Human adenovirus capsid protein IX (pIX) is involved in stabilizing the virion but has also been developed as a platform for presentation of various polypeptides on the surface of the virion. Whether such modifications affect the ability of pIX to stabilize the virion is unknown. We show that addition of large polypeptides to pIX can reduce both the DNA packaging capacity and the heat stability of the virion, which provides important guidance for the design of pIX-modified vectors.
KEYWORDS: adenoviruses, protein IX, vectors, virion structure
ABSTRACT
The human adenovirus (HAdV) protein IX (pIX) is a minor component of the capsid that acts in part to stabilize the hexon-hexon interactions within the mature capsid. Virions lacking pIX have a reduced DNA packaging capacity and exhibit thermal instability. More recently, pIX has been developed as a platform for presentation of large polypeptides, such as fluorescent proteins or large targeting ligands, on the viral capsid. It is not known whether such modifications affect the natural ability of pIX to stabilize the HAdV virion. In this study, we show that addition of large polypeptides to pIX does not alter the natural stability of virions containing sub-wild-type-sized genomes. However, similar virions containing wild-type-sized genomes tend to genetically rearrange, likely due to selective pressure caused by virion instability as a result of compromised pIX function.
IMPORTANCE Human adenovirus capsid protein IX (pIX) is involved in stabilizing the virion but has also been developed as a platform for presentation of various polypeptides on the surface of the virion. Whether such modifications affect the ability of pIX to stabilize the virion is unknown. We show that addition of large polypeptides to pIX can reduce both the DNA packaging capacity and the heat stability of the virion, which provides important guidance for the design of pIX-modified vectors.
INTRODUCTION
The human adenovirus (HAdV) protein IX (pIX) is a minor component of the virion that associates with the hexons that make up the facets of the icosahedron (1–4). Although involved in stabilizing the capsid, pIX is not essential for virion formation. However, capsids that are deficient of pIX are heat labile (5) and can only accommodate subgenomic-size DNA molecules (6, 7). pIX also appears to be a multifunctional protein. pIX has been implicated as a transcriptional activator (8, 9), although viruses deficient in pIX show no significant defect in growth or early or late gene expression (10, 11). pIX has also been implicated in sequestering promyelocytic leukemia protein within the host cell, which may function to enhance HAdV proliferation (12).
Most studies of pIX have involved HAdV-2 or HAdV-5. Within the HAdV virion, 240 copies of pIX form a continuous hexagonal network on the outer surface of the capsid that act to stabilize the virion (13–16). The N-terminal region of three pIX interact to form a triskelion structure that sits within the hexagonal bases of neighboring hexons in the group-of-nine (GON) hexon structure. The N-terminal regions of pIX interact strongly with hexon and likely perform a “cementing” function to stabilize the GON structure (17). The C-terminal region of four pIX interact through a coiled-coil structure, also known as a four-helix bundle, with each α-helix originating from a different triskelion structure. Interestingly, three of the four C-terminal α-helices come from the same facet of the virion and lay in the same orientation, whereas the fourth originates from an adjacent facet and interacts antiparallel to the other three (13). This tetrameric C-terminal structure sits at the boundaries of adjacent GON hexons, tangential to the virion surface (4). In this manner, the interconnected pIX essentially form a mesh-like structure around the entire capsid and laces together adjacent facets, thus providing structural stability.
Many research groups have used pIX as a platform for presentation of small or large polypeptides on the surface of the HAdV virion. The human, canine, and bovine pIX were shown to tolerate the addition of large polypeptides, including red, yellow, or green fluorescent proteins (18–23). pIX can also accommodate large polypeptides that can aid in determining virus biodistribution, such as human metallothionein, herpes simplex virus thymidine kinase, or firefly luciferase protein (24–26). pIX has served as a site for addition of a biotin acceptor peptide for metabolic biotinylation of the HAdV capsid, allowing for the subsequent avidin-mediated purification of the virus (27), or addition of avidin-linked targeting ligands (28). Addition of a reactive cysteine residue at the C terminus of pIX allowed for subsequent covalent addition of targeting ligands (29), whereas fusion of a leucine zipper domain to the C terminus of pIX allows for the noncovalent addition of proteins also containing a leucine zipper (30, 31). Several studies have shown that pIX can be used as a cell attachment protein, provided suitable cell binding ligands are attached to the protein (23, 28, 32–38). Finally, pIX has been used for the presentation of proteins and peptides for vaccines (39–43). Taken together, these observations suggest that pIX is an effective platform for addition of large polypeptides to the HAdV capsid.
As discussed above, pIX is essential to optimal virion stability and is also required for packaging of near-wild-type-size genomes. Viral genomes of >35 kb can only be packaged in virions containing pIX, whereas those of <35 kb can be accommodated in capsids containing or lacking pIX (5, 6, 10). Given the importance of pIX self-association at both the amino and the carboxy termini of the protein for proper function, there is the concern that elaborate modification of pIX may affect its natural ability to function (17). Indeed, thermal instability has been observed in some studies involving modification of pIX (33, 41). In this study, we show that the addition of fluorescent proteins to pIX reduces the DNA packaging capacity of the HAdV virion, suggesting that caution is required when using pIX as a platform for virion modification.
RESULTS AND DISCUSSION
Fusion of small polypeptides to pIX does not alter virion stability for viruses with small genomes.
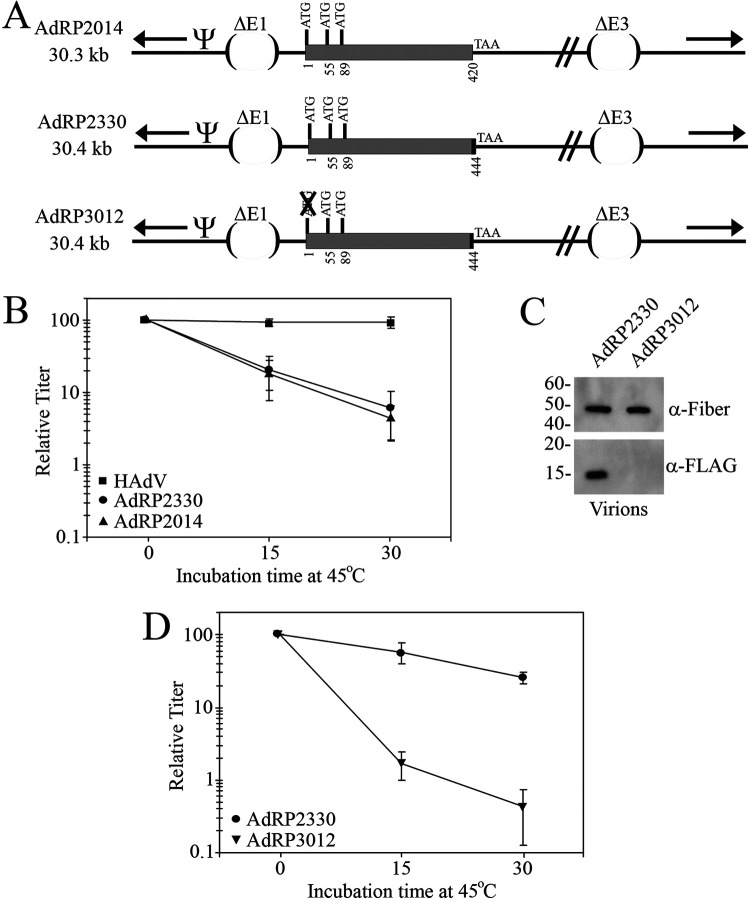
For convenience, we will refer to viruses with genomes less than 35 kb in size as “small genome” and those with genomes above 35 kb as “large genome.” We first examined whether addition of a small polypeptide, the 8 amino acid FLAG epitope tag, altered pIX function. We generated an E1/E3-deleted virus that was 30.4 kb in size, designated AdRP2330 (Fig. 1A), and examined its heat stability relative to an otherwise identical virus lacking the FLAG tag on pIX (AdRP2014), and compared these viruses to wild-type HAdV. As shown in Fig. 1B, both AdRP2330 and AdRP2014 had identical heat inactivation kinetics, suggesting the presence of the FLAG tag did not alter the function of pIX in these viruses containing small genomes. That both viruses showed ca. 5- and 10-fold decreases in virus stability relative to HAdV after heating for 15 and 30 min, respectively, was not surprising. In a previous study, we showed that the HAdV DNA genome itself contributes to the stability of HAdV and that virions containing less than ∼90% of the HAdV genome size showed a fairly dramatic and progressive loss of heat stability, which correlated with the actual genome length (44, 45). However, our analysis of these two viruses raises the question of whether the reduced stability is in fact due to an inability of pIX to perform its virion-stabilizing function for small genome viruses or whether the instability is strictly due to the reduced genome size. To address this possibility, we generated another virus, AdRP3012, which was identical in genome length to AdRP2330 but contained a series of base pair changes to mutate the pIX start codon and thus inactivate expression of the FLAG-tagged pIX (Fig. 1A). Analysis of virions from this virus, AdRP3012, showed that they did not contain a FLAG-tagged pIX (Fig. 1C). AdRP3012 clearly exhibited dramatically reduced heat stability relative to AdRP2330 (Fig. 1D), indicating that pIX contributes to virion stability for viruses containing small genomes. Furthermore, the presence of a small epitope tag does not adversely affect pIX function, at least for virions with small genomes.
FIG 1.
HAdV vectors with small genomes and small polypeptide additions to pIX are stable. (A) AdRP2014 is an E1/E3-deleted vector and expresses native pIX. AdRP2330 is also deleted of E1 and E3 and encodes pIX with a C-terminal FLAG epitope tag. AdRP3012 is similar to AdRP2330 but is mutated for the native pIX start codon. “Ψ” represents the packaging element, the inverted terminal repeats are indicated by black arrows, and the FLAG epitope tag is indicated by a black rectangle at the end of the pIX coding region. (B) HAdV, AdRP2330, and AdRP2014 were subjected to heating at 45°C for 0, 15, or 30 min, and the quantity of virus was determined by plaque assay on 293 cells. The normalized titer relative to unheated samples is shown, and the data represent the averages and standard deviations of n = 2 experiments performed in duplicate. (C) Protein samples of purified virions of AdRP2330 or AdRP3012 were separated by SDS-PAGE and subjected to immunoblot for FLAG and fiber (loading control). (D) AdRP2330 and AdRP3012 were subjected to heating at 45°C for 0, 15, or 30 min, and the quantity of virus was determined by plaque assay on 293 cells. The normalized titer relative to unheated samples is shown, and the data represent the averages and standard deviations of n = 2 experiments performed in duplicate.
Fusion of a large polypeptide to pIX does not alter virion stability for viruses with small genomes.
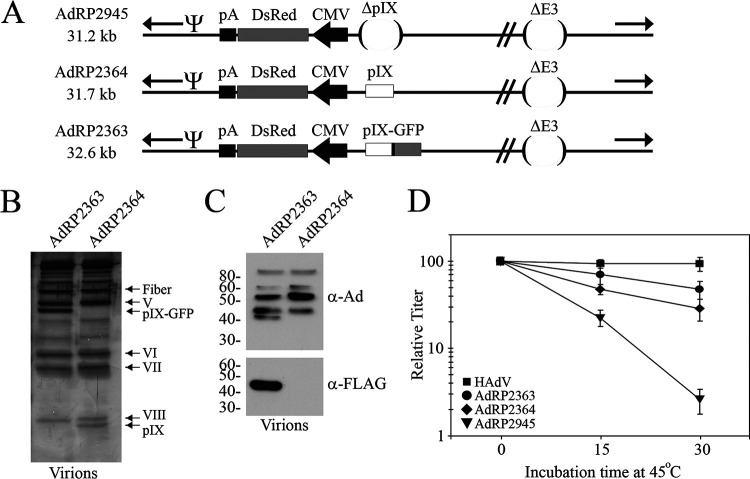
We next sought to determine whether addition of a larger peptide on pIX compromised its function, initially addressing this question using viruses with small genomes. AdRP2364 and AdRP2363 are similar in size (31.7 and 32.6 kb, respectively) but encode either native pIX or a pIX-GFP fusion protein, under regulation by the endogenous pIX promoter (Fig. 2A). The pIX and GFP coding regions are separated by an 18-amino-acid linker comprised primarily of the FLAG epitope tag and a flexible pentaglycine sequence (Table 1). Analysis of protein content of purified virions by SDS-PAGE and silver staining showed that each contained the expected pIX species (Fig. 2B). For AdRP2363, native pIX was not present in the virions, and pIX-GFP was evident as a unique protein species of ∼45 kDa on the silver stain gel and also evident by immunoblotting with antibody directed to all HAdV capsid proteins or FLAG (Fig. 2C). Native pIX was evident in the silver stain gel of AdRP2364 and, as expected, no FLAG signal was present for this virus. Importantly, the level of pIX-GFP incorporation in the virion appeared to be similar to native pIX (Fig. 2B). These two viruses were subjected to heat inactivation at 45°C for 0, 15, or 30 min, and the recovery was compared to HAdV and a virus containing an identical reporter construct, CMV-DsRed, but deleted of the entire pIX coding sequence, designated AdRP2945. Although both AdRP2363 and AdRP2364 showed reduced stability compared to HAdV, both viruses were considerably more stable than AdRP2945. This observation suggests that both pIX and pIX-GFP are able to function to stabilize HAdV capsids containing small genomes.
FIG 2.
HAdV vectors with small genomes and large polypeptide additions to pIX are stable. (A) AdRP2945 is deleted of E1, pIX, and E3 but contains a CMV-DsRed expression cassette replacing the E1 region (expression directed leftward). AdRP2364 is similar to AdRP2945 but contains an intact native pIX gene. AdRP2363 is also similar to AdRP2945 but encodes pIX in-frame with enhanced GFP. AdRP2363 has a FLAG epitope tag located between pIX and GFP, as indicated by a black rectangle. “Ψ” represents the packaging element, and the inverted terminal repeats are indicated by black arrows. (B) Purified virions of AdRP2363 and AdRP2364 were subjected to SDS-PAGE, and the resulting gel was silver stained to visualize capsid proteins. (C) Purified virions of AdRP2363 and AdRP2364 were subjected to SDS-PAGE, and the resulting gel was analyzed by immunoblotting with antibody raised to all HAdV capsid proteins and FLAG. (D) HAdV, AdRP2363, AdRP2364, and AdRP2945 were subjected to heating at 45°C for 0, 15, or 30 min, and the quantity of virus was determined by plaque assay on 293 cells. The normalized titer relative to unheated samples is shown, and the data represent the averages and standard deviations of n = 2 experiments performed in duplicate.
TABLE 1.
Protein sequence of pIX constructs
| pIX construct | Sequencea |
|---|---|
| HAdV-5 | …NAV1 |
| AdRP2330 | …NAV1 DYKDDDDK2 |
| AdRP3012 | …NAV1 DYKDDDDK2 |
| AdRP2363 | …NAV1 DYKDDDDK2 LREFGGGGGA3 SKG4… |
| AdRP3159 | …NAV1 DYKDDDDK2 LREFGGGGGA3 SKG4… |
| AdRP3160 | …NAV1 DYKDDDDK2 LREFGGGGGA3 SKG4… |
| AdRP2384 | …NAV1 DYKDDDDK2 LREFGGGGGA3 SKG4… |
| AdNP101ex | …NAV1 DYKDDDDK2 LSRITSYSIHYTKLCGP3 SKG4… |
| AdRP3064ex | …NAV1 DYKDDDDK2 LSRITSYSIHYTKLCGP3 SKG4… |
| AdCC110 | …NAV1 DYKDDDDK2 LREFGGGGGASAA3 MAS5… |
| AdRP2942 | …NAV1 DYKDDDDK2 LREFGGGGGASAA3 MAS5… |
Superscript numbers: 1, pIX; 2, FLAG epitope tag; 3, linker region; 4, GFP; 5, RFP.
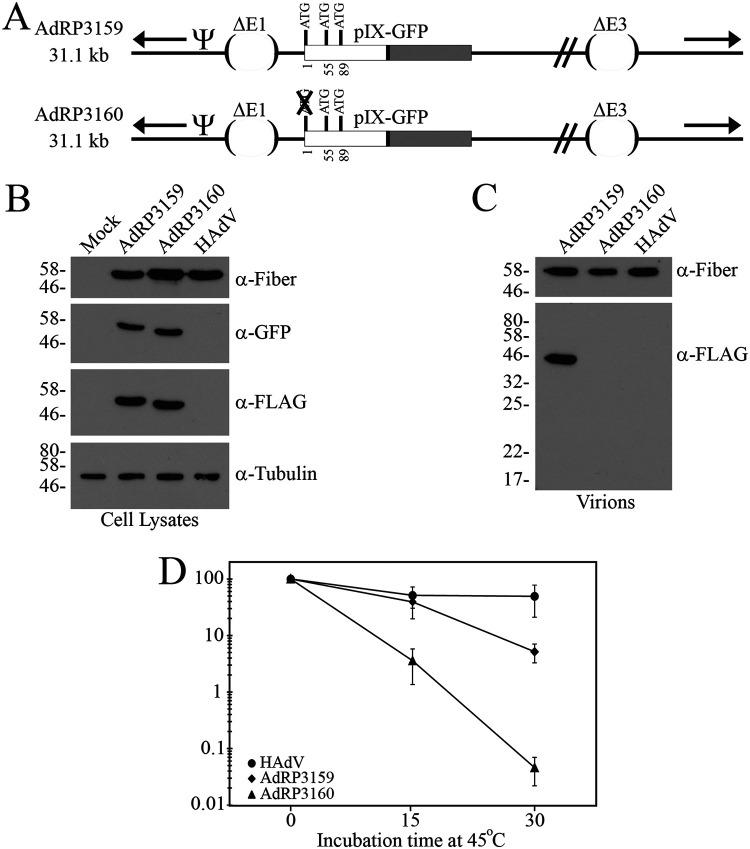
The heat inactivation curves shown in Fig. 2D resemble our previous data showing that small differences in the size of the HAdV genome can influence stability of the HAdV capsid (44, 45). Indeed, from a genome size perspective, HAdV > AdRP2363 > AdRP2364 > AdRP2945, which directly correlates to the viruses with the greatest to least thermal stability. To control for genome size effects, we constructed two viruses of identical genome size, but one expressed pIX-GFP (AdRP3159) and one was mutated at the natural start codon of pIX-GFP (AdRP3160) (Fig. 3A). The viruses were easily rescued and grown on 293 cells. Although pIX is required for recovery and stabilization of virions containing genome sizes greater than ∼35 kb, the relatively small genome size of AdRP3160 meant that the virions did not actually dependent on pIX for formation. To examine protein production from these viruses, we infected 293 cells at a multiplicity of infection (MOI) of 3 and examined fiber expression, a viral late protein, and surrogate marker for viral replication, as well as green fluorescent protein (GFP) and FLAG. Both viruses produced similar levels of fiber protein, suggesting that they each replicated at a similar efficiency. Somewhat surprisingly, the infected cell lysates from both AdRP3159 and AdRP3160 showed a positive signal for FLAG and GFP, although the protein band for AdRP3160 appeared slightly smaller (Fig. 3B). Thus, mutation of the native pIX start codon likely results in initiation of translation at one of the two in-frame methionine amino acid residues downstream of the native start codon. However, consistent with previous work showing that the N terminus of pIX is essential for incorporation of the protein into the virion (9), analysis of protein composition of purified virions showed that the N-terminal truncated pIX-GFP from AdRP3160 was not incorporated into the virion (Fig. 3C). Analysis of heat stability of the two viruses clearly showed that virions lacking pIX-GFP were considerably less stable than the virions containing pIX-GFP (Fig. 3D). Thus, at least in the context of HAdV containing small genomes, the addition of large polypeptides such as GFP does not appear to alter the ability of pIX to stabilize the HAdV virion.
FIG 3.
Addition of a large polypeptide on pIX does not compromise virion thermal stability for HAdV vectors with small genomes. (A) AdRP3159 is deleted of E1 and E3 and encodes pIX fused to GFP. Also shown is the native pIX start codon and two additional in-frame, downstream ATG codons. AdRP3160 is similar in structure to AdRP3159 but is mutated for the native start codon of pIX. Both AdRP3159 and AdRP3160 encode a FLAG epitope tag between pIX and GFP, as indicated by a black rectangle. “Ψ” represents the packaging element, and the inverted terminal repeats are indicated by black arrows. (B) 293 cells were infected with an MOI of 3 of AdRP3159, AdRP3160, or HAdV (or mock infected). After 24 h, crude protein extracts were prepared and subjected to immunoblot analysis for fiber protein, GFP, FLAG, and tubulin (loading control). (C) Purified virions of AdRP3159, AdRP3160, or HAdV were subjected to SDS-PAGE, and immunoblot analysis was performed for fiber (loading control) or FLAG. (D) HAdV, AdRP3159, and AdRP3160 were subjected to heating at 45°C for 0, 15, or 30 min, and the quantity of virus was determined by plaque assay on 293 cells. The normalized titer relative to unheated samples is shown, and the data represent the averages and standard deviations of n = 2 experiments performed in duplicate.
Fusion of a large polypeptide to pIX leads to instability for viruses with large genomes.
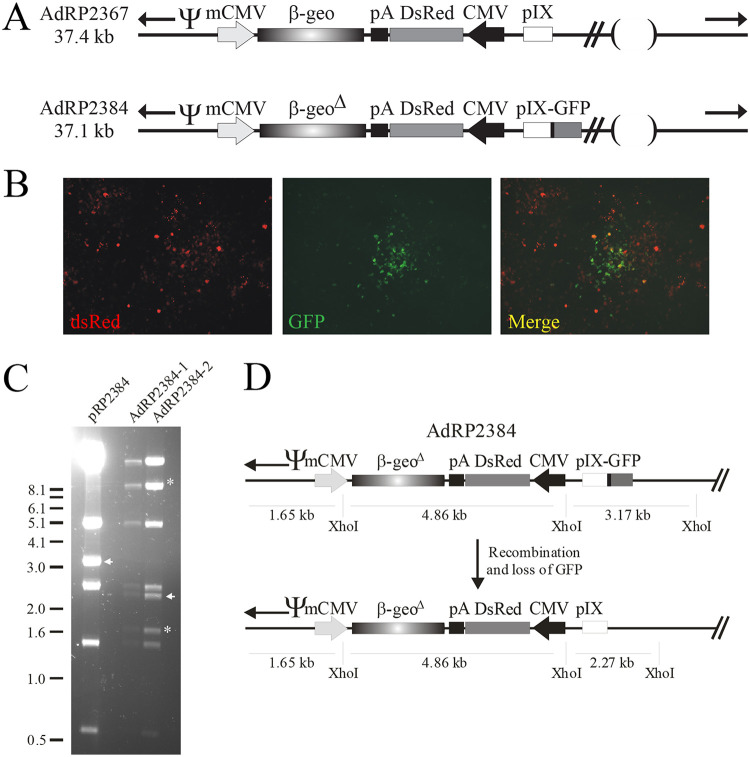
Since deficiency in pIX function leads to a reduced DNA packaging capacity in virions (6), we next addressed whether viable virus could be recovered that expressed pIX-GFP and had a large genome size. We designed a pair of HAdV vectors that contained large genomes (∼37 kb) and expressed either native pIX or pIX-GFP, AdRP2367 and AdRP2384, respectively (Fig. 4A). As expected, AdRP2367 was efficiently recovered and demonstrated normal growth and gene expression (data not shown). In contrast, AdRP2384 was difficult to recover and required several serial passages to yield a significant amount of virus for our studies. AdRP2384 was constructed to express both DsRed and pIX-GFP, but visual inspection by fluorescence microscopy of cells infected with the recovered virus clearly showed that the majority of the DsRed-expressing cells no longer expressed GFP (Fig. 4B). These data suggested that the recovered AdRP2384 had undergone spontaneous rearrangement or recombination to remove GFP from pIX. To gain insight into the genome structure of AdRP2384, we generated two independent large-scale preparations of this virus (i.e., each preparation originated from an independent transfection of pRP2384) and performed restriction digestion on the DNA recovered from the purified virions. As shown in Fig. 4C, both preparations of AdRP2384 had identical structure, which was notably different from pRP2384, the plasmid used to generate this virus. Of note, an expected 3.17-kb XhoI fragment containing the pIX-GFP gene was absent in the recovered viral DNA, and a smaller fragment of ∼2.2 kb was observed. This restriction pattern was consistent with a loss of GFP (Fig. 4D), likely through homologous recombination with HAdV-5 sequences contained in the 293 cell line during vector propagation.
FIG 4.
HAdV vectors with large genomes and large polypeptide additions to pIX show genome instability. (A) AdRP2367 is deleted of E1 and E3, contains a CMV-DsRed and murine CMV (mCMV) β-galactosidase/neomycin (β-geo) expression cassette replacing the E1 deletion, and encodes native pIX. AdRP2384 is similar in structure to AdRP2367 but contains a pIX-GFP fusion gene and has a deletion within the β-geo gene to maintain the size of the vector genome within the limits of packaging within the HAdV virion. AdRP2384 encodes a FLAG epitope tag between pIX and GFP as indicated by a black rectangle. “Ψ” represents the packaging element, and the inverted terminal repeats are indicated by black arrows. (B) A purified stock of AdRP2384 was serially diluted and used to infect 35-mm dishes of 293 cells. The cells were visualized 7 days later using a Zeiss Axiovert 200M microscope, with images collected using the 20× objective. (C) Genomic DNA was extracted from purified virions from two independent stocks of AdRP2384 (designated AdRP2384-1 and AdRP2384-2), digested with XhoI, and subjected to agarose gel electrophoresis. The bacterial plasmid used to generate the virus, pRP2384, was also digested and applied to the gel. For pRP2384, the inverted terminal repeats are contained in a large ∼12.2-kb fragment also containing the bacterial origin of replication and an ampicillin resistance gene, whereas in AdRP2384 the terminal regions of the virus are ∼1.7 and 8.5 kb and are indicated by asterisks. The fragment of pRP2384 containing the pIX-GFP gene is indicated by a white arrow at 3.2 kb, and the new band of ∼2.3 kb in AdRP2384 is also indicated by a white arrow. (D) Restriction fragment sizes of AdRP2384 and a putative recombinant in which the pIX-GFP gene is replaced by native pIX.
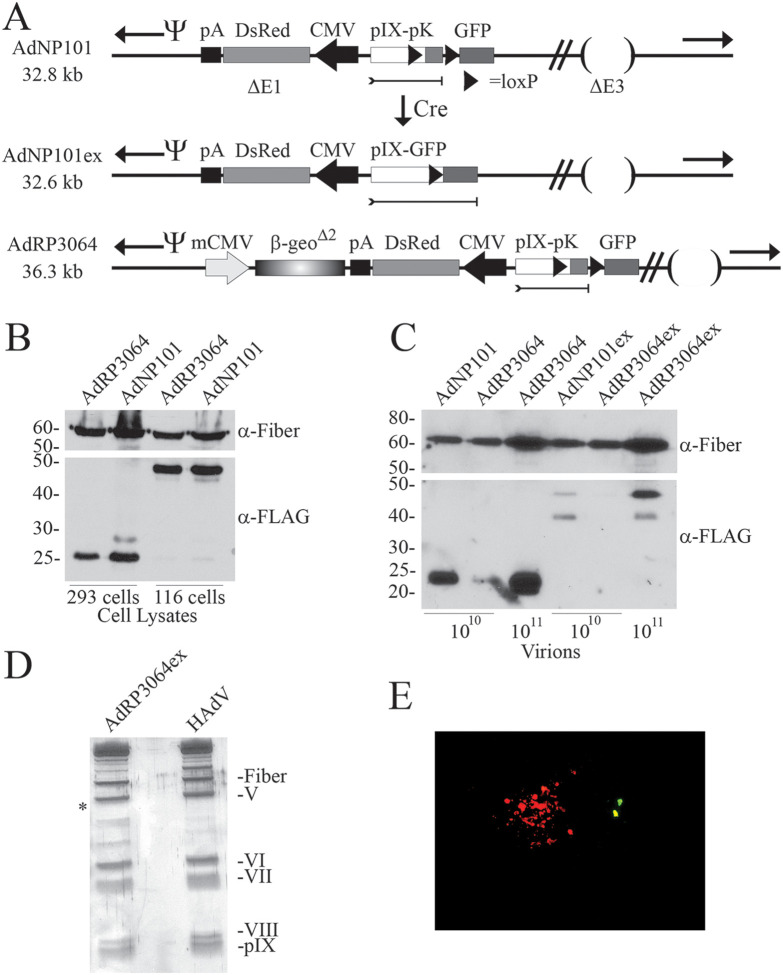
Previously, we used a Cre/loxP strategy to “swap” the targeting polypeptide present on the C terminus of pIX (36). In that study, a loxP-flanked sequence encoding polylysine (pK; binds heparan sulfate proteoglycans) was engineered onto the 3′ terminus of pIX, and the resulting fusion protein could be used for routine propagation of a “detargeted” virus mutated for the ability to bind the native HAdV-5 coxsackie adenovirus receptor. However, propagation of the virus on Cre-expressing cells resulted in Cre-mediated removal of the pK-coding sequence and simultaneously placement of a single-domain antibody coding sequence in frame with pIX for redirecting infection to cancer cells. We used a similar approach in an attempt to generate a large genome virus with pIX-GFP. We designed two viruses, AdNP101 (32.8 kb) and AdRP3064 (36.3 kb), that contained a pIX-loxP-pK-loxP with a GFP gene located downstream (Fig. 5A). Growth of these viruses in 293 cells resulted in exclusive production of the pIX-pK species, whereas growth of the two viruses in 116 cells (i.e., 293 cells expressing Cre recombinase) resulted in almost exclusive production of pIX-GFP (Fig. 5B). An analysis of the protein content of purified virions from virus grown on 293 cells showed that AdRP3064 had an apparent reduced incorporation of the pIX-pK polypeptide relative to AdNP104 (Fig. 5C). Similarly, for Cre-recombined viruses, AdRP3064ex showed reduced incorporation of pIX-GFP relative to AdNP101ex. Silver stain analysis of AdRP3064ex virions showed that the virions contained a small pIX-sized protein that was present at a similar level as native pIX in HAdV virions (Fig. 5D). Since this pIX-sized species in AdRP3064ex virions does not contain a FLAG tag (Fig. 5C), this suggests that, similar to AdRP2384, AdRP3064ex had undergone rearrangement to contain and express a wild-type pIX. Indeed, visual inspection of 293 cells infected with AdRP3064ex showed that DsRed-positive plaques formed without GFP signal, and GFP-positive cells did not spread into plaques (Fig. 5E). Thus, virus that truly expresses pIX-GFP appears unable to generate viable progeny virus to spread to adjacent cells. These data suggest that large polypeptide addition to pIX compromises the stability of viruses with large genomes.
FIG 5.
Cre-mediated swapping of polypeptides on pIX in HAdV vectors. (A) AdNP101 is deleted of E1 and E3 and contains a CMV-DsRed expression cassette replacing the E1 deletion. AdNP101 also encodes pIX fused to a polylysine peptide, which itself is flanked by loxP sites. Propagation of AdNP101 in 293 cells results in expression and incorporation of pIX-pK into the virion. However, propagation of AdNP101 on a Cre-expressing 293 cell line results in excision of the pK-encoding element and simultaneous placement of the GFP coding sequence in frame with pIX, designated AdNP101ex. Although not shown in the schematic, both pIX-pK and pIX-GFP also have a FLAG epitope tag at the 3′ end of the pIX coding region. AdRP3064 is similar in structure to AdNP101 but also contains an internally deleted mCMV-β-geo expression cassette within the E1 deletion. Propagation of AdRP3064 in Cre-expressing 293 cells generates AdRP3064ex. “Ψ” represents the packaging element, inverted terminal repeats are indicated by black arrows, and loxP sites are indicated by black triangles. (B) 293 or 116 cells, a 293-based cell line that expresses the Cre recombinase, were infected with AdRP3064 or AdNP101 at an MOI of 3; at 24 h postinfection, crude cellular protein extracts were prepared and analyzed by immunoblotting for FLAG or fiber. (C) Proteins from purified virions of AdNP101, AdRP3064, AdRPNP101ex, and AdRP3064ex were separated by SDS-PAGE and subjected to immunoblotting for FLAG or fiber (loading control). The number of virions analyzed in each lane is indicated. (D) Protein from purified virions of AdRP3064ex and HAdV were separated by SDS-PAGE and visualized by silver staining. The position where pIX-GFP should migrate is indicated by an asterisk. (E) A purified stock of AdRP3064ex was serially diluted and used to infect 35-mm dishes of 293 cells. The cells were visualized 7 days later using a Zeiss Axiovert 200M microscope, with images collected using a 20× objective.
HAdV vectors preferentially incorporate pIX with small rather than large polypeptide additions.
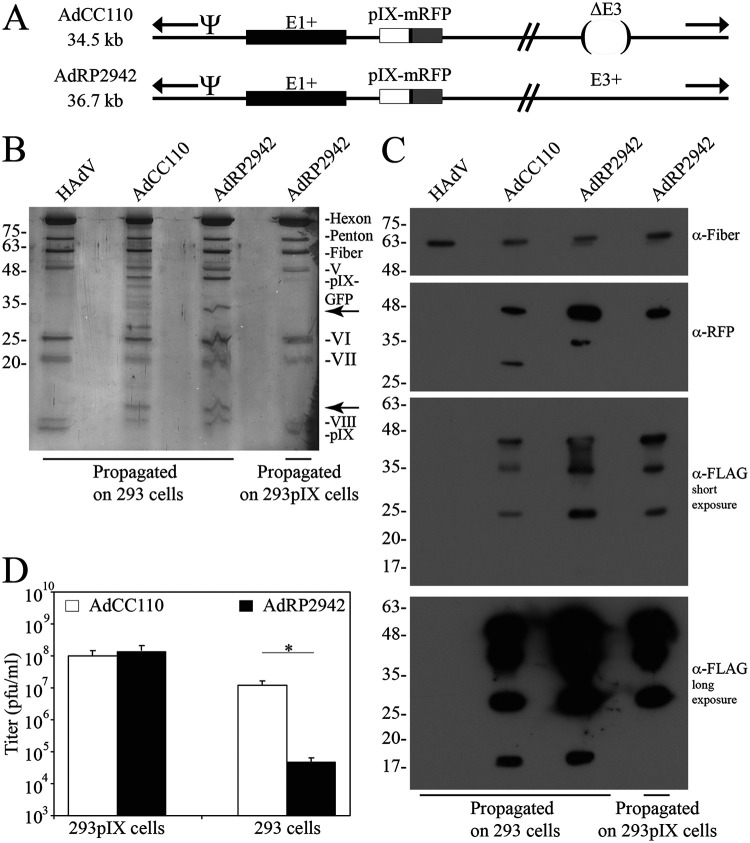
We next tested whether our observations with pIX-GFP were recapitulated with pIX-RFP. We designed two vectors that encoded pIX fused to monomeric red fluorescent protein (mRFP) but differed in the absence or presence of E3, giving rise to vectors of 34.5 kb (AdCC110) and 36.7 kb (AdRP2942), respectively (Fig. 6A). These vectors were rescued and propagated on both 293 and 293pIX cells. An analysis of the virion protein content on silver stain gels yielded two interesting observations. First, AdCC110 and AdRP2942 grown on 293 cells showed incorporation of the ∼45-kDa pIX-RFP protein and no incorporation of native pIX; however, these virions also showed prominent protein bands at ∼17 and ∼35 kDa which were not present in wild-type virions. Second, AdRP2942 grown on 293pIX cells incorporated little pIX-RFP, but similar levels of native pIX as observed in wild-type virions, suggesting a preference for incorporation of native protein. To determine the nature of the ∼17- and ∼35-kDa proteins, we tested whether these proteins were perhaps derived from pIX-RFP. Proteins from virions of HAdV and AdCC110 grown on 293 cells, and AdRP2942 grown on either 293 or 293 pIX cells were subjected to immunoblotting for RFP and FLAG. As shown in Fig. 6C, immunoblotting for RFP showed that in addition to the ∼45-kDa full-length pIX-RFP polypeptide, we also observed other proteins of ∼30 and 35 kDa. The immunoblot for FLAG yielded proteins of ∼45, 35, and 25 kDa, as well as 17 kDa on long exposure. These data suggest that multiple pIX-RFP species are produced in infected cells, either due to premature termination or proteolytic degradation, and that some of the smaller species are preferentially incorporated into virions at near wild-type-levels as native pIX. Different polypeptides likely differ in their translational efficiency or resistance to degradation which, ironically, may mean that virus is less likely to be recovered if the polypeptide addition has higher inherent stability. Monomeric RFP (mRFP) appears to be more prone to instability in HAdV-infected cells than GFP (compare Fig. 3C to 6C).
FIG 6.
Native pIX is preferentially incorporated into HAdV virions rather than pIX-RFP. (A) AdCC110 is deleted of E3 and encodes pIX fused to a cDNA for monomeric red fluorescent protein (mRFP). AdRP2942 is similar in structure to AdCC110 but encodes an intact E3 region. Both AdCC110 and AdRP2942 contain a sequence encoding a FLAG epitope tag between pIX and mRFP, as indicated by the black rectangle. “Ψ” represents the packaging element and inverted terminal repeats are indicated by black arrows. (B) Proteins from purified virions of HAdV, AdCC100, and AdRP2942 propagated on 293 cells and AdRP2942 propagated on 293pIX cells were separated by SDS-PAGE and visualized by silver staining. Bands of unknown identity are indicated by a black arrow. (C) Proteins from purified virions of HAdV, AdCC100, and AdRP2942 propagated on 293 cells and AdRP2942 propagated on 293pIX cells were separated by SDS-PAGE and subjected to immunoblotting for fiber (loading control), mRFP, and FLAG. (D) Stocks of AdCC110 and AdRP2942 that had both been propagated on 293pIX cells were used to infect 35-mm dishes of 293 or 293pIX cells at an MOI of 1; after 48 h, the cells were harvested into the medium. After freeze/thawing to aid in release of the virus, the samples were diluted, and titers were determined on 293pIX cells. The data show averages and standard deviations of n = 2 experiments performed in duplicate.
Finally, our data suggest that there is strong pressure on HAdV encoding pIX-GFP or -RFP that limits their fitness, resulting in the virus exploiting genome and protein instabilities to permit better propagation. We thus evaluated the burst size for these two viruses in the presence or absence of supplemented native pIX. AdCC110 and AdRP2942 that had both been propagated on 293pIX cells were used to infect either 293 or 293pIX cells at an MOI of 1; the infected cells were harvested into the medium and freeze-thawed, and virus titers were determined on 293pIX cells. As shown in Fig. 6D, both AdCC110 an AdRP2942 gave rise to similar bursts of virus when grown on 293pIX cells. However, on 293 cells, both viruses showed a reduction in virus recovery, with recovery of AdRP2942 reduced 3 logs compared to AdCC110. Taken together, our data suggest that for HAdV with genomes close to the wild-type size, the addition of large polypeptides to pIX compromises the ability of the protein to effectively stabilize the virion.
An interesting finding from our study is that pIX with a large polypeptide addition can effectively act to stabilize virions containing small genomes but appears not capable of supporting growth and stabilization of virions containing near-wild-type genome lengths. HAdV DNA is packaged into the capsid under pressure, estimated to be ∼30 atm (46). Presumably, virions with small genomes have lower internal pressure. Thus, a main function of pIX may be to counteract the outward force imposed by the compacted DNA (3). The addition of GFP or RFP to the C terminus of pIX likely does not affect formation of the N-terminal triskelion structure and thus may not affect the stabilization of individual GON. However, such large polypeptide additions likely affect the proper formation of the C-terminal tetrameric coiled-coil structure, weakening the interaction between adjacent GON. The reduced internal pressure in virions with small genomes may allow the virions to tolerate aberrant, or lack of, formation of the C-terminal four-helix bundle and yet still maintain thermal stability. However, pIX tolerability may be directly related to the size and nature of the polypeptide, as well as the size of the genome. For example, addition of GFP to pIX allowed for rescue of small-genome viruses that retained thermal stability, whereas other polypeptides may not. Incorporation of a longer linker region between pIX and the added polypeptide may allow pIX to form its natural stabilizing interconnections (32). Based on our study, we would predict that large polypeptide additions can be accommodated on pIX as long as the genome size is less than 35 kb, since this size reflects the upper limit for DNA packaging in virions lacking pIX stabilizing function (6, 7). Such virions would be stable at normal working temperatures and would only reveal thermal instability at elevated temperatures that are not typically used in normal research settings.
MATERIALS AND METHODS
Cell culture.
All cell culture media and reagents were obtained from Invitrogen (Burlington, Ontario, Canada). 293 (47), 293N3S (48), and A549 (human lung carcinoma, ATCC CCL 185) cells were grown in monolayer in minimum essential medium supplemented with 100 U of penicillin per ml, 100 μg of streptomycin per ml, 0.25 μg of amphotericin B (Fungizone) per ml, and 10% fetal bovine serum (complete medium). The 293-derived cell line that stably expresses the HAdV-5 pIX, 293pIXc4 (10), was grown in complete medium supplemented with 0.4 mg/ml hygromycin and is referred to here as 293pIX cells. The 116 cells, described previously (49), were kindly provided by Philip Ng (Baylor College of Medicine) and were maintained in complete medium supplemented with 0.4 mg/ml hygromycin.
HAdV vector cloning and culture.
All viruses used in this study were based on HAdV-5 (the wild-type virus is referred to as HAdV throughout the present study) and were constructed by using a combination of traditional cloning and RecA+ bacterium-mediated recombination (50). Additional details for cloning are available upon request. In all constructs, pIX expression is under regulation by the endogenous regulator region (i.e., either the intact pIX upstream regulatory region found in the wild-type virus or the remainder of the pIX upstream regulatory region contained in pΔE1sp1A [51]), and the native pIX polyadenylation sequence. Methods for propagation of HAdV vectors in 293 cells have been previously described (52).
AdRP2014, an early region 1 (E1) and E3-deleted vector, with no foreign transgene, was described previously (11, 53). AdRP2330 is similar to AdRP2014 but contains a FLAG epitope tag immediately following the C terminus of pIX (23) (Table 1). To remove the start codon of pIX, pRP2328, a pΔE1Sp1A-based shuttle plasmid encoding a C-terminal FLAG-tagged pIX (19), was PCR amplified with the synthetic oligonucleotides 5′-CGCCCTAGGGCACCAACTCGTTTGATGGAAGCATTG and 5′-GCGCCTAGGCGGCGGCGGCTGCTGCAAAACAGATAC, which amplify in opposite directions around the plasmid and change the native sequence at the start codon (CCATGA to CCTAGG), while also introducing an AvrII restriction site. The resulting linear PCR fragment was digested with AvrII and recircularized, and the resulting plasmid was sequenced to confirm fidelity and designated pRP3009. pRP3009 was recombined with the E1/E3-deleted HAdV genomic plasmid pRP2014, generating pRP3012, and the resulting virus AdRP3012. AdRP3159 and AdRP3160 are both E1/E3-deleted HAdV that contains a pIX-GFP fusion in the native pIX position; however, AdRP3160 is mutated in the native pIX start codon, with an identical sequence as described for AdRP3012.
pRP2306, a pΔE1Sp1A-derived plasmid encoding the pIX-GFP fusion protein, was described previously (19). This construct contains an 18-amino-acid linker between pIX and GFP, comprised primarily of the FLAG epitope tag and a flexible pentaglycine sequence (Table 1). A DsRed (Discosoma sp. red fluorescent protein) expression cassette, controlled by the human cytomegalovirus (CMV) immediate early enhancer/promoter, was cloned into the multiple cloning site of pRP2306, generating pRP2361. pRP2361 was recombined with the pRP2014 genomic plasmid to generate pRP2363 and the resulting virus AdRP2363. pRP2364 is similar in structure to pRP2363 but encodes an unmodified pIX gene. pRP2367 is similar in structure to pRP2364 (i.e., it encodes an unmodified pIX gene) but encodes both the DsRed and a murine CMV (mCMV)-β-geo (β-galactosidase-neomycin resistance gene fusion) expression cassette within the E1 deletion. pRP2384 is similar in structure to pRP2367 but encodes the pIX-GFP fusion protein and is deleted of ∼1.2 kb of β-geo coding sequence in order to maintain the size of the resulting virus below the absolute limit for HAdV DNA packaging (54).
Previously, we used the Cre/loxP recombination system to “swap” targeting motifs present on pIX (36). We used a similar system to create vectors that initially express pIX with a small C-terminal polylysine addition but could be induced to switch to expressing pIX-GFP in Cre-expressing cells. pRP2499 (36) was digested with NotI (filled in with Klenow polymerase) and BstEII and ligated to a 1.9-kb NheI (filled in)/BstEII fragment from pRL16 (23), generating pRP2990. Thus, pRP2990 is a pΔE1sp1A-derived plasmid containing a floxed polylysine-encoding sequence fused to the 3′ end of pIX, with the sequence for GFP downstream of the 3′ loxP site. Cre-mediated excision of the pK coding sequence results in fusion of the GFP coding sequence in-frame with pIX. A 428-bp HindIII/SpeI fragment from pRP2990 containing the floxed pK coding sequence was used to replace a HindIII/SpeI fragment in pRP2361, generating pNP100. pNP100 was recombined with the HAdV genomic plasmid pRP2014, generating pNP101. The amino acid sequence between pIX and GFP is shown in Table 1. A similar virus was constructed containing an additional transgene (a fragment from a mCMV-β-geo expression cassette) to increase the size of the virus genome closer to that of wild-type HAdV-5, designated AdRP3064. In these viruses, the pK motif does not serve any particular function relevant to this study.
HAdV-based vectors expressing a pIX-mRFP (monomeric red fluorescent protein [55]) fusion protein were generated as follows. mRFP was PCR amplified from pRP2483 (23) with the synthetic oligonucleotides 5′-GCGGCTAGCGCCGCCATGGCCTCCTCCGAGGACGTCATC and 5′-GCGACGCGTGGATCCTTAGGCGCCGGTGGAGTGGCGGC; the resulting PCR product was digested with NheI/MluI and cloned into NheI/MluI-digested pRP2288 (23), generating pRP2388. A BamHI fragment containing the pIX-mRFP cassette was cloned into BglII-digested pΔE1sp1AΔpIX (23) and designated pRFPpIX. An SphI fragment from pRFPpIX was used to replace the SphI fragment in pXC1 (56), generating a left-end shuttle plasmid with an intact E1 region and pIX-mRFP, designated pCC106. pCC106 was recombined with the HAdV genomic plasmids pRP2014 (E3–) and pRP2015 (E3+), generating pCC110 and pRP2942, respectively, and the corresponding viruses. The amino acid sequence between pIX and RFP in these constructs is shown in Table 1.
Analysis of virion DNA.
Purified virus (50 μl) was combined with 200 μl of SDS-proteinase K buffer (10 mM Tris-HCl [pH 7.4], 10 mM EDTA, 1% SDS [wt/vol], 1 mg/ml proteinase K). After an overnight incubation at 37°C, the DNA was phenol-chloroform extracted, ethanol precipitated, and resuspended in 50 μl of TE (10 mM Tris-HCl [pH 7.4], 1 mM EDTA). Then, 10 μl of the resulting DNA was digested overnight with XhoI and separated on a 1.0% agarose gel cast in TAE buffer (40 mM Tris [pH 8.3], 20 mM acetic acid, 1 mM EDTA) containing ethidium bromide.
Virus heat inactivation curves.
To determine the effect of heating on virus viability, an aliquot of each virus (108 PFU) was diluted 1/10 in phosphate-buffered saline and heated for 0, 15, or 30 min at 45°C. The virus was then diluted and titers were determined by plaque assay on 293 cells using standard methods.
Growth of viruses.
We examined the burst size of AdCC110 (34.5 kb) and AdRP2942 (36.7 kb), both of which encode pIX-RFP, on 293pIX and 293 cells as follows. Using stocks of both viruses that had been propagated on 293pIX cells, we infected duplicate 35-mm dishes of 293 or 293pIX cells at an MOI of 1; then, after 48 h, we harvested the cells into the medium, added sucrose to a final concentration of 4%, and subjected the crude cell lysate to one round of freeze/thaw. The virus samples were diluted, and titers were determined on 293pIX cells.
Immunoblot analysis.
To examine pIX, pIX-GFP or pIX-RFP incorporation into HAdV capsids by immunoblot analysis, 1010 virus particles, unless otherwise indicated, were mixed with an equal volume of SDS-PAGE protein loading buffer (62.5 mM Tris [pH 6.8], 10% glycerol, 2% SDS, 0.1% bromophenol blue, 5% 2-mercaptoethanol), boiled for 10 min, and separated by electrophoresis on a 15% SDS-polyacrylamide gel. The separated proteins were transferred to a polyvinylidene difluoride membrane (Millipore). The membrane was blocked with 5% milk in TBST (Tris-buffered saline containing 0.2% Tween 20 [Thermo Fisher Scientific]) and probed with antibodies diluted in 5% milk solution. The following primary antibodies were used: HAdV-5 fiber (1/10,000 dilution; Neomarkers, catalog no. MS-1027-P0), antibody raised to all HAd5 capsid proteins (1:10,000; Abcam, catalog no. ab6982), GFP (1:2,000; Invitrogen), RFP (1/5,000; Abcam, catalog no. ab62341), tubulin (1/5,000; Calbiochem, catalog no. CP06), and actin (1/10,000; Sigma-Aldrich, catalog no. A1978). The membranes were washed three times in TBST and incubated with the appropriate secondary antibodies conjugated to horseradish peroxidase (HRP). Blots were visualized by chemiluminescence reaction (Amersham ECL Plus or Immobilon Classico Western HRP substrate [Millipore]) and autoradiography. All immunoblot data are representative of two or more independent experiments.
Silver stain of polyacrylamide gel.
To examine the protein composition of the various HAdV capsids, 2.5 × 108 particles of virus were mixed with equal volume of SDS-PAGE protein loading buffer, boiled for 10 min, and separated by electrophoresis on a 12% SDS-polyacrylamide gel. The resulting gel was incubated in fix (50% ethanol, 5% acetic acid) for 30 min and washed for 10 min in 50% ethanol, followed by a final wash for 10 min in water. The gel was incubated for 2 min in sensitizer (0.02% thiosulfate) and washed twice in water for 5 min. The gel was incubated in 0.1% silver nitrate for 30 min, followed by two rapid washes in developer reagent (0.04% formaldehyde, 2% sodium carbonate). Finally, the gel was incubated in developer and, when sufficient color had developed, the reaction was stopped by incubating the gel in 5% acetic acid.
Immunofluorescence analysis.
Dishes (35 mm) of 293 cells were seeded at a density of 0.6 × 106 cells. The next day, the cells were infected with various dilutions of virus stocks for 1 h, and fresh medium was replaced. The cells were visualized 7 days later using a Zeiss Axiovert 200M microscope. Images were taken using a 20× objective and compiled in Adobe Photoshop CS4.
ACKNOWLEDGMENTS
Funding was provided by grants to R.J.P. from the Canadian Institutes of Health Research (CIHR; MOP-136898 and MOP-142316) and the Natural Sciences and Engineering Research Council (RGPIN-2014-04810 and RGPIN-2019-04786). E.R.M. was supported by an Ontario Graduate Scholarship from the Ontario Provincial Government and a scholarship from the CIHR.
REFERENCES
- 1.Boulanger P, Lemay P, Blair GE, Russell WC. 1979. Characterization of adenovirus protein IX. J Gen Virol 44:783–800. doi: 10.1099/0022-1317-44-3-783. [DOI] [PubMed] [Google Scholar]
- 2.Parks RJ. 2005. Adenovirus protein IX: a new look at an old protein. Mol Ther 11:19–25. doi: 10.1016/j.ymthe.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 3.Flint SJ. 2017. Viral moulds and cement: how interactions among human adenovirus hexons and their protein IX cement may buttress human adenovirus particles. J Mol Biol 429:2752–2754. doi: 10.1016/j.jmb.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 4.Reddy VS. 2017. The role of hexon protein as a molecular mold in patterning the protein IX organization in human adenoviruses. J Mol Biol 429:2747–2751. doi: 10.1016/j.jmb.2017.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colby WW, Shenk T. 1981. Adenovirus type 5 virions can be assembled in vivo in the absence of detectable polypeptide IX. J Virol 39:977–980. doi: 10.1128/JVI.39.3.977-980.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghosh-Choudhury G, Haj-Ahmad Y, Graham FL. 1987. Protein IX, a minor component of the human adenovirus capsid, is essential for the packaging of full length genomes. EMBO J 6:1733–1739. doi: 10.1002/j.1460-2075.1987.tb02425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McVey D, Zuber M, Ettyreddy D, Reiter CD, Brough DE, Nabel GJ, King CR, Gall JG. 2010. Characterization of human adenovirus 35 and derivation of complex vectors. Virol J 7:276. doi: 10.1186/1743-422X-7-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lutz P, Rosa-Calatrava M, Kedinger C. 1997. The product of the adenovirus intermediate gene IX is a transcriptional activator. J Virol 71:5102–5109. doi: 10.1128/JVI.71.7.5102-5109.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosa-Calatrava M, Grave L, Puvion-Dutilleul F, Chatton B, Kedinger C. 2001. Functional analysis of adenovirus protein IX identifies domains involved in capsid stability, transcriptional activity, and nuclear reorganization. J Virol 75:7131–7141. doi: 10.1128/JVI.75.15.7131-7141.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sargent K, Ng P, Evelegh C, Graham FL, Parks RJ. 2004. Development of a size restricted pIX deleted helper virus for amplification of helper dependent adenovirus vectors. Gene Ther 11:504–511. doi: 10.1038/sj.gt.3302107. [DOI] [PubMed] [Google Scholar]
- 11.Sargent K, Meulenbroek RA, Parks RJ. 2004. Activation of adenoviral gene expression by protein IX is not required for efficient virus replication. J Virol 78:5032–5037. doi: 10.1128/jvi.78.10.5032-5037.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosa-Calatrava M, Puvion-Dutilleul F, Lutz P, Dreyer D, de Thé H, Chatton B, Kedinger C. 2003. Adenovirus protein IX sequesters host-cell promyelocytic leukaemia protein and contributes to efficient viral proliferation. EMBO Rep 4:969–975. doi: 10.1038/sj.embor.embor943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu H, Jin L, Koh SB, Atanasov I, Schein S, Wu L, Zhou ZH. 2010. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science 329:1038–1043. doi: 10.1126/science.1187433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu X, Veesler D, Campbell MG, Barry ME, Asturias FJ, Barry MA, Reddy VS. 2017. Cryo-EM structure of human adenovirus D26 reveals the conservation of structural organization among human adenoviruses. Sci Adv 3:e1602670. doi: 10.1126/sciadv.1602670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai X, Wu L, Sun R, Zhou ZH. 2017. Atomic structures of minor proteins VI and VII in human adenovirus. J Virol 91:e00850-17. doi: 10.1128/JVI.00850-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kundhavai Natchiar S, Venkataraman S, Mullen TM, Nemerow GR, Reddy VS. 2018. Revised crystal structure of human adenovirus reveals the limits on protein IX quasi-equivalence and on analyzing large macromolecular complexes. J Mol Biol 430:4132–4141. doi: 10.1016/j.jmb.2018.08.011. [DOI] [PubMed] [Google Scholar]
- 17.Matteson NL, Barry MA, Reddy VS. 2018. Structure-based assessment of protein-protein interactions and accessibility of protein IX in adenoviruses with implications for antigen display. Virology 516:102–107. doi: 10.1016/j.virol.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 18.Zakhartchouk A, Connors W, van Kessel A, Tikoo SK. 2004. Bovine adenovirus type 3 containing heterologous protein in the C terminus of minor capsid protein IX. Virology 320:291–300. doi: 10.1016/j.virol.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 19.Meulenbroek RA, Sargent KL, Lunde J, Jasmin BJ, Parks RJ. 2004. Use of adenovirus protein IX to display large polypeptides on the virion: generation of fluorescent virus through incorporation of pIX-GFP. Mol Ther 9:617–624. doi: 10.1016/j.ymthe.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 20.Le LP, Everts M, Dmitriev IP, Davydova JG, Yamamoto M, Curiel DT. 2004. Fluorescently labeled adenovirus with pIX-EGFP for vector detection. Mol Imaging 3:105–116. doi: 10.1162/1535350041464874. [DOI] [PubMed] [Google Scholar]
- 21.Le LP, Li J, Ternovoi VV, Siegal GP, Curiel DT. 2005. Fluorescently tagged canine adenovirus via modification with protein IX-enhanced green fluorescent protein. J Gen Virol 86:3201–3208. doi: 10.1099/vir.0.80968-0. [DOI] [PubMed] [Google Scholar]
- 22.Tang Y, Le LP, Matthews QL, Han T, Wu H, Curiel DT. 2008. Derivation of a triple mosaic adenovirus based on modification of the minor capsid protein IX. Virology 377:391–400. doi: 10.1016/j.virol.2008.04.023. [DOI] [PubMed] [Google Scholar]
- 23.Poulin KL, Lanthier RM, Smith AC, Christou C, Risco Quiroz M, Powell KL, O’Meara RW, Kothary R, Lorimer IA, Parks RJ. 2010. Retargeting of adenovirus vectors through genetic fusion of a single-chain or single-domain antibody to capsid protein IX. J Virol 84:10074–10086. doi: 10.1128/JVI.02665-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, Le L, Sibley DA, Mathis JM, Curiel DT. 2005. Genetic incorporation of HSV-1 thymidine kinase into the adenovirus protein IX for functional display on the virion. Virology 338:247–258. doi: 10.1016/j.virol.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 25.Matthews QL, Sibley DA, Wu H, Li J, Stoff-Khalili MA, Waehler R, Mathis JM, Curiel DT. 2006. Genetic incorporation of a herpes simplex virus type 1 thymidine kinase and firefly luciferase fusion into the adenovirus protein IX for functional display on the virion. Mol Imaging 5:510–519. [PMC free article] [PubMed] [Google Scholar]
- 26.Liu L, Rogers BE, Aladyshkina N, Cheng B, Lokitz SJ, Curiel DT, Mathis JM. 2014. Construction and radiolabeling of adenovirus variants that incorporate human metallothionein into protein IX for analysis of biodistribution. Mol Imaging 13:7290.2014.00022. doi: 10.2310/7290.2014.00022. [DOI] [PubMed] [Google Scholar]
- 27.Campos SK, Parrott MB, Barry MA. 2004. Avidin-based targeting and purification of a protein IX-modified, metabolically biotinylated adenoviral vector. Mol Ther 9:942–954. doi: 10.1016/j.ymthe.2004.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campos SK, Barry MA. 2006. Comparison of adenovirus fiber, protein IX, and hexon capsomeres as scaffolds for vector purification and cell targeting. Virology 349:453–462. doi: 10.1016/j.virol.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 29.Corjon S, Wortmann A, Engler T, van Rooijen N, Kochanek S, Kreppel F. 2008. Targeting of adenovirus vectors to the LRP receptor family with the high-affinity ligand RAP via combined genetic and chemical modification of the pIX capsomere. Mol Ther 16:1813–1824. doi: 10.1038/mt.2008.174. [DOI] [PubMed] [Google Scholar]
- 30.Garas MN, Tillib SV, Zubkova OV, Rogozhin VN, Ivanova TI, Vasilev LA, Logunov DY, Shmarov MM, Tutykhina IL, Esmagambetov IB, Gribova IY, Bandelyuk AS, Naroditsky BS, Gintsburg AL. 2014. Construction of a pIX-modified adenovirus vector able to effectively bind to nanoantibodies for targeting. Acta Naturae 6:95–105. [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z, Yu B, Wang B, Yan J, Feng X, Wang Z, Wang L, Zhang H, Wu H, Wu J, Kong W, Yu X. 2016. A novel capsid-modified oncolytic recombinant adenovirus type 5 for tumor-targeting gene therapy by intravenous route. Oncotarget 7:47287–47301. doi: 10.18632/oncotarget.10075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vellinga J, Rabelink MJ, Cramer SJ, van den Wollenberg DJ, Van der Meulen H, Leppard KN, Fallaux FJ, Hoeben RC. 2004. Spacers increase the accessibility of peptide ligands linked to the carboxyl terminus of adenovirus minor capsid protein IX. J Virol 78:3470–3479. doi: 10.1128/jvi.78.7.3470-3479.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dmitriev IP, Kashentseva EA, Curiel DT. 2002. Engineering of adenovirus vectors containing heterologous peptide sequences in the C terminus of capsid protein IX. J Virol 76:6893–6899. doi: 10.1128/jvi.76.14.6893-6899.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vellinga J, de Vrij J, Myhre S, Uil T, Martineau P, Lindholm L, Hoeben RC. 2007. Efficient incorporation of a functional hyper-stable single-chain antibody fragment protein-IX fusion in the adenovirus capsid. Gene Ther 14:664–670. doi: 10.1038/sj.gt.3302908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Vrij J, Uil TG, van den Hengel SK, Cramer SJ, Koppers-Lalic D, Verweij MC, Wiertz EJ, Vellinga J, Willemsen RA, Hoeben RC. 2008. Adenovirus targeting to HLA-A1/MAGE-A1-positive tumor cells by fusing a single-chain T-cell receptor with minor capsid protein IX. Gene Ther 15:978–989. doi: 10.1038/gt.2008.26. [DOI] [PubMed] [Google Scholar]
- 36.Poulin KL, Tong G, Vorobyova O, Pool M, Kothary R, Parks RJ. 2011. Use of Cre/loxP recombination to swap cell binding motifs on the adenoviral capsid protein IX. Virology 420:146–155. doi: 10.1016/j.virol.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 37.Kurachi S, Koizumi N, Sakurai F, Kawabata K, Sakurai H, Nakagawa S, Hayakawa T, Mizuguchi H. 2007. Characterization of capsid-modified adenovirus vectors containing heterologous peptides in the fiber knob, protein IX, or hexon. Gene Ther 14:266–274. doi: 10.1038/sj.gt.3302859. [DOI] [PubMed] [Google Scholar]
- 38.de Vrij J, Dautzenberg IJ, van den Hengel SK, Magnusson MK, Uil TG, Cramer SJ, Vellinga J, Verissimo CS, Lindholm L, Koppers-Lalic D, Hoeben RC. 2012. A cathepsin-cleavage site between the adenovirus capsid protein IX and a tumor-targeting ligand improves targeted transduction. Gene Ther 19:899–906. doi: 10.1038/gt.2011.162. [DOI] [PubMed] [Google Scholar]
- 39.Vujadinovic M, Khan S, Oosterhuis K, Uil TG, Wunderlich K, Damman S, Boedhoe S, Verwilligen A, Knibbe J, Serroyen J, Schuitemaker H, Zahn R, Scheper G, Custers J, Vellinga J. 2018. Adenovirus based HPV L2 vaccine induces broad cross-reactive humoral immune responses. Vaccine 36:4462–4470. doi: 10.1016/j.vaccine.2018.06.024. [DOI] [PubMed] [Google Scholar]
- 40.Vujadinovic M, Vellinga J. 2018. Progress in adenoviral capsid-display vaccines. Biomedicines 6:81. doi: 10.3390/biomedicines6030081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salisch NC, Vujadinovic M, van der Helm E, Spek D, Vorthoren L, Serroyen J, Kuipers H, Schuitemaker H, Zahn R, Custers J, Vellinga J. 2017. Antigen capsid-display on human adenovirus 35 via pIX fusion is a potent vaccine platform. PLoS One 12:e0174728. doi: 10.1371/journal.pone.0174728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matthews QL, Farrow AL, Rachakonda G, Gu L, Nde P, Krendelchtchikov A, Pratap S, Sakhare SS, Sabbaj S, Lima MF, Villalta F. 2016. Epitope capsid-incorporation: new effective approach for vaccine development for Chagas disease. Pathog Immun 1:214–233. doi: 10.20411/pai.v1i2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Farrow AL, Peng BJ, Gu L, Krendelchtchikov A, Matthews QL. 2016. A novel vaccine approach for Chagas disease using rare adenovirus serotype 48 vectors. Viruses 8:78. doi: 10.3390/v8030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kennedy MA, Parks RJ. 2009. Adenovirus virion stability and the viral genome: size matters. Mol Ther 17:1664–1666. doi: 10.1038/mt.2009.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith AC, Poulin KL, Parks RJ. 2009. DNA genome size affects the stability of the adenovirus virion. J Virol 83:2025–2028. doi: 10.1128/JVI.01644-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ortega-Esteban A, Condezo GN, Pérez-Berná AJ, Chillón M, Flint SJ, Reguera D, San Martín C, de Pablo PJ. 2015. Mechanics of viral chromatin reveals the pressurization of human adenovirus. ACS Nano 9:10826–10833. doi: 10.1021/acsnano.5b03417. [DOI] [PubMed] [Google Scholar]
- 47.Graham FL, Smiley J, Russell WC, Nairn R. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol 36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 48.Graham FL. 1987. Growth of 293 cells in suspension culture. J Gen Virol 68:937–940. doi: 10.1099/0022-1317-68-3-937. [DOI] [PubMed] [Google Scholar]
- 49.Palmer D, Ng P. 2003. Improved system for helper-dependent adenoviral vector production. Mol Ther 8:846–852. doi: 10.1016/j.ymthe.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 50.Chartier C, Degryse E, Gantzer M, Dieterle A, Pavirani A, Mehtali M. 1996. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J Virol 70:4805–4810. doi: 10.1128/JVI.70.7.4805-4810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bett AJ, Haddara W, Prevec L, Graham FL. 1994. An efficient and flexible system for construction of adenovirus vectors with insertions or deletions in early regions 1 and 3. Proc Natl Acad Sci U S A 91:8802–8806. doi: 10.1073/pnas.91.19.8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ross PJ, Parks RJ. 2009. Construction and characterization of adenovirus vectors. Cold Spring Harbor Protoc 2009:pdb.prot5011. doi: 10.1101/pdb.prot5011. [DOI] [PubMed] [Google Scholar]
- 53.Christou C, Parks RJ. 2011. Rational design of murine secreted alkaline phosphatase for enhanced performance as a reporter gene in mouse gene therapy preclinical studies. Hum Gene Ther 22:499–506. doi: 10.1089/hum.2010.171. [DOI] [PubMed] [Google Scholar]
- 54.Bett AJ, Prevec L, Graham FL. 1993. Packaging capacity and stability of human adenovirus type 5 vectors. J Virol 67:5911–5921. doi: 10.1128/JVI.67.10.5911-5921.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. 2002. A monomeric red fluorescent protein. Proc Natl Acad Sci U S A 99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McKinnon RD, Bacchetti S, Graham FL. 1982. Tn5 mutagenesis of the transforming genes of human adenovirus type 5. Gene 19:33–42. doi: 10.1016/0378-1119(82)90186-X. [DOI] [PubMed] [Google Scholar]