We screened rabbit MHC class I RLA-A1-restricted peptides from the capsid protein VP60 of rabbit hemorrhagic disease virus (RHDV) and determined the structures of RLA-A1 complexed with three peptides, VP60-1, VP60-2, and VP60-10. From the structures, we found that the peptide binding motifs of RLA-A1 are extremely constraining. Thus, there is a generally restricted peptide selection for RLA-A1 compared to that for human HLA-A*0201. In addition, uncommon residues Gly53, Val55, and Glu56 of RLA-A1 are located between the 310 helix and α1 helix, which makes the steric position of the 310 helix in RLA-A1 much closer to the α2 helix than that found in other mammalian MHC class I molecules. This special conformation between the 310 helix and α1 helix plays a pivotal role in rabbit MHC class I assembly. Our results provide new insights into MHC class I molecule assembly and peptide presentation of domestic mammals. Furthermore, these data also broaden our knowledge on T-cell immunity in rabbits and may also provide useful information for vaccine development to prevent infectious diseases in rabbits.
KEYWORDS: rabbit, RHDV, MHC class I, RLA-A1, structure, peptide
ABSTRACT
Rabbits are pivotal domestic animals for both the economy and as an animal model for human diseases. A large number of rabbits have been infected by rabbit hemorrhagic disease virus (RHDV) in natural and artificial pandemics in the past. Differences in presentation of antigenic peptides by polymorphic major histocompatibility complex (MHC) molecules to T-cell receptors (TCR) on T lymphocytes are associated with viral clearance in mammals. Here, we screened and identified a series of peptides derived from RHDV binding to the rabbit MHC class I molecule, RLA-A1. The small, hydrophobic B and F pockets of RLA-A1 capture a peptide motif analogous to that recognized by human class I molecule HLA-A*0201, with more restricted aliphatic anchors at P2 and PΩ positions. Moreover, the rabbit molecule is characterized by an uncommon residue combination of Gly53, Val55, and Glu56, making the 310 helix and the loop between the 310 and α1 helices closer to the α2 helix. A wider A pocket in RLA-A1 can induce a special conformation of the P1 anchor and may play a pivotal role in peptide assembly and TCR recognition. Our study broadens the knowledge of T-cell immunity in domestic animals and also provides useful insights for vaccine development to prevent infectious diseases in rabbits.
IMPORTANCE We screened rabbit MHC class I RLA-A1-restricted peptides from the capsid protein VP60 of rabbit hemorrhagic disease virus (RHDV) and determined the structures of RLA-A1 complexed with three peptides, VP60-1, VP60-2, and VP60-10. From the structures, we found that the peptide binding motifs of RLA-A1 are extremely constraining. Thus, there is a generally restricted peptide selection for RLA-A1 compared to that for human HLA-A*0201. In addition, uncommon residues Gly53, Val55, and Glu56 of RLA-A1 are located between the 310 helix and α1 helix, which makes the steric position of the 310 helix in RLA-A1 much closer to the α2 helix than that found in other mammalian MHC class I molecules. This special conformation between the 310 helix and α1 helix plays a pivotal role in rabbit MHC class I assembly. Our results provide new insights into MHC class I molecule assembly and peptide presentation of domestic mammals. Furthermore, these data also broaden our knowledge on T-cell immunity in rabbits and may also provide useful information for vaccine development to prevent infectious diseases in rabbits.
INTRODUCTION
The European rabbit, Oryctolagus cuniculus, is found both in the wild and as domestic animals (1). Paleontological data show that this genus has undergone limited morphological diversification since its appearance at the end of the Miocene (1). Two species of rabbits were recognized from fossils, Oryctolagus lacosti in southern France and northwestern Italy and Oryctolagus laynensis in the Iberian Peninsula, with the latter being regarded as the species of origin of extant O. cuniculus (1).
Rabbits provide a very important animal model for human disease, especially immune-related diseases. Although rats are the most widely used animal model for osteoporosis research, rabbits show several advantages over rats for the modeling of osteoporosis, such as active Haversian remodeling analogous to that observed in humans, bone turnover faster than that of other models, and development of significant bone loss (2). In addition, rabbits can serve as a useful small-animal model for the study of acute and chronic hepatitis E virus (HEV) pathogenesis (3, 4). Previous work showed that rabbit HEV could cross the species barrier to infect humans (5). As the animal model of HEV, it is used not only for evaluating vaccine efficacy for humans but also for investigating the management of zoonotic transmission (6). Rabbits are also commonly used as animal models for several other infectious diseases (7), yet despite this, our knowledge of the adaptive immune system of Oryctolagus cuniculus remains limited.
Rabbit hemorrhagic disease virus (RHDV) is a calicivirus of the genus Lagovirus that causes rabbit hemorrhagic disease (RHD) in adult rabbits. This virus was first discovered in China in 1984. In less than 1 year, it killed about 140 million domestic rabbits in China and spread over 50,000 km2, which brought about large economic losses (8). RHDV subsequently spread around the world rapidly (9). In Australia and New Zealand, rabbits are considered to be an important agricultural pest which poses a threat to the endemic flora and fauna (10). RHDV was introduced to control rabbit numbers (11), resulting in a reduction by more than 95% of the wild rabbit population in some areas over several months. However, approximately 3% of the population survived after RHDV infection. Most of these surviving rabbits were 3- to 7-week-old rabbits with strong anti-RHDV antibodies (12). The role of the cellular immune response in these surviving rabbits was not evaluated. In most regions around the world, Oryctolagus cuniculus is mainly present as a domestic or industrial animal. RHDV infection continues to cause important economic losses, and the infection still has a significant negative ecological impact among wild rabbit populations and thus indirectly on their dependent predators (13, 14).
The capsid protein VP60 of RHDV is the main structural protein, and as the major target for the host immune defense against RHDV, it plays a pivotal role in virus diagnosis and vaccine design (15). Rabbits immunized with VP60 expressed by several heterologous systems showed full protection against challenge with lethal doses of RHDV (16–21). Studies showed that VP60 could induce strong neutralizing antibodies in rabbits after immunization (22, 23). In addition, VP60 could assemble virus-like particles (VLP), which are ideal candidate vectors for antigenic epitope delivery (24). Previous work showed that RHDV-VLP were essential for priming optimal cytotoxic T-lymphocyte (CTL) responses, which was very helpful for target antigen induction of cytotoxic killing of infected cells by CD8+ T cells in vivo (25). Cellular immunologic responses may also play an important role in RHDV clearance after immunization with VP60 vaccines in rabbits (26). However, the cellular immune responses in rabbits after viral infection and vaccination are as yet largely unknown.
Major histocompatibility complex class I (MHC-I) molecules play a vital role in activating CTLs by presenting antigen peptides to the surface of antigen-presenting cells (APCs) (27, 28). Rabbit MHC class I (RLA) was cloned in 1985 and comprises only one gene locus that can be transcribed and expressed in one RLA haplotype (29), which is different from human and mouse MHC class I genes which express multiple MHC class I antigens. Thus far, there are only six cDNA sequences of RLA deposited in the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/). The molecular basis of peptide presentation by RLA molecules remains largely unknown, which hinders the understanding of MHC class I-related adaptive immunity in rabbits.
In this study, we screened RLA-A1-restricted peptides from the capsid protein VP60 of RHDV and determined the structure of RLA-A1 complexed with three peptides, VP60-1, VP60-2, and VP60-10. This study expands our understanding of the T-cell immune response mediated by MHC class I in rabbits and may also provide useful information for vaccine development.
RESULTS
Overall structure of peptides binding to RLA-A1.
Thirteen peptides from capsid protein VP60 of RHDV were predicted in silico (http://www.cbs.dtu.dk/services/NetMHCpan/) to potentially bind to RLA-A1 and were verified by co-refolding in vitro (Table 1). Six of these 13 peptides bound to RLA-A1 (Fig. 1). Although VP60-8 and VP60-11 show weak binding to RLA-A1 in co-refolding, no soluble monomer complex with these peptides could be purified using Resource-Q anion-exchange chromatography (GE Healthcare), which was used for the other peptides that bound to RLA-A1 (data not shown). This indicates that VP60-8 and VP60-11 may have only transient binding to RLA-A1. The structures of three peptides, VP60-1, VP60-2, and VP60-10, presented by RLA-A1 were determined at the resolutions of 2.2 Å, 2.3 Å, and 2.6 Å, respectively (Table 2). There is only one RLA-A1 complex in one asymmetric unit for all three complex structures.
TABLE 1.
Potential binding peptides
| Namea | Position | Sequence | Scoreb | Refoldingc |
|---|---|---|---|---|
| VP60-1 | 542–550 | TLIDLTELI | 0.52206 | + |
| VP60-2 | 505–513 | ALMPGQFFV | 0.59905 | + |
| VP60-3 | 259–267 | GQIVGLQPV | 0.54241 | − |
| VP60-4 | 81–89 | SVADAPGSI | 0.46156 | + |
| VP60-5 | 136–44 | AVIPPGIEI | 0.43602 | + |
| VP60-6 | 487–495 | GSQPLPVTI | 0.48336 | − |
| VP60-7 | 536–544 | GTGASTTLI | 0.44076 | − |
| VP60-8 | 316–324 | WYANAGSAI | 0.47207 | − |
| VP60-9 | 191–199 | VLSVYNNLI | 0.51299 | + |
| VP60-10 | 341–349 | FVPFNSPNI | 0.44008 | + |
| VP60-11 | 517–525 | TFASGFMEI | 0.50784 | − |
| VP60-12 | 341–349 | GFMEIGLSV | 0.46530 | − |
| VP60-13 | 412–420 | NQNPTGLFV | 0.38125 | − |
All proteins were derived from major capsid protein VP60 of rabbit hemorrhagic disease virus (GenBank accession number DQ205345.1).
bValues were calculated by using http://www.cbs.dtu.dk/services/NetMHCpan/.
Peptides that can (+) or cannot (−) help the RLA-A1 heavy chain renature with human β2m.
FIG 1.
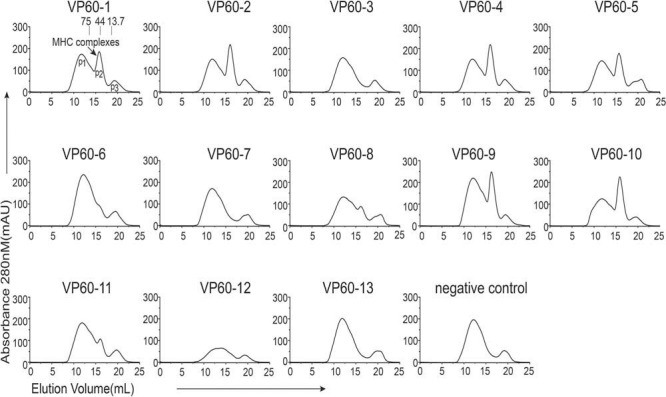
Screening of RHDV-derived peptides. Thirteen peptides from the capsid protein VP60 of RHDV were predicted in silico to potentially bind to RLA-A1 and were verified by refolding in vitro. RLA-A1 and hβ2m co-refolding without any peptides was used as a negative control. Peak 1 (P1), peak 2 (P2), and peak 3 (P3) represent the aggregated heavy chain, the correctly refolded heterotrimer RLA-A1/peptide complex, and the hβ2m, respectively.
TABLE 2.
X-ray data processing and refinement statistics
| Parameter | Value for:a
|
||
|---|---|---|---|
| RLA-A1/VP 60-1 | RLA-A1/VP 60-2 | RLA-A1/VP 60-10 | |
| PDB accession no. | 6M2J | 6M24 | 6M2K |
| Data processing | |||
| Space group | P212121 | P212121 | P212121 |
| Cell dimensions | |||
| a, b, c (Å) | 49.63, 74.36, 155.67 | 49.58, 76.22, 155.08 | 49.61, 74.30, 152.19 |
| α, β, γ (˚) | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 |
| Wavelength (Å) | 0.97911 | 1.03853 | 1.03923 |
| Resolution (Å) | 50.0–2.2 (2.28-2.2) | 50.0–2.3 (2.38–2.3) | 50.0–2.6 (2.69–2.6) |
| Total no. of reflections | 276,797 | 346,034 | 214,930 |
| No. of unique reflections | 29,903 | 27,014 | 18,060 |
| Completeness (%) | 99.7 (98.8) | 99.9 (100.0) | 99.9 (100.0) |
| Redundancy | 7.3 (9.3) | 12.2 (12.8) | 11.9 (12.2) |
| Rmerge (%)b | 17.4 (95.5) | 10.1 (85.7) | 11.2 (75.8) |
| I/σ | 2.41 | 2.75 | 2.78 |
| Refinement | |||
| Rwork (%)c | 20.2 | 19.4 | 21.5 |
| Rfree (%) | 25.6 | 24.6 | 29.4 |
| RMSD | |||
| Bond length (Å) | 0.009 | 0.008 | 0.009 |
| Bond angle (˚) | 1.250 | 1.200 | 1.270 |
| Avg B factor (Å2) | 38.27 | 36.77 | 40.34 |
| Ramachandran plot quality (%) | |||
| Favored (%) | 97.88 | 97.88 | 95.23 |
| Allowed (%) | 2.12 | 2.12 | 4.77 |
| Outliers (%) | 0.00 | 0.00 | 0.00 |
Numbers in parentheses represent the highest-resolution shell.
, where Ii refers to the observed intensity and is the average intensity of multiple observations of symmetry related reflections.
, where Rfree is calculated for a randomly chosen 5% of reflections, and Rwork is calculated for the remaining 95% of reflections used for structure refinement.
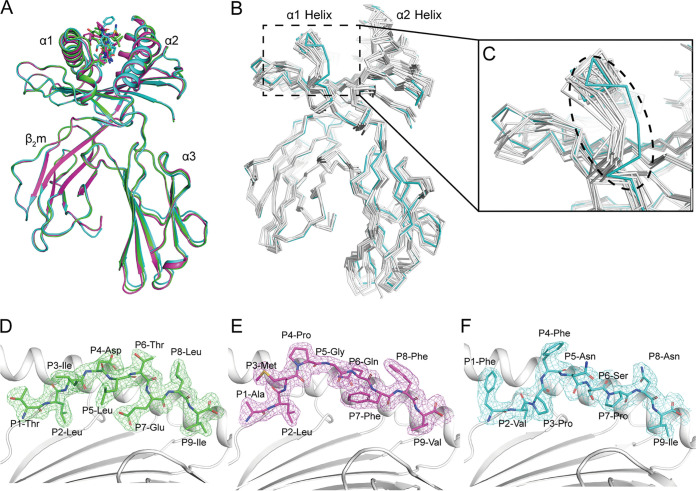
Overall, the structure of the domain arrangements and topologies of RLA-A1 displayed the common characteristics of classical MHC-I molecule structures determined thus far in other mammals. The extracellular region of the RLA-A1 heavy (H) chain was composed of the α1/α2 and α3 domains. The α1/α2 domains formed a peptide-binding groove (PBG) with two antiparallel α1 and α2 helices and eight-stranded β-sheets. Peptides presented by RLA-A1 lie along the PBG (Fig. 2A). The α3 of the H chain and β2-microglobulin (β2m) were typical immunoglobulin (Ig) domains under the PBG to stabilize the complex. The all-atom superimposition of RLA-A1/VP60-1, RLA-A1/VP60-2, and RLA-A1/VP60-10 showed similar overall conformations, with root mean square deviations (RMSDs) of 0.235 Å to 0.335 Å (Fig. 2A). The RMSDs of all atoms were between 0.581 Å and 1.213 Å for the superimposition of RLA-A1/VP60-1 onto other mammal MHC class I molecules, such as HLA-A*0201, macaque Mamu-A*01, equine Eqca-N*00601, canine DLA-88*50801, murine H2-Kd, feline FLA-E*01801, swine SLA-1*0401, and bovine N*01801 (Fig. 2B). The most distinct difference between RLA-A1 and other mammalian MHC class I molecules was located at the N terminus of the α1 helix, leading to a closer distance between the α1 helix of RLA-A1 and the N terminus of the α2 helix (Fig. 2B and C). The electron densities of VP60-1, VP60-2, and VP60-10 presented by RLA-A1 clearly exhibited the presenting conformations of the peptides with two major anchoring residues at the P2 and PΩ (P9 for the 9-mer) positions (Fig. 2D to F).
FIG 2.
Overall RLA-A1/peptide structures. (A) Structural alignment of RLA-A1/VP60-2 (magenta) and RLA-A1/VP60-10 (cyan) with RLA-A1/VP60-1 (green). The heavy chain of RLA-A1 and human β2m are shown as cartoons. Peptides are shown as sticks. (B) The superimposition of RLA-A1 (cyan) with other mammalian MHC-I molecules: human HLA-A*0201 (PDB code 3MGT), macaque Mamu-A*01 (PDB code 1ZVS), equine Eqca-N*00601 (PDB code 4ZUW), canine DLA-88*50801 (PDB code 5F1I), murine H2-Kd (PDB code 5GR7), feline FLA-E*01801 (PDB code 5XMF), swine SLA-1*0401 (PDB code 3QQ4), and bovine N*01801 (PDB code 3PWV) (all in white). (C) The main differences between RLA-A1 and other mammalian MHC-I molecules are located at the N terminus of the PBG, which is circled by a black dashed line. The authentic conformations of peptides VP60-1 (green) (D), VP60-2 (magenta) (E), and VP60-10 (cyan) (F) in the RLA-A1 PBG are shown in the 2Fo − Fc (where Fo and Fc are the observed and calculated structure factor amplitudes, respectively) electron groove density maps contoured at 1.0 s.
The small B pocket of RLA-A1 leads to a strict P2 anchor selection.
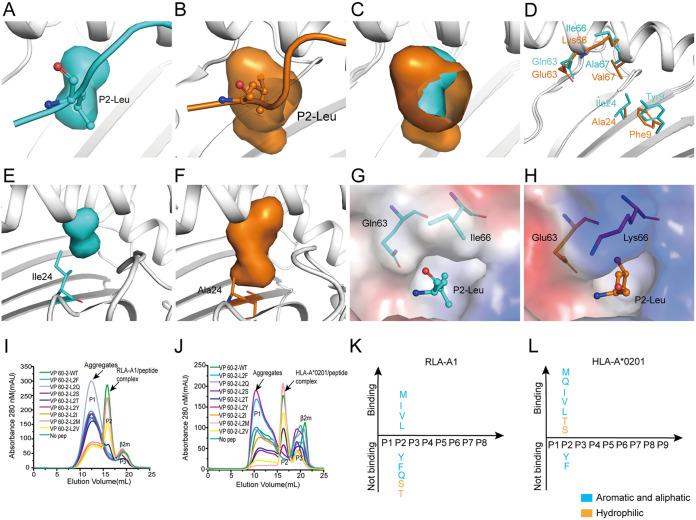
Previous studies reported that HLA-A*0201 has a hydrophobic B pocket which mainly accommodates aliphatic residues (Ala, Ser, Thr, Val, Leu, Ile, and Met) (30). Here, we found that the B pocket of RLA-A1 was also hydrophobic but much smaller than that of HLA-A*0201 (Fig. 3A to C). Further analysis indicated that the side chain of Ala24 in HLA-A*0201 lay at the bottom of the B pocket. However, in RLA-A1, an Ile24 residue at the same position made a much shallower pocket than that of HLA-A*0201 (Fig. 3E and F). Compared with HLA-A*0201 sequence, five amino acid mutations, F9Y, A24I, E63Q, K66I, and V67A, occurred in the B pocket of RLA-A1 (Fig. 3D). The inner side of the B pockets in both RLA-A1 and HLA-A*0201 were composed of hydrophobic amino acids and so could have anchors of aliphatic acids (Val, Leu, Ile, and Met) within both B pockets (Fig. 3I to L; Table 3). The B pocket entrance of HLA-A*0201 had an acidic amino acid, Glu63, and a basic amino acid, Lys66, which may be related to the accommodation of two hydrophilic amino acids, Ser and Thr, at the P2 anchor of the peptides (Fig. 3H). However, the two aliphatic acids, Gln63 and Ile66, at the same positions may prevent Ser and Thr anchoring in the B pocket of RLA-A1 (Fig. 3G). We then mutated P2-Leu of VP60-2 to P2-Phe, P2-Gln, P2-Ser, P2-The, P2-Tyr, P2-Ile, P2-Met, and P2-Val and refolded these mutated peptides with RLA-A1 and HLA-A*0201. Peptides VP60-2-L2M, VP60-2-L2Q, VP60-2-L2I, VP60-2-L2V, VP60-2-L2T, and VP60-2-L2S were presented by HLA-A*0201, while VP60-2-L2Y and VP60-2-L2F could not bind to HLA-A*0201. However, RLA-A1 bound to peptides VP60-2-L2M, VP60-2-L2Q, VP60-2-L2I, and VP60-2-L2V very well but could not bind to VP60-2-L2T, VP60-2-L2S, VP60-2-L2Y, and VP60-2-L2F (Fig. 3I to L; Table 3), indicating that the small and strong hydrophobic B pocket of RLA-A1 was much stricter in the choice of anchor P2 residue in peptides.
FIG 3.
The small and hydrophobic B pocket of RLA-A1. (A) Pocket B of RLA-A1 is shown as cyan cavities. P2-Leu of VP60-2 is shown as cyan sticks and spheres. The backbone of RLA-A1 is shown as a cartoon in white. (B) The B pocket of HLA-A*0201 is shown as orange cavities. P2-Leu of the peptide is shown as orange sticks and spheres. The backbone of HLA-A*0201 is shown as a cartoon in white. (C) Structure comparison of the B pocket between RLA-A1 and HLA-A*0201. (D) The different amino acids comprising the B pocket between RLA-A1 (cyan) and HLA-A*0201 (orange) are shown as sticks. (E) The B pocket of RLA-A1 is shown as cyan cavities. Ile24 of RLA-A1 is shown as cyan sticks. (F) The B pocket of HLA-A*0201 is shown as orange cavities. Ala24 of HLA-A*0201 is shown as orange sticks. (G and H) Vacuum electrostatic surface potential of the B pocket of RLA-A1 (G) and HLA-A*0201 (H). P2 residues of peptides and residues 63 and 66 of RLA-A1 and HLA-A*0201 are shown as sticks. The capacity of VP60-2 and its P2-Leu substitutions by small and aliphatic and aromatic amino acids in the second position for binding to RLA-A1 (I) and HLA-A*0201 (J) were evaluated by in vitro refolding. The binding motifs of the B pockets of RLA-A1 (K) and HLA-A*0201 (L) were validated by the renaturation method in vitro. RLA-A1 binds to peptides with aliphatic amino acids (Met, Leu, Ile, and Val). HLA-A*0201 binds not only to aliphatic amino acids (Met, Leu, Ile and Val) but also to hydrophilic amino acids (Ser and Thr).
TABLE 3.
P2-Leu substitutions of VP60-2 and PΩ-Ile substitutions of VP60-10
| Namea | Sequenceb | Refoldingc |
|
|---|---|---|---|
| RLA-A1 | HLA-A*0201 | ||
| VP60-2-WT | ALMPGQFFV | + | + |
| VP60-2-L2F | AFMPGQFFV | − | − |
| VP60-2- L2Q | AQMPGQFFV | − | + |
| VP60-2-L2S | ASMPGQFFV | − | + |
| VP60-2-L2T | ATMPGQFFV | − | + |
| VP60-2-L2Y | AYMPGQFFV | − | − |
| VP60-2-L2I | AIMPGQFFV | + | + |
| VP60-2-L2M | AMMPGQFFV | + | + |
| VP60-2-L2V | AVMPGQFFV | + | + |
| VP60-10-WT | FVPFNSPNI | + | + |
| VP60-10-I9F | FVPFNSPNF | − | − |
| VP60-10-I9Q | FVPFNSPNQ | − | − |
| VP60-10-I9Y | FVPFNSPNY | − | − |
| VP60-10-I9L | FVPFNSPNL | + | + |
| VP60-10-I9M | FVPFNSPNM | − | + |
| VP60-10-I9V | FVPFNSPNV | + | + |
WT, wild type.
The substitutions in the mutant peptides are underlined.
Peptides that can (+) or cannot (−) help the RLA-A1 heavy chain renature with human β2m.
F pocket and PΩ anchor restriction and orientation.
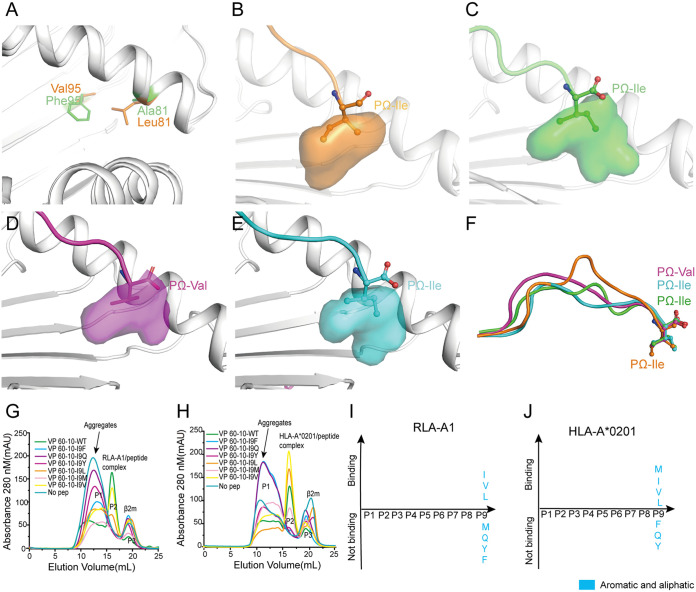
In addition to the B pocket, the F pocket of RLA-A1 is another primary anchor site for peptide binding, as in the MHC-I molecules of other mammals. The differing amino acids that make up the F pockets of RLA-A1 and HLA-A*0201 are located at positions 81 and 95 (Fig. 4). In RLA-A1, residue 81 was a small hydrophobic amino acid, Ala, and residue 95 was a large hydrophobic amino acid, Phe. In contrast, in HLA-A*0201, residue 81 was a large hydrophobic amino acid, Leu, and residue 95 was a small hydrophobic amino acid, Val (Fig. 5A). These amino acids retain the hydrophobicity of the F pockets of RLA-A1, similar to HLA-A*0201, but with a different orientation (Fig. 5B to E). When the structure of RLA-A1/peptide complexes was superimposed onto the previously determined structure of HLA-A*0201/peptide KI10 (PDB code 3MGT), we found the PΩ anchor of RLA-A1 to protrude in a different direction from that of the PΩ anchor of HLA-A*0201 (Fig. 5F).
FIG 4.
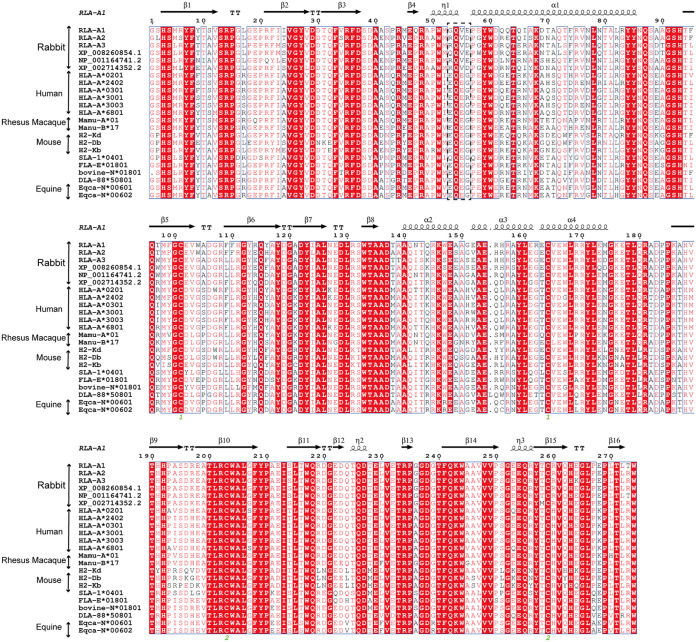
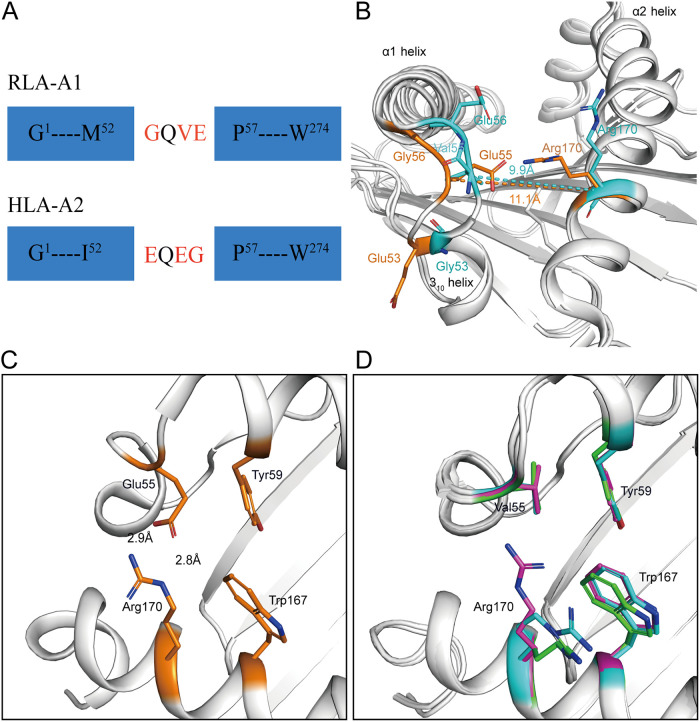
Sequence alignments with mammals. Structure-based sequence alignment of MHC-I molecules in rabbits and other representative mammals is shown (MHC I molecules covering the residues from positions 1 to 274 in RLA-A1). The coil and arrow above the sequence represent the α-helices and β-sheet, respectively. Residues in red indicate complete conservation, and residues boxed in blue represent 80% conservation, with the common amino acids shown in red. The residues at positions 53, 55, and 56 are shown in the black dotted box, and the residues at these three sites are more specific in rabbits; especially the residues at position 55 are completely conserved. The multiple sequence alignment was generated with ClustalX and ESPript.
FIG 5.
The hydrophobic F pocket of RLA-A1 with a different orientation. (A) The structure comparison of key discrepant residues of the F pocket, 81 and 95, between RLA-A1 (green) and HLA-A*0201 (orange). The residues are shown as sticks. (B) The F pocket of HLA-A*0201 is shown as an orange cavity. PΩ-Ile of the peptide is shown as orange sticks and spheres. The backbone of HLA-A*0201 is shown as a cartoon in white. The F pocket of RLA-A1/VP60-1 is shown in green (C), RLA-A1/VP60-2 is shown in magenta (D), and RLA-A1/VP60-10 is shown in cyan (E). PΩ-Ile residues of VP60-1, VP 60-2, and VP60-10 are shown as green, magenta, and cyan sticks and spheres, respectively. The backbone of RLA-A1 is shown as a cartoon in white. (F) The comparison of peptides presented by HLA-A*0201 and RLA-A1. H5-HA205-214 presented by HLA-A*0201 (PDB code 3MGT) is shown in orange. VP60-1, VP60-2, and VP60-10 presented by RLA-A1 are shown in green, magenta, and cyan, respectively. PΩ residues of VP60-1, VP60-2, VP60-10, and H5-HA205-214 are shown as sticks and spheres. The binding motifs of the F pocket of RLA-A1 (G) and HLA-A*0201 (H) were evaluated through a series of substitution mutations at the PΩ position. According to the results of renaturation in vitro, RLA-A1 bound to peptides with small and aliphatic amino acids (Leu, Ile, and Val) (I) at the PΩ position, and HLA-A*0201 bound to peptides with amino acids Met, Leu, Ile, and Val (J) at this position.
Among all the RLA-A1-binding peptides tested, VP60-10 bound well to both RLA-A1 and HLA-A*0201 (Fig. 5G and J). To evaluate restrictions imposed by the F pocket on RLA-A1-binding peptide motifs, we constructed a series of mutated P9-Ile peptides from VP60-10 (VP60-10-I9F, VP60-10-I9Q, VP60-10-I9Y, VP60-10-I9M, VP60-10-I9L, and VP60-10-I9V) (Table 3). VP60-10-I9F and VP60-10-I9Y could bind to neither RLA-A1 nor HLA-A*0201 (Fig. 5G to J); VP60-10-I9M was presented by HLA-A*0201 but not RLA-A1 (Fig. 5G to J); VP60-10-I9L and VP60-10-I9V, together with the wild-type VP60-10, bound both RLA-A1 and HLA-A*0201 (Fig. 5G to J). These data indicate that the F pocket of RLA-A1 is much more restrictive for PΩ anchoring of peptides than that of HLA-A*0201.
Uncommon residues Gly53, Val55, and Glu56 form the narrow, closed N-terminal PBG.
To analyze the gene diversity of RLA, all six RLA allele genes were aligned with typical mammalian MHC-I molecules. The alignment showed that residues Gly53, Val55, and Glu56 at the N terminus of the PBG of RLA-A1 amino acids were peculiar to rabbit MHC-I molecules; they differed from the corresponding residues in the sequences of the other mammals we compared them to. Most mammalian MHC-I molecules that we compared possessed Glu at position 53 of the H chain, with the exception of Gln53 in SLA-1*0401. Position 55 is a highly conserved Glu in the H chain of other mammalian MHC-I molecules, while position 56 is Gly for most mammals but Arg56 in human HLA-A*3001 (Fig. 4 and 6A). Previous work showed that the 310 helix (residues 49 to 53) located in the α1 domain plays a pivotal role in peptide assembly (31). Here, Gly53 was located in the 310 helix of RLA-A1. Val55 and Glu56 are key residues in the loop between the 310 helix and α1 helix. In RLA-A1, the conformation of the 310 helix and the loop between the 310 and α1 helices are much closer to the α2 helix than is found in HLA-A*0201 (Fig. 6B). When we compared the distance of the loop between the 310 and α1 helices and the α2 helix, the 310 helix and the loop between the 310 and α1 helices (9.9 Å between residue Val55 and Arg170) were found to be much closer to the α2 helix (11.1 Å between Glu55 and Arg170) in RLA-A1 than in HLA-A*0201 (Fig. 6B).
FIG 6.
The uncommon 310 helix of RLA-A1. (A) Schematic diagram of the construction of the uncommon residues Gly53, Val55, and Glu56 of RLA-A1 compared with common residues Glu53, Glu55, and Gly56, represented by HLA-A*0201. (B) The superimposition of RLA-A1 with HLA-A*0201. Residues Gly53, Val55, Glu56, and R170 of RLA-A1 are shown as sticks and labeled in cyan. Residues Glu53, Glu55, Gly56, and R170 represented by HLA-A*0201 are shown as sticks and labeled in orange. The 310 helix and the loop between the 310 helix and α1 helix are much closer to the α2 helix than to that of HLA-A*0201. (C and D) Two locked doors at the N terminus of HLA-A*0201 (C) and corresponding positions of RLA-A1 (D). Residues Tyr59 and Trp167 form the first locked door, shown as light orange in HLA-A*0201. Glu55 and Arg170 interact through a salt bridge which resembles a second locked door in HLA-A*0201. In RLA-A1, conserved residues Tyr59 and Trp167 also form the first locked door. However, reside Glu55 is replaced with Val55 which cannot interact with Arg170, so the second locked door in RLA-A1 is open.
In most typical mammalian MHC-I molecules, including HLA-A*0201, Tyr59 and Trp167 are amino acids with large side chains which form the N-terminal wall of the A pocket. This constructs a “locked door” at the N terminus of the PBG of these MHC-I molecules. In addition, a salt bridge between Glu55 and Arg170 can be recognized as a second locked door also at the N terminus of the PBG (Fig. 6C). However, in RLA-A1, residue 55 is a hydrophobic amino acid, Val, which cannot form a salt bridge with Arg170. In this situation, the second locked door is open, with the side chain of Arg170 pointing in another direction (Fig. 6D). The salt bridge between Glu55 and Arg170 in HLA-A*0201 may act as a lifting jack, bracing the N terminus of the PBG. However, RLA-A1 does not possess this second locked door.
The open second door at the N terminus of PBG affects peptide assembly.
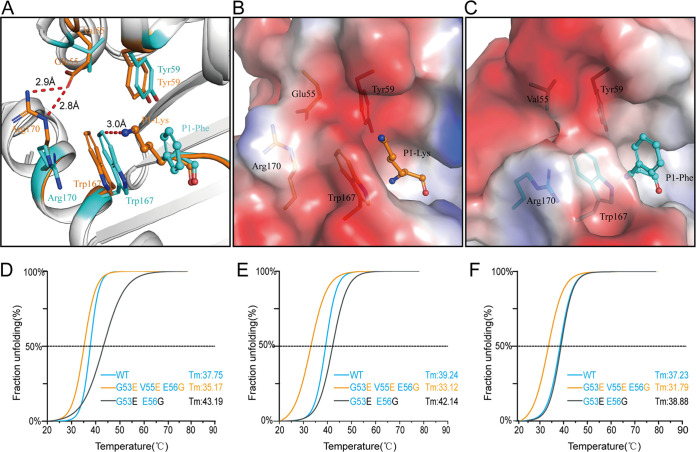
Superimposition of RLA-A1/VP60-10 onto HLA-A*0201 showed that when the second door at the N terminus of PBG in RLA-A1 was open, the conformation of Trp167 was slightly shifted toward the N terminus of the PBG (Fig. 7A). The side chain of P1-Phe of the binding peptide VP60-10 protruded toward Trp167 of RLA-A1, while the side chain of P1-Lys of the HLA-A*0201-binding peptide pointed in the other direction to avoid colliding with Trp167 of HLA-A*0201 (Fig. 7A). Thus, in most mammalian MHC-I molecules such as HLA-A*0201, there are two closed doors (residues Glu55 and Arg170 and residues Tyr59 and Trp167) at the N terminus of the PBG (Fig. 7B), while in RLA-A1, the open second door (residues Val55 and Arg170) at the N terminus forms a narrow N terminus which may indirectly change the conformation of the P1 residue (Fig. 7C).
FIG 7.
The second opened door at the N terminus of PBG affects peptide assembly. (A) The narrow N terminus of the PBG forces a conformational shift of Trp167. Trp167 of HLA-A*0201 is shown as an orange stick, and Trp167 of RLA-A1 is shown as a cyan stick. The superimposition of RLA-A1 over HLA-A*0201 shows that the conformation of Trp167 predisposes formation of the A pocket of the PBG. The distance of Trp167 in RLA-A1 to P1-Lys in HLA-A*0201 is 3.0 Å, meaning that the conformation shift of Trp167 in RLA-A1 could push the P1 residue of the peptide to the C terminus of the PBG. (B and C) Vacuum electrostatic surface potentials of the N terminus of the PBG of HLA-A*0201 (B) and RLA-A1 (C). Residues Glu55, Tyr59, Trp167, and Arg170 of HLA-A*0201 are shown as orange sticks under the vacuum electrostatic surface. Residues Val55, Tyr59, Trp167, and Arg170 of RLA-A1 are shown as cyan sticks under the vacuum electrostatic surface. P1-Lys of HLA-A*0201 and P1-Phe of RLA-A1 are shown as orange and cyan sticks, respectively. The thermostabilities of RLA-A1 wild type, its substitution mutations at positions 53 and 55, and its substitution mutations at positions 53, 55, and 56 binding to VP60-1 (D), VP60-2 (E), and VP60-10 (F) were tested by circular dichroism spectroscopy.
To investigate the potential impact of this special conformation of RLA-A1 on peptide binding, two mutants of RLA-A1 were constructed. One mutant, RLA-A1-Mu2, possessed the HLA-A*0201 residues Glu53 and Gly56 (Gly53Glu and Glu56Gly), while the second, RLA-A1-Mu3 (Gly53Glu, Val55Glu, and Glu56Gly), had all three uncommon residues of RLA-A1 mutated to the mammalian consensus of Glu53, Glu55, and Gly56, as found in HLA-A*0201. For RLA-A1-Mu2, peptides could still stabilize and even increase peptide binding compared with levels for RLA-A1 (Fig. 7D to F). The midpoint transition temperature (Tm) of RLA-A1-Mu2 complexed to peptides VP60-1, VP60-2, and VP60-10 were 43.19°C, 42.14°C, and 38.88°C, respectively, while the Tm values of wild-type RLA-A1 with peptides VP60-1, VP60-2, and VP60-10 were 37.75°C, 39.24°C, and 37.23°C, respectively. In contrast, RLA-A1-Mu3 had a lower binding capacity with peptides VP60-1, VP60-2, and VP60-10 (Tm values of 35.17°C, 33.12°C, and 31.79°C, respectively), which was lower than that of the corresponding wild-type RLA-A1 with peptides VP60-1, VP60-2, and VP60-10 (Tm values of 37.75°C, 39.24°C, and 37.23°C, respectively) (Fig. 7D to F).
Strong hydrophilic D pocket repels hydrophobic amino acids.
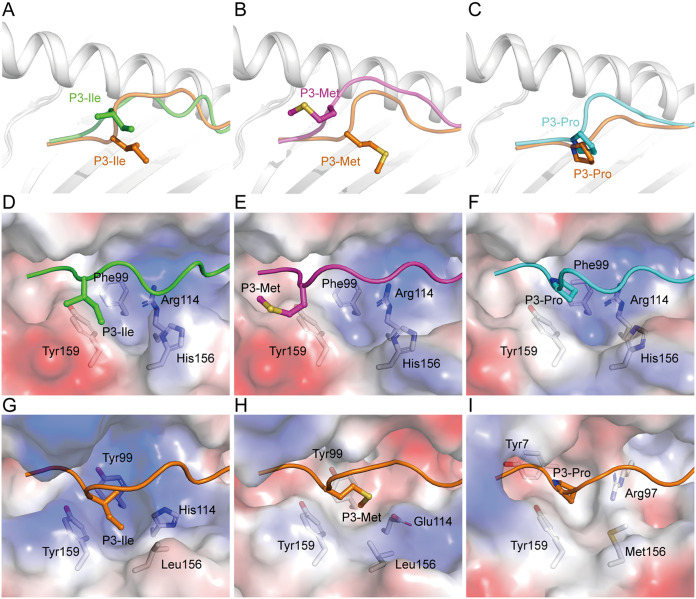
The P3 residues of peptides in our three RLA-A1/peptide complexes showed a repelled conformation outside the D pocket compared with that of MHC class I complexes (HLA-A*0201, HLA-A*3001, and Mamu-A*01) with the same amino acids at P3 (Fig. 8A to C). P3-Met of VP60-2 in the RLA-A1 complex was repelled out of the D pocket (Fig. 8B). Further analysis showed that the D pocket of RLA-A1 was consistent with Phe99, Arg114, His156, and Tyr159 (Fig. 8D to F). Consistent with three other structures, HLA-A*0201, HLA-A*3001, and Mamu-A*01, RLA-A1 residues 99 and 159 were hydrophobic amino acids, Phe and Tyr. But residue 114 was hydrophilic amino acids His and Glu in HLA-A*0201 and HLA-A*3001, respectively, and residue 97 was the hydrophilic amino acid Arg (Fig. 8G to I). The main difference between RLA-A1 and the other three structures that were used for comparison was at the amino acid found at position 156. In the three comparison mammalian structures, residue 156 was a hydrophobic amino acid Leu or Met, while in RLA-A1, residue 156 was the hydrophilic amino acid His (Fig. 8G to I). This difference makes the D pocket of RLA-A1 much more hydrophilic than the pockets of the other three MHC class I molecules. So, the strong hydrophilicity of the D pocket of RLA-A1 repels hydrophobic amino acids inserted into the pocket, with His156 being the key residue.
FIG 8.
The conformations of the P3 residue of peptides presented by RLA-A1. The structural comparison of the P3 residue of VP60-1 (A; green), VP60-2 (B; magenta), and VP60-10 (C; cyan) with the P3 residues of peptides in HLA-A*0201/AMPD2 (PDB code 4NO3), HLA-A*3003/MTB (PDB code 6J29), and Mamu-A*01/Tat-T18 (PDB code 1ZVS) complex, which is the same residue at the P3 position of the peptides, respectively. Peptides used for comparison are labeled in orange. The P3 residue of each peptide is shown as sticks and spheres. The backbones of MHC class I molecules are shown as cartoons in white. Vacuum electrostatic surface potentials of D pockets of RLA-A1/VP60-1 (D), RLA-A1/VP60-2 (E), RLA-A1/VP60-10 (F), HLA-A*0201/AMPD2 (G), HLA-A*3003/MTB (H), and Mamu-A*01/Tat-T18 (I) complexes are shown. Red represents negatively charged residues, blue represents positively charged residues, and gray indicates noncharged residues. Residues comprising the D pocket are shown as sticks under the vacuum electrostatic surface. The P3 residue of each peptide is shown as sticks and spheres.
DISCUSSION
The domestic rabbit is an important animal model for medical research in humans, especially immunological studies. Meanwhile, rabbits are also a pivotal economic animal. RHDV infection threatens the lives of rabbits and causes important economic losses. Rabbit T-cell immunity against virus infection or vaccination remains largely unknown. In the present study, we screened RLA-A1-restricted peptides derived from the capsid protein VP60 of RHDV. Identification of these peptides will benefit further studies of RLA-A1-mediated anti-RHDV responses and vaccine development. The binding motifs of RLA-A1 were determined, providing a good reference to predict CD8+ T-cell epitopes with the RLA-A1 restriction.
Determining the progress of peptide loading to the MHC class I molecule during biosynthetic maturation is crucial for understanding the general rules that govern the interaction of MHC class I molecules with peptides. MHC class I molecules assembled with β2m are stabilized by a suitable peptide in the endoplasmic reticulum (ER) (32, 33). In the ER, new MHC class I molecules are held by chaperones in a peptide-receptive transition state (34, 35). Available peptides processed by the proteasome are exchanged for chaperones and assemble with MHC class I molecules in the ER and then proceed to the cell surface in their mature conformations (36). Previous work showed that the 310 helix (residues 49 to 53) located in the α1 domain of the PBG plays vital roles in peptide assembly (31). The 310 helix is exposed in the open position in the ER transition state. When MHC-I assembles with a suitable peptide and matures, the 310 helix is closed, with buried hydrophobic side chains in the peptide-loaded form. In the bat MHC class I molecule Ptal-N*01:01, there is a 3-amino-acid insertion located at the 310 helix which facilitates residues Asp59, Asp65, and P1-Asp to form a hydrogen bond network. The hydrogen bond network of the A pocket could stabilize peptides with P1-Asp during peptide processing and exchange (37). In RLA-A1, uncommon residues G53, V55, and E56 are located between the 310 helix and α1 helix, which makes the conformation of the 310 helix in RLA-A1 much closer to the α2 helix than that in other mammalian MHC class I molecules. When we constructed the mutations G53 to E53 and E56 to G56, the stabilization of RLA-A1/VP60-1, RLA-A1/VP60-2, and RLA-A1/VP60-10 was not changed or even increased, based on a circular dichroism (CD) assay. However, when G53, V55, and E56 were mutated to E53, E55, and G56, stabilization of RLA-A1/VP60-1, RLA-A1/VP60-2, and RLA-A1/VP60-10 was clearly decreased. Arg167 may form a salt bridge with residue 55 of the RLA-A1 mutants and then close the second door at the N terminus of the PBG. These findings indicate that the uncommon residues G53, V55, and E56 of RLA-A1 play a pivotal role in MHC class I assembly in the rabbit.
Similar to other typical mammalian MHC class I molecules, the B and F pockets in the PBG of RLA-A1 accommodate the primary anchors of the binding peptides. However, uncommon conformational features of RLA-A1-loaded B and F pockets were identified through our structural investigations. The rabbit-specific residue Ile24 inserts into the B pocket of RLA-A1 to occupy its space, making a shallow B pocket. Furthermore, most amino acids comprising the B pocket of RLA-A1 are hydrophobic, thus forming a highly hydrophobic pocket. All of these features make the B pocket of RLA-A1 very restrictive for P2 anchoring. Meanwhile, the F pocket of RLA-A1 is much more restrictive than that of HLA-A*0201. Together with these features, the peptide binding motifs of RLA-A1 are extremely constraining. Thus, there is a generally restricted peptide selection for RLA-A1. Rabbits with RLA-A1 may be susceptible to pathogens whose sequences contain an RLA-A1-incompatible motif.
The variety of peptides presented by MHC class I reflect the capacity of T-cell responses to infectious diseases in some degree. For a chicken MHC class I molecule, BF2*2101, an unusually large central cavity in the binding groove offers substantial conformational flexibility to the crucial residue Arg9, allowing remodeling of key peptide-binding sites. This special feature allows it to bind to peptides with conspicuously different sequences, leading to chickens with BF2*2101 having strong resistance to Marek’s disease virus (38). In contrast, another chicken MHC class I molecule, BF2*0401, has a remarkably narrow PBG which limits the number of epitope peptides binding to it. The stringent restriction of BF2*0401 for peptide selection makes chickens with this allele susceptible to Marek’s disease virus (39). In this study, the restrictive binding motif of RLA-A1 may also limit T-cell responses during particular infectious diseases. As mentioned above, only six cDNA sequences of RLA have been published to date in the NCBI. RLA-A1 is one of these six genes. The survival of this genotype through evolution may be due to selective breeding or be the consequence of some other selective pressure, such as resistance to important pathogens for rabbits in the wild.
In most cases, the P3 residue of peptides inserts into the D pocket of the PBG of MHC-I molecules. The residue at position 3 acts as an anchor residue and is observed in the murine H-2Dd and macaque Mamu-A*01 (40, 41). In RLA-A1, P3-Ile and P3-Pro are repelled out of the D pocket, unlike other MHC class I complexes with the same amino acids in the binding peptides. P3-Met even protrudes out of the D pocket of the PBG. As we know, the exposed position of the MHC-I/peptide complex facilitates recognition by TCRs. The influenza virus peptide H1-P25 (LYKKLKREITF) with HLA-A*2402 restriction is conserved between H1N1 and H5N1 but has a dominant mutation with substitutions at position 9 from Ile to Met (I9M) (named peptide H7-P25) in H7N9 and H9N2. Study of the structure showed that Ile9 inserts into the E pocket while Met protrudes from the PBG. Functional study showed that H1-P25 induces stronger cross-reactivity than peptide H7-P25 only in pH1N1-specific T cells (42). In another case, the secondary anchor Ile5 of peptide 37-1 (YYSIIPHSI) drives a conformation shift of Trp73 on the α1 helix of H-2Kd to become exposed outside the PBG. The specific T-cell response to 37-1 is obviously decreased after mutation to I5A (YYSIAPHSI). The structure of H-2Kd/I5A also shows that the conformation of Trp73 shifts back to PBG (43). So, the uncommon shift of P3 in RLA-A1 may also be associated with T-cell recognition.
In conclusion, the present study demonstrates novel features of rabbit MHC class I molecules through structural and functional investigations. Our results provide new insights into MHC class I molecule assembly and peptide presentation. Furthermore, these data also broaden our knowledge on T-cell immunity in rabbits and may also provide useful information for vaccine development to prevent infectious diseases in rabbits.
MATERIALS AND METHODS
Preparation of expression constructs.
The cDNA coding for the heavy chain (residues 1 to 274) of rabbit MHC class I RLA-A1 (GenBank accession no. K02441.1) was synthesized (Genewiz, Suzhou, China) and cloned into a pET-21a(+) vector. Mutation constructs RLA-A1 G53E E56G and RLA-A1 G53E V55E E56G were constructed based on wild-type RLA-A1 by PCR and also cloned into a pET-21a(+) vector. An Escherichia coli vector expressing human β2m (residues 1 to 98) was constructed previously in our laboratory (44).
Synthesis of RHDV-derived peptides.
The RHDV-derived peptides were predicted by the NetMHCpan, version 4.0, server (http://www.cbs.dtu.dk/services/NetMHCpan/). A total of 13 peptides with high potential binding scores was selected (45, 46). Then peptides were synthesized and purified by reverse-phase high-performance liquid chromatography (HPLC) and mass spectrometry (SciLight Biotechnology, Beijing, China) with a purity of >90%. The peptides were lyophilized and stored at −80°C until use, when they were reconstituted in dimethyl sulfoxide (DMSO).
Expression, refolding, and purification of RLA-A1/peptide complexes.
RLA-A1 and human β2m (hβ2m) were overexpressed as inclusion bodies in the Escherichia coli strain BL21(DE3), and purified inclusion bodies were dissolved with 6 M guanidine-HCl buffer to a concentration of 30 mg/ml. Then, hβ2m, peptide, and RLA-A1 were injected and diluted at a molar ratio of 1:3:1 in refolding buffer (100 mM Tris-HCl, pH 8.0, 2 mM EDTA, 400 mM l-Arg, 0.5 mM oxidized glutathione, and 5 mM reduced glutathione) successively at 4°C for at least 8 h (47). After protein refolding, the RLA-A1 complexes were concentrated and resuspended in protein buffer with 20 mM Tris-HCl (pH 8.0) and 50 mM NaCl and then purified by a Superdex 200 Increase 10/300 GL column and Resource-Q anion-exchange chromatography (GE Healthcare, Beijing, China).
Crystallization and data collection.
As described previously, sitting-drop vapor diffusion technology was used for crystallization (48, 49). RLA-A1/peptide complex crystals were screened through a Crystal Screen I/II kit, Index Screen kit, and PEGRxI/II kit (all, Hampton Research, Aliso Viejo, CA) and kept in a stable environment of 4°C and 18°C for about 1 month. RLA-A1/VP60-10 crystals were observed in 0.1 M Bis-Tris (pH 6.5), and 20% (wt/vol) polyethylene glycol monomethyl ether 5000. The RLA-A1/VP60-1 crystals were grown in 0.1 M imidazole (pH 7.0) and 12% (wt/vol) polyethylene glycol 2000. The single crystal of RLA-A1/VP60-2 was grown in 0.2 M magnesium chloride hexahydrate, 0.1 M sodium citrate tribasic, pH 5.0, and 10% (wt/vol) polyethylene glycol 20000. RLA-A1/VP60-10 was grown in 0.1 M Bis-Tris, pH 6.5, and 20% (wt/vol) polyethylene glycol monomethyl ether 5000. These crystals were transferred to cryoprotective solutions containing 20% glycerol. The X-ray diffraction data were collected at 100 K at beamline BL19U of the Shanghai Synchrotron Radiation Facility (Shanghai, China).
Structure determination and refinement.
The collected data were processed using the Denzo program and the HKL2000 software package (HKL Research). The structures of RLA-A1/peptide complexes were determined using molecular replacement with the program MOLREP in CCP4 (50). The structure of PDB code 5DEG was used as the replacement model (51). Restrained refinement was performed using REFMAC5. The extensive model building was performed using COOT (52). The stereochemical quality of the final model was assessed with the program Refine. PyMOL (http://www.pymol.org/) and COOT were used to generate all structure-related figures.
Thermostabilities of proteins were evaluated by CD.
The thermostabilities of RLA-A1/peptide complexes and HLA-A2/peptide complexes were evaluated by circular dichroism (CD), as previously reported (53). Proteins were adjusted to 0.2 mg/ml with protein buffer. The CD spectra at 218 nm were tested on a Chirascan spectrometer (Applied Photophysics, Letterhead, UK) using a thermostatically controlled cuvette at temperature intervals of 0.2°C at an ascending rate of 1°C/min between 20 and 90°C. The proportion of unfolding protein was represented by the standard method: fraction unfolded (percent) = (θ − θN)/(θU − θN), where θ, θN, and θU represent the mean residue ellipticity, the mean residue ellipticity in the fully folded state, and the mean residue ellipticity in the completely unfolded state, respectively. The denaturation curve was formed by fitting with OriginPro, version 8.0 (OriginLab, Northampton, MA) (54), and the Tm was calculated by using data from the denaturation, curve as mentioned before.
Data availability.
The structures of the peptides in complex with RLA-A1 determined in this study are available in the Protein Data Bank under accession numbers 6M2J, 6M24, and 6M2K.
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (NSFC) (grant number 81971501) and the National Key Research and Development Program of China (grant number 2017YFC1200202). W.J.L. is supported by the Excellent Young Scientist Program of the NSFC (grant number 81822040). The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
We thank Jianhui Li (Institute of Biophysics, Chinese Academy of Sciences) for contributions during the work of circular dichroism spectroscopy.
We declare that we have no conflicts of interest.
REFERENCES
- 1.Branco M, Ferrand N, Monnerot M. 2000. Phylogeography of the European rabbit (Oryctolagus cuniculus) in the Iberian Peninsula inferred from RFLP analysis of the cytochrome b gene. Heredity (Edinb) 85 Pt 4:307–317. doi: 10.1046/j.1365-2540.2000.00756.x. [DOI] [PubMed] [Google Scholar]
- 2.Wanderman NR, Mallet C, Giambini H, Bao N, Zhao C, An KN, Freedman BA, Nassr A. 2018. An ovariectomy-induced rabbit osteoporotic model: a new perspective. Asian Spine J 12:12–17. doi: 10.4184/asj.2018.12.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu P, Bu Q, Wang L, Han J, Du R, Lei Y, Ouyang Y, Li J, Zhu Y, Lu F, Zhuang H. 2013. Transmission of hepatitis E virus from rabbits to cynomolgus macaques. Emerg Infect Dis 19:559–565. doi: 10.3201/eid1904.120827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han J, Lei Y, Liu L, Liu P, Xia J, Zhang Y, Zeng H, Wang L, Wang L, Zhuang H. 2014. SPF rabbits infected with rabbit hepatitis E virus isolate experimentally showing the chronicity of hepatitis. PLoS One 9:e99861. doi: 10.1371/journal.pone.0099861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abravanel F, Lhomme S, El CH, Schvartz B, Peron J-M, Kamar N, Izopet J. 2017. Rabbit hepatitis E virus infections in humans, France. Emerg Infect Dis 23:1191–1193. doi: 10.3201/eid2307.170318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L, Xia J, Wang L, Wang Y. 2017. Experimental infection of rabbits with genotype 3 hepatitis E virus produced both chronicity and kidney injury. Gut 66:561–562. doi: 10.1136/gutjnl-2016-312023. [DOI] [PubMed] [Google Scholar]
- 7.Bottagisio M, Coman C, Lovati AB. 2019. Animal models of orthopaedic infections. A review of rabbit models used to induce long bone bacterial infections J Med Microbiol 68:506–537. doi: 10.1099/jmm.0.000952. [DOI] [PubMed] [Google Scholar]
- 8.Xu WY. 1991. Viral haemorrhagic disease of rabbits in the people’s republic of china: epidemiology and virus characterisation. Rev Sci Tech 10:393–408. [PubMed] [Google Scholar]
- 9.Abrantes J, van-der-Loo W, Le-Pendu J, Esteves PJ. 2012. Rabbit haemorrhagic disease (RHD) and rabbit haemorrhagic disease virus (RHDV): a review. Vet Res 43:12. doi: 10.1186/1297-9716-43-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fenner F. 2010. Deliberate introduction of the European rabbit, Oryctolagus cuniculus, into Australia. Rev Sci Tech 29:103–111. doi: 10.20506/rst.29.1.1964. [DOI] [PubMed] [Google Scholar]
- 11.Cooke BD. 2002. Rabbit haemorrhagic disease: field epidemiology and the management of wild rabbit populations. Rev Sci Tech 21:347–358. doi: 10.20506/rst.21.2.1337. [DOI] [PubMed] [Google Scholar]
- 12.Mutze G, Cooke B, Alexander P. 1998. The initial impact of rabbit haemorrhagic disease on rabbit populations in South Australia. J Wildl Dis 34:221–227. doi: 10.7589/0090-3558-34.2.221. [DOI] [PubMed] [Google Scholar]
- 13.Gregg DA, House C, Meyer R, Berninger M. 1991. Viral haemorrhagic disease of rabbits in Mexico: epidemiology and viral characterization. Rev Sci Tech 10:435–451. doi: 10.20506/rst.10.2.556. [DOI] [PubMed] [Google Scholar]
- 14.Mitro S, Krauss H. 1993. Rabbit hemorrhagic disease: a review with special reference to its epizootiology. Eur J Epidemiol 9:70–78. doi: 10.1007/BF00463093. [DOI] [PubMed] [Google Scholar]
- 15.Li ZX, Hu WD, Li BC, Li TY, Zhou XY, Zhang Z. 2014. Egg yolk IgY against RHDV capsid protein VP60 promotes rabbit defense against RHDV infection. Vet Immunol Immunopathol 157:97–104. doi: 10.1016/j.vetimm.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Bertagnoli S, Gelfi J, Petit F, Vautherot JF, Rasschaert D, Laurent S, Le Gall G, Boilletot E, Chantal J, Boucraut-Baralon C. 1996. Protection of rabbits against rabbit viral haemorrhagic disease with a vaccinia-RHDV recombinant virus. Vaccine 14:506–510. doi: 10.1016/0264-410x(95)00232-p. [DOI] [PubMed] [Google Scholar]
- 17.Bertagnoli S, Gelfi J, Le Gall G, Boilletot E, Vautherot JF, Rasschaert D, Laurent S, Petit F, Boucraut-Baralon C, Milon A. 1996. Protection against myxomatosis and rabbit viral hemorrhagic disease with recombinant myxoma viruses expressing rabbit hemorrhagic disease virus capsid protein. J Virol 70:5061–5066. doi: 10.1128/JVI.70.8.5061-5066.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer L, Le-Gros FX, Mason PW, Paoletti E. 1997. A recombinant canarypox virus protects rabbits against a lethal rabbit hemorrhagic disease virus (RHDV) challenge. Vaccine 15:90–96. doi: 10.1016/s0264-410x(96)00102-8. [DOI] [PubMed] [Google Scholar]
- 19.Castañón S, Marín MS, Martín-Alonso JM, Boga JA, Casais R, Humara JM, Ordás RJ, Parra F. 1999. Immunization with potato plants expressing VP60 protein protects against Rabbit hemorrhagic disease virus. J Virol 73:4452–4455. doi: 10.1128/JVI.73.5.4452-4455.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernández-Fernández MR, Mouriño M, Rivera J, Rodríguez F, Plana-Durán J, García JA. 2001. Protection of rabbits against rabbit hemorrhagic disease virus by immunization with the VP60 protein expressed in plants with a potyvirus-based vector. Virology 280:283–291. doi: 10.1006/viro.2000.0762. [DOI] [PubMed] [Google Scholar]
- 21.Pérez-Filgueira DM, Resino-Talaván P, Cubillos C, Angulo I, Barderas MG, Barcena J, Escribano JM. 2007. Development of a low-cost, insect larvae-derived recombinant subunit vaccine against RHDV. Virology 364:422–430. doi: 10.1016/j.virol.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 22.Jiang Q, Yu Z, Liu J, Kong D, Guo D, Quan CS, Li BT, Hu XL, Qu L. 2018. Recombinant canine adenovirus type 2 expressing rabbit hemorrhagic disease virus VP60 protein provided protection against RHD in rabbits. Vet Microbiol 213:15–20. doi: 10.1016/j.vetmic.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 23.Wang L, Xia T, Guo T, Ru Y, Jiang Y, Cui W, Cui H, Qiao X, Tang L, Xu Y, Li Y. 2019. Recombinant lactobacillus casei expressing capsid protein VP60 can serve as vaccine against rabbit hemorrhagic disease virus in rabbits. Vaccines (Basel) 7:172. doi: 10.3390/vaccines7040172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen M, Song Y, Fan Z, Jiang P, Hu B, Xue J, Wei H, Wang F. 2014. Immunogenicity of different recombinant rabbit hemorrhagic disease virus-like particles carrying CD8+ T cell epitope from chicken ovalbumin (OVA). Virus Res 183:15–22. doi: 10.1016/j.virusres.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 25.Peacey M, Wilson S, Perret R, Ronchese F, Ward VK, Young V, Young SL, Baird MA. 2008. Virus-like particles from rabbit hemorrhagic disease virus can induce an anti-tumor response. Vaccine 26:5334–5337. doi: 10.1016/j.vaccine.2008.07.074. [DOI] [PubMed] [Google Scholar]
- 26.Deng Z, Geng Y, Wang K, Yu Z, Yang P, Yang Z, He C, Huang C, Yin L, He M, Tang L, Lai W. 2019. Adjuvant effects of interleukin-2 co-expression with VP60 in an oral vaccine delivered by attenuated Salmonella typhimurium against rabbit hemorrhagic disease. Vet Microbiol 230:49–55. doi: 10.1016/j.vetmic.2019.01.008. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Zhou M, Han J, Zhu X, Dong T, Gao GF, Tien P. 2005. Generation of murine CTL by a hepatitis B virus-specific peptide and evaluation of the adjuvant effect of heat shock protein glycoprotein 96 and its terminal fragments. J Immunol 174:195–204. doi: 10.4049/jimmunol.174.1.195. [DOI] [PubMed] [Google Scholar]
- 28.Turner SJ, Kedzierska K, Komodromou H, La Gruta NL, Dunstone MA, Webb AI, Webby R, Walden H, Xie W, McCluskey J, Purcell AW, Rossjohn J, Doherty PC. 2005. Lack of prominent peptide-major histocompatibility complex features limits repertoire diversity in virus-specific CD8+ T cell populations. Nat Immunol 6:382–389. doi: 10.1038/ni1175. [DOI] [PubMed] [Google Scholar]
- 29.Marche PN, Tykocinski ML, Max EE, Kindt TJ. 1985. Structure of a functional rabbit class I MHC gene: similarity to human class I genes. Immunogenetics 21:71–82. doi: 10.1007/BF00372243. [DOI] [PubMed] [Google Scholar]
- 30.Niu L, Cheng H, Zhang S, Tan S, Zhang Y, Qi J, Liu J, Gao GF. 2013. Structural basis for the differential classification of HLA-A*6802 and HLA-A*6801 into the A2 and A3 supertypes. Mol Immunol 55:381–392. doi: 10.1016/j.molimm.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mage M, Dolan M, Wang R, Boyd L, Revilleza MJ, Robinson H, Natarajan K, Myers NB, Hansen TH, Margulies DH. 2012. The peptide-receptive transition state of MHC class I molecules: insight from structure and molecular dynamics. J Immunol 189:1391–1399. doi: 10.4049/jimmunol.1200831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otten GR, Bikoff EK, Ribaudo RK, Kozlowski S, Margulies DH, Germain RN. 1992. Peptide and β2-microglobulin regulation of cell surface MHC class I conformation and expression. J Immunol 148:3723–3732. [PubMed] [Google Scholar]
- 33.Saunders PM, van-Endert P. 2011. Running the gauntlet: from peptide generation to antigen presentation by MHC class I. Tissue antigens 78:161–170. doi: 10.1111/j.1399-0039.2011.01735.x. [DOI] [PubMed] [Google Scholar]
- 34.Harris MR, Yu YY, Kindle CS, Hansen TH, Solheim JC. 1998. Calreticulin and calnexin interact with different protein and glycan determinants during the assembly of MHC class I. J Immunol 160:5404–5409. [PubMed] [Google Scholar]
- 35.Neefjes JJ, Hämmerling GJ, Momburg F. 1993. Folding and assembly of major histocompatibility complex class I heterodimers in the endoplasmic reticulum of intact cells precedes the binding of peptide. J Exp Med 178:1971–1980. doi: 10.1084/jem.178.6.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chapman DC, Williams DB. 2010. ER quality control in the biogenesis of MHC class I molecules. Semin Cell Dev Biol 21:512–519. doi: 10.1016/j.semcdb.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 37.Lu D, Liu K, Zhang D, Yue C, Lu Q, Cheng H, Wang L, Chai Y, Qi J, Wang LF, Gao GF, Liu WJ. 2019. Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats. PLoS Biol 17:e3000436. doi: 10.1371/journal.pbio.3000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koch M, Camp S, Collen T, Avila D, Salomonsen J, Wallny H-J, van Hateren A, Hunt L, Jacob JP, Johnston F, Marston DA, Shaw I, Dunbar PR, Cerundolo V, Jones EY, Kaufman J. 2007. Structures of an MHC Class I Molecule from B21 chickens illustrate promiscuous peptide binding. Immunity 27:885–899. doi: 10.1016/j.immuni.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J, Chen Y, Qi J, Gao F, Liu Y, Liu J, Zhou X, Kaufman J, Xia C, Gao GF. 2012. Narrow groove and restricted anchors of MHC Class I molecule BF2*0401 plus peptide transporter restriction can explain disease susceptibility of B4 chickens. J Immunol 189:4478–4487. doi: 10.4049/jimmunol.1200885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Achour A, Persson K, Harris RA, Sundback J, Sentman C, Lindqvist Y, Schneider G, Karre K. 1998. The crystal structure of H-2Dd MHC class I complexed with the HIV-1-derived peptide P18-I10 at 2.4 Å resolution: implications for T cell and NK cell recognition. Immunity 9:199–208. doi: 10.1016/S1074-7613(00)80602-0. [DOI] [PubMed] [Google Scholar]
- 41.Chu F, Lou Z, Chen Y, Liu Y, Gao B, Zong L, Khan A, Bell JI, Rao Z, Gao GF. 2007. First glimpse of the peptide presentation by rhesus macaque MHC class I: crystal structures of Mamu-A*01 complexed with two immunogenic SIV epitopes and insights into CTL escape. J Immunol 178:944–952. doi: 10.4049/jimmunol.178.2.944. [DOI] [PubMed] [Google Scholar]
- 42.Zhao M, Liu K, Luo J, Tan S, Quan C, Zhang S, Chai Y, Qi J, Li Y, Bi Y, Xiao H, Wong G, Zhou J, Jiang T, Liu W, Yu H, Yan J, Liu Y, Shu Y, Wu G, Wu A, Gao GF, Liu WJ. 2018. Heterosubtypic protections against human-infecting avian influenza viruses correlate to biased cross-T-Cell responses. mBio 9:e01408-18. doi: 10.1128/mBio.01408-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu WJ, Lan J, Liu K, Deng Y, Yao Y, Wu S, Chen H, Bao L, Zhang H, Zhao M, Wang Q, Han L, Chai Y, Qi J, Zhao J, Meng S, Qin C, Gao GF, Tan W. 2017. Protective T cell responses featured by concordant recognition of middle east respiratory syndrome coronavirus-derived CD8+ T cell epitopes and host MHC. J Immunol 198:873–882. doi: 10.4049/jimmunol.1601542. [DOI] [PubMed] [Google Scholar]
- 44.Zhang S, Liu J, Cheng H, Tan S, Qi J, Yan J, Gao GF. 2011. Structural basis of cross-allele presentation by HLA-A*0301 and HLA-A*1101 revealed by two HIV-derived peptide complexes. Mol Immunol 49:395–401. doi: 10.1016/j.molimm.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 45.Morten N, Claus L, Thomas B, Kasper L, Mikkel H, Sune J, Gustav RD, Bjoern P, Alessandro S, Ole L. 2007. NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS One 2:e796. doi: 10.1371/journal.pone.0000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoof I, Peters B, Sidney J, Pedersen LE, Sette A, Lund O, Buus S, Nielsen M. 2009. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics 61:1–13. doi: 10.1007/s00251-008-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao J, Xiang WZ, Zhang YL, Peng WY, Zhao M, Niu L, Chai Y, Qi JX, Wang F, Qi P, Pan C, Han L, Wang M, Kaufman J, Gao GF, Liu WJ. 2018. An invariant arginine in common with MHC class II allows extension at the C-terminal end of peptides bound to chicken MHC class I. J Immunol 201:3084–3095. doi: 10.4049/jimmunol.1800611. [DOI] [PubMed] [Google Scholar]
- 48.Liu K, Tan S, Chai Y, Chen D, Song H, Zhang C-H, Shi Y, Liu J, Tan W, Lyu J, Gao S, Yan J, Qi J, Gao GF. 2017. Structural basis of anti-PD-L1 monoclonal antibody avelumab for tumor therapy. Cell Res 27:151–153. doi: 10.1038/cr.2016.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tan S, Liu KF, Chai Y, Zhang WH, Gao S, Gao GF, Qi J. 2018. Distinct PD-L1 binding characteristics of therapeutic monoclonal antibody durvalumab. Protein Cell 9:135–135. doi: 10.1007/s13238-017-0412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. 1998. Crystallography & NMR system: new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 51.Loll B, Fabian H, Huser H, Hee C-S, Ziegler A, Uchanska-Ziegler B, Ziegler A. 2016. Increased conformational flexibility characterizes HLA-B*27 subtypes associated with ankylosing spondylitis. Arthritis Rheumatol 68:1172–1182. doi: 10.1002/art.39567. [DOI] [PubMed] [Google Scholar]
- 52.Emsley P, Lohkamp B, Scott W, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu S, Liu K, Chai Y, Wu Y, Lu D, Xiao W, Cheng H, Zhao Y, Ding C, Lyu J, Lou Y, Gao GF, Liu WJ. 2019. Divergent peptide presentations of HLA-A*30 alleles revealed by structures with pathogen peptides. Front Immunol 10:1709. doi: 10.3389/fimmu.2019.01709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tobita T, Oda M, Morii H, Kuroda M, Yoshino A, Azuma T, Kozono H. 2003. A role for the P1 anchor residue in the thermal stability of MHC class II molecule I-Ab. Immunol Lett 85:47–52. doi: 10.1016/S0165-2478(02)00206-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The structures of the peptides in complex with RLA-A1 determined in this study are available in the Protein Data Bank under accession numbers 6M2J, 6M24, and 6M2K.