Abstract
Background
Circular RNA UBE2D2 (circ_UBE2D2) has been found to be involved in the progression of breast cancer. Exosomes are critical mediators of intercellular communication, however, the function of exosomal circ_UBE2D2 in breast cancer remains vague.
Material/Methods
Cell viability was measured by Cell Counting Kit-8 assay. Western blot was used to detect the levels of estrogen receptor alpha (ERα), E-cadherin, vimentin, CD9, and CD63. Migrated and invaded cells were examined using Transwell assay. Circ_UBE2D2 and microRNA (miR)-200a-3p levels were detected using quantitative real-time polymerase chain reaction. Exosomes were isolated by ultracentrifugation method. The interaction between circ_UBE2D2 and miR-200a-3p was confirmed by dual-luciferase reporter assay and RNA immunoprecipitation assay. Murine xenograft model was established to conduct in vivo experiments.
Results
We found that circ_UBE2D2 was upregulated in breast cancer tamoxifen-resistant tissues and cell lines, and circ_UBE2D2 deletion mitigated tamoxifen resistance in breast cancer cells. Circ_UBE2D2 was also significantly loaded in exosomes isolated from resistant cells and could be transferred to parental cells. MiR-200a-3p was a target of circ_UBE2D2, and we demonstrated that exosomes mediated transfer of circ_UBE2D2 interacted with miR-200a-3p to enhance tamoxifen resistance of breast cancer cells by regulating cell viability, metastasis, and the level of ERα in vivo and in vitro.
Conclusions
Exosomes mediated transfer of circ_UBE2D2 reinforced tamoxifen resistance in breast cancer by binding to miR-200a-3p, providing new insights into the boost of the effectiveness of tamoxifen on breast cancer patients.
MeSH Keywords: Breast Neoplasms, Male; Cell Migration Assays; Cell Survival
Background
Breast cancer is the most common threat to the health of women worldwide. Currently, around 70% to 75% of breast cancer is reported to highly express estrogen receptor alpha (ERα), and the most ERα-positive breast cancer depends on estrogen signaling, which makes these cancers respond well initially to endocrine therapies [1–3]. Tamoxifen, an estrogen antagonist, is the most successful endocrine therapy for ERα-positive breast cancer [4,5]. Unfortunately, although remarkable benefits of tamoxifen in these treatment settings have been reported, many cases eventually acquired resistance to tamoxifen, thus resulting in relapse and death [6,7]. Therefore, a better understanding on the mechanism of tamoxifen resistance may contribute to the development of new approaches to overcome tamoxifen resistance and improve overall survival of breast cancer.
Exosomes are one of the extracellular vesicles that can be secreted by numerous cell types such as cancer cells [8]. Exosomes can transfer and exchange coding and non-coding RNAs, proteins and lipids, and are considered as potential modes of intercellular communication [9,10]. Recent studies have reported that exosomes participate in the modulation of chemoresistance of recipient cells and may be useful in cancer therapies [11]. Circular RNAs (circRNAs) are a class of non-coding transcripts that contain covalently closed loop structures [12]. Emerging evidence has revealed that circRNAs act as critical modulators in cancer progression by regulating various cellular processes through affecting a wide range of aspects of protein, DNA, and RNA expression as well as interactions [13,14]. In addition, they also are suggested to be involved in the regulation of chemoresistance of many types of cancers, including breast cancer. For example, circRNA-MTO1 was found to inhibit cell viability and enhance monastrol sensitivity by modulating TRAF4/Eg5 axis in breast cancer [15]. CircRNA-Cdr1 interacts with miR-1270 to upregulate the level of SCAI to sensitize ovarian cancer cells to cisplatin [16]. CircRNA ubiquitin-conjugating enzyme E2 D2 (circ_UBE2D2) is a newly identified RNA molecule. A recent study indicated that circ-UBE2D2 was elevated in breast cancer, and deletion of circ-UBE2D2 could inhibit the tumorigenesis of breast cancer cells via regulating miR-1236 or miR-1287 [17].
In this study we investigated whether exosomal circ-UBE2D2 was involved in chemoresistance in breast cancer. This study attempted to reveal the functions of circ-UBE2D2 in tamoxifen resistance in breast cancer, and explored whether exosome-transmitted circ-UBE2D2 was implicated in the regulation of tamoxifen resistance, as well as explore the potential molecular mechanism underlying the regulation in breast cancer.
Material and Methods
Patients and specimens
Breast cancer tissues were obtained from a total of 54 patients who underwent surgical resection at Shenzhen University General Hospital (Shenzhen University Clinical Medical Academy) and were immediately stored at −80°C until used. All patients were diagnosed by histopathological examination and only received tamoxifen (TAM)-based neo-adjuvant chemotherapy prior to surgery. The patients were divided into a tamoxifen-resistant (Resistant, n=25) group and a tamoxifen-sensitive (Sensitive, n=29) group depending on the sensitivity to tamoxifen. This study was permitted by the Ethics Committee of Shenzhen University General Hospital (Shenzhen University Clinical Medical Academy) and written informed consents were collected from all study participants.
Cell culture
Human breast cancer cell lines MCF-7 and T47D, and 293T cells were purchased from Shanghai Academy of life Science (Shanghai, China) and grown in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Carlsbad, CA, USA) containing 10% fetal bovine serum at 37°C with 5% CO2. Tamoxifen was obtained from Sigma (St. Louis, MO, USA) and dissolved in sterile water. Tamoxifen-resistant cells named MCF-7/TAM-R and T47D/TAM-R were generated through exposing parental MCF7 (MCF7/Par) and T47D (T47D/Par) cells to increasing concentrations of tamoxifen up to 5 μM over a period of 6 months. Tamoxifen-resistant cells were cultured in the same media supplemented with 1 μM tamoxifen. For exosome co-cultures, 2 μg/mL of exosomes were incubated with the MCF7/Par and T47D/Par cells (6×105) in the exosomes depleted culture medium.
Cell viability assay
Cells were seeded on 96-well plates at a density of 50 000 cells/mL for overnight, and then were treated with increasing concentrations of tamoxifen (0, 10, 20, 30, and 40 μM/L) for additional 48 hours. After the addition of 10 μL Cell Counting Kit-8 (CCK-8) solution in per well for another 4 hours, the optical density (OD) was measured by a microplate reader at 450 nm. Besides that, IC50 value of tamoxifen was calculated according to the relative survival curve.
Western blot
Proteins were extracted using RNA immunoprecipitation (RIP) assay lysis buffer (Beyotime, Beijing, China) and quantified by bicinchoninic acid (BAC) method, then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and shifted onto a polyvinylidene fluoride (PVDF) membrane. Immunoblotting used antibodies against CD9 (1: 5000, ab68418, Abcam, Cambridge, MA, USA), CD63 (1: 2000, ab68418, Abcam), E-cadherin (E-cad; 1: 1000, ab15148, Abcam), vimentin (1: 5000, ab92547, Abcam), ERα (1: 3000, ab13504, Abcam), GAPDH (1: 10 000, ab181602, Abcam) and the secondary horseradish peroxidase (HRP)-conjugated antibody (1: 1000, ab9482, Abcam). The protein bands were visualized using electrochemiluminescence (ECL).
Cells migration and invasion assays
For invasion assays, cells in serum-free DMEM were planted in the top chambers pre-coated with the Matrigel. Then 500 μL DMEM mixed with 10% FBS was added into the lower chambers. After 24 hours, cells on the lower face of the membranes were fixed and stained. Finally, 5 random fields were selected to count invaded cells with a microscope. For migration assays, the membranes of top chambers without the Matrigel were used and other methods of measurement were similar to the steps of cell invasion assays.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Whole-RNA extracts were prepared using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and total RNA from exosomes was isolated with the exoRNeasy Midi Kit (Qiagen, Valencia, CA, USA) according to the standard procedure. Then extracted RNA was interacted with Rnase R (Epicentre, Madison, WI, USA), followed by incubation with RNeasy MinElute Cleanup Kit (Qiagen). RNA samples were reversely transcribed into cDNA using a High Capacity cDNA Reverse Transcription Kit (Qiagen), and cDNA amplification was carried out with SYBR Premix Ex Taq (Qiagen). Glyceraldehyde 3-phosphate dehydrogenase (GADPH) and U6 small nuclear B noncoding RNA (U6) were used as internal references to normalize the fold changes using 2−ΔΔCt method. The specific primer sequences were listed as follows:
-
circ_UBE2D2:
forward 5′-AATGGCAGCATTTGTCTTGA-3′,
reverse 5′-GCCCCTGTGAGTAAGCTACG-3′;
-
miR-200a-3p:
forward 5′-GGCTAACACTGTCTGGTAACGATG-3′,
reverse 5′-GTGCAGGGTCCGAGGT-3′.
-
GADPH:
forward 5′-GATATTGTTGCCATCAATGAC-3′,
reverse 5′-TTGATTTTGGAGGGATCTCG-3′;
-
U6:
forward 5′-GCTTCGGCAGCACATATACTAAAAT-3′,
reverse 5′-CGCTTCACGAATTTGCGTGTCAT-3′.
Cell transfection
The mimic of miR-200a-3p, negative control (miR-NC), small interfering RNA (siRNA) targeting circ_UBE2D2 covalent closed junction (si-circ_UBE2D2), and the same vector harboring a scrambled sequence (si-NC) were synthesized by Genepharma (Shanghai, China). The transfection of oligonucleotides was conducted using Lipofectamine™ 3000 (Invitrogen).
Exosome (exo) isolation
Exosomes were isolated from MCF-7/TAM-R and MCF-7/Par cells culture media by ultracentrifugation method. Cell culture fluid was centrifuged at 3000 g for 30 minutes at 4°C to remove cellular debris/dead cells. Then the resulting supernatant was further centrifuged at 100 000 g for 70 minutes at 4°C, and followed by filtering using 0.22 μm filtration to concentrate the exosome-containing solution. Subsequently, the exosome containing pellet was washed with phosphate-buffered saline (PBS), and centrifuged at 100 000 g for 70 minutes again. Finally, cell exosomes were collected. The exosomes isolated from MCF-7/TAM-R cells were named as MCF-7/TAM-R-exo, while that from MCF-7/Par cells were termed as MCF-7/Par-exo.
Transmission electron microscopy (TEM)
The 10 mL exosome pellets were dropped on the carbon-coated copper grid and incubated for 5 minutes at 37°C, and then subjected to 2% phosphotungstic acid solution for 2 minutes, followed by washing with PBS for 3 times. Finally, the prepared samples were examined by using a transmission electron microscope (JEOL, Akishima, Japan).
Dual-luciferase reporter assay
The binding sequence of miR-200a-3p in circ_UBE2D2 and its mutated sequence were cloned into the pmiR-RB-Report (Promega, Shanghai, China), respectively. Afterwards, 293T were co-transfected with pmiR-RB-Report luciferase vectors containing wild-type or mutant circ_UBE2D2 and miR-200a-3p mimics or miR-NC using LipofectamineTM 3000 (Invitrogen). Lastly, a dual luciferase assay kit (Promega) was employed to analyze the luciferase activity.
RNA immunoprecipitation (RIP) assay
MCF7/Par and T47D/Par cells were lysed using RIP assay buffer (Millipore, Billerica, MA, USA), and then incubated with magnetic beads coated with anti-Ago2. Normal mouse IgG was served as a negative control. Finally, purified RNA was subjected to qRT-PCR analysis to determine the presence of the binding targets.
Xenograft experiments in vivo
Nine pathogen-free female BALB/c nude mice (5 weeks old) were purchased to establish xenograft models in line with the guidelines permitted by the Animal Research Committee of Shenzhen University General Hospital (Shenzhen University Clinical Medical Academy). All mice were maintained in specific pathogen-free conditions. The mice were randomly divided into 3 groups of 3 mice each, and the first group was subcutaneously inoculated with MCF-7/TAM-R cells, the second and third groups were subcutaneously injected with MCF-7/Par cells. When the volume of xenografts reached about 100 mm3, the first group was intratumorally injected with PBS, and the second and third groups were intratumorally injected with PBS and MCF-7/TAM-R-exo, and the all groups were gastrointestinally injection with tamoxifen (6 mg/kg) every day. Tumor volume was recorded every 5 days. At day 37, the mice were killed, and tumor masses were weighed and used for further molecular analysis.
Statistical analysis
Numerical results were expressed as the mean ± standard deviation (SD). The statistical difference between each group was analyzed using Student’s t-test or one-way analysis of variance (ANOVA) on GraphPad Prism 7 software. P values <0.05 or 0.01 suggested statistically significant.
Results
Characterization of tamoxifen-resistant breast cancer cell lines
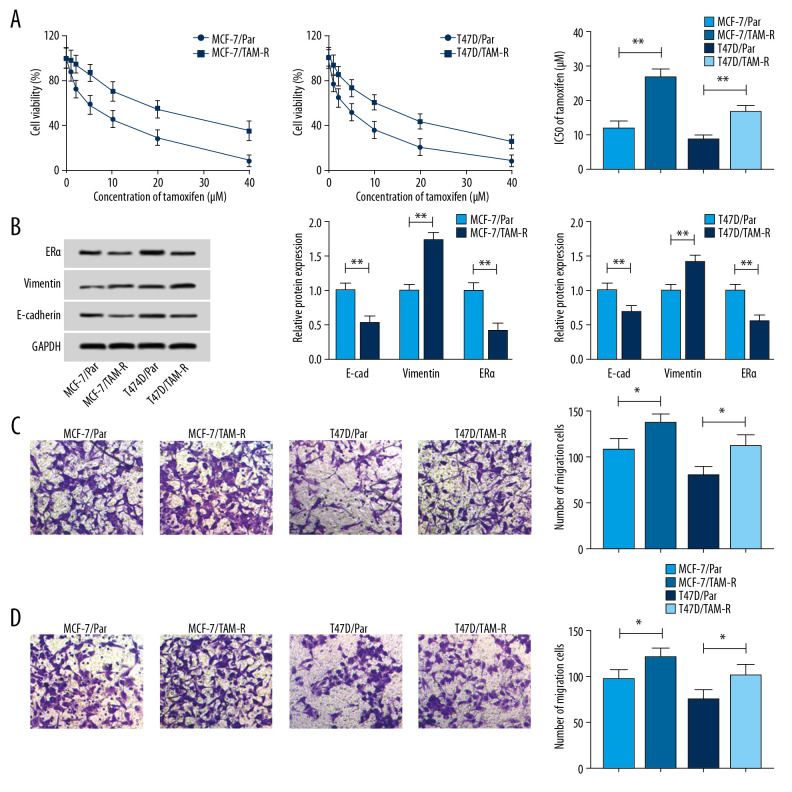
To explore underlying regulatory mechanism of tamoxifen resistance, 2 tamoxifen-resistant cells (MCF-7/TAM-R and T47D/TAM-R) derived from the parental cell lines MCF-7/Par and T47D/Par were established. As illustrated in Figure 1A, MCF-7/TAM-R and T47D/TAM-R cells showed increasing cell viability when incubated with increasing concentrations of tamoxifen (0, 10, 20, 30, and 40 μM/L) for 48 hours in contrast to the parental cells. Moreover, cell viability assay also indicated the IC50 values of tamoxifen were higher in MCF-7/TAM-R and T47D/TAM-R cells compared with the parental cells (Figure 1A), indicating resistant cells had a poor response to tamoxifen. Subsequently, western blot demonstrated an increase of vimentin protein expression and a decrease of E-cad protein expression in MCF-7/TAM-R and T47D/TAM-R cells compared with parental cells (Figure 1B); moreover, a markedly increased number of tamoxifen-resistant cells were observed to migrate and invade compared with parental cells (Figure 1C, 1D), suggesting tamoxifen resistance induced high metastasis in resistant cells. The target of tamoxifen in vivo is the ER, and the level of ER is the best predictor of benefit from tamoxifen, and loss of ER expression could confer resistance to therapy [5]. ERα is the major form of ER in breast. Thus, the level of ERα was detected; more importantly, tamoxifen resistance cells had a low level of ERα relative to parental cells (Figure 1B), thus resulting in tamoxifen resistance.
Figure 1.
Characterization of tamoxifen-resistant breast cancer cell lines. (A) CCK-8 analysis of cell viability and IC50 values in parent and resistant cells treated with different concentrations of tamoxifen. (B) Western blot analysis of the level of ERα, vimentin, and E-cad in parent and resistant cells incubated with 10 μM tamoxifen. (C, D) Transwell analysis of the migration and invasion of parental and resistant cells treated with 10 μM tamoxifen. * P<0.05, ** P<0.01. CCK-8 – Cell Counting Kit-8; ERα – estrogen receptor alpha.
Circ_UBE2D2 deletion mitigates tamoxifen resistance in breast cancer cells
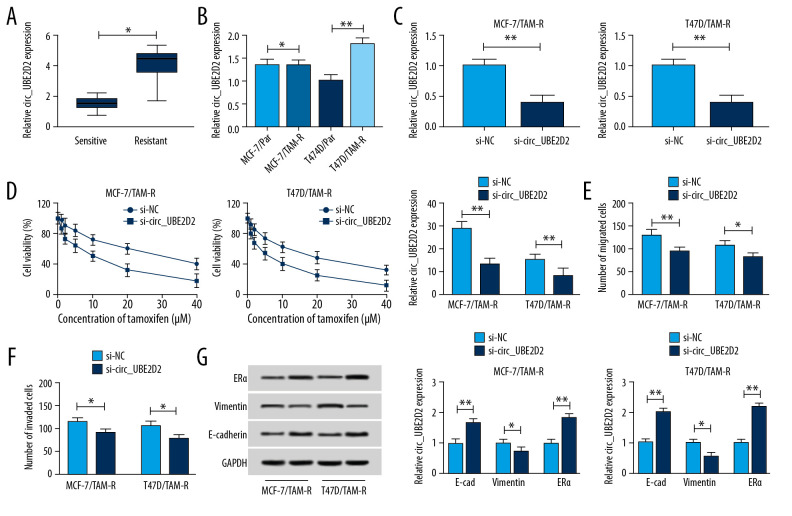
We found the expression of circ_UBE2D2 was significantly upregulated in tamoxifen-resistant breast cancer tissues and cell lines compared with the tamoxifen sensitive group and parental cells (Figure 2A, 2B). Then the biological functions of circ_UBE2D2 in tamoxifen resistance of breast cancer were investigated. Circ_UBE2D2 was interfered in MCF-7/TAM-R and T47D/TAM-R cells using siRNA sequences, as expected, circ_UBE2D2 downregulation in MCF-7/TAM-R and T47D/TAM-R cells were observed (Figure 2C). After that, CCK-8 assay showed that cell viability and IC50 value were markedly decreased by circ_UBE2D2 deletion (Figure 2D), indicating circ_UBE2D2 downregulation led MCF-7/TAM-R and T47D/TAM-R cells sensitive to tamoxifen. Meanwhile, Transwell assay indicated that knockdown of circ_UBE2D2 repressed the migration and invasion of MCF-7/TAM-R and T47D/TAM-R cells (Figure 2E, 2F). Additionally, western blot analysis found that circ_UBE2D2 silence increased the level of E-cad, but decreased the level of vimentin in MCF-7/TAM-R and T47D/TAM-R cells to suppress resistant cells EMT (Figure 2G). Moreover, circ_UBE2D2 silence increased the expression of ERα to attenuate the resistance to tamoxifen (Figure 2G). Taken together, circ_UBE2D2 deletion sensitized breast cancer cells to tamoxifen.
Figure 2.
Circ_UBE2D2 deletion mitigates tamoxifen resistance in tamoxifen-resistant breast cancer cells. (A, B) qRT-PCR analysis on the expression of circ_UBE2D2 in tamoxifen-resistant and tamoxifen-sensitive breast cancer patients, as well as in parent and resistant breast cancer cells was conducted. MCF-7/TAM-R and T47D/TAM-R cells were transfected with si-circ_UBE2D2 or si-NC. (C) The expression of circ_UBE2D2 in MCF-7/TAM-R and T47D/TAM-R cells was examined using qRT-PCR to analyze the interference efficiency. (D) The viability and IC50 values of MCF-7/TAM-R and T47D/TAM-R cells treated with different concentrations of tamoxifen were detected by CCK-8 assay. (E, F) The migration and invasion analysis of parental and resistant cells treated with 20 μM tamoxifen was performed using Transwell assay. (G) The levels of ERα, vimentin, and E-cad in parent and resistant cells treated with 20 μM tamoxifen were detected using western blot. * P<0.05, ** P<0.01. qRT-PCR – real-time polymerase chain reaction; CCK-8 – Cell Counting Kit-8; ERα – estrogen receptor alpha; E-cad – E-cadherin.
Upregulation of circ_UBE2D2 in exosomes from tamoxifen-resistant breast cancer cells
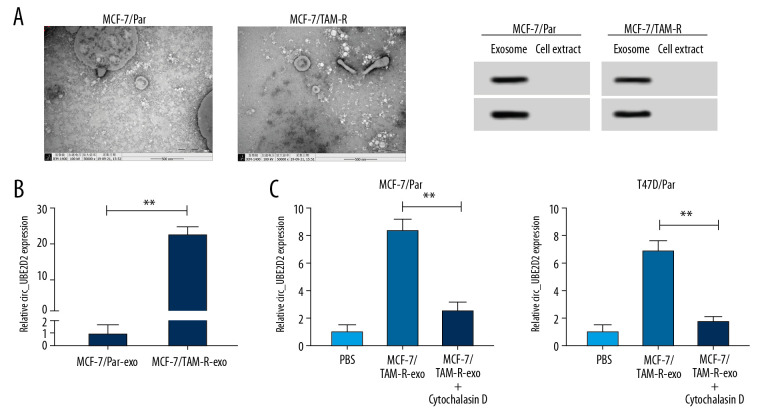
Cell exosomes were extracted from tamoxifen-resistant and parental breast cancer cells. The morphology (round-shaped vesicles) and size of exosomes were imaged using transmission electron microscopy (TEM) (Figure 3A). Meanwhile, western blot showed strong signals of exosomal markers CD63 and CD9 in MCF-7/TAM-R and MCF-7/Par exosomes compared with cell extract (Figure 3A). These data indicated the successful extraction of exosomes. Subsequently, the level of circ_UBE2D2 was detected in exosomes isolated from MCF-7/TAM-R (MCF-7/TAM-R-exo) and MCF-7/Par (MCF-7/Par-exo). As presented in Figure 3B, circ_UBE2D2 expression in MCF-7/TAM-R-exo was about 20-fold higher than that in MCF-7/Par-exo. After that, MCF-7/Par and T47D/Par cells were pretreated with PBS, and MCF-7/TAM-R-exo with or without cytochalasin D, an inhibitor of exosome receptor, and results showed the expression of circ_UBE2D2 was elevated after treatment with MCF-7/TAM-R-exo compared with treated with PBS, while this increase was inhibited by the cytochalasin D treatment (Figure 3C), indicating the increase of circ_UBE2D2 in parental cells was produced by the treatment of MCF-7/TAM-R-exo.
Figure 3.
Upregulation of circ_UBE2D2 in exosomes from tamoxifen-resistant breast cancer cells. Cell exosomes were extracted from tamoxifen-resistant and parental breast cancer cells. (A) The image of exosomes was captured by TEM, and the expression levels of exosomal markers CD9 and CD63 were measured by western blot. (B) Circ_UBE2D2 expression in MCF-7/TAM-R-exo and MCF-7/Par-exo was detected by qRT-PCR. (C) qRT-PCR analysis of circ_UBE2D2 was conducted in the MCF-7/TAM-R-exo co-cultured parental cells untreated with or treated with cytochalasin D. * P<0.05, ** P<0.01. TEM – transmission electron microscopy; qRT-PCR – real-time polymerase chain reaction.
Intercellular transfer of circ_UBE2D2 by exosomes enhances tamoxifen resistance in breast cancer cells in vitro
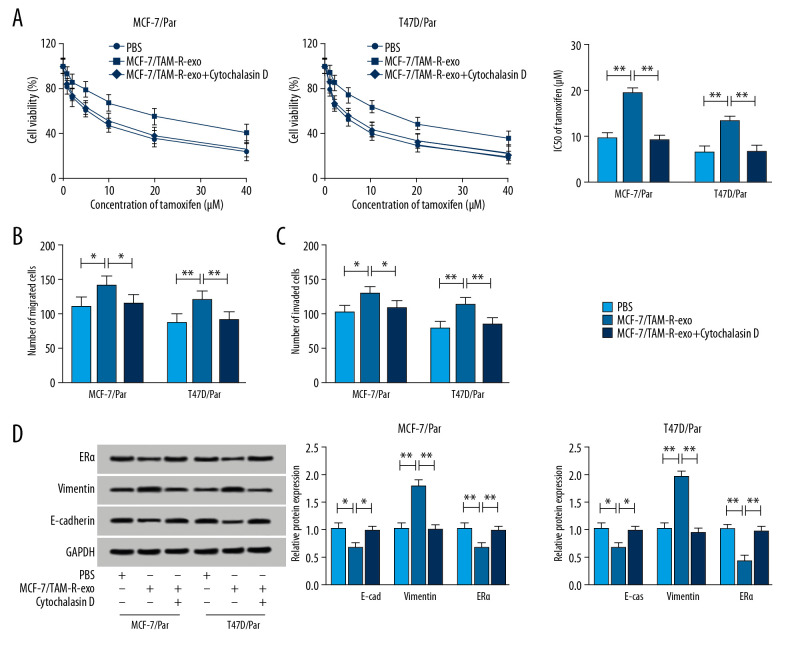
After treatment with MCF-7/TAM-R-exo, we detected whether exosome-transferred circ_UBE2D2 could regulate chemoresistance in sensitive cells of breast cancer. By performing a CCK-8 assay, we found MCF-7/TAM-R-exo enhanced the viability and IC50 values of MCF-7/Par and T47D/Par cells after tamoxifen treatment (Figure 4A). Then Transwell assay showed MCF-7/TAM-R-exo inhibited tamoxifen-induced migration and invasion suppression of sensitive cells (Figure 4B, 4C). What is more, western blot showed MCF-7/TAM-R-exo overturned tamoxifen-induced promotion on E-cad and ERα level and inhibition on vimentin expression in MCF-7/Par and T47D/Par cells (Figure 4D). However, these effects induced by MCF-7/TAM-R-exo could be reversed by cytochalasin D treatment (Figure 4A–4D). These results indicated intercellular transfer of circ_UBE2D2 by exosomes promoted tamoxifen resistance in vitro.
Figure 4.
Intercellular transfer of circ_UBE2D2 by exosomes enhances tamoxifen resistance in breast cancer cells in vitro. (A) Cell viability and IC50 values were detected using CCK-8 assay in MCF-7/Par and T47D/Par cells treated with tamoxifen at different concentrations. (B, C) The migration and invasion of parental cells treated with 10 μM tamoxifen were examined using Transwell assay. (D) Levels analysis of ERα, vimentin, and E-cad in parental cells treated with 10 μM tamoxifen were carried out using western blot. * P<0.05, ** P<0.01. ERα – estrogen receptor alpha; E-cad – E-cadherin.
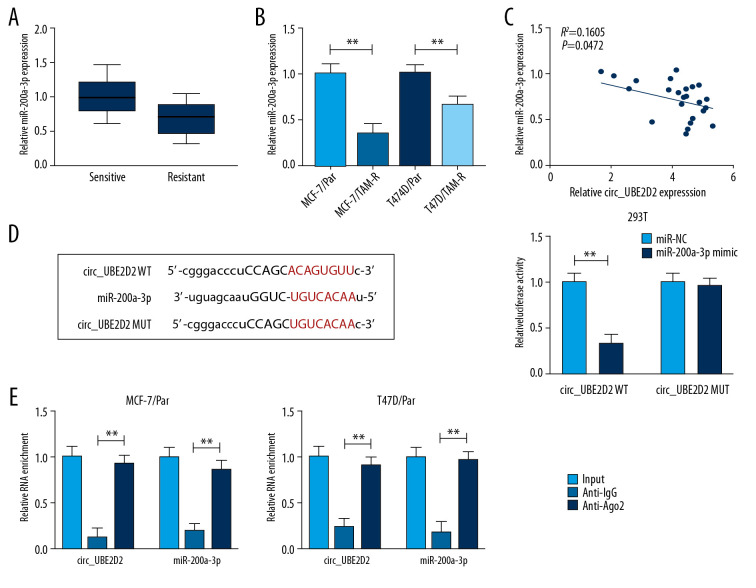
MiR-200a-3p is target of circ_UBE2D2
The expression of miR-200a-3p was measured and we found miR-200a-3p was downregulated in tamoxifen-resistant breast cancer tissues and cell lines (Figure 5A, 5B), and a negative regulation between miR-200a-3p and circ_UBE2D2 expression was observed (Figure 5C). After that, using the bioinformatics analysis of starBase 3.0 program, we found miR-200a-3p contained the putative binding sites of circ_UBE2D2 (Figure 5D). Then, the analysis of luciferase activity showed miR-200a-3p reduced the luciferase activity in 293T cells transfected with circ_UBE2D2 WT (Figure 5D). Furthermore, the RIP assay further verified the direct interaction between circ_UBE2D2 and miR-200a-3p with the significant enrichment of circ_UBE2D2 and miR-200a-3p expression in MCF-7/Par and T47D/Par cells after Ago2 RIP (Figure 5E). Therefore, circ_UBE2D2 directly bound to miR-200a-3p in breast cancer.
Figure 5.
MiR-200a-3p is target of circ_UBE2D2. (A, B) qRT-PCR analysis of the expression of miR-200a-3p in tamoxifen-resistant and -sensitive tissues, as well as in parent and resistant cells in breast cancer was conducted. (C) The correlation between miR-200a-3p and circ_UBE2D2 was analyzed using Pearson correlation analysis. (D) The putative binding site between miR-200a-3p and circ_UBE2D2 was presented. (D, E) The interaction between miR-200a-3p and circ_UBE2D2 was confirmed using the dual-luciferase reporter assay and RIP assay. * P<0.05, ** P<0.01. qRT-PCR – real-time polymerase chain reaction; RIP – RNA immunoprecipitation.
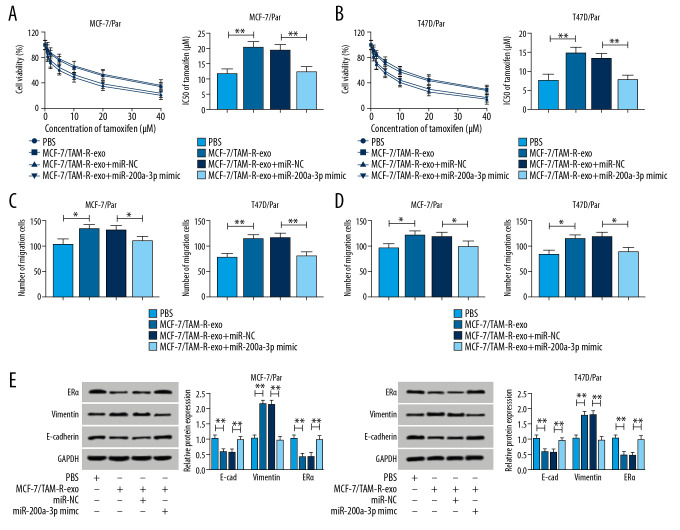
Exosomes mediated transfer of circ_UBE2D2 increases tamoxifen resistance in breast cancer cells by binding to miR-200a-3p
We further explored whether miR-200a-3p involved in the activity of exosomal circ_UBE2D2 on tamoxifen resistance. Results showed miR-200a-3p mimic transfection partially reversed MCF-7/TAM-R-exo induced promotion of cell viability (Figure 6A, 6B), elevation of IC50 values (Figure 6A, 6B), as well as increase of migrated and invaded cells (Figure 6C, 6D) in MCF-7/Par and T47D/Par cells in the treatment of tamoxifen. Synchronously, western blot assay also suggested that miR-200a-3p overexpression attenuated MCF-7/TAM-R-exo mediated increase of vimentin, and decrease of E-cad and ERα in MCF-7/Par and T47D/Par cells treated with 10 μM tamoxifen (Figure 6E). Collectively, exosomal circ_UBE2D2 enhanced the resistance of breast cancer cells to tamoxifen by binding to miR-200a-3p.
Figure 6.
Exosomes mediated transfer of circ_UBE2D2 increases tamoxifen resistance in breast cancer cells by binding to miR-200a-3p. Parental cells co-cultured with MCF-7/TAM-R-exo were transfected with miR-NC or miR-200a-3p mimic. (A, B) The viability and IC50 values in MCF-7/Par and T47D/Par cells treated with different concentrations of tamoxifen were detected using CCK-8 assay. (C, D) The migrated and invaded parental cells treated with 10 μM tamoxifen were evaluated by Transwell assay. (E) Western blot was used to measure the levels of ERα, vimentin, and E-cad in parental cells treated with 10 μM tamoxifen. * P<0.05, ** P<0.01. CCK-8 – Cell Counting Kit-8; ERα – estrogen receptor alpha; E-cad – E-cadherin.
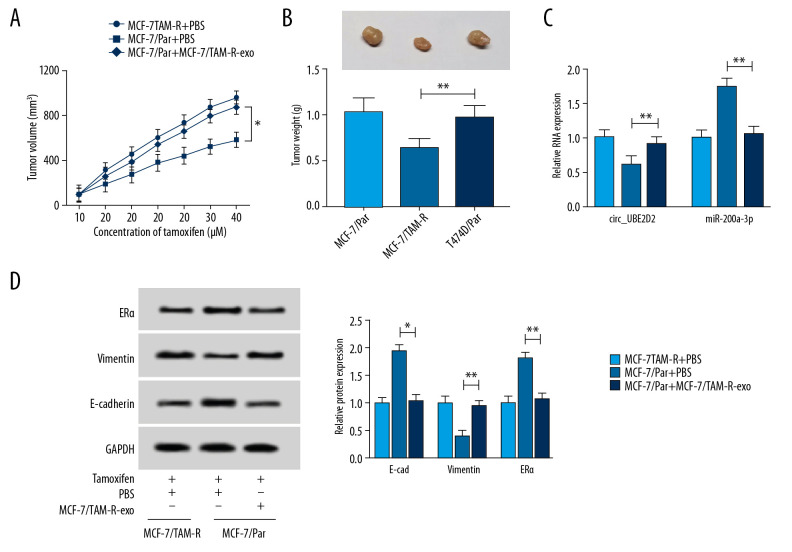
Exosomal circ_UBE2D2 accelerates tamoxifen resistance in vivo
We further elaborated the regulatory effects of exosomes mediated transfer of circ_UBE2D2 on xenograft mice. As presented in Figure 7A and 7B, MCF-7/TAM-R-exo treatment reversed tamoxifen-induced inhibition on the tumor growth reflected by the increase of tumor volume and weight in MCF-7/TAM-R-exo treatment group. Immediately, qRT-PCR analysis showed circ_UBE2D2 was highly expressed and miR-200a-3p was a low expression in MCF-7/TAM-R-exo treatment group (Figure 7C). Furthermore, western blot analysis showed MCF-7/TAM-R-exo abated tamoxifen-induced suppression of EMT and increase of ERα level in vivo (Figure 7D). Altogether, exosomal circ_UBE2D2 accelerated tamoxifen resistance in vivo via regulating miR-200a-3p.
Figure 7.
Exosomal circ_UBE2D2 accelerates tamoxifen resistance in vivo. (A) Tumor volume was assessed every 5 days. (B) Tumor weight was examined after 37 days. (C) The expression of circ_UBE2D2 and miR-200a-3p was detected by qRT-PCR in isolated tumor masses. (D) Protein expression of ERα, vimentin, and E-cad was evaluated tumor masses by western blot. * P<0.05, ** P<0.01. qRT-PCR – real-time polymerase chain reaction; ERα – estrogen receptor alpha; E-cad – E-cadherin.
Discussion
It has been documented that exosomes can act as a mediator of communication to transfer and exchange biomolecules between tumor cells and other cell types or the microenvironment in many cancers, thus affecting the development, progression and drug resistance of cancer [11,18]. For example, exosomes mediated transfer of miR-100-5p increased cisplatin resistance of recipient cells in lung cancer [19]. Exosome-transmitted long noncoding RNA (lncRNA) ARSR conferred sunitinib resistance in renal cancer cells through facilitating AXL and c-MET expression by sponging miR-34/miR-449 [20]. The anti-estrogen tamoxifen is the mainstay treatment of breast cancer with ERα-positive, while the resistance to tamoxifen has been investigated in the majority of breast cancer [2]. Recently, exosomes were reported to involve in the transferring of hormone/metformin resistance in breast cancer cells [21]. Besides that, Xu et al. found that exosomes transmitted lncRNA UCA1 increased cell tamoxifen resistance in breast cancer [22]. Wei et al. discovered that exosomes mediated transfer of miR-221/222 resulted in the enhancement of tamoxifen resistance in ER-positive breast cancer cells [23]. Furthermore, circRNA also were found to regulate cell tamoxifen resistance of breast cancer [24]. Thus, it is of great clinical to uncover the exosomes mediated transfer of circRNAs in the tamoxifen resistance of breast cancer.
In this study, circ_UBE2D2 was increased in breast cancer tamoxifen-resistant tissues and cell lines, and circ_UBE2D2 deletion mitigated tamoxifen resistance in breast cancer by suppressing resistant cell viability, metastasis and the inactivation of ERα, indicating the involvement of circ_UBE2D2 in the regulation of tamoxifen resistance. Subsequently, cell exosomes were extracted from tamoxifen-resistant and parental breast cancer cells, and the vesicles displayed a round shape with bilayered membranes, and the diameter from 40 nm to 250 nm under a TEM. Next, we found circ_UBE2D2 was also significant enrichment in exosomes driven from resistant cells and could be transferred to parental cells to enhance tamoxifen resistance of recipient cells. More importantly, xenograft analysis displayed that exosomal circ_UBE2D2 also enhanced tamoxifen resistance in vivo.
CircRNAs have various biological functions, among which the most studied is as a molecular sponge of microRNAs (miRNAs) [25]. Herein, through using bioinformatics analysis, we found miR-200a-3p was a target of circ_UBE2D2. MicroRNAs (miRNA) have been investigated to participate in the pathological process of many diseases by regulating cell metabolism, proliferation, migration, apoptosis, and drug resistance [26,27]. MiR-200a-3p is a functional miRNA and has been demonstrated to inhibit cell tumorigenesis in many types of cancers, such as papillary thyroid carcinoma [28], gastric cancer [29], renal cancer [30], and so on. In addition, miR-200a-3p also was implied to involve in the regulation of drug resistance. For instance, miR-200a-3p interacted with DUSP6 to enhance 5-fluorouracil resistance in hepatocellular carcinoma cells [31]. MiR-200a-3p involved in therapeutic response to promote temozolomide- sensitivity in glioblastoma multiforme [32]. Additionally, Zhang et al. found miR-200a-3p served as a target of LINC00894-002 to participate in LINC00894-002-mediated contribution to tamoxifen resistance in MCF-7/TAM-R cells [33]. This study displayed that miR-200a-3p was decreased in tamoxifen-resistant tissues and cell lines in breast cancer, subsequent experiments showed that miR-200a-3p reversed the facilitating action of exosomal circ_UBE2D2 on tamoxifen resistance in vivo and in vitro. Thus, a circ_UBE2D2/miR-200a-3p axis in the regulation of tamoxifen resistance in breast cancer cells was identified.
Conclusions
Our study demonstrated that circ_UBE2D2 was significantly loaded not only in tamoxifen-resistant breast cancer cells, but also in exosomes driven from resistant cells. Exosomes mediated transfer of circ_UBE2D2 markedly enhanced tamoxifen resistance in ER-positive breast cancer by binding to miR-200a-3p, indicating a promising biomarker and therapeutic target for drug-resistant in breast cancer.
Footnotes
Source of support: Departmental sources
References
- 1.Piva M, Domenici G, Iriondo O, et al. Sox2 promotes tamoxifen resistance in breast cancer cells. EMBO Mol Med. 2014;6:66–79. doi: 10.1002/emmm.201303411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergamaschi A, Katzenellenbogen BS. Tamoxifen downregulation of miR-451 increases 14-3-3zeta and promotes breast cancer cell survival and endocrine resistance. Oncogene. 2012;31:39–47. doi: 10.1038/onc.2011.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen L, Yang G, Dong H. Everolimus reverses palbociclib resistance in ER+ human breast cancer cells by inhibiting phosphatidylinositol 3-kinase (PI3K)/Akt/mammalian target of rapamycin (mTOR) pathway. Med Sci Monit. 2019;25:77–86. doi: 10.12659/MSM.912929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aldossary M, Alquraish F, Alazhri J. A case of locally advanced breast cancer in a 59-year-old man requiring a modified approach to management. Am J Case Rep. 2019;20:531–36. doi: 10.12659/AJCR.915377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jordan VC, O’Malley BW. Selective estrogen-receptor modulators and antihormonal resistance in breast cancer. J Clin Oncol. 2007;25:5815–24. doi: 10.1200/JCO.2007.11.3886. [DOI] [PubMed] [Google Scholar]
- 6.Mills JN, Rutkovsky AC, Giordano A. Mechanisms of resistance in estrogen receptor positive breast cancer: Overcoming resistance to tamoxifen/aromatase inhibitors. Curr Opin Pharmacol. 2018;41:59–65. doi: 10.1016/j.coph.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ring A, Dowsett M. Mechanisms of tamoxifen resistance. Endocr Relat Cancer. 2004;11:643–58. doi: 10.1677/erc.1.00776. [DOI] [PubMed] [Google Scholar]
- 8.Cocucci E, Meldolesi J. Ectosomes and exosomes: Shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015;25:364–72. doi: 10.1016/j.tcb.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 9.Abels ER, Breakefield XO. Introduction to extracellular vesicles: Biogenesis, RNA cargo selection, content, release, and uptake. Cell Mol Neurobiol. 2016;36:301–12. doi: 10.1007/s10571-016-0366-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–84. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang HD, Jiang LH, Hou JC, et al. Exosome: A novel mediator in drug resistance of cancer cells. Epigenomics. 2018;10:1499–509. doi: 10.2217/epi-2017-0151. [DOI] [PubMed] [Google Scholar]
- 12.Rybak-Wolf A, Stottmeister C, Glazar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870–85. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 13.Lux S, Bullinger L. Circular RNAs in cancer. Adv Exp Med Biol. 2018;1087:215–30. doi: 10.1007/978-981-13-1426-1_17. [DOI] [PubMed] [Google Scholar]
- 14.Zhao ZJ, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2017;14:514–21. doi: 10.1080/15476286.2015.1122162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Dong Y, Zhao L, et al. Circular RNAMTO1 suppresses breast cancer cell viability and reverses monastrol resistance through regulating the TRAF4/Eg5 axis. Int J Oncol. 2018;53:1752–62. doi: 10.3892/ijo.2018.4485. [DOI] [PubMed] [Google Scholar]
- 16.Zhao Z, Ji M, Wang Q, et al. Circular RNA Cdr1 as upregulates SCAI to suppress cisplatin resistance in ovarian cancer via miR-1270 suppression. Mol Ther Nucleic Acids. 2019;18:24–33. doi: 10.1016/j.omtn.2019.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Wang Y, Li J, Du C, et al. Upregulated circular RNA circ-UBE2D2 predicts poor prognosis and promotes breast cancer progression by sponging miR-1236 and miR-1287. Transl Oncol. 2019;12:1305–13. doi: 10.1016/j.tranon.2019.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruivo CF, Adem B, Silva M, et al. The biology of cancer exosomes: Insights and new perspectives. Cancer Res. 2017;77:6480–88. doi: 10.1158/0008-5472.CAN-17-0994. [DOI] [PubMed] [Google Scholar]
- 19.Qin X, Yu S, Zhou L, et al. Cisplatin-resistant lung cancer cell-derived exosomes increase cisplatin resistance of recipient cells in exosomal miR-100-5p-dependent manner. Int J Nanomedicine. 2017;12:3721–33. doi: 10.2147/IJN.S131516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qu L, Ding J, Chen C, et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell. 2016;29:653–68. doi: 10.1016/j.ccell.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 21.Semina SE, Scherbakov AM, Vnukova AA, et al. Exosome-mediated transfer of cancer cell resistance to antiestrogen drugs. Molecules. 2018;23(4):829. doi: 10.3390/molecules23040829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu CG, Yang MF, Ren YQ, et al. Exosomes mediated transfer of lncRNA UCA1 results in increased tamoxifen resistance in breast cancer cells. Eur Rev Med Pharmacol Sci. 2016;20:4362–68. [PubMed] [Google Scholar]
- 23.Wei Y, Lai X, Yu S, et al. Exosomal miR-221/222 enhances tamoxifen resistance in recipient ER-positive breast cancer cells. Breast Cancer Res Treat. 2014;147:423–31. doi: 10.1007/s10549-014-3037-0. [DOI] [PubMed] [Google Scholar]
- 24.Sang Y, Chen B, Song X, et al. circRNA_0025202 regulates tamoxifen sensitivity and tumor progression via regulating the miR-182-5p/FOXO3a axis in breast cancer. Mol Ther. 2019;27:1638–52. doi: 10.1016/j.ymthe.2019.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17:79. doi: 10.1186/s12943-018-0827-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tutar Y. miRNA and cancer; Computational and experimental approaches. Curr Pharm Biotechnol. 2014;15:429. doi: 10.2174/138920101505140828161335. [DOI] [PubMed] [Google Scholar]
- 27.Mishra S, Yadav T, Rani V. Exploring miRNA-based approaches in cancer diagnostics and therapeutics. Crit Rev Oncol Hematol. 2016;98:12–23. doi: 10.1016/j.critrevonc.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 28.Wu DM, Wang S, Wen X, et al. LncRNA SNHG15 acts as a ceRNA to regulate YAP1-Hippo signaling pathway by sponging miR-200a-3p in papillary thyroid carcinoma. Cell Death Dis. 2018;9:947. doi: 10.1038/s41419-018-0975-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia C, Zhang Y, Xie Y, et al. MiR-200a-3p plays tumor suppressor roles in gastric cancer cells by targeting KLF12. Artif Cells Nanomed Biotechnol. 2019;47:3697–703. doi: 10.1080/21691401.2019.1594857. [DOI] [PubMed] [Google Scholar]
- 30.Ding M, Sun X, Zhong J, et al. Decreased miR-200a-3p is a key regulator of renal carcinoma growth and migration by directly targeting CBL. J Cell Biochem. 2018;119:9974–85. doi: 10.1002/jcb.27326. [DOI] [PubMed] [Google Scholar]
- 31.Lee H, Kim C, Kang H, et al. MicroRNA-200a-3p increases 5-fluorouracil resistance by regulating dual specificity phosphatase 6 expression. Exp Mol Med. 2017;49:e327. doi: 10.1038/emm.2017.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berthois Y, Delfino C, Metellus P, et al. Differential expression of miR200a-3p and miR21 in grade II-III and grade IV gliomas: Evidence that miR200a-3p is regulated by O(6)-methylguanine methyltransferase and promotes temozolomide responsiveness. Cancer Biol Ther. 2014;15:938–50. doi: 10.4161/cbt.28920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X, Wang M, Sun H, et al. Downregulation of LINC00894-002 Contributes to tamoxifen resistance by enhancing the TGF-beta signaling pathway. Biochemistry (Mosc) 2018;83:603–11. doi: 10.1134/S0006297918050139. [DOI] [PubMed] [Google Scholar]