and their human orthologs were grouped into eight functional groups including 1) HR/double-strand break (DSB) repair, 2) other DNA repair pathways, 3) DNA damage checkpoint, 4) chromatin assembly, 5) chromatin remodeling, 6) chromosome cohesion, 7) nuclear pore, and 8) others (Fig. 1B). The biochemical mechanisms underlying each of these SL effects caused by the combined losses of function are mostly unknown; however, genetic evidence suggests that increased formation of DSBs during DNA replication in RAD27-mutant cells is likely to cause dependency on HR/DSB repair, checkpoint activation, and chromatin remodeling to protect the genome (14, 19, 20, 26–29). In contrast to RAD27, the RAD27 family members showed far fewer SL interactions with GIS genes [BioGRID database version 3.5.168 (21); Dataset S1]: EXO1 ranked 200th on the GIS gene SL list (five SL partners: DNA2, POL3, RAD5, RAD27, RFA1), YEN1 (human GEN1) ranked 680th on the GIS gene SL list (two SL partners: MMS4, DNA2), and RAD2 (human XPG) had no SL interactions with GIS genes, which include most genes that function in HR, DNA repair, DNA damage response, or other DNA metabolism pathways (3, 23–25).
Fig. 1.
RAD27 shares the greatest number of known SL interactions with S. cerevisiae GIS genes. (A) The number of reported SL interactions of genes with the 266 S. cerevisiae GIS and eGIS1 genes. (A, Inset) The entire distribution for all interactors. (B) RAD27 SL interactors (Sc genes) fall into common pathways, which are often conserved in humans (Hs genes).
FEN1 Inhibitors Selectively Kill BRCA1/BRCA2-Defective Cell Lines.
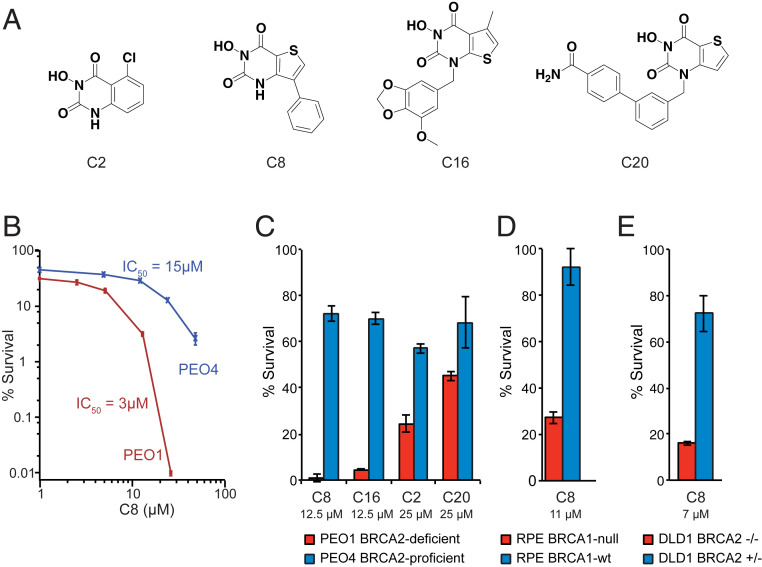
To determine if the SL relationships between RAD27 and HR defects are conserved in human cells, we synthesized four previously reported N-hydroxy pyrimidinedione-based FEN1 inhibitors (30) (C2, C8, C16, and C20; Fig. 2A), which also likely have some activity on FEN1 family members such as XPG, EXO1, and GEN1 (30, 31), and tested their ability to kill PEO1 and PEO4 ovarian cancer cells as assessed by clonogenic survival assay. PEO1 is a BRCA2-defective cell line derived from an ovarian tumor, and PEO4 is derived from a cisplatin-resistant relapse tumor from the same patient, in which the initial BRCA2 mutation was reverted (32, 33). We found that the BRCA2-mutant PEO1 cells were ∼5-fold more sensitive to killing by the FEN1 inhibitor C8 than the BRCA2-revertant PEO4 cells (Fig. 2B). Compared with PEO4 cells, PEO1 cells were also more sensitive to killing by the other three FEN1 inhibitors, with the relative sensitivity C8 > C16 >> C2, C20 at 12.5 μM (C8, C16) and 25 μM (C2, C20) (Fig. 2C). We used C8 in all subsequent experiments because, in comparison with the next most potent inhibitor (C16), C8 had a greater half-life in plasma after intraperitoneal (i.p.) administration at 5 mg/kg in mice (1.6 h half-life, maximum concentration (Cmax) 16 μM vs. no measurable exposure for C16; SI Appendix, Fig. S1). In addition, when C8 was administered daily to mice i.p. for 7 d, the concentration of C8 in plasma was essentially identical on days 1 and 7 (concentrations were determined 4 h after administration) and was comparable to the levels observed in the initial exposure experiment (SI Appendix, Fig. S1); furthermore, the plasma levels observed were proportional to the dose administered, indicating that higher effective concentrations of C8 are achieved at doses of 20 and 40 mg/kg than at 5 mg/kg. There was no evidence of weight loss or gross toxicity during this experiment, supporting the use of C8 as an in vivo tool compound.
Fig. 2.
BRCA-deficient cancer cell lines are sensitive to FEN1 inhibition. (A) Structures of the synthesized FEN1 inhibitors. (B) Clonogenic survival of BRCA2-deficient PEO1 and BRCA2-revertant PEO4 cells demonstrates that PEO1 cells are more sensitive to a 3-d exposure to the C8 FEN1 inhibitor. (C) Clonogenic survival demonstrates that PEO1 cells are more sensitive than PEO4 cells to a 3-d exposure to all of the FEN1 inhibitors, with C8 and C16 causing the greatest reduction in percent survival. C8 and C16 were tested at 12.5 μM and C2 and C20 were tested at 25 μM. (D) RPE1-hTERT p53−/− BRCA1-KO cells are more sensitive to a 3-d exposure to 11 μM C8 than the parental BRCA1–wild-type (wt) cells. (E) DLD1 BRCA2−/− cells are more sensitive to a 3-d exposure to 7 μM C8 than the parental BRCA2-proficient cells. Error bars represent the standard deviation.
Two additional pairs of matched cell lines were examined for their sensitivity to C8: 1) hTERT-immortalized and p53-null retinal pigmental epithelial (RPE) cells and a derivative in which BRCA1 was inactivated with CRISPR (34); and 2) DLD1 colorectal tumor cells and a derivative in which the wild-type copy of BRCA2 was inactivated by gene disruption (purchased from Horizon Discovery). For each pair, the BRCA-deficient derivative was more sensitive to killing by C8 than the matched BRCA-proficient parental cell line (Fig. 2 D and E).
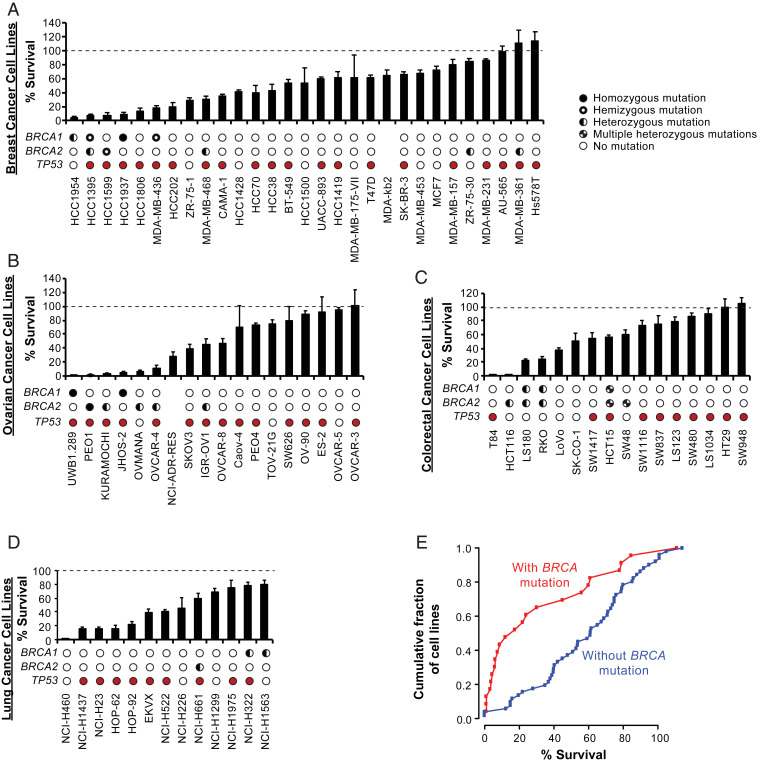
The ability of C8 to kill a panel of breast (Fig. 3A), ovarian (Fig. 3B), colorectal (Fig. 3C), and lung (Fig. 3D) cancer cell lines was then assessed in clonogenic survival assays after treating cells with C8 for 3 d at 12.5 μM, which was fourfold higher than or close to the half-maximal inhibitory concentration (IC50) values for PEO1 and PEO4 cells, respectively (Fig. 2B). For cell lines derived from each tumor type, the response to C8 treatment ranged from highly sensitive to completely resistant (Fig. 3). When the data for all of the cancer cell lines tested were combined, those with reported BRCA mutations tended to be more sensitive to C8 treatment than those without reported BRCA mutations (Fig. 3E; P = 0.0015, Kolmogorov–Smirnov test). The sensitive cell lines included breast and ovarian cancer cell lines with mutations in BRCA1 [HCC1954 (35), MDA-MB-436 (36), UWB1.289 (37), JHOS-2 (38), and the olaparib-resistant BRCA1-mutant cell line HCC1937 (36, 39–42)] or BRCA2 [HCC1395 (43), PEO1 (32), Kuramochi (44), Ovmana (44), IGR-OV1 (38), OVCAR-4 (38), and the olaparib-resistant BRCA2-mutant cell line HCC1599 (38, 42)]. Despite this association, we found that the reported BRCA mutation status was not always predictive of sensitivity to killing by C8. BRCA-mutant cell lines without C8 sensitivity typically contained heterozygous mutations and/or missense mutations of unknown significance [e.g., ZR-75-30 (36, 38), MDA-MB-361 (36, 38)], or they may contain mutations that do not substantially affect HR in these cell lines. On the other hand, BRCA–wild-type cell lines with C8 sensitivity may contain other genetic or epigenetic defects that are synthetic lethal with the loss of FEN1. Among these latter C8-sensitive cell lines were the breast cancer cell lines HCC1806, which is sensitive to some PARP inhibitors (45), and MDA-MB-468, which is heterozygous for an unverified BRCA2 missense mutation and is weakly sensitive to killing by C8 and PARP inhibitors (45).
Fig. 3.
Sensitivity of breast, ovarian, colorectal, and lung cancer cell lines to the C8 FEN1 inhibitor. (A–D) Clonogenic survival of breast cancer cell lines (A), ovarian cancer cell lines (B), colorectal cancer cell lines (C), and lung cancer cell lines (D) treated with 12.5 μM C8 for 3 d are displayed as bars. The BRCA1 and BRCA2 mutation status information is from the Broad Institute Cancer Cell Line Encyclopedia (CCLE) (38); zygosity was inferred from mutant vs. wild-type read counts and copy-number information. Homozygous mutations are displayed as solid black circles, hemizygous mutations are displayed as black circles with a central white spot, heterozygous mutations are displayed as half-filled circles, multiple heterozygous mutations are displayed as split filled circles, and no mutations are displayed as empty circles. Cell lines with TP53 mutations are shown with filled red circles and those without TP53 mutations are shown with empty circles. Cell lines with no data in the CCLE are shown without circles. (E) Cumulative percent survival of cell lines with and without BRCA mutations after 12.5 μM C8 for 3 d; P = 0.0015, Kolmogorov–Smirnov test. Error bars represent the standard deviation.
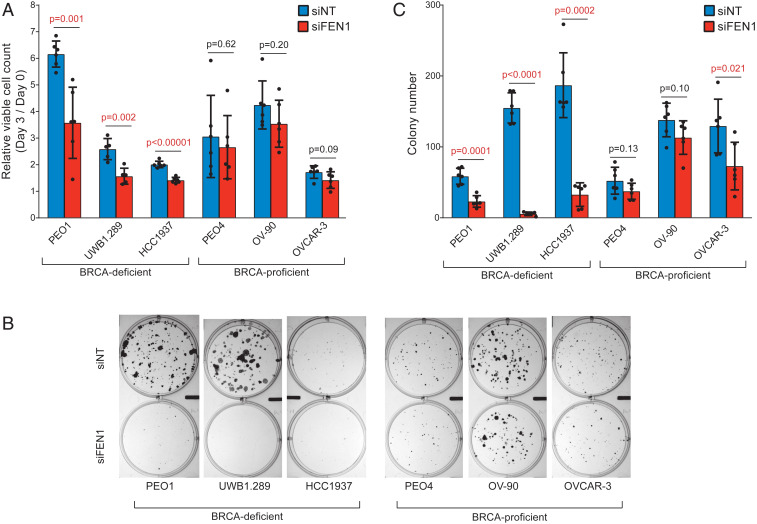
To confirm that the effect of C8 treatment on BRCA-deficient cell lines was due to FEN1 inhibition, siRNA-mediated knockdown of FEN1 was examined in two BRCA1-deficient (UWB1.289, HCC1937), one BRCA2-deficient (PEO1), and three BRCA-proficient (PEO4, OV-90, OVCAR-3) cancer cell lines. Three days after addition of a pool of four FEN1-targeting siRNAs (siFEN1s) or a pool of four nontargeting control siRNAs (siNTs), siFEN1 treatment caused FEN1 protein levels to be reduced by 50 to 80% in both BRCA-deficient and BRCA-proficient cell lines (SI Appendix, Fig. S2) and the number of viable trypan blue-excluding cells to be significantly reduced for the BRCA-deficient cell lines but not for the BRCA-proficient cell lines (Fig. 4A). To more directly compare the effect of siFEN1 with that of C8 treatment on clonogenic survival, six cultures of each cell line were transfected with siFEN1 or siNT, propagated with media changed every 3 d for 15 to 21 d, and then fixed and stained for the measurement of colony numbers (Fig. 4 B and C) and colony sizes (SI Appendix, Fig. S3). In comparison with the siNT control, transfection with siFEN1 resulted in a significant decrease in colony numbers for the BRCA-deficient cell lines (PEO1, UWB1.289, HCC1937), whereas only one (OVCAR-3) of the three BRCA-proficient cell lines (PEO4, OV-90, OVCAR-3) showed a small decrease in colony number that was of borderline significance (P = 0.021) (Fig. 4C). Of the colonies that survived siFEN1, we found a reproducible reduction of colony size among BRCA-defective cell lines but little if any decrease in colony size in BRCA-proficient cell lines (SI Appendix, Fig. S3). The short-term effect of siFEN1 on cell viability (determined at day 3 after transfection; Fig. 4A) was not as large as that seen in the longer-term clonogenic survival assay (Fig. 4B), suggesting that many of the siFEN1-transfected BRCA-deficient cells that were viable after 3 d either died or could not undergo additional cell divisions over time.
Fig. 4.
Treatment of BRCA-deficient cell lines with siFEN1 causes increased cell death and reduced clonogenic survival. (A) The number of viable cells is reduced by 3 d of siFEN1 treatment relative to 3 d of control siNT treatment for BRCA-deficient cell lines but not BRCA-proficient cell lines (n = 6, except for UWB1.289, where n = 5). P values were calculated using the two-tailed t test. (B) Representative images of the clonogenic survival assay of BRCA-deficient and BRCA-proficient cell lines treated with siNT or siFEN1. (C) The number of colonies in the clonogenic survival assay 15 to 21 d posttransfection was reduced upon siFEN1 treatment relative to siNT treatment in BRCA-deficient cell lines and OVCAR-3 but not PEO4 or OV-90 (n = 6). P values were calculated using the two-tailed t test. Error bars represent the standard deviation.
FEN1 Inhibition Blocks DNA Replication and Induces DNA Damage.
To determine the effect of FEN1 inhibition on DNA replication, cells were treated with 25 μM C8 for 3 d and then given a 45-min pulse of BrdU (5-bromo-2′-deoxyuridine), followed by FACS (fluorescence-activated cell sorting) analysis of DNA content and BrdU incorporation. C8 treatment of the BRCA-defective PEO1, UWB1.289, and HCC1937 cells reduced BrdU incorporation by varying degrees with a concomitant increase in the G2/M fraction. In PEO1 and HCC1937 cells, C8 increased the sub-G1 but not the G1 fraction, whereas with UWB1.289 cells, C8 significantly increased the G1 fraction but not the sub-G1 fraction (Fig. 5). The substantial inhibition of BrdU incorporation by C8 was also observed with other C8-sensitive cell lines, with and without BRCA defects, including HCT116, Ovmana, Kuramochi, NCI-H460, OVCAR-4, and JHOS-2 (SI Appendix, Fig. S4). In contrast, C8 treatment of the BRCA-proficient and C8-resistant PEO4, MDA-MB-231, OVCAR-3, and MCF7 cells either had no effect or a much lower effect on BrdU incorporation, and the G2/M or the sub-G1 fractions, when compared with vehicle-treated cells (Fig. 5 and SI Appendix, Fig. S4). These results suggest that FEN1 inhibitor-sensitive cell lines have reduced capacity for DNA replication and undergo cell-cycle arrest in G1 and/or G2, and possibly fragment their DNA.
Fig. 5.
Treatment of BRCA-deficient cell lines with C8 inhibits DNA replication. FACS profiles of total DNA (x axis) and incorporated BrdU (y axis) of cells treated with 25 μM C8 for 3 d and then pulse-labeled with BrdU for 45 min. Percentages of the number of cells in different phases of the cell cycle are shown. Rectangles depict the gates used to determine percentages.
To determine if the inhibition of DNA replication requires continuous treatment with C8, we tested the ability of BRCA2-mutant PEO1 cells and BRCA2-revertant PEO4 cells to recover BrdU incorporation after the removal of C8. In this experiment, cells were treated with 25 μM C8 for 3 d followed by incubation in either C8-free or C8-containing medium for an additional 3 d; the ability of cells to incorporate BrdU was analyzed by FACS at each step (SI Appendix, Fig. S5). In the BRCA2-proficient PEO4 cells, C8 treatment for 3 d reduced BrdU incorporation by 30% but, after drug removal, BrdU incorporation returned to levels found in untreated cultures (SI Appendix, Fig. S5). With the matched BRCA2-deficient PEO1 cells, C8 treatment for 3 d reduced BrdU incorporation by 30 to 50% (Fig. 5 and SI Appendix, Fig. S5), and this negative effect on DNA synthesis not only persisted but also increased to 90% inhibition at day 3 after drug removal (SI Appendix, Fig. S5). Moreover, the sub-G1 fraction also continued to increase in the C8-treated PEO1 cultures even after C8 removal (SI Appendix, Fig. S5). These results showed that FEN1 inhibition transiently interfered with DNA synthesis in BRCA-proficient cells but permanently disrupted the capacity of BRCA-deficient cells and other C8-sensitive cells to replicate their DNA even after drug removal.
Studies in S. cerevisiae have suggested that loss of Rad27 can cause increased levels of DSBs during DNA replication (14, 19, 20, 26–29); therefore, we measured the effect of C8 on the levels of the DNA damage markers γH2AX and 53BP1 foci (46) in PEO1 and PEO4 cells. In both cell lines, C8 treatment caused time-dependent increases in the levels of histone γH2AX (SI Appendix, Fig. S6A), in the number of cells with >25 histone γH2AX foci per nucleus (SI Appendix, Fig. S6 B and C), and in the number of cells with >25 53BP1 foci per nucleus (SI Appendix, Fig. S6 D and E) relative to vehicle-treated controls. PEO1 cells, however, had more pronounced increases of these DNA damage markers after C8 treatment than PEO4 cells (SI Appendix, Fig. S6). Induction of the DNA damage response including accumulation of histone γH2AX has been observed with other, related FEN1 inhibitors (31, 47). These results suggest that FEN1 inhibition results in increased DNA damage including DSBs in both BRCA-proficient and BRCA-deficient cells; the higher levels of 53BP1 foci in PEO1 cells compared with PEO4 cells are consistent with reduced HR in PEO1 cells lacking functional BRCA2.
Because the majority of the C8-sensitive cancer cell lines are p53-defective (Fig. 3), C8-induced DNA damage is likely to activate p53-independent death pathways (48). We found that C8 treatment led to a time- and concentration-dependent increase in DEVDase activity, which was greater in PEO1 cells relative to PEO4 cells (SI Appendix, Fig. S7A). Addition of the pan-caspase inhibitor ZVAD-fmk (49) reduced the DEVDase activity induced by either staurosporine [a known activator of apoptosis (50)] or by C8 in PEO1 cells (SI Appendix, Fig. S7B). However, ZVAD-fmk did not promote the clonogenic survival of C8-treated PEO1 or PEO4 cells (SI Appendix, Fig. S7C). Together, these results suggest that the replication defects and DNA damage caused by FEN1 inhibition can activate caspases, but the loss of viability still occurs in the absence of caspase activity.
Differential Sensitivity to C8 Is Recapitulated in Mice.
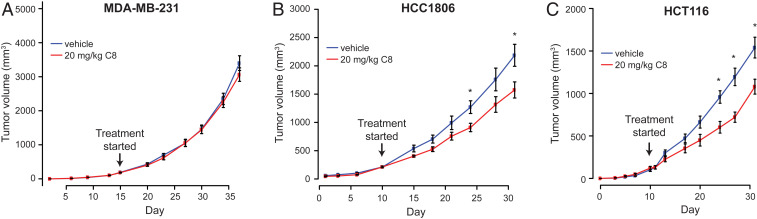
Because of its pharmacokinetic profile, C8 can be dosed in mice such that its plasma levels exceed the IC50 for tumor cell lines that are sensitive to killing by C8 (SI Appendix, Fig. S1); these pharmacokinetic data indicate that at a dose of 20 mg/kg, the plasma concentration of C8 should exceed the IC50 for C8-sensitive tumor cell lines for 4.5 to 6 h. Therefore, its tumor-inhibitory activity was evaluated for three cell lines in an immune-compromised mouse xenograft model (Fig. 6). Because PEO1 and the other BRCA-deficient cancer cell lines characterized in the cell-based studies do not form tumors in vivo, xenografts were established for two tumor cell lines that were sensitive to C8 (HCC1806 and HCT116) (Fig. 3) and one cell line that was relatively resistant to C8 (MDA-MB-231) (Fig. 3). The mice were administered vehicle or C8 at 20 mg/kg i.p. every 12 h, and the volume of the tumors was monitored with time. In the case of the C8-resistant MDA-MB-231 breast cancer cell line, tumor growth was similar in mice treated with C8 or vehicle (Fig. 6A). With the C8-sensitive HCC1806 and HCT116 cells, a significant reduction in tumor growth was observed in mice treated with C8 when compared with those treated with vehicle (Fig. 6 B and C). Although detailed evaluation of the in vivo efficacy of FEN1 inhibition will require the development of more potent and more stable FEN1 inhibitors, these results showed that the FEN1 inhibitor C8 can reach the site of tumors in mice and inhibit their growth in a manner that parallels that seen after treatment of cell lines in vitro.
Fig. 6.
C8 reduces growth rates of C8-sensitive but not C8-resistant cancer cell lines in mouse tumor models. Tumor volumes in mouse xenograft models established using C8-resistant cancer cells (MDA-MB-231) (A) and C8-sensitive cancer cells (HCC1806 and HCT116) (B and C). Compared with vehicle, twice-daily treatment with 20 mg/kg C8 significantly reduced tumor volumes in the HCC1806 (P = 0.03) and HCT116 (P = 0.02) mouse models but showed no effect in the MDA-MB-231 mouse model. Mean tumor volumes for each group are plotted, and error bars represent the SEM. An asterisk indicates tumor volume distributions that are significantly different with P < 0.05.
Discussion
There has been a growing interest in using SL to identify therapeutic targets for cancers with specific genetic defects. RNA interference, shRNA, and CRISPR have been used to perform SL screens in mammalian cells; however, due to methodological problems including off-target effects, context dependency, and poor reproducibility and because the readout of these methods is reduced growth rates, it has been difficult to identify strong SL interactions that are clinically actionable with high confidence (9, 10). PARP inhibitors that were developed for the treatment of cancers with DSB repair defects, such as BRCA1- and BRCA2-defective cancers, have been considered an example of applied SL (4, 51–53); however, recent studies have suggested that these inhibitors act through production of a trapped PARP1 complex on DNA and thus may be acting via a gain-of-function mechanism (34, 54). Moreover, acquired resistance to PARP inhibitors is a significant problem in the clinic (55–61), indicating the need for additional inhibitors for treatment of BRCA1- and BRCA2-defective cancers and cancers with HR defects. In the present study, we used validated SL relationships in S. cerevisiae to identify therapeutic targets for BRCA1- and BRCA2-defective cancers. Our bioinformatic analysis identified a number of such SL genes, of which RAD27/FEN1 was particularly specific for genes associated with DNA repair, including HR and chromatin metabolism. Consistent with this, our studies using FEN1 inhibitors and siRNA knockdown have demonstrated that FEN1 defects are SL with BRCA1 and BRCA2 defects; other studies have used CRISPR/shRNA dropout screens (62) or siRNA/shRNA knockdown in conjunction with FEN1 inhibitors (47) to provide evidence that FEN1 has SL relationships with BRCA2, ATM, MRE11A, FANCD2, and UBC13.
Using a pair of matched BRCA2-mutant (PEO1) and BRCA2-revertant (PEO4) ovarian cancer cell lines derived from tumors from the same patient (32), we found that four previously described FEN1 inhibitors selectively killed the BRCA2-mutant PEO1 cells in clonogenic survival assays relative to BRCA2-revertant PEO4 cells. However, the inhibitors exhibited a broad range of potencies in this cell-based assay, which could reflect differences in target interaction in cells or uptake/stability. The most potent of the inhibitors, C8, selectively killed a BRCA1-knockout (KO) mutant RPE clone and a BRCA2-knockout mutant DLD1 clone compared with BRCA-proficient matched parental cell lines, indicating that the sensitivity to killing observed was due to the BRCA1 and BRCA2 defects and not to some other property of the cancer cell lines studied. Analysis of different cancer cell line panels (breast, ovarian, colorectal, and lung) identified cell lines ranging from sensitive to resistant to killing by C8; among the sensitive cell lines were the BRCA1- and BRCA2-mutant cell lines, including an olaparib-resistant BRCA1-mutant line, HCC1937 (39–42), and an olaparib-resistant BRCA2-mutant line, HCC1599 (38, 42), and several cell lines that were wild-type for BRCA1 and BRCA2 or contained heterozygous BRCA1 or BRCA2 mutations. Finally, experiments with a subset of the BRCA1- or BRCA2-mutant cell lines and BRCA-proficient C8-resistant cell lines demonstrated that BRCA1- and BRCA2-mutant cell lines but not C8-resistant cell lines were sensitive to FEN1 siRNA knockdown. In aggregate, these results indicate that BRCA1 and BRCA2 mutants are sensitive to killing by FEN1 inhibition and that genetic or epigenetic alterations that result in resistance of BRCA mutants to PARP inhibitors do not necessarily cause resistance to FEN1 inhibition. Further analysis of which mechanisms underlying PARP inhibitor resistance cause or do not cause FEN1 inhibitor resistance would be particularly relevant for establishing the possible clinical utility of FEN1 inhibitors. Finally, given that several particularly C8-sensitive cell lines, most notably HCT116, T84, and NCI-H460, were wild-type for BRCA1 and BRCA2 or only contained a heterozygous BRCA2 mutation, our results also indicate that defects other than those in BRCA1 and BRCA2 can cause sensitivity to FEN1 inhibition in human cells. Moreover, since HCT116 is an HR-proficient cell line (63, 64), these results suggest that some of these other defects affect pathways other than HR. This result, which is predicted from S. cerevisiae SL networks, indicates that it would be interesting to establish the identity of the other defects that cause FEN1-inhibitor sensitivity in human cells. It should be noted that the FEN1 inhibitor we used in the present studies could have activity on FEN1 family members such as XPG (scRAD2), EXO1, and GEN1 (scYEN1) (30, 31), although the S. cerevisiae homologs of these genes have few (EXO1, YEN1) or no (RAD2) SL relationships with genes encoding components of HR, DSB repair, or the DNA damage response [BioGRID database version 3.5.168 (21); Dataset S1].
RAD27/FEN1 is a nonessential gene in S. cerevisiae (14). FEN1 is required for embryonic development in mice (65), whereas FEN1 mutations that eliminate some but not all of the FEN1 nuclease activities are tolerated in mice (66). In contrast, FEN1 is nonessential and can be deleted in chicken DT40 cells (67). Our results clearly show that DNA damage is induced upon FEN1 inhibition in human cells, as evidenced by the accumulation of histone γH2AX and 53BP1 foci, regardless of BRCA1 and BRCA2 status, and also suggest that FEN1 inhibition activates S-phase checkpoint pathways. However, many cell lines appear to be resistant to killing by FEN1 inhibitors and can recover once FEN1 inhibition ends. In contrast, BRCA1- and BRCA2-mutant cell lines as well as other C8-sensitive cell lines cannot recover from FEN1 inhibition; this lack of recovery correlates with an inability to properly replicate DNA after FEN1 inhibition. Rad27/FEN1 plays multiple roles in DNA metabolism, most notably by cleaving single-stranded flaps that occur during Okazaki fragment processing in DNA replication and during long-patch base-excision repair (18). Studies in S. cerevisiae have led to the proposal that in the absence of Rad27/FEN1, long single-stranded flaps accumulate during DNA replication and are processed to DSBs, which are either repaired by HR or misrepaired to yield mutations and chromosomal rearrangements (14, 19, 20, 26–29); the flaps could also result in stalled replication forks whose repair/restart requires HR (68). These mechanisms are consistent with the observed SL relationships between RAD27 and genes encoding components of HR and the DNA damage checkpoints in S. cerevisiae. These mechanisms can also explain the observed sensitivity of HR-defective BRCA1- and BRCA2-mutant cells to FEN1 inhibition and would explain the inability of these cells to recover the ability to replicate their DNA after the removal of FEN1 inhibitors.
FEN1 is an abundant protein in human cells. U2OS cells contain 1.4 × 105 FEN1 molecules per cell, suggesting the nuclear concentration of FEN1 is on the order of 1 µM; this is similar to the abundance of PARP1 (69). The experiments performed here utilized inhibitor concentrations that were above the IC50 for FEN1 and on the order of 10-fold higher than the concentration of FEN1; however, these conditions do not guarantee that FEN1 was fully inhibited under these experimental conditions. In this regard, siRNA knockdown of FEN1 was not complete yet was lethal to BRCA1/2-mutant cancer cell lines in our studies. One possible explanation for these effects is that cells require extremely high levels of FEN1 activity and that it is not necessary to inactivate all of the FEN1 to cause a phenotype. For example, given the small size of an Okazaki fragment (18) and the large size of the human diploid genome, there could be as many as 108 Okazaki fragments that require processing by FEN1 during each S phase. As a consequence, only partially perturbing FEN1 could cause replication defects, induction of DNA damage, and activation of cell-cycle checkpoints, all of which could contribute to cell death in BRCA1/2-mutant cancer cell lines seen in our studies. In addition, FEN1 interacts with other replication proteins during Okazaki fragment processing and similarly partially perturbing the FEN1 pool may disrupt lagging-strand replication protein complexes resulting in the same types of defects. It is also possible that FEN1 inhibitors could result in trapped FEN1–substrate complexes, not unlike that seen with PARP inhibitors (34, 54), although this would not account for the effects of siRNA knockdown of FEN1. Further studies will be required to better understand the mechanisms underlying the effects of FEN1 inhibition and depletion.
Together, these results suggest that FEN1 inhibitors could be possible therapeutic agents for treating cancers that are defective for HR and other DNA repair and DNA damage checkpoint pathways. The compounds tested here are unlikely to have clinical utility due to their low potencies and pharmacokinetic profiles. However, more suitable compounds could be developed using our cell-based assays for evaluating FEN1 inhibitors. The observation that normal cells can recover from FEN1 inhibition, whereas sensitive cells such as BRCA1 and BRCA2 mutants cannot, suggests that cycles of inhibitor treatment and recovery might be an effective approach for the use of FEN1 inhibitors.
Materials and Methods
Identification of the S. cerevisiae GIS Gene Interaction Network.
SL interactions, except for those that were conditional (e.g., increased sensitivity due to DNA damage agents) or involved three or more mutations (also called “genetic complex” interactions), were extracted from BioGRID database version 3.5.168 (21). The number of SL interactions of each gene with the 266 S. cerevisiae GIS gene was counted. Genes were ranked based on this count of the number of SL partners that were GIS genes (Fig. 1A and Dataset S1).
FEN1 Inhibitors.
FEN1 inhibitors were synthesized by Sundia Meditech as previously described (30). The compounds synthesized were compound 2 (called C2), 5-chloro-3-hydroxyquinazoline-2,4(1H,3H)-dione; compound 8 (called C8; laboratory inventory no. SMD154), 3-hydroxy-7-phenylthieno[3,2-d]pyrimidine-2,4(1H,3H)-dione; compound 16 (called C16), 3-hydroxy-1-((7-methoxybenzo[d][1,3]dioxol-5-yl)methyl)-5-methylthieno[2,3-d]pyrimidine-2,4(1H,3H)-dione; and compound 20 (called c20), 3′-((3-hydroxy-2,4-dioxo-3,4-dihydrothieno[3,2-d]pyrimidine-1(2H)-yl)methyl)-[1,1′-biphenyl]-4-carboxamide. Except for the mouse experiments, the compounds were dissolved in dimethyl sulfoxide (DMSO) and added directly to tissue-culture media. Formulations for use in the mouse experiments are indicated for each individual experiment.
Human Cell Lines.
RPE1-hTERT p53−/−, RPE1-hTERT p53−/− Cas9, and RPE1-hTERT p53−/− BRCA1-KO Cas9-null cells (34) were from Daniel Durocher, Lunenfeld-Tanenbaum Research Institute, Toronto, Canada. BRCA2-mutant PEO1 and BRCA2-revertant PEO4 cells (32) were obtained from Toshi Taniguchi, Fred Hutchinson Cancer Research Center, Seattle, WA. DLD1 BRCA2−/− cells (HD 105-007) were purchased from Horizon Discovery. All other cell lines were purchased from the American Type Culture Collection, National Cancer Institute (NCI), as part of the NCI-60 cell collection, RIKEN BioResource Research Center, or Japanese Health Science Research Resources Bank. Cells were propagated using the conditions and medium recommended by the source and stored in a liquid-nitrogen freezer.
Clonogenic Survival Assays.
PEO1 or PEO4 cells growing in log phase were plated in 12-well plates at 5 × 104 cells per well (n = 3) and then incubated at 37 °C and 5% CO2 in a humidified chamber. For experiments with the panels of ovarian, breast, colorectal, and lung cancer cell lines, cells growing in log phase were plated in 12-well plates at 5 × 103 cells per well (n = 3) and then incubated at 37 °C and 5% CO2 in a humidified chamber. After 24 h, the cells were treated with the indicated concentrations of FEN1 inhibitor or DMSO for an additional 3 d. The cells were then washed with 1 mL of phosphate-buffered saline (PBS) and trypsinized by the addition of 100 µL per well of 0.25% trypsin-ethylenediaminetetraacetate (EDTA) (Thermo Fisher Scientific). After incubation for 5 min at 37 °C, 900 µL of media was added per well, the cells were gently resuspended, and ∼125 and 500 cells were replated in 2 mL media per well in six-well plates. The plates were then incubated for 1 to 2 wk with medium replenishment after 3 d (2 mL per well fresh media added to existing media) and medium replacement (entire media replaced with 2 mL per well fresh media) after 6 d, with this schedule of replenishment/replacement repeated for the duration of the experiment. Resulting colonies were 1) washed with 2 mL per well PBS, 2) fixed with PBS containing 10% acetic acid and 10% methanol for 5 min, 3) washed with PBS, and 4) stained with 0.5 mL per well crystal violet (1% in methanol) for 5 min. The crystal violet solution was then aspirated, and the plates were washed by consecutive immersion in three containers of 1 L water each and dried for 24 h. Then the stained colonies containing >50 cells were counted.
To measure the clonogenic survival of the RPE1 and DLD1 matched pairs of cell lines, cells were plated at low density (900 to 3,000 per well) in six-well plates. At 24 h postseeding, cells were treated for 3 d with either DMSO or the indicated concentration of C8. On day 3 after drug addition, the media were replaced with fresh media, and the cells were incubated for a further 3 to 6 d with medium replacement every 3 d. Colonies were fixed and stained as described above. The stain was extracted using 10% acetic acid solution (1 mL per well), and the A570 of each sample was measured in duplicate using a Tecan Spark Multimode plate reader. Experiments were repeated three to eight times, and data were analyzed using GraphPad Prism.
To measure clonogenic survival after siRNA treatment, cells were seeded at low density in six-well plates and transfected after 24 h with either a 50 nM nontargeting siRNA pool or 50 nM FEN1 ON-TARGETplus SMARTpool (Dharmacon) according to the manufacturer’s instructions. At 24 h posttransfection, the transfection media were replaced with 2 mL per well fresh media, and an additional 2 mL per well of media was added 72 h posttransfection. The plates were incubated for 15 to 21 d posttransfection, with medium replenishment after 3 d (2 mL per well fresh media added to existing media) and medium replacement (entire media replaced with 2 mL per well fresh media) after 6 d, with this schedule of replenishment/replacement repeated for the duration of the experiment, and then the colonies were stained as described above.
Colony Number and Size Determination for siRNA Experiments.
Colony numbers and sizes were determined using Fiji (70). To provide a standard for calibrating Fiji, colonies were first counted by eye (n = 8 wells per cell line), regardless of colony size. To use Fiji, plates were first imaged using the AlphaImager HP System (ProteinSimple). The background for each image was then subtracted and the image was inverted. Thresholding was applied to the image to distinguish pixels belonging to colonies from those belonging to the background, and the watershed function was applied to the segmented image to separate overlapping objects; thresholding was optimized for each cell line so that the final automated colony counts reflected counts determined by eye. Colony counts and sizes were then calculated; this step was also optimized for each cell line based on size and circularity. The data were displayed using GraphPad Prism software.
Viable Cell Count Assays.
To measure viable cell counts after siRNA treatment, cells were seeded at 1x105 cells/well in 6-well plates and transfected after 24 h with either 50 nM non-targeting pool or 50 nM FEN1 ON-TARGETplus SMARTpool (Dharmacon), with fresh media added 24 h post-transfection. Viable cells were counted at the time of transfection and 3 d post-transfection. To determine the number of viable cells, cells were trypsinized and equal volumes of cell suspension and trypan blue solution (0.4% in 0.81% sodium chloride and 0.06% potassium phosphate dibasic solution, Bio-Rad) were mixed, and the samples counted using a TC20 Automated Cell Counter (Bio-Rad). Each biological replicate was counted 3 times.
BrdU Incorporation Assay.
DNA synthesis was quantified by analyzing incorporated BrdU using a BrdU Flow Kit (BD Pharmingen) according to the manufacturer’s instructions. Briefly, PEO1 or PEO4 cells (2 × 104 cells per milliliter, 5 mL per dish) were treated with or without 25 μM C8, pulse-labeled with BrdU (10 μM) for 45 min at the end of treatment, and then fixed and permeabilized. The subsequent double staining of cells with fluorescein isothiocyanate-conjugated anti-BrdU antibody and 7-aminoactinomycin D (7-AAD), which binds to total DNA, allowed the determination of BrdU incorporation at each step of the cell cycle. Stained cells were analyzed with the LSRII flow cytometer (BD Biosciences), and the results were analyzed using FlowJo software.
Immunoblotting.
Cell pellets were lysed in RIPA buffer with protease and phosphatase inhibitor mixtures (Cell Signaling Technology), sonicated with five pulses at 50% duty cycle (Ultrasonics), and cleared by centrifugation at 17,900 × g for 10 min at 4 °C, and the protein concentration was quantified using a modified Lowry protein assay (Bio-Rad; DC Protein Assay). Total cell lysates were separated by sodium dodecyl sulfate/polyacrylamide gel electrophoresis on 4 to 15% gradient gels (Bio-Rad; TGX) and transferred to polyvinylidene fluoride membranes (Bio-Rad) using a Trans-Blot Turbo System (Bio-Rad). Blots were incubated with primary antibodies against FEN1 (Abcam; ab462; 1:1,000), phosphoSer139-histone H2A.X (Cell Signaling Technology; 2577; 1:1,000), or GAPDH (Cell Signaling Technology; 2118S; 1:1,000), following the manufacturer’s instructions, washed in 1× TBST (Tris-buffered saline with 0.1% Tween 20), and incubated with horseradish peroxidase (HRP)-conjugated rabbit or mouse secondary antibodies (Cell Signaling Technology). Blots were washed in 1× TBST, developed using chemiluminescent HRP substrates (Thermo Fisher Scientific), and detected using a ChemiDoc MP imager (Bio-Rad).
Immunofluorescence Microscopy.
PEO1 and PEO4 cells (1 × 105 cells per well in six-well plates) were plated on sterile poly-l-lysine–coated coverslips (Thermo Fisher Scientific) and left to grow overnight. Following treatment with DMSO or 12.5 or 25 µM C8 for the indicated times, the cells were fixed with 4% paraformaldehyde for 15 min, permeabilized with 0.1% Triton-X/0.5% bovine serum albumin (BSA) for 15 min, and blocked with 2% BSA for 1 h, all at room temperature. After incubation with primary antibodies against histone γH2AX (05-636; Millipore Sigma) or 53BP1 (NB100-304; Novus Biologicals) at concentrations of 1:1,000 overnight at 4 °C, secondary antibodies at concentrations of 1:1,000 (Alexa Fluor 594 for γH2AX; Alexa Fluor 488 for 53BP1; Thermo Fisher Scientific) were applied for 1 h at room temperature. Coverslips were mounted using ProLong Gold Antifade Mountant with DAPI (Life Technologies). Images were collected using a Nikon ECLIPSE TE2000-S fluorescence microscope at 60×, and processed using Fiji software.
Caspase Activation Assays.
PEO1 and PEO4 cells (5 × 104 cells per well in 12-well plates) were washed with PBS and lysed in a buffer containing 50 mM Hepes (pH 7.5), 100 mM NaCl, 2 mM EDTA, 0.1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate, and 10% sucrose, and the protein concentration was measured. Next, 30 µg of each sample was incubated with 20 µg/mL of Ac-DEVD-AMC caspase-3 fluorogenic substrate (556449; BD Pharmingen) in lysis buffer supplemented with 10 mM dithiothreitol for 1 h at 37 °C. Fluorescent AMC liberated from Ac-DEVD-AMC was detected using a fluorometer with excitation at 380 nm and emission at 440 nm.
Mouse Pharmacokinetics Assays and Maximum Tolerated Dose Studies.
Pharmacokinetic profiling was performed by WuXi AppTec. C8 and C16 were dissolved in a vehicle composed of 70% polyethylene glycol (PEG) 400 in saline and administered at a dose of 5 mg/kg i.p. to groups of three male CD-1 mice. At different time points, plasma samples were obtained and the amount of either C8 or C16 present was determined by liquid chromatography-tandem mass spectrometry. Then the Cmax and T1/2 were determined.
A maximum tolerated dose study was also performed by WuXi AppTec. C8 was dissolved at different concentrations in a vehicle composed of 5% DMSO, 60% PEG400, and 35% water and administered at doses of 5, 20, or 40 mg/kg i.p. to groups of five female athymic nude mice for 7 d. Plasma concentrations of C8 were determined on days 1 and 7 at 4 h postdose. Weights of the mice were recorded every day until the end of day 8 of the experiment.
Xenograft Assay.
Xenograft experiments were performed in conjunction with Explora BioLabs. Female athymic nude mice aged 4 to 6 wk (n = 8 to 10 per group for C8 and vehicle) were injected with 2 to 3 × 106 cells subcutaneously into the mammary fat pad (MDA-MB-231 and HCC1806) or the right flank (HCT116). When the average tumor volume per group reached 150 mm3, the mice were administered i.p. injections of C8 (20 mg/kg of body weight) or vehicle (5% DMSO, 60% PEG400, 35% water) twice daily for 35 d. Body weight and tumor volume were measured twice per week. Two-tailed t tests of equal variance were used to compare tumor volumes in C8- and vehicle-treated mice, and P values were determined.
Supplementary Material
Acknowledgments
We thank Andy Shiau (Ludwig Institute for Cancer Research Small Molecule Discovery Program) for advice and comments on the manuscript and Dan Durocher (Lunenfeld-Tanenbaum Research Institute, Toronto) and Toshi Taniguchi (Fred Hutchinson Cancer Research Center) for providing cell lines. These studies were supported by the Ludwig Institute for Cancer Research and NIH Grant GM26017 (to R.D.K.).
Footnotes
Competing interest statement: R.D.K. and J.H.J.P. are both listed as authors on an upcoming review article based on meeting proceedings of talks presented at the 10th Weinman Symposium held in 2018. R.D.K. and J.H.J.P. did not directly collaborate on work presented at the symposium that is described in the review article.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009237117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.O’Neil N. J., Bailey M. L., Hieter P., Synthetic lethality and cancer. Nat. Rev. Genet. 18, 613–623 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Zinovyev A., Kuperstein I., Barillot E., Heyer W. D., Synthetic lethality between gene defects affecting a single non-essential molecular pathway with reversible steps. PLoS Comput. Biol. 9, e1003016 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Putnam C. D., Kolodner R. D., Pathways and mechanisms that prevent genome instability in Saccharomyces cerevisiae. Genetics 206, 1187–1225 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lord C. J., Ashworth A., PARP inhibitors: Synthetic lethality in the clinic. Science 355, 1152–1158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mateo J. et al., A decade of clinical development of PARP inhibitors in perspective. Ann. Oncol. 30, 1437–1447 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pujade-Lauraine E. et al.; SOLO2/ENGOT-Ov21 Investigators , Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): A double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 18, 1274–1284 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Hartwell L. H., Szankasi P., Roberts C. J., Murray A. W., Friend S. H., Integrating genetic approaches into the discovery of anticancer drugs. Science 278, 1064–1068 (1997). [DOI] [PubMed] [Google Scholar]
- 8.Kaelin W. G., Jr., The concept of synthetic lethality in the context of anticancer therapy. Nat. Rev. Cancer 5, 689–698 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Mair B., Moffat J., Boone C., Andrews B. J., Genetic interaction networks in cancer cells. Curr. Opin. Genet. Dev. 54, 64–72 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen J. P., Ideker T., Synthetic lethal networks for precision oncology: Promises and pitfalls. J. Mol. Biol. 430, 2900–2912 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rykowski M. C., Wallis J. W., Choe J., Grunstein M., Histone H2B subtypes are dispensable during the yeast cell cycle. Cell 25, 477–487 (1981). [DOI] [PubMed] [Google Scholar]
- 12.Bender A., Pringle J. R., Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 11, 1295–1305 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costigan C., Gehrung S., Snyder M., A synthetic lethal screen identifies SLK1, a novel protein kinase homolog implicated in yeast cell morphogenesis and cell growth. Mol. Cell. Biol. 12, 1162–1178 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tishkoff D. X., Filosi N., Gaida G. M., Kolodner R. D., A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell 88, 253–263 (1997). [DOI] [PubMed] [Google Scholar]
- 15.Morrison A., Johnson A. L., Johnston L. H., Sugino A., Pathway correcting DNA replication errors in Saccharomyces cerevisiae. EMBO J. 12, 1467–1473 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morrison A., Sugino A., The 3′→5′ exonucleases of both DNA polymerases delta and epsilon participate in correcting errors of DNA replication in Saccharomyces cerevisiae. Mol. Gen. Genet. 242, 289–296 (1994). [DOI] [PubMed] [Google Scholar]
- 17.Tran H. T., Gordenin D. A., Resnick M. A., The 3′→5′ exonucleases of DNA polymerases delta and epsilon and the 5′→3′ exonuclease Exo1 have major roles in postreplication mutation avoidance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 2000–2007 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balakrishnan L., Bambara R. A., Flap endonuclease 1. Annu. Rev. Biochem. 82, 119–138 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Symington L. S., Homologous recombination is required for the viability of rad27 mutants. Nucleic Acids Res. 26, 5589–5595 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loeillet S. et al., Genetic network interactions among replication, repair and nuclear pore deficiencies in yeast. DNA Repair 4, 459–468 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Oughtred R. et al., The BioGRID interaction database: 2019 update. Nucleic Acids Res. 47, D529–D541 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mullen J. R., Kaliraman V., Ibrahim S. S., Brill S. J., Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157, 103–118 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Putnam C. D. et al., Bioinformatic identification of genes suppressing genome instability. Proc. Natl. Acad. Sci. U.S.A. 109, E3251–E3259 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Putnam C. D. et al., A genetic network that suppresses genome rearrangements in Saccharomyces cerevisiae and contains defects in cancers. Nat. Commun. 7, 11256 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Srivatsan A. et al., Essential Saccharomyces cerevisiae genome instability suppressing genes identify potential human tumor suppressors. Proc. Natl. Acad. Sci. U.S.A. 116, 17377–17382 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Debrauwère H., Loeillet S., Lin W., Lopes J., Nicolas A., Links between replication and recombination in Saccharomyces cerevisiae: A hypersensitive requirement for homologous recombination in the absence of Rad27 activity. Proc. Nat. Acad. Sci. U.S.A. 98, 8263–8269 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C., Kolodner R. D., Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat. Genet. 23, 81–85 (1999). [DOI] [PubMed] [Google Scholar]
- 28.Kokoska R. J. et al., Destabilization of yeast micro- and minisatellite DNA sequences by mutations affecting a nuclease involved in Okazaki fragment processing (rad27) and DNA polymerase delta (pol3-t). Mol. Cell. Biol. 18, 2779–2788 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freudenreich C. H., Kantrow S. M., Zakian V. A., Expansion and length-dependent fragility of CTG repeats in yeast. Science 279, 853–856 (1998). [DOI] [PubMed] [Google Scholar]
- 30.Tumey L. N. et al., The identification and optimization of a N-hydroxy urea series of flap endonuclease 1 inhibitors. Bioorg. Med. Chem. Lett. 15, 277–281 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Exell J. C. et al., Cellularly active N-hydroxyurea FEN1 inhibitors block substrate entry to the active site. Nat. Chem. Biol. 12, 815–821 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakai W. et al., Functional restoration of BRCA2 protein by secondary BRCA2 mutations in BRCA2-mutated ovarian carcinoma. Cancer Res. 69, 6381–6386 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakai W. et al., Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature 451, 1116–1120 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zimmermann M. et al., CRISPR screens identify genomic ribonucleotides as a source of PARP-trapping lesions. Nature 559, 285–289 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun J. X. et al., A computational approach to distinguish somatic vs. germline origin of genomic alterations from deep sequencing of cancer specimens without a matched normal. PLoS Comput. Biol. 14, e1005965 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elstrodt F. et al., BRCA1 mutation analysis of 41 human breast cancer cell lines reveals three new deleterious mutants. Cancer Res. 66, 41–45 (2006). [DOI] [PubMed] [Google Scholar]
- 37.DelloRusso C. et al., Functional characterization of a novel BRCA1-null ovarian cancer cell line in response to ionizing radiation. Mol. Cancer Res. 5, 35–45 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Ghandi M. et al., Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomlinson G. E. et al., Characterization of a breast cancer cell line derived from a germ-line BRCA1 mutation carrier. Cancer Res. 58, 3237–3242 (1998). [PubMed] [Google Scholar]
- 40.Yazinski S. A. et al., ATR inhibition disrupts rewired homologous recombination and fork protection pathways in PARP inhibitor-resistant BRCA-deficient cancer cells. Genes Dev. 31, 318–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drew Y. et al., Therapeutic potential of poly(ADP-ribose) polymerase inhibitor AG014699 in human cancers with mutated or methylated BRCA1 or BRCA2. J. Natl. Cancer Inst. 103, 334–346 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Lehmann B. D. et al., Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Invest. 121, 2750–2767 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schröder-Heurich B. et al., Functional deficiency of NBN, the Nijmegen breakage syndrome protein, in a p.R215W mutant breast cancer cell line. BMC Cancer 14, 434 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ihnen M. et al., Therapeutic potential of the poly(ADP-ribose) polymerase inhibitor rucaparib for the treatment of sporadic human ovarian cancer. Mol. Cancer Ther. 12, 1002–1015 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hassan S., Esch A., Liby T., Gray J. W., Heiser L. M., Pathway-enriched gene signature associated with 53BP1 response to PARP inhibition in triple-negative breast cancer. Mol. Cancer Ther. 16, 2892–2901 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rothkamm K. et al., DNA damage foci: Meaning and significance. Environ. Mol. Mutagen. 56, 491–504 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Ward T. A., McHugh P. J., Durant S. T., Small molecule inhibitors uncover synthetic genetic interactions of human flap endonuclease 1 (FEN1) with DNA damage response genes. PLoS One 12, e0179278 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roos W. P., Kaina B., DNA damage-induced cell death by apoptosis. Trends Mol. Med. 12, 440–450 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Slee E. A. et al., Benzyloxycarbonyl-Val-Ala-Asp (OMe) fluoromethylketone (Z-VAD.FMK) inhibits apoptosis by blocking the processing of CPP32. Biochem. J. 315, 21–24 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bertrand R., Solary E., O’Connor P., Kohn K. W., Pommier Y., Induction of a common pathway of apoptosis by staurosporine. Exp. Cell Res. 211, 314–321 (1994). [DOI] [PubMed] [Google Scholar]
- 51.Farmer H. et al., Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Lord C. J., Ashworth A., Targeted therapy for cancer using PARP inhibitors. Curr. Opin. Pharmacol. 8, 363–369 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Bryant H. E. et al., Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Murai J. et al., Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 72, 5588–5599 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.D’Andrea A. D., Mechanisms of PARP inhibitor sensitivity and resistance. DNA Repair (Amst.) 71, 172–176 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Edwards S. L. et al., Resistance to therapy caused by intragenic deletion in BRCA2. Nature 451, 1111–1115 (2008). [DOI] [PubMed] [Google Scholar]
- 57.Gogola E. et al., Selective loss of PARG restores PARylation and counteracts PARP inhibitor-mediated synthetic lethality. Cancer Cell 33, 1078–1093.e12 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Gupta R. et al., DNA repair network analysis reveals shieldin as a key regulator of NHEJ and PARP inhibitor sensitivity. Cell 173, 972–988.e23 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lheureux S. et al., Somatic BRCA1/2 recovery as a resistance mechanism after exceptional response to poly (ADP-ribose) polymerase inhibition. J. Clin. Oncol. 35, 1240–1249 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Park P. H. et al., Amplification of the mutation-carrying BRCA2 allele promotes RAD51 loading and PARP inhibitor resistance in the absence of reversion mutations. Mol. Cancer Ther. 19, 602–613 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pettitt S. J. et al., Genome-wide and high-density CRISPR-Cas9 screens identify point mutations in PARP1 causing PARP inhibitor resistance. Nat. Commun. 9, 1849 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mengwasser K. E. et al., Genetic screens reveal FEN1 and APEX2 as BRCA2 synthetic lethal targets. Mol. Cell 73, 885–899.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bunz F. et al., Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282, 1497–1501 (1998). [DOI] [PubMed] [Google Scholar]
- 64.Yáñez R. J., Porter A. C., Therapeutic gene targeting. Gene Ther. 5, 149–159 (1998). [DOI] [PubMed] [Google Scholar]
- 65.Kucherlapati M. et al., Haploinsufficiency of flap endonuclease (Fen1) leads to rapid tumor progression. Proc. Natl. Acad. Sci. U.S.A. 99, 9924–9929 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng L. et al., Fen1 mutations result in autoimmunity, chronic inflammation and cancers. Nat. Med. 13, 812–819 (2007). [DOI] [PubMed] [Google Scholar]
- 67.Asagoshi K. et al., FEN1 functions in long patch base excision repair under conditions of oxidative stress in vertebrate cells. Mol. Cancer Res. 8, 204–215 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee M. et al., Rad52/Rad59-dependent recombination as a means to rectify faulty Okazaki fragment processing. J. Biol. Chem. 289, 15064–15079 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Beck M. et al., The quantitative proteome of a human cell line. Mol. Syst. Biol. 7, 549 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schindelin J. et al., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.