Abstract
Background
Gastric cancer (GC) is a worldwide malignancy and the molecular mechanism of the GC carcinogenesis has not been fully elucidated. Our previous study suggested CDCA5 played a role in GC development via regulating cell proliferation, migration, and apoptosis in GC cells.
Material/Methods
Here, we first carried out bioinformatics analysis and found cyclin-dependent kinase 1 (CDK1) was possibly associated with CDCA5 using STRING. Then, the expression levels of CDK1 and CDCA5 in cancer tissues were estimated through Oncomine and The Cancer Genome Atlas (TCGA) database. After that, functional experiments were exerted to detect the association of CDK1 and CDCA5. Finally, cell proliferation assay, colon formation assay, cell scratch assay, transwell migration and invasion assays were applied to explore the roles of CDK1 and CDCA5 in GC cells MGC-803.
Results
CDK1 and CDCA5 were both upregulated and co-expressed in GC tissues. The expression of CDK1 and CDCA5 in MGC-803 was positively related. CDK1 or CDCA5 inhibition can suppress the proliferation, colon formation, migration, and invasion abilities of GC cells.
Conclusions
Co-expression of CDK1 and CDCA5 might confer cell proliferation, migration, and invasion abilities in GC cells, and this can provide some clues for further therapies of gastric tumors.
MeSH Keywords: CDC2 Protein Kinase, Cell Cycle Proteins, Cell Migration Assays, Cell Proliferation, Neoplasm Invasiveness, Stomach Neoplasms
Background
Gastric cancer (GC) is a digestive tract malignant tumor and ranks the third cause among cancer-associated deaths in China [1]. Though many advances have been made in GC treatment, the 5-year survival rate is still low due to low early diagnosis rate, extensive invasion, and high lymphatic metastasis rate [2]. Therefore, studies about the molecular mechanisms in GC progression is still needed for better therapies.
CDCA5 (also named Sororin and with a full name of cell division cycle associated 5) is a conserved protein with 252 amino acids and the molecular weight is ~27.6 kDa. It plays a role in mitosis stabilizing cohesion complex association with chromatin [3]. Recently, CDCA5 has been found to be aberrantly expressed in many tumor tissues and associated with poor prognosis in some cancer types [4–6]. Our previous research demonstrated CDCA5 overexpression enhanced cell proliferation and migration, while CDCA5 inhibition can arrest cell cycle in G2/M phase in GC cells MGC-803 [7]. In order to find possible interaction proteins of CDCA5, we used the online free functional protein association network STRING (https://string-db.org/cgi/input.pl), and found CDK1, AURKB, and BIRC5 may interact with CDCA5. Then, we searched the gene expression levels of these genes using the Oncomine (https://www.oncomine.org/) and The Cancer Genome Atlas (TCGA) database (https://www.cancer.gov/). The results showed CDK1 was abnormally expressed in cancer tissues. CDK1 is a family member of the cyclin-dependent kinases (CDKs) which are important drivers in cell cycle regulation [8]. Research showed CDK1 was an essential regulator for cell cycle at G1/S and G2/M checkpoints [9,10] and it can also be regulated by certain microRNAs to inhibit tumor cell proliferation in cancers [11,12]. Until now, the role of CDK1 in GC is not clear. Herein, given that CDK1 may interact with CDCA5 in GC, we aimed to explore the relationship between CDCA5 and CDK1 and their roles in MGC-803 looking to identify potential new targets in GC treatment.
Material and Methods
Bioinformatics analysis
STRING is a well-known protein-protein interaction database [13]. We first input CDCA5 and found CDK1 may be a potential interaction protein using STRING. Then, we analyzed the mRNA expression levels of CDCA5 and found CDK1 in the TCGA [14] and Genotype-Tissue Expression (GTEx) projects on the GEPIA website (http://gepia.cancer-pku.cn/) which containing more than 9000 tumor tissues and 8000 normal tissues [15]. Next, based on the gene expression data, overall survival (OS) or disease-free survival (DFS) analysis were performed using Log-rank test for hypothesis test. Co-expression of CDK1 with CDCA5 in GC samples was also assessed from the Oncomine dataset under standardized instructions on the website [16–19].
Cell lines
Human GC cell line MGC-803 [20] was obtained from the Cell Bank of Sun Yat-Sen University (Guangzhou, China) and cultured in RPMI-1640 medium (Gibco BRL, Grand Island, NY, USA) supplement with 10% fetal bovine serum (FBS, Gibco BRL, Grand Island, NY, USA). The cells were incubated at 37°C with 5% CO2.
Small interfering RNAs and transfection
To knockdown CDCA5 and CDK1 in GC cell line, specific small interfering RNAs (siRNA) against CDCA5 and CDK1, and scramble siRNA not targeting any annotated human genes (control) were designed and purchased from Shanghai Genechem (Genechem, Shanghai, China). The siRNAs against CDCA5 were as follows:
-
si-CDCA5, sense: 5′-CGGAAAGUUUCCUCGCGUA-3′ and
antisense: 5′-UACGCGAGGAAACUUUCCG-3′.
Three siRNAs targeting CDK1 were designed as follows:
-
si-CDK1-1, sense: 5′-GGAACUUCGUCAUCCAAAU-3′ and
antisense: 5′-AUUUGGAUGACGAAGUUCC-3′;
-
si-CDK1-2, sense: 5′-GGUUAUAUCUCAUCUUUGA-3′ and
antisense: 5′-UCAAAGAUGAGAUAUAACC-3′;
-
si-CDK1-3, sense: 5′-GUACUGCAAUUCGGGAAAU-3′ and
antisense: 5′-AUUUCCCGAAUUGCAGUAC-3′.
For transfections, cells were first seeded onto 6-well plates. When the cell confluency got about 75%, the indicated siRNAs were transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) under the instruction manual.
RNA extraction and real-time quantitative polymerase chain reaction (RT-qPCR)
After the transfection, the cells were harvested 24 hours later and the total RNAs were extracted by TRIzol (Invitrogen, USA). The first-strand cDNA was synthesized by reverse transcription using 250 ng total RNAs in a volume of 10.0 μL using a first strand cDNA synthesis kit (Roche, Basel, Switzerland). Then the cDNA was diluted in sterile water and the quantitative real-time polymerase chain reaction (RT-qPCR) was carried out with a mixture as follows: 1.0 μL cDNA, 0.4 μL forward primer, 0.4 μL reverse primer, 5.0 μL SYBR® Select Master mix (Roche, Basel, Switzerland) and 3.2 μL deionized water. The amplification was exerted using a PikoReal™ Real-Time PCR system (Thermo Fisher Scientific, Waltham, MA, USA). The primers were obtained by Shanghai Generay Co., Ltd. (Shanghai, China) and the sequences were as follows:
-
CDCA5-f 5′-CGTAAGAAGAAGAAAATGCC-3′ and
CDCA5-r 5-ACAGGACAGGAGGGAGAG-3′;
-
CDK1-f 5-GGATGTGCTTATGCAGGATTCC-3′and
CDK1-r 5′-CATGTACTGACCAGGAGGGATAG-3′;
-
β-actin-f 5′-GGGAAATCGTGCGTGACATTAAGG-3′ and
β-actin-r 5′-CAGGAAGGAAGGCTGGAAGAGTG-3′.
β-actin was used as an endogenous control. The experiments were exerted in triplicate and every experiment was repeated 3 times independently. The 2−ΔΔCt method was applied for data analysis [21].
Western blot
Cells were washed 3 times with cold, sterile phosphate-buffered saline (PBS), and then lysed by addition of 1 mL of radioimmunoprecipitation assay (RIPA) lysis buffer (Solarbio, Beijing, China) supplemented with protease inhibitor cocktail (Sigma, St. Louis, MO, USA). Cell lysates were collected into tubes and centrifuged at 12 000 g for 15 minutes at 4°C. The supernatants and protein loading buffer (SDS-PAGE gel, CWBIO, Beijing, China) were mixed at a ratio of 4: 1 according to the instructions and then divided into 2 aliquots. One aliquot was fractionated by SDS-PAGE, and the other was stored at −80°C. All the western blotting procedures were performed according to the manuals. In this study, the primary antibodies used were as follows: a rabbit anti-human CDCA5 (ab192237, 1: 1000, Abcam, Cambridge, MA, USA), a rabbit anti-human CDK1 antibody (ab32094, 1: 1500, Abcam, Cambridge, MA, USA), a rabbit anti-human β-actin antibody (ab8227, 1: 1000; Abcam, Cambridge, MA, USA). An HRP-labeled goat anti-rabbit antibody (A0208, 1: 1000, Beyotime, China) was used as the second antibody. The band signals were detected using chemiluminescence (Millipore, MA, USA) by infrared imaging system (LI-COR Biosciences, Lincoln, NE, USA). The target protein was normalized to β-actin through a comparison of the gray scale values, and the analysis was performed by Quantity One (version 4.6.2).
Cell proliferation experiment
About 24 hours after the transfection, the cells were harvested and re-cultured in a 96-well microplate with about 3000 cells/well. Each group had 5-wells. After 24 hours, each well was added with 100 μL mixture containing of 10 μL CCK-8 (Cell Counting Kit-8, Dojindo Laboratories, Kumamoto, Japan) and 90 μL RPMI-1640, and the cells were continued cultured for 3 hours at 37°C with 5% CO2. The value of CCK-8 was measured at 450 nm by microplate reader (Thermo Fisher Scientific, USA) at the time point of 24, 48, and 72 hours. The experiments were performed for 3 independent times with each had 3 replicates.
Colon formation experiment
About 24 hours after the transfection, the cells were collected and re-cultured in 6-well plates with about 1.0×103 cells/well. The medium was switched to fresh Dulbecco’s Modified Eagle Medium (DMEM) medium with 10% FBS every 72 hours. Generally, 1–2 weeks later, the cells were fixed by pre-cold methanol for 20 minutes and then stained with 0.2% purple crystal at 25°C for 25–35 minutes. For the cell number counting, the colonies including more than 50 cells were included. The experiments were performed for 3 independent times with each had 3 replicates.
Cell scratch test
Forty-eight hours after cell transfection, when the confluence of the cells in the 6-well plate reached 100%, a single scratch wound at the same position in each well was generated by scratching the cell monolayer with a 10 μL sterile pipette tip. Then, the remaining cells washed 3 times with use fetal bovine serum (FBS)-free 1640 culture basic culture. Then the cells were filled with RPMI-1640 medium contained 2.5% FBS and incubated with 5% CO2 at 37°C. Spontaneous cellular migration of the cells at the edge of the scratch was observed and photographed at time 0, 24, and 48 hours using a light microscope (Olympus BX41, Olympus Corporation, Tokyo, Japan). The experiments were performed for 3 independent times with each had 3 replicates.
Transwell migration experiment
Twenty-four hours after the transfection, the cells were collected and re-suspended with blank RPMI-1640 medium. Then 200 μL suspensions (about 2.5×105 cells) were plated on the upper chamber of a Matrigel coated Transwell (200 μL/well), and the lower chambers contained 600 μL RPMI-1640 supplemented with 10% FBS. The chambers were placed in an incubator with 5% CO2 at 37°C. After 48 hours, the cells on the lower chambers were fixed with cold methanol for about 30 minutes, next the cells were stained with 0.1% crystal violet for about 15 minutes at 25°C. Finally, 5 random fields were photographed and counted using a light microscope. The experiments were performed for 3 independent times with each had 3 replicates.
Transwell invasion assay
The Matrigel matrix (Corning, Inc., Corning, NY, USA) and the RPMI-1640 medium without fetal bovine serum were mixed in a ratio of 1: 5, and then 60 μL above mixture was added into the upper chamber of Transwell for 4 hours at 37°C. Next, the Transwell upper chamber were added with 200 μL cell suspension including about 2.5×105 cells, and the lower chamber were added with 600 μL RPMI-1640 medium supplemented with 10% FBS. After that, the chambers were placed at 37°C with 5% CO2. Forty-eight hours after the incubation, the non-migrating cells were gently removed from the upper chamber with a cotton swab, and the remaining fixed and stained steps were similar with that in the aforementioned migration assay. The experiments were performed for 3 independent times with each had 3 replicates.
Statistical analysis
The data in this study were analyzed by SPSS 25.0 and GraphPad Prism 8. The data were indicated as the mean±standard deviation (SD). Independent sample t-test was applied for the comparison between 2 groups, and one-way ANOVA was exerted for comparisons between multiple groups. P<0.05 was thought to be statistically significant.
Results
Bioinformatics analysis suggested co-expression of CDCA5 and CDK1 in GC tissues
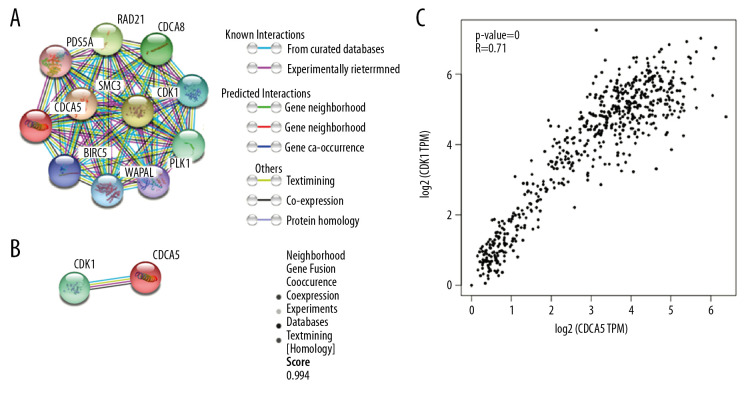
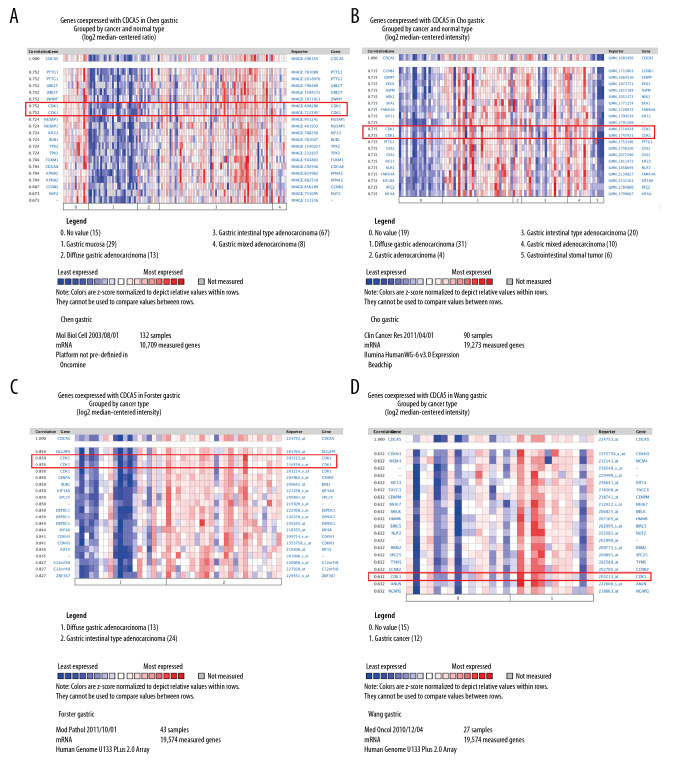
In the functional protein association network constructed by STRING, the interaction between CDCA5 and CDK1 was evaluated from 5 aspects: genomic context predictions, high-throughput laboratory experiments, co-expression, automated text mining, and previous knowledge in databases. The results showed a high score between the association of CDCA5 and CDK1 (Figure 1A, 1B). Data from the TCGA expression data suggested a positive correlation between CDCA5 and CDK1 expression using a pairwise gene expression correlation analysis (Figure 1C). Co-expression analysis of the clinical specimens from Oncomine database demonstrated CDCA5 was highly correlated with CDK1 in GC (Figure 2). These results confirmed an association of CDCA5 and CDK1.
Figure 1.
Co-expression between CDCA5 and CDK1. (A) Functional proteins association network of CDCA5 and CDK1 and other co-expressed genes in the STRING database. (B) Scoring of various aspects of the interaction between CDCA5 and CDK1. (C) Analysis of the TCGA gastric cancer dataset by the GEPIA website to determine the correlation between CDCA5 and CDK1 expression (* P<0.05)
Figure 2.
Co-expression analysis of CDCA5 and CDK1. Co-expression analysis of CDCA5 and CDK1 in clinical specimens of gastric cancer showed high correlation between the 2. (A) R=0.752, n=132; (B) R=0.715, n=90; (C) R=0.850, n=43; (D) R=0.632, n=27.
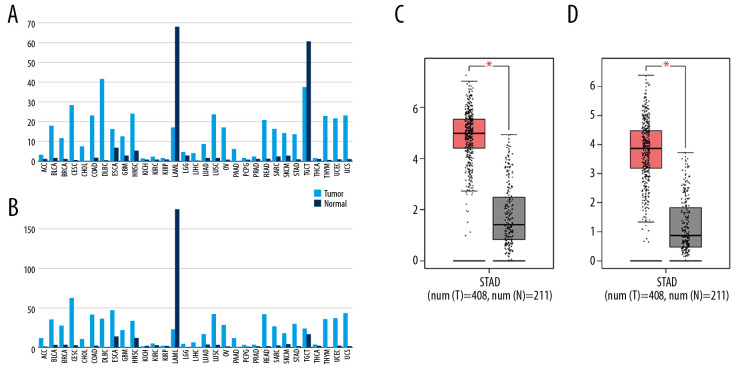
Then, the mRNA expression levels of CDCA5 and CDK1 were analyzed from TCGA and GTEx project containing a total of 9736 tumor samples and 8726 normal samples. The result presented both CDCA5 and CDK1 expressions were upregulated in most tumor tissues including GC compared with the matched normal tissues (Figure 3), which suggested CDCA5 and CDK1 had a similar expression pattern in GC tissues.
Figure 3.
Upregulation of CDCA5 and CDK1 in gastric cancer. (A) The gene expression profile of CDCA5 across all tumor samples and matched normal tissues, based on the expression of CDCA5 from the The Cancer Genome Atlas (TCGA) various types of cancer research cohort data sets, generated by the GEPIA website, the height of bar represents the median expression of certain tumor type or normal tissue. (B) The gene expression profile of CDK1 across all tumor samples and paired normal tissues. (C) Analysis of TCGA gastric cancer tissue data(n=408) and match TCGA normal and GTEx data (n=211) through the GEPIA website, to obtain CDCA5 expression levels. (D) Analysis of TCGA gastric cancer tissue data (n=408) and match TCGA normal and GTEx data (n=211) through the GEPIA website, to obtain CDK1 expression levels (Log2FC=1, P-value=0.01).
The mRNA expression levels of CDCA5 and CDK1 in 3 GC lines and selection of efficient siRNA fragments against CDCA5 and CDK1
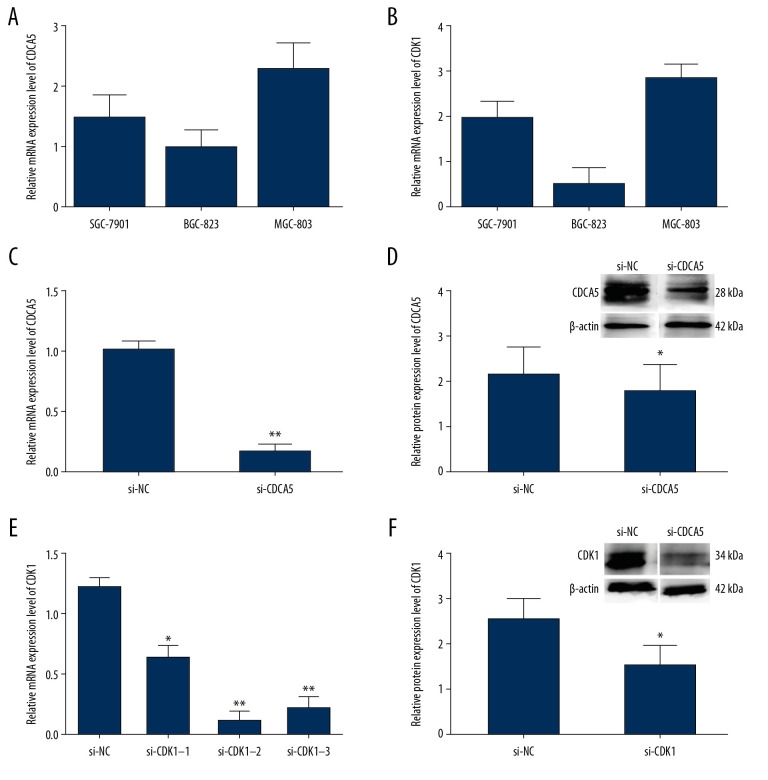
The relative mRNA expression levels of CDCA5 and CDK1 in 3 GC cell lines SGC-7901, BGC-823 and MGC-803 were tested by RT-qPCR, respectively. Figure 4A showed CDCA5 mRNA was the highest in MGC-803. Interesting, CDK1 had a similar expression pattern as CDCA5 in 3 cell lines (Figure 4B). Therefore, MGC-803 was chosen for the cell line in the following experiments.
Figure 4.
The relationship of CDCA5 and CDK1 in MGC-803 cells. The relative CDCA5 mRNA levels (A) and CDK1 mRNA levels (B) were analyzed in 3 different GC cell lines SGC-7901, BGC-823 and MGC-803 using RT-qPCR. Among which, MGC-803 cells could effectively be expressed in both CDCA5 and CDK1, and thus was chosen as the research cell line in this study. (C) Cells were assigned for 2 groups and transfected with the control siRNA (si-NC) and si-CDCA5, respectively. The CDCA5 mRNA level was reduced significantly compared with the control group (** P<0.01). (D) The CDCA5 protein level was reduced significantly compared with the control (* P<0.05). The results suggested the si-CDCA5 fragment was effective. (E) Cells transfected with three different CDCA5-specific siRNAs were named as si-CDK1–1, si-CDK1–2 and si-CDK1–3, respectively. Group transfected with scramble siRNA was named as si-NC and worked as a control. Compared with the si-NC control group, the mRNA level of CDK1 in the 3 siRNA interfering groups were all reduced, among which, the si-CDK1–2 group reduced most significantly (* P<0.05 and ** P<0.01). Thus, we selected si-CDK1–2 as the specific siRNA fragment against CDK1 for the following experiments and renamed si-CDK1. (F) The protein expression levels of CDK1 in the si-CDK1 group and the control. Consistent with the result in (E), the protein expression level reduced significantly in the si-CDK1 group (* P<0.05).
To explore the function of CDCA5 and CDK1, we applied siRNA interfering method to knockdown gene expression and investigate the effects on GC cell. For the CDCA5 inhibition experiments, compared with si-NC control group (transfecting siRNAs not targeting any gene), the mRNA and protein expression levels of CDCA5 in si-CDCA5 group (transfecting siRNAs targeting CDCA5) were significantly decreased (Figure 4C, 4D). In detail, the mRNA expression level was markedly reduced by 83.3% (Figure 4C), and the protein level was significantly reduced by 30.2% (Figure 4D).
For the CDK1 inhibition experiments, there were 3 siRNA fragments (named si-CDK1–1, si-CDK1–2, and si-CDK1–3) designed, and the RT-qPCR results showed all the 3 were efficient to knockdown CDK1, among which, the most efficient one was si-CDK1–2 (Figure 4E). In the group transfecting si-CDK1–2, compared with the control, the mRNA level was significantly reduced by 88.5% (Figure 4E), while the CDK1 protein level reduced by 40.8% (Figure 4F). Thus, the fragment of si-CDK1–2 was selected for subsequent experiments.
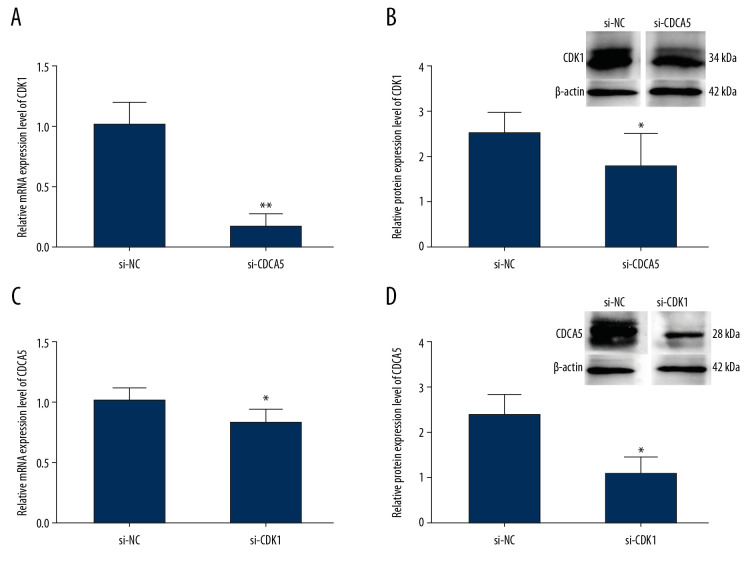
The expressions of CDCA5 and CDK1 in MGC-803 were positively related
To explore the relationship between CDCA5 and CDK1, we first detected the expression change of CDK1 after transfecting si-CDCA5 in MGC-803. CDCA5 protein has 252 amino acids with the molecular weight of about 27 kDa. CDK1 protein has 297 amino acids with the molecular weight of about 34 kDa. Compared with that of the si-NC group, the mRNA of CDK1 was significantly reduced by 83.5% (Figure 5A) and the protein of CDK1 reduced about 28% (Figure 5B). Then we detected the mRNA and protein expression levels of CDCA5 after transfecting si-CDK1 in MGC-803 cells. Compared with that of the si-NC group, the mRNA of CDCA5 was significantly reduced by 20% (Figure 5C) and the protein of CDK1 reduced about 58% (Figure 5D). These results confirmed a correlation between CDCA5 and CDK1 in GC cells.
Figure 5.
Relationship between CDK1 expression and CDCA5 expression. To explore the relationship between CDK1 and CDCA5, cells were knocked down one gene and checked the mRNA and protein expression level changes of the other gene. (A) Relative mRNA expression levels of CDK1 in cells transfected with si-NC and si-CDCA5 by quantitative real-time polymerase chain reaction (RT-qPCR) (** P<0.01). (B) Relative protein expression levels of CDK1 in cells transfected with si-NC and si-CDCA5 by western blot (* P<0.05). (C) Relative mRNA expression levels of CDCA5 in cells transfected with si-NC and si-CDK1 by RT-qPCR (* P<0.05). (D) Relative protein expression levels of CDCA5 in cells transfected with si-NC and si-CDK1 by western blot (* P<0.05).
Effects of CDCA5 and CDK1 in MGC-803 cells
To determine the effects of CDCA5 and CDK1 in GC cells, CDCA5 and CDK1 were knockdown by siRNA interfering, respectively. CCK-8 assay and colony formation experiments were performed to assess the proliferation ability; cell scratch assay and Transwell migration experiments were carried out to detect the migration ability; and Transwell assay was applied to detect the invasion ability.
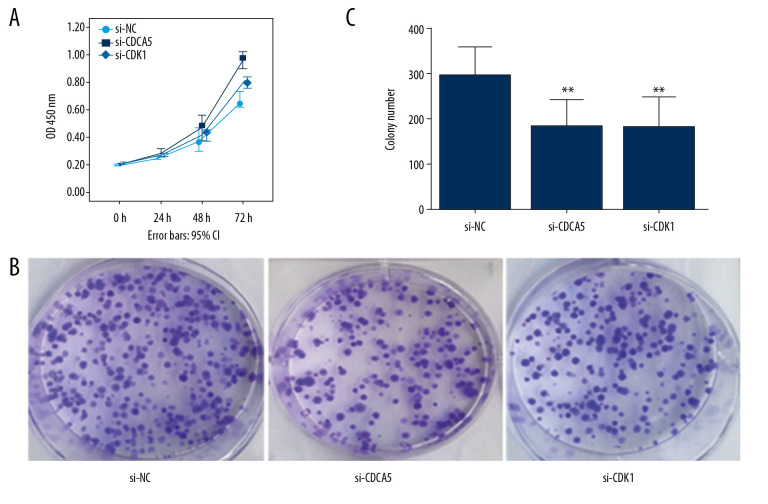
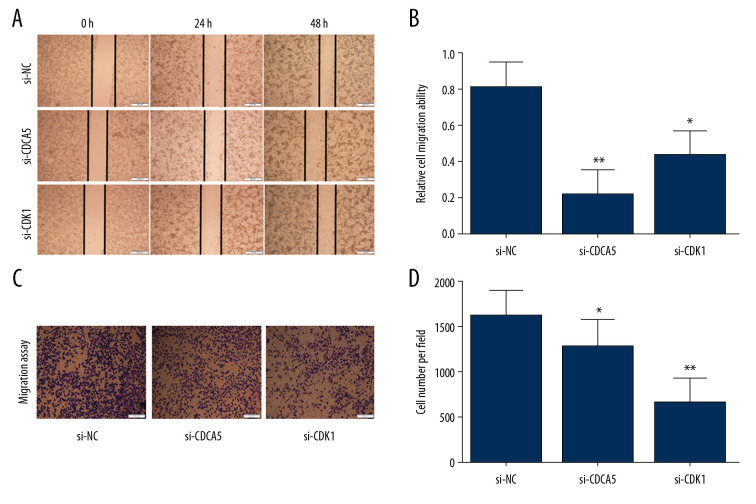
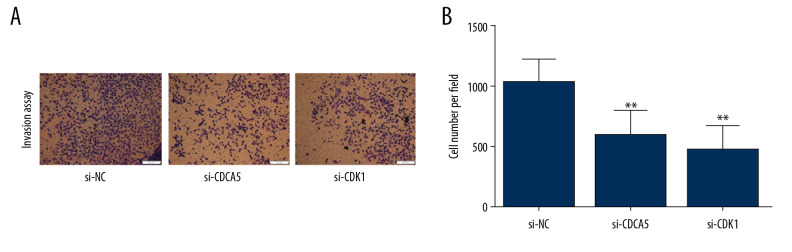
As a result, the CCK-8 assay and the plate colony formation experiment showed that both CDCA5 inhibition and CDK1 inhibition reduced the proliferation and colony forming ability of MGC-803 cells (Figure 6A–6C). Cell scraping experiments suggested both CDCA5 inhibition and CDK1 inhibition reduced cell migration ability of MGC-803 cells (Figure 7A, 7B). Similarly, the results of Transwell cell migration analysis indicated that CDCA5 inhibition or CDK1 inhibition suppressed the migration ability of MGC-803 cells (Figure 7C, 7D). The Transwell cell invasion tests also suggested that CDCA5 inhibition or CDK1 inhibition reduced the cell invasion capacity of MGC-803 cells (Figure 8A, 8B). These results showed CDK1 played a similar role as CDCA5 in GC cell progression.
Figure 6.
CDK1 or CDCA5 knockdown reduced cell proliferation in MGC-803. (A) The CCK-8 measurement results for the cells transfected with si-NC, si-CDCA5 and si-CDK1 at 0, 24, 48, and 72 hours, respectively. (B) The cell colon formation results for cells transfected with si-NC, si-CDCA5 and si-CDK1. (C) The cell colony number for cells transfected with si-NC, si-CDCA5 and si-CDK1 (** P<0.01).
Figure 7.
CDK1 or CDCA5 knockdown suppressed cell migration in MGC-803. (A) The cell scratch assay results for the cells transfected with si-NC, si-CDCA5 and si-CDK1 at 0, 24, 48 hours, respectively. (B) Relative cell migration ability of cells transfected with si-NC, si-CDCA5 and si-CDK1 at 48 hours (* P<0.05 and ** P<0.01). (C) The Transwell migration assay for cells transfected with si-NC, si-CDCA5 and si-CDK1, respectively. (D) The cell number per field for cells transfected with si-NC, si-CDCA5 and si-CDK1 (* P<0.05 and ** P<0.01).
Figure 8.
CDK1 or CDCA5 knockdown inhibited cell invasion in MGC-803. (A) The Transwell invasion assay for cells transfected with si-NC, si-CDCA5 and si-CDK1, respectively. (B) The cell number per field for cells transfected with si-NC, si-CDCA5 and si-CDK1 (** P<0.01).
Discussion
In this study, we first found the association of CDK1 and CDCA5 using STRING, Oncomine and TCGA database. Then we performed functional experiments to evaluate their relationship in gastric cells, and the results implicated the two proteins were positively regulated with each other, both can increase cell proliferation, migration, and invasion abilities of GC MGC-803 cells.
The CDCA5 protein is also named Sororin, which is a key regulator in sister chromatid condensation [22,23]. Recently, CDCA5 has also been found to be upregulated in many human tumor cases and its transcriptional level is significantly correlated with high pathological stage and overall survival rate, such as lung cancer [5], liver cancer [24], and oral squamous cell carcinoma [25]. In our previous study, we demonstrated the role of CDCA5 in GC cell proliferation, migration, and cell cycle changes [7]. Cyclin-dependent kinases (CDKs) are important drivers of cell cycle regulation and are thought to be potential targets for cancer therapy [26]. Among which CDK1 is a key regulator at G2/M checkpoint during the cell cycle. Knockdown of CDK1 had been proven to induce G2/M blockade and the uncontrolled mitotic activity may enhance genomic instability, which may lead to abnormal proliferation of malignant cells [10]. In fact, upregulation of CDK1 was often been observed in many advanced tumors [27–29] and dysregulation of CDK1 contributed to the failure of traditional chemotherapy drugs to eradicate cancer [30]. All these results suggested CDCA5 and CDK1 were cancer-related genes.
CDCA5 and CDK1 were positively related with each other and had similar roles in GC cell progression. Through knockdown assay by siRNA interfering, we found the mRNA and protein expression levels of CDCA5 were downregulated owing to CDK1 inhibition, and the same expression patterns were observed about the expression of CDK1 response to CDCA5 inhibition. Our results were consistent with Shen et al.’s study. They had also analyzed the relationship of CDCA5 and CDK1 in hepatocellular carcinoma and found CDCA5 inhibition suppressed cell survival as well as cell cycle and cell apoptosis by downregulation of CDK1 and CyclinB1. Moreover, CDCA5 overexpression upregulated the expression of CDK1 and CyclinB1 [24]. Our study suggested CDK1 inhibition or CDCA5 inhibition can suppress GC cell proliferation, migration, and invasion. Zhang et al. had also analyzed the function of CDCA5 inhibition in GC cells MKN45 and SGC7901, and they found the cell proliferation, migration, and invasion abilities were reduced, which were similar results with ours in MGC-803 cells [30]. Gao et al. investigated the effect of CDK1 in GC cells SGC-7901, and discovered downregulation of CDK1 inhibited the cell proliferation as in our study [31]. To our knowledge, this was the first comprehensively analysis of the proliferation, migration, and invasion abilities of CDK1 inhibition in GC cells.
CDCA5 and CDK1 can influence the expression of each other in GC cells, but it seemed that CDK1 had a bigger impact because CDCA5 protein reduced by about 58% (Figure 5D) while CDK1 reduced it by only about 28% (Figure 5B). It may be interpreted that CDCA5 was phosphorylated by CDK1 in GC cells. CDK1 is a protein kinase and the full consensus site for CDK1 phosphorylation is [S/T]Px[K/R] [32]. A previous study speculated that the phosphorylation and dephosphorylation processes of CDCA5 were involved in the regulation of cohesion, which was a necessary process of cell cycle, and the antagonism of a series of kinases such as CDK1 and protein phosphatase 2A played a unique role in this regulation [33,34]. Then, CDCA5 was found to be co-regulated with known cell cycle genes including CDK1 and CYCL1NB [35]. Next, CDCA5 phosphorylation during the mitosis process was confirmed and CDK1 was reported to play an important role in the complex mitosis involved in the regulation of CDCA5 protein [36]. Further, Zhang et al. analyzed CDCA5 protein primary structure and identified 3 putative CDK1 phosphorylation sites as S21PTK, S75PRR, and T159PGR by in vitro phosphorylation of CDCA5 by CDK1 using mass spectrometry. Especially, Ser21 conferred 70% of the total CDCA5 phosphorylation while Thr159 conferred 20%. Thus, they concluded Ser21 and Thr159 of CDCA5 were CDK1 phosphorylation sites [37]. Based on the previous studies, it can be speculated that CDCA5 was first phosphorylated by CDK1 and then influenced the malignant behaviors in GC cells. However, whether CDCA5 was phosphorylated by CDK1 on Ser21 and Thr159 or not in MGC-803 cells was not clear because Zhang et al. performed these experiments using HeLa cells, and thus more experiments are needed in the future.
Conclusions
Taken together, our work showed CDK1 and CDCA5 were positively correlated with each other, and both can promote cell proliferation, migration, and invasion in GC cells. CDCA5 and CDK1 may have the potential as diagnostic markers and therapeutic targets in GC therapy.
Footnotes
Conflict of interests
None.
Source of support: This work was supported by the Natural Science Foundation of Guangdong Province, China (No. 2016A030313683), and the National Natural Science Foundation of China (81200082)
References
- 1.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. Cancer J Clin. 2016;66(2):115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Zheng G, Xiong Y, Xu W, et al. A two-microRNA signature as a potential biomarker for early gastric cancer. Oncol Lett. 2014;7(3):679–84. doi: 10.3892/ol.2014.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang N, Pati D. Sororin is a master regulator of sister chromatid cohesion and separation. Cell Cycle. 2012;11(11):2073–83. doi: 10.4161/cc.20241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J, Xia C, Pu M, et al. Silencing of CDCA5 inhibits cancer progression and serves as a prognostic biomarker for hepatocellular carcinoma. Oncol Rep. 2018;40(4):1875–84. doi: 10.3892/or.2018.6579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen MH, Koinuma J, Ueda K, et al. Phosphorylation and activation of cell division cycle associated 5 by mitogen-activated protein kinase play a crucial role in human lung carcinogenesis. Cancer Res. 2010;70(13):5337–47. doi: 10.1158/0008-5472.CAN-09-4372. [DOI] [PubMed] [Google Scholar]
- 6.Xu T, Ma M, Dai J, et al. Gene expression screening identifies CDCA5 as a potential therapeutic target in acral melanoma. Human Pathol. 2018;75:137–45. doi: 10.1016/j.humpath.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 7.Chen T, Huang Z, Tian Y, et al. Role of triosephosphate isomerase and downstream functional genes on gastric cancer. Oncol Rep. 2017;38(3):1822–32. doi: 10.3892/or.2017.5846. [DOI] [PubMed] [Google Scholar]
- 8.Santamaría D, Barrière C, Cerqueira A, et al. Cdk1 is sufficient to drive the mammalian cell cycle. Nature. 2007;448(7155):811–15. doi: 10.1038/nature06046. [DOI] [PubMed] [Google Scholar]
- 9.Lohberger B, Leithner A, Stuendl N, et al. Diacerein retards cell growth of chondrosarcoma cells at the G2/M cell cycle checkpoint via cyclin B1/CDK1 and CDK2 downregulation. BMC Cancer. 2015;15:891. doi: 10.1186/s12885-015-1915-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Syn NL, Lim, Kong LR, et al. Pan-CDK inhibition augments cisplatin lethality in nasopharyngeal carcinoma cell lines and xenograft models. Signal Transduc Target Ther. 2018;3:9. doi: 10.1038/s41392-018-0010-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi Q, Zhou Z, Ye N, et al. MiR-181a inhibits non-small cell lung cancer cell proliferation by targeting CDK1. Cancer Biomark. 2017;20(4):539–46. doi: 10.3233/CBM-170350. [DOI] [PubMed] [Google Scholar]
- 12.Li L, Zhang Z, Yang Q, Ning M. Lycorine inhibited the cell growth of non-small cell lung cancer by modulating the miR-186/CDK1 axis. Life Sci. 2019;231:116528. doi: 10.1016/j.lfs.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 13.Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–13. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu W, Xu M, Wang L, et al. Integrative analysis of DNA methylation and gene expression identified cervical cancer-specific diagnostic biomarkers. Signal Transduct Target Ther. 2019;4:55. doi: 10.1038/s41392-019-0081-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang Z, Li C, Kang B, et al. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(Web Server issue):W98–102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X, Leung SY, Yuen ST, et al. Variation in gene expression patterns in human gastric cancers. Mol Biol Cell. 2003;14(8):3208–15. doi: 10.1091/mbc.E02-12-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho JY, Lim JY, Cheong JH, et al. Gene expression signature-based prognostic risk score in gastric cancer. Clin Cancer Res. 2011;17(7):1850–57. doi: 10.1158/1078-0432.CCR-10-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forster S, Gretschel S, Jons T, et al. THBS4, a novel stromal molecule of diffuse-type gastric adenocarcinomas, identified by transcriptome-wide expression profiling. Modern Pathol. 2011;24(10):1390–403. doi: 10.1038/modpathol.2011.99. [DOI] [PubMed] [Google Scholar]
- 19.Wang Q, Wen YG, Li DP, et al. Upregulated INHBA expression is associated with poor survival in gastric cancer. Med Oncol. 2012;29(1):77–83. doi: 10.1007/s12032-010-9766-y. [DOI] [PubMed] [Google Scholar]
- 20.Huang Z, Lin B, Pan H, et al. Gene expression profile analysis of ENO1 knockdown in gastric cancer cell line MGC-803. Oncol Lett. 2019;17(4):3881–89. doi: 10.3892/ol.2019.10053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta C(T)) method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Rankin S, Ayad NG, Kirschner MW. Sororin, a substrate of the anaphase-promoting complex, is required for sister chromatid cohesion in vertebrates. Mol Cell. 2005;18(2):185–200. doi: 10.1016/j.molcel.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 23.Nishiyama T, Ladurner R, Schmitz J, et al. Sororin mediates sister chromatid cohesion by antagonizing Wapl. Cell. 2010;143(5):737–49. doi: 10.1016/j.cell.2010.10.031. [DOI] [PubMed] [Google Scholar]
- 24.Shen Z, Yu X, Zheng Y, et al. CDCA5 regulates proliferation in hepatocellular carcinoma and has potential as a negative prognostic marker. OncoTargets Ther. 2018;11:891–901. doi: 10.2147/OTT.S154754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tokuzen N, Nakashiro K, Tanaka H, et al. Therapeutic potential of targeting cell division cycle associated 5 for oral squamous cell carcinoma. Oncotarget. 2016;7(3):2343–53. doi: 10.18632/oncotarget.6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fischer PM, Gianella-Borradori A. CDK inhibitors in clinical development for the treatment of cancer. Expert Opin Investig Drugs. 2003;12(6):955–70. doi: 10.1517/13543784.12.6.955. [DOI] [PubMed] [Google Scholar]
- 27.Pan XW, Chen L, Hong Y, et al. EIF3D silencing suppresses renal cell carcinoma tumorigenesis via inducing G2/M arrest through downregulation of Cyclin B1/CDK1 signaling. Int J Oncol. 2016;48(6):2580–90. doi: 10.3892/ijo.2016.3459. [DOI] [PubMed] [Google Scholar]
- 28.Ira G, Pellicioli A, Balijja A, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431(7011):1011–17. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thorenoor N, Faltejskova-Vychytilova P, Hombach S, et al. Long non-coding RNA ZFAS1 interacts with CDK1 and is involved in p53-dependent cell cycle control and apoptosis in colorectal cancer. Oncotarget. 2016;7(1):622–37. doi: 10.18632/oncotarget.5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z, Shen M, Zhou G. Upregulation of CDCA5 promotes gastric cancer malignant progression via influencing cyclin E1. Bioch Biophys Res Commun. 2018;496(2):482–89. doi: 10.1016/j.bbrc.2018.01.046. [DOI] [PubMed] [Google Scholar]
- 31.Gao SY, Li J, Qu XY, et al. Downregulation of CDK1 and cyclinB1 expression contributes to oridonin-induced cell cycle arrest at G2/M phase and growth inhibition in SGC-7901 gastric cancer cells. Asian Pac J Cancer Prev. 2014;15(15):6437–41. doi: 10.7314/apjcp.2014.15.15.6437. [DOI] [PubMed] [Google Scholar]
- 32.Ubersax JA, Woodbury EL, Quang PN, et al. Targets of the cyclin-dependent kinase CDK1. Nature. 2003;425(6960):859–64. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- 33.Zhang N, Panigrahi AK, Mao Q, Pati D. Interaction of Sororin protein with polo-like kinase 1 mediates resolution of chromosomal arm cohesion. J Biol Chem. 2011;286(48):41826–37. doi: 10.1074/jbc.M111.305888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGuinness BE, Hirota T, Kudo NR, et al. Shugoshin prevents dissociation of cohesion from centromeres during mitosis in vertebrate cells. PLoS Biol. 2005;3(3):e86. doi: 10.1371/journal.pbio.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walker MG. Drug target discovery by gene expression analysis: Cell cycle genes. Curr Cancer Drug Targets. 2001;1(1):73–83. doi: 10.2174/1568009013334241. [DOI] [PubMed] [Google Scholar]
- 36.Dreier MR, Bekier ME, Taylor WR. Regulation of Sororin by CDK1-mediated phosphorylation. J Cell Sci. 2011;124:2976–87. doi: 10.1242/jcs.085431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang N, Panigrahi AK, Mao Q, Pati D. Interaction of Sororin protein with polo-like kinase 1 mediates resolution of chromosomal arm cohesion. J Biol Chem. 2011;286(48):41826–3. doi: 10.1074/jbc.M111.305888. [DOI] [PMC free article] [PubMed] [Google Scholar]