Abstract
Microbial biochemicals have been indicated as the primary stimulators of innate immunity, the first line of the body’s defence against infections. However, the influence of topological features on a microbe’s surface on immune responses remains largely unknown. Here we demonstrate the ability of TiO2 microparticles decorated with nanospikes (spiky particles) to activate and amplify the immune response in vitro and in vivo. The nanospikes exert mechanical stress on the cells, which results in potassium efflux and inflammasome activation in macrophages and dendritic cells during phagocytosis. The spiky particles augment antigen-specific humoral and cellular immune responses in the presence of monophosphoryl lipid A and elicit protective immunity against tumour growth and influenza viral infection. The study offers insights into how surface physical cues can tune the activation of innate immunity and provides a basis for engineering particles with increased immunogenicity and adjuvanticity.
Microbial danger signals, collectively defined as pathogen-associated molecular patterns (PAMPs), alert the immune system of the presence of an invader and initiate an adaptive immune response1. PAMPs comprise a variety of biochemical cues or microbial materials, such as lipopolysaccharide (LPS), CpG DNA, viral RNA and so on, sensed by corresponding receptors in the host2,3. Although the extensive research on the effect that PAMPs have on the innate and adaptive immune response has yielded to the development of vaccine adjuvants, such as Toll-like receptor (TLR) agonists1,4,5, the role of physical cues on the microbes’ surface in triggering the immune system remains under investigated. Many pathogens have spike-like nanostructures on their surface, which are known to be crucial for their adhesion and infection6. For example, the influenza virus is decorated by nanospikes that comprise envelope proteins7, and some yeasts8 and bacteria are covered by hairy but rigid pilus or fimbriae9,10. The challenge to separate the surface structural cues on the microbe surface from the biochemical components, however, has hindered the research efforts aimed at uncovering how the nanotopographical cues alone influence the immune system.
So far, a few studies have been carried out to determine whether artificial micro-and/ or nanopatterned features or nanopillar structures attached to a planar substrate could influence immune cells11–13. It was found that nanofeatured substrates trigger the release of inflammatory factors11, modulate cell phenotypes12 and affect cellular events such as phagocytosis13. However, these studies relied on nanostructures fabricated on a planar substrate, with the cells seeded on top of the nanotopographical substrate. This two-dimensional (2D) approach is not amenable for solution-based delivery into the host and does not recapitulate the in vivo situation14, where microbes interact with host cells in 3D.
Here we fabricate TiO2 microparticles decorated with nanospikes and investigate their ability to activate innate immunity in vitro and in vivo. We found that macrophages actively take up spiky particles without the loss of viability or altered expression of genes in association with inflammation or antigen presentation. The spiky particles specifically activate inflammasomes by stimulating the K+ efflux during phagocytosis, probably due to a nanospike-mediated mechanical stress on the cell membrane. The injectable spiky particles upregulate the expression of co-stimulatory molecules like CD40 on dendritic cells (DCs) in an inflammasome-dependent manner and bolster antigen-specific humoral and cellular immune responses in vivo against tumour growth or influenza virus infection when combined with monophosphoryl lipid A (MPL), an agonist of TLR4. The study sheds lights on the significance of nanostructural cues in the regulation of innate immune responses and provides a basis for engineering more potent vaccines and adjuvants with physical cues for immune system activation.
Characterization of spiky particles and cell interfaces
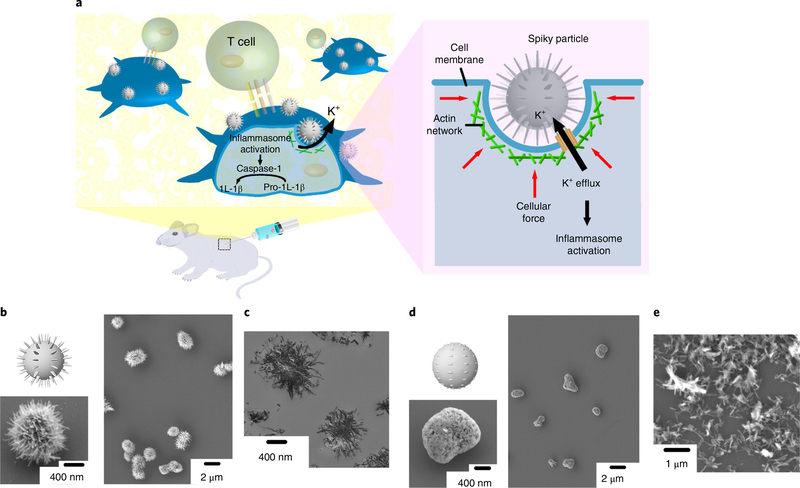
We aimed to address whether spiky particles could activate the innate immune cells in addition to chemical or biological cues, as illustrated in Fig. 1a. To separate the physical cues from chemical or biological ones, we fabricated TiO2 microparticles decorated with nanospikes by a two-step hydrothermal approach, whereby 1D nanostructural bundles were first generated from TiO2 powders, followed by the assembly of the bundle into spiky microparticles15. TiO2 is biologically inert and widely used as an additive in the food, cosmetic and pharmaceutical industries16,17. As revealed by scanning electron microscopy (SEM), the particles were covered with nanospikes (Fig. 1b) that resembled the surface spiky morphology of some bacteria or yeasts (Supplementary Section 1)8,9. The average particle size was 1.8 ± 0.3 μm and the surface spikes were ∼20 nm in diameter and 419 ± 83 nm in length. Transmission electron microscopy (TEM) confirmed that the nanospike structures protruded from the particle core (Fig. 1c). To generate particles with a rough topography devoid of sharp nanospikes for comparison, the spiky particles were sonicated to remove their nanospikes (Fig. 1d). The average diameter of the resultant rough particle was 1.3 ± 0.3 μm and the sonicated-off nanospikes (nanorods) are shown in Fig. 1e.
Fig. 1 |. Schematic and particle fabrication.
a, Schematic of spiky particles applied to activate immune cells and amplify immune responses in vivo. b,c, SEM (b) and TEM (c) images of spiky particles. d,e, SEM images of rough particles (d) and the sonicated-off nanospikes (nanorods) (e).
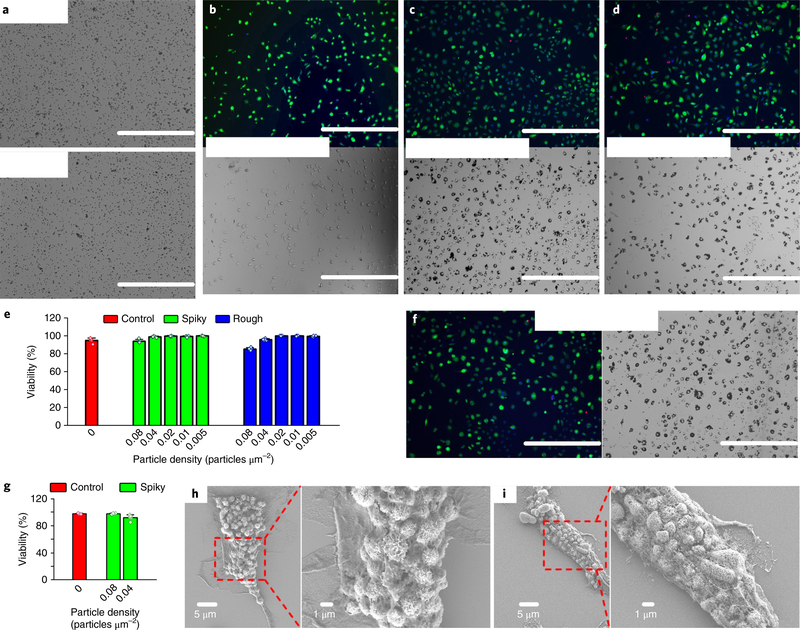
When bone-marrow-derived macrophages (BMMs) were incubated with different doses of microparticles for 48 hours, the cells survived at >90% in all the particle doses tested up to 0.08 particles μm−2 in the presence of either spiky or rough particles (Fig. 2a–e and Supplementary Fig. 2). Prolonged incubation with spiky particles up to 96 hours did not result in any overt cytotoxicity either (Fig. 2f,g). The metabolic activity of BMMs was not altered significantly, nor was apoptosis of the cells, even after their incubation with spiky particles for 72 hours (Supplementary Figs. 3 and 4). The non-cytotoxic feature of the spiky particles was extended to other primary cells, except for a slight decline (<20%) of the metabolic activity in bone-marrow-derived dendritic cells (BMDCs) at the highest dose tested (Supplementary Fig. 3). Although they display no adverse effect on cell viability, both spiky and rough particles were internalized vigorously by BMMs after 24 hours of incubation, as revealed by SEM (Fig. 2h,i).
Fig. 2 |. BMM viability and cell–particle interface study.
a, Optical images of spiky (top) and rough (bottom) particles at a typical dose of 0.04 particles μm−2. Scale bars, 200 μm. b-d, Fluorescence (top) and optical (bottom) images of BMMs after incubation without particles (b), or with spiky particles (c) or rough particles (d) at 0.04 particles μm−2 for 48h. The assay stained live cells with calcein (green), dead cells with ethidium bromide (red) and cell nuclei with Hoechst (blue). Scale bars, 400 μm. e, Macrophage viability after incubation with the indicated particles at different doses for 48h. n = 5 biologically independent samples. f, Fluorescent (left) and optical (right) images of BMMs after incubation with spiky particles at 0.04 particles μm−2 for 96 h. Scale bars, 400 μm. g, Macrophage viability after incubation with spiky particles at the indicated doses for 96h. n = 5 biologically independent samples. h,i, SEM images that show BMMs interfaced with spiky particles (h) and rough particles (i). The particles were observed to be engulfed or fully internalized. All the experiments were repeated twice with similar results. Statistical data presented as the mean± s.d.
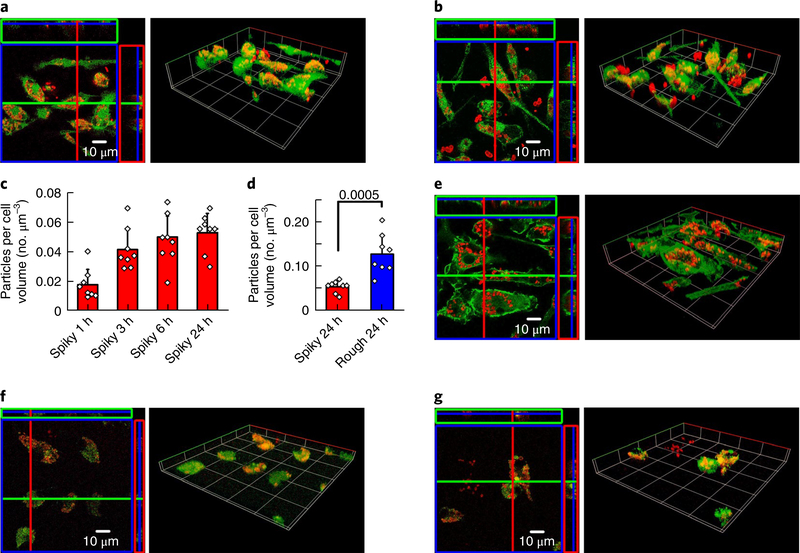
The cell-particle interface was examined with confocal fluorescence microscopy, and the fluorescence images were reconstructed with both orthographic and 3D views (Supplementary Section 5). The particles were conjugated with Alexa Fluor 660 dye (red) and incubated with calcein-labelled BMMs (green) for 24 hours (Fig. 3a,b). Both spiky and rough particles were observed to either accumulate within a cell or adhere on the cell surface. The amount of spiky particle uptake increased rapidly in the first three hours and reached a plateau thereafter (Fig. 3c), but the internalized spiky particles were about twofold less than in rough particles, which suggests that surface nanospikes hindered the microparticle uptake by BMMs (Fig. 3d). As for the interaction of the particles with cells, we observed a large number of particles proximal to or in association with the actin network, which was labelled with actin-specific Alexa Fluor 488 phalloidin (Fig. 3e), consistent with the localized assembly of actin filaments to facilitate particle phagocytosis and internalization18. After internalization, the particles appeared to enter endosomes, as evidenced by co-localization of endosome-specific green fluorescent protein and the particles (red), which corroborates that the particles were entrapped within intracellular compartments (Fig. 3f,g).
Fig. 3 |. BMM-particle interface study via confocal fluorescence microscopy.
a,b, BMMs interfaced with spiky particles (a) or rough particles (b) for 24h with cytosol staining: green, cell cytosol; red, particles. The confocal fluorescence images were reconstructed with both the orthographic view (left) and the 3D view (right). c,d, Quantitative analysis of spiky particle uptake at different time points (1, 3, 6 and 24h) (c), and comparison of particle uptake after 24 h of incubation (d). n = 8 independent cells. The significance was calculated by a two-tailed f-test. e, The interface between the BMM actin networks and spiky particles after 24 h of incubation. Green, cell actin network; red, particles. f,g, Spiky particles (f) or rough particles (g) entrapped in cell endosomes after 24h of particle incubation. Green, cell endosomes; red, particles. All the experiments were repeated twice with similar results. Statistical data presented as the mean±s.d.
Inflammasome activation by spiky particles
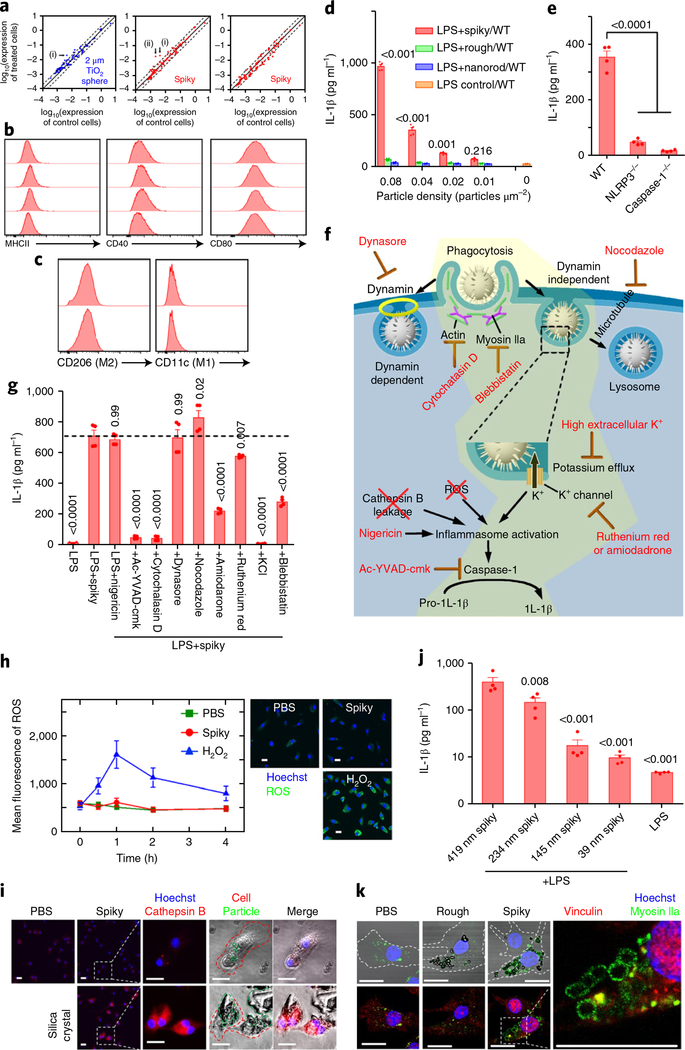
The impact of spiky particles (endotoxin free (Supplementary Section 6)) on gene expressions was investigated by a real-time reverse transcription polymerase chain reaction (RT-PCR) after BMMs were incubated with spiky particles for either 12 or 48 hours (Fig. 4a and Supplementary Fig. 7). Spiky particles alone exhibited little influence (mostly less than twofold) over the expression of 89 genes analysed in association with inflammatory responses and antigen presentation when compared with unstimulated controls or cells cultured with TiO2 microspheres, except for CXCL2, which is associated with inflammasome activation (Fig. 4a)19. No significant change was observed on the cell surface markers important for macrophage activation, such as CD40 and CD80, with the exception of a slight upregulation of major histocompatibility complex class II (MHCII), presumably due to phagocytosis (Fig. 4b)20. Spiky particles did not polarize macrophages either, because the expressions of the M1 marker CD11c and the M2 marker CD206 was unaltered (Fig. 4c)21. Similarly, expressions of M1 marker genes (Tnf, Il6, Il12b and Nos2) and M2 genes (Mrc1, Fizz1, Ym1 and Arg1) in the treated macrophages were unchanged as well (Supplementary Fig. 8)22.
Fig. 4 |. Spiky particles activate inflammasomes.
Particle concentrations were 0.04 particles μm−2 in all the studies except for d. a, Expression levels of 89 genes, which include cytokines, chemokines, cytokine/chemokine receptors and activation markers, were analysed by quantitative real-time RT-PCR after BMMs were cultured with control particles (2 μm TiO2 microspheres) (left) or spiky particles (centre and right) for 12 (left and centre) or 48 h (right). Solid lines indicate no significant changes between the control and treated cells, whereas the dashed lines indicate twofold up- or downregulation. The genes CD28 (i) and CXCL2 (ii) were altered by spiky particle treatments, but CD28 was also slightly upregulated by the control particle treatment. b, Cell activation markers CD40, CD80 and MHCII were analysed by flow cytometry after the BMMs were incubated with spiky (second row) or rough (third row) particles or nanorods (bottom row) for 12 h (the top row is the control). c, Cell differentiation into CD11c+ (M1 marker) or CD206+ (M2 marker) was analysed in BMMs incubated with spiky particles for 12 h (bottom; top is the control). d, The release of IL-1β owing to inflammasome activation was analysed by ELISA. n = 4 biologically independent samples. BMMs from the WT were primed with LPS for 3 h followed by incubation of the cells with the indicated doses of spiky particles, rough particles or nanorods for another 18 h. e, Spiky particle-induced inflammasome activations of BMMs from WT, caspase-1−/− or NLRP3−/− mice were analysed by ELISA. n = 4 biologically independent samples. f, Illustration of possible inflammasome activation mechanisms. The yellow region indicates a feasible mechanism supported by the experiments. g, Elucidation of the mechanism of the underlying inflammasome activation evoked by spiky particles. BMMs were pretreated with the indicated inhibitors for 0.5 h and treated with LPS and spiky particles. The dotted line indicates the mean of the LPS+spiky group. n = 4 biologically independent samples. h, ROS levels after incubation with spiky particles, or with H2O2 as the positive control. ROS levels at different time points are shown (left) and representative images were obtained 1 h after particle incubation (right). Green fluorescence indicates ROS production (blue is Hoechst). Scale bars, 20 μm. n = 10 images. i, The release of cathepsin B (red fluorescence) indicated a frustrated phagocytosis and lysosome disruption. BMMs were incubated for 3 h with spiky particles, or with silica crystals as the positive control. The red dashed lines indicate a cell border, and the green dashed lines indicate particles or silica crystals. Scale bars, 20 μm. j, The particles with different spike lengths (419 ± 83, 234 ±56,145 ± 42 and 39 ± 17nm) were cultured with BMMs in the presence of LPS as in d. IL-1β levels were measured by ELISA. n = 4 biologically independent samples. k, Recruitment of myosin IIa is shown by green fluorescence around the phagocytized particles. BMMs were cultured with rough or spiky particles for 5 h, after which vinculin was labelled with red fluorescence. The dashed line indicates a cell border. Scale bars, 20 μm. Data are presented as the mean± s.e.m. All the experiments were repeated three times with similar results. The significance was calculated by one-way ANOVA compared to the LPS control in d, to the WT mice in e, to LPS+spiky in g or to 419nm spiky+LPS in j.
Inflammasome activation is an important immune mechanism in response to microbes or environmental stress23. Among many events downstream of inflammasome activation, the cleavage of pro-caspase-1 into a mature caspase 1 that facilitates the maturation of pro-interleukin (IL)-iβ into IL-iβ is one of the key events. To increase the pro-IL-iβ availability in the cells, BMMs were primed with LPS and then incubated for 18 h with spiky or rough particles or nanorods at concentrations that ranged from 0.01 to 0.08 particles μm−2. Only spiky particles efficiently triggered the release of IL-iβ in a dose-dependent fashion, and raised IL-iβ levels as high as 17-fold over controls at 0.08 particles μm−2 (Fig. 4d). Under similar conditions, neither rough particles nor nanorods significantly stimulated IL-iβ release over that of controls at all the doses tested. As expected, spiky-particle-mediated release of IL-iβ depended on caspase-1 and nucleotide-binding oligomerization domain, leucine rich repeat and pyrin domain containing 3 (NLRP3) and deletion of either one abrogated the function of spiky particles (Fig. 4e). Vigorous inflammasome activation has been reported to cause pyroptosis24. Indeed, a significant loss of cell viability was found in a caspase-1- and NLRP3-dependent manner when a high amount (0.08 particles μm−2) of spiky particles was incubated with BMMs in the presence of LPS (Supplementary Fig. 9a). In marked contrast, nanorods and rough particles did not induce any pyroptosis significantly, in agreement with their failure to activate inflammasomes. Spiky particles also worked synergistically with MPL, a non-toxic form of LPS and a FDA-approved vaccine adjuvant to induce IL-1β, albeit at a slightly lower efficiency (Supplementary Fig. 9b). Unlike LPS, however, MPL in combination with spiky particles (spiky+MPL) did not impair cell viability in all the doses tested, which argues for a potential in vivo application of spiky+ MPL (Supplementary Fig. 9a). The finding that spiky particles activate inflammasomes only in the presence of biological cues like LPS or MPL suggests that the innate immune system can distinguish microbes from non-biological particles and tailor the immune response to the former accordingly.
Various inhibitors were next employed to interfere selectively with a specific pathway to elucidate the underlying mechanism (Fig. 4f and Supplementary Sections 10 and 11). As shown in Fig. 4g, spiky particles activated the inflammasome at a similar level as that of nigericin, a well-known inflammasome activator from bacteria. The inflammasome activation provoked by spiky particles was not impeded by the dynamin inhibitor, dynasore, which rules out the involvement of dynamin-dependent endocytosis25. In contrast, the caspase-1 inhibitor, Ac-YVAD-cmk, or a phagocytosis inhibitor, cytochalasin D, completely abolished the activation, which corroborates that uptake of spiky particles was mediated by phagocytosis26. Microtubules have been shown to promote the activation of inflammasomes by driving the spatial arrangement of the mitochondria27. This was, however, not the case for inflammasome activation by spiky particles because a microtubule-disrupting drug, nocodazole, did not affect the activation (Fig. 4g). Intracellular reactive oxygen species (ROS) are another well-documented activating factor for inflammasomes28, but ROS were not induced by spiky particles, whereas the cells treated with H2O2 in a parallel study generated abundant ROS (Fig. 4h). Some mineral crystals, such as silica crystals, can activate inflammasomes via a frustrated phagocytosis and trigger cathepsin B release into the cytosol via a damaged endosome membrane, which is also shown in Fig. 4i29. In comparison, spiky particles did not stimulate the release of cathepsin B in the parallel study (Fig. 4i).
It has been reported that nanocones attached to a planar surface could induce membrane curvature30 and activate mechanosensitive ion channels31. A number of mechanosensitive K+ or non-selective cation channels are located in the cell membrane32,33, among which K+ efflux is one of the common mechanisms associated with inflammasome activation34. The nanospikes might potentially introduce a higher mechanical stress on the cell membrane than non-spiky particles by the calculation detailed in Supplementary Section 12. The higher mechanical stress may facilitate the opening of the K+ channels and result in inflammasome activation during phagocytosis. In accordance with this, a graduated decline of spiky-particle-induced IL-1β production was correlated proportionally with the corresponding lengths of the nanospikes (Fig. 4j and Supplementary Fig. 13). Moreover, the inhibition of K+ efflux with a high extracellular K+ concentration completely abrogated the inflammasome activation induced by spiky particles (Fig. 4g). Inhibition of K+ channels with a K+ channel inhibitor, amiodarone or to a less extent, ruthenium red, also significantly blunted the release of IL-1β from BMMs treated with spiky particles (Fig. 4g). These results suggest that the spiky-particle-trigged efflux of K+ is channel dependent and likely to be driven by myosin IIa, a key contractile motor protein that generates mechanical forces during phagocytosis35. In accordance with this, a significant recruitment of myosin IIa (green) was observed around the phagocytized spiky particles, whereas such recruitment was negligible during the phagocytosis of rough particles (Fig. 4k). In addition, a blockade on Myosin IIa recruitment by a specific inhibitor blebbistatin sufficiently blunted the inflammasome activation provoked by spiky particles (Fig. 4g). These findings are in agreement with recent studies demonstrating that membrane mechanical stress could activate the NLRP3 inflammasome via a mechanosensitive channel-mediated K+ efflux32, or alter the physiological process of the cells30,36,37.
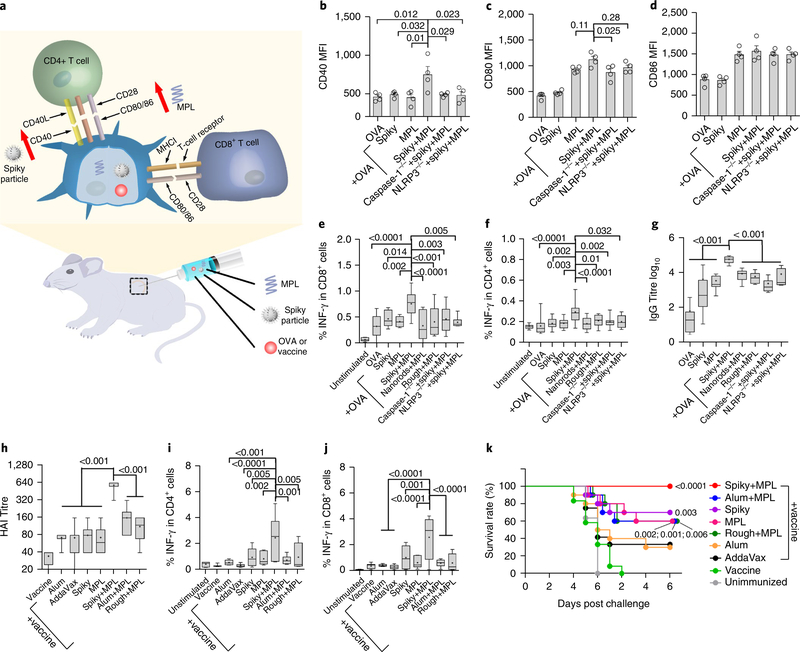
Spiky-particle-mediated promotion of DC maturation
The ability of spiky particles to promote DC maturation was subsequently substantiated in vitro and in vivo in light of an importance of inflammasome activation in DC maturation38. In this regard, we primed BMDCs with MPL and then incubated the cells with spiky particles for 12 hours, followed by analysis of the DC maturation markers CD40, CD80 and CD8639. Spiky particles combined with MPL significantly enhanced the CD40 expression on BMDCs compared to either alone, but such an enhancement was not attained with the CD80 or CD86 expression (Fig. 5a). Under similar conditions, neither rough particles nor nanorods augmented the CD40 expression in MPL-primed BMDCs (Fig. 5b and Supplementary Fig. 14a). Moreover, nanospike-mediated CD40 upregulation was blunted in BMDCs that lacked the two key genes (MyD88 and TRIF) in the TLR4-mediated signalling pathway (Fig. 5c and Supplementary Fig. 14b). CD40 expression was not augmented by spiky+MPL in BMDCs that lacked the caspase-1 or NLRP3 needed for spiky particles to activate the inflammasome (Fig. 5c). CD40 is an important co-stimulatory molecule that transduces ‘licensing’ signals to DCs from CD4+ T cells, by which DCs cross-present antigens into the MHCI pathway that is critical for CD8+ cytotoxic T-cell responses against intracellular pathogens and cancer cells40. To address the potential of spiky+MPL-stimulated DCs in CD8+ T-cell activation, BMDCs were incubated with ovalbumin (OVA) together with SIINFEKL peptide, a specific epitope for OT-I CD8+ T cells, in the presence of different stimuli and then co-cultured with CD8+ T cells isolated from OT-I transgenic mice. As shown in Fig. 5d, spiky+ MPL stimulated the highest amount of interferon gamma (IFN-γ) production compared to other stimuli, a strong indicator of the activation of CD8+ T cells. The T-cell activation again relied on the expression of caspase-1 in BMDCs. To confirm the in vitro observation in in vivo settings, the ability of these activated DCs to inhibit tumour growth was assessed using EG7 tumour cells expressing OVA. Mice were subcutaneously inoculated with EG7 and administered 3 and 7 days later with OVA-primed BMDCs activated by MPL alone or its combination with spiky particles, rough particles or nanorods. The injection of spiky+MPL-activated DCs almost completely blocked tumour growth, whereas MPL alone or with rough particles or nanorods was much less effective (Fig. 5e,f). The immunotherapeutic benefit of spiky+MPL-activated DCs was evident as early as at 15 days after tumour inoculation and continued throughout the entire experimental period, whereas such an immunotherapeutic benefit was observed only on the 21st day in mice that received MPL-activated DCs (Fig. 5e). As expected, spiky+MPL- treated DCs that lacked caspase-1 failed to control tumour growth (Fig. 5e,f), consistent with previous observations that the activation of the NLRP3 inflammasome in DCs conferred adaptive immunity against tumours41.
Fig. 5 |. Spiky particles enhance DC maturation and DC-meditated cancer immunotherapy.
a, Upregulation of the activation markers CD40, CD80 and CD86 on DCs. BMDCs were incubated with spiky particles, MPL or a combination of both for 12h. b, BMDCs were incubated with MPL plus spiky particles, rough particles or nanorods for 12 h, and the expression of the CD40 marker was analysed by flow cytometry. n = 4 biologically independent samples. MFI, mean fluorescence intensity. Representative flow cytometric profiles are given in Supplementary Fig. 14a. c, BMDCs from WT, caspase-1−/−, MyD88/ TRIF−/− and NLRP3−/− mice were cultured with spiky+MPL, and the expression of the CD40 marker was analysed by flow cytometry as in Supplementary Fig. 14b. n = 6 biologically independent samples. The particle dose was 0.04 particles μm−2 in b and c. d, In vitro CD8 T-cell activation assay. BMDCs from WT or caspase-1−/− mice were stimulated with antigen alone (OVA plus SIINFEKL peptide) or coupled with spiky particles, MPL alone or combinations. Activated DCs were subsequently incubated with T cells from OT-I mice for 3 d. The IFN-γ secreted from the T cells was measured by ELISA. n = 4 biologically independent samples. e, DC-mediated cancer immunotherapy. Mice were subcutaneously injected with 5×105 EG7 tumour cells and BMDCs, as in d, were administered 3 and 7d later. The tumour volumes were monitored every 3d. n = 8 tumours. f, The significance between the indicated groups on days 15–21 was calculated by two-way ANOVA and is shown as a heat map. All the experiments were repeated twice with similar results. The significance between the indicated groups in b-d was calculated by one-way ANOVA. Data are presented as the mean± s.e.m. NS, not significant.
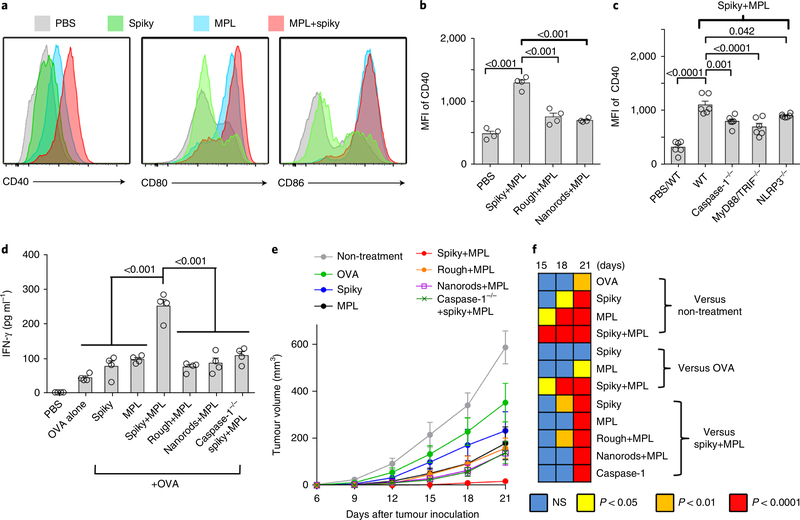
The study described above demonstrated clearly that spiky particles could work synergistically with MPL to promote DC maturation in vitro in an inflammasome-dependent manner. The synergistic effects were subsequently verified in vivo by immunization of wild-type (WT), NLRP3−/− or caspase 1−/− mice with OVA alone or together with MPL, spiky particles or spiky+MPL, as illustrated in Fig. 6a. Analysis of CD40 expression on DCs in draining lymph nodes 36 hours postimmunization revealed a significant upregulation of CD40 only in WT mice that received spiky+MPL, but not in mice that lacked either caspase-1 or NLRP3 (Fig. 6b). Unlike CD40, the expressions of CD80 and CD86 on DCs were enhanced by MPL alone and the enhancement of CD80, but not CD86, could be furthered by spiky particles (Fig. 6c,d).
Fig. 6 |. Spiky particles coupled with MPL as a potent adjuvant.
a, Schematic of a working model for the combination of spiky particles and MPL. b-d, WT, NLRP3−/− or caspase 1−/− mice received subcutaneous injections of OVA with or without spiky particles, MPL or spiky+MPL. The draining lymph nodes were collected 36 h later, and the expressions of CD40 (b), CD80 (c) and CD86 (d) markers on the gate of the DCs were analysed by flow cytometry (gating strategy is given in Supplementary Fig. 15). MFI is presented as the mean ± s.e.m. n = 4 mice. e,f, Cellular immune responses including IFN-γ-secreting CD8+ (e) and CD4+ (f) T cells were measured 7d after immunization. CD4+ and CD8+ T cells that expressed the indicated cytokine were analysed in the gate of CD3+ T cells, as detailed in Supplementary Fig. 16a. From left to right, n = 6, 14, 8, 8, 20, 12, 12, 12 and 6 mice. g, Serum IgG antibody titres were determined 14d after immunization. From left to right, n = 8, 8, 8, 6, 6, 6, 6 and 5 mice. h-k, Influenza vaccination and challenges. h, Mice were immunized on day 0 and day 14 with 2009 H1N1 monovalent influenza vaccine with the indicated adjuvants, including AddaVax (50μl per mouse), alum (1 mg per mouse), spiky (1 mg per mouse), MPL (10 μg per mouse) or spiky+MPL (1 mg / 10 μg per mouse). Sera were collected 7 d after the final immunization and measured for HAI titre. n = 6 mice, except for the vaccine and AddaVax for which n = 8. i,j, Cellular immune responses were also measured 5 d after the final immunization (gating strategy is given in Supplementary Fig. 16b). n = 6 mice, except for vaccine and AddaVax for which n = 8. k, The immunized mice were challenged with 20xLD50 A/California/7/2009 H1N1 influenza virus 14d after the final immunization. Survival rates were monitored for an additional 12d. n = 10 mice, except for vaccine and AddaVax for which n = 12. Survival curves are compared with vaccine group by the Gehan-Breslow-Wilcoxon test. All the experiments were repeated twice with similar results. The significance between the indicated groups in b-j was calculated by one-way ANOVA. Immune response data are presented as box and whiskers, and the mean is shown by a + sign. Body weight data are presented as the mean±s.e.m.
We went on to examine humoral and T-cell immunity in the mice after immunization with OVA in the presence or absence of various stimuli, as above. Mice that received spiky+MPL had the highest percentages of OVA-specific CD8+ and CD4+ T cells that produced IFN-γ. Under similar conditions, neither MPL nor spiky particles elicited a stronger CD8+ T cell response than OVA alone (Fig. 6e,f). There were no significant differences in IL-4-producing CD4+ (IL-4+CD4+) T cells among all the groups tested (Supplementary Fig. 17a), which confirms that spiky particles plus MPL function as a type 1 T helper cell (Th1)-skewed adjuvant similar to MPL. The spiky+MPL also augmented IgG production at levels 1,000-fold higher than OVA alone or 20-fold higher than MPL alone and the augment required caspase-1 or NLRP3 (Fig. 6g). The duo also evoked the highest level of IgG2c production among all the stimuli tested, an indicator of Th1 immune responses, but did not further enhance IgG1-predominant Th2 responses over MPL alone (Supplementary Fig. 18). In marked contrast, MPL coupled with either rough particles or nanorods did not exhibit any significant adjuvant effect relative to MPL alone, which underlines that the nanospikes had the needed surface topography to enhance in vivo immune responses. Furthermore, when CD40-CD40L interaction was transiently blocked by the anti-CD40L antibody injected right after immunization, the adjuvant effect of spiky+MPL on the CD8+ T-cell and humoral responses, and to a much less extent on the CD4+ T-cell response, was significantly blunted (Supplementary Fig. 19). Hence, upregulation of CD40 is likely to be one of the mechanisms behind spiky+MPL adjuvant, in agreement with the importance of CD40 in immune responses42.
Spiky particles coupled with MPL as a potent adjuvant
To demonstrate the clinical relevance of the finding, mice were given two intramuscular vaccinations with the 2009 H1N1 influenza vaccine alone or together with the MF59-like adjuvant (AddaVax), aluminium hydroxide (alum), spiky, MPL, spiky+MPL, alum+MPL or rough particles plus MPL. Alum and MF59 are clinically used as adjuvants in influenza vaccination, and alum+MPL is approved for HPV vaccines in the past decade and also tested in influenza vaccines5,43. Influenza vaccination with spiky+MPL induced haemagglutination inhibition (HAI) antibody titres 20-fold higher than the vaccine alone, eightfold higher than MPL, AddaVax or alum and four-fold higher than alum+MPL (Fig. 6h). The superior adjuvant effect of spiky+MPL was also attained in T-cell immune responses evidenced by the highest level of IFN-γ-producing CD4+ and IFN-γ-producing CD8+ T cells among all the adjuvants tested either alone or in combination (Fig. 6i,j and Supplementary Fig. 20). The duo adjuvants did not boost the level of IL-4+CD4+ T cells significantly (Supplementary Fig. 17b), in contrast to alum which gave rise to the highest level of IL-4+CD4+ T cells, because it is a Th2-biased adjuvant44. The immunized mice were subsequently challenged with a lethal dose (20 × LD50) influenza virus, as previously described45,46. As shown in Fig. 6k, mice that receive the influenza vaccine together with MPL+spiky were fully protected from the viral challenge without any overt body-weight loss, whereas all the other groups of mice had varying levels of significant weight loss (Supplementary Fig. 21). Moreover, all the mice in the spiky+MPL survived group (100%), but the survival rate diminished to 60–70% in mice that received the influenza vaccine combined with spiky, MPL, rough+MPL or alum+MPL, further down to 30–40% in mice immunized with the vaccine and alum or AddaVax and to 0% of mice immunized with the vaccine alone (Fig. 6k). Notably, the same amount of rough particles in combination with MPL was inferior to spiky+MPL in the induction of adaptive immunity and protection against viral challenges.
Conclusions
In summary, TiO2 microparticles decorated with nanospikes were fabricated and explored for their interaction with the innate immune system in vitro and in vivo. The spiky particles introduce mechanical stress on the cell membrane during phagocytosis, which leads to activation of the K+ efflux and inflammasomes in a caspase-1- and NLRP3-dependent fashion. The sensing of the nanospike structures coupled with activation of the TLR4 pathway synergistically promotes DC maturation and significantly bolsters the T-cell and humoral immune responses and the efficacies of DC-mediated cancer immunotherapy and influenza vaccination. The physical cue activates the innate immune system only in the presence of a biological cue, such as MPL or LPS, which may serve as an important check to tailor the immune response specifically to control microbial infections over non-contagious nano- and microparticles. Scientists have been puzzling for a long time as to whether surface topography, like spiky nanostructures, on bacteria and viruses participates in the activation of the innate immune system during microbe invasion. The current study not only sheds light on the potential roles for these nanostructures in the activation of innate immunity, but also forms a basis to engineer spiky surfaces to improve the immunogenicity or adjuvanticity of microparticle-based cancer immunotherapy and prophylactic vaccines.
Methods
Spiky particle fabrication
All the procedures were performed in an endotoxin-free environment. TiO2 (Degussa P25) powder (1 g) was dispersed in 40 ml of NaOH aqueous solution (5 M), stirred for 30 min and transferred into a Teflon-lined stainless-steel autoclave and heated at 120 °C for 24 h. The precipitate was obtained by centrifugation, washed with deionized water to a pH of ∼10.5 and then the precipitate was dried at 60 °C in vacuum. The resultant powder was 1D nanostructure bundles of sodium titanate. Then 0.1 g of the sodium titanate powder was dispersed in a mixture of 19 ml of NaOH aqueous solution (1 M) and 1.25 ml of H2O2 (30%) solution. The suspension was stirred magnetically, ultrasonicated for 30 min and then transferred into a Teflon-lined stainless-steel autoclave to heat at 150 °C for 8 h. The precipitate was attained by centrifugation, washed with deionized water to a pH of ~7 and dried at 60 °C in vacuum to generate sodium titanate spiky particles. The spiky particles were then treated with excess HNO3 solution (0.05 M) with agitation to produce hydrogen titanate spiky particles. The hydrogen titanate spiky particles were then washed with deionized water five times and calcined at 400 °C for 1 h to produce the TiO2 spiky particles. The well-dispersed particles were collected and separated from the aggregated ones based on the different sediment rates of the particles. The TiO2 spiky particles were resuspended in 1 ml of sterile water and added on top of 9 ml of sterile water in a 15 ml centrifuge tube, and the particles allowed to sediment for 30 min. The spiky particles gradually sank to the bottom at different rates dependent on their size. The upper portion of 4 ml that contained the spiky particles of smaller size was collected.
The spiky particles in water were sonicated (Bandelin Sonorex) for 0, 5, 10 or 20 h to produce nanospikes of length 419 ± 97 nm, 234 ± 56 nm, 145 ± 42 nm or 39 ± 17 nm, respectively. Spiky particles in water were sonicated for 16 h to remove the nanospikes on the surface to produce rough particles. The sonicated-off nanospikes were collected in the upper solution as nanorod samples after centrifugation at 5,000 revolutions per minute for 2 min. The precipitated rough particles were washed with sterile water five times prior to use. The spiky particles, rough particles and nanorods were sterilized by autoclaving and stored at 4 °C. The particles were resuspended in a cell culture medium before they were added to the cells or injection into mice.
Particle characterization
The particles were deposited on a Si wafer substrate and sputter-coated with gold-palladium for SEM imaging (Zeiss SUPRA 60). The particles were dispersed in 100% ethanol, embedded in fresh Epon, cut into ultrathin sections and subjected to TEM imaging (FEI Tecnai). To calibrate a particle dose, particles at a series of dilutions were added to a 96-well plate and allowed to sediment for 6 h. The particles were counted under an optical microscope to obtain a number of particles per area. The doses of particles are expressed as the unit of particle number per area (particles μm−2). When placed in a well of the 96-well plate, the dose of spiky particles was converted by 0.04 particles μm−2, equal to 58 μgml−1. A dose of nanorods was determined by the dose of particles used to generate the nanorods. For example, nanorods at a dose of 0.04 particles μm−2 meant that these nanorods were produced from spiky particles at 0.04 particles μm−2.
Animals
WT C57BL/6 mice were obtained from Jackson Laboratories and the Animal Facility of Sun Yat-Sen University and housed on a 12-h day/12-h night schedule (lights on from 19:00 to 07:00) at a constant temperature (22 ± 1 °C) and humidity (60%). Caspase-1 knockout mice (caspase-1−/−) and NLRP3 knockout mice (NLRP3−/−) were obtained from Jackson Laboratories and MyD88/TRIF double knockout mice (MyD88/TRIF−/−) provided by R. Medzhitov (Yale University). Studies were approved by the Animal Care and Use Committee at the Sun Yat-Sen University and Massachusetts General Hospital. All the animals received humane care and every effort was taken to minimize suffering in compliance with institutional and National Institutes of Health guidelines.
BMM and BMDC cultures
BMMs were prepared by culturing mouse bone-marrow cells isolated from WT C57BL/6, caspase-1−/− or NLRP3−/− mice with 100 ng ml−1 macrophage colony-stimulating factor (Biolegend) in PRMI 1640 medium plus 10% fetal bovine serum (FBS) for 7 d. BMDCs were prepared by culturing bone-marrow cells from WT, caspase-1−/− or MyD88/TRIF−/− mice with 10 ng ml−1 of granulocyte macrophage colony-stimulating factor (Biolegend) in PRMI 1640 medium plus 10% FBS for 7 d. BMDCs were also sorted on a highspeed cell sorter based on DC marker CD11c on BD FACSAria. All the procedures and experiments were performed in an endotoxin-free environment.
BMM live/dead assays
To determine the cell viability, BMMs were incubated with spiky particles or rough particles at a dose of 0.005, 0.01, 0.02, 0.04 or 0.08 particles μm−2. After the indicated times of particle treatment, cells were stained with Hoechst to label the cell nuclei, calcein AM to identify live cells and ethidium homodimer to exclude dead cells. The results were analysed by fluorescent microscopy (Mshot and Zeiss Axio).
XTT assays
BMMs, BMDCs and mouse embryonic fibroblasts were cultured with spiky particles at 0.02, 0.04 or 0.08 particles μm−2 for 72 h. To the cells, XTT (Sigma) and phenazine methosulfate (Sigma) were added and incubated for 2 h at 37 °C to measure the metabolic activities of the cells. The A450 values were recorded on a plate reader (Molecular Devices).
Cell–particle interface studies
Cells were cultured on a glass slide for 24 h and incubated with spiky particles or rough particles (0.04 particles μm−2) for an additional 24 h. The cells were fixed with 2% glutaraldehyde for 5 min, gradually dehydrated with ethanol of increasing concentrations (0, 30, 50, 70, 80, 90, 95 and 100% three times) and then prepared by critical point drying (Samdri). The cells were then sputter-coated with gold-palladium for SEM imaging. In addition, the spiky and rough particles were conjugated with red fluorescent dye for confocal imaging. To label the particles, they were first functionalized with (3-aminopropyl)triethoxysilane (APTES, Sigma) by dispersing the particles in ethanol that contained 5% APTES and heating at 60 °C for 3 h. The particles were then washed with ethanol and water, re-dispersed in water and incubated with 1 mg ml−1 Alexa Fluor 660 NHS Ester (succinimidyl ester) for 30 min to conjugate to the dye covalently.
BMMs were cultured onto a glass-bottom plate for 24 h and then the dye-conjugated particles were added and incubated with cells for the indicated periods in the experiments. The cell cytosol was stained with calcein AM and imaged with a confocal fluorescence microscope (Olympus FV-10i or Zeiss LSM). The images were reconstructed with ZEN Lite to generate orthographic and 3D views. For actin staining, the BMMs were fixed with paraformaldehyde and rinsed, and the actin was stained with ActinGreen 488 ReadyProbes a per the manufacturer’s instruction (Thermal Scientific). For endosome staining, the BMMs were transfected with a CellLight Early Endosomes-GFP BacMam plasmid (Thermal Scientific) for 24 h before particle addition to locate the endosomes. To determine the particle uptake, the confocal fluorescence images were analysed to quantify the green fluorescence volume (cell) and the red fluorescence volume (particles) in each cell. The single-particle volume was estimated on the basis of the particle diameter determined from SEM images. The particle number was estimated by the ratio of red fluorescence volume to single-particle volume.
Gene profiling
BMMs were incubated with spiky or control particles (∼2 μm TiO2 microsphere) at 0.04 particles μm−2 or left untreated for 12 or 48 h. RNAs were extracted from the treated cells by a RNeasy Mini Kit (Qiagen) according to the manufacturers manual. Expressions of 89 genes were analysed by quantitative real-time RT-PCR using the Dendritic and Antigen Presenting Cell PCR Array (Qiagen). Genes related to M1 and M2 macrophage subtypes were analysed by real-time RT-PCR (Roche) using the indicated primers in Supplementary Table 1.
Cell-surface marker analysis
Cell surface markers were analysed by flow cytometry (BD FACSAria). To this end, BMMs were incubated with spiky particles or left untreated for 12 h. BMDCs were cultured with MPL alone or along with spiky particles, rough particles or nanorods for 12 h. The cells were collected and stained by anti-MHCII (25–9-17, 1:200), anti-CD40 (3/23, 1:100), anti-CD80 (16–10A1, 1:100) and anti-CD86 (GL-1, 1:100) antibodies. Subtype markers of the macrophages were recognized by anti-CD11c (N418, 1:200) and anti-CD206 (C068C2, 1:100) antibodies. All the antibodies were obtained from Biolegend. The stained cells were fixed, quantified by flow cytometry and analysed with Flowjo software.
Inflammasome activation
BMMs were primed with 1 μgml−1 LPS or 1 μgml−1 MPL for 3 h and then cultured with spiky particles, rough nanoparticles or nanorods at 0.02–0.08 particles μm−2 for another 18 h. The IL-1β concentration in the culture medium was measured with an enzyme-linked immunosorbent assay (ELISA) kit (eBioscience). To elucidate the mechanism that underlies the inflammasome activation, an inhibitor or activator, such as nigericin (3 μM), Ac-YVAD-cmk (50 μM), cytochalasin D (2 μM), dynasore (80 μM), nocodazole (10 μM), ruthenium red (2 μM), amiodarone (10 μM), KCl (130 mM) or blebbistatin (50 μM), was added to the culture of BMMs 0.5 h prior to particle treatment.
ROS assays
BMMs were pre-incubated with 5 μM CM-H2DCFDA (Life Technologies) for 30 min and stained with Hoechst dye (Enzo Lifesciences) for another 10 min. The BMMs were then cultured in fresh medium that contained spiky particles at 0.04 particles μm−2 or 0.1% H2O2 as the positive controls. ROS levels were quantified by the fluorescent intensity of CM-H2DCFDA and recorded in real time with confocal fluorescence microscopy.
Cathepsin B assays
A release of cathepsin B into cytosol was assessed with a CV-cathepsin B detection kit (Enzo Lifesciences) as per the manufacturer’s instruction. Briefly, BMMs were cultured with spiky particles at 0.04 particles μm−2 or silica crystals at 500 μgml−1 for 3 h, and subsequently incubated with a CV-(RR)2 substrate for 1 h, followed by staining the cell nuclei with Hoechst dye. Cathepsin B that leaked into the cytosol was indicated by the fluorescence of the CV-(RR)2 substrate in cytosol imaged with confocal fluorescence microscopy.
Immunofluorescence staining
Spiky particles or rough particles (both at 0.04 particles μm−2) or PBS were added to the BMM culture in a glass-bottom dish pretreated with poly-L-lysine for 4 h. After the particle treatment, BMMs were washed, fixed with 4% paraformaldehyde and permeabilized by 0.1% Triton X-100. The permeabilized cells were incubated with rabbit anti-myosin Ila (3403, Cell Signaling Technology, 1:50) or mouse anti-vanculin (728526, R&D systems, 1:50) antibody and stained with Qdot525-conjugated Donkey anti-Rabbit IgG (Q-11441MP, Life Technologies, 1:100) or Alexa Fluor 594-conjugated Goat antiMouse IgG (Poly4053, Biolegend, 1:100). The stained cells were washed thoroughly and covered by ProLong Gold Antifade Mountant with 4,6-diamidino-2-phenylindole (Life Technologies). To mark active caspase-3, BMMs were cultured with spiky particles at 0.08 particles μm−2 for 72 h, fixed by 4% paraformaldehyde and then stained with 10 μgml−1 rabbit anti-active caspase-3 antibody (Abcam, ab2302), followed by secondary staining with AF647-conjugated Goat anti-Rabbit antibody (Abcam, ab5694, 1:200). The stained cells were washed thoroughly and covered by ProLong Gold Antifade Mountant with 4,6-diamidino-2-phenylindole (Life Technologies) and subjected to imaging with confocal fluorescence microscopy (FV-1000, Olympus).
OT-I-stimulation assay
OT-I cells were stimulated by BMDCs as previously described47,48. In brief, immature WT or caspase 1−/− BMDCs were incubated with 200 μgml−1 OVA overnight and then with 10 ng ml−1 SIINFEKL peptide plus MPL, spiky particles, rough particles, nanorods or their combinations for another 6 h. The antigen-primed DCs were washed thoroughly before co-culturing with OT-I cells. MPL was used at a dose of 1 μgml−1 and all the particles were used at a dose of 0.04 particles μm−2. T cells were isolated from OT-I mice via magnetic beads (Miltenyi Biotec) and incubated with BMDCs at a 1:1 ratio for 3 d. The IFN-γ concentration in the supernatant was quantified by an ELISA kit (Thermo Fisher).
DC-mediated cancer immunotherapy
DC-mediated cancer immunotherapy was carried out as previously described49,50. In brief, mice were subcutaneously (s.c.) injected with 5 × 105 EG7 tumour cells on the right flank. EG7 cells were obtained from and authenticated by ATCC, and tested negative for mycoplasma. OVA-primed BMDC (5 × 105) that received the different stimulations above were s.c. injected into mice 3 and 7 d after tumour inoculation. Tumour sizes were monitored by a quality calibrator every 3 d. Tumour volumes were calculated as ½ × a × b2, in which a and b are the long and short diameters of the tumour, respectively.
Immunization and analysis of immune responses
C57BL/6 or caspase 1−/− mice were immunized with 20 μg of OVA together with 10 μg of MPL,+ 1 mg of spiky particles, 10 μg of MPL + 1 mg spiky particles, 10 μg of MPL + 1 mg of rough particles or 10 μg of MPL + 1 mg of nanorods via s.c. injection on the lower back. The particles and nanorods were pretreated with 3-aminopropyltriethoxysilane and washed thoroughly before mixing with MPL. Some mice received two intraperitoneal injections of 200 μg anti-CD40L antibodies (MR-1, BioXcell) right after and 2 d after immunization to transiently block the CD40-CD40L interaction. To evaluate DC maturation in vivo, some mice were euthanized and the inguinal lymph nodes collected 36 h after immunization to analyse the CD40, CD80 and CD86 expressions. Other mice were euthanized 1 wk later, and splenocytes were prepared and stimulated overnight with OVA323_339 (ISQAVHAAHAEINEAGR) peptide, OVA257.264 (SIINFEKL) peptide and OVA, each at 10 μgml−1 in the presence of anti-CD28 antibody (37.51, BD Pharmingen, 4 μgml−1). Golgi-Plug (BD Pharmingen, 1:1,000) was added to the cell culture in the final 5 h. Cells cultured with anti-CD28 antibody and Golgi-Plug in the absence of antigens (unstimulated) served as a negative control. Cells were suspended in PBS with 10% FBS, stained by anti-CD4 (GK1.5, 1:100) and anti-CD8 (53–6.7, 1:100) antibodies, fixed and permeabilized by a Fix/Perm kit (BD Pharmingen) and then intracellularly stained with anti-IL-4 (11B11, 1:100) or anti-IFN-γ (XMG1.2, 1:100) antibody. All the antibodies were obtained from Biolegend unless otherwise indicated. The results were analysed with BD FACSAria and Flowjo software.
Humoral immune responses were measured 2 wk after immunization. Serum IgG, IgG1 and IgG2c titres were measured by ELISA with OVA at 1 μgml−1 as the coating antigen. Sera were serially diluted and incubated in the plate at room temperature for 2 h. After washing, horseradish peroxidase-conjugated anti-mouse IgG (Southern Biotec, 1:4,000), IgG1 (Southern Biotec, 1:4,000) or IgG2c (Southern Biotec, 1:4,000) antibody was added into the plates and incubated for 1 h at room temperature. The titres of specific antibody subtypes were quantified by using SIGMAFAS OPD as the substrate and reading the reaction at A490 on a plate reader (Molecular Devices).
Influenza vaccination and challenge
Mice were immunized on days 0 and 14 with the 2009 H1N1 monovalent clinical influenza vaccine (100 ng haemagglutinin (HA) content (Sanofi Pasteur)) alone or together with alum (1 mg, Thermo Fisher), spiky particles (1 mg), MPL (10 μg), AddaVax (50 μl, Invivogen), spiky+MPL, alum+MPL or rough+MPL. Five days after the final immunization, splenocytes were stimulated overnight with the influenza virus at 1 μgml−1 in the presence of anti-CD28 antibody (37.51, BD Pharmingen, 4 μgml−1) followed by incubation with Golgi-Plug (BD Pharmingen, 1:1,000) for another 5 h. Cells that received only anti-CD28 and Golgi-Plug (without antigen stimulation) served as negative controls. T cells that expressed IFN-γ or IL-4 were analysed as above. Sera were collected 7 d after the final immunization and measured for HAI titres. Serum samples were incubated with receptor-destroying enzyme (Denka Seiken) at 37 °C for 20 h followed by heat inactivation at 56 °C for 30 min. The resultant serum samples were incubated with four HA units of 2009 H1N1 influenza virus at 37 °C for 1 h after serial dilutions, and then with 0.5% chicken red blood cells (LAMPIRE Biological Laboratories) at room temperature for 30 min. The HAI titre was defined as the reciprocal of the highest dilution that inhibited four HA units. Immunized mice were intranasally challenged with 20 × LD50 A/California/7/2009 H1N1 influenza virus 14 d after the final immunization. Body-weight loss and survival rates were monitored for an additional 12 d.
Statistics and reproducibility
A two-tailed t-test was used to analyse the difference between two groups, and a one-way analysis of variance (ANOVA) among multiple groups with a confidence interval of 95%. All the data are presented as the mean ± s.d. or the mean ± s.e.m. as indicated in the experiments, except for antibody titres and T-cell responses, which are presented as box and whiskers plots, in which the whiskers are the minimum to maximum. The three lines of the boxes are the 25th percentile, the median and the 75th percentile, respectively. P values were calculated by PRISM software (GraphPad) or Origin and regarded significant if less than 0.05. Sample sizes were determined on the basis of preliminary experiments to give a statistical power of 90%. No animals were excluded from the analysis. Photoanalysis and tumour volume measurements were conducted in an investigator-blind fashion, but the investigators were not blinded to the other experiments, which were, however, carried out under highly standardized and predefined conditions.
Data availability
The data that support the plots within this paper and other findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgements
This work is supported in part by the National Natural Science Foundation of China (grant nos 61771498, 51705543 and 31530023) to X.X., J.T. and T.H., and by National Institutes of Health grants AI089779 and AI 113458 and department funds to M.X.W. X.X. thanks the Youth 1000 Talents Program of China and 100 Talents Program of Sun Yat-Sen University (76120-18821104). J.W. thanks the 100 Talents Program of Sun Yat-Sen University. The authors thank the Wellman Center Photopathology Core for their help in the histology analysis during this project.
Footnotes
Competing interests
The authors declare no competing interests.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, statements of data availability and associated accession codes are available at https://doi.org/10.1038/s41565-018-0274-0.
Supplementary information is available for this paper at https://doi.org/10.1038/s41565-018-0274-0.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Iwasaki A & Medzhitov R Control of adaptive immunity by the innate immune system. Nat. Immunol. 16, 343–353 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brubaker SW, Bonham KS, Zanoni I & Kagan JC Innate immune pattern recognition: a cell biological perspective. Annu. Rev. Immunol. 33, 257–290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Medzhitov R Toll-like receptors and innate immunity. Nat. Rev. Immunol. 1, 135–145 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Petrovsky N & Aguilar JC Vaccine adjuvants: current state and future trends. Immunol. Cell Biol. 82, 488–496 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Reed SG, Orr MT & Fox CB Key roles of adjuvants in modern vaccines. Nat. Med. 19, 1597–1608 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Kline KA, Falker S, Dahlberg S, Normark S & Henriques-Normark B Bacterial adhesins in host-microbe interactions. Cell Host Microbe 5, 580–592 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Harris AK et al. Structure and accessibility of HA trimers on intact 2009 H1N1 pandemic influenza virus to stem region-specific neutralizing antibodies. Proc. Natl Acad. Sci. USA 110, 4592–4597 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Duin D, Cleare W, Zaragoza O, Casadevall A & Nosanchuk JD Effects of voriconazole on Cryptococcus neoformans. Antimicrob. Agents Chemother. 48, 2014–2020 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waksman G & Hultgren SJ Structural biology of the chaperone-usher pathway of pilus biogenesis. Nat. Rev. Microbiol. 7, 765–774 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Purcell BK, Pruckler J & Clegg S Nucleotide sequences of the genes encoding type 1 fimbrial subunits of Klebsiella pneumoniae and Salmonella typhimurium. J. Bacteriol. 169, 5831–5834 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Padmanabhan J et al. Engineering cellular response using nanopatterned bulk metallic glass. ACS Nano 8, 4366–4375 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luu TU, Gott SC, Woo BWK, Rao MP & Liu WF Micro- and nanopatterned topographical cues for regulating macrophage cell shape and phenotype. ACS Appl Mater. Interfaces 7, 28665–28672 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christo SN et al. The role of surface nanotopography and chemistry on primary neutrophil and macrophage cellular responses. Adv. Healthcare Mater. 5, 956–965 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Xie X & Melosh NA Fabrication of sub-cell size ‘spiky’ nanoparticles and their interfaces with biological cells. J. Mater. Chem. B 3, 5155–5160 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Li J et al. Nanotube-based hierarchical titanate microspheres: an improved anode structure for Li-ion batteries. Chem. Commun. 48, 389–391 (2012). [DOI] [PubMed] [Google Scholar]
- 16.EFSA ANS panel. Scientific opinion on the re-evaluation of titanium dioxide (E171) as a food additive. EFSA J. 14, 4545 (2016). [Google Scholar]
- 17.Farrell TP & Magnuson B Absorption, distribution and excretion of four forms of titanium dioxide pigment in the rat. J. Food. Sci. 82, 1985–1993 (2017). [DOI] [PubMed] [Google Scholar]
- 18.May RC & Machesky LM Phagocytosis and the actin cytoskeleton. J. Cell Sci. 114, 1061–1077 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Neudecker V et al. Myeloid-derived miR-223 regulates intestinal inflammation via repression of the NLRP3 inflammasome. J. Exp. Med. 214, 1737–1752 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalupahana RS, Mastroeni P, Maskell D & Blacklaws BA Activation of murine dendritic cells and macrophages induced by Salmonella enterica serovar Typhimurium. Immunology 115, 462–472 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shahid M et al. IEX-1 deficiency induces browning of white adipose tissue and resists diet-induced obesity. Sci. Rep. 6, 24135 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jablonski KA et al. Novel markers to delineate murine M1 and M2 macrophages. PLoS One 10, e0145342 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Broz P & Dixit VM Inflammasomes: mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 16, 407–420 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Bergsbaken T, Fink SL & Cookson BT Pyroptosis: host cell death and inflammation. Nat. Rev. Microbiol. 7, 99–109 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patino T, Soriano J, Barrios L, Ibanez E & Nogues C Surface modification of microparticles causes differential uptake responses in normal and tumoral human breast epithelial cells. Sci. Rep. 5, 11371 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hornung V et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 9,847–856 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Misawa T et al. Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat. Immunol. 14,454–460 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Tschopp J & Schroder K NLRP3 inflammasome activation: the convergence of multiple signalling pathways on ROS production? Nat. Rev. Immunol. 10, 210–215 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Celada A & Maki RA The expression of I-A correlates with the uptake of interferon-gamma by macrophages. Eur. J. Immunol. 19, 205–208 (1989). [DOI] [PubMed] [Google Scholar]
- 30.Galic M et al. External push and internal pull forces recruit curvaturesensing N-BAR domain proteins to the plasma membrane. Nat. Cell Biol. 14, 874–U212 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bavi O et al. Influence of global and local membrane curvature on mechanosensitive ion channels: a finite element approach. Membranes 6,14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Compan V et al. Cell volume regulation modulates NLRP3 inflammasome activation. Immunity 37, 487–500 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Brohawn SG, Su Z & MacKinnon R Mechanosensitivity is mediated directly by the lipid membrane in TRAAK and TREK1 K+ channels. Proc. Natl Acad. Sci. USA 111, 3614–3619 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munoz-Planillo R et al. K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity 38, 1142–1153 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fenix AM et al. Expansion and concatenation of nonmuscle myosin IIA filaments drive cellular contractile system formation during interphase and mitosis. Mol. Biol. Cell 27, 1465–1478 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galic M et al. Dynamic recruitment of the curvature-sensitive protein ArhGAP44 to nanoscale membrane deformations limits exploratory filopodia initiation in neurons. eLife 3, e03116 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao W et al. Nanoscale manipulation of membrane curvature for probing endocytosis in live cells. Nat Nanotech. 12, 750–756 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kool M et al. Alum adjuvant stimulates inflammatory dendritic cells through activation of the NALP3 inflammasome. J. Immunol. 181, 3755–3759 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Chen X, Zeng Q & Wu MX Improved efficacy of dendritic cell-based immunotherapy by cutaneous laser illumination. Clin. Cancer Res. 18, 2240–2249 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurts C, Robinson BWS & Knolle PA Cross-priming in health and disease. Nat. Rev. Immunol. 10, 403–414 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Ghiringhelli F et al. Activation of the NLRP3 inflammasome in dendritic cells induces IL-1^-dependent adaptive immunity against tumors. Nat. Med. 15, 1170–1178 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Murugaiyan G, Martin S & Saha B Levels of CD40 expression on dendritic cells dictate tumour growth or regression. Clin. Exp. Immunol. 149, 194–202 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.MacLeod MKL et al. Vaccine adjuvants aluminum and monophosphoryl lipid A provide distinct signals to generate protective cytotoxic memory CD8 T cells. Proc. Natl. Acad. Sci. USA 108, 7914–7919 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marrack P, McKee AS & Munks MW Towards an understanding of the adjuvant action of aluminium. Nat. Rev. Immunol. 9, 287–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J, Shah D, Chen X, Anderson RR & Wu MX A micro-sterile inflammation array as an adjuvant for influenza vaccines. Nat. Commun. 5, 4447 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J, Li B & Wu MX Effective and lesion-free cutaneous influenza vaccination. Proc. Natl Acad. Sci. USA 112, 5005–5010 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Helft J et al. GM-CSF mouse bone marrow cultures comprise a heterogeneous population of CD11c+ MHCII+ macrophages and dendritic cells. Immunity 42, 1197–1211 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Popovic ZV et al. The proteoglycan biglycan enhances antigen-specific T cell activation potentially via MyD88 and TRIF pathways and triggers autoimmune perimyocarditis. J. Immunol. 187, 6217–6226 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Met O et al. The effect of a therapeutic dendritic cell-based cancer vaccination depends on the blockage of CTLA-4 signaling. Cancer Lett. 231, 247–256 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Kato M et al. Enhanced anti-tumor immunity by superantigen-pulsed dendritic cells. Cancer Immunol. Immunother. 60, 1029–1038 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the plots within this paper and other findings of this study are available from the corresponding author upon reasonable request.