Abstract
The G protein-coupled receptor 182 (GPR182) is an orphan GPCR, the expression of which is enriched in embryonic endothelial cells (ECs). However, the physiological role and molecular mechanism of action of GPR182 are unknown. Here, we show that GPR182 negatively regulates definitive hematopoiesis in zebrafish and mice. In zebrafish, gpr182 expression is enriched in the hemogenic endothelium (HE), and gpr182–/– display an increased expression of HE and hematopoietic stem cell (HSC) marker genes. Notably, we find an increased number of myeloid cells in gpr182–/– compared to wild-type. Further, by time-lapse imaging of zebrafish embryos during the endothelial-to-hematopoietic transition, we find that HE/HSC cell numbers are increased in gpr182–/– compared to wild-type. GPR182–/– mice also exhibit an increased number of myeloid cells compared to wild-type, indicating a conserved role for GPR182 in myelopoiesis. Using cell-based small molecule screening and transcriptomic analyses, we further find that GPR182 regulates the leukotriene B4 (LTB4) biosynthesis pathway. Taken together, these data indicate that GPR182 is a negative regulator of definitive hematopoiesis in zebrafish and mice, and provide further evidence for LTB4 signaling in HSC biology.
Keywords: G protein-coupled receptor, GPR182, hematopoietic stem cell, definitive hematopoiesis, myelopoiesis, Leukotriene B4
G-protein coupled receptors (GPCRs) are the most tractable class of proteins, with ∼30–40% of all drugs currently on the market targeting their activity.1−3 To date, many GPCRs remain categorized as “orphan” GPCRs, sparking much interest and investment to discover selective modulators of their activity for the development of novel therapeutics. As such, it is critical to define the function and molecular mechanism of these orphan receptors to understand the physiological impact of their inhibition. GPCRs constitute the largest receptor family and are involved in a variety of physiological processes that range from sensing external signals including light, odor, taste, and touch to mediating signal transduction pathways, such as in the autonomic nervous system and during inflammation.4 However, the role of GPCRs in hematopoiesis remains poorly characterized.
Historically, zebrafish have been recognized as an excellent genetic model system to study hematopoiesis because of a high level of similarity with mammals.5 Namely, zebrafish and mammals share all major types of blood cells, and these cells are produced via similar processes called primitive and definitive hematopoiesis.6 In zebrafish, primitive hematopoiesis occurs in the anterior lateral plate mesoderm, which gives rise to myeloid cells, and in the posterior lateral plate mesoderm, which gives rise to primitive erythrocytes.5 Definitive hematopoiesis produces hematopoietic stem cells (HSCs) capable of self-renewing and contributing to all blood lineages.7 HSCs first appear at approximately 30–32 h post fertilization (hpf) from hemogenic endothelial cells located at the ventral wall of the dorsal aorta (VDA), which is functionally equivalent to the aorta-gonad-mesonephros (AGM) region in amniotes.8,9 HSCs (marked by runx1 and c-myb expression) migrate to the caudal hematopoietic tissue (CHT) where they expand and further develop before moving to the kidney, which is the zebrafish equivalent to the mammalian bone marrow.10
GPR182 is a class A orphan GPCR. Initially, it was thought that GPR182 was a putative adrenomedullin receptor; however, it was later shown that adrenomedullin signals through a different GPCR complex.11Gpr182 is highly expressed in developing mouse and zebrafish endothelium and enriched in mammary tumor endothelium compared to normal mammary endothelium.12−14 Interestingly, gpr182 expression is significantly altered in a zebrafish model of myeloid leukemia.15 These reports suggest that Gpr182 functions in hematopoiesis in healthy and disease conditions.
Here, we show that GPR182 plays a negative function in definitive hematopoiesis. We found that gpr182–/– zebrafish embryos exhibit increased HE/HSC formation. In addition, we observed that loss of GPR182 in zebrafish and mice leads to an increase in the number of myeloid cells. Furthermore, we found, via drug screening and transcriptome analysis, that GPR182 regulates the leukotriene biosynthesis pathway. Overall, these data indicate that GPR182 negatively regulates definitive hematopoiesis and myelopoiesis in part through modulation of leukotriene biosynthesis.
Results
gpr182 Is Highly Expressed in Endothelial Cells
To understand the role of gpr182 during development, we first examined its expression pattern by WISH using zebrafish embryos at different stages (Figure 1A–I). gpr182 appears to be specifically expressed in blood vessels in embryos at 30, 48, and 60 hpf (Figure 1A, D, G). Interestingly, gpr182 exhibits a heterogeneous expression pattern between different vascular beds and different stages. To pinpoint the spatiotemporal expression pattern of gpr182, we performed WISH for gpr182 and immunofluorescent (IF) staining for enhanced green fluorescent protein (EGFP) on Tg(fli1a: EGFP) embryos which express EGFP specifically within ECs.16 As observed in whole embryos (Figure 1A–I) as well as on sections (Figure 1J–Q), at 30 and 48 hpf, gpr182 appears strongly expressed in the vicinity of the VDA (red arrows) and PCV (yellow arrows) in the trunk (Figure 1B, E, J, K, L, M) and in the caudal vein plexus (CVP) in the tail (Figure 1C, F, N, O, P, Q). By 60 hpf, the expression level of gpr182 is decreased in the trunk (Figure 1H) and maintained in the tail (Figure 1I). Using another EC reporter line, TgBAC(etsrp:EGFP), we sorted ECs and non-ECs and performed quantitative reverse transcription (qPCR) for gpr182 (Figure 1R). Consistent with WISH data, gpr182 is specifically expressed in ECs (Figure 1S).
Figure 1.
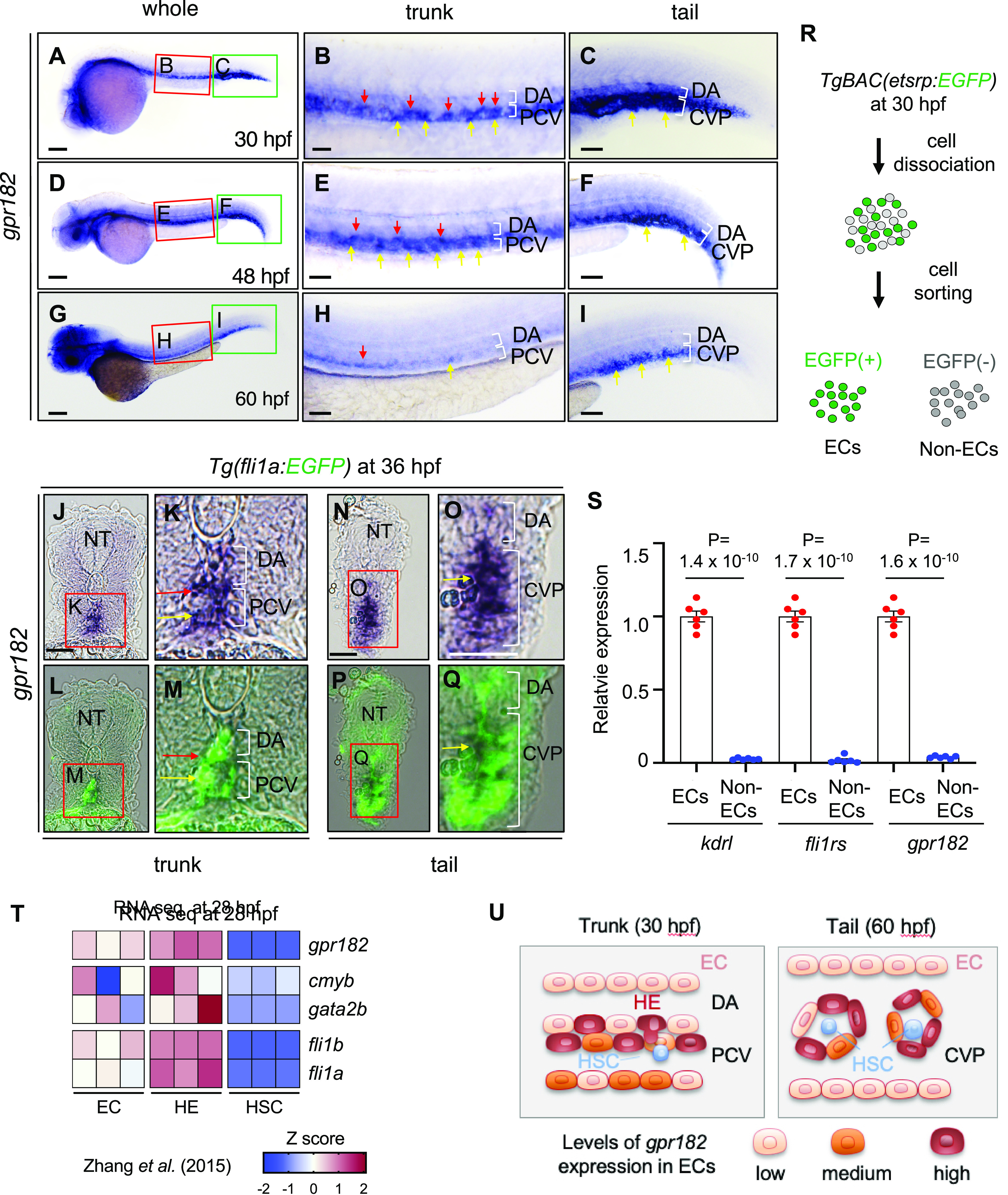
gpr182 is highly expressed in endothelial cells in zebrafish. (A–I) Brightfield images of whole-mount in situ hybridization (WISH) for gpr182 expression at 30 (A–C), 48 (D–F), and 60 (G–I) hpf. The red and green boxes in the left panels (A, D, G) are enlarged in the middle (B, E, H) and right (C, F, L) panels, respectively. Red and yellow arrows point to cells exhibiting strong expression of gpr182 in the ventral part of the DA (red) and in the PCV (yellow), respectively. Anterior to the left, dorsal to the top. (J–Q) Images of sectioned embryos after WISH for gpr182 expression in 36 hpf Tg(fli1a:EGFP) animals in the trunk (J–M) and tail (N–Q) region. The red boxes in panels J, K, N, and O are enlarged in their respective bottom panels (L, M, P, Q). Red and yellow arrows point to cells exhibiting strong expression of gpr182 in the ventral part of the DA (red) and in the PCV (yellow), respectively. (R) Schematic representation of EC sorting from 30 hpf TgBAC(etsrp:EGFP) embryos. (S) qPCR analysis of kdrl, fli1rs, and gpr182 mRNA expression levels in isolated ECs and non ECs from 30 hpf TgBAC(etsrp:EGFP) embryos. N = 6 biologically independent samples. A delta delta Ct (ΔΔCt) analysis was performed and EC expression levels were set at 1. Data are mean ± s.d., and a two-tailed Student’s t test was used to calculate P values. The threshold cycle (Ct) values are in Table S2. (T) Heatmap analysis of gpr182 expression in nonhemogenic ECs (kdrl+/runx1–), specified HECs (HE, kdrl+/runx1+), and potential HSCs (kdrl–/runx1+) sorted from 28 hpf Tg(kdrl:mCherry/runx1:EGFP) embryos.18 Heatmap was generated according to z-score of reads per kilobase per million reads (RPKMs) of each gene in multiple samples. RPKM and z-scores are summarized in Figure S1(A). (U) Schematic illustration showing gpr182 expression in ECs in the trunk (30 hpf) and tail (60 hpf) region. Scale bars, 200 μm (A, D, G), 50 μm (B, C, E, F, H, I, J–Q). CVP, caudal vein plexus; DA, dorsal aorta; ISV, intersegmental vessel; NC, notochord; NT, neural tube; PCV, posterior cardinal vein.
The HE emerges within the VDA around 30 hpf and undergoes a process called endothelial to hematopoietic transition (EHT) to give rise to HSCs.17 In this process, HE cells extrude from the VDA and migrate to the CHT. On the basis of the expression pattern of gpr182 in the trunk and tail, we hypothesized that gpr182 was highly enriched in the HE. To test this hypothesis, we used previously reported RNA seq data to assess the levels of gpr182 expression in the HE (kdrl+/runx1+), nonhemogenic EC (kdrl+/runx1-), and HSC (kdrl-/runx1+) sorted from Tg(kdrl:mCherry);Tg(runx1:EGFP) embryos at 28 hpf.18 Interestingly, gpr182 is highly enriched in the HE (Figure 1T, U, Figure S1A). These data support a role for gpr182 in definitive hematopoiesis during zebrafish development.
Zebrafish gpr182 Mutant Embryos Exhibit Wild-Type-Like Morphology, Vascular Development, and Blood Circulation
To understand the role of GPR182 in zebrafish, we generated gpr182 mutants (gpr182–/–) using the CRISPR-Cas9 technology.19,20 We designed guide RNAs (gRNAs) targeting the region that encodes the seventh transmembrane domain of Gpr182 (Figure 2A) and identified an allele carrying an 11 nucleotide deletion and predicted to encode a protein lacking the seventh transmembrane and intracellular domains (Figure 2B, Figure S1B). High resolution melt analysis confirmed the genotype of gpr182–/– zebrafish (Figure 2C).
Figure 2.
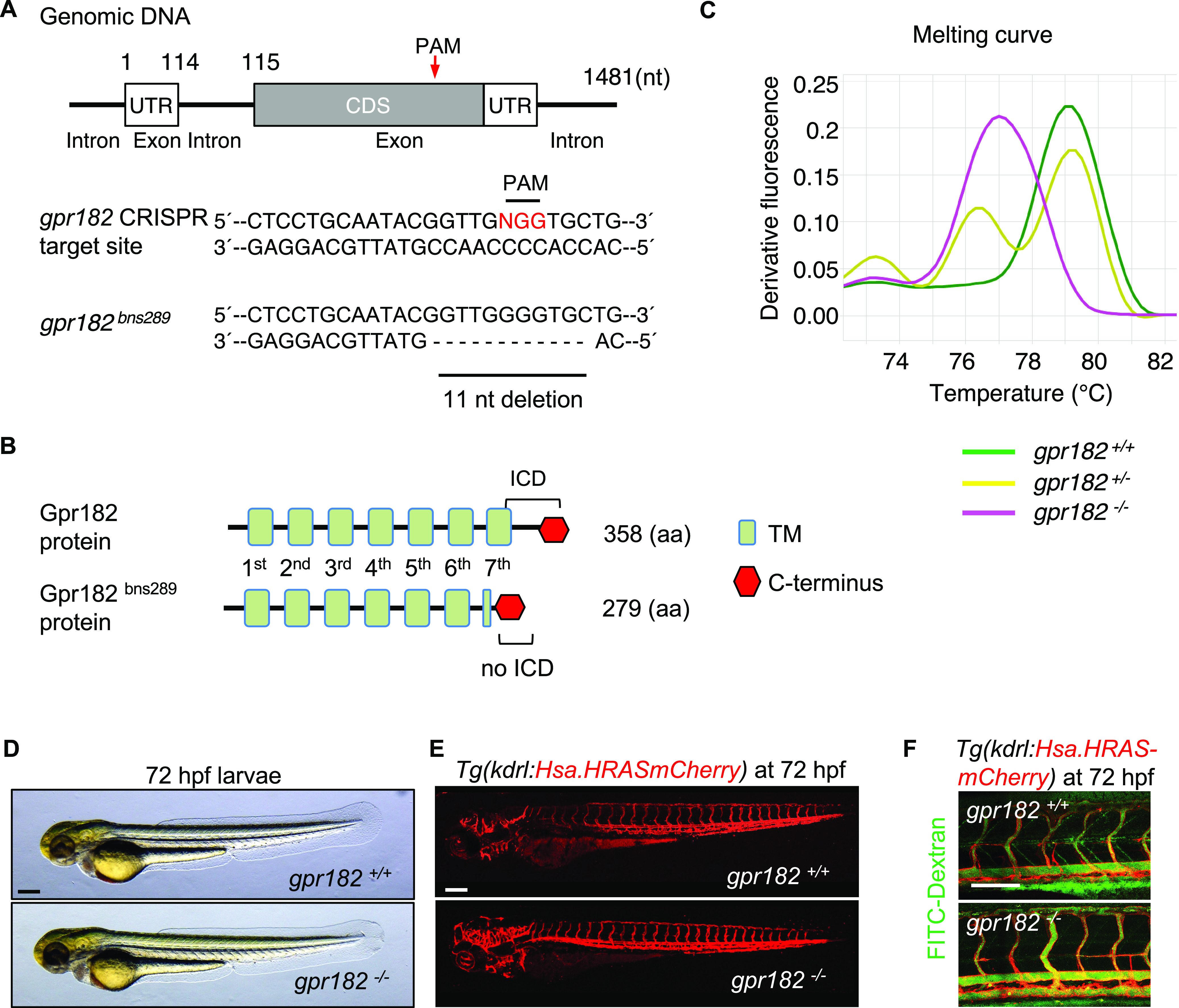
Zebrafish gpr182 mutant embryos exhibit wild-type-like vascular development. (A) Partial DNA sequence of the gpr182–/– allele (bns289) generated for this study. Red arrow points to the mutated region. (B) Schematic representation of wild-type and mutant Gpr182. Green boxes indicate the transmembrane domains (TM). Red hexagon indicates C-terminus. (C) High-resolution melt analysis (HRMA) of gpr182+/+, gpr182+/−, and gpr182–/– DNA. (D) Representative brightfield images of 72 hpf wild-type and gpr182–/– larvae. (E) Confocal images of 72 hpf Tg(kdrl:Hsa.HRASmCherry) wild-type and gpr182–/– larvae. (F) Microangiography of 72 hpf Tg(kdrl:Hsa.HRASmCherry) wild-type and gpr182–/– larvae injected intravascularly with 2000 kDa FITC-dextran; lateral views. Scale bars, 200 μm (D, F), 50 μm (F). Anterior to the left, dorsal to the top.
gpr182–/– embryos exhibit a wild-type morphology and develop into adulthood without any obvious defects (Figure 2D, Figure S1C). Using confocal microscopy on an endothelial reporter line, Tg(kdrl:Hsa.HRASmCherry), we found that gpr182–/– larvae display a wild-type like vascular morphology (Figure 2E). Considering that blood flow is critical for definitive hematopoiesis,21 we next examined blood circulation in gpr182–/– embryos using microangiography by injecting FITC-dextran into blood vessels. No circulation defect was observed in gpr182 mutants compared to wild-type (Figure 2F).
Gpr182 Mediates Developmental Hematopoiesis via Regulation of HE/HSC Formation and Myeloid Cell Differentiation
Since gpr182 is highly expressed in the HE at 30 hpf (red arrows in Figure 1B, E, L, M, T), we investigated its role by comparing the transcriptome of wild-type versus gpr182–/–Tg(kdrl:Hsa.HRASmCherry) positive ECs at 30 hpf (Figure 3A, B). For this analysis, we used the Database for Annotation, Visualization and Integrated Discovery (DAVID) bioinformatics resource, which allowed us to delineate the biological significance of gene changes due to loss of Gpr182 function.22 From this analysis, we found that gpr182–/– ECs exhibit an increased gene signature for the innate immune response, immune system processes, stress response, and chemotaxis. Importantly, gpr182–/–ECs also exhibited an increased expression of genes associated with definitive hematopoiesis and myeloid cell differentiation compared to wild-type ECs (Figure 3B). Conversely, genes related to blood circulation, regeneration, and cardiac conduction were down-regulated in gpr182–/– ECs compared to wild-type ECs (Figure 3B).
Figure 3.
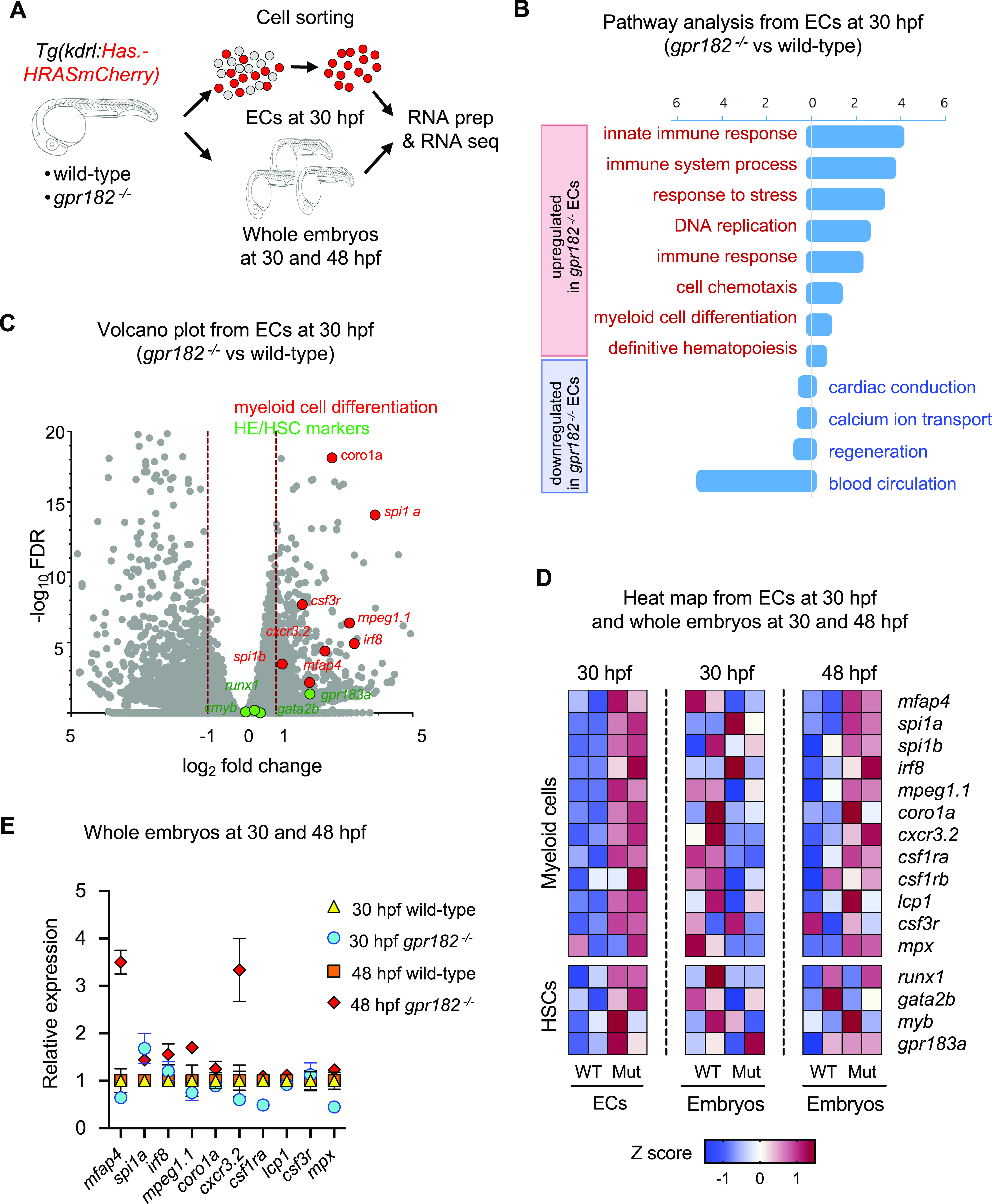
Transcriptomic analysis suggests an increase in definitive myelopoiesis in zebrafish gpr182 mutant embryos. (A) Schematic representation of the transcriptomic analysis. (B, C) Pathway analysis (B) and volcano plot (C) from RNA seq analysis of sorted ECs from 30 hpf wild-type and gpr182–/– embryos. (D) Heatmap analysis of ECs from 30 hpf wild-type and gpr182–/– embryos, and from whole embryos at 30 and 48 hpf. Heatmap was generated according to z-score of reads per kilobase per million reads (RPKMs) of each gene in multiple samples. RPKM and z-scores are summarized in Table S3. (E) Relative expression of myeloid markers in wild-type and gpr182–/– embryos at 30 and 48 hpf.
Considering that gpr182 expression is enriched in the HE, we were particularly interested in gpr182–/– EC-specific hematopoiesis-related gene signatures, especially those relating to myeloid cell differentiation and definitive hematopoiesis. We found that gpr182–/– ECs exhibited an increased expression of macrophage markers such as mfap4, mpeg1.1, cxcr3.2, and csf3r as well as of genes encoding transcription factors that regulate myeloid differentiation such as spi1a, spi1b, and irf8 compared to wild-type ECs (Figure 3C, D). Further, gpr182–/–ECs exhibited an increased expression of HE/HSC markers such as runx1, gata2b, and cmyb compared to wild-type ECs (Figure 3C, D). Altogether, these data suggest that Gpr182 regulates myeloid cell differentiation and HE/HSC formation during zebrafish development.
Interestingly, while we did not observe a difference in myeloid marker gene expression between wild-type and gpr182–/– embryos at 30 hpf, we did observe an increase at 48 hpf in gpr182–/– embryos compared to wild-type (Figure 3D, E). These data, together with the EC-specific transcriptomic data, suggest that ECs from gpr182–/– embryos, especially those within the HE, exhibit an increased potential to differentiate into myeloid cells at 48 hpf.
Zebrafish gpr182 Mutant Embryos Exhibit an Increased Number of HE/HSCs in the Ventral Wall of the Dorsal Aorta
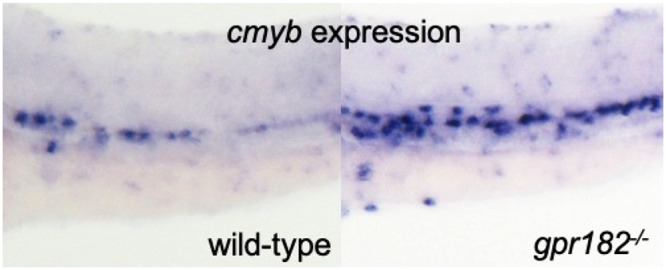
Next, we wanted to examine Gpr182 regulation of HE/HSC formation in more detail. First, we visualized HE/HSCs by performing WISH for the HE/HSC marker cmyb. We found that gpr182–/– embryos exhibit an increase in cmyb expression in the trunk compared to wild-type (white arrows, Figure 4A). In addition, consistent with the WISH data, we observed a 1.36 fold increase in cmyb mRNA expression levels in gpr182–/– embryos compared to wild-type using qPCR (Figure 4B). Next, we performed confocal imaging using a HE/HSC reporter line, Tg(cd41:EGFP); Tg(kdrl:Hsa.HRASmCherry), at 50 hpf. We found that gpr182–/– embryos exhibit a 1.75 fold increase in the number of cd41/kdrl double-positive HE/HSC cells (WT:22.1; gpr182–/–: 38.84) in the trunk (white arrows, Figure 4C, D) compared to wild-type. Since HE/HSCs form in the VDA and migrate to the CHT after an endothelial-to-hematopoietic transition (EHT),7,23 we examined HSCs in the CHT after 72 hpf and observed an increase in the number of HSC in the CHT of gpr182–/– compared to wild-type using both WISH (Figure 4E) and confocal imaging (Figure 4F, G). These data suggest that gpr182 negatively regulates HE/HSC formation during development.
Figure 4.
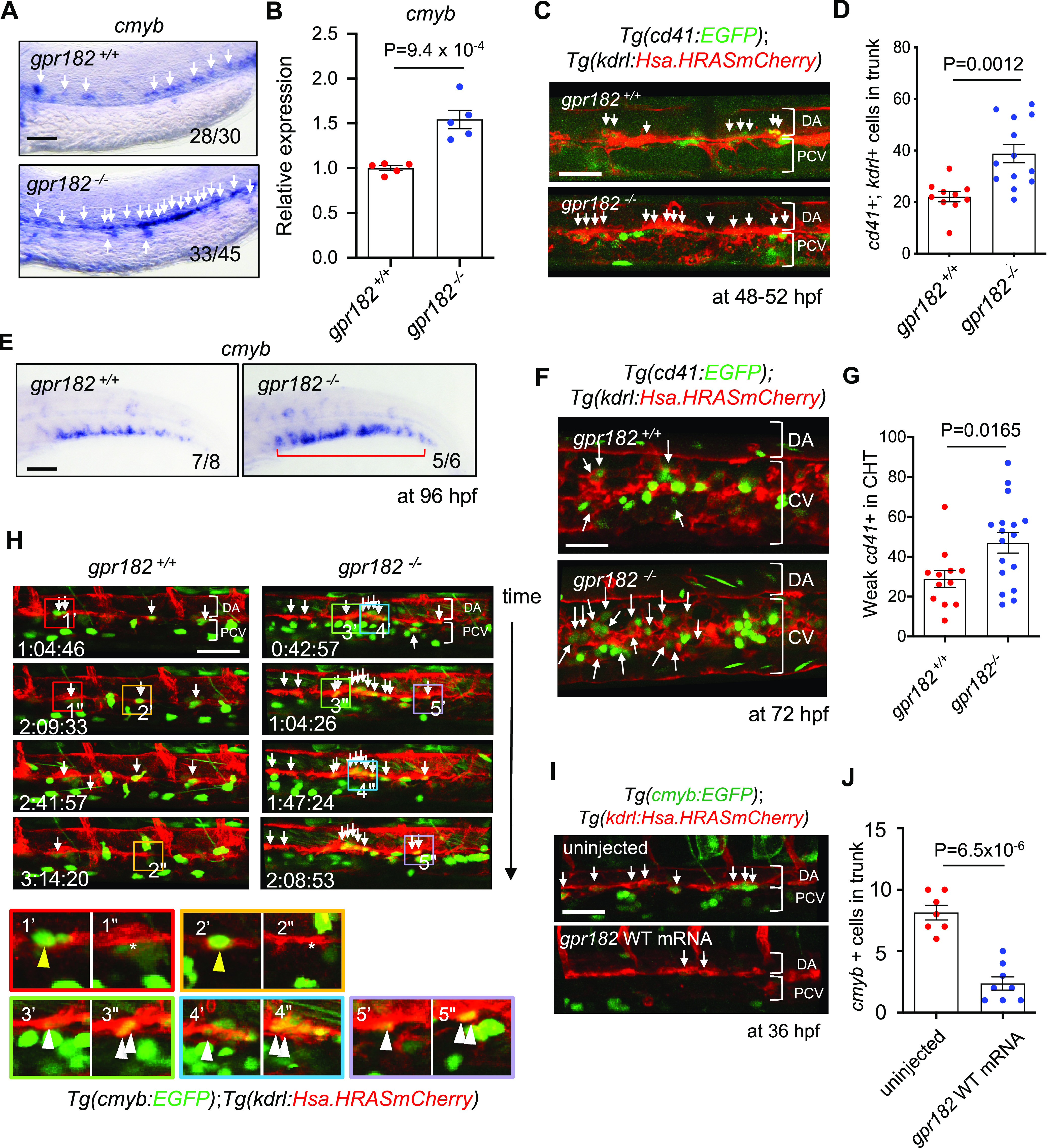
Zebrafish gpr182 mutant embryos exhibit increased HE and HSC formation. (A) Brightfield images of WISH for cmyb expression in wild-type and gpr182–/– embryos at 36 hpf. White arrows point to cmyb positive cells in the trunk. N/N, number of embryos showing representative phenotype/total number of embryos examined. Two independent experiments were performed with similar results. (B) qPCR analysis of cmyb mRNA levels from wild-type and gpr182–/– embryos at 36 hpf. N = 5 biological replicates. A delta delta Ct (ΔΔCt) analysis was performed and wild-type expression levels were set at 1. Data are mean ± s.d., and a two-tailed Student’s t test was used to calculate P values. (C) Confocal images of Tg(cd41:EGFP); Tg(kdrl:Hsa.HRASmCherry) wild-type and gpr182–/– embryos in the trunk at 48–52 hpf. White arrows point to cd41/kdrl double-positive cells in the trunk. (D) Number of cd4l/kdrl double-positive cells in the trunk (six somites). Wild-type N = 10, gpr182–/–N = 13. N obtained from three independent clutches. (E) Brightfield images of WISH for cmyb expression in wild-type and gpr182–/– larvae at 96 hpf. N/N, number of embryos showing representative phenotype/total number of embryos examined. Two independent experiments were performed with similar results. (F) Confocal images of Tg(cd41: EGFP); Tg(kdrl: Hsa.HRASmCherry) wild-type and gpr182–/– larvae in the tail at 72 hpf. White arrows point to weak EGFP positive HSCs in the tail. (G) Number of weak cd41:EGFP positive HSCs in the tail (4 somites) of wild-type and gpr182–/– embryos. Wild-type (N = 12) and gpr182–/– (N = 17), from three independent clutches. (H) Time-lapse confocal images of Tg(cmyb:GFP); Tg(kdrl:Hsa.HRASmCherry) wild-type and gpr182–/– embryos at 36 hpf in the trunk. White arrows point to cmyb/kdrl double-positive HE/HSCs. The red, orange, green, blue and purple boxes in the above panels are enlarged in the bottom panels, respectively. Yellow (1′–2′′) and white (3′–5′′) arrowheads in the bottom panels point to HE/HSCs of the wild-type and gpr182–/– embryo, respectively. (I) Confocal images of Tg(cmyb:GFP); Tg(kdrl:Hsa.HRASmCherry) uninjected and gpr182 mRNA injected embryos at 36 hpf. White arrows point to cmyb/kdrl double positive cells in the trunk. (J) Quantification of cmyb/kdrl double-positive cells in the trunk (six somites). Uninjected embryos (N = 7) and gpr182 wild-type mRNA injected embryos (N = 8), from three independent clutches. Data are mean ± s.d.. A two-tailed Student’s t test was used to calculate p-values. Scale bars, 50 μm. Anterior to the left, dorsal to the top. DA, dorsal aorta; CV, caudal vein; PCV, posterior cardinal vein; VDA, ventral wall of DA.
Since we observed an increase in the number of cd41/kdrl double-positive cells in the VDA during EHT in gpr182–/– embryos compared to wild-type (white arrows, Figure 4C, D), we were interested in investigating how Gpr182 regulates HE/HSC formation at the cellular level. Thus, we performed time-lapse confocal imaging using another HSC reporter line, Tg(cmyb:EGFP); Tg(kdrl:Hras-mChrry), starting at 36 hpf. Notably, we observed within the VDA during EHT an increased number of HE/HSCs in gpr182–/– embryos (Figure 4H (3′–5′′), compared to wild-type (Figure 4H (1′–2′′)). Furthermore, we observed a significant reduction in HE/HSC number in the trunk of embryos injected with gpr182 mRNA compared to uninjected embryos (Figure 4 I, J). Taken together, these data support the hypothesis that Gpr182 negatively regulates definitive hematopoiesis by inhibiting HE/HSC formation.
Zebrafish gpr182 Mutant Embryos Exhibit an Increased Number of Myeloid Cells
Due to the increase in myeloid differentiation marker gene expression observed in gpr182–/– ECs (Figure 3B, C, D), we investigated whether gpr182–/– embryos had an increase in myeloid cell numbers compared to wild-type. First, using the Tg(mpeg1:mCherry) macrophage reporter line, we found at 60 hpf a 1.59 fold increase in average macrophage numbers (wild-type, 31; gpr182–/–, 49.33) in gpr182–/– embryos compared to wild-type (white arrows, Figure 5A, B). Second, using the Tg(lyz:EGFP) neutrophil reporter line, we found at 72 hpf a 1.5 fold increase in average neutrophil numbers (wild-type, 51; gpr182–/–, 76.69) in gpr182–/– larvae compared to wild-type (white arrows, Figure 5C, D). Taken together, these data, along with the observation of increased myeloid marker gene expression in gpr182–/– embryos at 48 but not 30 hpf (Figure 3D, E), support the hypothesis that gpr182 negatively regulates myeloid cell differentiation.
Figure 5.
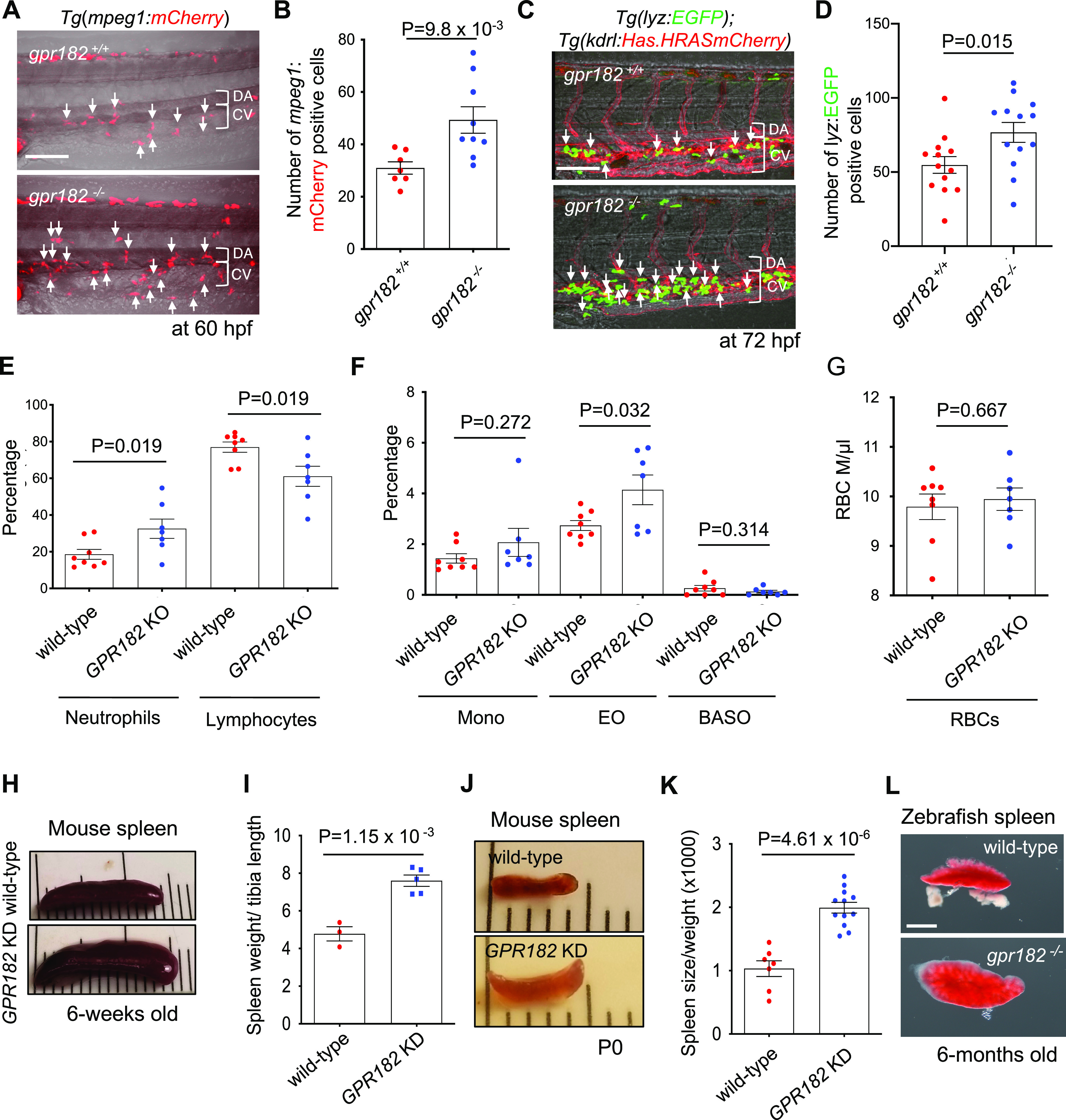
Zebrafish and mouse gpr182 mutants exhibit an increased number of myeloid cells. (A) Confocal images of 60 hpf Tg(mpeg1:mCherry) wild-type and gpr182–/– embryos in the tail. White arrows point to mpeg1:mCherry positive cells. (B) Number of mpeg1:mCherry positive cells in tail (six somites). Wild-type (N = 7), gpr182–/–(N = 9), from three independent clutches. (C) Confocal images of 72 hpf Tg(lyz:EGFP); Tg(kdrl:Hsa.HRASmCherry) wild-type and gpr182–/– larvae in the tail. White arrows point to lyz:EGFP positive cells. (D) Number of lyz:EGFP positive cells in the tail (six somites). Wild-type embryos (N = 13), gpr182–/– embryos (N = 13), from three independent clutches. (E–G) Whole blood analysis of 6-week old C57/B6 wild-type (N = 8) and GPR182 KO (N = 7) mice. (H) Bright-field images of 6-weeks old wild-type and GPR182 KD mouse spleens. (I) Number of wild-type (N = 3) and GPR182 KD (N = 5) mouse spleen sizes. (J) Bright-field images of P0 wild-type and GPR182 KD mouse spleens. (K) Quantification of P0 wild-type (N = 7) and GPR182 KD (N = 11) mouse spleens. Spleen size was normalized by body weight and P0 wild-type spleen size was set at 1. Data are mean ± s.d. and a two-tailed Student’s t test was used to calculate p-values. (L) Bright-field images of 6-months old adult wild-type and gpr182–/– zebrafish spleen (see Figure S1D). Scale bars, 50 μm (A, C), 1 mm (D). BASO, basophils; CHT, caudal hematopoietic tissue; CV, caudal vein; DA, dorsal aorta; EO, Eosinophils; KD, knockdown; KO, knockout; MONO, monocytes; PCV, posterior cardinal vein; RBCs, red blood cells.
GPR182 Regulation of Hematopoiesis Is Also Observed in Mice
Next, we wanted to investigate whether the physiological role of Gpr182 in hematopoiesis is conserved in higher vertebrates. To this end, we first used a previously described genetically engineered mouse model that has a complete loss of GPR182 function (GPR182 KO).24 On the basis of our zebrafish gpr182–/– data (Figure 3B–E, 5A–D), we hypothesized that GPR182 regulates myeloid cell differentiation, and predicted that the proportion of myeloid cells in the whole blood of wild-type mice would be significantly different than that in GPR182 KO mice. As predicted, we found an increase in myeloid cells, especially neutrophils, in GPR182 KO adult mice compared to wild-type (WT, 18.57%; N = 8; GPR182 KO, 32.52%; N = 7). Conversely, we found a decrease in lymphocytes in GRP182 KO mice compared to wild-type (WT, 76.98%; N = 8; GPR182 KO, 61.1%; N = 7) (Figure 5E). While we also observed subtle changes in the number of monocytes, basophils, and RBCs in GPR182 KO mice compared to wild-type, these differences were not significant (Figure 5F, G, Table S4). Taken together, these data support a role for GPR182 in the differentiation of myeloid cells in adult mice. Furthermore, these data indicate that the physiological role of Gpr182 in definitive hematopoiesis is conserved between zebrafish and mice.
Previously, Kelchele et al. reported that GPR182 KO adult mice exhibit an enlarged spleen.24 Thus, considering the role of GPR182 in hematopoiesis and the fact that the spleen is a major hematopoietic organ, we examined the spleen from GPR182 knockdown (KD) mice, which have an 85% reduction in Gpr182 mRNA, as well as from gpr182 mutant zebrafish. As Kelchele et al. observed, we found that adult and P0 GPR182 KD mice exhibit an increase in spleen size compared to wild-type (adult spleen: 1.6 fold; wild-type. N = 3; KD, N = 5. P0 pup spleen: 1.93 fold; wild-type, N = 7; KD, N = 11) (Figure 5H–K). Consistent with these observations in mice, 6-months old gpr182–/– adult zebrafish also exhibit an increase in spleen size compared to wild-type (Figure 5L and Figure S1D). These data further support the conclusion that the physiological role of GPR182 in hematopoiesis is conserved between zebrafish and mice.
GPR182 Regulates Hematopoiesis via Induction of the Leukotriene Biosynthesis Pathway
After finding a physiological role for GPR182 in definitive hematopoiesis, we next sought to define the molecular mechanism for the GPR182 function. Considering that GPR182 is an orphan receptor, we sought to modulate GPR182 activity by treating a stable cell line expressing a tagged version of human GPR182 (TANGO system) with a small molecule library (Figure 6A, Figure S2A,B).25 Of the 820 bioactive lipid compounds screened in duplicate (trial 1 and 2), we selected 11 potential hits that activated the hGPR182-TANGO system (Figure 6B, Figure S2C). These 11 hits were rescreened using the TANGO assay, from which, two compounds, MS-275 (HDAC inhibitor) and ibuprofen (cyclooxygenase inhibitor), were found to consistently activate the hGPR182-TANGO system (Figure 6B, Figure S2D).
Figure 6.
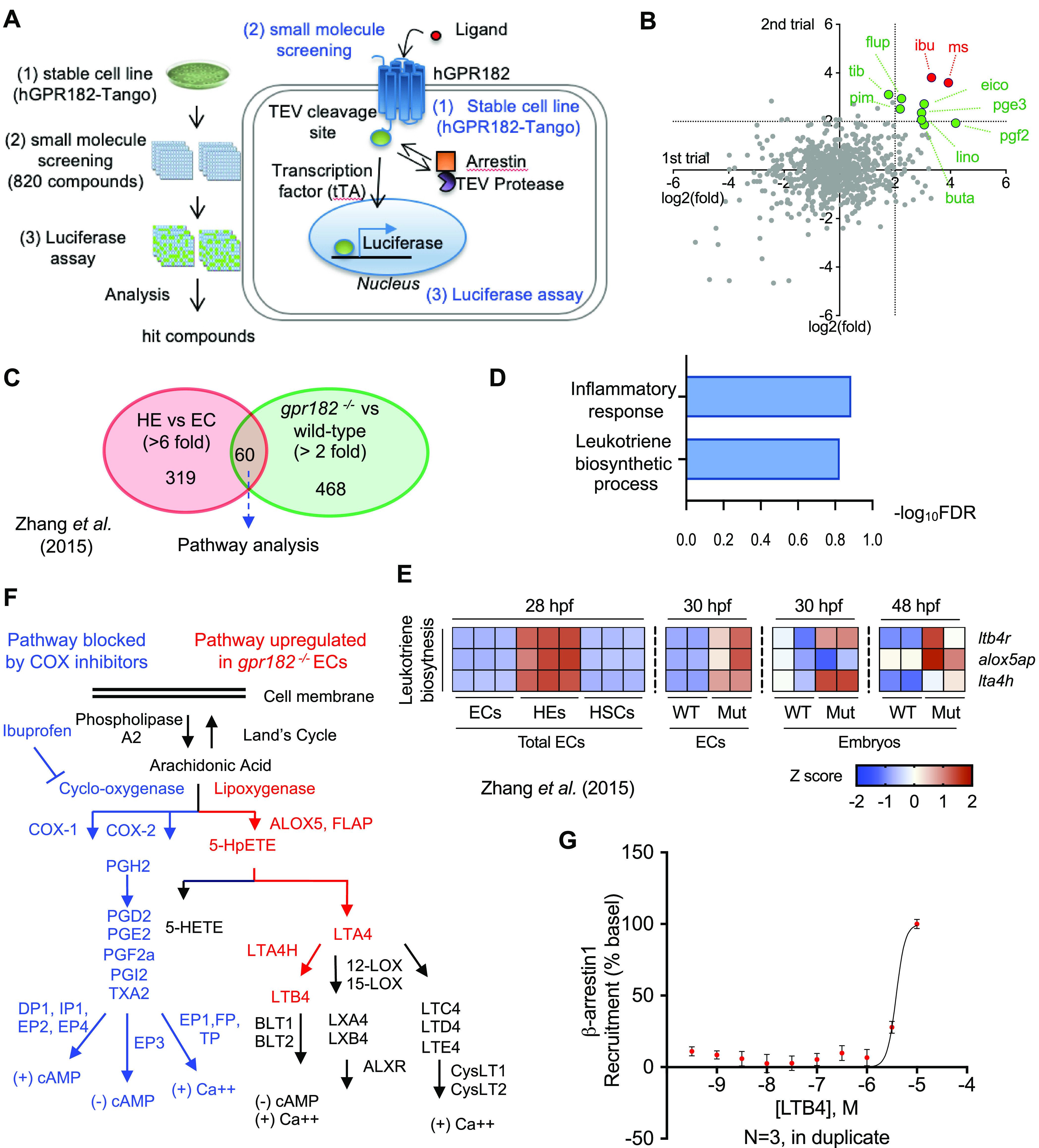
Activation of the leukotriene biosynthesis pathway by GPR182. (A) Schematic illustration of the human GPR182-TANGO (hGPR182-TANGO) system and associated small molecule screening. (B) Differential hGPR182-TANGO activation by small molecules tested during the primary screen. A total of 820 compounds from bioactive lipid small-molecule libraries were screened in duplicate. Dashed lines indicate the 2-fold (log2) ratio. Negative control (1% DMSO). (C) Venn diagram showing genes highly expressed in HE (red circle) and gpr182–/– ECs (green circle). (D) Pathway analysis of selected genes from panel C. (E) Heat-map analysis of leukotriene biosynthesis pathways in wild-type and gpr182–/– embryos at 30 and 48 hpf. (F) Schematic illustration of prostaglandin and leukotriene biosynthesis pathways. Pathways blocked by ibuprofen marked in blue; pathways upregulated in gpr182–/– ECs marked in red. (G) β-arrestin-1 recruitment assay. Best fit calculated by a nonlinear regression with four parameters and variable slope, ± S.E.M., N = 3 biological replicates. Curves and statistical significance were determined by nonlinear regression with a comparison of fits (F-test).
Since ibuprofen is known to inhibit cyclooxygenase (COX), we wanted to investigate how ibuprofen activates hGPR182. To address this question, we treated our hGPR182-TANGO stable cell line with a broad COX inhibitor, indomethacin, which resulted in the induction of reporter signal (>7 fold) compared to DMSO-treated control cells (Figure S2E,F). These data show that blocking COX activity with indomethacin leads to the activation of the hGPR182-TANGO reporter.
To understand why COX inhibition resulted in hGPR182-TANGO activation, we returned to our zebrafish gpr182–/– model. Specifically, we examined signaling pathways that were upregulated in gpr182–/– ECs compared to wild-type ECs. We hypothesized that the signaling pathways upregulated in gpr182–/– ECs were upregulated to compensate for the loss of GPR182 function. To narrow down the list of candidates, we first selected highly enriched genes (319 genes, >6 fold) in HEs compared to ECs and selected 60 overlapping genes among those (468 genes, >2 fold) that were also found to be increased in gpr182–/– ECs (Figure 6C).18 Next, using DAVID pathway analysis,22,26 we identified the upregulation of the leukotriene biosynthesis pathway in zebrafish gpr182–/– ECs (Figure 6D, E, Figure S3A, B).
Consistent with a conserved role between zebrafish and mice, we found an increase in Lta4h mRNA in the bone marrow and spleen isolated from GPR182 KD mice compared to wild-type (data not shown). Interestingly, both the cyclooxygenase and leukotriene biosynthesis (lipoxygenase) pathways are downstream of arachidonic acid metabolism and are inextricably tied to the inflammatory response (Figure 6F).27 These results link GPR182 cellular functions to arachidonic acid metabolism whereby COX inhibition promotes GPR182-TANGO activation, and loss of GPR182 function promotes upregulation of leukotriene biosynthesis (Figure 6F, Figure S2D, E). In addition, LTB4 treatment of HEK293T cells overexpressing GPR182 resulted in β-arrestin-1 recruitment at high concentrations (>1 μM), suggesting that LTB4 activates GPR182 signaling via a yet to be identified autocrine and/or paracrine mechanism (Figure 6G). Taken together, these data suggest that GPR182 regulates hematopoiesis and the inflammatory response through modulation of leukotriene biosynthesis. Future work will investigate the molecular mechanisms that govern GPR182 regulation of the leukotriene biosynthesis pathway.
Discussion
Previous reports have shown that GPR182 is expressed in endothelial cells in mice and humans. In this study, we validate these results and also find gpr182 expression to be enriched in the HE during zebrafish development (Figure 1B, E, T, U). Since the physiological role of GPR182 during development was unknown, we isolated gpr182–/– ECs using the Tg(kdrl:HsaHRAS-mCherry) EC reporter line at 30 hpf and performed transcriptomic analysis (Figure 3A). We found that the expression of genes related to definitive hematopoiesis and myelopoiesis were increased in gpr182–/– ECs compared to wild-type (Figure 3B–D). Further, due to an observed increase in myeloid marker gene expression in ECs, we hypothesized that the loss of GPR182 function increases the potential for myeloid cells to differentiate from HE/HSCs. Consistent with this hypothesis, we observed an increase in myeloid marker gene expression in zebrafish gpr182–/– embryos at 48 hpf (Figure 3D, E). These data suggest that GPR182 regulates myeloid differentiation in an HE/HSC dependent manner.
Importantly, we show that the role of GPR182 in hematopoiesis is conserved between zebrafish and mice. Specifically, whole blood isolated from GPR182 KO mice exhibited an increase in neutrophils and a decrease in lymphocytes compared to wild-type (Figure 5E). These data support a model suggesting that GPR182 promotes HSC differentiation into myeloid rather than lymphoid cells. It was recently reported that sinusoidal endothelial cells are important for HSC differentiation during regeneration in vivo(28) as well as HSC expansion in vitro.29 Interestingly, GPR182 is known to be specifically expressed in sinusoidal endothelial cells in the spleen, lymph node, and the bone marrow in humans.30 These data indicate that GPR182 expressed in sinusoidal endothelial cells of the bone marrow regulates HSC differentiation, and it will be interesting to investigate further the underlying mechanisms.
In this study, we also establish a novel link between GPR182 and inflammation. Specifically, we found that loss of GPR182 in zebrafish ECs and mouse bone marrow resulted in the increased expression of inflammatory signals (Figures 3B and 6E, data not shown). Inflammatory signals, such as TNF-α and interferons, are essential for hematopoiesis in the adult bone marrow and in zebrafish embryos.31−35 However, it is unknown how HE/HSCs recognize inflammatory signals to maintain homeostasis. To this end, we identified a novel link between GPR182 and the leukotriene biosynthesis pathway (Figure 6). Leukotriene B4 (LTB4) is a lipid metabolite produced by the leukotriene biosynthesis pathway that mediates inflammatory signals in response to bacterial infection and/or inflammatory conditions.36,37 Importantly, LTB4 has been previously linked to the regulation of hematopoiesis;38 however, the underlying molecular mechanisms are unknown. Here, we provide evidence for a novel link between GPR182 expressed in HE/HSCs and LTB4 synthesis. Considering this link between GPR182, hematopoiesis, and inflammation, future studies might investigate the molecular mechanisms by which GPR182 signaling regulates leukotriene biosynthesis as well as the role GPR182 plays in acute versus chronic models of inflammation.
In summary, we have generated a zebrafish gpr182 mutant and characterized the role of Gpr182 in HE/HSC formation as well as in myeloid cell differentiation. Importantly, we confirmed that the physiological function of GPR182 in hematopoiesis is conserved between zebrafish and mice. This observation underscores the importance of GPR182 in HSC formation and hematopoiesis. Further, we show that GPR182 functions as a negative regulator of definitive hematopoiesis to maintain inflammatory homeostasis via regulation of leukotriene biosynthesis. Identification of a role for GPR182 in definitive hematopoiesis and inflammation highlights it as a putative therapeutic target for the treatment of blood related pathologies including leukemia.
Material and Methods
Study Approval
All zebrafish (Danio rerio) husbandry was performed under standard conditions in accordance with institutional (Max Planck Gesellschaft) and national ethical and animal welfare guidelines approved by the ethics committee for animal experiments at the Regierungspräsidium Darmstadt, Germany. In addition, all animal experiments performed at the University of North Carolina at Chapel Hill were approved by the UNC Institutional Animal Care and Use Committee. Animals were housed in an AAALAC-accredited facility in compliance with the Guide for the Care and Use of Laboratory Animals guide as detailed on protocols.io (dx.doi.org/10.17504/protocols.io.baenibde).
Zebrafish
AB, Tg(fli1a:EGFP)y1 (ref (16)), TgBAC(etsrp:EGFP)ci1 (ref (39)), Tg(kdrl:Hsa.HRASmCherry)s896 (ref (40)), Tg(cd41:EGFP)la2 (ref (41)), Tg(cmyb:GFP)zf169 (ref (42)), Tg(mpeg:mCherry)ump2 (ref (43)), Tg(mpeg:EGFP)gl22 (ref (44)), and Tg(lyz:EGFP)nz117 (ref (44)) fish were used in this study. Embryos were staged by hpf at 28.5 °C.45
Mouse
The Gpr182tm2a(KOMP)Wtsi/+ (knockout first/promoter driven) mice used in this study were created from an embryonic stem (ES) cell clone (EPD0365_4_C08) obtained from the National Center for Research Resources-NIH-supported Knockout Mouse Project (KOMP) repository and generated by the CHORI, Sanger Institute, and UC Davis (CSD) Consortium for the NIH-funded KOMP.46 The CSD-targeted allele has been previously published.51,52 To generate GPR182 knockout (KO) mice, Gpr182tm2a(KOMP)Wtsi/+ mice were crossed with the B6.C-Tg(CMV-cre)1Cgn/J Tg mouse line expressing Cre recombinase ubiquitously (The Jackson Laboratory; stock no. 006054). To generate GPR182 knockdown (KD) mice, heterozygous Gpr182lacZ/+ mice were incrossed. Homozygous Gpr182 lacZ/lacZ mice showed the reduction of Gpr182 mRNA level compared to wild-type, as described in a previous publication.24 Thus, we used homozygous Gpr182LacZ/LacZ mice as GPR182 KD mice. All Gpr182-associated mouse lines were maintained on an isogenic C57BL/6 background. The genotyping primers used are listed in Table S1.
Generation of Zebrafish gpr182 Mutants
The gpr182 mutant line (gpr182bns289) was generated using the CRISPR-Cas9 system as previously described.19,20 pT7-gRNA and pT3TS-nlsCas9nls vectors were purchased from Addgene. A gRNA was designed to target the gpr182 exon using the CRISPR design tool (http://crispr.mit.edu/). Cas9 mRNA (100 pg) and a gRNA targeting the gene of interest (50 pg) were coinjected into zebrafish embryos at the one-cell stage. Mutant alleles were identified by high-resolution melt analysis (HRMA) of PCR products, allowing one to distinguish between heterozygous, WT, and homozygous mutants.
RNA Sequencing
RNA was isolated from sorted endothelial cells of 30 hpf wild-type sibling and gpr182–/–Tg(kdrl:Hsa.HRASmCherry) embryos as well as from 30 and 48 hpf wild-type and gpr182–/– embryos using the miRNeasy Micro Kit (Qiagen). To avoid contamination with genomic DNA, the samples were treated by on-column DNase digestion (DNase-Free DNase Set, Qiagen). RNA and library preparation integrity were verified with LabChip Gx Touch 24 (PerkinElmer). The RNA amount was adjusted on the number of isolated cells by FACS and approximately 4 ng of total RNA was used as input for SMARTer Stranded Total RNA-Seq Kit-Pico Input Mammalian (Takara Clontech). Sequencing was performed on the NextSeq500 instrument (Illumina) using v2 chemistry, resulting in an average of 27 M reads per library with 1 × 75 bp single end setup. The resulting raw reads were assessed for quality, adapter content, and duplication rates with FastQC.47 Reaper version 13–100 was employed to trim reads after a quality drop below a mean of Q20 in a window of 10 nucleotides.48 Only reads between 30 and 150 nucleotides were cleared for further analyses. Trimmed and filtered reads were aligned versus the Ensembl Zebrafish genome version DanRer10 (GRCz10.87) using STAR 2.4.0a with the parameter “-out Filter Mismatch Nover Lmax 0.1” to increase the maximum ratio of mismatches to mapped length to 10%.49 The number of reads aligning to genes was counted with the feature Counts 1.4.5-p1 tool from the Subread package.50 Only reads mapping at least partially inside exons were admitted and aggregated per gene. Reads overlapping multiple genes or aligning to multiple regions were excluded. The Ensemble annotation was enriched with UniProt data (release 06.06.2014) based on Ensemble gene identifiers.51
RNA-Seq Analysis
RNA-seq data were downloaded from the published paper. RSEM upper-quantile-normalized values from Illumina HiSeq_RNASeqV2 from 28 hpf gpr182–/– and wild-type tissue were log2 transformed. Samples with an expression value of 3 or lower were indistinguishable from background values and were thus considered a value of 0. The Bioconductor package edgeR version 3.26.8 was used to compute RPKM.52 Differentially expressed genes between two groups were determined using the R-package DEseq2 and edgeR with a criterion P-value cutoff of 0.05 and fold change cutoff of 2.53 Functional and pathway enrichment analyses were performed using the Database for Annotation Visualization and Integrated Discovery (DAVID; https://david.ncifcrf.gov/).26 The heatmap was generated using the R-package heatmap.2 function in the gplots package according to z-score of RPKMs (reads per kilobase per million reads) of each gene in multiple samples.
Whole-Mount In Situ Hybridization
For whole-mount in situ hybridization (WISH), zebrafish embryos and larvae were fixed in 4% PFA overnight at 4 °C and subsequently dehydrated in methanol and stored at −20 °C until required. Before hybridization, embryos were rehydrated to PBS/0.1% Tween and then digested in 10 mg/mL Proteinase K (Roche) followed by fixation in 4% PFA in PBS/0.1% Tween. Embryos were washed in PBS/0.1% Tween, preincubated in hybridization buffer at 70 °C for 4 h, and then incubated with Dig-labeled RNA probes in hybridization buffer at 70 °C overnight. After washing, the hybridized probes were detected with alkaline-phosphatase conjugated, alkaline-phosphatase-labeled antidigoxigenin antibody (11093274910, Roche, dilution 1:1,000) at 4 °C overnight, and the signal was visualized with BM purple (1144207001, Roche). Probes for gpr182 and cmyb were amplified from cDNA synthesized from total RNA extracted from 24 to 48 hpf zebrafish embryos. Primer information is in Table S1.
In Vivo Imaging and Image Processing
Pigmentation of embryos and larvae was inhibited by 1-phenyl-2-thiourea (Sigma). The embryos were treated with 100 mg/mL tricaine (Sigma), mounted in a drop of 1.0–1.5% low melting agarose in egg water and placed onto a glass-bottom Petri dish (MatTek Corporation, Ashland, Ma). Fluorescence images were obtained using an LSM800 confocal laser scanning microscope (Zeiss), an Olympus Fluoview FV1000 confocal laser scanning microscope (Olympus) or high-end stereoscopic microscopes (Nikon SMZ25). Three-dimensional-rendered z-stack images and three-dimensional surface-rendered images and movies were analyzed and assembled using the IMARIS software (BITPLANE).
Small Molecule Screening
To screen for agonists of GPR182, we used the Tango assay as previously described,24 and the Cayman Bioactive Lipid I Screening Library (reference 10506). Stable HTLA cells expressing hGPR182-TANGO HTLA were seeded on gelatin-coated 96-well plates at 50 000 cells per well in complete DMEM medium containing FBS. After 1 day, the medium was replaced with 100 μL of serum-free DMEM medium containing antibiotics for 4 h. Compounds to be tested, including DMSO controls, were then added directly to the wells at a final concentration of 10 μM, and the cells were further incubated for 16 h at 37 °C–5% CO2. After stimulation, the supernatant was removed and replaced by 100 μL of assay reagent per well (HBSS, Gibco 14025-092; 20 mM HEPES, Gibco 15630056; BrighGloReagentTM, Promega E2620; pH7.4 at room temperature). The plates were incubated in the dark for 20 min at room temperature on an orbital shaker at 400 rpm The emitted luminescence was then measured using the Flexstation 3 device (Molecular Probes).
Whole Blood Analysis
Whole blood samples were obtained by incising the right submandibular vein of anesthetized mice with a sterile 4 mm lancet. Anesthesia was induced by placing each mouse in an inhalation chamber with 4% isoflurane (FORANE, Baxter Healthcare). The volume of each blood sample was approximately 300 μL. For collecting blood, we used the tubes containing EDTA to prevent clotting. After, a complete blood count (CBC) test was performed at the Animal Histopathology and Laboratory Medicine Core (University of North Carolina, Chapel Hill). The results of the CBC test are summarized in Table S4.
β-Arrestin Recruitment Assays
To assay for β-arrestin1 recruitment, HEK293T cells were seeded in 10 cm2 dish and grown overnight. The following day, cells were transfected using calcium phosphate precipitation with GPR182-rLuc (1 μg), β-arrestin-1-YFP (5 μg), and GRK (4 μg). The next day, cells were seeded into a 96-well plate. After 24 h, the media was removed and 80 μL of PBS was added to each well. Then, 10 μL of Coelenterazine h was added to each well and incubated for 10 min in the dark. Finally, titrated concentrations of LTB4 were added to the plate, and the luminescence and fluorescence were read after 30 min. Data were analyzed with a nonlinear curve fit with a variable slope for either log(agonist) or log(antagonist).
Statistics
Statistical analysis was performed using GraphPad Prism 8.2.1 (GraphPad Software), and all data are represented as the mean ± SEM. Statistical significance for paired samples and for multiple comparisons was determined by Student’s t test and one-way analysis of variance with Tukey’s test, respectively. A P-value of less than 0.05 was considered statistically significant.
Acknowledgments
We thank Caron lab members for discussions and/or critical reading of the manuscript; Dr. Jiangdong Liu, Hans-Martin Maischein, Sharon Meaney-Gardian, Dr. Celia Shiau, Marianne Ploch for kind help; Michelle Altemara, Liz Blakeney, and the staff at the zebrafish and mouse facility of the Max Planck Institute for Heart and Lung Research and of UNC at Chapel Hill for excellent the assistance. This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2016R1A6A3A03007406) to H.-B.K.; a grant from the Excellence Cluster Cardio-Pulmonary System (ECCPS) to H.-B.K. and R.B.; the Global Core Research Center Program (GCRC, 2011-0030001), the Midcareer Researcher Program (2018R1A2B6001590) funded by the National Research Foundation of Korea (NRF) to K.-W.K.; by an NIH Grant (F32-HL134279) to D.I.M.; by an NCI Cancer Cell Biology Predoctoral T32 Training Program (T32CA071341) to D.S.S.; by the S.O. and NIH Grants (RO1-DK099156, RO1-HD060860, and R01-HL129086) to K.M.C.; an American Heart Association Innovator Award (16IRG27260077) to K.M.C.; and the Max Planck Society to D.Y.R.S.
Glossary
Abbreviations
- BASO
basophils
- CHT
caudal hematopoietic tissue
- CV
caudal vein
- CVP
caudal vein plexus
- DA
dorsal aorta
- EGFP
green fluorescent protein
- EHT
endothelial hematopoietic transition
- EO
Eosinophils
- GPCR
G-protein coupled receptors
- GPR182
G-protein coupled receptor 182
- hpf
hour post fertilization
- HSC
hematopoietic stem cells
- ICD
intracellular domain
- ISV
intersegmental vessel
- KD
knockdown
- KO
knockout
- LTB4
leukotriene B4
- MONO
monocytes
- NC
notochord
- NT
neural tube
- PCV
posterior cardinal vein
- qPCR
quantitative reverse transcription
- RBCs
red blood cells
- VDA
ventral wall of the dorsal aorta
- WISH
whole-mount in situ hybridization
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsptsci.0c00020.
Primers used for qPCR, genotyping, and WISH probe synthesis; cycle threshold (Ct) expression values of candidate genes obtained via qPCR; expression levels of myeloid and HE/HSC marker genes from the RNA seq data set; results of complete blood count test; additional figures as described in the text (PDF)
Author Contributions
H.-B.K., K.M.C., and D.Y.R.S. designed experiments; H.-B.K., K.-W.K, S.O., K.M.C., and D.Y.R.S. analyzed data; H.-B.K., D.I.M, R.B., A.L, C.S.M.H, T.S., S.G., and D.S.S. conducted experiments; and H.-B.K., K.M.C, and D.Y.R.S. wrote the paper with feedback from all authors.
The authors declare no competing financial interest.
Supplementary Material
References
- Sriram K.; Insel P. A. (2018) G Protein-Coupled Receptors as Targets for Approved Drugs: How Many Targets and How Many Drugs?. Mol. Pharmacol. 93 (4), 251–258. 10.1124/mol.117.111062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland S. (2013) Are GPCRs Still a Source of New Targets?. J. Biomol. Screening 18 (9), 947–966. 10.1177/1087057113498418. [DOI] [PubMed] [Google Scholar]
- Venkatakrishnan A. J.; Deupi X.; Lebon G.; Tate C. G.; Schertler G. F.; Babu M. M. (2013) Molecular Signatures of G-Protein-Coupled Receptors. Nature 494 (7436), 185–194. 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- Julius D.; Nathans J. (2012) Signaling by Sensory Receptors. Cold Spring Harbor Perspect. Biol. 4 (1), a005991 10.1101/cshperspect.a005991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway J. L.; Zon L. I. (2003) Ontogeny of Hematopoiesis: Examining the Emergence of Hematopoietic Cells in the Vertebrate Embryo. Curr. Top. Dev. Biol. 53, 139–158. 10.1016/S0070-2153(03)53004-6. [DOI] [PubMed] [Google Scholar]
- Jagannathan-Bogdan M.; Zon L. I. (2013) Hematopoiesis. Development (Cambridge, U. K.) 140 (12), 2463–2467. 10.1242/dev.083147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin S. H.; Zon L. I. (2008) Hematopoiesis: An Evolving Paradigm for Stem Cell Biology. Cell 132 (4), 631–644. 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand J. Y.; Chi N. C.; Santoso B.; Teng S.; Stainier D. Y.; Traver D. (2010) Haematopoietic Stem Cells Derive Directly from Aortic Endothelium during Development. Nature 464 (7285), 108–111. 10.1038/nature08738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissa K.; Herbomel P. (2010) Blood Stem Cells Emerge from Aortic Endothelium by a Novel Type of Cell Transition. Nature 464 (7285), 112–115. 10.1038/nature08761. [DOI] [PubMed] [Google Scholar]
- Tamplin O. J.; Durand E. M.; Carr L. A.; Childs S. J.; Hagedorn E. J.; Li P.; Yzaguirre A. D.; Speck N. A.; Zon L. I. (2015) Hematopoietic Stem Cell Arrival Triggers Dynamic Remodeling of the Perivascular Niche. Cell 160 (1–2), 241–252. 10.1016/j.cell.2014.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapas S.; Catt K.; Clark A. (1995) Cloning and Expression of CDNA Encoding a Rat Adrenomedullin Receptor. J. Biol. Chem. 270 (43), 25344–25347. 10.1074/jbc.270.43.25344. [DOI] [PubMed] [Google Scholar]
- Xiao L.; Harrell J. C.; Perou C. M.; Dudley A. C. (2014) Identification of a Stable Molecular Signature in Mammary Tumor Endothelial Cells That Persists in Vitro. Angiogenesis 17 (3), 511–518. 10.1007/s10456-013-9409-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takase H.; Matsumoto K.; Yamadera R.; Kubota Y.; Otsu A.; Suzuki R.; Ishitobi H.; Mochizuki H.; Kojima T.; Takano S.; Uchida K.; Takahashi S.; Ema M. (2012) Genome-Wide Identification of Endothelial Cell-Enriched Genes in the Mouse Embryo. Blood 120 (4), 914–923. 10.1182/blood-2011-12-398156. [DOI] [PubMed] [Google Scholar]
- Sumanas S.; Jorniak T.; Lin S. (2005) Identification of Novel Vascular Endothelial-Specific Genes by the Microarray Analysis of the Zebrafish Cloche Mutants. Blood 106 (2), 534–541. 10.1182/blood-2004-12-4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alghisi E.; Distel M.; Malagola M.; Anelli V.; Santoriello C.; Herwig L.; Krudewig A.; Henkel C. V.; Russo D.; Mione M. C. (2013) Targeting Oncogene Expression to Endothelial Cells Induces Proliferation of the Myelo-Erythroid Lineage by Repressing the Notch Pathway. Leukemia 27 (11), 2229–2241. 10.1038/leu.2013.132. [DOI] [PubMed] [Google Scholar]
- Lawson N. D.; Weinstein B. M. (2002) In Vivo Imaging of Embryonic Vascular Development Using Transgenic Zebrafish. Dev. Biol. 248 (2), 307–318. 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- Paik E. J.; Zon L. I. (2010) Hematopoietic Development in the Zebrafish. Int. J. Dev. Biol. 54 (6–7), 1127–1137. 10.1387/ijdb.093042ep. [DOI] [PubMed] [Google Scholar]
- Zhang P.; He Q.; Chen D.; Liu W.; Wang L.; Zhang C.; Ma D.; Li W.; Liu B.; Liu F. (2015) G Protein-Coupled Receptor 183 Facilitates Endothelial-to-Hematopoietic Transition via Notch1 Inhibition. Cell Res. 25 (10), 1093–1107. 10.1038/cr.2015.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vejnar C. E.; Moreno-Mateos M. A.; Cifuentes D.; Bazzini A. A.; Giraldez A. J. (2016) Optimized CRISPR-Cas9 System for Genome Editing in Zebrafish. Cold Spring Harb Protoc 2016 (10), 86850 10.1101/pdb.prot086850. [DOI] [PubMed] [Google Scholar]
- Wang H.; La Russa M.; Qi L. S. (2016) CRISPR/Cas9 in Genome Editing and Beyond. Annu. Rev. Biochem. 85, 227–264. 10.1146/annurev-biochem-060815-014607. [DOI] [PubMed] [Google Scholar]
- North T. E.; Goessling W.; Peeters M.; Li P.; Ceol C.; Lord A. M.; Weber G. J.; Harris J.; Cutting C. C.; Huang P.; Dzierzak E.; Zon L. I. (2009) Hematopoietic Stem Cell Development Is Dependent on Blood Flow. Cell 137 (4), 736–748. 10.1016/j.cell.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.; Sherman B.; Lempicki R. (2009) Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat. Protoc. 4 (1), 44–57. 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Chen A. T.; Zon L. I. (2009) Zebrafish Blood Stem Cells. J. Cell. Biochem. 108 (1), 35–42. 10.1002/jcb.22251. [DOI] [PubMed] [Google Scholar]
- Kechele D. O.; Blue R. E.; Zwarycz B.; Espenschied S. T.; Mah A. T.; Siegel M. B.; Perou C. M.; Ding S.; Magness S. T.; Lund P. K.; Caron K. M. (2017) Orphan Gpr182 Suppresses ERK-Mediated Intestinal Proliferation during Regeneration and Adenoma Formation. J. Clin. Invest. 127 (2), 593–607. 10.1172/JCI87588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeze W. K.; Sassano M. F.; Huang X.-P.; Lansu K.; McCorvy J. D.; Giguère P. M.; Sciaky N.; Roth B. L. (2015) PRESTO-Tango as an Open-Source Resource for Interrogation of the Druggable Human GPCRome. Nat. Struct. Mole. Biol. 22 (5), 362 10.1038/nsmb.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W.; Sherman B. T.; Tan Q.; Kir J.; Liu D.; Bryant D.; Guo Y.; Stephens R.; Baseler M. W.; Lane H. C.; Lempicki R. A. (2007) DAVID Bioinformatics Resources: Expanded Annotation Database and Novel Algorithms to Better Extract Biology from Large Gene Lists. Nucleic Acids Res. 35, W169–W175. 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricciotti E.; FitzGerald G. A. (2011) Prostaglandins and Inflammation. Arterioscler., Thromb., Vasc. Biol. 31 (5), 986–1000. 10.1161/ATVBAHA.110.207449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper A. T.; Butler J. M.; Nolan D. J.; Kranz A.; Iida K.; Kobayashi M.; Kopp H.-G.; Shido K.; Petit I.; Yanger K.; James D.; Witte L.; Zhu Z.; Wu Y.; Pytowski B.; Rosenwaks Z.; Mittal V.; Sato T. N.; Rafii S. (2009) Engraftment and Reconstitution of Hematopoiesis Is Dependent on VEGFR2-Mediated Regeneration of Sinusoidal Endothelial Cells. Cell Stem Cell 4 (3), 263–274. 10.1016/j.stem.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.; Pei H.; Xie X.; Wang S.; Jia Y.; Zhang B.; Fan Z.; Liu Y.; Bai Y.; Han Y.; He L.; Nan X.; Yue W.; Pei X. (2019) Liver Sinusoidal Endothelial Cells Promote the Expansion of Human Cord Blood Hematopoietic Stem and Progenitor Cells. Int. J. Mol. Sci. 20 (8), 1985. 10.3390/ijms20081985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid C.; Schledzewski K.; Mogler C.; Waldburger N.; Kalna V.; Marx A.; Randi A.; Géraud C.; Goerdt S.; Koch P.-S. (2018) GPR182 Is a Novel Marker for Sinusoidal Endothelial Differentiation with Distinct GPCR Signaling Activity in Vitro. Biochem. Biophys. Res. Commun. 497 (1), 32–38. 10.1016/j.bbrc.2018.01.185. [DOI] [PubMed] [Google Scholar]
- Li Y.; Esain V.; Teng L.; Xu J.; Kwan W.; Frost I. M.; Yzaguirre A. D.; Cai X.; Cortes M.; Maijenburg M. W.; Tober J.; Dzierzak E.; Orkin S. H.; Tan K.; North T. E.; Speck N. A. (2014) Inflammatory Signaling Regulates Embryonic Hematopoietic Stem and Progenitor Cell Production. Genes Dev. 28 (23), 2597–2612. 10.1101/gad.253302.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espin-Palazon R.; Weijts B.; Mulero V.; Traver D. (2018) Proinflammatory Signals as Fuel for the Fire of Hematopoietic Stem Cell Emergence. Trends Cell Biol. 28 (1), 58–66. 10.1016/j.tcb.2017.08.003. [DOI] [PubMed] [Google Scholar]
- Espín-Palazón R.; Stachura D. L.; Campbell C. A.; García-Moreno D.; Del Cid N.; Kim A. D.; Candel S.; Meseguer J.; Mulero V.; Traver D. (2014) Proinflammatory Signaling Regulates Hematopoietic Stem Cell Emergence. Cell 159 (5), 1070–1085. 10.1016/j.cell.2014.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamiphak S.; Kontarakis Z.; Stainier D. Y. (2014) Interferon Gamma Signaling Positively Regulates Hematopoietic Stem Cell Emergence. Dev. Cell 31 (5), 640–653. 10.1016/j.devcel.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q.; Zhang C.; Wang L.; Zhang P.; Ma D.; Lv J.; Liu F. (2015) Inflammatory Signaling Regulates Hematopoietic Stem and Progenitor Cell Emergence in Vertebrates. Blood 125 (7), 1098–1106. 10.1182/blood-2014-09-601542. [DOI] [PubMed] [Google Scholar]
- Kwon S.-Y.; Ro M.; Kim J.-H. (2019) Mediatory Roles of Leukotriene B4 Receptors in LPS-Induced Endotoxic Shock. Sci. Rep. 9 (1), 5936. 10.1038/s41598-019-42410-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki F.; Yokomizo T. (2019) The Leukotriene Receptors as Therapeutic Targets of Inflammatory Diseases. Int. Immunol. 31, 607. 10.1093/intimm/dxz044. [DOI] [PubMed] [Google Scholar]
- Chung J.; Kim G.-Y.; Mun Y.-C.; Ahn J.-Y.; Seong C.-M.; Kim J.-H. (2005) Leukotriene B4 Pathway Regulates the Fate of the Hematopoietic Stem Cells. Exp. Mol. Med. 37 (1), 45–50. 10.1038/emm.2005.6. [DOI] [PubMed] [Google Scholar]
- Proulx K.; Lu A.; Sumanas S. (2010) Cranial Vasculature in Zebrafish Forms by Angioblast Cluster-Derived Angiogenesis. Dev. Biol. 348 (1), 34–46. 10.1016/j.ydbio.2010.08.036. [DOI] [PubMed] [Google Scholar]
- Chi N. C.; Shaw R. M.; De Val S.; Kang G.; Jan L. Y.; Black B. L.; Stainier D. Y. (2008) Foxn4 Directly Regulates Tbx2b Expression and Atrioventricular Canal Formation. Genes Dev. 22 (6), 734–739. 10.1101/gad.1629408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H.-F.; Traver D.; Zhu H.; Dooley K.; Paw B. H.; Zon L. I.; Handin R. I. (2005) Analysis of Thrombocyte Development in CD41-GFP Transgenic Zebrafish. Blood 106 (12), 3803–3810. 10.1182/blood-2005-01-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North T. E.; Goessling W.; Walkley C. R.; Lengerke C.; Kopani K. R.; Lord A. M.; Weber G. J.; Bowman T. V.; Jang I.-H. H.; Grosser T.; Fitzgerald G. A.; Daley G. Q.; Orkin S. H.; Zon L. I. (2007) Prostaglandin E2 Regulates Vertebrate Haematopoietic Stem Cell Homeostasis. Nature 447 (7147), 1007–1011. 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elks P. M.; Brizee S.; van der Vaart M.; Walmsley S. R.; van Eeden F. J.; Renshaw S. A.; Meijer A. H. (2013) Hypoxia Inducible Factor Signaling Modulates Susceptibility to Mycobacterial Infection via a Nitric Oxide Dependent Mechanism. Plos Pathog. 9 (12), e1003789 10.1371/journal.ppat.1003789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellett F.; Pase L.; Hayman J. W.; Andrianopoulos A.; Lieschke G. J. (2010) Mpeg1 Promoter Transgenes Direct Macrophage-Lineage Expression in Zebrafish. Blood 117 (4), e49 10.1182/blood-2010-10-314120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel C. B.; Ballard W. W.; Kimmel S. R.; Ullmann B.; Schilling T. F. (1995) Stages of Embryonic Development of the Zebrafish. Dev. Dyn. 203 (3), 253–310. 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- UC Davis . Knockout Mouse Project (KOMP) Respository. https://www.komp.org (accessed 2016-11-30, n.d.)
- Babraham Bioinformatics . FastQC: A Quality Control Tool for High Throughput Sequence Data, http://www.bioinformatics.babraham.ac.uk/projects/fastqc. 2010.
- Davis M. P. A.; van Dongen S.; Abreu-Goodger C.; Bartonicek N.; Enright A. J. (2013) Kraken: A Set of Tools for Quality Control and Analysis of High-Throughput Sequence Data. Methods 63 (1), 41–49. 10.1016/j.ymeth.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A.; Davis C. A.; Schlesinger F.; Drenkow J.; Zaleski C.; Jha S.; Batut P.; Chaisson M.; Gingeras T. R. (2013) STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 29 (1), 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y.; Smyth G. K.; Shi W. (2014) FeatureCounts: An Efficient General-Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 30 (7), 923–930. 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Consortium U. (2013) Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res. 42 (Database issue), D191–8. 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D.; McCarthy D. J.; Smyth G. K. (2010) EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 26 (1), 139–140. 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I.; Huber W.; Anders S. (2014) Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 15 (12), 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


