Improved viral enrichment methods conclusively demonstrate EBV type 1 to be more prevalent in eBL patients than in geographically matched healthy controls, which previously underrepresented the prevalence of EBV type 2. Genome-wide association analysis between cases and controls identifies six eBL-associated nonsynonymous variants in EBNA1, EBNA2, BcLF1, and BARF1 genes. Analysis of population structure reveals that EBV type 2 exists as two genomic subgroups and was more commonly found in female than in male eBL patients.
KEYWORDS: genome sequencing, genetic variation, endemic Burkitt lymphoma, EBV type 1, EBV type 2, Epstein-Barr virus
ABSTRACT
Endemic Burkitt lymphoma (eBL), the most prevalent pediatric cancer in sub-Saharan Africa, is distinguished by its inclusion of Epstein-Barr virus (EBV). In order to better understand the impact of EBV variation in eBL tumorigenesis, we improved viral DNA enrichment methods and generated a total of 98 new EBV genomes from both eBL cases (n = 58) and healthy controls (n = 40) residing in the same geographic region in Kenya. Using our unbiased methods, we found that EBV type 1 was significantly more prevalent in eBL patients (74.5%) than in healthy children (47.5%) (odds ratio = 3.24, 95% confidence interval = 1.36 to 7.71, P = 0.007), as opposed to similar proportions in both groups. Controlling for EBV type, we also performed a genome-wide association study identifying six nonsynonymous variants in the genes EBNA1, EBNA2, BcLF1, and BARF1 that were enriched in eBL patients. In addition, viruses isolated from plasma of eBL patients were identical to their tumor counterparts consistent with circulating viral DNA originating from the tumor. We also detected three intertypic recombinants carrying type 1 EBNA2 and type 2 EBNA3 regions, as well as one novel genome with a 20-kb deletion, resulting in the loss of multiple lytic and virion genes. Comparing EBV types, viral genes displayed differential variation rates as type 1 appeared to be more divergent, while type 2 demonstrated novel substructures. Overall, our findings highlight the complexities of the EBV population structure and provide new insight into viral variation, potentially deepening our understanding of eBL oncogenesis.
IMPORTANCE Improved viral enrichment methods conclusively demonstrate EBV type 1 to be more prevalent in eBL patients than in geographically matched healthy controls, which previously underrepresented the prevalence of EBV type 2. Genome-wide association analysis between cases and controls identifies six eBL-associated nonsynonymous variants in EBNA1, EBNA2, BcLF1, and BARF1 genes. Analysis of population structure reveals that EBV type 2 exists as two genomic subgroups and was more commonly found in female than in male eBL patients.
INTRODUCTION
Epstein-Barr virus (EBV) infects more than 90% of the world’s population and typically persists as a chronic asymptomatic infection (1). Although most individuals endure a lifelong infection with minimal effect, EBV is associated with ∼1% of all human malignancies worldwide. EBV was first isolated from an endemic Burkitt lymphoma (eBL) tumor; this is the most prevalent pediatric cancer in sub-Saharan Africa (2). Repeated Plasmodium falciparum infections during childhood appear to drive this increased incidence (3). Malaria causes polyclonal B-cell expansion and increased expression of activation-induced cytidine deaminase (AID)-dependent DNA damage, leading to the hallmark translocation of the MYC gene under the control of the constitutively active immunoglobulin enhancer (4–6). How EBV potentiates eBL is incompletely understood; however, the clonal presence of this virus in almost every eBL tumor suggests a necessary role.
EBV strains are categorized into two types based on the high degree of divergence in the EBNA2 and EBNA3 genes (7–9). This longstanding evolutionary division is also present in orthologous primate viruses (10), and yet it remains unexplained. Although EBV type 1 has been extensively studied (11, 12), because it causes acute infectious mononucleosis and other diseases in the developed world, type 2 virus studies have not kept pace since infected individuals are less common and are found primarily in sub-Saharan Africa. This view is changing as several recent studies have reported a significant prevalence of type 2 circulating in western countries, suggesting a greater role for type 1 and greater potential for interactions between the EBV types worldwide (13, 14). To understand endemic Burkitt lymphoma (eBL), the African context provides a direct opportunity to examine viral variation because types 1 and 2 are found in both eBL patients and healthy individuals (8, 15, 16). Viral variation has been shown to impact differential transformation and growth, as well as the capacity to block apoptosis or immune recognition (7, 17, 18). However, studies focusing on only certain genomic regions or proteins potentially miss the disease associations of other loci (19, 20). Although new studies have been conducted (21, 22), genome-wide examinations in case-control studies are few and often do not type the virus. A recent study which investigated whole EBV genomes for variant associations with nasopharyngeal carcinoma among Chinese patients has discovered two variants associated with increased disease risk (23). Similarly, another study investigated genome-wide variants of HIV genomes in quest of finding the associations with drug resistance (24). However, to the best of our knowledge, such a viral genome-wide variant association for EBV and eBL remains to be explored. To address this shortfall and to provide a proof of concept to the field, we sequenced a set of EBV genomes in a disease/control setting.
Whole-genome sequencing of EBV is now attainable from tumor, blood, or saliva using targeted viral DNA capture methods (25–30). However, studying EBV from the blood of healthy individuals remains challenging due to the low viral abundance relative to human DNA (1 to 10 EBV copy/ng of blood DNA). In addition, EBV’s GC-rich genome is inefficiently amplified using conventional library preparation methods. Here, we present improved methods for EBV genome enrichment that allow us to sequence viruses directly from eBL patients and healthy children. Leveraging these samples, we sought to define the viral population structure and characterize viral subtypes collected from children hailing from the same region of western Kenya. In addition, we performed the first genome-wide association study to identify viral variants that correlate with eBL pathogenesis.
RESULTS
Study participant characteristics.
The objective of this study was to examine EBV genetic variation in a region of western Kenya with a high incidence of eBL (31) and determine whether any variants are associated with eBL pathogenesis. We leveraged specimens from eBL patients and healthy children residing in the same geographic area (Fig. 1C) (31). We sequenced the virus isolated from 58 eBL cases and 40 healthy Kenyan children, as controls. Patients between 1 and 13 years of age were predominantly male (74%), consistent with the sex ratio of eBL (Table 1) (31). Healthy controls had similar levels of malaria exposure based on previous epidemiologic studies (32). Control subjects ranged in age from 1 to 6 years. This difference in age was necessarily due to the finding that younger, healthy yet malaria-exposed children have higher average viral loads compared to older children who have developed immune control over this chronic viral infection (33). Therefore, it was not feasible to age-match controls with the eBL cases, who tend to be older. We made the assumption that children are infected with the same herpesvirus throughout life, and therefore genomes from younger children reflect EBV genomes that would be found in older children.
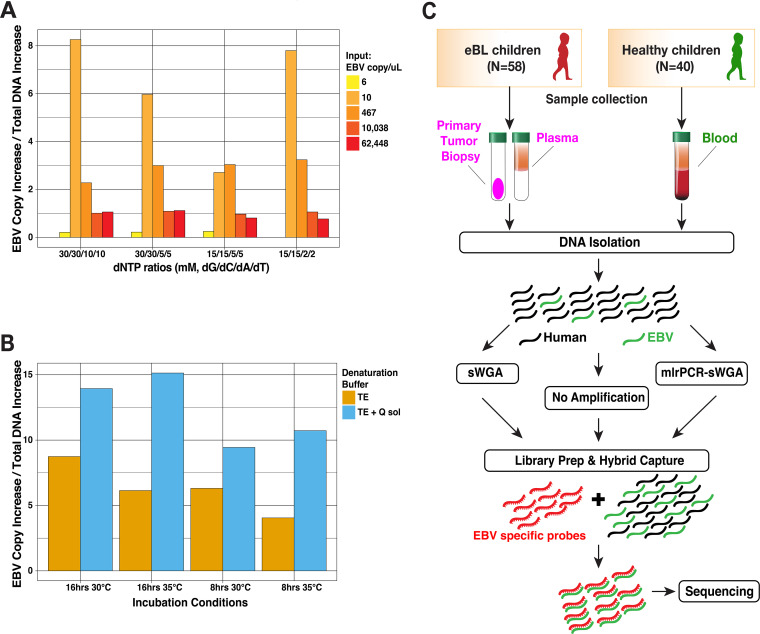
FIG 1.
Optimized EBV genome sequencing from tumors and primary clinical samples. (A) Optimization results of various dNTP concentrations in mlrPCR-sWGA reaction measured as the EBV copy increase normalized by the overall DNA increase. (B) Incubation buffer (TE [Tris-EDTA] or Q solution), time (8 or 16 h), and temperature optimization for better EBV copy increase. (C) Overview of sample collection and methods for sequencing virus from Kenyan children diagnosed with eBL and healthy children as controls. Hybrid capture was universally performed along with additional amplification and enrichment steps to overcome small amounts of virus and input DNA. mlrPCR-sWGA, multiplexed long-range PCR–specific whole-genome amplification.
TABLE 1.
Characteristics of children included in EBV sequencing analysis
| Characteristic | No. (%) |
|
|---|---|---|
| eBL patients (n = 58) | Healthy controls (n = 40) | |
| Age (yr) at collection | ||
| <6 | 16 (27.6) | 39 (97.5) |
| 7–13 | 42 (72.4) | 1 (2.5) |
| Female/male | 15/43 (25.9/74.1) | 20/20 (50.0/50.0) |
| Specimen type | ||
| Tumor biopsy | 41 (41.8) | |
| Blood | 40 (100.0) | |
| Plasma | 14 (14.2) | |
| New cultured eBL | 3 (3.0) | |
Sequencing and assembly quality.
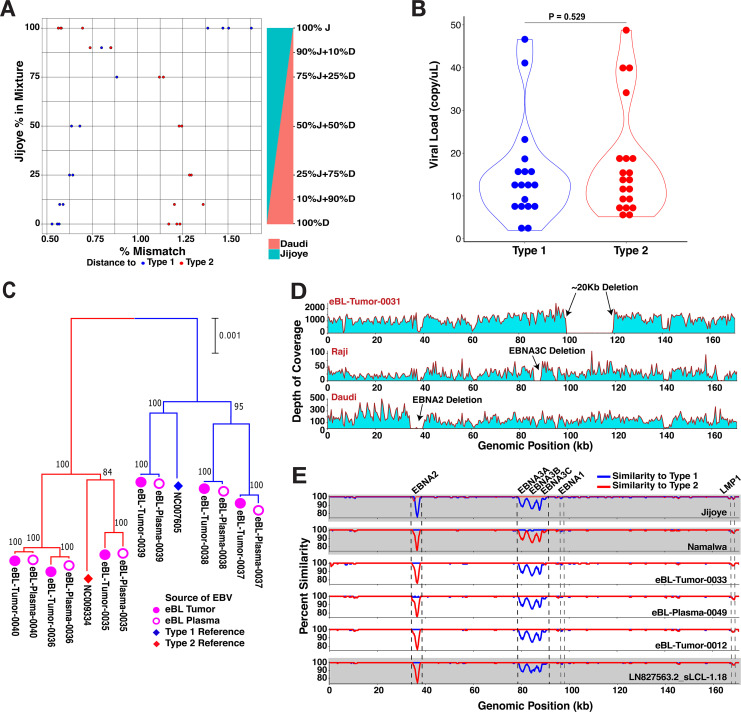
EBV is a large GC-rich double-stranded DNA virus with a 172-kb genome, ∼20% of which is a repetitive sequence. For the majority of eBL patients, we prepared sequencing libraries directly from tumor DNA, followed by hybrid capture enrichment. For low-copy-number viral samples, such as eBL plasma and healthy control blood, we designed and implemented additional viral whole-genome amplification and enrichment prior to library preparation and sequencing (Fig. 1C). We generated a study set of 114 genomes, including replicates from cell lines and primary clinical samples, representing 98 cases and controls. In addition, we sequenced 20 technical replicates for quality control purposes such as estimation of resequencing error or specific whole-genome amplification (sWGA) bias and sensitivity of detection of mixed infections. The baseline resequencing error rate was limited to ∼1.1 × 10−5 bases when our assemblies were compared to high-quality known strain genomes (34) (Table 2). The mean error rate was ∼2.1 × 10−5 bases for sWGA as determined by a GenomiPhi V2 DNA amplification kit (Sigma-Aldritch), while it was ∼1.1 × 10−4 bases when we used more sensitive multiplex long-range PCR amplification (mlrPCR) combined with sWGA (mlrPCR-sWGA; see Materials and Methods). We obtained an average of ∼5 million reads, resulting in an average 9,688 depth of coverage across assemblies (Table S3). De novo sequence assembly created large scaffolds covering nonrepetitive regions, except for three isolates with low coverage, yielded a median of 137,887-bp genomes (range, 47,534 to 146,920 bp). We determined the types of each isolate by calculating the nucleotide distance to both reference types in addition to read mapping rates against type-specific regions. Despite our ability to experimentally detect mixed types at levels as low as 10% (Fig. 2A), we found no evidence of mixed infections in our cases and controls. Also, to ensure that our sample inclusion was unbiased when selecting healthy individuals with high enough viremias to sequence, we quantified the baseline viral loads with biplex qPCR using primers for viral BALF5 and human β-actin gene (see the supplemental material). We compared the viral loads and found no significant difference between types 1 and 2 (P = 0.529, Fig. 2B).
TABLE 2.
Estimated sequencing error rates based on replicates and controlsa
| Prior amplification | Control expt assembly | Reference assembly | No. of substitution errors | No. of correct bases | Error rate (per base) | Mean error rate (per base) |
|---|---|---|---|---|---|---|
| No amplification | Jijoye | Jiyoye assembly* | 1 | 134,118 | 7.46E−06 | 1.13E−05 |
| Daudi | Daudi assembly* | 0 | 132,780 | 0.00E+00 | 1.13E−05 | |
| Raji_Rep1 | Raji assembly* | 0 | 130,560 | 0.00E+00 | 1.13E−05 | |
| Raji_Rep2 | Raji assembly* | 5 | 132,736 | 3.77E−05 | 1.13E−05 | |
| GenomiPhi-WGA | Raji_wga | Raji assembly* | 2 | 132,836 | 1.51E−05 | 2.13E−05 |
| eBLtumor-01_EBV_type1_wga | eBLtumor-01_EBV_type1# | 1 | 140,069 | 7.14E−06 | 2.13E−05 | |
| eBLtumor-02_EBV_type2_wga | eBLtumor-02_EBV_type2# | 4 | 142,932 | 2.80E−05 | 2.13E−05 | |
| eBLtumor-03_EBV_type2_wga | eBLtumor-03_EBV_type2# | 1 | 142,154 | 7.03E−06 | 2.13E−05 | |
| eBLtumor-04_EBV_type1_wga | eBLtumor-04_EBV_type1# | 5 | 141,643 | 3.53E−05 | 2.13E−05 | |
| eBLtumor-05_EBV_type1_wga | eBLtumor-05_EBV_type1# | 5 | 141,643 | 3.53E−05 | 2.13E−05 | |
| mlrPCR-sWGA | Raji_Rep1_mlrPCR-sWGA | Raji assembly* | 6 | 98,507 | 6.09E−05 | 1.09E−04 |
| Raji_Rep2_mlrPCR-sWGA | Raji assembly* | 11 | 130,538 | 8.43E−05 | 1.09E−04 | |
| Jijoye_Rep1_mlrPCR-sWGA | Jiyoye assembly* | 13 | 132,045 | 9.84E−05 | 1.09E−04 | |
| Daudi_mlrPCR-sWGA | Daudi assembly* | 25 | 130,160 | 1.92E−04 | 1.09E−04 |
*, Reference assemblies are from Palser et al. (34) (Jijoye, LN827800; Daudi, LN827545; and Raji, KF717093). #, The indicated isolates were sequenced without any amplification. Preprocess denotes whether the sample DNA was amplified prior to sequencing library preparation. The numbers of substitutions were determined by pairwise whole-genome alignments of control and reference assemblies. Error rates refer to the average mismatches to reference assemblies after normalizing to total covered genomic regions. GenomiPhi-WGA, whole-genome amplification using EBV-specific protected hexamers; mlrPCR-sWGA, preamplification with PCR primer pools followed by sWGA.
FIG 2.
Sequencing and detection quality, plasma-tumor pairs, and atypical genome isolates. (A) Controls for putative mixed infections and sampling bias against EBV types. The sensitivity of EBV genome typing approach measured by accurate type assignments of in-lab mixtures with predefined ratios. Each mixture of Daudi (type 1) and Jijoye (type 2) at various ratios was prepared in replicates. After the genome assembly, the type of the major strain was determined from the distance to both reference viral genomes. (B) Comparison of viral load levels of individuals carrying different EBV types (P = 0.529, t test). The nonsignificant difference suggests unbiased sampling among either type regardless of viral loads. We quantified viral loads with biplex qPCR using primers for viral BALF5 and human β-actin gene. (C) Comparison of virus from paired tumor (filled pink circles) and plasma samples (hollow pink circles) at diagnosis shows that viral DNA circulating in the peripheral blood represents the virus in the tumor. The neighbor-joining tree is scaled (0.001 substitutions per site) and includes standard reference genomes for type 1 (NC_007605, blue diamond) and type 2 (NC_009334, red diamond). (D) The depth of coverage showing an absence of reads from approximately 100 to 120 kb is indicative of a large deletion in the virus from an eBL tumor (top panel). In the middle and lower panels, although we did not detect any in our tumor or control viruses, we detected the deletions previously described in tumor lines, including EBNA3C deletion in Raji and EBNA2 deletion in Daudi strains. (E) Three intertypic viruses were detected by scanning across the genomes for percent identity in 1-kb windows to both type 1 and type 2 references (NC_007605 and NC_009334, respectively). The top two graphs (gray) represent the controls, Jijoye and Namalwa, followed by three intertypic viruses from this study and one publicly available intertypic virus (LN827563.2_sLCL-1.18 in gray).
Equivalence of tumor and plasma viral DNA in eBL cases.
Plasma EBV load has been studied to show its potential as a biomarker or as a prognostic marker in various lymphomas, including BL (35, 36). We included plasma specimens, along with the tumor biopsy specimens, from eBL patients in the whole-genome sequencing set to compare and contrast the two counterparts at the sequence level. Following the separate sequencing and genome assembly of six pairs of plasma- and tumor-associated viruses from six patients, we confirmed that viral DNA in the plasma was representative of the virus in the tumor cells (Fig. 2C). Accounting for the sequencing errors, the pairs appeared to be identical (Fig. S1). Out of these pairs, we further confirmed the subtypes of three EBV isolates (eBL-Tumor-0035, -0037, and -0038) from the plasma and tumor biopsy specimens using type-specific PCRs (see the methods in the supplemental material) in addition to five other samples (eBL-Tumor-0003, -0019, -0022, -0029, and -0030). Overall, these findings demonstrate that viral DNA isolated from eBL patient plasma represents the tumor virus and reflects its genome sequence to the circulating system. This further ensures the potential of plasma DNA for prognostic tools in disease monitoring.
Structural variation and intertypic recombinants.
First, we looked for large deletions within our viral genomes but did not detect any of the previously described deletions in EBNAs, even though we were able to detect, as positive controls, EBNA3C deletion in Raji and the EBNA2 deletion in Daudi cell lines. However, in one sample we did detect a novel 20-kb deletion, spanning from 100 to 120 kb in the genome (Fig. 2D), which appears as the lack of sequencing read coverage while the rest of the genome, even high-GC regions, show a high sequencing depth (>6,000× on average). This deleted region normally encodes multiple lytic phase genes, e.g., BBRF1/2, BBLF1/3, BGLF1/2/3/4/5, and BDLF2/3/4. Interestingly, none of the latent genes were affected by this deletion.
Next, we interrogated our isolates by comparing the pairwise similarities of each genome against EBV type 1 and type 2 references. By traversing through the genome with a window, we were able to delineate regions that were more similar to one type over the other (Fig. 2E). As expected, Jijoye, a type 2 strain, displayed less similarity against type 1 reference around its EBNA2 and EBNA3 genes, the most divergent region between types, whereas Namalwa as a type 1 strain shows the same pattern of dissimilarity against type 2 reference around the same regions. Interestingly, we found three patient-derived genomes—eBL-Tumor-0012, eBL-Tumor-0033, and eBL-Plasma-0049—with mixed similarity trends. Similar to a previously detected recombinant strain (LN827563.2_sLCL-1.18) (34), all of the intertypic isolates carried type 1 EBNA2 and type 2 EBNA3 genes. Although these new intertypic hybrids were all isolated from eBL patients only as opposed to healthy controls, this finding does not reach statistical significance (P = 0.268, chi square).
Genomic population structure is driven by type differences with distinct substructures in type 2 viruses.
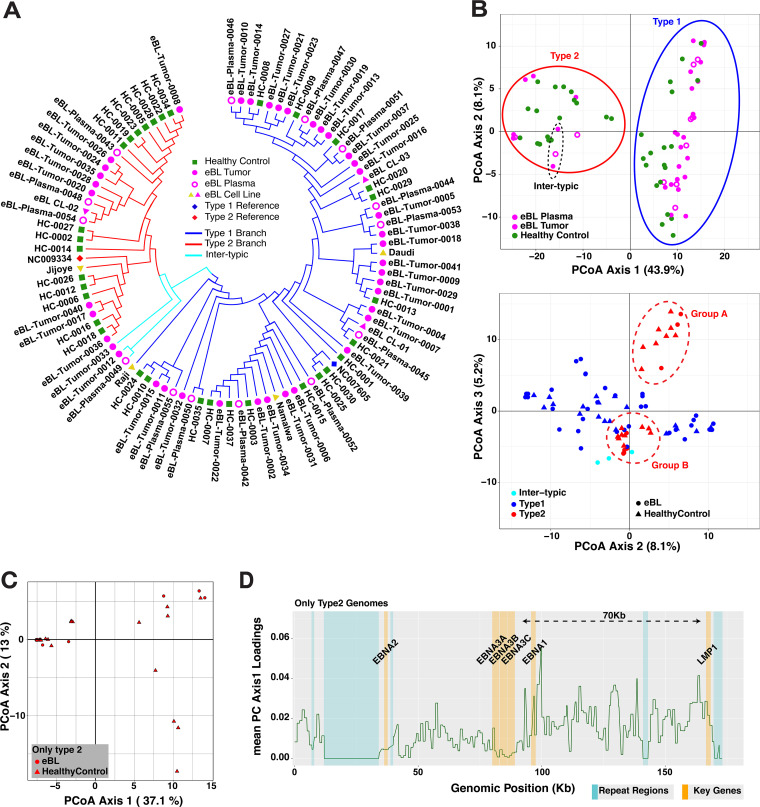
Our samples present a unique opportunity to study population structure of EBV types and their coevolution within a geographically defined region. As expected, the major bifurcation within the phylogenetic tree based on the entire genome occurs between type 1 and type 2 viruses (Fig. 3A). Viruses from both eBL patients and healthy controls appeared to be intermixed almost randomly within the type 1 branch. Interestingly, within type 2 genomes eight eBL-associated isolates formed a unique subcluster. The hybrid genomes are clustered with type 2s, which is consistent with type 2 EBNA3s representing a greater amount of sequence than the type 1 EBNA2 region.
FIG 3.
Diversity and phylogenetic analysis of EBV genomes in Kenyan population. (A) Phylogenetic tree of the western Kenya EBV genomes demonstrating the major type 1 and type 2 demarcation (blue and red branches, respectively). Pairwise distance calculations were based on Jukes-Cantor nucleotide substitution model, and the tree was constructed with the simple neighbor-joining method. Genomes are colored based on sample type: healthy children blood (green squares), eBL tumors (full pink circles), plasma of eBL children (hollow pink circles), and new and previous cell lines (pink and yellow triangles, respectively). Low-coverage genomes are excluded. (B) PCoA plots of nucleotide variations among whole-genome sequences with first and second axes (upper plot, colored by sample type) and second and third axes (lower plot, colored by EBV subtype and shapes represent case and control), which separates type 2 genomes into groups A and B (dashed red ellipses). The color coding is the same as in panel A. (C) First and second axes of PCoA using only type 2 genomes showing the separation of two groups. (D) Absolute loading values of axis 1 from PCoA with all variants are plotted throughout the genome. Values are averaged across the 1-kb window. A dashed arrow marks the region for sequence variations that predominantly drives the separation in the PCoA.
We further explored viral population structure with principal coordinate analysis (PCoA) of variation across the genome. While the first three components cumulatively explain 57.2% of the total variance, the first component, which solely accounted for 43.9% of the variance, separates genomes based on type 1 and type 2 (Fig. 3B, upper plot). Similar to the phylogenetic tree, intertypic genomes are positioned more closely to type 2s. Interestingly, the second and predominantly third components separate type 2 viruses into two distinct clusters, groups A and B (Fig. 3B, lower plot). These clusters were reflected, although not as distinctly, in the structure of the phylogenetic tree in the Fig. 3A as well. The PCoA loading values, which account for 37.1% of the variance between the type 2 groups, are predominantly driven by correlated variation spanning 70 kb upstream of EBNA3C (Fig. 3C and D). Together, these findings suggest that there are two EBV type 2 strains circulating within this population. We also examined viral variation from the perspective of LMP1. Interestingly, the vast majority of viruses were grouped into Alaskan and Mediterranean strains (see Fig. S2 in the supplemental material). The majority of genomes that carry Alaskan LMP1 are type 2 genomes, whereas Mediterranean strain LMP1s are mostly from type 1 genomes. For all available LMP1 type 2 sequences, group A and group B correlated with Mediterranean and Alaskan strains, respectively.
EBV type 2 is less diverse than EBV type 1.
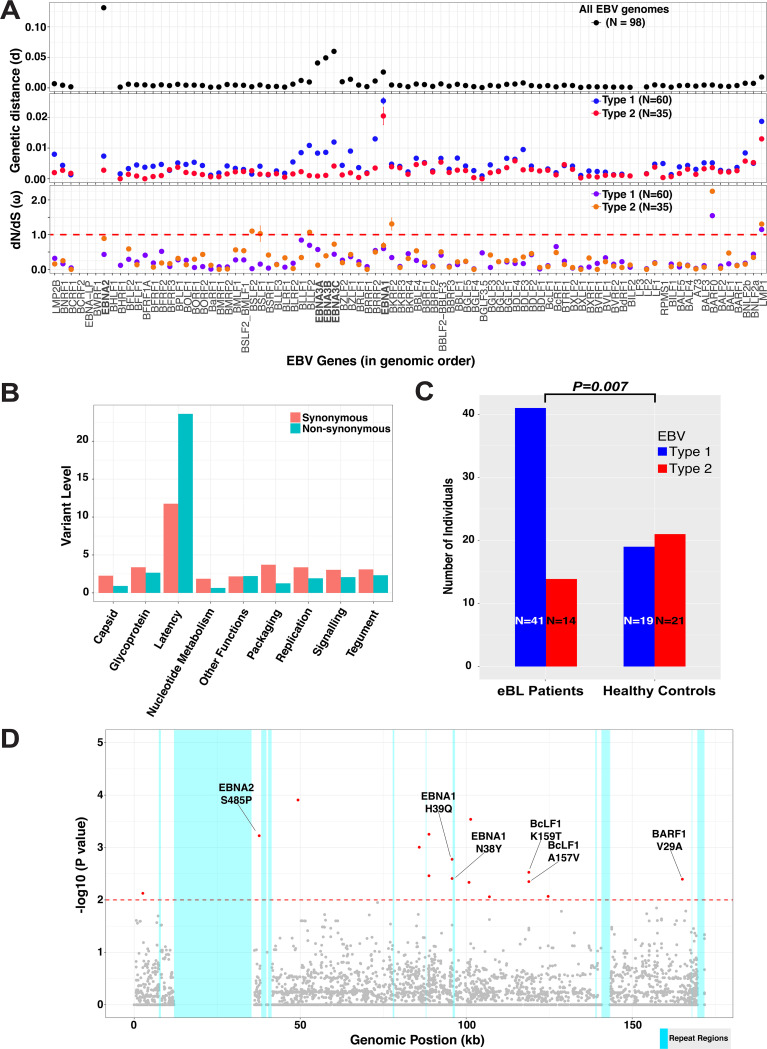
We further explored the pattern and nature of genomic variation across the genome comparing and contrasting EBV type 1 and type 2. Examining the pairwise divergence of coding genes for all viral genomes, we found that the divergence was highest in the type-specific EBNA genes (EBNA2 and EBNA3s), in particular, with EBNA2 showing the greatest divergence (d = 0.1313 ± 0.0023) (Fig. 4A, upper panel). Investigating each type separately, the diversity within types was low for EBNA2 and EBNA3Cs, consistent with type 1 and 2 being separated by many fixed differences (Fig. 4A, middle panel). In both types, intratype divergence was greatest for EBNA1 and LMP1. Most remarkable was the fact that type 2 generally showed lower levels of divergence across the genome (d = 0.0047 ± 0.0037 and d = 0.0025 ± 0.0027 for type 1 and type 2, respectively). We observed the same trend even with the balanced sample sizes through random downsampling (Fig. S3A and B). Overall, these measures suggest that EBV gene evolutionary rates differ by types.
FIG 4.
Diversity in EBV coding genes and significant associations of EBV type 1 genomes and single nucleotide variants with eBL. (A) Genetic distance metrics of each EBV gene calculated based on the Kimura two-parameter method averaged across all genomes (upper panel) or type 1/type 2 separately (middle panel). Lower panel shows nonsynonymous-to-synonymous change (dN/dS) ratios of viral protein coding genes averaged across all pairwise comparisons within each group separately. Error bars represent standard errors of the mean. (Three intertypic genomes are excluded). (B) Average synonymous and nonsynonymous variants in genes are summarized as functional categories of genes. Variant level represents the number of variants per gene normalized by gene length in kilobases. (C) The frequency of type 1 and type 2 genomes identified from eBL patients and healthy control children (excluding the three intertypic hybrid genomes) is significantly different (P = 0.007, Fisher exact test). (D) Manhattan plot for genome-wide associations of all single nucleotide variants tested for frequency differences between cases and controls controlling for type-specific variants. The significance of each locus association is represented by an empirical P value (negative log10 scale) that was calculated by 1 million permutations with random label swapping. Permutations were stratified for EBV genome type and adjusted for the missing genotypes due to lack of coverage. All significant variants associated with eBL cases are indicated in red (P < 0.01). Nucleotide positions are presented according to the type 1 reference genome.
To explore signatures of evolutionary selection, we examined the dN/dS ratios within coding sequences (Fig. 4A, lower panel). Overall, most genes showed signals of purifying selection, as indicated by ω < 1.0, except for LMP1, BARF0, and BKRF2 (only type 2). Interestingly, with dN/dS measures, EBNA2, BSLF1, BSLF2, and BLLF2 genes had relatively higher rates in type 2 compared to type 1 (P < 0.001). Having significantly different ω values for multiple genes (P < 0.001, t test) can suggest the existence of differential evolutionary pressure on these two divergent types. This can be interpreted as an ongoing adaptation process of type 2 genomes (through certain genes) in the population in contrast to fixed functions of these genes for type 1 genomes. Overall, the magnitude of average nonsynonymous and synonymous changes per gene, normalized by gene length, reflect the high-level diversity accumulated in certain genes (Fig. S4). Latency-associated genes generally have the highest nonsynonymous variant rates, but they also have the highest synonymous rates, consistent with longstanding divergence (Fig. 4B). Other functional categories, including lytic genes, have relatively low levels of nonsynonymous mutations, suggesting stronger purifying selection (see Table S4 for functional categories).
Global context of Kenyan viruses.
To more broadly contextualize our viral population from western Kenya, we examined the phylogeny of the Kenyan viruses along with other publicly available genomes from across the world (Table S5). Among all isolates, the most polymorphic genomic regions appeared to be around EBNA2 and EBNA3 genes (Fig. S5A). Phylogenetic tree shows that the major types, type 1 and type 2, are the main demarcation point regardless of the source or geographic location. The three intertypic genomes from our sample set neatly cluster with the previously isolated intertypic hybrid, sLCL-1.18 (Fig. S5B). Type 1 genomes from our study were split into two groups, with one forming a sub-branch only with Kenyan type 1, including Mutu, Daudi, and several Kenyan lymphoblastoid cell lines (LCLs). The second group interspersed with other African (Ghana, Nigeria, and North Africa) and non-African isolates. In addition, a few of our genomes from healthy carriers clustered with a group of mainly Australian isolates; however, none of them clustered with the South Asian group. Our Kenyan EBV type 2s generally intermixed with other type 2 genomes.
Viral genomic variants and associations with eBL.
Previous studies, which examined viral sequence variants with relative frequencies, often lack properly controlled disease association analysis in genome-wide context (14, 37). After excluding the intertypic hybrids, we compared type frequencies of EBV genomes isolated from eBL patients and healthy controls. We observed a significant difference in frequencies with 74.5% of eBLs carrying type 1, whereas only 25.5% carried type 2 infections. In contrast, 47.5% versus 52.5% of types 1 and 2, respectively, were found in healthy controls. EBV type 1 was associated with eBL (odds ratio [OR] = 3.24, 95% confidence interval [CI] = 1.36 to 7.71, P = 0.007 [Fisher exact test]) (Fig. 4C), independent of age and gender (all P > 0.05, Fig. S6). The type 1 prevalence was still ∼70% (and 30% type 2) among eBL children who are within the equivalent age range of their healthy counterparts (1 to 6 years old). The breakdown of the subtype frequencies based on gender revealed that the female eBL patients most frequently carried type 2 (type 1 n = 14 versus type 2 n = 2) while their healthy counterparts showed the opposite trend (type 1 n = 7 versus type 2 n = 26) (Table 3). On the other hand, male individuals carried both subtypes with roughly equivalent frequencies regardless of their disease status (n = 27 versus n = 26 and n = 12 versus n = 16, type 1 and type 2 among eBL and healthy control groups, respectively). We then expanded the association analysis to all 6,191 synonymous and nonsynonymous single nucleotide variations across the entire genome (Fig. 4D and Table S6). We conducted an initial association test for each nonsynonymous variant and detected 133 significant associations (Table S7; see also Materials and Methods). The vast majority of these variants were located within the type 1/type 2 region given the highly correlated nature of this region (Fig. S7). We then stratified by type to detect variants independent of viral type. This yielded six variants solely associated with the disease (Table 4). Variant 37668T>C represents a serine residue change to a proline at the C terminus of EBNA2 (S485P) which is carried by 24/54 eBL cases, whereas this variant was present in only 2/36 healthy controls. Two variants in EBNA1 at 95773A>T and 95778T>G (N38Y and H39Q, respectively) were both observed in 3/57 eBL isolates, while their corresponding frequencies were 11/36 and 12/37 among healthy controls. These two variants fall into one of the two chromosome binding domains of the EBNA1 protein, which plays a bridging role for tethering the viral episome to the host chromosome (38, 39). Other two significant variants we detected are within the BcLF1 gene which encodes viral major capsid protein. This protein is the most essential component of the self-assembly structures for the viral capsid (40). Elevated substitution rates in viruses of healthy controls as opposed to eBL-associated viruses comply with their role in capsid formation and pathogenesis. The BARF1 variant with higher frequencies in eBL-associated viruses that replaces the valine residue with an alanine might provide a fitness advantage with its role as a soluble form of CSF-1 receptor that neutralizes effects of human CSF-1. With this residual change in the protein, BARF1 increases its sequence identity to human CD80 since both share the same amino acid, alanine, at position 29 (41).
TABLE 3.
Characteristics of children included in our study and viral genome subtypes
| Group | Gender | Typea | Mean age (yr) | Count | % |
|---|---|---|---|---|---|
| eBL | Female | 1 | 7.57 | 14 | 14 |
| 2 | 4 | 2 | 2 | ||
| Male | 1 | 6.81 | 27 | 28 | |
| 2 | 7.08 | 26 | 27 | ||
| 1/2 | 8.33 | 9 | 9 | ||
| Healthy control | Female | 1 | 3.36 | 7 | 7 |
| 2 | 3.58 | 26 | 27 | ||
| Male | 1 | 3.3 | 12 | 12 | |
| 2 | 3.74 | 16 | 16 |
EBV subtypes are represented as 1, 2, and 1/2 for intertypic.
TABLE 4.
Single nucleotide variants associated with eBLa
| Gene | Position | Ref | Alt | AA change | eBLs |
Healthy controls |
P | OR | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Genotypes | Alt count | Genotypes | Alt count | |||||||
| EBNA2 | 37668 | T | C | S485P | 54 | 24 | 36 | 2 | 0.000328 | 0.1 |
| EBNA1 | 95773 | A | T | N38Y | 57 | 3 | 36 | 11 | 0.001322 | 6.67213 |
| EBNA1 | 95778 | T | G | H39Q | 57 | 3 | 37 | 12 | 0.000538 | 7.16129 |
| BcLF1 | 124703 | T | G | K159T | 56 | 1 | 34 | 7 | 0.003178 | 12.7377 |
| BcLF1 | 124709 | G | A | A157V | 56 | 1 | 34 | 7 | 0.003092 | 12.7377 |
| BARF1 | 165131 | T | C | V29A | 57 | 36 | 36 | 10 | 0.004082 | 0.349462 |
A single nucleotide variant association test results with P < 0.01 after type stratification. The table summarizes the statistically significant single nucleotide variant associations and their effects in the coding regions. Reference (Ref) refers to the genotype based on the consensus of all genomes in the sequencing set, and the variant position denotes the projection to the type 1 reference genome (NC_007605). An association test was performed for every variant position comparing the frequency of reference and alternative (minor allele) bases among eBL patient and healthy control children (Fisher exact test). Empirical P values are based on one million permutations. For the genotypes, genomes with missing data (Ns, lack of coverage) were excluded. Ref, reference allele; Alt, alternative/variant allele; AA, amino acid; OR, odds ratio.
Nucleotide variants in noncoding and promoter regions can affect regulation of viral gene expression and activity within host cells. BZLF1 is a regulator gene of lytic reactivation and classified based on its promoter as prototype Zp-P (B95-8) and Zp-V3 (M81 strain). Zp-V3 variant of the promoter has recently been found to enhance lytic activity and overrepresented in EBV-positive BLs (42). Therefore, we sought to find out whether our data set can validate the association. We determined variants at seven positions in the upstream promoter region of BZLF1 (Table S8). Interestingly, all of the Kenyan viruses carried C at both positions –525 and –274 (as in Zp-P) regardless of promoter type. We also found that –532 and –524 are variable in our isolates, while these two are not variant in both promoter types. Our results show that only 12.5% (5/40) type 1 promoter sequences fully resembled Zp-V3 in eBL group as opposed to 22% (2/9) healthy genomes, whereas all of the type 2 genomes, without exception, carried Zp-V3 type promoter regardless of disease status.
DISCUSSION
In this study, we investigated the genomic diversity of EBV by sampling viruses from children in western Kenya, where eBL incidence is high (32). Our improved methods allowed us to sequence asymptomatically infected healthy controls with relatively low peripheral blood viral loads and thereby examine the virus in the population at large (33). We performed the first association study comparing viral genomes from eBL patients and geographically matched controls, without the need for viral propagation in LCLs, thus showing that type 1 EBV, as well as potentially several non-type-specific variants, is associated with eBL. Furthermore, as the first study that characterized significant numbers of EBV type 2, we were able to compare and contrast both types and explore the viral population, thus discovering novel differences, including in population substructures and female-to-male frequencies, in EBV type 2. An extended cohort is required to further validate our results.
Our sequencing data demonstrated that EBV from plasma is representative of the tumor virus in eBL patients. This is consistent with the premise that peripheral EBV DNA originates from apoptotic tumor cells given that cell-free EBV DNA in eBL patients are mostly unprotected against DNase (43), as opposed to being encapsidated during lytic reactivation, and that plasma EBV levels are associated with tumor burden and stage (36). These findings support the use of plasma viremia as a surrogate biomarker for tumor burden and the development of plasma-based prognostic tests with predictive models that could be used during clinical trials (36). The lack of mixed infections observed in our healthy controls could be due to the limit of detection in blood compared to viruses isolated from saliva (14). Further studies are needed to extrapolate and understand the coevolution and dynamics of both EBV types.
In addition, we detected three intertypic recombinant EBV genomes solely found within our eBL patients, findings similar to those previously described in other cancers (44). It is unclear whether the intertypic genomes represent a common event with subsequent mutation and recombination or multiple independent events. If the latter is true, it supports more frequent mixed-type infections given that both parents have to be present in the same cell (45–47). It is interesting that all four intertypic viruses observed to date carry the same type EBNA2/EBNA3 combinations with the type 2 genes being so closely related (Fig. S8). Thus, if multiple events have generated these viruses, it suggests that certain strains may have a greater proclivity to recombine. Further studies will be needed to better define the intertypic population, their origins, and their association with disease.
Importantly, we were able to explore EBV population genetics and compare and contrast type 1 and type 2 because of their coprevalence in Africa. As already well described, the major differentiation in terms of genetic variability was the variation correlated with type 1 and type 2 viruses. These viral types showed distinct population characteristics with type 1 harboring greater diversity especially in functionally important latent genes. Combined with the observed nucleotide diversity, latency genes appear to have long-standing divergence that has accumulated significant synonymous changes (as opposed to recent sweeps on nonsynonymous changes that would erase synonymous variants). Global phylogenetic analysis emphasizes this diversity by providing two main subgroups for type 1 genomes in our sequencing set. One group represents core local Kenyan viruses, while the second group is a mixture of viruses from across the globe, with the exception of South Asian viruses that group separately. While previously sequenced type 2 viruses intermingle with western Kenya isolates, the majority of these originated from East Africa, with only a few from West Africa. Interestingly, intermingling is also true for type 2, as we observed two distinct groups. This is more apparent in PCoA, where type 2 virus forms two clusters. Based upon examination by PCoA, the loading values are determined by a broad stretch of the genome from the end of EBNA3C to LMP1, where Mediterranean and Alaskan designations correlate. It remains to be determined whether this substructure might be due to the introduction of previously geographically isolated viruses or distinct evolutionary trajectories within the population. Further study is needed with broader samplings to understand its significance, but our findings suggest that there may be significant epistasis potentially including LMP1.
By sequencing the virus directly from healthy controls, we were able to address the question of relative tumorigenicity between EBV types 1 and 2. We evaluated the long-standing presumption that type 1 virus is more strongly associated with eBL than is type 2. Our work was able to more definitely answer this question since we were not reliant on LCLs from healthy controls, where type 1 bias in transformation might explain the lack of previous associations. We earlier demonstrated, by mutational profiling of EBV-positive and -negative eBL tumors, that the virus, especially type 1, might mitigate the necessity for certain driver mutations in the host genome (16). In addition, our genome-wide results controlling for viral type substantiates investigations of non-type-associated variation that could also impart oncogenic risk, since we found suggestive trends for several nonsynonymous variants as well. Supporting the putative existence of EBV substrains that have increased oncogenic potential, we observed subcliques of solely eBL or control isolates within the type 2 genomes. Although these subgroups were formed with only seven or eight members, the significance of this observation will be deciphered with more extensive cohorts. On the other hand, only a small subset of type 1 viruses from eBL patients carried the BZLF1 promoter variant, which leads to a gain of function (42), while all type 2 viruses carried this variant, suggesting that this promoter might be beneficial for type 2 but makes it unlikely to be a driver of oncogenesis. It is essential to remember that the suggestive associations we uncovered require further validation with independent cohorts and should be treated cautiously.
Overall, our population-based study provides the groundwork to unravel the complexities of EBV genome structure and insight into viral variation that influences oncogenesis. Genomic and mutational analysis of BL tumors identified key differences based on viral content, suggesting new avenues for the development of prognostic molecular biomarkers and the potential for antiviral therapeutic interventions.
MATERIALS AND METHODS
Ethical approval and sample collection.
For this study, we recruited children between 2009 and 2012 with suspected eBL, between 2 and 14 years of age, undergoing initial diagnosis at Jaramogi Oginga Odinga Teaching and Referral Hospital (Kisumu, Kenya), which is a regional referral hospital for pediatric cancer in western Kenya (31). We also enrolled healthy age-matched children residing in the same regions of malaria endemicity in Kenya as controls. We obtained written informed consent from parents to enroll their child in this study. Ethical approval was obtained from the Institutional Review Board at the University of Massachusetts Medical School and the Scientific and Ethical Review Unit at the Kenya Medical Research Institute. From eBL patients, tumor biopsy specimens were collected using fine-needle aspirates (FNAs) and transferred into RNAlater at the bedside, prior to the induction of chemotherapy. Peripheral blood samples were collected from all children and fractionated by centrifugation prior to freezing into plasma and cell pellets. All samples were stored at –80°C prior to nucleic acid extraction.
Cell cultures and controlled mixtures.
The BL cultured cell lines Namalwa, Daudi, Raji, and Jijoye were grown in complete growth medium, RPMI 1640 (Life Technologies), with 2 mM l-glutamine adjusted to contain 1.5 g/liter sodium bicarbonate, 4.5 g/liter glucose, 10 mM HEPES, 1.0 mM sodium pyruvate, and 7.5% fetal bovine serum. We used Jijoye and Daudi as representative genomes of type 1 and type 2 strains. For mixing experiments, we created relative Jijoye/Daudi ratios of 10:90, 25:75, 75:25, and 90:10 in addition to sequencing each strain individually.
Improved enrichment of GC-rich EBV in low-abundance samples.
We used an Allprep DNA/RNA/protein minikit (Qiagen) for DNA isolations from FNAs and a QIAamp DNA kit for blood and plasma. We developed an improved multistep amplification and enrichment process for the GC-rich EBV genome, particularly in samples with low viral copies. We used EBV-specific whole-genome amplification (sWGA) to provide sufficient material and targeted enrichment with hybridization probes after the library preparation. For this, we designed 3′-protected oligonucleotides according to the instructions of Leichty and Brisson (48). For low-viral-load samples, we added a multiplex long-range PCR amplification (mlrPCR) step comprising two sets of nonoverlapping EBV-specific primers tiling across the genome (49). To increase viral DNA content in low abundant specimens, we applied an initial amplification with long-range PCR using a strategy consisting of two multiplexed sets of primers which combine tiled the viral genome, as designed by Kwok et al. (49). To this, we added EBV type 2-specific primers. Following the initial mlrPCRs, we mixed two independent reactions and then performed sWGA using phi29 polymerase with EBV-specific oligonucleotides. The overall DNA quality and quantities were assessed using NanoDrop and Picogreen and purified with 2× XP-Ampure magnetic beads. We prepared two reaction solutions with separate primer pools (2 μl of 10 μM each), using 2.5 μl of 10× long-range PCR buffer Mg2+ (Qiagen), 1.25 μl of deoxynucleoside triphosphate (dNTP; 10 mM each), 0.15 μl of long-range PCR enzyme mix (Qiagen), and 5 μl of 5× Q-solution (Qiagen), to which we added 10 ng of input DNA in 14 μl. The reaction conditions involved initial denaturation at 95°C for 3 min, followed by 20 cycles of 95°C for 30 s, gradient annealing at 58 to 49°C for 15 s each time, and extension at 72°C for 7 min, with a final extension at 68°C for 10 min. We then mixed two independent reactions, denatured at 95°C for 3 min, and then added sWGA reaction buffer that contains 7 μl of 10× phi29 reaction buffer (NEB), 3 μl of dNTP mix (final concentrations, 30 mM dGTP and dCTP and 10 mM dATP and dTTP), 7 μl of EBV-specific protected oligonucleotide mix (10 μM each), 2 μl of phi29 polymerase (10 U/μl; NEB), 0.7 μl of bovine serum albumin (0.1 μg/μl), and 0.3 μl of H2O. For samples with higher viral loads that did not require PCR amplification prior to sWGA, we denatured DNA using the same conditions but replaced the reaction buffer with Tris-EDTA (TE) or TE-Q-solution mix. We incubated the sWGA at 30°C for 16 h, followed by incubation at 65°C for 15 min to stop the reaction. Instead of random hexamers for the MDA (multiple strand displacement amplification) reaction, we used EBV-specific hexamers with 3′-end modification to protect against phi29 exonuclease activity (see the supplemental material for the primer sequences). For WGA with a GenomiPhi v2 kit, we followed the manufacturer’s instructions modified by adding extra 2× dGTP and dCTP. For hybrid capture, we followed the MyBaits (Arbor Biosciences) protocol in accordance with the manufacturer’s recommendations. After incubation at 65°C for 72 h, we purified the hybridization products with streptavidin beads and used Kapa HiFi to amplify the captured library. We quantified viral content with biplex qPCR using primers for viral BALF5 and human β-actin gene (33). For validation of EBV subtypes, we used primers spanning EBNA3C gene producing 153- and 246-bp products for types 1 and 2, respectively (see Table S1 in the supplemental material for all primers).
We improved the amplification yield by adding extra 2× dGTP/dCTP to the amplification buffers, especially for low EBV inputs (10 EBV copies/μl) (Table 5). We also tested the effect of Q-solution (Qiagen) on the sWGA yield and found that EBV yields were almost doubled (Fig. 1A and B). In addition, we found that a prolonged sWGA incubation time (16 h) improved amplification yield compared to a relatively shorter time (8 h). Combining the methods described above allowed for adequate input for hybrid capture even from low-viral-load healthy controls.
TABLE 5.
Optimization of mlrPCR-sWGA and whole-genome amplification reactions
| Variable | EBV input copies for sWGAa | Denaturation buffer | Avg EBV input (genome copy/μl) | Avg human input (β-actin/μl) | dNTP composition (G/C/T/A ratio) | Whole-genome amplification incubation time (h), temp (°C) | EBV DNA output (copy/ng) | Human DNA output (β-actin/ng) | EBV enrichmentb | Human enrichmentc | EBV enrichment/human enrichment |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Input EBV copies, dNTP composition | 10,000 | TE | 62,448 | 3,456 | 30/30/10/10 | 16 h, 30°C | 4,573 | 7 | 1.06 | 0.03 | 35.33 |
| 1,000 | TE | 10,038 | 13,911 | 30/30/10/10 | 16 h, 30°C | 505 | 59 | 1.01 | 0.08 | 12.63 | |
| 100 | TE | 467 | 10,883 | 30/30/10/10 | 16 h, 30°C | 55 | 70 | 2.28 | 0.12 | 19.00 | |
| 10 | TE | 10 | 11,326 | 30/30/10/10 | 16 h, 30°C | 4 | 68 | 8.26 | 0.12 | 68.83 | |
| 1 | TE | 6 | 19,483 | 30/30/10/10 | 16 h, 30°C | 0 | 55 | 0.21 | 0.06 | 3.50 | |
| 0 | TE | 11,551 | 30/30/10/10 | 16 h, 30°C | 76 | 0.23 | 0.00 | ||||
| 10,000 | TE | 62,448 | 3,456 | 30/30/5/5 | 16 h, 30°C | 4,847 | 2 | 1.12 | 0.01 | 112.00 | |
| 1,000 | TE | 10,038 | 13,911 | 30/30/5/5 | 16 h, 30°C | 544 | 75 | 1.09 | 0.11 | 9.91 | |
| 100 | TE | 467 | 10,883 | 30/30/5/5 | 16 h, 30°C | 72 | 65 | 3 | 0.12 | 25.00 | |
| 10 | TE | 10 | 11,326 | 30/30/5/5 | 16 h, 30°C | 3 | 65 | 5.97 | 0.11 | 54.27 | |
| 1 | TE | 6 | 19,483 | 30/30/5/5 | 16 h, 30°C | 0 | 81 | 0.22 | 0.09 | 2.44 | |
| 0 | TE | 11,551 | 30/30/5/5 | 16 h, 30°C | 71 | 0.21 | 0.00 | ||||
| 10,000 | TE | 62,448 | 3,456 | 15/15/5/5 | 16 h, 30°C | 3,483 | 16 | 0.81 | 0.07 | 11.57 | |
| 1,000 | TE | 10,038 | 13,911 | 15/15/5/5 | 16 h, 30°C | 488 | 84 | 0.97 | 0.12 | 8.08 | |
| 100 | TE | 467 | 10,883 | 15/15/5/5 | 16 h, 30°C | 73 | 109 | 3.04 | 0.19 | 16.00 | |
| 10 | TE | 10 | 11,326 | 15/15/5/5 | 16 h, 30°C | 1 | 80 | 2.71 | 0.14 | 19.36 | |
| 1 | TE | 6 | 19,483 | 15/15/5/5 | 16 h, 30°C | 0 | 103 | 0.25 | 0.11 | 2.27 | |
| 0 | TE | 11,551 | 15/15/5/5 | 16 h, 30°C | 118 | 0.35 | 0.00 | ||||
| 10,000 | TE | 62,448 | 3,456 | 15/15/2/2 | 16 h, 30°C | 3,324 | 12 | 0.77 | 0.05 | 15.40 | |
| 1,000 | TE | 10,038 | 13,911 | 15/15/2/2 | 16 h, 30°C | 533 | 77 | 1.06 | 0.11 | 9.64 | |
| 100 | TE | 467 | 10,883 | 15/15/2/2 | 16 h, 30°C | 78 | 105 | 3.24 | 0.19 | 17.05 | |
| 10 | TE | 10 | 11,326 | 15/15/2/2 | 16 h, 30°C | 4 | 105 | 7.8 | 0.18 | 43.33 | |
| 1 | TE | 6 | 19,483 | 15/15/2/2 | 16 h, 30°C | 0 | 110 | 0 | 0.12 | 0.00 | |
| 0 | TE | 11,551 | 15/15/2/2 | 16 h, 30°C | 108 | 0.32 | 0.00 | ||||
| Denaturation buffer, incubation time, and temperature | 10 | TE | 10 | 11,326 | 30/30/5/5 | 8 h, 30°C | 2 | 40 | 6.31 | 0.14 | 45.07 |
| 10 | TE | 10 | 11,326 | 30/30/5/5 | 16 h, 30°C | 2 | 40 | 8.74 | 0.14 | 62.43 | |
| 10 | TE | 10 | 11,326 | 30/30/5/5 | 8 h, 35°C | 1 | 22 | 4.05 | 0.08 | 50.63 | |
| 10 | TE | 10 | 11,326 | 30/30/5/5 | 16 h, 35°C | 2 | 22 | 6.14 | 0.08 | 76.75 | |
| 10 | TE+Q sol | 10 | 11,326 | 30/30/5/5 | 8 h, 30°C | 2 | 41 | 9.44 | 0.14 | 67.43 | |
| 10 | TE+Q sol | 10 | 11,326 | 30/30/5/5 | 16 h, 30°C | 3 | 60 | 13.94 | 0.21 | 66.38 | |
| 10 | TE+Q sol | 10 | 11,326 | 30/30/5/5 | 8 h, 35°C | 3 | 28 | 10.72 | 0.1 | 107.20 | |
| 10 | TE+Q sol | 10 | 11,326 | 30/30/5/5 | 16 h, 35°C | 4 | 49 | 15.13 | 0.17 | 89.00 |
Total EBV genomes = 20 μl of input sWGA.
Calculated as “(EBVpost/EBVpre)/(DNApost/DNApre).”
Calculated as “(humanpost/humanpre)/(DNApost/DNApre).”
Sequence design for RNA baits.
Capture bait sequences were designed using in-house scripts to target both types 1 and 2. In addition to type 1 and type 2 references, we also designed against other available complete genomes, including Mutu I, Akata, GD1, and GD2, to ensure the capture of divergent regions. Specifically, the design consisted of overlapping 120-nucleotide probes tiling every 30 bases (4× overlapping tiling) across the genomic sequences with increased probes for regions with elevated GC content (>65%). Additional probes were added based on the sequential analysis of additional genomes, when current probes were >5% divergent or there was a gap in coverage for a specific region (Table S2).
Sequencing library preparation and hybrid capture enrichment.
Illumina sequencing library preparation steps consisted of DNA shearing, blunt-end repair (Quick Blunting kit; NEB), 3′-adenylation (Klenow fragment 3′ to 5′ exo-; NEB), and ligation of indexed sequencing adaptors (Quick Ligation kit; NEB). We PCR amplified libraries to a final concentration with 10 cycles using KAPA HiFi HotStart ReadyMix and quantified them using a bioanalyzer. We then pooled sample libraries, balancing them according to their EBV content, and proceeded to target enrichment hybridization using custom EBV-specific biotinylated RNA probes (MyBaits; Arbor Biosciences). We performed sequencing using Illumina MiSeq, HiSeq 2000, and NextSeq 500 platforms and 1×75bp, 2×100bp, and 2×150bp, respectively.
Sequence preprocessing and de novo genome assembly.
We checked the sequence quality using FastQC (v0.10.1) after trimming residual adapter and low-quality bases (<20) using cutadapt (v1.7.1) (50) and prinseq (v0.20.4) (51), respectively. After removing reads that mapped to the human genome (hg38), we de novo assembled the remaining reads into contigs with VelvetOptimiser (v2.2.5) (52) using a kmer search ranging from 21 to 149 to maximize N50. We then ordered and oriented the contigs guided by the reference genomes (NC_007605.1 for type 1 and NC_009334.1 for type 2) using ABACAS, extended with read support using IMAGE (53), and merged the overlapping contigs to form larger scaffolds (using in-house scripts). By aligning reads back to scaffolds, we assessed contig quality requiring support from ≥5 unique reads. We created a final genome by demarcating repetitive and missing regions due to low coverage with sequential ambiguous “N” nucleotides. We excluded minor variants (<5% of reads) in final assemblies.
Diversity and variant association analysis.
We used Mafft (v7.215) (54) to generate multiple sequence alignment (msa) of genomes and masked the repetitive regions predefined in the EBV reference genome, NC_007605, in addition to repeat regions larger than 100 nucleotides detected by miropeat (55). For analyses based on viral genes, we extracted the coding region sequences from msa of assemblies according to the reference genome GenBank annotations. The substitutions in coding regions were translated in protein sequences based on a standard genetic code. The genetic distance between the sequences were calculated using the Kimura two-parameter method based on transition and transversion frequencies. We calculated dN/dS rates per gene based on pairwise Nei-Gojobori algorithm using the Python functions provided at https://github.com/a1ultima/hpcleap_dnds/ after excluding frameshift insertions and ambiguous bases. We constructed whole-genome phylogenetic trees based on neighbor-joining method and protein sequence trees based on the maximum-likelihood method with a Jukes-Cantor substitution model using MEGA (v6.0) (56). We determined variant sites of each isolate in reference to the EBV reference genome, NC_007605, based on msa using snp-sites (v2.3.2) (57). For PCoA, we used the R package dartR (v1.0.5) (58). We performed the variant association analysis using the “v-assoc” function from PSEQ/PLINK (59). To control for multiple testing, we calculated empirical P values with one million permutations (pseq proj v-assoc –phenotype eBL –fix-null –perm 1000000) with EBV type stratification, which permutes within types (–strata EBVtype).
Data availability.
Deposited genomes can be accessed in the European Nucleotide Archive (ENA) database under accession no. PRJEB38735 (study accession no. ERP122181), and raw reads can be downloaded from the Sequence Read Archive (SRA) database under BioProject accession number PRJNA552587 (study accession no. SRP212943).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by U.S. National Institutes of Health, National Cancer Institute, grants R01 CA134051 and R01 CA189806 (A.M.M., J.A.B., C.I.O., and Y.K.); The Thrasher Research Fund 02833-7 (A.M.M.); UMCCTS Pilot Project Program U1 LTR000161-04 (Y.K., J.A.B., and A.M.M.); and the Turkish Ministry of National Education Graduate Study Abroad Program (Y.K.). This publication was approved by the Director of KEMRI.
We thank the Kenyan children and their families who participated in this study. We thank Patrick Marsh for help with the EBV genotyping assays and Mercedeh Movassagh for sharing genotyping primers.
Y.K., C.I.O., and O.A. designed and performed experiments. Y.K. and C.I.O. analyzed and interpreted results. Y.K. made the figures. Y.K., J.A.B., and A.M.M. designed the research and wrote the paper. C.I.O., J.A.O., J.M.O., and A.M.M. organized clinical sample acquisition.
We declare there are no competing financial interests.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Young LS, Rickinson AB. 2004. Epstein-Barr virus: 40 years on. Nat Rev Cancer 4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 2.Crawford DH. 2001. Biology and disease associations of Epstein-Barr virus. Philos Trans R Soc Lond B Biol Sci 356:461–473. doi: 10.1098/rstb.2000.0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moormann AM, Bailey JA. 2016. Malaria: how this parasitic infection aids and abets EBV-associated Burkitt lymphomagenesis. Curr Opin Virol 20:78–84. doi: 10.1016/j.coviro.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Torgbor C, Awuah P, Deitsch K, Kalantari P, Duca KA, Thorley-Lawson DA. 2014. A multifactorial role for P falciparum malaria in endemic Burkitt’s lymphoma pathogenesis. PLoS Pathog 10:e1004170. doi: 10.1371/journal.ppat.1004170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simone O, Bejarano MT, Pierce SK, Antonaci S, Wahlgren M, Troye-Blomberg M, Donati D. 2011. TLRs innate immunereceptors and Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) CIDR1α-driven human polyclonal B-cell activation. Acta Trop 119:144–150. doi: 10.1016/j.actatropica.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robbiani DF, Deroubaix S, Feldhahn N, Oliveira TY, Callen E, Wang Q, Jankovic M, Silva IT, Rommel PC, Bosque D, Eisenreich T, Nussenzweig A, Nussenzweig MC. 2015. Plasmodium infection promotes genomic instability and AID-dependent B cell lymphoma. Cell 162:727–737. doi: 10.1016/j.cell.2015.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen JI, Wang F, Mannick J, Kieff E. 1989. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc Natl Acad Sci U S A 86:9558–9562. doi: 10.1073/pnas.86.23.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rowe M, Young LS, Cadwallader K, Petti L, Kieff E, Rickinson AB. 1989. Distinction between Epstein-Barr virus type A (EBNA 2A) and type B (EBNA 2B) isolates extends to the EBNA 3 family of nuclear proteins. J Virol 63:1031–1039. doi: 10.1128/JVI.63.3.1031-1039.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dambaugh T, Hennessy K, Chamnankit L, Kieff E. 1984. U2 region of Epstein-Barr virus DNA may encode Epstein-Barr nuclear antigen 2. Proc Natl Acad Sci U S A 81:7632–7636. doi: 10.1073/pnas.81.23.7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho YG, Gordadze AV, Ling PD, Wang F. 1999. Evolution of two types of rhesus lymphocryptovirus similar to type 1 and type 2 Epstein-Barr virus. J Virol 73:9206–9212. doi: 10.1128/JVI.73.11.9206-9212.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zimber U, Adldinger HK, Lenoir GM, Vuillaume M, Knebel-Doeberitz MV, Laux G, Desgranges C, Wittmann P, Freese UK, Schneider U. 1986. Geographical prevalence of two types of Epstein-Barr virus. Virology 154:56–66. doi: 10.1016/0042-6822(86)90429-0. [DOI] [PubMed] [Google Scholar]
- 12.Apolloni A, Sculley TB. 1994. Detection of A-type and B-type Epstein-Barr virus in throat washings and lymphocytes. Virology 202:978–981. doi: 10.1006/viro.1994.1422. [DOI] [PubMed] [Google Scholar]
- 13.Sixbey JW, Shirley P, Chesney PJ, Buntin DM, Resnick L. 1989. Detection of a second widespread strain of Epstein-Barr virus. Lancet 2:761–765. doi: 10.1016/s0140-6736(89)90829-5. [DOI] [PubMed] [Google Scholar]
- 14.Correia S, Palser A, Elgueta Karstegl C, Middeldorp JM, Ramayanti O, Cohen JI, Hildesheim A, Fellner MD, Wiels J, White RE, Kellam P, Farrell PJ. 2017. Natural variation of Epstein-Barr virus genes, proteins and pri-miRNA (revised). J Virol 91:e00375-17. doi: 10.1128/JVI.00375-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young LS, Yao QY, Rooney CM, Sculley TB, Moss DJ, Rupani H, Laux G, Bornkamm GW, Rickinson AB. 1987. New type B isolates of Epstein-Barr virus from Burkitt’s lymphoma and from normal individuals in endemic areas. J Gen Virol 68:2853–2862. doi: 10.1099/0022-1317-68-11-2853. [DOI] [PubMed] [Google Scholar]
- 16.Kaymaz Y, Oduor CI, Yu H, Otieno JA, Ong’echa JM, Moormann AM, Bailey JA. 2017. Comprehensive transcriptome and mutational profiling of endemic Burkitt lymphoma reveals EBV type-specific differences. Mol Cancer Res 15:563–576. doi: 10.1158/1541-7786.MCR-16-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lucchesi W, Brady G, Dittrich-Breiholz O, Kracht M, Russ R, Farrell PJ. 2008. Differential gene regulation by Epstein-Barr virus type 1 and type 2 EBNA2. J Virol 82:7456–7466. doi: 10.1128/JVI.00223-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaye KM, Izumi KM, Kieff E. 1993. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci U S A 90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wohlford EM, Asito AS, Chelimo K, Sumba PO, Baresel PC, Oot RA, Moormann AM, Rochford R. 2013. Identification of a novel variant of LMP-1 of EBV in patients with endemic Burkitt lymphoma in western Kenya. Infect Agents Cancer 8:34. doi: 10.1186/1750-9378-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang CM, Yu KJ, Mbulaiteye SM, Hildesheim A, Bhatia K. 2009. The extent of genetic diversity of Epstein-Barr virus and its geographic and disease patterns: a need for reappraisal. Virus Res 143:209–221. doi: 10.1016/j.virusres.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiara M, Manzari C, Lionetti C, Mechelli R, Anastasiadou E, Chiara Buscarinu M, Ristori G, Salvetti M, Picardi E, D’Erchia AM, Pesole G, Horner DS. 2016. Geographic population structure in Epstein-Barr virus revealed by comparative genomics. Genome Biol Evol 8:3284–3291. doi: 10.1093/gbe/evw226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou L, Chen J-N, Qiu X-M, Pan Y-H, Zhang Z-G, Shao C-K. 2017. Comparative analysis of 22 Epstein-Barr virus genomes from diseased and healthy individuals. J Gen Virol 98:96–107. doi: 10.1099/jgv.0.000699. [DOI] [PubMed] [Google Scholar]
- 23.Xu M, Yao Y, Chen H, Zhang S, Cao S-M, Zhang Z, Luo B, Liu Z, Li Z, Xiang T, He G, Feng Q-S, Chen L-Z, Guo X, Jia W-H, Chen M-Y, Zhang X, Xie S-H, Peng R, Chang ET, Pedergnana V, Feng L, Bei J-X, Xu R-H, Zeng M-S, Ye W, Adami H-O, Lin X, Zhai W, Zeng Y-X, Liu J. 2019. Genome sequencing analysis identifies Epstein-Barr virus subtypes associated with high risk of nasopharyngeal carcinoma. Nat Genet 51:1131–1136. doi: 10.1038/s41588-019-0436-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Power RA, Davaniah S, Derache A, Wilkinson E, Tanser F, Gupta RK, Pillay D, de Oliveira T. 2016. Genome-wide association study of HIV whole-genome sequences validated using drug resistance. bioRxiv https://www.biorxiv.org/content/10.1101/076216v1. [DOI] [PMC free article] [PubMed]
- 25.Depledge DP, Palser AL, Watson SJ, Lai I-C, Gray ER, Grant P, Kanda RK, Leproust E, Kellam P, Breuer J. 2011. Specific capture and whole-genome sequencing of viruses from clinical samples. PLoS One 6:e27805. doi: 10.1371/journal.pone.0027805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwok H, Wu CW, Palser AL, Kellam P, Sham PC, Kwong DLW, Chiang A. 2014. Genomic diversity of Epstein-Barr virus genomes isolated from primary nasopharyngeal carcinoma biopsy samples. J Virol 88:10662–10672. doi: 10.1128/JVI.01665-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, Yang W, Pan Y, Ji J, Lu Z, Ke Y. 2016. Genome-wide analysis of Epstein-Barr virus (EBV) isolated from EBV-associated gastric carcinoma (EBVaGC). Oncotarget 7:4903–4914. doi: 10.18632/oncotarget.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S, Xiong H, Yan S, Wu N, Lu Z. 2016. Identification and characterization of Epstein-Barr virus genomes in lung carcinoma biopsy samples by next-generation sequencing technology. Sci Rep 6:26156. doi: 10.1038/srep26156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lei H, Li T, Li B, Tsai S, Biggar RJ, Nkrumah F, Neequaye J, Gutierrez M, Epelman S, Mbulaiteye SM, Bhatia K, Lo S-C. 2015. Epstein-Barr virus from Burkitt Lymphoma biopsies from Africa and South America share novel LMP-1 promoter and gene variations. Sci Rep 5:16706. doi: 10.1038/srep16706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parras-Moltó M, López-Bueno A. 2018. Methods for enrichment and sequencing of oral viral assemblages: saliva, oral mucosa, and dental plaque viromes. Methods Mol Biol 1838:143–161. doi: 10.1007/978-1-4939-8682-8_11. [DOI] [PubMed] [Google Scholar]
- 31.Buckle G, Maranda L, Skiles J, Ong’echa JM, Foley J, Epstein M, Vik TA, Schroeder A, Lemberger J, Rosmarin A, Remick SC, Bailey JA, Vulule J, Otieno JA, Moormann AM. 2016. Factors influencing survival among Kenyan children diagnosed with endemic Burkitt lymphoma between 2003 and 2011: a historical cohort study. Int J Cancer 139:1231–1240. doi: 10.1002/ijc.30170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rainey JJ, Mwanda WO, Wairiumu P, Moormann AM, Wilson ML, Rochford R. 2007. Spatial distribution of Burkitt’s lymphoma in Kenya and association with malaria risk. Trop Med Int Health 12:936–943. doi: 10.1111/j.1365-3156.2007.01875.x. [DOI] [PubMed] [Google Scholar]
- 33.Moormann AM, Chelimo K, Sumba OP, Lutzke ML, Ploutz-Snyder R, Newton D, Kazura J, Rochford R. 2005. Exposure to holoendemic malaria results in elevated Epstein-Barr virus loads in children. J Infect Dis 191:1233–1238. doi: 10.1086/428910. [DOI] [PubMed] [Google Scholar]
- 34.Palser AL, Grayson NE, White RE, Corton C, Correia S, Ba Abdullah MM, Watson SJ, Cotten M, Arrand JR, Murray PG, Allday MJ, Rickinson AB, Young LS, Farrell PJ, Kellam P. 2015. Genome diversity of Epstein-Barr virus from multiple tumor types and normal infection. J Virol 89:5222–5237. doi: 10.1128/JVI.03614-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muncunill J, Baptista M-J, Hernandez-Rodríguez Á, Dalmau J, Garcia O, Tapia G, Moreno M, Sancho J-M, Martínez-Picado J, Feliu E, Mate J-L, Ribera J-M, Navarro J-T. 2019. Plasma Epstein-Barr virus load as an early biomarker and prognostic factor of human immunodeficiency virus-related lymphomas. Clin Infect Dis 68:834–843. doi: 10.1093/cid/ciy542. [DOI] [PubMed] [Google Scholar]
- 36.Westmoreland KD, Montgomery ND, Stanley CC, El-Mallawany NK, Wasswa P, van der Gronde T, Mtete I, Butia M, Itimu S, Chasela M, Mtunda M, Kampani C, Liomba NG, Tomoka T, Dhungel BM, Sanders MK, Krysiak R, Kazembe P, Dittmer DP, Fedoriw Y, Gopal S. 2017. Plasma Epstein-Barr virus DNA for pediatric Burkitt lymphoma diagnosis, prognosis and response assessment in Malawi. Int J Cancer 140:2509–2516. doi: 10.1002/ijc.30682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Correia S, Bridges R, Wegner F, Venturini C, Palser A, Middeldorp JM, Cohen JI, Lorenzetti MA, Bassano I, White RE, Kellam P, Breuer J, Farrell PJ. 2018. Sequence variation of Epstein-Barr virus: viral types, geography, codon usage, and diseases. J Virol 92:e01132-18. doi: 10.1128/JVI.01132-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sears J, Ujihara M, Wong S, Ott C, Middeldorp J, Aiyar A. 2004. The amino terminus of Epstein-Barr virus (EBV) nuclear antigen 1 contains at hooks that facilitate the replication and partitioning of latent EBV genomes by tethering them to cellular chromosomes. J Virol 78:11487–11505. doi: 10.1128/JVI.78.21.11487-11505.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanda T, Horikoshi N, Murata T, Kawashima D, Sugimoto A, Narita Y, Kurumizaka H, Tsurumi T. 2013. Interaction between basic residues of Epstein-Barr virus EBNA1 protein and cellular chromatin mediates viral plasmid maintenance. J Biol Chem 288:24189–24199. doi: 10.1074/jbc.M113.491167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henson BW, Perkins EM, Cothran JE, Desai P. 2009. Self-assembly of Epstein-Barr virus capsids. J Virol 83:3877–3890. doi: 10.1128/JVI.01733-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tarbouriech N, Ruggiero F, de Turenne-Tessier M, Ooka T, Burmeister WP. 2006. Structure of the Epstein-Barr virus oncogene BARF1. J Mol Biol 359:667–678. doi: 10.1016/j.jmb.2006.03.056. [DOI] [PubMed] [Google Scholar]
- 42.Bristol JA, Djavadian R, Albright ER, Coleman CB, Ohashi M, Hayes M, Romero-Masters JC, Barlow EA, Farrell PJ, Rochford R, Kalejta RF, Johannsen EC, Kenney SC. 2018. A cancer-associated Epstein-Barr virus BZLF1 promoter variant enhances lytic infection. PLoS Pathog 14:e1007179. doi: 10.1371/journal.ppat.1007179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mulama DH, Bailey JA, Foley J, Chelimo K, Ouma C, Jura WG, Otieno J, Vulule J, Moormann AM. 2014. Sickle cell trait is not associated with endemic Burkitt lymphoma: an ethnicity and malaria endemicity-matched case-control study suggests factors controlling EBV may serve as a predictive biomarker for this pediatric cancer. Int J Cancer 134:645–653. doi: 10.1002/ijc.28378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cho S-G, Lee W-K. 2000. Analysis of genetic polymorphisms of Epstein-Barr virus isolates from cancer patients and healthy carriers. J Microbiol Biotechnol 10:620–627. [Google Scholar]
- 45.Burrows JM, Khanna R, Sculley TB, Alpers MP, Moss DJ, Burrows SR. 1996. Identification of a naturally occurring recombinant Epstein-Barr virus isolate from New Guinea that encodes both type 1 and type 2 nuclear antigen sequences. J Virol 70:4829–4833. doi: 10.1128/JVI.70.7.4829-4833.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yao QY, Tierney RJ, Croom-Carter D, Cooper GM, Ellis CJ, Rowe M, Rickinson AB. 1996. Isolation of intertypic recombinants of Epstein-Barr virus from T-cell-immunocompromised individuals. J Virol 70:4895–4903. doi: 10.1128/JVI.70.8.4895-4903.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skare J, Farley J, Strominger JL, Fresen KO, Cho MS, Zur Hausen H. 1985. Transformation by Epstein-Barr virus requires DNA sequences in the region of BamHI fragments Y and H. J Virol 55:286–297. doi: 10.1128/JVI.55.2.286-297.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leichty AR, Brisson D. 2014. Selective whole genome amplification for resequencing target microbial species from complex natural samples. Genetics 198:473–481. doi: 10.1534/genetics.114.165498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwok H, Tong AHY, Lin CH, Lok S, Farrell PJ, Kwong DLW, Chiang A. 2012. Genomic sequencing and comparative analysis of Epstein-Barr virus genome isolated from primary nasopharyngeal carcinoma biopsy. PLoS One 7:e36939. doi: 10.1371/journal.pone.0036939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet j 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 51.Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Victorian Bioinformatics Consortium. 2012. VelvetOptimiser. http://www.vicbioinformatics.com/software.velvetoptimiser.shtml.
- 53.Swain MT, Tsai IJ, Assefa SA, Newbold C, Berriman M, Otto TD. 2012. A post-assembly genome-improvement toolkit (PAGIT) to obtain annotated genomes from contigs. Nat Protoc 7:1260–1284. doi: 10.1038/nprot.2012.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parsons JD. 1995. Miropeats: graphical DNA sequence comparisons. Comput Appl Biosci 11:615–619. doi: 10.1093/bioinformatics/11.6.615. [DOI] [PubMed] [Google Scholar]
- 56.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Page AJ, Taylor B, Delaney AJ, Soares J, Seemann T, Keane JA, Harris SR. 2016. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb Genom 2:e000056. doi: 10.1099/mgen.0.000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gruber B, Unmack PJ, Berry OF, Georges A. 2018. dartr: an r package to facilitate analysis of SNP data generated from reduced representation genome sequencing. Mol Ecol Resour 18:691–699. doi: 10.1111/1755-0998.12745. [DOI] [PubMed] [Google Scholar]
- 59.PLINK/SEQ. 2014. PLINK/SEQ: a library for the analysis of genetic variation data. https://atgu.mgh.harvard.edu/plinkseq/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Deposited genomes can be accessed in the European Nucleotide Archive (ENA) database under accession no. PRJEB38735 (study accession no. ERP122181), and raw reads can be downloaded from the Sequence Read Archive (SRA) database under BioProject accession number PRJNA552587 (study accession no. SRP212943).