Abstract
COVID-19 has spread to most countries in the world. However, there are differences in the rate of infection in different countries. Specifically, high incidence was reported in specific areas in China (Wuhan) and Italy (Lombardy). These differences may be related to different Human Leucocyte Antigen (HLA) patterns in various geographic areas. We suggest HLA spreading between Italy and China is related to the travels of Marco Polo through the Silk Road as a potential historic explanation to COVID-19 spreading.
Keywords: COVID-19, infection rate, HLA, Marco Polo, silk road
Introduction
Coronavirus disease 2019 (COVID-19) became a worldwide pandemic disease as of December 2019, with more than 5.5 million people infected, of whom more than 350,000 have died resulting in a crude fatality ratio [CFR] of 3.8%. The overall CFR varies by location and intensity of transmission and was reported as high as 5.8% in Wuhan vs. 0.7% in other areas in China. After China COVID-19 spread throughout the world, but became very aggressive in Italy, especially in Lombardy, causing high rates of infectivity and mortality.
Why COVID-19 infection rate and mortality were high in Wuhan and Lombardy, while in other European countries like Germany and Sweden the numbers were much lower? We suggest that the reason relates to Human Leucocyte Antigen (HLA) typing and that historical facts may lay behind the variability of COVID-19 severity and intensity of transmission in these areas. We suggest that HLA spreading from Italy through the travels of Marco Polo to China, travelling through the Silk Road and passing Iran-also a high area with high COVID-19 CFR- is the reason. Our hypothesis may also explain the high rate of infection in New York and London where there are many Italian and Chinese immigrants.
Historical background
Wuhan is the capital of Hubei province in central China with a population of over 11 million habitants. As of early 2020, Wuhan is most recognized as the origin of COVID-19 outbreak. Wuhan lies on the confluence of the Yangtze River and its largest tributary, the Han River, that divides the city into the three districts.
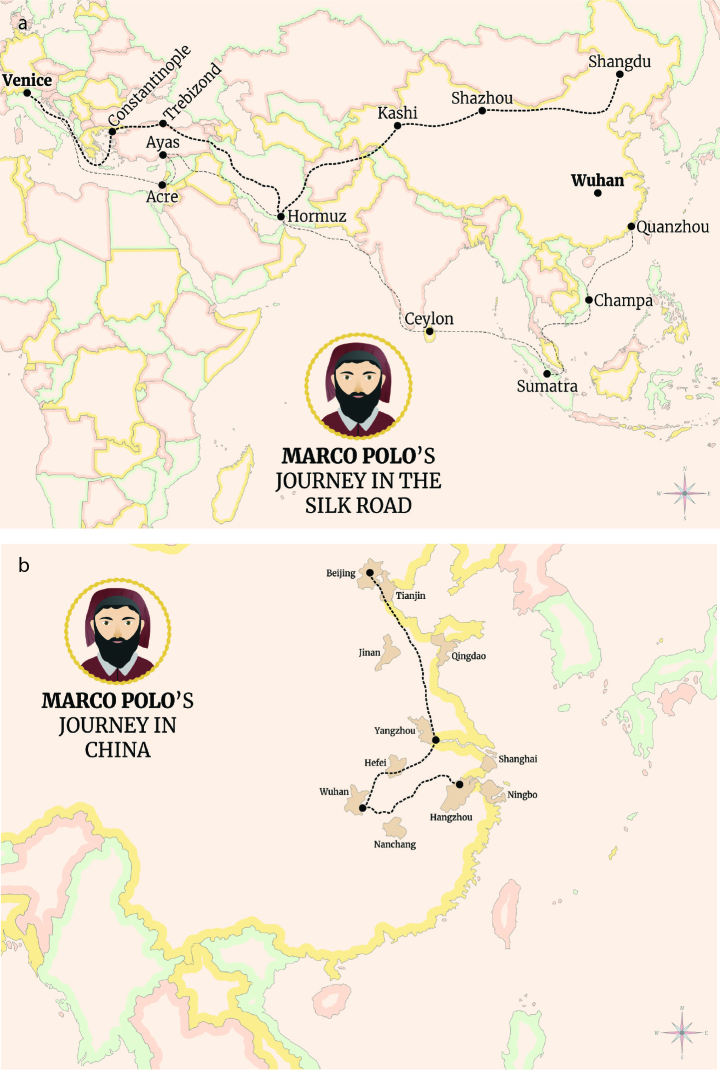
Marco Polo was born in in 1254 in Venice, the capital of the Veneto region in northeastern Italy, which is very close to the district of Lombardy (Figure 1a). He travelled through Asia along the Silk Road between 1271 and 1295, and is known as a merchant and explorer who was the first to reach China and at the time of the rich Mongol Empire in the Yuan Dynasty (1, 2). Marco learned the mercantile trade from his father and his uncle, and the three of them explored many places along the Silk Road-travelling through Persia (Iran of today), Afghanistan and the Pamir Mountains. Then, they cut across the vast Gobi Desert to Beijing in China. They were received by the Mongol emperor Kublai Khan, and Marco was appointed to serve as Khan’s foreign emissary to India and Burma, and was sent on many diplomatic missions throughout the empire. Marco immersed himself in Chinese culture, learning the language and customs and often visiting the court of Kublai Khan in Xanadu, the summer capital of the Yuan dynasty (Figure 1b). It is clear that the long travels in these roads took many days and young Marco got close acquaintance with the local population. During his 17 years of service in China, Marco traveled extensively inside the country and even accompanied the Mongol princess Kököchin to Persia in a two-year-trip around 1290 to 1293. After leaving the princess, he travelled to Constantinople (İstanbul) and then to Venice, returning to Italy after 24 years.
Figure 1. a, b.
Marco Polo travels from Italy through the Silk Road (a) and in China reaching Wuhan (b).
Suggested connection: HLA type associated with failure to eliminate COVID-19 virus
HLA genes are important components of immune system regulation. These genes code for glycoproteins that bind peptides and present them to T cell receptors. If the bound peptide is a viral pathogen it will result in cellular and humoral responses aimed to eliminate the pathogen. Therefore, HLA loci are the prototypical candidates for genetic susceptibility to infectious diseases, and different HLA haplotypes are associated with distinct disease susceptibilities (3). Indeed, the susceptibility to various infectious diseases such as tuberculosis, leprosy, HIV, HPV, hepatitis B, and influenza is associated with specific HLA haplotypes (4–6). Accordingly, it seems less advantageous to have HLA molecules with increased binding specificities to the SARS-CoV-2 virus peptides on the cell surface of an antigen-presenting cells. Moreover, geographic variation in HLA type may explain the different rates of susceptibility to COVID-19 in various autoimmune disease (7), and the possibility that specific alleles such as HLA-DRB1*16:02 allele may serve as a risk to some autoimmune diseases and as a protective factor in others (8).
Previous studies have reported the association of HLA-B*4601 with the severity of severe acute respiratory syndrome (SARS) infection in Taiwanese patients (9), and association of HLA class I (B*0703) and class II (DRB1*0301) genotypes with susceptibility and resistance to SARS in Chinese patients (10). Similarly, severity of SARS was shown to be significantly associated with HLA-B*4601 (11). In accordance, it seems rational to suggest that specific HLA loci will be associated with the development of anti-COVID-19 immunity as well as with induction of protective immunity. Indeed, Nguyen et al. (12) performed in-silico analysis of viral peptides for MHC class I binding affinity across 145 HLA -A, -B, and -C genotypes for all COVID-19 peptides. They reported that HLA-B*46:01 allele had the fewest predicted binding peptides for COVID-19. These findings suggest that individuals with this allele are less vulnerable to the disease. In their work the authors presented a series of world maps displaying the estimated global distributions and frequencies across 145 different HLA alleles, as well as the distribution of predicted SARS-CoV-2 peptide binding and global haplotype frequencies across all haplotypes containing that HLA allele. Looking at these maps and at geographical HLA comparison maps (www.igdawg.org/software/browser-beta.html), we suggest additional HLA alleles such as HLA-A*0201, HLA-DPA1*0103, HLA-B*07:04, HLA-B*13:02, HLA-B*14:03, HLA-C*01:01, and HLA-C*05:01, that are common between both the Italian and Chinese populations and therefore support the Marco Polo hypothesis of HLA geographic spreading.
Conclusion
We suggest that specific HLA loci associated with susceptibility to COVID-19 infection, will be similar between patients from Wuhan China, patients from Lombardy region in Italy and patients from Italian or Chinese origin in New York and London, as these alleles have spread to vulnerable populations through the historical travels of Marco Polo.
We suggest that studies using innovative technologies like RNAseq to construct detailed HLA portraits of vulnerable (Italy) and resistant (Sweden, Germany) populations will enable better understanding of how natural selection has acted and identify variants that increase the risk of COVID-19 infection.
Acknowledgement
The authors thank Mrs. Netta Kasher for the figure illustrations.
Footnotes
Conflict of Interest: The authors have no conflict of interest to declare.
Financial Disclosure: The authors declared that this study has received no financial support.
References
- 1.Latham RE. The Travels by Marco Polo. 30th ed. London: Penguin Books; 1958. [Google Scholar]
- 2.Bergreen L. Marco Polo: From Venice to Xanadu. New York, NY: Knopf Doubleday Publishing Group; 2007. [Google Scholar]
- 3.Meyer D, Aguiar VR, Bitarello BD, Brandt DY, Nunes K. A genomic perspective on HLA evolution. Immunogenetics. 2018;70:5–27. doi: 10.1007/s00251-017-1017-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackwell JM, Jamieson SE, Burgner D. HLA and infectious diseases. Clin Microbiol Rev. 2009;22:370–85. doi: 10.1128/CMR.00048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amiot L, Vu N, Samson M. Immunomodulatory properties of HLA-G in infectious diseases. J Immunol Res. 2014;2014 doi: 10.1155/2014/298569. 298569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joosten SA, Sullivan LC, Ottenhoff TH. Characteristics of HLA-E restricted T-cell responses and their role in infectious diseases. J Immunol Res. 2016;2016 doi: 10.1155/2016/2695396. 2695396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ehrenfeld M, Tincani A, Andreoli L, Cattalini M, Greenbaum A, Kanduc D, et al. Covid-19 and autoimmunity. Autoimmun Rev. 2020;19:102597. doi: 10.1016/j.autrev.2020.102597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y, Li S, Huang R, Zhang Z, Petersen F, Zheng J, et al. Comprehensive meta-analysis reveals an association of the HLA-DRB1*1602 allele with autoimmune diseases mediated predominantly by autoantibodies. Autoimmun Rev. 2020;19:102532. doi: 10.1016/j.autrev.2020.102532. [DOI] [PubMed] [Google Scholar]
- 9.Lin M, Tseng HK, Trejaut JA, Lee HL, Loo JH, Chue CC, et al. Association of HLA class I with severe acute respiratory syndrome coronavirus infection. BMC Med Genet. 2003;4:9. doi: 10.1186/1471-2156-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi Y, Wang Y, Shao C, Huang J, Gan J, Huang X, et al. COVID-19 infection: The perspectives on immune responses. Cell Death Differ. 2020;27:1451–4. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ng MH, Lau KM, Li L, Cheng SH, Chan WY, Hui PH, et al. Association of human-leukocyte-antigen class I (B*0703) and class II (DRB1*0301) genotypes with susceptibility and resistance to the development of severe acute respiratory syndrome. J Infect Dis. 2004;190:515–8. doi: 10.1086/421523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen A, David JK, Maden SK, Wood MS, Weeder BR, Nellore A, et al. Human leukocyte antigen susceptibility map for SARS-CoV-2. J Virol. 2020;94:e00510–20. doi: 10.1128/JVI.00510-20. [DOI] [PMC free article] [PubMed] [Google Scholar]