Abstract
Antimicrobial resistance (AMR) in Escherichia coli (E. coli) poses a public health concern worldwide. Wild birds and rodents, due to their mobility, are potential vehicles for transmission of AMR bacteria to humans. Ninety-six wild birds’ faecal samples and 135 rodents’ droppings samples were collected and analysed in 2017. Forty-six E. coli isolates from wild birds and rodents were subjected to AMR phenotypic and genotypic characterisation. The proportion of E. coli isolates resistant to at least one of the antimicrobials tested from wild birds (80.8%) was significantly higher than that of isolates from rodents (40.0%). The proportion of E. coli isolates resistant to each antimicrobial class for wild birds was 3.8% to 73.1% and that for rodents was 5.0% to 35.0%. Six out of 26 E. coli isolates from wild birds (23.1%) and two out of 20 (10.0%) isolates from rodents were multi-drug resistant (MDR) strains. These MDR E. coli isolates were detected with various antimicrobial resistance genes such as blaTEM-1B and qnrS1 and could be considered as part of the environmental resistome. Findings in this study suggested that wild birds and rodents could play a role in disseminating antimicrobial resistant E. coli, and this underscores the necessity of environment management and close monitoring on AMR bacteria in wild birds and rodents to prevent spreading of resistant organisms to other wildlife animals and humans.
Keywords: Escherichia coli (E. coli), wild birds, rodents, multi-drug resistant (MDR), resistance genes, antimicrobial susceptibility testing, whole genome sequencing
1. Introduction
Escherichia coli (E. coli) is a commensal bacterium found in the guts of animals [1,2]. It can be pathogenic and cause gastroenteritis, bacteraemia and urinary tract infections [2,3,4]. The bacterium is known to be susceptible to selection pressure [2] and has the high competency to pick up and transfer antibiotic resistance genes to and from other bacterial strains [2,5]. Resistance to antimicrobials in clinical and veterinary medicine has been increasingly reported in E. coli and this has become a public health concern worldwide [4]. The World Health Organization (WHO) has categorized Enterobacteriaceae (including E. coli) as pathogens of critical priority for antimicrobial resistance (AMR) investigation [6]. AMR in E. coli has been investigated in clinical settings and retail cooked food in Singapore [7,8]. However, to our knowledge, there is limited information on AMR in the environment such as in wild birds and rodents in Singapore. Wild birds and rodents, due to their potential interactions with humans and mobility [9], are potential reservoirs and vectors for transmission of antimicrobial resistant bacteria to humans, through pathways such as contact of faecal materials and contamination of food items [4,9,10]. This study aimed to investigate the occurrence and AMR of E. coli from wild birds and rodents in Singapore. The data from the study would enhance our understanding of AMR transmission in the environment and allow subsequent mitigation measures.
2. Materials and Methods
2.1. Sample Collection, Isolation and Identification
Sample collection: A total of 96 faecal samples from wild birds and 135 samples of rodents’ droppings were conveniently collected in 2017, as part of zoonotic disease surveillance programmes. The wild birds were collected by the Environmental Health Institute of National Environment Agency, where the wild birds’ carcasses were collected from urban areas and recreational parks. An approximate 1 g of wild bird faecal matter was collected after dissection of each wild bird. Samples of rodents’ droppings (rodent species were unidentified) were collected by Rodent Control Unit, of National Environment Agency Central Regional Office. These rodents’ droppings were found at bin chute, drain and kitchen areas.
Sample isolation was carried out according to the method as follows: 1–10 g of wild bird faecal or rodent droppings samples were incubated in 9 mL of Universal Pre-Enrichment Broth (Acumedia, Lansing, MI, USA) under aerobic conditions at 37 ± 1 °C for 16–18 h. A 10 μL loopful of enriched broth was streaked onto Eosine Methylene Blue agar (Acumedia, Lansing, MI, USA) and incubated under the same conditions. Single colonies were streaked on MacConkey agar (Acumedia, Lansing, MI, USA) and incubated under the same conditions for further isolation. The purified colonies were then streaked onto Tryptic Soy Agar (Acumedia, Lansing, MI, USA) and incubated under the same condition. E. coli confirmation was performed with an indole test. A pure colony from Tryptic Soy Agar was inoculated into Peptone water (Acumedia, Lansing, MI, USA) and incubated under the same conditions. Five drops (0.5 mL) of Remel™ Kovacs Indole Reagent (Thermo Scientific, Lenexa, KS, USA) was dispensed to the enriched peptone water. A pink ring interfaced between peptone water and indole reagent was observed for E. coli isolate. Next, randomly selected separate E. coli isolates (one colony per sample) were stored in Brain Heart Infusion broth with 15% glycerol until further usage.
DNA extraction of the presumptive E. coli isolates was carried out using DNeasy Blood & Tissue Kits (Qiagen, Hilden, Germany). Confirmation of E. coli isolates was performed by 16S ribosomal RNA polymerase chain reaction using forward primer 27f (5′-AGAGTTTGATCCTGGCTCAG-3′) and reverse primer 1492r (5′-GGTTACCTTGTTACGACTT-3′). Polymerase chain reaction conditions are described as follows [11]: A 50 μL reaction mix consisted of 10 μL of 5x Phusion High-Fidelity Buffer (Thermo Scientific, Vilnius, Lithuania), 1 μL of dNTP mix (1st BASE, Seri Kembangan, Malaysia), 0.5 μL (10 µM) of each primer (Integrated DNA Technologies, Singapore), 0.5 μL of Phusion Hot Start II DNA Polymerase (Thermo Scientific, Vilnius, Lithuania), 5 μL of DNA template and 32.5 μL of molecular grade water was used. PCR was conducted using thermocycler (Applied Biosystems, Waltham, MA, USA) with conditions as follows: initial denaturation at 98 °C for 30 s, 35 cycles consisting of denaturation at 98 °C for 10 s, annealing at 50 °C for 30 s and extension at 72 °C for 30 s and a final extension at 72 °C for 10 min. The amplified fragments were visualised at 2% agarose gel. Those isolates with an expected band size of 1465 bp were sequenced using BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Waltham, MA, USA). Raw sequences were assembled using BioEdit version 7.2.6.1 software. Assembled reads were uploaded to nucleotide BLAST database (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE = BlastSearch) (National Centre for Biotechnology Information, United States) for E. coli identification (>90%).
2.2. Antimicrobial Susceptibility Testing of E. coli Isolates
Susceptibilities to 12 antimicrobial agents of nine different classes were determined by disk diffusion method and interpreted according to the Clinical and Laboratory Standards Institute guideline 2020 [12]. The antimicrobials agents were: Penicillins (Ampicillin 10 µg), Aminoglycosides (Amikacin 30 µg and Gentamicin 10 µg), Beta-lactam/Beta-lactamase Inhibitor Combinations (Amoxicillin/Clavulanic acid 30 µg), Phenicols (Chloramphenicol 30 µg), Quinolones (Ciprofloxacin 5 µg, Nalidixic acid 30 µg, Norfloxacin 30 µg), Cephalosporins (Ceftriaxone 30 µg), Carbapenems (Meropenem 10 µg), Folate Pathway Synthesis (Sulphamethoxazole/Trimethoprim 25 µg) and Tetracyclines (Tetracycline 30 µg) (Oxoid, Basingstoke, UK). Escherichia coli ATCC® 25922 strain was used as the quality control strain. Isolates that expressed resistance or intermediate phenotypes were classified as resistant. Isolates that were resistant to three or more classes of antimicrobials were considered multi-drug resistant (MDR) [13].
2.3. Extended Spectrum Beta-Lactamases (ESBL) Testing for E. coli Isolates
Resistance to Ceftriaxone by disk diffusion was confirmed for ESBL production, as previously described [7].
2.4. Genotypic Characterisation by Whole Genome Sequencing
Based on phenotypic resistance results, eight MDR E. coli isolates (indicated in Table 1) were selected and subjected to genotypic analysis by whole genome sequencing. DNA extraction, library preparation and sequencing were performed as previously described [7]. Raw sequence data was deposited into Genbank under Bioproject accession number PRJNA625931. The raw reads were assembled using SPAdes version 3.11.0, with “-careful, -k auto and -cov-cutoff as off” parameters [14]. The genome data were analysed with reference to the ResFinder 3.1 database (https://cge.cbs.dtu.dk/services/ResFinder/) to identify antimicrobial resistance genes and chromosomal point mutations based on the following parameters: minimum length coverage of 60% and minimum identity of 90% (Centre for Genomic Epidemiology, Denmark) [15].
Table 1.
Antimicrobial susceptibility profiles of E. coli isolates from wild birds and rodents (#: Multi-drug resistant strain; S: Susceptible; I: Intermediate; R: Resistant. Antimicrobial agents used for testing were AK: Amikacin; AMC: Amoxicillin/Clavulanic acid; AMP: Ampicillin; C: Chloramphenicol; CIP: Ciprofloxacin; CN: Gentamicin; CRO: Ceftriaxone; MEM: Meropenem; NA: Nalidixic acid; NOR: Norfloxacin; SXT: Sulphamethoxazole/Trimethoprim; TE: Tetracycline).
| Sample ID | Sample Type | Source | AK30 | AMC30 | AMP10 | CRO30 | C30 | CIP5 | CN10 | MEM10 | NA30 | NOR10 | SXT25 | TE30 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C1776# | Wild Bird | Crow | S | S | R | S | R | R | S | S | R | R | R | R |
| C1775 | Wild Bird | Myna | S | S | R | S | S | S | S | S | S | S | S | S |
| 8657-0407-0411-1 | Rodent | Rodent | S | S | S | S | S | S | S | S | S | S | S | S |
| 8657-0948-0411 | Rodent | Rodent | S | S | I | S | S | S | S | S | S | S | S | S |
| 8645-0135# | Rodent | Rodent | S | S | R | S | R | S | S | S | S | S | R | R |
| C1797# | Wild Bird | Crested Goshawk | S | S | I | S | R | R | S | S | R | R | S | R |
| C1809-1 | Wild Bird | Hooded Pitta | S | S | I | S | S | S | S | S | S | S | S | S |
| C1789 | Wild Bird | Crow | S | S | I | S | S | S | S | S | S | S | S | S |
| C1798 | Wild Bird | Black-naped oriole | S | S | I | S | S | S | S | S | S | S | S | S |
| HHK-M04-060417-1 | Rodent | Rodent | S | S | S | S | S | S | S | S | S | S | S | S |
| HHK-M04-060417-3 | Rodent | Rodent | S | S | I | S | S | S | S | S | S | S | S | S |
| 8642-0159-0410-1 | Rodent | Rodent | S | S | S | S | S | S | S | S | S | S | S | S |
| C1742# | Wild Bird | Yellow Bittern | S | I | R | S | R | S | S | S | S | S | S | R |
| C1779 | Wild Bird | Crow | S | S | S | S | S | S | S | S | S | S | S | S |
| C1805-1# | Wild Bird | Crow | S | I | R | S | R | S | S | S | S | S | S | R |
| 8657-0146-0224 | Rodent | Rodent | S | S | S | S | S | S | S | S | S | S | S | S |
| NHA-M02-230117 | Rodent | Rodent | S | S | S | S | S | S | S | S | S | S | S | S |
| 8657-0258-0411-1 | Rodent | Rodent | S | S | S | S | S | S | S | S | S | S | S | S |
| 7574-0224-0224 | Rodent | Rodent | S | S | I | S | S | S | S | S | S | S | S | S |
| C1802-1 | Wild Bird | Crow | S | S | S | S | S | S | R | S | S | S | S | R |
| 8646-0251-0301 | Rodent | Rodent | S | S | I | S | S | S | S | S | S | S | S | S |
| HHK-M05-160317 | Rodent | Rodent | S | S | S | S | S | S | S | S | S | S | S | S |
| C1743E | Wild Bird | Black Bittern | S | S | S | S | S | S | S | S | S | S | S | S |
| C1781 | Wild Bird | Black Bittern | S | S | I | S | S | S | S | S | S | S | S | S |
| HHK-M07-060417-1 | Rodent | Rodent | S | I | R | S | S | S | S | S | S | S | S | R |
| C1806-1 | Wild Bird | Pied Fantail | S | S | S | S | S | S | S | S | S | S | S | S |
| C1736 | Wild Bird | Brahmin Kite | S | S | S | S | S | S | S | S | S | S | S | S |
| 8645-0205-0411-1 | Rodent | Rodent | S | S | S | S | S | S | S | S | S | S | S | S |
| C1795 | Wild Bird | Sparrow Hawk | S | S | I | S | S | S | S | S | R | S | S | S |
| C1794 | Wild Bird | Grey Heron | S | I | R | S | S | S | S | S | I | S | S | R |
| NRS-M02-150317 | Rodent | Rodent | S | S | S | S | S | S | S | S | S | S | S | S |
| C1783 | Wild Bird | Scops Owl | S | S | S | S | S | S | S | S | S | S | S | S |
| C1770 | Wild Bird | Myna | S | S | S | S | S | S | S | S | S | S | S | R |
| C1758# | Wild Bird | Black Bittern | S | I | R | S | S | S | S | S | S | S | R | R |
| C1757 | Wild Bird | Yellow Bittern | S | S | I | S | S | S | S | S | S | S | S | S |
| C1750 | Wild Bird | Sparrow Hawk | S | I | R | S | S | S | S | S | S | S | S | S |
| C1737 | Wild Bird | Crow | S | S | I | S | S | S | S | S | S | S | S | S |
| C1740 | Wild Bird | Crow | S | R | R | S | S | S | S | S | S | S | S | S |
| C1803-2 | Wild Bird | Crow | S | S | I | S | S | S | S | S | S | S | S | S |
| 8656-0339-0224 | Rodent | Rodent | S | S | S | S | S | S | S | S | S | S | S | S |
| TAH-M01-210317 | Rodent | Rodent | S | S | S | S | S | S | S | S | R | S | S | S |
| HHK-M04-060417-1A | Rodent | Rodent | S | S | S | S | S | S | S | S | S | S | S | S |
| 8655-0114# | Rodent | Rodent | S | R | R | S | S | R | S | S | R | R | S | R |
| C1738 | Wild Bird | Crow | S | S | I | S | S | S | S | S | S | S | S | S |
| C1722# | Wild Bird | Crow | S | R | R | R | R | I | R | S | R | S | R | R |
| HHK-M06-060417-1 | Rodent | Rodent | S | S | S | S | S | S | S | S | S | S | S | S |
2.5. Statistical Analysis
The 95% confidence intervals of proportions were calculated using http://vassarstats.net/prop1.html. Z-scores for two population proportions were calculated using https://www.socscistatistics.com/tests/ztest/default2.aspx.
3. Results
3.1. Occurrence of E. coli in Wild Birds and Rodents
Of the 96 wild birds’ faecal samples, E. coli was detected in 26 (27.1%) [95% CI: 19.2–36.7%] faecal samples from 12 different types of wild birds, with details shown in Table 2. Of 135 rodent droppings, 20 (14.8%) [95% CI: 9.8–21.8%] tested positive for E. coli (Table 2).
Table 2.
Occurrence of E. coli in wild birds and rodents.
| Types of Samples | Percentage of Samples Positive for E. coli | Sample Name (Scientific Name) | No. of E. coli Isolates |
|---|---|---|---|
| Wild Birds | 27.1% (26/96) [95% CI: 19.2–36.7%] | Crow (Corvus spp.) | Ten |
| Black Bittern (Ixobrychus flavicollis) | Three | ||
| Myna (Acridotheres spp.) | Two | ||
| Sparrow Hawk (Accipiter spp.) | Two | ||
| Yellow Bittern (Ixobrychus sinensis) | Two | ||
| Black-naped oriole (Oriolus chinensis) | One | ||
| Brahminy Kite (Haliastur indus) | One | ||
| Crested Goshawk (Accipiter trivirgatus) | One | ||
| Grey Heron (Ardea cinerea) | One | ||
| Hooded Pitta (Pitta sordida) | One | ||
| Pied Fantail (Rhipidura javanica) | One | ||
| Scops Owl (Otus spp.) | One | ||
| Rodents | 14.8% (20/135) [95% CI: 9.8–21.8%] | Black Rat/Brown Rat/House Mouse (Rattus rattus/Rattus norvegicus/Mus musculus) | 20 |
3.2. Antimicrobial Resistance in E. coli Isolated from Wild Birds and Rodents
Antimicrobial susceptibility test results for all the E. coli isolates are shown in Table 1. There were 80.8% (21/26) of E. coli isolates from wild birds and 40.0% (8/20) of E. coli from rodents being resistant to at least one of the antimicrobials tested in the study (Table 3). The proportion of E. coli isolates resistant to at least one of the antimicrobials tested from wild birds (80.8%, Z-score 2.8, p < 0.05) was significantly higher than that of isolates from rodents (40.0%) (Table 3).
Table 3.
Percentage of E. coli isolates from wild birds and rodents resistant to at least one antimicrobial.
| Wild Birds | Rodents | Z-Score |
|---|---|---|
| 80.8% (21/26) [95% CI: 62.1–91.5%] |
40.0% (8/20) [95% CI: 21.9–61.3%] |
2.8 (p < 0.05) |
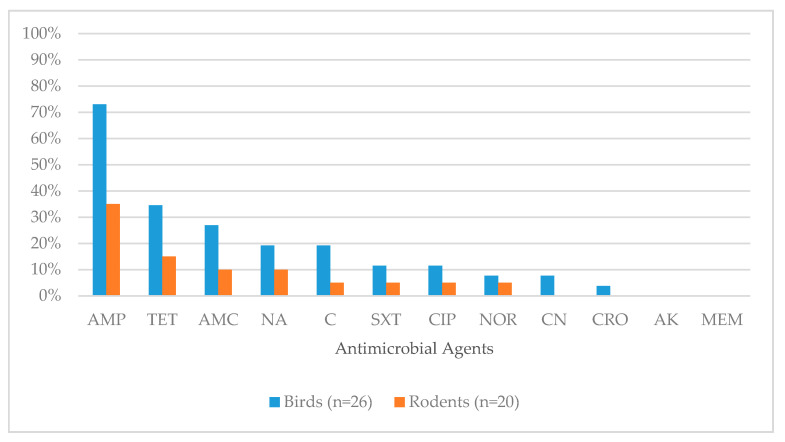
The proportions of E. coli isolates resistant to each antimicrobial class for wild birds (3.8% to 73.1%) and rodents (5.0% to 35.0%) are shown in Figure 1 and Table 4.
Figure 1.
Percentage of antimicrobial resistance in E. coli isolates from wild birds and rodents (Corresponding to Table 4). Antimicrobial agents used for testing were AK: Amikacin; AMC: Amoxicillin/Clavulanic acid; AMP: Ampicillin; C: Chloramphenicol; CIP: Ciprofloxacin; CN: Gentamicin; CRO: Ceftriaxone; MEM: Meropenem; NA: Nalidixic acid; NOR: Norfloxacin; SXT: Sulphamethoxazole/Trimethoprim; TE: Tetracycline.
Table 4.
Percentage of antimicrobial resistance in E. coli isolates from wild birds and rodents (Corresponding to Figure 1).
| Antimicrobial Class | Antimicrobial Agent Tested in the Study | Percentage of Isolates Showing Resistant Phenotype (n) | |
|---|---|---|---|
| Wild Birds (26) | Rodents (20) | ||
| Penicillins | Ampicillin | 73.1% (19/26) | 35.0% (7/20) |
| Tetracyclines | Tetracycline | 34.6% (9/26) | 15.0% (3/20) |
| Beta-lactam/beta-lactamase Inhibitor Combinations | Amoxicillin/Clavulanic acid | 26.9% (7/26) | 10.0% (2/20) |
| Quinolones | Nalidixic acid (Quinolone) | 19.2% (5/26) | 10.0% (2/20) |
| Phenicols | Chloramphenicol | 19.2% (5/26) | 5.0% (1/20) |
| Folate Pathway Synthesis | Sulphamethoxazole/Trimethoprim | 11.5% (3/26) | 5.0% (1/20) |
| Quinolones | Ciprofloxacin (Fluoroquinolone) | 11.5% (3/26) | 5.0% (1/20) |
| Quinolones | Norfloxacin (Fluoroquinolone) | 7.7% (2/26) | 5.0% (1/20) |
| Aminoglycosides | Gentamicin | 7.7% (2/26) | 0.0% |
| Third Generation Cephalosporins | Ceftriaxone | 3.8% (1/26) | 0.0% |
| Aminoglycosides | Amikacin | 0.0% | 0.0% |
| Carbapenems | Meropenem | 0.0% | 0.0% |
For isolates from wild birds, the most common phenotypic resistance exhibited was against Penicillins (Ampicillin), followed by Beta-lactam/Beta-lactamase Inhibitor Combinations (Amoxicillin/Clavulanic Acid), Tetracyclines (Tetracycline), Quinolones (Nalidixic acid) and Phenicols (Chloramphenicol). Lower resistance rates (less than or equal to 15.0%) were found for the other remaining five antimicrobial classes (Table 4). No resistance was found for Aminoglycosides (Amikacin) and Carbapenems (Meropenem).
For isolates from rodents, the most common phenotypic resistance exhibited was against Penicillins (Ampicillin). Lower resistance rates (less than or equal to 15.0%) were observed for other remaining seven antimicrobial classes (Table 4). No resistance was found for Aminoglycosides (Gentamicin), Third Generation Cephalosporins (Ceftriaxone), Aminoglycosides (Amikacin) and Carbapenems (Meropenem).
Six out of 26 E. coli isolates from wild birds (23.1%) and two out of 20 (10.0%) isolates from rodents were resistant to three or more antimicrobial classes and considered as multi-drug resistant (MDR) strains. One MDR E. coli isolate (C1722) recovered from wild birds tested positive for ESBL production.
3.3. Distribution of Resistance Genes in Eight MDR E. coli Isolates from Wild Birds and Rodents
At least one antimicrobial resistance gene was detected in all eight MDR E. coli isolates from wild birds and rodents (Table 5). An isolate (C1722) from wild bird had 17 resistance genes detected, the highest among the eight MDR E. coli isolates. It is worth noting that this isolate was ESBL-producing and harboured blaCTM-X-65 gene and tet(X) gene.
Table 5.
Distribution of resistance genes in the eight multi-drug resistant E. coli isolates.
| Isolate ID | Sample Source | Sample Description | Resistance Genes (n) | Aminoglycoside | ESBL | Quinolone | Fosfomycin | MLS a | Phenicol | Sulphonamide | Tetracycline | Trimethoprim | Chromosomal Point Mutations |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C1722 | Wild bird | Crow | 17 | aac(3)-IV, aadA2, aph(3′)-Ia, aph(3″)-Ib, aph(4)-Ia, aph(6)-Id, | blaCTX-M-65, blaTEM-1B | - | fosA3 | mdf(A) | cmlA1, floR | sul2, sul3 | tet(A), tet(X) | dfrA12 | gyrA (Ser83Leu) |
| C1742 | Wild bird | Yellow Bittern | 7 | aph(3′)-Ia | blaTEM-176 | qnrS1 | - | mdf(A) | floR | - | tet(A) | dfrA14 | - |
| C1758 | Wild bird | Black Bittern | 4 | aph(6)-Id | qnrS1 | - | mdf(A) | - | - | tet(A) | - | - | |
| C1776 | Wild bird | Crow | 11 | aadA1, aadA2, aph(3″)-Ib, aph(6)-Id | blaTEM-1B | oqxB | - | mdf(A), mph(A) | cmlA1 | sul3 | tet(A) | - | gyrA, (Ser83Leu), gyrA(Asp87Asn) & parC (Ser80Ile). |
| C1797 | Wild bird | Crested Goshawk | 3 | - | - | - | - | mdf(A) | catA1 | - | tet(B) | - | gyrA (Ser83Leu), gyrA(Asp87Asn) & parC (Ser80Ile). |
| C1805-1 | Wild bird | Crow | 7 | aadA1 | blaTEM-1B | qnrS1 | - | mdf(A) | floR | sul3 | tet(A) | - | - |
| 8645-0135 | Rodent | Rodent | 11 | aadA1, aadA2 | blaTEM-1B | qnrS1 | - | mdf(A) | cmlA1, floR | sul2, sul3 | tet(A) | dfrA12 | - |
| 8655-0114 | Rodent | Rodent | 1 | - | - | - | - | mdf(A) | - | - | - | - | - |
MLS a—Macrolide, Lincosamide and Streptogramin B.
Macrolide, Lincosamide and Streptogramin B (MLS) resistance gene (mdf(A)) was found in all MDR E. coli isolates. An isolate from wild bird (C1776) was detected with additional MLS resistance gene mph(A). Six out of eight (75.0%) MDR E. coli isolates were detected with Tetracycline resistance gene (tet(A)) while another MDR E. coli isolate from wild bird (C1797) was detected with Tetracycline resistance gene tet(B). Aminoglycoside resistance genes were detected in 75.0% (6/8) of MDR E. coli isolates. The most common Aminoglycoside resistance genes were aadA1 (37.5%, 3/8), aadA2 (37.5%, 3/8) and aph (6)-Id (37.5%, 3/8). Phenicol resistance genes were detected in 75.0% (6/8) of MDR E. coli isolates. floR (50.0%, 4/8) was the most frequently detected Phenicol resistance gene, followed by cmlA1 (37.5%, 3/8) and catA1 (12.5%, 1/8). ESBL resistance genes were detected in 62.5% (5/8) of the MDR E. coli isolates, which comprised of blaTEM-1B (50.0%, 4/8), blaTEM-176 (12.5%, 1/8) and blaCTM-X-65 (12.5%, 1/8). Plasmid Mediated Quinolone Resistance (PMQR) genes were found in 62.5% (5/8) of MDR E. coli isolates, with the common PMQR gene being qnrS1 (50.0%, 4/8). Another PMQR gene oqxB was found in an isolate from wild bird (C1776). Sulphonamide resistance genes were present in 50% (4/8) of MDR E. coli isolates, which include sul3 (50.0%, 4/8) and sul2 (25.0%, 2/8) resistance genes. Trimethoprim resistance genes (dfrA12, dfrA14) were found in 37.5% (3/8) of MDR E. coli. Fosfomycin (fosA3) resistance gene was detected in one isolate from wild bird (C1722).
Chromosomal point mutations in Quinolone resistant determining regions (QRDR) of gyrA and parC genes were observed in 37.5% (3/8) of the MDR E. coli isolates from wild birds. A gyrA mutation for amino acid substitution from Serine to Leucine at 83th position (Ser83Leu) was found in an isolate (C1722), which displayed resistance to Nalidixic Acid and Ciprofloxacin (Table 6). Two isolates (C1776 and C1797) carried double gyrA mutations responsible for amino acid change from Serine to Leucine at 83th position (Ser83Leu) and aspartic acid to asparagine at 87th position (Asp87Asn) and parC mutation for change from Serine to Isoleucine at 80th position (Ser80Ile). These two isolates were resistant to Nalidixic acid, Ciprofloxacin and Norfloxacin (Table 6).
Table 6.
Comparison of antimicrobial resistance phenotype and genotype of the eight multi-drug resistant E. coli isolates for the selected antimicrobial agents (AMC: Amoxicillin/Clavulanic acid; AMP: Ampicillin; C: Chloramphenicol; CIP: Ciprofloxacin; CRO: Ceftriaxone; NA: Nalidixic acid; NOR: Norfloxacin; TE: Tetracycline).
| Isolate ID | Sample Source | Sample Description | Beta-lactams | Quinolones | Phenicols | Tetracyclines | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Phenotype | Genotype | Phenotype | Genotype | Phenotype | Genotype | Phenotype | Genotype | |||
| C1722 | Wild bird | Crow | AMC *, AMP, CRO | blaCTX-M-65, blaTEM-1B | CIP, NA | Chromosomal mutations gyrA (Ser83Leu) | C | cmlA1, floR | TE | tet(A), tet(X) |
| C1742 | Wild bird | Yellow Bittern | AMC *, AMP | blaTEM-176 | - | qnrS1 | C | floR | TE | tet(A) |
| C1758 | Wild bird | Black Bittern | AMC *, AMP | - | - | qnrS1 | - | - | TE | tet(A) |
| C1776 | Wild bird | Crow | AMP | blaTEM-1B | CIP, NA, NOR | oqxB, Chromosomal mutations gyrA (Ser83Leu) gyrA (Asp87Asn) & parC (Ser80Ile) | C | cmlA1 | TE | tet(A) |
| C1797 | Wild bird | Crested Goshawk | AMP | - | CIP, NA, NOR | Chromosomal mutations gyrA (Ser83Leu) gyrA (Asp87Asn) & parC (Ser80Ile) | C | catA1 | TE | tet(B) |
| C1805_1 | Wild bird | Crow | AMP, AMC * | blaTEM-1B | - | qnrS1 | C | floR | TE | tet(A) |
| 8645_0135 | Rodent | Rodent | AMP | blaTEM-1B | - | qnrS1 | C | cmlA1, floR | TE | tet(A) |
| 8655_0114 | Rodent | Rodent | AMC *, AMP | - | CIP, NA, NOR | - | - | - | TE | - |
* Denotes a combination of two different classes of antimicrobial agents being used in the antimicrobial susceptibility testing.
3.4. Comparison between Phenotypic and Genotypic Characteristics of MDR E. coli Isolates
Comparison of antimicrobial phenotype and whole genome sequencing data of MDR E. coli isolates (n = 6) from wild birds and rodents (n = 2) was performed for Beta-lactams, Quinolones, Phenicols and Tetracyclines. The remaining five antimicrobial classes were not performed for comparison between antimicrobial resistance phenotype and genotype due to limitations of the antimicrobial susceptibility testing as follows: (1) Macrolides and Fosfomycin were not tested; (2) Many antimicrobial classes of Aminoglycosides (e.g., Kanamycin, Streptomycin) were excluded; (3) A combination of Sulphamethoxazole/Trimethoprim was used and there was no testing for individual antimicrobial classes of Sulphonamides and Trimethoprim.
In general, there was good agreement between phenotypic and genotypic characteristics for Phenicols and Tetracyclines. Discrepancies between phenotypic and genotypic resistance traits were detected in MDR E. coli isolates (Table 6). For instance, Quinolones resistance genes were detected in four MDR E. coli isolates (C1742, C1758, C1805_1 and 8645_0135) which were, however, phenotypically susceptible to the Quinolones included in this study. Isolate 8655_0114 was phenotypically resistant to Quinolones with no corresponding resistance gene. Three MDR E. coli (C1758 and C1797, 8655_0114) were phenotypically resistant to Penicillins and Beta-lactam/Beta-lactamase Inhibitor Combinations but there was no ESBL resistance gene detected.
4. Discussion
To our knowledge, this is the first report on the occurrence and antimicrobial resistant phenotype and genotype in E. coli isolates from wild birds and rodents in Singapore.
This study revealed that the occurrence of E. coli in wild birds (27.1%) in Singapore was relatively lower than that reported in other countries such as Switzerland (53.7%) and Saudi Arabia (93.0%) [16,17]. Similarly, the occurrence of E. coli in rodents (14.8%) was relatively lower than it is reported in other countries such as Trinidad and Tobago (83.8%) and Canada (62.7%) [18,19]. One possible reason for relatively lower occurrences on both wild birds and rodents could be due to differences in sampling and laboratory methods used in respective studies (e.g., the convenient collection of samples used in this study) that rendered comparison of occurrence data between studies challenging. Furthermore, the occurrence data could be affected by storage conditions of the collected samples. Another limitation of our study was that we were unable to identify the rodent species despite it being known that common rodent species found in Singapore include Black Rat (Rattus norvegicus), Brown Rat (Rattus rattus) and House Mouse (Mus musculus) [20]. The occurrence of E. coli could hypothetically originate from these pools of common rodent species. Our study provided an insight into the occurrence of E. coli isolates from wild birds and rodents in Singapore, which enhances our understanding of the local epidemiology of E. coli and could guide future epidemiological studies.
The proportion of E. coli isolates resistant to at least one of the antimicrobials tested from wild birds was significantly higher than that of isolates from rodents. Although different in behavior, diet and migration potential with regard to species, wild birds generally have a higher movement pattern than rodents and could have higher exposure to antimicrobial resistance determinants in the ecological niches. This could lead to a higher probability for wild birds in disseminating AMR determinants to humans or other wildlife animals [21]. Our results differ from a previous study in Singapore which indicated there was no phenotypic antimicrobial resistance detected in Salmonella isolates recovered from wild birds [22]. The difference in phenotypic resistance observed for E. coli and Salmonella isolates could be due to the fact that E. coli has a greater ability to acquire resistance than Salmonella, making E. coli more susceptible to antimicrobial selection pressure than Salmonella for the tested antimicrobials [23,24]. This implies the importance of using multiple bacteria organisms (both commensal and pathogenic) as AMR indicators in surveillances for better understanding of the distribution of resistant organisms or resistance determinants in the environment.
The antimicrobial resistance rate among E. coli isolates could be related to the usage of antimicrobials. E. coli isolates from both wild birds and rodents were phenotypically resistant to Penicillins, whereas isolates recovered from wild birds also displayed resistance to Beta-lactam/Beta-lactamase Inhibitor Combinations and Tetracyclines. These antimicrobials are commonly used in clinical and agricultural sectors [25,26,27,28,29,30]. This is of public health concern as these antimicrobials are the first-line drugs of choice for empirical treatment of infections caused by E. coli. The widespread resistance to these antimicrobials will render these antimicrobials ineffective for the treatment of infections and will increase the need for an alternative antimicrobial therapy option.
The lower percentage (less than or equal to 15.0%) of resistance observed in E. coli isolates from wild birds to antimicrobial classes of Aminoglycosides and Third Generation Cephalosporins suggested that there could be a relatively lower selection pressure of these antimicrobial classes as compared to other commonly used antimicrobials in wild birds. As these antimicrobial classes are not commonly used in food animal production [4,31], nor in clinical settings as a first-line drug for E. coli infection but as a drug of choice for invasive/resistant infections [4,32,33], the level of antimicrobial residual pollution in the environment is expected to be lower, and consequently lower exposure and selection pressure for wild birds [34]. Nevertheless, further monitoring of efficacy for these antimicrobials remains necessary.
Our study detected E. coli isolates from wild birds and rodents that were resistant to Quinolones (Nalidixic Acid), which is an indicator of reduced susceptibility for Fluoroquinolones (e.g., Ciprofloxacin and Norfloxacin). Indeed, Nalidixic Acid resistant E. coli isolates from wild birds (n = 5) also showed resistance to Ciprofloxacin (60.0%, 3/5) and Norfloxacin (40.0%, 2/5). A similar observation was found in rodents where Nalidixic Acid-resistant E. coli isolates (n = 2) were resistant to Ciprofloxacin (50.0%, 1/2) and Norfloxacin (50.0%, 1/2). These findings indicated the possible increasing trend for Fluroquinolones-resistant organisms present in the environment. It is important to implement continuous monitoring of Quinolone and Fluroquinolone resistance in E. coli from wild birds and rodents in order to prevent the spread of the resistance determinants into environmental niches.
All MDR isolates in our study were detected with MLS resistance gene mdf(A). A study showed that this resistance gene is expressed constitutively in E. coli [35] and it encodes for a multidrug efflux pump [36]. Another study has shown that the expression of mdf(A) in E. coli would confer multidrug resistance [37]. Isolate C1776 (from wild bird) harboured another MLS resistance gene mph(A), which encodes for enzymes capable of inactivating Erythromycin (a type of Macrolides) [38].
Our observation of tet(A) being the most common tetracycline resistance gene in all MDR isolates was in agreement with other studies in birds and rodents [26,39,40]. Isolate C1797 from wild bird was detected with the tet(B) gene—another gene which codes for efflux pump to transport Tetracycline out of bacterial cell [7]. Isolate C1722 from wild bird harboured tet(X), which is the first described tetracycline resistance gene that encodes for an enzyme which inactivates Tetracycline. The tet(X) gene was previously identified in environmental bacteria from soil, sewage plants and human clinical samples [41].
Aminoglycosides resistance genes encode for enzymes such as acetyltransferases (such as aac(3)-IV detected in our study), nucleotidyltransferases (such as aadA1 and aadA2 found in this study) or phosphotransferases (such as aph(3′)-Ia and aph (6)-Id identified in our study) which inactivate Aminoglycosides. The most common Aminoglycoside genes detected were aadA1, aadA2 and aph (6)-Id. Genes aadA1 and aadA2 confer resistance to Aminoglycosides (e.g., Spectinomycin and Streptomycin) [42] and they were also found in a study on birds from Australia [43]. The gene aph (6)-Id was reported as the most commonly detected gene in E. coli clinical isolates from Egypt [44].
Both Phenicol resistance genes floR and cml1a that encode for efflux transporter [45] were detected in our study. The floR gene was reported in Klebsiella pneumoniae from clinical isolates [46] and E. coli from cattle [47], while cml1a was found in E. coli from poultry [48]. Another gene catA1 was found in isolate C1797, which encodes for an enzyme that inactivates Phenicols [45] and it was previously also reported in birds [43].
Two types of ESBL resistance genes (blaTEM and blaCTX-M) were detected in MDR E. coli isolates and both resistance genes encode for Amber Class A Beta-lactamases [45]. The most commonly detected ESBL resistance gene blaTEM-1B, encodes for TEM-1 Beta-lactamase that hydrolyses Penicillins and First Generation Cephalosporins [7], and was reported in another study on birds [49]. The gene blaTEM-176, was reported in birds such as gull and rook [9]. On the other hand, blaCTX-M-65 which encodes for CTX-M Beta-lactamase that hydrolyses Penicillins and First to Third Generation Cephalosporins [50], was reported in clinical isolates [51] and raw retail chicken [52].
Qnrs1 encodes for a protein which binds to and protects both DNA gyrase and topoisomerase IV from inhibition by Quinolones [53]. Qnrs1 represented the most commonly detected PMQR gene among MDR isolates in this study and this is in agreement with other studies on E. coli and Enterobacteriaceae from birds [54,55]. Another PMQR gene oqxB that was detected in this study, was shown to encode for multidrug efflux pump [56]. This gene was found in Enterobacteriaceae from rooks throughout the European continent [55] and human clinical isolates from Korea [56]. Sulphonamide resistance genes sul2 and sul3 were identified, which is in line with other reports on birds [43,49] and rodents [40].
This study detected three MDR E. coli isolates with chromosomal point mutations in QRDR. Mechanisms of resistance to Quinolones include target gene mutations, active efflux pumps, decreased permeability for outer membrane and acquisition of resistance genes such as qnrS1 [30,57]. Target gene mutations include alteration of QRDR in DNA gyrase subunit A (gyrA) and topoisomerase IV subunit C (parC) [58]. In this study, detection of gyrA and parC mutations was found in 37.5% of the MDR E. coli isolates. These gyrA and parC mutations were identical as reported previously for Quinolone-resistant E. coli strains [57]. One ESBL-producing and MDR E. coli isolate (C1722) which displayed resistance to Nalidixic Acid and Ciprofloxacin was found to have a single gyrA mutation. Our finding differed from the report which indicated that a single gyrA mutation would result in resistance to Nalidixic Acid and an additional mutation in gyrA or parC would be required for resistance in Ciprofloxacin [57]. The different phenotypic resistance observed in the C1722 isolate could be due to the resistance mechanism for co-existence of the ESBL resistance gene (such as blaCTM-X-65) and single gyrA mutation, which was reported in a clinical study from China [51].
Our data, supported by other studies, suggest that the antimicrobial resistance genotype does not always correspond well with phenotypic expression and vice versa [59,60]. It is known that there are multiple complex mechanisms that can lead to bacteria becoming resistant to antimicrobials [59,60]. In the absence of corresponding resistance genes that encode for proteins responsible for enzymatic degradation of antimicrobials and alteration of bacterial proteins targeted by antimicrobials, isolates can exhibit resistance due to other mechanisms such as porin loss and efflux pumps [61]. For example, the detection of mdf(A) in MDR E. coli isolate from rodent (8655_0114), which encodes for a multidrug efflux pump, could possibly explain the observation that this isolate was conferred resistance to many antimicrobials but had no detectable corresponding resistance genes.
In our study, PMQR genes were detected in four MDR E. coli isolates (C1742, C1758, C1805_1 and 8645_0135) which were, however, phenotypically susceptible to the Quinolones tested. This is congruent with a study on E. coli from environmental samples in pig farms, which reported that PMQR genes alone are insufficient to confer resistance to Quinolones [62]. As discussed in the previous paragraph, further mechanisms such as mutations in QRDR regions would confer resistance to Quinolones. Despite the absence of phenotypic resistance traits, isolates harbouring antimicrobial resistance genes may be subjected to a transfer of resistance determinants to other bacterial isolates/species via horizontal gene transfer [63]. Thus, risks posed by susceptible isolates carrying resistance determinants should not be underestimated. Our findings suggested that the bacterial isolates should be characterised for both phenotypic and genotypic resistance traits, for a holistic interpretation and more thorough risk assessment. To have a more comprehensive antimicrobial resistance gene portfolio of the E. coli isolates obtained in this study, further genotypic screening could be carried out for the remaining antimicrobial resistant E. coli isolates.
Our study reports antimicrobial resistant E. coli isolates from wild birds and rodents in Singapore. In addition, genotypic characterisation by whole genome sequencing revealed the diversity of resistance genes in eight MDR E. coli isolates, which demonstrated the value of whole genome sequencing as an epidemiological tool for further understanding of antimicrobial resistance gene profiles in bacterial isolates. Once wild birds and rodents acquire antimicrobial resistant bacteria, these bacteria could continue to colonise and infect the hosts [64]. Therefore, wild birds and rodents could play a role for the dissemination of antimicrobial resistant bacteria and/or genes across different wildlife species and environmental sectors, perhaps via their faecal materials, as supported by other E. coli studies on wild birds and rodents [10,16,21,40,65]. An increasing number of cities today are undergoing urban rewilding, which transforms dense urban areas into green cities with nature assimilated. While the increased biodiversity in cities brings many benefits, it could also facilitate the crossovers of antimicrobial resistance pathogens or genes between urban and wildlife ecosystems. Hence, a close monitoring programme on the antimicrobial resistant bacteria in wildlife, especially in those animals that are in close proximity to human habitats, is recommended to complement surveillance systems in food animals, food and humans.
5. Conclusions
This study provides baseline data of occurrence and antimicrobial resistance characteristics of E. coli in wild birds and rodents representing a part of the environment in Singapore. Wild birds and rodents could play a contributing role to further spread antimicrobial resistance to other wildlife and environmental sectors through faecal contamination. The findings of our study highlight the importance of (i) environment management; (ii) close-monitoring on AMR bacteria, particularly in the potential reservoirs such as wild bird and rodents, and (iii) a deeper understanding of AMR transmission in our environment.
Acknowledgments
The authors thank the Rodent Control Unit, of the Central Regional Office, National Environment Agency for the collection of rodent droppings. In addition, the authors thank collaborators from the National Parks Board Singapore, and the Wildlife Reserves Singapore who provided the wild birds samples (carcasses). Lastly, the authors thank Singapore Centre for Environmental Life Sciences Engineering, Nanyang Technological University for providing whole genome sequencing services.
Author Contributions
Conceptualization, L.C.N. and K.T.A.; Data curation, J.Y.Q., Z.X.L., S.A., M.H., C.C., K.L.G.S., S.G. and M.Y.F.T.; Funding acquisition, J.S. and L.C.N.; Investigation, J.Y.Q., Z.X.L., S.A., M.H., C.C., K.L.G.S., S.G. and M.Y.F.T.; Methodology, L.C.N. and K.T.A.; Project administration, J.S., L.C.N. and K.T.A.; Resources, M.H. and C.C.; Supervision, J.S., L.C.N. and K.T.A.; Validation, J.Y.Q., Z.X.L., S.A., M.H., C.C., K.L.G.S., S.G. and M.Y.F.T.; Visualization, K.H.O.; Writing—original draft preparation, K.H.O.; Writing—review and editing, K.H.O., W.C.K., S.A., M.H., C.C., K.L.G.S., S.G., M.Y.F.T., J.S., L.C.N. and K.T.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Environment Agency, Singapore and Nanyang Technological University, Singapore.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Van den Bogaard A.E., Stobberingh E.E. Epidemiology of resistance to antibiotics: Links between animals and humans. Int. J. Antimicrob. Agents. 2000;14:327–335. doi: 10.1016/S0924-8579(00)00145-X. [DOI] [PubMed] [Google Scholar]
- 2.Marinho C.M., Santos T., Gonçalves A., Poeta P., Igrejas G. A Decade-Long Commitment to Antimicrobial Resistance Surveillance in Portugal. Front. Microbiol. 2016;7:1650. doi: 10.3389/fmicb.2016.01650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jindal A.K., Pandya K., Khan I.D. Antimicrobial resistance: A public health challenge. Med. J. Armed Forces Ind. 2015;71:178–181. doi: 10.1016/j.mjafi.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poirel L., Madec J.Y., Lupo A., Schink A.K., Kieffer N., Nordmann P., Schwarz S. Antimicrobial Resistance in Escherichia coli. Microbiol. Spectr. 2018;6:4:1–4:27. doi: 10.1128/microbiolspec.ARBA-0026-2017. [DOI] [PubMed] [Google Scholar]
- 5.Rasheed M.U., Thajuddin N., Ahamed P., Teklemariam Z., Jamil K. Antimicrobial Drug Resistance in Strains of Escherichia coli isolated from Food Sources. Rev. Inst. Med. Trop. São Paulo. 2014;56:341–346. doi: 10.1590/S0036-46652014000400012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organisation Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics 2017. [(accessed on 14 May 2020)]; Available online: http://apps.who.int/medicinedocs/documents/s23171en/s23171en.pdf.
- 7.Guo S., Tay M.Y.F., Aung K.T., Seow K.L.G., Ng L.C., Purbojati R.W., Drautz-Moses D.I., Schuster S.C., Schlundt J. Phenotypic and genotypic characterization of antimicrobial resistant Escherichia coli isolated from ready-to-eat food in Singapore using disk diffusion, broth microdilution and whole genome sequencing methods. Food Control. 2019;99:89–97. doi: 10.1016/j.foodcont.2018.12.043. [DOI] [Google Scholar]
- 8.Hsu L.-Y., Tan T.-Y., Jureen R., Koh T.-H., Krishnan P., Lin R.T.-P., Tee N.W.-S., Tambyah P.A. Antimicrobial Drug Resistance in Singapore Hospitals. Emerg. Infect. Dis. J. 2007;13:1944. doi: 10.3201/eid1312.070299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J., Ma Z.-B., Zeng Z.-L., Yang X.-W., Huang Y., Liu J.-H. The role of wildlife (wild birds) in the global transmission of antimicrobial resistance genes. Zool. Res. 2017;38:55–80. doi: 10.24272/j.issn.2095-8137.2017.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho P.-L., Lo W.-U., Lai E.L., Law P.Y., Leung S.M., Wang Y., Chow K.-H. Clonal diversity of CTX-M-producing, multidrug-resistant Escherichia coli from rodents. J. Med. Microbiol. 2015;64:185–190. doi: 10.1099/jmm.0.000001. [DOI] [PubMed] [Google Scholar]
- 11.Lane D.J. 16S/23S rRNA sequencing in nucleic acid techniques. In: Stackebrandt E., Goodfellow M., editors. Nucleic Acid Techniques in Bacterial Systematics. John Wiley and Sons; New York, NY, USA: 1991. pp. 115–148. [Google Scholar]
- 12.Clinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. CLSI; Wayne, PA, USA: 2020. CLSI Supplement M100. [Google Scholar]
- 13.Exner M., Bhattacharya S., Christiansen B., Gebel J., Goroncy-Bermes P., Hartemann P., Heeg P., Ilschner C., Kramer A., Larson E., et al. Antibiotic resistance: What is so special about multidrug-resistant Gram-negative bacteria? GMS Hyg. Infect. Control. 2017;12:Doc05. doi: 10.3205/dgkh000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., Aarestrup F.M., Larsen M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zurfluh K., Albini S., Mattmann P., Kindle P., Nüesch-Inderbinen M., Stephan R., Vogler B.R. Antimicrobial resistant and extended-spectrum β-lactamase producing Escherichia coli in common wild bird species in Switzerland. Microbiol. Open. 2019;8:e845. doi: 10.1002/mbo3.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shobrak M.Y., Abo-Amer A.E. Role of wild birds as carriers of multi-drug resistant Escherichia coli and Escherichia vulneris. Braz. J. Microbiol. 2014;45:1199–1209. doi: 10.1590/S1517-83822014000400010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nkogwe C., Raletobana J., Stewart-Johnson A., Suepaul S., Adesiyun A. Frequency of Detection of Escherichia coli, Salmonella spp., and Campylobacter spp. in the Faeces of Wild Rats Rattus spp.) in Trinidad and Tobago. Vet. Med. Int. 2011;2011:686923. doi: 10.4061/2011/686923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Himsworth C.G., Zabek E., Desruisseau A., Parmley E.J., Reid-Smith R., Jardine C.M., Tang P., Patrick D.M. Prevalence and Characteristics of Escherichia Coli and Salmonella spp. in the Feces of Wild Urban Norway and Black Rats (Rattus Norvegicus and Rattus Rattus) from an Inner-City Neighborhood of Vancouver, Canada. J. Wildl. Dis. 2015;51:589–600. doi: 10.7589/2014-09-242. [DOI] [PubMed] [Google Scholar]
- 20.National Environment Agency What is the Difference Between a Rat and a Shrew? Should the Public Be Worried about the Presence of Shrews? [(accessed on 14 July 2020)];2016 Available online: https://www.nea.gov.sg/media/nea-vox/index/what-is-the-difference-between-a-rat-and-a-shrew-should-the-public-be-worried-about-the-presence-of-shrews.
- 21.Arnold K.E., Williams N.J., Bennett M. ‘Disperse abroad in the land’: The role of wildlife in the dissemination of antimicrobial resistance. Biol. Lett. 2016;12:20160137. doi: 10.1098/rsbl.2016.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aung K.T., Chen H.J., Chau M.L., Yap G., Lim X.F., Humaidi M., Chua C., Yeo G., Yap H.M., Oh J.Q., et al. Salmonella in Retail. Food and Wild Birds in Singapore-Prevalence, Antimicrobial Resistance, and Sequence Types. Int. J. Environ. Res. Public Health. 2019;16:4235. doi: 10.3390/ijerph16214235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varga C., Rajić A., McFall M.E., Reid-Smith R.J., Deckert A.E., Pearl D.L., Avery B.P., Checkley S.L., McEwen S.A. Comparison of antimicrobial resistance in generic Escherichia coli and Salmonella spp. cultured from identical fecal samples in finishing swine. Can. J. Vet. Res. 2008;72:181–187. [PMC free article] [PubMed] [Google Scholar]
- 24.Mathew A., Jackson F., Saxton A. Effects of antibiotic regimens on resistance of Escherichia coli and Salmonella serovar Typhimurium in swine. J. Swine Health Prod. 2002;10:7–13. [Google Scholar]
- 25.Chopra I., Roberts M. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 2001;65:232–260. doi: 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diren Sigirci B., Celik B., Başaran Kahraman B., Bagcigil A.F., Ak S. Tetracycline Resistance of Enterobacteriaceae Isolated from Feces of Synanthropic Birds. J. Exot. Pet Med. 2019;28:13–18. doi: 10.1053/j.jepm.2017.12.003. [DOI] [Google Scholar]
- 27.Nessim N. Antibiotics in 2019: Everything You Need to Know. [(accessed on 14 March 2020)]; Available online: https://www.solvhealth.com/blog/antibiotics-everything-you-need-to-know.
- 28.Pozzi S.P., Ben-David Z. Use of amoxicillin + clavulanic acid combination in Veterinary Medicine and possible antibiotic-resistance in human pathogens; A world-wide overview. Isr. J. Vet. Med. 2002;57:95. [Google Scholar]
- 29.Iredell J., Brown J., Tagg K. Antibiotic resistance in Enterobacteriaceae: Mechanisms and clinical implications. Bmj. 2016;352:h6420. doi: 10.1136/bmj.h6420. [DOI] [PubMed] [Google Scholar]
- 30.Zwe Y.H., Tang V.C.Y., Aung K.T., Gutiérrez R.A., Ng L.C., Yuk H.-G. Prevalence, sequence types, antibiotic resistance and, gyrA mutations of Salmonella isolated from retail fresh chicken meat in Singapore. Food Control. 2018;90:233–240. doi: 10.1016/j.foodcont.2018.03.004. [DOI] [Google Scholar]
- 31.Hornish R.E., Kotarski S.F. Cephalosporins in veterinary medicine—Ceftiofur use in food animals. Curr. Top. Med. Chem. 2002;2:717–731. doi: 10.2174/1568026023393679. [DOI] [PubMed] [Google Scholar]
- 32.Fritzenwanker M., Imirzalioglu C., Herold S., Wagenlehner F.M., Zimmer K.P., Chakraborty T. Treatment Options for Carbapenem- Resistant Gram-Negative Infections. Dtsch Arztebl Int. 2018;115:345–352. doi: 10.3238/arztebl.2018.0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chua K.Y.L., Stewardson A.J. Individual and community predictors of urinary ceftriaxone-resistant Escherichia coli isolates, Victoria, Australia. Antimicrob. Resist. Infect. Control. 2019;8:36. doi: 10.1186/s13756-019-0492-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng G., Ning J., Ahmed S., Huang J., Ullah R., An B., Hao H., Dai M., Huang L., Wang X., et al. Selection and dissemination of antimicrobial resistance in Agri-food production. Antimicrob. Resist. Infect. Control. 2019;8:158. doi: 10.1186/s13756-019-0623-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar R., Bibi E. A single membrane-embedded negative charge is critical for recognizing positively charged drugs by the Escherichia coli multidrug resistance protein MdfA. EMBO J. 1999;18:822–832. doi: 10.1093/emboj/18.4.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sutcliffe J.A., Leclercq R. Mechanisms of Resistance to Macrolides, Lincosamides, and Ketolides. In: Schönfeld W., Kirst H.A., editors. Macrolide Antibiotics. Birkhäuser Basel; Basel, Switzerland: 2002. pp. 281–317. [Google Scholar]
- 37.Edgar R., Bibi E. MdfA, an Escherichia coli multidrug resistance protein with an extraordinarily broad spectrum of drug recognition. J. Bacteriol. 1997;179:2274–2280. doi: 10.1128/JB.179.7.2274-2280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noguchi N., Takada K., Katayama J., Emura A., Sasatsu M. Regulation of transcription of the mph(A) gene for macrolide 2′-phosphotransferase I in Escherichia coli: Characterization of the regulatory gene mphR(A) J. Bacteriol. 2000;182:5052–5058. doi: 10.1128/JB.182.18.5052-5058.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Handrova L., Kmet V. ; Kmet, V. Antibiotic resistance and virulence factors of Escherichia coli from eagles and goshawks. J. Environ. Sci. Health Part B. 2019;54:605–614. doi: 10.1080/03601234.2019.1608103. [DOI] [PubMed] [Google Scholar]
- 40.Huy H., Koizumi N., Ung T., Le T., Nguyen H., Hoang P., Nguyen C., Khong T., Hasebe F., Haga T., et al. Antibiotic-resistant Escherichia coli isolated from urban rodents in Hanoi, Vietnam. J. Vet. Med. Sci. 2020;82:653–660. doi: 10.1292/jvms.19-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leski T.A., Bangura U., Jimmy D.H., Ansumana R., Lizewski S.E., Stenger D.A., Taitt C.R., Vora G.J. Multidrug-resistant tet(X)-containing hospital isolates in Sierra Leone. Int. J. Antimicrob. Agents. 2013;42:83–86. doi: 10.1016/j.ijantimicag.2013.04.014. [DOI] [PubMed] [Google Scholar]
- 42.Clark N.C., Olsvik O., Swenson J.M., Spiegel C.A., Tenover F.C. Detection of a streptomycin/spectinomycin adenylyltransferase gene (aadA) in Enterococcus faecalis. Antimicrob. Agents Chemother. 1999;43:157–160. doi: 10.1128/AAC.43.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marcelino V., Wille M., Hurt A., González-Acuña D., Klaassen M., Eden J.-S., Shi M., Iredell J., Sorrell T., Holmes E. Meta-transcriptomics reveals a diverse antibiotic resistance gene pool in avian microbiomes. BMC Biol. 2019;17:31:1–31:11. doi: 10.1186/s12915-019-0649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hassan R., Tantawy M., Gouda N.A., Elzayat M.G., Gabra S., Nabih A., Diab A.A., El-Hadidi M., Bakry U., Shoeb M.R., et al. Genotypic characterization of multiple drug resistant Escherichia coli isolates from a pediatric cancer hospital in Egypt. Sci. Rep. 2020;10:4165. doi: 10.1038/s41598-020-61159-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Hoek A.H.A.M., Mevius D., Guerra B., Mullany P., Roberts A.P., Aarts H.J.M. Acquired antibiotic resistance genes: An overview. Front. Microbiol. 2011;2:203. doi: 10.3389/fmicb.2011.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu J., Zhang J., Xu L., Liu Y., Li P., Zhu T., Cheng C., Lu S., Xu T., Yi H., et al. Spread of the florfenicol resistance floR gene among clinical Klebsiella pneumoniae isolates in China. Antimicrob. Resist. Infect. Control. 2018;7:127. doi: 10.1186/s13756-018-0415-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cloeckaert A., Baucheron S., Flaujac G., Schwarz S., Kehrenberg C., Martel J.-L., Chaslus-Dancla E. Plasmid-Mediated Florfenicol Resistance Encoded by the floR Gene in Escherichia coli Isolated from Cattle. Antimicrob. Agents Chemother. 2000;44:2858–2860. doi: 10.1128/AAC.44.10.2858-2860.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olawale O.A., Obasola E.F., Yvonne A. Antibiotic resistance and resistance genes in Escherichia coli from poultry farms, southwest Nigeria. J. Infect. Dev. Ctries. 2014;8:1103–1112. doi: 10.3855/jidc.4222. [DOI] [PubMed] [Google Scholar]
- 49.Aeksiri N., Toanan W., Sawikan S., Suwannarit R., Pungpomin P., Khieokhajonkhet A., Niumsup P.R. First Detection and Genomic Insight into mcr-1 Encoding Plasmid-Mediated Colistin-Resistance Gene in Escherichia coli ST101 Isolated from the Migratory Bird Species Hirundo rustica in Thailand. Microb. Drug Resist. 2019;25:1437–1442. doi: 10.1089/mdr.2019.0020. [DOI] [PubMed] [Google Scholar]
- 50.Chen Y., Shoichet B., Bonnet R. Structure, function, and inhibition along the reaction coordinate of CTX-M beta-lactamases. J. Am. Chem. Soc. 2005;127:5423–5434. doi: 10.1021/ja042850a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi H., Sun F., Chen J., Ou Q., Feng W., Yong X., Xia P. Epidemiology of CTX-M-type extended-spectrum beta-lactamase (ESBL)-producing nosocomial-Escherichia coli infection in China. Ann. Clin. Microbiol. Antimicrob. 2015;14:1–5. doi: 10.1186/s12941-015-0063-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park H., Kim J., Ryu S., Jeon B. Predominance of blaCTX-M-65 and blaCTX-M-55 in extended-spectrum β-lactamase-producing Escherichia coli from raw retail chicken in South Korea. J. Glob. Antimicrob. Resist. 2019;17:216–220. doi: 10.1016/j.jgar.2019.01.005. [DOI] [PubMed] [Google Scholar]
- 53.Jacoby G.A. Mechanisms of resistance to quinolones. Clin. Infect. Dis. 2005;41(Suppl. 2):S120–S126. doi: 10.1086/428052. [DOI] [PubMed] [Google Scholar]
- 54.Oh J.Y., Kwon Y.K., Tamang M.D., Jang H.K., Jeong O.M., Lee H.S., Kang M.S. Plasmid-Mediated Quinolone Resistance in Escherichia coli Isolates from Wild Birds and Chickens in South Korea. Microb. Drug Resist. 2016;22:69–79. doi: 10.1089/mdr.2015.0090. [DOI] [PubMed] [Google Scholar]
- 55.Literak I., Micudova M., Tausova D., Cizek A., Dolejska M., Papousek I., Prochazka J., Vojtech J., Borleis F., Guardone L., et al. Plasmid-mediated quinolone resistance genes in fecal bacteria from rooks commonly wintering throughout Europe. Microb. Drug Resist. 2012;18:567–573. doi: 10.1089/mdr.2012.0075. [DOI] [PubMed] [Google Scholar]
- 56.Kim H.B., Wang M., Park C.H., Kim E.-C., Jacoby G.A., Hooper D.C. oqxAB Encoding a Multidrug Efflux Pump in Human Clinical Isolates of Enterobacteriaceae. Antimicrob. Agents Chemother. 2009;53:3582–3584. doi: 10.1128/AAC.01574-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruiz J. Mechanisms of resistance to quinolones: Target alterations, decreased accumulation and DNA gyrase protection. J. Antimicrob. Chemother. 2003;51:1109–1117. doi: 10.1093/jac/dkg222. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Z., Meng X., Wang Y., Xia X., Wang X., Xi M., Meng J., Shi X., Wang D., Yang B. Presence of qnr, aac(6′)-Ib, qepA, oqxAB, and mutations in gyrase and topoisomerase in nalidixic acid-resistant Salmonella isolates recovered from retail chicken carcasses. Foodborne Pathog. Dis. 2014;11:698–705. doi: 10.1089/fpd.2014.1736. [DOI] [PubMed] [Google Scholar]
- 59.Gow S.P., Waldner C.L., Harel J., Boerlin P. Associations between Antimicrobial Resistance Genes in Fecal Generic Escherichia coli Isolates from Cow-Calf Herds in Western Canada. Appl. Environ. Microbiol. 2008;74:3658. doi: 10.1128/AEM.02505-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lou Y., Liu H., Zhang Z., Pan Y., Zhao Y. Mismatch between antimicrobial resistance phenotype and genotype of pathogenic Vibrio parahaemolyticus isolated from seafood. Food Control. 2016;59:207–211. doi: 10.1016/j.foodcont.2015.04.039. [DOI] [Google Scholar]
- 61.Fernández L., Hancock R.E.W. Adaptive and mutational resistance: Role of porins and efflux pumps in drug resistance. Clin. Microbiol. Rev. 2012;25:661–681. doi: 10.1128/CMR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kindle P., Zurfluh K., Nuesch-Inderbinen M., von Ah S., Sidler X., Stephan R., Kummerlen D. Phenotypic and genotypic characteristics of Escherichia coli with non-susceptibility to quinolones isolated from environmental samples on pig farms. Porcine Health Manag. 2019;5:9. doi: 10.1186/s40813-019-0116-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nguyen D.T., Kanki M., Nguyen P.D., Le H.T., Ngo P.T., Tran D.N., Le N.H., Dang C.V., Kawai T., Kawahara R., et al. Prevalence, antibiotic resistance, and extended-spectrum and AmpC beta-lactamase productivity of Salmonella isolates from raw meat and seafood samples in Ho Chi Minh City, Vietnam. Int. J. Food Microbiol. 2016;236:115–122. doi: 10.1016/j.ijfoodmicro.2016.07.017. [DOI] [PubMed] [Google Scholar]
- 64.Argudín M.A., Deplano A., Meghraoui A., Dodémont M., Heinrichs A., Denis O., Nonhoff C., Roisin S. Bacteria from Animals as a Pool of Antimicrobial Resistance Genes. Antibiotics. 2017;6:12. doi: 10.3390/antibiotics6020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dolejska M., Literak I. Wildlife is overlooked in the epidemiology of medically important antimicrobial resistant bacteria. Antimicrob. Agents Chemother. 2019;63:1167-19. doi: 10.1128/AAC.01167-19. [DOI] [PMC free article] [PubMed] [Google Scholar]