The NF-κB pathway plays key roles in HIV-1 replication and reactivation of HIV-1 latency. A regulator inhibiting NF-κB activation may be a promising therapeutic strategy against HIV-1. Recently, NF-κB-interacting long noncoding RNA (NKILA) was identified to suppress the development of different human cancers by inhibiting IκB kinase (IKK)-induced IκB phosphorylation and NF-κB pathway activation, whereas the relationship between NKILA and HIV-1 replication is still unknown. Here, our results show that NKILA inhibits HIV-1 replication and reactivation by suppressing HIV-1 long terminal repeat (LTR)-driven transcription initiation. Moreover, NKILA inhibited the replication of HIV-1 clones with different coreceptor tropisms. This project may reveal a target for the development of novel anti-HIV drugs.
KEYWORDS: lncRNA, NKILA, NF-κB, antiviral activity, HIV-1 replication, HIV-1 latency
ABSTRACT
NF-κB-interacting long noncoding RNA (NKILA) was recently identified as a negative regulator of NF-κB signaling and plays an important role in the development of various cancers. It is well known that NF-κB-mediated activation of human immunodeficiency virus type 1 (HIV-1) long terminal repeat (LTR)-driven gene expression is required for HIV-1 transcription and reactivation of latency. However, whether NKILA plays essential roles in HIV-1 replication and latency is unclear. Here, by ectopic expression and silencing experiments, we demonstrate that NKILA potently inhibits HIV-1 replication in an NF-κB-dependent manner by suppressing HIV-1 LTR promoter activity. Moreover, NKILA showed broad-spectrum inhibition on the replication of HIV-1 clones with different coreceptor tropisms as well as on LTR activity of various HIV-1 clinical subtypes. Chromatin immunoprecipitation (ChIP) assays revealed that NKILA expression abolishes the recruitment of p65 to the duplicated κB binding sites in the HIV-1 LTR. NKILA mutants disrupting NF-κB inhibition also lost the ability to inhibit HIV-1 replication. Notably, HIV-1 infection or reactivation significantly downregulated NKILA expression in T cells in order to facilitate viral replication. Downregulated NKILA was mainly due to reduced acetylation of histone K27 on the promoter of NKILA by HIV-1 infection, which blocks NKILA expression. Knockdown of NKILA promoted the reactivation of latent HIV-1 upon phorbol myristate acetate (PMA) stimulation, while ectopic NKILA suppressed the reactivation in a well-established clinical model of withdrawal of azidothymidine (AZT) in vitro. These findings improve our understanding of the functional suppression of HIV-1 replication and latency by NKILA through NF-κB signaling.
IMPORTANCE The NF-κB pathway plays key roles in HIV-1 replication and reactivation of HIV-1 latency. A regulator inhibiting NF-κB activation may be a promising therapeutic strategy against HIV-1. Recently, NF-κB-interacting long noncoding RNA (NKILA) was identified to suppress the development of different human cancers by inhibiting IκB kinase (IKK)-induced IκB phosphorylation and NF-κB pathway activation, whereas the relationship between NKILA and HIV-1 replication is still unknown. Here, our results show that NKILA inhibits HIV-1 replication and reactivation by suppressing HIV-1 long terminal repeat (LTR)-driven transcription initiation. Moreover, NKILA inhibited the replication of HIV-1 clones with different coreceptor tropisms. This project may reveal a target for the development of novel anti-HIV drugs.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) is a severe health threat worldwide. Although combined antiretroviral therapy (cART) greatly revolutionized the treatment of HIV-1 infection, changing it from a fatal acute disease to a chronic condition, the viral reservoir that persists in resting memory CD4+ T cells and other myeloid lineages is a major barrier to HIV-1 eradication (1–6). A deeper understanding of the transcriptional events underlying HIV-1 replication and latency will facilitate the development of a strategy that permits the manipulation of key cellular and viral transcription factors.
Previous studies have discovered that factors leading to transcriptional inhibition, such as the presence of nuc1, sequestration of nuclear factor kappa B (NF-κB) in the cytoplasm or of positive transcription elongation (P-TEFb) in the inactive 7SK snRNP, and insufficiency of the transactivator Tat encoded by HIV-1, play dominant roles in HIV-1 replication and the establishment and maintenance of HIV-1 latency (7–12). As an important transcription factor, NF-κB/RelA promotes HIV-1 replication and influences HIV-1 latency by binding to the duplicate κB enhancer in the HIV-1 long terminal repeat (LTR) (11, 13–16).
NF-κB transcription factors are dimers produced by the combination of five different monomers (RelA [p65], NF-κB1 [p50], NF-κB2 [p52], c-Rel, and RelB) for dimerization, DNA binding, and nuclear translocation. NF-κB signaling is involved in various biological processes, including immunity, inflammation, cancer, and viral infection (11, 17–22). The p50/p65 heterodimer interacts with the NF-κB sites in the HIV LTR and recruits histone acetyltransferase (HAT) to facilitate HIV transcription upon activation (23), while HIV-1 employs its viral accessory protein Nef to boost NF-κB activation or Vpu to downmodulate NF-κB-dependent expression of interferon-stimulated genes (ISGs) (24). One NF-κB agonist, a compound called 5HN, promoted NF-κB induction and led to activation of latent HIV-1 without specific T-cell activation (25). Protein kinase C (PKC) agonists, including prostratin, bryostatin, and ingenol, activate NF-κB and increase the levels of cyclin-dependent kinases (CDKs), P-TEFb, and other related proteins, which are required for HIV-1 replication and reactivation (26–28). Williams et al. showed that efficient formation of elongated HIV-1 transcripts requires sustained activation of NF-κB, which increases de novo synthesis of Tat (29). Upon activation, NF-κB complexes (e.g., p50/p65 heterodimers) replace p50 homodimers to bind to κB sites in the LTR and to recruit the cellular histone acetyltransferase p300 which drives localized histone acetylation and promotes transcription initiation (29–31). Therefore, the NF-κB pathway has positive effects on HIV-1 replication and latency and may be a promising target for the development of new antiviral drugs.
Recently, NF-κB-interacting long noncoding RNA (NKILA), which is 2,570 bp in length and is located at chromosomal region 20q13, was initially identified as a tumor suppressor by its abrogation of NF-κB signaling (32–36); however, whether NKILA regulates HIV-1 replication or latency has not been characterized. Here, we investigated the potential role of NKILA in HIV-1 replication and reactivation of latent HIV-1. The results showed that NKILA potently inhibits the replication of various subtypes of HIV-1 and might regulate HIV-1 latency through NF-κB signaling. Our study discovered the regulatory function of a long noncoding RNA (lncRNA), NKILA, on HIV-1 by targeting NF-κB signaling, which provides important insight for the development of new therapeutic tools against HIV-1 infection.
RESULTS
NKILA potently inhibits HIV-1 replication.
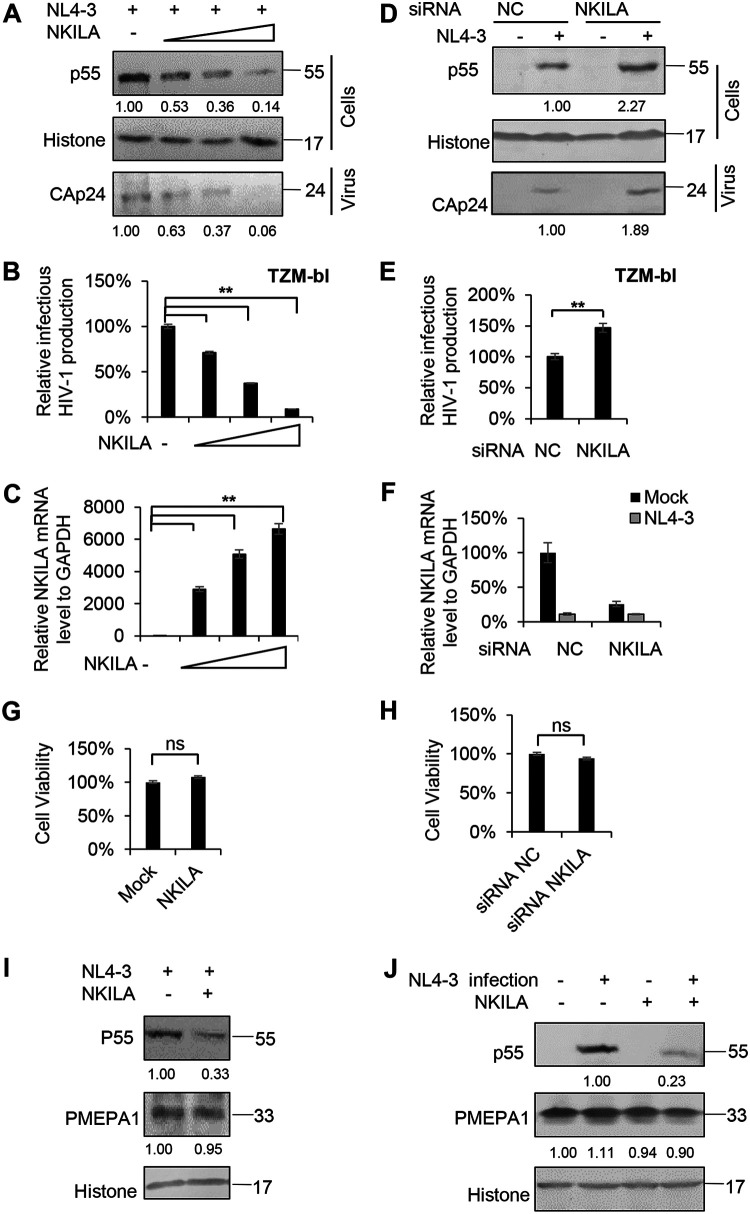
As a transcription factor, NF-κB plays an important role in HIV-1 transcription and replication. To investigate whether NF-κB-interacting lncRNA (NKILA), which represses NF-κB signaling (33), affects HIV-1 replication, we first transfected HEK293T cells with the pNL4-3 expression vector plus the negative-control vector VR1012 or the NKILA expression vector and then harvested cells 48 h later for immunoblotting and reverse transcription-quantitative PCR (qRT-PCR) analysis. With increasing levels of NKILA, Gagp55 expression in the cell lysate and capsid p24 (CAp24) expression in the viral supernatant from cells were decreased in a dose-dependent manner (Fig. 1A), and the infectious HIV-1 production was greatly decreased when TZM-bl cells were used as infection indicator cells (Fig. 1B), indicating that NKILA suppresses HIV-1 replication. The mRNA levels of NKILA were determined by qRT-PCR (Fig. 1C).
FIG 1.
NKILA inhibits HIV-1 replication. (A to C) Overexpression of NKILA inhibits HIV-1 replication in a dose-dependent manner. (A) Multiple dose amounts of NKILA expression vector (100 ng, 300 ng, and 900 ng) or negative-control vector were transfected with the pNL4-3 viral expression vector into HEK293T cells. After 48 h, cells and supernatants were harvested and analyzed by immunoblot (IB) analysis. The densities of bands from representative immunoblotting (IB) analyses were analyzed with ImageJ software to calculate the values, for cells relative to that for histone. (B) Infectious HIV-1 production was decreased with increasing NKILA expression, as indicated in TZM-bl cells. (C) The expression levels of NKILA mRNA were measured by qRT-PCR. The mRNA level of endogenous NKILA was set as 100%. (D to F) Knockdown of NKILA increased HIV-1 replication. (D) pNL4-3 or negative-control vector was cotransfected with siRNA NKILA or siRNA NC into HEK293T cells for 48 h. Cells and supernatants were harvested for IB analysis, and the densities of bands from representative IB analyses were analyzed as described for panel A. (E) NKILA increased the infectious HIV-1 production, as indicated in TZM-bl cells. The infectious HIV-1 production of siRNA NC was set as 100%. (F) The expression levels of NKILA in cells with NKILA knockdown were measured by qRT-PCR and normalized to GAPDH expression. Overexpression (G) or knockdown (H) of NKILA had no effect on cell viability by CCK-8 detection. (I)The inhibitory effect of NKILA on HIV-1 production was not associated with altered endogenous expression of the PMEPA1 protein. NKILA or negative-control vector was cotransfected with the pNL4-3 viral vector into HEK293T cells. Forty-eight hours after transfection, cell extracts were harvested and subjected to IB analysis with anti-PMEPA1 antibody to detect the PMEPA1 protein. (J) PMEPA1 protein expression was not affected by HIV-1 infection or NKILA expression. NKILA or negative-control vector was nucleofected to Jurkat cells. Forty-eight hours posttransfection, the cells were infected with the supernatant containing NL4-3 viral particles or an equal amount of medium. After 48 h of infection, cells were harvested for IB analysis. The densities of bands were analyzed as described for panel A. All results are presented as the means ± SDs from three independent experiments. ns, not significant; **, P < 0.01.
In contrast, compared to transfection with short interfering RNA negative control (siRNA NC), knockdown of NKILA with siRNA in HEK293T cells led to increased levels of Gagp55 in the cell lysate and CAp24 in the viral supernatant (Fig. 1D, 2nd and 4th lanes). The infectious HIV-1 production in the supernatant produced by NKILA siRNA cells was increased (Fig. 1E). The mRNA levels of NKILA are shown in Fig. 1F. A cell proliferation assay by Cell Counting Kit 8 (CCK-8) confirmed that ectopic expression or silencing of NKILA had no effect on cell viability (Fig. 1G and H), indicating that NKILA itself but not the cellular effects of NKILA accounts for HIV-1 inhibition. The PMEPA1 gene, which encodes a protein correlated with several tumors and participates in immune pathways, runs antisense to NKILA (37). Thus, we examined whether NKILA expression affects PMEPA1 expression. We cotransfected NKILA with or without the pNL4-3 expression plasmid into HEK293T cells and harvested the cells for immunoblot analysis. We observed that NKILA expression strongly inhibited Gagp55 expression in the cell lysate, while it had no effect on PMEPA1 expression (Fig. 1I). Moreover, PMEPA1 expression was not affected before or after HIV-1 infection (Fig. 1J), indicating that the inhibitory effect of NKILA on HIV-1 is not associated with PMEPA1 expression.
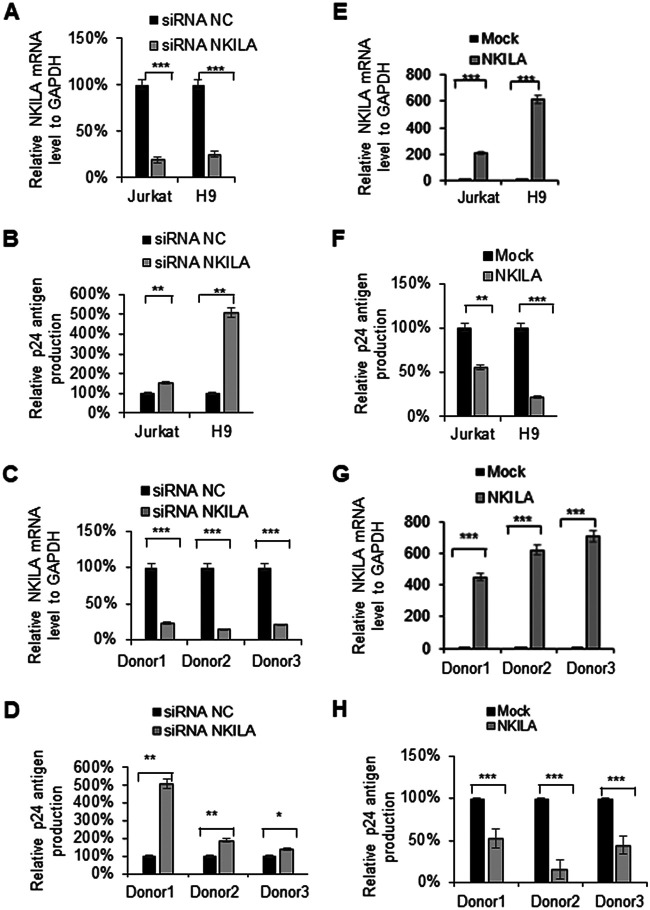
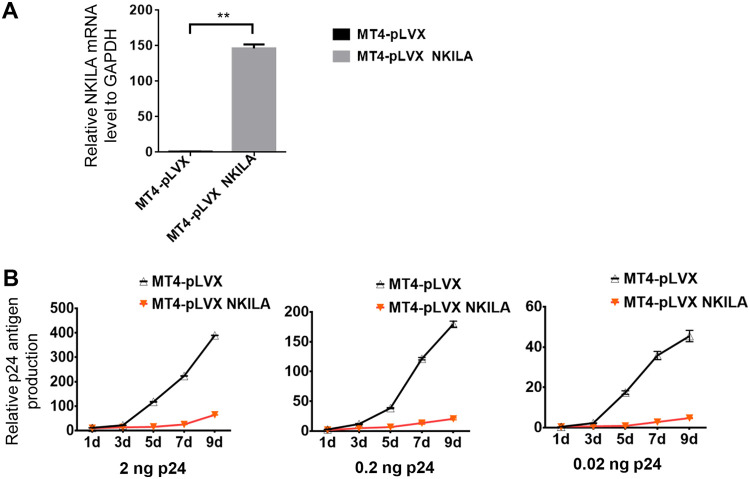
To further confirm whether the infectious HIV-1 production in target cells is affected by NKILA, we silenced NKILA in different T cells, such as Jurkat, H9, and CD4+ primary cells, using siRNA pools targeting NKILA or transfecting these cells with siRNA NC (Fig. 2A and C). Knockdown of NKILA increased the levels of p24 from ∼1.5- to 5-fold in the Jurkat and H9 cell lines (Fig. 2B) as well as in CD4+ T cells from three healthy donors (Fig. 2D). Accordingly, overexpression of NKILA in Jurkat, H9, or CD4+ primary cells by nucleofection (Fig. 2E and G) suppressed p24 antigen production (Fig. 2F and H). We also examined the lasting repressive function of stable expressing NKILA in MT4 cells on HIV-1 replication and confirmed that NKILA potently inhibited HIV-1 replication at different doses of HIV-1 infection compared to that of negative-control cell lines over time (Fig. 3B). NKILA stable expression in MT4 cells was confirmed by qRT-PCR (Fig. 3A). Collectively, these data suggest that NKILA strongly inhibits HIV-1 replication.
FIG 2.
NKILA inhibits HIV-1 replication in T cells. (A to D) Knockdown of NKILA in T cells promoted infectious HIV-1 production. The pNL4-3 viral vector was transfected into HEK293T cells, and supernatants containing HIV-1 NL4-3 virus were harvested for infection of Jurkat cells, H9 cells, or primary CD4 T cells from donors for 48 h which had been transfected with siRNA NKILA or siRNA NC for 24 h. (A and C) Total RNA was extracted from the cells, and the mRNA level of NKILA was measured by qRT-PCR. Cells transfected with siRNAs were infected with NL4-3 virus for 4 h, washed twice with PBS buffer, and cultured in medium (RPMI 1640, 10% FBS). (B and D) After 48 h, the supernatants were harvested, and CAp24 expression was measured by ELISA. (E to H) Overexpression of NKILA suppressed infectious HIV-1 production. Jurkat, H9, or primary CD4+ T cells which had been transfected with NKILA or negative-control vector for 24 h were then were infected with HIV-1 NL4-3 viruses. (E and G) Forty-eight hours postinfection, cells were harvested, and the mRNA level of NKILA was measured by qRT-PCR. (F and H) The supernatants were harvested, and CAp24 expression was measured by ELISA. Expression data were normalized to those of the siRNA NC or negative vector group. All results are presented as the means ± SDs from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
FIG 3.
NKILA inhibits HIV spreading replication. (A) Relative NKILA mRNA expression in MT4 cells expressing NKILA. MT4 cells were infected with lentiviruses containing NKILA or a scramble control, and at 48 h postinfection, puromycin (1 g/ml) was added to the medium for selection. (B) Two nanograms, 0.2 ng, or 0.02 ng p24 antigen of NL4-3 viral particles was added to infect the NKILA MT4 overexpression or control cell lines. Cells were washed with PBS three times after 4 h of infection. The supernatant p24 release amounts were analyzed by ELISA at 1 day, 3 days, 5 days, 7 days, and 9 days.
NKILA suppresses the activity of the HIV-1 LTR in a dose-dependent manner.
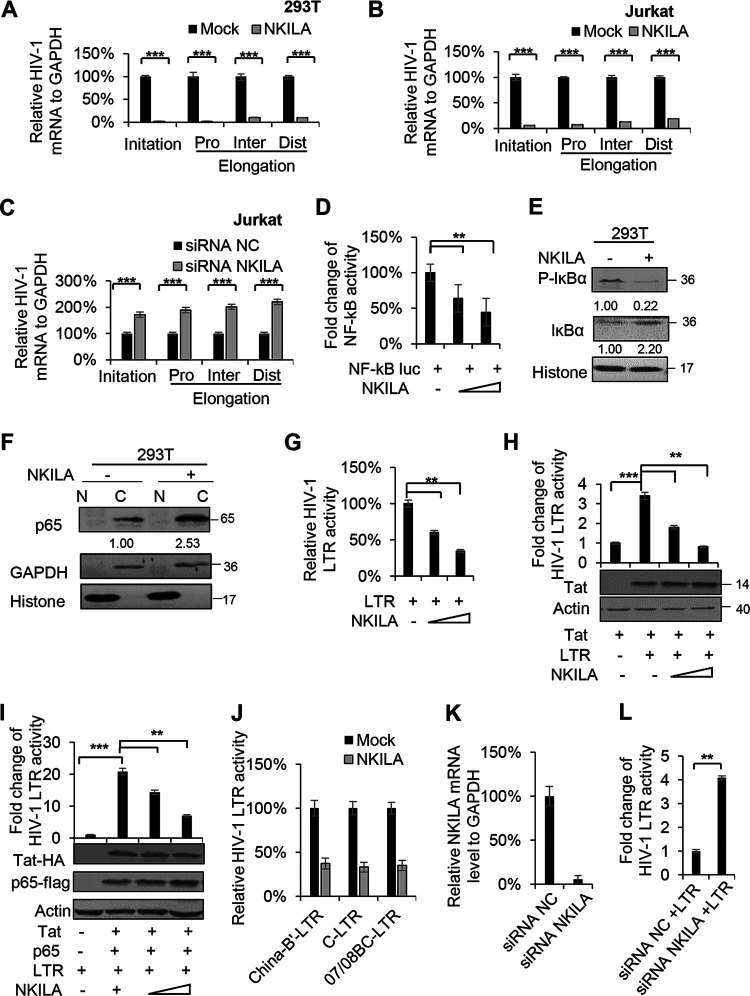
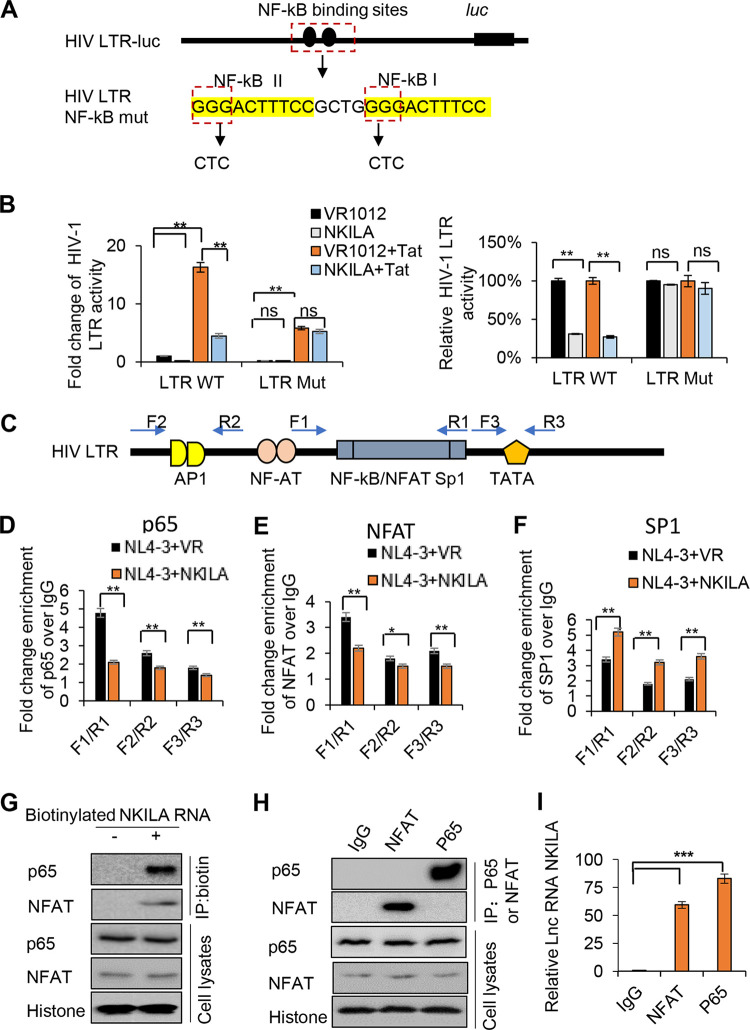
The HIV-1 LTR drives the transcription initiation and elongation of HIV-1 transcripts, and NF-κB induces the initiation of HIV-1 LTR-driven gene transcription (29, 32); thus, we hypothesized that NKILA might inhibit HIV-1 transcription initiation by suppressing NF-κB signaling. To investigate this possibility, we assessed HIV-1 transcription initiation and elongation by measuring the mRNA levels of the proximal (Pro), intermediate (Int), and distal (Dis) transcripts by qPCR in the presence or absence of NKILA. The results showed that NKILA significantly suppressed HIV-1 transcription initiation and then inhibited the transcription elongation (Fig. 4A and B). Accordingly, knockdown of NKILA increased the transcription initiation of HIV-1 and then increased the transcription elongation (Fig. 4C). We further confirmed that NKILA inhibited NF-κB-driven luciferase activity and induced a decrease in lκBα phosphorylation as well as a reduction in p65 translocation into the nucleus as evidenced by nuclear/cytoplasmic fractionation (Fig. 4D to F), consistent with the results of a previous study (33). NF-κB initially induces only a single round of RelA and RNA polymerase II (Pol II) binding, whereas Tat efficiently stimulates transcription elongation (29). We observed that NKILA inhibited not only LTR activity but also LTR activity induced by Tat alone or by both p65 and Tat (Fig. 4G to I). In addition, to investigate the effect of NKILA on LTR activities from different clinical HIV isolates, we transfected NKILA or a negative vector plus expression vector containing LTR from the China HIV-1 subtypes B′, C, and CRF07/08 BC and found a similar reduction of LTR-driven gene expression when NKILA was overexpressed (Fig. 4J). HIV-1 LTR activity was significantly increased when NKILA was depleted using siRNA in HEK293T cells (Fig. 4L). The mRNA expression levels of NKILA were confirmed by qPCR (Fig. 4K). Taken together, these data suggest active competition between NKILA and p65 for controlling HIV-1 LTR promoter activity.
FIG 4.
NKILA suppresses HIV-1 LTR-driven gene expression in a dose-dependent manner. (A) NKILA inhibits the transcription initiation and then inhibits the transcription elongation of HIV-1 in HEK293T cells. pNL4-3 was cotransfected with NKILA or negative-control vector into HEK293T cells, and cells were harvested 48 h posttransfection for quantitation of HIV-1 gene transcription initiation and elongation with special primers by qRT-PCR. (B and C) Effect of NKILA overexpression or knockdown on HIV 5′ LTR-driven transcription initiation and elongation in Jurkat cells. Jurkat cells transfected with NKILA vectors or siRNA NKILA were infected with HIV-luc/VSV-G for 2 days, and mRNA was extracted and analyzed by qRT-PCR. (D) NKILA inhibited NF-κB activity. Increasing amounts of NKILA or negative-control vector were cotransfected with pNF-κB-luciferase and pRenilla-luciferase reporter plasmids into HEK293T cells for 48 h. Cells were harvested for assessment of reporter gene expression by a dual luciferase reporter assay. (E) NKILA expression influenced the total and phosphorylated IĸBα protein levels, as evidenced by IB analysis, and the densities of bands were analyzed with ImageJ software to calculate the values relative to that for histone. (F) The localization of p65 protein in the cytoplasm was increased with NKILA overexpression. HEK293T cells transfected with NKILA or negative-control vector were harvested for chromatin fractionation, and protein expression was then analyzed by immunoblotting. The densities of bands were analyzed with ImageJ software to calculate the values relative to that for GAPDH. (G) Overexpression of NKILA inhibited LTR-driven gene expression. Increasing amounts of NKILA or negative-control vector were cotransfected with pHIV-1-LTR-luciferase and pRenilla-luciferase into HEK293T cells for 48 h. Cells were harvested for assessment of reporter gene expression by a dual-luciferase reporter assay. NKILA also decreased LTR-driven gene expression mediated by Tat (H) or by Tat combined with p65 (I). pHIV-1 5′ LTR plasmids were cotransfected with Tat expression vector or with both Tat and p65 expression vectors. Forty-eight hours later, cell extracts were harvested for detection of protein expression by IB analysis and quantitation of reporter activity by a dual-luciferase reporter assay. (J) NKILA repressed LTR-driven gene expression of different clinical HIV-1 strains with different NF-κB binding sites. HEK293T cells were cotransfected with the LTRs of different clinical HIV-1 strains found in China and with pRenilla for 48 h. Reporter gene expression was assessed by a luciferase assay. (K and L) Knockdown of NKILA increased LTR-driven gene expression. pHIV-1 5′ LTR plasmids were cotransfected with siRNA NKILA or siRNA NC into HEK293T cells. Forty-eight hours posttransfection, the cells were harvested and analyzed by qRT-PCR to measure the mRNA level of NKILA and were subjected to a dual-luciferase reporter assay to measure the reporter activity. The corresponding value of the control was set as 1 or 100% as appropriate. All results are representative of three independent experiments. The data are presented as the means ± SDs. **, P < 0.01; ***, P < 0.001.
The NF-κB binding site in HIV-1 LTR is required for HIV-1 inhibition by NKILA.
The HIV-1 5′ LTR usually contains a TATA box, three SP1 binding sites, and two NF-κB binding sites (16, 38), although HIV-1 subtype C viruses contain an additional NF-κB site in the enhancer region of HIV-1 LTR (39, 40). Chaudhary et al. identified critical nucleotides in the NF-κB binding sites in the HIV-1 5′ LTR that affect NF-κB function (14). To further determine whether NKILA-mediated inhibition of HIV-1 is NF-κB dependent, we constructed an HIV-1 LTR NF-κB mutant in a luciferase reporter plasmid as indicated in Fig. 5A. As expected, NKILA inhibited Tat-mediated wild-type (WT) LTR activity, while NKILA lost the ability to affect the activity of the LTR NF-κB mutant (Fig. 5B). We also observed that Tat-mediated activation was lower in the LTR mutant group than in the LTR WT group, which is consistent with the fact that NF-κB induces efficient HIV-1 transcription initiation. We next investigated the effect of NKILA on the in vivo binding of p65 to the LTR promoter by chromatin immunoprecipitation (ChIP) analysis. qRT-PCR was carried out with the primer pairs F1/R1, F2/R2, and F3/R3, as indicated in Fig. 5C, which encompassed the region occupied by p65 on the LTR promoter. In addition to acting as an NF-κB binding site, LTR also serves as the binding site for other transcription factors, including the nuclear factor of activated T cells (NFAT) and the specificity protein (SP1). F1/R1 includes NF-κB/NFAT and SP1 (nucleotides [nt] −143 to +14), while F2/R2 and F3/R3 contain the regions between nt −453 and −349 and +75 and +180 in the LTR, respectively. We observed significant enrichment using an anti-p65 antibody over that obtained using IgG with F1/R1, F2/R2, and F3/R3, while NKILA obviously reduced the recruitment of p65 to F1/R1 and slightly reduced the recruitment of p65 to F2/R2 and F3/R3 in HEK293T cells cotransfected with NKILA and pNL4-3 compared to that in cells transfected with pNL4-3 alone (Fig. 5D). We also examined the effect of NKILA on the binding of LTR to NFAT and SP1 and observed a similar influence of NKILA on NFAT as for NF-κB, while NKILA mainly increased the recruitment of SP1 to F1/R1. For lower binding of SP1 with F2/R2 and F3/R3 regions of LTR as well as the increases caused by NKILA, we speculated that the occupied place released by NF-κB increased the binding of LTR with SP1 (Fig. 5E and F); the phenomenon of functional incompatibility between the NF-κB motif and a unique SP1 had been reported previously (41). Interestingly, a biotinylated NKILA pulldown assay showed NFAT also interacts with NKILA as p65 (Fig. 5G), and reverse co-immunoprecipitation (co-IP) assays further verified the interaction between NKILA and NFAT (Fig. 5H to I), suggesting that NKILA competitively binding to NFAT with LTR caused reduced interaction between NFAT and LTR. Thus, our results indicate that NKILA inhibits p65 and NFAT recruitment to the NF-κB enhancer region in the LTR promoter.
FIG 5.
NKILA directly inhibits NF-κB-mediated HIV-1 transcription by obstructing p65 recruitment to the LTR promoter. (A) Representation of the HIV-LTR with mutations at NF-κB binding sites. (B) The effect of NKILA on the HIV-1 LTR promoter is specifically mediated by NF-κB binding sites. pHIV-1 5′ LTR WT or pHIV-1 5′ LTR mutant plasmids were cotransfected with NKILA or negative-control vector, and cells were harvested for a reporter assay 48 h posttransfection. (C) Schematic of the HIV-1 LTR primers (F1/R1, F2/R2, and F3/R3) used in the ChIP assays. (D) p65 recruitment to the LTR promoter at the NF-κB-SP1 enhancer region was disrupted by NKILA in HIV-1 NL4-3 virus-infected Jurkat cells. Jurkat cells transfected with NKILA or negative-control vector for 24 h were infected with HIV-1 NL4-3 virus for another 5 days. Fixed and isolated chromatin from HIV-1-infected Jurkat cells was immunoprecipitated with anti-p65 antibody or IgG as the negative control and was analyzed by qRT-PCR with the indicated primers spanning three different regions in the LTR promoter. The effect of NKILA on the recruitment of NFAT (E) or SP1 (F) to HIV-1 LTR. The experimental procedure is the same as for panel D. Fixed and isolated chromatin was immunoprecipitated with anti-NFAT or SP1 antibody or IgG as the negative control and then was analyzed as described for panel D. (G) The interaction of p65 and NFAT proteins with NKILA according to RNA pulldown assay. (H) NKILA interacted with NFAT and p65 proteins according to the co-IP assay. Jurkat cells were immunoprecipitated with anti-NFAT- or p65 antibody-conjugated agarose beads. NFAT and P65 proteins were detected by immunoblotting analysis. (I) The relative binding ability between NFAT or P65 with NKILA. The binding between NKILA and IgG was set as 1. **, P < 0.01; ***, P < 0.001.
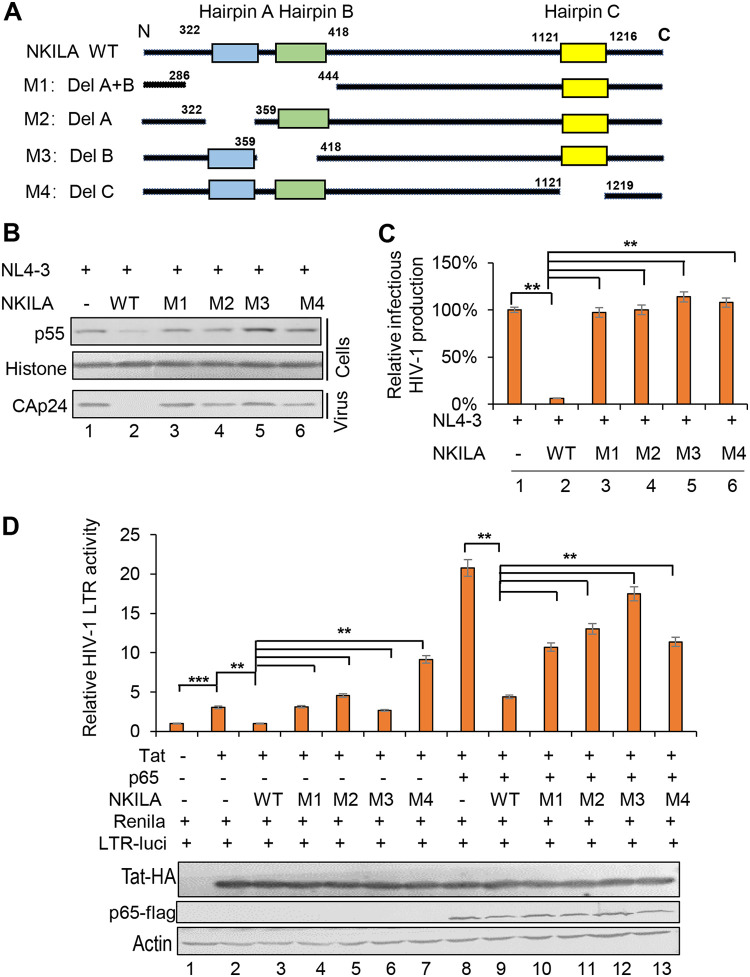
Functional domains of NKILA required for NF-κB interference are also required for HIV-1 restriction.
A previous study demonstrated that three hairpins of NKILA are essential for NF-κB signaling suppression through binding to the p65-p50-lκBα complex via p65 and physically masking the phosphorylation sites on lκBα (33). Hairpin A (nt 322 to 359) and hairpin B (nt 395 to 418) within the region encompassing nt 300 to 500, as well as hairpin C (nt 1121 to 1216), were predicted previously (33). To determine whether these three hairpins are required for HIV-1 inhibition, we constructed NKILA mutants as indicated in Fig. 6A. The results showed that compared to WT NKILA, all four mutants lost the ability to inhibit HIV-1 replication, as evidenced by measurement of the Gagp55 expression in the cell lysate and capsid p24 expression in the supernatant. In addition, the infectious HIV-1 NL4-3 production was also not affected by these mutants (Fig. 6B and C). The HIV-1 LTR luciferase assay showed that compared to WT NKILA, M1, M2, and M4 mutants could not inhibit Tat-mediated LTR activity, only NKILA M3 mutant showed considerable inhibitory effect. In Tat-p65-mediated LTR activity, all NKILA mutants showed a weaker inhibitory effect than WT NKILA (Fig. 6D). Taken together, functional domains in NKILA essential for NF-κB suppression are also necessary for HIV-1 restriction, indicating that the inhibitory effect of NKILA on HIV-1 is closely associated with its function in suppressing NF-κB signaling.
FIG 6.
NKILA mutants fail to abolish HIV-1 transcription and LTR promoter activity. (A) Schematic representation of NKILA with mutations at each of the hairpins (hairpins A, B, and C). (B) NKILA WT, the indicated mutants, or negative-control vector were cotransfected with pNL4-3ΔEnv and pVSV-G plasmids into HEK293T cells. Cells and partial supernatants were harvested for measurement of HIV-1 gene expression by IB analysis. (C) Supernatants containing virus-like particles (VLPs) were collected and used to infect TZM-bl cells. Viral infectious yield was measured by a luciferase reporter assay. (D) NKILA mutation completely abolished the ability to inhibit Tat with or without p65-mediated LTR promoter activity. NKILA WT or the indicated mutants were cotransfected with Tat or with both Tat and p65 plus pHIV-1-luciferase plasmids. Forty-eight hours posttransfection, reporter gene expression was analyzed by a luciferase reporter assay, and the expression of proteins was assessed by IB analysis. The corresponding value in the absence of Tat and p65 was set as 1 or 100% as appropriate. All results are representative of three independent experiments. The data are presented as the means ± SDs. **, P < 0.01; ***, P < 0.001.
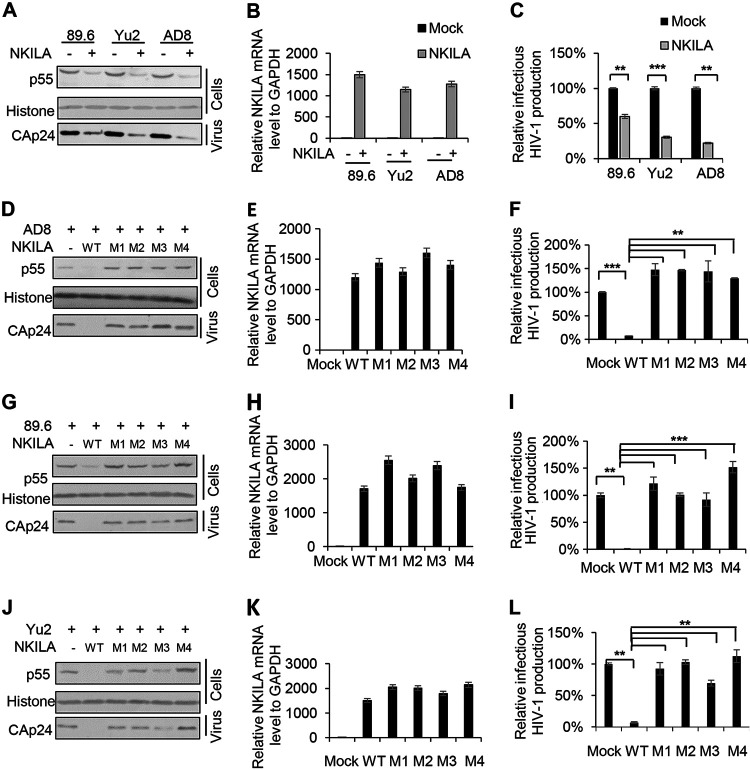
NKILA potently inhibits the replication of HIV-1 clones with different coreceptor tropisms.
To investigate the inhibitory effect of NKILA on HIV-1 clones with different coreceptor tropisms, we transfected cells with VR1012 or NKILA plus the 89.6 (R5X4-tropic), Yu2 (brain-derived HIV-1), or AD8 (macrophage-tropic) expression vectors into HEK293T cells and then harvested the cells and supernatants for immunoblot or qRT-PCR analysis. The results showed that Gagp55 expression in the cell lysate and capsid p24 (CAp24) expression in the viral supernatant from cells were obviously decreased (Fig. 7A), and the infectious production of these HIV-1 clones was greatly decreased (Fig. 7C). We also observed that compared to WT NKILA, the NKILA mutants lost the ability to inhibit the replication and the infectious production of these HIV-1 clones (Fig. 7D, F, G, I, J, and L). The expression of NKILA was determined by qRT-PCR analysis (Fig. 7B, E, H, and K). Combined with the data in Fig. 4J, these data indicate that NKILA can inhibit various HIV-1 clones by inhibiting LTR activity.
FIG 7.
NKILA has the ability to inhibit the replication of HIV-1 clones with different coreceptor tropisms. (A) Overexpression of NKILA suppressed the replication of HIV-1 clones with different coreceptor tropisms. The NKILA or negative-control vector was cotransfected with expression plasmids for the HIV-1 clones 89.6, Yu2, or AD8. Forty-eight hours posttransfection, cells and supernatants were harvested for measurement of HIV-1 gene expression by IB analysis (A) and were analyzed by qRT-PCR to measure NKILA mRNA levels (B). (C) TZM-bl cells were infected with viral supernatants for 48 h and harvested for measurement of infectious HIV-1 yield by a luciferase assay. (D to L) NKILA mutants lost their inhibition ability toward various HIV-1 clones. NKILA WT or the indicated mutants were cotransfected with expression vectors for the HIV-1 clones AD8 (D, E, and F), 89.6 (G, H, and I), or Yu2 (J, K, and L). Cells were harvested for detection of HIV-1 gene expression by IB analysis (D, G, and J) and were analyzed by qRT-PCR to measure NKILA mRNA levels (E, H, and K). Supernatants were used to infect TZM-bl cells for measurement of the infectious yield of various HIV-1 clones (F, I, and L). The corresponding value of the control was set as 1 or 100% as appropriate. All results are representative of three independent experiments. The data are presented as the means ± SDs. **, P < 0.01; ***, P < 0.001.
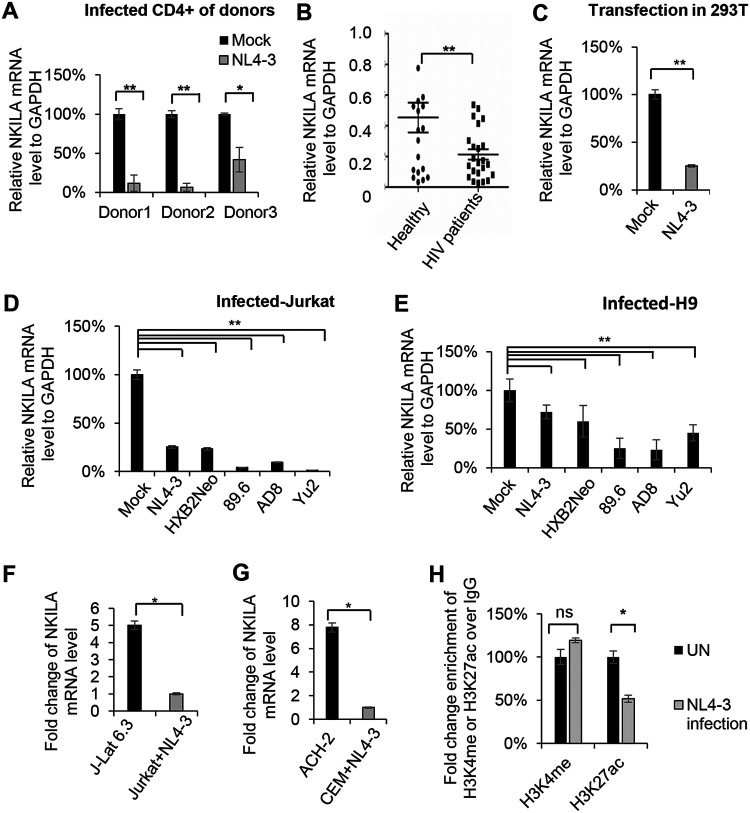
HIV-1 replication downregulates NKILA expression.
The above-described data showed that NKILA inhibits HIV-1 replication (Fig. 1 to 3), and we next examined whether HIV-1 infection or replication regulates NKILA expression. Primary CD4+ T cells from three healthy donors were infected with HIV-1 NL4-3 virus produced from the supernatant of HEK293T cells transfected with the NL4-3 expression vector. In cells infected with HIV-1, the mRNA levels of NKILA were greatly decreased (Fig. 8A) compared with that in uninfected CD4+ T cells. Moreover, NKILA expression was obviously downregulated in the CD4+ T cells from 23 HIV-1 infected patients compared with that in cells from 17 healthy donors (Fig. 8B). Even transfection with the NL4-3 expression vector in HEK293T cells caused a reduction in NKILA expression (Fig. 8C). Moreover, in addition to HIV-1 NL4-3 infection, the infection caused by the other HIV-1 clones HXB2Neo, 89.6, AD8, and Yu2 also induced a decrease in NKILA mRNA levels in the Jurkat and H9 cell lines (Fig. 8D and E). In latently infected J-Lat 6.3 and ACH-2 cells, we further observed that the mRNA levels of NKILA were higher than those in acutely infected Jurkat and CEM cells infected with NL4-3 viruses (Fig. 8F and G). In order to investigate the reason why NKILA was downregulated by HIV-1 infection, we examined the epigenetic modification in the promoter of NKILA, histone 3 K4 methylation (H3K4me) and histone 3 K27 acetylation (H3K27ac) according to the prediction by UCSC Genome Browser analysis. A ChIP assay showed that HIV-1 infection decreased H3K27 acetylation and had no effect on H3K4 methylation, which might be the reason why NKILA was downregulated (Fig. 8H).
FIG 8.
NKILA expression is inversely correlated with HIV-1 infection and production. HIV-1 infection downregulated NKILA expression in primary CD4+ T cells isolated from healthy donors (donors 1 to 3) (A), Jurkat cells (D), and H9 cells (E). Each HIV-1 strain expression vector was transfected into HEK293T cells. After 48 h, supernatants were harvested for infection of the indicated cells. The mRNA levels of NKILA in the mock- and HIV-1 NL4-3-infected primary CD4+ T (A), Jurkat (D), and H9 (E) cells were measured by qRT-PCR and normalized to the level of GAPDH. (B) The mRNA level of NKILA was decreased in primary CD4+ T cells from HIV-infected patients compared to those from healthy donors. (C) The mRNA level of NKILA was decreased by HIV-1 production. pNL4-3 or negative-control vector was transfected into HEK293T cells, and cells were harvested for analysis of NKILA expression by qRT-PCR. (F) J-Lat 6.3 cells are Jurkat cells with latent HIV-1 infection. NKILA expression in Jurkat cells with acute or latent HIV-1 infection was measured by qRT-PCR. (G) Acute infection with NL4-3 decreased the NKILA mRNA level compared to that in HIV-1 latently infected CEM cells (ACH-2), as evidenced by qRT-PCR. (H) HIV-1 infection decreased the H3K27 acetylation mark on the NKILA promoter, as revealed by anti-H3K4me and anti-H3K27ac ChIP assays in Jurkat cells showing the level of H3K27ac on the NKILA promoter following NL4-3 infection or when uninfected (UN). All results are representative of three independent experiments. The data are presented as the means ± SDs. ns, not significant; *, P < 0.05; **, P < 0.01.
NKILA is negatively correlated with HIV-1 reactivation.
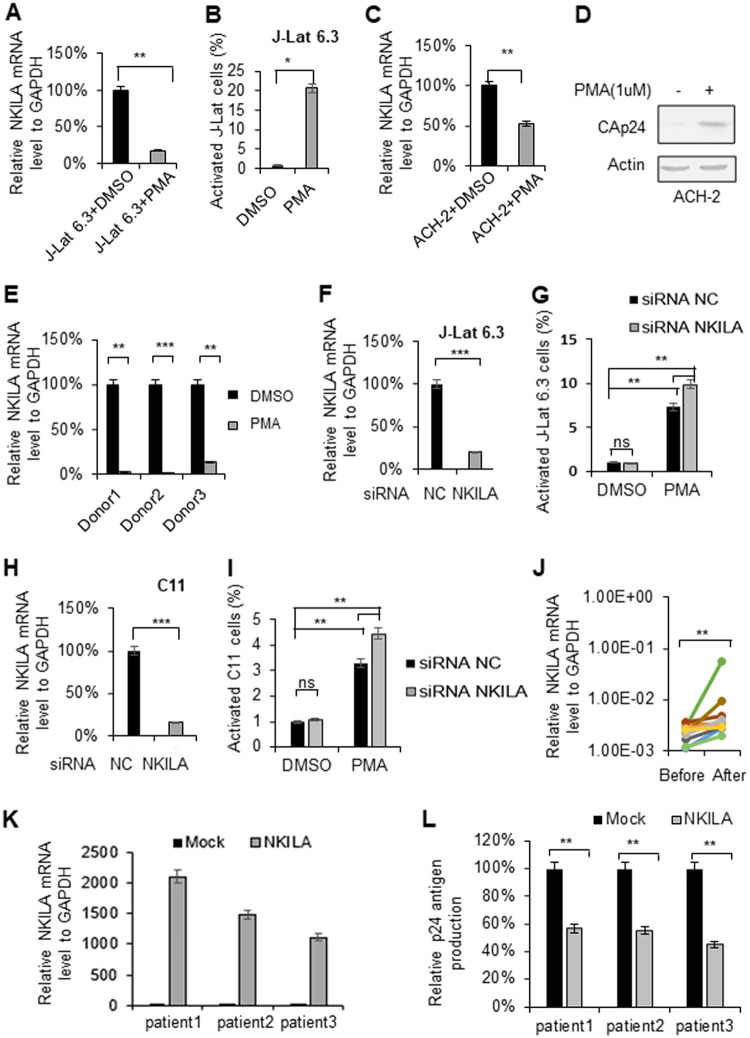
NF-κB signaling plays an important role in the reactivation of HIV-1 latency (11, 13, 14). To understand the function of NKILA-mediated transcriptional repression of HIV-1 by targeting the NF-κB pathway, we examined the mRNA levels of NKILA when the latently HIV-1-infected human T cell lines J-Lat 6.3 and ACH-2 were activated by phorbol myristate acetate (PMA). Treatment of J-Lat 6.3 and ACH-2 cells with PMA (1 μM) for 48 h decreased the mRNA levels of NKILA (Fig. 9A and C), while PMA stimulated HIV-1 transcription and replication, as reported (Fig. 9B and D). Moreover, CD4+ T cells obtained from HIV-1-infected patients with cART treatment and treated with PMA (1 μM) exhibited lower mRNA levels of NKILA than those without PMA treatment (Fig. 9E), suggesting that the physiological levels of NKILA might be associated with HIV-1 latency.
FIG 9.
NKILA is downregulated when latent cells are activated. Reactivation of latent HIV-1 increased NKILA expression. HIV-1 latently infected J-Lat 6.3 Jurkat cells and CD4+ CEM cells (ACH-2) were utilized to measure the change in NKILA expression during viral reactivation. J-Lat 6.3 (A) or ACH-2 (C) cells were treated with or without PMA (1 μM) for 48 h and were then harvested. mRNA was extracted from the cells for measurement of NKILA expression by qRT-PCR, and the expression levels were normalized to that of GAPDH. (B) HIV-1 reactivation in J-Lat 6.3 cells was measured by detecting GFP-positive cells by flow cytometry. The corresponding value of GFP-positive cells treated with dimethyl sulfoxide (DMSO) was set as 1 or 100% as appropriate. (D) HIV-1 reactivation of ACH-2 cells was measured by IB analysis with anti-CAp24. (E) PMA treatment decreased the NKILA levels in primary CD4+ T cells from HIV-1 infected patients with cART treatment. (F to I) Knockdown of NKILA increased HIV reactivation from latency. siRNA NKILA or siRNA NC was transfected into HIV-1 latently infected C11 and J-Lat 6.3 cells. Forty-eight hours posttransfection, cells were harvested for assessment of NKILA expression by qRT-PCR (F and H) and were stimulated with PMA for 48 h for detection of the GFP-positive cells by flow cytometry (G and I). The corresponding value of GFP-positive cells treated with DMSO was set as 1 or 100% as appropriate. (J) Successful cART treatment increased NKILA expression in PBMCs of HIV-1 infected patients. PBMCs were collected from HIV-1-infected individuals pretherapy and after 48 weeks of cART therapy. mRNA was extracted from PBMCs for measurement of NKILA expression levels by qRT-PCR. (K and L) Overexpression of NKILA in CD4+ T cells caused inhibition of reactivation of HIV-1. The CD4+ cells from three patients who had undergone HAART treatment were nucleofected with NKILA or control vector. Forty-eight hours posttransfection, NKILA mRNA levels were analyzed by qRT-PCR (K), and phytohemagglutinin M (PHA-M, 5 ng/ml) was added to the cell supernatants for 7 days. (L) The supernatant p24 antigen was analyzed by ELISA. The results are representative of three independent repeats. The qRT-PCR results in panels A, C, E, F, H, J, K, and L are shown as the corresponding values, with the value in the mock group set as 100%. The data are presented as the means ± SDs. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (unpaired t test).
To further explore the effect of NKILA on HIV reactivation, we transfected siRNA NKILA or siRNA NC into latent J-Lat 6.3 or C11 cells. Twenty-four hours posttransfection, the mRNA levels of NKILA in the cells were measured by qRT-PCR (Fig. 9F and H), and cells were stimulated with PMA for 48 h and harvested for detection of viral reactivation by flow cytometry. We observed that knockdown of NKILA increased the reactivation of HIV transcription from latency (Fig. 9G and I). Accordingly, overexpression of NKILA in CD4+ T cells from three HIV-1-infected individuals who had undergone highly active antiretroviral therapy (HAART) (Fig. 9K) decreased the rebound of latent HIV-1 upon stimulation by phytohemagglutinin (PHA), as determined by detected p24 amount in the supernatant (Fig. 9L). To examine the clinical association between HIV-1 replication and NKILA expression, we isolated peripheral blood mononuclear cells (PBMCs) from HIV-1-infected patients at pretherapy and posttherapy time points. After 48 weeks of cART therapy, the plasma viral load was suppressed below the level of detection (<50 copies), while the mRNA levels of NKILA were higher than that pretherapy (Fig. 9J), indicating that NKILA is negatively correlated with HIV-1 replication.
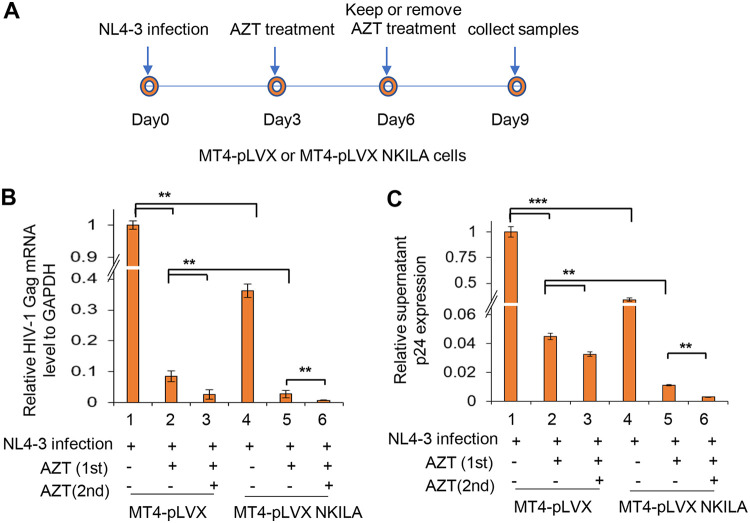
In order to substantiate the effect of NKILA on HIV-1 reactivation, we used a well-established clinical consequence of withdrawal of azidothymidine (AZT) treatment in vitro (42). MT4 cells stably expressing NKILA or negative-control vector were infected with HIV-1 NL4-3 for 3 days and then treated with AZT for 3 days (first treatment), washed, and placed back in culture in AZT-free or AZT-containing medium (second treatment) for another 3 days (Fig. 10A). HIV-1 Gag mRNA levels in cells and p24 amount in the supernatant were detected by qRT-PCR and enzyme-linked immunosorbent assay (ELISA) after the first and second treatments, respectively. The results showed that HIV-1 was effectively inhibited in cells expressing NKILA (Fig. 10B, lanes 1 and 4), and AZT treatment obviously enhanced the suppression (Fig. 10B and C, lanes 2, 3, 5, and 6). However, removing AZT after the first treatment in MT4 cells expressing NKILA led to weaker rebound of HIV-1 than in the negative-control group (Fig. 10B and C, lanes 2 and 5). Overall, these data indicate that NKILA is an important inhibitor of HIV-1 replication by suppressing NF-κB and might contribute to the maintenance of HIV-1 latency.
FIG 10.
NKILA overexpression maintains HIV-1 suppression after AZT withdrawal. (A) Experimental design for the analysis of HIV-1 replication in MT4 cells expressing NKILA or control cells. The cell lines were infected with 2 ng p24 antigen of NL4-3 viral particles. Three days postinfection, cells were treated with AZT (20 μM) or equal amounts of DMSO for 3 days and then switched to AZT-free or 20 μM AZT-containing medium for an additional 3 days. On day 9, the cells were harvested and analyzed for HIV Gag mRNA by qRT-PCR (B) and the amount of p24 antigen in the supernatant by ELISA (C).
DISCUSSION
Numerous studies have demonstrated that the host transcription factor NF-κB is utilized by HIV-1 to mediate transcriptional initiation and achieve high expression levels of the RNA genome (43–45). The enhancer region in the HIV-1 5′ LTR usually contains two identical κB binding sites located at nt −104 to −80, and HIV-1 viruses containing mutated LTR-κB sequences infect cells but fail to produce de novo transcripts (46). Moreover, p50/RelA also stimulates the early rounds of RNA Pol II elongation, although the conversion of RNA Pol II from the poorly processive enzymatic form to a highly processive form is transient (29, 31, 45, 47).
Accumulating evidence has demonstrated that lncRNAs are involved in numerous biological and pathological processes, including HIV-1 viral infection (48, 49). Recent studies identified that some lncRNAs, such as NRON, uc002yug.2, and MALAT1, significantly influence HIV-1 replication and latency (7, 36, 50, 51). Liu et al. (33) and other research groups (11, 36, 52) demonstrated that NKILA, as a negative regulator of NF-κB, inhibits cell proliferation, invasion, or metastasis in multiple cancers, including breast cancer, melanoma, non-small cell lung cancer, and hepatocellular carcinoma. However, whether NKILA affects HIV-1 replication and reactivation has not been investigated.
In the present study, we demonstrate that NKILA plays a key role in the regulation of HIV-1 transcription and replication. Beginning with transfection of an NKILA overexpression vector or siRNA NKILA into HEK293T cells, we found that NKILA strongly inhibited HIV-1 replication and infectious HIV-1 production in a dose-dependent manner (Fig. 1A to C) and that depletion of NKILA in HEK293T cells increased HIV-1 replication and infectious HIV-1 production (Fig. 1D to F). Moreover, NKILA suppressed HIV-1 infection in HIV-1-targeted cells, such as different T cell lines and primary CD4+ T cells (Fig. 2).
NKILA has been demonstrated to form a stable ternary complex with NF-κB/IκB via binding to p65 and retaining NF-κB in the cytoplasm, thus causing the suppression of NF-κB signaling (32, 33). The HIV-1 5′ LTR harbors κB binding sites for NF-κB interaction, and we observed that NKILA not only inhibited NF-κB activity but also inhibited the activation of HIV-1 LTR-driven gene expression mediated by Tat alone or even by p65 and Tat combined (Fig. 4). However, the promoter activity of HIV-1 LTR mutants harboring κB binding site mutations was not affected by NKILA expression compared to that of the WT LTR (Fig. 5). The ChIP assay showed significant enrichment using an anti-p65 antibody over that obtained using IgG with F1/R1 and less enrichment with F2/R2 and F3/R3 in the HIV-1 LTR, while NKILA obviously reduced the recruitment of p65 to F1/R1 (which contains the NF-κB-SP1 region located at nt −143 to +14) as well to F2/R2 or F3/R3 (which contain the regions between nt −453 to −349 and nt +75 to +180 in the LTR, respectively) (Fig. 5D). Interestingly, NKILA also had a similar influence on NFAT binding to LTR due to the interaction between NFAT and NKILA, which was confirmed in Fig. 5I, but had opposite effect on SP1 binding to HIV-1 LTR. Moreover, the functional domains in NKILA responsible for p65 binding as well as masking κB phosphorylation are also required for HIV-1 inhibition (Fig. 6). Considering the highly conserved κB binding sites in the 5′ LTR region among the majority of the HIV isolates (53), we observed that NKILA also potently inhibited the replication of HIV-1 clones with different coreceptor tropisms, such as the macrophage-tropic AD8 strain, the brain-derived Yu2 strain, and the R5X4-tropic 89.6 strain (Fig. 7). In the present study, we further examined the role of NKILA in simian immunodeficiency virus (SIV) infection and found that the replication of SIVmac239 was not affected by NKILA (data not shown). Despite the sequence conservation of the NF-κB binding site in the LTR of HIV and SIV, Ilyinskii et al. (54) and Pohlmann et al. (55) showed that SIV replicates efficiently in the absence of NF-κB binding, indicating that the requirement of NF-κB binding for viral replication was a prerequisite for NKILA to exert its antiviral activity.
Accumulating studies demonstrated that viruses frequently evolve a strategy to antagonize host restriction (56–58); for example, Vif, Vpu, and Vpx encoded by HIV mediate the degradation of APOBEC3, tetherin, and SAMHD1, respectively (59–64). Interestingly, we found that NKILA expression was downregulated in acute HIV-1 infection or HIV-1 reactivation from latency (Fig. 8 and 9). More importantly, we confirmed that NKILA expression was lower in HIV-1-infected patients than in healthy people, indicating that HIV-1 viruses try to overcome the inhibitory effect of NKILA to successfully replicate and survive in cells (Fig. 8B). Further investigation showed that HIV-1 downregulates the expression of NKILA by decreasing H3K27 acetylation in the promoter of NKILA (Fig. 8H). Anyway, it is hard to exclude the possibility that an indirect effect by HIV-1 infection causes NKILA downregulation. It is noteworthy that upregulated NKILA expression was observed in cytotoxic T lymphocytes (CTLs) and type 1 T helper (TH1) cells activated by PHA through STAT1 signaling (32); thus, different cell types or additional HIV-1 infection might be responsible for this opposite result.
Multiple mechanisms including transcriptional interference can establish HIV-1 latency. Among them, the NF-κB pathway required for HIV-1 transcription plays a critical role in latency reversal, as demonstrated by HSP70 binding protein 1 (HspBP1) and HSP90, which involve in NF-κB regulation (11, 14). By knockdown or overexpression of NKILA in latent HIV-1 cell lines or CD4+ T cells from three HIV-1-infected individuals who had undergone HAART, as well as in a well-established clinical model of withdrawal of AZT treatment, we demonstrated that NKILA decreased the rebound of latent HIV-1 (Fig. 9 and 10), suggesting that NKILA suppression of NF-κB activity might regulate HIV-1 latency.
To summarize, our novel finding that the NKILA restricts HIV-1 replication by inhibiting NF-κB-mediated activation of HIV-1 viral gene expression establishes that the NKILA is a restriction factor for HIV-1 replication. The information in this study might be useful for developing novel therapeutic strategies against HIV-1.
MATERIALS AND METHODS
Ethics statement.
This study was approved by the Ethics Review Committee of the First Hospital of Jilin University. All research participants signed informed consent forms. HIV-1-infected patients and healthy donors were recruited at the Changchun Center for Disease Control and Prevention, Jilin, China.
Plasmid construction.
The plasmids pNL4-3, pNL4-3 ΔEnv, pHXB2Neo, pYu2, p89.6, and pAD8 were obtained from the AIDS Research and Reference Reagents Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH). pVSV-G, pRenilla-luciferase, pTat-hemagglutinin (HA), and pHIV-1-LTR-luciferase were constructed as previously described (36, 65–67). The full-length NKILA fragment was amplified from the total cDNA of human peripheral blood mononuclear cells (PBMCs) and subcloned into the VR1012 vector, which was generously provided by Vical (San Diego, CA), via the SalI and BglII sites. Mutants (M1 to M4) of NKILA were generated from pNKILA by PCR-based site-directed mutagenesis (Shanghai Generay Biotech Company, China). The p65 expression vector with a C-terminal Myc-flag tag was cloned into the pCMV6-Entry vector via the SgfI and MluI sites (OriGene Technologies, Inc., USA). The plasmids pChina-B′-LTR, pC′-LTR, and p07/08-BC-LTR were kindly provided by Jianhua Wang (University of Chinese Academy of Sciences, China).
RNA extraction and real-time qRT-PCR.
RNA was isolated with TRIzol reagent according to the manufacturer's instructions (Invitrogen, Carlsbad, CA, USA). Before reverse transcription, RQ-1 DNase (Promega, Madison, WI) was used to treat RNA samples. cDNA synthesis was performed using a Transcriptor First Strand cDNA Synthesis kit (Roche, Basel, Switzerland) according to the manufacturer’s instructions. A total of 250 to 1,000 ng of total RNA was used as a template for each cDNA synthesis reaction, and samples containing only H2O or no reverse transcriptase were included as blank samples. cDNA was stored at −80°C until use. The qRT-PCR assay was performed with FastStart Universal SYBR green Master (Roche) on a Roche 480 instrument. Blank samples without reverse transcriptase was included in the NKILA qPCRs to exclude contamination by genomic DNA. qRT-PCR amplification of the target fragment was carried out as follows: initial activation at 95°C for 2 min, followed by 45 cycles at 95°C for 15 s, 57°C for 15 s, and 68°C for 20 s. To minimize the bias that may be caused by house genes, the geometric mean of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin gene expression was used to characterize the expression stability, and fold changes were calculated using the comparative threshold cycle (2−ΔΔCT) method according to the MIQE guidelines (68). The primers used for real-time qRT-PCR are listed below. The GAPDH forward sequence was CGTGCGTGACAT, the GAPDH reverse sequence was GTCAGGCAGCTCGTAGCTCTT, the NKILA forward sequence was AACCAAACCTACCCACAACG, the NKILA reverse sequence was ACCACTAAGTCAATCCCAGGTG, the β-actin forward sequence was ACCGAGCGCGGCTACAG, the β-actin reverse sequence was CTTAATGTCACGCACGATTTCC, the forward sequence of initial primers targeting base pairs 10 to 59 of the HIV-1 transcript was GTTAGACCAGATCTGAGCCT, the initial reverse sequence was GTGGGTTCCCTAGTTAGCCA, the forward sequence of proximal primers targeting base pairs 29 to 180 of the HIV-1 transcript was TGGGAGCTCTCTGGCTAACT, the proximal reverse sequence was TGCTAGAGATTTTCCACACTGA, the forward sequence of intermediate primers targeting base pairs 836 to 1015 of the HIV-1 transcript was GTAATACCCATGTTTTCAGCATTATC, the intermediate reverse sequence was TCTGGCCTGGTGCAATAGG, the forward sequence of distal primers targeting base pairs 2341 to 2433 of HIV-1 transcript was GAGAACTCAAGATTTCTGGGAAG, and the distal reverse sequence was AAAATATGCATCGCCCACAT. The HIV-1 Gag forward sequence was CTGAAGCGCGCACGGCAA and the HIV-1 Gag reverse sequence was GACGCTCTCGCACCCATCTC. For ChIP-NKILA promoter primers, the forward sequence was CGTTTTCGTCGTCGTCGTC, and the reverse sequence was CTCGACGAAAATTAACGCAAC.
Cells and antibodies.
HEK293T (catalog no. CRL-11268) and TZM-bl (catalog no. PTA-5659) cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; Gibco BRL, Grand Island, NY, USA), 100 μg/ml penicillin-streptomycin, and 1 mM Na pyruvate. H9 (catalog no. HTB-176; ATCC) and Jurkat (catalog no. TIB-152; ATCC) cells were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). The Jurkat C11 cell line was a gift from H. Z. Zhu (The College of Life Science, Fudan University). J-Lat 6.3 (catalog no. 9846), ACH-2 (catalog no. 349), MT4 (catalog no. 120), and CEM (catalog no. 117) cells were obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. H9, Jurkat, J-Lat 6.3, Jurkat C11, MT4, and CEM cells were maintained in RPMI 1640 (HyClone) containing 10% FBS, l-glutamine, and penicillin-streptomycin. ACH-2 cells were cultured in RPMI 1640 supplemented with 10% heat-inactivated FBS, 1% HEPES, 1% l-glutamine, and penicillin-streptomycin. PBMCs were isolated from blood samples of research donors through Ficoll gradient centrifugation, and the CD4+ T lymphocytes were then purified from PBMCs with anti-CD4-specific antibody-coated microbeads (Miltenyi Biotec, Germany) according to the manufacturer's instructions. Primary CD4+ T cells were maintained with 1/1,000 recombinant human interleukin 2 (IL-2) to maintain proliferation in RPMI 1640 supplemented with 1% penicillin-streptomycin, 1% l-glutamine, and 10% FBS. All cell lines were cultured at 37°C in a humidified atmosphere containing 5% CO2.
The following antibodies were used: mouse anti-HA monoclonal antibody (MAb) (901514; BioLegend, San Diego, CA, USA), mouse anti-myc MAb (05-724; Millipore, Burlington, MA, USA), mouse anti-β-actin MAb (A00702-100; Genscript, Piscataway, NJ, USA), goat anti-human histone polyclonal antibody (PAb) (A0150240; Genscript, Piscataway, NJ, USA), mouse anti-p24 MAb (catalog no. 1513; AIDS Research and Reference Reagents Program [ARRRP], USA), rabbit polyclonal to histone H3 (trimethyl K4) antibody (Abcam, 8580), rabbit polyclonal to histone H3 (acetyl K27) antibody (Abcam, 4729), rabbit polyclonal to SP1 antibody(Abcam, 227383), mouse-anti-NFAT monoclonal antibody (Abcam, 2722), and rabbit anti-PMEPA1 polyclonal antibody (Proteintech, 16521-1-ap).
Transfection and immunoblot analysis.
A total of 3 × 105 cells of adherent cell lines were seeded in 12-well plates and were then transfected using Lipofectamine 2000 (Invitrogen), while TZM-bl cells were transfected using Lipofectamine 3000 (Invitrogen) as recommended by the manufacturer. Human CD4+ T cells or T cell lines were nucleofected using an Amaxa Human T Cell Nucleofector kit with the U-014 program or kit V with the X-001 program (Lonza, Switzerland).
Cells were harvested 48 h after transfection or stimulation. Cell samples were boiled in 1× loading buffer (0.08 M Tris [pH 6.8], 2.0% SDS, 10% glycerol, 0.1 M dithiothreitol, and 0.2% bromophenol blue) and subsequently separated on a 12% polyacrylamide gel. Proteins were transferred to nitrocellulose membranes, and membranes were probed with various primary antibodies against the proteins of interest. Primary antibodies were diluted with 1% milk in phosphate-buffered saline (PBS), and incubation with primary antibodies was followed by incubation with a corresponding alkaline phosphatase (AP)-conjugated secondary antibody (Jackson ImmunoResearch, West Grove, PA, USA). Protein staining was carried out with the substrates 5-bromo-4-chloro-3-indolylphosphate (BCIP) and nitroblue tetrazolium (NBT) obtained from Sigma (St. Louis, MO, USA).
Chemical synthesis of short interfering RNA.
To knock down NKILA, chemically synthesized siRNA and nonspecific control siRNA were purchased from RiboBio (Guangzhou, China). RNAi against NKILA was carried out using a pool of three duplexed short interfering RNAs. The siRNA sequences were as follows (F indicates forward and R indicates reverse): the duplex 1 (siG151105032412) F1 sequence was GCCAGAAACUCUCCAAAUA-deoxyribosylthymine (dT)-dT, the duplex 1 R1 sequence was dTdTUAUUUGGAGAGUUUCUGGC, the duplex 2 (siG151105032422) F2 sequence was CAGGAGUGCUACAAGAACAdTdT, the duplex 2 R2 sequence was dTdTUGUUCUUGUAGCACUCCUG, the duplex 3 (siG151105032434) F3 sequence was CGCUGCAACUUAAGAGAAAdTdT, and the duplex 3 R3 sequence was dTdTUUUCUCUUAAGUUGCAGCG.
Cell proliferation assay.
A total of approximately 1 × 103 cells were plated in 96-well plates. Cell proliferation was assessed using Cell Counting Kit 8 (Transgen Biotech, Beijing, China) according to the manufacturer’s protocol. Absorbance at 450 nm was recorded using an iMark microplate reader (Bio-Rad, Hercules, CA) at 48 h.
HIV-1 production, infection, and reactivation.
HIV-1 virus was produced by transfecting pNL4-3 plasmids into HEK293T cells with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Forty-eight hours later, supernatants were collected, and HIV-1 virus was quantified using an HIV-1 p24 ELISA kit (Hebei Medical University, China).
TZM-bl cells containing an integrated HIV-1 LTR promoter, which were derived from HeLa cells, were used to assess infectious HIV-1 production. TZM-bl cells were seeded in a 24-well plate overnight and were then infected with supernatant containing HIV-1 viral particles. Forty-eight hours postinfection, the infected cells were harvested, and luciferase activity was measured with a GloMax 20/20 luminometer (Promega).
A total of 2 × 106 CD4+ T cells isolated from healthy donors or T cell lines were nucleofected with the NKILA or control vector. Forty-eight hours posttransfection, cells were infected with an equivalent volume of supernatant containing 2 ng HIV-1 p24 antigen for 4 h. Cells were washed 3 times with PBS and maintained in RPMI 1640 medium supplemented with IL-2 (10 ng/ml). Cell supernatants were collected on various days postinfection and analyzed using the HIV-1 p24 ELISA kit according to the manufacturer’s instructions.
To measure viral reactivation, HIV-1 latently infected J-Lat 6.3 and ACH-2 cells were nucleofected with siRNA NKILA or treated with the latent reactivation agent PMA (1 μM) as appropriate for 48 h. The green fluorescent protein (GFP)-positive J-Lat 6.3 cells were analyzed by flow cytometry (FACSCalibur; BD, Franklin Lakes, NJ, USA) for green fluorescence (FL1, 488 nm), while p24 expression in ACH-2 cells was measured by immunoblotting.
Purified CD4+ T cells isolated from HIV-1-infected individuals who had undergone HAART were nucleofected with the NKILA or control vector, and then the cells were cultured for another 2 days. Cells were collected for qRT-PCR or stimulated with phytohemagglutinin M (PHA-M) (5 ng/ml; Sigma-Aldrich) for 7 days. HIV-1 activation was detected by measuring the released p24 amount in the supernatants by ELISA.
Lentiviral production, transduction, and infection.
The full-length human NKILA sequence was cloned into the lentiviral expression vector pLVX-IRES-neo (Clontech Laboratories Inc., San Francisco, CA). Using a four-plasmid transient-cotransfection method, replication-defective vesicular stomatitis virus G protein-pseudotyped viral particles were packaged in HEK293T cells. Lentiviruses were harvested to infect MT4 cell lines. At 48 h postinfection, puromycin (1 g/ml) was added to the medium for selection. In order to remove the dead cells, the cell culture medium was replaced every 2 days. Five to 7 days later, expression of NKILA was detected by qRT-PCR. Two nanograms, 0.2 ng, or 0.02 ng HIV-1 p24 antigen was used to infect NKILA-overexpressing MT4 cells for 4 h. Cells were washed 3 times with PBS and maintained in RPMI 1640 medium. The supernatants were harvested and production of p24 antigen was measured by ELISA at 1 day, 3 days, 5 days, 7 days, and 9 days after infection.
Luciferase assay.
HEK293T cells were precultured in a 12-well cell culture plate and transfected using Lipofectamine 2000 according to the manufacturer’s instructions. Forty-eight hours posttransfection, cells were collected, and a dual luciferase reporter assay was performed using the Promega dual-luciferase reporter assay system (Promega) according to the manufacturer’s instructions with the GloMax 20/20 luminometer (Promega).
ChIP-qPCR.
Chromatin immunoprecipitation (ChIP) was performed with a chromatin immunoprecipitation kit (Millipore, 17-371). Approximately 4 × 106 cells were used for each immunoprecipitation (IP). Briefly, Jurkat cells were transfected with the NKILA or negative-control vector for 24 h and infected with supernatants containing HIV-1 viral particles for 48 h. For cross-linking proteins to DNA, the infected cells were treated with 1% formaldehyde (Sigma-Aldrich) for 10 min at room temperature. Unreacted formaldehyde was quenched by 125 mM glycine for 5 min at room temperature and centrifugation at 700 × g for 5 min at 4°C. The cell pellets were washed with cold PBS three times and resuspended in 1 ml SDS lysis buffer containing 1× protease inhibitor cocktail II. The cell lysates were sonicated on wet ice with 5 sets of 10-s pulses using a Cole-Parmer instrument to acquire the appropriate DNA fragment size between 150 and 900 bp and were centrifuged at 12,000 × g for 10 min at 4°C. The supernatants were collected and precleared by incubation with 60 μl of protein G agarose for 1 h at 4°C with rotation. The agarose was discarded by centrifugation at 5,000 × g for 1 min at 4°C, and the supernatants were transferred to new tubes for immunoprecipitation. Normal rabbit IgG (CST, 2729) and anti-p65 antibody (10745-1-AP) were separately added to the NKILA and negative vector groups, respectively. Immunoprecipitation was carried out overnight at 4°C with rotation with normal rabbit IgG or rabbit anti-p65 antibody, and incubation was continued with 60 μl of protein G agarose for another 2 h at 4°C with rotation. Low-salt immune complex wash buffer, high-salt immune complex wash buffer, and LiCl immune complex wash buffer were used sequentially one time each to wash the agarose, and Tris-EDTA (TE) was then used for two washes on a rotating platform, with brief centrifugation at 5,000 × g for 1 min. Then, 100 μl of elution buffer was added to each antibody/agarose complex for 15 min at room temperature, and the supernatants were collected by centrifugation at 3,000 × g for 1 min. The supernatants were incubated first with 8 μl of 5 M NaCl at 65°C for 4 h to reverse the DNA-protein cross-links and then with RNase A to remove RNA. Five volumes of bind reagent A were added to 1 volume of the supernatants. The supernatant/bind reagent A mixture was transferred to a filter in a collection tube and was then centrifuged at 12,000 × g for 30 s. Wash reagent B was used to wash the spin filter in the collection tube by centrifugation at 12,000 × g two times for 30 s each, and 50 μl of elution buffer C was added directly to the center of the spin filter membrane. The eluate was collected by centrifugation at 12,000 × g for 30 s and was then analyzed immediately by qRT-PCR. The quantitation regions targeted by ChIP primers are listed below. The HIV-1 LTR-F1 sequence was GGAGTACTACAAAGACTGCT, the LTR-R1 sequence was TAACCAGAGAGACCCAGTA, the LTR-F2 sequence was TGGAAGGGCTAATTTGGTC, the LTR-R2 sequence was CTGGCCCTGGTGTGTAGTTC, the LTR-F3 sequence was TAAAGCTTGCCTTGAGTGCT, and the LTR-R3 sequence was TGCTAGAGATTTTCCACACTGA. The NKILA promoter forward sequence was CGTTTTCGTCGTCGTCGTC, and NKILA promoter reverse sequence was CTCGACGAAAATTAACGCAAC. All ChIP-qPCR DNA signals were normalized to the negative vector group cellular DNA precipitated with IgG.
Co-immunoprecipitation.
Jurkat cells stably expressing NKILA were harvested and disrupted with lysis buffer (PBS containing 1% Triton X-100 and complete protease inhibitor cocktail [Roche]) at 4°C for 1 h. Cell lysates were clarified by centrifugation at 10,000 × g for 30 min at 4°C. Precleared cell lysates were mixed with anti-IgG antibody-conjugated agarose beads, anti-NFAT antibody-conjugated agarose beads, or anti-p65 antibody-conjugated agarose beads and incubated at 4°C for 4 h on an end-over-end rocker. The reaction mixtures were then washed six times with cold wash buffer (20 mM Tris-HCl [pH 7.5], 100 mM NaCl, 0.1 mM EDTA, 0.05% Tween 20) and subsequently analyzed by Western blotting or RNA was extracted for qRT-PCR analysis.
RNA pulldown assay.
RNA of NKILA was transcribed with a MEGAscriptTM T7 kit (Ambion, Austin, TX, USA), and RNA purified with a MEGA clear kit (Ambion) was annealed with the DNA probe (5′-TCCAGTTAAATTGAGATATACTTAC-3′) labeled with biotin by a Biotin 3′ End DNA Labeling kit (catalog no. 89818; Thermo). To be specific, 4 μg biotinylated RNA was heated to 90°C for 2 min to disrupt the secondary structure and placed on ice for 2 min to form the proper secondary structure for the pulldown assay. Then, 5 × 106 cells were lysed in lysis buffer as described for the co-IP assay supplemented with 20 U Protector RNase inhibitor (Roche) and centrifuged to get the clear cell lysate. Then, the lysate was precleared with 50 μl streptavidin-Sepharose beads (BioVision, Milpitas, CA, USA) by rotating at 4°C for 30 min. Then, folded RNA was added to the precleared cell lysate and rotated at 4°C for 3 h. One hundred microliters of beads was added to the reaction mixture and rotated at 4°C for 1 h followed by five washes using the wash buffer described for the co-IP assay supplemented with 20 U Protector RNase inhibitor (Roche). The proteins on beads were detected by Western blotting.
Statistical analysis.
All data represent the results from three independent experiments and are presented as the means ± standard deviations (SDs). Statistical significance was calculated using Student's t tests.
ACKNOWLEDGMENTS
We thank C. Y. Dai for the critical reagents. We thank the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), for critical reagents. We thank H. Z. Zhu (Fudan University School of Life Sciences) for the gift of latent cell line C11.
This work was supported in part by funding from the National Natural Science Foundation of China (81672004 and 31270202 to W.Z. and 81801993 to H.W.), the Jilin University Science and Technology Innovative Research Team (JLUSTIRT 2017TD-05 to W.Z.), the Science and Technology Department of Jilin Province (20190101003JH to W.Z. and 20190201272JC to H.W.), the Key Laboratory of Molecular Virology, Jilin Province (20102209 to W.Z.), the China Postdoctoral Science Foundation (2018M631869 to H.W.), and the Chinese Ministry of Science and Technology (2018ZX10302104-001-010 to B.-S.Z.).
We declare no conflict of interest.
REFERENCES
- 1.Brooks DG, Kitchen SG, Kitchen CM, Scripture-Adams DD, Zack JA. 2001. Generation of HIV latency during thymopoiesis. Nat Med 7:459–464. doi: 10.1038/86531. [DOI] [PubMed] [Google Scholar]
- 2.Courouce AM. 1987. Latency preceding seroconversion in sexually transmitted HIV infection. Lancet 330:1025. doi: 10.1016/S0140-6736(87)92587-6. [DOI] [PubMed] [Google Scholar]
- 3.Marcello A. 2006. Latency: the hidden HIV-1 challenge. Retrovirology 3:7. doi: 10.1186/1742-4690-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCune JM. 1995. Viral latency in HIV disease. Cell 82:183–188. doi: 10.1016/0092-8674(95)90305-4. [DOI] [PubMed] [Google Scholar]
- 5.Pomerantz RJ, Trono D, Feinberg MB, Baltimore D. 1990. Cells nonproductively infected with HIV-1 exhibit an aberrant pattern of viral RNA expression: a molecular model for latency. Cell 61:1271–1276. doi: 10.1016/0092-8674(90)90691-7. [DOI] [PubMed] [Google Scholar]
- 6.Saah AJ. 1987. Latency preceding seroconversion in sexually transmitted HIV infection. Lancet 2:1402. doi: 10.1016/s0140-6736(87)91296-7. [DOI] [PubMed] [Google Scholar]
- 7.Cary DC, Fujinaga K, Peterlin BM. 2016. Molecular mechanisms of HIV latency. J Clin Invest 126:448–454. doi: 10.1172/JCI80565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coiras M, López-Huertas MR, Pérez-Olmeda M, Alcamí J. 2009. Understanding HIV-1 latency provides clues for the eradication of long-term reservoirs. Nat Rev Microbiol 7:798–812. doi: 10.1038/nrmicro2223. [DOI] [PubMed] [Google Scholar]
- 9.Donahue DA, Kuhl BD, Sloan RD, Wainberg MA. 2012. The viral protein Tat can inhibit the establishment of HIV-1 latency. J Virol 86:3253–3263. doi: 10.1128/JVI.06648-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donahue DA, Wainberg MA. 2013. Cellular and molecular mechanisms involved in the establishment of HIV-1 latency. Retrovirology 10:11. doi: 10.1186/1742-4690-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson I, Low JS, Weston S, Weinberger M, Zhyvoloup A, Labokha AA, Corazza G, Kitson RA, Moody CJ, Marcello A, Fassati A. 2014. Heat shock protein 90 controls HIV-1 reactivation from latency. Proc Natl Acad Sci U S A 111:E1528–E1537. doi: 10.1073/pnas.1320178111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Y, Karijolich J, Glaunsinger B, Zhou Q. 2016. Pseudouridylation of 7SK snRNA promotes 7SK snRNP formation to suppress HIV-1 transcription and escape from latency. EMBO Rep 17:1441–1451. doi: 10.15252/embr.201642682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan JK, Greene WC. 2011. NF-kappaB/Rel: agonist and antagonist roles in HIV-1 latency. Curr Opin HIV AIDS 6:12–18. doi: 10.1097/COH.0b013e32834124fd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaudhary P, Khan SZ, Rawat P, Augustine T, Raynes DA, Guerriero V, Mitra D. 2016. HSP70 binding protein 1 (HspBP1) suppresses HIV-1 replication by inhibiting NF-kappaB mediated activation of viral gene expression. Nucleic Acids Res 44:1613–1629. doi: 10.1093/nar/gkv1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heusinger E, Kirchhoff F. 2017. Primate lentiviruses modulate NF-kappaB activity by multiple mechanisms to fine-tune viral and cellular gene expression. Front Microbiol 8:198. doi: 10.3389/fmicb.2017.00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nabel G, Baltimore D. 1987. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature 326:711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- 17.Canton J, Fehr AR, Fernandez-Delgado R, Gutierrez-Alvarez FJ, Sanchez-Aparicio MT, García-Sastre A, Perlman S, Enjuanes L, Sola I. 2018. MERS-CoV 4b protein interferes with the NF-kappaB-dependent innate immune response during infection. PLoS Pathog 14:e1006838. doi: 10.1371/journal.ppat.1006838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramasamy S, Saez B, Mukhopadhyay S, Ding D, Ahmed AM, Chen X, Pucci F, Yamin R, Wang J, Pittet MJ, Kelleher CM, Scadden DT, Sweetser DA. 2016. Tle1 tumor suppressor negatively regulates inflammation in vivo and modulates NF-kappaB inflammatory pathway. Proc Natl Acad Sci U S A 113:1871–1876. doi: 10.1073/pnas.1511380113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tietjen I, Williams DE, Read S, Kuang XT, Mwimanzi P, Wilhelm E, Markle T, Kinloch NN, Naphen CN, Tenney K, Mesplede T, Wainberg MA, Crews P, Bell B, Andersen RJ, Brumme ZL, Brockman MA. 2018. Inhibition of NF-kappaB-dependent HIV-1 replication by the marine natural product bengamide A. Antiviral Res 152:94–103. doi: 10.1016/j.antiviral.2018.02.017. [DOI] [PubMed] [Google Scholar]
- 20.Vento-Tormo R, Rodríguez-Ubreva J, Lisio LD, Islam ABMMK, Urquiza JM, Hernando H, López-Bigas N, Shannon-Lowe C, Martínez N, Montes-Moreno S, Piris MA, Ballestar E. 2014. NF-kappaB directly mediates epigenetic deregulation of common microRNAs in Epstein-Barr virus-mediated transformation of B-cells and in lymphomas. Nucleic Acids Res 42:11025–11039. doi: 10.1093/nar/gku826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wullaert A, Bonnet MC, Pasparakis M. 2011. NF-kappaB in the regulation of epithelial homeostasis and inflammation. Cell Res 21:146–158. doi: 10.1038/cr.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, Guo H, Su J, Rui Y, Zheng W, Gao W, Zhang W, Li Z, Liu G, Markham RB, Wei W, Yu XF. 2017. Inhibition of Vpx-mediated SAMHD1 and Vpr-mediated host helicase transcription factor degradation by selective disruption of viral CRL4 (DCAF1) E3 ubiquitin ligase assembly. J Virol 91:e00225-17. doi: 10.1128/JVI.00225-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stroud JC, Oltman A, Han A, Bates DL, Chen L. 2009. Structural basis of HIV-1 activation by NF-kappaB–a higher-order complex of p50:RelA bound to the HIV-1 LTR. J Mol Biol 393:98–112. doi: 10.1016/j.jmb.2009.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sauter D, Hotter D, Van Driessche B, Sturzel CM, Kluge SF, Wildum S, Yu H, Baumann B, Wirth T, Plantier JC, Leoz M, Hahn BH, Van Lint C, Kirchhoff F. 2015. Differential regulation of NF-kappaB-mediated proviral and antiviral host gene expression by primate lentiviral Nef and Vpu proteins. Cell Rep 10:586–599. doi: 10.1016/j.celrep.2014.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang HC, Xing S, Shan L, O'Connell K, Dinoso J, Shen A, Zhou Y, Shrum CK, Han Y, Liu JO, Zhang H, Margolick JB, Siliciano RF. 2009. Small-molecule screening using a human primary cell model of HIV latency identifies compounds that reverse latency without cellular activation. J Clin Invest 119:3473–3486. doi: 10.1172/JCI39199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang G, Mendes EA, Kaiser P, Sankaran-Walters S, Tang Y, Weber MG, Melcher GP, Thompson GR III, Tanuri A, Pianowski LF, Wong JK, Dandekar S. 2014. Reactivation of HIV latency by a newly modified Ingenol derivative via protein kinase Cdelta-NF-kappaB signaling. AIDS 28:1555–1566. doi: 10.1097/QAD.0000000000000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pérez M, de Vinuesa AG, Sanchez-Duffhues G, Marquez N, Bellido ML, Muñoz-Fernandez MA, Moreno S, Castor TP, Calzado MA, Muñoz E. 2010. Bryostatin-1 synergizes with histone deacetylase inhibitors to reactivate HIV-1 from latency. Curr HIV Res 8:418–429. doi: 10.2174/157016210793499312. [DOI] [PubMed] [Google Scholar]
- 28.Reuse S, Calao M, Kabeya K, Guiguen A, Gatot JS, Quivy V, Vanhulle C, Lamine A, Vaira D, Demonte D, Martinelli V, Veithen E, Cherrier T, Avettand V, Poutrel S, Piette J, de Launoit Y, Moutschen M, Burny A, Rouzioux C, De Wit S, Herbein G, Rohr O, Collette Y, Lambotte O, Clumeck N, Van Lint C. 2009. Synergistic activation of HIV-1 expression by deacetylase inhibitors and prostratin: implications for treatment of latent infection. PLoS One 4:e6093. doi: 10.1371/journal.pone.0006093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams SA, Kwon H, Chen LF, Greene WC. 2007. Sustained induction of NF-kappa B is required for efficient expression of latent human immunodeficiency virus type 1. J Virol 81:6043–6056. doi: 10.1128/JVI.02074-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerritsen ME, Williams AJ, Neish AS, Moore S, Shi Y, Collins T. 1997. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc Natl Acad Sci U S A 94:2927–2932. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. 2001. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol Cell 8:327–337. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- 32.Huang D, Chen J, Yang L, Ouyang Q, Li J, Lao L, Zhao J, Liu J, Lu Y, Xing Y, Chen F, Su F, Yao H, Liu Q, Su S, Song E. 2018. NKILA lncRNA promotes tumor immune evasion by sensitizing T cells to activation-induced cell death. Nat Immunol 19:1112–1125. doi: 10.1038/s41590-018-0207-y. [DOI] [PubMed] [Google Scholar]
- 33.Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, Lin L, Yao H, Su F, Li D, Zeng M, Song E. 2015. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 27:370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 34.Weidle UH, Birzele F, Kollmorgen G, Ruger R. 2017. Long non-coding RNAs and their role in metastasis. Cancer Genomics Proteomics 14:143–160. doi: 10.21873/cgp.20027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ke S, Li RC, Meng FK, Fang MH. 2018. NKILA inhibits NF-kappaB signaling and suppresses tumor metastasis. Aging (Albany NY) 10:56–71. doi: 10.18632/aging.101359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huan C, Li Z, Ning S, Wang H, Yu XF, Zhang W. 2018. Long noncoding RNA uc002yug.2 activates HIV-1 latency through regulation of mRNA levels of various RUNX1 isoforms and increased Tat expression. J Virol 92:e01844-17. doi: 10.1128/JVI.01844-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dijkstra JM, Alexander DB. 2015. The “NF-k B interacting long noncoding RNA” (NKILA) transcript is antisense to cancer-associated gene PMEPA1. F1000Res 4:96. doi: 10.12688/f1000research.6400.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia JA, Ou SH, Wu F, Lusis AJ, Sparkes RS, Gaynor RB. 1992. Cloning and chromosomal mapping of a human immunodeficiency virus 1 “TATA” element modulatory factor. Proc Natl Acad Sci U S A 89:9372–9376. doi: 10.1073/pnas.89.20.9372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blackard JT, Renjifo B, Fawzi W, Hertzmark E, Msamanga G, Mwakagile D, Hunter D, Spiegelman D, Sharghi N, Kagoma C, Essex M. 2001. HIV-1 LTR subtype and perinatal transmission. Virology 287:261–265. doi: 10.1006/viro.2001.1059. [DOI] [PubMed] [Google Scholar]
- 40.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73:152–160. doi: 10.1128/JVI.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verma A, Rajagopalan P, Lotke R, Varghese R, Selvam D, Kundu TK, Ranga U. 2016. Functional incompatibility between the generic NF-kappaB motif and a subtype-specific Sp1III element drives the formation of the HIV-1 subtype C viral promoter. J Virol 90:7046–7065. doi: 10.1128/JVI.00308-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chao TC, Zhang Q, Li Z, Tiwari SK, Qin Y, Yau E, Sanchez A, Singh G, Chang K, Kaul M, Karris MAY, Rana TM. 2019. The long noncoding RNA HEAL regulates HIV-1 replication through epigenetic regulation of the HIV-1 promoter. mBio 10:e02016-19. doi: 10.1128/mBio.02016-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan JK, Greene WC. 2012. Dynamic roles for NF-kappaB in HTLV-I and HIV-1 retroviral pathogenesis. Immunol Rev 246:286–310. doi: 10.1111/j.1600-065X.2012.01094.x. [DOI] [PubMed] [Google Scholar]
- 44.Perkins ND, Edwards NL, Duckett CS, Agranoff AB, Schmid RM, Nabel GJ. 1993. A cooperative interaction between NF-kappa B and Sp1 is required for HIV-1 enhancer activation. EMBO J 12:3551–3558. doi: 10.1002/j.1460-2075.1993.tb06029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.West MJ, Lowe AD, Karn J. 2001. Activation of human immunodeficiency virus transcription in T cells revisited: NF-kappaB p65 stimulates transcriptional elongation. J Virol 75:8524–8537. doi: 10.1128/jvi.75.18.8524-8537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alcami J, Lain de Lera T, Folgueira L, Pedraza MA, Jacque JM, Bachelerie F, Noriega AR, Hay RT, Harrich D, Gaynor RB. 1995. Absolute dependence on kappa B responsive elements for initiation and Tat-mediated amplification of HIV transcription in blood CD4 T lymphocytes. EMBO J 14:1552–1560. doi: 10.1002/j.1460-2075.1995.tb07141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams SA, Chen LF, Kwon H, Ruiz-Jarabo CM, Verdin E, Greene WC. 2006. NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J 25:139–149. doi: 10.1038/sj.emboj.7600900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. 2009. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A 106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lazar DC, Morris KV, Saayman SM. 2016. The emerging role of long non-coding RNAs in HIV infection. Virus Res 212:114–126. doi: 10.1016/j.virusres.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qu D, Sun WW, Li L, Ma L, Sun L, Jin X, Li T, Hou W, Wang JH. 2019. Long noncoding RNA MALAT1 releases epigenetic silencing of HIV-1 replication by displacing the polycomb repressive complex 2 from binding to the LTR promoter. Nucleic Acids Res 47:3013–3027. doi: 10.1093/nar/gkz117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bian D, Gao C, Bao K, Song G. 2017. The long non-coding RNA NKILA inhibits the invasion-metastasis cascade of malignant melanoma via the regulation of NF-kB. Am J Cancer Res 7:28–40. [PMC free article] [PubMed] [Google Scholar]
- 53.Burnett JC, Lim KI, Calafi A, Rossi JJ, Schaffer DV, Arkin AP. 2010. Combinatorial latency reactivation for HIV-1 subtypes and variants. J Virol 84:5958–5974. doi: 10.1128/JVI.00161-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ilyinskii PO, Simon MA, Czajak SC, Lackner AA, Desrosiers RC. 1997. Induction of AIDS by simian immunodeficiency virus lacking NF-kappaB and SP1 binding elements. J Virol 71:1880–1887. doi: 10.1128/JVI.71.3.1880-1887.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pohlmann S, Floss S, Ilyinskii PO, Stamminger T, Kirchhoff F. 1998. Sequences just upstream of the simian immunodeficiency virus core enhancer allow efficient replication in the absence of NF-kappaB and Sp1 binding elements. J Virol 72:5589–5598. doi: 10.1128/JVI.72.7.5589-5598.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sauter D, Kirchhoff F. 2018. Multilayered and versatile inhibition of cellular antiviral factors by HIV and SIV accessory proteins. Cytokine Growth Factor Rev 40:3–12. doi: 10.1016/j.cytogfr.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 57.Heigele A, Kmiec D, Regensburger K, Langer S, Peiffer L, Sturzel CM, Sauter D, Peeters M, Pizzato M, Learn GH, Hahn BH, Kirchhoff F. 2016. The potency of Nef-mediated SERINC5 antagonism correlates with the prevalence of primate lentiviruses in the wild. Cell Host Microbe 20:381–391. doi: 10.1016/j.chom.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jia X, Zhao Q, Xiong Y. 2015. HIV suppression by host restriction factors and viral immune evasion. Curr Opin Struct Biol 31:106–114. doi: 10.1016/j.sbi.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HC, Rice GI, Christodoulou E, Walker PA, Kelly G, Haire LF, Yap MW, de Carvalho LP, Stoye JP, Crow YJ, Taylor IA, Webb M. 2011. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480:379–382. doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- 60.Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J. 2011. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474:658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kluge SF, Sauter D, Kirchhoff F. 2015. SnapShot: antiviral restriction factors. Cell 163:774–774.e1. doi: 10.1016/j.cell.2015.10.019. [DOI] [PubMed] [Google Scholar]
- 62.Sheehy AM, Gaddis NC, Malim MH. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat Med 9:1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- 63.Simon V, Bloch N, Landau NR. 2015. Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nat Immunol 16:546–553. doi: 10.1038/ni.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 65.Wei W, Guo H, Ma M, Markham R, Yu XF. 2016. Accumulation of MxB/Mx2-resistant HIV-1 capsid variants during expansion of the HIV-1 epidemic in human populations. EBioMedicine 8:230–236. doi: 10.1016/j.ebiom.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang W, Du J, Evans SL, Yu Y, Yu XF. 2011. T-cell differentiation factor CBF-beta regulates HIV-1 Vif-mediated evasion of host restriction. Nature 481:376–379. doi: 10.1038/nature10718. [DOI] [PubMed] [Google Scholar]
- 67.Lu Z, Li Y, Wang J, Che Y, Sun S, Huang J, Chen Z, He J. 2017. Long non-coding RNA NKILA inhibits migration and invasion of non-small cell lung cancer via NF-kappaB/Snail pathway. J Exp Clin Cancer Res 36:54. doi: 10.1186/s13046-017-0518-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bustin SA, Benes V, Garson J, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley G, Wittwer CT, Schjerling P, Day PJ, Abreu M, Aguado B, Beaulieu J-F, Beckers A, Bogaert S, Browne JA, Carrasco-Ramiro F, Ceelen L, Ciborowski K, Cornillie P, Coulon S, Cuypers A, De Brouwer S, De Ceuninck L, De Craene J, De Naeyer H, De Spiegelaere W, Deckers K, Dheedene A, Durinck K, Ferreira-Teixeira M, Fieuw A, Gallup JM, Gonzalo-Flores S, Goossens K, Heindryckx F, Herring E, Hoenicka H, Icardi L, Jaggi R, Javad F, Karampelias M, Kibenge F, Kibenge M, Kumps C, Lambertz I, Lammens T, et al. 2013. The need for transparency and good practices in the qPCR literature. Nat Methods 10:1063–1067. doi: 10.1038/nmeth.2697. [DOI] [PubMed] [Google Scholar]