The HSV-1 gI is required for viral cell-to-cell spread within the host, but the molecular mechanisms of how gI exactly works have remained poorly understood. Here, we report a novel property of this molecule, namely, induction of rod-shaped structures, which appeared to represent a higher-order form of gI. We further mapped the critical residues and showed that the ability of gI to induce rod-shaped structures correlated well with the capability of HSV-1 to induce cell fusion in the UL24syn background, suggesting that the two events may have an intrinsic link. Our results shed light on the biological properties of HSV-1 gI and may have important implications in understanding viral pathogenesis.
KEYWORDS: rod-shaped structures, cell-cell fusion, cell-to-cell spread, glycoprotein I (gI), herpes simplex virus, membrane tubulation
ABSTRACT
The envelope glycoprotein I (gI) of herpes simplex virus 1 (HSV-1) is a critical mediator of virus-induced cell-to-cell spread and cell-cell fusion. Here, we report a previously unrecognized property of this molecule. In transfected cells, the HSV-1 gI was discovered to induce rod-shaped structures that were uniform in width but variable in length. Moreover, the gI within these structures was conformationally different from the typical form of gI, as a previously used monoclonal antibody mAb3104 and a newly made peptide antibody to the gI extracellular domain (ECD) (amino acids [aa] 110 to 202) both failed to stain the long rod-shaped structures, suggesting the formation of a higher-order form. Consistent with this observation, we found that gI could self-interact and that the rod-shaped structures failed to recognize glycoprotein E, the well-known binding partner of gI. Further analyses by deletion mutagenesis and construction of chimeric mutants between gI and gD revealed that the gI ECD is the critical determinant, whereas the transmembrane domain served merely as an anchor. The critical amino acids were subsequently mapped to proline residues 184 and 188 within a conserved PXXXP motif. Reverse genetics analyses showed that the ability to induce a rod-shaped structure was not required for viral replication and spread in cell culture but rather correlated positively with the capability of the virus to induce cell fusion in the UL24syn background. Together, this work discovered a novel feature of HSV-1 gI that may have important implications in understanding gI function in viral spread and pathogenesis.
IMPORTANCE The HSV-1 gI is required for viral cell-to-cell spread within the host, but the molecular mechanisms of how gI exactly works have remained poorly understood. Here, we report a novel property of this molecule, namely, induction of rod-shaped structures, which appeared to represent a higher-order form of gI. We further mapped the critical residues and showed that the ability of gI to induce rod-shaped structures correlated well with the capability of HSV-1 to induce cell fusion in the UL24syn background, suggesting that the two events may have an intrinsic link. Our results shed light on the biological properties of HSV-1 gI and may have important implications in understanding viral pathogenesis.
INTRODUCTION
Herpesviruses within the family Herpesviridae pose a huge threat to the health of both humans and animals. This class of large DNA viruses can be divided into three subfamilies, including Alphaherpesvirinae, Betaherpesvirinae, and Gammaherpesvirinae (1). Of them, herpes simplex virus 1 (HSV-1) is an alphaherpesvirus that can cause a variety of human diseases, including cold sore, ocular keratitis, genital herpes, herpes encephalitis, neonatal herpes, and Alzheimer’s diseases (2–5). Upon infection within a host, this virus can disseminate rapidly and efficiently from infected to neighboring uninfected cells through lateral cell-cell junctions, a mode of transmission that is termed “cell-to-cell spread” (CCS) (6). Notably, CCS is also critical for HSV latent infection and reactivation (7, 8). HSV-1 initiates infections in skin and mucosal epithelial cells and then spreads to peripheral sensory neurons to establish a latent infection. Similarly, virions produced following reactivation of latent infections are transmitted to epithelial cells via the same junctional connections (6, 7, 9, 10). The process for HSV-1 CCS is complex and requires many viral proteins, including viral core fusion complex gB, gD, and gH/gL, as well as other accessory proteins that are not necessary for virus entry, such as heterodimer gE/gI (9–18), US9 (14, 15, 19, 20), gK (19, 20), and so on.
The focus of this report is HSV-1 envelope glycoprotein gI, a type I transmembrane protein that contains a signal peptide (SP), an extracellular domain (ECD), a membrane-spanning region (TM), and a cytoplasmic tail (CT) (21). HSV gI is notable for size variations in its extracellular domain, particularly near the transmembrane region among different HSV strains (22, 23). In addition, it interacts with gE to form a heterodimer (gE/gI) (24–27). Past studies have suggested that gE/gI serves as a multifunctional executor during infection. It can function as the Fc receptor of antibodies (24), in which gI itself does not bind to IgG, but its interaction with gE can dramatically increase the affinity of gE with antibodies (21). The Fc receptor may interfere with antibody-related host defense (28–31). Antibodies specific for HSV-1 antigens can be simultaneously bound at the surface of infected cells to gE/gI via their Fc region and to a cell surface antigen by their antigen-binding fragments (Fabs) (25, 28, 29). This process, known as antibody bipolar bridging (ABB), may be a strategy to prevent the host from utilizing anti-HSV-1 antibodies in immune responses (25, 28, 29). The gE/gI complex is also involved in secondary envelopment (32, 33). Deletion of either gI or gE does not affect significantly viral replication, but simultaneous deletion of gE and gD leads to a severe defect in virion biogenesis (33). The third function of the gE/gI heterodimer is its requirement for HSV-induced cell fusion, leading to generation of giant multinucleated cells (34–36). In cell cultures, wild-type (WT) HSV-1 rarely induces syncytia, while mutations in genes encoding gB (UL27), gK (UL56), UL20, or UL24 can give rise to this property (37–40). However, it is becoming increasingly clear that gE and gI behave a little bit differently for syncytial formation. Although gE is required for all types of syncytial formation, gI is needed only in the context of UL24syn background (40). Last, most importantly, gE/gI promotes the sorting nascent virions to cell junctions and facilitates the viral spread across cell junction (7, 9–11, 13–16, 18). In mice, viral mutants lacking gI or gE produce smaller or barely perceptible punctate lesions, are cleared rapidly from the inoculation site, and reach low and transient titers in sensory ganglia (16, 41–43).
Despite considerable efforts in the past, we still do not know much about the biology of gI and how gI executes its function during infection. Our study here began with the exploration of the intracellular distribution of gI in transfected cells. We quickly discovered a novel phenotype of gI, namely, the induction of rod-shaped structures. We subsequently probed the nature and molecular determinants of the gI rods and then explored their significance in viral replication, spread, and cell-cell fusion.
RESULTS
Induction of rod-shaped structures by HSV-1 gI.
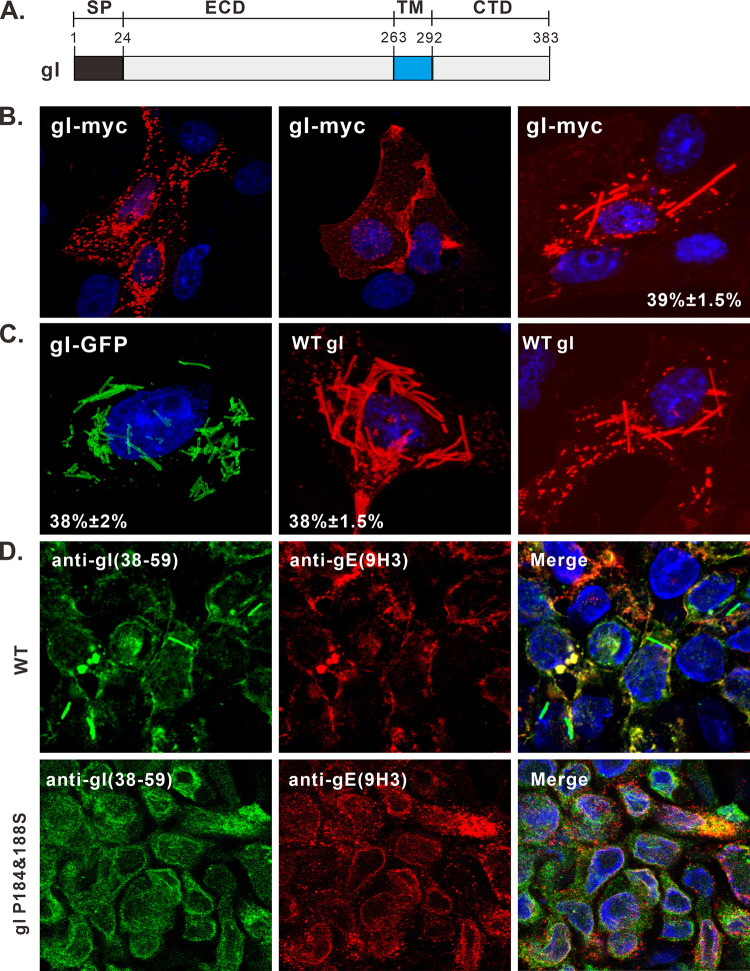
To understand the biological properties of gI, we started with analyzing its subcellular localization in transfected cells by using HSV-1 KOS strain gI as a model protein that has a size of 383 amino acids (aa) (Fig. 1A). To facilitate staining, gI was tagged with a c-Myc epitope at its C terminus to create a construct gI-myc. In transfected Vero cells, the immunofluorescence staining by anti-Myc antibodies revealed three different distribution patterns (Fig. 1B). The first represented the typical localization pattern in which gI was stained on the nuclear membrane and in the cytoplasm as speckles (Fig. 1B, left). In some cells, gI accumulated at cell junctions (Fig. 1B, middle). Unexpectedly, gI was found to induce some rod-shaped structures in the cytoplasm (Fig. 1B, right), a property that was not described before, and about 40% of cells expressing gI showed this phenotype.
FIG 1.
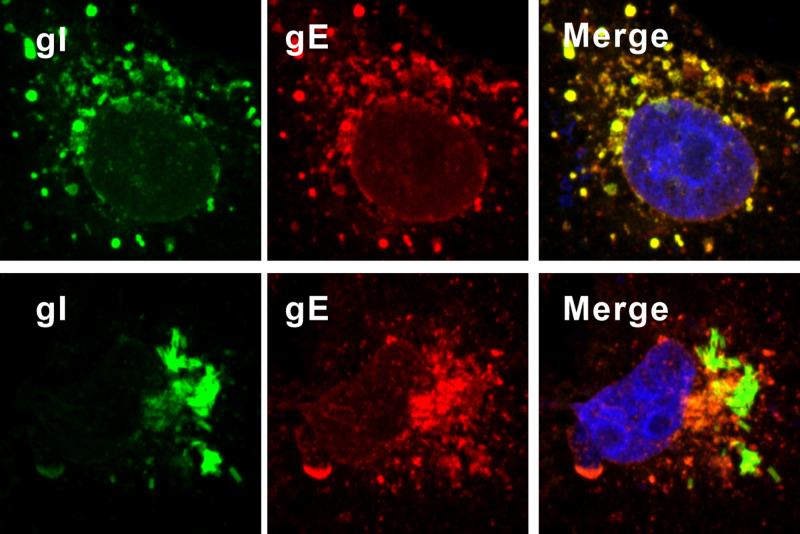
Induction of rod-shaped structures by HSV-1 gI. (A) Diagram of domain organization of HSV-1 KOS strain glycoprotein gI. (B and C) Vero cells were transfected to express HSV-1 gI-Myc, gI-GFP, and untagged gI. At 18 to 24 h posttransfection, the cells were fixed, permeabilized, and revealed by antibodies specific for the Myc tag, gI antigen, and HA tag or by GFP fluorescence. DAPI was used to stain cell nuclei (blue). The representative images were captured with a Leica confocal microscope and processed by using ImageJ. The percentage of cells showing rod-shaped structures relative to total gI-positive cells is also shown in the pictures. (D) HaCaT cells grown on coverslips were infected with WT HSV or the mutant gI P184&188S at an MOI of 0.1 at 37°C. After 1 h of incubation, the cells were rinsed with PBS and supplemented with fresh infection medium. At 24 h postinfection, the cells were fixed and stained with rabbit polyclonal antibodies against gI and mouse monoclonal antibodies against gE (red) as indicated. The rest of the steps were the same as panels B and C.
This observation was further verified by another construct, gI-green fluorescent protein (GFP), in which the GFP was fused to the cytoplasmic tail of gI (Fig. 1C, left). To rule out any adverse effect of foreign tags on the protein structure, the untagged version of gI was transiently expressed and probed with gI polyclonal antibodies (UP1725) to the extracellular domain. Again, we made the same observation (Fig. 1C, right). This finding was also extended to BHK-21 and HeLa cells (data not shown). As a control, the pseudorabies virus glycoprotein I did not show this property but rather displayed the typical distribution pattern of a viral glycoprotein (data not shown). Thus, the induction of rod-shaped structures by HSV-1 gI is virus specific, irrelevant to the foreign tags, and independent of cell types.
To investigate whether the rod structure can be induced in virus-infected cells, the HSV-1 KOS strain was used to infect HaCaT and Vero cells at a multiplicity of infection (MOI) of 0.1. The gI rod structures could be observed in infected HaCaT cells but with a low frequency of about 4.5% (Fig. 1D, top). Interestingly, they were rarely seen in Vero cells (data not shown). These results are not surprising, as the ability of gI to induce the rod-shaped structures is probably subject to regulation by viral factors during infection, a condition that is much more complex than expression alone in transfected cells.
Analysis of the nature and dimensions of the gI rod structures.
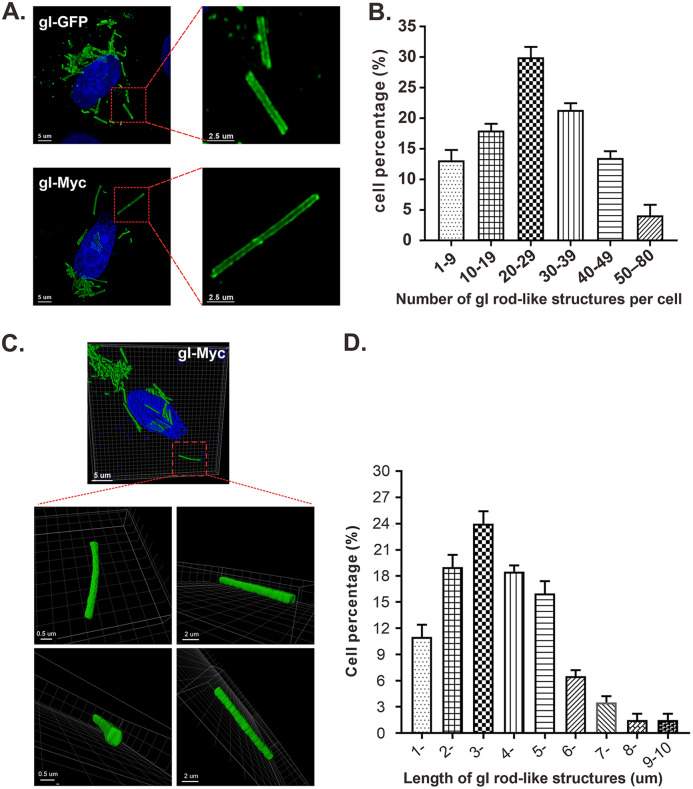
The gI rod-shaped structures were quite uniform in width but variable in length; some were very short rods, while some were long enough to cross the cytoplasm of a eukaryotic cell (Fig. 1). Examination at higher magnification revealed the structural details of the rods that were heavily stained on both sides but less stained in the middle region (Fig. 2A, left), while the information on both ends of the rods was not clear, which awaits to be further explored by superresolution microscopy in the future. Quantification analysis revealed that the number per cell of gI rod-shaped structures ranged from 1 to 80 with an average number of 24.5 and that there existed an approximately normal distribution of the rod number per cell versus percentage of cells having rods (Fig. 2B). Further analysis by three-dimensional reconstruction showed that the rod-shaped structures are not cylinders but, rather, cuboids (Fig. 2C). The gI rods varied from 1 to 10 μm in length with an average size of about 3.6 μm, while in most cells, they ranged from 2 to 5 μm (Fig. 2D). On the other hand, the width and height were quite uniform with an estimated dimensional parameter of 0.5 ± 0.1 μm (width) by 0.3 ± 0.1 μm (height). Thus, the gI rods appear to be quite regular structures.
FIG 2.
Analysis of the number, size, and dimensions of HSV-1 gI rod-shaped structures. Vero cells were transfected to express gI-Myc or gI-GFP. At 18 to 24 h posttransfection, the cells were fixed, permeabilized, and stained with antibodies to Myc epitope tag (for cells expressing gI-myc). DAPI was used to stain cell nuclei (blue). The representative images were captured with a Leica confocal microscope and processed by using ImageJ and Imaris. (A) Images of gI-GFP and gI-myc at higher magnification. (B) Statistical analysis of the number per cell of gI rods. The y axis indicates the percentage of cells having rod structures, and the x axis indicates the number range of rods per cell. (C) Three-dimensional reconstruction of gI rod structures by Imaris software. The size bars are indicated. (D) Quantification analysis of the length of rod structures. The y axis indicates the percentage of cells having rod structures, and the x axis indicates the length range of rods.
The gI within the rod-shaped structures exhibits a conformational difference from the typical form of gI.
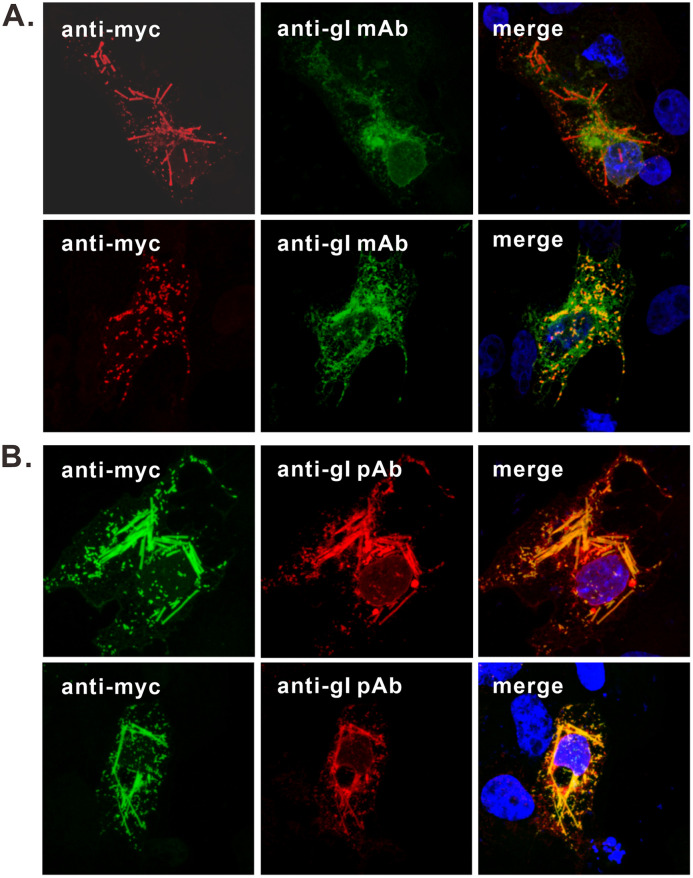
To probe the nature of the gI rods, we used a mouse monoclonal antibody, mAb3014, that was previously used for studying gI (21, 24). To our surprise, this antibody gave only a typical staining pattern of gI (Fig. 3A); it was not able to label the long rod-shaped structures, but rather the very short ones that showed a good colocalization relationship with that stained by anti-Myc antibody (Fig. 3A). In contrast, the gI polyclonal antibodies (UP1725) against the gI extracellular domain could label the rod-shaped structures that colocalized well with those by anti-Myc antibody (Fig. 3B). Thus, there exists a structural difference between the gI within the rods and the typical gI.
FIG 3.
The gI within the rod-shaped structures is structurally different from the typical form of gI. Vero cells were transfected to express gI-Myc. At 18 to 24 h posttransfection, the cells were fixed, permeabilized, and doubly stained with antibodies to Myc epitope tag and gI monoclonal antibody mAb3104 (A) or rabbit polyclonal antibodies to gI extracellular domain (UP1725) (B). DAPI was used to stain cell nuclei (blue). The representative images were captured with a Leica confocal microscope and processed using ImageJ.
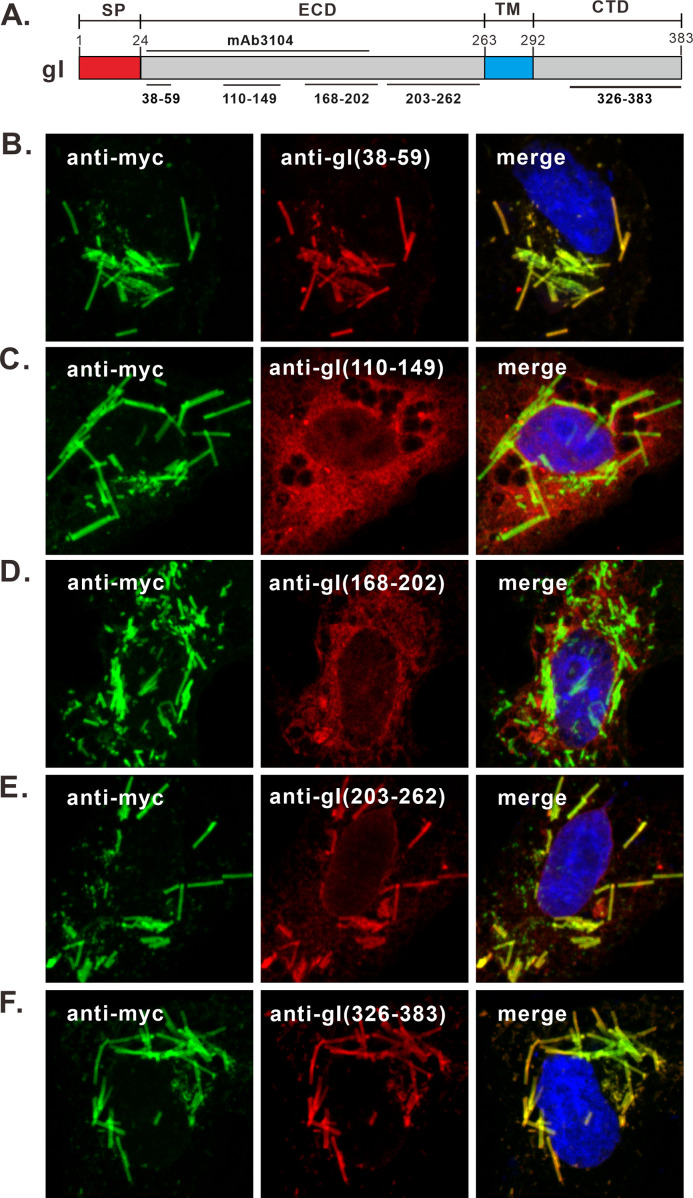
The sequence that gI mAb3104 recognizes is unclear, but previous studies suggest that the binding site is located within the gI region aa 43 to 200 and is probably conformation dependent (21). To further probe the structural difference, we made 5 rabbit polyclonal peptide antibodies against 5 different regions of gI (Fig. 4A), including aa 38 to 59, aa 110 to 149, aa 168 to 202, aa 203 to 262, and aa 326 to 383. These five polyclonal antibodies (pAbs) were subsequently named anti-gI(38-59), anti-gI(110-149), anti-gI(168-202) and anti-gI(203-262), and anti-gI(326-383). The probing results showed that three antibodies, including anti-gI(38-59), anti-gI(203-262), and anti-gI(326-383), could recognize rod-shaped structures that colocalized with those by the anti-Myc antibody (Fig. 4B, E, and F). In contrast, the other two antibodies against the middle region of the gI extracellular domain, including anti-gI(110-149) and anti-gI(168-202), could only recognize the typical form of gI (Fig. 4C and D). This result is consistent with that of mAb3104.
FIG 4.
The gI region aa 110 to 202 in the rod-shaped structures is not exposed outside. (A) Diagram of gI showing the recognition sites of gI antibodies. (B to F) Vero cells were transfected to express gI-Myc. At 18 to 24 h posttransfection, the cells were fixed, permeabilized, and doubly stained with antibodies specific for Myc tag (green) and gI peptide antibodies (red) as indicated. DAPI was used to stain cell nuclei (blue). The representative images were captured with a Leica confocal microscope and processed using ImageJ.
gI is a well-known binding partner of gE, and the interaction region for gE has been previously mapped to the middle region (aa 128 to 145) of the gI extracellular domain (21). According to the above results, this region of gI appears to be buried in the rod-shaped structures, perhaps driving their assembly. Hence, we predicted that the rod-shaped structures will not be able to interact with gE. Indeed, within the Vero cells coexpressing gE and gI, gE formed the typical speckles that resembled structures of ER or Golgi and colocalized well with the typical form of gI, but not the rod-shaped structures (Fig. 5). Thus, this result provides further evidence of the conformational change of gI as a result of formation of the rod-shaped structures, in which the antigenic epitopes with the middle region of gI (aa 110 to 202) are probably buried inside, while the regions aa 38 to 59 and aa 203 to 262 are exposed outside.
FIG 5.
The gI within rod-shaped structures does not colocalize gE. Vero cells were transfected to coexpress gI-myc and gE-HA. At 18 to 24 h posttransfection, the cells were fixed, permeabilized, and costained with antibodies specific for Myc tag (green) and HA antigen (red). The representative images were captured with a Leica confocal microscope and processed using ImageJ.
Evidence for gI self-interaction.
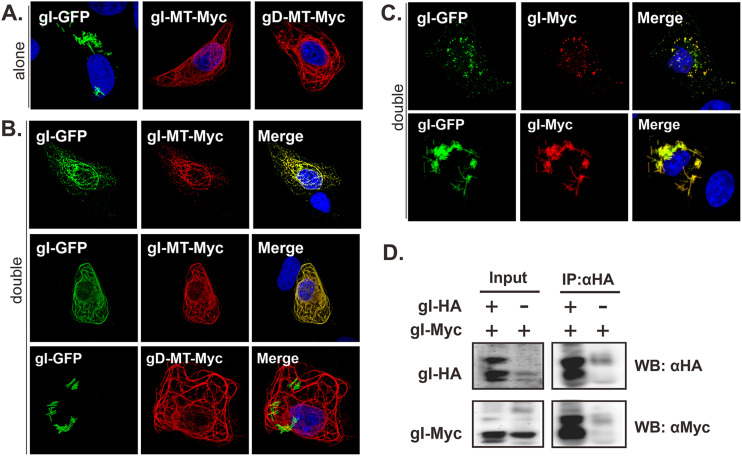
The gI rod-shaped structures resemble the morphology of tobacco mosaic virus (TMV) in which a single capsid protein homo-oligomerizes into higher-order structures. The very regularity of gI rods suggests that HSV-1 gI may have the potential to self-interact, a property that has not been reported before. To test this hypothesis, we performed a relocalization assay in which the cytoplasmic tail of gI is tagged with a microtubule localization signal (MT) to make a construct gI-MT-myc. In transfected cells, gI was targeted to form microtubule-like filaments (Fig. 6A). When coexpressed, we found that gI-GFP followed the same pattern of gI-MT-myc and became colocalized well with gI-MT-myc (Fig. 6B), while gI-GFP did not show this pattern when expressed alone (Fig. 6A). As a control, gI-GFP did not respond to gD-MT-myc (Fig. 6B, bottom), suggesting the interaction specificity and that the MT signal is not the factor for interaction. Consistent with this, gI-GFP colocalized well with gI-myc and could be assembled into the same rod-shaped structures (Fig. 6C). We also performed a coimmunoprecipitation assay (co-IP) and found that gI-myc could be coimmunoprecipitated by antibodies to the HA epitope targeting gI-HA (Fig. 6D). Thus, we provided two lines of evidence for gI self-interaction.
FIG 6.
Analysis of HSV-1 gI self-interaction. Vero cells were transfected to either singly express (A) or coexpress gI-GFP and gI-MT-myc, or gD-MT-myc (B) or gI-myc (C). The singly expressed proteins served as a control. At 18 to 24 h posttransfection, the cells were fixed, permeabilized, and stained with antibodies specific for Myc tag (red). The representative images were captured with a Leica confocal microscope and processed by using ImageJ. (D) Vero cells were transfected to express gI-HA and gI-myc or to singly express gI-myc. At 24 h posttransfection, the cells were lysed, and gI was immunoprecipitated with antibodies to HA epitope. The proteins bounded to the beads were separated by SDS-PAGE, followed by Western blotting with antibodies to either HA or c-myc epitope.
Analysis of the origin of gI rod-shaped structures.
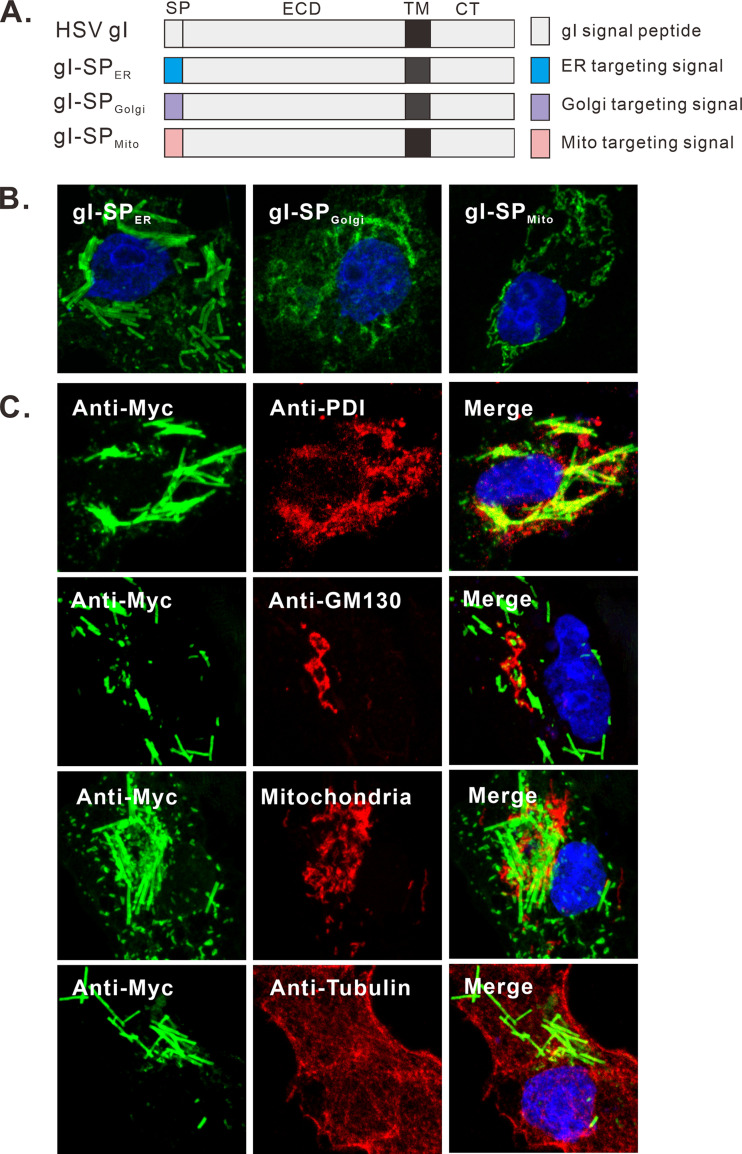
The fact that gI is a membrane protein strongly suggests that it is able to tubulate intracellular membranes as it assembles into rods. To probe the membrane origin, we first investigated the importance of the gI signal peptide. The N-terminal signal peptide sequence of gI was replaced with various subcellular organelle targeting signals (Fig. 7A), which have been shown to target GFP to the respective cellular organelles (44). We found that replacement with a similar ER targeting signal (gI-SPER) did not affect the formation of the rod-shaped structures, but not that with Golgi or mitochondria targeting signals, suggesting that the ER targeting signal is important for rod formation (Fig. 7B). We also investigated the colocalization relationship between rod-shaped structures and the makers of subcellular organelles in transfected Vero cells, but the rod structures did not colocalize with any markers of the endoplasmic reticulum (ER), Golgi, or microtubule (Fig. 7C). These results indicate the complex nature of the biogenesis of the rod-shaped structures, which warrants further investigation in the near future.
FIG 7.
Analysis of the membrane origin of gI rod-shaped structures. (A) Schematic diagrams of chimeric gI-myc constructs in which the gI signal peptide was replaced with either ER, Golgi, or mitochondrial targeting signal. (B) Vero cells were transfected to express the gI mutants and stained with antibodies to Myc epitope. (C) Vero cells were transfected to express gI-Myc. At 18 to 24 h posttransfection, the cells were fixed, permeabilized, and costained with antibodies to Myc tag (green) and other subcellular markers (red). The cell nuclei were stained with DAPI (blue). The representative images were captured with a Leica confocal microscope and processed by using ImageJ.
The extracellular domain of gI is a critical determinant for induction of rod-shaped structures.
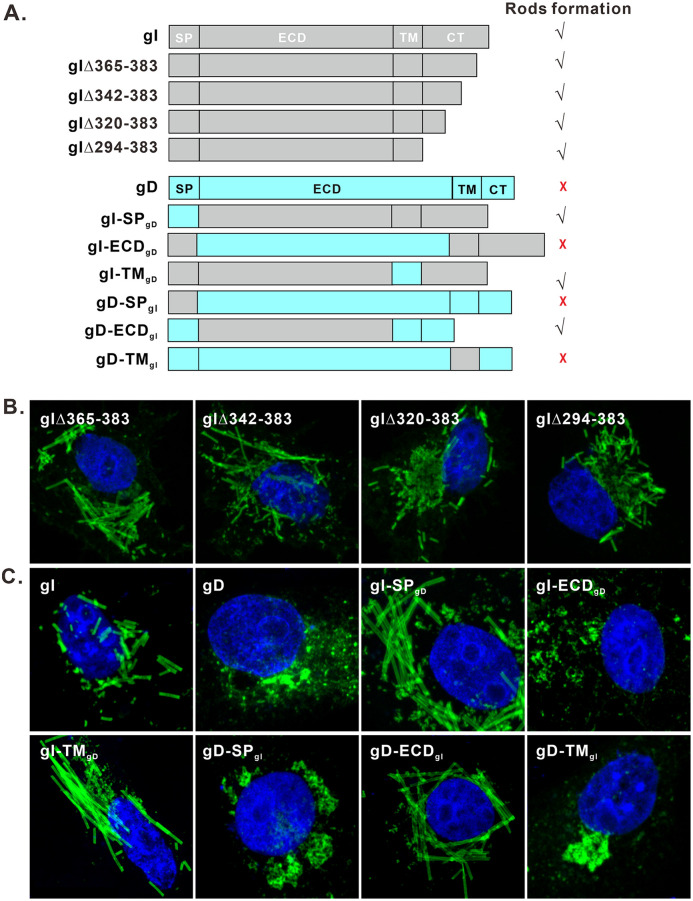
To identify the molecular determinant for forming rod-shaped structures, we first investigated the role of the gI cytoplasmic tail (aa 297 to 383), as it contains a putative amphipathic helix, two SH3-binding motifs, and an arginine-rich region, and all of these have the possibility to make gI polymerize. Accordingly, we made several truncation mutants of the gI cytoplasmic tail, including gIΔ294-383, gIΔ320-383, gIΔ342-383, and gIΔ365-383 (Fig. 8A, top). All the mutants retained the ability to induce rod structures (Fig. 8B). Thus, the gI cytoplasmic tail is not necessary for the rod formation.
FIG 8.
The gI extracellular domain is the critical determinant for inducing rod-shaped structures. (A) Diagram of gI/gD chimeric mutants and gI CT truncated mutants. The corresponding SP, ECD, and TM regions were swapped in a reciprocal manner. (B and C) Mutants were transiently expressed in Vero cells and were with antibodies to Myc and HA epitopes, followed with Alexa Fluor 488-conjugated secondary antibodies. The cell nuclei were stained with DAPI (blue). The representative images were captured with a Leica confocal microscope and processed by using ImageJ.
We next investigated the other three domains (SP, ECD, and TM) by taking the strategy of constructing chimeric mutants (Fig. 8A). The HSV-1 gD was chosen as the donor and acceptor, as it is also a type I transmembrane protein and is unable to induce the rod-shaped structures formation (Fig. 8C, top). A series of chimeras was constructed by swapping the respective SP, ECD, and the predicated TM regions in a reciprocal manner, and the resulted constructs were named gI-SPgD and gD-SPgI, gI-ECDgD and gD-ECDgI, and gI-TMgD and gD-TMgI, respectively (Fig. 8A, bottom). In the loss-of-function test, substitutions with gD SP or TM (gI-SPgD and gI-TMgD) did not affect the formation of the rod-shaped structures, but replacement with the ECD domain of gD (gI-ECDgD) could completely abolish this phenotype of gI (Fig. 8C), indicating a critical role of gI ECD domain. In the gain-of-function test, exchange with the gI ECD domain (gD-ECDgI) enabled gD to gain the ability to generate rod-shaped structures, but not those with SP or TM domains (gD-SPgI and gD-TMgI) (Fig. 8C).
Together, the above results indicate that the gI extracellular domain is the key determinant for inducing rod-shaped structures, and SP and TM are only needed for membrane insertion and anchoring, while the cytoplasmic tail seems nonessential.
Identification of the key residues or motifs for the formation of gI rod-shaped structures.
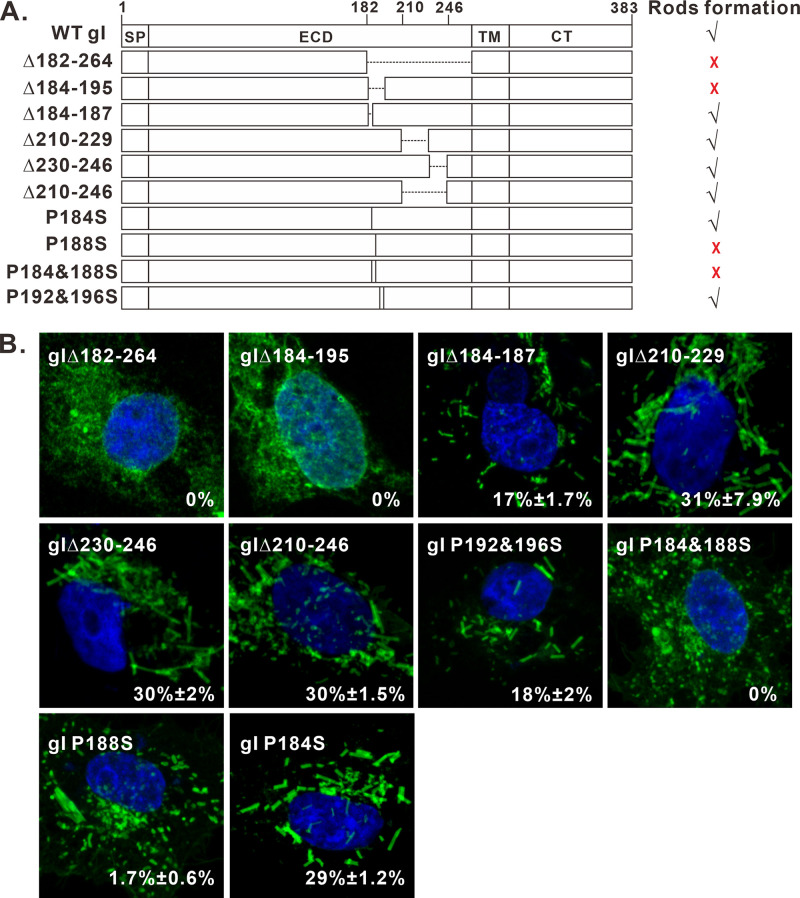
Bioinformatics analysis by online programs Phyre2 (45) and GlobPlot (46) suggest that the gI extracellular domain can be divided into two parts: aa 23 to 168 and aa 169 to 263. The region aa 23 to 168 contains 10 predicted beta sheets, suggesting its highly structured nature. In contrast, the region aa 169 to 263 has been reported to be a proline-rich domain (PRD) (22, 23). In addition, genetic mutations, insertions, and deletions often occur to this PRD region (22, 23). We started with this region by constructing a total of 6 deletion mutants (Fig. 9A), including gIΔ182-264, gIΔ184-195, gIΔ184-187, gIΔ196-209, gIΔ210-229 and gIΔ230-246, and gIΔ210-246, respectively. Of them, the mutant gIΔ196-209 failed to be expressed at a detectable level for unknown reasons (data not shown), while others were well expressed. The IFA results showed that deletion of either aa 182 to 264 or aa 184 to 195 blocked the formation of rod-shaped structures (Fig. 9B, top, left), while other deletions did not have an effect (Fig. 9B). We also introduced the same deletions into the chimeric construct gD-ECDgI, and the results were the same (data not shown). Together, these results suggest that the gI region aa 184 to 195 is a key determinant for rod formation.
FIG 9.
Mapping the key residues for formation of the gI rod-shaped structures. (A) Mutagenesis analysis of the gI extracellular domain. The deleted region is shown as a dashed line and point mutation as a vertical line with the relative positions indicated in the name of individual mutants. The capacity of inducing rod-shaped structures for each mutant is shown on the right. √, capable; ×, incapable. (B) Immunofluorescence staining of some of the mutants in transfected Vero cells with antibodies to Myc epitope. The images were obtained by confocal microscopy.
The sequence aa 184 to 196 contains a proline-rich motif (P184XXXP188XXXP192XXXP196). It forms a very regular pattern, and the four proline residues are evenly separated by three amino acids. To investigate whether these proline residues play a role in the rod structure formation, we performed site-directed mutagenesis by changing them to serine in different combinations in the background of gI-myc to generate two mutants: gI P184&188S and gI P192&196S (Fig. 9A). The mutant gI P184&188S showed the typical distribution pattern of a glycoprotein (Fig. 9B, middle), while gI P192&196S retained the ability to induce rods (Fig. 9B, middle), suggesting a crucial role in rod formation for prolines at positions 184 and 188 (P184 and P188). To further delineate the importance of these two residues, we mutated P184 and P188 individually to serine to generate two additional mutants: P184S and P188S (Fig. 9A). In transfected Vero cells, the mutant P184S retained the phenotype (Fig. 9B, bottom), while the mutation at position 188 (P188S) severely crippled the ability of gI to induce rods, and the percentage of cells expressing gI but showing rods was decreased to 1.7% (Fig. 9B, bottom), suggesting that P188 is a key residue. However, it took the double mutations (P184&188S) to completely block the induction of rod structures (Fig. 9B), suggesting that the two residues somehow work cooperatively to regulate the formation of rods. We also introduced the same mutations into the background of gD-ECDgI, and the results were the same (not shown). Thus, the residue P188 proline is the major determinant for the rod formation, but both P184 and P188 contribute.
The P184&188S mutation did not affect HSV-1 replication and spread in cell culture.
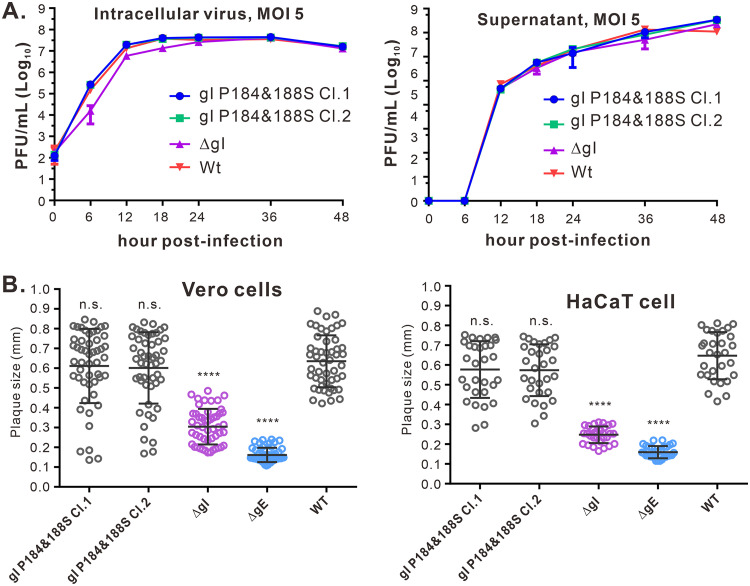
To analyze the importance of the gI ability to induce rods for viral replication and spread, we introduced the P184S and P188S mutations (gI P184&188S) into the HSV-1 KOS strain BAC clone by the galK selection system as described previously (47, 48). As a control, we created a gI-null virus (ΔgI) by replacing the start codon of gI with two stop codons. The correct clones were all verified by HindIII digestion and DNA sequencing. To examine the mutational effect, we analyzed growth properties in HaCaT cells infected with indicated viruses at an MOI of 5 (Fig. 10A). The results showed that the mutant gI P184&188S had kinetics similar to the parental virus (WT KOS) and reached a plateau around 24 h postinfection (hpi) within cells and at 36 hpi in the medium (Fig. 10A). In agreement with previous reports, ΔgI behaved like the WT virus (34, 40), despite a little decrease at an early time (Fig. 10A). In addition, we found that the mutant virus gI P184&188S failed to induce rod-shaped structures in HaCaT cells (Fig. 1D). To measure the mutational effect on viral spread, we analyzed plaque sizes in both Vero and HaCaT cells using plaque assay conditions. In Vero cells, the mutant gI P184&188S had plaque sizes similar to those of the WT and larger than ΔgI and ΔgE. Likewise, similar results were obtained with HaCaT cells infected with each of these viruses (Fig. 10B). Thus, we conclude that the ability of gI to induce rod-shaped structures is dispensable for viral replication and cell-free spread in epithelial cell culture.
FIG 10.
Effect of gI P184&188S mutation on viral replication and spread in cell culture. (A) Growth curve of WT HSV-1 and gI mutants in HaCaT cells at an MOI of 5. The intracellular virus and supernatant were harvested at indicated time points for titer determination by standard viral plaque assay on Vero cells. The data shown are representative of results from three independent experiments. (B) Quantitative analysis of the plaque sizes in Vero and HaCaT cells. Vero (left) and HaCaT (right) cells were infected with HSV-1 WT and recombinant viruses under plaque assay conditions. The cells were fixed at 5 days postinfection and plaques were visualized by staining with crystal violet. The diameters of 50 (Vero cells, left) or 30 (HaCaT cells, right) single plaques for each of the indicated viruses were measured. The horizontal bars indicate the mean for each group. ****, P < 0.001; n.s., not significant.
The P184&188S mutation affects the ability of HSV-1 to induce cell fusion in the UL24syn background.
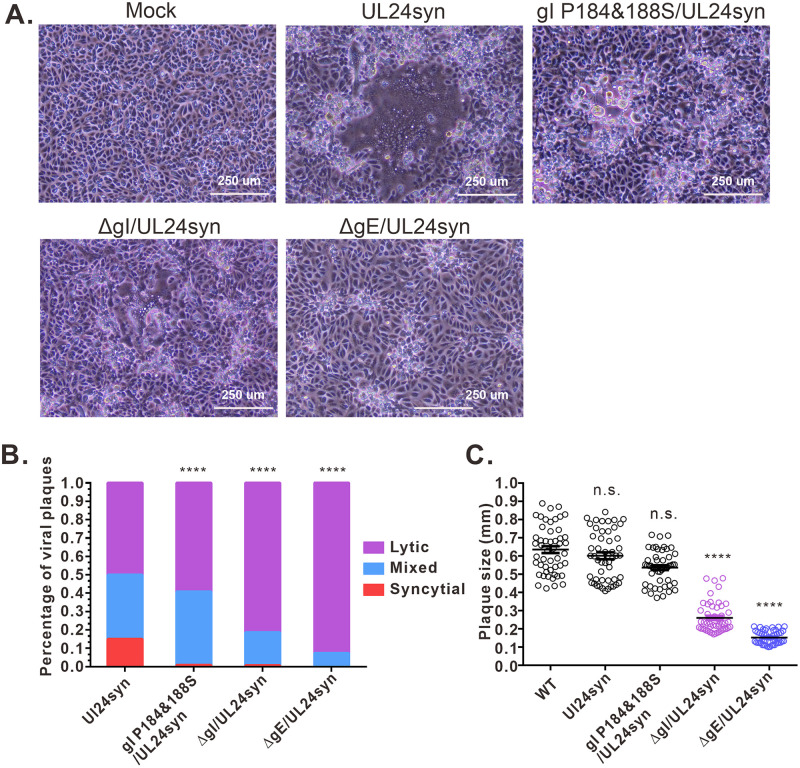
A recent study has revealed that there exist differential requirements for accessory proteins (e.g., gE, gI, UL16, etc.) among syncytial variants and that gI is only required for UL24syn-induced cell-cell fusion (40). To test the effect of P184&188S mutation on syncytial formation, we first converted the HSV KOS strain into a syncytial virus by introducing a Syn mutation (G121A) into the UL24 gene to generate the mutant virus UL24syn and then introduced the corresponding mutation into the UL24syn background to generate the mutant virus gI P184&188S/UL24syn. At the same, deletion mutants lacking gI (ΔgI/UL24syn) or gE (ΔgE/UL24syn) served as positive controls. The quantitative analysis showed that the mutant UL24.G121A (UL24syn) displayed a mixed phenotype that contained fully syncytial plaques (15%) and mixed (35%) and lytic (50%) plaques (Fig. 11A and B), which is a little different from the previous report that the UL24syn was fully fusogenic (40). As positive controls, deletion of gE completely blocked the fully syncytial phenotype (Fig. 11A and B), and the gI-null virus also showed the significantly crippled ability to induce syncytium (Fig. 11A and B). Interestingly, the introduction of the P184&188S mutation also significantly decreased the percentage of fully syncytial phenotype from 15% to 0.8% (Fig. 11B). We chose two independent clones, and they showed the same effect. On the other hand, the P184&188S mutation did not affect the viral plaque sizes on Vero cells (Fig. 11C). Thus, the ability of gI to induce rods appears to have a positive correlation with the capacity of the virus to induce syncytium in the UL24syn background in cell culture.
FIG 11.
Effect of gI P184&188S mutations on syncytium formation in the UL24syn background. (A) Virus-induced CPE. Vero cells were infected by the virus as indicated at an MOI of 0.01. At 36 hpi, the virus-induced CPE was taken by a Nikon inverted microscope. The data showed the representative pictures. (B) Quantitative analysis of virus induced-syncytium. Vero cells were infected at an MOI of 0.01. After 1 h of incubation, the cells were rinsed with PBS overlaid with DMEM containing 0.6% methylcellulose and incubated for 36 h. Afterward, the cells were fixed and stained with antibody to VP5, and the cell nuclei were stained with DAPI. The infection focus was scored as syncytial, mixed, or lytic according to the methods in Materials and Methods. (C) Quantitative analysis of the plaque sizes in Vero cells. The diameters of 50 single plaques for each of the indicated viruses were measured. The horizontal bars indicate the mean for each group. ****, P < 0.001; n.s., not significant.
DISCUSSION
Alphaherpesviruses are very adept at moving from one infected cell to an adjoining uninfected cell, specifically at sites of epithelial cell or neuronal junctions (6–8). For HSV-1, the role of the gE/gI heterodimer in viral cell-to-cell spread has been well documented (7, 9–11, 13–16, 18, 41, 42), but much is still unknown about how this protein pair works during infection. The past efforts have mainly focused on gE, leaving gI largely unexplored. In this report, we described a previously unrecognized property of HSV-1 gI. This molecule was able to induce rod-shaped structures within eukaryotic cells when expressed alone. The antibody probing revealed that the gI within these structures exhibited a conformational difference from the typical gI and likely represents a higher-order form. Further mutagenesis identified the gI extracellular domain as the critical determinant for inducing the rod-shaped structures, while the TM domain merely served as a membrane anchor. Finally, the reverse genetics analysis showed that the ability of gI to induce rods was not required for viral replication or spread in cell culture but rather positively correlated with the capacity of the virus to induce cell fusion in the UL24syn background.
The observation that HSV-1 gI could induce rod-shaped structures is unexpected. This fortuitous discovery stemmed from our approach to visualize gI intracellular distribution by using an epitope tag instead of a previously used gI mouse monoclonal antibody mAb3104 (21, 24). The rod-shaped structures were quite striking, as they were regular in width but variable in length, ranging from 1 to 10 μm. Interestingly, only about 40% of the transfected cells expressing gI showed this phenotype. The reasons why gI did not induce rod structures in some cells are currently not clear, but they may be related to cell physiological status and may require a host protein as a trigger, rather than the protein expression level, as gI was expressed well in both situations (Fig. 1). In HSV-1 infected cells, the frequency of cells having rods was quite low (Fig. 1). This result is not surprising, as all the other viral proteins, including all that interact with gI, are present, and they can potentially regulate this property of gI. We actually have some preliminary evidence showing that expression of gE could reduce the ability of gI to induce rod-shaped structures (data not shown). The rod formation is unlikely due to overexpression and protein aggregation, as the rod formation can essentially be eliminated by a single amino acid substitution. Moreover, the mutants with more drastic deletion mutations continued to make rods, even though they would be more likely to be misfolded. Finally, the very regularity of the width of the rods is inconsistent with a random protein aggregation but, rather, a likely result of protein polymerization.
Our finding represents the first example in virology and cell biology that a viral protein can induce such large and ordered structures at the micrometer level. The molecular mechanisms regarding the rod biogenesis remain unresolved. Since gI is a type I membrane protein, the formation of the rods required the transmembrane domain, and that the rods were heavily stained on both sides, it is possible that HSV-1 gI possesses the ability to induce membrane curvature and is able to tubulate cellular membranes to form such highly ordered structures. In retrospect, the vaccinia virus A17 protein was also found to induce tubule-like structures (49), but these structures are too small (at the nanometer level) to be seen by a conventional fluorescence microscope.
The morphology of HSV-1 gI rods is reminiscent of tobacco mosaic virus (TMV) and tubules formed by cellular membrane curvature-inducing cellular proteins (50, 51). Polymerization or homo-oligomerization of viral or cellular proteins into a higher order is often the key to assemble such very regular structures (52–58). This is likely the case for gI rod formation; the polymerization of gI may lead to the formation of such beautiful rod-shaped structures. This will also explain the conformational difference between the two forms of gI, as certain antibodies [mAb3104, anti-gI(110-149), and anti-gI(168-202)] could recognize the typical gI but fail to pick up the rod-shaped structures. In agreement with this scenario, we provided evidence that HSV-1 gI can self-interact (Fig. 6). More experiments are clearly required to investigate whether gI can polymerize. Our mutagenesis studies also identified a proline-rich motif (PRD) (P184XXXP188) critical for rod formation. Interestingly, in the dynamin protein superfamily, the PRD provides a platform to interact with SH3 motif-containing proteins to promote protein oligomerization (59). In addition, many proteins, such as P53 and HIV-1 Vif, often contain a proline-rich motif for protein multimerization (60, 61). Whether HSV-1 gI uses the similar mechanism and how gI exactly induces rod formation are currently not clear, which will be of great interest to pursue.
The rod-shaped structures most likely contain the full-length gI, as they could be recognized by antibodies against both the N terminus and cytoplasmic tail of gI. Although several antibodies failed to stain the region aa 110 to 202 of the rods, it is unlikely that this region is not included in the protein. We did the deep sequencing of HSV-1-infected cells and did not find any spliced mRNA of gI. In addition, small deletions or in-frame insertions of a hemagglutinin (HA)-epitope tag within this region could disrupt the formation of rod-shaped structures (data not shown).
We also tried to investigate whether the ability of gI to induce rod-shaped structures is important for viral replication and spread. We found that the introduction of the mutation gI P184&188S into the HSV-1 KOS did not affect viral replication or the viral plaque size. In contrast, the gI- or gE-negative mutant produced smaller plaques in both Vero and HaCaT cells, as expected. Because the induction of rod-shaped structures was not frequently seen in virus-infected cells, we hypothesized that the ability to induce membrane curvature by gI is well regulated under normal conditions (WT HSV-infected cells). This will be similar to the situation for gD, gB, and gH/gL, which can cause massive cell fusions when they are coexpressed in the absence of other viral proteins (62) but tightly regulated by other viral proteins in WT HSV-infected cells, unless the syn mutations are introduced into one of the genes encoding gB, gK, UL20, or UL24 to tip this balance (37–40). Therefore, we examined the membrane curvature-inducing ability of gI in this out-of-control situation. Fortunately, a recent study showed that gI is only required for UL24syn-induced cell fusion (40). A syncytial mutant UL24syn was constructed by introducing a substitution G121A into the WT UL24. Interestingly, the phenotype of UL24syn had a mixed phenotype, containing a little percentage of 15% fully syncytial plaques, 35% mixed, and 50% lytic plaques, which is slightly different from the previous report that the UL24syn was fully fusogenic (40). The reasons for the difference are currently not clear, but it may be due to a difference in the systems, such as the cells we used. Nevertheless, the G121A mutant still can induce a substantial percentage of syncytium. As a positive control, either deletion of either gE or gI blocked the formation of pure syncytium. Interestingly, the P184&188S mutation also severely crippled the syncytium formation (0.8%). Thus, the ability of gI to induce membrane curvature seems to positively correlate with the capacity of the virus to induce cell fusion in the UL24syn background in cell culture, suggesting a likely intimate relationship between curvature induction and virus-induced cell-cell fusion. Since gI is important for HSV anterograde transmission from neuronal cells to epithelial cells or within neuronal tissues, it will be of great interest to test in the future how the mutant P184&188S behaves compared to the gI-null virus (ΔgI) in the Campenot chamber culture system and in a mouse model. For the latter, because the KOS strain used in this study is a lab-adapted strain and does not infect BALB/c mouse efficiently, it is necessary to construct mutant viruses based on other pathogenic HSV-1 strains to further explore the functional importance of this novel property of gI. Nevertheless, the studies here further advanced our understanding of the biological properties of HSV-1 gI and may have important implications in understanding viral pathogenesis.
MATERIALS AND METHODS
Cells and virus strains.
Vero cells (ATCC CCL-81 and E6), HaCaT cells, BHK-21 cells, and HeLa cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS), penicillin (65 μg/ml), and streptomycin (131 μg/ml). All viruses used in this study were derived from the HSV-1 KOS strain, the genome of which has been cloned into a bacterial artificial chromosome (BAC) (47). For infection assays, Vero and HaCaT cells were grown in DMEM supplemented with 2% FBS, penicillin (65 μg/ml), and streptomycin (131 μg/ml).
Antibodies.
Rabbit anti-Myc polyclonal antibody (pAb) (catalog no. C3956), mouse anti-Myc monoclonal antibody (MAb) (catalog no. M4439), mouse anti-HA MAb (catalog no. H3663), and Mito Red (catalog no. 53271) were all purchased from Sigma-Aldrich (St. Louis, MO, USA). Mouse anti-HSV1 gE MAb (catalog no.ab6510) was purchased from Abcam. Rabbit anti-PDI MAb (product no. 3501), rabbit anti-GM130 MAb (product no. 12480), and mouse anti-α-tubulin MAb (product no. 3873) were purchased from Cell Signaling Technology (Danvers, MA, USA). Alexa Fluor 488-conjugated goat anti-rabbit IgG(H+L) F(ab′)2 fragment and Alexa Fluor 488-conjugated goat anti-mouse IgG(H+L) F(ab′)2 fragment were purchased from Molecular Probes (Invitrogen, CA, USA). Mouse anti-HSV1 gI MAb (24) was a generous gift by David C. Johnson (Oregon Health & Science University), and rabbit anti-HSV1 gI pAb (UP1725) (41) was a generous gift from Harvey Friedman (University of Pennsylvania).
The rabbit peptide antibodies were raised against different regions of gI, including gI aa 38 to 59, aa 110 to 149, aa 168 to 202, aa 203 to 262, and aa 326 to 383, which were expressed as a glutathione S-transferase (GST) fusion protein at its C terminus. The recombinant proteins were expressed and purified from Escherichia coli BL21 cells. The reactivities and specificities of the respective antibodies were tested by immunofluorescence under conditions of transfection. All the antibodies showed good reactivity and specificity (data not shown).
Construction of gI mutants.
All gI alleles were cloned into the multiple cloning site of pEGFP-N2 (Clontech), thereby producing vectors that encode wild-type gI and gI-GFP fusions. The mammalian expression plasmid for gI-Myc was created by adding a Myc tag to the C terminus of gI. The construct for gE-HA or gD-HA was generated by adding a HA epitope tag to the C terminus. Then, the signal peptides (SP), extracellular domain (ECD), and transmembrane domain (TM) between gI and gD were swapped by overlapping PCR. The gI signal peptide was replaced with either ER (MLLSVPLLLGLLGLAVA in the N terminus and retention signal KDEL in the C terminus), Golgi (MFSLFILVFLLFAVICVW), or mitochondrial (MSVLTPLLLRGLTGSARRLPVPRAKIHSL) targeting signals (44, 63). The expression plasmids of gI-MT-myc and gD-MT-myc were constructed by adding a microtubule localization signal (GIRINLAASSQPLDPEGPIAVTPRPPIRPSSGRYRGPGIHRSPPRRTASRPTSRSATT) to the gI C terminus plus an myc epitope tag. The molecular cloning was facilitated through homologous recombination instead of the conventional ligation strategy by using the ClonExpress II one step cloning kit (Vazyme, Nanjing, China). Then, the corresponding deletion and site-directed mutants originated from gI-Myc or chimeric gD-ECDgI-HA were made by standard molecular biological techniques. All created plasmids are under the control of the cytomegalovirus (CMV) promoter, and a Kozak core sequence is also added to allow optimal expression, as well as confirmed by sequencing.
Confocal microscopy.
Vero E6, BHK-21, and HeLa Cells grown on coverslips in 6-well plates were transfected when 60% to 70% confluent with 2 to 3 μg plasmid DNA per well using Lipofectamine 2000 DNA transfection reagent (Invitrogen, CA, USA) in Opti-MEM. After incubation at room temperature (RT) for 15 min, the DNA-Lipofectamine complexes were added directly to cells in an antibiotic-free culture medium. HaCaT cells cultured on coverslips in six-well plates were infected with the HSV-1 KOS strain at an MOI of 0.1. At 18 to 24 h posttransfection or postinfection, the immunofluorescence assay was protocoled as previously documented. Generally, the cells were fixed with 3.7% paraformaldehyde for 10 min at RT, then washed with phosphate-buffered saline (PBS) three times (5 min for each wash), permeabilized with 0.1% Triton X-100 in PBS-2% bovine serum albumin (BSA) for 10 min, and blocked with PBS-2% BSA for 30 min at RT. The cells were then incubated with the proper primary antibodies for 1 h in a humid chamber and then washed with PBS three times (5 min for each wash). After that, the cells were incubated with the appropriate secondary antibodies, including Alexa Fluor 488-conjugated goat anti-rabbit IgG(H+L) F(ab′)2 fragment and Alexa Fluor 488-conjugated goat anti-mouse IgG(H+L) F(ab′)2 fragment, for another 1 h and then washed once with PBS. All the primary and secondary antibodies were used at a dilution of 1:1,000. Nuclear DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI) (Molecular Probes) for 5 min. After three rinses, the samples were mounted and examined with a Leica confocal microscope and processed using ImageJ software. The final three-dimensional reconstruction, including surface rendering, was generated using Imaris (Bitplane, Switzerland).
Coimmunoprecipitation.
Vero cells were transfected to express gI-HA and gI-myc or to singly express gI-myc. At 24 h posttransfection, the cells were harvested and lysed in NP-40 lysis buffer (0.5% NP-40, 150 mM NaCl, 50 mM Tris-HCl, pH 8.0), and protease inhibitor cocktail (Sigma; catalog no. P8340) and clarified by centrifugation at 12,000 rpm for 20 min. The cell supernatants were precleared with protein A/G-Sepharose beads (Santa Cruz Biotechnology; catalog no. sc-2003) and then incubated with 5 μl mouse anti-HA MAb and 25 μl protein A/G-Sepharose beads overnight at 4°C with gentle rotation. The beads were washed five times with the NP-40 lysis buffer, and the proteins bounded to the beads were separated by SDS-PAGE, followed by Western blotting with proper antibodies.
Western blot analysis.
The protein samples were separated by SDS-PAGE, transferred onto polyvinylidene difluoride (PVDF) membranes, blocked with PBST (PBS with 0.05% Tween 20) containing 5% milk or BSA for 1.5 h (for detection of phosphorylated proteins, BSA was used instead of milk), and then probed with primary antibodies to HA or c-Myc epitope at RT. The membranes were then washed with PBST and incubated with proper horseradish peroxidase (HRP)-conjugated secondary antibodies with a dilution ratio of 1:10,000. The membrane was again washed and then developed with the Pierce ECL Western blot substrate (Thermo Fisher; catalog no. 32209).
Construction of mutant viruses.
The HSV-1 KOS strain BAC was used to make mutants using BAC recombineering system as previously described (47). The gE-null mutant (ΔgE) was described (64). The gI-null mutant (ΔgI) was made by replacing the gI start codons with two stop codons. Recombinant viruses HSV gI P184&188S, encoding gI with serine, substituted for proline-184 and proline-188. To convert wild-type KOS into a syncytial strain, a G121A mutant was introduced into the UL24 gene to generate a syncytial virus UL24syn. In the background of this virus, mutants gI P184&188S/UL24syn, ΔgI/UL24syn, and ΔgE/UL24syn were subsequently generated. All clones were verified by HindIII digestion, PCR analysis, and DNA sequencing of the relevant region. The resulting BAC plasmids were purified from E. coli (SW102) and transfected into Vero cells using Lipofectamine 2000 as previously described. After 3 to 4 days posttransfection, when cytopathic effects (CPE) appeared, the whole-cell culture was harvested, and this transfection stock was used to infect new Vero monolayers. Cells and media were harvested 2 to 3 days postinfection, subjected to 3 freeze-thaw rounds, and sonicated to create a virus stock.
Viral growth kinetics and plaque assays.
Six-well plates of HaCaT cells were infected with the P1-specified viruses at an MOI of 5. After 1 h of incubation at 37°C, the cells were first washed with acid buffer (135 Mm NaCl, 10 Mm KCl, 40 Mm citric acid, pH 3.0), then once with DMEM, and overlaid with 1 ml DMEM containing 2% FBS. At the indicated time points postinfection, infected cells and media were harvested separately and processed as follows. Cells were scraped off the plates, and media were cleared of cells by centrifugation at 12,000 rpm for 1 min and frozen at −80°C. Cells were washed three times with DMEM and freeze-thawed 3 times to release cell-associated viruses. Each sample was titrated on Vero cells using a standard viral plaque assay. For plaque size analyses, plaque areas were calculated using ImageJ software.
Quantitative analysis of syncytium form.
To measure and score syncytium formation, Vero cells were infected with the viruses at an MOI of 0.01 at 37°C. After 1 h of incubation, the cells were rinsed with PBS and overlaid with DMEM medium containing 0.6% methylcellulose. At the indicated times postinfection, the immunofluorescence assay was protocoled as previously documented. The images were captured under Nikon Eclipse Ci fluorescence microscope and processed by using ImageJ software. Each individual syncytium was assigned a phenotypic score as in the previous report (40). A score of syncytium indicated that the entire area of infection consisted of multinucleated cells. A score of mixed meant that the area of infection contained a syncytium with at least 5 nuclei directly juxtaposed to lytically infected cells. Finally, a score of lytic meant that no fused cells were apparent.
Statistical analysis.
Statistical analyses were performed using a one-way analysis of variance (ANOVA) in GraphPad Prism (version 6). P values of <0.05 were considered statistically significant; P values of <0.001 were considered extremely significant.
ACKNOWLEDGMENTS
We sincerely thank David C. Johnson, Harvey Friedman, and David Leib for providing reagents.
This work was supported by the National Natural Science Foundation of China (31770171) and a Chinese One Thousand Young Talents award (1051-21986001) to J.H. and by NIH grant R01 AI071286 to J.W.W. The funders were not involved in study design, data collection or interpretation, or the decision to submit the work for publication.
We declare no conflicts of interest.
REFERENCES
- 1.Davison AJ, Eberle R, Ehlers B, Hayward GS, McGeoch DJ, Minson AC, Pellett PE, Roizman B, Studdert MJ, Thiry E. 2009. The order Herpesvirales. Arch Virol 154:171–177. doi: 10.1007/s00705-008-0278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.James SH, Kimberlin DW. 2015. Neonatal herpes simplex virus infection. Infect Dis Clin North Am 29:391–400. doi: 10.1016/j.idc.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 3.Stahl JP, Mailles A. 2019. Herpes simplex virus encephalitis update. Curr Opin Infect Dis 32:239–243. doi: 10.1097/QCO.0000000000000554. [DOI] [PubMed] [Google Scholar]
- 4.Groves MJ. 2016. Genital herpes: a review. Am Fam Physician 93:928–934. [PubMed] [Google Scholar]
- 5.Harris SA, Harris EA. 2018. Molecular mechanisms for herpes simplex virus type 1 pathogenesis in Alzheimer's disease. Front Aging Neurosci 10:48. doi: 10.3389/fnagi.2018.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson DC, Huber MT. 2002. Directed egress of animal viruses promotes cell-to-cell spread. J Virol 76:1–8. doi: 10.1128/jvi.76.1.1-8.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith G. 2012. Herpesvirus transport to the nervous system and back again. Annu Rev Microbiol 66:153–176. doi: 10.1146/annurev-micro-092611-150051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kramer T, Enquist LW. 2013. Directional spread of alphaherpesviruses in the nervous system. Viruses 5:678–707. doi: 10.3390/v5020678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson DC, Webb M, Wisner TW, Brunetti C. 2001. Herpes simplex virus gE/gI sorts nascent virions to epithelial cell junctions, promoting virus spread. J Virol 75:821–833. doi: 10.1128/JVI.75.2.821-833.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howard PW, Wright CC, Howard T, Johnson DC. 2014. Herpes simplex virus gE/gI extracellular domains promote axonal transport and spread from neurons to epithelial cells. J Virol 88:11178–11186. doi: 10.1128/JVI.01627-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dingwell KS, Johnson DC. 1998. The herpes simplex virus gE-gI complex facilitates cell-to-cell spread and binds to components of cell junctions. J Virol 72:8933–8942. doi: 10.1128/JVI.72.11.8933-8942.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wisner T, Brunetti C, Dingwell K, Johnson DC. 2000. The extracellular domain of herpes simplex virus gE is sufficient for accumulation at cell junctions but not for cell-to-cell spread. J Virol 74:2278–2287. doi: 10.1128/jvi.74.5.2278-2287.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Polcicova K, Goldsmith K, Rainish BL, Wisner TW, Johnson DC. 2005. The extracellular domain of herpes simplex virus gE is indispensable for efficient cell-to-cell spread: evidence for gE/gI receptors. J Virol 79:11990–12001. doi: 10.1128/JVI.79.18.11990-12001.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snyder A, Polcicova K, Johnson DC. 2008. Herpes simplex virus gE/gI and US9 proteins promote transport of both capsids and virion glycoproteins in neuronal axons. J Virol 82:10613–10624. doi: 10.1128/JVI.01241-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howard PW, Howard TL, Johnson DC. 2013. Herpes simplex virus membrane proteins gE/gI and US9 act cooperatively to promote transport of capsids and glycoproteins from neuron cell bodies into initial axon segments. J Virol 87:403–414. doi: 10.1128/JVI.02465-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saldanha CE, Lubinski J, Martin C, Nagashunmugam T, Wang L, van Der Keyl H, Tal-Singer R, Friedman HM. 2000. Herpes simplex virus type 1 glycoprotein E domains involved in virus spread and disease. J Virol 74:6712–6719. doi: 10.1128/jvi.74.15.6712-6719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang F, Tang W, McGraw HM, Bennett J, Enquist LW, Friedman HM. 2005. Herpes simplex virus type 1 glycoprotein E is required for axonal localization of capsid, tegument, and membrane glycoproteins. J Virol 79:13362–13372. doi: 10.1128/JVI.79.21.13362-13372.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang F, Zumbrun EE, Huang J, Si H, Makaroun L, Friedman HM. 2010. Herpes simplex virus type 2 glycoprotein E is required for efficient virus spread from epithelial cells to neurons and for targeting viral proteins from the neuron cell body into axons. Virology 405:269–279. doi: 10.1016/j.virol.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saied AA, Chouljenko VN, Subramanian R, Kousoulas KG. 2014. A replication competent HSV-1(McKrae) with a mutation in the amino-terminus of glycoprotein K (gK) is unable to infect mouse trigeminal ganglia after cornea infection. Curr Eye Res 39:596–603. doi: 10.3109/02713683.2013.855238. [DOI] [PubMed] [Google Scholar]
- 20.David AT, Baghian A, Foster TP, Chouljenko VN, Kousoulas KG. 2008. The herpes simplex virus type 1 (HSV-1) glycoprotein K(gK) is essential for viral corneal spread and neuroinvasiveness. Curr Eye Res 33:455–467. doi: 10.1080/02713680802130362. [DOI] [PubMed] [Google Scholar]
- 21.Basu S, Dubin G, Nagashunmugam T, Basu M, Goldstein LT, Wang L, Weeks B, Friedman HM. 1997. Mapping regions of herpes simplex virus type 1 glycoprotein I required for formation of the viral Fc receptor for monomeric IgG. J Immunol 158:209–215. [PubMed] [Google Scholar]
- 22.Norberg P, Bergström T, Rekabdar E, Lindh M, Liljeqvist JA. 2004. Phylogenetic analysis of clinical herpes simplex virus type 1 isolates identified three genetic groups and recombinant viruses. J Virol 78:10755–10764. doi: 10.1128/JVI.78.19.10755-10764.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Norberg P, Olofsson S, Tarp MA, Clausen H, Bergstrom T, Liljeqvist JA. 2007. Glycoprotein I of herpes simplex virus type 1 contains a unique polymorphic tandem-repeated mucin region. J Gen Virol 88:1683–1688. doi: 10.1099/vir.0.82500-0. [DOI] [PubMed] [Google Scholar]
- 24.Johnson DC, Frame MC, Ligas MW, Cross AM, Stow ND. 1988. Herpes simplex virus immunoglobulin G Fc receptor activity depends on a complex of two viral glycoproteins, gE and gI. J Virol 62:1347–1354. doi: 10.1128/JVI.62.4.1347-1354.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sprague ER, Wang C, Baker D, Bjorkman PJ. 2006. Crystal structure of the HSV-1 Fc receptor bound to Fc reveals a mechanism for antibody bipolar bridging. PLoS Biol 4:e148. doi: 10.1371/journal.pbio.0040148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chapman TL, You I, Joseph IM, Bjorkman PJ, Morrison SL, Raghavan M. 1999. Characterization of the interaction between the herpes simplex virus type I Fc receptor and immunoglobulin G. J Biol Chem 274:6911–6919. doi: 10.1074/jbc.274.11.6911. [DOI] [PubMed] [Google Scholar]
- 27.Rizvi SM, Raghavan M. 2001. An N-terminal domain of herpes simplex virus type Ig E is capable of forming stable complexes with gI. J Virol 75:11897–11901. doi: 10.1128/JVI.75.23.11897-11901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ndjamen B, Farley AH, Lee T, Fraser SE, Bjorkman PJ. 2014. The herpes virus Fc receptor gE-gI mediates antibody bipolar bridging to clear viral antigens from the cell surface. PLoS Pathog 10:e1003961. doi: 10.1371/journal.ppat.1003961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frank I, Friedman HM. 1989. A novel function of the herpes simplex virus type 1 Fc receptor: participation in bipolar bridging of antiviral immunoglobulin G. J Virol 63:4479–4488. doi: 10.1128/JVI.63.11.4479-4488.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hook LM, Huang J, Jiang M, Hodinka R, Friedman HM. 2008. Blocking antibody access to neutralizing domains on glycoproteins involved in entry as a novel mechanism of immune evasion by herpes simplex virus type 1 glycoproteins C and E. J Virol 82:6935–6941. doi: 10.1128/JVI.02599-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagashunmugam T, Lubinski J, Wang L, Goldstein LT, Weeks BS, Sundaresan P, Kang EH, Dubin G, Friedman HM. 1998. In vivo immune evasion mediated by the herpes simplex virus type 1 immunoglobulin G Fc receptor. J Virol 72:5351–5359. doi: 10.1128/JVI.72.7.5351-5359.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DuRaine G, Wisner TW, Howard P, Williams M, Johnson DC. 2017. Herpes simplex virus gE/gI and US9 promote both envelopment and sorting of virus particles in the cytoplasm of neurons, two processes that precede anterograde transport in axons. J Virol 91:e00050-17. doi: 10.1128/JVI.00050-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farnsworth A, Goldsmith K, Johnson DC. 2003. Herpes simplex virus glycoproteins gD and gE/gI serve essential but redundant functions during acquisition of the virion envelope in the cytoplasm. J Virol 77:8481–8494. doi: 10.1128/jvi.77.15.8481-8494.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balan P, Davis-Poynter N, Bell S, Atkinson H, Browne H, Minson T. 1994. An analysis of the in vitro and in vivo phenotypes of mutants of herpes simplex virus type 1 lacking glycoproteins gG, gE, gI or the putative gJ. J Gen Virol 75:1245–1258. doi: 10.1099/0022-1317-75-6-1245. [DOI] [PubMed] [Google Scholar]
- 35.Chatterjee S, Koga J, Whitley RJ. 1989. A role for herpes simplex virus type 1 glycoprotein E in induction of cell fusion. J Gen Virol 70:2157–2162. doi: 10.1099/0022-1317-70-8-2157. [DOI] [PubMed] [Google Scholar]
- 36.Davis-Poynter N, Bell S, Minson T, Browne H. 1994. Analysis of the contributions of herpes simplex virus type 1 membrane proteins to the induction of cell-cell fusion. J Virol 68:7586–7590. doi: 10.1128/JVI.68.11.7586-7590.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gage PJ, Levine M, Glorioso JC. 1993. Syncytium-inducing mutations localize to two discrete regions within the cytoplasmic domain of herpes simplex virus type 1 glycoprotein B. J Virol 67:2191–2201. doi: 10.1128/JVI.67.4.2191-2201.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dolter KE, Ramaswamy R, Holland TC. 1994. Syncytial mutations in the herpes simplex virus type 1 gK (UL53) gene occur in two distinct domains. J Virol 68:8277–8281. doi: 10.1128/JVI.68.12.8277-8281.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarfo A, Starkey J, Mellinger E, Zhang D, Chadha P, Carmichael J, Wills JW. 2017. The UL21 tegument protein of herpes simplex virus 1 is differentially required for the syncytial phenotype. J Virol 91:e01161-17. doi: 10.1128/JVI.01161-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carmichael JC, Wills JW. 2019. Differential requirements for gE, gI, and UL16 among herpes simplex virus 1 syncytial variants suggest unique modes of dysregulating the mechanism of cell-to-cell spread. J Virol 93:e00494-19. doi: 10.1128/JVI.00494-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGraw HM, Awasthi S, Wojcechowskyj JA, Friedman HM. 2009. Anterograde spread of herpes simplex virus type 1 requires glycoprotein E and glycoprotein I but not Us9. J Virol 83:8315–8326. doi: 10.1128/JVI.00633-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dingwell KS, Doering LC, Johnson DC. 1995. Glycoproteins E and I facilitate neuron-to-neuron spread of herpes simplex virus. J Virol 69:7087–7098. doi: 10.1128/JVI.69.11.7087-7098.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Diefenbach RJ, Miranda-Saksena M, Douglas MW, Cunningham AL. 2008. Transport and egress of herpes simplex virus in neurons. Rev Med Virol 18:35–51. doi: 10.1002/rmv.560. [DOI] [PubMed] [Google Scholar]
- 44.De Giorgi F, Ahmed Z, Bastianutto C, Brini M, Jouaville LS, Marsault R, Murgia M, Pinton P, Pozzan T, Rizzuto R. 1999. Targeting GFP to organelles. Methods Cell Biol 58:75–85. doi: 10.1016/s0091-679x(08)61949-4. [DOI] [PubMed] [Google Scholar]
- 45.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Linding R, Russell RB, Neduva V, Gibson TJ. 2003. GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res 31:3701–3708. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gierasch WW, Zimmerman DL, Ward SL, Vanheyningen TK, Romine JD, Leib DA. 2006. Construction and characterization of bacterial artificial chromosomes containing HSV-1 strains 17 and KOS. J Virol Methods 135:197–206. doi: 10.1016/j.jviromet.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 48.Carmichael JC, Starkey J, Zhang D, Sarfo A, Chadha P, Wills JW, Han J. 2019. Glycoprotein D of HSV-1 is dependent on tegument protein UL16 for packaging and contains a motif that is differentially required for syncytia formation. Virology 527:64–76. doi: 10.1016/j.virol.2018.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Erlandson KJ, Bisht H, Weisberg AS, Hyun SI, Hansen BT, Fischer ER, Hinshaw JE, Moss B. 2016. Poxviruses encode a reticulon-like protein that promotes membrane curvature. Cell Rep 14:2084–2091. doi: 10.1016/j.celrep.2016.01.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gulati NM, Pitek AS, Steinmetz NF, Stewart PL. 2017. Cryo-electron tomography investigation of serum albumin-camouflaged tobacco mosaic virus nanoparticles. Nanoscale 9:3408–3415. doi: 10.1039/c6nr06948g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fu J, Zhang S, Wu J, Chen Y, Zhong Y, Zhou Y, Wang J, Chen S. 2020. Structural characterization of a polysaccharide from dry mycelium of Penicillium chrysogenum that induces resistance to Tobacco mosaic virus in tobacco plants. Int J Biol Macromol 156:67–79. doi: 10.1016/j.ijbiomac.2020.04.050. [DOI] [PubMed] [Google Scholar]
- 52.Rao Y, Ma Q, Vahedi-Faridi A, Sundborger A, Pechstein A, Puchkov D, Luo L, Shupliakov O, Saenger W, Haucke V. 2010. Molecular basis for SH3 domain regulation of F-BAR-mediated membrane deformation. Proc Natl Acad Sci U S A 107:8213–8218. doi: 10.1073/pnas.1003478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rao Y, Haucke V. 2011. Membrane shaping by the Bin/amphiphysin/Rvs (BAR) domain protein superfamily. Cell Mol Life Sci 68:3983–3993. doi: 10.1007/s00018-011-0768-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ayton GS, Lyman E, Krishna V, Swenson RD, Mim C, Unger VM, Voth GA. 2009. New insights into BAR domain-induced membrane remodeling. Biophys J 97:1616–1625. doi: 10.1016/j.bpj.2009.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Praefcke GJ, McMahon HT. 2004. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol 5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 56.Bian X, Klemm RW, Liu TY, Zhang M, Sun S, Sui X, Liu X, Rapoport TA, Hu J. 2011. Structures of the atlastin GTPase provide insight into homotypic fusion of endoplasmic reticulum membranes. Proc Natl Acad Sci U S A 108:3976–3981. doi: 10.1073/pnas.1101643108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barbot M, Jans DC, Schulz C, Denkert N, Kroppen B, Hoppert M, Jakobs S, Meinecke M. 2015. Mic10 oligomerizes to bend mitochondrial inner membranes at cristae junctions. Cell Metab 21:756–763. doi: 10.1016/j.cmet.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 58.Shibata Y, Voss C, Rist JM, Hu J, Rapoport TA, Prinz WA, Voeltz GK. 2008. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J Biol Chem 283:18892–18904. doi: 10.1074/jbc.M800986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Okamoto PM, Herskovits JS, Vallee RB. 1997. Role of the basic, proline-rich region of dynamin in Src homology 3 domain binding and endocytosis. J Biol Chem 272:11629–11635. doi: 10.1074/jbc.272.17.11629. [DOI] [PubMed] [Google Scholar]
- 60.Ruaro EM, Collavin L, Del Sal G, Haffner R, Oren M, Levine AJ, Schneider C. 1997. A proline-rich motif in p53 is required for transactivation-independent growth arrest as induced by Gas1. Proc Natl Acad Sci U S A 94:4675–4680. doi: 10.1073/pnas.94.9.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bernacchi S, Mercenne G, Tournaire C, Marquet R, Paillart JC. 2011. Importance of the proline-rich multimerization domain on the oligomerization and nucleic acid binding properties of HIV-1 Vif. Nucleic Acids Res 39:2404–2415. doi: 10.1093/nar/gkq979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Turner A, Bruun B, Minson T, Browne H. 1998. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J Virol 72:873–875. doi: 10.1128/JVI.72.1.873-875.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Omura T. 1998. Mitochondria-targeting sequence, a multi-role sorting sequence recognized at all steps of protein import into mitochondria. J Biochem 123:1010–1016. doi: 10.1093/oxfordjournals.jbchem.a022036. [DOI] [PubMed] [Google Scholar]
- 64.Han J, Chadha P, Starkey JL, Wills JW. 2012. Function of glycoprotein E of herpes simplex virus requires coordinated assembly of three tegument proteins on its cytoplasmic tail. Proc Natl Acad Sci U S A 109:19798–19803. doi: 10.1073/pnas.1212900109. [DOI] [PMC free article] [PubMed] [Google Scholar]