Abstract
Cell–cell communication via endogenous peptides and their receptors is vital for controlling all aspects of human physiology and most peptides signal through G protein-coupled receptors (GPCRs). Disordered peptides bind GPCRs through complex modes for which there are few representative crystal structures. The disordered peptide neurotensin (NT) is a neuromodulator of classical neurotransmitters such as dopamine and glutamate, through activation of neurotensin receptor 1 (NTS1). While several experimental structures show how NT binds NTS1, details about the structural dynamics of NT during and after binding NTS1, or the role of peptide dynamics on receptor activation, remain obscure. Here saturation transfer difference (STD) NMR revealed that the binding mode of NT fragment NT10-13 is heterogeneous. Epitope maps of NT10-13 at NTS1 suggested that tyrosine 11 (Y11) samples other conformations to those observed in crystal structures of NT-bound NTS1. Molecular dynamics (MD) simulations confirmed that when NT is bound to NTS1, residue Y11 can exist in two χ1 rotameric states, gauche plus (g+) or gauche minus (g–). Since only the g+ Y11 state is observed in all the structures solved to date, we asked if the g– state is important for receptor activation. NT analogues with Y11 replaced with 7-OH-Tic were synthesized to restrain the dynamics of the side chain. P(OH-TIC)IL bound NTS1 with the same affinity as NT10-13 but did not activate NTS1, instead acted as an antagonist. This study highlights that flexibility of Y11 in NT may be required for NT activation of NTS1.
Keywords: neurotensin, neurotensin receptor 1 (NTS1), NMR, peptide dynamics
Introduction
Neurotensin (NT) is a 13-residue peptide that is expressed in the central nervous, gastro-intestinal, and cardiovascular systems. NT is a neuromodulator of classical neurotransmitters such as dopamine and glutamate, primarily through activation of its major receptor, neurotensin receptor 1 (NTS1).1 NT and its receptors are implicated in nociception2 and inflammation,3 and in a variety of diseases and disorders including schizophrenia,4 substance abuse,5 cancer,6 Parkinson’s disease,1 eating disorders,7 and high blood pressure.8
Eight crystal structures of thermostabilized variants of rat neurotensin receptor 1 (rNTS1) have been solved in complex with the strong affinity 8-13 fragment of NT (NT8-13).9−12 Using Cryo-EM, a structure of a hNTS1–Arrestin 2 complex13 and two structures of a hNTS1–Gαi1β1γ2 complex14 were solved in the presence of NT8-13 and the synthetic peptide JMV449(KKPYIL), respectively. In all NTS1 structures, the agonist peptide adopts an extended conformation, with the C-terminal four residues (PYIL) buried deeply in the orthosteric site of the receptor. This conformation agrees with solid-state NMR experiments showing that the NT8-13 backbone torsional angles are consistent with a β-strand arrangement when in complex with NTS1.15 NMR studies of peptide-GPCR interactions indicate the retention of peptide flexibility after receptor binding.16,17 Wuthrich and co-workers16 showed that the N- and C-termini of dynorphin, a linear disordered neuropeptide, remain highly mobile even when bound to its cognate GPCR, the kappa opioid receptor. Furthermore, a recent molecular dynamics (MD) study of NT bound to NTS1 revealed a coordination between conformational changes in NT and the activation state of the receptor.18 Thus, the question remains: how ordered is neurotensin in the assumed orthosteric binding pose at NTS1?
Saturation transfer difference (STD)-NMR is a ligand-based detection technique that does not require isotopic labeling of the ligand or receptor. STD-NMR is widely used to probe weak-affinity ligand binding to soluble proteins,19 and has been used to probe the binding of small molecules to a number of proteins including several GPCR complexes.20−25 STD-NMR experiments require relatively low concentrations of protein compared to protein-detected NMR experiments, and a 20- to 100-fold molar excess of ligand over protein. As the intensity loss of the STD-NMR signal has an r–6 distance dependence, the experiment allows the determination of ligand binding modes via group epitope mapping (GEM),26 and can therefore be employed to interpret the proximities of ligand protons to the receptor protein. STD-NMR-based GEM is well established for mapping binding epitopes of small molecules, including peptides,27−34 to protein targets.
In this work, we combined STD-NMR-based GEM and molecular dynamics (MD) to probe the solution binding mode of a NT peptide (NT10-13) to a thermostabilized NTS1 variant (enNTS1).35 Together, the NMR and MD data revealed that the NT peptide adopts multiple conformations in the bound state. Tyrosine 11 (Y11) exists in two conformations, namely χ1 gauche plus (g+) and χ1 gauche minus (g–). Of these two conformations, only g+ has been observed in crystal and Cyro-EM structures, although a recent molecular dynamics study suggested that conformational changes of Y11 in NT may be important for NTS1 inactive–active state transitions.18 To probe this hypothesis, we designed a conformationally restrained synthetic peptide based on the NT10-13 sequence (PYIL) in which Y11 of NT10-13 was replaced with 7-hydroxy-3,4-dihydro-1h-isoquinoline-3 (7-OH-TIC). P(OH-TIC)IL bound rNTS1 with the same affinity as NT10-13 but unlike NT10-13 was unable to activate rNTS1-induced cell signaling. These results suggest that NT samples multiple conformations when bound to NTS1 and that the g– conformation of Y11 may be important for triggering inactive–active state transitions in NTS1 and hence cell signaling. Knowledge of such molecular complexity will be vital for developing novel NTS1 modulators.
Results and Discussion
Detection of NT-NTS1 Binding Heterogeneity with STD-NMR
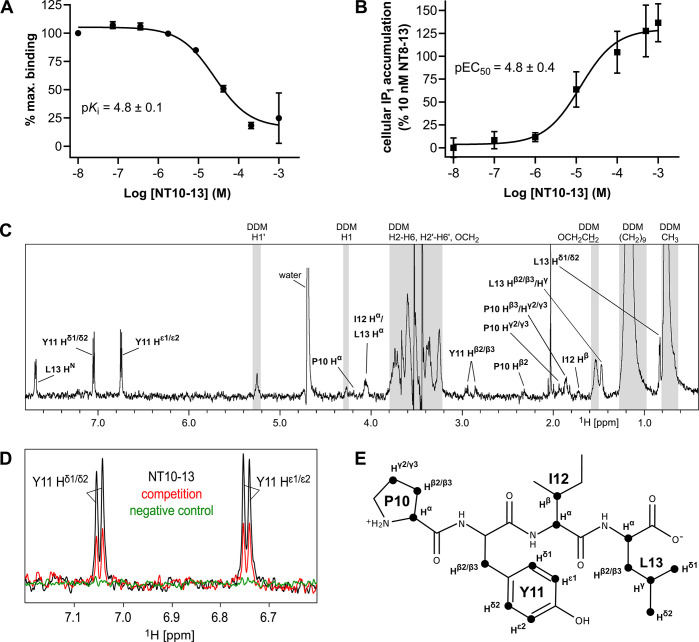
Compared to most other peptide–GPCRs pairs, the structural details of how NT8-13 interacts with NTS1 in its final bound state is well characterized. While NT8-13 is disordered in solution36,37 it adopts a β-strand conformation when bound to NTS1.9−11,15 To understand how NT transitions from disordered to ordered upon binding NTS1, and if any intermediate states exist, we aimed to map the average binding epitope of a neurotensin derivative to NTS1 using STD-NMR. To detect the interaction of neurotensin peptide derivatives with NTS1 using STD NMR, a sample of purified, correctly folded NTS1 protein is required. We made use of the thermostabilized NTS1 variant enNTS1, which can be purified with relative ease, retains ligand binding properties of wild-type NTS1 in solution, remains stable for many hours in detergent, and when expressed in human cells is signaling competent.35 Rapid transfer of magnetization from protein-bound ligand molecules to the free ligand population in solution is required for binding detection by STD-NMR. Ligands with slow off-rates, such as NT (sequence ZLYENKPRRPYIL, where Z is pyro-glutamate38) and the C-terminal hexapeptide NT8-13, which bind enNTS1 with subnanomolar affinities35 are not suitable.19 The C-terminal tetrapeptide NT10-13, which contains the key activating residues (PYIL), binds NTS1 with weaker affinity (reported Ki > 4 μM).39 Despite this, NT10-13 is a full agonist at NTS1, albeit with weaker potency (EC50 > 5 μM) compared to NT8-13 (EC50 = 25 nM).39 Using fluorescence-based saturation and inhibition binding assays, we determined the KD of NT8-13 and the Ki of NT10-13 at the MBP-enNTS1-muGFP construct to be 0.83 nM35 and 15.8 μM (pKi = 4.8 ± 0.1), respectively (Figure 1A). Using an inositol monophosphate (IP1) assay, NT10-13 activated Gαq/11 signaling at wild-type rNTS1 (Figure 1B) and enNTS1 stably expressing human embryonic kidney (HEK-293T) cells with an EC50 of 4.2 μM (pEC50 = 4.8 ± 0.4) compared to an EC50 of 3 nM for NT8-13 determined at enNTS1 in parallel (Figure S1). The binding data suggested that NT10-13 would be suitable for STD-NMR experiments.
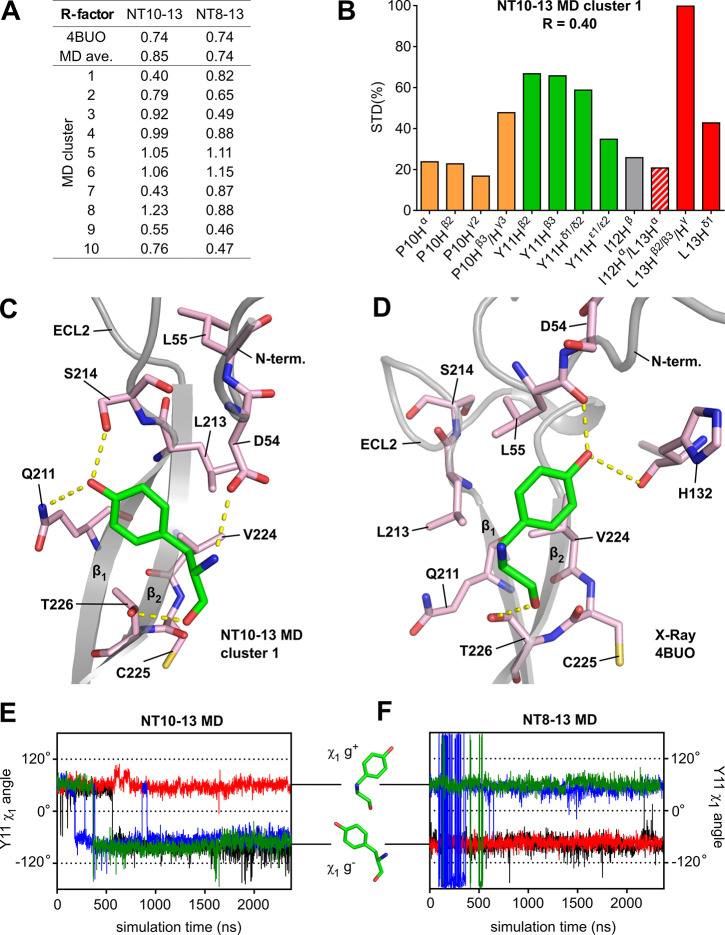
Figure 1.
The weak affinity peptide NT10-13 used for STD-NMR. (A) Inhibition of TAMRA-NT8-13 binding to MBP-enNTS1-muGFP using varying concentrations of NT10-13. Values reported represent the mean and standard deviation (SD) of three independent experiments measured in triplicate (n = 3) of the TAMRA fluorescence to the muGFP ratio calculated for each measurement, plotted against NT10-13 concentration. The −log dissociation constant (pKi) of NT10-13 is reported with the standard error (SE) calculated from separate measurements. (B) IP1 dose response curve of NT10-13 at HEK-293T cells expressing wild-type rNTS1. Values reported represent the mean and standard error of the mean (SE) of two independent experiments measured in triplicate (n = 2). (C) STD-NMR spectrum of NT10-13 interacting with cleaved enNTS1 showing visible peptide hydrogen resonances. Hydrogens overlapping with DDM signals (gray areas) were omitted for clarity. The DDM signals correspond to H1′ (5.28 ppm), H1 (4.30 ppm), the maltoside moiety (H2–H6 and H2′–H6′) and OCH2 of the n-dodecyl group (3.94–3.15 ppm), OCH2CH2 (1.50 ppm), (CH2)9 (1.19 ppm), and CH3 (0.78 ppm), which were assigned based on previously published data.70 (D) Close-up of the NT10-13 Y11 Hδ1/δ2 and Hε1/ε2 hydrogen region of the STD-NMR experiment containing 5 μM cleaved enNTS1 and 500 μM NT10-13 (black) overlaid with a competition STD-NMR experiment containing five μM cleaved enNTS1, 500 μM NT10-13, and 20 μM NT8-13 (red), and a control STD-NMR experiment containing 5 μM α1A-AR and 500 μM NT10-13 (green). (E) Skeletal formula of NT10-13 with hydrogen atoms used for epitope mapping marked with black discs.
The superior stability of enNTS1 in n-dodecyl-β-d-maltopyranoside (DDM) micelles allowed us to acquire a series of STD-NMR spectra on single samples for more than 24 h without significant loss of signal. The STD-NMR spectra for NT10-13 binding to cleaved enNTS1 are shown in Figure 1C and Figure S2B. Observable NT10-13 protons (Figure 1E) were assigned via a 2D 1H–1H TOCSY experiment (Figure S3; and Table S1) and most 1D 1H counterparts could be identified in 1H reference and STD-NMR spectra. In the presence of 20 μM NT8-13 (>99.9% binding saturation), the signal intensity of NT10-13 was decreased by up to 70%, as illustrated for the Y11 Hδ and Hε STD signals (Figure 1D), which is indicative of competitive behavior. The nearly absent signals for NT10-13 in control experiments using a thermostabilized α1A–adrenergic receptor (α1A-AR)21 (Figure S2D) suggested that the STD-NMR signals observed for enNTS1 were specific.
To epitope map the binding of NT10-13 to enNTS1, STD build-up curves of NT10-13 binding enNTS1 were obtained by acquiring STD-NMR spectra at nine different saturation times (0.5 to 5 s) (Figure S4A–C). For each of these spectra, the peak intensities were transformed to obtain STD amplification factors (STDAF) using eq 1 and the resultant curves were fitted using eq 2. The saturation that ligand protons receive when bound to protein is affected by their longitudinal relaxation times (T1) in the free state, which can lead to misinterpretations of large STD effects at long saturation times.19 Thus, the initial rate of magnetization transfer, STDFIT, was determined from each build-up curve (Figure S4D and Table S2). We subsequently compared the STDFIT method to the Group Epitope Mapping Considering Relaxation of the Ligand (GEM-CRL) method,40 whereby STD enhancements obtained using long saturation times are corrected using experimentally derived T1 relaxation times. GEM-CRL was performed using a saturation time of 3 s with T1 correction using eq 4. Since the STDFIT and STDGEM-CRL values were generally in good agreement (Figure S4D), the less time-intensive GEM-CRL method was used for competition STD experiments. Saturation transfer competition difference (STCD) spectra were obtained by subtracting STD-NMR spectra from receptor samples containing NT10-13 and 20 μM NT8-13 competitor from STD-NMR spectra of NT10-13, thus removing noncompetable signals (Figure S5). By applying GEM-CRL to the competition STD-NMR (STCD) approach, we could differentiate the signals derived from NT10-13 binding to the orthosteric binding site of enNTS1 from signals derived from interactions with the DDM surfactant and secondary, nonorthosteric interactions of the peptide with the receptor (Figure 2A). Table S4 lists the remaining signal intensities for each proton after subtraction of the competition experiment, suggesting that all ligand protons received significant magnetization outside of the orthosteric binding site. The signal for P10 Hα was completely abolished while I12 Hβ, I12 Hα/L13 Hα, and L13 Hδ1 were reduced to 33%, 35%, and 41% of the original signal indicating that particularly those protons are involved in nonspecific peptide-detergent and/or peptide-secondary receptor interactions.
Figure 2.
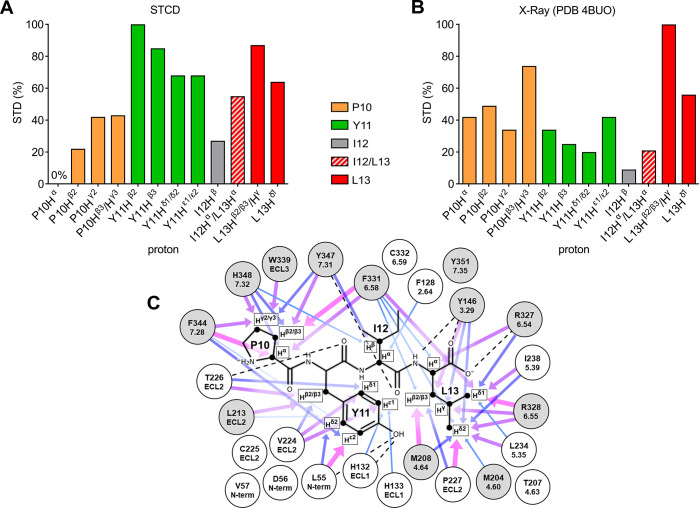
Comparison of experimental and crystal structure-derived epitope maps. (A) Experimental epitope map based on the STCD experiment. The corresponding values are P10 Hα, 0%; P10 Hβ2, 22%; P10 Hγ2, 42%; P10 Hβ2/γ3, 43%; Y11 Hβ2, 100%; Y11 Hβ3, 85%; Y11 Hδ1/δ2, 68%; Y11 Hε1/ε2, 68%; I12 Hβ, 27%; I12 Hα/L13 Hα, 55%; L13 Hβ2/β3/Hγ, 87%; and L13 Hδ1, 64%. (B) Epitope map generated using theoretical STD enhancements calculated based on a NTS1 crystal structure (PDB 4BUO, chain A). The corresponding values are P10 Hα, 42%; P10 Hβ2, 49%; P10 Hγ2, 34%; P10 Hβ3/γ3, 74%; Y11 Hβ2, 34%; Y11 Hβ3, 25%; Y11 Hδ1/δ2, 20%; Y11 Hε1/ε2, 42%; I12 Hβ, 9%; I12 Hα/L13 Hα, 21%; L13 Hβ2/β3/Hγ, 100%; and L13 Hδ1, 56%. (C) Schematic representation of residues (sequential and Ballesteros-Weinstein numbering) within 5 Å of protons used to generate the theoretical 4BUO (chain A) epitope map (Figure 2B). Arrows represent individual contributions to STD enhancements as a function of interproton distances ranging from 2.27–3 Å (thick arrows, magenta) to 4–4.5 Å (thin arrows, lightblue). Dotted lines represent hydrogen bonds. Gray circles represent residues resulting in loss of NT8-13 binding affinity when mutated to other residue types.41−43 Data sets for epitope mapping (panels A and B) were normalized to the largest value of each set.
As expected, key differences were observed between the NT10-13 STCD epitope map and an epitope map calculated from the crystal structure of NT8-13 bound to NTS1 (PDB 4BUO, Figure 2B,C). Peptide binding to GPCRs is a highly dynamic process16,44 and the contributions of multiple binding modes at the orthosteric binding site will be averaged in the STCD epitope map, whereas the crystal structure represents a single possible binding mode. The most obvious difference between the STCD epitope map and the crystal structure was the increased apparent binding intimacy of Y11 in the NMR approach. Y11 of NT is known to be important for binding NTS1, with substitution to other amino acids reducing binding affinity.45,46 Thus, to gain insight into the dynamics of the system, molecular dynamics (MD) simulations of enNTS1 bound to both NT10-13 and NT8-13 were conducted.
MD Simulations of NT10-13 and NT8-13 Binding
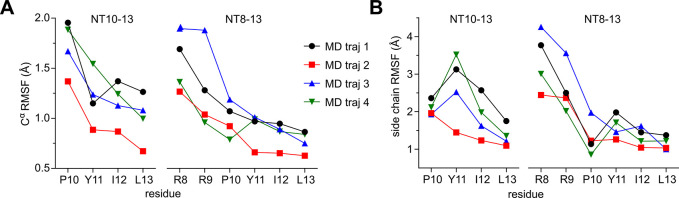
MD simulations were employed to assess if the dynamics of NT10-13 and NT8-13 while bound to enNTS1 would explain the increased Y11 binding observed with STD NMR. We built homology models of enNTS1 (enNTS1model) bound to NT10-13 and NT8-13 and ran four independent simulations of each complex. Each simulation had a duration of more than 2.3 μs. In the simulations, the protein structure deviated little from the starting conformation; over all simulations, the protein transmembrane Cα atoms had average RMSD values of 1.4 Å with respect to the starting structures. (Table S5). Analysis of the bound peptides showed that the C-termini are tightly bound and that the N-termini are relatively more flexible (see Figure 3A,B). The NT10-13 peptide showed larger fluctuations than NT8-13.
Figure 3.
Average RMSF per MD trajectory and residue. (A) Cα RMSF plots of NT10-13 and NT8-13 peptides bound to enNTS1model from MD trajectories. Peptide residues and Cα RMSF (Å) are shown on the x and y axis, respectively. (B) Side-chain RMSF plots of NT10-13 and NT8-13 peptides from MD trajectories. Peptide residues and side chain RMSF (Å) are shown on the x and y axis, respectively.
Each set of MD trajectories was clustered into 10 groups and representative structures were chosen from the center of each cluster (Table S6). Superimpositions of the 10 cluster representatives demonstrate the variability of ECL2 observed in the RMSF plots (Figures S6A,B). The bound ligand conformations closely align with the crystal structure conformation (Figures S6C,D), although the NT10-13 conformations are slightly more heterogeneous than the conformations observed for the same residues in the NT8-13 cluster frames. The backbone Cα atoms of the peptides were aligned (Figures S6C,D and Figure S7), and the torsion angles were compared between cluster frames (Supplementary Tables 7 and 8). Generally the backbone conformation of both NT8-13 and NT10-13 largely retained the extended conformation of the crystal structure. The Y11 side chain, however, adopted two distinct conformations in both complexes characterized by a gauche plus (g+) or gauche minus (g–) χ1 torsion angle. The presence of two distinct Y11 side chain conformations in simulations of both NT peptides suggests that the Y11 dynamics are not due to NT10-13 being a shorter fragment than NT8-13. Side chain RMSF plots indicate that the Y11 side chain is the most mobile of the C-terminal four residues in both peptides (Figure 3B). The Y11 side chain in the crystal structures of NTS1-bound NT8-13 has a χ1 g+ conformation, predominantly stabilized by hydrogen bonds between the phenolic OH of Y11 and the peptide carbonyls of L55 of the receptor N-terminus and H132 in ECL1 (Table S9). However, in the MD simulations the Y11 side chain also occupies a χ1 g– conformation stabilized by hydrogen bonds of the phenolic OH of Y11 to the Q211 Nε2 and S214 Oγ of β-strand 1 in ECL2 (Table S10). Generally, we observe a looser fit for NT10-13 in the orthosteric binding pocket compared to NT8-13, where the positively charged arginine side chains complement the net-negative charge of the receptor extracellular vestibule.
STD Enhancement Analysis
Theoretical STD enhancements (STDMD; eq 6) for the ligand protons were calculated using the cluster representatives from the MD simulations of NT10-13 and NT8-13. The STDMD values were weighted by cluster population to obtain average STDMD enhancements for the protons observed in our STD experiments (Figures S6E,F). The averaged STDMD enhancements for both NT10-13 and NT8-13 were reduced for all P10 protons as well as the two Y11 Hβ protons but are otherwise in good agreement with the theoretical enhancements determined for the crystal structure as indicated by the agreement factor R (using eq 7) of 0.38 and 0.23, respectively (Table S11). The agreement factor is a straightforward method to evaluate the correspondence between observed and calculated values where a lower R-factor indicates better and R = 0 perfect agreement.
The calculated STD enhancements of NT10-13 based on the X-ray structure of NT8-13 poorly matched the experimental STCD enhancements (R = 0.74) as did the averaged STDMD enhancements for both NT10-13 (R = 0.85) and NT8-13 (R = 0.74). We thus extended the fitting procedure to the individual cluster representatives (Figure 4A) and identified NT10-13 cluster 1 to correlate better with the STCD experiments (R = 0.40) than other cluster representatives (Figures S8, S9 and Table S11). NT10-13 cluster 1 includes 1225 out of 9479 frames (13%) of the combined NT10-13 trajectories. The most noticeable difference between NT10-13 MD cluster representative 1 and the other cluster representatives is the greater enhancement of all Y11 protons (Hβ2/β3, 67%/66%; Hδ1/δ2, 59%; and Hε1/ε2, 35%) (Figure S8) that best approaches the high enhancements of the Y11 protons observed in the STCD epitope map (Hβ2, 100%; Hβ3, 85%; Hδ1/δ2, 68%; and Hε1/ε2, 68%) shown in Figure 2A. Our fitting procedure identified another cluster, NT10-13 cluster 7, to correlate almost as well with the STCD experiment (R = 0.43) when compared to NT10-13 cluster 1. NT10-13 cluster 7 is less populated (568 frames) and the relative enhancements of the Y11 protons (Hβ2/β3, 55%/54%; and Hδ1/δ2, 48%) except Y11 Hε1/ε2 (36%) are slightly reduced (Figure S8). Because of the absence of stereospecific assignments for P10 Hγ2/γ3, Y11 Hβ2/β3, and L13 Hδ1/δ2, we also calculated the R-factor ratios for possible alternative assignments of those protons (Table S12). NT10-13 cluster 1 remained the best ranking cluster for any of the alternative assignment combinations with mostly improved R-factor ratios. Swapping of P10 Hγ2 with P10 Hγ3 and L13 Hδ1 with L13 Hδ2 resulted in R-factor ratios of 0.38 and 0.37, respectively, while interchanging both proton pairs concomitantly yielded an R-factor ratio of 0.35.
Figure 4.
Alternative Y11 χ1 angle rotamer supported by epitope mapping. (A) R-factor ratios comparing epitope maps based on theoretical STD enhancements and the experimental STCDAF-CRL epitope map. Theoretical epitope maps were generated based on the NTS1 crystal structure (PDB 4BUO); based on the averaged STD enhancements over four MD trajectories (MDav) of NT10-13 and NT8-13 bound to enNTS1model; and based on the individual cluster representatives (clusters 1–10) extracted from the NT10-13 and NT8-13 bound MD trajectories. (B) Epitope map based on STD enhancements of NT10-13 MD cluster representative 1 normalized to the maximum value. This cluster showed the lowest R-factor (R = 0.40) among the theoretical epitope maps generated. The corresponding values are P10 Hα, 22%; P10 Hβ2, 23%; P10 Hγ2, 17%; P10 Hβ2/γ3, 48%; Y11 Hβ2, 66%; Y11 Hβ3, 65%; Y11 Hδ1/δ2, 56%; Y11 Hε1/ε2, 40%; I12 Hβ, 26%; I12 Hα/L13 Hα, 21%; L13 Hβ2/β3/Hγ, 100%; and L13 Hδ1, 48%. (C) Close-up of the ECL2 region of NT10-13 MD cluster representative 1 showing that the Y11 side chain adopts a χ1 g– conformation. (D) Close-up of the ECL2 region of 4BUO showing the Y11 side chain in a χ1 g+ conformation. Close-ups have been reduced to residues interacting with Y11 for clarity (two-dimensional ligand maps showing all receptor residues interacting with NT10-13 given in Figure S12). (E and F) Rotamer χ1 angle of Y11 in four independent MD simulations of NT10-13 and NT8-13, respectively, bound to enNTS1model.
The conformation of NT10-13 in cluster 1 is consistent with the conformation of NT residues 10–13 in the crystal structure, except for the I12 and Y11 side chain conformations (Figure S10). As the resonances of most I12 protons are poorly resolved, we are unable to comment on possible alternative conformations of the I12 side chain compared to the crystal structure. Analysis of the Y11 side chain conformation revealed that it adopts a g– χ1 rotamer in NT10-13 MD cluster representative 1 (Figure 4C), stabilized by hydrogen bonds of the phenolic OH of Y11 to the Q211 Nε2 and S214 Oγ, which are part of β-strand 1 in ECL2. The peptide NH of Y11 forms a hydrogen bond to the D54 Oγ1 in the receptor N-terminus. In the crystal structure, the phenolic OH of Y11 forms hydrogen bonds with the peptide carbonyls of L55 of the receptor N-terminus and H132 in ECL1, which requires the Y11 χ1 angle to adopt a g+ rotamer (Figure 4D). Both conformations share a hydrogen bond between the peptide carbonyl of Y11 and T266 OHγ1 located in β-strand 2 which acts as an anchor point of Y11 in this highly structured part of ECL2. Analysis of the Y11 χ1 angles over the whole trajectories (Figures 4E,F) revealed that both the g+ and g– conformations tend to be stable throughout the simulations. For the NT8-13 bound enNTS1model MD simulations the final Y11 χ1 angle was maintained in the production runs: that is a χ1 g– conformation in trajectories 1 and 2, and a χ1 g+ conformation in trajectories 3 and 4. NT8-13 trajectory 3, however, has fluctuations at the start of the simulation (between 87 and 360 ns) whereby the Y11 side chain does not toggle to the other side of ECL2 but is pushed into the pocket toward ECL1 to form transient interactions with H133. The NT10-13 bound enNTS1model MD simulations all started with a Y11 χ1 g+ conformation. This conformation was maintained throughout trajectory 2 but switched to a g– conformation in the other three trajectories (after 184 ns in trajectory 3, 365 ns in trajectory 4, and 564 ns in trajectory 1). Once flipped, the χ1 angle remained g–, with only one transition from g– to g+ and back again observed within 50 ns (from 863 to 913 ns in trajectory 3).
In summary, in some of the MD simulations, Y11 was observed to be stable in the χ1 g+ conformation, whereas in others Y11 was stable in the χ1 g– conformations. Both Y11 conformations are consistent with previously reported mutagenesis and binding studies of NTS1.45−47 Substitution of Y11 with Phe, Trp, and the bicyclic aromatic amino acid 3-(1-naphthyl)-l-alanine (Nal) reduces binding affinity 5, 2, and 10-fold, respectively.45,46 Phe, Trp, and Nal retain the aromatic function at position 11 and the ability to activate NTS1 but remove the capability of the Tyr OH group to engage in hydrogen bonds. Taken together, our NMR and MD findings suggest that hydrogen bonding of the Y11 OH is more diverse than suggested by crystal structures.
Design and Characterization of Synthetic, Conformationally Restrained NT Analogues
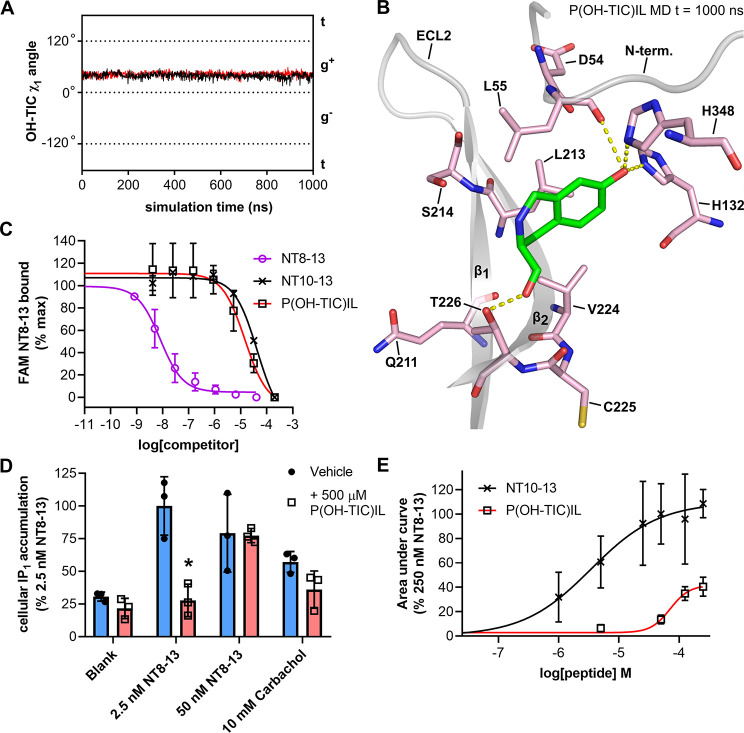
A recent molecular dynamics study suggested that conformational changes of Y11 in NT may be important for NTS1 inactive–active state transitions.18 Thus, to explore the relevance of Y11 dynamics to NTS1 activation we sought a peptide analogue where, upon binding, the Y11 side chain of neurotensin would be restrained to some degree. Hydroxy-3,4-dihydro-1h-isoquinoline-3-carboxylic acid (TIC) is an unnatural amino acid that has been used as a conformationally restrained phenylalanine and tyrosine replacement in peptides and thus was explored as a candidate for replacing Y11 in NT10-13. Several GPCR-acting peptides containing TIC substitutions have been synthesized to probe structure–activity relationships,48−51 including neurotensin.52,53 TIC-containing peptides have also been used to validate the biological relevance of tyrosine χ1 angles seen in crystal structures.54 Compared to phenylalanine, TIC has an additional six-membered ring formed from a bridging methylene group between the ortho carbon of the benzene ring and the peptide nitrogen (Figure S11A). This bridge restrains TIC so it can only sample χ1 g– and g+, with the trans conformation being excluded. We synthesized an NT10-13 analogue in which Y11 was replaced with 7-hydroxy-3,4-dihydro-1h-isoquinoline-3-carboxylic acid, termed P(OH-TIC)IL. Docking and MD simulations revealed that P(OH-TIC)IL can still bind enNTS1model, but with the χ1 angle remaining stable in g+ (Figure 5A,B), possibly due to the additional bulk of 7-OH-TIC sterically excluding the χ1 g– NTS1-bound conformation. In the laboratory P(OH-TIC)IL bound purified MBP-enNTS1-muGFP (Figure S11B) and wild-type (WT) rNTS1 expressed in HEK 293-T cells (Figure 5C), with comparable affinity to NT10-13. In a WT rNTS1 signaling assay, however, 500 μM P(OH-TIC)IL was unable to stimulate IP1 accumulation (Figure 5D), whereas NT10-13 is a full agonist in IP1 assays (Figure 1B). In fact, when WT rNTS1 expressing cells were treated with both 500 μM P(OH-TIC)IL and 2.5 nM NT8-13, P(OH-TIC)IL blocked the NT8-13 IP1 response (Figure 5D). Higher concentrations of NT8-13 could overcome the antagonism of WT rNTS1 by P(OH-TIC)IL (Figure 5D). P(OH-TIC)IL had no significant effect on IP1 accumulation stimulated by endogenously expressed muscarinic acetylcholine receptors with carbachol (Figure 5D), indicating that the inhibition of IP1 accumulation was a WT rNTS1-specific effect.
Figure 5.
Conformationally restrained P(OH-TIC)IL peptide. (A) Rotamer χ1 angle of Y11 in MD simulations of P(OH-TIC)IL (black trace) and RRP(OH-TIC)IL (red trace) bound to enNTS1model. (B) Snapshot taken at 1000 ns from the MD trajectory of P(OH-TIL)IL bound to enNTS1model showing OH-TIC. Other ligand residues and enNTS1 residues not interacting with OH-TIC or Y11 in the χ1 g+ conformation (Figure 4D) were omitted for clarity. (C) FAM-NT8-13 competition binding to HEK 293-T cells expressing WT rNTS1. Four nM FAM-NT binding was competed with increasing concentrations of NT8-13 (purple open circles), NT10-13 (black crosses), and P(OH-TIC)IL (black open squares). Values reported represent the mean and standard deviation (SD) from three independent experiments (n = 3). (D) IP1 accumulation assay on HEK-293T cells expressing WT rNTS1. Cells were treated with either vehicle (blank), 2.5 nM NT8-13, 50 nM NT8-13, or 10 mM Carbachol in the absence (blue bars and solid black circles) or presence (red bars and open black squares) of 500 μM P(OH-TIC)IL. Values reported represent the mean and standard deviation (SD) of three independent experiments. (E) xCELLigence dose response curves for NT10-13 (black crosses and black curve) and P(OH-TIC)IL (black open squares and red curve) at HEK 293T cells transfected with WT hNTS1. xCELLigence assays measure the impedance changes (cell index or CI) that cells impart over time on gold electrode-embedded microplates when treated with ligands. Area under the curve values were extracted from xCELLigence CI traces such as those in Figure S14 and normalized to the area under the curve given by 250 nM NT8-13 (100%). Values reported represent the mean and standard deviation (SD) of three independent experiments (n = 3).
We further characterized the pharmacology of P(OH-TIC)IL on WT human NTS1 (WT hNTS1) expressing cells due to the availability of more direct receptor activation assays. Activation of NTS1 leads to the recruitment of β-arrestin 1 and β-arrestin 2, which is important for receptor desensitization.55 Using a bioluminescent resonance energy transfer (BRET) assay, NT8-13 and NT10-13 were both able to stimulate the recruitment of human β-arrestin 2 to WT hNTS1-nanoluciferase (NanoLuc) fusion protein expressed in HEK 293T cells (Figure S12A,B). P(OH-TIC)IL, on the other hand, failed to significantly stimulate β-arrestin 2 recruitment at two concentrations (Figure S12A–D). When cells were pretreated with 250 μM P(OH-TIC)IL for 30 min, the NT8-13-induced β-arrestin 2 recruitment to WT hNTS1–NanoLuc was significantly reduced (Figure S13C,D), consistent with P(OH-TIC)IL being a weak antagonist.
A hNTS1-induced G protein activation assay was used to further explore the action of P(OH-TIC)IL. Using this BRET assay, NT8-13 robustly stimulates, in a dose dependent manner, the dissociation of a Gβγ-Venus fusion from Gα, allowing it to associate with a membrane anchored GRK3-NanoLuc fusion protein, which is detected through BRET (Figure S13A,B). P(OH-TIC)IL at 500 μM was unable to significantly inhibit hNTS1 induced Gβγ release by 1 nM NT8-13 (Figure S13C,D). There was a trend for 500 μM P(OH-TIC)IL to stimulate Gβγ release, although this was not significant (Figure S13C–D).
We have previously shown that the impedance-based xCELLigence system provides a highly sensitive assay for NTS1 cellular activation35 and thus was suited to determine if P(OH-TIC)IL was a low efficacy agonist. As expected, treatment of HEK 293-T cells transfected with WT hNTS1 with saturating concentrations of NT8-13, induced robust xCELLigence responses (Figure S14A and B). High concentrations (50, 100, and 250 μM) of NT10-13 induced similar xCELLigence responses to NT8-13, validating that NT10-13 is a full agonist at NTS1 (Figure S14A and Figure 5E). P(OH-TIC)IL treatment at the same concentrations (50, 100, and 250 μM), did induce significant xCELLigence responses through WT hNTS1 stimulation (Figure S14B and Figure 5E); however, the maximum response induced by P(OH-TIC)IL stimulation was significantly reduced at each concentration compared the NT10-13 and NT8-13. While these data suggest that P(OH-TIC)IL is a weak partial agonist at hNTS1, the lack of efficacy in the other signaling assays above makes unambiguous classification of P(OH-TIC)IL as a partial agonist impossible.
Overall these data suggest that the conformationally restricted OH-TIC residue prevents P(OH-TIC)IL from fully activating NTS1, even though it binds with the same affinity as the full agonist NT10-13. This, in turn, suggests that transitions between the g+ and g– χ1 conformations of Y11 may be important for NT-stimulated transition of NTS1 into active states after initial NT binding through the crystallographic pose. This is consistent with a recent MD study of NT and NTS1, in which NT Y11 χ1 oscillations between g+ and g– were observed and postulated to be a means by which receptor activation is triggered.18 The authors observed that the NT Y11 χ1 angle correlated to the side chain flipping of W3216.48 in TM6 of the receptor, even though W3216.48 is distant from the peptide binding site and is in fact part of the CWxP motif (rotamer toggle switch) known to transmit agonist binding-induced changes in the orthosteric site to the microswitches that control receptor conformational states.18 Thus, by disallowing NT Y11 χ1 oscillation with P(OH-TIC)IL, this conformational transition in W3216.48 may not be triggered. In addition to all inactive-state NTS1 crystal structures,10 active-like NTS1 crystal structures9,11,12 and active-state NTS1 effector protein complexes14,56 also show Y11 of the agonist peptide in the g+ conformation. Our data suggests that intermediate states with Y11 of the agonist peptide in the g– conformation may be the missing link to fully understand NTS1 activation. It will be interesting to see if Y11 populates the g– conformation when an NT-bound NTS1 active state (Gq-complex) structure is solved in the future. Overall, this study has demonstrated the power of combining solution-based NMR techniques with MD to gain insight into the dynamics of peptides when bound to GPCRs and how this knowledge can be applied to design novel GPCR-modulating ligands.
Methods
Protein Expression and Purification
A detailed description of the enNTS1 mutant, how it compares to WT rNTS1, and how it is expressed and purified to >90% homogeneity has been published previously.35 Briefly, enNTS1 was expressed in the cytoplasmic membrane of E. coli C43(DE3) cells using a pQE-30-derived vector with a N-terminal maltose binding protein (MBP) and a C-terminal monomeric ultrastable green fluorescent protein (muGFP)57 sequence flanking enNTS1. The cells were disrupted by sonication in 100 mM HEPES, 400 mM NaCl, and 20% glycerol, and the membranes were solubilized by adding 1.6% n-decyl-β-d-maltopyranoside (DM), 0.12% cholesterol hemisuccinate (CHS), and 0.6% CHAPS-hydrate (CHAPS). The fusion protein was captured using immobilized metal affinity chromatography (IMAC) and the buffer exchanged to 25 mM HEPES, 300 mM NaCl, 10% glycerol, 0.05% DDM pH 8. Protein from this step (MBP-enNTS1-muGFP) was used for the NT10-13 and P(OH-TIC)IL competition binding assays. The fusion proteins were removed via HRV 3C protease cleavage followed by reverse IMAC as both fusion proteins but not enNTS1 contained a His10-tag. The flowthrough containing cleaved enNTS1 was exchanged into phosphate buffer (50 mM potassium phosphate, 100 mM NaCl, 0.02% DDM, pH 7.4) and further purified by size exclusion chromatography (SEC). The enNTS1 containing SEC fractions were pooled and 86 μL aliquots at 29 μM receptor containing 20% d5-glycerol were snap frozen in liquid nitrogen and stored at −80 °C. Stabilized α1A-AR21, used as negative controls, was prepared as described for cleaved enNTS1.
NT10-13 Competition Binding Assay
Competition binding experiments were performed as described previously,35 using 0.5 nM 5-carboxytetramethylrhodamine (TAMRA) labeled NT8-13 (TAMRA-NT8-13, 5-TAMRA-Ser-Ala-Arg-Arg-Pro-Tyr-Ile-Leu) incubated with varying concentrations of unlabeled NT10-13. NT8-13 and TAMRA-NT8-13 were synthesized by GL Biochem (Shanghai) and dissolved to 20 μM in H2O. Purified MBP-enNTS1-muGFP fusion protein was immobilized on Streptavidin T1 Dynabeads (Life Technologies) as described previously,58 and resuspended in 50 mM potassium phosphate buffer, 100 mM NaCl, 0.02% DDM, pH 7.4. Receptor-bound beads were distributed into 96-well kingfisher plates, with approximately 20 pmol receptor per well. To determine nonspecific binding, paired samples were exposed to the same concentrations of TAMRA-NT8-13, but in the presence of 1 μM unlabeled NT8-13. Beads were washed for 5 min in buffer then transferred to black 96-well plates for measurement in a fluorescence plate reader (Omega polarstar, BMG Labtech). Each concentration point was measured in triplicate with the ratio of TAMRA fluorescence to muGFP calculated for each well and plotted against TAMRA-NT8-13 concentration. The data were analyzed using Graphpad Prism. Values reported represent the mean and standard error (SE) of three pKi values calculated from separate measurements.
IP1 Assay
Gαq/11 signaling assays were carried out using the IP-One HTRF Assay Kit (Cisbio Bioassays, France) measuring inositol monophosphate (IP1) using the manufacturers protocol on the wild-type rNTS1 or enNTS1 stably expressing HEK-293T cells described in Bumbak et al., 2018.35 The assay was conducted using 10 000 cells per well in a HTRF 365 well white plate and 7 μL of each treatment with stimulation for 1 h and 30 min at 37 °C. Time-resolved fluorescence measurements were conducted using a POLARstar Omega Microplate reader (BMG Labtech, Ortenburg, Germany). Samples were measured in triplicate, and the experiment was repeated at least twice. Data were analyzed using Graphad Prism.
NMR Sample Preparation
All peptides were synthesized by Genscript or GL Biochem and prepared at >95% purity. Thawed aliquots of purified enNTS1 were complemented with ligand, topped up with STD-NMR buffer (50 mM potassium phosphate, 100 mM NaCl, 0.05 or 0.02% (w/v) DDM, 10% D2O, pH 7.4) to 500 μL and added to a 5 mm NMR tube 1 h prior to measurement. The ligands used were kept as frozen stocks at concentrations of 223 mM NT10-13 in 100% d6-DMSO and 9.4 mM NT8-13 in 100% D2O. NT10-13 was added to final concentrations of 500 μM, and NT8-13 was used as the competitor at a final concentration of 20 μM.
NMR Experiments and Proton Assignments
NMR spectra were acquired at 25 °C on a 700 MHz Bruker Avance III HD spectrometer equipped with a triple-resonance TCI CryoProbe. Chemical shifts were referenced to internal DSS (0 ppm) for assignment purposes and in initial competition and control experiments. External referencing was employed for data collection samples. Samples for STD-NMR contained 500 μM NT10-13 in the presence of 5 μM cleaved enNTS1.
STD-NMR
Single saturation time STD-NMR spectra were acquired as described previously.19,26 Saturation was achieved using a train of 60 Gaussian shaped 50 ms pulses for saturation with a B1 field of 130 Hz, separated by 4 μs delays. The saturation time was 3 s and an additional relaxation delay between transients was set to 0.1 s. For the initial competition STD experiments with NT10-13 and the control experiment with cleaved α1A-AR, the on-resonance frequency was set to −1 ppm. For all other experiments the on-resonance frequency was set to 10 ppm, as excitation of the ligand or DDM was minimal at this frequency. The off-resonance frequency was set to 55 ppm. A T1ρ relaxation filter using a 40 ms spin-lock pulse was applied to suppress protein background signals. Excitation sculpting was employed for solvent suppression. Either 512 or 2048 transients were averaged over 32 K data points and a spectral width of 16 ppm. The data were multiplied by an exponential function with 2 Hz line-broadening and zero-filled once prior to Fourier transformation. STD-NMR build-up spectra were recorded analogous to single saturation time spectra but with saturation times set to 0.5, 0.75, 1, 1.5, 2, 2.5, 3, 4, and 5 s. An additional relaxation delay between transients for all build-up experiments was set so that the total time of saturation and relaxation delay equaled 5.1 s. Build-up curves were generated by plotting STD amplification factors (STDAF) calculated for each proton at each saturation time measured as proposed by Mayer and Meyer:26
| 1 |
where I0 is the intensity of a proton signal in the off-resonance spectrum and Isat is the corresponding intensity in the on-resonance spectrum. The STDAF allows direct comparison of experiments using different ligand and/or protein concentrations. The build-up curves were then fitted to a rising exponential function:59
| 2 |
where, as defined in eq 1, STDAF was determined for a given proton at saturation time t, STDMax is the maximal obtainable STD intensity at long saturation times, and ksat is the observed saturation rate constant. The initial slope 0 s saturation (STDFIT) is obtained by multiplying ksat with STDMax:
| 3 |
This slope (STDFIT) corresponds to the STD intensity in the absence of T1 correction. For simplicity and comparability STDFIT values are usually normalized by referencing the proton with the largest STDFIT values.59
Group Epitope Mapping Considering Relaxation of the Ligand (GEM-CRL) was applied as proposed by Kemper et al.40 The STD enhancements were measured using a single saturation time significantly longer than the T1 relaxation time of any ligand proton analyzed. The inversion–recovery method was used to measure 1H T1 relaxation times of a sample containing 500 μM NT10-13 in the presence of 5 μM enNTS1. The experimentally derived STD enhancements for each proton at 3 s saturation time were then modified as follows:
| 4 |
where T1 is the longitudinal relaxation time of the corresponding proton as determined by inversion recovery and STDMax is the equilibrium corrected STDAF value (eq 2) which was introduced for resonances (particularly Y11 Hδ1/δ2 and Y11 Hε1/ε2) that do not reach equilibrium at a saturation time of 3 s. STDMax values were calculated by rearranging eq 2:
| 5 |
where STDAF is the experimentally determined STD enhancement at 3 s saturation time, ksat was extracted from the exponential fit of the build-up data of the corresponding proton and t was set to 3 s. STDGEM-CRL values were determined in four individual STD-NMR experiments and normalized to the largest value of each experiment (Table S3) prior to averaging across the four experiments.
STD-NMR spectra were processed in Topspin 3.2 (Bruker); the peak intensities for each saturation time were extracted using Mnova NMR (Mestrelab), and STDAF were calculated in Microsoft Excel. STDAF were plotted and fitted using Graphpad Prism 6 to calculate the initial build-up rates (k0) for each peak. Statistical analysis was performed using one-way ANOVA with statistical significance valued at P < 0.05.
2D Total Correlation Spectroscopy (TOCSY) and Proton Assignments
1H resonances of NT10-13 were assigned using total correlation spectroscopy (TOCSY) with 60 ms mixing time, acquired under the same conditions as the STD-NMR experiments and in the presence of enNTS1. The TOCSY spectrum was processed using NMRPipe using a 90° shifted squared-sinebell window function in both dimensions and polynomial baseline correction after Fourier transformation. Peaks were assigned using the NMRFAM distribution of SPARKY.50 The Y11 HN was not apparent in any of the NMR spectra, probably due to fast exchange at pH 7.4. I12 HN (8.05 ppm) was observed in the 1H–1H TOCSY and in 1D 1H reference spectra but not in STD-NMR spectra. The detergent resonances dominated the STD-NMR spectra and partially masked ligand proton signals of P10 Hδ2/δ3 (3.28/3.26 ppm), I12 Hδ1 (0.76 ppm), I12 Hγ12/γ13 (1.34/1.04 ppm), I12 Hγ2 (0.80 ppm), and L13 Hδ1/δ2 (0.80 ppm), which were excluded from epitope mapping. P10 Hβ2 (2.33 ppm) and P10 Hβ3 (1.89 ppm) were stereospecifically assigned based on previously published work.60 P10 Hβ3 hence overlaps with the upfield resonance assigned to P10 Hγ2/γ3 (1.95/1.89 ppm). The resonance at 1.89 ppm was thus treated as the sum of the two protons P10 Hβ3 and either P10 Hγ2/γ3. Y11 Hβ2/β3 (2.94/2.86 ppm) and L13 Hδ1/δ2 (0.83/0.80 ppm) were clearly resolved in the TOCSY experiment but not assigned stereospecifically. I12 Hα (4.07 ppm) and L13 Hα (4.08 ppm) partially overlap and were thus treated as ambiguous. L13 Hβ2/β3 overlap with L13 Hγ and the corresponding peak (1.45 ppm) is thus assumed to represent the sum of the three protons. L13 HN was excluded due to possible solvent exchange processes. Our assignments are in good agreement with previously published assignments of free NT in the presence of dodecylphosphocholine (DPC) micelles.61 For convenience and representation, in the subsequent text downfield resonances are labeled P10 Hγ2 (1.95 ppm), Y11 Hβ2 (2.94 ppm), and L13 Hδ1 (0.83 ppm), and upfield resonances were labeled P10 Hγ3 (1.89 ppm), Y11 Hβ3 (2.86 ppm), and L13 Hδ2 (0.80 ppm).
Model System Construction
The homology model of the NTS1 mutant used in the present study (termed enNTS1model) was derived from the crystal structure of the NTS1–H4 mutant10 (PDB code 4BWB). NTS1–H4 was chosen because it represents the crystallized NTS1 mutant with the smallest number of substituted residues when compared with the enNTS1 mutant used herein. RosettaBackrub62 was used to generate enNTS1model, substituting D113S, L125V, L167R, A201S, I202L, and A260I. The nine N-terminal residues of enNTS1 (TSESDTAGP) were truncated in the construct used to generate the NTS1–H4 crystal structure because they were not resolved in the NTS1 GW5-T4L structure (PDB 4GRV).11 Furthermore, residues R91 to L95 (RKKSL) of ICL1 were not resolved while residues E273 to T290 of ICL3 were not present in the crystal structure. The free termini of the missing loop residues were joined together similar to previous work63 without perturbation of either TM1, TM2, TM5 or TM6. The backbone conformation of the lowest-energy homology model generated was effectively identical to the template structure 4BWB. The ligand that was carried over from 4BWB was replaced with NT8-13 in the conformation derived from 4BUO (chain C), since only residues NT8-12 were resolved in 4BWB. Residues R8 and R9 were removed manually for NT10-13 bound simulations. The enNTS1model–peptide complex was embedded into a rhombic dodecahedron cell containing a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) bilayer (40 lipids in each leaflet) and solvated with 4668 (NT10-13) and 4704 (NT8-13) simple point charge (SPC) water molecules.64 Twenty-three sodium and 30 chloride ions were added to obtain neutral systems at approximately 150 mM NaCl. The complete systems contained approximately 21370 atoms. The systems were checked for clashes between the receptor and POPC molecules in VMD, and clashes were resolved manually prior to relaxation. Each protein–bilayer system was relaxed over 100 ns, as outlined below.
MD Simulations with NT10-13 and NT8-13 peptides
Simulations were performed using Gromacs version 5.0.665 with the GROMOS 54a7 united-atom force field66 under periodic boundary conditions and using heavy-hydrogen atoms.67 The neighbor list and Lennard-Jones interaction cut-offs were set to 0.9 nm. The particle mesh Ewald (PME) algorithm68 was utilized for long-range interactions beyond these cut-offs. Four independent MD simulations using different random seeds for each NT10-13 and NT8-13 bound enNTS1 were performed. All systems were first subjected to energy minimization, followed by 500 ps of heating to 298 K with position restraints on the protein and the POPC headgroup phosphorus atoms under constant volume conditions. This was followed by 5 ns of MD under constant pressure with restraints on the protein atoms. A third equilibration phase with the V-rescale thermostat69 (310 K) and the Parrinello–Rahman barostat70 was performed for 1 ns, using a 2 fs time step and no position restraints on any atoms. The production runs were performed for at least 2.3 μs per run using the same conditions as the final equilibration phase, but a 5 fs time step. The MD frames for analysis were obtained by saving 1 frame for every nanosecond. All simulations were carried out on the Victorian Life-Science Super Computing Initiative (VLSCI) platform.
MD Simulations with OH-TIC Constrained NT Peptides
From unbiased simulations of the NT8-13 and NT10-13 peptides in complex with enNTS1model, one snapshot of the system coordinates with the Y11 χ1 angle in the g+ position was captured. To protonate the new complexes with OPLS3e atom types, we used the Protein Preparation Wizard71 tool of Maestro.72 The Y11 χ1 angles of each peptide (NT8-13 or NT10-13) were constrained to the adjacent amide in the g+ via the Y11 amide on the peptide backbone. A heavy atom restrained minimization was then performed to alleviate potential clashes using the Macromodel tool of Maestro, using implicit73 VGSB solvent and harmonic position constraints on all heavy atoms, with a force constant (k) of 0.1 kcal Å–1 mol–1. Subsequently, protein–ligand complexes were centered in an orthorhombic periodic cell74,75 and fit with OPLS-3e76 parameters using Desmond.77 These periodic boundary conditions were constructed such that the minimum distance of protein–ligand complex from the “face” of the cell was 10 Å. The system was then solvated with SPC water to a density of 1 g mL–1. To neutralize the charge on the system and to bring the salt concentration of the system to 150 mM, water(s) were replaced with Na+ or Cl– counterion(s).
System Equilibration
Using Desmond,77 an unconstrained, steepest descent energy minimization was performed on the system over a maximum of 2000 steps, using a convergence threshold of 0.1 kcal mol–1. The minimized system was then equilibrated and relaxed using the default Desmond relaxation protocol,78 which involves a series of minimizations and short MD simulations, to bring the system from its original state, to NVT (constant temperature) conditions and then to NPT (constant pressure) conditions. The final equilibration phase was conducted using the RESPA integrator79 with a 2 fs time step, the Nose-Hoover thermostat80 with τT = 1 ps to maintain a temperature of 300 K, and the Martyna-Tobias-Klein barostat81 using τP = 2 ps to sustain an average pressure of 1.0 bar. Short range Coulombic and vdW interactions were calculated with a 9.0 Å cutoff. Finally, the 1.0 μs MD production run was performed using the same NPT conditions as the last equilibration phase described above. Structures were outputted to the trajectory at a rate of 1 frame per nanosecond.
MD Data Analysis
Hydrogen-bond occupancies were calculated using the “hbond” tool of the MDTraj analysis software package.82 The Baker-Hubbard83 criteria were used: hydrogen bonds are between 2.5 and 4.0 Å long with angles (D–H–A) between 120° and 180°. Structures from all four MD trajectories were clustered with MSMBuilder 3.3.084 based on all ϕ, ψ, and χ1 angles of the receptor and the peptide with the -k-means method85 used and with the cluster number set to 10. The frame closest to each cluster center was selected for further analysis. Nonpolar hydrogen atoms, which are implicitly treated in the united-atom force field, were added to ligand and protein using ICM-Pro (MolSoft L.L.C., San Diego) without further minimization. The distances (r) between ligand and receptor protons within 5 Å, excluding exchangeable hydroxyl protons, were extracted from MD frames and the NTS1 crystal structure (4BUO) using a custom script for ICM-Pro. Interproton distances shorter than the sum of the van der Waals radii were manually set to the sum of the respective atomic radii.86 This manual correction was necessary, as distances less than 2 Å introduce a strong bias due to the r–6 relationship of the NOE effect. Theoretical STD enhancements (STDMD) for each frame were calculated from the measured ligand–protein distances using the equation:87
| 6 |
where i is a ligand proton and j are the receptor protons within 5 Å of i. The STDMD of equivalent methyl, diastereotopic P10 Hγ2/3 methylene and the stereochemically ambiguous Y11 Hδ1/2 and Hε1/2 protons of the ligand that were observed as a single peak in the STD-NMR experiments were averaged. To calculate the average STDMD values across the 10 clusters, each cluster representative was weighted according to the cluster size. The averaged STDMD values were then normalized to the largest value. The agreement of normalized STDMD sets with normalized experimental STDAF sets was assessed via the agreement factor R:88
| 7 |
where % STDMD,k and % STDAF,k represent the normalized theoretical and experimental STD enhancements, respectively, for proton k. MDTraj was run using the Anaconda Python distribution (Python 3.5.1, Continuum Analytics) in combination with the Jupyter notebook server 4.1.0 (www.ipython.org). RMSF data were calculated using the g_rmsf utility of Gromacs, and peptide torsion angles were measured using WhatIF.89 Further analysis of the MD trajectories and cluster representative frames was carried out using VMD 1.9.2.90 Representations of molecules were generated using PyMol 1.8.91
Cell-Based Competition Binding Assays
HEK 293-T cells stable expressing WT rNTS135 were resuspended into Freestyle 293 expression medium (ThermoFisher Scientific) at a concentration of around 1 000 000 cells per mL. Competition binding concentration series were setup in V bottom 96 well plates, with each well containing 50 μL of Freestyle 293 expression medium, 8 nM FAM-NT, and 2× the desired concentration of each particular competitor; 50 μL of the cell solution was added to each ligand well, resulting in a mixture of 4 nM FAM-NT, 50 000 cells, and various concentrations of competitor. The 96 well plates were incubated at 20 °C for 1 h before being measured with flow cytometry on a LSR Fortessa flow cytometer (BD biosciences). For each experiment, particular competitor concentrations samples were measured in duplicate, with 5000 cells recorded, and the mean FAM fluorescence intensity of each replicate determined. Experiments were repeated at least three times for each competitor. Data were analyzed in Graphpad Prism, and competition curves were fitted to a one site, fit Ki equation using the known Kd of FAM-NT on these cells (0.4 nM). Ki values from each experiment were combined to determine mean Ki ± SD for each competitor.
β-Arrestin Recruitment Assays
HEK 293-T cells were seeded in 6-well plates at 600 000 cells per well and cotransfected the following day with a construct encoding WT hNTS1 with a C-terminal NanoLuc tag (50 ng) and another encoding human β-arrestin 2 with a C-terminal Venus tag (200 ng). The cells were harvested 24 h later in phenol-red-free-DMEM containing 10% FBS and 40 000 cells in 80 μL per well plated out in a white 96-well plate (Nunc) and incubated at 37 °C under 5% CO2 for 24 h. For the BRET assays 10 μL of phenol-red-free-DMEM containing 10% FBS and 100 μM Furimazine were added to the wells and incubated in the dark for 30 min. At this stage any preincubated ligands (10 μL) were also added at the relevant concentration (i.e., 2.5 mM P(OH-TIC)IL for experiment in Figure S13C,D). To initiate the β-arrestin recruitment assay, 10 μL (or 11.1 μL for wells with preincubated ligands) of a 10 times concentrated agonist (or vehicle) was added to the well, and luminescent signals were immediately measured in a PHERAstar Omega plate reader (BMG LABTECH, Germany) prewarmed to 37 °C. Emitted light was measured at 410–490 nm for NanoLuc and 520–550 nm for Venus simultanueously. Signals were measured from each well every 30–40 s for up to 1 h. Ligand-induced BRET was calculated by subtracting the ratio of emission through the acceptor wavelength window (Venus) over emission through the donor wavelength window (NanoLuc) for a vehicle-treated cell sample vs peptide treated cells. For each experiment, duplicate treatments were measured simultaneously, and the resulting signals were averaged. For presented data, each experiment was repeated three times, and the averages from each experiment per treatment and time point were pooled. Data were analyzed in Graphpad prism, including area under the curve calculations (done on experimental replicates, not the pooled data) and one-way ANOVA with Tukey multiple comparisons test.
G Protein Activation Assays
HEK 293-T cells were seeded in 6-well plates at 600 000 cells per well and cotransfected the following day with the following plasmids: (1) a construct encoding WT hNTS1 (50 ng); (2) a construct encoding human Gαq (100 ng); and (3) a custom vector encoding Venus fragment (1–155) fused to human Gγ2, Venus fragment (156–239) fused to human Gβ1, and masGRK3 C-terminus fused to NanoLuc (50 ng). A sequence encoding a self-cleaving P2A peptide separates the human Gγ2 and the Gβ1 encoding fusions in plasmid (3). The split-Venus G protein fusions and masGRK3-luciferase designs in plasmid (3) are based on work by Hollins et al.92 The cells were harvested 24 h later in phenol-red-free-DMEM containing 10% FBS and 40 000 cells in 80 μL per well plated out in a white 96-well plate (Nunc) and incubated at 37 °C under 5% CO2 for 24 h. For the BRET assays 10 μL phenol-red-free-DMEM containing 10% FBS and 100 μM Furimazine were added to the wells and incubated in the dark for 30 min. At this stage any preincubated ligands (10 μL) were also added at the relevant concentration (i.e., 1 nM NT8-13 + 5 mM P(OH-TIC)IL in Figure S14C,D). To initiate the G protein activation assay, 10 μL (or 11.1 μL for wells with preincubated ligands) of a 10 times concentrated agonist (or vehicle) was added to the well and luminescent signals were immediately measured in a PHERAstar Omega plate reader (BMG LABTECH, Germany) prewarmed to 37 °C. Emited light was measured at 410–490 nm for NanoLuc and 520–550 nm for splitVenus simultanueously. Signals were measured from each well every 30–40 s for up to 1 h. Ligand-induced BRET was calculated by subtracting the ratio of emission through the acceptor wavelength window (Venus) over emission through the donor wavelength window (NanoLuc) for a vehicle-treated cell sample vs peptide treated cells. For each experiment, duplicate treatments were measured simultaneously, and the resulting signals were averaged. For the presented data, each experiment was repeated three times, and the averages from each experiment per treatment and time point were pooled. Data were analyzed in Graphpad prism, including area under the curve calculations (done on experimental replicates, not the pooled data) and one-way ANOVA with Tukey multiple comparisons test.
xCELLigence Assays
Label-free signaling assays were performed using the xCELLigence Single Plate Real Time Cell Analyzer (RTCA) (Acea Biosciences Inc.) as described previously.35 The xCELLigence device measures the impedance that cells impart over time on gold electrode-embedded microplates. Background impedance readings were initially recorded in 96-well E-Plates containing 50 μL of complete DMEM media per well before seeding each well with 18 000 HEK 293-T cells. Twenty-four hours later cells were transfected with a plasmid encoding WT hNTS1. After another 24 h the medium was aspirated and replaced with 90 μL of 0.5% FBS DMEM. Cells were then left for 4 h to establish a semiquiescent state for determining a baseline before stimulation. Stimulations were then conducted using an Eppendorf Research Pro multichannel pipet to quickly add 10 μL of treatment solutions per well. For each experiment, triplicate treatments were measured simultaneously and the resulting signals were averaged for each time point. For the presented data, each experiment was repeated three times. Data were analyzed in Graphpad prism, including area under the curve calculations (done on experimental replicates, not the pooled data). Dose response curves were generated by plotting the area under the cell impedance curves for 1 h after treatment with different doses of NT8-13 or P(OH-TIC)IL.
Acknowledgments
This research was supported by National Health and Medical Research Council of Australia project Grants 1100676 and 1043750 (R.A.D.B., P.R.G., and D.J.S.) and the Victorian Government Operational Infrastructure Support Program. R.A.D.B. is supported by an NHMRC Research Fellowship, and D.J.S. is supported by an NHMRC Dementia Fellowship. The authors thank Tania Ferraro and Sharon Layfield for technical assistance.
Glossary
Abbreviations
- GPCR
G protein-coupled receptors
- NT
neurotensin
- NTS1)
neurotensin receptor 1
- STD-NMR
saturation transfer difference nuclear magnetic resonance spectroscopy
- GEM
group epitope mapping
- MD
molecular dynamics
- DDM
n-dodecyl-β-d-maltopyranoside
- GEM-CRL
Group Epitope Mapping Considering Relaxation of the Ligand
- STCD
saturation transfer competition difference
- RMSD
root-mean-square deviation
- RMSF
root-mean-square fluctuation
- ECL
extracellular loop
- TIC
hydroxy-3,4-dihydro-1h-isoquinoline-3-carboxylic acid
- OH-TIC
7-hydroxy-3,4-dihydro-1h-isoquinoline-3-carboxylic acid
- WT
wild type
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsptsci.0c00026.
Supplementary data figures, tables and information (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Cáceda R.; Kinkead B.; Nemeroff C. B. (2006) Neurotensin: Role in psychiatric and neurological diseases. Peptides 27, 2385–2404. 10.1016/j.peptides.2006.04.024. [DOI] [PubMed] [Google Scholar]
- Al-Rodhan N. R. F.; Richelson E.; Gilbert J. A.; McCormick D. J.; Kanba K. S.; Pfenning M. A.; Nelson A.; Larson E. W.; Yaksh T. L. (1991) Structure-antinociceptive activity of neurotensin and some novel analogues in the periaqueductal gray region of the brainstem. Brain Res. 557, 227–235. 10.1016/0006-8993(91)90139-M. [DOI] [PubMed] [Google Scholar]
- Zhao D.; Keates A. C.; Kuhnt-Moore S.; Moyer M. P.; Kelly C. P.; Pothoulakis C. (2001) Signal Transduction Pathways Mediating Neurotensin-stimulated Interleukin-8 Expression in Human Colonocytes. J. Biol. Chem. 276, 44464–44471. 10.1074/jbc.M104942200. [DOI] [PubMed] [Google Scholar]
- Boules M. M.; Fredrickson P.; Muehlmann A. M.; Richelson E. (2014) Elucidating the role of neurotensin in the pathophysiology and management of major mental disorders. Behav. Sci. 4, 125–153. 10.3390/bs4020125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro L.; Tiozzo Fasiolo L.; Beggiato S.; Borelli A. C.; Pomierny-Chamiolo L.; Frankowska M.; Antonelli T.; Tomasini M. C.; Fuxe K.; Filip M. (2016) Neurotensin: A role in substance use disorder?. J. Psychopharmacol. 30, 112–127. 10.1177/0269881115622240. [DOI] [PubMed] [Google Scholar]
- Carraway R. E.; Plona A. M. (2006) Involvement of neurotensin in cancer growth: evidence, mechanisms and development of diagnostic tools. Peptides 27, 2445–2460. 10.1016/j.peptides.2006.04.030. [DOI] [PubMed] [Google Scholar]
- Li J.; Song J.; Zaytseva Y. Y.; Liu Y.; Rychahou P.; Jiang K.; Starr M. E.; Kim J. T.; Harris J. W.; Yiannikouris F. B.; Katz W. S.; Nilsson P. M.; Orho-Melander M.; Chen J.; Zhu H.; Fahrenholz T.; Higashi R. M.; Gao T.; Morris A. J.; Cassis L. A.; Fan T. W. M.; Weiss H. L.; Dobner P. R.; Melander O.; Jia J.; Evers B. M. (2016) An obligatory role for neurotensin in high-fat-diet-induced obesity. Nature 533, 411–415. 10.1038/nature17662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Gelais F.; Jomphe C.; Trudeau L. E. (2006) The role of neurotensin in central nervous system pathophysiology: What is the evidence?. J. Psych. Neurosci. 31, 229–245. [PMC free article] [PubMed] [Google Scholar]
- Krumm B. E.; White J. F.; Shah P.; Grisshammer R. (2015) Structural prerequisites for G-protein activation by the neurotensin receptor. Nat. Commun. 6, 7895. 10.1038/ncomms8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff P.; Hillenbrand M.; Klenk C.; Batyuk A.; Heine P.; Balada S.; Schlinkmann K. M.; Scott D. J.; Schutz M.; Pluckthun A. (2014) Structure of signaling-competent neurotensin receptor 1 obtained by directed evolution in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 111, E655–E662. 10.1073/pnas.1317903111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. F.; Noinaj N.; Shibata Y.; Love J.; Kloss B.; Xu F.; Gvozdenovic-Jeremic J.; Shah P.; Shiloach J.; Tate C. G.; Grisshammer R. (2012) Structure of the agonist-bound neurotensin receptor. Nature 490, 508–513. 10.1038/nature11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm B. E.; Lee S.; Bhattacharya S.; Botos I.; White C. F.; Du H.; Vaidehi N.; Grisshammer R. (2016) Structure and dynamics of a constitutively active neurotensin receptor. Sci. Rep. 6, 38564. 10.1038/srep38564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W.; Li Z.; Jin M.; Yin Y.-L.; de Waal P. W.; Pal K.; Yin Y.; Gao X.; He Y.; Gao J.; Wang X.; Zhang Y.; Zhou H.; Melcher K.; Jiang Y.; Cong Y.; Zhou X. E.; Yu X.; Xu H. E. (2019) A complex structure of arrestin-2 bound to a G protein-coupled receptor. Cell Res. 29, 822957. 10.1038/s41422-019-0256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H. E.; Zhang Y.; Hu H.; Suomivuori C.-M.; Kadji F. M. N.; Aoki J.; Krishna Kumar K.; Fonseca R.; Hilger D.; Huang W.; Latorraca N. R.; Inoue A.; Dror R. O.; Kobilka B. K.; Skiniotis G. (2019) Conformational transitions of a neurotensin receptor 1–Gi1 complex. Nature 572, 80–85. 10.1038/s41586-019-1337-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca S.; White J. F.; Sohal A. K.; Filippov D. V.; van Boom J. H.; Grisshammer R.; Baldus M. (2003) The conformation of neurotensin bound to its G protein-coupled receptor. Proc. Natl. Acad. Sci. U. S. A. 100, 10706–10711. 10.1073/pnas.1834523100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor C.; White K. L.; Doncescu N.; Didenko T.; Roth B. L.; Czaplicki G.; Stevens R. C.; Wuthrich K.; Milon A. (2015) NMR structure and dynamics of the agonist dynorphin peptide bound to the human kappa opioid receptor. Proc. Natl. Acad. Sci. U. S. A. 112, 11852–11857. 10.1073/pnas.1510117112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inooka H.; Ohtaki T.; Kitahara O.; Ikegami T.; Endo S.; Kitada C.; Ogi K.; Onda H.; Fujino M.; Shirakawa M. (2001) Conformation of a peptide ligand bound to its G-protein coupled receptor. Nat. Struct. Biol. 8, 161–165. 10.1038/84159. [DOI] [PubMed] [Google Scholar]
- Cong X.; Fiorucci S.; Golebiowski J. (2018) Activation dynamics of the neurotensin G protein-coupled receptor 1. J. Chem. Theory Comput. 14, 4467–4473. 10.1021/acs.jctc.8b00216. [DOI] [PubMed] [Google Scholar]
- Mayer M.; Meyer B. (1999) Characterization of ligand binding by saturation transfer difference NMR spectroscopy. Angew. Chem., Int. Ed. 38, 1784–1788. . [DOI] [PubMed] [Google Scholar]
- Bartoschek S.; Klabunde T.; Defossa E.; Dietrich V.; Stengelin S.; Griesinger C.; Carlomagno T.; Focken I.; Wendt K. U. (2010) Drug Design for G-Protein-Coupled Receptors by a Ligand-Based NMR Method. Angew. Chem., Int. Ed. 49, 1426–1429. 10.1002/anie.200905102. [DOI] [PubMed] [Google Scholar]
- Yong K. J.; Vaid T. M.; Shilling P. J.; Wu F.-J.; Williams L. M.; Deluigi M.; Plückthun A.; Bathgate R. A. D.; Gooley P. R.; Scott D. J. (2018) Determinants of ligand subtype-selectivity at α1A-adrenoceptor revealed using Saturation Transfer Difference (STD) NMR. ACS Chem. Biol. 13, 1090–1102. 10.1021/acschembio.8b00191. [DOI] [PubMed] [Google Scholar]
- Assadi-Porter F. M.; Tonelli M.; Maillet E.; Hallenga K.; Benard O.; Max M.; Markley J. L. (2008) Direct NMR detection of the binding of functional ligands to the human sweet receptor, a heterodimeric family 3 GPCR. J. Am. Chem. Soc. 130, 7212–7213. 10.1021/ja8016939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igonet S.; Raingeval C.; Cecon E.; Pučić-Baković M.; Lauc G.; Cala O.; Baranowski M.; Perez J.; Jockers R.; Krimm I.; Jawhari A. (2018) Enabling STD-NMR fragment screening using stabilized native GPCR: A case study of adenosine receptor. Sci. Rep. 8, 8142. 10.1038/s41598-018-26113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira A.; Pfeifer T. A.; Grigliatti T. A.; Andersen R. J. (2009) Functional Cell-Based Screening and Saturation Transfer Double-Difference NMR Have Identified Haplosamate A as a Cannabinoid Receptor Agonist. ACS Chem. Biol. 4, 139–144. 10.1021/cb800264k. [DOI] [PubMed] [Google Scholar]
- Burger J. L.; Jeerage K. M.; Bruno T. J. (2016) Direct nuclear magnetic resonance observation of odorant binding to mouse odorant receptor MOR244–3. Anal. Biochem. 502, 64–72. 10.1016/j.ab.2016.03.006. [DOI] [PubMed] [Google Scholar]
- Mayer M.; Meyer B. (2001) Group Epitope Mapping by Saturation Transfer Difference NMR To Identify Segments of a Ligand in Direct Contact with a Protein Receptor. J. Am. Chem. Soc. 123, 6108–6117. 10.1021/ja0100120. [DOI] [PubMed] [Google Scholar]
- Johnson M. A.; Pinto B. M. (2004) Saturation-transfer difference NMR studies for the epitope mapping of a carbohydrate-mimetic peptide recognized by an anti-carbohydrate antibody. Bioorg. Med. Chem. 12, 295–300. 10.1016/j.bmc.2003.09.041. [DOI] [PubMed] [Google Scholar]
- Neffe A. T.; Bilang M.; Meyer B. (2006) Synthesis and optimization of peptidomimetics as HIV entry inhibitors against the receptor protein CD4 using STD NMR and ligand docking. Org. Biomol. Chem. 4, 3259–3267. 10.1039/b605380g. [DOI] [PubMed] [Google Scholar]
- Tong W.; Gagnon M.; Sprules T.; Gilbert M.; Chowdhury S.; Meerovitch K.; Hansford K.; Purisima E. O.; Blankenship J. W.; Cheung N. K.; Gehring K.; Lubell W. D.; Saragovi H. U. (2010) Small-molecule ligands of GD2 ganglioside, designed from NMR studies, exhibit induced-fit binding and bioactivity. Chem. Biol. 17, 183–194. 10.1016/j.chembiol.2010.01.012. [DOI] [PubMed] [Google Scholar]
- Gimeno A.; Santos L. M.; Alemi M.; Rivas J.; Blasi D.; Cotrina E. Y.; Llop J.; Valencia G.; Cardoso I.; Quintana J. R.; Arsequell G.; Jimenez-Barbero J. (2017) Insights on the interaction between transthyretin and Aβ in solution. A saturation transfer difference (STD) NMR analysis of the role of iododiflunisal. J. Med. Chem. 60, 5749–5758. 10.1021/acs.jmedchem.7b00428. [DOI] [PubMed] [Google Scholar]
- Meinecke R.; Meyer B. (2001) Determination of the binding specificity of an integral membrane protein by saturation transfer difference NMR: RGD peptide ligands binding to integrin alpha(IIb)beta(3). J. Med. Chem. 44, 3059–3065. 10.1021/jm0109154. [DOI] [PubMed] [Google Scholar]
- Calvanese L.; Focà A.; Sandomenico A.; Focà G.; Caporale A.; Doti N.; Iaccarino E.; Leonardi A.; D’Auria G.; Ruvo M.; Falcigno L. (2017) Structural insights into the interaction of a monoclonal antibody and Nodal peptides by STD-NMR spectroscopy. Bioorg. Med. Chem. 25, 6589–6596. 10.1016/j.bmc.2017.10.036. [DOI] [PubMed] [Google Scholar]
- Melikian M.; Eluard B.; Bertho G.; Baud V.; Evrard-Todeschi N. (2017) Model of the Interaction between the NF-κB Inhibitory protein p100 and the E3 ubiquitin ligase β-TrCP based on NMR and Docking Experiments. J. Chem. Inf. Model. 57, 223–233. 10.1021/acs.jcim.5b00409. [DOI] [PubMed] [Google Scholar]
- Johnson M. A.; Jaseja M.; Zou W.; Jennings H. J.; Copié V.; Pinto B. M.; Pincus S. H. (2003) NMR Studies of Carbohydrates and Carbohydrate-mimetic Peptides Recognized by an Anti-Group B Streptococcus Antibody. J. Biol. Chem. 278, 24740–24752. 10.1074/jbc.M301846200. [DOI] [PubMed] [Google Scholar]
- Bumbak F.; Keen A. C.; Gunn N. J.; Gooley P. R.; Bathgate R. A. D.; Scott D. J. (2018) Optimization and 13CH3 methionine labeling of a signaling competent neurotensin receptor 1 variant for NMR studies. Biochim. Biophys. Acta, Biomembr. 1860, 1372–1383. 10.1016/j.bbamem.2018.03.020. [DOI] [PubMed] [Google Scholar]
- Nieto J. L.; Rico M.; Santoro J.; Herranz J.; Bermejo F. J. (1986) Assignment and conformation of neurotensin in aqueous solution by 1H NMR. Int. J. Pept. Protein Res. 28, 315–323. 10.1111/j.1399-3011.1986.tb03261.x. [DOI] [PubMed] [Google Scholar]
- Xu G. Y.; Deber C. M. (1991) Conformations of neurotensin in solution and in membrane environments studied by 2-D NMR spectroscopy. Int. J. Pept. Protein Res. 37, 528–535. 10.1111/j.1399-3011.1991.tb00771.x. [DOI] [PubMed] [Google Scholar]
- Myers R. M.; Shearman J. W.; Kitching M. O.; Ramos-Montoya A.; Neal D. E.; Ley S. V. (2009) Cancer, chemistry, and the cell: molecules that interact with the neurotensin receptors. ACS Chem. Biol. 4, 503–525. 10.1021/cb900038e. [DOI] [PubMed] [Google Scholar]
- KITABGI P.; POUSTIS C.; GRANIER C.; VAN RIETSCHOTEN J.; RIVIER J.; MORGAT J.-L.; FREYCHET P. (1980) Neurotensin Binding to Extraneural and Neural Receptors: Comparison with Biological Activity and Structure—Activity Relationships. Mol. Pharmacol. 18, 11–19. [PubMed] [Google Scholar]
- Kemper S.; Patel M. K.; Errey J. C.; Davis B. G.; Jones J. A.; Claridge T. D. W. (2010) Group epitope mapping considering relaxation of the ligand (GEM-CRL): Including longitudinal relaxation rates in the analysis of saturation transfer difference (STD) experiments. J. Magn. Reson. 203, 1–10. 10.1016/j.jmr.2009.11.015. [DOI] [PubMed] [Google Scholar]
- Labbe-Jullie C.; Barroso S.; Nicolas-Eteve D.; Reversat J. L.; Botto J. M.; Mazella J.; Bernassau J. M.; Kitabgi P. (1998) Mutagenesis and modeling of the neurotensin receptor NTR1. Identification of residues that are critical for binding SR 48692, a nonpeptide neurotensin antagonist. J. Biol. Chem. 273, 16351–16357. 10.1074/jbc.273.26.16351. [DOI] [PubMed] [Google Scholar]
- Barroso S.; Richard F.; Nicolas-Etheve D.; Reversat J. L.; Bernassau J. M.; Kitabgi P.; Labbe-Jullie C. (2000) Identification of residues involved in neurotensin binding and modeling of the agonist binding site in neurotensin receptor 1. J. Biol. Chem. 275, 328–336. 10.1074/jbc.275.1.328. [DOI] [PubMed] [Google Scholar]
- Botto J. M.; Chabry J.; Nouel D.; Paquet M.; Seguela P.; Vincent J. P.; Beaudet A.; Mazella J. (1997) Identification in the rat neurotensin receptor of amino-acid residues critical for the binding of neurotensin. Mol. Brain Res. 46, 311–317. 10.1016/S0169-328X(97)00006-5. [DOI] [PubMed] [Google Scholar]
- Sanfelice D.; Temussi P. A. (2014) The conformation of enkephalin bound to its receptor: an “elusive goal” becoming reality. Front. Mol. Biosci. 1, 1–8. 10.3389/fmolb.2014.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y. P.; Cusack B.; Groshan K.; Richelson E. (1996) Proposed ligand binding site of the transmembrane receptor for neurotensin(8–13). J. Biol. Chem. 271, 15060–15068. 10.1074/jbc.271.25.15060. [DOI] [PubMed] [Google Scholar]
- Cusack B.; Mccormick D. J.; Pang Y. P.; Souder T.; Garcia R.; Fauq A.; Richelson E. (1995) Pharmacological and Biochemical Profiles of Unique Neurotensin-8–13 Analogs Exhibiting Species Selectivity, Stereoselectivity, and Superagonism. J. Biol. Chem. 270, 18359–18366. 10.1074/jbc.270.31.18359. [DOI] [PubMed] [Google Scholar]
- Granier C.; van Rietschoten J.; Kitabgi P.; Poustis C.; Freychet P. (1982) Synthesis and characterization of neurotensin analogues for structure/activity relationship studies. Acetyl-neurotensin-(8--13) is the shortest analogue with full binding and pharmacological activities. Eur. J. Biochem. 124, 117–124. 10.1111/j.1432-1033.1982.tb05913.x. [DOI] [PubMed] [Google Scholar]
- Hruby V. J.; Cody W. L.; de Lauro Castrucci A. M.; Hadley M. E. (1988) Conformational and biological analysis of α-MSH fragment analogues with sterically constrained amino acid residues. Collect. Czech. Chem. Commun. 5, 2549–2573. 10.1135/cccc19882549. [DOI] [Google Scholar]
- Procházka Z.; Ancans J. E.; Slaninová J.; Machová A.; Barth T.; Lebl M. (1990) Synthesis and biological properties of vasopressin analogues containing 1,2,3,4-tetrahydroisoquinoline-3-carboxylic acid. Collect. Czech. Chem. Commun. 55, 1099–1105. 10.1135/cccc19901099. [DOI] [Google Scholar]
- Gibson S. E.; Guillo N.; Jones J. O.; Buck I. M.; Kalindjian S. B.; Roberts S.; Tozer M. J. (2002) CCK2 receptor antagonists containing the conformationally constrained phenylalanine derivatives, including the new amino acid Xic. Eur. J. Med. Chem. 37, 379–389. 10.1016/S0223-5234(02)01351-X. [DOI] [PubMed] [Google Scholar]
- Ballet S.; Feytens D.; Wachter R. D.; Vlaeminck M. D.; Marczak E. D.; Salvadori S.; Graaf C. d.; Rognan D.; Negri L.; Lattanzi R.; Lazarus L. H.; Tourwé D.; Balboni G. (2009) Conformationally constrained opioid ligands: The Dmt-Aba and Dmt-Aia versus Dmt-Tic scaffold. Bioorg. Med. Chem. Lett. 19, 433–437. 10.1016/j.bmcl.2008.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyl D. L.; Sefler A. M.; He J. X.; Sawyer T. K.; Wustrow D. J.; Akunne H. C.; Davis M. D.; Pugsley T. A.; Heffner T. G.; Corbin A. E.; Cody W. L. (1994) Structure-Activity and Conformational Studies of a Series of Modified C-Terminal Hexapeptide Neurotensin Analogs. Int. J. Pept. Protein Res. 44, 233–238. 10.1111/j.1399-3011.1994.tb00165.x. [DOI] [PubMed] [Google Scholar]
- Eiselt E.; Gonzalez S.; Martin C.; Chartier M.; Betti C.; Longpré J.-M.; Cavelier F.; Tourwè D.; Gendron L.; Ballet S.; Sarret P. (2019) Neurotensin Analogues Containing Cyclic Surrogates of Tyrosine at Position 11 Improve NTS2 Selectivity Leading to Analgesia without Hypotension and Hypothermia. ACS Chem. Neurosci. 10, 4535–4544. 10.1021/acschemneuro.9b00390. [DOI] [PubMed] [Google Scholar]
- Pfizenmayer A. J.; Vera M. D.; Ding X.; Xiao D.; Chen W.-C.; Joullié M. M.; Tandon D.; Toogood P. L. (1998) Synthesis and biological activity of [Tic5] didemnin B. Bioorg. Med. Chem. Lett. 8, 3653–3656. 10.1016/S0960-894X(98)00662-3. [DOI] [PubMed] [Google Scholar]
- Besserer-Offroy É.; Brouillette R. L.; Lavenus S.; Froehlich U.; Brumwell A.; Murza A.; Longpré J.-M.; Marsault É.; Grandbois M.; Sarret P.; Leduc R. (2017) The signaling signature of the neurotensin type 1 receptor with endogenous ligands. Eur. J. Pharmacol. 805, 1–13. 10.1016/j.ejphar.2017.03.046. [DOI] [PubMed] [Google Scholar]
- Huang W.; Masureel M.; Qianhui Q.; Janetzko J.; Inoue A.; Kato H. E.; Robertson M. J.; Nguyen K. C.; Glenn J. S.; Skiniotis G.; Kobilka B. K. (2020) Structure of the neurotensin receptor 1 in complex with β-arrestin 1. Nature 597, 303–308. 10.1038/s41586-020-1953-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D. J.; Gunn N. J.; Yong K. J.; Wimmer V. C.; Veldhuis N. A.; Challis L. M.; Haidar M.; Petrou S.; Bathgate R. A. D.; Griffin M. D. W. (2018) A Novel Ultra-Stable, Monomeric Green Fluorescent Protein For Direct Volumetric Imaging of Whole Organs Using CLARITY. Sci. Rep. 8, 667. 10.1038/s41598-017-18045-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D. J.; Kummer L.; Egloff P.; Bathgate R. A.; Pluckthun A. (2014) Improving the apo-state detergent stability of NTS1 with CHESS for pharmacological and structural studies. Biochim. Biophys. Acta, Biomembr. 1838, 2817–2824. 10.1016/j.bbamem.2014.07.015. [DOI] [PubMed] [Google Scholar]
- Mayer M.; James T. L. (2004) NMR-based characterization of phenothiazines as a RNA binding scaffold. J. Am. Chem. Soc. 126, 4453–4460. 10.1021/ja0398870. [DOI] [PubMed] [Google Scholar]
- Pogliani L.; Ellenberger M.; Valat J. (1975) A NMR investigation of proline and its derivatives. II—Conformational implications of the 1H NMR spectrum of L-proline at different pH. Org. Magn. Reson. 7, 61–71. 10.1002/mrc.1270070202. [DOI] [Google Scholar]
- Coutant J.; Curmi P. A.; Toma F.; Monti J. P. (2007) NMR solution structure of neurotensin in membrane-mimetic environments: Molecular basis for neurotensin receptor recognition. Biochemistry 46, 5656–5663. 10.1021/bi602567p. [DOI] [PubMed] [Google Scholar]
- Smith C. A.; Kortemme T. (2008) Backrub-like backbone simulation recapitulates natural protein conformational variability and improves mutant side-chain prediction. J. Mol. Biol. 380, 742–756. 10.1016/j.jmb.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T.; Fang Y.; Yuriev E.; Chalmers D. K. (2016) Ligand Binding Pathways of Clozapine and Haloperidol in the Dopamine D2 and D3 Receptors. J. Chem. Inf. Model. 56, 308–321. 10.1021/acs.jcim.5b00457. [DOI] [PubMed] [Google Scholar]
- Berendsen H. J. C., Postma J. P. M., van Gunsteren W. F., and Hermans J. (1981) Interaction Models for Water in Relation to Protein Hydration, In Intermolecular Forces: Proceedings of the Fourteenth Jerusalem Symposium on Quantum Chemistry and Biochemistry Held in Jerusalem, Israel, April 13–16, 1981 (Pullman B., Ed.) pp 331–342, Springer Netherlands, Dordrecht.
- Abraham M. J.; Murtola T.; Schulz R.; Páll S.; Smith J. C.; Hess B.; Lindahl E. (2015) GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25. 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- Schmid N.; Eichenberger A. P.; Choutko A.; Riniker S.; Winger M.; Mark A. E.; Gunsteren W. F. (2011) Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur. Biophys. J. 40, 843–856. 10.1007/s00249-011-0700-9. [DOI] [PubMed] [Google Scholar]
- Feenstra K. A.; Hess B.; Berendsen H. J. C. (1999) Improving efficiency of large time-scale molecular dynamics simulations of hydrogen-rich systems. J. Comput. Chem. 20, 786–798. . [DOI] [PubMed] [Google Scholar]
- Darden T.; York D.; Pedersen L. (1993) Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092. 10.1063/1.464397. [DOI] [Google Scholar]
- Bussi G.; Donadio D.; Parrinello M. (2007) Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- Parrinello M.; Rahman A. (1981) Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 52, 7182–7190. 10.1063/1.328693. [DOI] [Google Scholar]
- (2011) Protein preparation wizard, Epik version 2, Schrödinger, LLC.
- (2014) MacroModel, version 10.3, Schrödinger, LLC, New York, NY. [Google Scholar]
- Wang J.; Tan C.; Tan Y.-H.; Lu Q.; Luo R. (2008) Poisson-Boltzmann solvents in molecular dynamics simulations. Commun. Comput. Phys. 3, 1010–1031. [Google Scholar]
- Williams-Noonan B. J.; Yuriev E.; Chalmers D. K. (2018) Free Energy Methods in Drug Design: Prospects of “Alchemical Perturbation” in Medicinal Chemistry. J. Med. Chem. 61, 638–649. 10.1021/acs.jmedchem.7b00681. [DOI] [PubMed] [Google Scholar]
- Dubin D. H. (1990) First-order anharmonic correction to the free energy of a coulomb crystal in periodic boundary conditions. Phys. Rev. A: At., Mol., Opt. Phys. 42, 4972. 10.1103/PhysRevA.42.4972. [DOI] [PubMed] [Google Scholar]
- Roos K.; Wu C.; Damm W.; Reboul M.; Stevenson J. M.; Lu C.; Dahlgren M. K.; Mondal S.; Chen W.; Wang L.; Abel R.; Friesner R. A.; Harder E. D. (2019) OPLS3e: Extending Force Field Coverage for Drug-Like Small Molecules. J. Chem. Theory Comput. 15, 1863–1874. 10.1021/acs.jctc.8b01026. [DOI] [PubMed] [Google Scholar]
- (2014) Desmond Molecular Dynamics System, version 3.7, DE Shaw Research, New York, NY.
- Wang L.; Deng Y.; Knight J. L.; Wu Y.; Kim B.; Sherman W.; Shelley J. C.; Lin T.; Abel R. (2013) Modeling local structural rearrangements using FEP/REST: application to relative binding affinity predictions of CDK2 inhibitors. J. Chem. Theory Comput. 9, 1282–1293. 10.1021/ct300911a. [DOI] [PubMed] [Google Scholar]
- Leimkuhler B.; Margul D. T.; Tuckerman M. E. (2013) Stochastic, resonance-free multiple time-step algorithm for molecular dynamics with very large time steps. Mol. Phys. 111, 3579–3594. 10.1080/00268976.2013.844369. [DOI] [Google Scholar]
- Evans D. J.; Holian B. L. (1985) The nose–hoover thermostat. J. Chem. Phys. 83, 4069–4074. 10.1063/1.449071. [DOI] [Google Scholar]
- Martyna G. J.; Tobias D. J.; Klein M. L. (1994) Constant pressure molecular dynamics algorithms. J. Chem. Phys. 101, 4177–4189. 10.1063/1.467468. [DOI] [Google Scholar]
- McGibbon R. T.; Beauchamp K. A.; Harrigan M. P.; Klein C.; Swails J. M.; Hernandez C. X.; Schwantes C. R.; Wang L.-P.; Lane T. J.; Pande V. S. (2015) MDTraj: A Modern Open Library for the Analysis of Molecular Dynamics Trajectories. Biophys. J. 109, 1528–1532. 10.1016/j.bpj.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker E. N.; Hubbard R. E. (1984) Hydrogen-Bonding in Globular-Proteins. Prog. Biophys. Mol. Biol. 44, 97–179. 10.1016/0079-6107(84)90007-5. [DOI] [PubMed] [Google Scholar]
- Beauchamp K. A.; Bowman G. R.; Lane T. J.; Maibaum L.; Haque I. S.; Pande V. S. (2011) MSMBuilder2: Modeling Conformational Dynamics on the Picosecond to Millisecond Scale. J. Chem. Theory Comput. 7, 3412–3419. 10.1021/ct200463m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartigan J. A.; Wong M. A. (1979) Algorithm AS 136: A K-Means Clustering Algorithm. Journal of the Royal Statistical Society. Series C (Applied Statistics) 28, 100–108. 10.2307/2346830. [DOI] [Google Scholar]
- Seeliger D.; de Groot B. L. (2007) Atomic contacts in protein structures. A detailed analysis of atomic radii, packing, and overlaps. Proteins: Struct., Funct., Genet. 68, 595–601. 10.1002/prot.21447. [DOI] [PubMed] [Google Scholar]
- Seeger K.; Lein S.; Reuter G.; Berger S. (2005) Saturation Transfer Difference Measurements with SU(VAR)3–9 and S-adenosyl-l-methionine†. Biochemistry 44, 6208–6213. 10.1021/bi047749x. [DOI] [PubMed] [Google Scholar]
- Krishna N. R.; Agresti D. G.; Glickson J. D.; Walter R. (1978) Solution conformation of peptides by the intramolecular nuclear Overhauser effect experiment. Study of valinomycin-K+. Biophys. J. 24, 791–814. 10.1016/S0006-3495(78)85421-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriend G. (1990) WHAT IF: A molecular modeling and drug design program. J. Mol. Graphics 8, 52–56. 10.1016/0263-7855(90)80070-V. [DOI] [PubMed] [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. (1996) VMD: Visual molecular dynamics. J. Mol. Graphics 14, 33–38. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- (2015) The PyMOL Molecular Graphics System, version 1.8, Schrodinger, LLC.
- Hollins B.; Kuravi S.; Digby G. J.; Lambert N. A. (2009) The c-terminus of GRK3 indicates rapid dissociation of G protein heterotrimers. Cell. Signalling 21, 1015–1021. 10.1016/j.cellsig.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.