Abstract
Purpose
Keratoconus (KC) is a bilateral and noninflammatory disease, characterized by progressive thinning and anterior protrusion of the cornea and may result in severe visual impairment due to irregular astigmatism. Matrix metalloproteinases (MMP) are the main group of enzymes that degrade extracellular matrix proteins including collagens; Type IV collagen is found in the corneal stroma. MMP enzymatic activity is inhibited by tissue inhibitor of metalloproteinase-1 (TIMP-1). A decrease in TIMP-1 level is associated with the development of KC. In the present study, we investigated the impact of COL4A4 rs2228557 C/T and TIMP-1 rs4898 C/T (X-chromosome) variants on the odds of KC development in a sample of Iranian population.
Methods
This case–control study was conducted on 140 patients with KC and 150 healthy control subjects. We used modified methods of Nested-PCR and ARMS-PCR in combination (Nested-ARMS-PCR) and confirmed their validity with RFLP–PCR.
Results
Significant differences were noticed between KC patients and healthy individuals regarding the genotype TY or T allele frequencies of rs4898 in the male subjects (OR = 0.43, 95%CI: 0.20–0.92, P = 0.03), whereas no significant differences were identified in the female subjects (OR = 1.07, 95%CI: 0.52–2.20, P = 0.85). The rs2228557, T allele was associated with KC (OR = 0.69, 95% CI: 0.50–0.97, P = 0.035).
Conclusion
In the rs2228557 variant, T allele acts as a protective factor from the disease and decreases the risk of KC compared with the C allele. Also, in our investigation about rs4898, we found that TY genotype or T allele decreased the risk of KC compared with the C allele in males and was a protective factor for KC in our population
Keywords: Collagen, COL4A4, Keratoconus, Polymorphism, TIMP-1
INTRODUCTION
Keratoconus (KC) is defined as a bilateral, non-inflammatory, and progressive disease characterized by conical protrusion of the cornea. This disease may result in severe visual impairment due to irregular astigmatism and stromal scarring. KC eventually affects both eyes, although the involvement is usually asymmetric. The symptoms of KC-affected patients are different depending on the stage of the disease.[1,2,3] Glasses or contact lenses can provide useful vision in the early stage of the disease; nonetheless, corneal transplantation is mandatory for visual rehabilitation in 20% of the patients who are in advanced stage. Corneal thinning is considered as one of the identifying characteristics of KC. Central corneal thickness (CCT) is lower in KC patients by 75 µm as compared to normal controls.[4,5]
The incidence of KC is approximately 1 per 2,000, and its prevalence is 54.5 per 100,000. This disease occurs in both genders,[6] with different rates among different ethnic groups.[7] KC usually begins in teens, and its progression slows after the age of 30 years.[6]
KC is a multi-factorial disorder; environmental factors cause KC in genetically susceptible individuals. The environmental factors that may play roles in the pathogenesis of KC include eye rubbing, allergy, connective tissue dysfunction, and contact lens wear. Moreover, subjects with a family history of KC are more susceptible to this disorder.[8] Using family-based linkage, several case–control studies have determined various genes that increase the odds of KC development.[9] The gene candidates for KC include LOX,[10] VSX1,[11] GPX-1,[12] TGF-1,[13] COL4A3 and COL4A4 [14] (polymorphism or mutation), and TIMP-1,[15] MMP-2, MMP-9 [16] (gene expression). KC is still an enigmatic disease in many aspects, including inheritance, basic pathophysiology, prevention, associated risk factors, disease development, as well as therapeutic approaches.[17]
Type IV collagen is only present in basement membranes and constitutes their main structural component. Collagen type IV gene, alpha-4 (COL4A4), is located in the region 2q35–q37 with a gene span composed of 113 kb and 48 exons.[18,19] Type IV collagen is not expressed in cornea in the normal condition and its presence indicates a corneal pathology; therefore, type IV collagen can be a potential candidate in the pathogenesis of KC. In support of this theory, alterations in the expression level of collagen type IV (α-4 chains) were observed in corneas inflicted by KC.[20,21] Matrix metalloproteinases (MMP) are the major expressed metalloproteases in the cornea. It has been demonstrated that the proteolytic activity of MMP increases in KC. This finding suggests that abnormal MMP activity plays a role in the pathogenesis of KC.[16,22]
Four types of tissue inhibitor of metalloproteinase (TIMP1-4) have been detected; three of which, including TIMPs-1, 3, and 4, are nested within an intron in the genes of synapsins. TIMP inhibits collagenases and proteoglycanase called matrix metalloproteinase. All four types of TIMPs have various biological activities apart from their metalloproteinase inhibitory activity. These biological activities include the promotion of cell proliferation, cancer promotion, regulation of angiogenesis, as well as pro- and anti-apoptotic and synaptic flexibility activities, many of which are independent of metalloprotease inhibition. TIMP-1 is associated with synapsin 1 and its gene is located in X11p11.23–11.4 consisting of six exons. Mature TIMP-1 is a 28.5 kDa glycoprotein that consists of 184 amino acid residues. The natural precursor contains a signal peptide of 23 residues which are cleaved throughout the protein maturation.[23,24,25,26,27] TIMP-1 suppresses angiogenesis and controls the balance in the corneal tissue by inhibiting the action of matrix metalloproteinase to protect tissues from permanent damage.[28] Furthermore, it has been demonstrated that increased MMP and decreased TIMP levels are associated with the development of KC.[29]
COL4A4 gene rs2228557 (F1644F) (HGVM1660028) is located in chromosome 2 exon 48, NM_000092.4 region. Several studies examined the association between COL4A4 and KC and revealed that this gene is associated with KC; however, some other studies failed to find the same relationship in different populations.[14,22,30,31]
TIMP-1 gene rs4898 (+372C/T) (HGVM6380940) is located within the intron of the synapsin gene, and single nucleotide polymorphism (SNP) is located in exon 5, the region of NM_006950.3. Some studies investigated the association of rs4898 gene polymorphism with disorders including intracerebral hemorrhage,[32] systemic sclerosis,[33] and severe sepsis.[34]
The present study aimed to evaluate the possible association of TIMP-1 rs4898 C/T gene polymorphism and COL4A4 rs2228557 C/T gene polymorphism with the development of KC in a sample of Iranian population.
METHODS
Patients
The current retrospective case–control study was conducted at the Alzahra Eye Hospital, Zahedan University of Medical Sciences, Zahedan, Iran and recruited 140 unrelated Iranian patients with KC and 150 unrelated healthy controls. The patients were diagnosed with KC after a comprehensive ophthalmic examination using the following criteria: (1) clinical signs of KC (Munson sign, protrusion, Vogts striae, corneal thickness, scarring, Fleischer ring) and abnormal findings in corneal topography (Pentacam AXL, OCULUS INC); (2) the three quantitative videokeratographic indices used for the screening of KC were central corneal power 47.2 D, inferior–superior value 1.4 D, Sim-K astigmatism 1.5 D, and skewed radial axes 21°.[10,35] Patients with other ocular diseases were excluded from the study. Controls were sex- and age-matched healthy participants who were unrelated to the patients and were selected from a geographic area similar to that of KC subjects.
The Ethics Committee of Zahedan University of Medical Sciences, Zahedan, Iran approved the study protocol and informed consent was signed by all participants. This study complied with the tenets of the Declaration of Helsinki.
Analysis of the TIMP-1 (rs4898), COL4A4 (rs2228557) Polymorphisms
Blood samples were collected in EDTA-containing tubes and genomic DNA was extracted from the peripheral blood leukocytes using salting out method as previously described.[36] All procedures were performed under a standardized setting to avoid variation in DNA quality. SNP rs2228557 COL4A4 was evaluated by ARMS–PCR. For the detection of rs4898 TIMP-1, we used the combination of Nested-polymerase chain reaction (Nested-PCR)[37] and amplification refractory mutation system-PCR(ARMS-PCR)[38] (Nested-ARMS-PCR). The verification of these methods was accomplished using Restriction Fragment Length Polymorphism (RFLP-PCR).[39]
The rs4898 location was very challengeable for ARMS-PCR; therefore, for the detection of the SNP, we used Nested or hemi-Nested-PCR primers, as mentioned previously.[40] The advantages of this modification include elimination of non-specific products, protection of SNP position for the next steps, low cost, and short duration of the process.
PCR reactions were performed using PCR master mix (Ampliqon Taq 2x mastermix, Denmark) according to the manufacturer's instructions. For investigation of COL4A4 (rs2228557), amplification reaction was provided in 20 μL volume including: 1 μL template DNA (100 ng/μL), 1 μL of each primer (10 pmol/μL), 10 μL mastermix, and 7 μL DNase-free water. The PCR conditions were set as follow: 95°C for 5 min, 30 cycles of 95°C for 30 sec, 55°C for 35 sec, 72°C for 30 sec, and a final extension at 72°C for 5 min. PCR products were detected by electrophoresis on a 2% agarose gel staining by ethidium bromide (Figure 1A).
Figure 1.
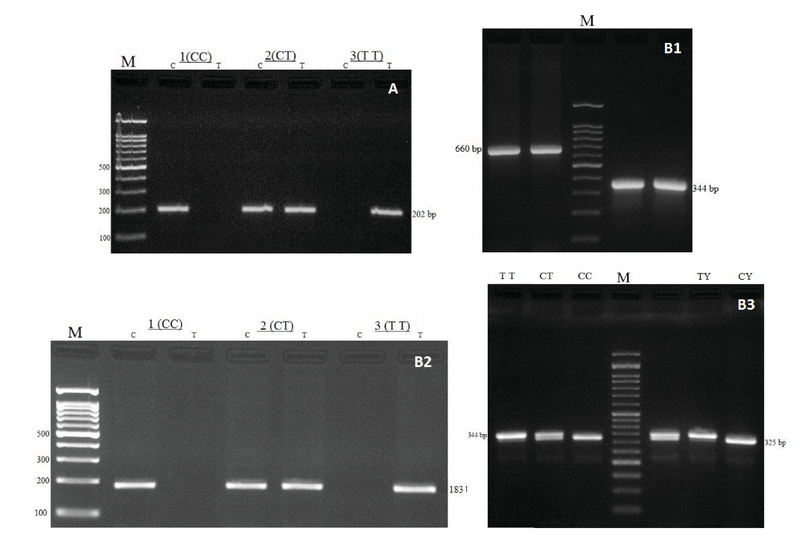
Electrophoresis pattern for the detection of SNPs in COL4A4 rs2228557 and TIMP-1 rs4898. (A) Arms-PCR products of COL4A4 rs2228557, M: DNA marker (100 bp). (B1) TIMP-1 rs4898, Nested-PCR products, M: DNA marker (100bp), product size (660 and 344bp), respectively. (B2) ARMS-PCR products, product size (202bp), (TT, TC, CC) demonstrate genotypes. (B3) RFLP-PCR products, M: DNA marker (50bp), product sizes were 344bp for TT, 344bp and 325bp for TC, 325bp for CC in female, 344bp for TY and 325bp for CY in male, (TT, TC, CC) demonstrate female and (TT = TY, CC = CY) demonstrate male genotypes, (Y stands for the Y-chromosome).
In the first stage of study for TIMP-1 rs4898, Nested-PCR reaction was performed in 20 μL volume including: 1 μL template DNA (100 ng/μL), 1 μL of each primer (10 pmol/μL), 10 μL mastermix, and 7 μL DNase-free water. The PCR conditions were set as follow: 95°C for 5 min, 30 cycles of 95°C for 30 sec, 64°C for 40 sec, 72°C for 30 sec, and a final extension at 72°C for 5 min. In the second phase, the PCR product obtained from the first stage (660 bp) was used as the template and diluted 1:50. Primers for ARMS-PCR were designed to detect the SNP (Table 1). In this step, ARMS-PCR reaction was performed in 20 μL volume including: 1 μL template (1:50 dilution), 1 μL of each primer (10 pmol/μL), 10 μL mastermix, and 7 μL DNase-free water. The PCR conditions were set as follow: 95°C for 5 min, 17 cycles of 95°C for 30 sec, 56°C for 30 sec, 72°C for 30 sec, and a final extension at 72°C for 5 min. PCR products were detected by electrophoresis on a 2% agarose gel staining by ethidium bromide (202bp product). Consequently, SNP (rs4898) TIMP-1 was successfully detected with the combination of two methods (Nested-PCR and ARMS-PCR) (Figures B1 & B2).
Table 1.
The list of primers and methods used for detection of Single Nucleotide Polymorphisms (SNPs) TIMP-1 rs4898 and COL4A4 rs2228557
|
| |||
| TIMP-1(rs4898) T/C | Primers(5'-3') | Product size | |
| Stage 1Nested-PCR | F | TGGGGACACCAGAAGTCAAC | 660 bp |
| R | TAAGCTCAGGCTGTTCCAGG | ||
| Stage 2ARMS-PCR | F Common | AGGCTCTGATGAGAATGGTCCCA | 202 bp |
| R (C allele) | CAGATTGTTCCAGGGAGCCAAG | ||
| R (T allele) | CAGATTGTTCCAGGGAGCCAAA | ||
| Stage 3RFLP-PCR | F | CCGCCATGGAGAGTGTCTGC | 344 bp |
| R* | AGGCTGTTCCAGGGAGTCGC | ||
| COL4A4(rs2228557 ) C/T | |||
| ARMS-PCR | F Common | TGTCTGAGCCCTAATTCTCT | 183 bp |
| R (C allele) | GAGCCAGAAGCTATACTTATTTGAG | ||
| R (T allele) | GAGCCAGAAGCTATACTTATTTGAA | ||
| F, forward; R, reverse; R*, altered reverse | |||
Table 2.
Genotype and allelic frequencies of COL4A4 rs2228557 and TIMP-1 rs4898 polymorphisms between keratoconus (KC) patients and healthy controls.
|
| ||||
| Variants | KC Patients n (%) | Controls n (%) | *OR (95% CI) | P -value |
| rs2228557,COL4A4 | ||||
| CC | 67 (47.8) | 61 (40.7) | Ref. | — |
| CT | 39 (27.9) | 37 (24.7) | 0.96 (0.54–1.69) | 0.887 |
| TT | 34 (24.3) | 52 (34.6) | 0.59 (0.34–1.03) | 0.067 |
| Allele | ||||
| C | 173 (61.8) | 159 (53) | Ref. | — |
| T | 107 (38.2) | 141 (47) | 0.69 (0.50–0.97) | 0.035 |
| rs4898,TIMP-1 | Male | Male | ||
| CY | 46 (75.4) | 37 (57) | Ref. | — |
| TY | 15 (24.6) | 28 (43) | 0.43 (0.20–0.92) | 0.038 |
| Female | Female | |||
| CC | 28 (47.5) | 31 (52.5) | Ref. | — |
| CT | 29 (49.2) | 30 (50.8) | 1.01 (0.46–2.19) | 0.97 |
| TT | 22 (47.8) | 24 (52.2) | 1.07 (0.52–2.20) | 0.854 |
| Allele | ||||
| C | 85 (53.8) | 92 (54.1) | Ref. | — |
| T | 73 (46.2) | 78 (45.9) | 1.01 (0.66–1.56) | 0.953 |
| Ref., reference; OR, odds ratio; CI, confidence interval; n, number *Adjusted for sex and age. (Y states for the Y-chromosome) | ||||
Table 3.
Correlation of clinical and keratometric parameters with COL4A4 (rs2228557) and TIMP-1(rs4898) in keratoconus patients.
|
| |||||
| Parameters evaluated | Patients n (%) | rs2228557 | P -value | rs4898 | P -value |
| Male | Female | ||||
| KC ocular | |||||
| OD | 42 (30.0) | 0.25 | 0.39 | 0.4 | |
| OS | 36 (25.7) | ||||
| OU | 62 (44.3) | ||||
| Level of KC | |||||
| KK 1 | 33 (23.6) | 0.81 | 0.014 | 0.97 | |
| KK 2 | 45 (32.1) | ||||
| KK 3 | 62 (44.3) | ||||
| CXL | |||||
| OD | 39 (27.9) | 0.58 | 0.71 | 0.37 | |
| OS | 40 (28.6) | ||||
| OU | 42 (30.0) | ||||
| Candidate | 19 (13.6) | ||||
| Correlation of clinical and keratometric parameters with COL4A4 (rs2228557) and TIMP-1 (rs4898) in keratoconus patients. KC, keratoconus; OD, right eye; OS, left eye; OU, both eyes; CXL, cross-linking surgery KK1, 2, 3 are phenotyping classification and show the level progress of keratoconus disease in patients | |||||
RFLP-PCR was applied to validate the method and results of screening of rs4898 polymorphism in TIMP-1. As the original sequence of the region surrounding the polymorphism does not make a restriction enzyme site, a site-directed mutagenesis PCR primer (R*) was designed. This primer differs from the referent sequence in two bases and lies close to the polymorphic spot to alter the sequence and provide a restriction enzyme site at product. Thus, the original sequence of the region is TT(C) GTGG, while our PCR product was in fact TT(C) GCGA. The C-variant is a palindrome which forms a site for the Bsp68I (NruI) restriction enzyme (Thermo scientific).[40]
Initially, we amplified PCR product (660bp) using the abovementioned conditions. For alteration in the restriction enzyme site, secondary primers were added to template 1:50 dilution of first stage using the following condition: 1 μL template (1:50 dilution), 1 μL of each primer (10 pmol/μL), 10 μL mastermix, and 7 μL DNase-free water. The PCR conditions were set as follow: 95°C for 5 min, 30 cycles of 95°C for 30 sec, 63°C for 30 sec, 72°C for 30 sec, and a final extension at 72°C for 5 min. PCR products were detected by electrophoresis on a 2% agarose gel with ethidium bromide (344bp product). For optimal results, PCR products were digested at 37°C for 5 h according to the manufacturer's instruction. The restriction of the C-allele PCR product resulted in 325bp and 19bp digest products, whereas the T allele (wild type) products remained unrestricted (Figure B3).
Statistical analysis
Statistical analyses were performed using the SPSS software version 19.0 (SPSS Inc., Chicago IL, USA). Frequencies were compared between the study groups using the Chi-square test. Association of gene polymorphisms with KC was investigated and compared between the groups using logistic regression analysis, estimation of odds ratio (OR), and 95% confidence intervals (CI), respectively. A p-value 0.05 was regarded as statistically significant.
RESULTS
A total of 140 patients (61 male and 79 female subjects), aged 28 12.5 years, included the KC group. The control group consisted of 150 healthy controls (65 male and 85 female subjects), aged 29.8 15.6 years.
There was no significant difference between the two groups regarding the age and gender (P = 0.20).
The Col4A4 rs2228557 C/T variant, T allele was associated with a decrease in the risk of KC development (OR = 0.69, 95% CI: 0.50–0.97, P = 0.035), as compared to C allele. Our results indicated that TT was not associated with KC as compared to CC (OR = 0.59, 95% CI = 0.34–1.03, P = 0.067) (Table 2).
Table 2 demonstrates the genotype and allelic frequencies of TIMP-1 (rs4898) gene polymorphism in each study group. Since TIMP-1 is an X-linked gene, the results were compared in male and female subjects separately. This analysis demonstrated that TY genotype or T allele decreased the risk of KC in male subjects as compared to the C allele (OR = 0.43, 95% CI: 0.20–0.92, P = 0.03). However, no significant association was found between TT genotype (OR = 1.07, 95% CI: 0.52–2.20, P = 0.854) or T allele (OR = 1.01, 95%CI: 0.66–1.56, P = 0.95) and KC in female subjects.
Table 3 illustrates the associations of COL4A4 (rs2228557) and TIMP-1 (rs4898) with KC severity. This polymorphism TIMP1 (rs4898 T/C) located at X chromosome exists in two alleles and their combination results in five possible genotypes. Because men lack a second X-chromosome, the possible genotypes are CY and TY hemizygotes (Y states for the Y-chromosome). As for women, there are CC and TT homozygotes and CT heterozygotes (Table 2). In fact, men just have allele and women have genotype and allele. For this reasons, calculation of TIMP1 (rs4898) distinctly separated in men and women was done. So, results in men and women can be different.
DISCUSSION
Keratoconus is an eye disorder characterized by bilateral, asymmetrical, noninflammatory, and progressive thinning of the cornea. The shape of cornea progressively alters from the normal round shape to a cone shaped one. Although the etiology of KC is not clear, defective cross-linking between adjacent collagen fibers may play an important role in its pathogenesis.[41] SNPs and gene variants suggest an intricate etiology or the convergence of multiple disease pathways.[42]
Biochemical investigations have suggested that the amount of collagen fibers decrease in KC. Also, the weight of KC corneas was found to be reduced; therefore, it can be assumed that collagenase enzymes might be involved.[43]
In the current study, we investigated the impact of COL4A4 and TIMP-1 variants on the risk of KC development in a sample of Iranian population. Our results showed that the T allele reduced the risk of disease development, as compared to the C allele. Results in the distribution of genotypes (CC, CT, TT) in rs2228557 of the COL4A4 gene between KC patients and controls in the Stabuc-Silih et al's study were different from our results.[14] Similar to our findings, Kokolakis et al demonstrated that the TT genotype was significantly over-represented in healthy individuals and suggested a protective role for this genotype in the KC development.[31]
The level of TIMP-1 significantly decreases in KC as compared to normal corneas. Given the fact that KC is not associated with extensive scarring or inflammatory infiltrates, substantial degradation should occur in the extracellular matrix. A decreased level of TIMP-1 increases gelatinase and collagenase activities and apoptosis which are characteristic phenomena in KC. Decreases in TIMP-1 might play a role in matrix degradation which is a characteristic feature of KC.[15,44] Furthermore, it has been recognized that increased MMP and decreased TIMP-1 levels are associated with the development of KC.[29]
The TIMP-1 gene is located in Xp11.3–p11.23 and has three types of polymorphism including TIMP-1 (19 C/T) in the 5'-UTR, TIMP-1 (261 C/T) in exon 4, and TIMP-1 (372 T/C) (rs4898) in exon 5. The TIMP-1 (rs4898) polymorphism is an important site which has been reported in other studies. This variation does not result in changes in the amino acid sequence (F124F). This polymorphism exists in two alleles and their combination results in five possible genotypes. Because male subjects lack a second X-chromosome, the possible genotypes are CY and TY hemizygotes. In female subjects, however, there are CC and TT homozygotes and CT heterozygote genotypes.[40]
Our data suggest that C or T allele is associated with TIMP-1 (rs4898) polymorphism in patients or controls. The C allele of the 372T/C polymorphism was more frequently found in female than male controls.[45] However, in other studies, the C allele was detected more frequently in male patients with an abdominal aortic aneurysm.[46] Meijer et al investigated the male subjects with inflammatory bowel disease carrying TIMP-1 (rs4898) T allele. They reported lower levels of TIMP-1 in surgically resected inflamed tissue, as compared to C allele carriers.[47] In addition, Indelicato et al reported that TIMP-1 (rs4898) C allele frequency increased in males but not females with systemic sclerosis, as compared to healthy individuals.[48] Along the same lines, Wei et al revealed that C allele carriers of TIMP-1 (rs4898) run a greater risk of developing ankylosing spondylitis disease.[49] Furthermore, it was found that among cirrhotic patients, males with TIMP-1 (372C/T) T allele developed cirrhosis at a younger age.[50]
Our findings indicated that TIMP-1 (rs4898) was associated with the clinical characteristics of KC only in our male sample population. Nevertheless, the analysis of genotype and allele frequencies revealed no significant differences in female patients as compared to female controls. The T allele decreased the risk of KC, as compared to the C allele in males which can be attributed to the location of TIMP-1 gene at Xp11.3–p11.23. Males only have one X-chromosome, and the functional difference of genetic polymorphism of TIMP-1 (rs4898) appears more obvious due to the lack of heterozygotes. We cannot compare our results with the literature because no previous study has evaluated the correlation between the TIMP-1 variants and the risk of KC development.
In conclusion, our study showed that in the COL4A4 rs2228557 C/T variant, the T allele acts as a protective factor against the disease and decreases the risk of KC. In addition, TIMP-1 rs4898 C/T the TY genotype or T allele in males can decrease the risk of KC in comparison with the C allele. Further studies with a larger sample size and different ethnicities are required to confirm these findings.
Financial Support and Sponsorship
This study was supported by a dissertation grant (M.Sc. thesis of Davood Yari, NO. 6077) from the Deputy for Research, Zahedan University of Medical Sciences.
Conflicts of Interests
There are no conflicts of interest.
References
- 1.Burdon KP, Vincent AL. Insights into keratoconus from a genetic perspective. Clin Exp Optom 2013;96:146–154. [DOI] [PubMed]
- 2.Gajecka M, Radhakrishna U, Winters D, Nath SK, Rydzanicz M, Ratnamala U, et al. Localization of a gene for keratoconus to a 5.6-Mb interval on 13q32. Invest Ophthalmol Vis Sci 2009;50:1531–1539. [DOI] [PMC free article] [PubMed]
- 3.Lee LR, Hirst LW, Readshaw G. Clinical detection of unilateral keratoconus. Aust N Z J Ophthalmol 1995;23:129–133. [DOI] [PubMed]
- 4.Li X, Bykhovskaya Y, Canedo AL, Haritunians T, Siscovick D, Aldave AJ, et al. Genetic association of COL5A1 variants in keratoconus patients suggests a complex connection between corneal thinning and keratoconus. Invest Ophthalmol Vis Sci 2013;54:2696–2704. [DOI] [PMC free article] [PubMed]
- 5.Grewal DS, Brar GS, Grewal SP. Assessment of central corneal thickness in normal, keratoconus, and post-laser in situ keratomileusis eyes using Scheimpflug imaging, spectral domain optical coherence tomography, and ultrasound pachymetry. J Cataract Refract Surg 2010;36:954–964. [DOI] [PubMed]
- 6.Rabinowitz YS. Keratoconus. Surv Ophthalmol 1998;42:297–319. [DOI] [PubMed]
- 7.Pearson AR, Soneji B, Sarvananthan N, Sandford-Smith JH. Does ethnic origin influence the incidence or severity of keratoconus? Eye 2000;14:625–628. [DOI] [PubMed]
- 8.Sahebjada S, Schache M, Richardson AJ, Snibson G, MacGregor S, Daniell M, et al. Evaluating the association between keratoconus and the corneal thickness genes in an independent Australian population. Invest Ophthalmol Vis Sci 2013;54:8224–8228. [DOI] [PubMed]
- 9.Bae HA, Mills RA, Lindsay RG, Phillips T, Coster DJ, Mitchell P, et al. Replication and meta-analysis of candidate loci identified variation at RAB3GAP1 associated with keratoconus. Invest Ophthalmol Vis Sci 2013;54:5132–5135. [DOI] [PMC free article] [PubMed]
- 10.Hasanian-Langroudi F, Saravani R, Validad MH, Bahari G, Yari D. Association of Lysyl oxidase (LOX) polymorphisms with the risk of Keratoconus in an Iranian population. Ophthalmic Genet 2014;0:1–6. [DOI] [PubMed]
- 11.Heon E, Greenberg A, Kopp KK, Rootman D, Vincent AL, Billingsley G, et al. VSX1: a gene for posterior polymorphous dystrophy and keratoconus. Hum Mol Genet 2002;11:1029–1036. [DOI] [PubMed]
- 12.Yari D, Saravani R, Saravani S, Ebrahimian K, Galavi HR. Genetic polymorphisms of catalase and glutathione peroxidase-1 in keratoconus. Iran J Public Health 2018;47:1567–1574. [PMC free article] [PubMed]
- 13.Guan T, Liu C, Ma Z, Ding S. The point mutation and polymorphism in keratoconus candidate gene TGFBI in Chinese population. Gene 2012;503:137–139. [DOI] [PubMed]
- 14.Stabuc-Silih M, Ravnik-Glavac M, Glavac D, Hawlina M, Strazisar M. Polymorphisms in COL4A3 and COL4A4 genes associated with keratoconus. Mol Vis 2009;15:2848–2860. [PMC free article] [PubMed]
- 15.Lee JE, Oum BS, Choi HY, Lee SU, Lee JS. Evaluation of differentially expressed genes identified in keratoconus. Mol Vis 2009;15:2480–2487. [PMC free article] [PubMed]
- 16.Fullerton J, Paprocki P, Foote S, Mackey DA, Williamson R, Forrest S. Identity-by-descent approach to gene localisation in eight individuals affected by keratoconus from north-west Tasmania, Australia. Hum Genet 2002;110:462–470. [DOI] [PubMed]
- 17.Patel D, McGhee C. Understanding keratoconus: what have we learned from the New Zealand perspective? Clin Exp Optom 2013;96:183–187. [DOI] [PubMed]
- 18.Boye E, Mollet G, Forestier L, Cohen-Solal L, Heidet L, Cochat P, et al. Determination of the genomic structure of the COL4A4 gene and of novel mutations causing autosomal recessive Alport syndrome. Am J Hum Genet 1998;63:1329–1340. [DOI] [PMC free article] [PubMed]
- 19.Mariyama M, Zheng K, Yang-Feng TL, Reeders ST. Colocalization of the genes for the alpha 3(IV) and alpha 4(IV) chains of type IV collagen to chromosome 2 bands q35-q37. Genomics 1992;13:809–813. [DOI] [PubMed]
- 20.Stachs O, Bochert A, Gerber T, Koczan D, Thiessen HJ, Guthoff RF. [The extracellular matrix structure in keratoconus]. Ophthalmologe 2004;101:384–389. [DOI] [PubMed]
- 21.Bochert A, Berlau J, Koczan D, Seitz B, Thiessen HJ, Guthoff R. [Gene expression in keratoconus. Initial results using DNA microarrays]. Ophthalmologe 2003;100:545–549. [DOI] [PubMed]
- 22.Nielsen K, Hjortdal J, Pihlmann M, Corydon TJ. Update on the keratoconus genetics. Acta Ophthalmol 2013;91:106–113. [DOI] [PubMed]
- 23.Brew K, Dinakarpandian D, Nagase H. Tissue inhibitors of metalloproteinases: evolution, structure and function. Biochim Biophys Acta 2000;1477:267–283. [DOI] [PubMed]
- 24.Brew K, Nagase H. The tissue inhibitors of metalloproteinases (TIMPs): an ancient family with structural and functional diversity. Biochim Biophys Acta 2010;1803:55–71. [DOI] [PMC free article] [PubMed]
- 25.Guedez L, Stetler-Stevenson WG, Wolff L, Wang J, Fukushima P, Mansoor A, et al. In vitro suppression of programmed cell death of B cells by tissue inhibitor of metalloproteinases-1. J Clin Invest 1998;102:2002–2010. [DOI] [PMC free article] [PubMed]
- 26.Usher PA, Sieuwerts AM, Bartels A, Lademann U, Nielsen HJ, Holten-Andersen L, et al. Identification of alternatively spliced TIMP-1 mRNA in cancer cell lines and colon cancer tissue. Mol Oncol 2007;1:205–215. [DOI] [PMC free article] [PubMed]
- 27.Akahane T, Akahane M, Shah A, Connor CM, Thorgeirsson UP. TIMP-1 inhibits microvascular endothelial cell migration by MMP-dependent and MMP-independent mechanisms. Exp Cell Res 2004;301:158–167. [DOI] [PubMed]
- 28.Johnson MD, Kim HR, Chesler L, Tsao-Wu G, Bouck N, Polverini PJ. Inhibition of angiogenesis by tissue inhibitor of metalloproteinase. J Cell Physiol 1994;160:194–202. [DOI] [PubMed]
- 29.Brown D, Chwa MM, Opbroek A, Kenney MC. Keratoconus corneas: increased gelatinolytic activity appears after modification of inhibitors. Curr Eye Res 1993;12:571–581. [DOI] [PubMed]
- 30.Saravani R, Hasanian-Langroudi F, Validad MH, Yari D, Bahari G, Faramarzi M, et al. Evaluation of possible relationship between COL4A4 gene polymorphisms and risk of keratoconus. Cornea 2015;34:318–322. [DOI] [PubMed]
- 31.Kokolakis NS, Gazouli M, Chatziralli IP, Koutsandrea C, Gatzioufas Z, Peponis VG, et al. Polymorphism analysis of COL4A3 and COL4A4 genes in Greek patients with keratoconus. Ophthalmic Genet 2014;35:226–228. [DOI] [PubMed]
- 32.Wang HX, Yang QD, Liu BQ, Zhang L, Ma MM, Hu ZY, et al. TIMP-1 polymorphisms in a Chinese Han population with intracerebral hemorrhage. Int J Neurosci 2014;124:61–67. [DOI] [PubMed]
- 33.Skarmoutsou E, D'Amico F, Marchini M, Malaponte G, Scorza R, Mazzarino MC. Association of TIMP-1 +372 SNP with digital ulcer manifestation in female systemic sclerosis patients. Hum Immunol 2012;73:950–953. [DOI] [PubMed]
- 34.Lorente L, Martin M, Plasencia F, Sole-Violan J, Blanquer J, Labarta L, et al. The 372 T/C genetic polymorphism of TIMP-1 is associated with serum levels of TIMP-1 and survival in patients with severe sepsis. Crit Care 2013;17:R94. [DOI] [PMC free article] [PubMed]
- 35.Mikami T, Meguro A, Teshigawara T, Takeuchi M, Uemoto R, Kawagoe T, et al. Interleukin 1 beta promoter polymorphism is associated with keratoconus in a Japanese population. Mol Vis 2013;19:845–851. [PMC free article] [PubMed]
- 36.Hashemi M, Amininia S, Ebrahimi M, Hashemi SM, Yousefi J, Eskandari-Nasab E, et al. Association between LAPTM4B gene polymorphism and breast cancer susceptibility in an Iranian population. Med Oncol 2014;31:111. [DOI] [PubMed]
- 37.Haff LA. Improved quantitative PCR using nested primers. PCR Methods Appl 1994;3:332–337. [DOI] [PubMed]
- 38.Newton CR, Graham A, Heptinstall LE, Powell SJ, Summers C, Kalsheker N, et al. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res 1989;17:2503–2516. [DOI] [PMC free article] [PubMed]
- 39.Dai S, Long Y. Genotyping Analysis Using an RFLP Assay. Methods Mol Biol 2015;1245:91–99. [DOI] [PubMed]
- 40.Naychov ZD, Hiyama E, Uchida N, Arihiro K, Nagao M, Takahashi S, et al. TIMP-1 c.T372C Genetic Polymorphism as a Possible Predictor for Acute Aortic Dissection. Hiroshima J Med Sci 2013;62:31–37.
- 41.Ormonde S. Refractive surgery for keratoconus. Clin Exp Optom 2013;96:173–182. [DOI] [PubMed]
- 42.Crawford A, Fassett RG, Geraghty DP, Kunde DA, Ball MJ, Robertson IK, et al. Relationships between single nucleotide polymorphisms of antioxidant enzymes and disease. Gene 2012;501:89–103. [DOI] [PubMed]
- 43.Collier SA. Is the corneal degradation in keratoconus caused by matrix-metalloproteinases? Clin Experiment Ophthalmol 2001;29:340–34. [DOI] [PubMed]
- 44.Kenney MC, Chwa M, Atilano SR, Tran A, Carballo M, Saghizadeh M, et al. Increased levels of catalase and cathepsin V/L2 but decreased TIMP-1 in keratoconus corneas: evidence that oxidative stress plays a role in this disorder. Invest Ophthalmol Vis Sci 2005;46:823–832. [DOI] [PubMed]
- 45.Krex D, Rohl H, Konig IR, Ziegler A, Schackert HK, Schackert G. Tissue inhibitor of metalloproteinases-1, -2, and -3 polymorphisms in a white population with intracranial aneurysms. Stroke 2003;34:2817–2821. [DOI] [PubMed]
- 46.Hinterseher I, Krex D, Kuhlisch E, Schmidt KG, Pilarsky C, Schneiders W, et al. Tissue inhibitor of metalloproteinase-1 (TIMP-1) polymorphisms in a Caucasian population with abdominal aortic aneurysm. World J Surg 2007;31:2248–2254. [DOI] [PubMed]
- 47.Meijer MJ, Mieremet-Ooms MA, van Hogezand RA, Lamers CB, Hommes DW, Verspaget HW. Role of matrix metalloproteinase, tissue inhibitor of metalloproteinase and tumor necrosis factor-alpha single nucleotide gene polymorphisms in inflammatory bowel disease. World J Gastroenterol 2007;13:2960–2966. [DOI] [PMC free article] [PubMed]
- 48.Indelicato M, Chiarenza V, Libra M, Malaponte G, Bevelacqua V, Marchini M, et al. Analysis of TIMP-1 gene polymorphisms in Italian sclerodermic patients. J Clin Lab Anal 2006;20:173–176. [DOI] [PMC free article] [PubMed]
- 49.Wei JC, Lee HS, Chen WC, Shiu LJ, Yang SF, Wong RH. Genetic polymorphisms of the matrix metalloproteinase-3 (MMP-3) and tissue inhibitors of matrix metalloproteinases-1 (TIMP-1) modulate the development of ankylosing spondylitis. Ann Rheum Dis 2009;68:1781–1786. [DOI] [PubMed]
- 50.Ikebuchi Y, Ishida C, Okamoto K, Murawaki Y. Association of TIMP-1 and TIMP-2 gene polymorphisms with progression of liver fibrosis in patients with type C chronic liver disease. Biochem Genet 2013;51:564–574. [DOI] [PubMed]
- 51.Saravani R, Yari D, Saravani S, Hasanian-Langroudi F. Correlation between the COL4A3, MMP-9, and TIMP-1 polymorphisms and risk of keratoconus. Jpn J Ophthalmol 2017;61:218–222. [DOI] [PubMed]


