Abstract
In humans, bone marrow (BM) failure syndromes, both constitutional and acquired, predispose to myeloid malignancies. We have modeled acquired immune aplastic anemia, the paradigmatic disease of these syndromes, in the mouse by infusing lymph node (LN) cells from specific pathogen free (SPF) CD45.1 congenic C57BL/6 (B6) donors into hybrid CByB6F1 recipients housed either in conventional (CVB) or SPF facilities. Severity of BM damage was reduced in CVB recipients; they also had reduced levels of CD44+CD62L− effector memory T cells, reduced numbers of donor-type CD44+ T cells, and reduced expansion of donor-type CD8 T cells carrying T cell receptor β variable regions (TCR-Vβ) 07, 11 and 17. Analyses of fecal samples through 16S rRNA amplicon sequencing revealed greater gut microbiota phylogenetic diversity in CVB mice relative to that of SPF mice. Thus, the presence of a broader spectrum of gut microorganisms in CVB-housed CByB6F1 could have primed recipient animal’s immune system leading to suppression of allogeneic donor T cell activation and expansion and attenuation of host BM destruction. These results suggest potential benefit of a diverse gut microbiota in patients receiving bone marrow transplants.
Keywords: Conventional housing, T cell activation-expansion, bone marrow failure, gut microbiota
1 |. INTRODUCTION
In human bone marrow (BM) failure syndromes, hematopoiesis is deficient, leading to marrow hypocellularity, peripheral blood cytopenias, and clinically in untreated severe cases, to death due to infection, bleeding, and anemia.1 In patients with acquired immune aplastic anemia (AA) who improve blood counts and survive after immunosuppressive therapy, about 15% “evolve”, with development over time to a myeloid neoplasm, myelodysplastic syndrome (MDS) and/or acute myeloid leukemia (AML).2 The risk of myeloid cancer is high also in constitutional BM failure, as in Fanconi anemia, in which basic cellular mechanisms such as DNA repair are genetically defective.3,4 The marrow failure environment and, in immune AA, inflammation appears to favor the development and selection of specific hematopoietic clones, with somatic mutations and/or chromosome aberrations, predisposing to MDS and AML.5 We have developed an animal model to examine the pathophysiology of immune AA and to test for plausible pathophysiologic mechanisms and therapeutic interventions.6,7 The model is also suitable to examine environmental features that influence hematopoiesis and the immune response.
In humans and animals, bacteria, fungi, protozoa, virus and other microorganisms form a microbial ecosystem that help to maintain health.8,9 Reduced microbiota diversity or imbalances between pathogenic and symbiotic microbial species can lead to disease.9–13 Mice housed under specific pathogen free (SPF) conditions have been shown to have reduced immune function relative to mice co-housed with pet-store animals.14 The spectrum of gut microbiota diversity is narrower in SPF-housed mice than in mice living in the wild,15 or in embryo-transferred “wilding” mice.16 Environmental influences (eg., gut-flora rich environments) have been shown to interact with host susceptibility factors to determine the frequency of myc translocations, ultimately leading to mouse plasma cell tumors.17
In the current work, we used our previously-established mouse model to address whether mice born and raised under conventional (CVB) conditions differ from mice born and raised under SPF conditions in their susceptibility to immune-mediated destruction of BM hematopoietic cells. Previous reports indicated that mice housed in germ-free (GF) environments without a normal microbial ecosystem,18,19 or mice treated with antibiotics to eradicate gut microorganisms,20 manifested hematopoietic dysfunction compared to mice housed in SPF environments. Bacteria and their metabolites affect T cell activation and immunity and have been implicated in prevention and alleviation of certain diseases.21,22 Here, we infused lymph node (LN) cells from inbred C57BL/6 (B6) donors into MHC-mismatched hybrid CByB6F1 recipients,6 in order to compare responses of recipients born and housed in CVB and SPF facilities. CByB6F1 CVB mice had higher gut microbiota diversity, reduced levels of BM damage mediated by the infused allogeneic LN cells, and restrained expansion/activation of donor T cells, relative to CByB6F1 SPF mice. Our observations affirm a positive role of the conventional housing environment in augmenting host immune reactivity to allogeneic LN cell-mediated BM damage.
2 |. MATERIALS AND METHODS
2.1 |. Mice and induction of BM failure
Inbred C57BL/6J, BALB/cByJ (BALB), and congenic B6.SJL-PtprcaPep3b/Boy (B6-CD45.1) mice were obtained from the Jackson Laboratory (Bar Harbor, ME) and housed in two different facilities at the National Institutes of Health: a SPF facility with barrier-control, covered cages, individual cage air ventilation, and a CVB facility with suspended shelf, and open-top cage. In both facilities, B6 and BALB mice were inbred to produce new mice and were also cross-bred to produce CByB6F1 hybrids. Animals in both facilities had free access to commercial rodent chow and acidified (pH 2.7–3.0) water, and were used at 2–4 months of age. All animal studies were pre-approved by animal care and use committees at both the National Heart, Lung, and Blood Institute and the National Cancer Institute.
Cervical, axillary and inguinal LNs were removed from B6-CD45.1 donors of the SPF facility. Donor LN cells were homogenized and washed in complete RPMI 1640 media (Invitrogen, CA), filtered through 90 μM nylon mesh (Small Parts, Miami Lake, FL), counted in a Vicell counter (Beckman Coulter, Miami, FL), and injected into hybrid CByB6F1 recipients in both CVB and SPF facilities through lateral tail vein at 28 × 106 BM cells/recipient. Recipient mice were bled and euthanized at day 17 after LN cell infusion for tissue collection and sample analyses.
2.2 |. Cell analyses and histology
Blood was collected from retro-orbital sinus into Eppendorf tubes in the presence of 5 mM ethylenediaminetetraacetic acid (EDTA, Sigma, St. Louis, MO). Complete blood counts (CBC) were performed using a HemaVet 950 analyzer (Drew Scientific, Inc., Miami Lake, FL). After euthanasia, mouse spleen and BM cells were extracted into RPMI 1640 media (Invitrogen, CA, supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 100 μg/ml glutamine), filtered through 90 μM nylon mesh (Small Parts, Miami Lake, FL), and counted in a Vicell counter (Beckman Coulter, Miami, FL). Spleen (SP) and BM cells were stained with antibody mixtures for flow cytometry analyses using a BD Canto II flow cytometer and associated FACS-diva software (Becton Dickson, San Diego, CA). Monoclonal antibodies for murine CD3e (clone 145–2C11), CD4 (Clone GK1.5), CD8a (Clone 53–6.7), CD11b (clone M1/70), CD44 (clone 1M7), CD45R (clone RA3–6B2), CD45.1 (Clone A20), CD48 (HM48–1), CD62L (MEL-14), CD117 (Clone 2B8), CD150 (Clone TC15–12F12.2), erythroid cells (Clone TER-119), granulocytes (Clone RB6–8C5), and stem cell antigen 1 (Sca1, Clone D7) were all from Biolegend (San Diego, CA) and were conjugated to fluorescein isothiocyanate (FITC), phycoerythrin (PE), PE-cyanin 5 (PE-Cy5), PE-cyanin 7 (PE-Cy7), allophycocyanin (APC) or brilliant violet 421 (BV421). FITC-conjugated antibodies for mouse T cell receptor β variable region (TCR-Vβ) 2 (Clone B20.6), 3 (Clone KJ25), 4 (Clone KT4), 5.1/5.2 (Clone MR9–4), 6 (Clone RR4–7), 7 (Clone TR310), 8.1/8.2 (Clone MR5–2), 8.3 (Clone 1B3.3), 9 (Clone MR10–2), 10b (Clone B21.5), 11 (Clone RR3–15), 12 (Clone MR11–1), 13 (Clone MR12–3), 14 (Clone 14–2) and 17a (Clone KJ23) were obtained from BD-Biosciences (San Diego, CA).
Sternums were fixed in 10% neutral buffered formalin, sectioned at 5 µM thickness, and stained with hematoxylin and eosin (VitroVivo Biotech, LLC., Rockville, MD). Slides were scanned and digital files were generated using a Leica Aperio ScanScope CS (Leica Biosystems, Buffalo Grove, IL). TIFF files were generated from the exact same relative area at the exact same magnification (20x or 40x) for each slide. Each slide was examined and confirmed to have adequate quantity and quality of tissue for review under an Olympus BX40 microscope (Olympus, Waltham, MA). Representative photomicrographs were generated from scanned images. Overall percent cellularity of each bone marrow slide was estimated, utilizing blinded-review, by a comparative veterinary pathologist and confirmed by general consensus among authors.
2.3 |. 16S rRNA amplicon sequencing
Fecal pellets were collected from CByB6F1 mice in CVB and SPF facilities at day −2 (N=12) and day 17 (N=8) of LN cell infusion. Fecal samples were stored at −80°C. Before analyses, pellets were loaded individually into wells of a 96-well plate for DNA extraction followed by 16S rRNA gene amplicon sequencing at the National Cancer Institute Microbiome and Genetics Core Facility. Microbiota diversity was determined using established methods for sequencing and analyses as previously documented.23
2.4 |. Data analyses
Data obtained from blood, BM cell counts and flow cytometry were analyzed by unpaired t-test or one-way ANOVA using GraphPad Prism statistical software (GraphPad Software, Inc., La Jolla, CA), or by two-way ANOVA with JMP statistical discovery software (SAS Institute Inc., Cary, NC). Data were presented as means with standard errors.
3 |. RESULTS
3.1 |. Attenuated BM destruction in CVB mice following allogeneic LN cell infusion
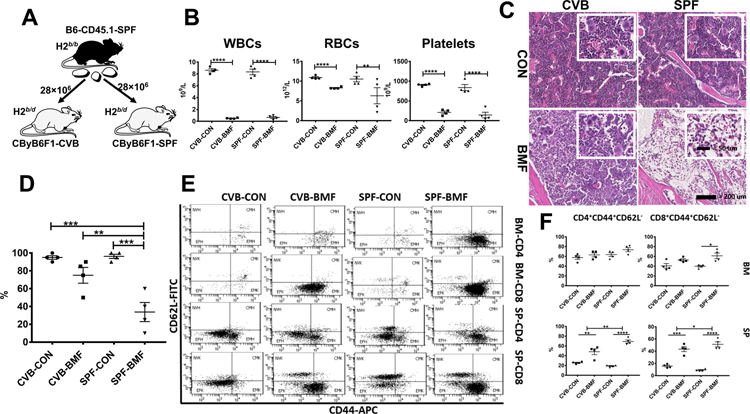
To determine if SPF versus CVB housing conditions affect immune-mediated BM failure, we employed a previously-established B6⇒CByB6F1 LN cell infusion model of BM failure by infusing 28 × 106 LN cells from B6-CD45.1 donors born and housed in SPF facility into CByB6F1 recipients born and housed in CVB and SPF facilities respectively (Figure 1A). At day 17 following LN cell infusion, CByB6F1 recipients from both CVB and SPF facilities developed full-blown BM failure (CVB-BMF and SPF-BMF) with severe declines in white blood cells (WBC), red blood cells (RBC) and platelets relative to control mice (CVB-CON and SPF-CON) without LN cell infusion (Figure 1B). We collected the sternum from CByB6F1 recipients and examined histological changes. Sections of sternum from CVB-CON and SPF-CON mice showed normal cellularity including the presence of mega-size megakaryocytes (Figure 1C). BM cellularity was notably reduced in CVB-BMF mice relative to CVB-CON controls but hematopoietic cells were still visible in the BM cavity (Figure 1C). In contrast, the BM cavities of SPF-BMF mice were essentially empty with no sign of megakaryocytes or other BM cells (Figure 1C). Quantitative analyses of sternum images using a scale of 0 (no depletion) to 4 (high) to score the degree of BM depletion revealed higher levels of BM depletion in SPF-BMF mice relative to mice from other groups (Figure 1D). Of the remaining BM cells, infiltrating CD45.1+ donor LN cells were present at lower proportions (12.2%) in CVB-BMF mice than that in SPF-BMF (20.4%) mice (Fig S1).
FIGURE 1.
Attenuated BM destruction and reduced BM T cell activation in CVB-housed CByB6F1 mice following infusion of allogeneic LN cells from SPF-housed B6-CD45.1 donors. A, LN cells from SPF-housed B6-CD45.1 donors were injected into CByB6F1 recipients in both CVB and SPF facilities at 28 × 106 LN cells/mouse. B, Recipient mice measured 17 days after LN cell infusion (BMF) showed drastic declines in WBCs, RBCs and platelets indicative of BM failure relative to control mice (CON) without LN cell infusion. C, Sterna from BMF and CON mice were collected at day 17, fixed in 10% formalin, decalcified, cut, hematoxylin & eosin stained. Representative 20× photomicrographs (and 40× insets) of sternal BM from mice representing each group: CVB-CON and CVB-BMF or SPF-CON and SPF-BMF. D, Overall percent cellularity of each BM slide was estimated, under blinded-review, by a comparative veterinary pathologist and confirmed by general consensus among authors. E, BM and spleen cells were stained with CD62L and CD44 antibodies to divide T cells into naïve (NV, CD44−CD62L+), center memory (CM, CD44+CD62L+), effector (EF, CD44−CD62L−), and effector memory (EM, CD44+CD62L−) subsets for both CD4 and CD8 cells, shown as representative dot plots. F, Proportions of EM-CD4 and EM-CD8 T cells were calculated and shown as means with standard errors. *, P<.05; **, P<.01; ***, P<.001; ****, P<.0001.
3.2 |. Suppressed T cell activation and reduced T cell expansion in CVB mice following LN cell infusion
Analyses of cellular changes in BM and spleen at day 17 following LN cell infusion revealed large proportions of effector memory (EM, CD44+CD62L−) CD4 and CD8 T cells in the BM and spleens of CVB-BMF and SPF-BMF mice relative to CVB-CON and SPF-CON controls (Figure 1E). In the BM, increases in the EM-CD4 T cell proportion were relatively small for both CVB-BMF/CVB-CON and SPF-BMF/SPF-CON comparisons. In contrast, the EM-CD8 T cell proportion was significantly increased in SPF-BMF relative to SPF-CON controls, but not in CVB-BMF relative to CVB-CON controls (Figure 1F). In the spleen, proportions of EM-CD4 T cells were 19% in SPF-CON, 69% in SPF-BMF, 26% in CVB-CON and 48% in CVB-BMF mice respectively, representing a 2.6-fold increase in SPF-BMF mice and a 0.9-fold increase in CVB-BMF mice (Figure 1F). Proportions of EM-CD8 T cells were 9% in SPF-CON, 51% in SPF-BMF, 16% in CVB-CON and 44% in CVB-BMF mice respectively, showing a 4.6-fold increase in SPF-BMF and a 1.8-fold increase in CVB-BMF mice relative to their respective controls (Figure 1F). Thus, increases in splenic EM-CD4 and EM-CD8 T cells were attenuated in CVB-BMF mice relative to SPF-BMF mice.
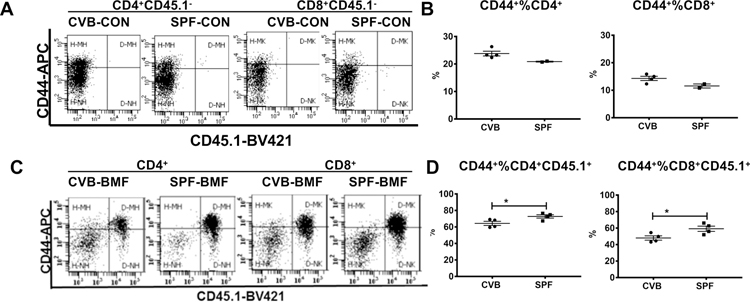
Donor T cell expansion and activation is an important pathophysiological change associated with immune-mediated BM failure in our mouse model;6 the proportional increase in activated T cells in SP and BM is the key cellular change orchestrating the damage to host BM cells.7,24 To distinguish residual host T cells from donor T cells we examined CD44 expression on T cells in SP, an organ that contains large numbers of T and B cells. In CVB-CON and SPF-CON mice, all splenic T cells were of the CD45.1− host type (Figure 2A); proportions of CD44+CD4 and CD44+CD8 T cells were higher in CVB-CON than in SPF-CON mice (Figure 2B). In CVB-BMF and SPF-BMF mice, on the other hand, splenic CD4 and CD8 T cells were mostly CD45.1+ donor-type, with high level expression of CD44 in donor but not host T cells, indicative of donor T cell activation (Figure 2C). Proportions of CD45.1+ donor-type CD44+CD4 and CD44+CD8 T cells were 64% and 48% in CVB-BMF mice and were 73% and 59% in SPF-BMF mice respectively, showing lower levels of donor T cell activation in the spleens of CVB-BMF than in SPF-BMF mice. (Figure 2D). We also measured CD44 expression on T cells in the BM. Proportions of CD44+%CD4+ and CD44+%CD8+ T cells were both high in CVB-CON and SPF-CON mice (Figure S2A, S2B). In CVB-BMF and SPF-BMF mice, T cell infiltration/expansion in the BM (Figures S2C, S2D) followed the same trend as seen in the spleen (Figures 2C, 2D), but the effects were less prominent.
FIGURE 2.
CVB housing increases host but reduces donor memory T cell proportions in the spleen following allogeneic LN cell infusion. Lymph node (LN) cells from SPF B6-CD45.1 donors were infused into CByB6F1 recipients in both CVB and SPF facilities at 28 × 106 cells/mouse. Animals were euthanized 17 days later and spleen cells were extracted and analyzed. A, Proportions of CD44+ memory CD4 and CD8 T cells in CVB-CON and SPF-CON control mice (CD45.1−) shown as representative dot plots. B, Proportions of CD44+ memory CD4 and CD8 T cells in CVB-CON and SPF-CON control mice shown as means with standard errors. C, Proportions of donor-type (CD45.1+) or host-type (CD45.1−) CD4+CD44+ and CD8+CD44+ memory T cells in CVB-BMF and SPF-BMF mice shown as representative dot plots. D, Proportions of donor-type (CD45.1+) CD4+CD44+ and CD8+CD44+ memory T cells in CVB-BMF and SPF-BMF mice shown as means with standard errors. *, P<.05.
3.3 |. Reduced clonal T cell expansion in CVB mice following LN cell infusion
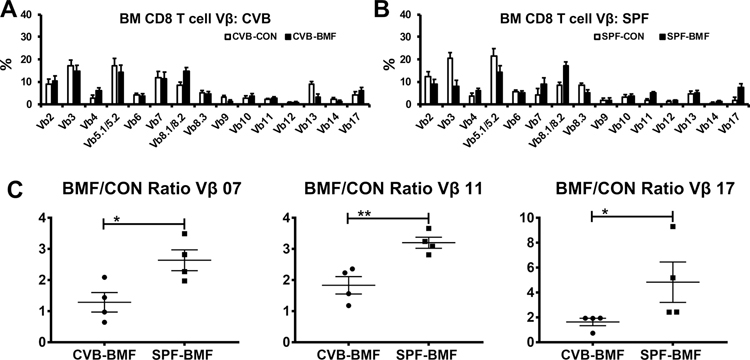
We monitored T cell clonal expansion in BM and Spleen by determining relative proportions of TCR Vβ subsets. Since the majority of BM T cells in BMF mice were of donor-origin (CD45.1) similar to those shown in Fig 2C, we focused on comparing donor-type T cells in BMF mice with T cells from CON mice. These analyses revealed no difference in TCR Vβ usage in CD4 T cells between CVB-BMF and SPF-BMF mice relative to their CVB-CON and SPF-CON controls in either BM or SP (Figure S3). In BM CD8 T cells, TCR Vβ representation patterns differed between donor-type T cells from CVB-BMF mice and host T cells from CVB-CON mice (Figure 3A), as well as between donor-type T cells from SPF-BMF mice and host T cells from SPF-CON mice (Figure 3B). A greater representation of TCR Vβ 8.1/8.2 in donor CD8 T cells was found in both CVB-BMF (Figure 3A) and SPF-BMF (Figure 3B) mice relative to the CD8 T cells of CVB-CON and SPF-CON mice. We calculated CVB-BMF/CVB-CON ratios and SPF-BMF/SPF-CON ratios for all 15 TCR Vβ subsets, and found the ratios of TCR Vβ 07, 11 and 17 to be significantly lower in CVB than in SPF mice (Figure 3C), indicating that expansion of CD8 T cells specific for these three TCR Vβ subsets was attenuated in CVB-BMF mice. In splenic CD8 T cells, TCR Vβ representation patterns also differed between donor-type T cells from CVB-BMF mice and host T cells from CVB-CON mice (Figure S4A), as well as between donor-type T cells from SPF-BMF mice and host T cells from SPF-CON mice (Figure S4B). Proportions of TCR Vβ11 CD8 T cells were higher in CVB-CON than in SPF-CON controls and were lower in CVB-BMF than in SPF-BMF mice (Figure S4C). Thus, the calculated BMF/CON ratio for TCR Vβ11 CD8 T cells was significantly lower in CVB than in SPF mice (Figure S4D), indicating an attenuation of TCR Vβ11 CD8 T cell expansion in CVB-BMF mice.
FIGURE 3.
Reduced donor CD8 T cell Vβ07, 11 and 17 expansion in CVB-BMF mice following allogeneic SPF donor LN cell injection. BM cells were collected from untreated (CON) or LN-cell-infused (BMF) mice from CVB and SPF facilities at day 17 following the infusion of SPF donor LN cells, and were stained with a panel of 15 antibodies specific for T cell receptor β variable regions (Vβ). A, Relative proportions of Vβ subsets in host CD8 T cells from CVB-CON mice and relative proportions of Vβ subsets in donor-type CD8 T cells from CVB-BMF mice. B, Relative proportions of Vβ subsets in host CD8 T cells from SPF-CON mice and relative proportions of Vβ subsets in donor-type CD8 T cells from SPF-BMF mice. C, Proportions of donor-type CD8 T cell Vβ in CVB-BMF and SPF-BMF mice over average proportion of host CD8 T cell Vβ in CVB-CON and SPF-CON mice were calculated as ratios, showing reduced expansion in Vβ 07, Vβ 11 and Vβ 17 CD8 T cells in CVT-BMF mice relative to SPF-BMF mice. Data shown as means with standard errors. *, P<.05; **, P<.01.
3.4 |. Gut microbiota diversity is increased in CVB mice
We collected fecal samples from CByB6F1 recipients in both CVB and SPF facilities and performed 16S rRNA amplicon sequencing to characterize gut microbial composition. Indeed, we found that gut microbiota phylogenetic diversity was higher in CVB mice with significantly more operational taxa unit (OTU) than in SPF mice (Figure 4A). The distribution of OTUs formed two clusters by Bray Curtis principal components analysis for CVB and SPF samples respectively (Fig.4B); no obvious differences were observed between samples collected before and after the induction of BM failure, with only one outlier sample from a CVB-BMF mouse showing a high level (~70%) of Enterobacteriaceae (Figure 4B). LefSe analysis shows the differentially abundant taxa for CVB and SPF mice (Figure 4C). Thus, mice born and housed in the CVB facility carry significantly higher gut microbiota diversity in general. Prevotellaceae, Rikenellaceae, Lactobacillaceae were abundant in the CVB mice and underrepresented in the SPF mice; similarly, Erysipelotrichaceae, Bifidobacteriaceae, Clostridiaceae were abundant in the SPF mice and underrepresented in the CVB mice.
FIGURE 4.
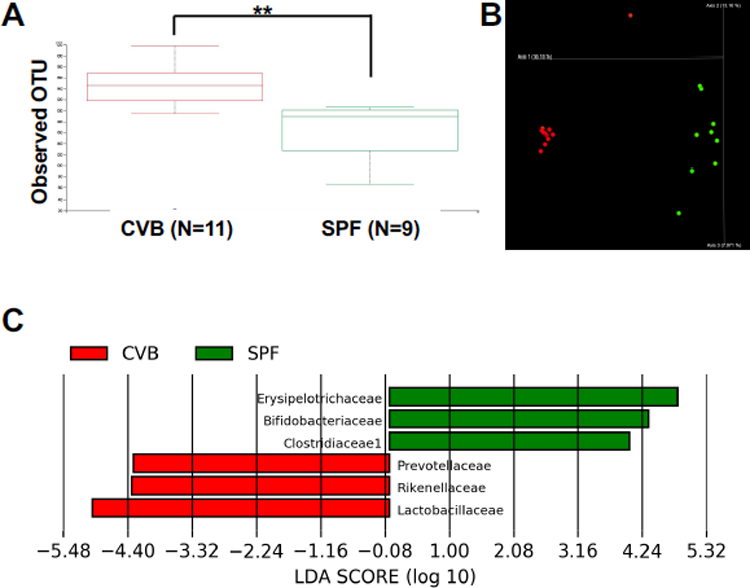
Gut microbiota for CVB and SPF mice. A, 16S rRNA amplicon sequencing was performed on fecal samples from CByB6F1 mice of CVB (red) and SPF (green) facilities and the alpha diversity boxplots represent the significant difference in number of operation taxa units (OTUs) observed across the two housing conditions. The Kruskal-Wallis test was applied for statistical analysis. B, Microbiota OTUs from CVB and SPF mice formed two clusters by bray_curtis_emperor principle components analysis. C,16S rRNA amplicon sequencing data were further processed using the LefSe application to show highly differentially abundant taxa across the two housing conditions. These taxa were detected with very high LDA scores of > 4. **, P<.01.
Correlation analyses were performed between hematopoietic cell population sizes and the levels of the six differentially represented microbial families (Fig. 4C) from fecal samples collected 17 days after LN cell infusion (Tables S1, S2). Surprisingly, fecal Bifidobacteriaceae (enriched in SPF samples) levels were negatively correlated with several blood cell measurements, especially red blood cells, hemoglobin and hematocrit, and positively correlated with the percentages of CD4 and CD8 T cells in both spleen and BM (Table S1). Measurements for hematopoietic populations showed weak but positive correlations with levels of fecal Rikenellaceae (enriched in CVB samples) (Table S2). There is a positive correlation between the percent of CD44/CD4 double positive cells and levels of Prevotellaceae (enriched in CVB samples). However, in general, correlations were not strong between hematopoietic populations and levels of the other microbial families (Tables S1, S2).
4 |. DISCUSSION
In our study, broad diversity in gut microbiota in recipient animals housed in a conventional environment was associated with enhanced host immunity, suppressed allogeneic donor T cell activation and expansion, and mitigation of immune-mediated damage to host BM hematopoietic cells. Observations in other animal models and in patients also indicate that microbial diversity influences immune responses and disease outcomes. Co-housing laboratory mice with pet-store mice enabled co-housed mice to have increased microbial exposure with increased immunity toward Listeria monocytogenes.14 In scurfy mice, deficiency of regulatory T cells resulted in gut microbial dysbiosis and autoimmunity while remodeling microbiota with Lactobacillus reuteri reduced multiorgan inflammation and prolonged survival.22 In the clinic, adult survivors of childhood acute lymphoblastic leukemia had reduced anal microbiota diversity in association with signs of immune dysregulation and chronic inflammation.25 In allogeneic hematopoietic cell transplantation recipients, treatment with fecal microbiota transplantation from third party donors was effective in restoring patient gut microbiota diversity and reducing non-commensal infection.26,27
Proportions of activated T cells (CD44+ CD4 and CD8), especially effector memory T cells (CD62L−CD44+ CD4 and CD8), were higher in the spleens of CVB-CON versus SPF-CON mice (without allogeneic LN cell infusion). These observations are consistent with earlier reports in which memory and effector memory T cells were proportionately higher in laboratory mice co-housed with pet-store mice, and correlated with increased immune activity against infection.14 Thus, the greater diversity of microbiota in control CVB mice may have augmented baseline levels of TCR Vβ07, 11 and 17 CD8 T cells in CVB mice, thereby limiting the expansion of SPF-B6-CD45.1 donor cells and reducing immune responses against host BM cells.
We found greater diversity in gut microbiota in CVB compared to SPF mice. Furthermore, members of the Prevotellaceae, Rikenellaceae and Lactobacillaceae were most abundant in CVB mice while members of the Erysipelotrichaceae, Bifidobacteriaceae, Clostridiaceae were more adundant in SPF mice. With observations of increased resistance to allogeneic LN infusion and reduced allogeneic LN cell infusion-induced immune responses in CVB mice, our findings are in accordance with previous observations. Postbiotic metabolites produced by six strains of Lactobacillus exhibited selective cytotoxicity against malignant cancer cells in a strain-specific and cancer cell-type specific manner while sparing normal cells.28 In a Rag1−/− mouse model, fecal transplantation increased the presence of Prevotellaceae and orchestrated recovery of hematopoietic stem and progenitor cells.29 In a model of experimental autoimmune encephalomyelitis, a specific strain of Lactobacillaceae was found to reduce inflammatory cell infiltration and to ameliorate overall disease.30 Human patients with Parkinson’s disease relative to healthy controls have lower abundance of Prevotellaceae,35 while low abundance of Rikenellaceae was found in patients with liver cirrhosis.36 Thus, the three bacterial families with increased representation in CVB mice are beneficial in the maintenance of normal cellular function/homeostasis and may help to prevent certain diseases/cancers.
In contrast, the three bacterial families with increased abundance in SPF mice are related to inflammatory responses in general. Patients with inflammatory bowel disease associated arthropathy or with rheumatoid arthritis were found to have increased abundance of Clostridiaceae in their stool samples.39 The importance of Erysipelotrichaceae in human health has been linked to their effect on host lipid metabolism and their association with inflammatory responses.31 Interestingly, patients with chronic human immunodeficiency virus infection following effective antiretroviral therapy had increased abundance of Erysipelotrichaceae (seen in our SPF mice) and reduced presence of Rikenellaceae (seen in our CVB mice) family members.37 Bifidobacteriaceae family members are the most dominant gut inhabitants in early life in humans and animals.32,33
The higher gut microbiota diversity in CVB mice relative to SPF mice echoes other comparisons between wild and domesticated mice,15,16 and between mice with and without antibiotic treatment.20,40–42 While antibiotic treatment reduces gut microbiota diversity in general, the effect may vary depending on baseline microbiota composition,42 as well as on the type of antibiotics used.43 One specific example is Vancomycin which causes no significant decrease in the overall bacterial biomass in mouse caecal content but alters the composition of microbiota by eliminating Anaeroplasmataceae, Rikenellaceae and Ruminococcaceae and expanding Lactobacillaceae.40 Future studies would help to determine whether antibiotic treatment could alter the gut microbiota and phenocopy the gut diversity of SPF mice.
The abundance of Bifidobacteriaceae family members in SPF mice was simultaneously negatively correlated with several hematopoietic cell populations and positively correlated with CD4/CD8 T cell percentages, suggesting a potential connection between Bifidobacteriaceae and augmented T cell expansion leading to deficits in mature blood cells. In contrast, Rikenellaceae abundance in CVB mice was positively correlated with some hematopoietic cell populations and negatively correlated with CD4 and CD8 T cell percentages suggesting the possibility of their protective role to mitigate immune responses and marrow damage. Future studies are needed to confirm such functional roles of Bifidobacteriaceae and Rikenellaceae in the regulation of immunity and hematopoiesis.
Conventionally housed mice exhibited increased microbiota phylogenetic diversity and enhanced immune defenses that effectively suppressed the activation and expansion of donor T cells following the infusion of allogeneic LN cells. Thus, microbiota diversity in the gut may modulate immune responses to alleviate immune-mediated BM destruction. Human studies have linked changes in intestinal flora to various clinical outcomes after allogeneic-hematopoietic stem cell transplant; in fact, relapse of primary malignancy is the major hurdle to survival after transplantation.44 Therefore our data in mice add further support to the interactions, reviewed in Andermann et al,45 between hematopoietic cell transplant outcomes and the gut microbiome.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Heart, Lung, and Blood Institute and the National Cancer Institute Intramural Research Programs. JL was supported by fellowships from Natural Science Foundation of Jiangsu Province (Project Number: BK20131034), Jiangsu Provincial Administration of traditional Chinese Medicine (Project number: YB201812) and Jiangsu Government Scholarship for Overseas Studies. Microbiome data are available on request from the authors.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- 1.Ishii K, Young NS. Anemia of Central Origin. Semin Hematol. 2015;52(4):321–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maciejewski JP, Selleri C. Evolution of clonal cytogenetic abnormalities in aplastic anemia. Leuk Lymphoma. 2004;45(3):433–440. [DOI] [PubMed] [Google Scholar]
- 3.Porter CC. Germ line mutations associated with leukemias. Hematology Am Soc Hematol Educ Program. 2016;2016(1):302–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alter BP, Giri N, Savage SA, et al. Malignancies and survival patterns in the National Cancer Institute inherited bone marrow failure syndromes cohort study. Br J Haematol. 2010;150(2):179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Young NS. Aplastic Anemia. N Engl J Med. 2018;379(17):1643–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bloom ML, Wolk AG, Simon-Stoos KL, Bard JS, Chen J, Young NS. A mouse model of lymphocyte infusion-induced bone marrow failure. Exp Hematol. 2004;32(12):1163–1172. [DOI] [PubMed] [Google Scholar]
- 7.Chen J, Lipovsky K, Ellison FM, Calado RT, Young NS. Bystander destruction of hematopoietic progenitor and stem cells in a mouse model of infusion-induced bone marrow failure. Blood. 2004;104(6):1671–1678. [DOI] [PubMed] [Google Scholar]
- 8.Cosorich I, Dalla-Costa G, Sorini C, et al. High frequency of intestinal TH17 cells correlates with microbiota alterations and disease activity in multiple sclerosis. Sci Adv. 2017;3(7):e1700492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marino E, Richards JL, McLeod KH, et al. Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat Immunol. 2017;18(5):552–562. [DOI] [PubMed] [Google Scholar]
- 10.Alexander JL, Wilson ID, Teare J, Marchesi JR, Nicholson JK, Kinross JM. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat Rev Gastroenterol Hepatol. 2017;14(6):356–365. [DOI] [PubMed] [Google Scholar]
- 11.Scott TA, Quintaneiro LM, Norvaisas P, et al. Host-Microbe Co-metabolism Dictates Cancer Drug Efficacy in C. elegans. Cell. 2017;169(3):442–456.e418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tronstad RR, Kummen M, Holm K, et al. Guanylate Cyclase C Activation Shapes the Intestinal Microbiota in Patients with Familial Diarrhea and Increased Susceptibility for Crohn’s Disease. Inflamm Bowel Dis. 2017;23(10):1752–1761. [DOI] [PubMed] [Google Scholar]
- 13.Al-Obaide MAI, Singh R, Datta P, et al. Gut Microbiota-Dependent Trimethylamine-N-oxide and Serum Biomarkers in Patients with T2DM and Advanced CKD. J Clin Med. 2017;6(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beura LK, Hamilton SE, Bi K, et al. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature. 2016;532(7600):512–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosshart SP, Vassallo BG, Angeletti D, et al. Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell. 2017;171(5):1015–1028 e1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosshart SP, Herz J, Vassallo BG, et al. Laboratory mice born to wild mice have natural microbiota and model human immune responses. Science. 2019;365(6452). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janz S. Genetic and environmental cofactors of Myc translocations in plasma cell tumor development in mice. J Natl Cancer Inst Monogr. 2008(39):37–40. [DOI] [PubMed] [Google Scholar]
- 18.Khosravi A, Yanez A, Price JG, et al. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe. 2014;15(3):374–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwamura C, Bouladoux N, Belkaid Y, Sher A, Jankovic D. Sensing of the microbiota by NOD1 in mesenchymal stromal cells regulates murine hematopoiesis. Blood. 2017;129(2):171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Josefsdottir KS, Baldridge MT, Kadmon CS, King KY. Antibiotics impair murine hematopoiesis by depleting the intestinal microbiota. Blood. 2017;129(6):729–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paynich ML, Jones-Burrage SE, Knight KL. Exopolysaccharide from Bacillus subtilis Induces Anti-Inflammatory M2 Macrophages That Prevent T Cell-Mediated Disease. J Immunol. 2017;198(7):2689–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He B, Hoang TK, Wang T, et al. Resetting microbiota by Lactobacillus reuteri inhibits T reg deficiency-induced autoimmunity via adenosine A2A receptors. J Exp Med. 2017;214(1):107–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Namasivayam S, Maiga M, Yuan W, et al. Longitudinal profiling reveals a persistent intestinal dysbiosis triggered by conventional anti-tuberculosis therapy. Microbiome. 2017;5(1):71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen J, Feng X, Desierto MJ, Keyvanfar K, Young NS. IFN-gamma-mediated hematopoietic cell destruction in murine models of immune-mediated bone marrow failure. Blood. 2015;126(24):2621–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chua LL, Rajasuriar R, Azanan MS, et al. Reduced microbial diversity in adult survivors of childhood acute lymphoblastic leukemia and microbial associations with increased immune activation. Microbiome. 2017;5(1):35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeFilipp Z, Hohmann E, Jenq RR, Chen YB. Fecal Microbiota Transplantation: Restoring the Injured Microbiome after Allogeneic Hematopoietic Cell Transplantation. Biol Blood Marrow Transplant. 2019;25(1):e17–e22. [DOI] [PubMed] [Google Scholar]
- 27.DeFilipp Z, Peled JU, Li S, et al. Third-party fecal microbiota transplantation following allo-HCT reconstitutes microbiome diversity. Blood Adv. 2018;2(7):745–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chuah LO, Foo HL, Loh TC, et al. Postbiotic metabolites produced by Lactobacillus plantarum strains exert selective cytotoxicity effects on cancer cells. BMC Complement Altern Med. 2019;19(1):114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwon O, Lee S, Kim JH, Kim H, Lee SW. Altered gut microbita composition in Rag1-deficient mice contributes to modulating hemostasis of hematopoietic stem and progenitor cells. Immune Netw. 2015;15(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He B, Hoang TK, Tian X, et al. Lactobacillus reuteri Reduces the Severity of Experimental Autoimmune Encephalomyelitis in Mice by Modulating Gut Microbiota. Front Immunol. 2019;10:385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaakoush NO. Insights into the Role of Erysipelotrichaceae in the Human Host. Front Cell Infect Microbiol. 2015;5:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mattarelli P, Brandi G, Calabrese C, et al. Occurrence of Bifidobacteriaceae in human hypochlorhydria stomach. Microb Ecol Health Dis. 2014;25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lugli GA, Milani C, Turroni F, et al. Comparative genomic and phylogenomic analyses of the Bifidobacteriaceae family. BMC Genomics. 2017;18(1):568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horie M, Miura T, Hirakata S, et al. Comparative analysis of the intestinal flora in type 2 diabetes and nondiabetic mice. Exp Anim. 2017;66(4):405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scheperjans F, Aho V, Pereira PA, et al. Gut microbiota are related to Parkinson’s disease and clinical phenotype. Mov Disord. 2015;30(3):350–358. [DOI] [PubMed] [Google Scholar]
- 36.Kajihara M, Koido S, Kanai T, et al. Characterisation of blood microbiota in patients with liver cirrhosis. Eur J Gastroenterol Hepatol. 2019. [DOI] [PMC free article] [PubMed]
- 37.Dinh DM, Volpe GE, Duffalo C, et al. Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J Infect Dis. 2015;211(1):19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonai M, Shigehisa A, Kigawa I, et al. Galacto-oligosaccharides ameliorate dysbiotic Bifidobacteriaceae decline in Japanese patients with type 2 diabetes. Benef Microbes. 2017;8(5):705–716. [DOI] [PubMed] [Google Scholar]
- 39.Muniz Pedrogo DA, Chen J, Hillmann B, et al. An Increased Abundance of Clostridiaceae Characterizes Arthritis in Inflammatory Bowel Disease and Rheumatoid Arthritis: A Cross-sectional Study. Inflamm Bowel Dis. 2019;25(5):902–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson CJ, Young VB. Antibiotic administration alters the community structure of the gastrointestinal micobiota. Gut Microbes. 2010;1(4):279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kennedy EA, King KY, Baldridge MT. Mouse Microbiota Models: Comparing Germ-Free Mice and Antibiotics Treatment as Tools for Modifying Gut Bacteria. Front Physiol. 2018;9:1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lavelle A, Hoffmann TW, Pham HP, Langella P, Guedon E, Sokol H. Baseline microbiota composition modulates antibiotic-mediated effects on the gut microbiota and host. Microbiome. 2019;7(1):111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun. 2009;77(6):2367–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peled JU, Jenq RR, Holler E, van den Brink MR. Role of gut flora after bone marrow transplantation. Nat Microbiol. 2016;1:16036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andermann TM, Peled JU, Ho C, et al. The Microbiome and Hematopoietic Cell Transplantation: Past, Present, and Future. Biol Blood Marrow Transplant. 2018;24(7):1322–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Microbiome data are available from the authors upon request.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.