Among 12 serotypes of fowl adenovirus (FAdV), FAdV-1, FAdV-4, and FAdV-10 all carry two fiber genes (i.e., fiber-1 and fiber-2), whereas other serotypes have only one. As important viral surface proteins, the fibers play vital roles in the infection and pathogenesis of FAdV. However, the importance of the fibers to the infection and pathogenesis of FAdV may be different from each other. Recent studies reveal that fiber-2 is identified as a determinant of virulence, but which fiber triggers the infection of FAdV-4 remains unknown. In this study, fiber-1 was identified as a key factor for directly mediating the infection of FAdV-4 through its shaft and knob domains, whereas fiber-2 did not play a role in triggering FAdV-4 infection. The results suggest that fiber-1 and its knob domain may serve as a target for identifying the receptor of FAdV-4 and developing efficient drugs or vaccines against FAdV-4.
KEYWORDS: FAdV-4, fiber-1, infection, knob domain, vaccine, hepatitis-hydropericardium syndrome
ABSTRACT
Recently, the disease of hepatitis-hydropericardium syndrome (HPS) caused by serotype 4 fowl adenovirus (FAdV-4) has spread widely and resulted in huge economic losses to the poultry industry. Although the genome of FAdV-4 has two fiber genes (fiber-1 and fiber-2), the exact role of the genes in the infection of FAdV-4 is barely known. In this study, through superinfection resistance analysis and an interfering assay, we found that fiber-1, but not fiber-2, was the key factor for directly triggering the infection of FAdV-4. The truncation analysis further revealed that both of the shaft and knob domains of fiber-1 were required for the infection. Moreover, the sera against the knob domain were able to block FAdV-4 infection, and the knob-containing fusion protein provided efficient protection against the lethal challenge of FAdV-4 in chickens. All the data demonstrated the significant roles of fiber-1 and its knob domain in directly mediating the infection of FAdV-4, which established a foundation for identifying the receptor of FAdV-4 and developing efficient vaccines against FAdV-4.
IMPORTANCE Among 12 serotypes of fowl adenovirus (FAdV), FAdV-1, FAdV-4, and FAdV-10 all carry two fiber genes (i.e., fiber-1 and fiber-2), whereas other serotypes have only one. As important viral surface proteins, the fibers play vital roles in the infection and pathogenesis of FAdV. However, the importance of the fibers to the infection and pathogenesis of FAdV may be different from each other. Recent studies reveal that fiber-2 is identified as a determinant of virulence, but which fiber triggers the infection of FAdV-4 remains unknown. In this study, fiber-1 was identified as a key factor for directly mediating the infection of FAdV-4 through its shaft and knob domains, whereas fiber-2 did not play a role in triggering FAdV-4 infection. The results suggest that fiber-1 and its knob domain may serve as a target for identifying the receptor of FAdV-4 and developing efficient drugs or vaccines against FAdV-4.
INTRODUCTION
Fowl adenovirus (FAdV) belongs to the genus Aviadenovirus (1). Based on the analyses with restriction enzyme and sera cross-neutralization, FAdV is currently clustered into 5 species (FAdV-A to FAdV-E) with 12 serotypes (FAdV-1 to -8a and -8b to -11) (2). The infection of FAdV mainly causes clinical symptoms such as inclusion body hepatitis (IBH), hepatitis-hydropericardium syndrome (HPS), and gizzard erosion and ulceration (GEU) (2–4). Notably, different serotypes of FAdV generally cause different clinical symptoms. IBH is generally induced by the infection of FAdV-8a, FAdV-8b, FAdV-2, and FAdV-11, HPS is mainly caused by FAdV-4, and adenoviral gizzard erosion (AGE) is closely related to the infection of FAdV-1 (5–10). Recently, the outbreak of HPS caused by FAdV-4 has caused huge economic losses to the poultry industry globally (11, 12). However, the molecular mechanism for the infection and pathogenesis of FAdV-4 are barely known.
Among the viral surface proteins, including fiber, hexon, and penton, the fiber proteins of FAdV not only effectively induce the virus-neutralizing antibodies but also mediate the viral attachment to the cellular receptor for entering host cells (13–15). Different from other serotypes of FAdV, serotypes FAdV-1, FAdV-4, and FAdV-10 have two fibers (fiber-1 and fiber-2). Zhang et al. recently reported that fiber-2 was the key determinant for the virulence of the highly pathogenic FAdV-4 endemic in China (16). Our previous study also demonstrated that a monoclonal antibody (MAb) against fiber-2 efficiently blocks the infection of FAdV-4 (11). However, the roles of fiber-1 in the viral infection and pathogenesis of FAdV-4 are less studied. In this study, we found that fiber-1, but not fiber-2, was the key factor for directly mediating the infection of FAdV-4. Moreover, the knob domain containing fusion protein derived from fiber-1 could be used as a subunit vaccine to provide efficient protection against the lethal challenge of FAdV-4 in chickens.
RESULTS
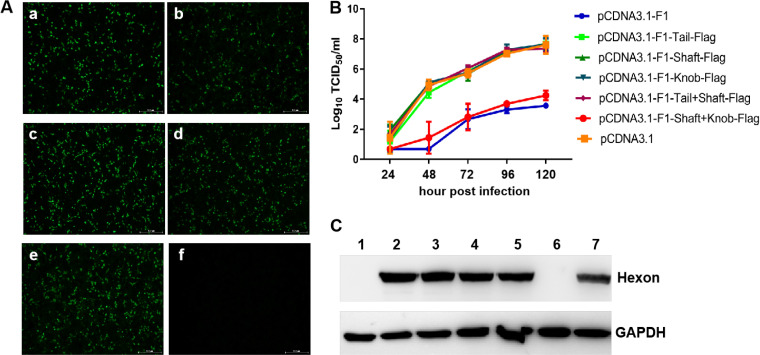
Fiber-1 efficiently conferred superinfection resistance against FAdV-4.
To identify which fiber protein directly mediates the infection of FAdV-4, we performed a superinfection resistance assay by the transfection of the FAdV-4-sensitive cell line LMH with expression plasmids of the fiber genes. The similar transfection efficacy and expression levels of fiber-1 and fiber-2 in the cells transfected with pcDNA3.1-F1 and pcDNA3.1-F2 were first confirmed as shown in Fig. 1A and B. The superinfection resistance assay showed that the viral titers of FAdV-4 in the cells transfected with pcDNA3.1-F1 were significantly lower than those in the cells transfected with pcDNA3.1-F2 or control vector pcDNA3.1 at different time points, as described in Fig. 1C. Notably, at 72 h postinfection (hpi), the virus titers in the cells transfected with pcDNA3.1-F1 were approximately 100 times less than those in the cells transfected with pcDNA3.1-F2 or control vector pcDNA3.1. Western blot analysis further confirmed this superinfection resistance activity by fiber-1. As shown in Fig. 1D, the strong band of viral hexon protein was found in the cells transfected with pcDNA3.1 or pcDNA3.1-F2 at 120 hpi, whereas the hexon protein could not be efficiently detected in the cells transfected with pcDNA3.1-F1. These data demonstrate that the fiber-1 protein, but not fiber-2, can efficiently block or inhibit the viral replication of FAdV-4 in the susceptible cell line LMH possibly by blocking the cell receptor for FAdV-4 infection, highlighting that the fiber-1 protein directly mediates the infection of FAdV-4.
FIG 1.
Fiber-1 efficiently conferred superinfection resistance against FAdV-4. (A) LMH cells transfected with pcDNA3.1-F1 (a), pcDNA3.1-F2 (b), and pcDNA3.1 (c) were analyzed by IFA using the chicken sera against FAdV-4. (B) LMH cells transfected with pcDNA3.1-F1 (lane 1), pcDNA3.1-F2 (lane 2), and pcDNA3.1 (lane 3) were analyzed by Western blotting using chicken sera against FAdV-4. (C) Supernatants from the infected LMH cells at different time points, which were transfected with pcDNA3.1-F1, pcDNA3.1-F2, and pcDNA3.1 in advance, were titrated with TCID50. (D) Infected LMH cells at 120 hpi, which were transfected with pcDNA3.1-F1 (lane 1), pcDNA3.1-F2 (lane 2), and pcDNA3.1 (lane 3) in advance, were analyzed by Western blotting using MAb 1B4 against the hexon protein of FAdV-4. These experiments were performed for three times with similar results.
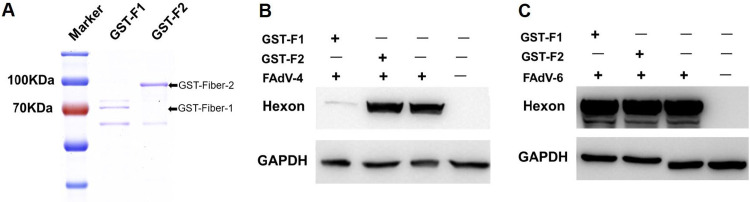
Purified GST-fiber-1 efficiently interfered with the infection of FAdV-4.
To confirm the superinfection resistance against FAdV-4 mediated by fiber-1, we performed an interfering assay for the infection of FAdV-4 using purified glutathione transferase (GST)-fiber-1 and GST-fiber-2. As shown in Fig. 2A, the GST fusion proteins GST-fiber-1 and GST-fiber-2 were purified. The interfering assay with the purified proteins revealed that GST-fiber-1, but not GST-fiber-2, efficiently interfered with the infection and replication of FAdV-4 in the LMH cells. As described in Fig. 2B, the hexon protein of FAdV-4 was hardly detected in the infected LMH cells with GST-fiber-1, whereas the strong band of the hexon protein was detected in the infected LMH cells with GST-fiber-2 or without treatment. To evaluate the specificity of the interfering activity of the GST-fiber-1 protein against the infection of FAdV-4, the serotype 6 fowl adenovirus (FAdV-6, from ATCC) was used as a control in the interfering assay. As described in Fig. 2C, the hexon protein of FAdV-6 was obviously detected in the infected LMH cells with GST-fiber-1, GST-fiber-2, or without treatment. The data clearly demonstrate that the interfering activity of GST-fiber-1 against the infection of FAdV-4 is specific to FAdV-4.
FIG 2.
Purified GST-fiber-1 efficiently interfered with the infection of FAdV-4. (A) SDS-PAGE analysis of the purified GST-fiber-1 and GST-fiber-2 fusion proteins. (B) LMH cells infected with FAdV-4 and treated with purified GST-fiber-1 or GST-fiber-2 were analyzed using MAb 1B4 against the hexon protein by Western blotting. (C) LMH cells infected with FAdV-6 and treated with purified GST-fiber-1 or GST-fiber-2 were analyzed using MAb against the Hexon protein by Western blotting. Experiments shown in panels B and C were performed three times with similar results.
Shaft and knob domains of fiber-1 contributed to the superinfection resistance.
To determine which domain of the fiber-1 protein contributes to the superinfection resistance conferred by fiber-1, a set of fiber-1 fragments with different domains carrying a Flag tag at the C terminus were constructed and designated pcDNA3.1-F1-tail-Flag, pcDNA3.1-F1-shaft-Flag, pcDNA3.1-F1-knob-Flag, pcDNA3.1-F1-tail+shaft-Flag, and pcDNA3.1-F1-shaft+knob-Flag. The transfection efficacy and expression of these fiber-1 fragments were confirmed by indirect immunofluorescence assay (IFA) using antibody against Flag. As described in Fig. 3A, all of these fiber-1 fragments were efficiently expressed with similar transfection efficacy. The superinfection resistance assay using these fiber-1 fragments showed that the virus titers of FAdV-4 in the cells transfected with pcDNA3.1-F1 or pcDNA3.1-F1-shaft+knob-Flag were significantly lower than those in the cells transfected with pcDNA3.1, pcDNA3.1-F1-tail-Flag, pcDNA3.1-F1-shaft-Flag, pcDNA3.1-F1-knob-Flag, and pcDNA3.1-F1-tail+shaft-Flag at 48, 96, and 120 hpi (h postinfection) (Fig. 3B). Notably, the virus titers in the cells transfected with pcDNA3.1-F1-tail-Flag, pcDNA3.1-F1-shaft-Flag, pcDNA3.1-F1-knob-Flag, and pcDNA3.1-F1-tail+shaft-Flag were very similar to those in cells transfected with pcDNA3.1 at different time points, indicating the FAdV-4 virus was not inhibited by transfection with pcDNA3.1-F1-tail-Flag, pcDNA3.1-F1-shaft-Flag, pcDNA3.1-F1-knob-Flag, and pcDNA3.1-F1-tail+shaft-Flag. Western blot analysis further confirmed this blocking activity against the infection of FAdV-4 conferred by shaft and knob domains of fiber-1. As shown in Fig. 3C, the viral hexon protein could not be efficiently detected in the cells transfected with pcDNA3.1-F1 or pcDNA3.1-F1-shaft+knob-Flag at 120 hpi, whereas the strong band of hexon protein was found in the cells transfected with pcDNA3.1, pcDNA3.1-F1-tail-Flag, pcDNA3.1-F1-shaft-Flag, pcDNA3.1-F1-knob-Flag, and pcDNA3.1-F1-tail+shaft-Flag. Together, the results demonstrate that both shaft and knob domains of fiber-1 are required for the superinfection resistance of the fiber-1 protein.
FIG 3.
The shaft and knob domains contributed to the superinfection resistance. (A) Expression of the truncated fragments of fiber-1 in LMH cells was detected by IFA using a monoclonal antibody against Flag. (a to f) LMH cells were transfected with pcDNA3.1-F1-tail-Flag, pcDNA3.1-F1-shaft-Flag, pcDNA3.1-F1-knob-Flag, pcDNA3.1-F1-tail+shaft-Flag, pcDNA3.1-F1-shaft+knob-Flag, and pcDNA3.1, respectively. (B) Supernatants from infected LMH cells at different time points, which were transfected with pcDNA3.1-F1, pcDNA3.1-F1-tail-Flag, pcDNA3.1-F1-shaft-Flag, pcDNA3.1-F1-knob-Flag, pcDNA3.1-F1-tail+shaft-Flag, pcDNA3.1-F1-shaft+knob-Flag, and pcDNA3.1 in advance, were titrated with TCID50. (C) Infected LMH cells at 120 hpi, which were transfected with pcDNA3.1-F1 (lane 1), pcDNA3.1-F1-tail-Flag (lane 2), pcDNA3.1-F1-shaft-Flag (lane 3), pcDNA3.1-F1-knob-Flag (lane 4), pcDNA3.1-F1-tail+shaft-Flag (lane 5), pcDNA3.1-F1-shaft+knob-Flag (lane 6), and pcDNA3.1 (lane 7) in advance, were analyzed by Western blotting using MAb 1B4 against the hexon protein of FAdV-4. These experiments were performed three times with similar results.
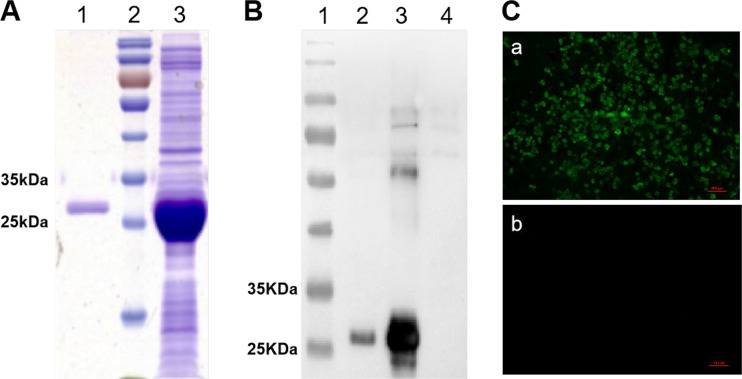
The antibodies against the knob domain efficiently block the infection of FAdV-4.
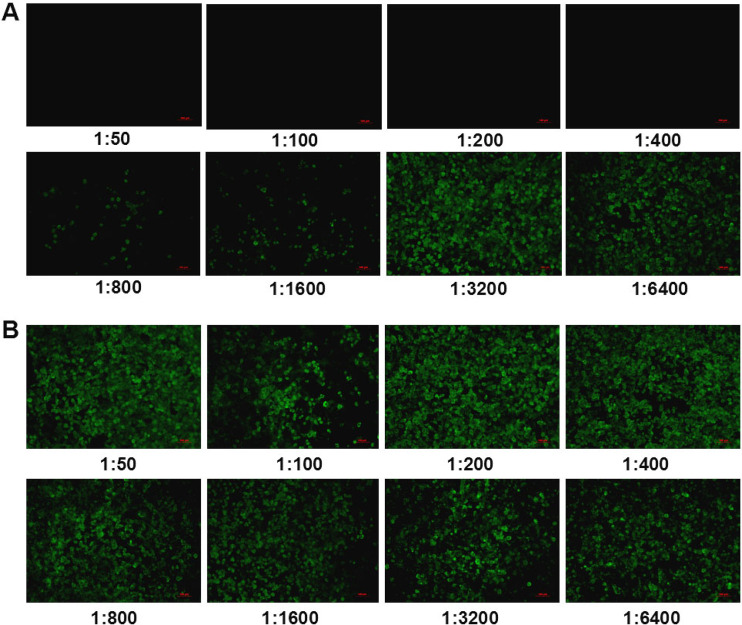
Since both shaft and knob domains play vital roles in directly mediating the infection of FAdV-4 in LMH cells, and the knob domain is located outside fiber-1, we speculated that the knob of fiber-1 might directly bind to the cellular receptor during the infection of FAdV-4; thus, the sera against the knob could efficiently block the infection of FAdV-4. To test this, the knob fusion protein His-knob was generated, followed by preparation of the sera against the knob domain in mice. As shown in Fig. 4A and B, the fusion protein containing the His-knob domain was highly expressed in pellets of Escherichia coli upon induction by isopropyl-β-d-thiogalactopyranoside (IPTG). Moreover, the purified fusion protein was well recognized by mouse sera against fiber-1. The specificity of the sera against the knob domain from mouse was confirmed by the IFA analysis (Fig. 4C). To investigate whether the mouse sera against the knob domain efficiently blocks the infection of FAdV-4, a neutralizing assay was performed. As shown in Fig. 5A, the sera against the fusion protein from mouse efficiently blocked the viral replication of FAdV-4 in LMH cells in a dose-dependent manner. The neutralizing titers of the mouse sera remained effective with the dilution of 1:400. Notably, only a few viruses were detected in the cells treated with the mouse sera against His-knob, even with the dilutions of 1:800 to 1:1,600; in contrast, many viruses were detected in the cells treated with the control sera only, with the different dilutions shown in Fig. 5B. The results demonstrate that the fusion protein His-knob is good in regard to antigenicity, and the mouse sera against the His-knob can effectively inhibit the infection of FAdV-4.
FIG 4.
Expression of knob domain-containing fusion protein and preparation of its polyclonal antibodies. (A) SDS-PAGE analysis on the expression and purification of the His-knob containing fusion protein. Lane 1, purified fusion protein containing His-knob domain; lane 2, prestained protein ladder; lane 3, unpurified fusion protein containing His-knob domain. (B) Western blot analysis for the expression and purification of the His-knob domain-containing fusion protein by using a mouse antibody against fiber-1. Lane 1, prestained protein ladder; lane 2, purified fusion protein containing His-knob domain; lane 3, unpurified fusion protein containing His-knob domain; lane 4, pCold-I prokaryotic expression vector control. (C) IFA analysis with the mouse sera against the knob domain of fiber-1. (a) Reaction of mouse sera against the knob domain with the LMH cells infected with FAdV-4. (b) Reaction of mouse sera against the knob domain with the noninfected LMH cells. Experiments in panels B and C were performed three times with similar results.
FIG 5.
Antisera against knob domain efficiently inhibited the infection of FAdV-4. (A) Serial dilutions (1:50 to 1:6,400) of mouse sera against the knob domain were mixed with FAdV-4, followed by infection of the LMH cells with the mixture, which was analyzed by IFA at day 4 postinfection. (B) Serial dilutions (1:50 to 1:6,400) of negative mouse sera were mixed with FAdV-4, followed by infection of the LMH cells with the mixture, which was analyzed by IFA at day 4 postinfection. These experiments were performed three times with similar results.
The knob-containing fusion protein provides efficient protection against FAdV-4.
To further evaluate whether the fusion protein containing His-knob could be used as a vaccine candidate to provide protection against FAdV-4, chickens were immunized with the purified fusion protein containing the His-knob domain and challenged with a lethal dose of FAdV-4 at day 28 postvaccination. Cloacal swab samples, livers, spleens, and kidneys were collected at designated time points and titrated with LMH cells. The clinical symptoms and mortality of the challenged chickens were monitored daily. After challenging, the chickens without vaccination exhibited signs of depression, loss of appetite, lethargy, and yellow-green and thin feces, and the mortality reached 45.5% (5/11) at day 5 postchallenge. In contrast, the chickens with vaccination did not exhibit any clinical symptoms, with no death. Moreover, hydropericardium syndrome, the typical lesions caused by FAdV-4 infection, were found in the hearts in the challenged chickens without vaccination (the control group) but not in the vaccinated group (Fig. 6A). In addition, the livers in the challenged chickens without vaccination (the control group) were enlarged and lighter in color than those in the vaccinated group (Fig. 6A). These clinical data were consistent with viral shedding in the swabs and viral loading in the tissues. As shown in Fig. 6B, the viruses were detected with high titer in the cloacal swab samples from the challenged chickens without vaccination at early time points postchallenge; in contrast, those in the swab samples were hardly detected in the challenged chickens with vaccination. Also, the viruses were detected with high titer in the livers, spleens, and kidneys from the challenged chickens without vaccination at day 3 postchallenge, whereas those from the challenged chickens with vaccination were hardly detected (Fig. 6C). Together, the results demonstrate that the fusion protein containing the His-knob domain may be used as a vaccine candidate to provide efficient protection against the lethal challenge of FAdV-4.
FIG 6.
The knob domain-containing fusion protein conferred efficient protection against FAdV-4. (A) Pathological changes in the chickens from the control group and the vaccinated group (these are ventral views of the heart and liver). (B) Cloacal swab samples (n = 4) from the control group and the vaccinated group were collected and titrated with TCID50 at different time points postchallenge. (C) Tissue samples (n = 3) (including liver, spleen, and kidney) were collected from the control group and the vaccinated group and titrated with TCID50 at day 3 postchallenge.
DISCUSSION
As the causative agent for the hepatitis-hydropericardium syndrome (HPS) that is recently endemic in global poultry industry, FAdV-4 has caused a huge economic loss. However, the molecular mechanisms for the infection and pathogenesis of FAdV-4 are largely unknown. As one of the surface proteins, the fiber protein of adenovirus plays vital roles in mediating viral infection and inducing neutralizing antibodies (14, 15). Notably, FAdV-A (serotype FAdV-1) and FAdV-C (serotypes FAdV-4 and FAdV-10) have two fiber proteins (fiber-1 and fiber-2), whereas other serotypes of FAdV only carry one fiber protein. Previous reports demonstrate that fiber-2 of CELO (FAdV-1) is essential for virus growth and assembly, while fiber-1 plays important roles in the binding to the coxsackievirus and adenovirus receptor (CAR) derived from human cells (17). The fiber-2 protein of FAdV-4 was also recently identified as a key virulent determinant for the highly pathogenic FAdV-4 and as an important factor involving in the infection of FAdV-4 (11, 16). Here, we identified fiber-1, but not fiber-2, as the key factor for directly mediating the infection of FAdV-4 through its shaft and knob domains by using a superinfection resistance assay and interfering assays. Moreover, we found that the sera against the knob domain efficiently neutralized the virus and blocked the infection of FAdV-4. These data demonstrate that the knob domain of fiber-1 may directly interact with the cell receptor that mediates the infection of FAdV-4. However, the knob domain alone did not confer the superinfection resistance, as the resistance was also dependent on the shaft domain. This is probably because the function of the knob is heavily dependent on its homotrimer structure (13). Since the shaft domain of the fiber-1 protein carries trimeric motifs, the knob domain in the shaft+knob fusion protein should be a functional homotrimer (13). The purified fusion protein containing the His-knob domain when delivered as a vaccine provided efficient protection against a lethal challenge of FAdV-4, which further confirmed the importance of fiber-1 and its knob domain in mediating the infection of the FAdV-4. In contrast, although previous reports show that fiber-2 plays a vital role in mediating the infection of FAdV-4 and can serve an efficient vaccine target (11, 15), we did not find the superinfection resistance effect and interfering activity of fiber-2 on the infection of FAdV-4 in the superinfection resistance assay and interfering assays. However, the molecular mechanism for these characteristics of fiber-2 need to be further elucidated. These results indicate that the role of fiber-2 in mediating the infection of FAdV-4 might be indirect. Since both fiber-1 and fiber-2 are thought to be linked with the penton protein on the viral particle, the antibodies induced by fiber-2 can inhibit or neutralize FAdV-4 possibly through its interference with the interaction between fiber-1 and the cell receptor during its binding to fiber-2. Notably, a paper published by Pan et al. during our revision for the manuscript also demonstrates that fiber-1, not fiber-2, directly binds to the cell receptor, chicken CAR homology, for FAdV-4 infection (18). The finding by Pan et al. is consistent with or confirms our data. However, Pan et al. did not identify which domain of fiber-1 is responsible for initiating the infection and did not use knob of fiber-1 as an immunogen to protect chickens.
In summary, this is the first report that fiber-1 directly mediates the infection of FAdV-4 through its shaft and knob domains, which is identified through superinfection resistance assay, interfering assay, and serum neutralizing assay. Although the chicken CAR homology has just been recently identified as a cell receptor for fiber-1 by Pan et al. (18), identifying the effective domain in fiber-1 for binding to its receptor and other potential host molecules for interacting with fiber-1 is critical to elucidate the molecular basis for initiating the infection of FAdV-4. The efficient protection against the lethal challenge of FAdV-4 in chickens conferred by the vaccination of the knob domain-containing fusion protein not only confirms the significant roles of the fiber-1 and its knob domain in mediating the infection of FAdV-4 but also highlight a promising application of knob domain-based subunit vaccines against FAdV-4 in the future.
MATERIALS AND METHODS
Viruses and cells.
FAdV-4 isolate SD2015 was isolated and maintained in our laboratory (11). The chicken liver cell line (LMH, from ATCC) was cultured in F12-Dulbecco’s modified Eagle medium (DMEM) (Gibco, NY, USA) with 10% fetal bovine serum (FBS) (Lonsera, Shanghai, China).
Antibodies.
The chicken sera against FAdV-4 and monoclonal antibody (MAb) 3C2 against fiber-2 of FAdV-4 were prepared in our laboratory (11). Mouse sera against fiber-1 were also prepared in our laboratory. The MAb 1B4 against hexon of FAdV-4 was a gift kindly given by Hongjun Chen (Shanghai Veterinary Research Institute).
Plasmids.
The plasmids pcDNA3.1-F1 and pcDNA3.1-F2 efficiently expressing the fiber-1 and fiber-2 proteins of FAdV-4, respectively, and the plasmids GST-F1 and GST-F2 efficiently expressing the GST-fiber-1 and GST-fiber-2 fusion proteins were all constructed and kept in our laboratory. To construct the fragments of fiber-1 with different domains, the domains of tail, shaft, knob, tail+shaft, and shaft+knob with Flag tags in the C termini were first amplified by PCR using the primers listed in Table 1 and then cloned into the linear vector pcDNA3.1 by the ClonExpress II One Step Cloning kit (Vazyme Nanjing China). The corresponding recombinant plasmids were designated pcDNA3.1-F1-tail-Flag, pcDNA3.1-F1-shaft-Flag, pcDNA3.1-F1-knob-Flag, pcDNA3.1-F1-tail+shaft-Flag, and pcDNA3.1-F1-shaft+knob-Flag, respectively. For the expression of the His-knob fusion protein, the knob domain of fiber-1 was cloned into the pCold-I prokaryotic expression vector, and the recombinant plasmid was named as pCold-I-F1-knob.
TABLE 1.
Primers for PCR amplification for the truncation of the fiber-1 gene
| PCR product | Sequence (5′→3′) |
|---|---|
| Knob | AGCTTGGTACCGAATGAGCCCCTTTGCGAC |
| ATATCTGCAGAATTTACTTATCGTCGTCATCCTTGTAATCGGGGCCCGGAGCAT | |
| Shaft | AGCTTGGTACCGAATGGGTCAGCAGATCGC |
| ATATCTGCAGAATTTACTTATCGTCGTCATCCTTGTAATCCAGATAGGTCGGAC | |
| Tail | AGCTTGGTACCGAATGTCGGCCCTAATCGC |
| ATATCTGCAGAATTTACTTATCGTCGTCATCCTTGTAATCTCCACCTCCCCCAC | |
| Shaft and knob | AGCTTGGTACCGAATGGGTCAGCAGATCGC |
| ATATCTGCAGAATTTACTTATCGTCGTCATCCTTGTAATCGGGGCCCGGAGCAT | |
| Tail and shaft | AGCTTGGTACCGAATGTCGGCCCTAATCGC |
| ATATCTGCAGAATTTACTTATCGTCGTCATCCTTGTAATCCAGATAGGTCGGAC |
Expression and purification of the fusion protein.
The recombinant plasmids, including GST-fiber-1, GST-fiber-2, and pCold-I-F1-knob were transformed into BL21 cells, and the expression of the fusion proteins was induced by 1 mM IPTG for 12 h at 16°C and determined by SDS-PAGE and Western blotting. The GST-fiber-1, GST-fiber-2, and His-knob fusion proteins were then purified from the BL21 cells.
Preparation of sera against the knob domain of fiber-1 from mice.
Six-week-old female BALB/c mice were immunized with 50 μg of the purified His-knob protein emulsified with 200 μl of complete Freund’s adjuvant by intraperitoneal injection, followed by immunizing the mice with equal amounts of His-knob protein emulsified with 200 μl of incomplete Freund's adjuvant 2 weeks later. The immunization of the mice was then boosted with 50 μg of the His-knob protein without the adjuvant twice at a 2-week interval.
Superinfection resistance assay.
LMH cells were first transfected with the different recombinant plasmids for 24 h, followed by infection with the FAdV-4 at a multiplicity of infection (MOI) of 0.01. The supernatants from the infected cells were then collected at 24, 48, 72, 96, and 120 h postinfection and titrated according to the 50% tissue culture infection dose (TCID50).
Interfering assay using purified fiber-1 and fiber-2 fusion proteins.
Purified fiber-1 or fiber-2 fusion proteins were first mixed with FAdV-4 or FAdV-6 at an MOI of 0.01, followed by inoculation into the LMH cells. After 2 h, the inoculated cells were washed twice with phosphate-buffered saline (PBS) and then cultured in F12-DMEM with 1% FBS. At day 4 postinoculation, the LMH cells were collected and analyzed using MAb 1B4 against the hexon of FAdV by Western blotting.
Indirect immunofluorescence assay.
LMH cells transfected with the plasmids individually for 48 h were fixed with 4% paraformaldehyde for 25 min at 37°C, followed by three washes with PBS, and then were treated with 2% Triton X-100 for 25 min at 37°C. After washing with PBS three times, the cells were blocked with 1% bovine serum albumin (BSA) for 30 min at 37°C. The cells were stained with the diluted antibodies against the designated proteins for 45 min at 37°C. After washing three times with PBS, the cells were stained with the diluted second antibody (goat anti mouse IgG-fluorescein isothiocyanate [FITC]) for another 45 min at 37°C. Again, after three washes with PBS, the cells were observed by inverted fluorescence microscopy.
Western blot assay.
LMH cells transfected with plasmids or infected with FAdVs were collected and lysed in lysis buffer (CST, MA, USA) with phenylmethylsulfonyl fluoride (PMSF; Beyotime, Shanghai China) and protease and phosphatase inhibitors (CST). The lysates were boiled in the loading buffer. After SDS-PAGE, the denatured proteins were transferred onto nitrocellulose (NC) membranes (GE Healthcare Life sciences, Freiburg, Germany). After blocking with 5% skim milk in PBS with Tween 20 (PBST) for 2 h at room temperature (RT), the membranes were reacted with the corresponding MAbs or sera for 2 h at RT. After three washes with PBST, the membranes were incubated with horseradish peroxidase (HRP)-labeled goat anti-mouse IgG or HRP-labeled rabbit anti-chicken IgG. After another three washes, the membranes were developed with chemiluminescent reagents and imaged with an automatic imaging system (Tanon 5200).
Neutralization test.
Different dilutions of the polyclonal antibody against the knob domain of the fiber-1 protein were first mixed with 200 TCID50 of FAdV-4 and incubated for 1 h at 37°C. Then, the mixtures were added to the 96-well plate with LMH cells and incubated for 2 h at 37°C. After washing once, the cells were cultured in F12-DMEM with 1% fetal bovine serum. After culturing for 96 h, the cells were fixed and subjected to IFA analysis by using MAb 3C2 against fiber-2 of FAdV-4 as previously described (11).
Animal experiment.
Twenty-two 2-week-old specific-pathogen-free (SPF) chickens were randomly divided into two groups. Chickens in group I (n = 11) were immunized with 150 μg of the purified His-knob-containing fusion protein mixed with polymeric adjuvant (Seppic, Castres, France) via a pectoral injection, whereas chickens in group II (n = 11) as a control were injected with polymeric adjuvant. At day 28 postinjection, all the chickens from both group I and group II were challenged with 106 TCID50/ml of FAdV-4 (SD2015 strain) via muscle injection at the dose of 200 μl. The cloacal swab samples were collected at days 1, 3, 5, 7, 9, and 11 postinjection and titrated with LMH cells. The livers, spleens, and kidneys of the chickens were also collected at day 3 postinjection and titrated with LMH cells. The clinical symptoms and mortality of the injected chickens were monitored daily.
Titration of viral titer in the organs and cloacal swab samples.
The livers, spleens, and kidneys of the chickens were collected and homogenized in PBS (1 g tissue in 1 ml PBS). Then, the homogenates were treated with penicillin and streptomycin for 0.5 h at 37°C and centrifuged to obtain the supernatant. The cloacal samples collected from the chickens were placed in 800 μl of PBS. After three cycles of repeated freeze-thawing and vortexing, the samples were also treated with penicillin and streptomycin for 0.5 h at 37°C and centrifuged to obtain the supernatant. The virus-containing supernatants were then serially diluted in F12-DMEM with 1% of FBS and were inoculated into the 96-well plates with LMH cells. Four days postinfection, the supernatants were titrated in LMH cells by IFA using MAb 3C2 against fiber-2, and TCID50s of those supernatants were calculated by the Reed-Muench method.
Statistical analysis.
All the results are presented as means ± standard deviations. The statistical analysis in this study was performed with one-way analysis of variance (ANOVA) using GraphPad 5 software. A P value of <0.05 was considered significant.
Ethics approval.
All animal experiments complied with institutional animal care guidelines and were approved by the Animal Care Committee of Yangzhou University. At the end of the experiment, all the chickens were euthanized by CO2.
Data availability.
The data sets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
ACKNOWLEDGMENTS
We thank Jianjun Zhang (Sinopharm Yangzhou VAC Biological Engineering Co. Ltd.) for kindly providing SPF chickens.
This study was supported by the National Key Research & Development (R&D) Plan (2018YFD0500106)and the Jiangsu Agricultural Science and Technology Innovation Fund [CX(19)3026], the Undergraduate Training Programs for Innovation and Entrepreneurship in Yangzhou University (C201811117020Y), the Key Laboratory of Prevention and Control of Biological Hazard Factors (Animal Origin) for Agrifood Safety and Quality (26116120), the Research Foundation for Talented Scholars in Yangzhou University, and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
We declare no competing interests.
W.W., J.Y., and A.Q. designed the project. W.W., Q.L., T.L., Q.X., and H.C. carried out the experiments. W.W., Q.L., H.C., H.S., and Z.W. analyzed the data. W.W., T.L., T.G., and J.Y. drafted the manuscript. J.Y. supervised all the experiments and participated in the data analysis. H.S., Z.W., H.C., T.G., and A.Q. discussed and prepared the final report. All of the authors read and approved the final manuscript.
REFERENCES
- 1.Adams MJ, Carstens EB, Lefkowitz EJ. 2012. Family – Adenoviridae, p 25–141. In King AMG, Adams MJ, Carstens EB, Lefkowitz EJ. (eds), Virus taxonomy. Elsevier, Amsterdam, Netherlands. doi: 10.1016/b978-0-12-384684-6.00009-4. [DOI] [Google Scholar]
- 2.Niczyporuk JS. 2016. Phylogenetic and geographic analysis of fowl adenovirus field strains isolated from poultry in Poland. Arch Virol 161:33–42. doi: 10.1007/s00705-015-2635-4. [DOI] [PubMed] [Google Scholar]
- 3.Mittal D, Jindal N, Tiwari AK, Khokhar RS. 2014. Characterization of fowl adenoviruses associated with hydropericardium syndrome and inclusion body hepatitis in broiler chickens. Virusdisease 25:114–119. doi: 10.1007/s13337-013-0183-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okuda Y, Ono M, Shibata I, Sato S. 2004. Pathogenicity of serotype 8 fowl adenovirus isolated from gizzard erosions of slaughtered broiler chickens. J Vet Med Sci 66:1561–1566. doi: 10.1292/jvms.66.1561. [DOI] [PubMed] [Google Scholar]
- 5.De Herdt P, Timmerman T, Defoort P, Lycke K, Jaspers R. 2013. Fowl adenovirus infections in Belgian broilers: a ten-year survey. Vlaams Diergeneeskundig Tijdschrift 82:125–132. [Google Scholar]
- 6.Kaján GL, Kecskeméti S, Harrach B, Benkő M. 2013. Molecular typing of fowl adenoviruses, isolated in Hungary recently, reveals high diversity. Vet Microbiol 167:357–363. doi: 10.1016/j.vetmic.2013.09.025. [DOI] [PubMed] [Google Scholar]
- 7.Marek A, Günes A, Schulz E, Hess M. 2010. Classification of fowl adenoviruses by use of phylogenetic analysis and high-resolution melting-curve analysis of the hexon L1 gene region. J Virol Methods 170:147–154. doi: 10.1016/j.jviromet.2010.09.019. [DOI] [PubMed] [Google Scholar]
- 8.Mazaheri A, Prusas C, Voss M, Hess M. 1998. Some strains of serotype 4 fowl adenoviruses cause inclusion body hepatitis and hydropericardium syndrome in chickens. Avian Pathol 27:269–276. doi: 10.1080/03079459808419335. [DOI] [PubMed] [Google Scholar]
- 9.Ojkic D, Martin E, Swinton J, Vaillancourt JP, Boulianne M, Gomis S. 2008. Genotyping of Canadian isolates of fowl adenoviruses. Avian Pathol 37:95–100. doi: 10.1080/03079450701805324. [DOI] [PubMed] [Google Scholar]
- 10.Schachner A, Marek A, Grafl B, Hess M. 2016. Detailed molecular analyses of the hexon loop-1 and fibers of fowl aviadenoviruses reveal new insights into the antigenic relationship and confirm that specific genotypes are involved in field outbreaks of inclusion body hepatitis. Vet Microbiol 186:13–20. doi: 10.1016/j.vetmic.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 11.Wang P, Zhang J, Wang W, Li T, Liang G, Shao H, Gao W, Qin A, Ye J. 2018. A novel monoclonal antibody efficiently blocks the infection of serotype 4 fowl adenovirus by targeting fiber-2. Vet Res 49:29. doi: 10.1186/s13567-018-0525-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye J, Liang G, Zhang J, Wang W, Song N, Wang P, Zheng W, Xie Q, Shao H, Wan Z, Wang C, Chen H, Gao W, Qin A. 2016. Outbreaks of serotype 4 fowl adenovirus with novel genotype, China. Emerg Microbes Infect 5:e50. doi: 10.1038/emi.2016.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harakuni T, Andoh K, Sakamoto RI, Tamaki Y, Miyata T, Uefuji H, Yamazaki KI, Arakawa T. 2016. Fiber knob domain lacking the shaft sequence but fused to a coiled coil is a candidate subunit vaccine against egg-drop syndrome. Vaccine 34:3184–3190. doi: 10.1016/j.vaccine.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 14.Henry LJ, Xia D, Wilke ME, Deisenhofer J, Gerard RD. 1994. Characterization of the knob domain of the adenovirus type 5 fiber protein expressed in Escherichia coli. J Virol 68:5239–5246. doi: 10.1128/JVI.68.8.5239-5246.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruan S, Zhao J, Yin X, He Z, Zhang G. 2018. A subunit vaccine based on fiber-2 protein provides full protection against fowl adenovirus serotype 4 and induces quicker and stronger immune responses than an inactivated oil-emulsion vaccine. Infect Genet Evol 61:145–150. doi: 10.1016/j.meegid.2018.03.031. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Liu R, Tian K, Wang Z, Yang X, Gao D, Zhang Y, Fu J, Wang H, Zhao J. 2018. Fiber2 and hexon genes are closely associated with the virulence of the emerging and highly pathogenic fowl adenovirus 4. Emerg Microbes Infect 7:199. doi: 10.1038/s41426-018-0203-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan PK, Michou AI, Bergelson JM, Cotten M. 2001. Defining CAR as a cellular receptor for the avian adenovirus CELO using a genetic analysis of the two viral fibre proteins. J Gen Virol 82:1465–1472. doi: 10.1099/0022-1317-82-6-1465. [DOI] [PubMed] [Google Scholar]
- 18.Pan Q, Wang J, Gao Y, Wang Q, Cui H, Liu C, Qi X, Zhang Y, Wang Y, Li K, Gao L, Liu A, Wang X. 2020. Identification of chicken CAR homology as a cellular receptor for the emerging highly pathogenic fowl adenovirus 4 via unique binding mechanism. Emerg Microbes Infect 9:586–596. doi: 10.1080/22221751.2020.1736954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets used and/or analyzed during the present study are available from the corresponding author on reasonable request.