Induction of autophagy through inhibition of MTOR has been shown to inhibit HIV replication. However, inhibition of the mechanistic target of rapamycin (MTOR) has cellular effects that may alter HIV infection through other mechanisms. Here, we examined the HIV-inhibitory effects of the MTOR-independent inducer of autophagy, trehalose. Of note, we identified that in addition to the inhibition of the intracellular replication of HIV by autophagy, trehalose decreased viral entry in human primary macrophages and CD4+ T cells through the downregulation of C-C motif chemokine receptor 5 (CCR5) in T cells and CD4 in both T cells and macrophages. Thus, we showed that trehalose uniquely inhibits HIV replication through inhibition of viral entry and intracellular degradation in the two most important target cells for HIV infection.
KEYWORDS: CCR5, CD4, HIV-1, MTOR, T cells, trehalose, autophagy, macrophages
ABSTRACT
Autophagy is a highly conserved recycling pathway that promotes cell survival during periods of stress. We previously reported that induction of autophagy through the inhibition of the mechanistic target of rapamycin (MTOR) inhibits HIV replication in human macrophages and CD4+ T lymphocytes (T cells). However, the inhibition of MTOR has modulatory effects beyond autophagy that might affect viral replication. Here, we examined the effect on HIV replication of trehalose, a nontoxic, nonreducing disaccharide that induces autophagy through an MTOR-independent mechanism. Treatment of HIV-infected macrophages and T cells with trehalose inhibited infection in a dose-dependent manner. Uninfected and HIV-infected macrophages and T cells treated with trehalose exhibited increased markers of autophagy, including LC3B lipidation with further accumulation following bafilomycin A1 treatment, and increased levels of LAMP1, LAMP2, and RAB7 proteins required for lysosomal biogenesis and fusion. Moreover, the inhibition of HIV by trehalose was significantly reduced by knockdown of ATG5. Additionally, trehalose downregulated the expression of C-C motif chemokine receptor 5 (CCR5) in T cells and CD4 in both T cells and macrophages, which reduced HIV entry in these cells. Our data demonstrate that the naturally occurring sugar trehalose at doses safely achieved in humans inhibits HIV through two mechanisms: (i) decreased entry through the downregulation of CCR5 in T cells and decreased CD4 expression in both T cells and macrophages and (ii) degradation of intracellular HIV through the induction of MTOR-independent autophagy. These findings demonstrate that cellular mechanisms can be modulated to inhibit HIV entry and intracellular replication using a naturally occurring, nontoxic sugar.
IMPORTANCE Induction of autophagy through inhibition of MTOR has been shown to inhibit HIV replication. However, inhibition of the mechanistic target of rapamycin (MTOR) has cellular effects that may alter HIV infection through other mechanisms. Here, we examined the HIV-inhibitory effects of the MTOR-independent inducer of autophagy, trehalose. Of note, we identified that in addition to the inhibition of the intracellular replication of HIV by autophagy, trehalose decreased viral entry in human primary macrophages and CD4+ T cells through the downregulation of C-C motif chemokine receptor 5 (CCR5) in T cells and CD4 in both T cells and macrophages. Thus, we showed that trehalose uniquely inhibits HIV replication through inhibition of viral entry and intracellular degradation in the two most important target cells for HIV infection.
INTRODUCTION
Antiretroviral therapy (ART) has significantly altered the course of human immunodeficiency type 1 (HIV) infection, and those receiving treatment can achieve sustained viral suppression. Current antiretrovirals target multiple stages of the HIV life cycle, including attachment, fusion, entry, reverse transcription, integration, and maturation (1). Whereas multiple FDA-approved drugs target HIV reverse transcription, integration, and maturation, only enfuvirtide (inhibiting fusion) and maraviroc (targeting CCR5) inhibit viral entry (2, 3). HIV entry is initiated through the attachment and fusion of the envelope protein gp120, gp41, the host cell CD4 receptor, and a coreceptor, usually either C-C motif chemokine receptor 5 (CCR5) or C-X-C motif chemokine receptor 4 (CXCR4) (4).
Macroautophagy (here referred to as autophagy) is a cellular pathway responsible for recycling amino acids from degraded long-lived proteins or damaged organelles to promote cellular homeostasis during times of stress or starvation (5). Initiation of autophagy is mediated by a complex consisting of Unc-51-like kinase 1 or 2 (ULK1/2), ATG13, and focal adhesion kinase family interacting protein of 200 kDa (FIP200) (6). The mechanistic target of rapamycin (MTOR) is a master regulator of cell growth and autophagy. Under nutrient-rich conditions, MTOR is associated with the ULK complex and remains inactivated through phosphorylation, inhibiting autophagy initiation. However, during cell stress, starvation, or sirolimus (rapamycin) treatment, MTOR dissociates from the ULK complex, becomes hypophosphorylated, and initiates autophagy (7). Following initiation, the nucleation complex, which consists of p150, ATG14, and Beclin1, is recruited to initiate the formation of a double membrane structure known as a phagophore. Subsequently, the ATG12-ATG5-ATG16L complex is recruited to mediate phagophore elongation (8). Concurrently, microtubule-associated protein 1 light chain 3 beta (LC3B-I, cytosolic) is conjugated to phosphatidylethanolamine (PE) through a process involving ATG4, ATG7, and ATG3 to form LC3B-II, which tethers to the growing phagophore. As the phagophore grows, it matures until it eventually sequesters a portion of the cytosol and cargo within a double-membrane vesicle, termed the autophagosome (8). The outer membrane of the autophagosome then fuses with the membrane of a lysosome to form the autolysosome, where the internal compartment is degraded and the contents are released back into the cytosol for reuse (8, 9). Recent studies suggest that autophagy plays an important role in the regulation of various cellular pathways, such as aging, inflammation, immune activation, immune response to microbial pathogens (10–12), and neurodegenerative diseases, such as Alzheimer’s, Parkinson’s, and Huntington’s disease (13, 14).
An increasing number of studies indicate that autophagy plays an important role in HIV pathogenesis. Our research and that of others has shown that upon initial entry into cells, HIV induces autophagy to promote its own replication. However, once HIV has established a permissive infection, viral proteins inhibit autophagy to promote cell survival and viral persistence. Induction of autophagy by rapamycin and other MTOR inhibitors inhibits HIV replication, and the overinduction of a Na+/K+-ATPase-dependent form of autophagy, termed autosis, can selectively kill latently HIV-infected resting memory T lymphocytes (15).
Although an abundance of research has focused on MTOR-dependent autophagy, MTOR-independent autophagy, including the inositol pathway and the calcium/calpain pathway, is also important for cellular homeostasis (16–19). Additionally, multiple inducers of MTOR-independent autophagy have been identified, including SMER28, spermidine, and trehalose (20–26). Of the MTOR-independent inducers of autophagy, trehalose has been studied most extensively. Trehalose is a natural sugar composed of two glucose units joined by an alpha-alpha-1 linkage. (27). It is the most common sugar found in the blood circulation of insects (28). Humans are unable to synthesize trehalose, but they possess trehalase, which can hydrolyze trehalose in the small intestine (29). Trehalose treatment promotes the clearance of mutant proteins associated with Alzheimer’s, Huntington’s, and prion diseases (26, 30–32). In addition, trehalose has been found to inhibit human cytomegalovirus (HCMV) in a variety of cell types (33). The mechanism of trehalose-induced autophagy remains controversial. One group has proposed that trehalose treatment inhibits several cellular glucose transporters (GLUT), which in turn decreases the cellular ATP level and activates the AMP-activated protein kinase (AMPK) and ULK1 phosphorylation to induce autophagy (34, 35). Additionally, trehalose induces autophagy in human foreskin fibroblasts (HEFs) by altering the levels of RAB7 and Rab11 proteins without affecting their cellular glucose uptake (36) and via lysosome-mediated transcription factor EB (TFEB) activation in a motor neuron degeneration model (37). Currently, trehalose is considered a “generally recognized as safe” (GRAS) substance by the U.S. Food and Drug Administration, which makes it an attractive potential therapeutic agent (27). In this report, we examined the effects of trehalose-mediated autophagy on HIV replication.
RESULTS
Trehalose inhibits HIV replication in human primary macrophages.
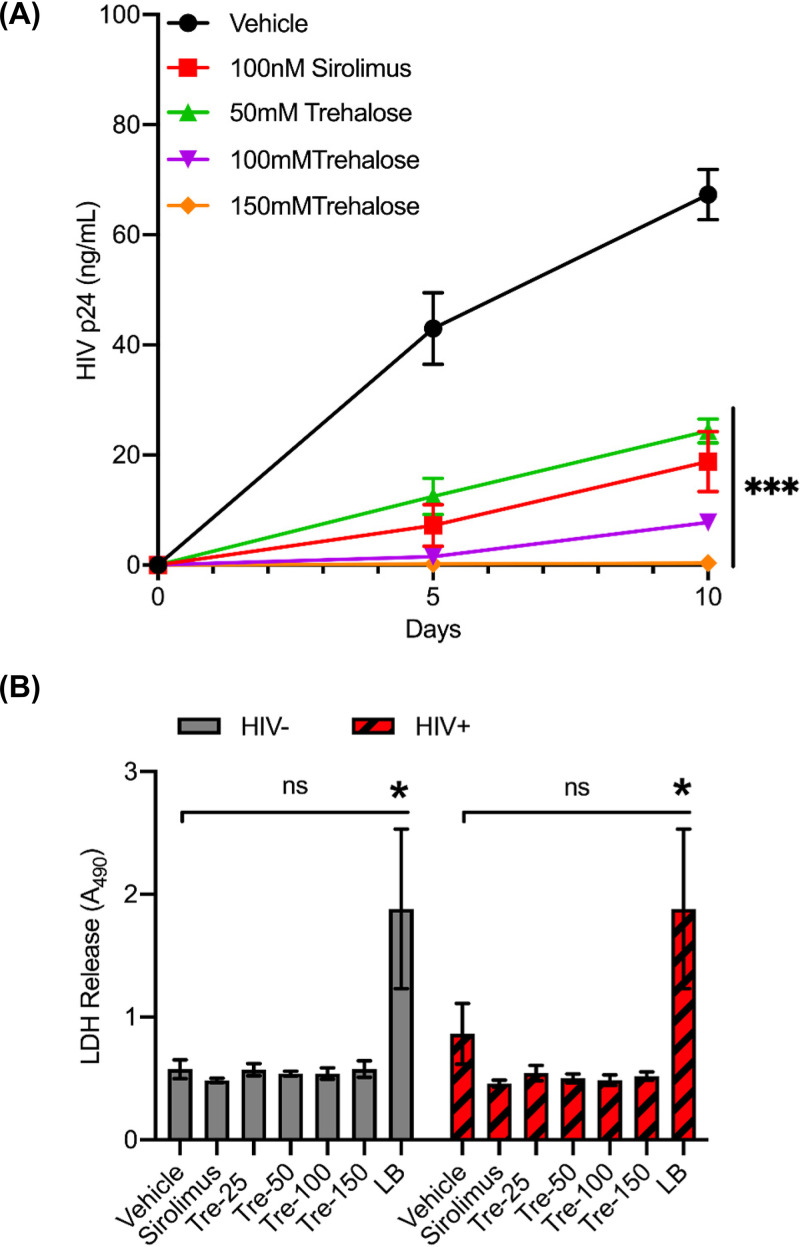
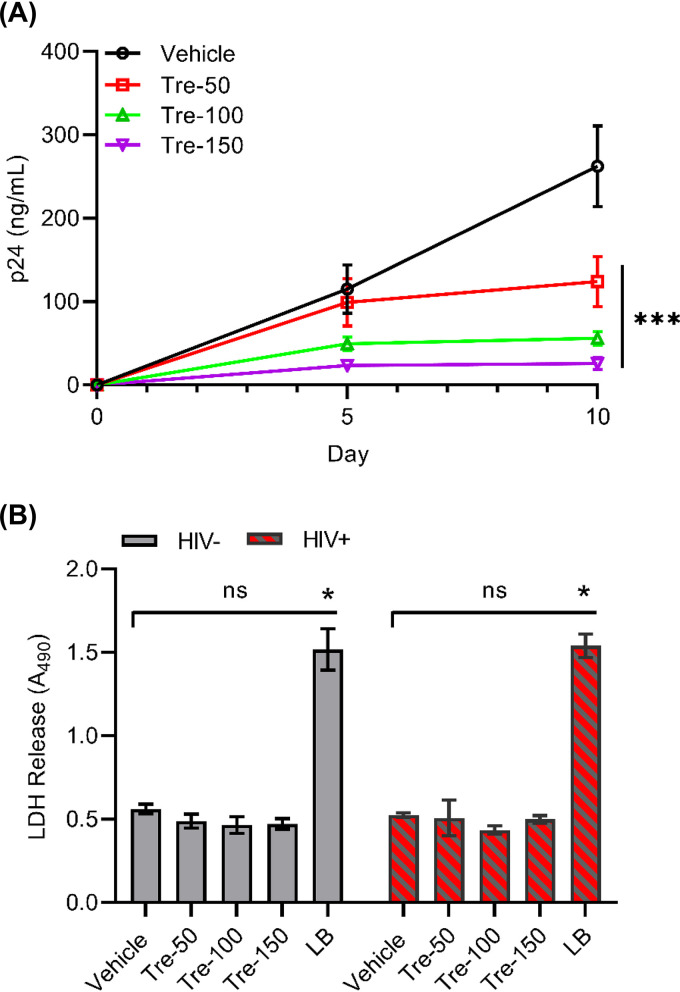
HIV modulates the autophagy pathway to its advantage by differentially regulating the early and late degradative stages to establish a permissive infection in human macrophages (38–40). We have shown earlier that chemical inducers of autophagy can inhibit HIV replication via induction of MTOR-dependent autophagy (41–43). In our current study, we first analyzed the effects of MTOR-independent autophagy on HIV replication in human macrophages. Untreated and trehalose-treated (50 mM to 150 mM) macrophages were infected with HIV (0.04 multiplicity of infection [MOI]) for 8 h and incubated for 10 days in medium in the presence or absence of trehalose. Culture supernatants were tested for p24 antigen production at day 5 and day 10 postinfection (p.i.) by enzyme-linked immunosorbent assay (ELISA). Sirolimus-pretreated (100 nM) infected macrophages were used as controls for HIV inhibition by autophagy. Trehalose pretreatment decreased the quantity of HIV p24 release in culture supernatants, resulting in a >70% reduction in p24 release at 50 mM and a >85% reduction in both 100-mM and 150- mM trehalose pretreatment groups by day 10 p.i. (P < 0.001; Fig. 1A).
FIG 1.
Trehalose is a noncytotoxic inhibitor of HIV replication in human macrophages. (A) Macrophages were pretreated with vehicle, 100 nM sirolimus, and 50 mM to 150 mM trehalose for 12 h prior to exposure to HIV (0.04 MOI). Cell culture supernatants were collected at days 5 and 10 p.i. for extracellular p24 antigen quantification by ELISA. Data are derived from four independent donors and presented as means ± the standard error of the mean (s.e.m.). (B) Uninfected and HIV-infected macrophages were incubated with vehicle, 100 nM sirolimus, or increasing concentrations of trehalose for 10 days. At day 10, cell culture supernatants were collected, and spectrophotometric measurement of extracellular LDH was used to determine cellular toxicity. Uninfected cells treated with 1× lysis buffer (LB) were used as a positive control. Data are derived from three independent donors and presented as means ± s.e.m. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Though trehalose is considered a GRAS substance and has been approved for human consumption at up to 50 g/day, it is unknown if it exerts any cytotoxicity of human macrophages that might result in what appears to be inhibition of HIV infection. Therefore, we also assessed the cytotoxicity profile of trehalose-treated macrophages by measuring the extracellular lactate dehydrogenase (LDH) release in the culture supernatants collected from macrophages treated with trehalose (25 mM to 150 mM) for 10 days alone or with HIV infection (Fig. 1B). Sirolimus-treated (100 nM) and vehicle-treated macrophages were used as controls. Trehalose-treated uninfected and HIV-infected macrophages showed minimal change in cell survival after 10 days of treatment at different concentrations of trehalose. Spectrophotometric measurements did not show any significant increase in LDH release in the culture supernatants in the presence of trehalose in both uninfected and HIV-infected macrophages (P > 0.1; Fig. 1B).
Trehalose increases autophagic flux in primary human macrophages by inducing TFEB nuclear translocation and activation of autophagy genes.
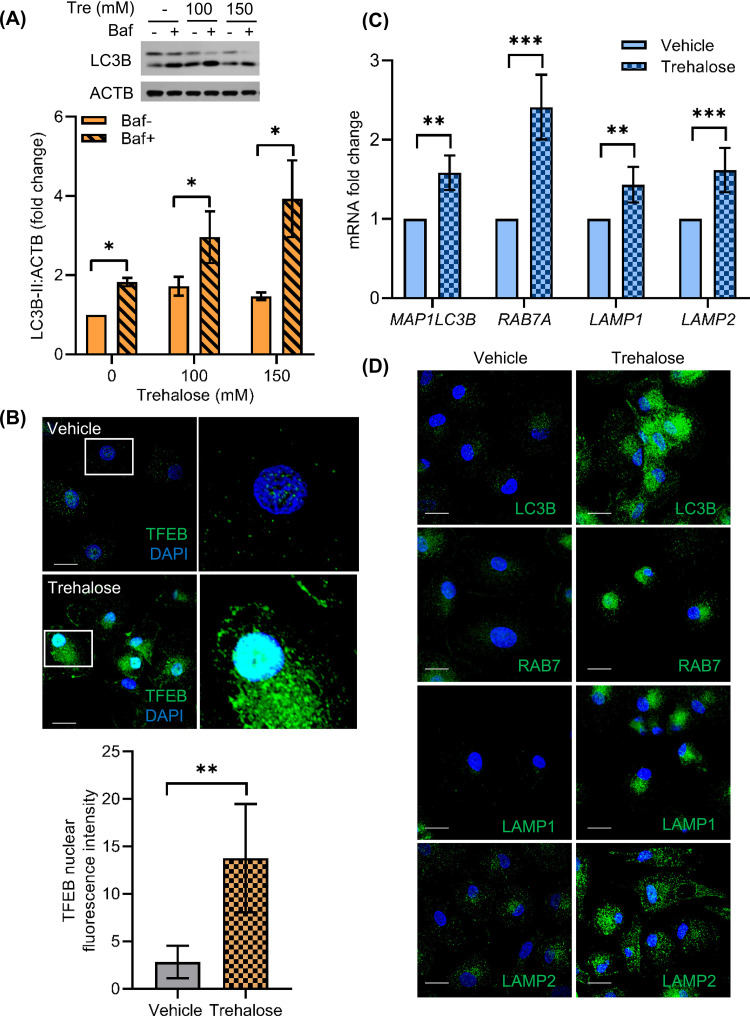
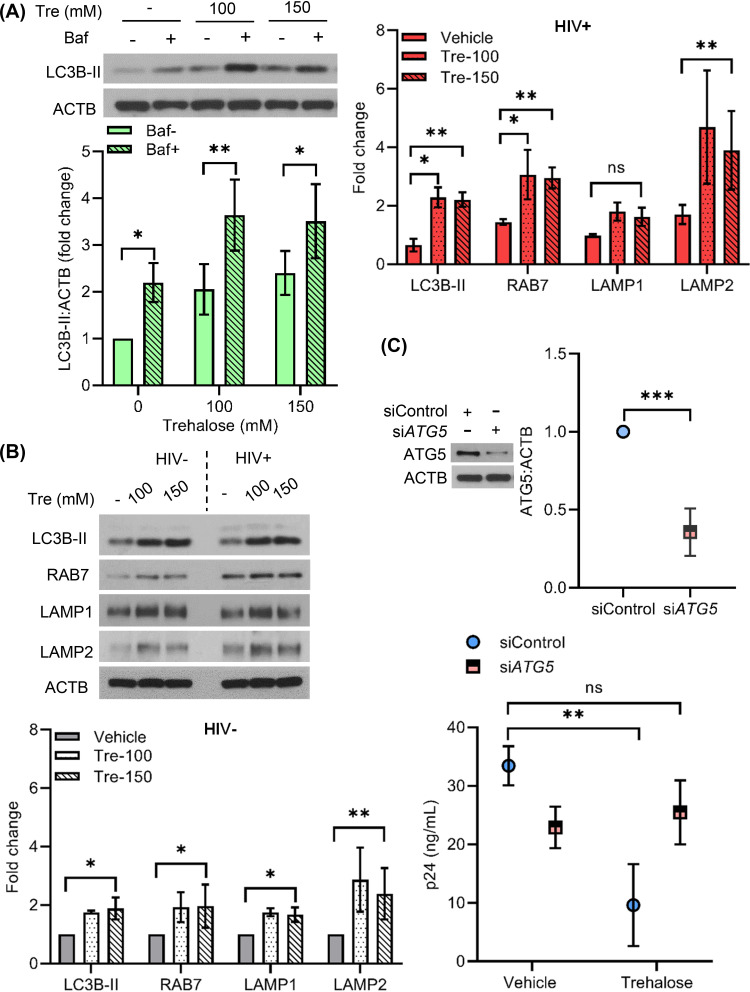
Trehalose has been identified to induce autophagy in several human cell types, including fibroblasts, keratinocytes, aortic endothelial cells, and neurons (26, 31, 33, 44). Therefore, we sought to confirm that autophagy induction was occurring in HIV-infected macrophages and was responsible for the inhibition of infection. The initial steps of autophagy include formation of autophagosomes, which involves the lipidation of LC3B-I to form LC3B-II. The mature autophagosome then fuses with lysosome to form the autolysosome, where autophagic cargo degradation and LC3B-II turnover takes place. Relative quantification of LC3B-II is therefore used as an important marker to assess autophagy in cells. To assess trehalose-mediated modulation of autophagic flux, we incubated macrophages in medium in the presence or absence of 100 mM and 150 mM trehalose for 12 to 24 h. Cells were harvested for lysate preparation and analyzed by Western blotting using antibody to LC3B. Macrophages treated with 100 mM and 150 mM trehalose exhibited 1.7- and 1.5-fold higher LCB-II levels, respectively, compared to untreated cells (P = 0.03 and P = 0.02, respectively; Fig. 2A). Increased LC3B-II expression can be due to an increase in autophagosome formation or their accumulation because of blockage of the autophagy pathway at the autophagosome and lysosome fusion step. SQSTM1 degradation is generally used as a marker to measure the completion of autophagic flux (45), but some recent studies, including one that specifically studied the effects of trehalose in keratinocytes, suggest that SQSTM1 degradation may not be directly correlated with increased LC3B lipidation (31, 46, 47). Therefore, in our experiments we included a 2-h cotreatment with bafilomycin A1 to prevent the turnover of LC3B-II by blocking the autolysosome acidification via inhibition of the vacuolar H+ ATPase. As such, if trehalose-treated cells exhibit a further increase in LC3B-II levels, it would indicate that autophagic flux was going to completion (43, 47–50). In the presence of bafilomycin A1, we observed 1.8- and 2.7-fold higher LC3B lipidation in macrophages incubated with 100 mM and 150 mM trehalose, respectively (P = 0.04 and P = 0.03, respectively; Fig. 2A). Thus, trehalose treatment induces autophagic flux in macrophages.
FIG 2.
Trehalose induces autophagy in human macrophages via activation of TFEB nuclear translocation. Macrophages were treated with trehalose (Tre 100 mM, 150 mM) for 12 h. Bafilomycin A1 (Baf) cotreatment was used to confirm the increase in autophagy flux in the presence of trehalose. Cells were harvested, lysed, and analyzed for LC3B lipidation by immunoblotting. (A) (Top) Representative immunoblot showing expression of LC3B-II and ACTB. (Bottom) Relative fold change (densitomeric analysis) in LC3B-II expression normalized to ACTB. Data are derived from three independent donors and presented as means ± s.e.m. (B) Macrophages were treated with vehicle or 100 mM trehalose for 24 h and analyzed for TFEB nuclear translocation by IF analysis using anti-TFEB (green) antibody and DAPI nuclear stain (blue). (Top) Representative immunofluorescent images showing TFEB localization in vehicle- or trehalose-treated macrophages. (Bottom) The bar graph represents the quantification of TFEB nuclear intensity in >20 cells for each sample analyzed. Scale bar = 10 μM (n = 3). (C) RT-qPCR analysis on the following mRNAs: MAP1LC3B, RAB7A, LAMP1, and LAMP2 in vehicle or trehalose-treated (100 mM) macrophages. The relative fold difference in the mRNA expression was determined using vehicle-treated cells as a control. Data are derived from three independent donors and presented as means ± s.e.m. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (D) Macrophages were treated with vehicle or 100 mM trehalose for 24 h, and IF analysis was performed with anti-LC3B (green), anti-RAB7 (green), LAMP1 (green), and LAMP2 (green) antibodies, and nuclei were stained with DAPI (blue). Scale bar = 10 μM (n = 3).
Trehalose has been shown to induce nuclear translocation of TFEB and upregulation of TFEB target genes, including lysosomal hydrolases, lysosomal membrane proteins (LAMP), and several other autophagy-related proteins, including BECN1 and LC3B (37, 51, 52). In the case of HCMV infection, trehalose treatment increased the levels of RAB7 protein, enhancing lysosomal biogenesis, vacuolation and acidification, and HCMV autophagic degradation in infected HFE cells (36). In order to understand the mechanism of trehalose-induced autophagy in macrophages, we first evaluated whether trehalose treatment modulated TFEB nuclear translocation in these cells. Macrophages were incubated in the absence or presence of 100 mM trehalose for 24 h, and the expression and localization of TFEB were analyzed by confocal immunofluorescence (IF) microscopy. Following 24 h of treatment with 100 mM trehalose, both untreated and trehalose-treated macrophages were fixed, permeabilized, stained with antibody to TFEB and DAPI (4′,6-diamidino-2-phenylindole) nuclear stain, and analyzed by confocal IF microscopy. Untreated macrophages expressed very low TFEB, which was mostly confined to the cytoplasm (Fig. 2B, top). Following 24-h treatment with 100 mM trehalose, TFEB expression and nuclear translocation were significantly increased in macrophages (Fig. 2B, top). In the presence of trehalose, macrophages exhibited 10-fold higher TFEB nuclear translocation (Fig. 2B, bottom; nuclear TFEB quantification P = 0.009). These data suggest that trehalose treatment induces both TFEB expression and nuclear translocation in human primary macrophages.
Having shown that trehalose induces TFEB nuclear translocation in macrophages, we next evaluated the expression of lysosomal biogenesis and autophagy-related genes LAMP1, LAMP2, MAP1LC3B, and RAB7A by reverse transcriptase quantitative PCR (RT-qPCR) (Fig. 2C) and confocal immunofluorescence microscopy (Fig. 2D). In the presence of trehalose, macrophages exhibited a ≥1.7-fold increase in mRNA expression of autophagy and lysosomal biogenesis-related genes MAP1LC3B, RAB7A, LAMP1, and LAMP2 compared to untreated cells (P < 0.01; Fig. 2C). IF images also confirmed that trehalose treatment not only increased TFEB expression and nuclear translocation (Fig. 2B), but also induced expression of autophagy and lysosomal biogenesis-related proteins (Fig. 2D). While untreated macrophages showed minimal LC3B, RAB7, LAMP1, and LAMP2 expression, following trehalose treatment, we observed increased punctated staining for LC3B in the cytoplasm and increased punctated staining for RAB7, LAMP1, and LAMP2 proteins in the perinuclear region (Fig. 2D). These data further support that trehalose treatment modulates autophagy in macrophages via TFEB activation and induction of autophagy-related gene expression.
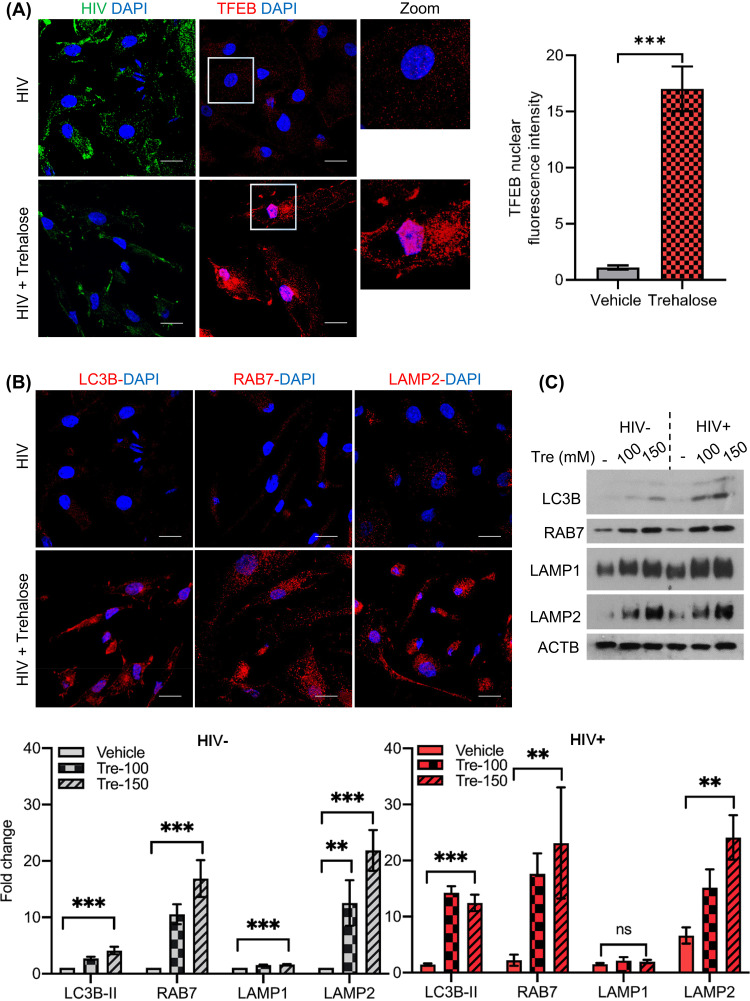
Having shown a central role for TFEB in the induction of autophagy in uninfected cells by trehalose, we next examined if TFEB-regulated autophagy was altered following trehalose treatment of infected macrophages. For these experiments, HIV-infected macrophages were incubated in the presence or absence of trehalose (100 mM) for 10 days, and expression of TFEB and its localization was compared in HIV-infected and trehalose-treated HIV-infected cells using confocal immunofluorescence microscopy as described previously (Fig. 2B). TFEB localized predominantly in the cytoplasm of HIV-infected macrophages (Fig. 3A). However, in the presence of trehalose, both TFEB expression and nuclear translocation are increased in infected macrophages (Fig. 3A, left). Trehalose-treated HIV-infected macrophages exhibited a >15-fold increase in TFEB nuclear accumulation compared to control infected cells (Fig. 3A, right; quantification, P = 0.008). IF images confirmed that trehalose treatment increased both TFEB nuclear translocation (Fig. 3A) and expression of autophagy and lysosomal biogenesis-related proteins in HIV-infected macrophages (Fig. 3B). Similar to what was observed in uninfected cells, trehalose treatment of HIV-infected macrophages also increased LC3B lipidation >9-fold (P < 0.001; Fig. 3C, bottom), RAB7 by >10-fold (P < 0.01; Fig. 3C), and LAMP2 >10-fold (P < 0.01; Fig. 3C, bottom), whereas LAMP1 was increased only 2-fold P > 0.05).
FIG 3.
Trehalose activates TFEB nuclear translocation and induces autophagy in HIV-infected macrophages. HIV-infected macrophages were treated with vehicle or trehalose (100 mM) on day 3 p.i. and incubated for 10 days. (A) IF analysis with anti-TFEB antibody (red) and DAPI (nucleus, blue). (B) IF analysis with anti-LC3B, anti-RAB7, and anti-LAMP2 (red) antibody and DAPI (nucleus, blue). (C) Uninfected and HIV-infected macrophages were treated with vehicle or trehalose (Tre; 100 mM, 150 mM) on day 3 p.i. and incubated for 10 days. At day 10, cells were harvested and lysates were analyzed by immunoblotting with anti-LC3B, anti-RAB7, anti-LAMP1, anti-LAMP2, and ACTB antibody. (Top) Representative immunoblots of LC3B, RAB7, LAMP1, and LAMP2. (Bottom) Densitometric analysis of immunoblots for uninfected (gray) and infected (red) cells. Data are derived from three independent donors and presented as means ± s.e.m. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Trehalose inhibits HIV entry in human primary macrophages.
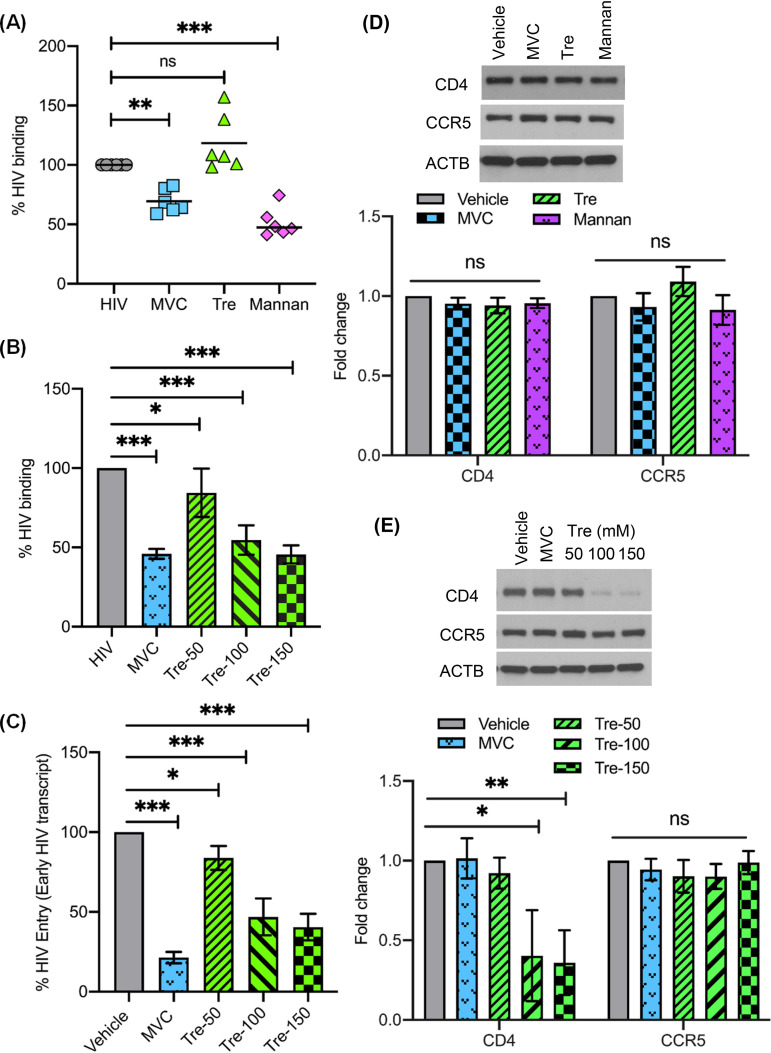
Although our findings to this point indicated that the induction of autophagy by trehalose is likely responsible for the intracellular inhibition of HIV in macrophages, it was important to determine if trehalose might affect HIV infection through viral entry. Therefore, our next series of experiments were designed to determine if trehalose altered viral binding or entry in macrophages. Macrophages were incubated with or without trehalose (100 mM) prior to exposure to HIV for 35 min as previously described (53). Maraviroc-pretreated (1 μM) and mannan-pretreated (16 mg/ml) macrophages were used as controls for inhibition of HIV binding to the surface of macrophages by blocking CCR5 or mannose receptor, respectively (3, 53). Following HIV exposure, cells were washed three times and lysed, and lysates were analyzed for p24 antigen by ELISA to measure HIV binding. Compared to untreated HIV-exposed macrophages, there was a 30% reduction in HIV binding in maraviroc-treated macrophages and approximately 50% reduction in mannan-treated macrophages (P < 0.01 and P < 0.001, respectively; Fig. 4A); however, no significant difference was observed in viral binding following 30 min of trehalose treatment (Fig. 4A). To assess if time of exposure to trehalose might alter binding and entry of HIV, we next treated macrophages with trehalose (50 mM to 150 mM) for 6 h followed by exposure to HIV for 35 min to assess binding (Fig. 4B) or for 3 h to assess entry (Fig. 4C). To assess entry, genomic DNA was extracted and HIV strong stop DNA (a measure of early HIV transcription or entry) was quantified by qPCR. In contrast to what had been observed following a 30-min exposure to trehalose, 6-h pretreatment resulted in a dose-dependent decrease in HIV binding (Fig. 4B). Additionally, similar to what was observed for HIV binding, a dose-dependent decrease in early HIV transcription was observed following a 6-h pretreatment of macrophages with trehalose, indicative of a decrease in viral entry (Fig. 4C).
FIG 4.
Trehalose inhibits HIV entry into human macrophages. (A) Macrophages pretreated with either 1 μM maraviroc (MVC), 100 mM trehalose (Tre), or 16 mg/ml mannan for 30 min were exposed to HIV for 35 min, after which cells were washed and lysates prepared to detect HIV p24 antigen by ELISA. (B) Macrophages pretreated with either 1 μM maraviroc (MVC) or 50 mM to 150 mM trehalose (Tre-50, Tre-100, Tre-150) for 6 h were exposed to HIV for 35 min, and lysates were assessed for HIV binding by ELISA. (C) Macrophages pretreated with either 1 μM maraviroc (MVC) or 50 mM to 150 mM trehalose (Tre-50, Tre-100, Tre-150) for 6 h were exposed to HIV for 3 h. Following 3 h of incubation, genomic DNA was prepared to measure early virus transcript by qPCR, and results are presented as percent HIV entry compared to vehicle-treated HIV-exposed macrophages. (D) (Top) Representative immunoblot showing expression of CD4, CCR5, and ACTB in cell lysates from vehicle and macrophages treated with MVC (1 μM), trehalose (100 mM), and mannan (16 mg/ml) (30-min treatment). (Bottom) Relative fold change (densitometric analysis) in CD4 and CCR5 protein normalized to ACTB. (E) (Top) Representative immunoblot showing expression of CD4, CCR5, and ACTB in cell lysates from vehicle and MVC-treated (1 μM) and trehalose-treated (50 mM to 150 mM) macrophages (6-h treatment). (Bottom) Relative fold change (densitometric analysis) in CD4 and CCR5 proteins normalized to ACTB. Data are derived from three independent donors and presented as means ± s.e.m. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
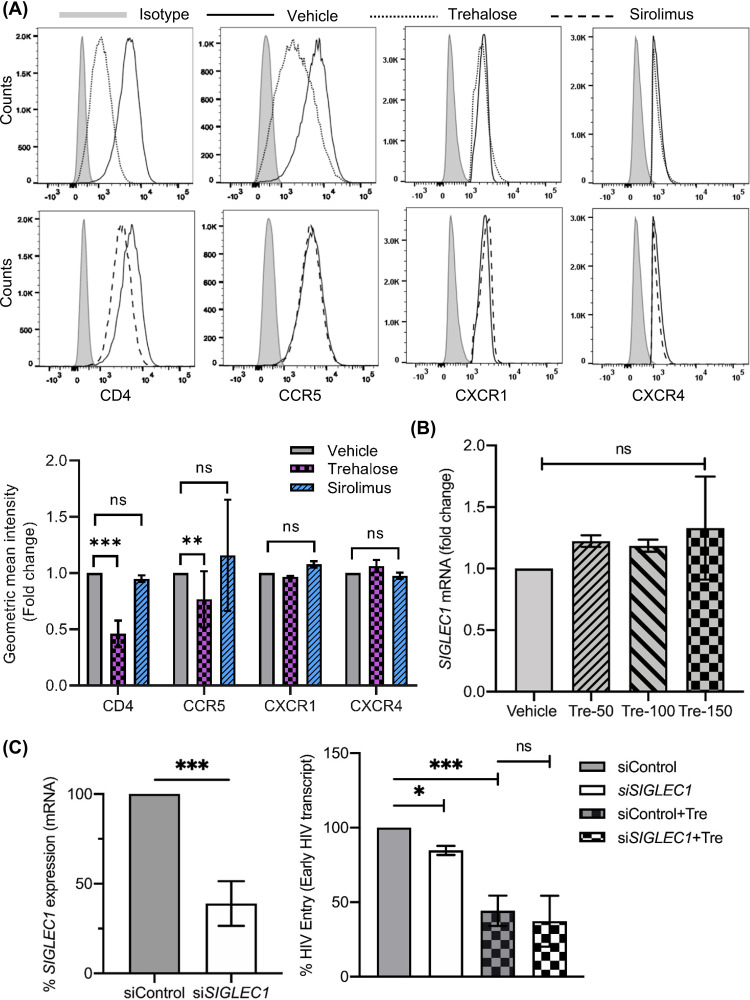
Having identified that a 6-h treatment with trehalose decreases viral entry, we hypothesized that the steady-state expression of CD4 or the major coreceptor on macrophages, CCR5, would be altered by trehalose. Therefore, we analyzed the expression of CD4 and CCR5 in our binding experiments following 30-min or 6-h pretreatment with vehicle, maraviroc, and trehalose by immunoblotting (Fig. 4D and E). Consistent with our binding and entry experiments, we did not observe any significant change in CD4 and CCR5 expression following 30 min of incubation (Fig. 4D). However, there was a dose-dependent decrease in CD4 expression following 6 h of incubation with trehalose (50 mM to 150 mM) (P < 0.001; Fig. 4E). These findings were further confirmed by flow cytometry (Fig. 5A, top left) that demonstrated a 60% reduction in CD4 surface expression (P < 0.001) and a 28% decrease in CCR5 on macrophages in the presence of trehalose (P = 0.002). In contrast, macrophages treated with sirolimus, a known MTOR-dependent inducer of autophagy, resulted in no significant change in CD4 or CCR5 expression (Fig. 5A). In total, these findings indicate that trehalose partially inhibits HIV binding and entry through the downregulation of CD4 and CCR5 expression on macrophages.
FIG 5.
Trehalose treatment reduces surface expression of CD4 and CCR5 in human macrophages. (A) Macrophages treated with vehicle, sirolimus, or trehalose for 12 h were analyzed for expression of CD4, CCR5, CXCR1, and CXCR4 by flow cytometry. (Top) Representative histogram showing surface expression of CD4, CCR5, CXCR1, and CXCR4 receptors. (Bottom) Geometric mean intensity fold change showing expression of CD4, CCR5, CXCR1, and CXCR4. Data are presented as means of four independent donors ± s.e.m. (B) Macrophages treated with vehicle or trehalose (50 mM to 150 mM) were analyzed for SIGLEC1 expression by qRT-PCR. (C) Macrophages transfected with nonspecific control siRNA (siControl) or SIGLEC1 siRNA were treated with 100 mM trehalose for 6 h followed by incubation with HIV (0.04 MOI) for 3 h. At 48 h post siRNA transfection, cells were harvested and analyzed for SIGLEC1 expression by qRT-PCR and presented as percent SIGLEC1 expression compared to control cells (siControl) (left). At 3 h post-HIV exposure, cells were trypsinized and washed, and genomic DNA was prepared and analyzed for early HIV transcript levels by qPCR. Results are presented as percent HIV entry compared to control siRNA transfected HIV-exposed cells (siControl). Data are derived from four independent donors and presented as means ± s.e.m. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Having shown a decrease in expression of CD4 and a modest decrease in CCR5 expression following trehalose treatment, we examined if there was a general decrease in cell surface receptor expression. Macrophages were treated as described above, and surface expression for two additional surface receptors, CXCR1 and CXCR4, was assessed by flow cytometry (Fig. 5A, top right). Of note, no significant change in CXCR1 and CXCR4 surface expression was observed (P > 0.5; Fig. 5A, bottom, quantification) following trehalose treatment. These results suggest that trehalose treatment specifically reduces the surface expression of CD4 and CCR5 receptors in macrophages.
The SIGLEC1 receptor is known to interact with sialic acid on the HIV envelope and is thought to mediate CD4-independent HIV binding and entry in macrophages (54, 55). To assess if trehalose treatment also affects SIGLEC1-mediated entry in macrophages, we analyzed the effect of trehalose on SIGLEC1 expression by qPCR and found no significant effect (Fig. 5B, right). Because SIGLEC1 expression is very low in adherent macrophages and cannot be reliably detected by immunoblotting (56), we used gene silencing (Fig. 5C, left) to assess the effect of trehalose on SIGLEC1-mediated HIV entry (Fig. 5C, right). Compared to control small interfering RNA (siRNA)-treated HIV-exposed macrophages, knockdown of SIGLEC1 had a modest effect on viral entry; however, no additional effect was observed on viral entry in trehalose-treated macrophages (Fig. 5C). In total, trehalose pretreatment did not show any direct effect on SIGLEC1 expression, and SIGLEC1 does not appear to play an important role in the trehalose-mediated decrease in viral entry.
Trehalose inhibits HIV infection and replication via reduction of CD4-dependent HIV entry and induction of autophagy.
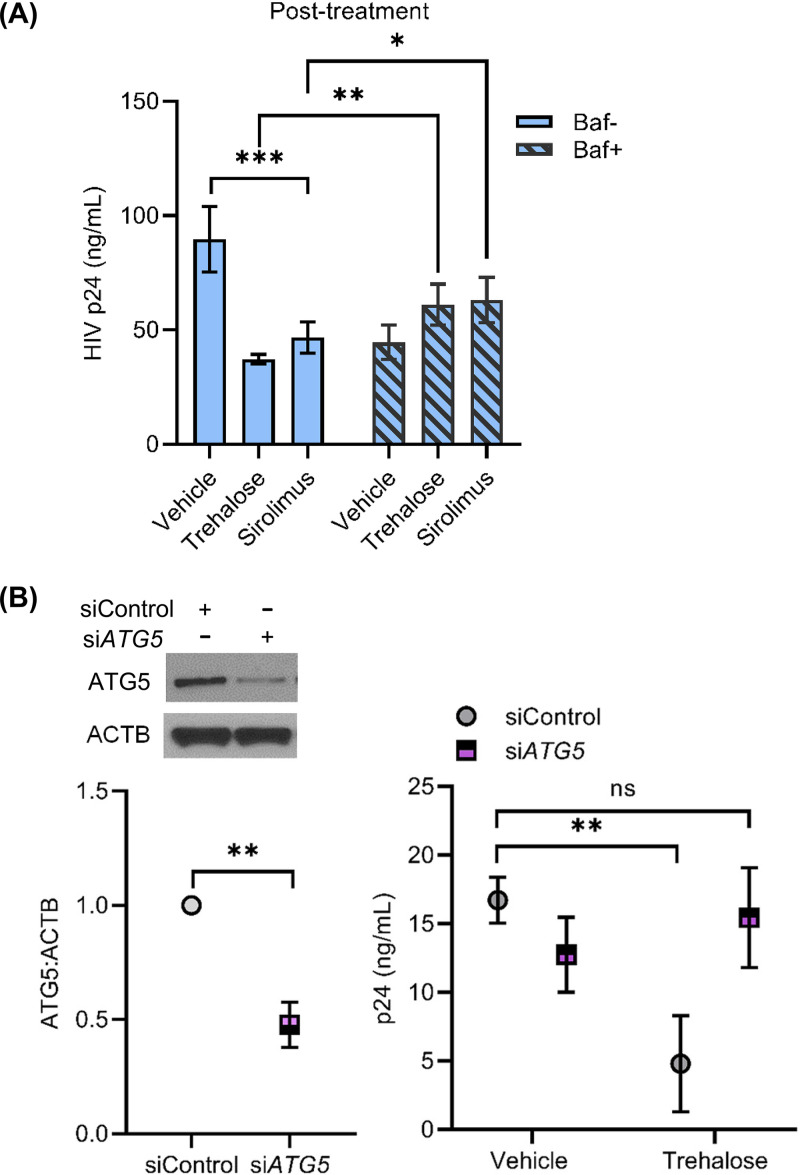
Having unexpectedly identified that trehalose decreases HIV entry into macrophages, it was important to establish our original hypothesis that the non-MTOR induction of autophagy by trehalose is important in HIV inhibition. To determine the role of autophagy in trehalose intracellular inhibition, macrophages were infected with HIV for 8 h and cultured for 72 h without any treatment, at which time trehalose (100 mM) was added to the culture medium. Cultures were maintained for an additional 7 days, at which time supernatants were collected (10 days p.i.) and p24 antigen was measured by ELISA (Fig. 6A). Trehalose-treated macrophages released 50% less p24 antigen in culture supernatants compared to untreated HIV-infected cells (P < 0.001; Fig. 6A). Bafilomycin A1 cotreatment partially blocked trehalose-mediated inhibition of HIV, as under these conditions, p24 release in the culture supernatants was increased by 1.6-fold compared to the trehalose-treated cells (P < 0.005; Fig. 6A). To further confirm the importance of autophagy in trehalose-mediated inhibition of HIV, we employed gene silencing of the autophagy elongation protein ATG5 (Fig. 6B, left). Following knockdown of ATG5 in HIV-infected macrophages, trehalose treatment failed to inhibit HIV replication (Fig. 6B, right). In total, these findings confirm that trehalose inhibits intracellular HIV in infected macrophages through induction of autophagy, which is consistent with recent findings on the effect of trehalose on mycobacteria/HIV coinfection of human macrophages (57).
FIG 6.
Trehalose inhibits HIV replication in HIV-infected macrophages by induction of autophagy. (A) HIV-infected macrophages were incubated for 72 h, at which time cells were treated with vehicle, trehalose (100 mM), or sirolimus 100 nM). Bafilomycin A1 (Baf) cotreatment was used to inhibit trehalose- or sirolimus-induced autophagic flux. At day 10 p.i., extracellular p24 antigen release was analyzed in the culture supernatants by ELISA. Data are derived from four independent donors and presented as means ± s.e.m. (B) HIV-infected macrophages transfected with nonspecific control siRNA (siControl) or ATG5 siRNA were treated with 100 mM trehalose for 3 days. At 48 h post-siRNA transfection, cells were harvested and analyzed for ATG5 expression by immunoblotting (left). At 72 h post-trehalose treatment, culture supernatants were collected and analyzed for p24 release by ELISA. Data are derived from three independent donors and presented as means ± s.e.m. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Trehalose inhibits HIV replication in human CD4+ T cells.
Although HIV infects macrophages, the virus predominantly infects CD4+ T cells. Having shown that trehalose inhibits HIV replication in human macrophages, we analyzed the effects of trehalose treatment in HIV-infected T cells (Fig. 7A) as described earlier for macrophages. Untreated and trehalose-treated (50 mM to 150 mM) T cells were infected with HIV for 6 h and incubated for 10 days with medium in the presence or absence of trehalose. Culture supernatants were tested for p24 antigen release at day 5 and day 10 p.i. by ELISA (Fig. 7A). Trehalose-treated HIV-infected T cells exhibited a dose-dependent decrease in HIV p24 release compared to untreated HIV-infected T cells (Fig. 7A). By day 10, we observed approximately 30%, 50%, and 70% inhibition in 50 mM, 100 mM, and 150 mM trehalose-treated cells, respectively (P < 0.001; Fig. 7A). Trehalose-treated uninfected and HIV-infected T cells showed little change in cell survival after trehalose treatment at all concentrations tested (Fig. 7B). We did not observe a significant increase in LDH release in the culture supernatants in the presence of trehalose in both uninfected and HIV-infected T cells (P > 0.1; Fig. 7B).
FIG 7.
Trehalose treatment inhibits HIV replication in human CD4+ T cells. (A) PHA-stimulated T cells (PHA-T cells) were pretreated with vehicle and increasing concentrations of trehalose (50 mM to 150 mM) for 6 h prior to exposure to HIV. Trehalose-pretreated HIV-infected T cells were incubated with vehicle or trehalose (Tre; 50 mM to 150 mM) for 10 days, and culture supernatants were collected at days 5 and 10 p.i. for extracellular p24 antigen quantification by ELISA. Data are derived from three independent donors and presented as means ± s.e.m. (B) Uninfected and HIV-infected T cells were incubated with vehicle or various concentrations of trehalose for 10 days. At day 10, cell culture supernatants were collected, and spectrophotometric measurement of extracellular LDH was used to determine cellular toxicity. Uninfected cells treated with 1× lysis buffer (LB) were used as a positive control. Data are derived from four independent donors and presented as means ± s.e.m. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Trehalose inhibits HIV entry in human CD4+ T cells.
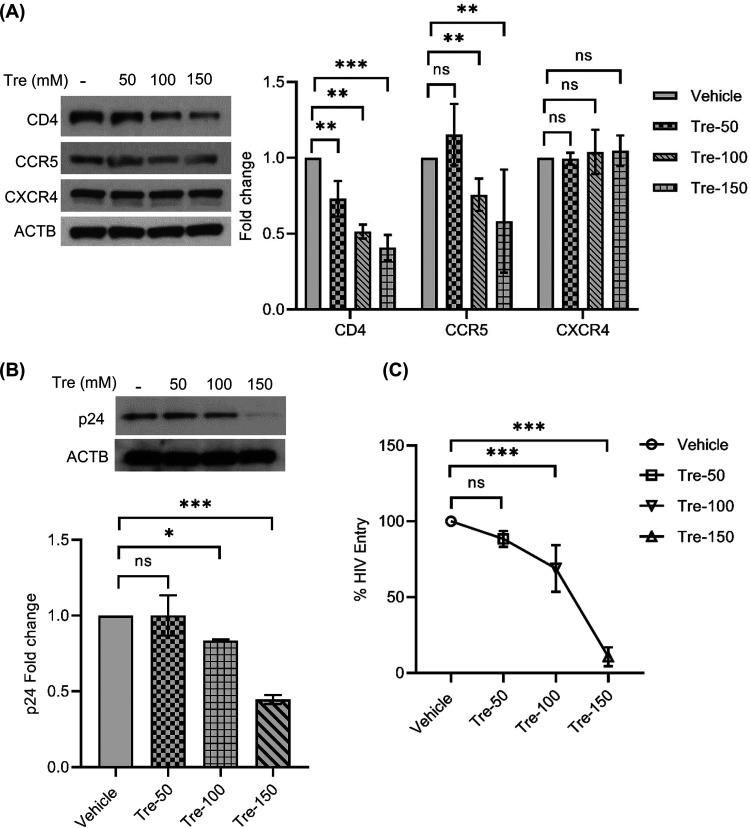
Having previously shown that trehalose inhibits HIV entry through the decreased expression of CD4 and CCR5 in macrophages, we sought to examine if viral entry was similarly reduced in CD4+ T cells. Phytohemagglutinin (PHA)-stimulated T cells were incubated with or without trehalose (50 mM to 150 mM) for 6 h, after which cells were collected, lysed, and analyzed for CD4, CCR5, and CXCR4 expression by immunoblotting (Fig. 8A). Trehalose treatment altered both CD4 and CCR5 expression in T cells. We observed 27%, 49%, and 60% decreases in CD4 expression on T cells treated with trehalose at 50 mM, 100 mM, and 150 mM, respectively (P < 0.05, P < 0.05, P < 0.001, respectively), and 25% and 42% reductions in CCR5 expression at the two highest trehalose concentrations (P < 0.01; Fig. 8A). No significant change was found in CXCR4 expression in T cells following trehalose treatment at all concentrations tested (P > 0.3; Fig. 8A).
FIG 8.
Trehalose treatment inhibits HIV entry into human CD4+ T cells by modulating the CD4 and CCR5 expression. (A) PHA-T cells were treated with vehicle or trehalose (50 to 150 mM) for 6 h and analyzed for expression of CD4, CCR5, and CXCR4 by immunoblotting. (Left) Representative immunoblot showing the expression of CD4, CCR5, CXCR4, and ACTB. (Right) Relative fold change (densitometric analysis) in CD4, CCR5, and CXCR4 protein normalized to ACTB. (B and C) Untreated and trehalose-pretreated (73) T cells were exposed to HIV (0.04 MOI) for 8 h. At 24 p.i., cell lysates were prepared to detect HIV antigen by immunoblotting (B) and ELISA (C). Data are derived from four independent donors and presented as means ± s.e.m. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To confirm that trehalose pretreatment decreased CD4 and CCR5 sufficiently to decrease HIV entry, T cells were exposed to HIV for 8 h, washed, and incubated for an additional 24 h in growth medium. At 24 h post-HIV infection, cells were collected and washed, and lysates were analyzed for p24 by immunoblotting and p24 antigen capture ELISA (Fig. 8B and C). Compared to controls, there was a 16.5% and 55% reduction in intracellular p24 expression in 100 mM and 150 mM trehalose-treated T cells by immunoblotting (P = 0.05 and P < 0.001, respectively; Fig. 8B). Similar to the immunoblotting data, a dose-dependent decrease in HIV entry in trehalose-treated T cells was also observed by ELISA (Fig. 8C). CD4+ T cells exhibited approximately 30% and 80% reductions in HIV entry following 100-mM and 150-mM trehalose treatments, respectively (P < 0.001; Fig. 8C). These results confirm that trehalose-mediated downregulation in CD4 and CCR5 expression also inhibits HIV entry in T cells.
Trehalose inhibits HIV infection in CD4+ T cells via inhibition of HIV entry and induction of autophagy.
Having demonstrated that trehalose inhibits HIV entry in CD4+ T cells, we next evaluated the role of autophagy in trehalose-mediated inhibition of HIV postentry. Following the methods described for macrophages, HIV-infected T cells treated with trehalose at 100 mM and 150 mM exhibited 2.0- and 2.4-fold higher LC3B-II levels, respectively, compared to untreated cells (P = 0.02 and P = 0.009, respectively; Fig. 9A). In the presence of bafilomycin A1, 100 mM and 150 mM trehalose-treated T cells exhibited 1.7- and 1.4-fold, respectively, higher LC3B lipidation (P = 0.002 and P = 0.03, respectively; Fig. 9A), confirming the induction of autophagic flux by trehalose. We next evaluated the expression of autophagy and lysosomal biogenesis-related proteins LC3B, RAB7, LAMP1, and LAMP2 in trehalose-treated T cells by immunoblotting (Fig. 9B). As observed previously with macrophages, trehalose treatment (100 mM and 150 mM) of infected T cells resulted in a significant increase in LC3B lipidation (2.2-fold; P = 0.007 and P < 0.001, respectively) and expression levels of RAB7 (3- and 2.9-fold; P = 0.007 and P = 0.001, respectively) and LAMP2 (4.6- and 3.9-fold, respectively; P < 0.001) (Fig. 9B). Though we observed 1.8-fold higher LAMP1 expression in infected T cells in the presence of trehalose (100 mM and 150 mM), this increase was not significant (P = 0.1 and P = 0.098, respectively; Fig. 9B). Importantly, following knockdown of the autophagy elongation protein ATG5 in infected T cells (P < 0.001; Fig. 9C, top), trehalose treatment failed to inhibit HIV replication (P = 0.28; Fig. 9C, bottom).
FIG 9.
Trehalose inhibits intracellular HIV replication in CD4+ T cells via induction of autophagy. (A) PHA-T cells were treated with trehalose (Tre; 100 mM, 150 mM) for 6 h. Bafilomycin cotreatment was included to confirm the increase in autophagy flux in the presence of trehalose. Cells were harvested, lysed, and analyzed for expression of LC3B lipidation by immunoblotting. (Top) Representative immunoblot showing expression of LC3B-II and ACTB. (Bottom) Relative fold change (densitometric analysis) in LC3B-II expression normalized to ACTB. (B) Uninfected and HIV-infected CD4+ T cells were treated with vehicle or trehalose (100 mM and 150 mM) on day 3 p.i. and incubated for 10 days. At day 10, cells were harvested and lysates were analyzed by immunoblotting with anti-LC3B, anti-RAB7, anti-LAMP1, and anti-LAMP2 and ACTB antibody. (Top) Representative immunoblots of LC3B, RAB7, LAMP1, and LAMP2. (Bottom) Densitometric analysis of uninfected CD4+ T cells (gray) and infected CD4+ T cells (upper right, red). (C) Infected T cells transfected with nonspecific control siRNA (siControl) or ATG5 siRNA were treated with 100 mM trehalose for 3 days. At 48 h post-siRNA transfection, cells were harvested and analyzed for ATG5 expression by immunoblotting (top). At 72 h post-trehalose treatment, culture supernatants were collected and analyzed for p24 release by ELISA (bottom). Data are derived from three independent donors and presented as means ± s.e.m. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
In total, these findings show that trehalose treatment inhibits HIV infection through two independent pathways. First, it downregulates the expression of CD4 in macrophages and both CD4 and CCR5 in T cells to reduce virus entry. Second, trehalose promotes the degradation of intracellular virus via induction of autophagy in both cell types.
DISCUSSION
Autophagy plays an important role in host-virus interactions and viral pathogenesis (58). Whereas some viruses use the proteins of autophagy to enhance replication, other viruses are inhibited by autophagy. Still others, including HIV, utilize the proteins of autophagy to enhance replication, but after establishing a permissive infection inhibit autophagy to maintain cell survival. In these cases, the induction of autophagy can lead to inhibition of the targeted virus, including HIV (39, 41, 42, 59–62).
Most research examining the effects of autophagy on viral infections has focused on the induction of autophagy through inhibition of MTOR. However, there is a growing literature on the induction of autophagy via MTOR-independent pathways and the effect on microbial pathogens (33, 63, 64). Of the non-MTOR inducers of autophagy, trehalose is among the most studied and has been shown to inhibit human cytomegalovirus, varicella zoster virus and Zika virus replication (33, 36; D. Spector, personal communication). Recently, trehalose was also shown to limit mycobacterial infection during HIV coinfection by reversing the autophagy block in HIV-infected peripheral blood mononuclear cells (PBMC) and macrophages (57). The α,α-trehalose derivatives containing guanidino groups have also been found to inhibit HIV replication through the disruption of HIV-1 tat-TAR interaction in human cell lines (65). In the present study, we found that trehalose is a potent inhibitor of HIV in human macrophages and T cells without inducing cytotoxicity. Of note, we have also observed that other non-MTOR inhibitors of autophagy, spermidine and SMER28, inhibit HIV replication (our unpublished data). Uniquely, however, we have identified that the inhibition of HIV infection by trehalose is dependent not only on the induction of MTOR-independent autophagy, but also via the inhibition of viral entry through decreased expression of CD4 in both T cells and macrophages and CCR5 in T cells. We have also shown that the decreased expression of CD4 and CCR5 by trehalose is specific to these receptors since trehalose had no effect on CXCR1, CXCR4, or SIGLEC1. Thus, trehalose differs from other anti-HIV compounds in that it affects HIV infection at entry and intracellularly.
Trehalose has been found to induce autophagy by distinct mechanisms in different disease model systems. In the case of HCMV infection, trehalose treatment increases levels of RAB7 promoting lysosomal biogenesis, fusion, and autophagic degradation of the virus (36). In multiple neurodegenerative disease (ND) model systems, trehalose-induced autophagy occurs via activation of TFEB. In these ND models, trehalose treatment increases TFEB nuclear translocation and gene expression of TFEB targets to activate degradative autophagy (37, 52, 66). In agreement with these studies, we also found that trehalose treatment increased the nuclear accumulation of TFEB and expression of lysosomal biogenesis and degradation proteins RAB7, LAMP1, and LAMP2 and LC3B lipidation. The finding that trehalose alters TFEB is of particular interest since in previous studies we have shown that during permissive infection, HIV Nef binds to BECN1, resulting in MTOR activation, TFEB phosphorylation and cytosolic sequestration, and the inhibition of autophagy (41). The TFEB-mediated inhibition of autophagy leads to enhanced cell survival and maintenance of HIV infection. Trehalose reverses this process and, working through TFEB, enhances autophagy and inhibits HIV intracellular replication.
The upregulation of RAB7 by trehalose is also of considerable interest given our finding that trehalose decreases the expression of CD4. HIV Nef downregulates CD4 and the major histocompatibility complex class I from the cell surface via endosomal trafficking pathways to establish continued viral fitness and persistence (67). Recently, Nef has been found to target serine incorporator 5 (SERINC5) for internalization and receptor-mediated endocytosis and relocalization of SERINC5 to Rab5+ early, RAB7+ late, and Rab11+ recycling endosomes. Endosomal sorting to the RAB7-dependent late endosome/lysosomal route leads to degradation, a similar strategy that has been found for Nef-mediated downregulation of CD4 (68, 69). Thus, the increase of RAB7 observed following trehalose treatment of CD4+ T cells and macrophages suggests that the decreased expression of CD4 observed in these cells could be related to the CD4 endosomal/lysosomal degradation.
In summary, we have shown that the nontoxic, naturally occurring sugar trehalose can uniquely inhibit HIV infection both at pre- and postentry. Preentry inhibition by trehalose occurs by decreasing cell expression of CD4 and CCR5 on T cells and CD4 on macrophages. Postentry, trehalose inhibits HIV infection through the induction of MTOR-independent autophagy mediated through TFEB activation. These findings demonstrate that cellular mechanisms can be modulated to inhibit HIV entry and intracellular replication using trehalose. Trehalose may be a useful, safe adjunct in the maintenance of patients who achieve an HIV functional cure.
MATERIALS AND METHODS
Ethics Statement.
Healthy HIV seronegative donors were enrolled for venous blood draw using a protocol that was reviewed and approved by the Human Research Protection Program of the University of California San Diego in accordance with the requirements of the Code of Federal Regulations on the Protection of Human Subjects (45 CFR 46 and 21 CFR 50 and 56) and was in full compliance with the principles expressed in the Declaration of Helsinki. Written informed consent was obtained from all the donors prior to their participation.
Cell Culture and Reagent Preparation.
Human peripheral blood mononuclear cells (PBMC) were isolated from whole blood or buffy coat of HIV-seronegative donors by density gradient centrifugation over Ficoll-Paque Plus (GE Healthcare). Monocyte-derived macrophages (macrophages) were derived from PBMCs using a monocyte isolation kit (Miltenyi Biotec) and cultured in RPMI 1640 medium with l-glutamine (Gibco) supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin (Gibco), 10% vol/vol heat-inactivated fetal bovine serum (FBS; Sigma-Aldrich), and 10 ng/ml macrophage colony-stimulating factor (MCSF; R&D Systems) at 5% CO2 and 37°C for 7 to 10 days with medium change every 3 days. Phytohemagglutinin (PHA, Sigma-Aldrich)-stimulated PBMC (PHA-PBMC) were prepared by incubating 2 × 106 PBMC ml−1 in expansion medium (RPMI 1640 supplemented with 10% [vol/vol] heat-inactivated FBS, 2 mM l-glutamine, 105 U liter−1 penicillin, 0.1 g liter−1 streptomycin with 1 mg/ml PHA) for 48 h in 5% CO2 at 37°C. CD4+ T cells were purified from PHA-PBMC by negative selection using the Miltenyi Biotech human CD4+ T cell isolation kit and cultured in RPMI 1640 supplemented with 10% (vol/vol) heat-inactivated FBS, 2 mM l-glutamine, 105 U liter−1 penicillin, 0.1 g liter−1 streptomycin, and 20 U ml−1 human interleukin-2 (IL-2) (47).
For autophagy induction, the culture medium was supplemented with different concentrations of trehalose (50 to 150 mM; Sigma-Aldrich) or sirolimus (rapamycin) (Sigma-Aldrich) at 100 nM concentration. The lysosomal degradation inhibitor, bafilomycin A1 (Enzo Life Sciences), was reconstituted in DMSO and used at a final concentration of 50 nM to measure autophagic flux. Maraviroc (Toronto Research Chemicals) was reconstituted in methanol and used at a final concentration of 1 μM.
Virus assay.
HIVBa-L was obtained through the AIDS Research and Reference Reagent Program, from Suzanne Gartner and Robert Gallo (49, 70). HIVBa-L virus stocks were prepared and the titer was determined as previously described (71). The 50% tissue culture infective dose (TCID50) was determined using the Spearmen-Karber method (72) using the Alliance HIVp24 antigen ELISA (Perkin Elmer). Cells were pretreated with an autophagy-inducing agent for 6 to 12 h and then infected with 105 TCID50/ml HIVBa-L per 5 × 105 cells unless otherwise stated.
For virus binding assessment, macrophages (2.5 × 105/well in a 48-well plate) were preincubated with 100 μl of medium (10% RPMI/FBS) or maraviroc (1 μM), mannan (16 mg/ml), or trehalose (100 mM) for 30 min at 37°C. Pretreated macrophages were further incubated with HIV for 35 min at 37°C (53). The cells were washed three times and then lysed in 200 μl of 1% TritonX100/PBS for 1 h at 37°C. The amount of bound viral p24 was measured by ELISA capture assay (ZeptoMetrix, Buffalo, NY).
For virus entry assessment, macrophages (0.5 × 105/well in a 24-well plate) were preincubated with 400 μl of medium (10% RPMI/FBS) or maraviroc (1 μM) or various concentrations of trehalose (50 mM to 150 mM) for 6 h. Pretreated macrophages were further incubated with HIV for 3 h at 37°C. After 3 h, cells were washed twice with 1× PBS and trypsinized to remove the virus from the surface and washed with PBS. Genomic DNA was extracted from cells using the Pure Link genomic DNA minikit (Life Technologies) and used to measure the level of HIV strong stop DNA by qPCR using TaqMan primer/probe sets (Applied Bioscience) as described previously (54). Briefly, TaqMan primer probe sets for HIV strong stop DNA (measure of early reverse transcript, virus entry) (forward primer [5′R]-5′AACTAGGGAACCCACTGCTTAA, reverse primer [3′U5]-5′TGAGGGATCTCTAGTTACCAGAGTCA, and R-probe [5′FAM]-CCTCAATAAAGCTTGCCTTGAGTGCTTCAA) were used for qPCR amplification of HIV early transcripts in genomic DNA samples. Predesigned GAPDH primer probe sets (Applied Biosciences) were used to both normalize the sample inputs and serve as a template integrity control. The cycling parameters used were as follows: 95°C for 10 min and 45 cycles at 95°C for 15 s and 60°C for 1 min. Normalized relative levels were calculated as log2-transformed fold difference from corresponding control (vehicle-treated) sample by 2-ΔΔCT, where ΔΔCT = ΔCT of treatment sample – ΔCT of vehicle-treated cells, and ΔCT = average cycle threshold (CT) of HIV target – average CT of corresponding GAPDH (54).
Virus entry was also assessed by measuring intracellular p24 levels in cell lysates using ELISA. For T cells, virus entry was assessed in vehicle or trehalose-pretreated (50 mM to 150 mM), HIV-infected T cells. HIV-exposed T cells were cultured for 24 h and washed 3 times with DPBS and lysed with 120 μl of cell lytic M cell lysis reagent (Sigma-Aldrich) as described above. Virus entry was analyzed by measuring the intracellular p24 levels in lysates using immunoblotting and ELISA.
Cytotoxicity assay.
Cytotoxicity and cell death were measured in vehicle and trehalose-treated macrophages with or without HIV infection using the LDH cytotoxicity detection kit (TaKaRa). Briefly, following 24 h of incubation with vehicle or trehalose, cells were infected with HIV and cultured in the presence of trehalose for 10 days. Uninfected cells were also cultured for 10 days in the presence of trehalose. At day 10, culture supernatants were collected, and LDH release was measured by ELISA.
Flow cytometry.
For cell surface expression analysis of CD4, CCR5, CXCR1, and CXCR4 by flow cytometry, macrophages were treated with trehalose for 12 h, collected, and stained with the following fluorescent-tagged antibodies according to the manufacturer’s instructions: allophycocyanin-CD4 (APC-CD4), phycoerythrin-cyanine7-CCR5 (PECy7-CCR5), fluorescein-5-isothiocyanate-CXCR1 (FITC-CXCR1), and phycoerythrin-CXCR4 (PE-CXCR4) (all from eBioscience) along with a live/dead fixable aqua dead cell stain (Molecular Probes). Stained cells were fixed with fixation buffer (BD Biosciences) and resuspended in stain buffer for flow cytometry analysis using BD FACS Canto RUO-ORANGE analyzer. Data were analyzed using FlowJo software v10.
RNA interference.
The TriFECT Dicer-substrate siRNA (DsiRNA) kit for ATG5 (hs.Ri.ATG5.13) and SIGLEC1 (hs.Ri.SIGLEC1.13) were purchased from Integrated DNA Technologies, Inc. (IDT, San Diego, CA) for silencing ATG5 in primary macrophages and T cells and SIGLEC1 in primary macrophages. Negative-control siRNA (DS NC1) was used as a nonspecific targeting control. DsiRNA transfections were performed using lipofectamine RNAi MAX reagent (Thermo Fisher) following the manufacturer’s instructions. Two days later, cells were analyzed for target gene silencing using RT-qPCR analysis and used in experiments.
RNA isolation and real-time qPCR.
Total RNA was isolated using an RNeasy minikit (Qiagen) according to the manufacturer’s protocol, and 200 to 500 ng RNA was used in 20 to 40 μl of reverse transcription reaction using a high-efficiency cDNA reverse transcription kit (Applied Biosystems). TaqMan gene expression assays (Applied Biosystems) were used for microtubule-associated protein 1 light chain 3 beta (MAP1LC3B), lysosomal-associated membrane protein 1 (LAMP1), lysosomal-associated membrane protein 2 (LAMP2), RAB7A, SIGLEC1, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene RT-qPCR analysis. The fold change in gene expression was calculated by the ΔΔCT method using GAPDH as an internal loading control. All RT-qPCR assays were performed in triplicate.
Immunoblotting.
Cell lysates were prepared using RIPA lysis buffer (Pierce) supplemented with 1× halt protease and phosphatase inhibitor cocktail (Thermo Scientific) according to the manufacturer’s instructions. Lysates were resolved on Bolt 12% Bis-Tris plus gels and transferred to polyvinylidene difluoride membrane (PVDF) or nitrocellulose membrane (Thermo Fisher Scientific). The primary antibodies in 5% dry milk in Tris-buffered saline with 0.1% Tween 20 (TBS-T) that were used were ACTB (Sigma-Aldrich), LC3B and CXCR4 (both from Novus Biologicals), LC3B, LAMP2, and RAB7 (all from Cell Signaling), LAMP1 (Santa Cruz Biotechnology), and CD4 and CCR5 (both from Abcam). Target proteins were detected using the WesternBreeze chemiluminescence kit (Invitrogen), and their relative band intensities were analyzed and compared with the ACTB bands using ImageJ (NIH).
Immunofluorescence microscopy.
Macrophages (untreated [uninfected or HIV-infected] and trehalose-treated [uninfected or HIV-infected]) were fixed and permeabilized using 4% (wt/vol) paraformaldehyde and 0.25% (vol/vol) TritonX-100 (Sigma-Aldrich). For immunofluorescence staining, cells were incubated with appropriate primary antibodies: anti-TFEB, anti-LAMP1, anti-LAMP2, anti-HIV gag/p24 (all from Abcam), and RAB7 (Cell Signaling) followed by incubation with Alexa Fluor-conjugated secondary antibodies (Molecular Probes). Cells were nuclear stained and mounted using Prolong Gold antifade mountant with 4′6′-diamidino-2-phenylindole (DAPI) (Molecular Probes). Stained cells were visualized using an Olympus FluoView FV-1000 confocal imaging system and minimally processed with Adobe Photoshop CS6.
Statistical analysis.
All results were assessed in GraphPad Prism 8.0 (GraphPad, La Jolla, CA). Statistical significance was determined by using two-tailed Student’s t test, analysis of variance (ANOVA), and Wilcoxon rank test as appropriate.
ACKNOWLEDGMENTS
This work was supported, in whole or in part, by the National Institute of Neurological Disorders and Stroke of the National Institutes of Health (RO1 NS084912 and RO1 NS104015 to S.A.S.) and the International Maternal Pediatrics Adolescent AIDS Clinical Trial Network (IMPAACT; impaactnetwork.org). Overall support for the IMPAACT Network was provided by the National Institute of Allergy and Infectious Diseases (NIAID of the NIH under award numbers UM1AI068632 [IMPAACT LOC], UM1AI068616 [IMPAACT SDMC], and UM1AI106716 [IMPAACT LC], with cofunding from the Eunice Kennedy Shriver National Institute of Child Health and Human Development; NICHD) and the National Institute of Mental Health; NIMH). The content here is solely the responsibility of the authors and does not represent the official views of the NIH. The funders had no role in study design, data collection, analysis and decision to publish, or preparation of the manuscript.
We thank Deborah Spector for insightful discussions, Jennifer Santini, UCSD Neuroscience Core and Light Microscopy Facility, the NINDS P30NS04710 core grant for confocal microscopy technical assistance and the Flow Cytometry Core at the UC San Diego Centre for AIDS Research (P30 AI036214), the VA San Diego Health Care System, and the San Diego Veterans Medical Research Foundation for flow cytometry technical support.
REFERENCES
- 1.Laskey SB, Siliciano RF. 2014. A mechanistic theory to explain the efficacy of antiretroviral therapy. Nat Rev Microbiol 12:772–780. doi: 10.1038/nrmicro3351. [DOI] [PubMed] [Google Scholar]
- 2.Adamson CS, Freed EO. 2009. Anti-HIV-1 therapeutics: from FDA-approved drugs to hypothetical future targets. Mol Interv 9:70–74. doi: 10.1124/mi.9.2.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuritzkes DR. 2009. HIV-1 entry inhibitors: an overview. Curr Opin HIV AIDS 4:82–87. doi: 10.1097/COH.0b013e328322402e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilen CB, Tilton JC, Doms RW. 2012. HIV: cell binding and entry. Cold Spring Harb Perspect Med 2:a006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jing K, Lim K. 2012. Why is autophagy important in human diseases? Exp Mol Med 44:69–72. doi: 10.3858/emm.2012.44.2.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parzych KR, Klionsky DJ. 2014. An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal 20:460–473. doi: 10.1089/ars.2013.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. 2010. mTOR regulation of autophagy. FEBS Lett 584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He C, Klionsky DJ. 2009. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klionsky DJ, Eskelinen EL, Deretic V. 2014. Autophagosomes, phagosomes, autolysosomes, phagolysosomes, autophagolysosomes… wait, I’m confused. Autophagy 10:549–551. doi: 10.4161/auto.28448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deretic V, Saitoh T, Akira S. 2013. Autophagy in infection, inflammation and immunity. Nat Rev Immunol 13:722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gozuacik D, Kimchi A. 2004. Autophagy as a cell death and tumor suppressor mechanism. Oncogene 23:2891–2906. doi: 10.1038/sj.onc.1207521. [DOI] [PubMed] [Google Scholar]
- 12.Rubinsztein DC, Marino G, Kroemer G. 2011. Autophagy and aging. Cell 146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 13.Levine B, Kroemer G. 2008. Autophagy in the pathogenesis of disease. Cell 132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nixon RA. 2013. The role of autophagy in neurodegenerative disease. Nat Med 19:983–997. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- 15.Zhang G, Luk BT, Hamidy M, Zhang L, Spector SA. 2018. Induction of a Na(+)/K(+)-ATPase-dependent form of autophagy triggers preferential cell death of human immunodeficiency virus type-1-infected macrophages. Autophagy 14:1359–1375. doi: 10.1080/15548627.2018.1476014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Renna M, Jimenez-Sanchez M, Sarkar S, Rubinsztein DC. 2010. Chemical inducers of autophagy that enhance the clearance of mutant proteins in neurodegenerative diseases. J Biol Chem 285:11061–11067. doi: 10.1074/jbc.R109.072181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarkar S. 2013. Regulation of autophagy by mTOR-dependent and mTOR-independent pathways: autophagy dysfunction in neurodegenerative diseases and therapeutic application of autophagy enhancers. Biochem Soc Trans 41:1103–1130. doi: 10.1042/BST20130134. [DOI] [PubMed] [Google Scholar]
- 18.Sarkar S, Floto RA, Berger Z, Imarisio S, Cordenier A, Pasco M, Cook LJ, Rubinsztein DC. 2005. Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol 170:1101–1111. doi: 10.1083/jcb.200504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams A, Sarkar S, Cuddon P, Ttofi EK, Saiki S, Siddiqi FH, Jahreiss L, Fleming A, Pask D, Goldsmith P, O’Kane CJ, Floto RA, Rubinsztein DC. 2008. Novel targets for Huntington’s disease in an mTOR-independent autophagy pathway. Nat Chem Biol 4:295–305. doi: 10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarkar S, Perlstein EO, Imarisio S, Pineau S, Cordenier A, Maglathlin RL, Webster JA, Lewis TA, O’Kane CJ, Schreiber SL, Rubinsztein DC. 2007. Small molecules enhance autophagy and reduce toxicity in Huntington’s disease models. Nat Chem Biol 3:331–338. doi: 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian Y, Bustos V, Flajolet M, Greengard P. 2011. A small-molecule enhancer of autophagy decreases levels of Abeta and APP-CTF via Atg5-dependent autophagy pathway. FASEB J 25:1934–1942. doi: 10.1096/fj.10-175158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buttner S, Broeskamp F, Sommer C, Markaki M, Habernig L, Alavian-Ghavanini A, Carmona-Gutierrez D, Eisenberg T, Michael E, Kroemer G, Tavernarakis N, Sigrist SJ, Madeo F. 2014. Spermidine protects against alpha-synuclein neurotoxicity. Cell Cycle 13:3903–3908. doi: 10.4161/15384101.2014.973309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LaRocca TJ, Gioscia-Ryan RA, Hearon CM Jr, Seals DR. 2013. The autophagy enhancer spermidine reverses arterial aging. Mech Ageing Dev 134:314–320. doi: 10.1016/j.mad.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madeo F, Eisenberg T, Buttner S, Ruckenstuhl C, Kroemer G. 2010. Spermidine: a novel autophagy inducer and longevity elixir. Autophagy 6:160–162. doi: 10.4161/auto.6.1.10600. [DOI] [PubMed] [Google Scholar]
- 25.Pietrocola F, Lachkar S, Enot DP, Niso-Santano M, Bravo-San Pedro JM, Sica V, Izzo V, Maiuri MC, Madeo F, Mariño G, Kroemer G. 2015. Spermidine induces autophagy by inhibiting the acetyltransferase EP300. Cell Death Differ 22:509–516. doi: 10.1038/cdd.2014.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. 2007. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem 282:5641–5652. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 27.Richards AB, Krakowka S, Dexter LB, Schmid H, Wolterbeek AP, Waalkens-Berendsen DH, Shigoyuki A, Kurimoto M. 2002. Trehalose: a review of properties, history of use and human tolerance, and results of multiple safety studies. Food Chem Toxicol 40:871–898. doi: 10.1016/s0278-6915(02)00011-x. [DOI] [PubMed] [Google Scholar]
- 28.Arrese EL, Soulages JL. 2010. Insect fat body: energy, metabolism, and regulation. Annu Rev Entomol 55:207–225. doi: 10.1146/annurev-ento-112408-085356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ouyang Y, Xu Q, Mitsui K, Motizuki M, Xu Z. 2009. Human trehalase is a stress responsive protein in Saccharomyces cerevisiae. Biochem Biophys Res Commun 379:621–625. doi: 10.1016/j.bbrc.2008.12.134. [DOI] [PubMed] [Google Scholar]
- 30.Aguib Y, Heiseke A, Gilch S, Riemer C, Baier M, Schatzl HM, Ertmer A. 2009. Autophagy induction by trehalose counteracts cellular prion infection. Autophagy 5:361–369. doi: 10.4161/auto.5.3.7662. [DOI] [PubMed] [Google Scholar]
- 31.Chen X, Li M, Li L, Xu S, Huang D, Ju M, Huang J, Chen K, Gu H. 2016. Trehalose, sucrose and raffinose are novel activators of autophagy in human keratinocytes through an mTOR-independent pathway. Sci Rep 6:28423. doi: 10.1038/srep28423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kruger U, Wang Y, Kumar S, Mandelkow EM. 2012. Autophagic degradation of tau in primary neurons and its enhancement by trehalose. Neurobiol Aging 33:2291–2305. doi: 10.1016/j.neurobiolaging.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 33.Belzile JP, Sabalza M, Craig M, Clark E, Morello CS, Spector DH. 2016. Trehalose, an mTOR-independent inducer of autophagy, inhibits human cytomegalovirus infection in multiple cell types. J Virol 90:1259–1277. doi: 10.1128/JVI.02651-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim J, Kundu M, Viollet B, Guan KL. 2011. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mardones P, Rubinsztein DC, Hetz C. 2016. Mystery solved: trehalose kickstarts autophagy by blocking glucose transport. Sci Signal 9:fs2. doi: 10.1126/scisignal.aaf1937. [DOI] [PubMed] [Google Scholar]
- 36.Clark AE, Sabalza M, Gordts P, Spector DH. 2017. Human cytomegalovirus replication is inhibited by the autophagy-inducing compounds trehalose and SMER28 through distinctively different mechanisms. J Virol 92. doi: 10.1128/JVI.02015-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rusmini P, Cortese K, Crippa V, Cristofani R, Cicardi ME, Ferrari V, Vezzoli G, Tedesco B, Meroni M, Messi E, Piccolella M, Galbiati M, Garre M, Morelli E, Vaccari T, Poletti A. 2019. Trehalose induces autophagy via lysosomal-mediated TFEB activation in models of motoneuron degeneration. Autophagy 15:631–621. doi: 10.1080/15548627.2018.1535292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Killian MS. 2012. Dual role of autophagy in HIV-1 replication and pathogenesis. AIDS Res Ther 9:16. doi: 10.1186/1742-6405-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kyei GB, Dinkins C, Davis AS, Roberts E, Singh SB, Dong C, Wu L, Kominami E, Ueno T, Yamamoto A, Federico M, Panganiban A, Vergne I, Deretic V. 2009. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J Cell Biol 186:255–268. doi: 10.1083/jcb.200903070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nardacci R, Ciccosanti F, Marsella C, Ippolito G, Piacentini M, Fimia GM. 2017. Role of autophagy in HIV infection and pathogenesis. J Intern Med 281:422–432. doi: 10.1111/joim.12596. [DOI] [PubMed] [Google Scholar]
- 41.Campbell GR, Rawat P, Bruckman RS, Spector SA. 2015. Human immunodeficiency virus type 1 Nef inhibits autophagy through transcription factor EB sequestration. PLoS Pathog 11:e1005018. doi: 10.1371/journal.ppat.1005018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Campbell GR, Spector SA. 2011. Hormonally active vitamin D3 (1alpha,25-dihydroxycholecalciferol) triggers autophagy in human macrophages that inhibits HIV-1 infection. J Biol Chem 286:18890–18902. doi: 10.1074/jbc.M110.206110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Campbell GR, Spector SA. 2013. Inhibition of human immunodeficiency virus type-1 through autophagy. Curr Opin Microbiol 16:349–354. doi: 10.1016/j.mib.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kimura S, Fujita N, Noda T, Yoshimori T. 2009. Monitoring autophagy in mammalian cultured cells through the dynamics of LC3. Methods Enzymol 452:1–12. doi: 10.1016/S0076-6879(08)03601-X. [DOI] [PubMed] [Google Scholar]
- 45.Bjorkoy G, Lamark T, Pankiv S, Overvatn A, Brech A, Johansen T. 2009. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol 452:181–197. doi: 10.1016/S0076-6879(08)03612-4. [DOI] [PubMed] [Google Scholar]
- 46.El-Khoury V, Pierson S, Szwarcbart E, Brons NH, Roland O, Cherrier-De Wilde S, Plawny L, Van Dyck E, Berchem G. 2014. Disruption of autophagy by the histone deacetylase inhibitor MGCD0103 and its therapeutic implication in B-cell chronic lymphocytic leukemia. Leukemia 28:1636–1646. doi: 10.1038/leu.2014.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, Adhihetty PJ, Adler SG, Agam G, Agarwal R, Aghi MK, Agnello M, Agostinis P, Aguilar PV, Aguirre-Ghiso J, Airoldi EM, Ait-Si-Ali S, Akematsu T, Akporiaye ET, Al-Rubeai M, Albaiceta GM, Albanese C, Albani D, Albert ML, Aldudo J, Algül H, Alirezaei M, Alloza I, Almasan A, Almonte-Beceril M, Alnemri ES, Alonso C, Altan-Bonnet N, Altieri DC, Alvarez S, Alvarez-Erviti L, Alves S, Amadoro G, Amano A, Amantini C, Ambrosio S, Amelio I, Amer AO, Amessou M, Amon A, An Z, et al. 2016. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barth S, Glick D, Macleod KF. 2010. Autophagy: assays and artifacts. J Pathol 221:117–124. doi: 10.1002/path.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gartner S, Markovits P, Markovitz DM, Kaplan MH, Gallo RC, Popovic M. 1986. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science 233:215–219. doi: 10.1126/science.3014648. [DOI] [PubMed] [Google Scholar]
- 50.Mizushima N, Yoshimori T, Levine B. 2010. Methods in mammalian autophagy research. Cell 140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dehay B, Bove J, Rodriguez-Muela N, Perier C, Recasens A, Boya P, Vila M. 2010. Pathogenic lysosomal depletion in Parkinson’s disease. J Neurosci 30:12535–12544. doi: 10.1523/JNEUROSCI.1920-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Palmieri M, Pal R, Nelvagal HR, Lotfi P, Stinnett GR, Seymour ML, Chaudhury A, Bajaj L, Bondar VV, Bremner L, Saleem U, Tse DY, Sanagasetti D, Wu SM, Neilson JR, Pereira FA, Pautler RG, Rodney GG, Cooper JD, Sardiello M. 2017. mTORC1-independent TFEB activation via Akt inhibition promotes cellular clearance in neurodegenerative storage diseases. Nat Commun 8:14338. doi: 10.1038/ncomms14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nguyen DG, Hildreth JE. 2003. Involvement of macrophage mannose receptor in the binding and transmission of HIV by macrophages. Eur J Immunol 33:483–493. doi: 10.1002/immu.200310024. [DOI] [PubMed] [Google Scholar]
- 54.Jobe O, Trinh HV, Kim J, Alsalmi W, Tovanabutra S, Ehrenberg PK, Peachman KK, Gao G, Thomas R, Kim JH, Michael NL, Alving CR, Rao VB, Rao M. 2016. Effect of cytokines on Siglec-1 and HIV-1 entry in monocyte-derived macrophages: the importance of HIV-1 envelope V1V2 region. J Leukoc Biol 99:1089–1106. doi: 10.1189/jlb.2A0815-361R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zou Z, Chastain A, Moir S, Ford J, Trandem K, Martinelli E, Cicala C, Crocker P, Arthos J, Sun PD. 2011. Siglecs facilitate HIV-1 infection of macrophages through adhesion with viral sialic acids. PLoS One 6:e24559. doi: 10.1371/journal.pone.0024559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jobe O, Kim J, Tycksen E, Onkar S, Michael NL, Alving CR, Rao M. 2017. Human primary macrophages derived in vitro from circulating monocytes comprise adherent and non-adherent subsets with differential expression of Siglec-1 and CD4 and permissiveness to HIV-1 infection. Front Immunol 8:1352. doi: 10.3389/fimmu.2017.01352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharma V, Makhdoomi M, Singh L, Kumar P, Khan N, Singh S, Verma HN, Luthra K, Sarkar S, Kumar D. 2020. Trehalose limits opportunistic mycobacterial survival during HIV co-infection by reversing HIV-mediated autophagy block. Autophagy doi: 10.1080/15548627.2020.1725374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chiramel AI, Brady NR, Bartenschlager R. 2013. Divergent roles of autophagy in virus infection. Cells 2:83–104. doi: 10.3390/cells2010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Borel S, Robert-Hebmann V, Alfaisal J, Jain A, Faure M, Espert L, Chaloin L, Paillart JC, Johansen T, Biard-Piechaczyk M. 2015. HIV-1 viral infectivity factor interacts with microtubule-associated protein light chain 3 and inhibits autophagy. AIDS 29:275–286. doi: 10.1097/QAD.0000000000000554. [DOI] [PubMed] [Google Scholar]
- 60.Campbell GR, Spector SA. 2012. Toll-like receptor 8 ligands activate a vitamin D mediated autophagic response that inhibits human immunodeficiency virus type 1. PLoS Pathog 8:e1003017. doi: 10.1371/journal.ppat.1003017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shoji-Kawata S, Sumpter R, Leveno M, Campbell GR, Zou Z, Kinch L, Wilkins AD, Sun Q, Pallauf K, MacDuff D, Huerta C, Virgin HW, Helms JB, Eerland R, Tooze SA, Xavier R, Lenschow DJ, Yamamoto A, King D, Lichtarge O, Grishin NV, Spector SA, Kaloyanova DV, Levine B. 2013. Identification of a candidate therapeutic autophagy-inducing peptide. Nature 494:201–206. doi: 10.1038/nature11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yakoub AM, Shukla D. 2015. Autophagy stimulation abrogates herpes simplex virus-1 infection. Sci Rep 5:9730. doi: 10.1038/srep09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yuan S, Zhang ZW, Li ZL. 2017. Cell death-autophagy loop and glutamate-glutamine cycle in amyotrophic lateral sclerosis. Front Mol Neurosci 10:231. doi: 10.3389/fnmol.2017.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang X, Chen S, Song L, Tang Y, Shen Y, Jia L, Le W. 2014. MTOR-independent, autophagic enhancer trehalose prolongs motor neuron survival and ameliorates the autophagic flux defect in a mouse model of amyotrophic lateral sclerosis. Autophagy 10:588–602. doi: 10.4161/auto.27710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang M, Xu Z, Tu P, Yu X, Xiao S, Yang M. 2004. Alpha,alpha-trehalose derivatives bearing guanidino groups as inhibitors to HIV-1 Tat-TAR RNA interaction in human cells. Bioorg Med Chem Lett 14:2585–2588. doi: 10.1016/j.bmcl.2004.02.073. [DOI] [PubMed] [Google Scholar]
- 66.Evans TD, Jeong SJ, Zhang X, Sergin I, Razani B. 2018. TFEB and trehalose drive the macrophage autophagy-lysosome system to protect against atherosclerosis. Autophagy 14:724–726. doi: 10.1080/15548627.2018.1434373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shi J, Xiong R, Zhou T, Su P, Zhang X, Qiu X, Li H, Li S, Yu C, Wang B, Ding C, Smithgall TE, Zheng YH. 2018. HIV-1 Nef antagonizes SERINC5 restriction by downregulation of SERINC5 via the endosome/lysosome system. J Virol 92. doi: 10.1128/JVI.00196-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Garcia JV, Miller AD. 1991. Serine phosphorylation-independent downregulation of cell-surface CD4 by nef. Nature 350:508–511. doi: 10.1038/350508a0. [DOI] [PubMed] [Google Scholar]
- 69.Pereira EA, daSilva LL. 2016. HIV-1 Nef: taking control of protein trafficking. Traffic 17:976–996. doi: 10.1111/tra.12412. [DOI] [PubMed] [Google Scholar]
- 70.Popovic M, Gartner S, Read-Connole E, Beaver B. 1988. Cell tropism and expression of HIV-1 isolates in natural targets, p 21–27. In Girard M, Valette L (ed), Retroviruses of human AIDS and related animal diseases, vol 3 Pasteur Vaccins, Marnes-La-Coquette, France. [Google Scholar]
- 71.Campbell GR, Loret EP, Spector SA. 2010. HIV-1 clade B Tat, but not clade C Tat, increases X4 HIV-1 entry into resting but not activated CD4+ T cells. J Biol Chem 285:1681–1691. doi: 10.1074/jbc.M109.049957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Japour AJ, Mayers DL, Johnson VA, Kuritzkes DR, Beckett LA, Arduino JM, Lane J, Black RJ, Reichelderfer PS, D’Aquila RT. 1993. Standardized peripheral blood mononuclear cell culture assay for determination of drug susceptibilities of clinical human immunodeficiency virus type 1 isolates. The RV-43 Study Group, the AIDS Clinical Trials Group Virology Committee Resistance Working Group. Antimicrob Agents Chemother 37:1095–1101. doi: 10.1128/AAC.37.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jaramillo M, Gomez MA, Larsson O, Shio MT, Topisirovic I, Contreras I, Luxenburg R, Rosenfeld A, Colina R, McMaster RW, Olivier M, Costa-Mattioli M, Sonenberg N. 2011. Leishmania repression of host translation through mTOR cleavage is required for parasite survival and infection. Cell Host Microbe 9:331–341. doi: 10.1016/j.chom.2011.03.008. [DOI] [PubMed] [Google Scholar]