Emerging coronaviruses, including SARS-CoV-2 and porcine CoVs, can infect enterocytes, cause diarrhea, and be shed in the feces. New approaches are needed to understand enteric pathogenesis and to develop vaccines and therapeutics to prevent the spread of these viruses. Here, we exploited a reverse genetic system for an enteric CoV, porcine epidemic diarrhea virus (PEDV), and outline an approach of genetically inactivating highly conserved viral factors known to limit the host innate immune response to infection. Our report reveals that generating PEDV with inactive versions of three viral interferon antagonists, nonstructural proteins 1, 15, and 16, results in a highly attenuated virus that does not cause diarrhea in animals and elicits a neutralizing antibody response in virus-infected animals. This strategy may be useful for generating live attenuated vaccine candidates that prevent disease and fecal spread of enteric CoVs, including SARS-CoV-2.
KEYWORDS: pathogenesis, porcine epidemic diarrhea virus, SARS-CoV-2, coronavirus, endoribonuclease, interferon antagonist, nsp1, nsp15, nsp16, vaccine
ABSTRACT
Coronaviruses (CoVs) have repeatedly emerged from wildlife hosts and infected humans and livestock animals to cause epidemics with significant morbidity and mortality. CoVs infect various organs, including respiratory and enteric systems, as exemplified by newly emerged severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The constellation of viral factors that contribute to developing enteric disease remains elusive. Here, we investigated CoV interferon antagonists for their contribution to enteric pathogenesis. Using an infectious clone of an enteric CoV, porcine epidemic diarrhea virus (icPEDV), we generated viruses with inactive versions of interferon antagonist nonstructural protein 1 (nsp1), nsp15, and nsp16 individually or combined into one virus designated icPEDV-mut4. Interferon-responsive PK1 cells were infected with these viruses and produced higher levels of interferon responses than were seen with wild-type icPEDV infection. icPEDV-mut4 elicited robust interferon responses and was severely impaired for replication in PK1 cells. To evaluate viral pathogenesis, piglets were infected with either icPEDV or icPEDV-mut4. While the icPEDV-infected piglets exhibited clinical disease, the icPEDV-mut4-infected piglets showed no clinical symptoms and exhibited normal intestinal pathology at day 2 postinfection. icPEDV-mut4 replicated in the intestinal tract, as revealed by detection of viral RNA in fecal swabs, with sequence analysis documenting genetic stability of the input strain. Importantly, icPEDV-mut4 infection elicited IgG and neutralizing antibody responses to PEDV. These results identify nsp1, nsp15, and nsp16 as virulence factors that contribute to the development of PEDV-induced diarrhea in swine. Inactivation of these CoV interferon antagonists is a rational approach for generating candidate vaccines to prevent disease and spread of enteric CoVs, including SARS-CoV-2.
IMPORTANCE Emerging coronaviruses, including SARS-CoV-2 and porcine CoVs, can infect enterocytes, cause diarrhea, and be shed in the feces. New approaches are needed to understand enteric pathogenesis and to develop vaccines and therapeutics to prevent the spread of these viruses. Here, we exploited a reverse genetic system for an enteric CoV, porcine epidemic diarrhea virus (PEDV), and outline an approach of genetically inactivating highly conserved viral factors known to limit the host innate immune response to infection. Our report reveals that generating PEDV with inactive versions of three viral interferon antagonists, nonstructural proteins 1, 15, and 16, results in a highly attenuated virus that does not cause diarrhea in animals and elicits a neutralizing antibody response in virus-infected animals. This strategy may be useful for generating live attenuated vaccine candidates that prevent disease and fecal spread of enteric CoVs, including SARS-CoV-2.
INTRODUCTION
Coronaviruses (CoVs) have repeatedly emerged from wildlife hosts and infected humans and livestock animals to cause epidemics with significant morbidity and mortality. The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 2019 and the rapid, global spread of infection in humans highlight the need for developing therapeutics and vaccines to limit coronavirus pandemics (1–3). By leveraging knowledge from studies of model systems of coronavirus infection and pathogenesis, we can develop and apply strategies that limit replication and spread of coronavirus infections in humans and other animals. Here, we describe the use of an enteric coronavirus that infects swine and our approach of inactivating highly conserved regions of this coronavirus with the goal of limiting virus replication in infected animals and eliciting neutralizing antibody responses.
Porcine epidemic diarrhea virus (PEDV) belongs to the family Coronaviridae in the order Nidovirales. The members of this family of viruses all have large (∼30-kb), positive-sense RNA genomes encapsidated by nucleocapsid (N) protein and enveloped by host membranes modified by viral structural proteins designated envelope (E), membrane (M), and spike (S). The spike protein gives the virus the typical crown-like appearance when visualized by electron microscopy (2). The spike protein engages the host receptor and mediates fusion of the viral and host membranes, allowing entry of the viral genomic RNA into the cytoplasm of the cell. CoV genomic RNA is translated to generate a large polyprotein that is processed into 15 or 16 nonstructural proteins (nsps) that assemble together to make the double membrane vesicle (DMV) replication complex (4, 5). These nsps were initially proposed to function exclusively in the replication and transcription of viral RNA (vRNA). However, recent studies revealed that many of these proteins are multifunctional and play important roles in limiting the host response to virus infection by acting as antagonists of the host type I and type III interferon (IFN) responses (reviewed in reference 6). Type I IFNs (alpha IFN [IFN-α] and IFN-β) and type III IFNs (IFN-λ) work in an autocrine fashion and in a paracrine fashion to induce an antiviral state by expressing interferon-stimulated genes (ISGs) that limit replication of coronaviruses (7). Studies performed by Channappanavar and coworkers documented that this virus-mediated delay in the host interferon response during infection contributes to more-severe disease (8, 9). Understanding the mechanisms used by viruses to delay the host innate immune response and inactivating these viral mechanisms may provide new targets for antiviral therapeutics and new approaches for generating vaccines.
We and others are developing and testing strategies of inactivating coronavirus interferon antagonists to reduce viral pathogenesis and generate candidate live attenuated virus vaccines (10–14). Menachery and coworkers showed that inactivating the highly conserved CoV 2′-O-methyltransferase (MTase) enzyme in nsp16 results in virus that activates the host interferon response, is attenuated in animals, and elicits a protective immune response to Middle East respiratory syndrome CoV (MERS-CoV) (12). Hou and coworkers showed that inactivating nsp16 in combination with a deletion in the spike glycoprotein in the PEDV-PC22A strain generated a virus that exhibited reduced pathogenesis but that still caused diarrhea in piglets (14). We wanted to extend these studies and determine if inactivating multiple antagonists would affect replication and pathogenicity of PEDV. Currently, it is unclear if inactivating multiple CoV antagonists in the context of an enteric infection would impact the disease and the host immune response to infection.
We also wanted to evaluate the role of a highly conserved replicase interferon antagonist, the endoribonuclease (EndoU) contained within nsp15. The rationale for investigating EndoU as a virulence factor stems from promising results obtained using the murine coronavirus, mouse hepatitis virus (MHV). We generated MHV with a mutation in a catalytic histidine residue of EndoU and found that this virus replicated as well as wild-type (WT) virus in interferon-nonresponsive cells, revealing that EndoU activity was not required for CoV replication. We found that EndoU-mutant murine coronavirus elicited a robust type I interferon response in interferon-responsive macrophages, caused no clinical disease, and elicited a protective immune response in mice (10, 15). These results documented the critical role of EndoU activity in the pathogenesis of the murine coronavirus. To determine if EndoU activity played a role as a virulence factor in other CoVs, we initiated studies using PEDV. We found that inactivating EndoU/nsp15 in PEDV was sufficient to elicit both type I and type III interferon responses from infected cells and to reduce clinical disease in infect piglets (11). However, the inactivation of this single interferon antagonist was not sufficient to fully attenuate PEDV, as the infected piglets still exhibited some diarrhea (11). In addition, the PEDV nsp1 (F44A) mutation was shown to reduce the ability of nsp1 to antagonize innate immune responses in an overexpression system in cultured cells (16, 17), but the effect of this mutation in the context of virus replication was unknown. We reasoned that inactivation of multiple interferon antagonists should be evaluated as an approach for attenuating enteric CoVs.
Here, we evaluate the role of three CoV interferon antagonists, nsp1, nsp15, and nsp16, as virulence factors in the replication and pathogenesis of PEDV. We used reverse genetics to generate isogenic viruses that contained mutations that inactivated residues known to be essential for interferon antagonism. We evaluated these mutant viruses for their ability to elicit type I and type III interferon responses in cell culture, and the most promising candidate was selected for an animal study to evaluate clinical signs of disease and neutralizing antibody response in piglets.
RESULTS
Generating interferon antagonist mutant strains of PEDV-Colorado.
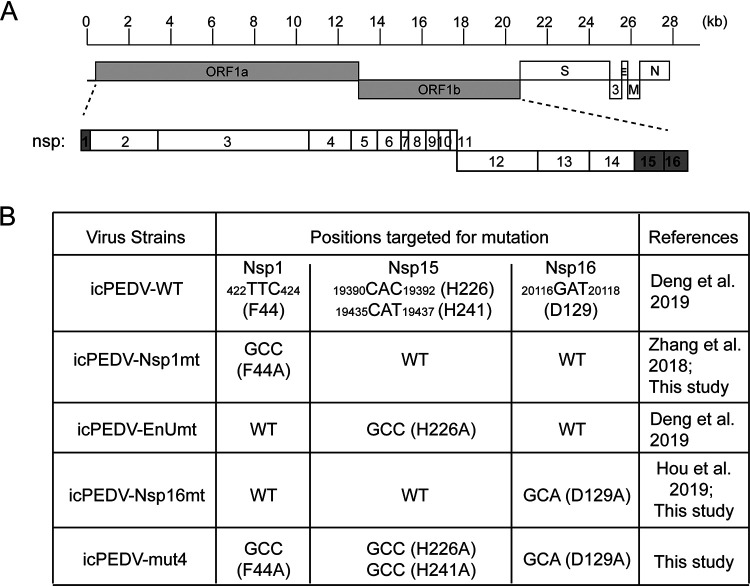
We previously documented our use of reverse genetics to generate an infectious clone of the wild-type PEDV-Colorado strain and a strain containing a mutation in the catalytic histidine residue (H226A) of endoribonuclease/nsp15 that inactivates enzyme activity. These viruses were designated icPEDV-WT and icPEDV-EnUmt (11). We found that icPEDV-EnUmt generated an earlier and more robust interferon response in cell culture than the wild-type virus and was attenuated in piglets. Using the same strategy of incorporating mutations to inactivate IFN antagonists, we generated three new viruses that contain mutations in nsp1 or nsp15 or nsp16 (Fig. 1). We incorporated the nsp1 F44A mutation that had previously been documented to inactivate IFN antagonism (16) and designated this virus icPEDV-Nsp1mt. Regarding the nsp16 mutation, Hou and coworkers working with porcine CoV 22A found that a single alanine substitution (underlined) in the KDKE catalytic site of the 2′-O-methyltransferase resulted in a replication-competent strain of PEDV, whereas substitution of all four catalytic site residues resulted in lower levels of PEDV replication in Vero cells (14). Here, we incorporated the single alanine substitution in the catalytic D129 site of nsp16 in the PEDV-Colorado strain, designated icPEDV-Nsp16mt. Finally, we incorporated the mutations in Nsp1 and Nsp16 with mutations that inactivate both catalytic histidine residues (H226A and H241A) of nsp15 into one virus and designated that virus icPEDV-mut4. We abbreviate the designations for these icPEDVs in the remaining figures in this paper as follows: WT, Nsp1mt, EnUmt, Nsp16mt, and mut4.
FIG 1.
Schematic diagram of the genome organization of PEDV and positions of nucleotide changes made in icPEDV mutant viruses. (A) Schematic diagram of the genome organization of PEDV, with nonstructural proteins 1, 15, and 16 highlighted in gray. (B) Location of nucleotide sequence targeted for mutagenesis. The resulting change in the amino acid sequence is listed. For reference, Zhang et al. reported that the mutation F44A in Nsp1 reduced interferon antagonism in an overexpression system (16), and Hou et al. reported that mutation of Nsp16 in PEDV strain PC22A was attenuating in piglets (14). (See also Deng et al. [11].)
Evaluating replication of IFN antagonist mutant viruses in Vero cells and PK1 cells.
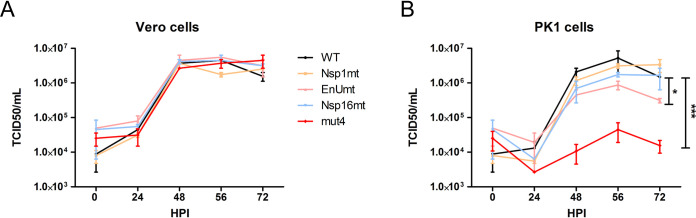
We previously reported that inactivating EndoU by mutating catalytic histidine residue H226A had no effect on virus replication in interferon-nonresponsive Vero cells but that replication was reduced in interferon-responsive PK1 cells (11). To determine whether the mutation(s) that we introduced had any effect on virus replication, we compared the replication kinetics of the viruses in Vero cells over 72 h. As expected, we found that all four of the mutant viruses replicated efficiently in Vero cells, with kinetics that were similar to those seen with WT PEDV (Fig. 2A). These results are consistent with the concept that interferon antagonists are not required for virus replication in interferon-nonresponsive cells. In contrast to the results in Vero cells, we found that the levels of replication of EnUmt and mut4 were significantly reduced in PK1 cells, with mut4 replication reduced by 1,000-fold compared to WT PEDV (Fig. 2B). Since PK1 cells are interferon responsive, we hypothesized that the mutant viruses were activating the expression of type I and type III interferons and interferon-responsive genes (ISGs), which limited the production of infectious virus progeny over time.
FIG 2.
Evaluating the growth kinetics of icPEDV wild-type and nsp-mutant viruses in Vero cells and PK1 cells. (A) Vero cells or (B) PK1 cells were infected with designated virus at a dose of 0.1 TCID50 per cell. Cell culture supernatants were collected at the indicated hours postinfection (HPI). The titer of infectious virus in the supernatant was determined in Vero cells using a TCID50 assay performed in triplicate, and the results show the means ± standard deviations (SD). Data sets at the same time point were analyzed with an unpaired t test. Asterisks indicate that the results of between-group comparisons of the data sets obtained at 48, 56, and 72 HPI were statistically significant as follows: *, P < 0.05; ***, P < 0.001.
Evaluating the kinetics of the IFN response in PEDV-infected PK1 cells.
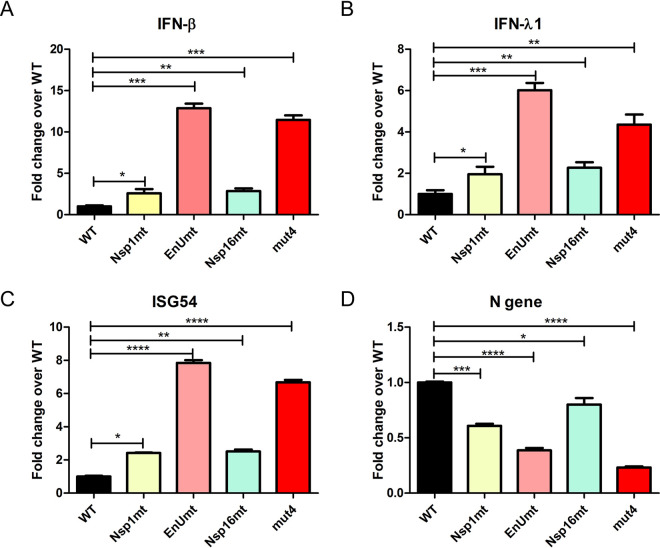
We showed that PK1 cells respond to PEDV infection by activating the transcription of interferons (type I IFN-β and type III IFN-λ) and interferon-stimulated genes, such as ISG54 (11). To determine if inactivating multiple CoV interferon antagonists affected the IFN response to infection, we infected PK1 cells with the designated virus, incubated the infected cells for 24 h, harvested RNA from the cells, and performed real-time reverse transcription-quantitative PCR (RT-qPCR) to evaluate the levels of expression of IFNs and ISG54 and the viral nucleocapsid (N) RNA. We compared the relative levels of expression of the target mRNA and of porcine GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA, assigned the level of expression in PEDV-WT-infected cells a value of 1, and determined the fold change in gene expression detected in the mutant virus-infected cells. We found that the mutant virus-infected cells all had higher levels of IFNs and ISG54 than the icPEDV-WT-infected cells (Fig. 3A to C). Cells infected with icPEDV-EnUmt and the icPEDV-mut4 had the highest levels of IFN and ISG54 expression, with greater than 5-fold change compared to wild-type PEDV infected cells. We also evaluated viral replication by monitoring levels of PEDV N gene mRNA (Fig. 3D). We found that viral mRNA levels were significantly reduced in the mutant virus-infected cells, with icPEDV-mut4 being the most impaired. We noted that there was a correlation of high levels of IFN expression with reduced levels of viral RNA expression, consistent with the concept that IFN expression in PK1 cells limits virus replication. Both icPEDV-EnUmt and icPEDV-mut4 activated significantly higher levels of IFNs than the wild-type virus, and significantly lower levels of infectious particles were produced from the infected cells (Fig. 2B). Since nsp1, nsp15, and nsp16 possess distinct functions in evading the host antiviral responses, it is possible that multiple mechanisms contribute to the overall attenuation of virus replication in PK1 cells.
FIG 3.
Evaluating the interferon responses to icPEDV infection in PK1 cells. PK1 cells were mock infected or infected with the indicated strain of PEDV at a dose of 0.1 TCID50 per cell. At 24 HPI, cells were lysed to collect total RNA for cDNA synthesis, and quantitative PCR was used to measure the relative levels of expression of the indicated mRNAs for IFN-β (A), IFN-γ1 (B), ISG54 (C), and the N gene (D). The level of gene expression in the wild-type-virus-infected samples was assigned a value of 1, and the fold change in expression is indicated relative to the PEDV wild-type-virus-infected sample. Values are presented as means ± SD and were analyzed by unpaired t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Evaluating the replication and pathogenesis of icPEDV-mut4 in piglets.
We previously reported that inoculation of piglets with icPEDV-WT resulted in significant disease and mortality in piglets and that inoculation with icPEDV-EnUmt was associated with lower levels of virus shedding and no mortality (11). Our in vitro studies indicated that icPEDV-mut4 induced levels of IFNs at 24 h postinfection similar to those induced by icPEDV-EnUmt but that the titer of infectious icPEDV-mut4 was significantly reduced in PK1 cells (Fig. 2B). Therefore, we wanted to evaluate the pathogenesis of icPEDV-mut4 in piglets and compare the results to those seen with piglets inoculated with icPEDV-WT. For these studies, 34 piglets from 3 sows were randomized into 3 groups (Fig. 4A). Piglets were orally inoculated with the designated virus (500 50% tissue culture infective doses [TCID50] per piglet) and monitored daily for signs of clinical disease. Fecal swabs were obtained from each piglet and PEDV RNA levels were determined by RT-qPCR. We found that the icPEDV-WT-infected piglets all had signs of diarrhea (ranging from soft stool to watery diarrhea), as documented in the clinical score (Fig. 4B). In contrast, similarly to the mock-infected animals, the icPEDV-mut4-infected animals showed no clinical signs of disease. By evaluating the shedding of viral RNA in the fecal swab, we found that there was a significant reduction in the level of shedding in the icPEDV-mut4-infected animals compared to the icPEDV-WT-infected animals (Fig. 4C). We detected an average of 1010 copies of PEDV RNA/ml in the fecal swabs from icPEDV-WT-infected animals at day 2 postinfection compared to 104 copies of PEDV RNA/ml in 1 of 11 piglets infected with icPEDV-mut4. The levels of virus shedding were consistently 100 to 1,000 times lower in the fecal swabs of the icPEDV-mut4-infected animals than in those of the icPEDV-WT-infected animals. The level of PEDV RNA in the fecal swab was below the limit of detection by day 18 postinfection in the icPEDV-mut4-infected animals.
FIG 4.
Experimental design and outcomes after PEDV infection. (A) A total of 34 piglets from 3 sows were randomly grouped to 3 groups. These piglets were either mock infected or infected with icPEDV wild-type or icPEDV-mut4 virus. Blood samples were drawn prior to infection and also at 21 days after infection. (B) Clinical symptoms were evaluated and scored daily. The score is based on the status of feces and the overall appearance of each animal. Individual values and geometric means are presented. (C) Evaluating shedding of viral RNA. RNA was isolated from rectal swab samples and subjected to quantitative PCR to determine the number of genomic RNA copies per milliliter of sample. Individual values and geometric means are presented and were analyzed with unpaired t tests used for comparisons between the groups at the same time point. *, P < 0.05.
icPEDV-mut4 sequence is maintained during virus replication in piglets.
To determine if the icPEDV-mut4 sequence was maintained during virus replication in the piglets, we generated amplicons that encompassed the 3 regions of interest with specific primers (Table 1) and compared the sequence to that of icPEDV-WT. The RNA was obtained from the fecal swabs taken on the last day that each of the 11 piglets was PCR positive, which ranged from day 8 to day 12 postinfection. Amplicons for nsp1, nsp15, and nsp16 were obtained from 7/11 fecal samples, while 4 samples did not produce a product in testing under the same conditions, suggesting that viral RNA was not present at sufficient quantities for efficient amplification of the replicase region. The amplicons were subjected to sequence analysis. We found that all four of the mutations engineered into the icPEDV-mut4 genome were maintained, even after 12 days of replication in the piglet. Data representative of results of this analysis of amplicons sequenced from one fecal sample are shown in Fig. 5. These results indicate that the mutations introduced into nsp1, nsp15, and nsp16 were maintained during virus replication in infected animals.
TABLE 1.
PCR primers
| Method | Targeted site | Primer sequence (5′–3′) |
|
|---|---|---|---|
| Forward | Reverse | ||
| Mutagenesis | Nsp1-F44A | TTTATGCAATGCCGTgcCGTGTCCTTCGATCT | AGATCGAAGGACACGgcACGGCATTGCATAAA |
| Nsp15-H226A | GATTACGGCTTTGAGgcCGTTGTGTATGGTGAT | ATCACCATACACAACGgcCTCAAAGCCGTAATC | |
| Nsp15-H241A | ACCCTTGGTGGTTTGgccCTACTAATTTCGCAG | CTGCGAAATTAGTAGggcCAAACCACCAAGGGT | |
| Nsp16-D129A | GATTTAGTTATATCTGcaATGTATGATGGTAAG | CTTACCATCATACATtgCAGATATAACTAAATC | |
| Mutation stability RT-PCR | Nsp1: PEDV-210F/-627R | AGACAAACAGCCTTCCTCCG | ATGTTACCACCACGACGACC |
| Nsp15: PEDV-19071F/-19731R | CGACTTTGAGGGTGACGTCT | CCACAACATCCACCTCCACA | |
| Nsp15: PEDV-18319F/-19712R | CCAGAATTTTCTGTGGTCTGTCG | GTCTGGAGTTTATGATCCTTACACCAC | |
| Nsp16: PEDV-19923F/-19712R | TGCGTGTGCTACATCTTGGT | GTCAATCACAGCGCCACTTG | |
| Mutation sequencing | Nsp1: PEDV-235F/-549R | GTCTGGGGGTTGTGTGGATA | CAAATGTTTTTGGGGCGGCT |
| Nsp15: PEDV-19218F/-19637R | TGGTGTCCCAGTTAACACACA | TCGTCAAGCAGGAGATCCAT | |
| Nsp16: PEDV-20028F/-20345R | GTGTGGATTACGTTAGCGATGC | AGAATGCCTCTGACGATGACG | |
Underlined sequence characters represent the codon of mutated amino acid; lowercase sequence characters represent the mutated nucleotides.
FIG 5.
Mutation stability in icPEDV-mut4-infected piglets late in infection. The nucleotide sequences of regions of icPEDV-WT were compared to the chromatograms and sequences generated for the icPEDV-mut4-infected piglets for each of the four mutations (nsp1-F44A; nsp15-H226A, nsp15-H226A, and nsp16-D129A). The sequence shown was derived from one of seven piglets with identical results.
Pathological evaluation of icPEDV-WT and icPEDV-mut4 replication in intestinal sites at day 2 postinfection.
Histopathological examination of the jejunum section (Fig. 6A) and ileum section (Fig. 6B) from the mock-infected control piglets and the icPEDV-mut4-infected piglets revealed no significant pathological lesions in either group. Intestinal villi were tall and lined by tall columnar epithelial cells. In contrast, piglets infected with icPEDV-WT exhibited classical microscopic lesions within sections of small intestine consisting of villus atrophy with degeneration and necrosis of villus tip enterocytes. Villus tip enterocytes were swollen, were rounded to flattened in appearance, and were often separated from the underlying lamina. The superficial lamina was congested and contained a few neutrophils. Immunohistochemistry (IHC) was utilized to detect PEDV nucleocapsid protein in all groups (Fig. 6, lower panels). Viral antigen was not detected by IHC within intestinal sections of the mock-infected control piglets or the icPEDV-mut4-infected piglets. In contrast, viral antigen was detected within mucosal epithelial cells of the icPEDV-WT-infected piglets. These results document the dramatic differences in pathology seen at day 2 postinfection, which is a time point typically used to document the rapid and robust infection and spread of PEDV in the intestinal epithelial cells of piglets. Overall, we report delayed and reduced fecal shedding and a remarkable reduction in tissue pathology seen after infection with icPEDV-mut4 compared to what is typically revealed at day 2 postinfection in piglets challenged with icPEDV-WT.
FIG 6.
Histology and IHC staining of uninfected control, icPEDV-WT-infected, and icPEDV-mut4-infected piglet jejunum and ileum. Piglets were euthanized at day 2 postinfection. Images show representative histological slides of (A) jejunum specimens (×10) and (B) ileum specimens (×4) showing H&E staining and immunohistochemistry (IHC) staining using mouse anti-PEDV-N antibody.
Infection with icPEDV-mut4 elicits virus-specific IgG serum antibodies with neutralizing activity.
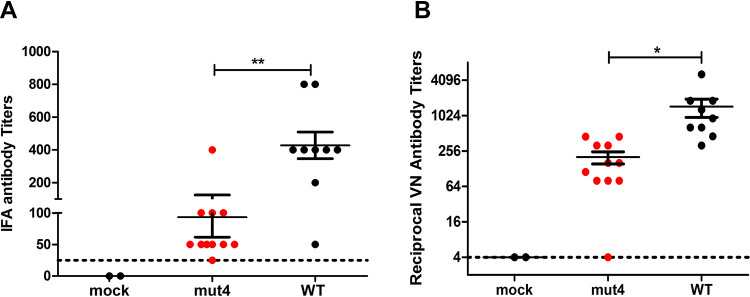
To determine if the icPEDV-mut4-infected piglets generated an antibody response to the infection, we obtained serum samples at day 21 postinfection and used an immunofluorescence assay (IFA) to evaluate the level of IgG in response to PEDV (18). We found that icPEDV-mut4-infected animals had significantly higher levels of virus-specific IgG than the mock-infected animals (Fig. 7A). As expected, we detected the highest titers of PEDV-specific IgG in the WT-infected piglets that exhibited clinical symptoms of disease. To determine the titers of neutralizing antibodies, we performed a virus-neutralizing (VN) assay as described previously (14, 19). Importantly, the icPEDV-mut4-infected animals generated a robust neutralizing antibody response, with an average titer of 1:256, whereas the VN titers of control sera were undetectable (Fig. 7B). The positive-control serum samples collected from the icPEDV-WT-infected piglets contained high titers (average, 1:1,024 dilution) of neutralizing antibody. Taken together, these results demonstrate that icPEDV-mut4 replicates in enterocytes and elicits a neutralizing antibody response in the piglets without causing clinical signs of disease.
FIG 7.
Virus-specific IgG titer and neutralizing antibody titer in sera collected from piglets infected with icPEDV-WT or icPEDV-mut4. (A) Fluorescence-linked immunosorbent assay (FLISA) was used to determine the PEDV-specific IgG titer. Briefly, PEDV-infected Vero cells were fixed with ethanol/acetone (1:1) and then incubated with serially diluted porcine sera. The highest dilution of serum that produced positive signal was determined as the FLISA titer. (B) Virus neutralizing (VN) antibody titers in sera collected 21 dpi. (See details in Materials and Methods). Each value and means ± standard errors of the means (SEM) are presented. *, P < 0.05; **, P < 0.01. Dashed horizontal lines represent the limit of detection.
DISCUSSION
Here, we report that inactivation of three independent coronavirus interferon antagonists is a rational approach for attenuating a highly pathogenic, enteric coronavirus. Starting with a pathogenic strain of PEDV, we incorporated mutations into known catalytic or essential sites of nonstructural proteins 1, 15, and 16 to generate icPEDV-mut4. We found that this mutant virus replicated as efficiently as wild-type virus in Vero cells but that it was highly impaired for replication in interferon-responsive porcine kidney (PK) epithelial cells. We administered the parental strain and the mutant strain to highly susceptible piglets and found that icPEDV-mut4-infected animals exhibited no clinical signs of disease (diarrhea). Importantly, we found that icPEDV-mut4 did replicate in the infected animals, as revealed by shedding of virus in the feces, and elicited an adaptive immune response, as revealed by detection of virus-specific IgG and neutralizing antibody in the serum at 21 day postinfection. Furthermore, the genetic signature of icPEDV-mut4 was maintained in the viral RNA shed from the infected animals. We conclude that inactivation of the three coronavirus interferon antagonists is a rational approach for generating a live attenuated coronavirus vaccine candidate strain.
Our findings revealing the delay in virus shedding until after day 2 postinfection (Fig. 4C) and the lack of detectable viral antigen in gut of icPEDV-mut4-infected animals (Fig. 6) were unexpected. Previously, we found that mutation of nsp15 alone reduced PEDV shedding and mortality and that virus antigen was detected in the gut at day 7. Here, we decided to look at day 2 postinfection, a time when viral antigen and pathology are generally readily detected (19, 20). Interestingly, we noted that only 1 of the 11 icPEDV-mut4-infected animals had a level of shedding that was above the limit of detection and that neither of the 2 animals euthanized for pathological studies had notable tissue pathology or detectable levels of viral antigen. These results document the profound attenuation of icPEDV-mut4 in piglets. The detection of viral RNA from day 3 to day 16 in the piglets provides evidence that the mutant virus does replicate in vivo, albeit at a relatively low level compared to the wild-type virus infection. This low level of virus replication is likely essential for generating the adaptive immune response that we detected as virus-specific IgG and neutralizing antibody responses (Fig. 7). Future studies will determine if the level of the adaptive response to icPEDV-mut4 infection is sufficient to protect from homologous or heterologous challenge with PEDV.
Recent advances in the ability to engineer live attenuated viruses and prevent reversion to virulence have stimulated renewed interest in the use of this classic type of vaccine (21). Here, we show that the mutations engineered into icPEDV-mut4 were maintained in the viral RNA shed in the feces at late times postinfection. The presence of four mutations in the genome likely reduces the potential for reversion to virulence, but this has yet to be formally tested. To reduce the potential for a CoV live attenuated vaccine to undergo recombination that could restore virulence, changes in the transcriptional regulatory sequences could be incorporated into the engineered vaccine strain that would eliminate the potential for successful recombination with endemic CoVs (22). These advances in engineering live attenuated vaccines offer an opportunity to generate designer vaccines that can efficiently induce mucosal immunity.
This approach of inactivating a viral interferon antagonist to attenuate pathogenic viruses has been successful for diverse viruses, including poxviruses (23, 24), influenza viruses (25, 26), and flaviviruses (27). This approach relies on the ability of the mutant virus to activate the host innate immune response, predominantly manifesting as type I and type III IFNs. The IFNs and interferon-stimulated genes (ISGs) generate a hostile environment that limits virus replication while still allowing sufficient virus replication to activate the adaptive immune response. The challenge in applying this approach to coronaviruses has been the difficulty of identifying a key antagonist, or a constellation of antagonists, to inactivate such that the virus replicates at a level that elicits a protective response without causing any clinical disease.
Our study focused on three conserved coronavirus interferon antagonists, nsp1, nsp15, and nsp16. These antagonists were selected because previous studies documented that mutating specific residues was sufficient to disrupt the antagonistic activity of the protein. Nsp1 is highly divergent among coronaviruses, and only alphacoronavirus and betacoronavirus strains encode it (28, 29). The nsp1 of alphacoronavirus is about 110 amino acids in length (versus the approximately 170 to 245 amino acids of betacoronavirus nsp1) and shares low sequence similarity with betacoronavirus nsp1 (30). Nsp1 is a multifunctional antagonist that blocks innate immune responses (reviewed previously [31]). Previous studies used overexpression of PEDV nsp1 to identify key residues required for disrupting interferon antagonism (16, 17), although the exact mechanism used by PEDV nsp1 has not yet been elucidated (31). Our studies demonstrated that the F44A mutation of PEDV nsp1 in the context of virus replication resulted in an elevated interferon response in comparison to the WT infection (Fig. 3). Thus, PEDV nsp1 is an interferon antagonist.
Nsp16 is a 2′-O-methyltransferase that is important for modifying the viral RNA to mimic the methylation found in host mRNA, thus evading sensing by host pattern recognition receptors (32). Leveraging previous work on PEDV nsp16 (14), we generated the nsp16 D129A mutant virus for our studies, as this change is sufficient to disrupt interferon antagonism activity, as shown in Fig. 3 and in previous studies (14, 32), without compromising efficient virus replication in Vero cells (Fig. 2A).
Our third target for inactivation of a viral interferon antagonist was nsp15. Nsp15 is a viral endoribonuclease that, as we recently reported (33), targets the poly uridine residues at the 5′ end of the negative-sense RNA. EndoU activity trims the viral negative-sense RNA such that it is not recognized by the MDA5 host pattern recognition receptor. The structure of CoV endoribonuclease has been solved, revealing two required histidine residues in the catalytic sites, and mutation of either of these histidine residues to alanine inactivates the enzyme (34–37). For this study, we targeted the two required catalytic histidine residues of nsp15 (H226A and H241A) and incorporated those mutations in combination with the mutations in nsp1 and nsp16. The combination mutant virus, icPEDV-mut4, replicated efficiently in Vero cells, elicited a robust type I and III IFN response in PK1 cells with limited virus replication, and was highly attenuated when administered to naive piglets. This mutant virus was able to replicate in enterocytes to a sufficient level to elicit an adaptive immune response, as documented by the presence of virus-specific IgG and neutralizing antibody in the serum.
The results of this study are in agreement with those of previous studies performed using mouse-adapted versions of SARS-CoV and MERS-CoV that documented the efficacy of combination attenuation strategies for generating live attenuated coronavirus vaccines (13, 38, 39). Those studies focused on the combination of inactivation of nsp16 (MTase) and ExoN (proofreading activity) as a strategy that could be leveraged as a platform for generating vaccines for emerging coronaviruses. Our studies complement and extend these studies by using an enteric CoV model system, the porcine coronavirus PEDV, and infection of piglets.
Our use of a model system where the coronavirus replicates in enterocytes and causes diarrhea will be informative for studying SARS-CoV-2, which has also caused symptoms of diarrhea and evidence of shedding in the feces (40–43). Recent studies documented the replication of SARS-CoV-2 in human gut enterocytes (44, 45). Furthermore, fecal shedding of SARS-CoV-2 can occur for many days, even after the virus is no longer detected in the upper respiratory tract of patients (46, 47). It is likely that SARS-CoV-2 emerged either directly or indirectly from a virus reservoir in bats (2), where the virus replicates in the enteric tract. Understanding the virulence factors that allow coronavirus replication in the enteric tract, such as nsp1, nsp15, and nsp16, is critical for generating effective candidate vaccines.
The results presented here describe a candidate vaccine strain that will have to undergo rigorous evaluation to determine if it elicits a protective response to challenge virus and to determine the genetic stability of the virus during passage in animals. Our future studies will be aimed at determining the dose of icPEDV-mut4 required to elicit responses that protect animals from homologous strains of PEDV and potentially from heterologous challenge from more-diverse strains of PEDV. In addition, it will be important to evaluate the genetic stability of icPEDV-mut4 during passage, particularly the potential for a virus with 4 mutations to revert to virulence during passage in animals. Finally, any candidate vaccine strains will need to be made resistant to RNA recombination with exiting strains. Graham and coworkers outlined a strategy that rewires the coronavirus transcriptional regulatory sequences that could be applied to any candidate live attenuated coronavirus vaccines (22). Using synthetic biology, any promising candidate vaccine can be rewired to significantly reduce the likelihood of recombination with existing coronaviruses. Overall, our studies revealed the critical role of coronavirus interferon antagonists as virulence factors in enteric disease. This appreciation of virulence factors that contribute to enteric disease is particularly important now that we are facing pandemic spread of SARS-CoV-2. We hope that the results presented here will contribute to rapid progress in efforts to limit the spread, morbidity, and mortality of existing and potentially emerging coronavirus infections.
MATERIALS AND METHODS
Ethics statement.
All animal work was performed according to Institutional Animal Care and Use Protocol guidelines (ACUP ARS-2017-603) at the National Animal Disease Center (NADC) in Ames, IA. Piglets were consistently monitored for signs of distress over the course of the experiments; those that showed such signs were removed and euthanized by the use of carbon dioxide inhalation to prevent unnecessary suffering.
Cells and virus.
Porcine kidney epithelial cells, LLC-PK1 (ATCC CL101), termed PK1 cells, were purchased from the ATCC and grown in growth medium containing modified Eagle medium (MEM) (Corning; catalog no. 10010CV) supplemented with heat-inactivated 5% fetal calf serum (FCS) (Atlanta Biological) and 1% penicillin/streptomycin (Pen/Strep) (HyClone). Vero cells (USDA Animal and Plant Health Inspection Agency, National Veterinary Services Laboratory [APHIS-NVSL]) were grown in growth media containing MEM (Gibco; catalog no. 41500-018) supplemented with 10% FCS, 0.5% lactalbumin enzymatic hydrolysate (Sigma; catalog no. 68458-87-7), and 1% Pen/Strep.
An infectious clone of the PEDV wild-type strain (icPEDV-WT) and a PEDV mutant expressing catalytic-inactive endoribonuclease (icPEDV-EnUmt) were generated in our previous study (11). A similar approach was used to engineer icPEDV-Nsp1mt, icPEDV-Nsp16mt, and icPEDV-mut4. icPEDV-Nsp1mt expresses a mutated Nsp1 that carries a Phe44-to-Ala substitution. icPEDV-Nsp16mt encodes a catalytic-inactive 2′-O-methyltransferase which harbors an Asp129-to-Ala mutation in Nsp16. icPEDV-mut4 has combined mutations in Nsp1 and Nsp16 in addition to two catalytic histidine mutations (His226-to-Ala and His241-to-Ala) of nsp15. Primers used in the site-directed mutagenesis are listed in Table 1. All these recombinant PEDVs were rescued in Vero cells and sequenced to confirm the engineered mutations. To make large stocks, these viruses were propagated once more in Vero cells with maintenance media containing FCS-free growth media, 0.15% Bacto tryptose phosphate broth (Becton Dickinson; catalog no. 260300) (29.5 g/liter), and 5 μg/ml trypsin (Sigma; catalog no. 59427C). The culture medium of infected cells was harvested when ∼90% cells showed cytopathic effect (CPE), titrated on Vero cell monolayers, and stored at –80°C.
Growth kinetics and titration of icPEDV-WT and mutant viruses.
Vero or PK1 cells were seeded into a 24-well (1.5 × 105 cells/well) plate and infected with either icPEDV-WT or the designated mutant virus at a dose of 0.1 TCID50 per cell in the presence of 5 μg/ml trypsin. After 1 h of incubation, the inoculum was removed and replaced with serum-free maintenance medium. Cell culture supernatant was collected at the indicated time points after infection and subjected to titration in Vero cells using a standard TCID50 assay as previously described (11).
Analysis of gene expression using RT-qPCR.
To measure mRNA levels in cells, 3 × 105 PK1 cells per well were plated in a 12-well plate, 16 h prior to infection. Cells were washed twice with phosphate-buffered saline (PBS) and infected with the indicated virus at a dose of 0.1 TCID50 per cell in the presence of 5 μg/ml trypsin (Sigma; catalog no. 59427C). Cells were harvested at different time points using the RLT buffer provided with an RNeasy minikit (Qiagen; catalog no. 74104), and total RNA was extracted as instructed by the manufacturer’s protocol. A 500-ng volume of RNA was used for cDNA synthesis performed with a RT2 HT first-strand kit (Qiagen; catalog no. 330411). Quantitative PCR (qPCR) was performed using RT2 SYBR green qPCR mix (Qiagen; catalog no. 330502) and a Bio-Rad CFX96 system. The thermocycler was set as follows: one step at 95°C (10 min); 40 cycles of 95°C (15 s) and 55°C (1 min) followed by plate reading; one step at 95°C (10 s); and a melt curve from 65°C to 95°C using increments of 0.5°C/0.05 s. Samples were assayed in triplicate, and data are representative of results from three independent experiments. The levels of mRNA were determined relative to β-actin mRNA levels and are expressed as 2−ΔCT [threshold cycle (ΔCT) = CT(gene of interest) − CT(GAPDH)]. The details of the PCR assay performed for the primers used in this study are listed in Table 2.
TABLE 2.
qPCR primer and probe sequences
| Method | Target | Primer sequence (5′–3′) |
|
|---|---|---|---|
| Forward | Reverse | ||
| SYBR green | Porcine IFN-β | AGCAGATCTTCGGCATTCTC | GTCATCCATCTGCCCATCAA |
| Porcine IFN-λ1 | ACTGTGATGCTGGACTTGG | GCATCCTTGGCTTTCTTGAAG | |
| Porcine GAPDH | ACCTCCACTACATGGTCTACA | ATGACAAGCTTCCCGTTCTC | |
| Porcine ISG54 | CTGGCAAAGAGCCCTAAGGA | CTCAGAGGGTCAATGGAATTCC | |
| Porcine N gene | CACTAACCTGGGTGTCAGAAA | CGTGAAGTAGGAGGTGTGTTAG | |
| Taqmana | PEDV N gene | GAATTCCCAAGGGCGAAAAT | TTTTCGACAAATTCCGCATCT |
The sequence of the N gene probe used in the Taqman assay was as follows: FAM-CGTAGCAGCTTGCTTCGGACCCA-BHQ (FAM, 6-carboxyfluorescein; BHQ, black hole quencher).
Evaluating pathogenesis of icPEDV-WT and icPEDV-mut4.
Six pregnant sows free of clinical disease and shown to be negative for PEDV antibodies via IFA were purchased from a commercial farm and transported to NADC prior to farrowing. Piglets were weaned from sows at between 3 and 6 days of age, given an iron injection, and ear tagged. Piglets were blocked by litter and assigned to 3 treatment groups: icPEDV-WT (n = 13), icPEDV-mut4 (n = 13), and controls (n = 8). Each group was housed in a separate biosafety level 2 animal room, fed a diet of milk replacer and starter, and given ad libitum access to water. Twenty-four hours after weaning, piglets were orally inoculated with 2 ml of virus solution at a titer of 500 TCID50/pig. Control pigs received 2 ml of medium inoculum orally. Piglets were rectally swabbed and given a clinical diarrhea score at 0 to 10, 12, 14, 18, and 21 days postinoculation (dpi). Clinical diarrhea scores were assigned by the following criteria: 0 = normal feces, 1 = soft stool, 2 = semiliquid stool, 3 = liquid feces, 4 = voluminous watery diarrhea. Rectal swabs were collected with a sterile polyester-tipped applicator (Puritan Medical Products, Guilford, ME) immersed in 3 ml of serum-free MEM. Blood was collected in a serum separation tube (BD Vacutainer, Franklin Lakes, NJ) and centrifuged to harvest serum at 0 and 21 dpi. Samples were stored at –80°C until time of testing.
Two piglets from each group were euthanized at 2 dpi for collection of jejunum and ileum tissue samples. Intestinal sections were fixed in 10% neutral buffered formalin and routinely processed. All remaining animals were euthanized on 21 dpi. Euthanasia was performed with an intravenous administration of barbiturate (Fatal Plus; Vortech Pharmaceuticals, Dearborn, MI) following the manufacturer-labeled dose.
PEDV RNA quantification using TaqMan real-time RT-PCR.
Viral RNA was quantified from rectal swabs as previously described (48). Briefly, RNA extraction was performed using a MagMAX pathogen RNA/DNA kit (catalog no. 4462359; Applied Biosystems) following the manufacturer’s recommendations for fecal samples. Viral RNA was eluted into 90 μl of elution buffer. Following extraction, 5-μl volumes of the nucleic acid templates were added to 20 μl of Path-ID multiplex one-step RT-PCR master mix (catalog no. 4442137; Applied Biosystems). Real-time reverse transcription-quantitative PCR (RT-qPCR) was performed on an ABI 7500 Fast instrument run in standard mode under the following conditions: reverse transcription at 45°C for 10 min and denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 45 s. The primer and the probe (listed in Table 2) were synthesized (Integrated DNA Technologies, Coralville, IA) and targeted a conserved region (nucleotides 941 to 1028) of the PEDV N gene with modifications specific to PEDV strain USA/Colorado/2013 (GenBank accession no. KF272920). PEDV genome copy numbers were calculated based on a standard RNA transcript overlapping the target region.
Evaluating icPEDVmut 4 sequence from fecal samples.
Viral RNA was purified from fecal swabs obtained from 11 piglets inoculated with icPEDV-mut4 and taken from the last day that viral RNA was detected in the quantitative real-time PCR (8 to 14 days), as described above. Three regions containing the four engineered mutations in the icPEDV-mut4 genome (19.5 μl vRNA) were first amplified using a SuperScript IV one-step RT-PCR system (Thermo Fisher Scientific, Waltham, MA) and specific primers (PEDV-210F/PEDV627R for nsp1; PEDV-19071F/PEDV-19731R or PEDV-18319F/PEDV-19712R for nsp15; PEDV-19923F/PEDV-20395R for nsp16) (Table 1). The amplicons were subjected to column purification, and a 1-μl volume of the product was further amplified using a FailSafe PCR system (Lucigen Corporation, Middleton, WI) with the same primers. PCR products from the nsp1 and nsp16 amplifications were purified and sent for sequencing using sequencing primers (PEDV-235F and PEDV-549R for nsp1; PEDV-20028F and PEDV-20345R for nsp16) (Table 1). Products from the nsp15 amplification were sent for sequencing using primers PEDV-19218F and PEDV-19637R or were cloned into pCR-Blunt II-TOPO vector and sequenced using primers PEDV-18319F and PEDV-19712R.
Immunofluorescence assay to determine the titer of virus-specific IgG.
In a 96-well plate, Vero cells were infected with PEDV at a dose of 0.1 TCID50 per cell in the presence of 2 μg/ml trypsin. At 16 h postinfection, cells were washed once with PBS and fixed with cold methanol/acetone (50%/50% [vol/vol]) for 15 min at –20°C. Fixed cells were then blocked with PBS containing 5% FCS for 30 min at 37°C and incubated with serially diluted pig serum for 1 h at 37°C. Cells were washed three times with PBS and incubated with fluorescein isothiocyanate (FITC)-conjugated goat anti-swine IgG (H+L) secondary antibody (catalog no. 6050-02; Southern Biotech). Subsequently, cells were washed three times with PBS before examination using a fluorescence microscope.
H&E staining and immunohistochemistry.
The details of this method were described previously (11). Briefly, tissues were fixed in neutral buffered formalin, processed, and embedded in paraffin. Sections (5 μm thick) were cut and stained with hematoxylin and eosin (H&E) stain utilizing a Tissue-Tek automated slide stainer (Sakura Finetek USA, Torrence, CA). A veterinary pathologist who was blind to the treatment groups evaluated sections of small intestine by light microscopy to identify location and subjectively assess villus atrophy and crypt hyperplasia. For immunohistochemistry, the tissue sections were mounted on positively charged glass slides and oven dried for 60 min at 60˚C. Slides were deparaffinized and then rinsed three times in deionized water, followed by soaking in Tris-buffered saline with Tween 20 for 5 min. Slides were placed in a Dako autostainer (Agilent, Santa Clara, CA) and run through a preprogrammed immunohistochemistry (IHC) protocol. The IHC protocol utilizes Protease XIV (Millipore Sigma, St. Louis, MO) for antigen retrieval, murine monoclonal antibody SD6-29 specific for the nucleocapsid of PEDV (18) at 1:1,000 dilution, Dako Envision+ horseradish peroxidase (HRP) (Agilent, Santa Clara, CA), and DAB (diaminobenzidine) substrate chromogen (Agilent, Santa Clara, CA). The slides were then counterstained in hematoxylin and placed beneath cover slips.
Viral neutralizing antibody determination.
The details of this method were described previously (14). Briefly, the collected sera were first subjected to heat inactivation at 56°C for 30 min and to serial 4-fold dilution with MEM. Diluted sera were then mixed with a same volume of virus solution containing 100 TCID50/25 μl of icPEDV-WT. After incubation for 90 min at 37°C, the mixture was used to infect Vero cells in 96-well plates with controls of mock and virus-only infections. After a 1-h infection, the inoculum was discarded. The cells were washed three times with PBS and cultured with the maintenance medium containing 5 μg/ml of trypsin. After 3 days of infection, the titers of neutralizing antibody were determined by the use of the Reed and Muench method (49). The VN titer of a serum sample is the reciprocal of its dilution that gives no CPE in 50% of the wells.
ACKNOWLEDGMENTS
We thank Jason Huegel, Justin Miller, Alyssa Dannen, Randy Leon, and Nate Horman for providing animal care; Deb Adolphson, Sarah Anderson, and Miranda Dietz for technical assistance (animal handling, RT-qPCR, nucleotide sequencing); and Matthew Hackbart and Amornrat O’Brien for comments and suggestions.
This work was funded by USDA-Loyola University Chicago cooperative agreement 59-5030-8-003B, NIH AI085089 (to S.C.B.), and USDA ARS CRIS Project 5030-32000-118-00D.
REFERENCES
- 1.Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, Hu Y, Tao Z-W, Tian J-H, Pei Y-Y, Yuan M-L, Zhang Y-L, Dai F-H, Liu Y, Wang Q-M, Zheng J-J, Xu L, Holmes EC, Zhang Y-Z. 2020. A new coronavirus associated with human respiratory disease in China. Nature 580. doi: 10.1038/s41586-020-2202-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–274. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gosert R, Kanjanahaluethai A, Egger D, Bienz K, Baker SC. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J Virol 76:3697–3708. doi: 10.1128/jvi.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knoops K, Kikkert M, van den Worm SHE, Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, Mommaas AM, Snijder EJ. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol 6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kindler E, Thiel V. 2014. To sense or not to sense viral RNA-essentials of coronavirus innate immune evasion. Curr Opin Microbiol 20:69–75. doi: 10.1016/j.mib.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mesev EV, LeDesma RA, Ploss A. 2019. Decoding type I and III interferon signalling during viral infection. Nat Microbiol 4:914–924. doi: 10.1038/s41564-019-0421-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Channappanavar R, Fehr AR, Vijay R, Mack M, Zhao J, Meyerholz DK, Perlman S. 2016. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe 19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Channappanavar R, Fehr AR, Zheng J, Wohlford-Lenane C, Abrahante JE, Mack M, Sompallae R, McCray PB, Meyerholz DK, Perlman S. 2019. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J Clin Invest 129. doi: 10.1172/JCI126363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng X, Hackbart M, Mettelman RC, O'Brien A, Mielech AM, Yi G, Kao CC, Baker SC. 2017. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc Natl Acad Sci U S A 114:E4251–E4260. doi: 10.1073/pnas.1618310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng X, van Geelen A, Buckley AC, O’Brien A, Pillatzki A, Lager KM, Faaberg KS, Baker SC. 2019. Coronavirus endoribonuclease activity in porcine epidemic diarrhea virus suppresses type I and type III interferon responses. J Virol 93:e02000-18. doi: 10.1128/JVI.02000-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menachery VD, Gralinski LE, Mitchell HD, Dinnon KH, Leist SR, Yount BL, Graham RL, McAnarney ET, Stratton KG, Cockrell AS, Debbink K, Sims AC, Waters KM, Baric RS. 2017. Middle East respiratory syndrome coronavirus nonstructural protein 16 is necessary for interferon resistance and viral pathogenesis. mSphere 2:e00346-17. doi: 10.1128/mSphere.00346-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menachery VD, Gralinski LE, Mitchell HD, Dinnon KH, Leist SR, Yount BL, McAnarney ET, Graham RL, Waters KM, Baric RS. 2018. Combination attenuation offers strategy for live attenuated coronavirus vaccines. J Virol 92:e00710-18. doi: 10.1128/JVI.00710-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou Y, Ke H, Kim J, Yoo D, Su Y, Boley P, Chepngeno J, Vlasova AN, Saif LJ, Wang Q. 2019. Engineering a live attenuated PEDV vaccine candidate via inactivation of the viral 2′-O methyltransferase and the endocytosis signal of the spike protein. J Virol 93:e00406-19. doi: 10.1128/JVI.00406-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Volk A, Hackbart M, Deng X, Cruz-Pulido Y, O’Brien A, Baker SC. 2020. Coronavirus Endoribonuclease and deubiquitinating interferon antagonists differentially modulate the host response during replication in macrophages. J Virol 94. doi: 10.1128/JVI.00178-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Q, Ke H, Blikslager A, Fujita T, Yoo D. 2018. Type III interferon restriction by porcine epidemic diarrhea virus and the role of viral protein nsp1 in IRF1 signaling. J Virol 92:e01677-17. doi: 10.1128/JVI.01677-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Q, Ma J, Yoo D. 2017. Inhibition of NF-κB activity by the porcine epidemic diarrhea virus nonstructural protein 1 for innate immune evasion. Virology 510:111–126. doi: 10.1016/j.virol.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okda F, Liu X, Singrey A, Clement T, Nelson J, Christopher-Hennings J, Nelson EA, Lawson S. 2015. Development of an indirect ELISA, blocking ELISA, fluorescent microsphere immunoassay and fluorescent focus neutralization assay for serologic evaluation of exposure to North American strains of porcine epidemic diarrhea virus. BMC Vet Res 11:180. doi: 10.1186/s12917-015-0500-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas JT, Chen Q, Gauger PC, Giménez-Lirola LG, Sinha A, Harmon KM, Madson DM, Burrough ER, Magstadt DR, Salzbrenner HM, Welch MW, Yoon KJ, Zimmerman JJ, Zhang J. 2015. Effect of porcine epidemic diarrhea virus infectious doses on infection outcomes in Naïve conventional neonatal and weaned pigs. PLoS One 10:e0139266. doi: 10.1371/journal.pone.0139266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung K, Annamalai T, Lu Z, Saif LJ. 2015. Comparative pathogenesis of US porcine epidemic diarrhea virus (PEDV) strain PC21A in conventional 9-day-old nursing piglets vs. 26-day-old weaned pigs. Vet Microbiol 178:31–40. doi: 10.1016/j.vetmic.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Te Yeh M, Bujaki E, Dolan PT, Smith M, Wahid R, Konz J, Weiner AJ, Bandyopadhyay AS, Van Damme P, De Coster I, Revets H, Macadam A, Andino R. 2020. Engineering the live-attenuated polio vaccine to prevent reversion to virulence. Cell Host Microbe 27:736–751.e8. doi: 10.1016/j.chom.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graham RL, Deming DJ, Deming ME, Yount BL, Baric RS. 2018. Evaluation of a recombination-resistant coronavirus as a broadly applicable, rapidly implementable vaccine platform. Commun Biol 1:179. doi: 10.1038/s42003-018-0175-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith GL, Talbot-Cooper C, Lu Y. 2018. How does vaccinia virus interfere with interferon? Adv Virus Res 100:355–378. doi: 10.1016/bs.aivir.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 24.Smith GL, Benfield CTO, Maluquer de Motes C, Mazzon M, Ember SWJ, Ferguson BJ, Sumner RP. 2013. Vaccinia virus immune evasion: mechanisms, virulence and immunogenicity. J Gen Virol 94:2367–2392. doi: 10.1099/vir.0.055921-0. [DOI] [PubMed] [Google Scholar]
- 25.Marazzi I, Garcia-Sastre A. 2015. Interference of viral effector proteins with chromatin, transcription, and the epigenome. Curr Opin Microbiol 26:123–129. doi: 10.1016/j.mib.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 26.Du Y, Shi Y, Zhang T-H, Dai L, Gong D, Brar G, Shu S, Luo J, Tseng Y-W, Wu T-T, Wang J, Sun R, Xin L, Bai H, Shu Y, Wu NC, Reiley W. 2018. Genome-wide identification of interferon-sensitive mutations enables influenza vaccine design. Science 359:290–296. doi: 10.1126/science.aan8806. [DOI] [PubMed] [Google Scholar]
- 27.Xu Y, Li X, Zhu B, Liang H, Fang C, Gong Y, Guo Q, Sun X, Zhao D, Shen J, Zhang H, Liu H, Xia H, Tang J, Zhang K, Gong S. 2020. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med 26:502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, Poon LLM, Guan Y, Rozanov M, Spaan WJM, Gorbalenya AE. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol 331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ziebuhr J. 2005. The coronavirus replicase. Curr Top Microbiol Immunol 287:57–94. doi: 10.1007/3-540-26765-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Connor RF, Roper RL. 2007. Unique SARS-CoV protein nsp1: bioinformatics, biochemistry and potential effects on virulence. Trends Microbiol 15:51–53. doi: 10.1016/j.tim.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Narayanan K, Ramirez SI, Lokugamage KG, Makino S. 2015. Coronavirus nonstructural protein 1: common and distinct functions in the regulation of host and viral gene expression. Virus Res 202:89–100. doi: 10.1016/j.virusres.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Züst R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, Siddell SG, Ludewig B, Thiel V. 2011. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol 12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hackbart M, Deng X, Baker SC. 2020. Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc Natl Acad Sci U S A 117:8094–8103. doi: 10.1073/pnas.1921485117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ricagno S, Egloff M-P, Ulferts R, Coutard B, Nurizzo D, Campanacci V, Cambillau C, Ziebuhr J, Canard B. 2006. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc Natl Acad Sci U S A 103:11892–11897. doi: 10.1073/pnas.0601708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ivanov KA, Hertzig T, Rozanov M, Bayer S, Thiel V, Gorbalenya AEA, Ziebuhr J. 2004. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc Natl Acad Sci U S A 101:12694–12699. doi: 10.1073/pnas.0403127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhardwaj K, Guarino L, Kao CC. 2004. The severe acute respiratory syndrome coronavirus nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J Virol 78:12218–12224. doi: 10.1128/JVI.78.22.12218-12224.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu X, Zhai Y, Sun F, Lou Z, Su D, Xu Y, Zhang R, Joachimiak A, Zhang XC, Bartlam M, Rao Z. 2006. New antiviral target revealed by the hexameric structure of mouse hepatitis virus nonstructural protein nsp15. J Virol 80:7909–7917. doi: 10.1128/JVI.00525-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Menachery VD, Mitchell HD, Cockrell AS, Gralinski LE, Yount BL, Graham RL, McAnarney ET, Douglas MG, Scobey T, Beall A, Dinnon K, Kocher JF, Hale AE, Stratton KG, Waters KM, Baric RS. 2017. MERS-CoV accessory ORFs play key role for infection and pathogenesis. mBio 8:e00665-17. doi: 10.1128/mBio.00665-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolles M, Deming D, Long K, Agnihothram S, Whitmore A, Ferris M, Funkhouser W, Gralinski L, Totura A, Heise M, Baric RS. 2011. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J Virol 85:12201–12215. doi: 10.1128/JVI.06048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, Zhao Y, Li Y, Wang X, Peng Z. 2020. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA 323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. 2020. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, Spitters C, Ericson K, Wilkerson S, Tural A, Diaz G, Cohn A, Fox L, Patel A, Gerber SI, Kim L, Tong S, Lu X, Lindstrom S, Pallansch MA, Weldon WC, Biggs HM, Uyeki TM, Pillai SK, Washington State 2019-nCoV Case Investigation Team. 2020. First case of 2019 novel coronavirus in the United States. N Engl J Med 382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brünink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C, Wendtner C. 2020. Virological assessment of hospitalized patients with COVID-2019. Nature 581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 44.Lamers MM, Beumer J, van derVaart J, Knoops K, Puschhof J, Breugem TJ, Ravelli RBG, van Schayck JP, Mykytyn AZ, Duimel HQ, van Donselaar E, Riesebosch S, Kuijpers HJH, Schippers D, van de Wetering WJ, de Graaf M, Koopmans M, Cuppen E, Peters PJ, Haagmans BL, Clevers H. 2020. SARS-CoV-2 productively infects human gut enterocytes. Science doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou J, Li C, Liu X, Chiu MC, Zhao X, Wang D, Wei Y, Lee A, Zhang AJ, Chu H, Cai J-P, Yip CC-Y, Chan I-Y, Wong KK-Y, Tsang O-Y, Chan K-H, Chan J-W, To KK-W, Chen H, Yuen KY. 2020. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat Med doi: 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
- 46.Cheung KS, Hung IFN, Chan PPY, Lung KC, Tso E, Liu R, Ng YY, Chu MY, Chung TWH, Tam AR, Yip CCY, Leung K-H, Fung AY-F, Zhang RR, Lin Y, Cheng HM, Zhang AJX, To KKW, Chan K-H, Yuen K-Y, Leung WK. 2020. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from the Hong Kong cohort and systematic review and meta-analysis. Gastroenterology doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. 2020. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology 158:1831–1833.e3. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller LC, Crawford KK, Lager KM, Kellner SG, Brockmeier SL. 2016. Evaluation of two real-time polymerase chain reaction assays for Porcine epidemic diarrhea virus (PEDV) to assess PEDV transmission in growing pigs. J Vet Diagn Invest 28:20–29. doi: 10.1177/1040638715621949. [DOI] [PubMed] [Google Scholar]
- 49.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]