Abstract
Introduction
Bacteriophage therapy can be developed to target emerging diarrhoeal pathogens, but doing so in the absence of microbiome disruption, which occurs with antibiotic treatment, has not been established.
Aim
Identify a therapeutic bacteriophage that kills diarrhoeagenic enteroaggregative Escherichia coli (EAEC) while leaving the human microbiome intact.
Methodology
Phages from wastewater in Portland, OR, USA were screened for bacteriolytic activity by overlay assay. One isolated phage, PDX, was classified by electron microscopy and genome sequencing. A mouse model of infection determined whether the phage was therapeutic against EAEC. 16S metagenomic analysis of anaerobic cultures determined whether a normal human microbiome was altered by treatment.
Results
Escherichia virus PDX, a member of the strictly lytic family Myoviridae, killed a case-associated EAEC isolate from a child in rural Tennessee in a dose-dependent manner, and killed EAEC isolates from Columbian children. A single dose of PDX (multiplicity of infection: 100) 1 day post-infection reduced EAEC recovered from mouse faeces. PDX also killed EAEC when cultured anaerobically in the presence of human faecal bacteria. While the addition of EAEC reduced the β-diversity of the human microbiota, that of the cultures with either faeces alone, faeces with EAEC and PDX, or with just PDX phage was not different statistically.
Conclusion
PDX killed EAEC isolate EN1E-0007 in vivo and in vitro, while not altering the diversity of normal human microbiota in anaerobic culture, and thus could be part of an effective therapy for children in developing countries and those suffering from EAEC-mediated traveller’s diarrhoea without causing dysbiosis.
Keywords: phage therapy, EAEC, Myoviridae, Escherichia virus, dysbiosis, antibiotic alternatives
Introduction
Diarrhoeagenic Escherichia coli is a major cause of infectious disease worldwide, and in particular for those living in developing countries due to decreased access to safe water and healthcare. Of the E. coli pathotypes, enteroaggregative E. coli (EAEC) is a heterogeneous category, causing both acute and persistent diarrhoea [1]. EAEC has been implicated in outbreaks [2, 3], and is also the second leading cause of traveller’s diarrhoea. This pathotype has been linked to persistent diarrhoea and malnutrition of children and patients with HIV/AIDS living in developing countries. In the USA, it was the most commonly isolated diarrhoeal pathogen in emergency rooms in a large-scale study in Maryland and Connecticut [3]. Thus, EAEC is an emerging pathogen, for those living both in the developing and more developed regions of the world.
Antibiotics are generally prescribed for traveller’s diarrhoea, but with the ever-increasing problem of drug resistance, treatment options are becoming more limited. The type of antibiotic given will depend on regional resistance patterns [4]. In developing countries, recurrent and persistent bouts of diarrhoea caused by these bacteria can result in nutrient and growth deficiencies, and even life-threatening fluid loss in children [5]. Though ultimately self-limiting, due to the persistent nature of EAEC disease, the syndrome is not effectively controlled by oral rehydration therapies alone. Thus, combined with the problem of drug resistance, alternative treatment strategies are urgently needed.
Microbiologists are generally familiar with the work of Felix d’Herelle, who discovered bacteriophages during an outbreak of dysentery among soldiers during the First World War, and the subsequent success of phage therapy against bacterial infections in eastern Europe in the subsequent decades [6, 7]. Today, even the general public is learning of phage therapy due to the prevalence of multiple-drug-resistant (MDR) bacteria. Recently, phages have been used successfully for the treatment of bacterial pathogens, including Pseudomonas aeruginosa and Staphylococcus spp., in Poland and Romania, respectively [8–10]. However, few phage therapy clinical trials have been conducted in Western countries. Two successful phage therapy applications, of single-dose phage cocktails, a mixture of bacteriophages that target different pathogenic bacteria, have been conducted in the USA. In one case, the phage cocktail was administered topically to burn victims in Queen Astrid military hospital in Brussels, Belgium [11]. The other successful phase II clinical trial of phage therapy was administered to patients suffering from chronic MDR P. aeruginosa ear infections in London, UK [12]. This study was terminated early as the treatment group exhibited such impressive recoveries that the physicians wanted to be able to deliver the phage preparation to the control group of patients to treat their infections. Three patients enrolled in the study experienced full resolution of symptoms as determined by both the study physicians and self-reporting. Although there are significant hurdles for developing mechanisms for US Food and Drug Administration (FDA) approval of the clinical use of bacteriophages [13, 14], the results of these studies strongly suggest that the efficacy of phage therapeutics is not the limiting factor in making such applications available.
A clear advantage of using phages as therapy against bacterial infections is that they are thought to only target specific pathogens, often only one species or strain, without disrupting the normal microbiota, or causing dysbiosis. This is in stark contrast to more traditional antibiotics, which are life-saving drugs, but are also guaranteed to cause dysbiosis due to the conserved nature of drug targets. Recent reports demonstrate that this type of disruption can be associated with a number of different deleterious health effects [15]. One such review presents data on the loss of microbiome diversity in children treated with antibiotics [16], which includes a reduction of α-diversity, a metric of biodiversity, of up to 40 % from ciprofloxacin, an antibiotic commonly given for EAEC infections. Only a few have shown that phage can be used to treat bacterial infections without causing dysbiosis in humans [17–19]. In one, a commercially available Russian phage cocktail was tested in human volunteers, without pathogen challenge, showing no adverse effects with oral phage exposure. The authors also found no evidence of potentially deleterious genes in the phage genomes [18].
In this study, using a sequential isolation technique to enrich for bacteriophages capable of infecting the desired hosts, we isolated a strictly lytic phage from wastewater in Portland, OR, USA that kills strains of EAEC bacteria. Using electron microscopy techniques, the PDX phage was tentatively identified as a member of the family Myoviridae that infect E. coli , and this was confirmed through functional genome bioinformatic and phylogenetic analysis. We characterized the growth and infectivity of the PDX phage, and showed that it kills a case-associated EAEC strain both in vitro and in vivo. In a mouse model, a single dose of the phage 1 day post-infection significantly reduced the number of bacteria recovered from faeces over a 5-day period. Lastly, we showed that PDX kills EAEC when cultured anaerobically in the presence of human faecal bacteria, without altering the α-diversity of the human microbiota.
Methods
Bacterial strains and growth conditions
The bacteria used for phage isolation and clinical EAEC isolates for host susceptibility testing are listed in Table 1. The EPEC clinical isolates for host susceptibility testing are listed in Table S1 (available in the online version of this article). All strains were grown in lysogeny broth (LB) at 37 °C with shaking at 225 r.p.m. or on LB agar at 37 °C. All phage lysates were stored in SM gel buffer [50 mM Tris-HCl (pH 7.5), 0.1 M NaCl, 8 mM MgSO4, 0.01 % gelatin] at 4 ˚C, and warmed to room temperature for all experiments unless otherwise stated.
Table 1.
E. coli strains used in this study
|
Strain |
Genotype/serotype |
Syndrome* |
Spot test result† |
Plaque formation‡ |
Source§ |
|---|---|---|---|---|---|
|
EN1E-0007 |
EAEC O86:H27 |
D |
+ |
+ |
[66] |
|
LRT9 |
EPEC O11:abH2 |
U |
+ |
+ |
[67] |
|
MC4100 |
F-araD139 Δ(argF-lac) U169 rpsL 150 StrR relA1 flhD5301 deoC1 ptsF25 rbsR |
NP |
+ |
+ |
[68] |
|
BC/Ae017.2 |
EAEC AMR, AMCR |
D |
− |
− |
UB|| |
|
BC/AaUIM099.2 |
EAEC |
D |
+ |
− |
UB |
|
BC/AaUIM171.3 |
EAEC AMR |
D |
− |
− |
UB |
|
BC/AaUIM206.1 |
EAEC |
D |
+ |
− |
UB |
|
BC/Ac277.1 |
EAEC |
D |
− |
− |
UB |
|
BC/Ac338.3 |
EAEC AMR, AMCR |
D |
+ |
+ |
UB |
|
BC/AaUIM363.2 |
EAEC |
D |
+ |
+ |
UB |
|
BC/Ac408.1 |
EAEC |
D |
+ |
− |
UB |
|
BC/Ac422.2 |
EAEC |
D |
− |
− |
UB |
|
BC/Ac482.1 |
EAEC AMR, SXTR |
D |
− |
− |
UB |
|
BC/AaUIM517.1 |
EAEC |
D |
− |
− |
UB |
|
BC/Ac512.2 |
EAEC |
D |
− |
+ |
UB |
*D, acute diarrhoea; U, details unknown; NP, non-pathogenic laboratory strain.
†For the spot test, phage in 5 µl drops were placed on streaks of the different clinical isolates, and then screened for clearing after overnight growth. A plus sign indicates that the strain was lysed by Escherichia virus PDX.
‡Plaque formation was assessed with serial dilutions of the PDX phage, propagated on different host strains using the pour plate method. In this case, individual plaques indicated reproductive infections [20]. A plus sign indicates PDX replicates (can form plaques on) the strain.
§Clinical isolate EN1E-0007 obtained from Tennessee, UB isolates obtained from Columbia
||Division of Pediatrics Infectious Diseases, University at Buffalo.
Phage isolation and propagation
Bacteriophage PDX was initially isolated from untreated (influent) sewage collected at the Portland Wastewater Treatment Plant on 1 October 2015 using standard techniques [20]. The raw sewage was filtered through a 0.2 µm filter (Corning, Corning, NY, USA) to remove bacteria. The filtered sewage was then added to host bacteria in liquid culture. Phage–host solutions were combined with 0.5 % LB soft agar and overlaid on LB agar plates. Plates were then observed for the presence of plaques after overnight growth at 37 ˚C. Clear, defined plaques of interest were isolated and transferred using a sterilized inoculating needle and agitation in 1 ml solution of lambda diluent (10 mM Tris, pH 7.5; 10 mM MgSO4). To obtain a high-titre phage preparation, a crude lysate was generated. A clear PDX plaque was isolated and suspended in 1 ml of sterile lambda diluent. The phage suspension was combined with overnight culture of the bacterial host and incubated at 37 ˚C with shaking at 225 r.p.m. overnight. Chloroform was added to kill any remaining bacteria. This solution was centrifuged at 4 ˚C at 4000 g for 10 min to remove bacteria. The supernatant was recovered and treated again with chloroform.
Phages were sequentially isolated from E. coli strain MC4100, EPEC strain LRT9 and EAEC clinical isolate EN1E-0007 using a sequential isolation technique to enrich for polyvalent antigen recognition [21]. EPEC LRT9 was used because it is also a laboratory strain that is amenable to P1 transduction [22]. Several clearly defined plaques from the final overlay were picked using a sterile inoculating needle and suspended in microcentrifuge tubes containing SM buffer. The suspension was incubated at room temperature for 15 minutes to allow the phages to diffuse through the medium, followed by filtration through a sterile 0.2 µm filter to remove any bacteria that may have been transferred from the overlay plate. The final phage suspension was then propagated on EAEC strain EN1E-0007 to make a purified high-titre lysate. Bacteria were removed from the culture by centrifugation at 6055 g, and then the supernatant was sterile-filtered with a 0.2 µm filter. The titre of the lysate was determined by serial dilutions and overlay assay as described. After overnight incubation, the plates were observed and the number of plaque-forming units per ml (p.f.u. ml−1) was calculated. The purified high-titre lysate in SM buffer was stored at 4 °C.
Host susceptibility assay
A spot test was used to determine the host range of PDX on clinical EAEC and EPEC clinical isolates (Tables 1 and S1). Each LB agar plate was streaked with three–four spatially separated bacterial strains using a sterilized inoculating loop. A 10 µl purified lysate sample was pipetted on top of the centre of each linear bacterial streak and incubated overnight at 37 ˚C. Host susceptibility was indicated by a clearing within the streak of a given bacterial strain. Clinical EAEC and EPEC isolates were also tested for the ability to support PDX replication using the overlay assay described above and observing phage plaques (Tables 1 and S1).
Electron microscopy
PDX was analysed using transmission electron microscopy (TEM). Purified high-titre lysate was precipitated in 25 % PEG 6000–8000 in 2.5M NaCl and stored overnight at 4 ˚C. Precipitated phages in solution were centrifuged at 4 ˚C, 4000 g for 10 min. The supernatant was removed and the pellet was suspended in 40 µl of 50 mM Tris, pH 7.4, 10 mM MgSO4. The phage suspension was placed on copper-coated formvar grids (Ted Pella, Inc., Redding, CA, USA) and negatively stained with 1 % uranyl acetate. Samples were examined under a FEI Tecnai Spirit TEM system at an operating voltage of 120 kV (FEI, Hillsboro, OR, USA). All TEM was performed at the Multi-Scale Microscopy Core (MMC) with technical support from the Oregon Health and Science University (OHSU)/FEI Living Lab and the OHSU Center for Spatial Systems Biomedicine (OCSSB).
Phage genome sequencing and bioinformatics
A high-titre lysate was purified using a cesium chloride gradient, and then genomic dsDNA was extracted with an Epicentre DNA isolation kit (Madison, WI, USA) according to the manufacturer’s instructions. All library preparation and Illumina next-generation sequencing was performed at the Massachusetts General Hospital (MGH) Genome Core (Cambridge, MA, USA). Raw reads were filtered and processed for quality using Galaxy [23]. The read adaptors were trimmed with Trimmomatic Galaxy (v 0.32.3) using the default settings [24]. The quality of the trimmed reads was processed with Fast QC (v 0.11.4) using default settings [25]. The reads were assembled with a de novo De Bruijn algorithm-based software SPAdes with the virus kingdom selected (v 3.7.1) [26]. Next the quality of the assembly was determined using the Quality Assessment Tool for genome assemblies (QUAST v 2.3) (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The assembled genome was annotated using ORF annotation (NCBI ORFfinder) and PROKKA with default settings and National Center for Biotechnology Information (NCBI) smart Basic Local Alignment Tool (blast) (v 1.12) [27]. Annotation outputs were compared in tandem and putative coding sequences (CDSs) and proteins with known functions above 95 % were selected using the default algorithm parameters.
Next, output CDSs from the genomic annotations were aligned for similarity to other sequences in the NCBI database using the blast (v. 2.3.1) suite, specifically using the blastp and tblastn tools with default settings [28, 29]. Finally, genes associated with structural components were analysed using the Virfam webserver, which uses a machine learning hierarchical agglomerative clustering method to construct a protein phylogeny between PDX and all other phages contained in the ACLAME database.
Accession number
The assembled genome sequence of the Escherichia virus PDX was deposited in GenBank under accession number MG963916.
One-step growth curves
As our target for therapy is EAEC, growth kinetics of PDX using the one-step growth curve method were performed on strain EN1E-0007 [30]. LB liquid cultures were grown to an OD600 of 0.6–0.7, mid- to late-exponential phase. The purified phage lysate was added to the liquid culture tubes to infect the bacteria with a multiplicity of infection (m.o.i.) of 0.01. The liquid bacterial culture and phage lysate were incubated at room temperature for 10 min. Following the incubation period, the solution was centrifuged at 8000 g for 10 min. The supernatant was discarded and the pellet was resuspended in LB to make the absorption mixture. Immediately after resuspension the absorption mixture was serially diluted and plated with the bacterial host. The phages were propagated using a standard plaque overlay [20]. This procedure was repeated at 20 min time intervals for a total of 120 min. After 24 h of incubation, the plaques on all the plates were recorded and the p.f.u. ml−1 was calculated. The titre over time (120 min) was plotted to obtain a one-step growth curve.
Time–kill assays
Bacterial density was measured over time to determine the course of phage infection [31]. Liquid cultures of EAEC strain EN1E-0007 were incubated with PDX. The in vitro lytic efficiency of the phage was examined at several m.o.i.s. The wells of a flat-bottomed 96-well microtitre plate were filled with 100 µl of inoculated double-strength LB and 100 µl of purified lysate dilutions in SM buffer. Bacterial cultures were grown to late-exponential phase and diluted to 106 colony-forming units (c.f.u.) ml−1. PDX lysate was diluted to 105, 106, 107 or 108 p.f.u. ml−1, corresponding to m.o.i.s of 0.1, 1, 10 and 100, respectively. Each phage–host combination at specific m.o.i.s was performed in triplicate wells, and each experiment was performed three times. Controls for plate sterility, phage suspension sterility and bacterial growth without phage addition were included. To prevent between-well contamination by aerosolizing of bacteria, an adhesive transparent plate cover was placed over the top of the plate. The plates were incubated at 37 °C with orbital shaking for 12 h and OD at 600 nm was measured using a Microtiter Plate Reader (Sunrise, Tecan Group Ltd, Austria) at 30 min intervals.
Murine model of intestinal colonization
C56BL/6 mouse models have been previously developed for EPEC [32] and EAEC [33, 34]. Although mice do not display robust histopathological or clinical signs of disease, they serve as an in vivo colonization model to extend findings from in vitro experiments. C57BL/6 J WT mice aged 6–8 weeks received ampicillin (1 g l−1) added to the drinking water for 2 days to reduce the gut microbiome [33]. After 2 days, ampicillin water was removed and replaced with sterile drinking water for the rest of the experiment. Mice were infected via oral gavage with 4.0×106 c.f.u. E. coli resuspended in 1× PBS, pH 7.4 [32]. Fifteen minutes prior to inoculation with bacteriophage PDX, mice were given 2.6 % sodium bicarbonate by oral gavage to neutralize stomach acid [35]. Bacteriophages were diluted to 4.0×109 p.f.u. ml−1 in SM buffer +0.01 % gelatin and were introduced by oral gavage for a dose of 4.0×108 p.f.u. EAEC strain EN1E-0007 was monitored by daily collection of faeces and enumeration of c.f.u. g−1 faeces. Faeces were resuspended and serially diluted in LB plus 1 % saponin to reduce bacterial clumping and plated on LB plates containing 100 µg ml−1 streptomycin. Streptomycin-resistant bacteria from the mice tested indole-positive, demonstrating that EAEC was successfully isolated, cultured and enumerated.
Ethics statement
C57BL/6 J WT mice (male and female) were purchased from Jackson Laboratories and housed at Oregon Health and Science University (OHSU) under specific pathogen-free conditions in an ABSL-2 facility in accordance with University and Federal guidelines. All experimental protocols were approved by OHSU’s Department of Comparative Medicine (DCM) in accordance with the University’s Institutional Animal Care and Use Committee (IACUC).
Anaerobic culture
Faecal cultures were prepared using methods adapted from Caldwell and Bryant’s 1966 Hungate roll tube method for non-selective growth of rumen bacteria [36]. In order to mimic the nutrient, mineral, pH and environmental conditions of human gut in vitro, M10 broth media was prepared and dispensed into sterile Hungate tubes with rubber stoppers. Anaerobic conditions were achieved by sparging with 100 % CO2 gas at 10 PSI for 15 min. Resazurin dye was used as a visual indicator of anaerobic conditions. In the presence of oxygen, resazurin is reduced to resorufin, a distinctly red compound. In the absence of oxygen (i.e. anaerobic conditions) resazurin turns M10 media colourless [36].
Faeces were collected from a 21-year-old male in Portland, OR, USA who had not received antibiotics for over 1 year. One gram of fresh faeces was immediately placed in a sterile Hungate tube and bubbled for 10 min at 10 PSI with 100 % CO2 gas. The faeces were suspended in 10 ml of sterile M10 medium, which was transferred aseptically and anaerobically via a sterile syringe and hypodermic needle from one stoppered Hungate tube to another. The resulting slurry was vortexed vigorously and serially diluted in stoppered Hungate tubes of M10 to a final concentration of 1.0×10−4 g faeces ml−1, and then grown anaerobically for 16 h at 37 °C using the Hungate method [36]. After a 16 h incubation period, the starting culture was used to inoculate subsequent faecal cultures as described below.
Streptomycin-resistant (strR) EAEC EN1E-0007 were grown anaerobically under 100 % CO2 with faecal slurry and PDX using the Hungate method in M10 medium [36]. Cultures were inoculated to final concentrations: 16 h faecal, M10 culture diluted 1 : 100, 4.0×103 c.f.u. ml−1 EAEC of isolate EN1E-0007 and 4.0×105 p.f.u. ml−1 phage for an m.o.i. of 100. As a therapeutic comparison, EAEC-challenged faecal cultures were incubated with or without 25 µg ml−1 ciprofloxacin. Cultures were incubated for 16 h in a stationary incubator at 37 °C. DNA from these cultures was isolated for 16S rDNA sequencing analysis after 16 h of incubation.
Immediately prior to DNA extraction, a sample from each culture was plated to enumerate the concentrations of strR EAEC in the anaerobic cultures. As a control, faecal samples were plated on LB agar with streptomycin, and no growth was observed. Serial dilutions were performed in a 96-well plate in sterile LB, followed by plating in triplicate (10 µL drops) of each concentration on LB agar containing 100 µg ml−1 streptomycin. Plates were incubated for 20 h at room temperature and colonies were counted using a dissection microscope.
DNA was isolated from faecal cultures using a GenElute Bacterial Genomic DNA kit (Sigma-Aldrich, St Louis, MO, USA) following the protocol for cultures containing Gram-positive and Gram-negative bacteria. Whole-genomic DNA was quantified using a Nanodrop 1000 (Thermo Fisher Scientific, Waltham, MA, USA).
16S metagenomic sequencing and analysis
Genomic DNA was sent to the Oregon State University Center for Genome Research and Biocomputing for multiplex sequencing analysis using the Illumina MiSeq sequencing platform, with paired-end forward and reverse reads that were 150 bp in length and a sequencing depth of coverage totalling 1 million reads. The V3–V4 region of the 16S rRNA gene was amplified and used to prepare a dual-index library using the Nextera XT library preparation kit. Sequences were quality-filtered and demultiplexed using FastQC, then processed in RStudio (v 1.1.383; RStudio, Inc., Boston, MA, USA) using the DADA2 package [37] to denoise and merge the forward and reverse reads. As per the DADA2 pipeline, chimeric sequences were removed and preprocessed reads were clustered into 100 % identical sequences representing amplicon sequence variants (ASVs) to capture the full extent of genetic diversity in the microbiome [38], and then read counts were converted into a count table. ASVs were assigned taxonomy to the level of genus using the silva ribosomal RNA gene database (release 132) as a reference [37, 39]. Non-bacterial taxa were removed from the dataset. ASV sequences were aligned using ClustalOmega (v 1.2.4) [40] and a 16S phylogenetic tree was constructed using FastTree (v 2.1.7) [41].
The count table, taxonomy table, metadata table and phylogenetic tree were used for quantitative and qualitative analysis using the R packages Phyloseq (v 1.24.2) [42] and DESeq2 (v 1.20.0 [43]). Chao1 and Shannon diversity indices were calculated in Phyloseq from unnormalized ASV counts as measures of alpha diversity using Tukey post-hoc HSD for statistical analysis. Chao1 is an estimated measure of species richness that adjusts for variable read depths across samples using the number of singletons in each sample [44, 45], while the Shannon index is an estimate of diversity accounting for both richness and evenness [46]. ASV counts were normalized across samples using a variance-stabilizing transformation (VST) implemented in DESeq2 prior to calculating beta diversity. The adjusted VST count table and phylogenetic tree were used to construct a weighted UniFrac distance matrix [47], and beta diversity was plotted using principal coordinate analysis (PCoA) as the ordination method. To test whether samples form significantly different clusters based on treatment groups, the Adonis2 tool from the Vegan package (v 2.5–2) [22] was used to run a PERMANOVA [48] for testing whether group centroids were significantly different. Finally, to test which taxa are differentially abundant across treatment groups, pairwise binomial Wald tests were performed on VST-adjusted data in DESeq2. Each statistical test described above was executed a second time on the dataset with ASVs representing the genus Escherichia filtered out to confirm that any microbiome community differences detected were not primarily due to counts of inoculated bacteria.
Results
Isolation and initial characterization of a Myoviridae phage
Sequential host isolation with MC4100, and then EPEC and EAEC strains LRT9 and EN1E-0007, respectively, allowed us to enrich the phages for polyvalent antigen recognition [21]. The lysates for Escherichia virus PDX had a titre of 8.92×1012 p.f.u. ml−1 when propagated on EAEC strain EN1E-0007, the host bacterial pathogen for this study. As a negative control, PDX phages were unable to lyse a strain of Bacillus subtilis (ATCC #6051).
To test how broadly PDX recognized EAEC bacteria we found that the PDX phages lysed 5 and formed plaques on 3 of 12 EAEC case-associated strains from children in Columbia (Table 1). One of these strains, BC/Ac338.3, exhibited multiple drug resistances, to ampicillin (AM) and amoxicillin clavulanate (AMC). PDX lysed 9 and formed plaques on 5 out of 20 clinical, diarrhoeal EPEC isolates from children living in the Seattle area (Table S1; [49]). These enteropathogenic isolates represented a range of serotypes and variety of syndromes, including acute, chronic and in one case, strain TB96A, serotype O75:HN, bloody diarrhoea. Thus, the Escherichia virus PDX replicated in, and formed plaques on, multiple case-associated EAEC and EPEC isolates.
To obtain preliminary classification of PDX, we employed TEM. TEM imaging revealed that the phage most likely belonged to the family Myoviridae, identified by their canonical morphology: a small-isometric non-enveloped head with a long contractile tail (Fig. 1; [50]). The head diameter of PDX was 76.4±4.34 nm and the tail length was 114.0±2.26 nm (n=13) (Fig. 1a,b,c). Based on morphological features and metrics obtained using TEM, we tentatively identified PDX to be a member of the family Myoviridae, which infect E. coli (Fig. 1d).
Fig. 1.
Transmission electron micrographs of bacteriophage Escherichia virus PDX. Samples were negatively stained with 1 % uranyl acetate solution, prepared on a carbon formvar grid, and were imaged at 120 kV on a FEI Tecan-i Spirit TEM system. The arrow in (a) indicates the base plate. The arrows in (b) identify three putative tail fibres. The image in (c) illustrates the small isometric, non-enveloped head with long contractile tail, specific to the family Myoviridae. The image in (d) illustrates PDX infecting EAEC, with a contractile tail, indicated by an arrow, penetrating the outer membrane and periplasmic space. Scale bars are as indicated.
Genomic sequence analysis
Therapeutic phages must be virulent, or lytic, so as to minimize the risks associated with horizontal gene transfer by temperate or lysogenic phages [51]. Thus, we used genome sequence analysis to confirm that PDX was a member of the strictly lytic family of Myoviridae phages. The genome of PDX is 138 828 bp dsDNA, which is typical of the family Myoviridae. The GC content is 43%, which is comparable to other lytic phages (FastQC Galaxy). We identified 206 putative CDSs (PROKKA, ORFfinder), but did not identify toxin-encoding or lysogenic genes, which are characterized as any gene containing the terms ‘integrase’, ‘excisionase’, ‘recombinase’, or ‘repressor’ within the genome when compared to the NCBI blast database. Additionally, in a standard 20 amino acid search, a total of 6 putative tRNAs were predicted with open reading frames (ORFs) ranging from 72 to 88 bp in length (tRNAscan-SE). Virfam software uses the translated amino acid of CDSs from a phage genome as input and searches for similarity to other structural phage protein sequences deposited in the Aclame database.
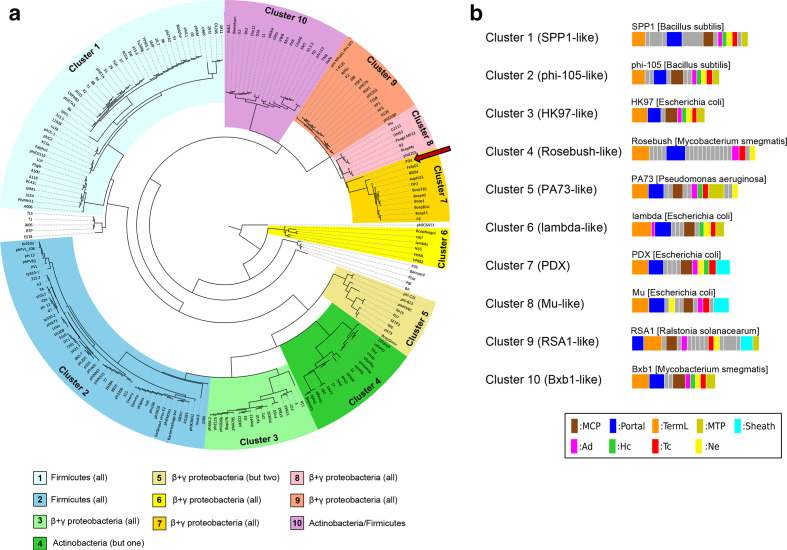
Based on the results of the Virfam analysis, the head–neck–tail structure genome organization in PDX belongs to the ‘neck type one – cluster 7’ category (Fig. 2a). Neck type one phages genomes contain the following proteins: portal protein, adaptor of type 1 (Ad1), head-closure of type 1(Hc1), neck protein of type 1 (Ne1), and tail-completion of type 1 (Tc1) (Fig. 2b). Cluster 7 phage genomes are reported in Virfam to be strictly Myoviridae with relatively small genome sizes and gene content (61–150 genes), although PDX contains 206 CDSs (Table 2).
Fig. 2.
Genome and structural protein classification analysis of PDX. (a) Tree representation of PDX (indicated by red arrow) classification among other Caudovirales phages, constructed from a hierarchical agglomerative clustering procedure applied to a matrix of similarity scores between pairs of phages (by combining HHsearch probabilities with the percentage identity) and present in the Aclame database. The different branches of the tree were sorted into 10 clusters and are highlighted by different background colors. The bacterial hosts of the phages are listed for each cluster in the legend at the bottom of the figure. (b) Gene organization of PDX and other phage genomes that belong to the neck one clusters. The legend at the bottom indicates which family a protein belongs to. In the legend, ‘but one’ and ‘but two’ indicate that all but one or two of the phages are lytic against the bacterial family indicated by the coloured boxes below the phylogeny.
Table 2.
Genomic analysis of PDX
|
Feature |
Quantity |
|---|---|
|
Size (bp) |
138 828 |
|
GC content (%) |
43 |
|
Coding domain sequences (CDSs) |
206 |
|
tRNAs |
6 |
By blast analysis, the three genomes with the highest whole-genome nucleotide identity match were E. coli O157 typing phage 4 (98 %), Escherichia phage JES-2013 (98%), and Escherichia phage Murica (Table S2). Murica is a Myoviridae phage that shares a 97 % nucleotide identity with PDX, and is strictly lytic against enterotoxigenic E. coli (ETEC) [52]. E. coli O157 typing phage 4 is lytic against E. coli O157, and also shares 99 % amino acid identity with the PDX tail fibre proteins [53]. Phage JES-2013 is lytic against Lactococcus lactis [54]. The putative tail fibre sequences from PDX had a 99 % amino acid identity with Myoviridae Escherichia phage V18 [55]. Overall, the phage that had the highest percentage identity scores with the PDX genome sequence and tail fibre amino acid sequence were all strictly lytic Myoviridae against various E. coli strains.
Phage PDX kills enteroaggregative E. coli both in vitro and in vivo
To characterize the phage life cycle, we performed a one-step growth curve of PDX (Fig. 3a). PDX infecting EAEC strain EN1E-0007 had a maximum relative virus count of 3.6×106 p.f.u. ml−1 after 100 min of infection. The bacteriolytic activity of PDX against EAEC strain EN1E-0007 was measured in a time–kill assay revealing a dose-dependent inhibition of bacterial growth. At an m.o.i. of 100, PDX suppressed the growth of EAEC (Fig. 3b). To demonstrate killing further, strain EN1E-0007 at 5×103 c.f.u. ml−1 was split into two cultures, one grown without and one with 5×105 p.f.u. ml−1 PDX phage, an m.o.i. of 100. After 5 h incubation, we found 1.8×106 c.f.u. ml−1 of strain EN1E-0007, while no bacteria were recovered from the culture containing bacteria and phage (the c.f.u. ml−1 values presented are the averages of an experiment performed in triplicate). Bacteria that were not treated with phage showed normal logistic growth curves that match the expected growth rate for this strain. A dose-dependent relationship between m.o.i. and point of infection was observed: at greater m.o.i.s, infection occurred earlier than for lower m.o.i.s (Fig. 3b).
Fig. 3.
One-step growth curve analysis and time–kill assays of EAEC infected with PDX. (a) Over the 120 min infection period, bacterial cultures were sampled, serially diluted and plated with a plaque overlay procedure (see the Methods section). After 24 h incubation, the plaques were counted and the titre (p.f.u. ml−1) was determined. Error bars represent the standard error of the mean. (b) Time–kill assay of EAEC strain EN1E-0007 treated with PDX. In a 96-well plate, exponential culture was inoculated with dilutions of PDX high-titre lysate for m.o.i.s of 0.1, 1, 10 and 100. The plates were incubated at 37 °C with orbital shaking for 12 h and the OD at 600 nm was measured using a Microtiter Plate Reader (Tecan). Two independent assays were performed in triplicate, with representative, averaged assay data presented. Error bars indicate ±1 standard deviation.
In many phage treatments, an increase in bacterial growth was observed after around 8–10 h of incubation. This is likely due to the emergence of mutant bacteria that are resistant to phage infection and has been observed in other similar studies [34, 56]. We isolated the bacteria that were resistant to phage infection. Phage induction, by treatment with mitomycin C, did not induce PDX plaques in the resistant, mutant EAEC. The lack of phage induction supported the conclusion that PDX is strictly virulent, and therefore appropriate for use as a therapeutic agent.
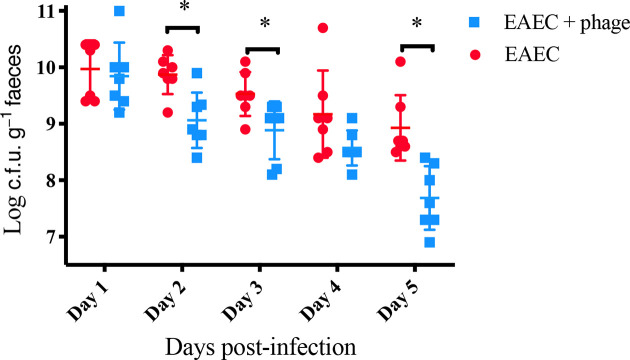
We next used a mouse colonization model to test the prediction that PDX can kill EAEC strain EN1E-0007 in vivo. A single dose of PDX caused a statistically significant decrease in EAEC colony counts at 2, 3, and 5 days post-infection (Fig. 4). The most pronounced decrease in EAEC occurred at 5 days post-infection, with mean 8.9 log c.f.u. g−1 faeces from EAEC-challenged mice and mean 7.7 log c.f.u. g−1 faeces from those treated with PDX (P=0.0016). As controls, we showed that the bacteria recovered from mouse faeces were streptomycin-resistant E. coli (by indole test) and that no bacteria from faecal pellets from uninfected mice grew on selective plates. We concluded that PDX reduced colonization of EAEC strain EN1E-0007 in the mouse model of infection.
Fig. 4.
A single dose of PDX decreased murine gut carriage of EAEC. Twenty-four hours after inoculation with EAEC, mice were gavaged with PDX phage at an m.o.i. of 100 or an equal volume of buffer control. No colonies were detected in phage-only control mice across the 5-day period. Statistical analysis was carried out to compare PDX-treated mice to control-treated mice at each indicated day (n=14 animals per day, day 2, P=0.0097; day 3, P=0.0214; day 5, P=0.0016). Each error bar is constructed using 1 standard deviation from the mean. Levels connected by an asterisk are statistically significantly different (P<0.05).
PDX phage kills EAEC strain EN1E-0007 without altering the diversity of human faecal bacteria when propagated anaerobically in vitro
To determine whether PDX could kill EAEC bacteria in the presence of the human gut microbiome, faeces were propagated anaerobically in M10 medium in the presence of the diarrhoeagenic EAEC strain EN1E-0007. Cultures lacking a PDX challenge grew from the 4.0×103 c.f.u. ml−1 inoculum to 9.0×107 c.f.u. ml−1 of EAEC in the absence of any additional agent (Table 3). In the cultures receiving EAEC and PDX phage the inoculum remained relatively unchanged, decreasing slightly to 1.7×103 c.f.u. ml−1 from the initial 4.0×103 c.f.u. ml−1. The cultures with ciprofloxacin, an antibiotic commonly given to treat traveller’s diarrhoea, were killed with no bacterial growth observed (Table 3). We concluded that PDX phage reduced growth of EAEC strain EN1E-0007 by nearly 5 log c.f.u. ml−1 in the presence of human microbiota when cultured anaerobically to mimic the human gut.
Table 3.
Colony counts of streptomycin-resistant EAEC strain EN1E-0007 in faecal culture after 16 h incubation at 37 °C
|
Treatment |
Faeces (c.f.u. ml−1) |
Faeces+EAECc (c.f.u. ml−1) |
|---|---|---|
|
Neat |
0 |
9.0×107 |
|
PDX a |
0 |
1.7×103 |
|
Ciprofloxacinb |
n/a |
0 |
Human faecal samples were collected and cultured anaerobically under the following regime: faeces only, faeces+EAEC, faeces+EAEC+ciprofloxacin, faeces+EAEC+PDX and faeces+PDX. Each challenge was cultured in triplicate.
a, Cultures were inoculated to 4.0x 105 p.f.u. ml−1 PDX for an m.o.i of 100.
b, Cultures were inoculated to 25 μg ml−1 ciprofloxacin.
c, Cultures were inoculated to 4.0x 10 c.f.u. ml−1 EAEC isolate EN1E-0007.
To test for any adverse effects of PDX on the human microbiota, genomic DNA was isolated from triplicate samples of the M10 anaerobic cultures presented in Table 3. High-throughput multiplex sequencing using Illumina MiSeq with 150 bp forward and reverse reads was used to analyse DNA from faeces only, faeces plus PDX, faeces plus EAEC, and faeces plus EAEC and PDX. As no DNA was recovered from ciprofloxacin-challenged cultures, no sequence data were generated. After processing and filtering steps, a total of 2303 unique ASVs were detected across all samples.
Alpha diversity (α-diversity), the mean ASV diversity of each sample, was calculated using the ‘estimate richness’ function in Phyloseq (Fig. 5). The Shannon diversity index was used to characterize diversity, considering the number of unique taxa (richness) and the evenness of the taxa present. The mean Shannon indices for faeces only (6.3), faeces+PDX (6.3) and faeces+EAEC+PDX (6.3) did not differ significantly from one another, but all differed significantly from faeces+EAEC (5.8), which showed the lowest α-diversity (P=0.0035). While the Shannon index utilizes the abundances of each taxa in its diversity calculation, the Chao1 estimator was selected as an additional non-parametric α-diversity estimator for the number of unique taxa present in each sample. The mean Chao1 estimates for faeces only (999.7), faeces+PDX (986.7) and faeces+EAEC+PDX (1014.2) did not differ significantly from one another, but all differed significantly from faeces+EAEC (682.5), which showed the lowest α-diversity (P=0.0081).
Fig. 5.
PDX does not alter the α-diversity of an in vitro human microbiome. Chao1 and Shannon indices were used to assess the α-diversity of cluster ASV reads. Statistical analyses were carried out to compare mean Chao1 estimates across challenges (n=12 cultures, faeces only; faeces+EAEC,*, P>0.0081; faeces+PDX; faeces+EAEC+ PDX). Statistical analyses were also carried out to compare mean Shannon indices across challenges (n=12 cultures, faeces only; faeces+EAEC,*, P>0.0035; faeces+PDX; faeces+EAEC+PDX).
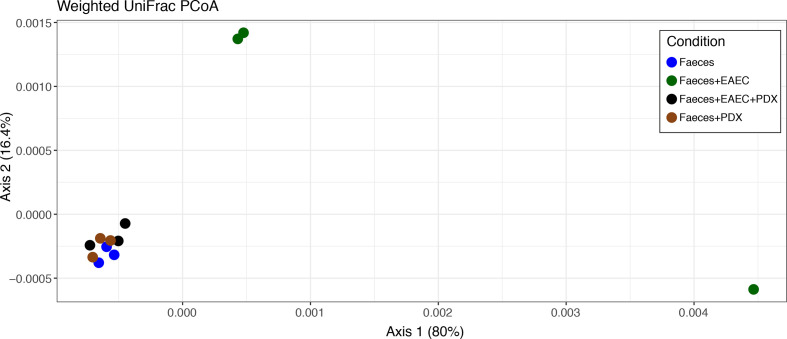
While alpha diversity metrics measure microbiome community within samples, beta diversity (β-diversity) is a measure of differences between samples. A weighted UniFrac dissimilarity matrix was used to plot samples accounting for the abundances and phylogenetic relationships of ASVs. PCoA revealed that all samples from faeces only, faeces+PDX and faeces+EAEC+PDX cluster closely together, while samples from faeces+EAEC are found far from this cluster (Fig. 6). Using Adonis2 to run PERMANOVA, we find that centroids from treatment groups are significant different from each other (F=2.83, P=0.014), which is primarily driven by the faeces+EAEC group. he dispersion of samples is not significantly different between groups (P=0.647), meeting the assumption of PERMANOVA.
Fig. 6.
PDX does not alter β-diversity of an in vitro human microbiome. β-diversity was assessed through PCoA for the ordination of a weighted UniFrac distance matrix, accounting for ASV abundance and phylogenetic relationships among taxa. Faeces, faeces+PDX and faeces+EAEC+PDX conditions cluster together, suggesting microbiome communities are unchanged with the addition of PDX, while the addition of EAEC alone alters the community significantly (PERMANOVA: F=2.83, P=0.014)
DESeq2 was used to identify specific taxa that have differential abundances between treatment groups. The only pairwise comparisons that yielded differentially abundant taxa (FDR<0.05) were faeces+EAEC vs faeces (305 taxa), faeces+EAEC vs faeces+PDX (261 taxa) and faeces+EAEC vs faeces+EAEC+PDX (232 taxa) (Fig. 7). For faeces+EAEC, we observed alteration of the abundance of taxa of all phyla (except a single representative of the phylum Verrucomicrobia that was unaffected) in the presence of EAEC isolate EN1E-0007. Taxa of the Bacterioidetes were reduced, as were a number of taxa of genera of the Firmicutes , and Acinetobacteria increased compared to the other groups. No comparisons between the faeces, faeces+PDX and faeces+EAEC+PDX groups revealed any differentially abundant taxa, even when the FDR cutoff was made less stringent at FDR<0.1.
Fig. 7.
Faeces with EAEC is the only condition that shows changes in specific taxa vs controls. Differential abundance analysis was performed via pairwise binomial Wald tests in DESeq2. Among all conditions, comparisons between (a) faeces+EAEC vs faeces, (b) faeces+EAEC vs faeces+PDX and (c) faeces+EAEC vs faeces+EAEC+PDX all yielded differentially abundant taxa (P<0.05), while no comparisons among faeces, faeces+PDX and faeces+EAEC+PDX showed any significant differences in ASV abundance, suggesting that the addition of PDX is highly targeted and does not affect the broader microbiome community.
All microbiome analyses were repeated with taxa in the genus Escherichia removed to test whether inoculated E. coli abundances were driving diversity patterns. However, the results remained largely unchanged for each test. Based on the collective microbiome results, we concluded that PDX phage killed EAEC isolate EN1E-0007 when cultured anaerobically in the presence of human gut microbiota without affecting bacterial community diversity.
Discussion
Using EAEC clinical isolate EN1E-0007 as our target EAEC pathotype, the Myoviridae Escherichia virus PDX killed the bacteria both in vitro and in vivo. EN1E-0007 is a case-associated isolate from a child living in rural Tennessee, and we also demonstrated that PDX formed plaques on case-associated EAEC isolates from children in Columbia (Tables 1 and S1). Laboratory E. coli strains, such as MC4100, have often lost some of the peripheral portions of their LPS or O-antigen repeats. Instead of possessing the more variable O-antigen repeats, the core polysaccharides of the LPS is left exposed, allowing for phage recognition of this and other more core antigens [57]. Due to the disparate location of these EAEC isolates, and the fact that PDX formed plaques on case-associated EPEC strains from children in the Seattle area, the results suggested that the sequential method [21] for isolating phages that recognized more core bacterial antigens was successful.
There is precedent in the literature for isolating therapeutic phage to target core antigens in order to broaden their host range. For example, for Salmonella enterica serovar Typhimurium, where the O-antigen repeat of the LPS is not produced via deletion of the gene rfbP, in order to expose the outer core of the LPS [58]. The authors found that the phage SSU5 was able to inhibit the growth of a number of other Salmonella strains, and also Cronobacter sakazakii , Shigella flexneri and E. coli , positing that, ‘an unknown common receptor for SSU5 might be shared among the bacteria that belong to the family Enterobacteriaciae.’ We have taken the same approach in order to broaden the host range of PDX, to target related bacteria from disparate locations. Although it is possible that EAEC strains exist in the Portland Metro area, to the best of our knowledge there have been no reports of clinical isolation of this pathotype within the region. While prevailing thought might indicate that phage cannot be isolated to target related bacterial pathogens from different locations, reports indicate otherwise [58], and our empirical data indicate that we can ‘train’ phage to recognize core antigens using domesticated laboratory strains of E. coli with altered LPS and kill related bacteria isolated, not only from patients living in different parts of the USA, but also those from different continents.
In order for phages to be used as therapeutics, they must be strictly lytic, and several lines of evidence indicate that PDX is a lytic phage. PDX was classified as a member of the strictly lytic bacteriophage family Myoviridae based on morphological characteristics from TEM imaging (Fig. 1). Neither the lambda suppressor CI nor CI-associated genes that are responsible for lysogeny in phage lambda were identified in the genome of PDX. It is noteworthy that the genome of PDX contains 206 CDSs, which is more than the other phages in this cluster in the current Aclame database (Table 2). Analysis of the head–neck structure CDSs in the Virfam suite identified PDX as belonging to neck type one – cluster 7 (Fig. 2a, b). Importantly, phage induction, by treatment with mitomycin C, did not induce PDX plaques in mutant derivatives of EAEC strain EN1E-0007 resistant to PDX infection. We concluded that the PDX phage was a member of the family Myoviridae with a strictly lytic lifestyle.
Other phages with high sequence identity to PDX are being investigated for use as therapeutics or for food contamination interventions. For example, the Myoviridae phage Murica (Table S2) is being researched as a potential treatment for contaminated food products [52]. Escherichia phage V18 was part of a successful prophylactic phage cocktail used in the prevention of traveller’s diarrhoea in mice infected with six bacterial strains, including E. coli, Shigella flexneri, Shigella sonnei , Salmonella enetica, Listeria monocytogenes or Staphylococcus aureus , and Lac (−) E. coli K-12 C600 [55]. Similar to PDX replicating in clinical isolates of both EAEC and EPEC pathotypes, the related V18 phage was cited as having a host range that included ETEC, EAEC and enterohaemorrhagic E. coli (EHEC). The ability of PDX to kill case-associated strains of two E. coli pathotypes, EAEC and EPEC, makes it an ideal candidate for therapeutic administration early, alone or in a cocktail, for patients when a bacterial infection is suspected but the species or strain is unknown, as is common in developing regions of the world. This is particularly the case since EAEC and EPEC bacteria remain a major cause of infantile diarrhoea [59]. While it is important to use phage cocktails, or multiple phages to combat phage-resistant bacterial mutants [60], in order to develop safe and effective therapies, it is also important to perform in vitro and in vivo characterization of individual phages [61, 62].
A clear advantage of using lytic phages as therapy against bacterial infections is specific targeting of pathogens without broad destruction of the normal microbiota, although this idea is not established for human infections in the literature. Here, by culturing normal human faeces anaerobically in vitro we showed that the PDX phage kills EAEC EN1E-0007 bacteria in this context, and using 16S rDNA analysis we showed that the α- and β-diversity of the microbiota were unaffected (Figs 5–7). Several points are worth noting. First, by ‘spiking in’ the EAEC strain into the faecal culture, the α-diversity is of course altered (Fig. 5), not unlike when humans are infected with enteric pathogens, and this has also been demonstrated in mice [63]. In contrast, the gut microbiome α-diversity was observed to be reduced by up to 40 % in patients given ciprofloxacin, a fluoroquinolone commonly prescribed for E. coli infections [16]. There have been many studies that indicate that antibiotic treatment has adverse effects on microbiota diversity [35]. In our study, the addition of ciprofloxacin to the anaerobic faecal culture not only killed EAEC strain EN1E-0007, but it obliterated the normal microbiota, such that no DNA was recoverable for the 16S analysis. While a recent study used an in vitro human gut culture, chosen to represent a limited bacterial community, to show the phage lyses a target laboratory strain of E. coli without significantly altering quantities of mutualistic gut bacteria [64], ours is the first study to our knowledge to address this question with a 16S metagenomic analysis from cultured normal human faeces. The development of antimicrobials begins with in vitro testing, and thus our approach demonstrates that therapeutic phage can be developed to target pathogens without human gut microbiome dysbiosis.
Dysbiosis from antibiotics leads to loss of keystone taxa, loss of diversity, shifts in metabolic capacity and blooms of pathogens [16]. Similarly, for children living in the developing regions of the world, repeated bouts of diarrhoea can lead to dysbiosis and serious health issues [59]. These children are at risk for malnutrition, and often suffer from environmental enteropathy: smaller villi, larger crypts, diminished absorption of nutrients and increased inflammation. During bouts of acute diarrhoea protective microbiome members, such as Bacterioidetes and Firmicutes , are reduced, creating dysbiosis. In our faecal culture analysis, consistently, in the presence of the EAEC clinical isolate and treatment with phage absent, we observed a reduction in he abundance of taxa in the genera belonging to the phylum Bacterioidetes, as well as reduction in a number of taxa in the genera of the phylum Firmicutes (Fig. 7). As EAEC is a major contributor to disease in these countries, and is thought to be an underestimated pathogen [1], PDX represents a potential therapeutic option, eliminating persistent diarrhoea, while helping to support a healthy microbiome to prevent environmental enteropathy in children.
The problem of treating MDR infections will only become more acute and alternative therapy ideas are urgently needed. Phage therapeutics can be one such option. For example, PDX and phages with similar characteristics would benefit travellers suffering from MDR, EAEC-mediated diarrhoea because it formed plaques on the case-associated, MDR strain BC/Ac338.3 (Table 1). While researchers have discussed the idea of compassionate phage therapy, few trials have occurred either in Europe or the USA. In the USA, phage therapy has mostly occurred for unmet medical needs, as experimental therapies when all other treatment options have been exhausted (https://www.sciencemag.org/news/2019/05/viruses-genetically-engineered-kill-bacteria-rescue-girl-antibiotic-resistant-infection), as phages are able to eliminate bacteria in a manner independent of the MDR phenotype [60, 65]. UC San Diego recently launched The Center for Innovative Phage Applications and Therapeutics (IPATH) to refine treatments and to bring phage therapies to market (http://www.sciencemag.org/news/2018/06/can-bacteria-slaying-viruses-defeat-antibiotic-resistant-infections-new-us-clinical). They hope to generate libraries of phages and to use cocktails for individual patients to treat MDR infections. Our efforts and those of other researchers will be essential for characterizing therapeutic phages to be used against specific pathogens while leaving the microbiome healthy and intact.
Supplementary Data
Funding information
This work was funded, in part, by Reed College Biology Undergraduate Research Project Funds.
Acknowledgements
The authors gratefully acknowledge Dr Oscar Gómez and Julio Guerra, MS, Division of Pediatrics Infectious Diseases, 875 Ellicott Street, University at Buffalo, The State University of New York University, Buffalo, NY 14203, for providing EAEC strain EN1E-0007 and clinical isolates from children suffering from diarrhoea in Columbia. We also acknowledge Katie McPherson for completion of control experiments and critical reading of the manuscript upon review.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: c.f.u., colony forming units; EAEC, Enteroaggregative E. coli; EPEC, Enteropathogenic E. coli; m.o.i., multiplicity of infection; p.f.u., plaque forming units.
Two supplementary tables are available with the online version of this article.
Escherichia virus PDX was deposited in GenBank under accession number MG963916.
References
- 1.Kaur P, Chakraborti A, Asea A. Enteroaggregative Escherichia coli: an emerging enteric food borne pathogen. Interdiscip Perspect Infect Dis. 2010;2010:254159. doi: 10.1155/2010/254159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, et al. Recent advances in understanding enteric pathogenic Escherichia coli . Clin Microbiol Rev. 2013;26:822–880. doi: 10.1128/CMR.00022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nataro JP, Mai V, Johnson J, Blackwelder WC, Heimer R, et al. Diarrheagenic Escherichia coli infection in Baltimore, Maryland, and new Haven, Connecticut. Clin Infect Dis. 2006;43:402–407. doi: 10.1086/505867. [DOI] [PubMed] [Google Scholar]
- 4.Infante RM, Ericsson CD, Jiang Z-D, Ke S, Steffen R, et al. Enteroaggregative Escherichia coli diarrhea in travelers: response to rifaximin therapy. Clin Gastroenterol Hepatol. 2004;2:135–138. doi: 10.1016/S1542-3565(03)00322-7. [DOI] [PubMed] [Google Scholar]
- 5.Opintan JA, Nataro JP, Gepi-Attee R, Newman MJ, Guerrant RL, et al. Pediatric diarrhea in southern Ghana: etiology and association with intestinal inflammation and malnutrition. Am J Trop Med Hyg. 2010;83:936–943. doi: 10.4269/ajtmh.2010.09-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abedon ST, Kuhl SJ, Blasdel BG, Kutter EM. Phage treatment of human infections. Bacteriophage. 2011;1:66–85. doi: 10.4161/bact.1.2.15845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dublanchet A, Bourne S. The EPIC of phage therapy. Can J Infect Dis Med Microbiol. 2007;18:15–18. doi: 10.1155/2007/365761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neguţ AC, Săndulescu O, Popa M, Streinu-Cercel A, Alavidze Z, et al. Experimental approach for bacteriophage susceptibility testing of planktonic and sessile bacterial populations - Study protocol. Germs. 2014;4:92–96. doi: 10.11599/germs.2014.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Negut AC, Chifiriuc MC, Sandulescu O. Bacteriophage-driven inhibition of biofilm formation in Staphylococcus strains from patients attending a Romanian reference center for infectious diseases. FEMS microbiology. 2016 doi: 10.1093/femsle/fnw193. [DOI] [PubMed] [Google Scholar]
- 10.Negut AC S-C. Bacteriophages–novel biotechnology tools available in clinical practice in Romania. Romanian. 2017 [Google Scholar]
- 11.Rose T, Verbeken G, Vos DD, Merabishvili M, Vaneechoutte M, et al. Experimental phage therapy of burn wound infection: difficult first steps. Int J Burns Trauma. 2014;4:66–73. [PMC free article] [PubMed] [Google Scholar]
- 12.Wright A, Hawkins CH, Anggård EE, Harper DR. A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin Otolaryngol. 2009;34:349–357. doi: 10.1111/j.1749-4486.2009.01973.x. [DOI] [PubMed] [Google Scholar]
- 13.Furfaro LL, Payne MS, Chang BJ. Bacteriophage therapy: clinical trials and regulatory hurdles. Front Cell Infect Microbiol. 2018;8:376. doi: 10.3389/fcimb.2018.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCallin S, Sacher JC, Zheng J, Chan DK. Current state of compassionate phage therapy. Viruses. 2019;11:343. doi: 10.3390/v11040343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keeney KM, Yurist-Doutsch S, Arrieta M-C, Finlay BB. Effects of antibiotics on human microbiota and subsequent disease. Annu Rev Microbiol. 2014;68:217–235. doi: 10.1146/annurev-micro-091313-103456. [DOI] [PubMed] [Google Scholar]
- 16.Vangay P, Ward T, Gerber JS, Knights D. Antibiotics, pediatric dysbiosis, and disease. Cell Host Microbe. 2015;17:553–564. doi: 10.1016/j.chom.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, et al. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun. 2009;77:2367–2375. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCallin S, Alam Sarker S, Barretto C, Sultana S, Berger B, et al. Safety analysis of a Russian phage cocktail: from metagenomic analysis to oral application in healthy human subjects. Virology. 2013;443:187–196. doi: 10.1016/j.virol.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 19.Sarker SA, McCallin S, Barretto C, Berger B, Pittet A-C, et al. Oral T4-like phage cocktail application to healthy adult volunteers from Bangladesh. Virology. 2012;434:222–232. doi: 10.1016/j.virol.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 20.Merabishvili M, Pirnay J-P, Verbeken G, Chanishvili N, Tediashvili M, et al. Quality-Controlled small-scale production of a well-defined bacteriophage cocktail for use in human clinical trials. PLoS One. 2009;4:e4944. doi: 10.1371/journal.pone.0004944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu P, Mathieu J, Li M, Dai Z, Alvarez PJJ. Isolation of polyvalent bacteriophages by sequential Multiple-Host approaches. Appl Environ Microbiol. 2016;82:808–815. doi: 10.1128/AEM.02382-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Legendre P, Minchin PR, O’hara RB. Package ‘vegan. Community ecology. 2013.
- 23.Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, et al. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol. 2010;Unit 19.10:1–21. doi: 10.1002/0471142727.mb1910s89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 28.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, et al. Gapped blast and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, et al. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 2005;272:5101–5109. doi: 10.1111/j.1742-4658.2005.04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manohar P, Tamhankar AJ, Lundborg CS, Ramesh N. Isolation, characterization and in vivo efficacy of Escherichia phage myPSH1131. PLoS One. 2018;13:e0206278. doi: 10.1371/journal.pone.0206278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forti F, Roach DR, Cafora M, Pasini ME, Horner DS, et al. Design of a broad-range bacteriophage cocktail that reduces Pseudomonas aeruginosa biofilms and treats acute infections in two animal models. Antimicrob Agents Chemother. 2018;62 doi: 10.1128/AAC.02573-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Savkovic SD, Villanueva J, Turner JR, Matkowskyj KA, Hecht G. Mouse model of enteropathogenic Escherichia coli infection. Infect Immun. 2005;73:1161. doi: 10.1128/IAI.73.2.1161-1170.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harrington SM, Sheikh J, Henderson IR. The PIC protease of enteroaggregative Escherichia coli promotes intestinal colonization and growth in the presence of mucin. infection and. 2009. [DOI] [PMC free article] [PubMed]
- 34.Park M, Lee J-H, Shin H, Kim M, Choi J, et al. Characterization and comparative genomic analysis of a novel bacteriophage, SFP10, simultaneously inhibiting both Salmonella enterica and Escherichia coli O157:H7. Appl Environ Microbiol. 2012;78:58–69. doi: 10.1128/AEM.06231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galtier M, Sordi LD, Maura D. Bacteriophages to reduce gut carriage of antibiotic resistant uropathogens with low impact on microbiota composition. Environmental. 2016 doi: 10.1111/1462-2920.13284. [DOI] [PubMed] [Google Scholar]
- 36.Caldwell DR, Bryant MP. Medium without rumen fluid for nonselective enumeration and isolation of rumen bacteria. Appl Microbiol. 1966;14:794–801. doi: 10.1128/AEM.14.5.794-801.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, et al. DADA2: high-resolution sample inference from illumina amplicon data. Nat Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Callahan BJ, McMurdie PJ, Holmes SP. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017;11:2639–2643. doi: 10.1038/ismej.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, et al. The Silva ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Price MN, Dehal PS, Arkin AP. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chao A. Nonparametric estimation of the number of classes in a population. Scandinavian Journal of statistics. 1984 [Google Scholar]
- 45.Chao A. Estimating the population size for capture-recapture data with unequal catchability. Biometrics. 1987;43:783–791. doi: 10.2307/2531532. [DOI] [PubMed] [Google Scholar]
- 46.Shannon CE. A mathematical theory of communication. Bell System Technical Journal. 1948;27:379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x. [DOI] [Google Scholar]
- 47.Lozupone C, Lladser ME, Knights D, Stombaugh J. UniFrac: an effective distance metric for microbial community comparison. The ISME. 2011 doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anderson MJ. Permutational Multivariate Analysis of Variance (PERMANOVA) Wiley StatsRef: Statistics Reference Online; 2014. [Google Scholar]
- 49.Bokete TN, Whittam TS, Wilson RA, Clausen CR, O'Callahan CM, et al. Genetic and phenotypic analysis of Escherichia coli with enteropathogenic characteristics isolated from Seattle children. J Infect Dis. 1997;175:1382–1389. doi: 10.1086/516470. [DOI] [PubMed] [Google Scholar]
- 50.Crowther RA. Viruses and the development of quantitative biological electron microscopy. IUBMB Life. 2004;56:239–248. doi: 10.1080/15216540412331279996. [DOI] [PubMed] [Google Scholar]
- 51.Vandenheuvel D, Lavigne R, Brüssow H. Bacteriophage therapy: advances in formulation strategies and human clinical trials. Annu Rev Virol. 2015;2:599–618. doi: 10.1146/annurev-virology-100114-054915. [DOI] [PubMed] [Google Scholar]
- 52.Wilder JN, Lancaster JC, Cahill JL, Rasche ES, Kuty Everett GF. Complete genome sequence of enterotoxigenic Escherichia coli myophage murica. Genome Announc. 2015;3:e01135-15. doi: 10.1128/genomeA.01135-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cowley LA, Beckett SJ, Chase-Topping M, Perry N, Dallman TJ, et al. Analysis of whole genome sequencing for the Escherichia coli O157:H7 typing phages. BMC Genomics. 2015;16:271. doi: 10.1186/s12864-015-1470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Samson JE, Bélanger M, Moineau S. Effect of the abortive infection mechanism and type III toxin/antitoxin system AbiQ on the lytic cycle of Lactococcus lactis phages. J Bacteriol. 2013;195:3947–3956. doi: 10.1128/JB.00296-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aleshkin AV, Rubalskii EO, Volozhantsev NV, Verevkin VV, Svetoch EA, et al. A small-scale experiment of using phage-based probiotic dietary supplement for prevention of E. coli traveler's diarrhea. Bacteriophage. 2015;5:e1074329. doi: 10.1080/21597081.2015.1074329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cairns BJ, Payne RJH. Bacteriophage therapy and the mutant selection window. Antimicrob Agents Chemother. 2008;52:4344–4350. doi: 10.1128/AAC.00574-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chibani-Chennoufi S, Sidoti J, Bruttin A, Dillmann M-L, Kutter E, et al. Isolation of Escherichia coli bacteriophages from the stool of pediatric diarrhea patients in Bangladesh. J Bacteriol. 2004;186:8287–8294. doi: 10.1128/JB.186.24.8287-8294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim M, Kim S, Park B, Ryu S. Core lipopolysaccharide-specific phage SSU5 as an auxiliary component of a phage cocktail for Salmonella biocontrol. Appl. Environ. Microbiol. 2014 doi: 10.1128/AEM.03494-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the global enteric multicenter study, GEMs): a prospective, case-control study. The Lancet. 2013;382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 60.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 61.Chan BK, Abedon ST, Loc-Carrillo C. Phage cocktails and the future of phage therapy. Future Microbiol. 2013;8:769–783. doi: 10.2217/fmb.13.47. [DOI] [PubMed] [Google Scholar]
- 62.Jeon J, Park J-H, Yong D. Efficacy of bacteriophage treatment against carbapenem-resistant Acinetobacter baumannii in Galleria mellonella larvae and a mouse model of acute pneumonia. BMC Microbiol. 2019;19:70. doi: 10.1186/s12866-019-1443-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bretin A, Lucas C, Larabi A, Dalmasso G, Billard E, et al. AIEC infection triggers modification of gut microbiota composition in genetically predisposed mice, contributing to intestinal inflammation. Sci Rep. 2018;8:12301. doi: 10.1038/s41598-018-30055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cieplak T, Soffer N, Sulakvelidze A, Nielsen DS. A bacteriophage cocktail targeting Escherichia coli reduces E. coli in simulated gut conditions, while preserving a non-targeted representative commensal normal microbiota. Gut Microbes. 2018;9:391–399. doi: 10.1080/19490976.2018.1447291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chattopadhyay MK, Chakraborty R, Grossart H-P, Reddy GS, Jagannadham MV. Antibiotic resistance of bacteria. Biomed Res Int. 2015;2015:501658. doi: 10.1155/2015/501658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Foster MA, Iqbal J, Zhang C, McHenry R, Cleveland BE, et al. Enteropathogenic and enteroaggregative E. coli in stools of children with acute gastroenteritis in Davidson County, Tennessee. Diagn Microbiol Infect Dis. 2015;83:319–324. doi: 10.1016/j.diagmicrobio.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ferreira GM, Spira B. The PST operon of enteropathogenic Escherichia coli enhances bacterial adherence to epithelial cells. Microbiology. 2008;154:2025–2036. doi: 10.1099/mic.0.2008/016634-0. [DOI] [PubMed] [Google Scholar]
- 68.Casadaban MJ. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.