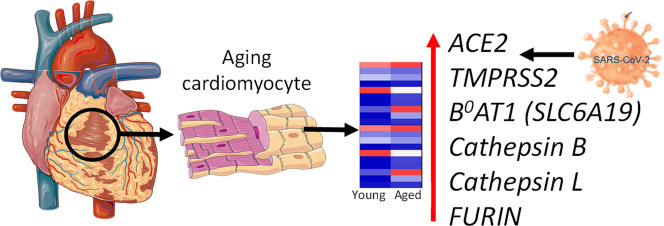
Graphical abstract
Keywords: Human cardiomyocytes, RNA-sequencing, Angiotensin, Apelin, Bradykinin, SARS-CoV-2, COVID-19
Abbreviations: ACE, angiotensin I converting enzyme; ACE2, angiotensin I converting enzyme 2; ADAM17, ADAM metallopeptidase domain 17; AGTR1, angiotensin II receptor type 1; APLNR, apelin receptor; BDKRB1, bradykinin receptor B1; BDKRB2, bradykinin receptor B2; CTSB, cathepsin B; CTSL, cathepsin L; FURIN, furin (paired basic amino acid cleaving enzyme); IFNAR1, interferon (alpha beta and omega) receptor 1; IFNAR2, interferon (alpha beta and omega) receptor 2 (IFNAR1and 2 form chains for interferons alpha and beta); IFNGR1, Interferon gamma receptor 1; IL6R, interleukin 6 receptor; IRF1, interferon regulatory factor 1 (regulates transcription of interferons); MAS1, MAS1 proto-oncogene G protein-coupled receptor; SLC6A19, solute carrier family 6 (neutral amino acid transporter) member 19 (aka B0AT1); TMPRSS2, transmembrane protease serine 2; TMPRSS11D, transmembrane protease serine 11D; TMPRSS11E, transmembrane protease serine 11E.
Introduction
Age (>70 years), case fatality rate (CFR,10.2%) and coexisting conditions, particularly cardiovascular disease (CFR,10.5%) and hypertension (CFR,6.0%), are independent predictors of adverse outcome for 45,000 COVID-19 patients in China [1]. A consensus has emerged that SARS-CoV-2 uses the same ‘receptor’ as SARS-CoV, the angiotensin converting enzyme 2 (ACE2), for initial binding to the host cell. This must be co-expressed with the serine protease TMPRSS2, that primes the spike protein S1 for endocytosis-mediated internalization of virus, employing the S2 domain for fusion to the host membrane (Fig. 1 [A]) [[2], [3], [4]]. A key difference in SARS-CoV-2 is a spike protein second site (S2’), proposed to be cleaved by the proteinase furin2. Once inside the cell cysteine proteases, cathepsin L and B, are thought critical for endosomal processing in certain cells [3,4].
Fig. 1.
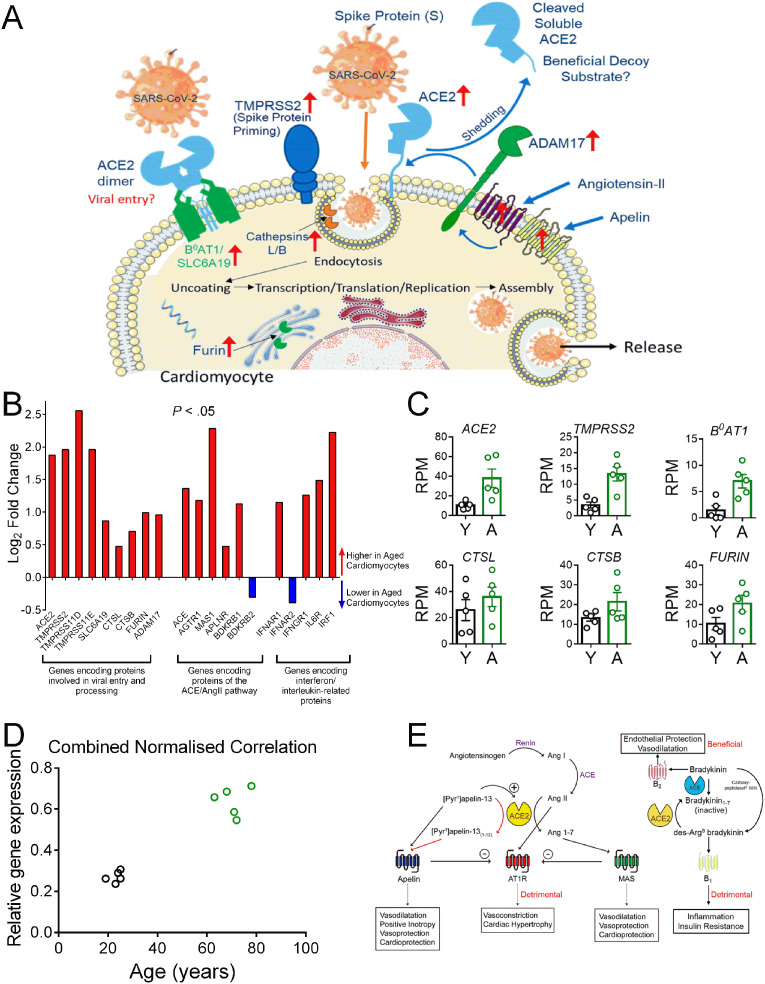
Genes Encoding Proteins Mediating Viral Entry are Upregulated with Age. See recent review [4] for a comprehensive list of references supporting the concepts outlined in this letter.
[A] Schematic diagram of the key proteins predicted from RNASeq data to be expressed by human cardiomyocytes. We propose SARS-CoV-2 binds initially to ACE2 (with the ACE2/B0AT1 complex as a potential second entry site). TMPRSS2 priming of the spike protein S1, together with further protease activation by cathepsins B and L, facilitates viral cell entry and internalization by endocytosis. Furin may also have a role in this process. Internalization of the virus with ACE2 inhibits ACE2 carboxypeptidase activity that normally hydrolyses Ang-II, apelin and des-Arg9-bradykinin. ADAM17, present on the cell surface, cleaves ACE2 to a soluble form that circulates in the plasma and could act as a decoy substrate for the virus. Levels of ADAM17 may be regulated by Ang-II and apelin acting via their respective G protein-coupled receptors [4]. ↑ - indicates genes with increased expression in aged cardiomyocytes vs young.
[B] Log2 fold change for expressed genes encoding SARS-CoV-2 entry proteins, peptide receptors in ACE/ACE2 pathway and IL-6 and interferon receptors in aged cardiomyocytes. A Chi Squared analysis for the selected gene panel shown in [B] showed significant enrichment for differentially expressed genes compared with the RNASeq data set as an entirety (P < 0.05, 95% confidence level).
[C] Scatter dot plot showing individual data points (with mean ± S.E.M) of key genes with higher expression in young (Y) verses aged (A) cardiomyocytes. Aged cardiomyocytes were double positive for ACE2 and TMPRSS2, critical for viral entry.
[D] Cardiomyocyte expression levels of the gene panel shown in [C] correlates positively with age. For each of the six targets presented in [C], the mean relative gene expression was calculated, normalized to the sample with the highest expression in RPM (non-parametric Spearman correlation, two-tailed, r = 0.86, P = 0.0025).
[E] Schematic diagram of genes encoding GPCRs that show higher expression in aged human cardiomyocytes compared with young. Activation of the renin-angiotensin system, with overproduction of Ang-II, contributes to acute respiratory distress syndrome following infection by coronoviruses. The Ang-II synthetic enzyme ACE and the ANG-II cognate receptor gene AGTR1 increased with age, together with B1 receptor gene, BDKBR1. This bradykinin receptor is selectively activated by des-Arg9-bradykinin (normally inactiveated by ACE2), representing a second deleterious pathway. Both pathways could be blocked with clinically approved drugs (ACE inhibitors, AT1 receptor antagonists and the B2 receptor antagonist Icatibant) for secondary treatment in pateints with COVID-19. Internalization of ACE2 by the virus is predicted to increase Ang II levels but reduce those of Ang1–7 but the gene encoding the proposed Ang1–7 receptor, MAS, was still detected in aged cardiomyocytes, as was the apelin receptor. The beneficial potent positive inotropic action of apelin in the heart as well as its anti-thrombotic and anti-diabetic properties, suggests its receptor is a promising therapeutic target for administered apelin and this strategy is currently under clinical investigation.
In the cardiovascular system, ACE2 protein [5] and mRNA [6] are present in cardiomyocytes. ACE2 normally functions as a carboxypeptidase cleaving single C-terminal amino acids thus hydrolysing Pro-Phe in Ang-II to Ang1 – 7, [Pyr1]-apelin-13 to [Pyr1]-apelin(1 − 12) and des-Arg9-bradykinin to inactive bradykinin(1 – 8) [4]. Internalization of ACE2 by virus potentially reduces the beneficial counter regulatory function of these peptide products to the RAAS pathway [2,4]. Conversely, the serine protease ADAM17 cleaves ACE2 to release its ectodomain and this is stimulated by Ang-II and, potentially, apelin acting via their respective G-protein coupled receptors [7]. Shed ACE2 binds SARS-CoV-2, a complex predicted not to internalize, and therefore circulating ACE2 could be exploited as a beneficial viral decoy substrate. Intriguingly, ACE2 is highly expressed in the GI tract where it is associated with B0AT1 (SLC6A19) that actively transports most neutral amino acids across the apical membrane of epithelial cells [4,8]. It is not yet known if B0AT1 and ACE2 are co-expressed in cardiomyocytes and represent an important mechanism of viral entry, but they can form a heterodimer, with the ACE2 capable of binding the spike protein S1 [8]. Interleukin 6 (IL-6), normally transiently produced, is elevated in serum and positively correlated with disease severity in COVID-19 patients [9].
We hypothesised that differential expression of genes encoding proteins in these pathways in aged cardiomyocytes could explain why the myocardium of older patients might be particularly vulnerable to the virus, manifesting as cardiovascular complications such as myocarditis. To selectively analyse cardiomyocyte gene expression, we generated strand-specific RNA-sequencing libraries from RNA isolated from flow-sorted cardiomyocyte nuclei from left ventricular tissue [10]. RNASeq data were compared between five young (19-25 yr) and five older (63-78 yr) Caucasian males, not on medication, with no evidence of cardiovascular disease post-mortem.
Viral entry, membrane fusion and endocytosis
The key finding (Fig. 1[B,C]) is that expression of genes encoding proteins hypothesised as important for viral entry, crucially including ACE2 and TMPRSS2, were upregulated in aged cardiomyocytes, as well as TMPRS11E and TMPRS11D, (where function is less well established), FURIN and cathepsins CTSL and CTSB. Importantly, while B 0 AT1/SLC6A19 was minimally expressed in young cardiomyocytes it was upregulated in aged cardiomyocytes. Moreover, combined relative cardiomyocyte expression of these genes correlated positively with age (Fig. 1[D]).
ACE/ACE2 regulation
The expression of Ang-II synthetic enzyme ACE and cognate receptor gene AGTR1 together with BDKBR1 increased with age, the latter normally not expressed in myocytes until induced by inflammation and selectively activated by des-Arg9-bradykinin (Fig. 1[B, E]) [9]. Both are potentially deleterious peptides, that we hypothesise would be increased by loss of cell surface ACE2 caused by viral internalization or ADAM17-mediated shedding. Expression of genes encoding receptors for peptides that normally mediate beneficial counter-regulatory effects to Ang-II in the heart, MAS1/Ang1 – 7, APLNR/apelin and BDKRB2/bradykinin were both up and downregulated.
Inflammatory mediators and endogenous antiviral strategies
Penetration of SARS-CoV-2 into lung alveoli, resulting in the ‘cytokine storm’ is a major determinant for COVID-19 patient intubation and mortality. IL-6 is a key mediator [4]: its receptor (IL6R) is expressed on cardiomyocytes, with a modest increase with age suggesting anti-IL-6R monoclonal antibodies, currently in trials [9] may prove cardioprotective.
Unlike most other cells, cardiomyocyte numbers remain stable, regenerating slowly. Although particularly vulnerable to viral infection they have evolved intrinsic mechanisms to combat cytotoxic effects and viral replication mainly by generating type 1 interferon responses. SARS-CoV-2 counterattacks by producing proteins that interfere with interferon pathways. While interferon receptor genes (IFNAR1/IFNAR2) were expressed in cardiomyocytes, no pattern emerged with age.
Our objective is to highlight SARS-CoV-2 related genes that have higher expression in aged compared with young adult cardiomyocytes. Nuclear RNA-seq measures transcriptional activity and therefore future studies will be needed to test the consequences of the detected changes in gene expression for cardiomyocyte function. In particular, it is not established whether direct viral infection results in injury to cardiomyocytes or if cardiac complications are predominantly mediated by cytokines. While older individuals have a worse outcome, it is not yet established if they are more vulnerable to viral infection. Selective enzyme inhibitors/antagonists are available as experimental compounds or clinically approved drugs to dissect these pathways (https://www.guidetopharmacology.org/coronavirus.jsp). Drugs to treat both acute and long-term effects of SARS-CoV-2 will need to focus on these three key stages of infection: preventing the virus entering cells, stopping viral replication and reducing resulting tissue damage, particularly in the hearts of aged individuals that are the most vulnerable members of society to COVID-19.
Funding
This work was supported by the Wellcome Trust 107715/Z/15/Z (UK), Addenbrooke's Charitable Trust (UK), British Heart Foundation Translational Award TG/18/4/33770 (A.P.D. & J.J.M.)(UK). Wellcome Trust PhD fellowship (Cardiovascular & Metabolic Disease)(UK), CVON EARLY-HFPEF consortium grant (Dutch Heart Foundation, Netherlands) and EMBO short-term fellowship (Ref 8471) (E.L.R). Research Foundation Flanders FWO (Fonds Wetenschappelijk Onderzoek) Odysseus Project Grant 90663, KULeuven Internal Funding (C1) (C14/17/088 (Belgium) and BBSRC Institute Strategic Program Grant (BBS/E/B/000C0400-403) (H.L.R.)(UK). The Center for Regenerative Therapies Dresden, the Karolinska Institutet, the Swedish Research Council, the Ragnar Söderberg Foundation, and the Åke Wiberg Foundation (O.B.)(Sweden).
Disclosures
None.
Acknowledgements
Heart samples were obtained from the KI Donatum, Karolinska Institutet, Stockholm, Sweden with ethical approval. Quality control and sequencing was carried out at The Babraham Institute Sequencing Facility, Cambridge (UK).
References
- 1.Sommerstein R. Preventing a covid-19 pandemic: ACE inhibitors as a potential risk factor for fatal Covid-19. BMJ. 2020;368 Doi: https://www.bmj.com/content/368/bmj.m810/rr-2. [Google Scholar]
- 2.Liu P.P., Blet A., Smyth D., Li H. The science underlying COVID-19: implications for the cardiovascular system. Circulation. 2020;142:68–78. doi: 10.1161/CIRCULATIONAHA.120.047549. [DOI] [PubMed] [Google Scholar]
- 3.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexander S.P.H., Armstrong J., Davenport A.P., Davies J., Faccenda E., Harding S.D. A rational roadmap for SARS-CoV-2/COVID-19 pharmacotherapeutic research and development. IUPHAR review 29. Br. J. Pharmacol. 2020:1–25. doi: 10.1111/bph.15094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hikmet F, Méar L, Edvinsson A, Micke P, Uhlén M, Lindskog C. The protein expression profile of ACE2 in human tissues bioRxiv 2020.03.31.016048; Doi: doi: 10.1101/2020.03.31.016048. [DOI] [PMC free article] [PubMed]
- 6.Tucker N.R., Chaffin M., Bedi K.C., Papangeli I., Akkad A.D., Arduini A. Myocyte specific upregulation of ACE2 in cardiovascular disease: implications for SARS-CoV-2 mediated myocarditis. Circ. 2020 doi: 10.1161/CIRCULATIONAHA.120.047911. Preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu J., Sriramula S., Xia H., Moreno-Walton L., Culicchia F., Domenig O. Clinical relevance and role of neuronal AT1 receptors in ADAM17-mediated ACE2 shedding in neurogenic hypertension. Circ. Res. 2017;121:43–55. doi: 10.1161/CIRCRESAHA.116.310509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toniati P., Piva S., Cattalini M., Garrafa E., Regola F., Castelli F. Tocilizumab for the treatment of severe COVID-19 pneumonia with hyperinflammatory syndrome and acute respiratory failure: a single center study of 100 patients in Brescia, Italy. Autoimmun. Rev. 2020;19 doi: 10.1016/j.autrev.2020.102568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thienpont B., Aronsen J.M., Robinson E.L., Okkenhaug H., Loche E., Ferrini A. The H3K9 dimethyltransferases EHMT1/2 protect against pathological cardiac hypertrophy. J. Clin. Invest. 2017;127:335–348. doi: 10.1172/JCI88353. [DOI] [PMC free article] [PubMed] [Google Scholar]