Abstract
NF-E2-related factor 2 (NRF2) is a basic leucine zipper transcription factor, a master regulator of redox homeostasis regulating a variety of genes for antioxidant and detoxification enzymes. NRF2 was, therefore, initially thought to protect the liver from oxidative stress. Recent studies, however, have revealed that mutations in NRF2 cause aberrant accumulation of NRF2 in the nucleus and exert the upregulation of NRF2 target genes. Moreover, among all molecular changes in hepatocellular carcinoma (HCC), NRF2 activation has been revealed as a more prominent pathway contributing to the progression of precancerous lesions to malignancy. Nevertheless, how its activation leads to poor prognosis in HCC patients remains unclear. In this review, we provide an overview of how aberrant activation of NRF2 triggers HCC development. We also summarize the emerging roles of other NRF family members in liver cancer development.
Keywords: NF-E2-related factor 2, transcription factor, redox homeostasis, oxidative stress, hepatocellular carcinoma
1. Introduction
Liver cancer is one of the most troublesome human malignancies, with an annual incidence of around 600,000 worldwide (https://doi.org/10.3322/canjclin.55.2.74). Among different types of liver cancer, hepatocellular carcinoma (HCC) is the sixth most common malignancy of liver (http://gco.iarc.fr/) and the eighth leading cause of cancer-related deaths in Europe (http://gco.iarc.fr/today). Chronic hepatitis B (HBV) or C (HCV) virus infection, alcohol intake, diabetes, fatty liver disease, and chronic liver injury cause permanent hepatocellular damage, hepatocyte regeneration, and inflammation, which are key risk factors for HCC. Infection with HBV or HCV and alcohol-induced hepatocarcinogenesis are associated with oxidative stress in the liver [1,2,3,4,5]. Oxidative stress also contributes to genomic instability, and the altered gene expression leads to HCC development [6,7,8]. Moreover, oxidative stress has been suggested to cause cancer-specific gene mutations in the cell cycle, apoptosis, and various processes of the regeneration cycle, which may lead to liver damage [9,10,11,12].
NF-E2-related factor 2 (NRF2) transcription factor is activated by oxidative stress, and recent studies have suggested that the aberrant activation of NRF2 triggers hepatomegaly and HCC development [13,14], however, this factor also acts to protect the liver from oxidative stress. The protein interaction of NRF2 and Kelch-like ECH-associated protein 1 (KEAP1) is known to orchestrate the NRF2-dependent oxidative stress response to maintain liver homeostasis. In the presence of oxidative stress, KEAP1 is degraded in the cytoplasm, and due to the degradation, NRF2 is released from KEAP1 [15,16]. Thereafter, NRF2 is phosphorylated and translocates into the nucleus, and induces a series of cytoprotective genes by binding to the antioxidant response element (ARE) after heterodimerization with small musculoaponeurotic fibrosarcoma oncogene homolog (Maf) proteins [17,18,19]. More importantly, several studies have demonstrated that somatic mutations occur in the coding region of NRF2 and are associated with poor prognoses and overall low survival rates in several cancers [20,21,22,23]. The mutations are mostly located in the DLG or ETGE motifs of NRF2 and have been reported to impair the NRF2 binding ability to KEAP1, which in turn leads to aberrant nuclear accumulation of NRF2 [21,23,24]. In this review, we describe how the aberrant transcriptional activation of NRF2 caused by its nuclear accumulation may develop HCC at molecular level.
2. Oxidative Stress-Dependent HCC Pathogenesis
Multiple genetic and epigenetic changes are involved in the development of HCC. Many studies have revealed that oxidative stress is one of the causes linked to tumor initiation and progression by disrupting the normal cellular redox homeostasis [25,26,27]. Oxidative stress also induces mitochondrial dysfunction, accelerates telomere shortening, causes DNA damage, and is associated with mutations of apoptosis-specific genes in HCC [28,29,30,31,32]. An increased level of reactive oxygen species (ROS), generated by Kupffer cells during hepatic inflammation, has been associated with the progression of liver pathologies [33]. Similarly, oxidative stress disrupts intracellular signaling pathways and contributes to HCC. Alterations in nuclear factor-kappa beta (NF-κB), peroxisome proliferator-activated receptor alpha (PPARα), mitogen-activated protein kinase (MAPK), extracellular signal-regulated kinases (ERK), and transforming growth factor β1 (TGF-β1) pathways due to oxidative stress are commonly associated with HCC, as all these pathways are involved in the activation of cellular proliferation and survival [34,35,36,37,38,39]. Moreover, HCV infection enhances NF-κB and TGF-β1 expression through the production of ROS and activation of p38-MAPK, ERK, and JNK, and thus promotes the development of hepatic fibrosis [35,37]. Rodent models of DEN/CCl4-induced hepatocarcinogenesis display a significant upregulation of liver-specific NF-κB and TGF-β1/Smad3 signaling [40]. In contrast, HepG2 HCC cells exposed to cadmium telluride quantum dots-induced oxidative stress exhibit decreased levels of Glutathione-SH and Bcl2 and increased expression of NRF2 together with apoptosis induction due to the activation of the MAPK-JNK pathway [36]. These studies suggest that multiple intracellular signaling pathways are activated by oxidative stress, and notably, either carcinogenic or anticarcinogenic pathways are triggered in a context-dependent manner.
3. Dysregulation of NRF2-KEAP1 Physical Interaction Triggers Several Types of Cancer
Several studies have indicated that the NRF2-KEAP1 signaling pathway functions as an oxidative stress sensor [16]. KEAP1 is an adapter protein for the E3 ubiquitin ligase complex that controls the stability and accumulation of NRF2 [41]. Under normal conditions, KEAP1 binds to NRF2 and directs it to Cullin 3-RING E3 (CUL3 E3) ligase for ubiquitination and subsequent proteasomal degradation [42,43]. Upon exposure to oxidative or electrophilic stresses, KEAP1-mediated proteasomal degradation of NRF2 is inhibited and leads to NRF2-mediated transcription of various genes in several types of tissue, including liver. NRF2 is a member of the Cap’n’Collar (CNC) subfamily of basic leucine zipper (bZIP) transcription factors that regulate a wide variety of genes for antioxidant and detoxification enzymes [44,45]. This activity relies on its transactivation capacity and its heterodimeric partner, small MAF transcription factor [46]. The NRF2 protein consists of seven conserved NRF2-ECH homology (Neh) domains. Neh1 contains CNC-bZIP domain via which NRF2 dimerizes with Maf [46]. The consensus binding site of NRF2 is the ARE sequence (TGACNNNGC) [47,48,49]. The N-terminal region contains the highly conserved Neh2 domain, which negatively regulates the transcriptional activity of NRF2. The Neh2 domain of NRF2 contains DLG and ETGE motifs, which are the binding sites for KEAP1 [50,51,52]. Alternatively, repression of Nrf2 is achieved by interactions of Neh7 with the DNA-binding domain of retinoic X receptor α [53]. Neh6 is target for E3 ubiquitin ligase β-TrCP leading to degradation [54]. The Neh3–5 domains are thought to bind to transcriptional system [55,56]. KEAP1 is a member of BTB-Kelch family of proteins. The BTB domain is N-terminally situated and is responsible for homodimerization of KEAP1 and interaction with CUL3 [57]. The C-terminal Kelch domain binds to the ETGE motif or DLG motif of NRF2 [51]. The Kelch domain forms a six-bladed β-propeller structure, and this domain is evolutionarily conserved among species. Moreover, among the six Kelch blades, four β-strands are conserved in each blade [58]. Recent studies have provided insight into how mutations disturb the structure of the BTB-Kelch domain that is responsible for NRF2 binding. It has been reported that one single-point mutation, a proline substitution for serine 383 (S383P) in KEAP1, significantly reduces the ability of the Kelch domain to bind with the Neh2 domain of NRF2 [58]. Moreover, KEAP1 substitution mutations of cysteine residues Cys273S/A and Cys288S/A do not affect the ability of KEAP1 to interact with NRF2, but they impair the KEAP1-mediated degradation of NRF2 [59,60]. Notably, many studies have revealed that the loss of interaction between NRF2 and KEAP1 causes tumor development in multiple cancer types. Furthermore, it has been reported that the deletion of Exon2 in NRF2, which reduces interaction with KEAP1, causes tumor development in lung and liver cancer [61]. Together, a tightly regulated balance of NRF2 and KEAP1 interaction is essential to protect cells or tissues from oxidative stress, and the failure of that mechanism (e.g., mutations of critical amino acids) triggers cancer development.
4. Mutations in NRF2 and KEAP1 Functional Domains Induce HCC Development
The International Cancer Genome Consortium (ICGC; https://icgc.org/) has identified somatic mutations in the NRF2 gene of human lung, liver, breast, head, and neck cancer patients. Notably, these mutations were mostly located within the DLG and ETGE motifs, which provides NRF2 with gain-of-function activity in different cancer types [21,23,62]. Therefore, we surveyed the ICGC database and compiled the HCC somatic mutations found in DLG and ETGE motifs (Table 1). KEAP1 mutations are also found within the BTB, IVR, and Kelch domains (Figure 1). Comprehensive genomic analyses have identified somatic mutations in the NRF2 and KEAP1 genes in various types of cancer [21,23,63]. Interestingly, mutations in KEAP1 and NRF2 are mutually exclusive and rarely occurred in the same cancer cell [64]. Whole-exome sequencing has identified 6.4% of the somatic mutations in NRF2 in HCC patients [24]. Somatic NRF2 and KEAP1 mutations were most often found in lung squamous cell carcinoma, esophageal carcinoma, uterine corpus endometrial carcinoma, lung adenocarcinoma, head and neck cancers, and HCC; the overlapping somatic NRF2 or KEAP1 mutations are associated with a sustained NRF2 activation phenotype [21,23]. In 995 lung cancer cases, 423 cases were estimated to have constitutive NRF2 activation, and 165 cases harbored either NRF2 or KEAP1 mutations with higher expression of NRF2 target genes, including Aldo-Keto Reductase Family 1 Member B10 (AKR1B10), Aldo-Keto Reductase Family 1 Member B15 (AKR1B15), Glutathione Peroxidase 2 (GPX2), Thioredoxin Reductase 1 (TXNRD1), Glutamate-Cysteine Ligase Modifier Subunit (GCLM), and Glutamate-Cysteine Ligase Catalytic Subunit Glutamate-Cysteine Ligase Catalytic Subunit (GCLC) [21]. It has been reported that NRF2 gain-of-function mutations are one of the possible triggers of HCC [65,66]. In an experimental rat model of hepatocarcinogenesis, it was found that the NRF2 gene was frequently mutated or activated during the early stage of the tumorigenic process [66]. This suggests that NRF2 plays a potent role for the initiation of HCC and is mandatory for the development of preneoplastic lesions. The role of constitutive NRF2 activation is well established in chemo- and radio resistance in various tumors [67,68]. In Lung Squamous Cell Carcinoma (LSCC) mice models developed by KEAP1 deletion, the persistent activation of NRF2 has been suggested to contribute to increased tumor formation, metastasis, and resistance to oxidative stress and irradiation [67]. The NRF2 DLG and ETGE motifs have been reported as driver mutations in several types of cancer including HCC [62,65,66]. A recent CRISPR/Cas9 genome-wide screening study demonstrated that KEAP1 depletion causes aberrant NRF2 transcriptional activity as well as high chemoresistance. Upregulation in NRF2-target gene expression including NAD(P)H-quinone oxidoreductase 1 (NQO1), GPX2, and TXNRD1 was also observed [69]. Moreover, microdeletion of NRF2 exon 2 (where the DLG and ETGE domains are located) in JHH cells is associated with increased NRF2 target gene expression and is similar to HCC cell lines containing KEAP1 mutations [61]. NRF2 DLG and ETGE mutations that demonstrate a loss of interaction with KEAP1, are localized to the nucleus and exert sustained target gene activation. In fact, NRF2 mutations occur frequently in the DLG or ETGE motifs and activate Nqo1, Gclc, and Gsta4 pathways. This suggests that NRF2 mutations are able to enhance NRF2 transcriptional activity [62]. Mutation in the DLG motif of NRF2 induces ARE-regulated PPP enzyme transcription, which is required for cell growth and proliferation [65,70]. NRF2 also translocates into the nucleus in HCV-infected hepatocytes and causes mouse double minute 2 homolog (MDM2)-mediated retinoblastoma protein (Rb) degradation. This subsequently induces HCC progression [71]. Overall, these findings suggest that either the NRF2 DLG/ETGE mutations or KEAP1 mutations induce aberrant NRF2 activity and may induce HCC through NRF2-ARE pathway activation. Moreover, methylation of the KEAP1 promoter has been reported to induce cancer development and chemo- and radio- resistance in multiple cancer types [72,73]. It has been determined that the most frequent somatic mutations found in HCC are telomerase reverse transcriptase (TERT) promoter mutations, identified in premalignant lesions in cirrhosis [74]. TERT is essential for telomere elongation and maintenance during cell division. TERT promoter mutations are associated with increased telomerase activity, which displays enhanced NRF2 expression and inhibited glycogen accumulation [75]. As such, it appears there are many possible pathways to trigger liver cancer via aberrant NRF2 transcriptional activity, and further phenotypic validation of the roles of these pathways in liver cancer development merits investigation.
Table 1.
NF-E2-related factor 2 (NRF2) mutations found in NRF2-ECH homology 2 (Neh2) domain ETGE and DLG motif of hepatocellular carcinoma (HCC) patients (International Cancer Genome Consortium (ICGC) database).
| Mutation ID | Genomic DNA Change | Type | Motif | Substitution to | Clinical Significance | Occurrence of Mutation |
|---|---|---|---|---|---|---|
| MU871836 | chr2:g.178098959T > C | single base substitution | DLG | G | Likely pathogenic | 5 |
| MU866686 | chr2:g.178098953C > T | single base substitution | DLG | E | ND | 2 |
| MU1327674 | chr2:g.178098960C > T | single base substitution | DLG | N | Likely pathogenic | 2 |
| MU29615597 | chr2:g.178098959T > G | single base substitution | DLG | A | ND | 1 |
| MU83818151 | chr2:g.178098954C > T | single base substitution | DLG | R | Pathogenic/Likely pathogenic | 1 |
| MU1324215 | chr2:g.178098960C > G | single base substitution | DLG | H | Likely pathogenic | 1 |
| MU830878 | chr2:g.178098956A > C | single base substitution | DLG | R | ND | 3 |
| MU1330977 | chr2:g.178098957G > A | single base substitution | DLG | F | ND | 1 |
| MU825005 | chr2:g.178098800T > C | single base substitution | ETGE | G | ND | 4 |
| MU7421282 | chr2:g.178098809T > C | single base substitution | ETGE | G | ND | 2 |
| MU29777568 | chr2:g.178098806G > A | single base substitution | ETGE | I | ND | 2 |
| MU29708787 | chr2:g.178098799T > G | single base substitution | ETGE | D | ND | 2 |
| MU1890585 | chr2:g.178098804C > A | single base substitution | ETGE | C | ND | 2 |
| MU1332094 | chr2:g.178098804C > T | single base substitution | ETGE | S | ND | 2 |
| MU3162037 | chr2:g.178098809T > A | single base substitution | ETGE | V | Likely pathogenic | 2 |
| MU128988244 | chr2:g.178098803C > G | single base substitution | ETGE | A | ND | 1 |
| MU1804262 | chr2:g.178098799T > A | single base substitution | ETGE | D | ND | 1 |
| MU41238347 | chr2:g.178098804C > G | single base substitution | ETGE | R | ND | 1 |
| MU1817004 | chr2:g.178098807T > G | single base substitution | ETGE | P | ND | 1 |
| MU112734927 | chr2:g.178098809T > G | single base substitution | ETGE | A | ND | 1 |
| MU871364 | chr2:g.178098803C > A | single base substitution | ETGE | V | ND | 1 |
| MU2689228 | chr2:g.178098800T > G | single base substitution | ETGE | A | ND | 1 |
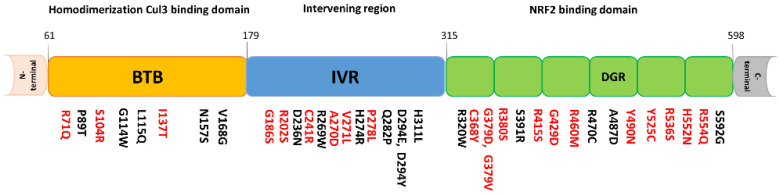
Figure 1.
Human Kelch-like ECH-associated protein 1 (KEAP1) mutations (liver cancer) identified by the International Cancer Genome Consortium. The amino acid positions of the identified mutations of KEAP1 are shown and the amino acid positions in red color indicate the location of mutations that are conserved among several species (Human, Mouse, Bovine, and Zebrafish). The 61BTB179 domain is required for homodimerization of Keap1 by interactions with the Cul3 based E3 ubiquitin ligase system. The 315DGR598 or 6 Kelch-repeat domain binds to NRF2 through Neh2 domain of NRF2. 180IVR314 domain between BTB and DGR domain important sensing oxidative stress and xenobiotic stimuli via modification of its four cysteine residues by electrophiles.
5. Aberrantly Activated NRF2 Targets Several Gene Expressions in HCC
As summarized in Table 2, it has been suggested that excessive NRF2 transcriptional activity promotes the development of liver cancer by regulating the expression of various genes. During oxidative stress, cells initiate an adaptive response that upregulates expression of a large array of cytoprotective genes. The battery of genes is regulated through NRF2 binding to the ARE consensus binding sequence, which includes glutamate-cysteine ligase (GCL), thioredoxin reductase 1, NAD(P)H-quinone oxidoreductase 1 (NQO1), and heme oxygenase-1 (HO-1) [76]. In healthy cells, HO-1 exhibits its cytoprotective effect through detoxification and ROS scavenging mechanisms that decrease the possibility of tumor initiation. However, in a tumor microenvironment, sustained NRF2 expression persists due to stress, which tightly regulates the expression of HO-1. Thus, in growing tumors, HO-1 plays a pro-tumorigenic role by increasing tumor cell proliferation and metastasis and blocking cell death [77,78].
Table 2.
List of NRF2 target genes and their effects on HCC development.
| NRF2 Target Genes | Effect of Gene Expression | ARE in Promoter |
|---|---|---|
| NAD(P)H dehydrogenase, quinone 1 (NQO1) and Hemeoxigenase 1 (HO-1) [92] | Constitutive activation of NRF2 by hepatotoxin contributes to the upregulation of NQO1 and HO-1. This promotes liver cancer cell growth. | YES |
| B-cell lymphoma-extra-large (Bcl-xL) [68] | Antioxidant stabilized NRF2 increases the expression of Bcl-xL gene which causes reduction in apoptosis, increase cell survival, and drug resistance in Hepa1-6 cells. | YES |
| Glutathione S-transferase A4 (GSTA4) [93]. Glutamate-cysteine ligase (GCLC) [91], [94] | Constitutive activation of NRF2 in preneoplastic lesions of HCC increases the expression of GSTA4 and GCLC and promotes HCC cell growth. | YES |
| Placental glutathione S-transferase (GST-P) [95,96] | NRF2/MafK heterodimer activates GST-P gene (a prominent tumor marker for hepatocarcinogenesis) through the binding with GPE1 enhancer during hepatocarcinogenesis. | YES |
| Matrix metalloproteinases-9 (MMP-9) [82], [97] | Up-regulation of NRF2 in HepG2 cells increases the MMP-9 expression which promotes the cell invasion ability of HCC. In response to ROS, NRF2 induces MMP-9 expression in HepG2 cells, which contributes to cancer cell migration and invasiveness. |
YES |
| Platelet-Derived Growth Factor-A (PDGFA) [98] | NRF2 promotes PDGFA transcription by interacting with SP1 thus promotes HCC proliferation. | NO (Through interaction with Sp1) |
| Prostaglandin reductase-1 (PTGR1) [99] | PTGR1 expression regulated by NRF2 regulates antioxidant responses to promote cell proliferation in HCC. PTGR1 overexpression in HCC increases cell proliferation and develop resistance to ROS-induced cell death. | YES |
| Peroxisome proliferator-activated receptor γ (PPARγ) [100] | Overexpression of NRF2 in HFD increases the expression of PPARγ and accumulates hepatic triglyceride which initiates NAFLD. | NO (Correlation was tested) |
| 26S proteasome non-ATPase regulatory subunit 10 (PSMD10) or Gankyrin [101] | Increased NRF2 activity up-regulates gankyrin expression in HCC. | YES |
| Mouse double minute 2 homolog (MDM2) [71,102] | During HCV infection, NRF2 translocates into the nucleus and induces MDM2-mediated retinoblastoma protein (Rb) degradation. This induces HCC progression. | YES |
The extracellular matrix metalloproteinase MMP-9 demonstrates a crucial role in HCC invasion and progression [79]. Many studies have suggested that MMP-9 is a prognostic biomarker to predict tumor invasiveness and recurrence in HCC patients [80,81]. Moreover, it was found that 98 samples displayed MMP-9 positive expression in 143 HCC tissue samples. It has been reported that the MMP-9 gene expression has a strong correlation with the metastatic potential of HCC cell lines (MHCC97-L, MHCC97-H, and HCCLM6) and its expression was significantly increased in an in vitro HCC invasion model [79]. This evidence suggests that MMP-9 plays a critical role in HCC invasiveness and metastasis. A positive correlation between NRF2 and MMP-9 expression in human HCC samples and HCC cell lines has been reported [82]. Moreover, NRF2-mediated induction of MMP-9 plays an important role in cell proliferation and invasion [82].
The presence of a strong correlation between NRF2 and the PI3K-Akt signaling pathway has been demonstrated in driving metabolic gene expression and increased cellular proliferation [83]. In many cancers, the PI3K-Akt pathway is constitutively active and may be responsible for increasing the nuclear levels of NRF2 by inhibiting the GSK3-mediated degradation of NRF2.
Peroxiredoxin 1 (PRDX1), a NRF2 target gene, is known to act as an oxidative stress sensor and promote liver cancer growth [84,85]. In HepG2 cells, a significant decrease in cell proliferation and upregulation of proapoptotic genes BAX and Caspase 3 were observed when PRDX1 was silenced, suggesting the prosurvival and tumorigenic roles of PRDX1. Furthermore, proteomic analysis has revealed changes in expression and oxidation of proteins involved in central metabolism and tumor growth, indicating that PRDX1 is one of metabolic reprogramming factors in cancer cells [86].
Methylenetetrahydrofolate dehydrogenase 1–like (MTHFD1L) is an enzyme, involved in the folate cycle which is also known as a target gene of NRF2. Transcriptome sequencing of HCC patients and The Cancer Genome Atlas (TCGA) data showed that MTHFD1L is significantly overexpressed in different cancers, including HCC. During HCC cell proliferation, the folate cycle provides nutrition to the cells by supplying metabolites for NADPH and DNA synthesis. The MTHFD1L promoter has three ARE sequence elements and is transcriptionally controlled by NRF2. Genetic knockdown (KD) of either NRF2 or MTHFD1L or inhibition of the folate cycle through antifolate drug can inhibit liver cancer cell proliferation by increasing oxidative stress, altering the metabolic program, and sensitizing HCC cells to sorafenib treatment [87].
In cancer cells, the aberrant activation of NRF2 helps their metabolic adaptations through regulating the key genes involved in glucose metabolism pathways. It has been reported that the effect of NRF2 on the regulation of glucose metabolism is partly through suppressing the transcription of miR-1 and miR-206. The miR-1 and miR-206 regulate their target genes of the PPP (pentose phosphate pathway) (Glucose-6-phosphate dehydrogenase (G6PD), Phosphogluconate Dehydrogenase (PGD), Transketolase (TKT), and Transaldolase 1 (TALDO1)) [88], suggesting that suppression of these miRNAs expression caused by NRF2 aberrant activation may enhance tumor development through glucose metabolism deficiency. Additionally, suppressed expression of miR-1 also has been proposed to be important in liver cancer cell growth [89]. On the other hand, in a resistant-hepatocyte rat model of HCC, microRNA profiling revealed an upregulation of miR-200a at the very early stage of tumorigenesis. miR-200a downregulates KEAP1 in several cancer types including HCC [90,91] and it promotes induced expression of NQO-1 and GSTA-4, and GCLC genes which are the direct target of NRF2 [91]. These findings suggest that NRF2 plays important roles in the promotion of liver cancer cell growth through the regulation of several genes and microRNAs.
6. Aberrant Activation of NRF2, a Critical Regulator of Lipid and Cholesterol Metabolism, Leads to HCC
Non-alcoholic fatty liver disease (NAFLD) is considered one of the risk factors of HCC. NAFLD includes a variety of liver pathologies including the accumulation of triglycerides in the hepatocytes, liver inflammation, and non-alcoholic steatohepatitis (NASH) that leads to cirrhosis and thereafter HCC [103,104,105]. The most important mechanism of NASH pathogenesis is increased hepatic iron accumulation, as well as oxidative DNA damage [106]. Highly proliferative cancer cells show strong affinity towards lipid and cholesterol metabolisms [107], and high levels of cholesterols and lipids are now considered hallmarks of many aggressive cancers [108,109,110,111,112]. It is also evident that hepatic lipid and fatty acid overload are related to the development of HCC [113]. The pathogenic role of NRF2 for the initiation and development of hepatic steatosis was described previously [114]. Additionally, enhanced NRF2 activity augments hepatic steatosis and increased lipid deposition in the liver has been reported. In leptin-deficient mice, constitutive activation of NRF2 via KEAP1-KD established insulin resistance, inhibited the accumulation of lipids in adipose tissue, and subsequently increased hepatic steatosis. [114]. Moreover, dysfunction in the starvation-induced hepatic lipid droplets (LDs) synthesis in liver-specific Atg5 (L-Atg5)-deficient mouse livers was associated with sustained NRF2 activation [115]. Though it has been reported that NRF2 activation may reduce cholesterol injury by regulating the lipid homeostasis, how the hyperactivation of this transcription factor in NAFLD leads to HCC needs to be further clarified.
NAFLD results from unbalanced lipid metabolism. Forkhead box protein A1 (FOXA1) is a triglyceride synthesis inhibitor, and it is well known to lower fatty acid uptake [116]. Thus, FOXA1 is thought to be an antisteatotic regulator in lipid metabolic pathways hepatocytes. Notably, it was found that excessive cholesterol synthesis causes the accumulation of NRF2. The accumulated NRF2 suppresses the expression of FOXA1 [117], and the downregulation of FOXA1 has been found in human and rat NAFLD [116]. In this regard, the disruption of lipid metabolism and oxidative stress have been reported as the main causes of NAFLD, and NRF2 is related to lipid homeostasis [118,119]. Peroxisome proliferator-activated receptor gamma (PPARγ) is also one of the most studied lipid metabolism regulators in hepatocytes and it contributes to the development of NAFLD [120]. Interestingly, it has been reported that PPARγ gene expression is regulated by NRF2 [121]. Moreover, it was found that liver-specific Nrf2-KO mice with high-fat diet (HFD) had less steatosis and inflammation with less hepatic triglyceride levels and decreased PPARγ activity [100]. Furthermore, constitutively activated NRF2 signaling in Keap1-KD mice fed a HFD exhibited greater lipogenic gene expression, inflammation, and increased hepatic steatosis [122]. These findings suggest that aberrant activation of NRF2 helps to trigger development of NASH or NAFLD, therefore, gain-of-function type mutations in NRF2 may initiate the development of HCC by inducing NASH or NAFLD. Nevertheless, since the NRF2-controlled gene network contributing to the promotion of HCC is not clear, further studies are needed to investigate how NRF2 induces NASH or NAFLD at the molecular level.
7. Emerging Mechanism of NRF2 Activation-Induced HCC
ROS is usually considered to be carcinogenic and several chemopreventive strategies for the usage of NRF2 have been proposed [123,124]. Since NRF2 is widely known to be a potent protector in anti-oxidative response, a question arises as to how NRF2 DLG and ETGE mutations lead to increased malignancy of HCC and trigger its resistance to chemotherapy. This issue has been discussed very intensively in several reviews [14,125] and an excellent hypothesis is proposed by Sporn and his group that the role of NRF2 can be altered depending on the stage of tumor progression [126]. They proposed a model in which enhancement of NRF2 activity can protect advanced tumors from the cytotoxic effects of ROS that are induced by oncogenic signaling whereas NRF2 activation acts as protective for tumor establishment in normal condition. Interestingly, oncogenic gene mutations such as K-RasG12D, B-RafV619E enhanced transcription of NRF2 with elevated NRF2 target gene expression and lowered intracellular ROS [127]. Therefore, it is assumed that aberrant transcriptional activity induced by high expression or mutation of NRF2 may lead to malignancy in combination with other factors (for example, mutation of oncogene leads cells to early cancer state or abnormality of metabolic state changes in cellular environment). In these conditions, the cells can be shifted to malignancy when they are induced to become HCC progenitor cells. Interestingly, it has been shown that elevated p62 levels aberrantly activate NRF2 transcriptional activity, which induces HCC pathogenesis by accelerating the survival of HCC-initiating cells [128]. p62, which is encoded by Sequestosome-1 (SQSTM1), is an autophagy adaptor. It activates NRF2 through inactivation of Keap1 [129]. Autophagy is an evolutionary conserved cellular mechanism that maintains cell homeostasis by targeting damaged organelles or mistranslated proteins for lysosomal degradation. Atg7 deletion mice develop hepatocellular adenoma accompanied by aberrant accumulation of p62 followed by NRF2 activation [130]. The study further elucidated the role of p62 in aberrant activation of NRF2 in HCC. The persistent activation of NRF2 is associated with p62 accumulation and the development of HCC in vitro [130]. Furthermore, high levels of p62 expression activates NRF2 and mTORC1 in HCC [128]. Consequently, this NRF2 activation spares HCC-initiating cells from oxidative stress-induced cell death [128]. This is supported by the fact that the kinase-dead mutation of p62 (S349A) in Human hepatoma cell line-1 (Huh-1) cells significantly reduces colony formation capacity with decreased NQO1 mRNA expression [131]. Moreover, a xenograft experiment using a nude mouse demonstrated that the tumor formation capacity of mutant Huh-1 cells (p62 KO and S349A) is reduced as compared to the wild-type. Additionally, p62-mediated NRF2 activation in HCC cells facilitates the glucuronate pathway and glutathione synthesis in HCV positive HCC [132]. NRF2 activation contributes to metabolic reprogramming in HCC harboring phosphorylated p62 [133]. This leads to increased cell proliferation and increases tolerance to anti-cancer drugs in HCC [132]. These findings clearly demonstrate that sustained activation of NRF2 by p62 activation is responsible for HCC pathology, suggesting that NRF2 and KEAP1 mutations, as well as aberrant p62 activation enhance the growth of HCC cells through metabolic dysregulation. NRF2 DLG and ETGE mutations lose the capacity to interact with KEAP1 and localize mainly to the nucleus. Accordingly, these mutations may activate NRF2 target gene expression and exhibit a similar phenotype to p62 activation in the liver. Future studies on the effect of NRF2 DLG and ETGE mutations in combination with autophagy and/or other cellular function in HCC are warranted.
8. Emerging Roles of the CNC Family of Transcription Factors in HCC
The CNC bZIP family of transcription factors, which comprises four closely related factors, NRF1, NRF2, NRF3, and p45 NF-E2, have developmental and homeostatic functions [134,135]. The CNC gene encodes different proteins, with evolutionary conservation between Drosophila CNC isoform and mammalian NRF1, NRF2, and NRF3 [136]. With similar binding and expression profiles, NRF1, NRF2, and NRF3 reside outside of the nucleus under normal conditions [137]. NRF2 resides in the cytoplasm and NRF1 and NRF3 in the endoplasmic reticulum [137,138]. All three transcription factors are essential for maintaining redox homeostasis and directing cellular stress responses. Much like NRF2, NRF1 contains the Neh2 domain and NRF3 does not [139]. NRF-encompassing amino acids 171–244 of Neh2 share 72% homology with the Neh2 domain of NRF2 [137]. In addition, NRF1 has conserved DLG and ETGE motifs within the Neh2 domain, which are essential for KEAP1–NRF1 interaction [140,141]. Moreover, a study indicated that NRF1 expression was significantly reduced in KEAP1-KO H1299 cells, suggesting that KEAP1 stabilizes NRF1 [142]. On the other hand, it has been reported that cytoplasmic localization of NRF1 is independent of KEAP1 whereas KEAP1 physically interacts with NRF1 [137]. Therefore, it is still not clear whether KEAP1 regulates NRF1 function and further study is needed. NRF1 and NRF2 have overlapping targets binding ARE-containing genes, but have distinctive roles [143]. It has also been reported that NRF1 and NRF2 simultaneously control the basal expression of ARE-containing genes in fibroblasts [144]. Likewise, NRF1-3 are known to regulate proteasome gene expression [145,146,147]. Moreover, the role of NRF1 and NRF3 has been indicated in human cancers, including HCC [148,149,150]. These results suggest that NRF1 and NRF3 mutations and aberrant gene expressions may trigger HCC. The molecular regulation and biological function of NRF3 in cancer cells have been elucidated [150,151,152]. When exposed to stress, NRF3 translocates to the nucleus, heterodimerizes with the small Maf proteins similarly to NRF2 via ARE, and activates U2AF homology motif kinase 1 (UHMK1) gene expression [150]. This study suggests that NRF3 functions as an inducible transcription factor in cancer progression. Moreover, analysis of TCGA data revealed that NRF3 was highly expressed in HCC tissues, and its expression was positively correlated with tumor grade and stage [149]. In addition, NRF3 deficiency has been revealed to predispose to T-cell lymphoblastic lymphoma when exposed to carcinogens [153]. Likewise, liver-specific inactivation of the Nrf1 gene in adult mice has been reported to trigger NASH [154], suggesting that the proper activity of NRF1 and NRF3 blocks carcinogenesis, including liver cancer. Therefore, the roles of NRF1 and NRF3 and their mutational effects in HCC merit investigation.
9. Conclusions
In this review, we discussed the current evidence on how aberrant NRF2 transcriptional activity causes HCC development (Figure 2). Moreover, we reviewed the mutations found in ICGC databases in the specific domain that is essential for the KEAP1-NRF2 interaction impact on HCC development. The aberrant activation of NRF2, the dark side of this protein expression, induces the transcription of a series of cytoprotective and xenobiotic-metabolizing genes. Furthermore, align with NRF2, other CNC family members, NRF1 and NRF3, are also dysregulated during HCC development. Hepatocytes are in a continuous struggle to maintain cellular homeostasis, owing to diverse physiological functions of the liver. In a diseased microenvironment, when NRF2 over-activation is induced through mutation, epigenetic changes, competition or other constitutive changes, and cellular responses are variable. Thus, how the hyperactivity of NRF2 leads to drug resistance and tumor development is the hotspot of future research. Further studies are needed to clarify the underlying mechanisms and investigate the role of NRF2 mutations in the development of liver cancer.
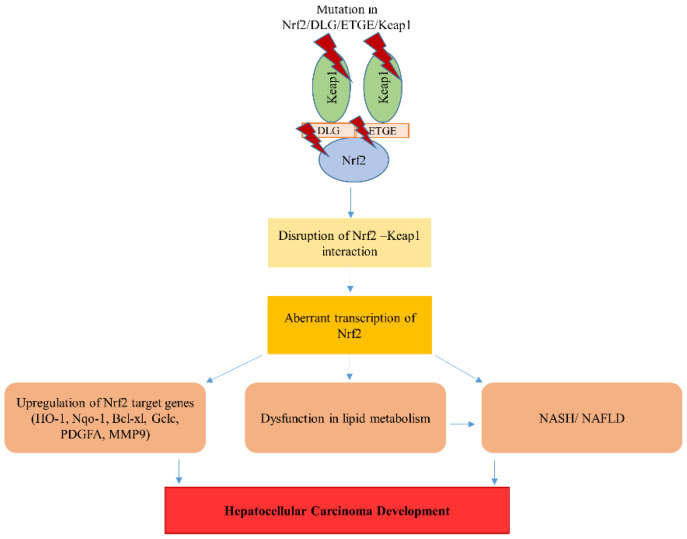
Figure 2.
Mutations in NRF2 or Keap1 cause aberrant accumulation of NRF2 in the nucleus that leads to an increase in NRF2 target genes. This aberrant activation of NRF2 dysregulates the lipid metabolism responsible for the non-alcoholic steatohepatitis (NASH)/non-alcoholic fatty liver disease (NAFLD) pathology. Consequently, these events lead to the development of HCC.
Abbreviations
| AKR1B10 | Aldo-Keto Reductase Family 1 Member B10 |
| AKR1B15 | Aldo-Keto Reductase Family 1 Member B15 |
| ARE | Antioxidant response element |
| Atg5 | Autophagy Related 5 |
| Atg7 | Autophagy Related 7 |
| BAX | BCL2 Associated X |
| Bcl-xL | B-cell lymphoma-extra-large |
| Bcl2 | B-cell lymphoma 2 |
| bZIP | Basic leucine zipper |
| CRISPR/Cas9 | Clustered regularly interspaced short palindromic repeats/ CRISPR-associated protein 9 |
| CUL3 E3 | Cullin 3-RING E3 |
| DEN/CCl4 | Diethylnitrosamine/Carbon tetrachloride |
| ERK | Extracellular signal-regulated kinases |
| FGF19 | Fibroblast growth factor 19 |
| FOXA1 | Forkhead box protein A1 |
| G6PD | Glucose-6-phosphate dehydrogenase |
| GCLC | Glutamate-Cysteine Ligase Catalytic Subunit |
| GCLM | Glutamate-Cysteine Ligase Modifier Subunit |
| GPX2 | Glutathione Peroxidase 2 |
| GSK3 | Glycogen synthase kinase 3 |
| GST-P | Placental glutathione S-transferase |
| GSTA4 | Glutathione S-transferase A4 |
| HBV | Hepatitis B virus |
| HCC | Hepatocellular carcinoma |
| HCV | Hepatitis C virus |
| HFD | High-fat diet |
| HO-1 | Heme oxygenase-1 |
| Huh-1 | Human hepatoma cell line-1 |
| ICGC | International Cancer Genome Consortium |
| JNK | c-Jun N-terminal kinases |
| KD | Knockdown |
| KEAP1 | Kelch-like ECH-associated protein 1 |
| KO | Knockout |
| LSCC | Lung Squamous Cell Carcinoma |
| Maf | Musculoaponeurotic fibrosarcoma |
| MAPK | Mitogen-activated protein kinase |
| Mdm2 | Mouse double minute 2 homolog |
| MMP-9 | Matrix metalloproteinase |
| MTHFD1L | Methylenetetrahydrofolate dehydrogenase 1–like |
| mTORC1 | Mammalian target of rapamycin complex 1 |
| NAFLD | Non-alcoholic fatty liver disease |
| NASH | Non-alcoholic steatohepatitis |
| NF-κB | Nuclear factor-kappa beta |
| NQO1 | NAD(P)H-quinone oxidoreductase 1 |
| NRF1 | Nuclear respiratory factor 1 |
| NRF2 | NF-E2-related factor 2 |
| NRF3 | Nuclear factor-like factor 3 |
| PDGFA | Platelet-Derived Growth Factor-A |
| PGD | Phosphogluconate Dehydrogenase |
| PI3K-Akt | Protein Kinase B Phosphoinositide-3-kinase |
| PPARα | Peroxisome proliferator-activated receptor alpha |
| PPARγ | Peroxisome proliferator-activated receptor γ |
| PPP | Pentose phosphate pathway |
| PRDX1 | Peroxiredoxin 1 |
| PSMD10 | 26S proteasome non-ATPase regulatory subunit 10 |
| PTGR1 | Prostaglandin reductase-1 |
| Rb | Retinoblastoma protein |
| ROS | Reactive oxygen species |
| SQSTM1 | Sequestosome-1 |
| Smad3 | SMAD family member 3 |
| TALDO1 | Transaldolase 1 |
| TCGA | The Cancer Genome Atlas |
| TERT | Telomerase reverse transcriptase |
| TGF-β1 | Transforming growth factor beta 1 |
| TKT | Transketolase |
| TXNRD1 | Thioredoxin Reductase 1 |
| UHMK1 | U2AF homology motif kinase 1 |
| β-TrCP | Beta-transducin repeats-containing proteins |
Author Contributions
E.H. and H.T. drafted the manuscript and M.R.K., A.S.T., M.Ś., P.L., D.W., A.G.A., and E.D.P. edited the final manuscript. All authors have read and approved the final manuscript.
Funding
This work was supported by the Polish National Science Center OPUS 13 (2017/25/B/NZ5/02762) for HT.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Abdalla M.Y., Ahmad I.M., Spitz D.R., Schmidt W.N., Britigan B.E. Hepatitis C Virus-Core and Non Structural Proteins Lead to Different Effects on Cellular Antioxidant Defenses. J. Med. Virol. 2005;76:489–497. doi: 10.1002/jmv.20388. [DOI] [PubMed] [Google Scholar]
- 2.Brault C., Levy P., Duponchel S., Michelet M., Salle A., Pecheur E.I., Plissonnier M.L., Parent R., Vericel E., Ivanov A.V., et al. Glutathione Peroxidase 4 is Reversibly Induced by HCV to Control Lipid Peroxidation and to Increase Virion Infectivity. Gut. 2016;65:144–154. doi: 10.1136/gutjnl-2014-307904. [DOI] [PubMed] [Google Scholar]
- 3.Higuchi H., Adachi M., Miura S., Gores G.J., Ishii H. The Mitochondrial Permeability Transition Contributes to Acute Ethanol-Induced Apoptosis in Rat Hepatocytes. Hepatology. 2001;34:320–328. doi: 10.1053/jhep.2001.26380. [DOI] [PubMed] [Google Scholar]
- 4.Sidharthan S., Kottilil S. Mechanisms of Alcohol-Induced Hepatocellular Carcinoma. Hepatol. Int. 2014;8:452–457. doi: 10.1007/s12072-013-9494-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsai S.M., Lin S.K., Lee K.T., Hsiao J.K., Huang J.C., Wu S.H., Ma H., Wu S.H., Tsai L.Y. Evaluation of Redox Statuses in Patients with Hepatitis B Virus-Associated Hepatocellular Carcinoma. Ann. Clin. Biochem. 2009;46:394–400. doi: 10.1258/acb.2009.009029. [DOI] [PubMed] [Google Scholar]
- 6.Zhou L., Yang Y., Tian D., Wang Y. Oxidative Stress-Induced 1, N6-Ethenodeoxyadenosine Adduct Formation Contributes to Hepatocarcinogenesis. Oncol. Rep. 2013;29:875–884. doi: 10.3892/or.2013.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka H., Fujita N., Sugimoto R., Urawa N., Horiike S., Kobayashi Y., Iwasa M., Ma N., Kawanishi S., Watanabe S., et al. Hepatic Oxidative DNA Damage is Associated with Increased Risk for Hepatocellular Carcinoma in Chronic Hepatitis C. Br. J. Cancer. 2008;98:580–586. doi: 10.1038/sj.bjc.6604204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartsch H., Nair J. Oxidative Stress and Lipid Peroxidation-Derived DNA-Lesions in Inflammation Driven Carcinogenesis. Cancer Detect. Prev. 2004;28:385–391. doi: 10.1016/j.cdp.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 9.Takaki A., Kawano S., Uchida D., Takahara M., Hiraoka S., Okada H. Paradoxical Roles of Oxidative Stress Response in the Digestive System before and After Carcinogenesis. Cancers. 2019;11:213. doi: 10.3390/cancers11020213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujinaga H., Tsutsumi T., Yotsuyanagi H., Moriya K., Koike K. Hepatocarcinogenesis in Hepatitis C: HCV Shrewdly Exacerbates Oxidative Stress by Modulating both Production and Scavenging of Reactive Oxygen Species. Oncology. 2011;81(Suppl. 1):11–17. doi: 10.1159/000333253. [DOI] [PubMed] [Google Scholar]
- 11.Bhargava A., Raghuram G.V., Pathak N., Varshney S., Jatawa S.K., Jain D., Mishra P.K. Occult Hepatitis C Virus Elicits Mitochondrial Oxidative Stress in Lymphocytes and Triggers PI3-Kinase-Mediated DNA Damage Response. Free Radic. Biol. Med. 2011;51:1806–1814. doi: 10.1016/j.freeradbiomed.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 12.Jungst C., Cheng B., Gehrke R., Schmitz V., Nischalke H.D., Ramakers J., Schramel P., Schirmacher P., Sauerbruch T., Caselmann W.H. Oxidative Damage is Increased in Human Liver Tissue Adjacent to Hepatocellular Carcinoma. Hepatology. 2004;39:1663–1672. doi: 10.1002/hep.20241. [DOI] [PubMed] [Google Scholar]
- 13.He F., Antonucci L., Yamachika S., Zhang Z., Taniguchi K., Umemura A., Hatzivassiliou G., Roose-Girma M., Reina-Campos M., Duran A., et al. NRF2 Activates Growth Factor Genes and Downstream AKT Signaling to Induce Mouse and Human Hepatomegaly. J. Hepatol. 2020;72:1182–1195. doi: 10.1016/j.jhep.2020.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raghunath A., Sundarraj K., Arfuso F., Sethi G., Perumal E. Dysregulation of Nrf2 in Hepatocellular Carcinoma: Role in Cancer Progression and Chemoresistance. Cancers. 2018;10:481. doi: 10.3390/cancers10120481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McMahon M., Itoh K., Yamamoto M., Hayes J.D. Keap1-Dependent Proteasomal Degradation of Transcription Factor Nrf2 Contributes to the Negative Regulation of Antioxidant Response Element-Driven Gene Expression. J. Biol. Chem. 2003;278:21592–21600. doi: 10.1074/jbc.M300931200. [DOI] [PubMed] [Google Scholar]
- 16.Itoh K., Wakabayashi N., Katoh Y., Ishii T., Igarashi K., Engel J.D., Yamamoto M. Keap1 Represses Nuclear Activation of Antioxidant Responsive Elements by Nrf2 through Binding to the Amino-Terminal Neh2 Domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang H.C., Nguyen T., Pickett C.B. Phosphorylation of Nrf2 at Ser-40 by Protein Kinase C Regulates Antioxidant Response Element-Mediated Transcription. J. Biol. Chem. 2002;277:42769–42774. doi: 10.1074/jbc.M206911200. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen T., Huang H.C., Pickett C.B. Transcriptional Regulation of the Antioxidant Response Element. Activation by Nrf2 and Repression by MafK. J. Biol. Chem. 2000;275:15466–15473. doi: 10.1074/jbc.M000361200. [DOI] [PubMed] [Google Scholar]
- 19.Itoh K., Chiba T., Takahashi S., Ishii T., Igarashi K., Katoh Y., Oyake T., Hayashi N., Satoh K., Hatayama I., et al. An Nrf2/small Maf Heterodimer Mediates the Induction of Phase II Detoxifying Enzyme Genes through Antioxidant Response Elements. Biochem. Biophys. Res. Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 20.Yamaguchi Y., Kamai T., Higashi S., Murakami S., Arai K., Shirataki H., Yoshida K.I. Nrf2 Gene Mutation and Single Nucleotide Polymorphism rs6721961 of the Nrf2 Promoter Region in Renal Cell Cancer. BMC Cancer. 2019;19:1137. doi: 10.1186/s12885-019-6347-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerins M.J., Ooi A. A Catalogue of Somatic NRF2 Gain-of-Function Mutations in Cancer. Sci. Rep. 2018;8:12846. doi: 10.1038/s41598-018-31281-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shibata T., Kokubu A., Saito S., Narisawa-Saito M., Sasaki H., Aoyagi K., Yoshimatsu Y., Tachimori Y., Kushima R., Kiyono T., et al. NRF2 Mutation Confers Malignant Potential and Resistance to Chemoradiation Therapy in Advanced Esophageal Squamous Cancer. Neoplasia. 2011;13:864–873. doi: 10.1593/neo.11750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shibata T., Ohta T., Tong K.I., Kokubu A., Odogawa R., Tsuta K., Asamura H., Yamamoto M., Hirohashi S. Cancer Related Mutations in NRF2 Impair its Recognition by Keap1-Cul3 E3 Ligase and Promote Malignancy. Proc. Natl. Acad. Sci. USA. 2008;105:13568–13573. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guichard C., Amaddeo G., Imbeaud S., Ladeiro Y., Pelletier L., Maad I.B., Calderaro J., Bioulac-Sage P., Letexier M., Degos F., et al. Integrated Analysis of Somatic Mutations and Focal Copy-Number Changes Identifies Key Genes and Pathways in Hepatocellular Carcinoma. Nat. Genet. 2012;44:694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J., Shao M., Liu M., Peng P., Li L., Wu W., Wang L., Duan F., Zhang M., Song S., et al. PKCalpha Promotes Generation of Reactive Oxygen Species Via DUOX2 in Hepatocellular Carcinoma. Biochem. Biophys. Res. Commun. 2015;463:839–845. doi: 10.1016/j.bbrc.2015.06.021. [DOI] [PubMed] [Google Scholar]
- 26.Gentric G., Maillet V., Paradis V., Couton D., L’Hermitte A., Panasyuk G., Fromenty B., Celton-Morizur S., Desdouets C. Oxidative Stress Promotes Pathologic Polyploidization in Nonalcoholic Fatty Liver Disease. J. Clin. Investig. 2015;125:981–992. doi: 10.1172/JCI73957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakurai T., He G., Matsuzawa A., Yu G.Y., Maeda S., Hardiman G., Karin M. Hepatocyte Necrosis Induced by Oxidative Stress and IL-1 Alpha Release Mediate Carcinogen-Induced Compensatory Proliferation and Liver Tumorigenesis. Cancer Cell. 2008;14:156–165. doi: 10.1016/j.ccr.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z., Li Z., Ye Y., Xie L., Li W. Oxidative Stress and Liver Cancer: Etiology and Therapeutic Targets. Oxid. Med. Cell. Longev. 2016;2016:7891574. doi: 10.1155/2016/7891574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koliaki C., Szendroedi J., Kaul K., Jelenik T., Nowotny P., Jankowiak F., Herder C., Carstensen M., Krausch M., Knoefel W.T., et al. Adaptation of Hepatic Mitochondrial Function in Humans with Non-Alcoholic Fatty Liver is Lost in Steatohepatitis. Cell Metab. 2015;21:739–746. doi: 10.1016/j.cmet.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 30.Malik A.N., Czajka A. Is Mitochondrial DNA Content a Potential Biomarker of Mitochondrial Dysfunction? Mitochondrion. 2013;13:481–492. doi: 10.1016/j.mito.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 31.Satapati S., Sunny N.E., Kucejova B., Fu X., He T.T., Mendez-Lucas A., Shelton J.M., Perales J.C., Browning J.D., Burgess S.C. Elevated TCA Cycle Function in the Pathology of Diet-Induced Hepatic Insulin Resistance and Fatty Liver. J. Lipid Res. 2012;53:1080–1092. doi: 10.1194/jlr.M023382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiemann S.U., Satyanarayana A., Tsahuridu M., Tillmann H.L., Zender L., Klempnauer J., Flemming P., Franco S., Blasco M.A., Manns M.P., et al. Hepatocyte Telomere Shortening and Senescence are General Markers of Human Liver Cirrhosis. FASEB J. 2002;16:935–942. doi: 10.1096/fj.01-0977com. [DOI] [PubMed] [Google Scholar]
- 33.Vrba J., Modriansky M. Oxidative Burst of Kupffer Cells: Target for Liver Injury Treatment. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech. Repub. 2002;146:15–20. doi: 10.5507/bp.2002.003. [DOI] [PubMed] [Google Scholar]
- 34.Mahmoudvand S., Shokri S., Taherkhani R., Farshadpour F. Hepatitis C Virus Core Protein Modulates several Signaling Pathways Involved in Hepatocellular Carcinoma. World J. Gastroenterol. 2019;25:42–58. doi: 10.3748/wjg.v25.i1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chusri P., Kumthip K., Hong J., Zhu C., Duan X., Jilg N., Fusco D.N., Brisac C., Schaefer E.A., Cai D., et al. HCV Induces Transforming Growth Factor beta1 through Activation of Endoplasmic Reticulum Stress and the Unfolded Protein Response. Sci. Rep. 2016;6:22487. doi: 10.1038/srep22487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen K.C., Willmore W.G., Tayabali A.F. Cadmium Telluride Quantum Dots Cause Oxidative Stress Leading to Extrinsic and Intrinsic Apoptosis in Hepatocellular Carcinoma HepG2 Cells. Toxicology. 2013;306:114–123. doi: 10.1016/j.tox.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 37.Lin W., Tsai W.L., Shao R.X., Wu G., Peng L.F., Barlow L.L., Chung W.J., Zhang L., Zhao H., Jang J.Y., et al. Hepatitis C Virus Regulates Transforming Growth Factor beta1 Production through the Generation of Reactive Oxygen Species in a Nuclear Factor kappaB-Dependent Manner. Gastroenterology. 2010;138:2509–2518.e1. doi: 10.1053/j.gastro.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gong G., Waris G., Tanveer R., Siddiqui A. Human Hepatitis C Virus NS5A Protein Alters Intracellular Calcium Levels, Induces Oxidative Stress, and Activates STAT-3 and NF-Kappa B. Proc. Natl. Acad. Sci. USA. 2001;98:9599–9604. doi: 10.1073/pnas.171311298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKillop I.H., Schmidt C.M., Cahill P.A., Sitzmann J.V. Altered Expression of Mitogen-Activated Protein Kinases in a Rat Model of Experimental Hepatocellular Carcinoma. Hepatology. 1997;26:1484–1491. doi: 10.1002/hep.510260615. [DOI] [PubMed] [Google Scholar]
- 40.Mahmoud A.M., Mohammed H.M., Khadrawy S.M., Galaly S.R. Hesperidin Protects Against Chemically Induced Hepatocarcinogenesis Via Modulation of Nrf2/ARE/HO-1, PPARgamma and TGF-beta1/Smad3 Signaling, and Amelioration of Oxidative Stress and Inflammation. Chem. Biol. Interact. 2017;277:146–158. doi: 10.1016/j.cbi.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 41.Zhang D.D., Lo S.C., Cross J.V., Templeton D.J., Hannink M. Keap1 is a Redox-Regulated Substrate Adaptor Protein for a Cul3-Dependent Ubiquitin Ligase Complex. Mol. Cell. Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Motohashi H., Yamamoto M. Nrf2-Keap1 Defines a Physiologically Important Stress Response Mechanism. Trends Mol. Med. 2004;10:549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 43.Kobayashi A., Kang M.I., Okawa H., Ohtsuji M., Zenke Y., Chiba T., Igarashi K., Yamamoto M. Oxidative Stress Sensor Keap1 Functions as an Adaptor for Cul3-Based E3 Ligase to Regulate Proteasomal Degradation of Nrf2. Mol. Cell. Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Basak P., Sadhukhan P., Sarkar P., Sil P.C. Perspectives of the Nrf-2 Signaling Pathway in Cancer Progression and Therapy. Toxicol. Rep. 2017;4:306–318. doi: 10.1016/j.toxrep.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moi P., Chan K., Asunis I., Cao A., Kan Y.W. Isolation of NF-E2-Related Factor 2 (Nrf2), a NF-E2-Like Basic Leucine Zipper Transcriptional Activator that Binds to the Tandem NF-E2/AP1 Repeat of the Beta-Globin Locus Control Region. Proc. Natl. Acad. Sci. USA. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Motohashi H., Katsuoka F., Engel J.D., Yamamoto M. Small Maf Proteins Serve as Transcriptional Cofactors for Keratinocyte Differentiation in the Keap1-Nrf2 Regulatory Pathway. Proc. Natl. Acad. Sci. USA. 2004;101:6379–6384. doi: 10.1073/pnas.0305902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hirotsu Y., Katsuoka F., Funayama R., Nagashima T., Nishida Y., Nakayama K., Engel J.D., Yamamoto M. Nrf2-MafG Heterodimers Contribute Globally to Antioxidant and Metabolic Networks. Nucleic Acids Res. 2012;40:10228–10239. doi: 10.1093/nar/gks827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taguchi K., Motohashi H., Yamamoto M. Molecular Mechanisms of the Keap1-Nrf2 Pathway in Stress Response and Cancer Evolution. Genes Cells. 2011;16:123–140. doi: 10.1111/j.1365-2443.2010.01473.x. [DOI] [PubMed] [Google Scholar]
- 49.Wasserman W.W., Fahl W.E. Functional Antioxidant Responsive Elements. Proc. Natl. Acad. Sci. USA. 1997;94:5361–5366. doi: 10.1073/pnas.94.10.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim J., Keum Y.S. NRF2, a Key Regulator of Antioxidants with Two Faces towards Cancer. Oxid. Med. Cell. Longev. 2016;2016:2746457. doi: 10.1155/2016/2746457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tong K.I., Katoh Y., Kusunoki H., Itoh K., Tanaka T., Yamamoto M. Keap1 Recruits Neh2 through Binding to ETGE and DLG Motifs: Characterization of the Two-Site Molecular Recognition Model. Mol. Cell. Biol. 2006;26:2887–2900. doi: 10.1128/MCB.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Katoh Y., Iida K., Kang M.I., Kobayashi A., Mizukami M., Tong K.I., McMahon M., Hayes J.D., Itoh K., Yamamoto M. Evolutionary Conserved N-Terminal Domain of Nrf2 is Essential for the Keap1-Mediated Degradation of the Protein by Proteasome. Arch. Biochem. Biophys. 2005;433:342–350. doi: 10.1016/j.abb.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 53.Wang H., Liu K., Geng M., Gao P., Wu X., Hai Y., Li Y., Li Y., Luo L., Hayes J.D., et al. RXRalpha Inhibits the NRF2-ARE Signaling Pathway through a Direct Interaction with the Neh7 Domain of NRF2. Cancer Res. 2013;73:3097–3108. doi: 10.1158/0008-5472.CAN-12-3386. [DOI] [PubMed] [Google Scholar]
- 54.Rada P., Rojo A.I., Chowdhry S., McMahon M., Hayes J.D., Cuadrado A. SCF/Beta-TrCP Promotes Glycogen Synthase Kinase 3-Dependent Degradation of the Nrf2 Transcription Factor in a Keap1-Independent Manner. Mol. Cell. Biol. 2011;31:1121–1133. doi: 10.1128/MCB.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katoh Y., Itoh K., Yoshida E., Miyagishi M., Fukamizu A., Yamamoto M. Two Domains of Nrf2 Cooperatively Bind CBP, a CREB Binding Protein, and Synergistically Activate Transcription. Genes Cells. 2001;6:857–868. doi: 10.1046/j.1365-2443.2001.00469.x. [DOI] [PubMed] [Google Scholar]
- 56.Nioi P., Nguyen T., Sherratt P.J., Pickett C.B. The Carboxy-Terminal Neh3 Domain of Nrf2 is Required for Transcriptional Activation. Mol. Cell. Biol. 2005;25:10895–10906. doi: 10.1128/MCB.25.24.10895-10906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cullinan S.B., Gordan J.D., Jin J., Harper J.W., Diehl J.A. The Keap1-BTB Protein is an Adaptor that Bridges Nrf2 to a Cul3-Based E3 Ligase: Oxidative Stress Sensing by a Cul3-Keap1 Ligase. Mol. Cell. Biol. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li X., Zhang D., Hannink M., Beamer L.J. Crystal Structure of the Kelch Domain of Human Keap1. J. Biol. Chem. 2004;279:54750–54758. doi: 10.1074/jbc.M410073200. [DOI] [PubMed] [Google Scholar]
- 59.Kobayashi A., Kang M.I., Watai Y., Tong K.I., Shibata T., Uchida K., Yamamoto M. Oxidative and Electrophilic Stresses Activate Nrf2 through Inhibition of Ubiquitination Activity of Keap1. Mol. Cell. Biol. 2006;26:221–229. doi: 10.1128/MCB.26.1.221-229.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang D.D., Hannink M. Distinct Cysteine Residues in Keap1 are Required for Keap1-Dependent Ubiquitination of Nrf2 and for Stabilization of Nrf2 by Chemopreventive Agents and Oxidative Stress. Mol. Cell. Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goldstein L.D., Lee J., Gnad F., Klijn C., Schaub A., Reeder J., Daemen A., Bakalarski C.E., Holcomb T., Shames D.S., et al. Recurrent Loss of NFE2L2 Exon 2 is a Mechanism for Nrf2 Pathway Activation in Human Cancers. Cell Rep. 2016;16:2605–2617. doi: 10.1016/j.celrep.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 62.Zavattari P., Perra A., Menegon S., Kowalik M.A., Petrelli A., Angioni M.M., Follenzi A., Quagliata L., Ledda-Columbano G.M., Terracciano L., et al. Nrf2, but Not Beta-Catenin, Mutation Represents an Early Event in Rat Hepatocarcinogenesis. Hepatology. 2015;62:851–862. doi: 10.1002/hep.27790. [DOI] [PubMed] [Google Scholar]
- 63.Fujimoto A., Furuta M., Totoki Y., Tsunoda T., Kato M., Shiraishi Y., Tanaka H., Taniguchi H., Kawakami Y., Ueno M., et al. Whole-Genome Mutational Landscape and Characterization of Noncoding and Structural Mutations in Liver Cancer. Nat. Genet. 2016;48:500–509. doi: 10.1038/ng.3547. [DOI] [PubMed] [Google Scholar]
- 64.Cancer Genome Atlas Research Network Comprehensive Genomic Characterization of Squamous Cell Lung Cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ngo H.K.C., Kim D.H., Cha Y.N., Na H.K., Surh Y.J. Nrf2 Mutagenic Activation Drives Hepatocarcinogenesis. Cancer Res. 2017;77:4797–4808. doi: 10.1158/0008-5472.CAN-16-3538. [DOI] [PubMed] [Google Scholar]
- 66.Orru C., Szydlowska M., Taguchi K., Zavattari P., Perra A., Yamamoto M., Columbano A. Genetic Inactivation of Nrf2 Prevents Clonal Expansion of Initiated Cells in a Nutritional Model of Rat Hepatocarcinogenesis. J. Hepatol. 2018;69:635–643. doi: 10.1016/j.jhep.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 67.Jeong Y., Hoang N.T., Lovejoy A., Stehr H., Newman A.M., Gentles A.J., Kong W., Truong D., Martin S., Chaudhuri A., et al. Role of KEAP1/NRF2 and TP53 Mutations in Lung Squamous Cell Carcinoma Development and Radiation Resistance. Cancer Discov. 2017;7:86–101. doi: 10.1158/2159-8290.CD-16-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Niture S.K., Jaiswal A.K. Nrf2-Induced Antiapoptotic Bcl-xL Protein Enhances Cell Survival and Drug Resistance. Free Radic. Biol. Med. 2013;57:119–131. doi: 10.1016/j.freeradbiomed.2012.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng A., Chevalier N., Calderoni M., Dubuis G., Dormond O., Ziros P.G., Sykiotis G.P., Widmann C. CRISPR/Cas9 Genome-Wide Screening Identifies KEAP1 as a Sorafenib, Lenvatinib, and Regorafenib Sensitivity Gene in Hepatocellular Carcinoma. Oncotarget. 2019;10:7058–7070. doi: 10.18632/oncotarget.27361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Patra K.C., Hay N. The Pentose Phosphate Pathway and Cancer. Trends Biochem. Sci. 2014;39:347–354. doi: 10.1016/j.tibs.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aydin Y., Chedid M., Chava S., Danielle Williams D., Liu S., Hagedorn C.H., Sumitran-Holgersson S., Reiss K., Moroz K., Lu H., et al. Activation of PERK-Nrf2 Oncogenic Signaling Promotes Mdm2-Mediated Rb Degradation in Persistently Infected HCV Culture. Sci. Rep. 2017;7:9223. doi: 10.1038/s41598-017-10087-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 72.Fabrizio F.P., Costantini M., Copetti M., La Torre A., Sparaneo A., Fontana A., Poeta L., Gallucci M., Sentinelli S., Graziano P., et al. Keap1/Nrf2 Pathway in Kidney Cancer: Frequent Methylation of KEAP1 Gene Promoter in Clear Renal Cell Carcinoma. Oncotarget. 2017;8:11187–11198. doi: 10.18632/oncotarget.14492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu X., Sun C., Liu B., Jin X., Li P., Zheng X., Zhao T., Li F., Li Q. Genistein Mediates the Selective Radiosensitizing Effect in NSCLC A549 Cells Via Inhibiting Methylation of the keap1 Gene Promoter Region. Oncotarget. 2016;7:27267–27279. doi: 10.18632/oncotarget.8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nault J.C., Mallet M., Pilati C., Calderaro J., Bioulac-Sage P., Laurent C., Laurent A., Cherqui D., Balabaud C., Zucman-Rossi J. High Frequency of Telomerase Reverse-Transcriptase Promoter Somatic Mutations in Hepatocellular Carcinoma and Preneoplastic Lesions. Nat. Commun. 2013;4:2218. doi: 10.1038/ncomms3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ahmad F., Dixit D., Sharma V., Kumar A., Joshi S.D., Sarkar C., Sen E. Nrf2-Driven TERT Regulates Pentose Phosphate Pathway in Glioblastoma. Cell Death Dis. 2016;7:e2213. doi: 10.1038/cddis.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kansanen E., Kuosmanen S.M., Leinonen H., Levonen A.L. The Keap1-Nrf2 Pathway: Mechanisms of Activation and Dysregulation in Cancer. Redox Biol. 2013;1:45–49. doi: 10.1016/j.redox.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gozzelino R., Jeney V., Soares M.P. Mechanisms of Cell Protection by Heme Oxygenase-1. Annu. Rev. Pharmacol. Toxicol. 2010;50:323–354. doi: 10.1146/annurev.pharmtox.010909.105600. [DOI] [PubMed] [Google Scholar]
- 78.Loboda A., Damulewicz M., Pyza E., Jozkowicz A., Dulak J. Role of Nrf2/HO-1 System in Development, Oxidative Stress Response and Diseases: An Evolutionarily Conserved Mechanism. Cell. Mol. Life Sci. 2016;73:3221–3247. doi: 10.1007/s00018-016-2223-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen R., Cui J., Xu C., Xue T., Guo K., Gao D., Liu Y., Ye S., Ren Z. The Significance of MMP-9 Over MMP-2 in HCC Invasiveness and Recurrence of Hepatocellular Carcinoma After Curative Resection. Ann. Surg. Oncol. 2012;19(Suppl. 3):S375–S384. doi: 10.1245/s10434-011-1836-7. [DOI] [PubMed] [Google Scholar]
- 80.Nart D., Yaman B., Yilmaz F., Zeytunlu M., Karasu Z., Kilic M. Expression of Matrix Metalloproteinase-9 in Predicting Prognosis of Hepatocellular Carcinoma After Liver Transplantation. Liver Transplant. 2010;16:621–630. doi: 10.1002/lt.22028. [DOI] [PubMed] [Google Scholar]
- 81.Zhang Q., Chen X., Zhou J., Zhang L., Zhao Q., Chen G., Xu J., Qian F., Chen Z. CD147, MMP-2, MMP-9 and MVD-CD34 are Significant Predictors of Recurrence After Liver Transplantation in Hepatocellular Carcinoma Patients. Cancer Biol. Ther. 2006;5:808–814. doi: 10.4161/cbt.5.7.2754. [DOI] [PubMed] [Google Scholar]
- 82.Zhang M., Zhang C., Zhang L., Yang Q., Zhou S., Wen Q., Wang J. Nrf2 is a Potential Prognostic Marker and Promotes Proliferation and Invasion in Human Hepatocellular Carcinoma. BMC Cancer. 2015;15:531. doi: 10.1186/s12885-015-1541-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mitsuishi Y., Taguchi K., Kawatani Y., Shibata T., Nukiwa T., Aburatani H., Yamamoto M., Motohashi H. Nrf2 Redirects Glucose and Glutamine into Anabolic Pathways in Metabolic Reprogramming. Cancer Cell. 2012;22:66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 84.Chorley B.N., Campbell M.R., Wang X., Karaca M., Sambandan D., Bangura F., Xue P., Pi J., Kleeberger S.R., Bell D.A. Identification of Novel NRF2-Regulated Genes by ChIP-Seq: Influence on Retinoid X Receptor Alpha. Nucleic Acids Res. 2012;40:7416–7429. doi: 10.1093/nar/gks409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sun Q.K., Zhu J.Y., Wang W., Lv Y., Zhou H.C., Yu J.H., Xu G.L., Ma J.L., Zhong W., Jia W.D. Diagnostic and Prognostic Significance of Peroxiredoxin 1 Expression in Human Hepatocellular Carcinoma. Med. Oncol. 2014;31:786. doi: 10.1007/s12032-013-0786-2. [DOI] [PubMed] [Google Scholar]
- 86.Aguilar-Melero P., Prieto-Alamo M.J., Jurado J., Holmgren A., Pueyo C. Proteomics in HepG2 Hepatocarcinoma Cells with Stably Silenced Expression of PRDX1. J. Proteom. 2013;79:161–171. doi: 10.1016/j.jprot.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 87.Lee D., Xu I.M., Chiu D.K., Lai R.K., Tse A.P., Lan Li L., Law C.T., Tsang F.H., Wei L.L., Chan C.Y., et al. Folate Cycle Enzyme MTHFD1L Confers Metabolic Advantages in Hepatocellular Carcinoma. J. Clin. Investig. 2017;127:1856–1872. doi: 10.1172/JCI90253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Singh A., Happel C., Manna S.K., Acquaah-Mensah G., Carrerero J., Kumar S., Nasipuri P., Krausz K.W., Wakabayashi N., Dewi R., et al. Transcription Factor NRF2 Regulates miR-1 and miR-206 to Drive Tumorigenesis. J. Clin. Investig. 2013;123:2921–2934. doi: 10.1172/JCI66353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Datta J., Kutay H., Nasser M.W., Nuovo G.J., Wang B., Majumder S., Liu C.G., Volinia S., Croce C.M., Schmittgen T.D., et al. Methylation Mediated Silencing of MicroRNA-1 Gene and its Role in Hepatocellular Carcinogenesis. Cancer Res. 2008;68:5049–5058. doi: 10.1158/0008-5472.CAN-07-6655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 90.Eades G., Yang M., Yao Y., Zhang Y., Zhou Q. MiR-200a Regulates Nrf2 Activation by Targeting Keap1 mRNA in Breast Cancer Cells. J. Biol. Chem. 2011;286:40725–40733. doi: 10.1074/jbc.M111.275495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Petrelli A., Perra A., Cora D., Sulas P., Menegon S., Manca C., Migliore C., Kowalik M.A., Ledda-Columbano G.M., Giordano S., et al. MicroRNA/gene Profiling Unveils Early Molecular Changes and Nuclear Factor Erythroid Related Factor 2 (NRF2) Activation in a Rat Model Recapitulating Human Hepatocellular Carcinoma (HCC) Hepatology. 2014;59:228–241. doi: 10.1002/hep.26616. [DOI] [PubMed] [Google Scholar]
- 92.Gan N., Sun X., Song L. Activation of Nrf2 by Microcystin-LR Provides Advantages for Liver Cancer Cell Growth. Chem. Res. Toxicol. 2010;23:1477–1484. doi: 10.1021/tx1001628. [DOI] [PubMed] [Google Scholar]
- 93.Huang Y., Li W., Kong A.N. Anti-Oxidative Stress Regulator NF-E2-Related Factor 2 Mediates the Adaptive Induction of Antioxidant and Detoxifying Enzymes by Lipid Peroxidation Metabolite 4-Hydroxynonenal. Cell Biosci. 2012;2:40. doi: 10.1186/2045-3701-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Levings D.C., Wang X., Kohlhase D., Bell D.A., Slattery M. A Distinct Class of Antioxidant Response Elements is Consistently Activated in Tumors with NRF2 Mutations. Redox Biol. 2018;19:235–249. doi: 10.1016/j.redox.2018.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ikeda H., Nishi S., Sakai M. Transcription Factor Nrf2/MafK Regulates Rat Placental Glutathione S-Transferase Gene during Hepatocarcinogenesis. Biochem. J. 2004;380:515–521. doi: 10.1042/bj20031948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Muramatsu M., Sakai M. Mechanisms of a Tumor Marker, Glutathione Transferase P, Expression during Hepatocarcinogenesis of the Rat. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2006;82:339–352. doi: 10.2183/pjab.82.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Endo H., Owada S., Inagaki Y., Shida Y., Tatemichi M. Glucose Starvation Induces LKB1-AMPK-Mediated MMP-9 Expression in Cancer Cells. Sci. Rep. 2018;8:10122. doi: 10.1038/s41598-018-28074-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu D., Zhang Y., Wei Y., Liu G., Liu Y., Gao Q., Zou L., Zeng W., Zhang N. Activation of AKT Pathway by Nrf2/PDGFA Feedback Loop Contributes to HCC Progression. Oncotarget. 2016;7:65389–65402. doi: 10.18632/oncotarget.11700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sanchez-Rodriguez R., Torres-Mena J.E., Quintanar-Jurado V., Chagoya-Hazas V., Del Castillo E.R., Del Pozo Yauner L., Villa-Trevino S., Perez-Carreon J.I. Ptgr1 Expression is Regulated by NRF2 in Rat Hepatocarcinogenesis and Promotes Cell Proliferation and Resistance to Oxidative Stress. Free Radic. Biol. Med. 2017;102:87–99. doi: 10.1016/j.freeradbiomed.2016.11.027. [DOI] [PubMed] [Google Scholar]
- 100.Li L., Fu J., Liu D., Sun J., Hou Y., Chen C., Shao J., Wang L., Wang X., Zhao R., et al. Hepatocyte-Specific Nrf2 Deficiency Mitigates High-Fat Diet-Induced Hepatic Steatosis: Involvement of Reduced PPARgamma Expression. Redox Biol. 2020;30:101412. doi: 10.1016/j.redox.2019.101412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang C., Tan Y.X., Yang G.Z., Zhang J., Pan Y.F., Liu C., Fu J., Chen Y., Ding Z.W., Dong L.W., et al. Gankyrin has an Antioxidative Role through the Feedback Regulation of Nrf2 in Hepatocellular Carcinoma. J. Exp. Med. 2016;213:859–875. doi: 10.1084/jem.20151208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.You A., Nam C.W., Wakabayashi N., Yamamoto M., Kensler T.W., Kwak M.K. Transcription Factor Nrf2 Maintains the Basal Expression of Mdm2: An Implication of the Regulation of p53 Signaling by Nrf2. Arch. Biochem. Biophys. 2011;507:356–364. doi: 10.1016/j.abb.2010.12.034. [DOI] [PubMed] [Google Scholar]
- 103.Gawrieh S., Dakhoul L., Miller E., Scanga A., deLemos A., Kettler C., Burney H., Liu H., Abu-Sbeih H., Chalasani N., et al. Characteristics, Aetiologies and Trends of Hepatocellular Carcinoma in Patients without Cirrhosis: A United States Multicentre Study. Aliment. Pharmacol. Ther. 2019;50:809–821. doi: 10.1111/apt.15464. [DOI] [PubMed] [Google Scholar]
- 104.Grohmann M., Wiede F., Dodd G.T., Gurzov E.N., Ooi G.J., Butt T., Rasmiena A.A., Kaur S., Gulati T., Goh P.K., et al. Obesity Drives STAT-1-Dependent NASH and STAT-3-Dependent HCC. Cell. 2018;175:1289–1306.e20. doi: 10.1016/j.cell.2018.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Margini C., Dufour J.F. The Story of HCC in NAFLD: From Epidemiology, Across Pathogenesis, to Prevention and Treatment. Liver Int. 2016;36:317–324. doi: 10.1111/liv.13031. [DOI] [PubMed] [Google Scholar]
- 106.Hoki T., Miyanishi K., Tanaka S., Takada K., Kawano Y., Sakurada A., Sato M., Kubo T., Sato T., Sato Y., et al. Increased Duodenal Iron Absorption through Up-Regulation of Divalent Metal Transporter 1 from Enhancement of Iron Regulatory Protein 1 Activity in Patients with Nonalcoholic Steatohepatitis. Hepatology. 2015;62:751–761. doi: 10.1002/hep.27774. [DOI] [PubMed] [Google Scholar]
- 107.Beloribi-Djefaflia S., Vasseur S., Guillaumond F. Lipid Metabolic Reprogramming in Cancer Cells. Oncogenesis. 2016;5:e189. doi: 10.1038/oncsis.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Abramczyk H., Surmacki J., Kopec M., Olejnik A.K., Lubecka-Pietruszewska K., Fabianowska-Majewska K. The Role of Lipid Droplets and Adipocytes in Cancer. Raman Imaging of Cell Cultures: MCF10A, MCF7, and MDA-MB-231 Compared to Adipocytes in Cancerous Human Breast Tissue. Analyst. 2015;140:2224–2235. doi: 10.1039/C4AN01875C. [DOI] [PubMed] [Google Scholar]
- 109.Accioly M.T., Pacheco P., Maya-Monteiro C.M., Carrossini N., Robbs B.K., Oliveira S.S., Kaufmann C., Morgado-Diaz J.A., Bozza P.T., Viola J.P. Lipid Bodies are Reservoirs of Cyclooxygenase-2 and Sites of Prostaglandin-E2 Synthesis in Colon Cancer Cells. Cancer Res. 2008;68:1732–1740. doi: 10.1158/0008-5472.CAN-07-1999. [DOI] [PubMed] [Google Scholar]
- 110.Guillaumond F., Bidaut G., Ouaissi M., Servais S., Gouirand V., Olivares O., Lac S., Borge L., Roques J., Gayet O., et al. Cholesterol Uptake Disruption, in Association with Chemotherapy, is a Promising Combined Metabolic Therapy for Pancreatic Adenocarcinoma. Proc. Natl. Acad. Sci. USA. 2015;112:2473–2478. doi: 10.1073/pnas.1421601112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Qiu B., Ackerman D., Sanchez D.J., Li B., Ochocki J.D., Grazioli A., Bobrovnikova-Marjon E., Diehl J.A., Keith B., Simon M.C. HIF2alpha-Dependent Lipid Storage Promotes Endoplasmic Reticulum Homeostasis in Clear-Cell Renal Cell Carcinoma. Cancer Discov. 2015;5:652–667. doi: 10.1158/2159-8290.CD-14-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yue S., Li J., Lee S.Y., Lee H.J., Shao T., Song B., Cheng L., Masterson T.A., Liu X., Ratliff T.L., et al. Cholesteryl Ester Accumulation Induced by PTEN Loss and PI3K/AKT Activation Underlies Human Prostate Cancer Aggressiveness. Cell Metab. 2014;19:393–406. doi: 10.1016/j.cmet.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Guri Y., Colombi M., Dazert E., Hindupur S.K., Roszik J., Moes S., Jenoe P., Heim M.H., Riezman I., Riezman H., et al. MTORC2 Promotes Tumorigenesis Via Lipid Synthesis. Cancer Cell. 2017;32:807–823.e12. doi: 10.1016/j.ccell.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 114.Xu J., Kulkarni S.R., Donepudi A.C., More V.R., Slitt A.L. Enhanced Nrf2 Activity Worsens Insulin Resistance, Impairs Lipid Accumulation in Adipose Tissue, and Increases Hepatic Steatosis in Leptin-Deficient Mice. Diabetes. 2012;61:3208–3218. doi: 10.2337/db11-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li Y., Chao X., Yang L., Lu Q., Li T., Ding W.X., Ni H.M. Impaired Fasting-Induced Adaptive Lipid Droplet Biogenesis in Liver-Specific Atg5-Deficient Mouse Liver is Mediated by Persistent Nuclear Factor-Like 2 Activation. Am. J. Pathol. 2018;188:1833–1846. doi: 10.1016/j.ajpath.2018.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Moya M., Benet M., Guzman C., Tolosa L., Garcia-Monzon C., Pareja E., Castell J.V., Jover R. Foxa1 Reduces Lipid Accumulation in Human Hepatocytes and is Down-Regulated in Nonalcoholic Fatty Liver. PLoS ONE. 2012;7:e30014. doi: 10.1371/journal.pone.0030014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Suzuki A., Shim J., Ogata K., Yoshioka H., Iwata J. Cholesterol Metabolism Plays a Crucial Role in the Regulation of Autophagy for Cell Differentiation of Granular Convoluted Tubules in Male Mouse Submandibular Glands. Development. 2019;146 doi: 10.1242/dev.178335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Huang J., Tabbi-Anneni I., Gunda V., Wang L. Transcription Factor Nrf2 Regulates SHP and Lipogenic Gene Expression in Hepatic Lipid Metabolism. Am. J. Physiol. Gastrointest. Liver Physiol. 2010;299:G1211–G1221. doi: 10.1152/ajpgi.00322.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mehta K., Van Thiel D.H., Shah N., Mobarhan S. Nonalcoholic Fatty Liver Disease: Pathogenesis and the Role of Antioxidants. Nutr. Rev. 2002;60:289–293. doi: 10.1301/002966402320387224. [DOI] [PubMed] [Google Scholar]
- 120.Souza-Mello V. Peroxisome Proliferator-Activated Receptors as Targets to Treat Non-Alcoholic Fatty Liver Disease. World J. Hepatol. 2015;7:1012–1019. doi: 10.4254/wjh.v7.i8.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pi J., Leung L., Xue P., Wang W., Hou Y., Liu D., Yehuda-Shnaidman E., Lee C., Lau J., Kurtz T.W., et al. Deficiency in the Nuclear Factor E2-Related Factor-2 Transcription Factor Results in Impaired Adipogenesis and Protects Against Diet-Induced Obesity. J. Biol. Chem. 2010;285:9292–9300. doi: 10.1074/jbc.M109.093955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.More V.R., Xu J., Shimpi P.C., Belgrave C., Luyendyk J.P., Yamamoto M., Slitt A.L. Keap1 Knockdown Increases Markers of Metabolic Syndrome After Long-Term High Fat Diet Feeding. Free Radic. Biol. Med. 2013;61:85–94. doi: 10.1016/j.freeradbiomed.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Farombi E.O., Shrotriya S., Na H.K., Kim S.H., Surh Y.J. Curcumin Attenuates Dimethylnitrosamine-Induced Liver Injury in Rats through Nrf2-Mediated Induction of Heme Oxygenase-1. Food Chem. Toxicol. 2008;46:1279–1287. doi: 10.1016/j.fct.2007.09.095. [DOI] [PubMed] [Google Scholar]
- 124.Panieri E., Santoro M.M. ROS Homeostasis and Metabolism: A Dangerous Liason in Cancer Cells. Cell Death Dis. 2016;7:e2253. doi: 10.1038/cddis.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Menegon S., Columbano A., Giordano S. The Dual Roles of NRF2 in Cancer. Trends Mol. Med. 2016;22:578–593. doi: 10.1016/j.molmed.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 126.Sporn M.B., Liby K.T. NRF2 and Cancer: The Good, the Bad and the Importance of Context. Nat. Rev. Cancer. 2012;12:564–571. doi: 10.1038/nrc3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.DeNicola G.M., Karreth F.A., Humpton T.J., Gopinathan A., Wei C., Frese K., Mangal D., Yu K.H., Yeo C.J., Calhoun E.S., et al. Oncogene-Induced Nrf2 Transcription Promotes ROS Detoxification and Tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Umemura A., He F., Taniguchi K., Nakagawa H., Yamachika S., Font-Burgada J., Zhong Z., Subramaniam S., Raghunandan S., Duran A., et al. P62, Upregulated during Preneoplasia, Induces Hepatocellular Carcinogenesis by Maintaining Survival of Stressed HCC-Initiating Cells. Cancer Cell. 2016;29:935–948. doi: 10.1016/j.ccell.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y.S., Ueno I., Sakamoto A., Tong K.I., et al. The Selective Autophagy Substrate p62 Activates the Stress Responsive Transcription Factor Nrf2 through Inactivation of Keap1. Nat. Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 130.Inami Y., Waguri S., Sakamoto A., Kouno T., Nakada K., Hino O., Watanabe S., Ando J., Iwadate M., Yamamoto M., et al. Persistent Activation of Nrf2 through p62 in Hepatocellular Carcinoma Cells. J. Cell Biol. 2011;193:275–284. doi: 10.1083/jcb.201102031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ichimura Y., Waguri S., Sou Y.S., Kageyama S., Hasegawa J., Ishimura R., Saito T., Yang Y., Kouno T., Fukutomi T., et al. Phosphorylation of p62 Activates the Keap1-Nrf2 Pathway during Selective Autophagy. Mol. Cell. 2013;51:618–631. doi: 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 132.Saito T., Ichimura Y., Taguchi K., Suzuki T., Mizushima T., Takagi K., Hirose Y., Nagahashi M., Iso T., Fukutomi T., et al. P62/Sqstm1 Promotes Malignancy of HCV-Positive Hepatocellular Carcinoma through Nrf2-Dependent Metabolic Reprogramming. Nat. Commun. 2016;7:12030. doi: 10.1038/ncomms12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ichimura Y., Komatsu M. Activation of p62/SQSTM1-Keap1-Nuclear Factor Erythroid 2-Related Factor 2 Pathway in Cancer. Front. Oncol. 2018;8:210. doi: 10.3389/fonc.2018.00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.An J.H., Blackwell T.K. SKN-1 Links C. Elegans Mesendodermal Specification to a Conserved Oxidative Stress Response. Genes Dev. 2003;17:1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Motohashi H., O’Connor T., Katsuoka F., Engel J.D., Yamamoto M. Integration and Diversity of the Regulatory Network Composed of Maf and CNC Families of Transcription Factors. Gene. 2002;294:1–12. doi: 10.1016/S0378-1119(02)00788-6. [DOI] [PubMed] [Google Scholar]
- 136.Grimberg K.B., Beskow A., Lundin D., Davis M.M., Young P. Basic Leucine Zipper Protein Cnc-C is a Substrate and Transcriptional Regulator of the Drosophila 26S Proteasome. Mol. Cell. Biol. 2011;31:897–909. doi: 10.1128/MCB.00799-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang W., Chan J.Y. Nrf1 is Targeted to the Endoplasmic Reticulum Membrane by an N-Terminal Transmembrane Domain. Inhibition of Nuclear Translocation and Transacting Function. J. Biol. Chem. 2006;281:19676–19687. doi: 10.1074/jbc.M602802200. [DOI] [PubMed] [Google Scholar]
- 138.Zhang Y., Lucocq J.M., Hayes J.D. The Nrf1 CNC/bZIP Protein is a Nuclear Envelope-Bound Transcription Factor that is Activated by t-Butyl Hydroquinone but Not by Endoplasmic Reticulum Stressors. Biochem. J. 2009;418:293–310. doi: 10.1042/BJ20081575. [DOI] [PubMed] [Google Scholar]
- 139.Fuse Y., Kobayashi M. Conservation of the Keap1-Nrf2 System: An Evolutionary Journey through Stressful Space and Time. Molecules. 2017;22:436. doi: 10.3390/molecules22030436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chenais B., Derjuga A., Massrieh W., Red-Horse K., Bellingard V., Fisher S.J., Blank V. Functional and Placental Expression Analysis of the Human NRF3 Transcription Factor. Mol. Endocrinol. 2005;19:125–137. doi: 10.1210/me.2003-0379. [DOI] [PubMed] [Google Scholar]
- 141.Kobayashi M., Itoh K., Suzuki T., Osanai H., Nishikawa K., Katoh Y., Takagi Y., Yamamoto M. Identification of the Interactive Interface and Phylogenic Conservation of the Nrf2-Keap1 System. Genes Cells. 2002;7:807–820. doi: 10.1046/j.1365-2443.2002.00561.x. [DOI] [PubMed] [Google Scholar]
- 142.Tian W., De la Vega M.R., Schmidlin C.J., Ooi A., Zhang D.D. Kelch-Like ECH-Associated Protein 1 (KEAP1) Differentially Regulates Nuclear Factor Erythroid-2-Related Factors 1 and 2 (NRF1 and NRF2) J. Biol. Chem. 2018;293:2029–2040. doi: 10.1074/jbc.RA117.000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Liu P., Kerins M.J., Tian W., Neupane D., Zhang D.D., Ooi A. Differential and Overlapping Targets of the Transcriptional Regulators NRF1, NRF2, and NRF3 in Human Cells. J. Biol. Chem. 2019;294:18131–18149. doi: 10.1074/jbc.RA119.009591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Leung L., Kwong M., Hou S., Lee C., Chan J.Y. Deficiency of the Nrf1 and Nrf2 Transcription Factors Results in Early Embryonic Lethality and Severe Oxidative Stress. J. Biol. Chem. 2003;278:48021–48029. doi: 10.1074/jbc.M308439200. [DOI] [PubMed] [Google Scholar]
- 145.Kobayashi A., Waku T. New Addiction to the NRF2-Related Factor NRF3 in Cancer Cells: Ubiquitin-Independent Proteolysis through the 20S Proteasome. Cancer Sci. 2020;111:6–14. doi: 10.1111/cas.14244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tsuchiya Y., Taniguchi H., Ito Y., Morita T., Karim M.R., Ohtake N., Fukagai K., Ito T., Okamuro S., Iemura S., et al. The Casein Kinase 2-nrf1 Axis Controls the Clearance of Ubiquitinated Proteins by Regulating Proteasome Gene Expression. Mol. Cell. Biol. 2013;33:3461–3472. doi: 10.1128/MCB.01271-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kapeta S., Chondrogianni N., Gonos E.S. Nuclear Erythroid Factor 2-Mediated Proteasome Activation Delays Senescence in Human Fibroblasts. J. Biol. Chem. 2010;285:8171–8184. doi: 10.1074/jbc.M109.031575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chen J., Wang M., Xiang Y., Ru X., Ren Y., Liu X., Qiu L., Zhang Y. Nrf1 is Endowed with a Dominant Tumor-Repressing Effect Onto the Wnt/beta-Catenin-Dependent and Wnt/beta-Catenin-Independent Signaling Networks in the Human Liver Cancer. Oxid. Med. Cell. Longev. 2020;2020:5138539. doi: 10.1155/2020/5138539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yu M.M., Feng Y.H., Zheng L., Zhang J., Luo G.H. Short Hairpin RNA-Mediated Knockdown of Nuclear Factor Erythroid 2-Like 3 Exhibits Tumor-Suppressing Effects in Hepatocellular Carcinoma Cells. World J. Gastroenterol. 2019;25:1210–1223. doi: 10.3748/wjg.v25.i10.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chowdhury A.M.M.A., Katoh H., Hatanaka A., Iwanari H., Nakamura N., Hamakubo T., Natsume T., Waku T., Kobayashi A. Multiple Regulatory Mechanisms of the Biological Function of NRF3 (NFE2L3) Control Cancer Cell Proliferation. Sci. Rep. 2017;7:12494. doi: 10.1038/s41598-017-12675-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Waku T., Nakamura N., Koji M., Watanabe H., Katoh H., Tatsumi C., Tamura N., Hatanaka A., Hirose S., Katayama H., et al. NRF3-POMP-20S Proteasome Assembly Axis Promotes Cancer Development Via Ubiquitin-Independent Proteolysis of p53 and Retinoblastoma Protein. Mol. Cell. Biol. 2020;40 doi: 10.1128/MCB.00597-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bury M., Le Calve B., Lessard F., Dal Maso T., Saliba J., Michiels C., Ferbeyre G., Blank V. NFE2L3 Controls Colon Cancer Cell Growth through Regulation of DUX4, a CDK1 Inhibitor. Cell Rep. 2019;29:1469–1481.e9. doi: 10.1016/j.celrep.2019.09.087. [DOI] [PubMed] [Google Scholar]
- 153.Chevillard G., Paquet M., Blank V. Nfe2l3 (Nrf3) Deficiency Predisposes Mice to T-Cell Lymphoblastic Lymphoma. Blood. 2011;117:2005–2008. doi: 10.1182/blood-2010-02-271460. [DOI] [PubMed] [Google Scholar]
- 154.Xu Z., Chen L., Leung L., Yen T.S., Lee C., Chan J.Y. Liver-Specific Inactivation of the Nrf1 Gene in Adult Mouse Leads to Nonalcoholic Steatohepatitis and Hepatic Neoplasia. Proc. Natl. Acad. Sci. USA. 2005;102:4120–4125. doi: 10.1073/pnas.0500660102. [DOI] [PMC free article] [PubMed] [Google Scholar]