The translation of picornaviral genome RNA mediated by the internal ribosomal entry site (IRES) is a crucial step for virus infections. Virus-host interactions play a critical role in the regulation of IRES-dependent translation, but the regulatory mechanism remains largely unknown. In this study, we identified an ITAF, hnRNP K, that negatively regulates FMDV replication by inhibiting viral IRES-mediated translation. In addition, we describe a novel translational regulation mechanism involving the proteolytic cleavage of hnRNP K by FMDV protease 3C. The cleavage of hnRNP K yields two cleavage products with opposite functions: the cleavage product hnRNP K1–364 retains a partial inhibitory effect on IRES activity, and the cleavage product hnRNP K364–465 becomes a positive regulator of FMDV replication. Our findings shed light on the effect of a novel ITAF on the translational regulation of picornavirus and provide new insights into translational control during viral infection.
KEYWORDS: foot-and-mouth disease virus, heterogeneous nuclear ribonucleoprotein K, internal ribosomal entry site, translational regulation, virus-host interactions
ABSTRACT
Cap-independent translation initiation on picornavirus mRNAs is mediated by an internal ribosomal entry site (IRES) in the 5′ untranslated region. The regulation of internal initiation requires the interaction of IRES-transacting factors (ITAFs) with the IRES. In this study, we identified a novel ITAF, heterogeneous nuclear ribonucleoprotein K (hnRNP K), which negatively regulates foot-and-mouth disease virus (FMDV) translation and viral replication. Further investigation revealed that the KH2 and KH3 domains of hnRNP K directly bind to domains II, III, and IV of the FMDV IRES, resulting in the inhibition of IRES-mediated translation by interfering with the recognition of another positive ITAF, polypyrimidine tract-binding protein (PTB). Conversely, hnRNP K-mediated inhibition was antagonized by the viral 3C protease through the cleavage of hnRNP K at the Glu-364 residue during FMDV infection. Interestingly, the N-terminal cleavage product, hnRNP K1–364, retained partial inhibitory effects on IRES activity, whereas the C-terminal cleavage product, hnRNP K364–465, became a positive regulator of FMDV replication. Our findings expand the current understanding of virus-host interactions concerning viral recruitment and the modulation of ITAFs, providing new insights into translational control during viral infection.
IMPORTANCE The translation of picornaviral genome RNA mediated by the internal ribosomal entry site (IRES) is a crucial step for virus infections. Virus-host interactions play a critical role in the regulation of IRES-dependent translation, but the regulatory mechanism remains largely unknown. In this study, we identified an ITAF, hnRNP K, that negatively regulates FMDV replication by inhibiting viral IRES-mediated translation. In addition, we describe a novel translational regulation mechanism involving the proteolytic cleavage of hnRNP K by FMDV protease 3C. The cleavage of hnRNP K yields two cleavage products with opposite functions: the cleavage product hnRNP K1–364 retains a partial inhibitory effect on IRES activity, and the cleavage product hnRNP K364–465 becomes a positive regulator of FMDV replication. Our findings shed light on the effect of a novel ITAF on the translational regulation of picornavirus and provide new insights into translational control during viral infection.
INTRODUCTION
Translational control is an important component of the regulation of gene expression. In eukaryotic cells, the majority of mRNAs initiate translation by a mechanism that depends on the recognition of the m7G(5′)ppp(5′)N structure (termed cap) located at the 5′ end of most mRNAs (1). In contrast to the general mechanism of cap-dependent translation initiation, a growing number of viral and cellular mRNAs are governed by internal ribosome entry site (IRES) elements (2–7). The IRES elements recruit ribosomes for translation initiation in a cap-independent manner, which is dependent on the structural organization of the IRES and its interactions with cellular proteins, including eukaryotic translation initiation factors (eIFs) and other noncanonical translation factors termed IRES-transacting factors (ITAFs) (8). This RNA-dependent translation mechanism was first discovered in the 5′ untranslated region (5′ UTR) of picornavirus (9, 10). Since the discovery of IRES elements, the regulation of IRES-dependent translation has been regarded as a critical step for some viral infections, with important effects on virulence, tissue tropism, and pathogenicity (11).
IRES-dependent translational control largely depends on ITAFs. To date, several cellular proteins, such as ITAFs, that modulate picornaviral IRES function have been identified, including polypyrimidine tract-binding protein (PTB) (12), poly(rC)-binding protein 2 (PCBP2) (13), FBP1 (14), FBP2 (15), gemin5 (16), autoantigen La (17), and upstream N-Ras protein (Unr) (18). These cellular ITAFs interact with various viral IRES elements to regulate their activity by affecting ribosome recruitment or modifying the structure of the IRES itself (8). Correspondingly, viruses have developed sophisticated strategies to regulate IRES activity by modifying host factors, allowing them to efficiently replicate in host cells. For example, picornaviral proteases can cleave eIF4G to inhibit host mRNA translation, thus facilitating viral translation and replication (19). In addition, hepatitis A virus (HAV) 3C protease (3Cpro) cleaves the positive ITAF PCBP2, resulting in the loss of RNA binding affinity and the downregulation of viral protein synthesis during the late stage of viral replication (20). PCBP2 can also be cleaved by poliovirus (PV) 3Cpro between the KH2 and KH3 domains, leading to the loss of SRp20 association and translation inhibition. Cleaved PCBP2 still binds to the cloverleaf structure, and PCBP2 can thus act as a switch toward genome replication (21, 22). However, the cleavage of ITAF is not always involved in the shutdown of translation. FBP1, a positive regulator of the IRES activity of enterovirus 71 (EV71), can be cleaved by 2Apro, and its N-terminal cleavage product, FBP11–371, additively promotes enterovirus A71 (EV-A71) IRES activity in coordination with full-length FBP1 (23). Several ITAFs that negatively regulate viral IRES activity, such as gemin5 and FBP2, which negatively regulate the FMDV and EV-A71 IRESs, respectively, are also cleaved during viral infection, resulting in the blockage of negative IRES activity regulation (24, 25). Therefore, an improved understanding of the virus-host interactions involved in IRES-driven translation could have important implications for the development of effective viral prevention or control strategies.
Foot-and-mouth disease (FMD) is an acute, systemic disease of domestic and wild cloven-hooved animal species that is caused by foot-and-mouth disease virus (FMDV), a member of the genus Aphthovirus within the Picornaviridae family (26–28). The highly contagious nature of FMDV and the associated productivity losses make it a primary animal health concern worldwide (29, 30). Currently, the disease remains prevalent in many regions of the world, resulting in significant economic losses. Similar to other picornaviruses, the FMDV genome lacks a 5′-cap structure, and its translation is controlled by an IRES located in the 5′ UTR (8, 31).
To gain insight into the host-virus interactions regulating translation during FMDV infection, we isolated eight IRES-associated cellular proteins, including heterogeneous nuclear ribonucleoprotein K (hnRNP K), using a biotinylated FMDV IRES RNA pulldown assay followed by liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS) analysis (32). These proteins are potentially involved in FMDV IRES-mediated translation. Here, we further investigated the interactions of cellular hnRNP K with viral IRES and its effect on FMDV replication. As a member of the hnRNP family, hnRNP K is an essential RNA- and DNA-binding protein that regulates transcription, translation, pre-mRNA splicing, RNA stability, chromatin remodeling, and signal transduction (33). Other members of this family, including PTBP1 (hnRNP I) (34), PCBP2 (hnRNP E) (35), AUF1 (hnRNP D) (36), hnRNP Q (37), and hnRNP A1 (38), have been shown to play key roles in modulating IRES activity and viral infection.
In this study, hnRNP K was shown to act as a novel ITAF of the picornavirus FMDV that negatively regulates viral replication by inhibiting viral IRES-dependent translation. Furthermore, the interaction between the FMDV IRES and hnRNP K was further confirmed by mapping the interaction regions in both the IRES element and the hnRNP K protein. Conversely, FMDV has developed a strategy in which the viral 3C protease cleaves hnRNP K in a protease activity-dependent manner to antagonize the restriction of hnRNP K. Interestingly, the function of hnRNP K is altered when its amino terminus is cleaved, generating a C-terminal cleavage product that positively regulates FMDV replication. Our results demonstrate a new function of hnRNP K in inhibiting viral infection and provide new insights into strategies for the control of viral infection.
RESULTS
hnRNP K is an FMDV IRES-binding protein.
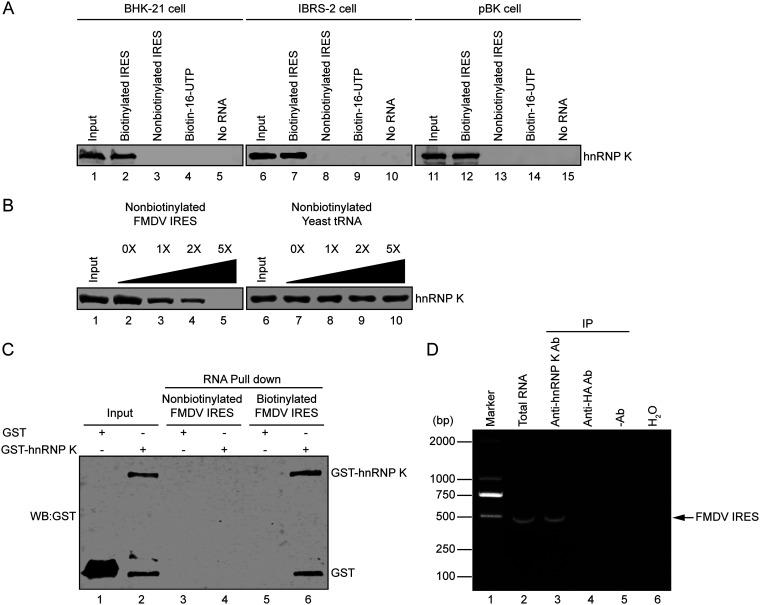
To further understand the mechanism of IRES-mediated translation initiation, the interaction of IRES-associated hnRNP K with FMDV IRES RNA was further verified by immunoblotting with an anti-hnRNP K antibody. The hnRNP K protein pulled down with biotin-IRES strongly interacted with the anti-hnRNP K antibody but was not pulled down by nonbiotinylated IRES RNA, biotin-16-UTP, or beads, used as negative controls (Fig. 1A). To determine whether the interaction between hnRNP K and the FMDV IRES occurred in a cell type-specific manner, lysates from pig-derived IBRS-2 cells and bovine-derived pBK cells were used in RNA pulldown assays, as these cell lines are susceptible to FMDV infection. The association of hnRNP K with the FMDV IRES occurred in all cell lines tested, indicating that the interaction between the FMDV IRES and hnRNP K was not restricted to one species or cell type (Fig. 1A). To further confirm the interaction between the FMDV IRES and hnRNP K, different amounts of nonbiotinylated FMDV IRES RNA or Saccharomyces cerevisiae yeast RNA were added to assess the competition for the IRES binding site using an RNA pulldown assay. The interaction was outcompeted by nonbiotinylated FMDV IRES RNA instead of yeast RNA, demonstrating that hnRNP K specifically interacts with FMDV IRES RNA (Fig. 1B). To exclude the possibility that the other host proteins in cells may participate in hnRNP K-FMDV IRES binding, unlabeled or biotin-labeled FMDV IRES RNA was incubated with Escherichia coli-expressed and purified glutathione S-transferase (GST)-hnRNP K or only GST. The result showed that GST-hnRNP K, but not GST, coprecipitated with biotin-labeled FMDV IRES RNA in vitro (Fig. 1C), indicating that hnRNP K directly binds to the FMDV IRES.
FIG 1.
Interaction of hnRNP K with the FMDV IRES. (A) The FMDV IRES interacts with hnRNP K in various cell lines. Extracts of BHK-21, IBRS-2, or pBK cells were incubated with biotinylated FMDV IRES, nonbiotinylated FMDV IRES, biotin-16-UTP, or no RNA. After the beads were washed, bound proteins were resolved using 12% SDS-PAGE. The hnRNP K protein was visualized by immunoblot analysis with an anti-hnRNP K antibody. The inputs of different cell extracts are shown in lanes 1, 6, and 11. (B) The specific association between hnRNP K and FMDV IRES was confirmed by competition assays. Various amounts of unlabeled RNA (FMDV IRES or yeast tRNA) were added to compete with the biotin-labeled FMDV IRES for binding hnRNP K in RNA-protein pulldown assays, and the eluted proteins were subjected to 12% SDS-PAGE and analyzed by Western blotting using an anti-hnRNP K antibody. The cell lysate inputs are shown in lanes 1 and 6. (C) Purified GST-hnRNP K or GST protein (50 μg) was incubated with unlabeled or biotinylated FMDV IRES, and the biotinylated RNA-protein complexes were precipitated with streptavidin beads and analyzed by Western blotting (WB) using an anti-GST antibody. (D) FMDV RNA was pulled down with hnRNP K from FMDV-infected cell extracts. BHK-21 cells were infected with FMDV at an MOI of 1 for 6 h before cell extract collection. Samples with mouse anti-hnRNP K, with mouse anti-HA, and without antibodies were incubated with 200 μg of FMDV-infected cell extracts, and protein G/A-agarose beads were then added to each sample to pull down the immune complexes. Following a washing step, the RNA was extracted and subjected to RT-PCR using FMDV IRES-specific primers. Total RNA extracted from FMDV-infected cell extracts was used as a positive control (lane 2) for RT-PCR, while H2O was used as a negative control (lane 6). The DNA was separated by agarose gel electrophoresis, and the expected 510-bp band is indicated by an arrow. Ab, antibody; IP, immunoprecipitation.
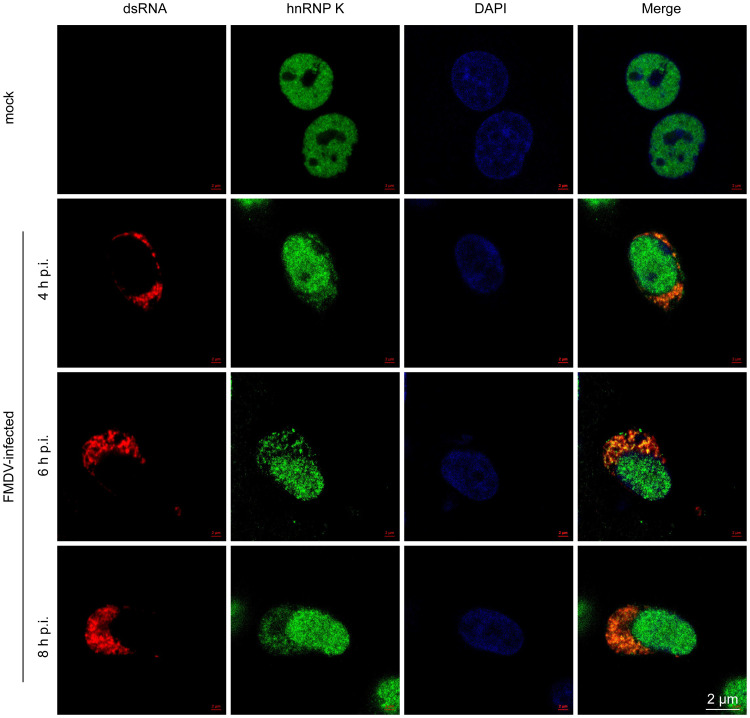
To validate the association of hnRNP K with FMDV IRES RNA in FMDV-infected cells, an RNA immunoprecipitation assay was performed. The result showed that a DNA band of the expected size (510 bp) was amplified from the immunoprecipitates pulled down by the anti-hnRNP K antibody but not from those obtained using the anti-HA antibody or the control without antibody, indicating that hnRNP K associates with the FMDV IRES in FMDV-infected cells (Fig. 1D). Additionally, the subcellular localization of hnRNP K and FMDV genomic RNA in FMDV-infected cells was examined. Endogenous hnRNP K localized in the nucleus of cells in the absence of FMDV infection, whereas hnRNP K was redistributed to the cytoplasm and colocalized with viral RNA in FMDV-infected cells (Fig. 2). These results indicate that during FMDV infection, hnRNP K is translocated from the nucleus to the cytoplasm, where it is retained and interacts with viral RNA.
FIG 2.
Cellular localization of hnRNP K and FMDV RNA. BHK-21 cells were mock infected or infected with FMDV at an MOI of 1 for 4, 6, or 8 h and then stained with antibodies against dsRNA (red), hnRNP K (green), and DAPI (blue). The samples were observed and imaged using a confocal microscope. p.i., postinfection.
Taken together, these results demonstrate that hnRNP K associates with FMDV IRES both in vitro and in vivo.
hnRNP K negatively regulates FMDV replication in infected cells.
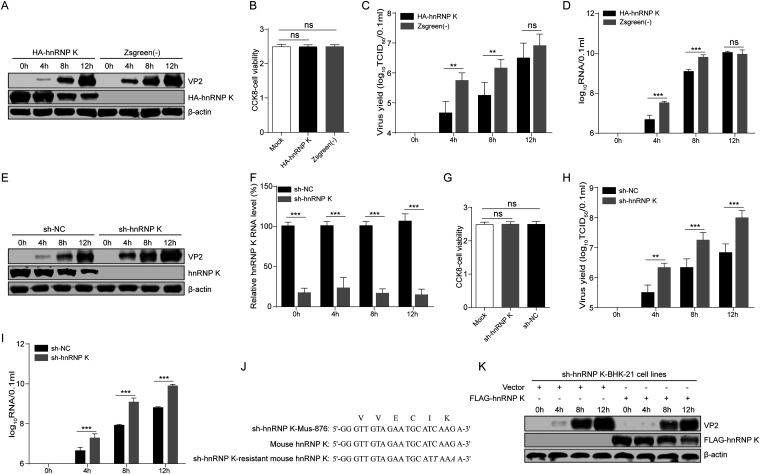
To address the biological role of hnRNP K binding to the FMDV IRES in the viral life cycle, BHK-21 cells were transduced with recombinant lentivirus overexpressing N-terminal hemagglutinin (HA)-tagged mouse hnRNP K (HA-hnRNP K) or green fluorescent protein (ZsGreen) as a control (Fig. 3A). Importantly, the overexpression of hnRNP K did not affect cell viability (Fig. 3B). The transduced cells were infected with FMDV at a multiplicity of infection (MOI) of 1.0. Compared to ZsGreen overexpression, hnRNP K overexpression resulted in 12.5- and 8.1-fold decreases in the production of infectious FMDV progeny at 4 and 8 h postinfection (hpi), respectively (Fig. 3C). The levels of the FMDV protein VP2 were also reduced in hnRNP K-overexpressing cells compared to those in the ZsGreen-transduced control cells, as measured by Western blotting (Fig. 3A). The levels of viral RNA detected by real-time reverse transcriptase (RT) quantitative PCR (qPCR) were also lower in the cells with hnRNP K overexpression than in the ZsGreen-transduced control cells (Fig. 3D). However, 12 h later, the viral VP2 protein and viral RNA levels and the viral titers in the hnRNP K-overexpressing cells increased to levels that were similar to those observed in the ZsGreen-transduced control cells (Fig. 3A, C, and D). These results strongly suggest that the overexpression of hnRNP K inhibits viral replication in the early stages of FMDV infection.
FIG 3.
hnRNP K negatively regulates FMDV replication. (A to D) Overexpression of hnRNP K reduces FMDV protein expression, the viral titer, and viral RNA production. hnRNP K-overexpressing or ZsGreen-transduced control BHK-21 cells were infected with FMDV at an MOI of 1, and the samples were harvested at the indicated time points postinfection. (A) The hnRNP K and viral VP2 levels in cells were detected by immunoblotting using anti-HA and anti-VP2 antibodies. (B) The proliferation of hnRNP K-overexpressing cells, ZsGreen-transduced control cells, and BHK-21 cells was detected by the CCK8 assay. (C) The viral titers in the supernatants were determined by TCID50 assays. (D) The viral RNA levels in cells were detected by RT-qPCR. (E to K) The reduction in hnRNP K expression enhances FMDV protein expression, viral titers, and viral RNA production. The knockdown effect of hnRNP K was detected at the protein level by immunoblotting with an anti-hnRNP K antibody (E) and at the RNA level by RT-qPCR using hnRNP K-specific primers (F). (G) The proliferation of hnRNP K-knockdown cells, cells transduced with sh-NC as a control, and BHK-21 cells was detected by the CCK8 assay. (E) The viral VP2 protein levels were determined by Western blotting assays. The viral titers (H) and viral RNA levels (I) in the culture supernatants were examined at different time points by the TCID50 assay and RT-qPCR. (J) The sequences of shRNAs targeting mouse hnRNP K (sh-hnRNP K-Mus-876), the targeted region of mouse hnRNP K, and sh-hnRNP K-resistant mouse hnRNP K are shown. The nucleotides that are different are underlined and shown in italic letters. The amino acids encoded by this region are also indicated at the top. (K) hnRNP K-knockdown BHK-21 cells transfected with a control vector (p3×FLAG-CMV) or a FLAG-tagged mouse hnRNP K expression plasmid (p3×FLAG-M-hnRNP K) were infected with FMDV at an MOI of 1. Cell lysates were harvested at 0, 4, 8, and 12 hpi and subjected to immunoblotting with antibodies against FMDV VP2, FLAG (for FLAG-tagged M-hnRNP K), and actin, as indicated. (B to D, F to I) The results are presented as the means ± SD from three independent experiments. The asterisks indicate significant differences between groups, as assessed by Student's t test (**, P < 0.01; ***, P < 0.001). ns, not significant.
The notion that hnRNP K inhibits FMDV replication was further verified by hnRNP K knockdown. The expression of hnRNP K in BHK-21 cells was knocked down by transduction with a lentivirus expressing a short hairpin RNA (shRNA) targeting hnRNP K mRNA (sh-hnRNP K-Mus-876), resulting in the BHK-sh-hnRNP K cell line. The results showed that hnRNP K expression was greatly decreased in BHK-sh-hnRNP K cells at the protein and RNA levels (Fig. 3E and F). Similarly, the knockdown of hnRNP K did not affect cell viability (Fig. 3G). These cells were then tested for their ability to support FMDV replication, and consistent with the hnRNP K overexpression results, the knockdown of hnRNP K resulted in significantly enhanced FMDV progeny production at 4, 8, and 12 hpi compared to that observed in the cells transduced with negative-control shRNA (sh-NC) as a control (Fig. 3H). Similarly, viral protein expression and RNA synthesis were accelerated in hnRNP K-knockdown cells (Fig. 3E and I).
To validate the specificity of hnRNP K knockdown on FMDV infection, we augmented the hnRNP K level in hnRNP K-knockdown cells by ectopic expression from an hnRNP K-expressing construct that carried two wobble mutations, rendering it resistant to shRNA targeting hnRNP K mRNA (sh-hnRNP K) without altering the amino acid sequence (Fig. 3J). hnRNP K-knockdown cells transfected with a vector (p3×FLAG-CMV) or a construct expressing FLAG-tagged hnRNP K (M-hnRNP K/p3×FLAG-CMV) were infected with FMDV at an MOI of 1. After reconstitution with hnRNP K, a higher level of FMDV VP2 protein expression was no longer observed in hnRNP K-knockdown cells compared to control cells (Fig. 3K). Taken together, these results demonstrate that hnRNP K inhibits FMDV replication, functioning as a host defense protein in FMDV-infected cells.
hnRNP K inhibits the IRES-mediated translation of FMDV.
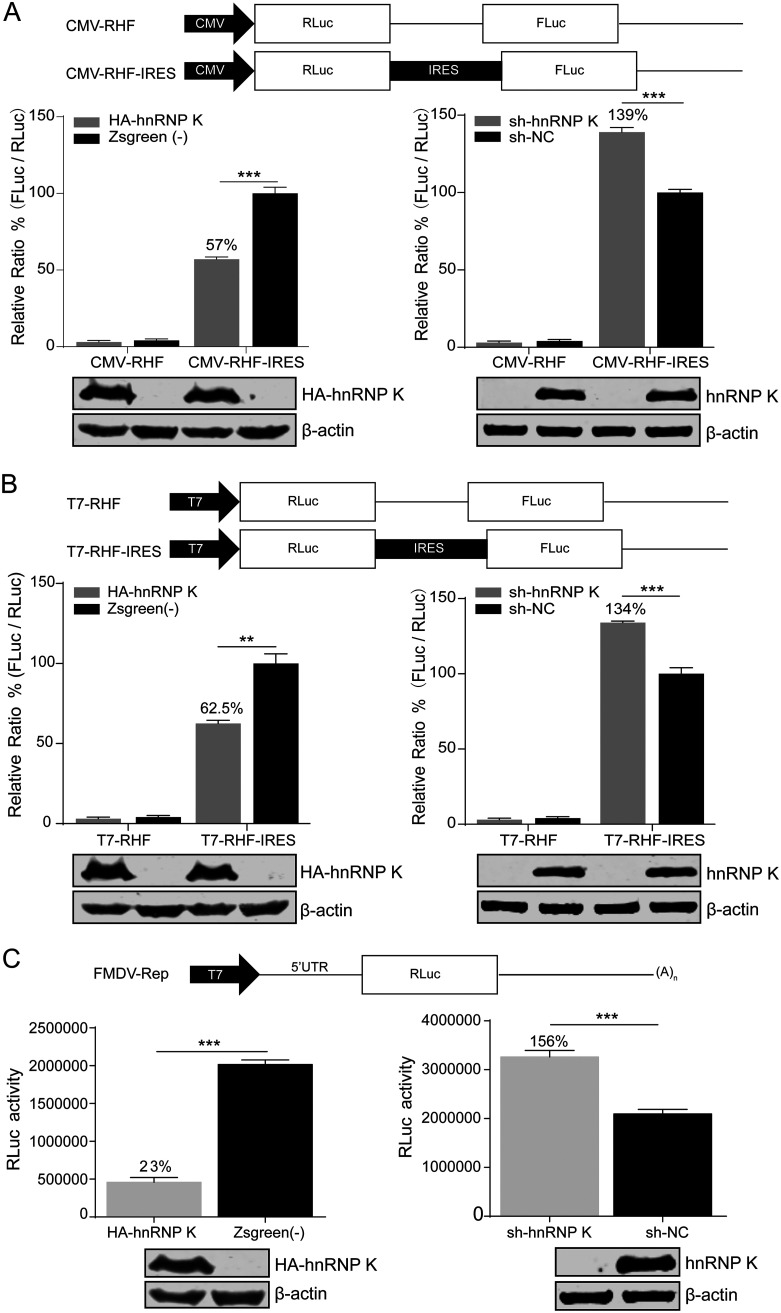
Since the interaction of hnRNP K with the FMDV IRES inhibits viral replication in the early stage of the FMDV life cycle, we further investigated the role of hnRNP K in IRES-mediated translation. A bicistronic reporter plasmid was used to evaluate FMDV IRES activity (Fig. 4A), as the translation of the first cistron (Renilla luciferase [RLuc]) is cap dependent, while the translation of the second cistron (firefly luciferase [FLuc]) is dependent on FMDV IRES activity. The relative IRES activity was reported as the ratio of FLuc expression to RLuc expression. The bicistronic reporter plasmid was transfected into hnRNP K-overexpressing and hnRNP K-knockdown cells. At 48 h posttransfection, cell lysates were collected to calculate the ratio of FLuc activity to RLuc activity. The FMDV IRES activity was significantly decreased in hnRNP K-overexpressing cells to 57% (P < 0.001) of that observed in the ZsGreen-transduced control cells (Fig. 4A, left). In contrast, the FMDV IRES activity was increased to 139% (P < 0.001) in the hnRNP K-knockdown cells compared to that in the cells transduced with sh-NC as a control (Fig. 4A, right). In addition, the direct transfection of bicistronic reporter mRNA containing the FMDV IRES was performed, revealing that FMDV IRES activity was reduced to 62.5% (P < 0.01) in hnRNP K-overexpressing cells and increased to 134% (P < 0.001) in hnRNP K-knockdown cells compared to that in the control cells (Fig. 4B). As shown in Table 1, the cap-dependent activity was not affected by the overexpression or knockdown of hnRNP K.
FIG 4.
hnRNP K inhibits FMDV IRES-mediated translation. (A) (Top) Schematic diagram of the bicistronic reporter plasmids pCMV-RHF and pCMV-RHF-IRES. The plasmids express bicistronic mRNA, consisting of an RLuc gene at the first cistron and the FMDV-IRES and FLuc genes at the second cistron (CMV, cytomegalovirus). (Middle) The bicistronic constructs pCMV-RHF or pCMV-RHF-IRES were transfected into hnRNP K-overexpressing or hnRNP K-knockdown BHK-21 cells. At 48 h posttransfection, the RLuc and FLuc activities in the cell lysates were analyzed. The bars in the histogram represent the FLuc/RLuc activity percentages. (Bottom) Western blot assays were performed to analyze the levels of hnRNP K expression. The luciferase activity of ZsGreen-transduced control cells or cells transduced with sh-NC as a control was set as 100%. (B) (Top) Schematic diagram of the bicistronic pT7-RHF and pT7-RHF-IRES reporter plasmids. (Middle) In vitro-transcribed T7-RHF or T7-RHF-IRES RNA was transfected into hnRNP K-overexpressing or hnRNP K-knockdown BHK-21 cells. At 48 h posttransfection, the RLuc and FLuc activities in the cell lysates were analyzed. (Bottom) Western blot assays were performed to analyze the levels of hnRNP K expression. The luciferase activity of ZsGreen-transduced control cells or cells transduced with sh-NC as a control was set as 100%. (C) (Top) Schematic diagram of the FMDV reporter replicon. The FMDV replicon was generated by replacing the P1 region of FMDV with the RLuc gene in the FMDV full-length infectious cDNA clone pYS, which has been reported previously (73). (Middle) hnRNP K-overexpressing or hnRNP K-knockdown BHK-21 cells were transfected with the FMDV-Rep replicon. At 12 h posttransfection, the cells were harvested, lysed, and assayed for RLuc activity. (Bottom) Western blot assays were performed to analyze the levels of hnRNP K expression. The RLuc activity of the ZsGreen-transduced control cells or cells transduced with sh-NC as a control was set as 100%. (A to C) The results are presented as the means ± SD from three independent experiments. The asterisks indicate significant differences between groups, as assessed by Student's t test (**, P < 0.01; ***, P < 0.001).
TABLE 1.
RLuc and FLuc values corresponding to Fig. 4
| Construct | FLuc/RLuc or RLuc value for cells transduced with: |
|||
|---|---|---|---|---|
| hnRNP K | ZsGreen | sh-hnRNP K | sh-NC | |
| CMV-RHF | 6,860/843,520 | 9,400/867,330 | 3,260/652,540 | 4,420/662,530 |
| 4,320/835,460 | 6,720/866,810 | 2,050/645,820 | 3,100/651,230 | |
| 9,200/825,430 | 11,310/811,450 | 4,180/633,510 | 5,410/655,220 | |
| CMV-RHF-IRES | 129,270/836,320 | 235,460/868,240 | 152,260/656,500 | 115,360/691,340 |
| 119,960/835,610 | 226,820/876,830 | 145,870/645,800 | 105,250/661,640 | |
| 137,890/845,360 | 246,880/885,360 | 149,070/663,600 | 112,030/678,230 | |
| T7-RHF | 7,110/905,680 | 9,680/924,600 | 6,390/893,100 | 8,630/904,500 |
| 6,160/1,156,800 | 8,060/1,008,000 | 3,220/908,260 | 5,050/950,650 | |
| 10,080/936,780 | 12,870/956,800 | 5,490/925,680 | 6,750/910,280 | |
| T7-RHF-IRES | 238,620/1,458,600 | 410,560/1,568,500 | 388,390/1,214,500 | 268,680/1,125,800 |
| 239,710/1,486,500 | 400,860/1,503,900 | 283,550/1,202,500 | 210,360/1,186,500 | |
| 254,340/1,465,400 | 408,650/1,518,600 | 245,220/1,223,600 | 180,460/1,215,600 | |
| FMDV-Rep | 431,000 | 208,0000 | 338,0000 | 2,180,000 |
| 415,000 | 201,0000 | 312,0000 | 2,110,000 | |
| 530,000 | 196,0000 | 329,0000 | 1,990,000 | |
Since use of the splice site or cryptic promoter in the IRES sequence of the bicistronic construct may interfere with the ratio of FLuc/RLuc activity, an in vitro-transcribed FMDV replicon RNA was transfected into the hnRNP K-overexpressing and hnRNP K-knockdown cell lines, and RLuc activity was measured at 12 h posttransfection. The FMDV IRES activity in the hnRNP K-overexpressing cells was reduced to 23% (P < 0.001) of that observed in the ZsGreen-transduced control cells, while in the hnRNP K-knockdown cells, the activity was increased to 156% (P < 0.001) of that observed in the negative-control cells (Fig. 4C). The lower panels in Fig. 4A to C show the overexpression and knockdown efficiency of hnRNP K. Taken together, these results indicate that hnRNP K negatively regulates the IRES-mediated translation of FMDV.
Regions of interaction between FMDV IRES and hnRNP K.
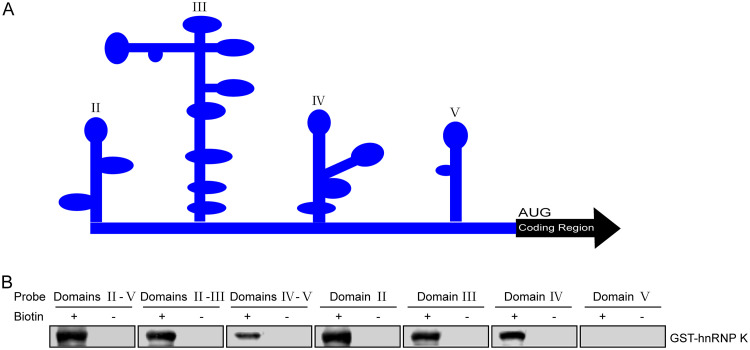
To better elucidate the interaction between FMDV IRES and hnRNP K, the binding regions involved in the interaction between the viral IRES and the hnRNP K protein were mapped. Considering the secondary structure of FMDV IRES, as predicted by use of the M-FOLD program (Fig. 5A), various truncated forms of FMDV IRES were synthesized by in vitro transcription and labeled with biotin. Subsequently, an RNA pulldown assay was performed to assess the interaction between hnRNP K and the truncated IRES constructs. The results showed that hnRNP K was pulled down with biotinylated IRES domain(s) II-V, II-III, IV-V, II, III, and IV but not with domain V (Fig. 5B), suggesting that hnRNP K interacts with domains II, III, and IV of the FMDV IRES.
FIG 5.
The regions of the FMDV IRES that interact with hnRNP K. (A) Predicted RNA secondary structure of the FMDV IRES. The M-FOLD program was used to predict the RNA secondary structure. (B) Map of the hnRNP K interaction region in the FMDV IRES. The purified recombinant protein GST-hnRNP K was mixed with RNA probes spanning different regions of the FMDV IRES. The RNA-protein complexes were purified with streptavidin beads, resolved by SDS-PAGE, and used in Western blot assays with an anti-GST antibody.
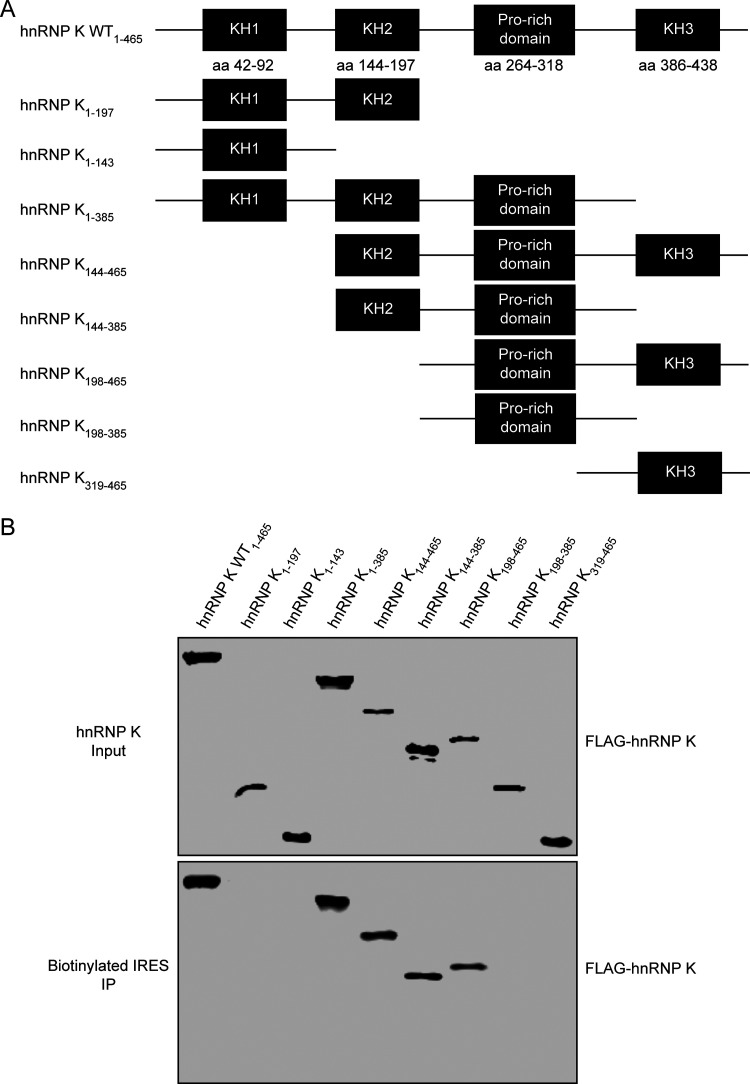
hnRNP K is an RNA-binding protein that was originally identified to be a component of the hnRNP complex and contains three KH domains (KH1, KH2, and KH3) and a proline-rich domain flanked by the KH2 and KH3 domains. The KH domain is one of the most common RNA-binding domains that directly contacts single-stranded RNA, while the proline-rich domain is important for protein-protein interactions (39, 40). To determine which functional domains of hnRNP K interact with the FMDV IRES, FLAG-tagged hnRNP K and its truncated forms (Fig. 6A) were expressed in HEK293T cells and purified by an anti-FLAG G1 affinity resin, and their ability to interact with FMDV IRES was assessed by an RNA pulldown assay. The FMDV IRES associated with full-length hnRNP K and four of the assayed truncations (hnRNP K constructs with amino acids [aa] 1 to 385, aa 144 to 465, aa 144 to 385, and aa 198 to 465) but not with hnRNP K constructs with aa 1 to 197, aa 198 to 385, and aa 319 to 465 (Fig. 6B). Considering that the hnRNP K truncations that bound to FMDV IRES depend on both the proline-rich domain and the KH2 or KH3 domain, our results suggest that hnRNP K interacts with the FMDV IRES through the KH2 and KH3 domains.
FIG 6.
hnRNP K protein domains that interact with the FMDV IRES. (A) Schematic diagram of hnRNP K and its truncated mutants. The hnRNP K protein contains three KH domains and a proline-rich domain. (B) Mapping of the regions in hnRNP K that interact with the FMDV IRES. Cell lysates prepared from HEK293T cells overexpressing FLAG-tagged wild-type hnRNP K or its truncated mutants were purified with an anti-FLAG G1 affinity resin and subjected to a biotin-RNA pulldown assay using the full-length FMDV IRES. Both the crude lysate (input) and proteins bound to the streptavidin beads (labeled RNA) were resolved by SDS-PAGE and detected by Western blotting using an anti-HA antibody. Unlabeled RNA was used as a negative control.
hnRNP K outcompetes PTB for binding to the FMDV IRES.
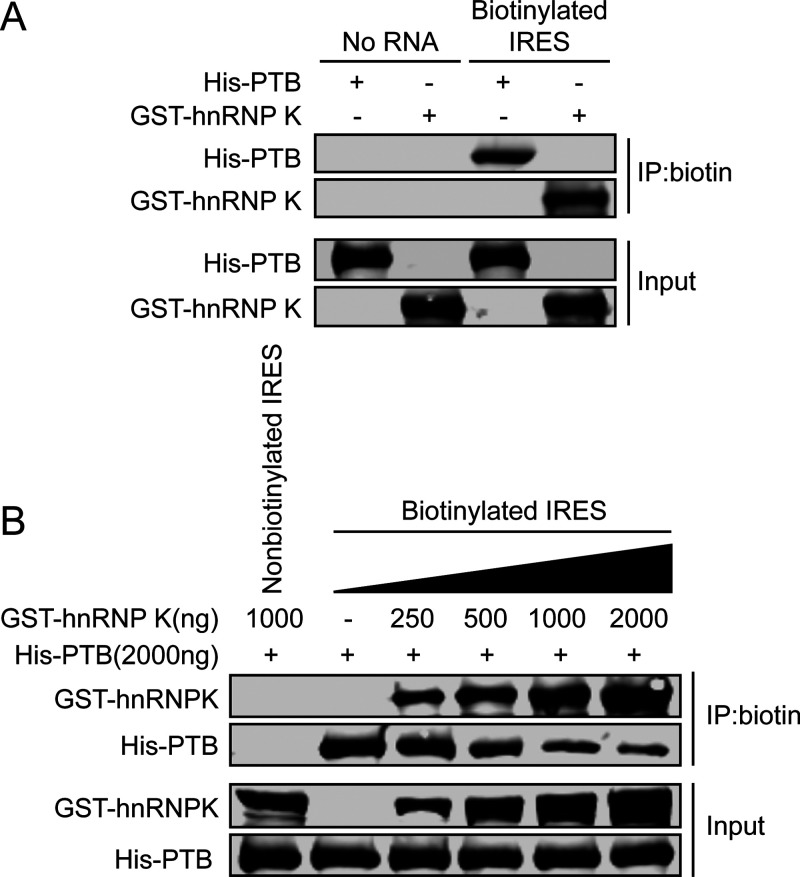
To gain insight into the mechanism by which hnRNP K serves as a negative regulator of FMDV IRES-mediated translation, we investigated whether hnRNP K affects the ability of the IRES to recruit other ITAFs. The cellular protein PTB is known to be required for the translation initiation of FMDV, during which PTB interacts with domains II, IV, and V of the viral IRES to positively regulate IRES-mediated translation (41–43). In this study, our results showed that the binding sites of hnRNP K are located in domains II, III, and IV of the FMDV IRES (Fig. 5B), where domains II and IV overlap the sites at which IRES binds PTB. Therefore, we speculated that hnRNP K may competitively bind to the FMDV IRES, which impairs the interaction between the FMDV IRES and PTB. We first confirmed that the biotinylated FMDV IRES could pull down hnRNP K and PTB by RNA pulldown assays (Fig. 7A). Subsequently, in vitro competitive binding assays were performed, revealing that the interaction between PTB and the FMDV IRES was significantly reduced with increasing hnRNP K protein levels (Fig. 7B). The nonbiotinylated FMDV IRES did not coprecipitate with PTB or hnRNP K, indicating that the interaction between the FMDV IRES and the protein is specific (Fig. 7B). These results demonstrated that hnRNP K outcompetes PTB for binding to the FMDV IRES, which suggests that hnRNP K inhibits FMDV IRES-mediated translation activity by interfering with the recognition of PTB as a positive regulator of translation.
FIG 7.
hnRNP K competes with PTB for the FMDV IRES binding site. (A) The interaction between hnRNP K or PTB and the FMDV IRES assessed by RNA pulldown assays. Purified GST-hnRNP K or His-PTB as a recombinant protein was incubated with biotinylated FMDV IRES RNA. No RNA was used as a negative control. Then, streptavidin beads were used to capture the biotinylated RNA, and the complexes were analyzed by Western blotting using anti-GST and anti-His antibodies. (B) Increased amounts of hnRNP K decreased the interaction between PTB and the FMDV IRES, according to the results of the RNA pulldown assay. Various amounts (in micrograms) of GST-hnRNP K were added to compete with His-PTB for interaction with the biotinylated FMDV IRES RNA. Nonbiotinylated RNA was used as a negative control. The eluted proteins were subjected to SDS-PAGE (12%), and antibodies against GST and His were then used in the Western blot analysis.
hnRNP K is cleaved by viral 3C protease during FMDV infection.
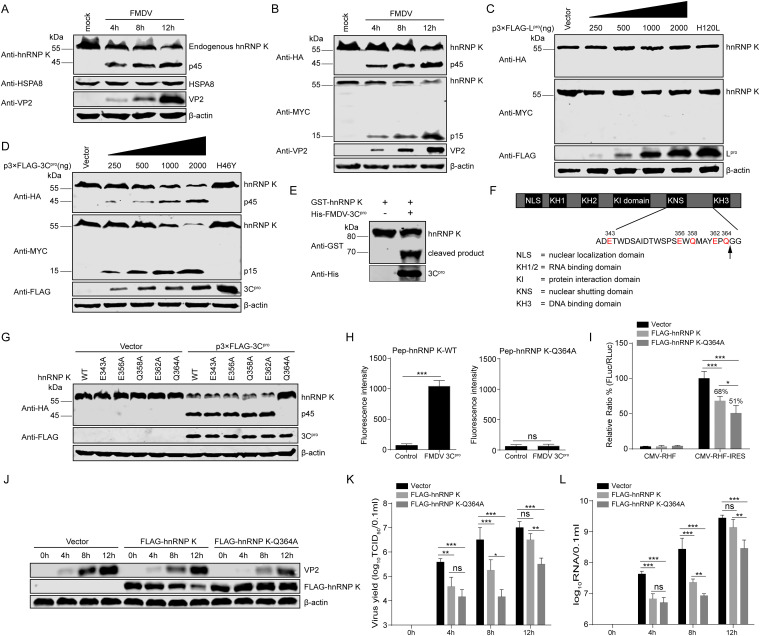
Our data indicated that hnRNP K acts as an ITAF that negatively regulates FMDV replication by downregulating IRES-mediated translation during the early stage of viral infection (Fig. 3 and 4). However, during the late stage of FMDV infection (12 hpi), the levels of viral protein expression, RNA synthesis, and virus production in the hnRNP K-overexpressing cells were increased to levels similar to those in the ZsGreen-transduced control cells (Fig. 3A, C, and D). In addition, hnRNP K was truncated in FMDV-infected BHK-21 cells compared to mock-infected BHK-21 cells (Fig. 8A), whereas the endogenous HSPA8 protein in BHK-21 cells was unaffected by FMDV infection (Fig. 8A), excluding the possibility that the cellular proteins were blocked by FMDV infection. These results indicate that FMDV has developed strategies to specifically antagonize the restriction of hnRNP K. To determine the possible cleavage products produced during FMDV infection, plasmids expressing N-terminal HA-tagged and C-terminal MYC-tagged hnRNP K were transfected into BHK-21 cells, after which the cells were infected with FMDV. The cleavage products were detected by Western blotting using anti-HA and anti-MYC antibodies. The generation of two stable cleavage products, p45 and p15, was observed at 4, 8, and 12 hpi, concomitant with a decrease in the expression of full-length HA-hnRNP K-MYC (p60) (Fig. 8B). These results indicate that hnRNP K is specifically cleaved during FMDV infection, yielding two cleavage products with apparent molecular weights of 45 and 15 kDa.
FIG 8.
FMDV 3Cpro cleaves hnRNP K in a protease activity-dependent manner. (A) The endogenous levels of hnRNP K or HSPA8 expression in mock-infected or FMDV-infected BHK-21 cells. The levels of hnRNP K and HSPA8 in FMDV-infected BHK-21 cells (or mock-infected cells) were detected at 4, 8, and 12 hpi by Western blotting. The viral VP2 protein was detected as a viral marker in these infected cells. (B) The cleavage of hnRNP K occurs in FMDV-infected cells. BHK-21 cells were transfected with a plasmid expressing HA-hnRNP K-MYC. At 24 h posttransfection, the cells were infected with FMDV, and the cell lysates were analyzed by Western blotting. (C) FMDV Lpro does not cleave hnRNP K. HEK293T cells were transfected with wild-type HA-hnRNP K-MYC (4 μg) together with increasing quantities of the FLAG-Lpro (0, 0.25, 0.5, 1, or 2 μg) or FLAG-Lpro H120L mutant (2 μg) plasmid. Cell lysates were prepared at 30 h posttransfection and analyzed by Western blotting. (D) hnRNP K is cleaved by FMDV 3Cpro in vivo. HEK293T cells were transfected with wild-type HA-hnRNP K-MYC (4 μg) together with increasing quantities of the FLAG-3Cpro (0, 0.25, 0.5, 1, or 2 μg) or FLAG-3Cpro H46Y mutant (2 μg) plasmid. Cell lysates were prepared at 30 h posttransfection and analyzed by Western blotting. (E) hnRNP K is cleaved by FMDV 3Cpro in vitro. The purified recombinant protein GST-hnRNP K was incubated with the purified recombinant protein His-FMDV 3Cpro, and the reaction products were analyzed by Western blotting with anti-GST and anti-His antibodies. (F) Schematic representation of wild-type hnRNP K and its potential 3C protease target sites. (G) FMDV 3Cpro cleaves hnRNP K at Q364. HEK293T cells were transfected with HA-tagged wild-type hnRNP K or its mutants, as indicated, together with FLAG-3Cpro or the empty vector. Cell lysates were prepared at 30 h posttransfection and analyzed by Western blotting. (H) FMDV 3Cpro cleaved hnRNP K-derived substrates in vitro. Two fluorogenic peptide substrates, Pep-hnRNP K-WT and Pep-hnRNP K-Q364A (DABCYL-MAYEPQ↓GGSGYE-Edans and DABCYL-MAYEPA↓GGSGYE-Edans), were introduced with His-FMDV 3Cpro in the FRET assay. (I) Effect of the cleavage-resistant mutant of hnRNP K on FMDV IRES activity. The plasmid pCMV-RHF or pCMV-RHF-IRES was transfected into BHK-21 cells overexpressing FLAG-hnRNP K, FLAG-hnRNP K-Q364A, or the FLAG vector control. At 48 h posttransfection, the RLuc and FLuc activities in the cell lysates were analyzed. The luciferase activity of the FLAG vector control was set as 100%. (J to L) Effect of the hnRNP K-Q364A mutant on FMDV replication. BHK-21 cells were transfected with FLAG-hnRNP K, FLAG-hnRNP K-Q364A, or the FLAG vector control and then infected with FMDV at an MOI of 1 at 24 h posttransfection. The cells and supernatants were harvested at 0, 4, 8, and 12 hpi. (J) The viral VP2 levels in the cells were detected by Western blotting. (K) The viral titers in the supernatants were determined by TCID50 assays. (L) The viral RNA levels in cells were detected by RT-qPCR. (H, I, K, and L) The results are presented as the means ± SD from at least three independent experiments. The asterisks indicate significant differences between groups, as assessed by Student's t test (**, P < 0.01; ***, P < 0.001).
In a previous study, a number of eIFs and ITAFs were shown to be proteolyzed in picornavirus-infected cells by proteases encoded in the viral genome (19, 20, 22–24). The FMDV genome is known to encode two functional proteases, the leader protease (Lpro) and the 3C protease (3Cpro). To determine which protease could be responsible for the observed cleavage of hnRNP K, HEK293T cells were cotransfected with a plasmid encoding HA-hnRNP K-MYC and increasing amounts of plasmids expressing wild-type (WT) Lpro or an inactive Lpro (H120L) mutant (44). The hnRNP K protein in the cell extracts was not cleaved by wild-type Lpro (Fig. 8C), suggesting that Lpro is not responsible for the proteolysis of hnRNP K during FMDV infection. Subsequently, the same experiment was performed to detect the catalytic activity of 3Cpro to cleave hnRNP K, revealing that hnRNP K was cleaved by wild-type 3Cpro but not the catalytically inactive mutant (H46Y) (45) of 3Cpro (Fig. 8D). Furthermore, the two cleavage products did not differ from those observed in FMDV-infected cells (Fig. 8D). To further confirm the cleaving effect of FMDV 3Cpro on hnRNP K in vitro, purified FMDV 3Cpro (His-FMDV 3Cpro) and GST-hnRNP K were incubated, and their reaction products were detected with anti-His and anti-GST antibodies. Purified GST-hnRNP K was also cleaved by FMDV 3Cpro in vitro (Fig. 8E). These results indicate that hnRNP K is cleaved by viral 3Cpro in a protease activity-dependent manner during FMDV infection.
To verify the properties of FMDV 3Cpro to cleave hnRNP K, we next examined potential 3Cpro cleavage sites in the amino acid sequence of hnRNP K. Previous investigations of 3Cpro substrate specificity recognized a partiality at the P1 position for both glutamine (Gln, Q) and glutamic acid (Glu, E) (46). Based on the sizes of the cleavage products, a series of hnRNP K mutants in which the invariant Gln or Glu was replaced by alanine (Ala, A) was constructed, and their cleavage by 3Cpro was examined (Fig. 8F). hnRNP K or its mutants were coexpressed with FMDV 3Cpro in HEK293T cells. The Q364A mutation blocked hnRNP K cleavage, yielding only the full-length hnRNP K protein in the presence of 3Cpro (Fig. 8G). In contrast, the E343A, E356A, Q358A, and E362A mutations had no impact on the 3Cpro-mediated cleavage of hnRNP K (Fig. 8G). To further determine whether Q364 of hnRNP K was the cleavage site of FMDV 3Cpro, we designed two fluorogenic peptide (Pep) substrates, wild-type Pep-hnRNP K (Pep-hnRNP K-WT) and Pep-hnRNP K-Q364A, which were derived from the hnRNP K-containing cleavage site, and fluorescence resonance energy transfer (FRET) assays were performed to explore whether FMDV 3Cpro could cleave these peptide substrates. When purified His-FMDV 3Cpro was incubated with the two fluorogenic peptide substrates in vitro, FMDV 3Cpro cleaved the Pep-hnRNP K-WT substrate instead of the Pep-hnRNP K-Q364A substrate (Fig. 8H). In addition, the cleavage-resistant mutant of hnRNP K inhibited IRES-mediated translation and viral replication better than wild-type hnRNP K (Fig. 8I to L). These data indicate that the viral 3C protease proteolytically cleaves hnRNP K at glutamine acid residue 364 (Q364) in vitro.
Effect of hnRNP K products cleaved by 3C protease on IRES-mediated translation and FMDV replication.
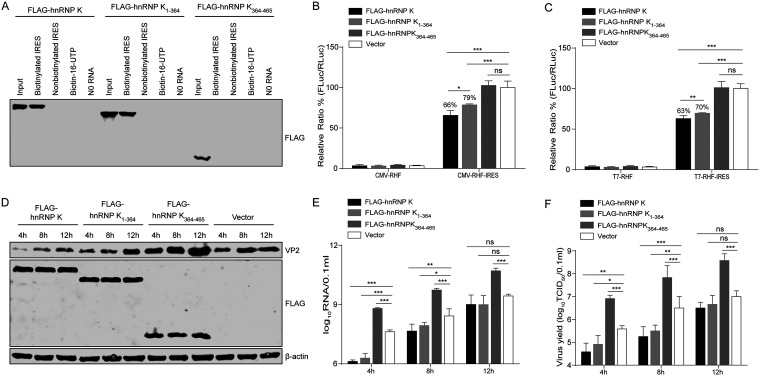
Our results showed that hnRNP K binds directly to the FMDV IRES element to act as a translation repressor. Therefore, we questioned whether the proteolytic products of hnRNP K retain the capacity to repress FMDV translation. To this end, we assessed the binding abilities of the hnRNP K cleavage products toward FMDV IRES RNA using an RNA pulldown assay. The full-length hnRNP K and its truncated form, hnRNP K1–364, bound to the FMDV IRES, but hnRNP K364–465 did not (Fig. 9A). Subsequently, the effects of hnRNP K1–364 and hnRNP K364–465 on IRES-mediated translation were examined by measuring the translation efficiency of bicistronic constructs harboring the FMDV IRES between the RLuc and FLuc reporter genes. Full-length hnRNP K and truncated hnRNP K1–364 decreased IRES activity to 66% and 79% (P < 0.001), respectively, whereas hnRNP K364–465 did not affect FMDV IRES activity (Fig. 9B). Similarly, direct transfection of bicistronic reporter mRNA was also performed. Consistent with the result described above, full-length hnRNP K and truncated hnRNP K1–364 reduced the FMDV IRES activity to 63% and 70% (P < 0.001), respectively, while hnRNP K364–465 had no effect on FMDV IRES activity (Fig. 9C). These results indicate that the N-terminal cleavage product hnRNP K1–364 retains its ability to inhibit IRES-mediated translation, but the repressive effect is attenuated compared to that exerted by the intact form of hnRNP K.
FIG 9.
Effect of hnRNP K products cleaved by 3C protease on IRES activity and FMDV replication. (A) Binding assay of hnRNP K and its truncated forms with FMDV IRES RNA. Lysates from HEK293T cells overexpressing FLAG-hnRNP K, FLAG-hnRNP K1–364, or FLAG-hnRNP K364–465 were purified by an anti-FLAG G1 affinity resin and incubated with biotinylated or nonbiotinylated FMDV IRES RNA, biotin-16-UTP, or no RNA. The protein-RNA complexes were pulled down with streptavidin beads and separated by SDS-PAGE. FLAG-tagged hnRNP K was detected by Western blotting. (B and C) Effects of hnRNP K and its truncated forms on FMDV IRES activity in vitro. BHK-21 cells were first transfected with FLAG-hnRNP K, FLAG-hnRNP K1–364, FLAG-hnRNP K364–465, or the FLAG vector control. After 24 h, the plasmid (pCMV-RHF or pCMV-RHF-IRES) (B) and the in vitro-transcribed RNA (T7-RHF or T7-RHF-IRES) (C) were transfected into BHK-21 cells. At 48 h posttransfection, the RLuc and FLuc activities in the cell lysates were analyzed. The luciferase activity of the FLAG vector control was set as 100%. (D to F) Effect of hnRNP K and its cleavage products (hnRNP K1–364 and hnRNP K364–465) on viral replication. BHK-21 cells were transfected with FLAG-hnRNP K, FLAG-hnRNP K1–364, FLAG-hnRNP K364–465, or the FLAG vector control and then infected with FMDV at an MOI of 1 at 24 h posttransfection. The cells and supernatants were harvested at 4, 8, and 12 hpi. (D) The full-length hnRNP K and hnRNP K truncations and viral VP2 levels in the cells were detected by Western blotting using anti-VP2, anti-FLAG, and antiactin antibodies, respectively. (E) The viral RNA levels in the cells were detected by RT-qPCR. (F) The viral titers in the supernatants were determined by TCID50 assays. (B, C, E, F) The results are presented as the means ± SD from at least three independent experiments. The asterisks indicate significant differences between groups, as assessed by Student's t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
To determine the effect of the hnRNP K proteolytic products on FMDV replication, FLAG-, FLAG-hnRNP K-, FLAG-hnRNP K1–364-, and FLAG-hnRNP K364–465-overexpressing BHK-21 cells were infected with FMDV at an MOI of 1. The level of FMDV VP2 protein expression in the cells overexpressing hnRNP K and hnRNP K1–364 was lower than that in the FLAG-transfected control cells (Fig. 9D). Interestingly, the level of VP2 protein expression in hnRNP K364-465-overexpressing cells was higher than that in the FLAG-transfected control cells (Fig. 9D). Similarly, the levels of RNA synthesis and viral production were decreased in the hnRNP K- and hnRNP K1–364-overexpressing cells compared with those in the FLAG-transfected control cells, whereas the viral RNA levels and titers of FMDV were increased in the hnRNP K364–465-overexpressing cells compared with those in the FLAG-transfected control cells (Fig. 9E and F). These results indicate that the overexpression of the two cleavage products of hnRNP K has different effects on FMDV replication: the N-terminal hnRNP K1–364 product partially retains inhibitory effects on viral replication, whereas the C-terminal hnRNP K364–465 product promotes viral replication.
DISCUSSION
The regulation of IRES-mediated translation is regarded as a critical step for picornavirus infection, as it has important effects on virulence, pathogenicity, and tissue tropism (11). To date, the regulatory mechanisms of viral IRES-mediated internal translation initiation remain poorly understood. In this study, we determined that hnRNP K is a novel ITAF of the picornavirus FMDV that negatively regulates FMDV replication by inhibiting viral IRES-mediated translation. Further investigation of the inhibitory mechanism of hnRNP K on IRES-mediated translation showed that hnRNP K competes with the positive ITAF PTB to bind the IRES and inhibit FMDV translation and replication. Interestingly, the FMDV 3C protease subverts the inhibitory effect of hnRNP K on viral replication by generating the fragment hnRNP K364–465, which promotes viral growth.hnRNP K is a polycytidine-binding protein that is involved in various cellular processes, including chromatin remodeling, transcriptional regulation, splicing, and RNA translation, through interactions with RNA, DNA, and multiple proteins (47–49). Lin et al. (50) showed that hnRNP K is enriched in the cytoplasm, in which EV71 replication occurs, to interact with the EV71 5′ UTR and promote viral replication. Our results indicated that the KH2 or KH3 domain of hnRNP K specifically binds to the highly structured domains II, III, and IV of the viral IRES in the cytoplasm and negatively regulates FMDV replication (Fig. 3, 5, and 6). Further functional analysis involving the knockdown and overexpression of hnRNP K in an in vitro translation assay underscored a novel function of the hnRNP K protein in translation control. The higher IRES-mediated translation efficiency in hnRNP K-knockdown cells and the corresponding lower efficiency of IRES activity in hnRNP K-overexpressing cells (Fig. 4) indicate that hnRNP K negatively regulates FMDV IRES-mediated translation.
The IRES-mediated initiation of translation depends on the structural organization of the IRES and its recruitment of numerous cellular proteins (eIFs and ITAFs), and RNA-binding proteins are able to interact with each other during this process (51–53). Thus, hnRNP K may recruit or interfere with another RNA-binding protein(s) to regulate translation. In this study, we observed that hnRNP K directly binds to a specific region of the FMDV IRES and outcompetes the positive ITAF PTB, which recognizes the same regions (Fig. 5 and 7). Several alternative but not mutually exclusive possibilities could explain how hnRNP K competes with PTB to bind the IRES and negatively regulate the IRES-mediated translation of FMDV. One possibility is the existence of similar recognition motifs in the competitor proteins or that the RNA-binding sites of the competitor proteins are closely located, leading to steric interference. In support of this model, our results showed that the sites on hnRNP K and PTB that bind to FMDV IRES overlap in domains II and IV (Fig. 5), which compete for the same IRES RNA-binding site, and this competition leads to the negative regulation of viral IRES-mediated translation. Another possibility is that the potential protein-protein interactions prevent the binding of ITAFs with the IRES element. PTB acts as an RNA chaperone to stabilize the IRES structure for FMDV translation by interacting with the FMDV IRES (34, 54), whereas the interaction of PTB and hnRNP K assists in the RNA-related biological processes of hnRNP K (55), suggesting that hnRNP K-induced interference in PTB binding to the IRES may be related to protein-protein interactions.
During coevolution with their hosts, viruses have developed strategies to actively counteract host antiviral responses, and the balance between host antiviral activity and viral antagonism may determine disease and pathogenesis outcomes. Many viruses have developed sophisticated strategies to evade host antiviral factors and replicate efficiently in host cells (56–58). Previous studies have shown that the proteases Lpro and 3Cpro of FMDV antagonize the host antiviral response via a repertoire of mechanisms during viral infection (59–62). In addition, some members of the hnRNP family, including PCBP, PTB, hnRNP M, AUF1, and hnRNP K, are also cleaved by different viral proteases to regulate viral infection through different mechanisms (21, 22, 63–66). Our results presented here demonstrate that FMDV antagonizes the inhibition of the cellular hnRNP K protein via cleavage by its own 3C protease, resulting in the generation of the two cleavage products with opposite functions during FMDV replication that ultimately determine the viral infection outcome. This important balance between host antiviral activity and viral antagonism is achieved during viral infection, and we speculate that the positive effect of the C-terminal cleavage product hnRNP K364–465 may overcome this inhibitory effect of the full-length hnRNP K and N-terminal cleavage product hnRNP K1–364 and therefore contribute to the balance established during FMDV cellular infection.hnRNP K1–364 is a cleavage product generated during FMDV infection that lacks the C-terminal KH3 domain. Our data presented in Fig. 9 clearly demonstrate that the direct binding of hnRNP K and hnRNP K1–364 to the FMDV IRES negatively regulates IRES-driven translation and viral replication, but the repressive effect of hnRNP K1–364 is attenuated compared to that exerted by the intact form of hnRNP K. However, it is unclear why hnRNP K1–364 retains its ability to inhibit IRES-mediated translation. To better survive in host cells, picornaviruses abolish unfavorable host factors through viral enzymatic cleavage to facilitate viral propagation (67). In this study, the cleavage of hnRNP K altered its RNA-binding ability in the FMDV IRES. The N-terminal cleavage product hnRNP K1–364 retained the KH2 domain but lacked the KH3 domain, capable of binding to the viral IRES, thereby reducing its inhibitory effect on IRES-mediated translation (Fig. 8 and 9). Similarly, the cellular mRNA decay protein AUF1, a negative ITAF for PV, can bind to stem-loop IV of the IRES to negatively regulate viral propagation (65). However, this host antiviral response is partly inhibited through the proteolytic cleavage of AUF1 by viral proteinase 3CD (65). Surprisingly, we observed that another cleavage product of hnRNP K, hnRNP K364–465, becomes a positive regulator of FMDV replication (Fig. 9). The KH3 domain is known to play a crucial role in RNA binding, as the isolated domain has been shown to bind to nucleic acids, albeit with a lower affinity than the full-length protein (68). Thus, we speculate that hnRNP K364–465 positively regulates FMDV replication by interacting with viral RNAs other than the IRES or protein(s). Recently, Cao et al. demonstrated that another member of the hnRNP family, hnRNP M, inhibits RNA virus-triggered innate immunity by antagonizing the RNA sensing of RIG-I-like receptors (69). hnRNP K and hnRNP M are known to have similar structures and functions (70). Structurally, they are modular proteins that consist of three RNA-binding domains [K(H/R)RM] connected by linker regions of various lengths. Functionally, they are nuclear proteins that induce nucleocytoplasmic shuttling during viral infection. Therefore, we also propose another possibility, which is that hnRNP K364–465 positively regulates FMDV replication by inhibiting virus-triggered innate immunity. However, the mechanism by which hnRNP K364–465 positively regulates FMDV replication will be investigated in future studies.
Collectively, our results revealed that hnRNP K is a novel ITAF for FMDV that negatively regulates FMDV replication by inhibiting viral IRES-mediated translation but that is antagonized by viral 3C protease. Interestingly, one of the cleavage products, hnRNP K364–465, becomes a positive regulator of viral replication. Our results broaden the knowledge of host-virus interactions and provide new insights into translational control during viral infection.
MATERIALS AND METHODS
Ethics statement.
The animal work for the preparation of primary fetal bovine kidney cells was carried out in strict accordance with Chinese regulations for laboratory animals (71) and the Laboratory Animal-Requirements of Environment and Housing Facilities (standard number GB 14925-2010; National Laboratory Animal Standardization Technical Committee) (72). Protocols for the animal studies were approved by the Committee on the Ethics of Animal Experiments of the Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences (protocol number 100515-01).
Plasmid construction.
Plasmids for in vitro transcription assays were constructed as follows: the entire FMDV IRES sequence was amplified by PCR from the FMDV full-length infectious cDNA clone pYS (73) using primers for IRES (forward primer 5′-CG GAATTC TAA TAC GAC TCA CTA TAG GGC ACG AAA CGC GCC GTC GCT TGA GGA GGA CT-3′ and reverse primer 5′-AAA GATATC TTA AAG ACA GTT GTT CGA AGG AAA GGT GCC GGC CTC-3′) containing the T7 promoter, and the fragment was cloned into the EcoRI and EcoRV sites of the vector pVAX1 (Invitrogen). Underlined nucleotides represent the restriction enzyme recognition sites, while italicized nucleotides represent the T7 promoter. Various truncated forms of the IRES were constructed via the amplification and insertion of truncation fragments into the EcoRI and EcoRV sites of the vector pVAX1.
The overexpression plasmids p3×FLAG-Lpro, p3×FLAG-3Cpro, p3×FLAG-Lpro (H120L), p3×FLAG-3Cpro (H46Y), p3×FLAG-hnRNP K, pHA-hnRNP K, and pHA-PTB were prepared as follows: the plasmid pYS was used as a template to PCR amplify Lpro (using forward primer 5′-CGC GAATTC AAT GGA GTT CAC ACT TCA CAA C-3′ and reverse primer 5′-CGC AGATCT TTA TCT GAG TCG TTT CTG AAC-3′ primers) and 3Cpro (using forward primer 5′-TTT GAATTC AAT GAG TGG TGC CCC ACC GAC T-3′ and reverse primer 5′-TTT AGATCT TTA CTC GTG GTG TGG TTC GGG-3′ primers), after which the fragments were cloned into the EcoRI and BglII sites of the vector p3×FLAG-CMV. The Lpro and 3Cpro, Lpro-H120L, and 3Cpro-H46Y mutants were constructed by overlap extension PCR using specific mutagenic primers. The cDNA from baby hamster kidney (BHK-21) cells was used as a template to amplify hnRNP K (using forward primer 5′-TTT GCGGCCGC GAT GGA GAC CGA ACA GCC AGA AGA AAC-3′ and reverse primer 5′-TCT GGATCC TTA GAA TCC TTC AAC ATC TGC-3′ primers), which was inserted into the NotI and BamHI sites of the vector p3×FLAG-CMV. The truncated forms of hnRNP K, hnRNP K-Q364A, hnRNP K1–364, and hnRNP K364–465 were constructed by PCR amplification using the p3×FLAG-hnRNP K plasmid as a template, and the fragments were inserted into the NotI and BamHI sites of the vector p3×FLAG-CMV. The various truncated forms of hnRNP K were constructed by PCR amplification and insertion of the truncation fragments into the vector p3×FLAG-CMV. The p3×FLAG-hnRNP K plasmid was used as a template to PCR amplify hnRNP K (using forward primer 5′-TGT GAATTC ATG GAG ACC GAA CAG CCA GAA GAA AC-3′ and reverse primer 5′-TTT GCTAGC TTA CAG ATC CTC TTC AGA GAT GAG TTT CTG CTC GAA TCC TTC AAC ATC TGC-3′ primers), which was cloned into the EcoRI and NheI sites of the vector pCAGGS-HA, which encodes an N-terminal HA tag and a C-terminal MYC tag (italicized nucleotides represent the MYC-tag sequence). The mutagenesis of hnRNP K (E343A, E356A, Q358A, E362A, and E364A) was performed by overlap extension PCR using specific mutagenic primers.
The GST-tagged recombinant hnRNP K protein was expressed in E. coli from the plasmid pGEX-hnRNP K. To construct this plasmid, pHA-hnRNP K was used as the template for PCR amplification using primers for hnRNP K (forward primer 5′-TGT GAATTC GAG ACC GAA CAG CCA GAA GAA AC-3′ and reverse primer 5′-TCT CTCGAG TTA GAA TCC TTC AAC ATC TGC-3′). The obtained PCR product was digested with BamHI/XhoI and subcloned into the vector pGEX-6p-1 (Amersham Pharmacia Biotech). The cDNAs of PTB and FMDV 3Cpro were also cloned into the pET-28a vectors to express recombinant proteins.
pT7-RHF was constructed by PCR amplifying the Renilla luciferase (RLuc) gene from the vector pGL4.75[hRLuc/CMV] (Promega, WI, USA) using primers for RLuc (forward primer 5′-AAC AGATCT TAA TAC GAC TCA CTA TAG ATG GCT TCC AAG GTG TAC GAC CCC GAG-3′ and reverse primer 5′-AAC AGATCT TTA CTG CTC GTT CTT CAG CAC GCG CT-3′), and the product was cloned into the BglII site of the vector pGL3-basic (Promega). Italicized nucleotides represent the T7 promoter. The bicistronic reporter plasmid pT7-RHF-IRES, which contains the FMDV IRES sequence between RLuc and FLuc, was constructed via the insertion of a BglII-FMDV IRES-NcoI fragment. The bicistronic reporter plasmids pCMV-RHF and pCMV-RHF-IRES were constructed by digesting the constructs pT7-RHF and pT7-RHF-IRES with SacI/XbaI to obtain the fragments T7-RHF and T7-RHF-IRES, which were cloned into the vector pGL4.75[hRLuc/CMV]. The FMDV replicon was generated by replacing the P1 region of FMDV with the RLuc gene in the FMDV full-length infectious cDNA clone of pYS, as reported previously (73).
Cells and viruses.
Baby hamster kidney cells (BHK-21 cells; ATCC CCL-10), porcine kidney cells (IBRS-2 cells; ATCC CRL-1835), human embryonic kidney 293T cells (HEK293T cells; CRL-11268), and primary fetal bovine kidney cells (pBK cells) were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) containing 10% fetal bovine serum (FBS; HyClone) and 2 mM l-glutamine at 37°C with 5% CO2 in a humidified atmosphere. The FMDV strain O/YS/CHA/05 (GenBank accession number HM008917) used in our study was generated from the infectious cDNA clone pYS (73). Viral titers were determined by 50% tissue culture infective dose (TCID50) assays with BHK-21 cells (74).
Protein expression and purification.
Escherichia coli strain BL21(DE3) was transformed with pGEX-hnRNP K or pGEX-6P-1 and then induced with 0.25 mM isopropyl-1-thio-β-d-galactopyranoside for 20 h at 16°C. Recombinant GST-hnRNP K and GST proteins were purified with glutathione-Sepharose (Amersham Pharmacia Biotech) and eluted with 10 mM reduced glutathione.
Construction of stable hnRNP K-overexpressing and hnRNP K-silenced cell lines.
The PCR-amplified hnRNP K gene from BHK-21 cells was ligated into the retroviral vector pLVX-IRES-ZsGreen1 (Clontech) to produce the recombinant plasmid pLVX-ZsGreen1-hnRNP K. For lentivirus preparation, HEK293T cells were cotransfected with pLVX-ZsGreen1-hnRNP K or pLVX-IRES-ZsGreen1 and two helper plasmids, psPAX2 and pMD2.G, using the Lipofectamine 2000 reagent (Invitrogen). The culture supernatants containing the viral particles were harvested and used to infect BHK-21 cells for 48 h. The transduced cells expressing hnRNP K-ZsGreen1 or ZsGreen1 were screened by flow cytometry using a MoFlo XDP high-speed cell sorter (Beckman Coulter).
The lentivirus vector pGLVU6/Puro, encoding short hairpin RNAs (shRNAs) targeting mouse hnRNP K (sh-hnRNP K-Mus-277 [5′-GGA GAC CGA ACA GCC AGA AGA-3′], targeting nucleotides [nt] 3 to 23 of the mouse hnRNP K mRNA; sh-hnRNP K-Mus-659 [5′-GCT GTG GAA TGC TTA AAT TAC-3′], targeting nt 385 to 405 of the mouse hnRNP K mRNA; sh-hnRNP K-Mus-876 [5′-GGG TTG TAG AAT GCA TCA AGA-3′], targeting nt 602 to 622 of the mouse hnRNP K mRNA; sh-hnRNP K-Mus-1093 [5′-GCC TCC TTC TAG AAG AGA TTA-3′], targeting nt 819 to 839 of the mouse hnRNP K mRNA) and the negative control (sh-hnRNP K-Mus-NC [5′-GTT CTC CCG AAC GTG TCA CGT-3′]) were obtained from Shanghai GenePharma Co. Ltd. (Shanghai, China). For lentivirus preparation, HEK293T cells were cotransfected with pGLVU6/Puro-sh-hnRNP K or pGLVU6/Puro and the helper plasmids, pGag/Pol, pRev, and pVSV-G using the Lipofectamine 2000 reagent (Invitrogen). The culture supernatants containing packaged lentiviruses were harvested. BHK-21 cells were transduced with sh-hnRNP K lentivirus for 48 h and selected with puromycin (2.5 μg/ml) to knock down mouse hnRNP K expression.
In vitro transcription.
The T7 promoter-FMDV IRES plasmids were linearized with EcoRV and purified by phenol-chloroform extraction. RNA transcripts were synthesized using a RiboMAX large-scale RNA production system (T7 kit; Promega) according to the manufacturer’s protocol. Biotinylated RNA was synthesized by adding 1.25 μl of 20 mM biotinylated protein biotin-16-UTP (Roche) to a final volume of 20 μl to constitute an in vitro transcription reaction mixture for RNA labeling. Synthesized RNAs were purified using an RNeasy minikit (Qiagen) and analyzed on 1% agarose gels.
RNA pulldown assay.
The RNA pulldown assay was performed based on a previously published method (15, 75, 76). Fresh cell lysates were lightly sonicated and centrifuged at 16,000 × g for 10 min at 4°C. Then, the supernatants were transferred to new tubes and the protein was quantified by measuring the absorbance at 280 nm. Egg white avidin (EMD Chemicals) and yeast RNA (Sigma) were added to block endogenous biotinylated proteins and nonspecific ribonucleoprotein complexes (RNPs), and the mixture was incubated on a rotating shaker at 4°C for 20 min. After blocking, the lysates were again centrifuged at 16,000 × g for 10 min at 4°C. The supernatants were transferred to new tubes with 200-U/ml RNasin (Promega). The biotinylated FMDV IRES RNA was heated to 90°C for 2 min in RNA folding buffer (10 mM Tris [pH 7], 0.1 M KCl, 10 mM MgCl2), and the mixture was shifted to room temperature for 20 min to allow proper secondary structure formation. For the biotinylated RNA-binding assay, a reaction mixture containing 200 μg of cell extract and 5 μg of biotinylated RNA was prepared. The mixture, at a final volume of 100 μl, was incubated in RNA mobility-shift buffer (5 mM HEPES [pH 7.1], 40 mM KCl, 2 mM MgCl2, 1 U RNasin, 0.25 mg/ml heparin) for 60 min at 30°C and then added to 100 μl of Dynabeads M-280 streptavidin (Invitrogen) and allowed to bind for 10 min at room temperature. The RNA-protein complexes were washed five times with RNA mobility-shift buffer without heparin. After the last wash, 30 μl of 1× SDS-PAGE sample buffer was added to the beads, and the captured proteins were analyzed by SDS-PAGE and immunoblotting assays.
Western blot analysis.
Cells were lysed with radioimmunoprecipitation assay (RIPA) buffer (Sigma) containing a cocktail of protease inhibitors (Roche). The protein concentrations were determined using a bicinchoninic acid protein assay kit (Pierce). Equal amounts of protein were loaded and separated by SDS-PAGE and then transferred to polyvinylidene difluoride (PVDF) membranes. The PVDF membranes were blocked with phosphate-buffered saline (PBS) containing 5% nonfat dry milk and subsequently incubated with primary antibodies against various proteins, including mouse anti-FMDV VP2 (77), mouse anti-hnRNP K (Santa Cruz Biotechnology), rabbit anti-hnRNP K (Proteintech), mouse anti-PTB (Proteintech), mouse anti-HSPA8 (Proteintech), mouse anti-FLAG (Sigma), mouse anti-GST (GenScript), mouse anti-HA (GenScript), and mouse anti-β-actin (GenScript). After being washed, the membranes were incubated with secondary IRDye 800CW goat anti-mouse IgG or IRDye 800CW goat anti-rabbit IgG antibodies, and signal detection was performed using a near-infrared fluorescence scanning imaging system (LiCor Odyssey).
RNA immunoprecipitation.
BHK-21 cells were infected with FMDV at a multiplicity of infection (MOI) of 1.0 for 6 h and lysed in RIPA buffer (Sigma). Cell lysates were precleared by incubation on ice for 1 h with protein A/G-agarose (GE Healthcare), after which the nonspecific complexes were pelleted by centrifugation at 10,000 × g for 10 min at 4°C, and the supernatants were used in immunoprecipitation assays. Two hundred micrograms of precleared lysate was mixed with 5 μg of mouse anti-hnRNP K, mouse anti-HA, or control mouse IgG antibody and incubated at 4°C for 4 h. Subsequently, prewashed protein A/G-agarose beads were added to each sample and the mixtures were incubated overnight at 4°C. The immune complexes were then washed three times with RNA mobility-shift buffer, and the RNA was extracted from the immunoprecipitated complexes with a Simply P total RNA extraction kit (BioFlux) according to the manufacturer’s instructions. cDNA was synthesized using PrimeScript reverse transcriptase (TaKaRa), and PCR analysis was performed by using primers specific for the FMDV IRES (forward primer 5′-GCA CGA AAC GCG CCG TCG CTT GAG G-3′ and reverse primer 5′-TTA AAG ACA GTT CGA AGG AAA GGT G-3′).
Confocal microscopy.
BHK-21 cells grown on glass coverslips were infected with FMDV at an MOI of 1. At 4, 6, or 8 h postinfection (hpi), the culture medium was removed and the cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100. After being blocked with 5% bovine serum albumin in PBS, the hnRNP K protein was detected using a rabbit anti-hnRNP K antibody (Proteintech), and FMDV RNA was detected using an anti-double-stranded RNA (anti-dsRNA) antibody (J2 monoclonal antibody; English & Scientific Consulting). Secondary Alexa Fluor 488-labeled goat anti-rabbit IgG (Beyotime) or Alexa Fluor 633-conjugated anti-mouse immunoglobulin (Invitrogen) antibodies were added sequentially for 1 h at room temperature. After the cells were washed with PBS, the nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI; Beyotime), and the specimens were observed with a confocal laser-scanning microscope (Zeiss model LSM880).
RNA extraction and RT-qPCR.
FMDV RNA levels were measured via real-time reverse transcriptase (RT) quantitative PCR (qPCR), as previously described (78). Briefly, total RNA was extracted from FMDV-infected BHK-21 cells using a Simply P total RNA extraction kit (BioFlux) according to the manufacturer’s instructions, and 200 ng of total RNA was subjected to cDNA synthesis using PrimeScript reverse transcriptase (TaKaRa). cDNA quantification was performed using an Mx3005P instrument (Agilent Technologies) with SYBR green real-time PCR master mix (Toyobo). To assess hnRNP K knockdown, total RNA was extracted from the cells, and RT-qPCR was performed using primers for hnRNP K (forward primer 5′-GCT GCC CTC ACT CCA CT-3′ and reverse primer 5′-TGG GAG ACT CGG AAA T-3′). GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as an internal control and was amplified with specific primers (forward primer 5′-ACA TGG CCT CCA AGG AGT AAG A-3′ and reverse primer 5′-GAT CGA GTT GGG GCT GTG ACT-3′). The mean values and standard deviations (SD) were derived from triplicate measurements. In addition, genomic DNA contamination was excluded by including non-reverse-transcribed RNA as a control for each sample.
Luciferase assays.
For the bicistronic expression assay, the bicistronic pCMV-RHF/pCMV-RHF-IRES or pT7-RHF/pT7-RHF-IRES reporter plasmids were transfected into hnRNP K-overexpressing or ZsGreen-transduced control cells and hnRNP K-knockdown cells or cells transduced with sh-NC. After 48 h, cell extracts were prepared in passive lysis buffer (Promega) and assayed for RLuc and FLuc activity on a PE EnVision reader using a dual-luciferase reporter assay system (Promega) according to the manufacturer’s instructions. For the FMDV luciferase replicon reporter assay, the replicon RNA was transcribed in vitro and transfected into hnRNP K-overexpressing or ZsGreen-transduced control cells, hnRNP K-knockdown cells, or cells transduced with sh-NC as a control. At 12 h posttransfection, the cells were lysed in passive lysis buffer, and the RLuc activity was measured using a Renilla luciferase assay system kit (Promega).
Expression of recombinant proteins and cleavage assay in vitro.
The expression, purification, and in vitro cleavage assay of proteins were performed as described previously (66). First, FMDV 3Cpro and hnRNP K were expressed in E. coli. The expressed His-FMDV 3Cpro protein was purified from clarified bacterial lysates by metal chelation chromatography. The expressed GST-hnRNP K protein was purified on a glutathione-agarose column. To examine hnRNP K cleavage in vitro, aliquots of recombinant His-FMDV 3Cpro and GST-hnRNP K were incubated in buffer A (50 mm Tris acetate, pH 8.5, 1 mm EDTA, 10% glycerol, 1 mm dithiothreitol [DTT]). After incubation at 30°C for 20 min, the reactions were terminated by the addition of 1× loading buffer and then subjected to Western blot analysis.
FRET-based assay for assessment of enzymatic characteristics.
Based on sites of FMDV 3Cpro-mediated hnRNP K cleavage, two fluorogenic peptide substrates, DABCYL-MAYEPQ↓GGSGYE-Edans and DABCYL-MAYEPA↓GGSGYE-Edans, which were derived from the hnRNP K-containing cleavage site and where DABCYL is N-[4-(4-dimethylamino)phenylazo]benzoic acid, were designed for artificial synthesis (GenScript). The donor and receptor fluorophores formed a quenching pair and showed fluorescence resonance energy transfer (FRET) within the peptide (79). The increases in fluorescence due to cleavage of the fluorogenic peptide substrates were monitored at 490 nm with excitation at 340 nm, using a fluorescence spectrophotometer (80). The expressed and purified His-FMDV 3Cpro was used in the FRET assays. All reactions were performed in a buffer containing 20 mM Tris-HCl (pH 7.4), 100 mM NaCl, and 5 mM DTT. The enzyme concentration used in the assay was 1 μM, and the substrate concentration was 10 μM.
Statistical analysis.
Data handling, analysis, and graphic representation were performed using Prism (version 5.0) software (GraphPad Software, San Diego, CA). Significant differences were determined using Student's t test. The data are presented as the means ± SD. P values of >0.05 were considered not significant.
ACKNOWLEDGMENTS
We thank Su Li (State Key Laboratory of Veterinary Biotechnology, Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Harbin, China) for critically reading and revising the manuscript and for providing very helpful comments and suggestions.
This work was supported by the National Natural Science Foundation of China (grants 31770173 and 31400138) and the National Key Research and Development Program of China (grant 2016YFD0501505).
We have no competing financial interests to declare.
REFERENCES
- 1.Sonenberg N, Dever TE. 2003. Eukaryotic translation initiation factors and regulators. Curr Opin Struct Biol 13:56–63. doi: 10.1016/s0959-440x(03)00009-5. [DOI] [PubMed] [Google Scholar]
- 2.Balvay L, Lopez Lastra M, Sargueil B, Darlix J-L, Ohlmann T. 2007. Translational control of retroviruses. Nat Rev Microbiol 5:128–140. doi: 10.1038/nrmicro1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martínez-Salas E, Pacheco A, Serrano P, Fernandez N. 2008. New insights into internal ribosome entry site elements relevant for viral gene expression. J Gen Virol 89:611–626. doi: 10.1099/vir.0.83426-0. [DOI] [PubMed] [Google Scholar]
- 4.Lukavsky PJ. 2009. Structure and function of HCV IRES domains. Virus Res 139:166–171. doi: 10.1016/j.virusres.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Filbin ME, Kieft JS. 2009. Toward a structural understanding of IRES RNA function. Curr Opin Struct Biol 19:267–276. doi: 10.1016/j.sbi.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baird SD, Turcotte M, Korneluk RG, Holcik M. 2006. Searching for IRES. RNA 12:1755–1785. doi: 10.1261/rna.157806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spriggs KA, Bushell M, Willis AE. 2010. Translational regulation of gene expression during conditions of cell stress. Mol Cell 40:228–237. doi: 10.1016/j.molcel.2010.09.028. [DOI] [PubMed] [Google Scholar]
- 8.Martínez-Salas E, Francisco-Velilla R, Fernandez-Chamorro J, Lozano G, Diaz-Toledano R. 2015. Picornavirus IRES elements: RNA structure and host protein interactions. Virus Res 206:62–73. doi: 10.1016/j.virusres.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 9.Jang SK, Kräusslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol 62:2636–2643. doi: 10.1128/JVI.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pelletier J, Sonenberg N. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 11.Komar AA, Hatzoglou M. 2015. Exploring internal ribosome entry sites as therapeutic targets. Front Oncol 5:233. doi: 10.3389/fonc.2015.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pilipenko EV, Pestova TV, Kolupaeva VG, Khitrina EV, Poperechnaya AN, Agol VI, Hellen CU. 2000. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev 14:2028–2045. [PMC free article] [PubMed] [Google Scholar]
- 13.Blyn LB, Towner JS, Semler BL, Ehrenfeld E. 1997. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J Virol 71:6243–6246. doi: 10.1128/JVI.71.8.6243-6246.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang PN, Lin JY, Locker N, Kung YA, Hung CT, Lin JY, Huang HI, Li ML, Shih SR. 2011. Far upstream element binding protein 1 binds the internal ribosomal entry site of enterovirus 71 and enhances viral translation and viral growth. Nucleic Acids Res 39:9633–9648. doi: 10.1093/nar/gkr682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin JY, Li ML, Shih SR. 2009. Far upstream element binding protein 2 interacts with enterovirus 71 internal ribosomal entry site and negatively regulates viral translation. Nucleic Acids Res 37:47–59. doi: 10.1093/nar/gkn901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pineiro D, Fernandez N, Ramajo J, Martinez-Salas E. 2013. Gemin5 promotes IRES interaction and translation control through its C-terminal region. Nucleic Acids Res 41:1017–1028. doi: 10.1093/nar/gks1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costa-Mattioli M, Svitkin Y, Sonenberg N. 2004. La autoantigen is necessary for optimal function of the poliovirus and hepatitis C virus internal ribosome entry site in vivo and in vitro. Mol Cell Biol 24:6861–6870. doi: 10.1128/MCB.24.15.6861-6870.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boussadia O, Niepmann M, Créancier L, Prats A-C, Dautry F, Jacquemin-Sablon H. 2003. Unr is required in vivo for efficient initiation of translation from the internal ribosome entry sites of both rhinovirus and poliovirus. J Virol 77:3353–3359. doi: 10.1128/jvi.77.6.3353-3359.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borman AM, Kirchweger R, Ziegler E, Rhoads RE, Skern T, Kean KM. 1997. elF4G and its proteolytic cleavage products: effect on initiation of protein synthesis from capped, uncapped, and IRES-containing mRNAs. RNA 3:186–196. [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang B, Seitz S, Kusov Y, Zell R, Gauss-Müller V. 2007. RNA interaction and cleavage of poly(C)-binding protein 2 by hepatitis A virus protease. Biochem Biophys Res Commun 364:725–730. doi: 10.1016/j.bbrc.2007.09.133. [DOI] [PubMed] [Google Scholar]
- 21.Perera R, Daijogo S, Walter BL, Nguyen JHC, Semler BL. 2007. Cellular protein modification by poliovirus: the two faces of poly(rC)-binding protein. J Virol 81:8919–8932. doi: 10.1128/JVI.01013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chase AJ, Daijogo S, Semler BL. 2014. Inhibition of poliovirus-induced cleavage of cellular protein PCBP2 reduces the levels of viral RNA replication. J Virol 88:3192–3201. doi: 10.1128/JVI.02503-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hung CT, Kung YA, Li ML, Brewer G, Lee KM, Liu ST, Shih SR. 2016. Additive promotion of viral internal ribosome entry site-mediated translation by far upstream element-binding protein 1 and an enterovirus 71-induced cleavage product. PLoS Pathog 12:e1005959. doi: 10.1371/journal.ppat.1005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pineiro D, Ramajo J, Bradrick SS, Martinez-Salas E. 2012. Gemin5 proteolysis reveals a novel motif to identify L protease targets. Nucleic Acids Res 40:4942–4953. doi: 10.1093/nar/gks172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen LL, Kung YA, Weng KF, Lin JY, Horng JT, Shih SR. 2013. Enterovirus 71 infection cleaves a negative regulator for viral internal ribosomal entry site-driven translation. J Virol 87:3828–3838. doi: 10.1128/JVI.02278-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grubman MJ, Baxt B. 2004. Foot-and-mouth disease. Clin Microbiol Rev 17:465–493. doi: 10.1128/cmr.17.2.465-493.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Domingo E, Baranowski E, Escarmís C, Sobrino F. 2002. Foot-and-mouth disease virus. Comp Immunol Microbiol Infect Dis 25:297–308. doi: 10.1016/s0147-9571(02)00027-9. [DOI] [PubMed] [Google Scholar]
- 28.Mason PW, Grubman MJ, Baxt B. 2003. Molecular basis of pathogenesis of FMDV. Virus Res 91:9–32. doi: 10.1016/s0168-1702(02)00257-5. [DOI] [PubMed] [Google Scholar]
- 29.Sáiz M, Núñez JI, Jimenez-Clavero MA, Baranowski E, Sobrino F. 2002. Foot-and-mouth disease virus: biology and prospects for disease control. Microbes Infect 4:1183–1192. doi: 10.1016/s1286-4579(02)01644-1. [DOI] [PubMed] [Google Scholar]
- 30.Sobrino F, Sáiz M, Jiménez-Clavero M, Núñez J, Rosas M, Baranowski E, Ley V. 2001. Foot-and-mouth disease virus: a long known virus, but a current threat. Vet Res 32:1–30. doi: 10.1051/vetres:2001106. [DOI] [PubMed] [Google Scholar]
- 31.Serrano P, Pulido MR, Sáiz M, Martínez-Salas E. 2006. The 3′ end of the foot-and-mouth disease virus genome establishes two distinct long-range RNA-RNA interactions with the 5′ end region. J Gen Virol 87:3013–3022. doi: 10.1099/vir.0.82059-0. [DOI] [PubMed] [Google Scholar]
- 32.Sun C, Liu M, Chang J, Yang D, Zhao B, Wang H, Zhou G, Weng C, Yu L. 2020. hnRNP L negatively regulates foot-and-mouth disease virus replication through inhibiting viral RNA synthesis by interacting with IRES in 5′ untranslated region. J Virol 94:e00282-20. doi: 10.1128/JVI.00282-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barboro P, Ferrari N, Balbi C. 2014. Emerging roles of heterogeneous nuclear ribonucleoprotein K (hnRNP K) in cancer progression. Cancer Lett 352:152–159. doi: 10.1016/j.canlet.2014.06.019. [DOI] [PubMed] [Google Scholar]
- 34.Luz N, Beck E. 1991. Interaction of a cellular 57-kilodalton protein with the internal translation initiation site of foot-and-mouth disease virus. J Virol 65:6486–6494. doi: 10.1128/JVI.65.12.6486-6494.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sean P, Nguyen JHC, Semler BL. 2009. Altered interactions between stem-loop IV within the 5′ noncoding region of coxsackievirus RNA and poly(rC) binding protein 2: effects on IRES-mediated translation and viral infectivity. Virology 389:45–58. doi: 10.1016/j.virol.2009.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paek KY, Kim CS, Park SM, Kim JH, Jang SK. 2008. RNA-binding protein hnRNP D modulates internal ribosome entry site-dependent translation of hepatitis C virus RNA. J Virol 82:12082–12093. doi: 10.1128/JVI.01405-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim JH, Paek KY, Ha SH, Cho S, Choi K, Kim CS, Ryu SH, Jang SK. 2004. A cellular RNA-binding protein enhances internal ribosomal entry site-dependent translation through an interaction downstream of the hepatitis C virus polyprotein initiation codon. Mol Cell Biol 24:7878–7890. doi: 10.1128/MCB.24.18.7878-7890.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin JY, Shih SR, Pan M, Li C, Lue CF, Stollar V, Li ML. 2009. hnRNP A1 interacts with the 5′ untranslated regions of enterovirus 71 and Sindbis virus RNA and is required for viral replication. J Virol 83:6106–6114. doi: 10.1128/JVI.02476-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siomi H, Matunis MJ, Michael WM, Dreyfuss G. 1993. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res 21:1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomonaga T, Levens D. 1995. Heterogeneous nuclear ribonucleoprotein K is a DNA-binding transactivator. J Biol Chem 270:4875–4881. doi: 10.1074/jbc.270.9.4875. [DOI] [PubMed] [Google Scholar]
- 41.Luz N, Beck E. 1990. A cellular 57 kDa protein binds to two regions of the internal translation initiation site of foot-and-mouth disease virus. FEBS Lett 269:311–314. doi: 10.1016/0014-5793(90)81182-n. [DOI] [PubMed] [Google Scholar]
- 42.Kolupaeva VG, Hellen CU, Shatsky IN. 1996. Structural analysis of the interaction of the pyrimidine tract-binding protein with the internal ribosomal entry site of encephalomyocarditis virus and foot-and-mouth disease virus RNAs. RNA 2:1199–1212. [PMC free article] [PubMed] [Google Scholar]
- 43.Niepmann M, Petersen A, Meyer K, Beck E. 1997. Functional involvement of polypyrimidine tract-binding protein in translation initiation complexes with the internal ribosome entry site of foot-and-mouth disease virus. J Virol 71:8330–8339. doi: 10.1128/JVI.71.11.8330-8339.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piccone ME, Zellner M, Kumosinski TF, Mason PW, Grubman MJ. 1995. Identification of the active-site residues of the L proteinase of foot-and-mouth disease virus. J Virol 69:4950–4956. doi: 10.1128/JVI.69.8.4950-4956.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grubman MJ, Zellner M, Bablanian G, Mason PW, Piccone ME. 1995. Identification of the active-site residues of the 3C proteinase of foot-and-mouth disease virus. Virology 213:581–589. doi: 10.1006/viro.1995.0030. [DOI] [PubMed] [Google Scholar]
- 46.Birtley JR, Knox SR, Jaulent AM, Brick P, Leatherbarrow RJ, Curry S. 2005. Crystal structure of foot-and-mouth disease virus 3C protease. New insights into catalytic mechanism and cleavage specificity. J Biol Chem 280:11520–11527. doi: 10.1074/jbc.M413254200. [DOI] [PubMed] [Google Scholar]
- 47.Bomsztyk K, Denisenko O, Ostrowski J. 2004. HnRNP K: one protein multiple processes. Bioessays 26:629–638. doi: 10.1002/bies.20048. [DOI] [PubMed] [Google Scholar]
- 48.Michelotti EF, Michelotti GA, Aronsohn AI, Levens D. 1996. Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol Cell Biol 16:2350–2360. doi: 10.1128/mcb.16.5.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Du Q, Melnikova IN, Gardner PD. 1998. Differential effects of heterogeneous nuclear ribonucleoprotein K on Sp1- and Sp3-mediated transcriptional activation of a neuronal nicotinic acetylcholine receptor promoter. J Biol Chem 273:19877–19883. doi: 10.1074/jbc.273.31.19877. [DOI] [PubMed] [Google Scholar]
- 50.Lin JY, Li ML, Huang PN, Chien KY, Horng JT, Shih SR. 2008. Heterogeneous nuclear ribonuclear protein K interacts with the enterovirus 71 5′ untranslated region and participates in virus replication. J Gen Virol 89:2540–2549. doi: 10.1099/vir.0.2008/003673-0. [DOI] [PubMed] [Google Scholar]
- 51.Andreev DE, Fernandez-Miragall O, Ramajo J, Dmitriev SE, Terenin IM, Martinez-Salas E, Shatsky IN. 2007. Differential factor requirement to assemble translation initiation complexes at the alternative start codons of foot-and-mouth disease virus RNA. RNA 13:1366–1374. doi: 10.1261/rna.469707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fernandez N, Fernandez-Miragall O, Ramajo J, Garcia-Sacristan A, Bellora N, Eyras E, Briones C, Martinez-Salas E. 2011. Structural basis for the biological relevance of the invariant apical stem in IRES-mediated translation. Nucleic Acids Res 39:8572–8585. doi: 10.1093/nar/gkr560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, Weng Z, Moore MJ. 2012. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell 151:750–764. doi: 10.1016/j.cell.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Song Y, Tzima E, Ochs K, Bassili G, Trusheim H, Linder M, Preissner KT, Niepmann M. 2005. Evidence for an RNA chaperone function of polypyrimidine tract-binding protein in picornavirus translation. RNA 11:1809–1824. doi: 10.1261/rna.7430405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim JH, Hahm B, Kim YK, Choi M, Jang SK. 2000. Protein-protein interaction among hnRNP shuttling between nucleus and cytoplasm. J Mol Biol 298:395–405. doi: 10.1006/jmbi.2000.3687. [DOI] [PubMed] [Google Scholar]
- 56.Simon V, Bloch N, Landau NR. 2015. Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nat Immunol 16:546–553. doi: 10.1038/ni.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang S, Chi X, Wei H, Chen Y, Chen Z, Huang S, Chen J-L. 2014. Influenza A virus-induced degradation of eukaryotic translation initiation factor 4B contributes to viral replication by suppressing IFITM3 protein expression. J Virol 88:8375–8385. doi: 10.1128/JVI.00126-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Z, Ning S, Su X, Liu X, Wang H, Liu Y, Zheng W, Zheng B, Yu X-F, Zhang W. 2018. Enterovirus 71 antagonizes the inhibition of the host intrinsic antiviral factor A3G. Nucleic Acids Res 46:11514–11527. doi: 10.1093/nar/gky840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Devaney MA, Vakharia VN, Lloyd RE, Ehrenfeld E, Grubman MJ. 1988. Leader protein of foot-and-mouth disease virus is required for cleavage of the p220 component of the cap-binding protein complex. J Virol 62:4407–4409. doi: 10.1128/JVI.62.11.4407-4409.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Los Santos T, Diaz-San Segundo F, Grubman MJ. 2007. Degradation of nuclear factor kappa B during foot-and-mouth disease virus infection. J Virol 81:12803–12815. doi: 10.1128/JVI.01467-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Falk MM, Grigera PR, Bergmann IE, Zibert A, Multhaup G, Beck E. 1990. Foot-and-mouth disease virus protease 3C induces specific proteolytic cleavage of host cell histone H3. J Virol 64:748–756. doi: 10.1128/JVI.64.2.748-756.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang D, Fang L, Li K, Zhong H, Fan J, Ouyang C, Zhang H, Duan E, Luo R, Zhang Z, Liu X, Chen H, Xiao S. 2012. Foot-and-mouth disease virus 3C protease cleaves NEMO to impair innate immune signaling. J Virol 86:9311–9322. doi: 10.1128/JVI.00722-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Back SH, Kim YK, Kim WJ, Cho S, Oh HR, Kim J-E, Jang SK. 2002. Translation of polioviral mRNA is inhibited by cleavage of polypyrimidine tract-binding proteins executed by polioviral 3C(pro). J Virol 76:2529–2542. doi: 10.1128/jvi.76.5.2529-2542.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jagdeo JM, Dufour A, Fung G, Luo H, Kleifeld O, Overall CM, Jan E. 2015. Heterogeneous nuclear ribonucleoprotein M facilitates enterovirus infection. J Virol 89:7064–7078. doi: 10.1128/JVI.02977-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cathcart AL, Rozovics JM, Semler BL. 2013. Cellular mRNA decay protein AUF1 negatively regulates enterovirus and human rhinovirus infections. J Virol 87:10423–10434. doi: 10.1128/JVI.01049-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jagdeo JM, Dufour A, Klein T, Solis N, Kleifeld O, Kizhakkedathu J, Luo H, Overall CM, Jan E. 2018. N-Terminomics TAILS identifies host cell substrates of poliovirus and coxsackievirus B3 3C proteinases that modulate virus infection. J Virol 92:e02211-17. doi: 10.1128/JVI.02211-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee KM, Chen CJ, Shih SR. 2017. Regulation mechanisms of viral IRES-driven translation. Trends Microbiol 25:546–561. doi: 10.1016/j.tim.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 68.Backe PH, Messias AC, Ravelli RBG, Sattler M, Cusack S. 2005. X-ray crystallographic and NMR studies of the third KH domain of hnRNP K in complex with single-stranded nucleic acids. Structure 13:1055–1067. doi: 10.1016/j.str.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 69.Cao P, Luo W-W, Li C, Tong Z, Zheng Z-Q, Zhou L, Xiong Y, Li S. 2019. The heterogeneous nuclear ribonucleoprotein hnRNPM inhibits RNA virus-triggered innate immunity by antagonizing RNA sensing of RIG-I-like receptors. PLoS Pathog 15:e1007983. doi: 10.1371/journal.ppat.1007983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Han SP, Tang YH, Smith R. 2010. Functional diversity of the hnRNPs: past, present and perspectives. Biochem J 430:379–392. doi: 10.1042/BJ20100396. [DOI] [PubMed] [Google Scholar]
- 71.Ministry of Science and Technology. Guidelines for the care of laboratory animals. Ministry of Science and Technology, Beijing, People’s Republic of China. [Google Scholar]
- 72.National Laboratory Animal Standardization Technical Committee. 2010. Laboratory animal-requirements of environment and housing facilities. National Laboratory Animal Standardization Technical Committee, Beijing, People’s Republic of China. [Google Scholar]
- 73.Sun C, Yang D, Gao R, Liang T, Wang H, Zhou G, Yu L. 2016. Modification of the internal ribosome entry site element impairs the growth of foot-and-mouth disease virus in porcine-derived cells. J Gen Virol 97:901–911. doi: 10.1099/jgv.0.000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zeng J, Wang H, Xie X, Li C, Zhou G, Yang D, Yu L. 2014. Ribavirin-resistant variants of foot-and-mouth disease virus: the effect of restricted quasispecies diversity on viral virulence. J Virol 88:4008–4020. doi: 10.1128/JVI.03594-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chien H-L, Liao C-L, Lin Y-L. 2011. FUSE binding protein 1 interacts with untranslated regions of Japanese encephalitis virus RNA and negatively regulates viral replication. J Virol 85:4698–4706. doi: 10.1128/JVI.01950-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harris D, Zhang Z, Chaubey B, Pandey VN. 2006. Identification of cellular factors associated with the 3′-nontranslated region of the hepatitis C virus genome. Mol Cell Proteomics 5:1006–1018. doi: 10.1074/mcp.M500429-MCP200. [DOI] [PubMed] [Google Scholar]
- 77.Yu Y, Wang H, Zhao L, Zhang C, Jiang Z, Yu L. 2011. Fine mapping of a foot-and-mouth disease virus epitope recognized by serotype-independent monoclonal antibody 4B2. J Microbiol 49:94–101. doi: 10.1007/s12275-011-0134-1. [DOI] [PubMed] [Google Scholar]
- 78.Reid SM, Ferris NP, Hutchings GH, Zhang Z, Belsham GJ, Alexandersen S. 2002. Detection of all seven serotypes of foot-and-mouth disease virus by real-time, fluorogenic reverse transcription polymerase chain reaction assay. J Virol Methods 105:67–80. doi: 10.1016/s0166-0934(02)00081-2. [DOI] [PubMed] [Google Scholar]
- 79.Matayoshi ED, Wang GT, Krafft GA, Erickson J. 1990. Novel fluorogenic substrates for assaying retroviral proteases by resonance energy transfer. Science 247:954–958. doi: 10.1126/science.2106161. [DOI] [PubMed] [Google Scholar]
- 80.Tian X, Lu G, Gao F, Peng H, Feng Y, Ma G, Bartlam M, Tian K, Yan J, Hilgenfeld R, Gao GF. 2009. Structure and cleavage specificity of the chymotrypsin-like serine protease (3CLSP/nsp4) of porcine reproductive and respiratory syndrome virus (PRRSV). J Mol Biol 392:977–993. doi: 10.1016/j.jmb.2009.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]