Human adenovirus type 55 (HAdV55) has recently emerged as a highly virulent respiratory pathogen and has been linked to severe and even fatal pneumonia in immunocompetent adults. However, the cellular receptors mediating the entry of HAdV55 into host cells remain unclear, which hinders the establishment of HAdV55-infected animal models and the development of antiviral approaches. In this study, we demonstrated that human desmoglein-2 (hDSG2) plays the major roles during HAdV55 infection. Human CD46 (hCD46) could also mediate the infection of HAdV55, but the efficiency was much lower than for hDSG2. We generated two transgenic mouse lines that express either hDSG2 or hCD46, both of which enabled HAd55 infection in otherwise nontransgenic mice. hDSG2 transgenic mice enabled more efficient HAdV55 infection than hCD46 transgenic mice. Our study adds to our understanding of HAdV55 infection and provides an animal model for evaluating HAdV55 vaccines and therapeutics.
KEYWORDS: human adenovirus type 55, human CD46, human desmoglein-2, receptor, transgenic mouse
ABSTRACT
Human adenovirus type 55 (HAdV55) represents an emerging respiratory pathogen and causes severe pneumonia with high fatality in humans. The cellular receptors, which are essential for understanding the infection and pathogenesis of HAdV55, remain unclear. In this study, we found that HAdV55 binding and infection were sharply reduced by disrupting the interaction of viral fiber protein with human desmoglein-2 (hDSG2) but only slightly reduced by disrupting the interaction of viral fiber protein with human CD46 (hCD46). Loss-of-function studies using soluble receptors, blocking antibodies, RNA interference, and gene knockout demonstrated that hDSG2 predominantly mediated HAdV55 infection. Nonpermissive rodent cells became susceptible to HAdV55 infection when hDSG2 or hCD46 was expressed, but hDSG2 mediated more efficient HAd55 infection than hCD46. We generated two transgenic mouse lines that constitutively express either hDSG2 or hCD46. Although nontransgenic mice were resistant to HAdV55 infection, infection with HAdV55 was significantly increased in hDSG2+/+ mice but was much less increased in hCD46+/+ mice. Our findings demonstrate that both hDSG2 and hCD46 are able to mediate HAdV55 infection but hDSG2 plays the major roles. The hDSG2 transgenic mouse can be used as a rodent model for evaluation of HAdV55 vaccine and therapeutics.
IMPORTANCE Human adenovirus type 55 (HAdV55) has recently emerged as a highly virulent respiratory pathogen and has been linked to severe and even fatal pneumonia in immunocompetent adults. However, the cellular receptors mediating the entry of HAdV55 into host cells remain unclear, which hinders the establishment of HAdV55-infected animal models and the development of antiviral approaches. In this study, we demonstrated that human desmoglein-2 (hDSG2) plays the major roles during HAdV55 infection. Human CD46 (hCD46) could also mediate the infection of HAdV55, but the efficiency was much lower than for hDSG2. We generated two transgenic mouse lines that express either hDSG2 or hCD46, both of which enabled HAd55 infection in otherwise nontransgenic mice. hDSG2 transgenic mice enabled more efficient HAdV55 infection than hCD46 transgenic mice. Our study adds to our understanding of HAdV55 infection and provides an animal model for evaluating HAdV55 vaccines and therapeutics.
INTRODUCTION
Human adenoviruses (HAdVs) are nonenveloped, double-stranded DNA viruses belonging to the family Adenoviridae. Up to 90 genotypes of HAdVs, divided into seven species (A to G), have been identified according to their hemagglutination activities, genomic sequences, and additional properties (1). Recently, severe pneumonia cases and deaths have been reported in outbreaks associated with human adenovirus type 55 (HAdV55) (2–11). It has been proposed that HAdV55 evolved from recombination between HAdV11 and HAdV14 (11, 12). The genome of HAdV55 has 97.6% nucleotide sequence identity with that of HAdV11 and 98.8% with that of HAdV14. Similar to HAdV11 and HAdV14, HAdV55 is also classified into species B based on genetic alignment (12). However, unlike HAdV11, which typically infects the kidney and urinary tract (13), HAdV55 is similar to HAdV14 and causes mainly respiratory infections (2, 4, 13, 14). The level of herd immunity to HAdV55 is 22.4%, much lower than that to other HAdVs, such as HAdV4 (58.4%), HAdV5 (77%), and HAdV7 (63.8%) (15–17). HAdV55 has exerted potential threats to public health, especially to crowded populations, including military recruits and school students (2–12). Currently, no prophylactic vaccines or antiviral drugs are available.
HAdV infection into host cells is initially mediated by the attachment of viral trimeric fiber protein to a cellular surface receptor (18–25). Most HAdVs, except those in species B and some genotypes in species D, utilize human coxsackie-adenovirus receptor (hCAR) as the primary receptor (18, 19, 25). As for species B HAdVs, two receptors have been reported, human CD46 (hCD46) and human desmoglein-2 (hDSG2). hCD46 is a ubiquitously expressed membrane cofactor protein that protects host cells from complement damage. hCD46 has been recognized as the cellular receptor for several human pathogens, including measles virus, human herpesvirus 6, and Neisseria, in addition to species B HAdVs (20, 26–28). hDSG2 is an integral component of desmosomes that contributes to the formation of cell-cell junctions (29). Loss of hDSG2 function may affect cardiomyocyte cohesion and may be associated with life-threatening cardiac diseases, such as sudden cardiac death (30, 31). It has been proposed that species B HAdVs have three groups based on the receptors to which they bind (19). Group 1 exclusively uses hCD46 as an attachment receptor, including HAdV16, -21, -35, and -50; group 2 predominantly engages hDSG2 for productive infection, including HAdV3, -7, and -14; and group 3 employs both hCD46 and hDSG2, such as HAdV11 (18–24). However, there are existing disputes regarding the receptor usage of species B HAdVs. Earlier studies showed that ectopic expression of hCD46 in rodent cells promoted infection with HAdV3, -11, -14, and -35 (21–23), and the short consensus repeat domains I and II were the common binding sites engaged by species B HAdVs (32, 33). There are also reports that hDSG2 is the major receptor for HAdV3, -7, and -14 (19, 34). However, one report has shown that hCD46 makes no significant contribution to HAdV3 infection (35). These discrepancies, according to several subsequent studies, might be attributed to the affinity of fiber protein for different cellular receptors (18, 33).
The cellular receptor that mediates HAdV55 infection has not been elucidated. Based on the genomic analysis, HAdV55 is phylogenetically close to HAdV14 in fiber and penton but phylogenetically close to HAdV11 in hexon (11, 12). It is important to note that the fiber knob (FK) domain of HAdV55 (Y16/SX/2011 strain) has a 139Val-to-139Ile mutation compared to that of prototype HAdV14 (de Wit strain) and has an insertion of two additional amino acids, 251Lys and 252Glu, compared to that of HAdV14a (CHN/GZ01/2010 strain) (12, 36). It remains unclear if these mutations alter the interaction of HAdV55 with cellular receptors and thereby lead to a different pattern of receptor usage. Defining the receptor usage of HAdV55, therefore, will add to our understanding of its infection process, facilitate the establishment of animal models supporting HAdV55 infection, and contribute to the development of HAdV55 vaccines and therapeutics.
In this study, we applied multiple loss-of-function and gain-of-function experiments to investigate the roles of hDSG2 and hCD46 in mediating HAdV55 attachment and infection. Since rodent cells and mice cannot be productively infected by HAdV55, we further generated transgenic mouse lines expressing hDSG2 or hCD46 for evaluating HAdV55 infection in otherwise nonpermissive mice.
RESULTS
Interaction of HAdV55 fiber with hDSG2 is required for HAdV55 infection.
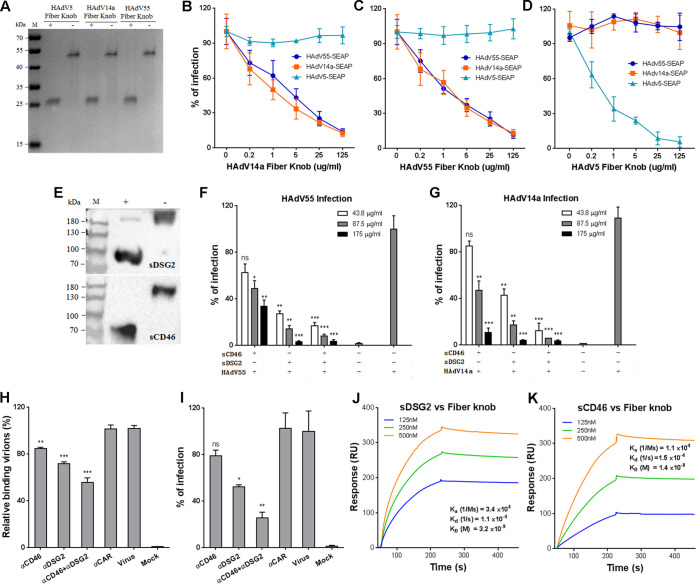
Given that HAdV55 fiber is phylogenetically close to HAdV14 fiber and since earlier studies showed that the interaction of fiber with hDSG2 or hCD46 is necessary for HAdV14 infection (19, 23, 36), we investigated if these interactions were also required for HAdV55 infection. We produced the fiber knobs of HAdV55, HAdV14a, and HAdV5 in Escherichia coli. All these fiber knobs showed a polymeric form that could be disrupted into a monomeric form by preheating under a reducing condition (Fig. 1A), similar to those reported by others (34, 36, 37). Human lung adenocarcinoma A549 cells were preincubated with fiber knobs and then infected with HAdV55-SEAP, HAdV14-SEAP, or HAdV5-SEAP expressing a reporter protein-secreted embryonic alkaline phosphatase (SEAP). HAdV14a fiber knob blocked the infection with HAdV14a and HAdV55 but not HAdV5 in a dose-dependent manner (Fig. 1B). HAdV55 fiber knob also dose dependently blocked infection with HAdV55 and HAdV14a but not HAdV5 (Fig. 1C). As a control, HAdV5 fiber knob only blocked infection with HAdV5 and not HAdV14a or HAdV55 (Fig. 1D). Therefore, HAdV55 fiber knob effectively disrupts the interaction of HAdV55 and HAdV14a with their receptors on host cells.
FIG 1.
Blocking the interaction of HAdV55 fiber with hDSG2 reduces HAdV55 binding and infection. (A) SDS-PAGE analysis of the fiber knobs of HAdV5, HAdV14a and HAdV55. +, the samples were preheated in the presence of β-mercaptoethanol; -, the samples were not preheated and β-mercaptoethanol was not added. (B) HAdV14a fiber knob competitively inhibits the infection of HAdV55 and HAdV14a. (C) HAdV55 fiber knob competitively inhibits the infection of HAdV55 and HAdV14a. (D) HAdV5 fiber knob competitively inhibits the infection of HAdV5 but not HAdV55 or HAdV14a. A549 cells were preincubated with the fiber knob of HAdV14a, HAdV55, or HAdV5 and then infected with HAdV14a-SEAP, HAdV55-SEAP, or HAdV5-SEAP at 200 vp/cell. At 24 h postinfection, the SEAP activity in the cultured media was measured. Percent infection was calculated as the percentage of SEAP activity in fiber knob-treated cells versus that in PBS-treated cells. (E) Western blot analysis of sCD46 and sDSG2 under reducing and nonreducing conditions. (F) sDSG2 and sCD46 competitively inhibit HAdV55 infection. (G) sDSG2 and sCD46 competitively inhibit HAdV14a infection. HAdV55-SEAP or HAdV14a-SEAP was incubated with sDSG2, sCD46, or both and then used to infect A549 cells (200 vp/cell). At 24 h postinfection, the SEAP activity in the cultured media was measured. Percent infection was calculated as the percentage of SEAP activity resulting from HAdV55-SEAP or HAdV14a-SEAP infection in the presence of sDSG2, sCD46, or both versus that in the absence of these competitors. (H) hDSG2 antibodies reduce HAdV55 binding to host cells. A549 cells were preincubated with hDSG2 antibodies (6D8 and 8E5), hCD46 antibodies (M177 and MEM258), or both and then incubated with HAdV55. The relative bound viruses were measured by qPCR and calculated using β-actin as a reference. Genome copies of HAdV55 bound to A549 cells were taken as 100%. (I) hDSG2 antibodies reduce HAdV55 infection. A549 cells preincubated with either hDSG2 antibodies or hCD46 antibodies were infected with HAdV55-SEAP at 50 vp/cell. At 24 h postinfection, the SEAP activity in the cultured media was measured. Percent infection was calculated as the percentage of SEAP activity resulting from HAdV55-SEAP infection in the presence of anti-hDSG2 antibodies, anti-sCD46 antibodies, or both versus that in the absence of these antibodies. (J) Kinetics of binding of HAdV55 fiber knob to sDSG2. (K) Kinetics of binding of HAdV55 fiber knob to sCD46. Data are representative of those from three independent experiments and presented as the means ± standard deviations (SD). Comparison between groups was performed by one-way analysis of variance (ANOVA; n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, no significance.
We next examined if HAdV55 infection could be competitively inhibited by soluble hDSG2 (sDSG2) or hCD46 (sCD46), two homodimers comprising the extracellular domains fused to human IgG Fc region (Fig. 1E). HAdV55-SEAP or HAdV14a-SEAP was incubated with sDSG2 or sCD46 and was then used to infect A549 cells. Both sDSG2 and sCD46 reduced HAdV55 infection in a dose-dependent manner (Fig. 1F). However, sDSG2 showed a much greater inhibitory effect than sCD46. The combination of sDSG2 and sCD46 moderately further reduced HAdV55 infection (Fig. 1F). Similarly, sDSG2 and sCD46 dose dependently inhibited HAdV14a infection, with sDSG2 showing a much greater inhibitory effect (Fig. 1G). This result suggests that sDSG2 and sCD46 are able to block the interaction of HAdV55 with its receptors, but sDSG2 has a significantly higher blocking efficacy than sCD46.
We employed monoclonal antibodies to further examine the interaction of HAdV55 with its possible receptors. A549 cells were preincubated with blocking monoclonal antibody cocktails against hDSG2 or hCD46 and then infected with HAdV55-SEAP. Both hDSG2 antibodies and hCD46 antibodies reduced the binding of HAdV55, and hDSG2 antibodies achieved a greater reduction than hCD46 antibodies (Fig. 1H). The combination of these antibodies resulted in the greatest reduction in viral binding (Fig. 1H). In contrast, hCAR antibodies had no blocking effects on HAdV55 binding. Accordingly, SEAP expression mediated by HAdV55-SEAP was reduced by hDSG2 antibodies and, to a lesser degree, by hCD46 antibodies. Concurrent incubation with both hDSG2 antibodies and hCD46 antibodies resulted in the most reduction (Fig. 1I). Therefore, the interaction of fiber with hDSG2 or hCD46 is required for HAdV55 binding and infection, but hDSG2 appears to play a greater role than hCD46. We then performed surface plasmon resonance (SPR) analysis to examine the interaction of HAdV55 fiber knob with hDSG2 and hCD46. HAdV55 fiber knob showed binding activity to both sDSG2 and sCD46 (Fig. 1J and K), suggesting that HAdV55 fiber knob directly interacts with hDSG2 and hCD46.
HAdV55 infection is correlated with the expression level of hDSG2 and, to a lesser degree, with that of hCD46 on different cell lines.
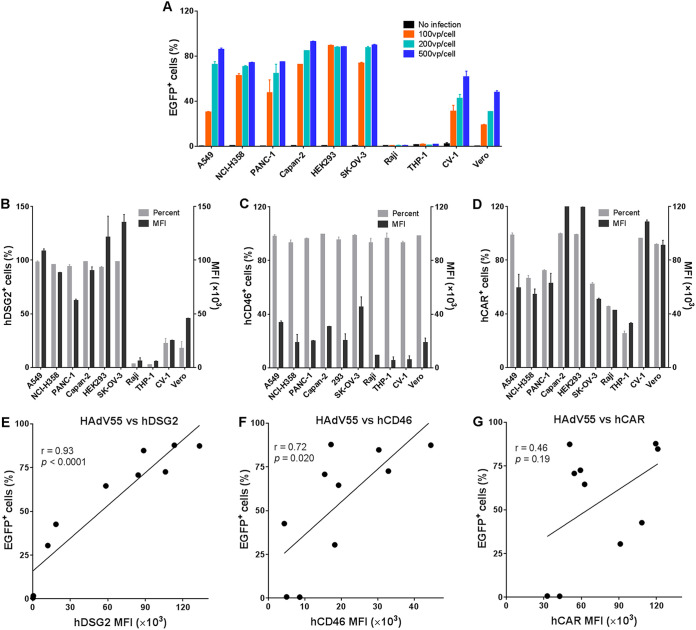
To determine if the infection efficiency of HAdV55 in a variety of cells is related to the expression level of hDSG2 and hCD46, we performed infection assays using 10 cell lines derived from humans and monkeys. Human epithelial-like cells, including A549, NCI-H358, PANC-1, Capan-2, HEK293, and SK-OV-3 cells, as well as monkey CV-1 and Vero cells, could be efficiently infected by HAdV55-enhanced green fluorescent protein (EGFP) (Fig. 2A). However, human hematopoietic cell lines Raji and THP-1 showed no HAdV55-EGFP infection (Fig. 2A). We examined the expression of hDSG2 and hCD46 on these cells by flow cytometry analysis using monoclonal antibodies. All human epithelial-like cells expressed a high level of hDSG2 and a somewhat lower level of hCD46. However, monkey CV-1 and Vero cells showed weak staining by hDSG2 antibody and moderate staining by hCD46 antibody (Fig. 2B and C). The amino acid sequences of hDSG2 and hCD46 and their counterparts in African green monkey (Chlorocebus sabaeus) have similarities of up to 96.3% and 86.2%, respectively. The DSG2 and CD46 expressed in these two cell lines could be recognized by anti-hDSG2 or anti-hCD46 antibodies in a Western blot assay (data not shown). The weak staining, therefore, most likely reveals a relatively low abundance of these two receptors on monkey cells. Notably, Raji cells and THP-1 cells showed weakly positive staining for hCD46 but were negative for hDSG2 (Fig. 2B and C). As a control, hCAR was detected in all human epithelial-like cells and monkey cells and, at a lesser abundance, in Raji cells and THP-1 cells (Fig. 2D). Regression analysis revealed that the infectivity of HAdV55 in these cell lines was strongly correlated with the expression of hDSG2 (r = 0.93 and P < 0.0001) but weakly correlated with the expression of hCD46 (r = 0.72 and P = 0.02). No correlation between HAdV55 infection and the expression of hCAR was observed (r = 0.46 and P = 0.19) (Fig. 2E to G). Thus, HAdV55 infection is most likely associated with hDSG2 and, to a lesser degree, with hCD46 but not hCAR on the cell surface.
FIG 2.
HAdV55 infection is positively correlated with the expression level of hDSG2. (A) Infection efficiency of HAdV55-EGFP in different cell lines derived from humans and monkeys. Each indicated cell line was infected with HAdV55-EGFP at 0, 100, 200, and 500 vp per cell. At 24 h postinfection, cells were examined using flow cytometry. The frequency of EGFP-positive cells is shown. Data are presented as the means ± SD. (B) Expression level of hDSG2 in different cell lines. (C) Expression level of hCD46 in different cell lines. (D) Expression level of hCAR in different cell lines. Each indicated cell line was stained with a FITC-labeled anti-hDSG2 antibody, a PE-labeled anti-hCD46 antibody, or a PE-labeled anti-hCAR antibody and examined using flow cytometry. The percentage of positive cells and the mean fluorescence intensity (MFI) of each cell line are shown. (E) Correlation of HAdV55 infectivity to the expression level of hDSG2. (F) Correlation of HAdV55 infectivity to the expression level of hCD46. (G) Correlation of HAdV55 infectivity to the expression level of hCAR. The infectivity of HAdV55 is presented as the frequency of EGFP-positive cells resulting from HAdV55-EGFP infection at 200 vp per cell. Representative results from three independent experiments are shown. The strength of the correlation was measured by the Pearson correlation coefficient.
Knockdown and knockout of hDSG2 decrease HAdV55 attachment and infection.
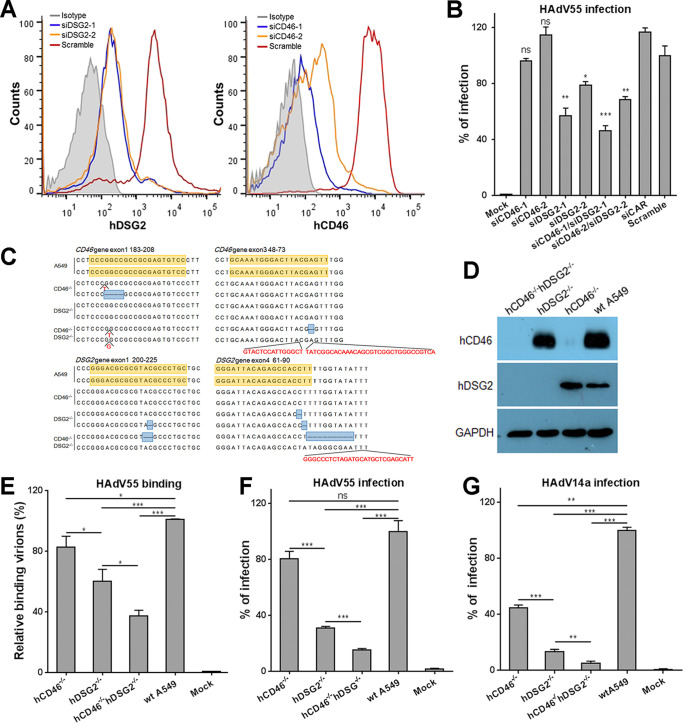
To test if reducing the expression of hDSG2 or hCD46 in host cells can affect HAdV55 infection, we knocked down the expression of hDSG2 or hCD46 in A549 cells using RNA interference (Fig. 3A). Knockdown of hDSG2 alone significantly reduced HAdV55 infection (Fig. 3A and B), whereas knockdown of hCD46 alone showed no significant reduction of HAdV55 infection (Fig. 3A and B). Concurrent downregulation of hDSG2 and hCD46 also reduced HAdV55 infection, but not lower than knockdown of hDSG2 alone (Fig. 3B). The small interfering RNA targeting hCAR and the scramble control showed no significant influence on HAdV55 infection (Fig. 3B).
FIG 3.
Knockdown or knockout of hDSG2 reduces HAdV55 binding and infection. (A) siRNAs downregulate the expression of hDSG2 and hCD46. A549 cells were transfected with siRNAs targeting hDSG2 or hCD46 or a scrambled control. At 60 h posttransfection, cells were stained with a FITC-labeled anti-hDSG2 antibody, a PE-labeled anti-hCD46 antibody, or an isotype control and examined by flow cytometry. One representative graph for hDSG2 (left) and hCD46 (right) knockdown is shown. (B) Downregulating the expression of hDSG2 reduces HAdV55 infection. A549 cells pretransfected with siRNAs were infected with HAdV55-SEAP at 200 vp/cell. At 24 h postinfection, the SEAP activity in the cultured media was measured. Percent infection was calculated as the percentage of the SEAP activity in cells transfected with siRNAs versus that in cells transfected with the scrambled control. (C) Alignment of the genomic sequence flanking the target sequence of the sgRNA in gene knockout cell lines with wild-type A549 cells. The deletions and the insertions in both alleles are marked. (D) Knockout of the hDSG2 and hCD46 genes eliminates their expression. The expression levels of hDSG2 and hCD46 in hCD46−/− cells, hDSG2−/− cells, hCD46−/− hDSG2−/− cells, and wild-type (wt) A549 cells were examined by Western blot analysis. GAPDH was examined in parallel as an internal control. (E) Knockout of hDSG2 decreases HAdV55 binding. Gene knockout cell lines as well as wild-type A549 cells were incubated with HAdV55 viral particles at 200 vp/cell, and the genome copies of the bound viral particles were measured by qPCR and calculated using β-actin as a reference. Genome copies of HAdV55 bound to wt A549 were taken as 100%. Uninfected A549 cells were used as negative controls. (F) Knockout of hDSG2 decreases HAdV55 infection. (G) Knockout of hDSG2 decreases HAdV14a infection. Gene knockout cell lines as well as wild-type A549 cells were infected with HAdV55-SEAP (F) or HAdV14a-SEAP (G) at 200 vp/cell. One day later, the SEAP activity in the cultured media was measured. Percent infection was calculated as the percentage of the SEAP activity in gene knockout cell lines versus that in wild-type A549 cells. Data are representative of those from three independent experiments and presented as the means ± SD. Comparison between groups was conducted by one-way ANOVA (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To rule out the possibility that residual hDSG2 and hCD46 on cell surface still mediate HAdV55 infection, we performed gene knockout in A549 cells using CRISPR-Cas9 technology to completely eliminate the expression of hDSG2 or hCD46 or both hDSG2 and hCD46. Three cell lines were constructed by introducing biallelic frameshift mutations into the exons of the DSG2 gene (hDSG2−/−) or CD46 gene (hCD46−/−) or both (hCD46−/− hDSG2−/−) (Fig. 3C). hDSG2−/− cells expressed hCD46 but not hDSG2, whereas hCD46−/− cells expressed hDSG2 but not hCD46 (Fig. 3D). hCD46−/− hDSG2−/− cells had no expression of either hCD46 or hDSG2 (Fig. 3D). hDSG2−/− cells showed significantly decreased binding to HAdV55 compared to that of wild-type A549 cells, whereas hCD46−/− cells showed only slightly decreased binding to HAdV55 compared to that of wild-type A549 cells (Fig. 3E). The binding of hCD46−/− hDSG2−/− cells to HAdV55 was further reduced (Fig. 3E). This result reveals that hDSG2 is the key contributor to HAdV55 binding.
To determine if complete knockout of hDSG2 or hCD46 or both hDSG2 and hCD46 reduces HAdV55 infection, we assessed the SEAP activity in the cultured media of these cells infected with HAdV55-SEAP. Compared to wild-type A549 cells, hDSG2−/− cells that expressed hCD46 but not hDSG2 showed a significant reduction of SEAP expression, whereas hCD46−/− cells that expressed hDSG2 but not hCD46 showed no significant reduction of SEAP expression (Fig. 3F). Concurrent knockout of hDSG2 and hCD46 further reduced SEAP expression (Fig. 3F). We also performed HAdV14a infection using these knockout cell lines. Knockout of hCD46 significantly reduced HAdV14a infection, to ∼40% (Fig. 3G), implying that hCD46 plays a slightly more important role in HAdV14a infection than in HAdV55 infection. Knockout of hDSG2 showed a much greater inhibition than knockout of hCD46 on HAdV14a infection (Fig. 3G), suggesting that similar to HAdV55, HAdV14a also uses hDSG2 as the major receptor. Together, these results show that hDSG2 has a dominating role in mediating the infection of HAd55 and HAdV14a, but hCD46 plays only a minor role in mediating the infection of these two HAdVs.
Expression of hDSG2 enables HAdV55 infection in nonpermissive rodent cells.
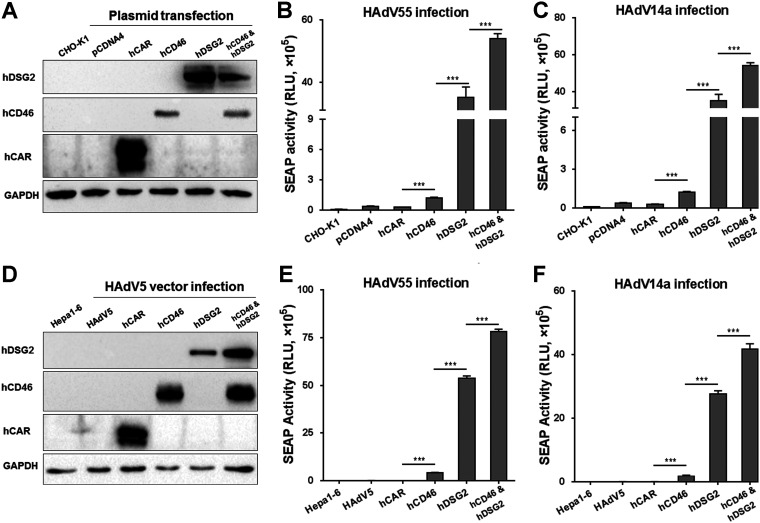
To investigate if ectopic expression of hDSG2 or hCD46 in rodent cells can enable HAdV55 infection, we transfected CHO-K1 cells with plasmids expressing either hDSG2 or hCD46 or both. Forty-eight hours later, cells were infected with HAdV55-SEAP. Cells expressing hDSG2 could be efficiently infected by HAdV55, as shown by high SEAP activity in the cultured medium (Fig. 4A and B). Cells expressing hCD46 could also be infected by HAdV55, but the efficiency was much lower than that in hDSG2-expressing cells. Cells expressing both hDSG2 and hCD46 were the most efficiently infected by HAdV55 (Fig. 4A and B). Infection with HAdV14a in CHO-K1 cells transiently expressing hDSG2 or hCD46 or both showed a pattern similar to that of infection with HAdV55 (Fig. 4C), revealing that the presence of hDSG2 effectively supports the infection of HAdV55 and HAdV14a in nonpermissive CHO-K1 cells.
FIG 4.
Transient expression of hDSG2 promotes HAdV55 infection in rodent cells. (A) Plasmid-mediated expression of hDSG2 and hCD46. CHO-K1 cells were transfected with either pCDNA4-hDSG2, pCDNA4-hCD46, or both and the resultant expression of hDSG2 or hCD46 was examined by Western blot analysis. Cells transfected with pCDNA4-hCAR or pCDNA4TO were used as negative controls. GAPDH was also examined as an internal control. The blots are from different gels with equal volumes of each sample loaded. (B) Transient expression of hDSG2 facilitates HAdV55 infection in CHO-K1 cells. (C) Transient expression of hDSG2 facilitates HAdV14a infection in CHO-K1 cells. Transfected cells were infected with HAdV55-SEAP or HAdV14a-SEAP at 200 vp/cell, and the SEAP activity in the cultured media was measured 24 h after infection. (D) HAdV5-mediated expression of hDSG2 and hCD46. Hepa1-6 cells were preinfected with either HAdV5-hDSG2, HAdV5-hCD46, or both, and the resultant expression of hDSG2 or hCD46 was examined by Western blot analysis. Cells preinfected with HAdV5-hCAR or empty HAdV5 were used as negative controls. GAPDH was also examined as an internal control. The blots are from different gels with equal volumes of each sample loaded. (E) HAdV5-mediated expression of hDSG2 facilitates HAdV55 infection in Hepa1-6 cells. (F) HAdV5-mediated expression of hDSG2 facilitates HAdV14a infection in Hepa1-6 cells. Cells preinfected with HAdV5-hDSG2 or HAdV5-hCD46 were infected with HAdV55-SEAP or HAdV14a-SEAP at 200 vp/cell. The SEAP activity in the cultured media was measured 24 h after infection. Data are representative of those from three independent experiments and presented as the means ± SD. Statistical analysis was conducted by one-way ANOVA (n = 3). ***, P < 0.001.
We then validated this result using another rodent cell line Hepa1-6. Hepa1-6 cells express CAR and thus can be infected by HAdV5, but they cannot be infected by species B HAdVs. The amino acid sequence identities of DSG2 and CD46 between mice and humans are only 76% and 49%, respectively (38–41). Hepa1-6 cells were preinfected with recombinant HAdV5 vectors expressing hDSG2 or hCD46 and then infected with HAdV55-SEAP. Similar to the results with CHO-K1 cells, Hepa1-6 cells expressing hDSG2 effectively supported HAdV55-SEAP infection (Fig. 4D and E). Hepa1-6 cells expressing hCD46 only weakly supported HAdV55-SEAP infection (Fig. 4D and E). Hepa1-6 cells expressing both hDSG2 and hCD46 most effectively supported HAdV55-SEAP infection (Fig. 4D and E). Notably, HAdV14a infection in these Hepa1-6 cells infected with HAdV5 vectors exhibited a pattern similar to that of HAdV55 infection (Fig. 4F). Therefore, rodent cells can be efficiently infected by HAdV55 when hDSG2 alone is expressed but can only be moderately infected by HAdV55 when hCD46 alone is expressed.
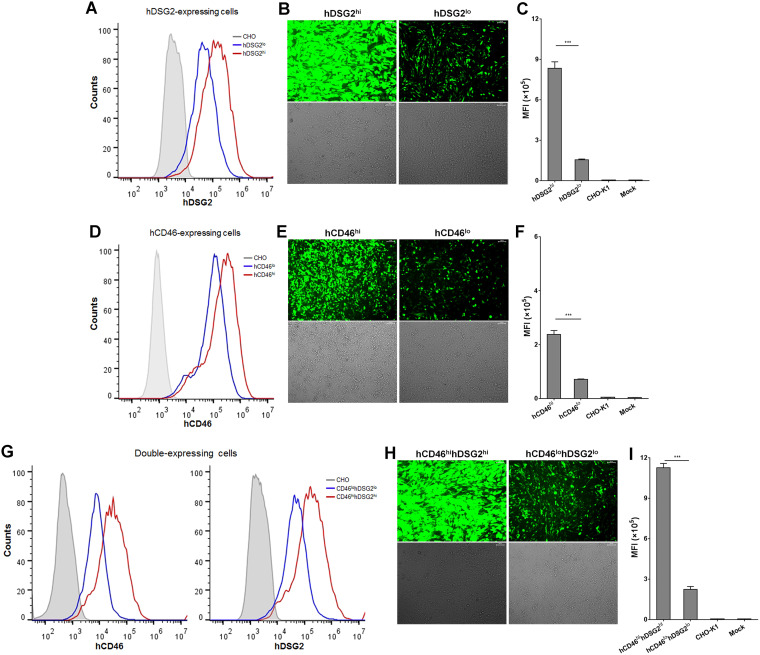
To further dissect the contribution of hDSG2 and hCD46 during HAdV55 infection, we constructed six CHO-K1-derived cell lines stably expressing hDSG2, hCD46, or both hDSG2 and hCD46 at either a low or a high level. hDSG2-expressing cells effectively supported HAdV55-EGFP infection, and the efficiency was positively correlated with the abundance of hDSG2 (Fig. 5A to C). Compared to hDSG2-expressing cells, hCD46-expressing cells were moderately susceptible to HAdV55-EGFP infection (Fig. 5D to F). Cells expressing both hDSG2 and hCD46 exhibited high susceptibility to HAdV55-EGFP infection (Fig. 5G to I). Consistent results from different cell lines suggest that both hDSG2 and hCD46 can mediate HAdV55 infection but that hDSG2 has much greater efficiency.
FIG 5.
CHO-K1-derived stable cell lines expressing hDSG2 are more susceptible to HAdV55 infection than hCD46-expressing cells. (A) CHO-K1-derived stable cell lines expressing hDSG2. (B) hDSG2-expressing cell lines are susceptible to HAdV55 infection. (C) MFI of cells infected with HAdV55-EGFP. (D) CHO-K1-derived stable cell lines expressing hCD46. (E) hCD46-expressing cell lines are moderately infected by HAdV55. (F) MFI of cells infected with HAdV55-EGFP. (G) CHO-K1-derived stable cell lines simultaneously expressing hDSG2 and hCD46. (H) Cell lines expressing both hDSG2 and hCD46 are susceptible to HAdV55 infection. (I) MFI of cells infected with HAdV55-EGFP. Each stable cell line was stained with a FITC-labeled anti-hDSG2 antibody or a PE-labeled anti-hCD46 antibody and examined by flow cytometry. The cell lines were then infected with HAdV55-EGFP at 200 vp/cell. At 24 h postinfection, cells were imaged under a fluorescence microscope, and the expression of EGFP was examined by flow cytometry. Data are representative of those from three independent experiments. The MFI is presented as the mean ± SD. Comparison between groups was conducted by one-way ANOVA (n = 3). ***, P < 0.001.
hDSG2 knock-in transgenic mice and hCD46 knock-in transgenic mice support HAdV55 infection.
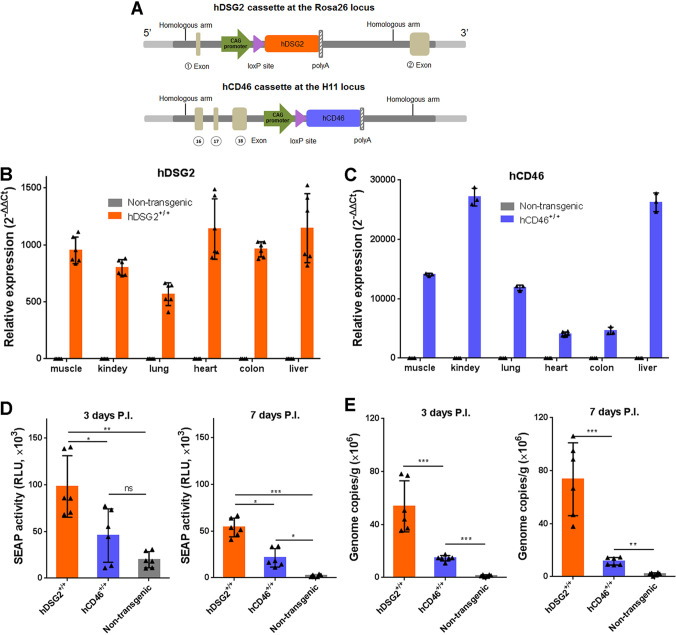
We attempted to infect wild-type BALB/c and C57BL/6 mice with HAdV55 but failed to observe significant infection. We thus generated an hDSG2 knock-in transgenic mouse line and an hCD46 knock-in transgenic mouse line based on C57BL/6 mice background. Using a site-specific integration strategy mediated by CRISPR-Cas9, we inserted the expression cassette for hDSG2 or hCD46 into the Rosa26 locus or the H11 locus, respectively (Fig. 6A). Expression of hDSG2 and hCD46 was detected in the muscle, kidney, lung, heart, colon, and liver of the respective transgenic mice (Fig. 6B and C). Wild-type mice, hDSG2+/+ mice, and hCD46+/+ mice were intravenously injected with the HAdV55-SEAP reporter virus. At 3 days postinjection, significantly higher SEAP activity was detected in the sera of hDSG2+/+ mice than in those of hCD46+/+ mice (Fig. 6D). At 7 days postinjection, SEAP activity could still be detected in the sera of hDSG2+/+ mice but was much lower in hCD46+/+ mice and not detectable in nontransgenic mice (Fig. 6D). Since SEAP activity in hDSG2+/+ mice was continuously higher than that in hCD46+/+ mice, hDSG2 likely plays a more important role than hCD46 in mediating HAdV55 infection in transgenic mice. At 3 days postinfection, HAdV55 genomes could be detected in the livers of hDSG2+/+ mice and hCD46+/+ mice, at a level much higher than in the livers of nontransgenic mice (Fig. 6E). Again, the level of HAdV55 genomes was much higher in the livers of hDSG2+/+ mice than in the livers of hCD46+/+ mice. Taken together, the results show that hDSG2+/+ mice appeared to be a more permissive model than hCD46+/+ mice in enabling the infection of HAdV55 in otherwise nonpermissive wild-type mice.
FIG 6.
hDSG2 transgenic mice support HAdV55 infection with a higher efficiency than hCD46 transgenic mice. (A) Schematic diagram of the expression cassettes for hDSG2 and hCD46 in the genome of transgenic mice. (B) The expression levels of hDSG2 in the indicated tissues were examined by reverse transcription-qPCR. (C) The expression levels of hCD46 in the indicated tissues were examined by reverse transcription-qPCR. The relative expression levels of hDSG2 and hCD46 were calculated using β-actin mRNA as a reference. Samples from nontransgenic mice were used as negative controls. Data are representative of those from three independent experiments and presented as the means ± SD (n = 3 to 6). (D) SEAP activity in the sera of transgenic and nontransgenic mice at 3 days (left) or 7 days (right) postinfection. Each group of mice (n = 6) was infected with 2 × 1010 vp of HAdV55-SEAP by an intravenous injection. The sera were collected at 3 days or 7 days postinjection, and the SEAP activity was examined. (E) Genome copies of HAdV55 in the liver of the infected mice at 3 days (left) or 7 days (right) postinfection. Data are representative of those from three independent experiments and presented as the means ± SD. Comparison between groups was performed using one-way ANOVA. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DISCUSSION
The emerging HAdV55 exhibits relatively high virulence due to its association with acute respiratory diseases and even deaths in immunocompetent adults (3, 42). Defining the potential cellular receptors is thus important for understanding the virological properties of HAdV55. Our results demonstrate for the first time that hDSG2 plays the major roles in mediating HAdV55 binding and infection (Fig. 1 to 5). hCD46 can also be engaged by HAdV55 but plays only minor or accessory roles during HAdV55 infection. This study, in addition with others, reveals that a group of species B HAdVs such as HAdV3, -7, -14, and -55 primarily recognize hDSG2 and to a lesser degree recognize hCD46 in cultured cells. This may add to the concept that many species B HAdVs and species D HAdVs can utilize more than one cellular receptor to achieve productive infection (18, 43). Based on these observations, we generated new transgenic mouse lines with the expression of hDSG2 or hCD46 in multiple organs (Fig. 6). We demonstrated that hDSG2 promotes HAdV55 infection in transgenic mice with a higher efficiency than hCD46 (Fig. 6). Since the reporter gene carried by recombinant HAdV55 can be efficiently expressed, hDSG2 transgenic mice may enable the assessment of vaccines based on recombinant HAdV55 vectors.
The fiber protein, in particular the knob domain, determines the receptor usage of an HAdV (44). Previously, HAdV7, -11, and -35 were reported to bind to hCD46 using similar sites on the fiber protein (21, 22, 32), and the common Arg279 amino acid in the fiber knob determines high binding affinity (33). An Arg279Gln mutation is observed in the fiber knob of both HAdV55 and HAdV14a, implying that their binding affinity for hCD46 should be relatively weak (33). Another group has shown that the fiber knobs of HAdV3 and HAdV7, both harboring the Arg279Gln mutation, have nearly undetectable affinity to hCD46 when its density is low but have significant avidity to hCD46 when its density is high (18). We show that HAdV55 fiber knob, in a polymeric form, directly binds to the dimeric hDSG2 and hCD46 (Fig. 1). Therefore, hCD46 may be bound by HAdV55 when it is highly expressed on the cellular surface, and hDSG2 might be preferably bound by HAdV55 when they are concurrently expressed.
We utilized multiple approaches, including fiber knob protein, soluble receptors, and specific antibodies, to competitively block the attachment of HAdV55. Unlike the fiber knobs that efficiently inhibit HAdV55 infection, soluble receptors and antibodies, used either alone or as a combination, only partially inhibited HAdV55 infection (Fig. 1). Similar results have been observed with HAdV3 and -7 (18, 19). We speculate that (i) HAdV55 can infect host cells through nonreceptor routes, especially at a high infection dose, and (ii) there are other, unknown receptors that can mediate HAdV55 infection. The results using gene knockout A549 cells support these speculations. Because the expression of hDSG2 or hCD46 was abrogated in hCD46−/− hDSG2−/− cells, the interaction of fiber with hDSG2 or hCD46 was eliminated (Fig. 3). However, we still observed significant HAdV55 infection in these cells, although at a sharply decreased level. It has been recognized that other cellular receptors, such as CD80 and CD86, which are two costimulatory molecules expressed on professional antigen-presenting cells, are able to be bound by species B HAdVs (45, 46). Further study is needed to dissect whether nonreceptor pathways or other unknown receptors mediate HAdV55 infection in hCD46−/− hDSG2−/− cells.
We have noted that the concentrations of fiber knobs, soluble receptors, and blocking antibodies used in our competition assays appeared to be higher than those used in other studies (19, 34). However, the experimental settings are quite different. Those two studies carried out competition assays on cells of the human cervical cancer cell line HeLa, whereas we performed these assays on human lung adenocarcinoma cell line A549 cells. The expression patterns of hCD46 and hDSG2 in these two cell lines may be different, which may result in different doses of reagents required to achieve an efficient inhibition of HAdV55 infection. In addition, those two studies used the expression of GFP as an indicator of HAdV infection, whereas our study used another reporter gene, SEAP, as an indicator of HAdV infection. The sensitivity of these two reporter genes may also add to the discrepancy between our and other studies. Nearly 80% HeLa cells can be infected by HAdV3-GFP in the presence of 25 μg/ml of HAdV3 fiber knob (34), whereas in our study the same concentration of HAdV14 FK and HAdV55 FK can reduce viral infection to about 30% (Fig. 1B and C). In another study which also used A549 cells, 5 μg/ml of sCD46 showed an extremely slight inhibition on the infection of HAdV3 and HAdV7 (18). In the same study, 40 μg/ml of anti-DSG2 antibodies reduced the binding of HAdV3 to ∼60%, similar to that observed in our studies (Fig. 1H). Thus, it may be reasonable that different concentrations of competitors are needed in assays based on different cell lines and different reporter HAdVs.
One significant finding of this study is that our hDSG2 transgenic mouse can potentially be infected by HAdV55. Although hDSG2 transgenic mice and hCD46 transgenic mice were previously reported (38, 39, 47, 48), the insertion of the transgenes into the genome of those mice was mediated by a random integration strategy. Since the native promoter and regulating elements were retained, the expression of hDSG2 and hCD46 might be widely diverse among tissues (39, 48). In this study, the expression cassettes of hDSG2 and hCD46 was controlled by a constitutive promoter and they were inserted into the Rosa26 locus and the H11 locus, respectively. These loci have been shown to support global expression of the knock-in transgene (49). The resultant transgenic mice, therefore, had significant expression of hDSG2 or hCD46 in multiple tissues (Fig. 6). An intravenous inoculation of HAdV55-SEAP led to significant expression of SEAP in the hDSG2+/+ mice and, to a lesser degree, in the hCD46+/+ mice but not in nontransgenic mice, indicating that hDSG2 is able to promote HAdV55 infection not only in cell lines but also in transgenic mice.
Our findings that HAdV55 infects hDSG2-expressing tumor cells may have intrinsic value for exploring oncolytics based on HAdV55. hDSG2 is overexpressed in several human epithelial tumors, including squamous cell carcinomas, gastric cancer, breast cancer, and bladder cancer (50, 51). hDSG2 is important for tumor cell proliferation and microenvironment modulation and thus affects the pathogenesis of a cancer (51). hDSG2-engaging HAdVs, such as HAdV3, have been developed as oncolytics and have shown great potency in killing tumor cells and reducing tumor growth (52, 53). However, the seroprevalences of neutralizing antibodies to HAdV3 and another hDSG2-engaging adenovirus, HAdV7, are relatively high in populations (60% or above) (17, 53), which may limit the oncolytic potency (54). Because the seroprevalence of neutralizing antibody to HAdV55 is significantly lower than those of neutralizing antibodies to HAdV3, -5, and -7 (16), we propose that recombinant HAdV55, upon modification to selectively replicate in tumor cells, might be developed as an alternative virotherapy.
In summary, we demonstrated that hDSG2 predominantly mediates HAdV55 infection and generated a new hDSG2 transgenic mouse line that effectively supports HAdV55 infection. Our findings shed light on the development of prophylactic and therapeutic approaches against HAdV55 infection.
MATERIALS AND METHODS
Plasmids and proteins.
Plasmids expressing hDSG2 (GenBank accession no. NP_758861.1), hCD46 (GenBank accession no. NP_001934.2), and hCAR (GenBank accession no. NM_001338.5) were constructed as follows. The cDNA sequences encoding hDSG2, hCD46, and hCAR (Genecopoeia) were cloned into pCDNA4TO (Addgene) by restricted digestion to obtain pCDNA4-hCD46, pCDNA4-hDSG2, and pCDNA4-hCAR, respectively. The coding sequences for the extracellular domain of hDSG2 (residues 50 to 609) and hCD46 (residues 35 to 328) were amplified and fused to the coding sequence for human IgG Fc at the 5′ terminus by PCR. Plasmids expressing sDSG2 and sCD46 were constructed by inserting hDSG2-Fc and hCD46-Fc fragments into the pEZ-M13 vector (Genecopoeia) to obtain pEZ-sDSG2 and pEZ-sCD46, respectively.
HAdV14a fiber knob (residues 118 to 323), HAdV55 fiber knob (residues 118 to 325), and HAdV5 fiber knob (residues 378 to 581) were produced in E. coli as described previously (55). sDSG2 and sCD46 were expressed in human embryonic kidney 293T (HEK293T) cells. In brief, HEK293T cells were transfected with pEZ-sDSG2 or pEZ-sCD46 using Lipofectamine 2000 (Thermo Fisher Scientific). At 3 days posttransfection, culture supernatants were harvested, centrifuged, and purified by protein A agarose (GE Healthcare). The proteins were then dialyzed into phosphate-buffered saline (PBS), filtered through 0.2-μm filters, and stored at –80°C.
Viruses.
HAdV14 and HAdV55 recombinants expressing EGFP or SEAP have been described previously (16). HAdV5 recombinants expressing hDSG2, hCD46, or hCAR were constructed according to previously described methods (56). In brief, the coding sequences of hDSG2, hCD46, and hCAR were released from the expression plasmids mentioned above and inserted into a pGA1 vector to obtain shuttle plasmids. Genomic plasmids were constructed by homologous recombination between linearized shuttle plasmids with pAd5ΔE1ΔE3. HAdV5-hDSG2, HAdV5-hCD46, and HAdV5-hCAR were then rescued, propagated in HEK293 cells, and purified in a manner similar to the production of recombinant HAdV55.
Cell lines.
Human lung carcinoma NCI-H358, human ovarian cancer SK-OV-3, human lymphoma Raji, African green monkey kidney CV-1, and Chinese hamster ovary CHO-K1 cells were purchased from the Cell Resource Center of Shanghai Institute of Life Sciences, Chinese Academy of Sciences. Human lung carcinoma A549, human pancreatic adenocarcinoma PANC-1 and Capan-2, HEK293, human monocytic THP-1, African green monkey kidney Vero, and murine hepatoma Hepa1-6 cells were kept in our lab. A549, PANC-1, Capan-2, HEK293, CV-1, Vero, and Hepa1-6 cells were cultured in Dulbecco’s modified Eagle medium (DMEM; Gibco) containing 10% fetal bovine serum (FBS; Gibco). SK-OV-3 cells were maintained in McCoy’s 5A (modified) medium containing 15% FBS. NCI-H358, Raji, and THP-1 cells were cultured in RPMI 1640 medium containing 10% FBS. CHO-K1 cells were cultured in Ham’s F-12K medium containing 10% FBS. All the culture media mentioned above were supplemented with 100 U/ml of penicillin and 100 μg/ml of streptomycin.
Gene knockout cell lines were constructed using CRISPR-Cas9 technology. The single guide RNAs (sgRNAs) targeting hDSG2 and hCD46 are listed in Table 1. The sense and antisense DNA sequences corresponding to these sgRNAs were synthesized, annealed, and cloned into plasmids under the control of the U6 promoter. A549 cells were cotransfected with pcDNA3.3-hCas9 (Addgene) and plasmids expressing either DSG2-sgRNAs, CD46-sgRNAs, or both. After selection for 7 days using 400 μg/ml of G418 (Gibco), single cells were isolated by limiting dilution in 96-well plates and screened through two strategies: (i) the fragments flanking the target sequences of sgRNAs in the cellular genome were amplified and sequenced, and (ii) the expression of hDSG2 and hCD46 was examined by Western blot analysis. Cell clones were propagated, frozen, and stored in liquid nitrogen.
TABLE 1.
sgRNAs, siRNAs, and primers used in this study
| Purpose | Name | Sequence (5′–3′) |
|---|---|---|
| Gene knockouta | CD46-sgRNA1 | GGACACTCGCGGCGGCCGGG |
| CD46-sgRNA2 | GCAAATGGGACTTACGAGTT | |
| DSG2-sgRNA1 | GCAGGGCGTACGCGCGTCCC | |
| DSG2-sgRNA2 | GGGATTACAGAGCCACCTTT | |
| Gene knockdown | siCD46-1 sense | GGAGCCACCAACAUUUGAATT |
| siCD46-1 antisense | UUCAAAUGUUGGUGGCUCCTT | |
| siCD46-2 sense | GCCUGUUAUAGAGAAACAUTT | |
| siCD46-2 antisense | AUGUUUCUCUAUAACAGGCTT | |
| siDSG2-1 sense | CCUCCAGUGUUCUACCUAATT | |
| siDSG2-1 antisense | UUAGGUAGAACACUGGAGGTT | |
| siDSG2-2 sense | CCAAUUGCCAAGAUACAUUTT | |
| siDSG2-2 antisense | AAUGUAUCUUGGCAAUUGGTT | |
| siCAR sense | GCCAGAAGUUUGAGUAUCATT | |
| siCAR antisense | UGAUACUCAAACUUCUGGCTT | |
| Scramble sense | UUCUCCGAAGGUGUCACGUTT | |
| Scramble antisense | ACGUGACACGUUCGGAGAATT | |
| Genome quantification | Q-Ad55 F | TGTTTTCCACTGAATGGCATAGG |
| Q-Ad55 R | GGAAACTTCGCCATAGATTGGC | |
| Gene expression | Q-CD46 F | TGTTTGAATGCGATAAGGGT |
| Q-CD46 R | TGAGACTGGAGGCTTGTAAG | |
| Q-DSG2 F | CTACACCCATTCCCATCAAGGTC | |
| Q-DSG2 R | GCTTGATCTATCCATGCTCTCG | |
| Q-βactin F | GACCCTGAAGTACCCCATTGA | |
| Q-βactin R | CTCAAACATGATCTGGGTCATCT | |
| CD46 F1 | GCCTCCAAGTCTTGACAGTAGATTA | |
| CD46 R1 | GCTTCTGTACACTGCCAGACATG | |
| CD46 R2 | CTATTGGCGTTACTATGGGAACA | |
| Transgenic mouse genotypingb | DSG2 F1 | GTTATCAGTAAGGGAGCTGCAGTG |
| DSG2 R1 | CTGCTTACATAGTCTAACTCGCGAC | |
| DSG2 R2 | GGCAGTTTACCGTAAATACTCCAC | |
| CAG F2 | CTGGTTATTGTGCTGTCTCATCAT | |
| DSG2 R3 | CTGGTAACATTCAGTTCTCCAGTATC | |
| CD46 R3 | GTGCATCTGATAACCAAACTCGTA |
The target sequences of sgRNAs for hCD46 and hDSG2 gene are shown. The sense and antisense DNAs corresponding to these targets were synthesized.
For mouse genotyping, two sets of primers for both hCD46 cassette and hDSG2 cassette were designed. F1-R1/R2 pairs were used to distinguish homozygous, heterozygous, and nontransgenic mice. CAG F2-R3 pairs were used to confirm the insertion of the cassette in mouse genome and validate the deletion of loxP-poly(A)-loxP sequence.
Stable cell lines expressing either hDSG2, hCD46, or both were constructed as follows. CHO-K1 cells were transfected with pCDNA4-hDSG2, pCDNA4-hCD46, or both. Cells transfected with pCDNA4-hCAR were used as controls. Two days after transfection, cells were cultured in the presence of 400 μg/ml of Zeocin (Thermo Fisher Scientific). Ten days later, single clones were picked out and propagated. Cell clones were examined for the expression of hDSG2 or hCD46 by flow cytometry. Finally, cell clones expressing different levels of hDSG2, hCD46, or both were propagated and subjected to infection assays.
Antibodies.
Mouse monoclonal antibodies to human β-actin, hDSG2 (6D8 and 8E5), or hCD46 (M177 and E4.3), phycoerythrin (PE)-labeled mouse anti-hCAR antibody (E1-1), fluorescein isothiocyanate (FITC)- or PE-labeled goat anti-mouse antibody, and mouse IgG1 isotype were purchased from Santa Cruz Biotechnology, Inc. A rabbit anti-hDSG2 antibody (EPR6768) was purchased from Abcam. A mouse anti-hCD46 antibody (MEM258) was obtained from AbD Serotec. A PE-labeled mouse anti-hCD46 antibody (MEM258) was purchased from Thermo Fisher Scientific. A FITC-labeled mouse anti-hDSG2 antibody was obtained from Lifespan Biosciences, Inc. A mouse anti-hCAR antibody (RmcB) was obtained from Merck Millipore.
Animals.
Transgenic mice expressing hDSG2 or hCD46 were constructed by CRISPR-Cas9 technology in a C57BL/6 background. In brief, the cassettes for hDSG2 or hCD46 consisted of a CAG promoter, a poly(A) signal flanked by two loxP sites, the coding region, and a poly(A) signal from the 5′ to 3′ terminus (Fig. 6A). The hDSG2 or hCD46 cassette was inserted between the homologous arms corresponding to the Rosa26 or H11 locus, respectively. These two loci have been shown to be able to incorporate large foreign genes (49, 57). These two fragments, as well as plasmids expressing Cas9 and the corresponding sgRNAs, were delivered into mouse embryos through pronuclear microinjection. After birth and sex maturation, the mice were crossed with Ella-Cre mice (Jackson Laboratory) to obtain transgenic mice with deleted transcription terminal signals. The resultant mice were then genotyped by PCR analysis of genomic DNA extracted from mouse tail tip samples. The primers are listed in Table 1. Experiments involving mice were in accordance with the guidelines set by the Association for the Assessment and Accreditation of Laboratory Animal Care and were approved by the Institutional Animal Care and Use Committee of Guangzhou Institutes of Biomedicine and Health (GIBH).
To test infection with HAdV55 in transgenic mice, 6-week-old hDSG2+/+ mice or hCD46+/+ mice or control nontransgenic mice were infected with HAdV55-SEAP at 2 × 1010 viral particles (vp) per mouse via intravenously injection. At 3 days and 7 days postinfection, serum samples were collected and subjected to SEAP activity examination. At the same time points, a proportion of infected animals were sacrificed and the livers were harvested and homogenized. Finally, the supernatants of the homogenates were subjected to DNA extraction and quantitative PCR (qPCR).
SPR analysis.
To assess the interaction of HAdV55 fiber knob with hDSG2 and hCD46, SPR analysis was performed on a Biacore X100 system (GE Healthcare). In brief, a CM5 chip was activated using 0.2 M 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide and 0.05 M N-hydroxysuccinimide in water and labeled with fiber knobs at 0.5 μM in 10 mM sodium acetate buffer (pH 4.5). Excess reactive groups were deactivated using 1 M ethanolamine HCl (pH 8.5). Subsequently, the sensor chip was conditioned three times with 50 mM NaOH. sDSG2 and sCD46 were injected at 2-fold-increasing concentrations from 125 nM to 500 nM in PBS at 30 μl/min. After each cycle, the chip was regenerated using 10 mM NaOH. The obtained sensorgrams were corrected by subtracting the data from blank buffer injections.
Virus binding assays.
To assess the blocking effects of anti-hDSG2 and anti-hCD46 antibodies, A549 cells were washed with PBS, resuspended in ice-cold adhesion buffer (DMEM supplemented with 2% FBS) containing monoclonal antibodies to hCAR (RmcB, 20 μg/ml), hDSG2 (20 μg/ml of 6D8 plus 20 μg/ml of 8E5), and/or hCD46 (20 μg/ml of M177 plus 20 μg/ml of MEM258), and then incubated on ice for 1 h. HAdV55-SEAP or HAdV14a-SEAP was then added at 200 vp/cell and incubated on ice for another 1 h. The cells were then harvested, washed with ice-cold PBS, and subjected to total DNA extraction using a QIAamp DNA minikit (Qiagen) according to the manufacturer’s protocol. Finally, viral DNA was analyzed by real-time qPCR. To assess the ability of HAdV55 to bind to gene knockout cells, hDSG2−/−, hCD46−/−, and hCD46−/− hDSG2−/− cells were incubated with HAdV55-SEAP (200 vp/cell) in ice-cold adhesion buffer. After incubation for 1 h, the bound viruses were processed and analyzed as described above.
Virus infection assays.
HAdV55-EGFP infection assays were performed as follows. Human-, monkey-, and CHO-K1 cell-derived stable cell lines were infected with HAdV55-EGFP at the desired doses at 37°C for 1 h. At 24 h postinfection, cells were imaged using a fluorescence microscopy or analyzed by flow cytometry.
HAdV55-SEAP and HAdV14a-SEAP infection assays were performed as follows. (i) A549 cells (3 × 104/well) in 96-well plates were transfected with siDSG2, siCD46, or both (10 pM each small interfering RNA [siRNA]; GenePharma), and siCAR and scrambled siRNA were used as controls. At 60 h posttransfection, a proportion of cells were subjected to flow cytometry analysis, and the other cells were infected with HAdV55-SEAP. (ii) CHO-K1 cells (1 × 105/well) in 96-well plates were transfected with pCDNA4-hDSG2, pCDNA4-hCD46, or both (0.2 μg of each plasmid), and pCDNA4TO and pCDNA4-hCAR were used as controls. At 48 h posttransfection, a proportion of transfected cells was subjected to Western blot analysis, and the other cells were infected with HAdV55-SEAP or HAdV14a-SEAP. (iii) Hepa1-6 cells (5 × 104/well) in 96-well plates were infected with HAdV5-hDSG2 or HAdV5-hCD46 or both (1000 vp of each virus per cell), and HAdV5-empty and HAdV5-hCAR were used as controls. At 36 h postinfection, a proportion of infected cells was subjected to Western blot analysis, and the other cells were infected with HAdV55-SEAP or HAdV14a-SEAP. (iv) Gene knockout cells (3 × 104/well) in 96-well plates were infected with HAdV55-SEAP. The dose of HAdV55-SEAP was 200 vp/cell. At 2 h postinfection, the infection mixtures were replaced with fresh culture media. At 24 h after HAdV55-SEAP or HAdV14a-SEAP infection, the culture media were harvested, and the SEAP activity was examined using a Phospha-Light system (Thermo Fisher Scientific). Relative light units (RLUs) were analyzed.
Virus infection inhibition assays.
For fiber knob competition, A549 cells were preincubated with HAdV55 fiber knob, HAdV14a fiber knob, or HAdV5 fiber knob at serially increasing concentrations (0.2, 1, 5, 25, and 125 μg/ml) at 4°C for 1 h. HAdV55-SEAP, HAdV14a-SEAP, or HAdV5-SEAP was added at 200 vp/cell and incubated at 4°C for another 1 h. Cells were then rinsed and incubated in phenol red-free DMEM containing 2% FBS at 37°C for 24 h. Finally, the SEAP activity in the cultured media was analyzed. For soluble receptor competition, HAdV55-SEAP or HAdV14a-SEAP (200 vp/cell) was preincubated with sDSG2, sCD46, or both at three concentrations (43.8, 87.5, and 175 μg/ml) in DMEM at 4°C for 1 h. The mixtures were then transferred to A549 cells and incubated at 4°C for another 1 h. After a thorough washing, cells were incubated at 37°C for 24 h. The SEAP activity in the cultured media was then assessed. For antibody competition, A549 cells were preincubated with anti-hDSG2 (20 μg/ml of 6D8 plus 20 μg/ml of 8E5), anti-hCD46 (20 μg/ml of M177 plus 20 μg/ml of MEM258), or anti-hCAR antibodies (RmcB; 40 μg/ml) at 4°C for 1 h. HAdV55-SEAP (50 vp/cell) was then added and incubated at 4°C for another 1 h. The cells were washed and incubated at 37°C for 24 h. The SEAP activity in the cultured media was then analyzed.
Western blot analysis.
Cells were treated with radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime) and subjected to SDS-PAGE. After transfer, membranes were blocked with 5% bovine serum albumin (BSA) and incubated with antibodies against hDSG2 (6D8), hCD46 (M177), hCAR (RmcB), or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in PBST (PBS supplemented with 0.05% Tween 20 and 5% nonfat milk). After incubation overnight at 4°C, membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies and developed with chemiluminescent HRP substrate (Merck).
Flow cytometry analysis.
To assess the expression levels of hDSG2 and hCD46 on the cellular surface, cells were detached, blocked with complete DMEM, and then washed three times with PBS. Subsequently, cells were incubated with a FITC-labeled anti-hDSG2 antibody and/or PE-labeled anti-hCD46 antibody at 1 μg/ml for 1 h. After being washed three times with PBS, cells were analyzed using an Accuri C6 flow cytometer (BD Biosciences). To assess the expression of EGFP, cells infected by HAdV55-EGFP were detached, washed three times with PBS, and analyzed using an Accuri C6 flow cytometer.
qPCR.
To assess the genome copies of HAdV55 bound to the cellular surface or in the liver of infected mice, HAdV55 genomes were extracted and subjected to qPCR using iTaq Universal SYBR green Supermix (Bio-Rad). The primers used are listed in Table 1. To quantify the binding virions on the cellular surface, the housekeeping β-actin gene was used as a reference. Relative binding virions were calculated by the threshold cycle (ΔΔCT) method, and the virions bund to A549 cells were taken as 100%. To quantify viral genome copies in mouse tissues, a standard curve was made using serially diluted HAdV55 genome extracted from viral stocks.
To assess the expression of hDSG2 and hCD46 in transgenic mice, total RNAs were extracted from mouse tissues using a total RNA miniprep kit (Axygen) and subjected to reverse transcription using an iScript cDNA synthesis kit (Bio-Rad) followed by qPCR using iTaq Universal SYBR green Supermix (Bio-Rad). RNA samples from nontransgenic mice were run in parallel as controls. The expression of the housekeeping β-actin gene was used as a reference. The CT values of each gene were normalized to that ofβ-actin, and 2−ΔΔCT values were used to compare mRNA levels among tissues.
Data processing and statistics.
The collection and analysis of flow cytometry data were performed with FlowJo software (version 7.6.2; Tree Star, Inc.). Statistical comparisons between groups were analyzed by one-way analysis of variance (ANOVA) and computed with SPSS version 13.0 (SPSS Inc.). P values less than 0.05 were considered statistically significant. Data graphs were constructed using GraphPad Prism version 7 (GraphPad Software). Figures were created using Photoshop version CS2 (Adobe Systems Inc.).
ACKNOWLEDGMENTS
We thank Cuie Li and Zhi Wang for their technical assistance.
This study was supported by the National Natural Science Foundation of China (31470892), the National Science and Technology Major Project (2017ZX10204401003 and 2018ZX10101003005), and the Guangzhou Health Care and Cooperative Innovation Major Project (201803040004).
X.L., C.Y., and S.G. are employees of Guangzhou nBiomed Ltd. The other authors declare no competing financial interests.
REFERENCES
- 1.Ismail AM, Lee JS, Lee JY, Singh G, Dyer DW, Seto D, Chodosh J, Rajaiya J. 2018. Adenoviromics: mining the human adenovirus species D genome. Front Microbiol 9:2178. doi: 10.3389/fmicb.2018.02178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao B, Huang GH, Pu ZH, Qu JX, Yu XM, Zhu Z, Dong JP, Gao Y, Zhang YX, Li XH, Liu JH, Wang H, Xu Q, Li H, Xu WB, Wang C. 2014. Emergence of community-acquired adenovirus type 55 as a cause of community-onset pneumonia. Chest 145:79–86. doi: 10.1378/chest.13-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun B, He H, Wang Z, Qu J, Li X, Ban C, Wan J, Cao B, Tong Z, Wang C. 2014. Emergent severe acute respiratory distress syndrome caused by adenovirus type 55 in immunocompetent adults in 2013: a prospective observational study. Crit Care 18:456. doi: 10.1186/s13054-014-0456-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X, Kong M, Su X, Zou M, Guo L, Dong X, Li L, Gu Q. 2014. An outbreak of acute respiratory disease in China caused by human adenovirus type B55 in a physical training facility. Int J Infect Dis 28:117–122. doi: 10.1016/j.ijid.2014.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chmielewicz B, Benzler J, Pauli G, Krause G, Bergmann F, Schweiger B. 2005. Respiratory disease caused by a species B2 adenovirus in a military camp in Turkey. J Med Virol 77:232–237. doi: 10.1002/jmv.20441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kajon AE, Dickson LM, Metzgar D, Houng HS, Lee V, Tan BH. 2010. Outbreak of febrile respiratory illness associated with adenovirus 11a infection in a Singapore military training camp. J Clin Microbiol 48:1438–1441. doi: 10.1128/JCM.01928-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan D, Zhu H, Fu Y, Tong F, Yao D, Walline J, Xu J, Yu X. 2016. Severe community-acquired pneumonia caused by human adenovirus in immunocompetent adults: a multicenter case series. PLoS One 11:e0151199. doi: 10.1371/journal.pone.0151199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salama M, Amitai Z, Amir N, Gottesman-Yekutieli T, Sherbany H, Drori Y, Mendelson E, Carmeli Y, Mandelboim M. 2016. Outbreak of adenovirus type 55 infection in Israel. J Clin Virol 78:31–35. doi: 10.1016/j.jcv.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 9.Yoo H, Gu SH, Jung J, Song DH, Yoon C, Hong DJ, Lee EY, Seog W, Hwang IU, Lee D, Jeong ST, Huh K. 2017. Febrile respiratory illness associated with human adenovirus type 55 in South Korea military, 2014–2016. Emerg Infect Dis 23:1016–1020. doi: 10.3201/eid2306.161848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heo JY, Noh JY, Jeong HW, Choe KW, Song JY, Kim WJ, Cheong HJ. 2018. Molecular epidemiology of human adenovirus-associated febrile respiratory illness in soldiers, South Korea. Emerg Infect Dis 24:1221–1227. doi: 10.3201/eid2407.171222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Z, Zhu Z, Tang L, Wang L, Tan X, Yu P, Zhang Y, Tian X, Wang J, Zhang Y, Li D, Xu W. 2009. Genomic analyses of recombinant adenovirus type 11a in China. J Clin Microbiol 47:3082–3090. doi: 10.1128/JCM.00282-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walsh MP, Seto J, Jones MS, Chodosh J, Xu W, Seto D. 2010. Computational analysis identifies human adenovirus type 55 as a re-emergent acute respiratory disease pathogen. J Clin Microbiol 48:991–993. doi: 10.1128/JCM.01694-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J, Nian QG, Zhang Y, Xu LJ, Hu Y, Li J, Deng YQ, Zhu SY, Wu XY, Qin ED, Jiang T, Qin CF. 2014. In vitro characterization of human adenovirus type 55 in comparison with its parental adenoviruses, types 11 and 14. PLoS One 9:e100665. doi: 10.1371/journal.pone.0100665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu QB, Tong YG, Wo Y, Wang HY, Liu EM, Gray GC, Liu W, Cao WC. 2014. Epidemiology of human adenovirus and molecular characterization of human adenovirus 55 in China, 2009–2012. Influenza Other Respir Viruses 8:302–308. doi: 10.1111/irv.12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun C, Zhang Y, Feng L, Pan W, Zhang M, Hong Z, Ma X, Chen X, Chen L. 2011. Epidemiology of adenovirus type 5 neutralizing antibodies in healthy people and AIDS patients in Guangzhou, southern China. Vaccine 29:3837–3841. doi: 10.1016/j.vaccine.2011.03.042. [DOI] [PubMed] [Google Scholar]
- 16.Zheng X, Rong X, Feng Y, Sun X, Li L, Wang Q, Wang M, Liu W, Li C, Yang Y, Zhou R, Lu J, Feng L, Chen L. 2017. Seroprevalence of neutralizing antibodies against adenovirus type 14 and 55 in healthy adults in southern China. Emerg Microbes Infect 6:e43. doi: 10.1038/emi.2017.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye X, Xiao L, Zheng X, Wang J, Shu T, Feng Y, Liu X, Su W, Wang Q, Li C, Chen L, Feng L. 2018. Seroprevalence of neutralizing antibodies to human adenovirus type 4 and 7 in healthy populations from southern China. Front Microbiol 9:3040. doi: 10.3389/fmicb.2018.03040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trinh HV, Lesage G, Chennamparampil V, Vollenweider B, Burckhardt CJ, Schauer S, Havenga M, Greber UF, Hemmi S. 2012. Avidity binding of human adenovirus serotypes 3 and 7 to the membrane cofactor CD46 triggers infection. J Virol 86:1623–1637. doi: 10.1128/JVI.06181-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang HJ, Li ZY, Liu Y, Persson J, Beyer I, Moller T, Koyuncu D, Drescher MR, Strauss R, Zhang XB, Wahl JK, Urban N, Drescher C, Hemminki A, Fender P, Lieber A. 2011. Desmoglein 2 is a receptor for adenovirus serotypes 3, 7, 11 and 14. Nat Med 17:96–104. doi: 10.1038/nm.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaggar A, Shayakhmetov DM, Lieber A. 2003. CD46 is a cellular receptor for group B adenoviruses. Nat Med 9:1408–1412. doi: 10.1038/nm952. [DOI] [PubMed] [Google Scholar]
- 21.Segerman A, Atkinson JP, Marttila M, Dennerquist V, Wadell G, Arnberg N. 2003. Adenovirus type 11 uses CD46 as a cellular receptor. J Virol 77:9183–9191. doi: 10.1128/jvi.77.17.9183-9191.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sirena D, Lilienfeld B, Eisenhut M, Kalin S, Boucke K, Beerli RR, Vogt L, Ruedl C, Bachmann MF, Greber UF, Hemmi S. 2004. The human membrane cofactor CD46 is a receptor for species B adenovirus serotype 3. J Virol 78:4454–4462. doi: 10.1128/jvi.78.9.4454-4462.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marttila M, Persson D, Gustafsson D, Liszewski MK, Atkinson JP, Wadell G, Arnberg N. 2005. CD46 is a cellular receptor for all species B adenoviruses except types 3 and 7. J Virol 79:14429–14436. doi: 10.1128/JVI.79.22.14429-14436.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tuve S, Wang HJ, Ware C, Liu Y, Gaggar A, Bernt K, Shayakhmetov D, Li ZY, Strauss R, Stone D, Lieber A. 2006. A new group B adenovirus receptor is expressed at high levels on human stem and tumor cells. J Virol 80:12109–12120. doi: 10.1128/JVI.01370-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bergelson JM, Cunningham JA, Droguett G, Kurt-Jones EA, Krithivas A, Hong JS, Horwitz MS, Crowell RL, Finberg RW. 1997. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 26.Dorig RE, Marcil A, Chopra A, Richardson CD. 1993. The human Cd46 molecule is a receptor for measles-virus (Edmonston strain). Cell 75:295–305. doi: 10.1016/0092-8674(93)80071-l. [DOI] [PubMed] [Google Scholar]
- 27.Santoro F, Kennedy PE, Locatelli G, Malnati MS, Berger EA, Lusso P. 1999. CD46 is a cellular receptor for human herpesvirus 6. Cell 99:817–827. doi: 10.1016/s0092-8674(00)81678-5. [DOI] [PubMed] [Google Scholar]
- 28.Kallstrom H, Gill DB, Albiger B, Liszewski MK, Atkinson JP, Jonsson AB. 2001. Attachment of Neisseria gonorrhoeae to the cellular pilus receptor CD46: identification of domains important for bacterial adherence. Cell Microbiol 3:133–143. doi: 10.1046/j.1462-5822.2001.00095.x. [DOI] [PubMed] [Google Scholar]
- 29.Broussard JA, Getsios S, Green KJ. 2015. Desmosome regulation and signaling in disease. Cell Tissue Res 360:501–512. doi: 10.1007/s00441-015-2136-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlipp A, Schinner C, Spindler V, Vielmuth F, Gehmlich K, Syrris P, Mckenna WJ, Dendorfer A, Hartlieb E, Waschke J. 2014. Desmoglein-2 interaction is crucial for cardiomyocyte cohesion and function. Cardiovasc Res 104:245–257. doi: 10.1093/cvr/cvu206. [DOI] [PubMed] [Google Scholar]
- 31.Zou Y, Zhang Q, Zhang JH, Chen XK, Zhou W, Yang ZZ, Yang Q, Yu H, Li LJ, He Y, Li CT, Zhang SH, Zhu SH, Luo B, Gao YZ. 2019. A common indel polymorphism of the desmoglein-2 (DSG2) is associated with sudden cardiac death in Chinese populations. Forensic Sci Int 301:382–387. doi: 10.1016/j.forsciint.2019.06.008. [DOI] [PubMed] [Google Scholar]
- 32.Fleischli C, Sirena D, Lesage G, Havenga MJE, Cattaneo R, Greber UF, Hemmi S. 2007. Species B adenovirus serotypes 3, 7, 11 and 35 share similar binding sites on the membrane cofactor protein CD46 receptor. J Gen Virol 88:2925–2934. doi: 10.1099/vir.0.83142-0. [DOI] [PubMed] [Google Scholar]
- 33.Persson BD, Muller S, Reiter DM, Schmitt BBT, Marttila M, Sumowski CV, Schweizer S, Scheu U, Ochsenfeld C, Arnberg N, Stehle T. 2009. An arginine switch in the species B adenovirus knob determines high-affinity engagement of cellular receptor CD46. J Virol 83:673–686. doi: 10.1128/JVI.01967-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang HJ, Li ZY, Yumul R, Lara S, Hemminki A, Fender P, Lieber A. 2011. Multimerization of adenovirus serotype 3 fiber knob domains is required for efficient binding of virus to desmoglein 2 and subsequent opening of epithelial junctions. J Virol 85:6390–6402. doi: 10.1128/JVI.00514-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hall K, Blair Zajdel ME, Blair GE. 2009. Defining the role of CD46, CD80 and CD86 in mediating adenovirus type 3 fiber interactions with host cells. Virology 392:222–229. doi: 10.1016/j.virol.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 36.Wang H, Tuve S, Erdman DD, Lieber A. 2009. Receptor usage of a newly emergent adenovirus type 14. Virology 387:436–441. doi: 10.1016/j.virol.2009.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Z, Tian X, Liu W, Xian Y, Chen W, Chen H, Zhou R. 2019. Development of two antigen-binding fragments to a conserved linear epitope of human adenovirus and their application in immunofluorescence. PLoS One 14:e0219091. doi: 10.1371/journal.pone.0219091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verhaagh S, de Jong E, Goudsmit J, Lecollinet S, Gillissen G, de Vries M, van Leuven K, Que I, Ouwehand K, Mintardjo R, Weverling GJ, Radosevic K, Richardson J, Eloit M, Lowik C, Quax P, Havenga M. 2006. Human CD46-transgenic mice in studies involving replication-incompetent adenoviral type 35 vectors. J Gen Virol 87:255–265. doi: 10.1099/vir.0.81293-0. [DOI] [PubMed] [Google Scholar]
- 39.Wang H, Beyer I, Persson J, Song H, Li Z, Richter M, Cao H, van Rensburg R, Yao X, Hudkins K, Yumul R, Zhang XB, Yu M, Fender P, Hemminki A, Lieber A. 2012. A new human DSG2-transgenic mouse model for studying the tropism and pathology of human adenoviruses. J Virol 86:6286–6302. doi: 10.1128/JVI.00205-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lei J, Jacobus EJ, Taverner WK, Fisher KD, Hemmi S, West K, Slater L, Lilley F, Brown A, Champion B, Duffy MR, Seymour LW. 2018. Expression of human CD46 and trans-complementation by murine adenovirus 1 fails to allow productive infection by a group B oncolytic adenovirus in murine cancer cells. J Immunother Cancer 6:55. doi: 10.1186/s40425-018-0350-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahoney MG, Simpson A, Aho S, Uitto J, Pulkkinen L. 2002. Interspecies conservation and differential expression of mouse desmoglein gene family. Exp Dermatol 11:115–125. doi: 10.1034/j.1600-0625.2002.110203.x. [DOI] [PubMed] [Google Scholar]
- 42.Xu W, Xu Z, Huang L, Qin EQ, Zhang JL, Zhao P, Tu B, Shi L, Li WG, Chen WW. 2019. Transcriptome sequencing identifies novel immune response genes highly related to the severity of human adenovirus type 55 infection. Front Microbiol 10:130. doi: 10.3389/fmicb.2019.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seiradake E, Henaff D, Wodrich H, Billet O, Perreau M, Hippert C, Mennechet F, Schoehn G, Lortat-Jacob H, Dreja H, Ibanes S, Kalatzis V, Wang JP, Finberg RW, Cusack S, Kremer EJ. 2009. The cell adhesion molecule “CAR” and sialic acid on human erythrocytes influence adenovirus in vivo biodistribution. PLoS Pathog 5:e1000277. doi: 10.1371/journal.ppat.1000277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baker AT, Greenshields-Watson A, Coughlan L, Davies JA, Uusi-Kerttula H, Cole DK, Rizkallah PJ, Parker AL. 2019. Diversity within the adenovirus fiber knob hypervariable loops influences primary receptor interactions. Nat Commun 10:741. doi: 10.1038/s41467-019-08599-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Short JJ, Pereboev AV, Kawakami Y, Vasu C, Holterman MJ, Curiel DT. 2004. Adenovirus serotype 3 utilizes CD80 (B7.1) and CD86 (B7.2) as cellular attachment receptors. Virology 322:349–359. doi: 10.1016/j.virol.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 46.Short JJ, Vasu C, Holterman MJ, Curiel DT, Pereboev A. 2006. Members of adenovirus species B utilize CD80 and CD86 as cellular attachment receptors. Virus Res 122:144–153. doi: 10.1016/j.virusres.2006.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu J, Boehme P, Zhang W, Fu J, Yumul R, Mese K, Tsoukas R, Solanki M, Kaufmann M, Lu R, Schmidtko A, Stewart AF, Lieber A, Ehrhardt A. 2018. Human adenovirus type 17 from species D transduces endothelial cells and human CD46 is involved in cell entry. Sci Rep 8:13442. doi: 10.1038/s41598-018-31713-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kemper C, Leung M, Stephensen CB, Pinkert CA, Liszewski MK, Cattaneo R, Atkinson JP. 2001. Membrane cofactor protein (MCP; CD46) expression in transgenic mice. Clin Exp Immunol 124:180–189. doi: 10.1046/j.1365-2249.2001.01458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tasic B, Hippenmeyer S, Wang C, Gamboa M, Zong H, Chen-Tsai Y, Luo L. 2011. Site-specific integrase-mediated transgenesis in mice via pronuclear injection. Proc Natl Acad Sci U S A 108:7902–7907. doi: 10.1073/pnas.1019507108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brennan D, Mahoney MG. 2009. Increased expression of Dsg2 in malignant skin carcinomas: a tissue-microarray based study. Cell Adh Migr 3:148–154. doi: 10.4161/cam.3.2.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Overmiller AM, Pierluissi JA, Wermuth PJ, Sauma S, Martinez-Outschoorn U, Tuluc M, Luginbuhl A, Curry J, Harshyne LA, Wahl JK III, South AP, Mahoney MG. 2017. Desmoglein 2 modulates extracellular vesicle release from squamous cell carcinoma keratinocytes. FASEB J 31:3412–3424. doi: 10.1096/fj.201601138RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hemminki O, Bauerschmitz G, Hemmi S, Lavilla-Alonso S, Diaconu I, Guse K, Koski A, Desmond RA, Lappalainen M, Kanerva A, Cerullo V, Pesonen S, Hemminki A. 2011. Oncolytic adenovirus based on serotype 3. Cancer Gene Ther 18:288–296. doi: 10.1038/cgt.2010.79. [DOI] [PubMed] [Google Scholar]
- 53.Su X, Tian X, Jiang Z, Ma Q, Liu Q, Lu X, Zhou R. 2016. Human adenovirus serotype 3 vector packaged by a rare serotype 14 hexon. PLoS One 11:e0156984. doi: 10.1371/journal.pone.0156984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alemany R. 2014. Oncolytic adenoviruses in cancer treatment. Biomedicines 2:36–49. doi: 10.3390/biomedicines2010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feng Y, Sun X, Ye X, Feng Y, Wang J, Zheng X, Liu X, Yi C, Hao M, Wang Q, Li F, Xu W, Li L, Li C, Zhou R, Chen L, Feng L. 2018. Hexon and fiber of adenovirus type 14 and 55 are major targets of neutralizing antibody but only fiber-specific antibody contributes to cross-neutralizing activity. Virology 518:272–283. doi: 10.1016/j.virol.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Y, Feng L, Li L, Wang D, Li C, Sun C, Li P, Zheng X, Liu Y, Yang W, Niu X, Zhong N, Chen L. 2015. Effects of the fusion design and immunization route on the immunogenicity of Ag85A-Mtb32 in adenoviral vectored tuberculosis vaccine. Hum Vaccin Immunother 11:1803–1813. doi: 10.1080/21645515.2015.1042193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soriano P. 1999. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]