Abstract
In spring 2013, a novel avian‐origin influenza A (H7N9) virus emerged in mainland China. The burden of H7N9 infection was estimated based on systematic review and meta‐analysis. The systematic search for available literature was conducted using Chinese and English databases. We calculated the pooled seroprevalence of H7N9 infection and its 95% confidence interval by using Freeman‐Tukey double arcsine transformation. Out of 16 890 records found using Chinese and English databases, 54 articles were included in the meta‐analysis. These included studies of a total of 64 107 individuals. The pooled seroprevalence of H7N9 infection among humans was 0.122% (95% CI: 0.023, 0.275). In high‐risk populations, the highest pooled seroprevalence was observed among close contacts (1.075%, 95% CI: 0.000, 4.357). The seroprevalence among general population was (0.077%, 95% CI: 0.011, 0.180). Our study discovered that asymptomatic infection of H7N9 virus did occur, even if the seroprevalence among humans was low.
Keywords: H7N9, influenza A, meta‐analysis, seroprevalence, systematic review
1. INTRODUCTION
In February and March 2013, a novel avian‐origin influenza A (H7N9) virus was identified, which caused more than 100 human cases in mainland China.1, 2 Up to September 5, 2018, a total of 1567 H7N9 human cases were reported, including more than 615 mortalities.3 The case fatality rate of H7N9 patients was close to 40%.3, 4 In March 2019, the cases of H7N9 infection re‐emerged in China after a period of 14 months.5 The majority of the H7N9 patients lived in mainland China. Hong Kong, Taiwan, Malaysia, and Canada also reported human cases of sporadic H7N9 infection, which were imported from mainland China.4
The patients with H7N9 infection who presented with severe clinical symptoms and showed a high case fatality rate have attracted global attention. The proportion of the population with mild or asymptomatic infection, that is, the iceberg below the sea level, was also worthy of attention. A number of serologic studies have been conducted on the seroprevalence of H7N9 infection among humans. However, it is difficult to estimate the extent of infection owing to different variables, such as study populations, test methods, and positive cutoff values in various studies. The purpose of our meta‐analysis was to examine the seroprevalence of influenza A (H7N9) infection among humans and to estimate the overall burden of H7N9 infection.
2. METHOD
2.1. Search strategy
A systematic search of the relevant literature was conducted for relevant articles published before October 22, 2018. The search terms were as follows: [“H7N9” OR “influenza A” OR “influenza A virus”] AND [“seroprevalence” OR “seropositive” OR “seronegative” OR “serologic” OR “serological” OR “seroepidemiology”]. Both Chinese and English databases were searched. Chinese databases consisted of the China National Knowledge Infrastructure, Chinese Science and Technology Periodical Database (VIP), and WanFang Database. English databases consisted of PubMed, Web of Science, and the Cochrane Library. Additionally, we searched the World Health Organization (WHO)'s website, regional health department's website, and reference lists of selected studies.
2.2. Inclusion and exclusion criteria
The inclusion criteria were as follows: (a) studies reporting the seroprevalence of H7N9 infection among humans and (b) cross‐sectional, retrospective, and cohort studies or routine surveillance.
The exclusion criteria were as follows: (a) study only examined the H7 subtype, (b) non‐H7N9 virus strains used in experiments, (c) study subjects were H7N9 patients or influenza patients, (d) sample size was too small (N < 10), (e) duplicated data, (f) study did not provide key data, that is, one‐third of the data (total number, the number of seropositive cases, and seroprevalence) were missing, and (g) conference papers.
Our aim was to summarize the antibody level against the H7N9 subtype. We excluded the serological studies that only examined H7 sole subtype or used non‐H7N9 virus strains in the test for the following reasons. First, the results of the tests only for the H7 subtype could only indicate previous infection with the H7 subtype, such as H7N1. Second, the hemagglutinin gene fragment of the influenza virus is constantly mutating. The H7 antigen of other subtypes is different from the H7N9 antigen reported in China because the H7 antigen can change.6, 7 Therefore, the results of the detection of other subtypes of H7 were not convincing. Although previous studies have shown cross‐reactivities between H7N9 and divergent H7 subtypic viruses,8, 9 the specific avian influenza A(H7N9) virus should be used in hemagglutinin inhibition (HI) or microneutralization (MN) assays.10
2.3. Quality assessment and data abstraction
Two researchers (QW and KX) independently reviewed and assessed each included article according to the following 10 criteria11: (a) whether it was a population‐based study, (b) whether the study time and location were provided, (c) whether the study population was ≥100 subjects, (d) whether the study population had avian exposure, (e) whether the characteristics of the study population were mentioned, (f) whether HI was carried out, (g) whether MN was carried out, (h) whether horse red blood cells were used in the HI assay, (i) whether the seropositive cutoff value was mentioned in the study, and (j) whether the seropositive cutoff value provided in the study referred to the WHO criteria. If the two researchers were in disagreement about the quality of a study, a third researcher (HJ) would make the final decision. “Yes” indicated a score of one, and “No” or “Not provided” indicated a score of zero; finally, we calculated the total score of the 10 items.
Similarly, data abstraction was carried out by two researchers (QW and KX). After extraction, data were checked by a third researcher (HJ). If there was a difference, the original literature would be reviewed for re‐extraction. The following data were extracted: first author, publication year, study type, population sample, study region, fieldwork dates, sample size, number of seropositive cases, seroprevalence, test method, seropositive cutoff value, HI test cell, and number of humans in each dilution titer (1:10‐1:640). For cohort studies, the number of people who showed seroconversion, the criteria of seroconversion, and follow‐up time was also extracted.
2.4. Data analysis
Excel and Stata software were used in this study. The data were subjected to Freeman‐Tukey double arcsine transformation, and we reported the pooled seroprevalence and its 95% confidence interval (CI) using the DerSimonian‐Laird random effects.12, 13 Analyses were conducted using the metaprop package in Stata software.14 We assessed the heterogeneity between the studies with the I 2 statistic. If the heterogeneity test result was I 2 < 50%, a fixed effect model was used; otherwise, a random effect model was used.
The WHO has suggested the criteria for confirming whether the results of H7N9 serologic tests are positive: for single‐serum samples, HI ≥ 1:160; for paired serum samples (acute and convalescent sera), a 4‐fold rise in HI titer.15 In single‐serum samples, sera with HI titer of 20‐80 should be confirmed by MN or WB assay.15 In addition to pooling seroprevalence according to the original study criteria, we re‐judged the seropositive results according to the WHO criteria to explore the influence of different thresholds on pooled seroprevalence.11 Based on the studies that reported the number of humans in each titer, we re‐judged the seropositive results in the included studies. The statistical significance of H7N9 seroprevalences that were calculated by the WHO criteria and original study criteria was assessed using the Wilcoxon rank sum test. We performed statistical tests for the included studies that provided number of humans in each titer to ensure comparability. For cohort studies, the incidence of seroconversion was analyzed after calculating data using the same standard unit (per person‐months), defined as follows: number of seroconverted humans in the cohorts divided by the number of person‐months of follow‐up. We further performed stratified subgroup and meta‐regression analyses.
3. RESULTS
3.1. Search results
A total of 16 890 records were obtained from Chinese and English databases according to the search terms mentioned before, of which 71 articles were reviewed in full‐text (Figure 1). Further, 17 articles were excluded on the basis of the exclusion criteria: 5 studies only examined H7 sole subtype, 1 study used other H7 subtype in the test, 1 study involved H7N9 patients who survived, 6 studies did not provide the data, 3 studies provided replicated data, and 1 study was a conference study. Finally, 54 studies were included in the analysis, consisting of a total of 64 107 individuals.
Figure 1.
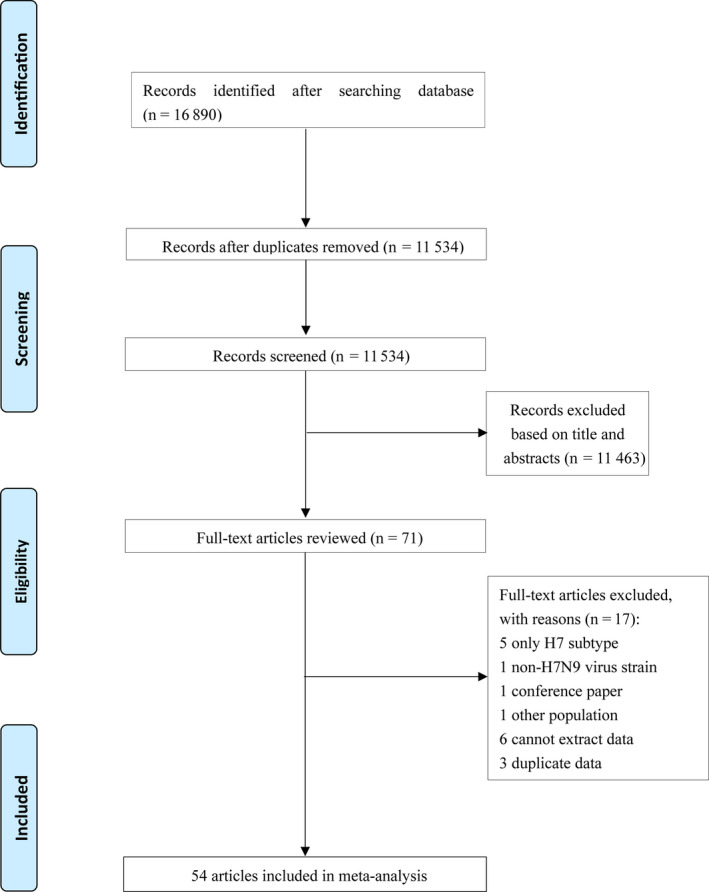
Flowchart of the literature search and study selection
3.2. Study characteristics and quality assessment
The 54 included studies were conducted in different regions and with different populations (Table S1). One study was from Taiwan, one from Hong Kong, and the rest were from mainland China. One study conducted in India was excluded because a non‐H7N9 virus strain was used in the experiments,16 and one study in Vietnam was excluded because it only examined the H7 subtype.17 There were 43 articles that involved poultry workers, 5 articles that involved swine workers, 8 articles that involved close contacts, and 14 articles that involved the general population. Some articles provided data on various study populations.
Of the 54 studies, 3 studies reported the seroprevalence before 2013, 13 studies reported it during the first epidemic wave (1/2013‐9/2013), 14 studies during the second epidemic wave (10/2013‐9/2014), and 4 studies during the third epidemic wave (10/2014‐9/2015). The other studies did not provide the study period or provided ambiguous dates that were difficult to classify; 23 studies followed the WHO criteria, 21 studies did not, and others did not specify the criteria. The scores of quality assessment ranged from 3 to 10 points, with an average of 7.2 ± 1.8 (Table S2).
3.3. Seroprevalence of influenza A (H7N9)
The pooled seroprevalence of H7N9 infection among humans was 0.122% (95% CI: 0.023, 0.275). The seroprevalence reported in the included studies ranged from 0.000% to 17.143%. In the 37 included studies that reported the number of people in each titer, the pooled seroprevalence was 0.046% (95% CI: 0.000, 0.193) according to the original study criteria, but was 0.003% (95% CI: 0.000, 0.081) according to the WHO criteria. However, the difference was not statistically significant (Z = −1.334, P = .182). Of the 37 articles, 14 were not in accordance with the WHO criteria. The pooled incidence of seroconversion in our study was 0.087% (95% CI: 0.007, 0.223) per person‐months.
The seroprevalence of the H7N9 virus varied widely in different regions (Table 1, Figure 2). We reported the seroprevalence in different regions of mainland China, including the eastern, central, and western regions.18 The seroprevalence in the eastern region was higher than that in the other two regions.
Table 1.
Seroprevalence in different groups a
| Variable | Ref (n) | n | Event | Seroprevalence (%) | 95% CI (%) | I 2 (%) | |
|---|---|---|---|---|---|---|---|
| Total | 54 | 64 107 | 410 | 0.122 | 0.023, 0.275 | 88.6 | |
| Different populations | Poultry workers | 43 | 27 383 | 317 | 0.254 | 0.041, 0.584 | 90.2 |
| Swine workers | 5 | 8596 | 5 | 0.005 | 0.000, 0.064 | 19.6 | |
| Close contacts | 8 | 793 | 21 | 1.075 | 0.000, 4.357 | 74.7 | |
| General population | 14 | 25 620 | 50 | 0.077 | 0.011, 0.180 | 71.1 | |
| Time | Before 2013 | 3 | 3089 | 0 | 0.000 | 0.000, 0.053 | 0.0 |
| First epidemic wave | 13 | 10 166 | 95 | 0.109 | 0.000, 0.670 | 90.3 | |
| Second epidemic wave | 14 | 22 550 | 166 | 0.441 | 0.101, 0.942 | 93.1 | |
| Third epidemic wave | 4 | 6005 | 4 | 0.000 | 0.000, 0.000 | 53.6 | |
| Region in mainland China | Eastern | 34 | 52 458 | 389 | 0.129 | 0.009, 0.346 | 92.0 |
| Central | 9 | 6819 | 8 | 0.000 | 0.000, 0.000 | 8.6 | |
| Western | 12 | 4514 | 0 | 0.000 | 0.000, 0.006 | 0.0 | |
| Seropositive value | HI titer ≥ 1:20 | 6 | 4942 | 13 | 0.007 | 0.000, 0.226 | 66.2 |
| HI titer ≥ 1:40 | 5 | 9200 | 93 | 1.056 | 0.247, 2.348 | 95.5 | |
| HI titer ≥ 1:80 | 7 | 18 698 | 94 | 0.147 | 0.001, 0.440 | 90.1 | |
| HI titer ≥ 1:160 | 18 | 7971 | 146 | 0.500 | 0.003, 1.501 | 92.8 | |
| HI titer ≥ 1:20 and MN titer ≥ 1:20 | 4 | 6975 | 13 | 0.000 | 0.000, 0.033 | 40.1 | |
| Test HI cell | Turkey red blood cell | 7 | 4910 | 33 | 0.013 | 0.000, 0.506 | 82.6 |
| Horse red blood cell | 36 | 47 961 | 303 | 0.158 | 0.026, 0.364 | 90.1 | |
| Chicken red blood cell | 4 | 3781 | 10 | 0.000 | 0.000, 0.229 | 66.5 | |
| Test method | HI | 40 | 39 263 | 329 | 0.203 | 0.026, 0.490 | 91.6 |
| HI an MN | 11 | 23 114 | 81 | 0.078 | 0.000, 0.245 | 79.3 | |
| Sample size | <500 | 46 | 18 659 | 212 | 0.169 | 0.005, 0.484 | 86.1 |
| ≥500 | 19 | 45 317 | 198 | 0.213 | 0.078, 0.398 | 91.2 |
HI: Hemagglutination inhibition test; MN: Microneutralization test.
Figure 2.
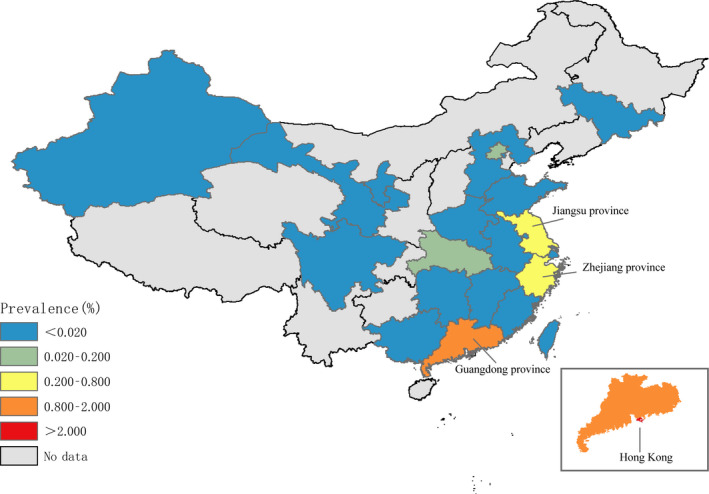
Seroprevalence of H7N9 virus among humans
The seroprevalence among close contacts was 1.075% (95% CI: 0.000, 4.357), ranging from 0.000% to 14.286%. The seroprevalence among close contacts was the highest in Jiangsu province (Table S3). The seroprevalence rates among poultry and swine workers were 0.254% (95% CI: 0.041, 0.584) and 0.005% (95% CI: 0.000, 0.064), respectively. The seroprevalence among poultry workers was the highest in Hong Kong and the lowest in some central and western provinces in mainland China. The seroprevalence among humans before 2013 was 0.000%. The seroprevalence was higher in the first two epidemic waves than in the third one.
3.4. Meta‐analysis regression
The results of univariate analysis showed that time and population significantly affected the heterogeneity of the meta‐analysis results (Table 2). We added region to the multivariate analysis because its adjusted R 2 was 2.55% in the univariate analysis. The results showed that the variables included in the regression were time and population, and the adjusted R 2 was 15.89%, which suggested that time and population can explain part of the heterogeneity.
Table 2.
The results of meta‐regression a
| Covariate | Coefficient | 95% CI | t | P | Adjusted R 2 (%) |
|---|---|---|---|---|---|
| Univariate analysis | |||||
| Time | |||||
| Other time | ‐ | ‐ | ‐ | ‐ | 4.25 |
| First and second epidemic wave | 0.068 | 0.009, 1.126 | 2.27 | .025 | |
| Region in mainland China | |||||
| Western | ‐ | ‐ | ‐ | ‐ | 2.55 |
| Eastern | 0.084 | −0.017, 0.185 | 1.65 | .102 | |
| Central | 0.017 | −0.095, 0.129 | 0.30 | .763 | |
| Population | |||||
| Swine worker | ‐ | ‐ | ‐ | ‐ | 4.24 |
| Close contacts | 0.220 | 0.054, 0.385 | 2.63 | .010 | |
| Poultry workers | 0.103 | 0.006, 0.200 | 2.09 | .038 | |
| General population | 0.024 | −0.082, 0.130 | 0.44 | .657 | |
| Test method | |||||
| HI and MN | ‐ | ‐ | ‐ | ‐ | −0.15 |
| HI | 0.039 | −0.026, 0.103 | 1.19 | .237 | |
| Test HI cell | |||||
| Chicken red blood cell | ‐ | ‐ | ‐ | ‐ | −2.24 |
| Turkey red blood cell | 0.011 | −0.148, 0.169 | 0.13 | .895 | |
| Horse red blood cell | 0.016 | −0.112, 0.144 | 0.25 | .805 | |
| Sample size | |||||
| n ≥ 500 | ‐ | ‐ | ‐ | ‐ | 0.06 |
| n < 500 | 0.039 | −0.020, 0.098 | 1.30 | .197 | |
| Multivariate analysis | 15.89 | ||||
| Time | |||||
| Other time | ‐ | ‐ | ‐ | ‐ | ‐ |
| First and second epidemic wave | 0.062 | 0.005, 0.119 | 2.17 | .032 | |
| Region in mainland China | |||||
| Western | ‐ | ‐ | ‐ | ‐ | ‐ |
| Eastern | 0.099 | −0.002, 0.201 | 1.94 | .055 | |
| Central | 0.022 | −0.088, 0.133 | 0.40 | .693 | |
| Population | |||||
| Swine worker | ‐ | ‐ | ‐ | ‐ | ‐ |
| Close contacts | 0.152 | −0.027, 0.332 | 1.68 | .095 | |
| Poultry workers | 0.151 | 0.057, 0.245 | 3.18 | .002 | |
| General population | 0.050 | −0.050, 0.150 | 0.98 | .327 | |
“‐”: the first line of every covariate represented reference; adjusted R 2 was used to indicate the degree of heterogeneity explained by study characteristics.
4. DISCUSSION
We performed this systematic review and meta‐analysis of the serological studies on influenza A (H7N9) to estimate the burden of this virus among humans. Strikingly, mild or asymptomatic human infection with H7N9 did exist, even if the proportion was small. Similar to influenza A (H5N1 and H9N2), the reported human cases were only a tip of the iceberg of a large number of infections.11, 19
High seroprevalence among poultry workers suggests that avian exposure is a risk factor for infection. A previous study on H5N1 and H9N2 infections suggested that age and chronic lung problems were consistent with elevated titers.20 Some case‐control studies on clinical H7N9 patients also found that patients with chronic obstructive lung disease (COPD) and those receiving immunosuppressive medications were highly susceptible to be infected with influenza virus infection.21, 22 Hence, avian exposure, chronic lung problems, and poor immune status may be associated with influenza A (H7N9) infection, in both clinical H7N9 cases and silent infections.
Poultry workers with intense avian exposure tend to have high risk of being infected with the influenza virus.20 However, the data were consistent with the same phenomenon seen with H5N1, where the virus was so poorly adapted to humans that most hosts could not be productively infected, leading to high exposure to the virus but low seroprevalence.23 Previous studies have provided evidence that the host gene plays an important role in susceptibility to infection and clinical outcomes.24, 25 The severe influenza cases are the result of rare genetic susceptibilities, attributable to the interferon‐induced transmembrane protein 3 (IFITM3) or other risk factors.23, 26, 27, 28 The possible association between glycine decarboxylase (GLDC), IFITM3, and toll‐like receptors 3 (TLR3) and the outcomes of influenza A (H7N9) infection was also examined by a few researchers.29, 30 Controversially, an epidemiological study analyzing clusters of H7N9 patients found that genetic susceptibility to H7N9 virus infection was limited.31 The association between susceptibility‐conferring genes and influenza A (H7N9) virus infection warrants further in‐depth studies.
The risk of infection among humans was high as per the pooled seroprevalence of close contacts. Close contacts were defined as healthcare workers who had not taken effective protective measures and family members who took care of patients during the treatment of suspected or confirmed infection; the staff who lived with the patients or had experienced other close contact situations from one day before the suspicion or confirmation of infection were placed in isolation.32 Articles providing information about close contacts were reviewed for further evidence. Of the 8 studies that reported the data, the seroprevalence in 5 studies was 0.000%; in the remaining 3 studies, it was 3.200%, 6.667%, and 14.286%. In the three studies that reported seropositivity, only one study reported that none were exposed to poultry, swine, or other animals.33 The seropositive close contacts in the study included healthcare workers and family members.33 Previous studies suggested that the transmissibility of H7N9 virus among persons cannot be ignored, even if it was limited.34, 35, 36 The close contacts of H7N9 patients require protective measures and more close attention.
Swine, the intermediate hosts that facilitate the reassortment of influenza viruses, were likely to cause infection in humans.37, 38 The animal could provide a suitable vessel for the influenza A (H7N9) virus to survive and evolve.39 Controversially, the seroprevalence of H7N9 among swine workers was low in our pooled results. On one hand, this might be associated with swine infected with H7N9. The previous serological reports did not provide evidence of swine involvement in H7N9 virus ecology.40, 41, 42 One study had shown that the H7N9 virus might not efficiently infect swine.43 On the other hand, the virus transmission ability from pigs to other mammals might be limited.44 The reasons mentioned above may explain the low seroprevalence among swine workers. More studies need to explore the difference between the adaption of H7N9 and other influenza A subtypes to swine.
With regard to time, cases of subclinical H7N9 infection were not reported before 2013. Compared with the first epidemic wave, mild or asymptomatic infection showed a downward trend during the third epidemic wave. The small number of studies on seroprevalence during this wave might have led to this finding. Besides, studies showed that antibody titers in people waned over time.45, 46 China experienced six waves of H7N9 epidemics.3 The highest number of humans cases was reported in the fifth epidemic wave.47 However, only three laboratory‐confirmed human cases were reported in the recent wave.3 The situation of asymptomatic infection with the H7N9 virus is unclear. Studies on the seroprevalence of the H7N9 virus among humans need to be performed, especially considering the sixth wave when cases of H7N9 infection were rarely reported.
Of note, there were some limitations to our study. First, cross‐sectional studies accounted for most of the included studies. The proportion rather than incidence was provided in these studies. The serologic results of a cross‐sectional study can be misleading because antibodies may wane over time. The risk factors for infection could not be explored exhaustively because the included studies provided inadequate information about the demographic, health, and exposure variables. Second, the different seropositive thresholds might have led to the overestimation of the seroprevalence among humans. Third, cross‐reactive immunity cannot be ignored despite excluding those articles. Fourth, the result of the meta‐regression did not explain the source of heterogeneity. Probably, the proportion was too small or even zero in most of the included studies.
5. CONCLUSION
Our study found that subclinical infection with the H7N9 virus did occur, even if the seroprevalence among humans was low. Be it the high‐risk group or general population, a certain degree of infection did exist. Stringent seropositive standards should be developed and observed to ensure that serology assays are reliable and convincing. Sensitive detection tests for the influenza A (H7N9) virus are need to be carried out to provide warnings before the evolvement and adaptation of the virus to the human body.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
AUTHOR CONTRIBUTIONS
QW and HJ designed the study. QW, KX, LQY, and WHX conducted the literature search and review. QW and KX reviewed citations and extracted data. QW, NYS, HYC, HDH, CJB, XFZ, and YLL analyzed the data. HJ and QW interpreted the results. All authors critically revised for important intellectual content. All authors approved the final version. Qiang Wang: Conceptualization (equal); data curation (lead); formal analysis (lead); funding acquisition (equal); methodology (equal); writing‐review & editing (lead). Ke Xu: Data curation (equal); formal analysis (equal); methodology (equal). Weihua Xie: Data curation (equal); methodology (equal). Liuqing Yang: Data curation (equal); methodology (equal). Haiyan Chen: Methodology‐Supporting. Naiyang Shi: Data curation (equal); methodology (equal). Changjun Bao: Formal analysis (equal); supervision (equal). Haodi Huang: Formal analysis (equal). Xuefeng Zhang: Formal analysis (equal); supervision (equal). Yilan Liao: Formal analysis (equal). Hui Jin: Conceptualization (equal); methodology (equal); supervision (lead); writing‐review & editing (equal).
Supporting information
Table S1
Table S2
Table S3
ACKNOWLEDGEMENTS
We thank all the editors and reviewers for their work and suggestions.
Wang Q, Xu K, Xie W, et al. Seroprevalence of H7N9 infection among humans: A systematic review and meta‐analysis. Influenza Other Respi Viruses. 2020;14:587–595. 10.1111/irv.12736
The peer review history for this article is available at https://publons.com/publon/10.1111/irv.12736
Funding information
This work was supported by Chinese National Natural Fund (Grant number: 81573258); Jiangsu Provincial Major Science & Technology Demostation Project (Grant numbers: BE2015714, and BE2017749); Jiangsu Provincial Six Talent Peak (Grant number: WSN‐002); Jiangsu Provincial Key Medical Discipline (Grant number: ZDXKA2016008).
REFERENCES
- 1. Gao RB, Cao B, Hu YW, et al. Human infection with a novel avian‐origin influenza A (H7N9) virus. N Engl J Med. 2013;368(20):1888‐1897. [DOI] [PubMed] [Google Scholar]
- 2. Li Q, Zhou L, Zhou MH, et al. Epidemiology of human infections with avian influenza A(H7N9) virus in China. N Engl J Med. 2014;370:520‐532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. World Health Organization . Human Infection with Avian Influenza A (H7N9) Virus – China: Update. https://www.who.int/csr/don/05‐september‐2018‐ah7n9‐china/en/. Accessed December 30, 2018. [Google Scholar]
- 4. Xiang NJ, Bai T, Kang K, et al. Sero‐epidemiologic study of influenza A (H7N9) infection among exposed populations, China 2013–2014. Influenza Other Respir Viruses. 2017;11(2):170‐176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yu DS, Xiang GF, Zhu WF, et al. The re‐emergence of highly pathogenic avian influenza H7N9 viruses in humans in mainland China, 2019. Eurosurveillance. 2019;24(21):13‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huang K‐YA, Rijal P, Jiang H, et al. Structure–function analysis of neutralizing antibodies to H7N9 influenza from naturally infected humans. Nat Microbiol. 2019;4(2):306‐315. [DOI] [PubMed] [Google Scholar]
- 7. Liu M, Song T, Hua S, Wu A, Jiang T. Computational analysis of antigenic epitopes of avian influenza A (H7N9) viruses. Sci Chin Life Sci. 2015;58(7):687‐693. [DOI] [PubMed] [Google Scholar]
- 8. Guo L, Wang D, Zhou H, et al. Cross‐reactivity between avian influenza A (H7N9) virus and divergent H7 subtypic‐ and heterosubtypic influenza A viruses. Sci Rep. 2016;6:22045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rudenko L, Isakova‐Sivak I, Donina S. H7N3 live attenuated influenza vaccine has a potential to protect against new H7N9 avian influenza virus. Vaccine. 2013;31(42):4702‐4705. [DOI] [PubMed] [Google Scholar]
- 10. OIE/FAO . Network of Expertise on Animal Influenza. Technical Brief: Implications for Pigs; 2013. http://www.offlu.net/index.php?id=302. Accessed April 19, 2013. [Google Scholar]
- 11. Khan SU, Anderson BD, Heil GL, Liang S, Gray GC. A systematic review and meta‐analysis of the seroprevalence of influenza A (H9N2) infection among humans. J Infect Dis. 2015;212(4):562‐569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Barendregt JJ, Doi SA, Lee YY, Norman RE, Vos T. Meta‐analysis of prevalence. J Epidemiol Commun Health. 2013;67(11): 974‐978. [DOI] [PubMed] [Google Scholar]
- 13. Freeman MF, Tukey JW. Transformations related to the angular and the square root. Ann Math Stat. 1950;21:607‐611. [Google Scholar]
- 14. Nyaga VN, Arbyn M, Aerts M. Metaprop: a Stata command to perform meta‐analysis of binomial data. Arch Public Health. 2014;72(1):39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. World Health Organization . Serological Detection of Avian Influenza A (H7N9) Infections by Horse Red Blood Cells Haemagglutination‐Inhibition Assay; 2013. http://www.who.int/influenza/gisrs_laboratory/cnic_serological_diagnosis_hai_a_h7n9_20131220.pdf. Accessed December 30, 2018. [Google Scholar]
- 16. Pawar SD, Tandale BV, Gurav YK, Parkhi YK, Kode SS. Immunity status against influenza A subtype H7N9 and other avian influenza viruses in a high‐risk group and the general population in India. J Infect Dis. 2014;210(1):160‐161. [DOI] [PubMed] [Google Scholar]
- 17. Boni MF, Chau NV, Dong N, et al. Population‐level antibody estimates to novel influenza A/H7N9. J Infect Dis. 2013;208(4):554‐558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. National Bureau of Statistics of China . Method of Dividing the East, West, Middle and Northeast Regions in China. http://www.stats.gov.cn/ztjc/zthd/sjtjr/dejtjkfr/tjkp/201106/t20110613_71947.htm. Accessed December 30, 2018. [Google Scholar]
- 19. Wang TT, Parides MK, Palese P. Seroevidence for H5N1 influenza infections in humans: meta‐analysis. Science. 2012;335:1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gomaa MR, Kayed AS, Elabd MA, et al. Avian influenza A(H5N1) and A(H9N2) seroprevalence and risk factors for infection among egyptians: a prospective, controlled seroepidemiological study. J Infect Dis. 2015;211(9):1399‐1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou L, Ren R, Ou J, et al. Risk factors for influenza A(H7N9) disease in China, a matched case control study, October 2014 to April 2015. Open Forum Infect Dis. 2016;3(3):ofw182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu B, Havers F, Chen EF, et al. Risk factors for influenza A(H7N9) disease‐China, 2013. Clin Infect Dis. 2014;59(6):787‐794. [DOI] [PubMed] [Google Scholar]
- 23. Morens DM, Taubenberger JK. How low is the risk of influenza A(H5N1) infection? J Infect Dis. 2015;211(9):1364‐1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Albright FS, Orlando P, Pavia AT, Jackson GG, Albright LAC. Evidence for a heritable predisposition to death due to influenza. J Infect Dis. 2008;197(1):18‐24. [DOI] [PubMed] [Google Scholar]
- 25. Telenti A, di Iulio J. Regulatory genome variants in human susceptibility to infection. Hum Genet. 2019. 10.1007/s00439-019-2091-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Everitt AR, Clare S, Pertel T, et al. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484(7395):519‐523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang YH, Zhao Y, Li N, et al. Interferon‐induced transmembrane protein‐3 genetic variant rs12252‐C is associated with severe influenza in Chinese individuals. Nat Commun. 2013;4:1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lin TY, Brass AL. Host genetic determinants of influenza pathogenicity. Curr Opin Virol. 2013;3(5):531‐536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou J, Wang D, Wong BHY, et al. Identification and characterization of GLDC as host susceptibility gene to severe influenza. Embo Mol Med. 2019;11(1):e9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee N, Cao B, Ke CW, et al. IFITM3, TLR3, and CD55 gene SNPs and cumulative genetic risks for severe outcomes in Chinese patients with H7N9/H1N1(pdm09) influenza. J Infect Dis. 2017;216(1):97‐104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang XL, Wu P, Pei Y, et al. Assessment of human‐to‐human transmissibility of avian influenza A(H7N9) virus across 5 waves by analyzing clusters of case patients in mainland China, 2013–2017. Clin Infect Dis. 2019;68(4):623‐631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. China National Health and Family Planning Commission . Chinese Guideline for Prevention and Control for Human Infection with A(H7N9) Avian Influenza (2014 edition), 3rd ed.; 2014. http://www.nhc.gov.cn/jkj/s3578/201401/ca44b7a218274b7d8335b34da7ed23da.shtml. Accessed December 30, 2018 [in Chinese]. [Google Scholar]
- 33. Ma MJ, Ma GY, Yang XX, et al. Avian influenza A (H7N9) virus antibodies in close contacts of infected persons, China, 2013–2014. Emerg Infect Dis. 2015;21(4):709‐711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Qi X, Qian YH, Bao CJ, et al. Probable person to person transmission of novel avian influenza A (H7N9) virus in Eastern China, 2013: epidemiological investigation. BMJ. 2013;347(3):f4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fang CF, Ma MJ, Zhan BD, et al. Nosocomial transmission of avian influenza A (H7N9) virus in China: epidemiological investigation. BMJ. 2015;351:h5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang JJ, Su N, Dong ZF, et al. The fifth influenza A (H7N9) epidemic: a family cluster of infection in Suzhou city of China, 2016. Int J Infect Dis. 2018;74:128‐135. [DOI] [PubMed] [Google Scholar]
- 37. Ma MJ, Wang GL, Anderson BD, et al. Evidence for cross‐species influenza A virus transmission within swine farms, China: a one health, prospective cohort study. Clin Infect Dis. 2018;66(4):533‐540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Smith GJ, Vijaykrishna D, Bahl J, et al. Origins and evolutionary genomics of the 2009 swine‐origin H1N1 influenza A epidemic. Nature. 2009;459(7250):1122‐1125. [DOI] [PubMed] [Google Scholar]
- 39. Xu LL, Bao LL, Deng W, et al. Rapid adaptation of avian H7N9 virus in pigs. Virology. 2014;452‐453:231‐236. [DOI] [PubMed] [Google Scholar]
- 40. Zhou P, Hong ML, Merrill MM, He HM, Sun LS, Zhang GH. Serological report of influenza a (H7N9) infections among pigs in Southern China. BMC Vet Res. 2014;10. Article number: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhao FR, Zhou DH, Lin T, et al. The first lack of evidence of H7N9 avian influenza virus infections among pigs in Eastern China. Microb Pathog. 2015;80:63‐66. [DOI] [PubMed] [Google Scholar]
- 42. Lan DS, Wei S, Hou ZZ. Serological investigation and analysis of swine influenza in Liaoning, 2012–2017. Chin J Zoonoses. 2019;35(1):91‐95. [Google Scholar]
- 43. Powell JD, Abente EJ, Torchetti MK, Killian ML, Vincent AL. An avian influenza virus A(H7N9) reassortant that recently emerged in the United States with low pathogenic phenotype does not efficiently infect swine. Influenza Other Respir Viruses. 2019;13(3):288‐291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhu H, Wang D, Kelvin DJ, et al. Infectivity, transmission, and pathology of human‐isolated H7N9 influenza virus in ferrets and pigs. Science. 2013;341(6142):183‐186. [DOI] [PubMed] [Google Scholar]
- 45. Toner ES, Adalja AA, Nuzzo JB, Inglesby TV, Henderson DA, Burke DS. Assessment of serosurveys for H5N1. Clin Infect Dis. 2013;56(9):1206‐1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Buchy P, Vong S, Chu S, et al. Kinetics of neutralizing antibodies in patients naturally infected by H5N1 virus. PLoS One. 2010;5(5):e10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang XL, Jiang H, Wu P, et al. Epidemiology of avian influenza A H7N9 virus in human beings across five epidemics in mainland China, 2013–17: an epidemiological study of laboratory‐confirmed case series. Lancet Infect Dis. 2017;17(8):822‐832. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Table S2
Table S3


