Abstract
Background
The Serious Outcomes Surveillance Network of the Canadian Immunization Research Network (CIRN SOS) has been performing active influenza surveillance since 2009 (ClinicalTrials.gov identifier: NCT01517191). Influenza A and B viruses are identified and characterized using real-time reverse-transcriptase polymerase chain reaction (RT-PCR), and multiplex testing has been performed on a subset of patients to identify other respiratory virus aetiologies. Since both methods can identify influenza A and B, a direct comparison was performed.
Methods
Validated real-time RT-PCRs from the World Health Organization (WHO) to identify influenza A and B viruses, characterize influenza A viruses into the H1N1 or H3N2 subtypes and describe influenza B viruses belonging to the Yamagata or Victoria lineages. In a subset of patients, the Seeplex RV15 One-Step ACE Detection assay (RV15) kit was also used for the detection of other respiratory viruses.
Results
In total, 1111 nasopharyngeal swabs were tested by RV15 and real-time RT-PCRs for influenza A and B identification and characterization. For influenza A, RV15 showed 98.0 % sensitivity, 100 % specificity and 99.7 % accuracy. The performance characteristics of RV15 were similar for influenza A subtypes H1N1 and H3N2. For influenza B, RV15 had 99.2 % sensitivity, 100 % specificity and 99.8 % accuracy, with similar assay performance being shown for both the Yamagata and Victoria lineages.
Conclusions
Overall, the detection of circulating subtypes of influenza A and lineages of influenza B by RV15 was similar to detection by real-time RT-PCR. Multiplex testing with RV15 allows for a more comprehensive respiratory virus surveillance in hospitalized adults, without significantly compromising the reliability of influenza A or B virus detection.
Keywords: influenza, multiplex PCR, subtype, lineage, validation
Introduction
Influenza virus infection is a leading infectious cause of morbidity and mortality in developed countries, and is of considerable public health concern [1–7]. Many vaccine formulations have been developed to reduce the burden of influenza illness, but the continued evolution of the viruses through antigenic drift requires that the vaccines be reformulated each year [8–10]. Monitoring the epidemiology and burden associated with circulating influenza viruses is important to make informed recommendations on vaccine use [8–10], and the currently circulating strains include influenza A virus subtypes H1N1 and H3N2 and the Yamagata and Victoria lineages of influenza B [11].
Since 2009, the Serious Outcomes Surveillance Network of the Canadian Immunization Research Network (CIRN SOS) has been conducting active surveillance for acute respiratory illness in hospitalized adults to monitor the burden of influenza illness and assess the effectiveness of seasonal influenza vaccines against laboratory-confirmed influenza [1–7]. The CIRN SOS Network comprises 15 to 45 acute care (depending on the year) hospitals across Canada, and influenza testing is performed using real-time reverse-transcriptase polymerase chain reaction (RT-PCR) methods derived from the World Health Organization (WHO) [12–14]. These methods allow the identification of influenza A and B, the discrimination of influenza A viruses into H1N1 and H3N2 subtypes, and the characterization of influenza B viruses into Yamagata or Victoria lineages [12–14]. These viruses were all circulating in Canada at the time of this study.
While WHO-based real-time RT-PCRs methods are often viewed as the reference standard for influenza virus detection, diagnostic laboratories and surveillance studies often test for other viral aetiologies of respiratory illness [15–28]. To avoid the high cost and labour associated with individual virus detection, multiplex RT-PCR technologies have been developed and are now commercially available [28]. Multiplex RT-PCR can detect influenza A and B, as well as non-influenza respiratory viruses (NIRVs), such as respiratory syncytial virus (RSV), coronaviruses, rhinoviruses, human metapneumovirus (hMPV), human parainfluenza viruses, adenovirus and enteroviruses [11, 15–28]. While the focus often lies on influenza, NIRVs can also be a significant causes of morbidity and mortality [29–36]. NIRVs can also co-circulate with influenza [11, 29], and can mirror the clinical presentations of influenza, or other viral or bacterial respiratory tract infections [30–36]. From a diagnostic perspective, rapid identification of viral respiratory viruses has potential benefits, such as a decrease in antibiotic prescriptions, a decrease in laboratory investigations and more judicious use of oseltamivir, and allows for the implementation of infection control practices, such as patient cohorting [31, 37, 38]. From a surveillance perspective, understanding the epidemiology of NIRVs can inform guidelines for patient management [9–11, 38] and help guide the development of new vaccines or therapeutics [31]. This study focuses on molecular detection of influenza viruses for the purpose of surveillance.
The CIRN SOS Network has performed testing for NIRVs on a subset of nasopharyngeal swabs collected for influenza surveillance. NIRV testing was performed using a Health Canada approved test, the Seeplex RV15 One-step ACE Detection assay (Seegene, Inc., Seoul, Republic of Korea) (RV15). This conventional multiplex RT-PCR uses dual-priming oligonucleotide (DPO) technology [39, 40] for the detection of influenza A and B, as well as 13 other respiratory viruses. The performance of RV15 has previously been compared against cell culture, direct immunofluorescence, real-time RT-PCR and other multiplex RT-PCRs for respiratory viruses [16–25, 36, 41]; however, to the best of our knowledge, the performance of RV15 for the detection of influenza A and B has yet to be compared against that of the WHO real-time RT-PCR reference methods. Given that RV15 showed variable performance for other viruses compared to other molecular methods [16, 17, 19, 36], an assessment of RV15 against the WHO reference methods for influenza A and B is well justified. Both RV15 and the WHO real-time RT-PCRs were used by the CIRN SOS Network, allowing a direct comparison of these methods.
Methods
Ethics
This study was approved by the research ethics boards (REBs) at each participating hospital (ClinicalTrials.gov identifier: NCT01517191). These included the William Osler Health System Research Ethics Board (Brampton, ON), the University of Edmonton Health Research Ethics Board (Edmonton, AB), the Capital Health Research Ethics Board (Halifax, NS), the Hamilton Health Sciences/McMaster Health Sciences Research Ethics Board (Hamilton, ON), the Horizon Health Network Research Ethics Board (Moncton, NB), the BMD Research Ethics Board MUHC – Montreal General Hospital (Montreal, QC), the Ottawa Health Science Network Research Ethics Board (Ottawa, ON), le Comité d'éthique de la recherche du CHU de Québec (Québec, QC), the Horizon Health Network Research Ethics Board (Saint John, NB), le Comité d'éthique de la recherche sur l’humain du Centre hospitalier de Sherbrooke (Sherbrooke, QC), the Health Sciences North Research Ethics Board (Sudbury, ON), the Mount Sinai Research Ethics Board (Toronto, ON), the North York General Research Ethics Board (Toronto, ON), the Toronto East General Hospital Research Ethics Board (Toronto, ON), le Comité d'éthique de la recherche de Trois-Rivières-Centre hospitalier affilié universitaire regional (Trois-Rivières, QC), the University of British Columbia Clinical Research Ethics Board (Vancouver, BC) and the University of Manitoba Health Research Ethics Board (Winnipeg, MB).
Active surveillance by the CIRN SOS Network
Active surveillance for influenza was performed across up to 45 hospitals in 5 Canadian provinces over consecutive influenza seasons starting in 2009, but the specimens from this study were collected between November 2011 and May 2013. On a daily basis, dedicated SOS Network surveillance monitors reviewed all adult admissions (aged ≥16 years) to identify patients with an acute respiratory illness, and patient demographics and outcomes were collected. Patient demographics and outcomes have been the subject of several CIRN SOS Network publications [1–7].
Specimen collection and processing
Within 7 days of onset of illness, consenting patients were enrolled and tested for influenza viruses A and B from NP swabs collected in universal transport media (UTM) (Copan Diagnostics). All swabs were divided into aliquots and archived at −80 °C for batch shipment on dry ice to the CIRN SOS Reference Laboratory at the Canadian Center for Vaccinology (CCfV) (Halifax, NS).
Real-time RT-PCR
The CIRN SOS Reference Laboratory retested all specimens from each CIRN site using validated real-time RT-PCR methods [12–14]. Total nucleic acids (TNAs) were extracted from 140 µl of NP swab material using a MagNaPure LC 2.0 instrument (Roche Diagnostics). The TNAs were eluted into 60 µl, and 5 µl served as a template for each 25 µl real-time RT-PCR reaction consisting of 0.5 µl SuperScript III RT/Platinum Taq Mix PCR enzyme mix (Invitrogen, Carlsbad, CA, USA), 1×PCR Master Mix, 0.8 µM primers and 0.2 µM probes (Table S1, available in the online version of this article). Initially, influenza strains were identified as A or B with a duplex real-time RT-PCR using primers targeting the matrix genes for each influenza type (Table S1). Subsequently, TNA that was positive for influenza A was subjected to a real-time RT-PCR subtyping assay targeting the haemagglutinin (HA) genes specific to either H1 or H3. TNA that was positive for influenza B was subjected to real-time RT-PCR characterization into Victoria or Yamagata lineages. Amplifications of all real-time RT-PCR assays were performed on an Applied Biosystems 7500 Fast Instrument (Life Technologies), under the following conditions: reverse transcription at 50 °C for 30 min; activation of the Taq DNA polymerase at 95 °C for 2 min; and 45 cycles of 95 °C for 15 s (denaturation) and 55 °C for 30 s (combined annealing and extension). Threshold cycle (C t) values were provided by the manufacturer’s software, and the C t cutoff for positivity was determined using previously validated values at defined thresholds (Table S2).
RV15 respiratory virus multiplex PCR
Following the same TNA extraction method as described for real-time RT-PCR analyses, TNA was extracted from a separate aliquot of NP swab material and 10 µl of TNA was used as a template for RV15 reactions, as recommended in the manufacturer’s instructions. Amplification was performed in a 96-well plate in a C1000 Touch Thermocycler (BioRad Laboratories Ltd, Mississauga, ON, Canada). Amplicons were resolved using 1.2 % (w/v) agarose gel electrophoresis with staining using 1.0 µg ml−1 ethidium bromide (final concentration), and visualized on a GelDoc XR+instrument with ImageLab software (version 5.1) (BioRad Laboratories).
Statistical analyses
Influenza A and B results from the RV15 or real-time RT-PCR assays were classified as positive or negative, and compared to a composite reference standard where concordant results between two of three methods were considered a true positive or negative result. Discrepant analyses were performed using real-time RT-PCR and/or sequencing by the National Microbiology Laboratory (Winnipeg, MB, Canada). Statistical Analysis Software (SAS) version 9.4 (SAS Institute, Cary, NC, USA) was used to assess significant differences between methods using 2×2 contingency tables and McNemar’s chi square test. A P value ≤0.05 was considered statistically significant. Sensitivity, specificity, accuracy, misclassification rates and kappa statistics were reported with 95 % confidence intervals (CIs).
Cloning of the matrix gene targets of influenza A and B and viral quantification
Viral loads in NP swabs were assessed based on standard curves generated by amplification of the matrix gene targets for influenza A and B using conventional RT-PCR, cloning of each target into plasmids and performance of real-time RT-PCR on serial dilutions of the quantified plasmids.
Analytical sensitivity
To directly compare the limits of detection (LoD) of RV15 and the WHO real-time RT-PCRs, parallel testing was performed using 10-fold serially diluted viruses. The viruses used were reference strains of the CIRN SOS Network that had been characterized by the National Microbiology Laboratory (Winnipeg, MB) as [A/California/7/2009 (H1N1)], [A/Perth/16/2009 (H3N2)], [B/Brisbane/60/2008 (Victoria lineage)] and [B/Wisconsin/1/2010 (Yamagata lineage)]. Triplicates values from three independent experiments were analysed, and the C t values obtained from real-time RT-PCR for influenza A and B were used to estimate viral concentrations (relative to the standard curves generated using plasmid controls). The LoD was estimated at a probability of 95 % by Probit analysis [36, 41] using StatPlus 2009 Professional version 5.7.8.
Results
Number of influenza A and B cases evaluated
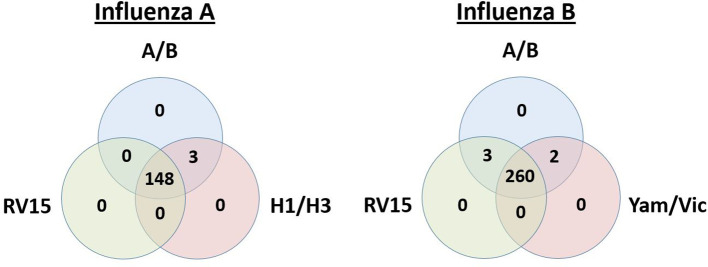
Laboratory data were available on 1111 adults hospitalized with acute respiratory illness, who had all 3 molecular tests of interest performed: RV15, influenza A/B duplex real-time RT-PCR for influenza A or B screening; and H1/H3 real-time RT-PCR subtyping for positive influenza A specimens or real-time RT-PCR Yamagata/Victoria lineage discrimination for positive influenza B. Of the 1111 patients, 151 were positive for influenza A (96 H1N1 and 55 H3N2) and 265 were positive for influenza B (184 Yamagata lineage and 81 Victoria lineage). Most of the results overlapped between testing methods, but some discrepant results were observed (Figs 1 and 2).
Fig. 1.
Overlap of positive influenza A or B results between RV15 and the WHO real-time RT-PCRs. Venn diagrams illustrating the overlap of positive results for influenza A or influenza B in patients tested by the Seegene multiplex PCR (RV15), the WHO real-time RT-PCRs for influenza A or B detection (A/B) and influenza A subtyping (H1/H3) or influenza B discrimination for Yamagata or Victoria (Yam/Vic) lineages.
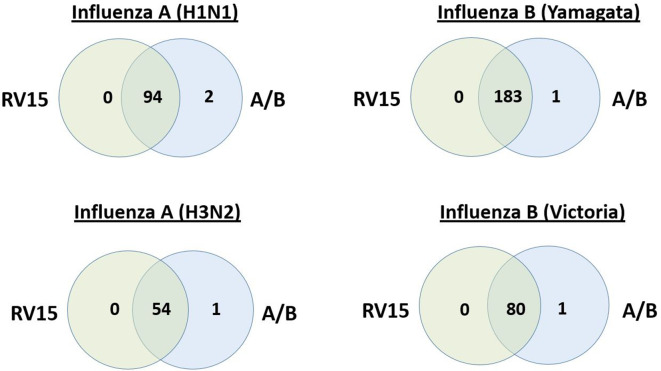
Fig. 2.
Overlap of results between RV15 and the WHO real-time RT-PCRs for characterized influenza A or B viruses. Venn diagram illustrating the overlap of positive results between RV15 and influenza A/B real-time RT-PCRs for characterized influenza A subtypes (H1N1 or H3N2) and influenza B lineages (Yamagata or Victoria).
Performance of RV15 for detection of influenza A and B
Compared to the composite reference standard, RV15 showed a sensitivity for influenza A of 98.0 % (148/151; 95 % CI: 95.8–98.0 %) and a specificity of 100.0 % (960/960; 95 % CI: 99.6–100.0 %). Discordant results were seen in three influenza A positive results (Table 1; Fig. 1). The accuracy of RV15 for influenza A was 99.7 % (1108/1111; 95 % CI: 99.1–99.7 %). Similar performance characteristics were noted for influenza A H1N1 and H3N2 (Table 1; Fig. 2). For influenza B, RV15 showed a sensitivity of 99.2 % (263/265; 95 % CI: 98.0–99.2 %) and a specificity of 100.0 % (846/846; 95 % CI: 99.6–100.0 %). Discordant results were seen in three influenza B positive results (Table 1; Fig. 1). The accuracy of RV15 for influenza B was 99.8 % (1109/1111; 95 % CI: 99.2–99.8 %). Similar performance characteristics were noted for the influenza B Yamagata and Victoria lineages (Table 1; Fig. 2). Overall, no significant differences were noted between real-time RT-PCR methods or RV15 and the composite reference standard, for all viruses, influenza A subtypes or influenza B lineages (with P values ranging from 0.248 to 1.000).
Table 1.
Assay performance against the composite reference standard
|
Assay |
Target |
Performance characteristics |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
TP |
TN |
FP |
FN |
Sensitivity (95 % CI) |
Specificity (95 % CI) |
Accuracy (95 % CI) |
Misclassification rate (95 % CI) |
Kappa (95 % CI) |
||
|
FluA/B real-time RT-PCR |
FluA (total) |
151 |
960 |
0 |
0 |
100.0 (98.1–100.0) |
100.0 (99.7–100.0) |
100.0 (99.5–100.0) |
0.0 (0.0–0.5) |
100.0 (97.8–100.0) |
|
FluA (H1N1) |
96 |
1015 |
0 |
0 |
100.0 (97.1–100.0) |
100.0 (99.7–100.0) |
100.0 (99.5–100.0) |
0.0 (0.0–0.5) |
100.0 (96.8–100.0) |
|
|
FluA (H3N2) |
55 |
1056 |
0 |
0 |
100.0 (94.9–100.0) |
100.0 (99.7–100.0) |
100.0 (99.5–100.0) |
0.0 (0.0–0.5) |
100.0 (94.7–100.0) |
|
|
FluB (Total) |
265 |
846 |
0 |
0 |
100.0 (98.9–100.0) |
100.0 (99.7–100.0) |
100.0 (99.5–100.0) |
0.0 (0.0–0.5) |
100.0 (98.6–100.0) |
|
|
FluB (Yamagata) |
184 |
927 |
0 |
0 |
100.0 (98.5–100.0) |
100.0 (99.7–100.0) |
100.0 (99.5–100.0) |
0.0 (0.0–0.5) |
100.0 (98.2–100.0) |
|
|
FluB (Victoria) |
81 |
1030 |
0 |
0 |
100.0 (96.5–100.0) |
100.0 (99.7–100.0) |
100.0 (99.5–100.0) |
0.0 (0.0–0.5) |
96.3 (96.3–100.0) |
|
|
RV15 |
FluA (total) |
148 |
960 |
0 |
3 |
98.0 (95.8–98.0) |
100.0 (99.6–100.0) |
99.7 (99.1–99.7) |
0.3 (0.3–0.9) |
98.8 (96.2–98.8) |
|
FluA (H1N1) |
94 |
1015 |
0 |
2 |
97.9 (94.9–97.9) |
100.0 (99.7–100.0) |
99.8 (99.2–99.8) |
0.2 (0.2–0.8) |
98.8 (95.1–98.8) |
|
|
FluA (H3N2) |
54 |
1056 |
0 |
1 |
98.2 (92.7–98.2) |
100.0 (99.7–100.0) |
99.9 (99.4–99.9) |
0.1 (0.1–0.6) |
99.0 (93.2–99.0) |
|
|
FluB (Total) |
263 |
846 |
0 |
2 |
99.2 (98.0–99.2) |
100.0 (99.6–100.0) |
99.8 (99.2–99.8) |
0.2 (0.2–0.8) |
99.5 (97.9–99.5) |
|
|
FluB (Yamagata) |
183 |
927 |
0 |
1 |
99.5 (97.8–99.5) |
100.0 (99.7–100.0) |
99.9 (99.4–99.9) |
0.1 (0.1–0.6) |
99.7 (97.7–99.7) |
|
|
FluB (Victoria) |
80 |
1030 |
0 |
1 |
98.8 (95.0–98.8) |
100.0 (99.7–100.0) |
99.9 (99.4–99.9) |
0.1 (0.2–0.6) |
99.3 (95.2–99.3) |
|
CIs, confidence intervals; FluA, influenza A; FluB, influenza B; FN, false negative; FP, false positive; TN, true negative; TP, true positive.
Discrepant analyses
For influenza A specimens not detected by RV15, the C t values obtained with influenza A/B real-time RT-PCR were 32.3, 34.2 and 34.5, which is near the previously established detection limit (i.e. C t cutoff of 36.4) (Table S2). Similarly, the C t values for influenza B real-time RT-PCR results in cases not detected by RV15 were 34.3 and 36.9, where the previously established C t cutoff for influenza B was 37.3 (Table S2). These results are consistent with specimens with low viral loads, spanning concentrations of 1.7, 2.0, and 4.9 copies/reaction for influenza A and 0.2 and 0.8 copies/reaction for influenza B. Upon retest of discrepant results by RV15, all results remained negative, whereas testing at the National Microbiology Microbiology (Winnipeg, MB) confirmed the positive results by RT-PCR and sequencing.
Analytical sensitivity
In this study, the estimated LoD of RV15 for each influenza A subtype and influenza B lineage was higher than that of the real-time RT-PCR for influenza A/B (Table 2), but below the detection limits of the real-time RT-PCRs for influenza A subtyping or B lineage characterization (Tables 2 and S3). This is consistent with previously validated cutoffs for positivity for the real-time RT-PCR methods (Table S2). At viral concentrations that were below the detectable limit of RV15 but detected using influenza A/B real-time RT-PCR, the C t values from the real-time RT-PCR spanned from 32.7 to 34.1 for influenza A H1N1, from 32.0 to 34.6 for influenza A H3N2, from 36.1 to 37.3 for influenza B Yamagata lineage and from 34.1 to 36.8 for influenza B Victoria lineage (data not shown). These C t values represent concentrations near the assay cutoff values for each target (Table 2 and S2).
Table 2.
Estimated LoD for detection of influenza A subtype or influenza B lineages
|
Virus |
LoD in copies/reaction* (95 % CI) |
|||
|---|---|---|---|---|
|
RV15 |
Real-time RT-PCR influenza A/B detection |
Real-time RT-PCR influenza A H1/H3 subtyping |
Real-time RT-PCR influenza B Yamagata/Victoria lineage |
|
|
FluA H1N1 |
4.0 (3.2–6.8) |
1.5 (1.2–2.5) |
9.5 (7.0–12.2) |
na |
|
FluA H3N2 |
4.2 (2.2–5.6) |
0.9 (0.7–1.3) |
0.9 (0.7–1.3) |
na |
|
FluB Yamagata |
5.2 (4.0–6.4) |
0.4 (0.1–0.6) |
na |
25.0 (22.6–26.6) |
|
FluB Victoria |
3.2 (3.0–3.4) |
1.5 (1.1–1.4) |
na |
3.2 (2.6–3.9) |
*The concentration that could reproducibly (at 95 % confidence) be detected by the test method is described as the LoD. The LoD was determined by Probit analysis by testing replicate aliquots of virus dilutions (see Table S3). For each virus aliquot, the C t values obtained by the WHO real-time RT-PCR were used to infer viral load. Concentrations of virus dilutions were estimated using standard curves generated with plasmids pFluA and pFluB from this study (see Supplementary Material).
CI, confidence intervals; FluA, influenza A; FluB, influenza B; LoD, limit of detection; na, not applicable.
Discussion
The CIRN SOS Network uses WHO-based real-time RT-PCRs to identify and characterize influenza A subtypes and influenza B lineages. Compared to the WHO reference methods, the RV15 assay had an accuracy of 99.7 and 99.8 % for the detection of influenza A and B, respectively.
While this was not statistically significant, RV15 was unable to detect influenza A or B in a small subset of specimens in which the viral loads were low. Conventional multiplex PCRs such as RV15 are often less sensitive than real-time RT-PCR, and this can be explained in part by the assay principle. Real-time RT-PCR detects fluorescence signals captured during PCR amplification cycles, and results are defined objectively using validated cutoffs for positivity. Conventional RT-PCR is an end-point detection of amplicons following electrophoresis and staining; a process where visualization of amplicons can be difficult and subjective when working with specimens with low concentrations of target [15–17, 21–23, 36, 41].
Whether conventional or real-time RT-PCR is used, the performance of these molecular methods can also vary, depending on factors such as genetic mismatches in the PCR target region, which could arise in influenza over time via antigenic drift. This emphasizes the need to verify the performance for characterized and circulating subtypes and lineages of influenza. However, for RV15, only a limited number of studies have assessed its ability to detect influenza A and B viruses, and none have specifically looked at its performance for the detection of influenza A virus subtypes or influenza B lineages [15, 19, 20, 23, 35]. Cho et al. [23] compared RV15 to a composite reference standard that included culture and a commercial real-time RT-PCR and demonstrated that RV15 had a sensitivity of 93.4 % (with 95 % CI: 88.8–93.4 %) for influenza A and one of 79.5 % (95 % CI: 79.0–88.6 %) for influenza B. Gharabaghi et al. [19] assessed the performance of RV15 against direct fluorescent antibody testing, virus culture and isolation, and three additional multiplex PCR methods. The specificities for influenza A and B were 98.8 and 100 %, and the sensitivities were 96.9 % (95 % CI: 91.9–96.9 %) and 100 % (92.6–100 %), respectively. In the present study, the performance of RV15 was verified for the detection of recent influenza A subtypes (H1N1 and H3N2) and influenza B lineages (Yamagata and Victoria), and it showed similar performance characteristics to the WHO real-time RT-PCR. Since this study used specimen collected prospectively from 2011 to 2013, it could be argued that the performance of the molecular methods assessed could vary with more recent circulating influenza strains, if primer or probe mismatches occur in the assay gene targets. However, the CIRN SOS laboratory participates in yearly quality assurance programmes proficiency testing for influenza A and B detection and characterization using all the assays from this study (all real-time RT-PCRs and the RV15 assay), and no differences in assay performance have been observed over time (data not shown). In this study, only a small subset of results in specimens with low viral loads were not detected by RV15, with this representing 0.3 and 0.2 % of the total tests for influenza A and B, respectively. While the lower sensitivity of multiplex PCRs such as RV15 was expected, the possibility of target gene sequence mismatches cannot be excluded.
It should be noted that viral loads were estimated using quantification relative to a standard curve generated using plasmid DNA controls. With influenza virus being an RNA virus, comparison to plasmid DNA does not account for the reverse transcription step. Regardless, all viruses were subjected to direct method comparison in the analytical analyses, which includes the reverse transcription step. Subsequently, the resulting C t values were subjected to comparison against the standard curve derived from plasmid DNA results, as plasmid DNA was more readily quantifiable. Overall, with this approach it remains valid to infer that the specimens not detected by RV15 had low viral loads, but the absolute quantity of virus should be considered to be an estimate (Tables 2, S2 and S3). Overall, this would have little impact on the conclusions from this study.
This study’s strengths include prospectively collected specimens from a defined patient population (adults hospitalized with acute respiratory illness), comparison of results against reference methods for influenza A and B detection, and analyses performed on influenza viruses characterized by subtyping or lineage determination. The main limitation of the study was that it was focused solely on influenza virus. However, this is justified because influenza viruses are the only respiratory viruses for which vaccines are currently available, and the performance for the detection of other viruses has been assessed previously [17, 19–23]. While assessing the performance of RV15 against validated real-time RT-PCRs for influenza is well justified, this study focused on the use of RV15 for population-based surveillance in hospitalized adults. These findings should not be extrapolated to other patient populations or for applications in clinical diagnostic testing where individual-level results are prioritized. The patients tested in this study were hospitalized adults, often presenting with co-morbidities and severe outcomes [1–7]. Further, respiratory virus testing is often performed using testing algorithms following initial screening methods for influenza A and B. As such, nucleic acids extracted from clinical specimens are sometimes inadvertently subjected to a freeze/thaw cycle prior to testing. The impact of freeze/thaw was not assessed in this study, as testing was performed following independent nucleic acid extraction on a different specimen aliquot. However, given that RV15 failed to detect a small subset of influenza A and B specimens at low viral loads, additional freeze/thaw cycles may further compromise influenza virus detection, and should thus be avoided. Finally, the workflow is relatively simple with RV15 for small numbers of specimens, but additional benefits could be afforded by using imaging software enabling automated amplicon detection if high-throughput specimen processing is required [15–17, 19–23]. Such automated analyses of RV15 amplicons could also reduce reduce result subjectivity compared to interpretations made from visual assessment of amplicons [15–17, 19–23]. Automated analyses were not evaluated in this study, but the technical staff performing RV15 testing were blinded to the real-time RT-PCR method results to avoid bias, and the RV15 results were remained unchanged with subsequent independent review by other blinded staff members.
Overall, the performance of RV15 was comparable to the WHO real-time RT-PCR standards for the detection of recently circulating subtypes of influenza A and lineages of influenza B, and it only missed a very small subset of influenza A and B results at low viral loads. Given that the performance characteristics of the RV15 multiplex PCR are not provided in the manufacturer kit insert, these data are of value for its users, which include several acute care hospitals and provincial public health laboratories in Canada [36, 41]. This study shows that the RV15 conventional multiplex PCR can be used for surveillance studies for respiratory viruses without significantly compromising detection of influenza A and B. The use of multiplex technologies such as RV15 can help better define the epidemiology of influenza and NIRVs, and these data are important for the development of novel therapeutics and vaccines.
Supplementary Data
Funding information
Influenza virus surveillance was funded by the Public Health Agency of Canada (PHAC) and the Canadian Institutes of Health Research (CIHR), and through a Collaborative Research Agreement between GlaxoSmithKline Biologicals SA and the CIRN SOS Network. CIRN and SM provided funding for RV15 testing. The authors received no financial support or other form of compensation related to the development of the manuscript, and were solely responsible for final manuscript content and data interpretation.
Acknowledgements
The authors would like to thank the patients and families whose participation made this study possible. The authors would also like to thank the many dedicated CIRN SOS Network surveillance monitors, the hospital staff and the CCfV staff, who were instrumental in the recruitment, collection, processing and archiving of specimens, and data collection.
Author contributions
All authors participated design and implementation or implementation or analysis, interpretation of the study, and the development of the manuscript. All authors had full access to the data and gave final approval before submission.
Conflicts of interest
J. L. received research grants from Merck for work outside the study, but no personal payments. T. F. H. and S. A. M. report payments to their institution from the GSK group of companies for the conduct of the study, and payments from Pfizer, Merck, Novartis and Sanofi‐Pasteur outside the submitted work. M. K. A. reports grants from the GSK group of companies, Pfizer and Sanofi, but no personal payments. J. M. reports payments to her institution from the GSK group of companies and Sanofi for her participation in advisory boards. A. P. reports payments from Actelion, Sanofi‐Pasteur and Genentech. J. P. reports payments from the GSK group of companies, Merck, Roche and Synthetic Biologics outside the submitted work. L. V. received research grants from the GSK group of companies, Pfizer, Optimer, Cubist and Merck, and personal fees from Merck, Optimer and Cubist. G. D. S. reports he was external consultant at Business and Decision Life Sciences (on behalf of GSK) at the time of the study, and is currently employed by the GSK group of companies and holds shares in the GSK group of companies. No other conflicts were declared.
Footnotes
Abbreviations: bp, base pairs; CCfV, Canadian Center for Vaccinology; CI, confidence intervals; CIHR, Canadian Institutes of Health Research; CIRN SOS, Serious Outcomes Surveillance Network of the Canadian Immunization Research Network; Ct, threshold cycle; DPO, dual priming oligonucleotide; HA, hemagglutinin; hMPV, humanmetapneumovirus; LoD, limits of detection; NIRVs, non-influenza respiratory viruses; NML, National Microbiology Laboratory; PHAC, Public Health Agency of Canada; REB, research ethics boards; RSV, respiratory syncytialvirus; RV15, Seeplex RV15 One-Step ACE Detection assay; SAS, Statistical Analysis Software; TNA, total nucleic acids; UTM, universal transport media; WHO, World Health Organization.
Three supplementary tables and supplementary materials are available with the online version of this article.
References
- 1.McNeil S, Shinde V, Andrew M, Hatchette T, Leblanc J, et al. Interim estimates of 2013/14 influenza clinical severity and vaccine effectiveness in the prevention of laboratory-confirmed influenza-related hospitalisation, Canada, February 2014. Euro Surveill. 2014;19:20729. doi: 10.2807/1560-7917.ES2014.19.9.20729. [DOI] [PubMed] [Google Scholar]
- 2.McNeil SA, Andrew MK, Ye L, Haguinet F, Hatchette TF, et al. Interim estimates of 2014/15 influenza vaccine effectiveness in preventing laboratory-confirmed influenza-related hospitalisation from the Serious Outcomes Surveillance Network of the Canadian Immunization Research Network, January 2015. Euro Surveill. 2015;20:21024. doi: 10.2807/1560-7917.ES2015.20.5.21024. [DOI] [PubMed] [Google Scholar]
- 3.Andrew MK, Shinde V, Hatchette T, Ambrose A, Boivin G, et al. Influenza vaccine effectiveness against influenza-related hospitalization during a season with mixed outbreaks of four influenza viruses: a test-negative case-control study in adults in Canada. BMC Infect Dis. 2017;17:805. doi: 10.1186/s12879-017-2905-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrew MK, Shinde V, Ye L, Hatchette T, Haguinet F, et al. The importance of frailty in the assessment of influenza vaccine effectiveness against influenza-related hospitalization in elderly people. J Infect Dis. 2017;216:405–414. doi: 10.1093/infdis/jix282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nichols MK, Andrew MK, Hatchette TF, Ambrose A, Boivin G, et al. Influenza vaccine effectiveness to prevent influenza-related hospitalizations and serious outcomes in Canadian adults over the 2011/12 through 2013/14 influenza seasons: A pooled analysis from the Canadian Immunization Research Network (CIRN) Serious Outcomes Surveillance (SOS Network) Vaccine. 2018;36:2166–2175. doi: 10.1016/j.vaccine.2018.02.093. [DOI] [PubMed] [Google Scholar]
- 6.Mulpuru S, Li L, Ye L, Hatchette T, Andrew MK, et al. Effectiveness of influenza vaccination on hospitalizations and risk factors for severe outcomes in hospitalized patients with COPD. Chest. 2019;155:69–78. doi: 10.1016/j.chest.2018.10.044. [DOI] [PubMed] [Google Scholar]
- 7.Nichols MK, Andrew MK, Ye L, Hatchette TF, Ambrose A, et al. The Impact of Prior Season Vaccination on Subsequent Influenza Vaccine Effectiveness to Prevent Influenza-related Hospitalizations Over 4 Influenza Seasons in Canada. Clin Infect Dis. 2018 doi: 10.1093/cid/ciy1009. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 8.Ambrose CS, Levin MJ. The rationale for quadrivalent influenza vaccines. Hum Vaccin Immunother. 2012;8:81–88. doi: 10.4161/hv.8.1.17623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Public Health Agency of Canada National Advisory Committee on Immunization (NACI) https://www.canada.ca/en/public-health/services/immunization/national-advisory-committee-on-immunization-naci.html January 3, 2019.
- 10.World health Organization (WHO) WHO recommendations on the composition of influenza virus vaccines. 2018 https://www.who.int/influenza/vaccines/virus/recommendations/en/ January 3, 2019.
- 11.Public Health Agency of Canada (PHAC) Respiratory Virus Detections in Canada. https://www.canada.ca/en/public-health/services/surveillance/respiratory-virus-detections-canada.html January 3, 2019.
- 12.World Health Organization (WHO) CDC Protocol of real-time RT-PCR for influenza A(H1N1) 2009 http://www.who.int/csr/resources/publications/swineflu/CDCRealtimeRTPCR_SwineH1Assay-2009_20090430.pdf January 3, 2019.
- 13.World Health Organization (WHO) WHO information for molecular diagnosis of influenza virus in humans - update. 2011 https://www.who.int/influenza/resources/documents/molecular_diagnosis_influenza_virus_humans_update_201108.pdf January 3, 2019.
- 14.Biere B, Bauer B, Schweiger B. Differentiation of influenza B virus lineages Yamagata and Victoria by real-time PCR. J Clin Microbiol. 2010;48:1425–1427. doi: 10.1128/JCM.02116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J, Lee HS, Cho YG, Choi SI, Kim DS, et al. Evaluation of allplex respiratory panel 1/2/3 multiplex real-time PCR assays for the detection of respiratory viruses with influenza A virus subtyping. Ann Lab Med. 2018;38:46–50. doi: 10.3343/alm.2018.38.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parrott G, Kinjo T, Nabeya D, Uehara A, Nahar S, et al. Evaluation of Anyplex™ II RV16 and RB5 real-time RT-PCR compared to Seeplex ® RV15 OneStep ACE and PneumoBacter ACE for the simultaneous detection of upper respiratory pathogens. J Infect Chemother. 2017;23:859–861. doi: 10.1016/j.jiac.2017.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho CH, Chulten B, Lee CK, Nam MH, Yoon SY, et al. Evaluation of a novel real-time RT-PCR using TOCE technology compared with culture and Seeplex RV15 for simultaneous detection of respiratory viruses. J Clin Virol. 2013;57:338–342. doi: 10.1016/j.jcv.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roh KH, Kim J, Nam MH, Yoon S, Lee CK, et al. Comparison of the Seeplex reverse transcription PCR assay with the R-mix viral culture and immunofluorescence techniques for detection of eight respiratory viruses. Ann Clin Lab Sci. 2008;38:41–46. [PubMed] [Google Scholar]
- 19.Gharabaghi F, Hawan A, Drews SJ, Richardson SE. Evaluation of multiple commercial molecular and conventional diagnostic assays for the detection of respiratory viruses in children. Clin Microbiol Infect. 2011;17:1900–1906. doi: 10.1111/j.1469-0691.2011.03529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bibby DF, McElarney I, Breuer J, Clark DA. Comparative evaluation of the Seegene Seeplex RV15 and real-time PCR for respiratory virus detection. J Med Virol. 2011;83:1469–1475. doi: 10.1002/jmv.22125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Radko S, Ian Stuart J, Zahariadis G. Evaluation of three commercial multiplex assays for the detection of respiratory viral infections. J Virol Methods. 2017;248:39–43. doi: 10.1016/j.jviromet.2017.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choudhary ML, Anand SP, Tikhe SA, Walimbe AM, Potdar VA, et al. Comparison of the conventional multiplex RT-PCR, real time RT-PCR and Luminex xTAG® RVP fast assay for the detection of respiratory viruses. J Med Virol. 2016;88:51–57. doi: 10.1002/jmv.24299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho CH, Lee CK, Nam MH, Yoon SY, Lim CS, et al. Evaluation of the AdvanSure™ real-time RT-PCR compared with culture and Seeplex RV15 for simultaneous detection of respiratory viruses. Diagn Microbiol Infect Dis. 2014;79:14–18. doi: 10.1016/j.diagmicrobio.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drews SJ, Blair J, Lombos E, DeLima C, Burton L, et al. Use of the Seeplex RV Detection kit for surveillance of respiratory viral outbreaks in Toronto, Ontario, Canada. Ann Clin Lab Sci. 2008;38:376–379. [PubMed] [Google Scholar]
- 25.Chen H, Weng H, Lin M, He P, Li Y, et al. The clinical significance of FilmArray Respiratory Panel in diagnosing community-acquired pneumonia. Biomed Res Int. 2017;2017:7320859. doi: 10.1155/2017/7320859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang YW, Gonsalves S, Sun JY, Stiles J, Gilhuley KA, et al. Clinical evaluation of the Luminex NxTAG Respiratory Pathogen Panel. J Clin Microbiol. 2016;54:1912–1914. doi: 10.1128/JCM.00482-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Popowitch EB, O'Neill SS, Miller MB. Comparison of the Biofire FilmArray RP, Genmark eSensor RVP, Luminex xTAG RVPv1, and Luminex xTAG RVP fast multiplex assays for detection of respiratory viruses. J Clin Microbiol. 2013;51:1528–1533. doi: 10.1128/JCM.03368-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahony JB, Blackhouse G, Babwah J, Smieja M, Buracond S, et al. Cost analysis of multiplex PCR testing for diagnosing respiratory virus infections. J Clin Microbiol. 2009;47:2812–2817. doi: 10.1128/JCM.00556-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stefanska I, Romanowska M, Donevski S, Gawryluk D, Brydak LB, et al. Co-infections with influenza and other respiratory viruses. Adv Exp Med Biol. 2013;756:291–301. doi: 10.1007/978-94-007-4549-0_36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Beek J, Veenhoven RH, Bruin JP, van Boxtel RAJ, de Lange MMA, et al. Influenza-like illness incidence is not reduced by influenza vaccination in a cohort of older adults, despite effectively reducing laboratory-confirmed influenza virus infections. J Infect Dis. 2017;216:415–424. doi: 10.1093/infdis/jix268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang JW, Lam TT, Zaraket H, Lipkin WI, Drews SJ, et al. Global epidemiology of non-influenza RNA respiratory viruses: data gaps and a growing need for surveillance. Lancet Infect Dis. 2017;17:e320–e326. doi: 10.1016/S1473-3099(17)30238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ackerson B, Tseng HF, Sy LS, Solano Z, Slezak J, et al. Severe morbidity and mortality associated with respiratory syncytial virus versus influenza infection in hospitalized older adults. Clin Infect Dis. 2019;69:197–203. doi: 10.1093/cid/ciy991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kodama F, Nace DA, Jump RLP. Respiratory syncytial virus and other noninfluenza respiratory viruses in older adults. Infect Dis Clin North Am. 2017;31:767–790. doi: 10.1016/j.idc.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nguyen C, Kaku S, Tutera D, Kuschner WG, Barr J, et al. Viral respiratory infections of adults in the intensive care unit. J Intensive Care Med. 2016;31:427–441. doi: 10.1177/0885066615585944. [DOI] [PubMed] [Google Scholar]
- 35.Hung IF, Zhang AJ, To KK, Chan JF, Zhu SH, et al. Unexpectedly higher morbidity and mortality of hospitalized elderly patients associated with rhinovirus compared with influenza virus respiratory tract infection. Int J Mol Sci. 2017;18:E259. doi: 10.3390/ijms18020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hatchette TF, Drews SJ, Grudeski E, Booth T, Martineau C, et al. Detection of enterovirus D68 in Canadian laboratories. J Clin Microbiol. 2015;53:1748–1751. doi: 10.1128/JCM.03686-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Charlton CL, Babady E, Ginocchio CC, Hatchette TF, Jerris RC, et al. Practical guidance for clinical microbiology laboratories: viruses causing acute respiratory tract infections. Clin Microbiol Rev. 2018;32:e00042–18. doi: 10.1128/CMR.00042-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schleihauf E, Fathima S, Pettipas J, LeBlanc JJ, Hatchette TF, et al. Respiratory outbreak investigations: How many specimens should be tested? Infect Control Hosp Epidemiol. 2015;36:1344–1347. doi: 10.1017/ice.2015.171. [DOI] [PubMed] [Google Scholar]
- 39.Kim SR, Ki CS, Lee NY. Rapid detection and identification of 12 respiratory viruses using a dual priming oligonucleotide system-based multiplex PCR assay. J Virol Methods. 2009;156:111–116. doi: 10.1016/j.jviromet.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma X, Xu H, Shi L, Yang P, Zhang L, et al. A multiplex PCR assay for the detection of five influenza viruses using a dual priming oligonucleotide system. BMC Infect Dis. 2015;15:93. doi: 10.1186/s12879-015-0818-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hatchette TF, Drews SJ, Bastien N, Li Y, German G, et al. Detection of influenza H7N9 virus: all molecular tests are not equal. J Clin Microbiol. 2013;51:3835–3838. doi: 10.1128/JCM.01808-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.