Highlights
-
•
Chloroquine/hydroxychloroquine is an old anti-malarial drug belongs to 4-aminoquinolines.
-
•
Chloroquine has additional anti-virus, anti-bacteria, anti-protozoan, anti-autoimmunity and anti-cancer effects.
-
•
Mechanism of action of chloroquine in malaria and other diseases is not understood well.
-
•
Chloroquine directly targets Plasmodium hemoglobin degradation pathway.
-
•
In addition, chloroquine may directly target host’s autophagy, innate and adaptive immunity in malaria.
Abstract
Due to the rapid onset and spread of the COVID-19 pandemic, the treatment of COVID-19 patients by hydroxychloroquine alone or in combination with other drugs has captured a great deal of attention and triggered considerable debate. Historically, the worldwide use of quinoline based-drugs has led to a spectacular reduction in death from malaria. Unfortunately, scientists have been forced to seek alternative drugs to treat malaria due to the emergence of chloroquine-resistant parasites in the 1960s. The repurposing of hydroxychloroquine against viral infections, various types of cancer and autoimmune diseases has been ongoing for more than 70 years, with no clear understanding of its mechanism of action (MOA). Here, we closely examine the MOA of this old but influential drug in and beyond malaria. Better insights into how chloroquine targets the host’s cellular and immune responses may help to develop applications against to new pathogens and diseases, and perhaps even restore the clinical utility of chloroquine against malaria.
Current Opinion in Immunology 2020, 66:98–107
This review comes from a themed issue on Host pathogens
Edited by Ashley L St. John and Thomas E Morrison
For a complete overview see the Issue and the Editorial
Available online 18th August 2020
https://doi.org/10.1016/j.coi.2020.07.005
0952-7915/© 2020 Elsevier Ltd. All rights reserved.
Introduction
Malaria is an ancient disease that co-evolved with human populations and their migratory spread over the globe. Not so long ago, only 100 years, in fact, 77% of the world population was suffering from malaria, this number was reduced to 48% due to a century long elimination effort [1]. Today, almost half a million people still die from malaria every year [2]. The ‘success’ against malaria is largely due to the quinoline-containing antimalarial drugs such as quinine, chloroquine and mefloquine [3].
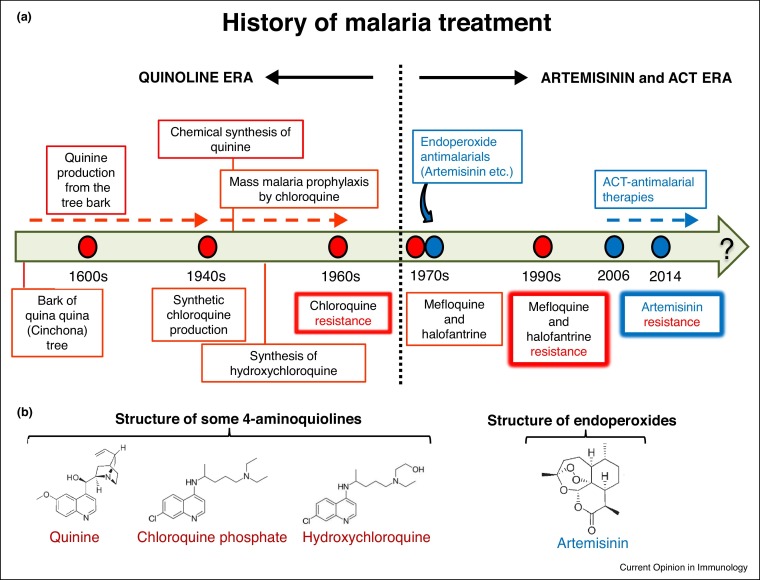
The history of the quinoline antimalarials dates back to 400 years ago. The Incas first extracted it from the bark of the quina-quina tree grown in the Andes (Figure 1 ). The tree later was widely came to be known as the ‘Cinchona tree’ after the Countess of Chinchon of Spain who was treated with the bark extract in the 17th century, and the tree bark was then brought to Europe [4]. The quinine was chemically extracted from these barks and used for centuries until the discovery of its synthetic analog chloroquine in 1934, which exhibited better tolerability and side effects. Although it took 10 years for chloroquine to come in to use in humans as a cheap, efficacious and affordable drug, it eventually came to be over used for protection from Plasmodium falciparum infections in many parts of the world, which resulted in the emergence of drug resistance and its withdrawal from P. falciparum treatment in South-East Asia, South America and Africa [3] (Figure 1). In addition, P. vivax-chloroquine resistant strains emerged in the 1990s in Southeast Asia, overall making more than 80% of worldwide wild parasite isolates proving resistant to chloroquine [5]. The chloroquine alternatives mefloquine and halofantrine were introduced in the 1970s and used for 30 years until parasite resistance appeared in these drugs towards P. falciparum strains.
Figure 1.
(a) History of malaria treatment. (b) Chemical structures of some quinoline drugs quinine, chloroquine phosphate and hydroxychloroquine and non-quinoline drug artemisinin.
The studies of the 2015 Nobel laureate Dr. Youyou Tu on Artemisia extracts since the beginning of 1970s led to the discovery of the artemisinin based drugs which do not belong to the quinoline class of drugs [6]. Since 2006 artemisinin combination therapies (ACTs) have been used to treat P. falciparum and complicated chloroquine-resistant P. vivax infections. The reason why artemisinin is used together with other agents such as quinoline-related drugs is due to the very short half-life of artemisinin, so the additional drugs help to prevent the recrudescence of the parasites [7]. Although recent studies have confirmed the signs of artemisinin resistance in P. falciparum [8], artemisinin and its derivatives have nevertheless provided a breakthrough treatment modality for malaria and rendered the quinoline drugs a secondary treatment option in most of the world. In the course of the recent coronavirus pandemic, treatment of COVID-19 patients with hydroxychloroquine has provoked a great deal of a debate. Chloroquine’s possible action on viral load and replication, lysosomal function and cellular immune responses has been vigorously discussed [9,10]. Therefore, I here summarize the current knowledge on the mechanisms of action of chloroquine against malaria. I wish to obtain novel insights into the effect of chloroquine on the host, rather than the parasite, which will facilitate its repurposing against various conditions, including viral infections, cancer and autoimmune diseases, and perhaps may even help to restore its clinical utility against malaria.
The mechanism of action of chloroquine on Plasmodium infected erythrocytes
Chloroquine generally refers to chloroquine phosphate (C18 H26 ClN3), a weak base drug that belongs to the first group of quinolone derivatives, the 4-aminoquinolines. Chloroquine’s hydroxyl derivative hydroxychloroquine (C18 H26 ClN3 O) that was developed in the 1950s presumably has a similar mechanism of action along with a higher safety profile. How chloroquine acts against malaria is still not well understood, although it is known that chloroquine affects only erythrocytic-stage parasites after diffusing across the erythrocyte and parasite membranes due to its small size and lipophilic characteristics. Two possibilities are suggested (Figure 2 ).
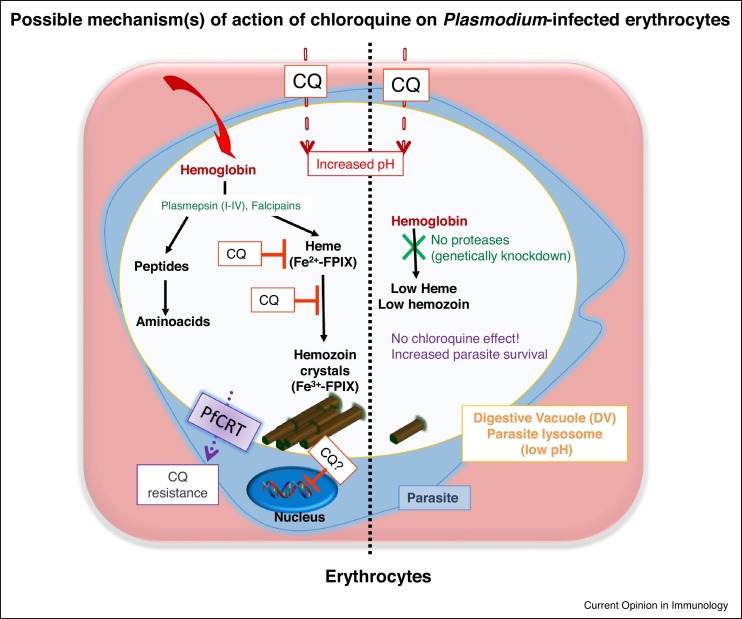
Figure 2.
Possible mechanism (s) of action of chloroquine during blood stage malaria infection. After invasion of erythrocytes, Plasmodium parasites form their own DV, a lysosome-like acidic compartment important for parasite metabolism and survival. In acidic DVs, the host-hemoglobin is degraded by parasite proteases for the vital needs, such as amino acids and the free-heme (Fe2+–protophorphyrin IX) is detoxified by converting it into insoluble crystals hemozoin (Fe3+–protophorphyrin IX). A weak base chloroquine accumulates in DVs, increases DV pH and binds heme and crystal surfaces, thereby blocks every steps of hemozoin formation which eventually leads heme toxicity and parasite death. In the absence of hemoglobin degrading proteases hemoglobin remains undigested and free heme is significantly diminished and the effect of chloroquine on parasites does not occur. Ineffective presence of chloroquine, on the other hand, may create the chloroquine-resistant parasites via a mutation in P. falciparum chloroquine resistance transporter (PfCRT) and possibly other genes.
First, chloroquine has long been known to bind to DNA and RNA. Early studies suggested it could inhibit DNA and RNA synthesis by binding to nucleic acids via electrostatic forces, hydrogen bonds, and van der Waals forces [11,12]. This may explain why chloroquine as well as hydroxychloroquine can inhibit replication of certain viruses in vitro, such as HIV, Zika virus, influenza A virus, herpes simplex virus, SARS-CoV, and chikungunya virus [13]. Hence, it is reasonable that chloroquine may interact in such a manner with the Plasmodium DNA/RNA machinery within erythrocytes. However, chloroquine-Plasmodium DNA interactions inside the parasite nucleus were found to require rather high concentrations of the drug (at a toxic level that exerts an inhibitory effect even on the growth of host cells), which would be far more than the concentration required for the clearance of parasites in vivo [4]. Therefore, this idea was found to be unfavorable.
The second is the effect of the weak base chloroquine on heme-like structures. During the life cycle of parasites in erythrocytes, the host hemoglobin is degraded by parasite proteases called Plasmepsins (I–IV) and Falcipains [14] for their own amino acid needs. The liberated free heme is subsequently polymerized into hemozoin, a black crystalloid metabolite and the hallmark of Plasmodium parasites, in a process that develops in the acidic lysosome-like parasite digestive vacuole (DV) residing inside the erythrocytes (Ref. [15] and Figure 2). The structural similarity between heme (monomeric) and hemozoin (dimeric) is well known and it is preserved at a low pH, but readily disassociates in alkaline solutions [16]. Thus, the accumulation of the weak base chloroquine in the DV perhaps naturally prevents crystallization dynamics due to the pH increase. Supporting this, recent studies have clearly shown that chloroquine binds heme (Ferriprotoporphyrin IX, Fe+2) as well as crystal surfaces with a strong affinity and thus ends up blocking hemozoin (Ferriprotoporphyrin IX, Fe+3 dimers) formation at every step of crystallization [17•,18•], thus allowing free-heme toxicity to parasites. On the other hand, there are other important facets of the mechanism of action of chloroquine against erythrocytic stage parasites. Recent studies showed that chemically labeled chloroquine molecules could be detected in DVs and on parasitic membranes, but not on red blood cell membranes [19,18•]. Chloroquine probably compromises the DV membrane and leads to an extrusion of DV proteases such as plasmepsin-IV which hydrolyze hemoglobin, in both in vitro and in vivo conditions [20]. A recent in vivo study using genetically modified mutant-parasite model confirmed this. In the absence of P. berghei hemoglobin degrading proteases (plasmepsin-IV and berghepain-2), hemoglobin was shown to be undigested and free heme release was significantly diminished. Therefore, no effect of chloroquine on parasites occurred, so no parasite death resulted [21]. This study also implies that the Plasmodium DNA/RNA machinery remains intact even in the presence of chloroquine.
However, ineffective presence of chloroquine has been linked to genetic pressure on the parasites resulting in chloroquine-resistant parasites. The main mechanism of resistance to chloroquine by P. falciparum parasites has been shown to involve mutation of the P. falciparum chloroquine resistance transporter (PfCRT), a transmembrane protein located in the DV membrane, which effluxes chloroquine into the cytosol [3] (Figure 2). However, other studies have indicated that this chloroquine-resistance phenotype does not involve all P. falciparum parasite strains, rather more multigenic involving other genetic loci [22,23]. Furthermore, chloroquine’s effect on the P. falciparum and P. vivax parasites seems to involve different mechanisms. While P. falciparum trophozoites with a single large food vacuole are fully susceptible to chloroquine, P. vivax trophozoites with many small vacuoles are not [24••,25]. Further studies are clearly needed to elucidate chloroquine’s pleiotropic effects on infected-erythrocytes and how Plasmodium parasites develop resistance to this drug. Because chloroquine resistance in P. falciparum seems to be decreasing in Africa after years of withdrawal [26], the opportunity for bringing back this cheap and safe drug to the field is growing.
Interestingly, the blood fluke Schistosoma mansoni similarly digests host hemoglobin and release free heme which is detoxified through hemozoin formation [27]. Based on this finding, in vivo experimental studies showed that chloroquine treatment decreased hemozoin formation, the viability of the worms and the severity of infection in S. mansoni-infected mice [27], confirming chloroquine’s pleiotropic effects on various pathogens.
Mechanisms of action of chloroquine: beyond infected erythrocytes
It is well known, mainly from empirical non-malarial usage of chloroquine in autoimmune diseases such as systemic lupus erythematosus (SLE) and rheumatoid arthritis (RA), that chloroquine has a wide range of different effects on immune cells and inflammation. Therefore, chloroquine may exert an effect on the immune system during malaria treatment, a possibility which has been largely overlooked. Early studies in animals and a few post-mortem studies in humans have shown that chloroquine accumulates at high concentrations in the eye (due to potent binding to melanin), lung, liver and spleen (possibly due to preferred accumulation in lysosomes), with low levels in muscle, brain tissue and bone, albeit only after several months of medication [12]. In fact, one of the reasons for the widespread investigation into the side effects of chloroquine and its derivatives is due to the prolonged treatment needs of SLE and RA patients (i.e. several months or even years). These studies have concluded that hydroxychloroquine is effective with fewer side effects than many other drugs for SLE treatment, even during pregnancy [10]. It is of note that the overall cumulative doses of chloroquine used for autoimmune disease treatment is at least 100 times greater (200−400 mg/day over weeks and years depending on the patient) than for prophylaxis (100−250 mg/week during and after stay in endemic area) or treatment of malaria (400 mg/day over 3 days) [12]. Chloroquine has been used safely for the treatment of pregnant women (chloroquine can easily cross the placenta with no apparent harmful effects to the fetus) as well as in lactating women and newborn/infants during cases of malaria [28]. In contrast, certain other quinolines, such as primaquine, may exert hemolytic effects in people with glucose-6-phosphate dehydrogenase deficiency (G6PD) [29]. The rare acute side effects of chloroquine during malaria treatment include gastrointestinal symptoms and itching, but it comparatively rarely induces neurological or cardiovascular symptoms, such as cardiac arrhythmia due to prolongation of cardiac repolarization (the QTc interval), and when such effects do occur, they are usually associated with high doses due to rapid intravenous infusion of the drug and/or high peak concentrations [10]. Importantly, children 4–8 years-old who are infected with chloroquine-resistant P. falciparum and treated with double or nearly triple-doses of the standard chloroquine protocol appear to tolerate the drug and are completely cured of malaria, albeit with prolonged QT intervals, but with no cardiac arrhythmias [30••]. Of note, hydroxychloroquine alone seems to be well tolerated if not given at a bolus concentration, and additional consideration is needed when it is given along with additional drugs such as digitoxin, tamoxifen, methotrexate and cyclosporine, as well as primaquine and azithromycin (the drug–chloroquine interactions are extensively summarized in Ref. [10]).
Similar to the case of the DV of Plasmodium parasites, chloroquine accumulates in any low-pH (i.e. a pH of less than 4–5) organelle such as lysosomes due to its weak base property and reduces the organelle’s acidification. Weak base chloroquine is usually uncharged and readily diffused into the lysosome, where it easily becomes protonated, allowing chloroquine to concentrate within compartments higher than its extracellular concentration and increasing the pH up to 6. The increase in the lysosomal pH in immune cells by chloroquine has three possible consequences during malaria.
Chloroquine and host autophagy during malaria
Autophagy (canonical or macroautophagy) is a cellular process that help cells to degrade and recycle their own components through an intracellular engulfment process which involves the formation of double membrane vesicles known as autophagosomes, which fuse with lysosomes for the final enzymatic digestion of components for their nutrient content [31,32] (Figure 3 ). This final acidic lysosomal activity is a key step in autophagy and its neutralization by weak alkaline chloroquine, or inhibition with bafilomycin A1 (that inhibits the lysosomal proton pump) disturbs lysosomal function, and either event leads to the failure of autophagy.
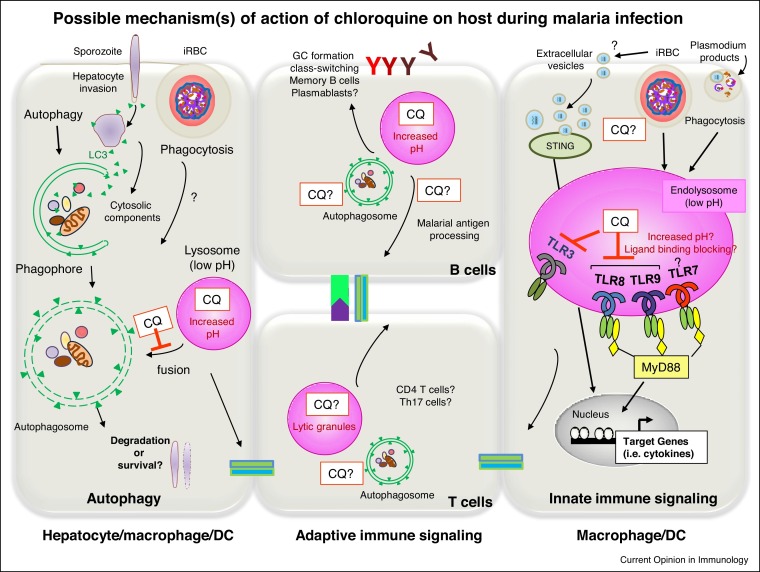
Figure 3.
Possible mechanism (s) of action of chloroquine on immune cells during malaria infection. Chloroquine has a clear anti-inflammation and anti-cytokine effect during malaria. It may accumulate in any low-pH organelle such as lysosomes in immune cells due to its weak base property and increases pH. This may affect autophagy of cells which require final acidic lysosomal activity that chloroquine can easily neutralize this process. Chloroquine may be affecting host autophagy machinery targeting sporozoites inside the hepatocytes or alternatively might have an effect on the accumulated CD8+ T cells, liver-resident T cells or activated T cells and B cells during infection. Chloroquine may have direct effect on pattern recognition receptors (PRRs), particularly TLR-mediated cytokine induction, because the acidic pH of endosome is a requirement for endosomal TLR activation. Therefore, chloroquine may interfere with the interaction of Plasmodium DNA, hemozoin and RNA with TLR9, TLR7 and TLR8 or inhibit the maturation of Plasmodium-containing phagosome. Chloroquine might modulate the extracellular vesicles (EVs) secreted from infected RBCs containing parasite small RNA and genomic DNA which activate cytosolic STING pathway. Overall, all these possible effects may modulate antigen presenting cells (APCs, mainly monocytes/macrophages and dendritic cells), lytic granule processing and modulate adaptive immune responses to malaria.
Several types of autophagy against pathogens have been described over the last two decades. In the course of selective autophagy (xenophagy), the autophagosome forms around the engulfed pathogens. Xenophagy helps eliminate pathogens, but in turn, may be hijacked by pathogens for their own survival. The recently described microtubule-associated protein 1 light chain 3 (LC3)-associated phagocytosis (LAP) involves only a few of the autophagy initiation complex proteins and does not result in autophagosome formation, but helps to eliminate pathogens via direct fusion with lysosomes [33]. During the pre-erythrocytic stages of malaria infection sporozoites invade hepatocytes and make a shield out of a membrane-bound parasitophorous vacuole (PV) and then replicate in it. The host autophagy machinery targets these PVs by decorating PV membranes with autophagy markers, including LC3. As a result, Plasmodium sporozoites either hijack the hepatocyte autophagy pathway and gain nutrients for their growth (elimination is avoided by PV transmembrane protein UIS3 [34]), or are degraded by a xenophagy-like mechanism. If the immune system is successfully activated (such as by the secretion of IFN-γ), approximately 50% of the intracellular sporozoites are cleared by a LAP-like process in hepatocytes [35].
Chloroquine has a direct effect on erythrocytic-stage but not sporozoite stage parasites. However, chloroquine has been used as an autophagy inhibitor in experimental studies investigating host autophagy in hepatocytes during sporozoite development [36]. On the other hand, the protection against Plasmodium parasites has been shown in animal and human experimental studies when individuals are immunized with infectious sporozoites under the cover of chloroquine chemoprophylaxis [37••,38,39•]. Although the role of chloroquine in these studies was to suppress following blood stage parasites, whether chloroquine induces autophagy inhibition or exerts an immunomodulatory effect has not been elucidated. For instance, chloroquine’s prophylactic effect during live sporozoite immunizations was compared with radiation-attenuated sporozoite (RAS) immunizations in humans and found to be 20 times more efficient, requiring only 45 mosquitoes bites versus 1000 bites, although the induction of the efficient parasite-specific CD8+ T cells response and IFN-γ production was comparable [40]. As it is believed that the presence of the liver stage parasite is a prerequisite for the induction of protective responses after sporozoite immunization (confirmed by concurrent primaquine treatment, which abrogated protection), chloroquine prophylaxis may need a different interpretation. The difference between live sporozoites and RAS sporozoites might be due to the sporozoites’ interaction with the hepatocyte autophagy machinery and chloroquine’s direct effect on it. Alternatively, chloroquine might have an effect on the accumulated CD8+ T cells or liver-resident T cells as autophagy helps to maintain liver-resident CD8+ T cells and their mitochondrial fitness [41]. These are open questions to be answered in the future.
There is very little information on the role of host autophagy during blood stage malaria infection, although Plasmodium parasites’ own autophagy in the blood stage has been studied [42,32]. Rapid acidification of phagosomes occurs when macrophages are stimulated with infected-erythrocytes in vitro, which may block the efficient signaling required for cytokines, as shown by the blocking of this this pathway by the acidification blocker Baf-A1 [43]. However, whether this actually occurs as a result of a suppression of acidification by chloroquine has not been reported.
Autophagy is known to be enhanced during T cell activation and proliferation [44], therefore whether chloroquine treatment inhibits the autophagic flux of activated T and/or B cells during malaria infection needs to be investigated.
Chloroquine and the innate immune system during malaria
In addition to its anti-parasitic effects, when chloroquine is given to P. falciparum-infected children it clearly exerts anti-inflammatory and anti-cytokine effect [30••]. Chloroquine’s suppression of cytokinesis during malaria is suspected to be due to its direct effect on pattern recognition receptors (PRRs), particularly TLR-mediated cytokine induction [45]. Although further studies are needed, there are a few possible explanations of the target molecule(s) for chloroquine on immune cells. During the blood stage, infected erythrocytes, ruptured merozoites and parasite products such as hemozoin are continuously phagocytized by monocytes/macrophages or DCs. TLR3, TLR7, TLR8 and TLR9 are the only TLRs among the 13 in the TLR family that are located on acidic organelle endosomes, and mainly recognize different classes of nucleic acids of either endogenous or exogenous origin. The acidic pH of endosomes is a requirement for endosomal TLR activation. However, chloroquine was found to have more activities than just this, for example directly interacting with cognate ligands (nucleic acids), changing the chemical environment and masking TLR ligand-binding epitopes [46]. CpG ODN-induced immune activation was inhibited by chloroquine via directly competing with CpG ODN for binding to the TLR9-ectodomain and changing its conformation. Moreover, poly (I:C) (dsRNA) interaction with its receptor TLR3 was affected similarly by chloroquine. In contrast, chloroquine’s inhibition of TLR8 was mostly due to the manipulation of endosomal pH by chloroquine [47]. Plasmodium DNA and RNA either alone or complexed with malarial products are ligands recognized by either endosomal TLR9 or TLR7 as well as cytosolic DNA sensing pathways such as STING (recently reviewed in Ref. [48]). Although malarial hemozoin’s recognition by TLR9 in DCs [49] has been extensively debated [48], this is the only study that has showed direct evidence that Plasmodium hemozoin-mediated cytokine activation was blocked by chloroquine treatment, and that hemozoin directly interacted with the TLR9-ectodomain protein via its heme molecule and thereby changed its confirmation [50]. Just recently, TLR8 was found to be involved in P. falciparum RNA recognition in human TLR8-expressing cells and this interaction was similarly blocked by chloroquine [51•]. Therefore, it is possible that chloroquine, in addition to its ability to inhibit hemozoin formation in erythrocytes, may also interfere with the interaction of Plasmodium hemozoin, DNA and RNA with TLR9, TLR7 and TLR8. Alternatively, it may inhibit the maturation of Plasmodium product-containing phagosomes, thereby inhibiting subsequent innate immune activation (Figure 3) and not have a direct effect on endocytic cell entry or the replication machinery, as in the case of viral infections. There is also the possibility that macrophage/monocyte and DC acidification mechanisms and levels may interfere with endosomal TLR recognition in different cells [43]. Nevertheless, the contribution of these various ligands from the Plasmodium parasite in the control of the immune system via the TLRs and chloroquine’s direct effect on innate immunity needs in vivo investigation.
A recent study showed that chronic accumulation of Plasmodium products in the bone marrow induces bone loss, but this pathology does not occur when either MyD88 (the adaptor protein for most TLR signaling) is lacking or mutant parasites lacking hemozoin (i.e. lacking Plasmepsin IV and berghepain 2) were used [52]. This study suggests that intact parasite DNA or RNA alone, although sensed via MyD88, have a limited capacity to induce pathology in the absence of hemozoin. Instead, hemozoin and the accumulation of other as yet unknown Plasmodium products activate cytokines via MyD88 are responsible for malaria-induced bone loss. Furthermore, repeated chloroquine treatment was shown to play a minimal role for the accumulation of hemozoin in the bone marrow which caused bone loss, suggesting the hemozoin crystals that form under chloroquine treatment remain intact and continuously induce immune activation and pathology even after parasite clearance. Clearly, further investigation of this bone pathology in vivo using animal models and in humans is needed to understand chloroquine’s long term effects during malaria infection.
Hydroxychloroquine was also shown to inhibit the activity of another nucleic acid sensor, cyclic GMP-AMP synthase (cGAS), by interfering with its binding to cytosolic DNA upstream of STING as well as by blocking the sorting of STING vesicles to lysosomes for STING degradation [53,54•]. During malaria, it is likely that extracellular vesicles (EVs) secreted from infected RBCs containing small RNA and genomic DNA of the parasite activate the cytosolic STING pathway [55•,56], although whether chloroquine directly targets these EVs has not been investigated. It is of note that recent studies have suggested another possibility that chloroquine can enhance the production of EVs [57,58]. Thus, how chloroquine interferes with Plasmodium-secreted EVs needs further investigation.
There is also a very likely possibility that chloroquine may block the interaction of cytosolic DNA as well as RNA with other sensing pathways such as the AIM2 or IPS1 (MAVS) [53]. This raises the possibility that recognition of Plasmodium RNA in hepatocytes [59] might be blocked by chloroquine, which has also not been investigated during sporozoite infection and vaccination.
Chloroquine and the adaptive immune system during malaria
The lysosomotropic properties of chloroquine may have an effect on antigen presenting cells (APCs, mainly monocytes/macrophages and dendritic cells) and modulate several innate signaling pathways, such as TLRs (mentioned above), proinflammatory cytokine production, and co-stimulatory molecule signaling, thus helping to elicit a proper adaptive immune responses. Furthermore, chloroquine’s long half-life (∼50 days) may also result in the modulation of several adaptive immune cell functions such as that results in impaired antigen processing, or suppression of antigen presentation to CD4+ T-cells, or inhibition of the differentiation into and secretion of cytokines from Th1 and Th17 cells [10,60,61], or inhibiting the perforin processing [62]. In contrast, several other studies have shown that chloroquine enhances DCs’ cross-presentation of soluble antigens, but not particulate antigens, to CD8+ T cells. This has been shown under both in vitro and in vivo conditions, for example during vaccination or in the course of viral infections, and is most probably due to the reduced degradation of antigens in the presence of weak base agents, resulting in a higher accumulation in endosomes and subsequent efficient export into the cytosol leading to efficient cross-presentation [63].
During malaria, in general, while antibodies are important in controlling blood-stage parasitemia, it is the CD8+ T-cell responses are critically involved in pre-erythrocytic immunity. Unexpectedly, RAS immunization under prophylaxis with or without chloroquine induced equally strong CD8+ T cells responses [40], suggesting that chloroquine’s effect was on other, as yet unknown mechanism(s) or on other cells. Interestingly, a recent study showed that humans immunized with live sporozoites under chloroquine chemoprophylaxis reacted to a broad repertoire of novel antigens in both breadth and magnitude, as assessed by a comprehensive P. falciparum protein microarray of blood [39•]. Furthermore, the direct effect of chloroquine/primaquine prophylaxis/treatment doses on vaccination using model antigens in mice was investigated and it was found that chloroquine clearly modulates antigen-specific B cell responses [64]. In fact, chloroquine together with an adjuvant (i.e. alum) increases the protective efficacy of whole-killed blood-stage vaccines via humoral immune responses due to an expansion of GC B cells and class‐switch recombination [65]. A synthetic analog of malarial hemozoin has been proposed to be a universal adjuvant [66]. It will be interesting to see in further studies investigating the effect of chloroquine on this particular malaria-derived adjuvant and whether it has the ability to bind directly to the hemozoin surface [18•].
Conclusions
Chloroquine has been one of the most affordable, relatively safe and widely used medications in the history of humankind, with pleiotropic functions under protozoan, viral, bacterial and inflammatory conditions. We owe a debt to chloroquine for its effectiveness in ameliorating the impact of malaria cases all over the world, but we still do not understand its mechanism of action, and how Plasmodium parasites develop resistance to it. Based on the information gained from non-malarial studies, we emphasize that more research is required to understand the host-mediated activity of chloroquine during malaria. We may repurpose chloroquine and its derivatives and facilitate its return to the shrinking list of antimalarials.
Authors’ contributions
Review was written and figures were prepared by CC.
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
The author acknowledges funding from the Japan Science and Technology Agency (JST-CREST to CC) and the International Joint Research Project of the Institute of Medical Science, the University of Tokyo (to CC).
References
- 1.Hay S.I., Guerra C.A., Tatem A.J., Noor A.M., Snow R.W. The global distribution and population at risk of malaria: past, present, and future. Lancet Infect Dis. 2004;4:327–336. doi: 10.1016/S1473-3099(04)01043-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coban C., Lee M.S.J., Ishii K.J. Tissue-specific immunopathology during malaria infection. Nat Rev Immunol. 2018;18:266–278. doi: 10.1038/nri.2017.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ross L.S., Fidock D.A. Elucidating mechanisms of drug-resistant Plasmodium falciparum. Cell Host Microbe. 2019;26:35–47. doi: 10.1016/j.chom.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foley M., Tilley L. Quinoline antimalarials: mechanisms of action and resistance and prospects for new agents. Pharmacol Ther. 1998;79:55–87. doi: 10.1016/S0163-7258(98)00012-6. [DOI] [PubMed] [Google Scholar]
- 5.Ginsburg H. Should chloroquine be laid to rest? Acta Trop. 2005;96:19–23. doi: 10.1016/j.actatropica.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 6.Wang J., Xu C., Wong Y.K., Li Y., Liao F., Jiang T., Tu Y. Artemisinin, the magic drug discovered from traditional Chinese medicine. Engineering. 2019;5:32–39. doi: 10.1016/j.eng.2018.11.011. [DOI] [Google Scholar]
- 7.Nosten F., White N.J. Artemisinin-based combination treatment of falciparum malaria. Am J Trop Med Hyg. 2007;77:181–192. doi: 10.4269/ajtmh.2007.77.181. [DOI] [PubMed] [Google Scholar]
- 8.Tilley L., Straimer J., Gnädig N.F., Ralph S.A., Fidock D.A. Artemisinin action and resistance in Plasmodium falciparum. Trends Parasitol. 2016;32:682–696. doi: 10.1016/j.pt.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu T.Y., Frieman M., Wolfram J. Insights from nanomedicine into chloroquine efficacy against COVID-19. Nat Nanotechnol. 2020;15:247–249. doi: 10.1038/s41565-020-0674-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schrezenmeier E., Dörner T. Mechanisms of action of hydroxychloroquine and chloroquine: implications for rheumatology. Nat Rev Rheumatol. 2020;16:155–166. doi: 10.1038/s41584-020-0372-x. [DOI] [PubMed] [Google Scholar]
- 11.Parker F.S., Irvin J.L. The interaction of chloroquine with nucleic acids and nucleoproteins. J Biol Chem. 1952;199:897–909. [PubMed] [Google Scholar]
- 12.Browning D.J. Hydroxychloroquine and Chloroquine Retinopathy. 2014. Pharmacology of chloroquine and hydroxychloroquine; pp. 35–63. [DOI] [Google Scholar]
- 13.Li C., Zhu X., Ji X., Quanquin N., Deng Y.Q., Tian M., Aliyari R., Zuo X., Yuan L., Afridi S.K. Chloroquine, a FDA-approved drug, prevents zika virus infection and its associated congenital microcephaly in mice. EBioMedicine. 2017;24:189–194. doi: 10.1016/j.ebiom.2017.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nasamu A.S., Polino A.J., Istvan E.S., Goldberg D.E. Malaria parasite plasmepsins: more than just plain old degradative pepsins. J Biol Chem. 2020;295:8425–8441. doi: 10.1074/jbc.rev120.009309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coban C., Yagi M., Ohata K., Igari Y., Tsukui T., Horii T., Ishii K.J., Akira S. The malarial metabolite hemozoin and its potential use as a vaccine adjuvant. Allergol Int. 2010;59:115–124. doi: 10.2332/allergolint.10-RAI-0194. [DOI] [PubMed] [Google Scholar]
- 16.Lee M.S.J., Igari Y., Tsukui T., Ishii K.J., Coban C. Current status of synthetic hemozoin adjuvant: a preliminary safety evaluation. Vaccine. 2016;34:2055–2061. doi: 10.1016/j.vaccine.2016.02.064. [DOI] [PubMed] [Google Scholar]
- 17•.Olafson K.N., Nguyen T.Q., Rimer J.D., Vekilov P.G. Antimalarials inhibit hematin crystallization by unique drug–surface site interactions. Proc Natl Acad Sci U S A. 2017;114:7531–7536. doi: 10.1073/pnas.1700125114. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows detailed molecular mechanism of quinoline drugs action that quinolines directly interact with crystal surfaces rather than soluble forms and suppress every steps of heme crystal growth.
- 18•.Kapishnikov S., Staalsø T., Yang Y., Lee J., Pérez-Berná A.J., Pereiro E., Yang Y., Werner S., Guttmann P., Leiserowitz L. Mode of action of quinoline antimalarial drugs in red blood cells infected by Plasmodium falciparum revealed in vivo. Proc Natl Acad Sci U S A. 2019;116:22946–22952. doi: 10.1073/pnas.1910123116. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows mode of action of quinoline drugs at the molecular level in vivo within Plasmodium-infected red blood cells.
- 19.Woodland J.G., Hunter R., Smith P.J., Egan T.J. Chemical proteomics and super-resolution imaging reveal that chloroquine interacts with Plasmodium falciparum multidrug resistance-associated protein and lipids. ACS Chem Biol. 2018;13:2939–2948. doi: 10.1021/acschembio.8b00583. [DOI] [PubMed] [Google Scholar]
- 20.Ch’ng J.H., Lee Y.Q., Gun S.Y., Chia W.N., Chang Z.W., Wong L.K., Batty K.T., Russell B., Nosten F., Renia L. Validation of a chloroquine-induced cell death mechanism for clinical use against malaria. Cell Death Dis. 2014;5:e1305. doi: 10.1038/cddis.2014.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin J. wen, Spaccapelo R., Schwarzer E., Sajid M., Annoura T., Deroost K., Ravelli R.B.G., Aime E., Capuccini B., Mommaas-Kienhuis A.M. Replication of Plasmodium in reticulocytes can occur without hemozoin formation, resulting in chloroquine resistance. J Exp Med. 2015;212:893–903. doi: 10.1084/jem.20141731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen N., Russell B., Fowler E., Peters J., Cheng Q. Levels of chloroquine resistance in Plasmodium falciparum are determined by loci other than pfcrt and pfmdr1. J Infect Dis. 2002;185:405–406. doi: 10.1086/338470. [DOI] [PubMed] [Google Scholar]
- 23.Reiling S.J., Krohne G., Friedrich O., Geary T.G., Rohrbach P. Chloroquine exposure triggers distinct cellular responses in sensitive versus resistant Plasmodium falciparum parasites. Sci Rep. 2018;8:11137. doi: 10.1038/s41598-018-29422-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24••.Kim A., Popovici J., Menard D., Serre D. Plasmodium vivax transcriptomes reveal stage-specific chloroquine response and differential regulation of male and female gametocytes. Nat Commun. 2019;10:371. doi: 10.1038/s41467-019-08312-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows at transcriptional level that chloroquine has little effect on trophozoite stages of P. vivax parasites, which is different than P. falciparum parasites, suggesting that chloroquine may act through a different mechanism in different parasite species.
- 25.Sá J.M., Kaslow S.R., Moraes Barros R.R., Brazeau N.F., Parobek C.M., Tao D., Salzman R.E., Gibson T.J., Velmurugan S., Krause M.A. Plasmodium vivax chloroquine resistance links to pvcrt transcription in a genetic cross. Nat Commun. 2019;10:4300. doi: 10.1038/s41467-019-12256-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conrad M.D., Rosenthal P.J. Antimalarial drug resistance in Africa: the calm before the storm? Lancet Infect Dis. 2019, 19: 338-351;19:338–351. doi: 10.1016/S1473-3099(19)30261-0. [DOI] [PubMed] [Google Scholar]
- 27.Oliveira M.F., d’Avila J.C.P., Tempone A.J., Corrêa Soares J.B.R., Rumjanek F.D., Ferreira-Pereira A., Ferreira S.T., Oliveira P.L. Inhibition of heme aggregation by chloroquine reduces Schistosoma mansoni infection. J Infect Dis. 2004;190:843–852. doi: 10.1086/422759. [DOI] [PubMed] [Google Scholar]
- 28.Saito M., Gilder M.E., McGready R., Nosten F. Antimalarial drugs for treating and preventing malaria in pregnant and lactating women. Expert Opin Drug Saf. 2018;17:1129–1144. doi: 10.1080/14740338.2018.1535593. [DOI] [PubMed] [Google Scholar]
- 29.Baird J.K., Valecha N., Duparc S., White N.J., Price R.N. Diagnosis and treatment of Plasmodium vivax malaria. Am J Trop Med Hyg. 2016;93:35–51. doi: 10.4269/ajtmh.16-0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30••.Ursing J., Rombo L., Eksborg S., Larson L., Bruvoll A., Tarning J., Rodrigues A., Kofoed P.E. High-dose chloroquine for uncomplicated Plasmodium falciparum malaria is well tolerated and causes similar QT interval prolongation as standard-dose chloroquine in children. Antimicrob Agents Chemother. 2020;64 doi: 10.1128/AAC.01846-19. e01846-19. [DOI] [PMC free article] [PubMed] [Google Scholar]; This important study investigated the safety and tolerability of 2–3 times the total standard dose of chloroquine used for treatment of children with uncomplicated P. falciparum malaria and found it safe and tolerable with no toxicity issues.
- 31.Lelliott P.M., Coban C. IFN-γ protects hepatocytes against Plasmodium vivax infection via LAP-like degradation of sporozoites. Proc Natl Acad Sci U S A. 2016;113:6813–6815. doi: 10.1073/pnas.1607007113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mizushima N. The ATG conjugation systems in autophagy. Curr Opin Cell Biol. 2020;63:1–10. doi: 10.1016/j.ceb.2019.12.001. [DOI] [PubMed] [Google Scholar]
- 33.Sanjuan M.A., Dillon C.P., Tait S.W.G., Moshiach S., Dorsey F., Connell S., Komatsu M., Tanaka K., Cleveland J.L., Withoff S. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 34.Real E., Rodrigues L., Cabal G.G., Enguita F.J., Mancio-Silva L., Mello-Vieira J., Beatty W., Vera I.M., Zuzarte-Luís V., Figueira T.N. Plasmodium UIS3 sequesters host LC3 to avoid elimination by autophagy in hepatocytes. Nat Microbiol. 2017;3:17–25. doi: 10.1038/s41564-017-0054-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boonhok R., Rachaphaew N., Duangmanee A., Chobson P., Pattaradilokrat S., Utaisincharoen P., Sattabongkot J., Ponpuak M. LAP-like process as an immune mechanism downstream of IFN-γ in control of the human malaria Plasmodium vivax liver stage. Proc Natl Acad Sci U S A. 2016;113:3519–3528. doi: 10.1073/pnas.1525606113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wacker R., Eickel N., Schmuckli-Maurer J., Annoura T., Niklaus L., Khan S.M., Guan J.L., Heussler V.T. LC3-association with the parasitophorous vacuole membrane of Plasmodium berghei liver stages follows a noncanonical autophagy pathway. Cell Microbiol. 2017;19:e12754. doi: 10.1111/cmi.12754. [DOI] [PubMed] [Google Scholar]
- 37••.Tran T.M., Bijker E.M., Haks M.C., Ottenhoff T.H.M., Visser L., Schats R., Venepally P., Lorenzi H., Crompton P.D., Sauerwein R.W. Whole-blood transcriptomic signatures induced during immunization by chloroquine prophylaxis and Plasmodium falciparum sporozoites. Sci Rep. 2019;9:8386. doi: 10.1038/s41598-019-44924-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study provides whole blood transcription profiling and related molecular signatures associated with protection of humans immunized with P. falciparum sprorozoites under chloroquine prophylaxis.
- 38.Moncunill G., Scholzen A., Mpina M., Nhabomba A., Hounkpatin A.B., Osaba L., Valls R., Campo J.J., Sanz H., Jairoce C. Antigen-stimulated PBMC transcriptional protective signatures for malaria immunization. Sci Transl Med. 2020;12:eaay8924. doi: 10.1126/scitranslmed.aay8924. [DOI] [PubMed] [Google Scholar]
- 39•.Obiero J.M., Campo J.J., Scholzen A., Randall A., Bijker E.M., Roestenberg M., Hermsen C.C., Teng A., Jain A., Davies D.H. Antibody biomarkers associated with sterile protection induced by controlled human malaria infection under chloroquine prophylaxis. mSphere. 2019;4 doi: 10.1128/mspheredirect.00027-19. e00027-19. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study provides humoral immune correlates of protection of humans immunized with P. falciparum sprorozoites under chloroquine prophylaxis.
- 40.Bijker E.M., Nganou-Makamdop K., Van Gemert G.J., Zavala F., Cockburn I., Sauerwein R.W. Studying the effect of chloroquine on sporozoite-induced protection and immune responses in Plasmodium berghei malaria. Malar J. 2015;14:130. doi: 10.1186/s12936-015-0626-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swadling L., Pallett L.J., Diniz M.O., Baker J.M., Amin O.E., Stegmann K.A., Burton A.R., Schmidt N.M., Jeffery-Smith A., Zakeri N. Human liver memory CD8+ T cells use autophagy for tissue residence. Cell Rep. 2020;30:687–698. doi: 10.1016/j.celrep.2019.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coppens I. How toxoplasma and malaria parasites defy first, then exploit host autophagic and endocytic pathways for growth. Curr Opin Microbiol. 2017;40:32–39. doi: 10.1016/j.mib.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 43.Wu X., Gowda N.M., Gowda D.C. Phagosomal acidification prevents macrophage inflammatory cytokine production to malaria, and dendritic cells are the major source at the early stages of infection: implication for malaria protective immunity development. J Biol Chem. 2015;290:23135–23147. doi: 10.1074/jbc.M115.671065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clarke A.J., Simon A.K. Autophagy in the renewal, differentiation and homeostasis of immune cells. Nat Rev Immunol. 2019;19:170–183. doi: 10.1038/s41577-018-0095-2. [DOI] [PubMed] [Google Scholar]
- 45.Hugosson E., Montgomery S.M., Premji Z., Troye-Blomberg M., Björkman A. Relationship between antipyretic effects and cytokine levels in uncomplicated falciparum malaria during different treatment regimes. Acta Trop. 2006;99:75–82. doi: 10.1016/j.actatropica.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 46.Kužnik A., Benčina M., Švajger U., Jeras M., Rozman B., Jerala R. Mechanism of endosomal TLR inhibition by antimalarial drugs and imidazoquinolines. J Immunol. 2011;186:4794–4804. doi: 10.4049/jimmunol.1000702. [DOI] [PubMed] [Google Scholar]
- 47.Figueroa-Lozano S., Valk-Weeber R.L., Akkerman R., Abdulahad W., van Leeuwen S.S., Dijkhuizen L., de Vos P. Inhibitory effects of dietary N-glycans from bovine lactoferrin on Toll-Like receptor 8; comparing efficacy with chloroquine. Front Immunol. 2020;11:790. doi: 10.3389/fimmu.2020.00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gowda D.C., Wu X. Parasite recognition and signaling mechanisms in innate immune responses to malaria. Front Immunol. 2018;9:3006. doi: 10.3389/fimmu.2018.03006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coban C., Ishii K.J., Kawai T., Hemmi H., Sato S., Uematsu S., Yamamoto M., Takeuchi O., Itagaki S., Kumar N. Toll-like receptor 9 mediates innate immune activation by the malaria pigment hemozoin. J Exp Med. 2005;201:19–25. doi: 10.1084/jem.20041836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coban C., Igari Y., Yagi M., Reimer T., Koyama S., Aoshi T., Ohata K., Tsukui T., Takeshita F., Sakurai K. Immunogenicity of whole-parasite vaccines against Plasmodium falciparum involves malarial hemozoin and host TLR9. Cell Host Microbe. 2010;7:50–61. doi: 10.1016/j.chom.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 51•.Coch C., Hommertgen B., Zillinger T., Daßler-Plenker J., Putschli B., Nastaly M., Kümmerer B.M., Scheunemann J.F., Schumak B., Specht S. Human TLR8 senses RNA from Plasmodium falciparum-infected red blood cells which is uniquely required for the IFN-γ response in NK cells. Front Immunol. 2019;10:371. doi: 10.3389/fimmu.2019.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows Plasmodium-derived natural hemozoin interacts with TLR9 in DCs and activates cytokines that are blocked by chloroquine treatment.
- 52.Lee M.S.J., Maruyama K., Fujita Y., Konishi A., Lelliott P.M., Itagaki S., Horii T., Lin J.-W., Khan S.M., Kuroda E. Plasmodium products persist in the bone marrow and promote chronic bone loss. Sci Immunol. 2017;2:eaam8093. doi: 10.1126/sciimmunol.aam8093. [DOI] [PubMed] [Google Scholar]
- 53.An J., Woodward J.J., Sasaki T., Minie M., Elkon K.B. Cutting edge: antimalarial drugs inhibit IFN-β production through blockade of cyclic GMP-AMP synthase–DNA interaction. J Immunol. 2015;194:4089–4093. doi: 10.4049/jimmunol.1402793. [DOI] [PubMed] [Google Scholar]
- 54•.Gonugunta V.K., Sakai T., Pokatayev V., Yang K., Wu J., Dobbs N., Yan N. Trafficking-mediated STING degradation requires sorting to acidified endolysosomes and can be targeted to enhance anti-tumor response. Cell Rep. 2017;21:3234–3242. doi: 10.1016/j.celrep.2017.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows cytosolic STING needs to be degraded via acidified endolysosomes.
- 55•.Sisquella X., Ofir-Birin Y., Pimentel M.A., Cheng L., Abou Karam P., Sampaio N.G., Penington J.S., Connolly D., Giladi T., Scicluna B.J. Malaria parasite DNA-harbouring vesicles activate cytosolic immune sensors. Nat Commun. 2017;8:1985. doi: 10.1038/s41467-017-02083-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows Plasmodium DNA containing EVs activate cytosolic STING sensor, which maybe degraded in endolysosomes.
- 56.Babatunde K.A., Yesodha Subramanian B., Ahouidi A.D., Martinez Murillo P., Walch M., Mantel P.Y. Role of extracellular vesicles in cellular cross talk in malaria. Front Immunol. 2020;11:22. doi: 10.3389/fimmu.2020.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heath N., Osteikoetxea X., De Oliveria T.M., Lázaro-Ibáñez E., Shatnyeva O., Schindler C., Tigue N., Mayr L.M., Dekker N., Overman R. Endosomal escape enhancing compounds facilitate functional delivery of extracellular vesicle cargo. Nanomedicine. 2019;14:2799–2814. doi: 10.2217/nnm-2019-0061. [DOI] [PubMed] [Google Scholar]
- 58.Ortega F.G., Roefs M.T., de Miguel Perez D., Kooijmans S.A., de Jong O.G., Sluijter J.P., Schiffelers R.M., Vader P. Interfering with endolysosomal trafficking enhances release of bioactive exosomes. Nanomed Nanotechnol Biol Med. 2019;20:102014. doi: 10.1016/j.nano.2019.102014. [DOI] [PubMed] [Google Scholar]
- 59.Liehl P., Zuzarte-Luís V., Chan J., Zillinger T., Baptista F., Carapau D., Konert M., Hanson K.K., Carret C., Lassnig C. Host-cell sensors for Plasmodium activate innate immunity against liver-stage infection. Nat Med. 2014;20:47–53. doi: 10.1038/nm.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang J., Yang X., Yang J., Li M. Hydroxychloroquine inhibits the differentiation of Th17 cells in systemic lupus erythematosus. J Rheumatol. 2018;45:818–826. doi: 10.3899/jrheum.170737. [DOI] [PubMed] [Google Scholar]
- 61.Schmidt R.L.J., Jutz S., Goldhahn K., Witzeneder N., Gerner M.C., Trapin D., Greiner G., Hoermann G., Steiner G., Pickl W.F. Chloroquine inhibits human CD4+ T-cell activation by AP-1 signaling modulation. Sci Rep. 2017;7:42191. doi: 10.1038/srep42191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Austin Taylor M., Bennett M., Kumar V., Schatzle J.D. Functional defects of NK cells treated with chloroquine mimic the lytic defects observed in perforin-deficient mice. J Immunol. 2000;165:5048–5053. doi: 10.4049/jimmunol.165.9.5048. [DOI] [PubMed] [Google Scholar]
- 63.Accapezzato D., Visco V., Francavilla V., Molette C., Donato T., Paroli M., Mondelli M.U., Doria M., Torrisi M.R., Barnaba V. Chloroquine enhances human CD8+ T cell responses against soluble antigens in vivo. J Exp Med. 2005;202:817–828. doi: 10.1084/jem.20051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Joseph H., Eriksson E., Schofield L. Early suppression of B cell immune responses by low doses of chloroquine and pyrimethamine: implications for studying immunity in malaria. Parasitol Res. 2019;118:1987–1992. doi: 10.1007/s00436-019-06335-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fu Y., Lu X., Zhu F., Zhao Y., Ding Y., Ye L., Guo B., Liu T., Xu W. Improving the immunogenicity and protective efficacy of a whole-killed malaria blood-stage vaccine by chloroquine. Parasite Immunol. 2020;42:e12682. doi: 10.1111/pim.12682. [DOI] [PubMed] [Google Scholar]
- 66.Lee M.S.J., Natsume-Kitatani Y., Temizoz B., Fujita Y., Konishi A., Matsuda K., Igari Y., Tsukui T., Kobiyama K., Kuroda E. B cell-intrinsic MyD88 signaling controls IFN-γ-mediated early IgG2c class switching in mice in response to a particulate adjuvant. Eur J Immunol. 2019;49:1433–1440. doi: 10.1002/eji.201848084. [DOI] [PubMed] [Google Scholar]