Abstract
Anti-apoptotic protein BCL-XL plays a key role in tumorigenesis and cancer chemotherapy resistance, rendering it an attractive target for cancer treatment. However, BCL-XL inhibitors such as ABT-263 cannot be safely used in the clinic because platelets solely depend on BCL-XL to maintain their viability. To reduce the on-target platelet toxicity associated with the inhibition of BCL-XL, we designed and synthesized PROTAC BCL-XL degraders that recruit CRBN or VHL E3 ligase because both of these enzymes are poorly expressed in human platelets compared to various cancer cell lines. We confirmed that platelet-toxic BCL-XL/2 dual inhibitor ABT-263 can be converted into platelet-sparing CRBN/VHL-based BCL-XL specific degraders. A number of BCL-XL degraders are more potent in killing cancer cells than their parent compound ABT-263. Specifically, XZ739, a CRBN-dependent BCL-XL degrader, is 20-fold more potent than ABT-263 against MOLT-4 T-ALL cells and has >100-fold selectivity for MOLT-4 cells over human platelets. Our findings further demonstrated the utility of PROTAC technology to achieve tissue selectivity through recruiting differentially expressed E3 ligases.
Keywords: BCL-XL, PROTAC, apoptosis, platelet, degradation
Graphical Abstract

1. Introduction
Apoptosis is a conserved and highly regulated biological process that plays a critical role in maintaining cellular homeostasis [1]. Resistance to apoptosis is a common feature in human malignancies and considered a key hallmark of cancer [2]. Therefore, targeting specific anti-apoptotic pathways is a promising cancer therapeutic strategy. Members of B-cell lymphoma 2 (BCL-2) protein family, consisting of both pro- and anti-apoptotic proteins, regulates the intrinsic apoptotic pathway. Several anti-apoptotic BCL-2 family proteins such as BCL-2, BCL-XL, and MCL-1 have been validated as anticancer targets [3–5]. Inhibition of these proteins with small-molecule inhibitors promotes Bax/Bak oligomerization and ultimately induces mitochondrial outer membrane permeabilization, followed by cytochrome c release and activation of caspases to execute apoptosis [1].
ABT-737 (Figure 1), the first potent BCL-2/BCL-XL dual inhibitor, was developed via fragment-based drug discovery using NMR [6,7]. Optimization of ABT-737 afforded navitoclax (ABT-263) (Figure 1) [8,9], an orally bioavailable BCL-2/BCL-XL dual inhibitor that has been in Phase II clinical trials for hematological malignancies and small cell lung cancer (SCLC). However, ABT-263 treatment leads to rapid and dose-dependent thrombocytopenia when dosed as a single agent [10], consistent with the studies showing the dependency of platelets on BCL- XL to maintain their viability [11,12]. To overcome this on-target, dose-limiting toxicity, a selective BCL-2 inhibitor, venetoclax (ABT-199) (Figure 1), was developed. Venetoclax has shown antileukemic activity without the induction of thrombocytopenia [13]. Subsequently, it was approved by the FDA for the treatment of chronic lymphocytic leukemia (CLL) and small lymphocytic lymphoma (SLL) as a single agent, and for acute myeloid leukemia (AML) in combination with low-intensity chemotherapy [14].
Figure 1.
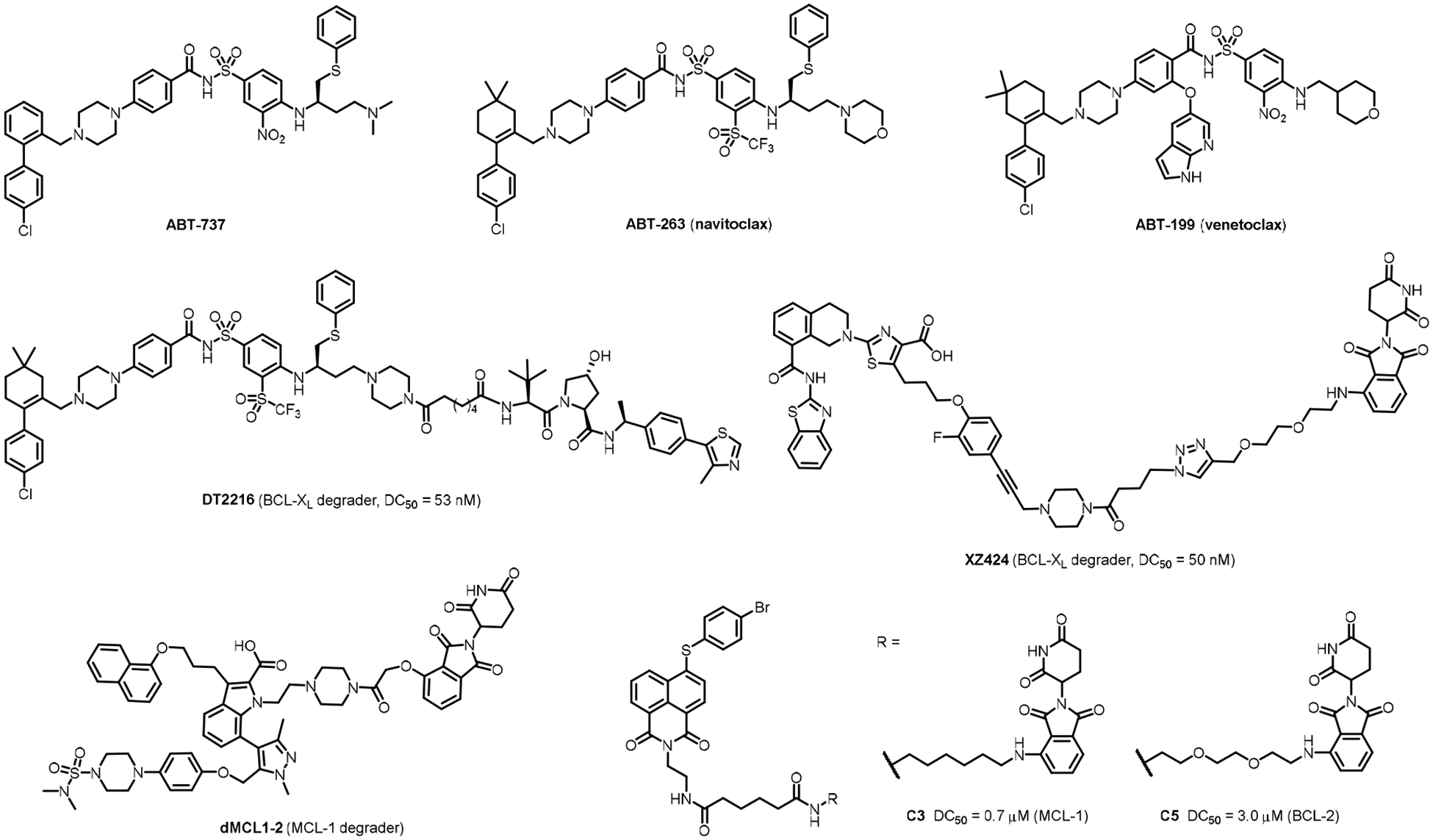
Chemical structures of representative BCl-2/BCL-XL inhibitors and BCL-2 family protein degraders.
The overall response rate of CLL patients to venetoclax is 71–79% but the complete remission rate (20%) is relatively low [15]. Upregulation of BCL-XL by microenvironmental survival signals has been identified as the major component accountable for the resistance, consistent with the high efficacy of BCL-2/BCL-XL dual inhibitor ABT-263 in killing venetoclax resistant CLL cells [16]. In addition, venetoclax has limited utility for the treatment of solid tumors [17,18] because BCL-2 is mainly associated with the survival of hematological malignancies and BCL-XL is the most common BCL-2 family member overexpressed in solid tumors, as well as in a subset of leukemia and lymphoma cells [19]. Bioinformatics analyses also reveal a strong correlation between the levels of BCL-XL expression and resistance to chemotherapies [20]. Further, it has been well established that inhibition of BCL-XL by ABT-263 is primarily responsible for the observed synergy with chemotherapies in solid tumors [21,22]. Taken together, BCL-XL is one of the most important validated cancer targets.
More recently, we and others discovered that ABT-263 and other BCL-XL inhibitors, such as A-1331852 and A-1155463 are potent senolytics, referring to small-molecules that can selectively kill senescent cells [23–26]. This is because BCL-XL is a key anti-apoptotic protein in many types of senescent cells. Subsequent studies on ABT-263 in mouse models have demonstrated that clearance of chemotherapy-induced senescent cells reduces several short- and long-term adverse effects of the chemotherapy, as well as cancer relapse and metastasis [27]. These findings suggest an added benefit of targeting BCL-XL for cancer treatment. Therefore, it is highly desirable to develop a strategy that can retain the versatility and efficacy of BCL-XL inhibitors, while reducing their on-target platelet toxicity.
Several strategies have been devised to minimize the on-target platelet toxicity associated with the inhibition of BCL-XL. One strategy is to combine ABT-263 with chemotherapy or targeted therapies to reduce the dose regimen of ABT-263 so that thrombocytopenia can be manageable [22]. In another strategy, prodrugs of BCL-2/BCL-XL dual inhibitors have been prepared to limit drug exposure to platelets [28], and the lead candidate, APG-1252, is currently in Phase I clinical trial [29]. Further, antibody-drug conjugates (ADCs) of BCL-XL inhibitors have been designed to deliver the inhibitors to cancer cells more specifically. For example, ABBV-155, a BCL-XL inhibitor ADC that targets B7H3 expressing cancer cells, is currently in Phase I clinical trial [30]. We have recently demonstrated that the proteolysis targeting chimera (PROTAC), an emerging therapeutic modality [31–33], is also a feasible solution to reduce platelet toxicity associated with BCL-XL inhibition [34]. This was based on the hypothesis that because PROTACs engage E3 ligases to induce protein degradation, they can achieve cell/tissue selectivity if the E3 ligase they recruit is differentially expressed in cells or tissues [34]. Von Hippel–Lindau (VHL) cullin-2 and cereblon (CRBN) cullin-4A RING E3 ligases, of which small-molecule ligands have been successfully employed in PROTAC design, were found to be minimally expressed in human platelets [35,36]. In a proof of concept study, we have shown that DT2216 (Figure 1), a VHL-based PROTAC derived from ABT-263, is more potent to a variety of BCL-XL dependent cancer cells with significantly less platelet toxicity than its parent compound ABT-263 [34]. In another study, through XZ424 (Figure 1), which is constructed by tethering BCL-XL specific inhibitor A-1155463 to CRBN ligand pomalidomide, we have demonstrated that CRBN could be an alternative E3 ligase to be recruited to build BCL-XL degraders with low platelet toxicity [37].
The discovery of PROTAC MCL-1 and BCL-2 degraders (Figure 1) have been reported recently [38,39]. Herein, we describe our efforts in the discovery of potent PROTAC BCL-XL degraders with low platelet toxicity. Emerging evidence suggests that it could be beneficial to parallelly develop two or more PROTAC series hijacking different E3 ligases [40–42]. Thus, we have evaluated both CRBN and VHL E3 ligases based BCL-XL degraders. The critical role of linkerology in degradation potency and efficacy of PROTACs is well estabolished [43–45]. Therefore, we have also systematically varied the length and composition of the linker unit.
2. Results and discussion
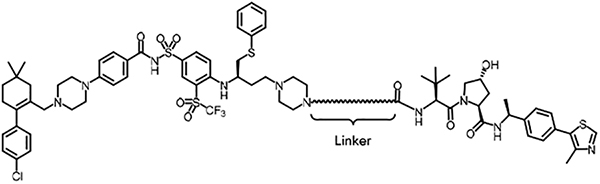
Cytotoxicity of the designed PROTACs and their ability in inducing BCL-XL degradation were primarily evaluated in MOLT-4, a T-cell acute lymphoblastic leukemia (T-ALL) cell line dependent on BCL-XL for survival. We focused on ABT-263 as the ‘warhead’ in our PROTAC design. The co-crystal structures of ABT-263 in complex with BCL-2/BCL-XL (PDB code 4LVT and 4QNQ, Figure 2) [13] indicate that the morpholine ring in ABT-263 is solvent-exposed, making it a suitable position for tethering to an E3 ligase binding unit through a linker. Accordingly, we initially converted ABT-263 to a ‘PROTAC-ready’ derivative by replacing the morpholine ring with a piperazine ring, followed by tethering it to a previously reported VHL ligand [46] through various linkers to yield compounds 1a-f, 2a-d, 3a-c, and 4a-b (Scheme 1, Table 1).
Figure 2.

X-ray crystal structures of ABT-263 (black) bound to (A) BCL-XL, available from PDB, code: 4QNQ, and (B) BCL-2, available from PDB, code: 4LVT. The local view of the key hydrogen bond between morpholine ring and protein is shown in the additional box.
Scheme 1.

Reagents and conditions: (a) i. MeNH2, MgSO4, THF, rt; ii. NaBH3CN, THF, rt; (b) Troc-Cl, TEA, DCM, rt; (c) TFA, DCM, rt; (d) 24, TEA, acetonitrile, reflux; (e) 26, EDCI, DMAP, DCM, rt; (f) Zn, HOAc, THF, rt.
Table 1.
BCL-XL Degraders Designed with Various Linkers Tethering through a Piperazine Ring to a VHL Ligand
 |
|||||
|---|---|---|---|---|---|
| Compd | Linker | IC50 (nM)a | Compd | Linker | IC50 (nM)a |
| ABT-263 | - | 230 | 2b | 82.0 | |
| 1a | 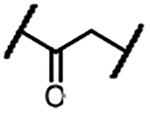 |
>2000 | 2c | 94.5 | |
| 1b | 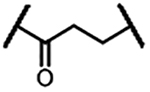 |
>2000 | 2d | 181 | |
| 1c |  |
>2000 | 3a | 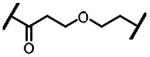 |
324 |
| 1d | 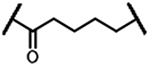 |
467 | 3b | 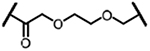 |
512 |
| 1e (DT2216) |  |
77.1 | 3c | 232 | |
| 1f |  |
108 | 4a | 151 | |
| 2a |  |
130 | 4b | 914 | |
IC50 values are the means of at least three independent experiments; reduction of MOLT-4 cell viability after 48 h treatment.
We first evaluated the effects of these compounds on the viability of MOLT-4 T-ALL cells, with ABT-263 as the positive control (Table 1), followed by examining their ability to BCL- 2/BCL-XL degradation in the same cell line using Western blotting (Figure 3). In the series with linkers containing an amide linkage and an alkane chain, we synthesized compounds 1a-f with 1–6 methylene groups in the alkane chain (Table 1) and determined the optimal linker length by comparing their potencies in the cell viability assay. DT2216 (1e) with five methylene groups in the linker was the most potent in this series and 3 times more potent than ABT-263. Shortening the linker in 1e afforded 1a-d, which exhibited largely decreased cellular potencies in the same assay. However, the analog (1f) with one methylene unit longer than that in 1e showed slightly reduced cytotoxicity against MOLT-4 cells. These results indicate that the optimal linker length is six carbon atoms. The cell viability assay data of 1a-f were well correlated with the protein degradation data (Figure 3), indicating that the cytotoxicity is mainly derived from BCL-XL protein degradation. Replacing the amide linkage in 1a-f with a C-N linkage yielded analogs 2a-d. The optimal linker length in this series is five carbon atoms (compound 2b) and appears to be one carbon atom shorter than the amide linkage containing series, which could be attributed to the increased flexibility of the C-N bond in comparison to the amide bond. Replacement of one methylene group in the linker of 1e with an oxygen atom afforded 3a, which was ~4 times less potent than 1e. In general, introduction of oxygen atoms into the linker unit is detrimental to cellular potency. None of the oxygen-containing PROTACs we synthesized showed better cytotoxicity to MOLT-4 cells in comparison to their corresponding alkane linker analogs (e.g. 3a vs 1e, 3b vs 1f, and 4a vs 2b). Interestingly, although 4b was slightly more potent in inducing BCL-XL degradation than 4a at 100 nM (Figure 3B), it was ~6 times less potent in reducing MOLT-4 cell viability. The cytotoxicity of the PROTACs is derived from both direct inhibition of BCL-XL and degradation of this protein. The relative high cellular potency of 4a could be due to its higher cell permeability than 4b despite its low efficiency in inducing BCL-XL protein degradation.
Figure 3.

Western blot analysis of the effect of VHL-based BCL-XL degraders on BCL-XL and BCL-2 protein levels in MOLT-4 T-ALL cells. Cells were treated with 100 nM of each compound for 16 h before harvesting. Degradation activity is reported as % of total protein remaining after compound treatment relative to the vehicle after normalization with β-actin as quantified by Image J software.
The salt bridge between Glu96 of BCL-XL and the protonated nitrogen atom on the morpholine ring of ABT-263 is one of the crucial interactions for high BCL-XL binding (Figure 2). However, the ring system is not required to maintain high binding affinity as the morpholine ring in ABT-263 can be replaced by a dimethylamino group as shown in ABT-737. Thus, we replaced the morpholine ring in ABT-263 with an N-methylamino group and tethered it to the same VHL binding moiety as compounds in Table 1 through an alkane chain to generate compounds 6a-f (Table 2). Interestingly, despite the smaller linkage unit (a single N atom vs. a piperazine ring), the optimal linker length for this series of compounds was also six carbon atoms; compound 6c was the most potent degrader among its analogs (Figure 3B) and exhibited a comparable cellular potency to 1e in reducing MOLT-4 cell viability. Compound 5, which was generated by replacing the C-N linkage in 6c with an amide bond, had no effect on MOLT-4 cell viability (IC50 > 2 μM) and weak BCL-XL degradation in MOLT-4 cells at 100 nM, confirming the importance of the salt bridge in maintaining high BCL-XL protein binding and the subsequent protein degradation. Incorporation of oxygen atoms into the linker unit of 6d caused small increases in both protein degradation and cellular potency (7a vs. 6d). In addition, PEG-linker containing PROTACs, i.e. 7a-c, are linker length independent in inducing BCL-XL protein degradation and cell death.
Table 2.
BCL-XL Degraders Designed with Various Linkers Tethering through an N-Methylamino Group to a VHL Ligand
 |
|||||
|---|---|---|---|---|---|
| Compd | Linker | IC50 (nM)a | Compd | Linker | IC50 (nM)a |
| 5 | >2000 | 6e | 139 | ||
| 6a |  |
1243 | 6f | 1058 | |
| 6b | 460 | 7a | 97.0 | ||
| 6c | 81.3 | 7b | 200 | ||
| 6d | 147 | 7c | 206 | ||
IC50 values are the means of at least three independent experiments; reduction of MOLT-4 cell viability after 48 h treatment.
Similar to DT2216 (1e) [34], none of these VHL-based PROTACs were able to degrade BCL-2 in MOLT-4 cells (Figure 3), further demonstrated the feasibility of achieving target specificity through conversion of non-selective inhibitors to PROTACs. Our studies with the most potent analog in this series, DT2216, have shown that BCL-XL, but not BCL-2, can form stable ternary complexes with DT2216 and VHL in live cells as determined by nanoBRET assay [47], which could contribute to the specific BCL-XL degradation [34].
We next replaced the VHL binding moiety in compounds 1–4 with CRBN binding moiety pomalidomide. It is well established that different E3 ligases require different linker length and/or composition for optimal cellular potency. Thus, we also conducted a systematic investigation of linker length and composition in the CRBN-based PROTACs. As shown in Table 3, compounds with an amide linkage were generally less potent in reducing MOLT-4 cell viability when compared with their corresponding C-N linkage containing analogs with a similar linker length. Among the alkane-linker containing PROTACs (8a-e and 9a-c), compounds with the shortest linkers, i.e. 8a and 9a, were the most potent. Compound 8a, which contains an amide linkage and three methylene groups, was 3 times more potent than the parent compound ABT-263. Switching the amide linkage to a C-N linkage resulted in a significant improvement on cellular potency; compound 9a, which contains three methylene groups in the linker, was 8 times more potent than ABT-263. Compounds with an amide linkage and a PEG-chain, 10a-c, appeared to have little dependence on linker length for their cellular potency. Replacing the amide linkage in 10a-c with a urea linkage, exemplified by 11a, had little effect on the ability to inhibit MOLT-4 growth. However, the thiourea analog 11b exhibited a ~6-fold decrease in potency compared to 11a, in the same assay. In contrast to 9a-c that contain a C-N linkage and an alkane-chain in their linkers, PEG-linker containing analogs 12a-c preferred longer linker length for optimal cellular potency. Compound 12c, which has the longest linker in this series with 11 atoms, had the highest cellular potency that was ~12 times more potent than ABT-263 in reducing MOLT-4 cell viability. These results further demonstrate the empirical nature of PROTAC design, which is at least partially attributable to the fact that potencies of reducing cell viability for inhibitor-derived PROTACs are a combination of protein degradation and direct protein inhibition, both of which are dependent on cell permeability. As a result, analogs with similar intrinsic protein degradation efficiency may exhibit completely different ability to induce protein degradation in cells.
Table 3.
BCL-XL Degraders Designed with Various Linkers Tethering through a Piperazine Ring to Pomalidomide
 |
|||||
|---|---|---|---|---|---|
| Compd | Linker | IC50 (nM)a | Compd | Linker | IC50 (nM)a |
| 8a |  |
82.0 | 10a | 161 | |
| 8b |  |
346 | 10b | 211 | |
| 8c |  |
518 | 10c | 126 | |
| 8d |  |
>2000 | 11a | 118 | |
| 8e | >2000 | 11b | 722 | ||
| 9a |  |
28.3 | 12a | 60.5 | |
| 9b |  |
43.0 | 12b | 41.4 | |
| 9c | 100 | 12c | 17.3 | ||
IC50 values are the means of at least three independent experiments; reduction of MOLT-4 cell viability after 48 h treatment.
We evaluated all the compounds in Table 3 for their ability to induce BCL-XL/BCL-2 protein degradation in MOLT-4 cells at 50 nM for 16 h. As shown in Figure 4, a good correlation between the ability to reduce MOLT-4 cell viability and inducing BCL-XL degradation was observed within each analog series. However, there was no obvious correlation among compounds from different series. For example, compounds 9a-c were more potent in the cell viability assay but much less effective in inducing protein degradation than 10a-c, indicating the cellular activity of 9a-c was more dependent on direct protein inhibitory effects and permeability. Similar to VHL-based PROTACs, no BCL-2 degradation was observed with these CRBN-based PROTACs.
Figure 4.
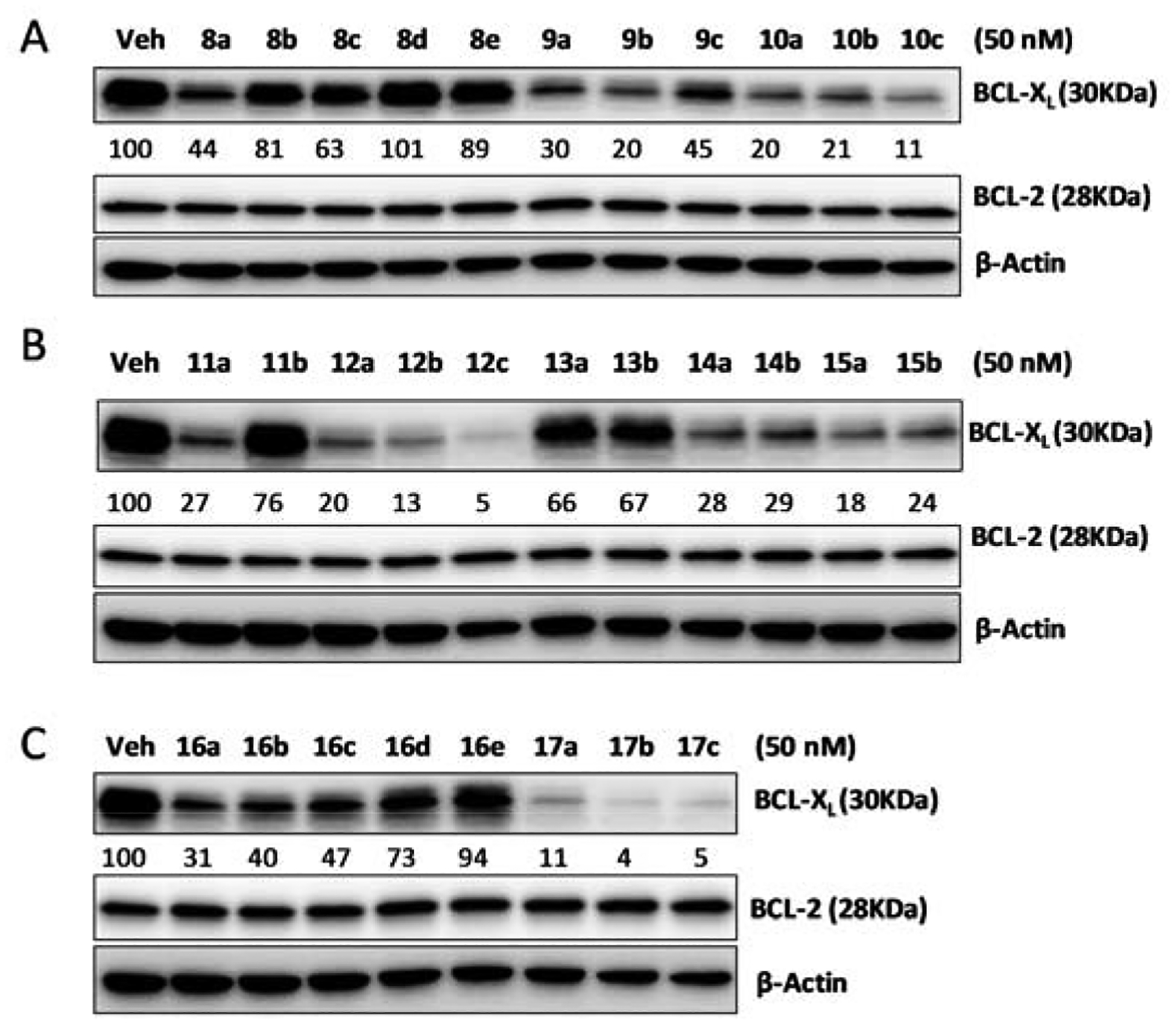
Western-blot analysis of BCL-XL and BCL-2 protein levels in MOLT-4 T-ALL cells after treating with CRBN-based BCL-XL degraders at 50 nM for 16 h. Degradation activity reported as % of total protein remaining after compound treatment relative to vehicle after normalization with β-actin as quantified by Image J software.
Subsequently, the N-methylamine precursor for analogs in Table 2 was employed for CRBN-based PROTACs and the cell viability data are shown in Table 4. Compounds 13a and 13b with an amide linkage to connect the 1,2,3-triazole containing linker and the precursor displayed moderate cytotoxicity against MOLT-4 cells. The extension of the alkane-chain portion of the linker resulted in an improvement in cellular activity (13a vs. 14a, 13b vs. 14b). The PEG-chain portion was more tolerated for modification, supported by the similar activity for 13a and 13b (or 14a and 14b). Switching the amide linkage in compounds 14a-b to C-N linkage, which resulted in 15a-b, significantly increased cytotoxicity in MOLT-4 cells. Protein degradation data indicated that 15a and 15b are better BCL-XL degraders than their amide analogs (Figure 4). Similar to the compounds in Table 2, we also synthesized both alkane-chain and PEG-chain containing PROTACs with a C-N linkage. Compared to the corresponding VHL-based PROTACs with the same linker (Table 2, Figure 3), the CRBN-based PROTACs displayed much higher cytotoxicity and BCL-XL degradation activity. Compound 17b (XZ739), which contains a PEG linker with linker length of 11 atoms, was the most potent BCL-XL degrader against MOLT-4 cells and was ~22 times more potent than ABT-263.
Table 4.
BCL-XL Degraders Designed with Various Linkers Tethering through an N-Methylamino Group to Pomalidomide
 |
|||||
|---|---|---|---|---|---|
| Compd | Linker | IC50 (nM)a | Compd | Linker | IC50 (nM)a |
| 13a |  |
770 | 16b |  |
38.7 |
| 13b |  |
769 | 16c | 60.9 | |
| 14a |  |
177 | 16d | 147 | |
| 14b |  |
179 | 16e | >2000 | |
| 15a | 29.2 | 17a | 35.0 | ||
| 15b | 35.0 | 17b (XZ739) | 10.1 | ||
| 16a |  |
32.5 | 17c | 19.3 | |
IC50 values are the means of at least three independent experiments; reduction of MOLT-4 cell viability after 48 h treatment.
We selected several potent BCL-XL degraders for further investigation. The DC50 values (the concentrations at which 50% protein has been degraded) were determined in MOLT-4 cells after treatment with each individual degrader for 16 h. As shown in Table 5 and Figure S2, the CRBN-based PROTACs 12c, 15a, 16a, and 17b (XZ739) were more potent in inducing BCL-XL degradation and reducing the viability of MOLT-4 cells than the VHL-based PROTACs DT2216, 2b, and 6c. Among these PROTACs, XZ739 was the most potent BCL-XL degrader with a DC50 value of 2.5 nM. Likely due to the reduced membrane permeability relative to their parent compound ABT-263, coupled with the lack of BCL-2 degradation, none of these ABT-263 derived BCL-XL degraders were significantly more potent than ABT-263 in reducing the viability of RS4;11, a B-cell ALL cell line that mainly depends on BCL-2 for survival [13]. The CRBN-based BCL-XL degraders were also more potent than the VHL-based degraders in H146 cells, a small cell lung cancer cell line that depends on both BCL-2 and BCL-XL for survival and has low VHL expression (Figure S1). Subsequently, the BCL-XL degraders in Table 5 were tested for their toxicity against human platelets and most displayed >100-fold selectivity for MOLT-4 cells over platelets. In contrast, ABT-263 showed no selectivity between these platelets and MOLT-4 cells.
Table 5.
Cellular Activity of Selected Analogs in Human Platelets and a Panel of Human Cancer Cell Lines
| Compd | IC50 (nM)a 48 h treatment | DC50 (nM)b | IC50 ratioc | |||
|---|---|---|---|---|---|---|
| MOLT-4 | RS4;11 | H146 | Platelets | |||
| ABT-263 | 227 | 49.0 | 43.8 | 242 | ND | 1.1 |
| DT2216 | 77.1 | 213 | 278 | >10,000 | 53 | >130 |
| 2b | 82.0 | 76 | 203 | >10,000 | 93 | >122 |
| 6c | 81.3 | 189 | 265 | >10,000 | 71 | >123 |
| 12c | 17.3 | 38.5 | 24.6 | 1,560 | 4.5 | 90 |
| 15a | 29.2 | 62.2 | 61.7 | 6,250 | 6.3 | 214 |
| 16a | 32.5 | 129 | 70.2 | 3,296 | 10.6 | 101 |
| 17b (XZ739) | 10.1 | 41.8 | 25.3 | 1,217 | 2.5 | 120 |
IC50 values are the means of at least three independent experiments;
in MOLT-4 cells, 16 h treatment;
IC50 ratio between human platelets and MOLT-4 cells.
Western blot analysis revealed that XZ739 dose-dependently induced BCL-XL degradation in MOLT-4 cells (Figure 5A). In addition, the BCL-XL degradation induced by XZ739 in MOLT-4 was rapid, starting within 2 h; and 8 h after drug treatment, more than 96% of the protein was degraded with 100 nM of XZ739 (Figure 5B). The effects of XZ739 on BCL-XL protein levels in MOLT-4 were long-lasting and also reversible, as indicated in the ‘washout’ experiment (Figure 5C). Further, XZ739-induced BCL-XL degradation can be abrogated by proteasome inhibitor MG-132 (Figure 5D) or excess competitive CRBN ligand pomalidomide (Figure 5E), indicating that the degradation depends on both proteasomes and the CRBN E3 ligase. To further confirm that the CRBN E3 ligase is involved in XZ739-induced BCL-XL degradation. We synthesized XZ739-NC, a negative control compound of XZ739, in which a methyl group is installed on the amino group in the pomalidomide moiety of XZ739 to block CRBN binding. Not surprisingly, XZ739-NC did not induce BCL-XL degradation in MOLT-4 cells (Figure 5F).
Figure 5.
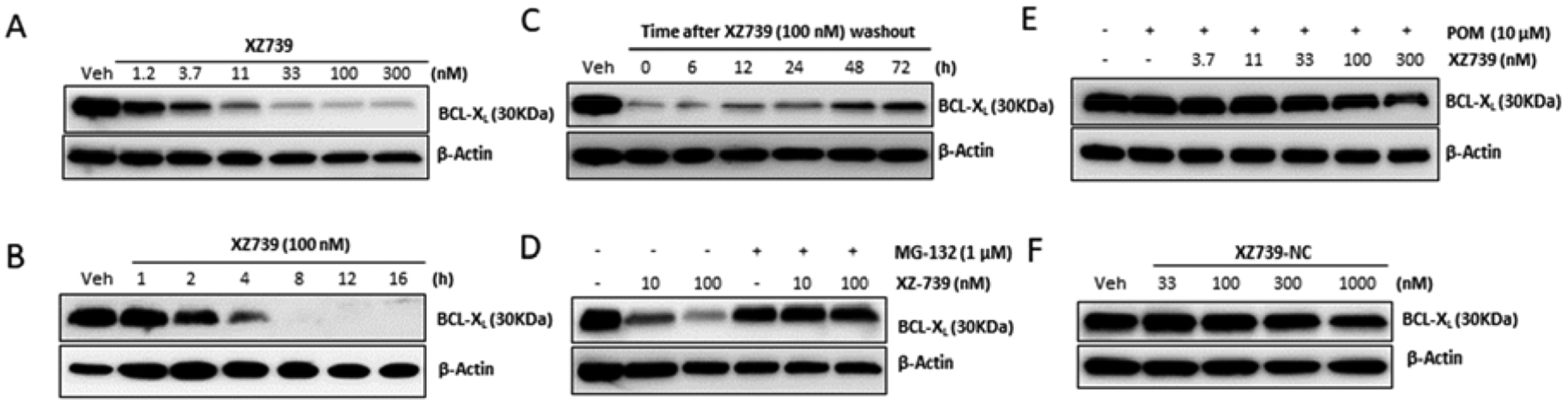
XZ739-induced BCL-XL degradation. (A) Western blots showing the BCL-XL protein levels in MOLT-4 cells treated with the indicated concentrations of XZ739 for 16 h. (B) BCL-XL protein levels in MOLT-4 cells treated with 100 nM of XZ739 at the indicated time points. (C) BCL-XL protein levels in MOLT-4 cells after incubation with 100 nM of XZ739 for 16 h followed by drug washout, resuspension and incubation of the cells for additional time as indicated in a drug-free medium. (D-E) Pretreatment with 10 μM pomalidomide (POM) or 1 μM MG-132 for 2 h blocked the degradation of BCL-XL induced by XZ739. (F) Western blot analysis of BCL-XL in MOLT-4 cells treated with XZ739-NC at indicated concentrations for 16 h. Data are representative of two independent experiments.
In human platelets, no significant changes in BCL-XL protein levels were observed after 16 h treatment with up to 1.0 μM of XZ739 (Figure 6A). The cytotoxicity of high concentrations of XZ739 to platelets most likely derived from BCL-XL inhibition rather than degradation as pre-incubation of platelets with pomalidomide did not affect the cytotoxicity of XZ739 to platelets while a ~27-fold shift in IC50 values was observed in MOLT-4 cells with pomalidomide pretreatment (Figure 6B).
Figure 6.
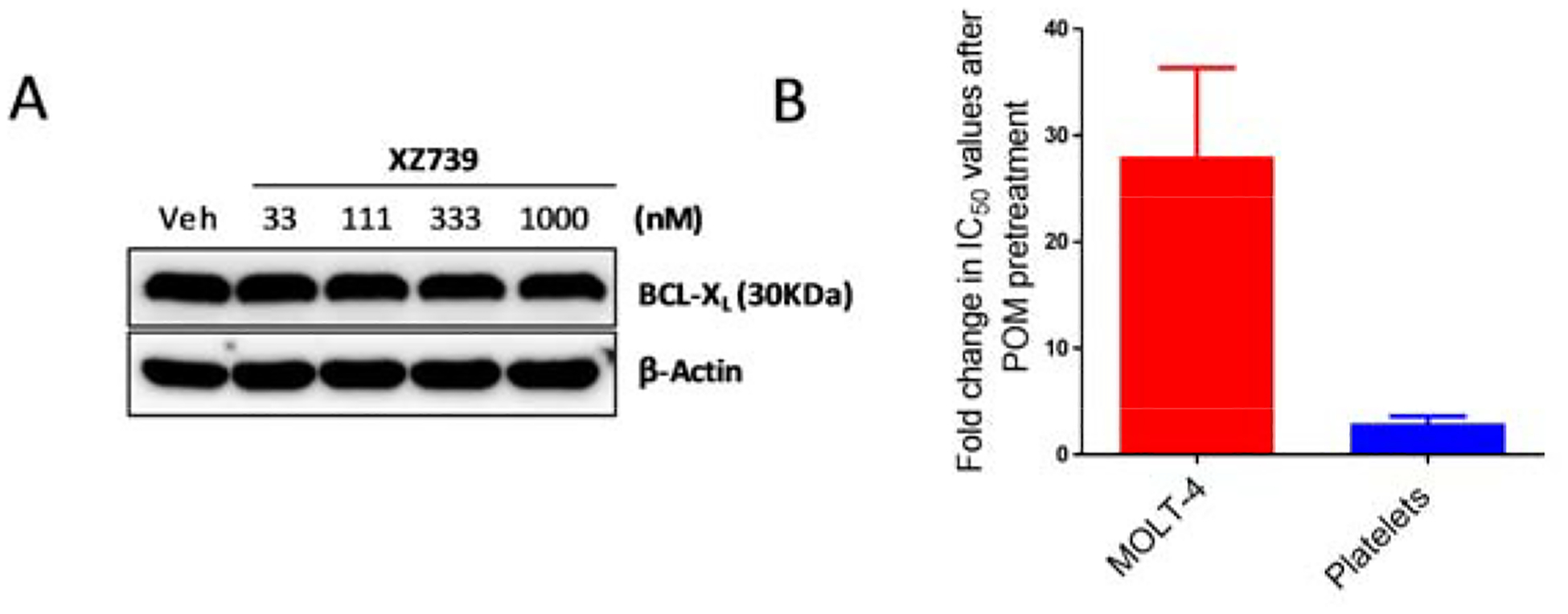
XZ739 shows improved selectivity because of its low activity in inducing BCL-XL degradation in human platelets, and the remaining cytotoxicity to human platelets mainly depends on BCL-XL inhibition. (A) Western blot analysis of BCL-XL levels after treatment of human platelets with indicated concentration of XZ739 for 16 h. (B) IC50 fold changes in MOLT-4 cells and human platelets after blocking with 10 μM pomalidomide (POM). IC50 values in MOLT-4 cells and platelets were normalized to 1.0.
We next assessed the impact of XZ739 on apoptosis. Western blot analysis showed that XZ739 dose-dependently increased poly (ADP-ribose) polymerase (PARP) and caspase-3 cleavage in MOLT-4 cells (Figure 7A), suggesting the apoptotic cell-death mechanism. Further, to confirm that XZ739 induces cell death through caspase-mediated apoptosis, we did flow cytometry analysis of apoptosis using Annexin-V and propidium iodide (PI) staining (Figure 7B). We found that, 10 nM of XZ739 treatment for 48 h significantly increased the percentage of Annexin-V-positive cells in MOLT-4 cells compared to the vehicle group. In comparison, ABT-263 showed similar effect at much higher concentration (100 nM). In addition, pretreatment with 10 μM of pan-caspase inhibitor Q-VD-OPh (QVD) for 2 h partially inhibited the XZ739-induced apoptosis, which confirms that XZ739 induces cell death through a caspase-dependent mechanism.
Figure 7.
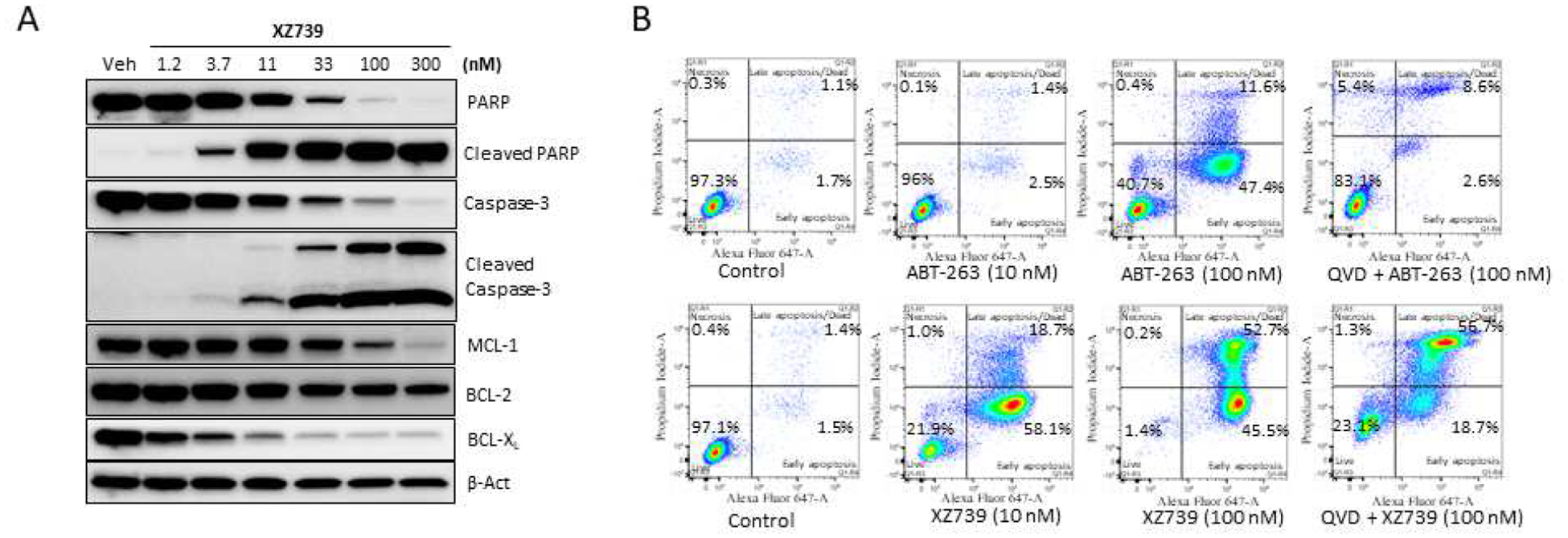
Characterization of XZ739-mediated apoptosis in MOLT-4 cells. (A) Western blot analysis of cleaved-PARP and cleaved-caspase-3 after 16 h treatment with indicated concentrations of XZ739. (B) Flow cytometry analysis of apoptosis using Annexin-V and PI staining. MOLT-4 cells were treated with XZ739 or ABT-263 at 10 and 100 nM for 48 h. XZ739 (100 nM) significantly increased the percentage of apoptotic cells, and QVD (10 μM) pretreatment for 2 h inhibited the apoptosis induced by XZ739. Data are representative of two independent experiments.
CRBN-targeting immunomodulatory drugs (IMiDs) such as thalidomide, pomalidomide and lenalidomide can induce degradation of a number of neo-substrate proteins through a C2H2 degron. PROTACs that are based on these IMiDs may have the potential to exert some off-target effects by degrading some of these C2H2 zinc finger proteins [48]. To test this possibility, we evaluated the effects of XZ739 on the expression of the Ikaros proteins that are well known targets of those CRBN E3 ligands, and found that IKZF1 and IKZF3 protein levels were not affected in MOLT-4 cells treated with 10 nM XZ739 but downregulated when the cells were treated with 100 nM XZ739 (Figure S6).
3. Chemistry
Precursor 18 that contains a piperazinyl moiety for linker attachment and aldehyde 20 were prepared as described in our recent publication [34]. The synthesis of methylamino group containing precursor 19 is outlined in Scheme 1. Briefly, aldehyde 20 was converted to amine 21 through a reductive amination reaction. The amino group in 21 was protected with 2,2,2-trichloroethoxycarbonyl (Troc) group by treating with trichloroethyl chloroformate (Troc-Cl) to give 22. Removal of the Boc protection group in 22 afforded 23, which was subjected to SNAr substitution with 24 to yield N-acylsulfonamide 25. Coupling of 25 with acid 26 in the presence of EDCI and DMAP afforded 27. Removal of the Troc group in 27 with Zn/HOAc gave 19.
The syntheses of VHL-based PROTACs are illustrated in Schemes 2 and 3. The VHL ligand motif 28 was prepared by following reported methods [46]. Coupling of 28 with acid 29a-f followed by hydrolysis with LiOH afforded 30a-f. Direct coupling between dicarboxylic acid 29g-i and 28 also worked smoothly in good yields (62–89%). PROTACs 1a-f, 3a-c, and 5 were obtained via coupling of precursor 18/19 with 30a-i (Scheme 2).
Scheme 2.
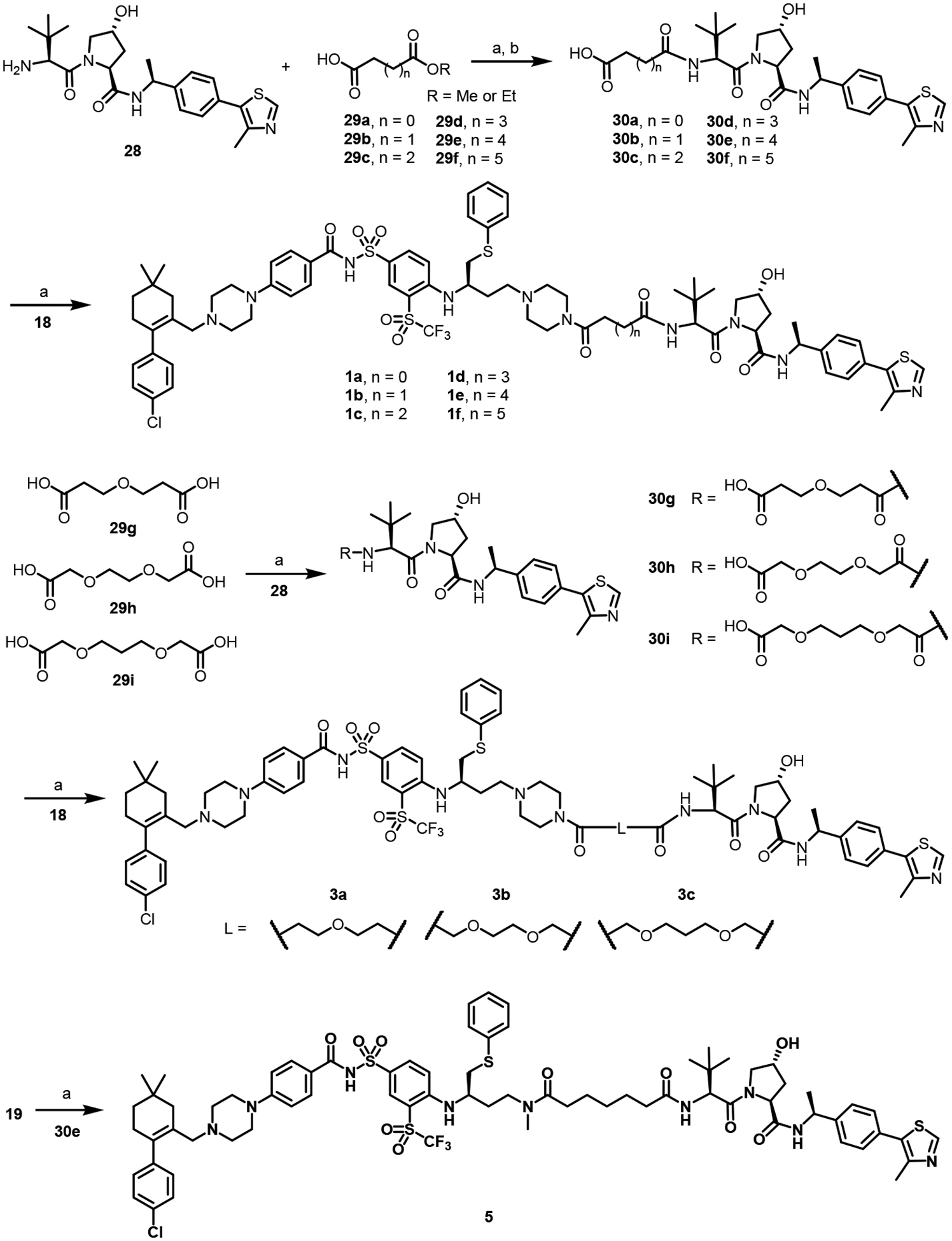
Reagents and conditions: (a) HATU, TEA, DCM, rt; (b) LiOH monohydrate, MeOH, H2O, rt.
Scheme 3.
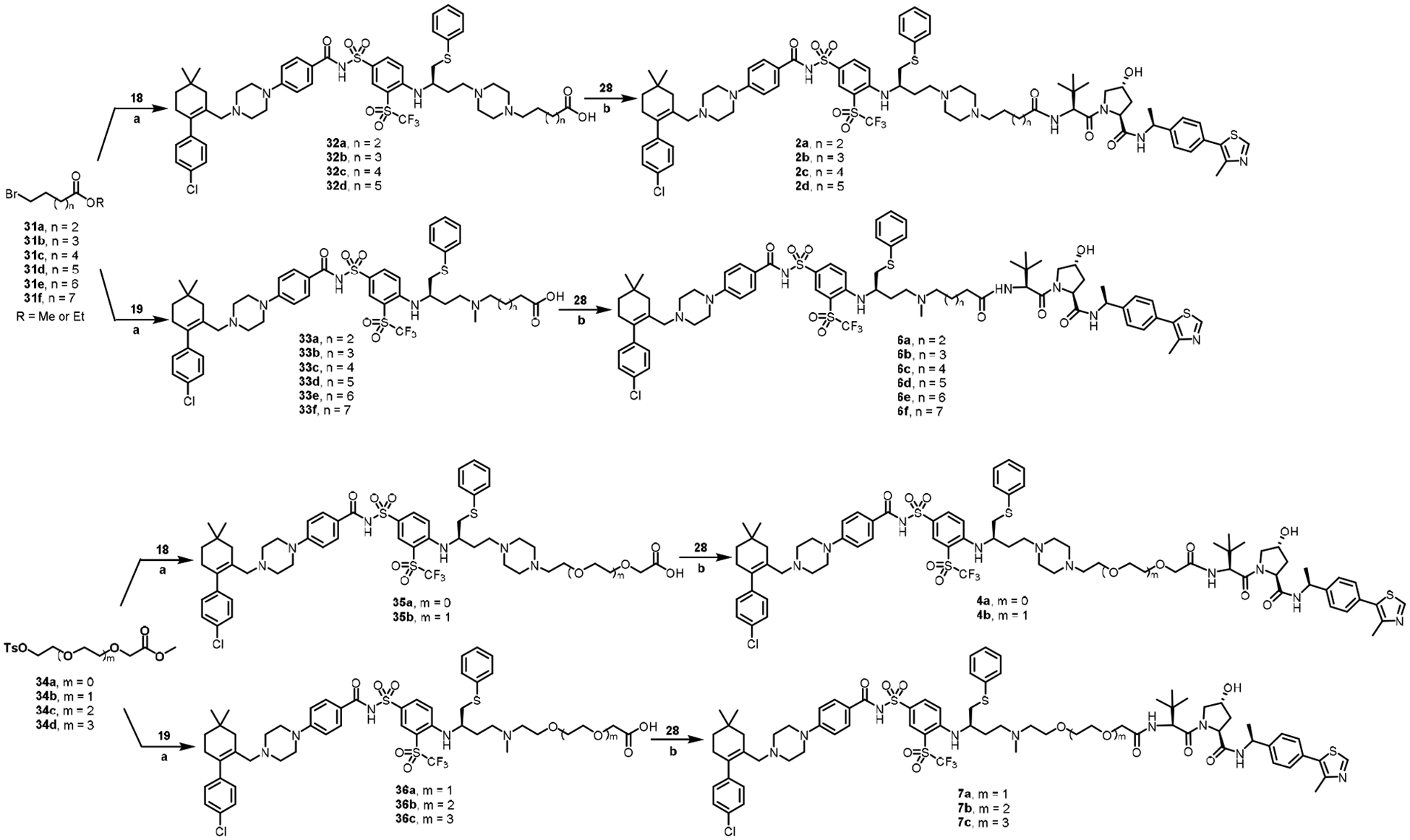
Reagents and conditions: (a) i. K2CO3, NaI, DMSO, 80 °C; ii. LiOH monohydrate, MeOH, THF, H2O, rt; (b) 28, HATU, TEA, DCM, rt.
The synthesis of the series with C-N linkage started from bromides 31a-f or tosylates 34a-d (Scheme 3). Nucleophilic substitution with amine 18/19 in the presence of K2CO3 and NaI at 80 °C, followed by ester hydrolysis, afforded acid intermediates 32a-d, 33a-f, 35a-b, and 36a-c, respectively, which were coupled with VHL ligand 28 to generate the corresponding final products 2a-d, 4a-b, 6a-f, and 7a-c.
The syntheses of CRBN-based PROTAC BCL-XL degraders are illustrated in Schemes 4–7. As shown in Scheme 4, amino acids 37a-e were coupled with 38 in the presence of DIPEA to afford intermediates 39a-e. Compound 8a-e were synthesized by amide coupling of 39a-e with 18. PEG linker containing PROTACs 10a-c were synthesized by initial coupling of amine 40a-c with 38, followed by removal of the tert-butyl on them to give intermediates 41a-c and amide coupling with 18.
Scheme 4.
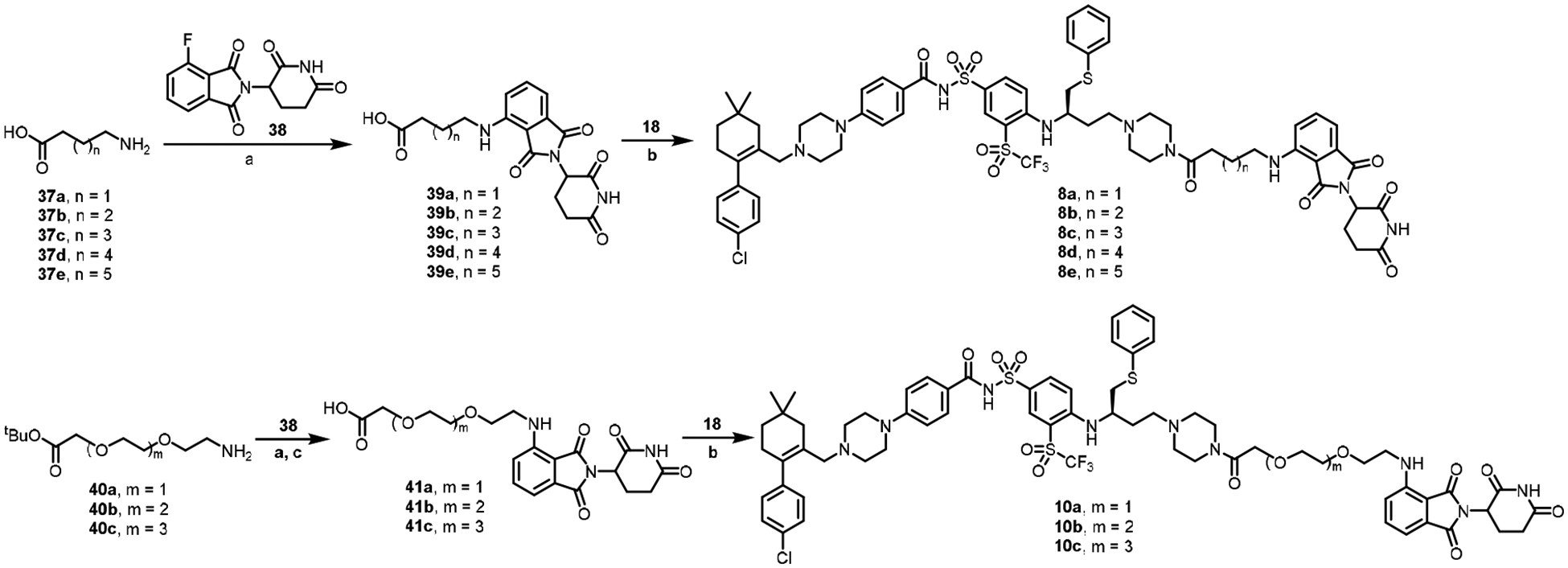
Reagents and conditions. (a) DIPEA, DMF, 90 °C; (b) 18, HATU, DIPEA, DCM, rt; (c) TFA, DCM, rt.
Scheme 7.

Reagents and conditions. (a) DIPEA, DMF, 90 °C; (b) i. COCl2, DMSO, DCM, −78 °C; ii. 50 or 53, DCM, −78 °C; iii. TEA, DCM, −78 °C then rt; (c) NaBH3CN, TEA, DCM, rt; (d) MsCl, TEA, DCM, rt; (e) DIPEA, NaI, 1,4-dioxane, 90 °C.
As shown in Scheme 5, compounds 13a-b and 14a-b were produced through coupling of azido acid 42a-b with precursor 19, followed by a click reaction with alkynes 44a/44b [37]. The synthesis of 15a-b started from a nucleophilic substitution of tosylate 45 with 19, followed by click reaction.
Scheme 5.

Reagents and conditions: (a) HATU, DIPEA, DCM, rt; (b) CuSO4·5H2O, sodium L-ascorbate, tBuOH, THF, H2O, 50 °C; (c) K2CO3, NaI, DMSO, 80 °C.
The synthesis of compounds 11a and 11b is shown in Scheme 6. The key intermediate 48 was synthesized by coupling of compounds 47 and 38, followed by removal of the Boc group by treating with TFA/DCM. Compound 48 was treated with carbonyldiimidazole or 1,1’-thiocarbonyldiimidazole and then with precursor 18 to afford compound 11a and 11b, respectively.
Scheme 6.
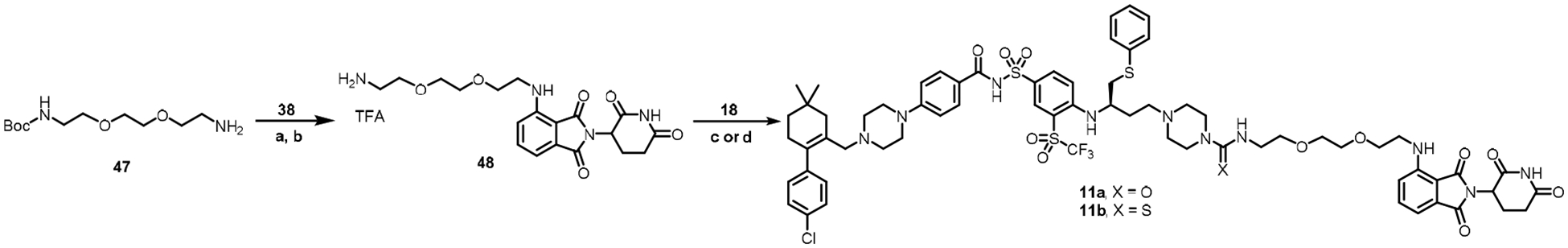
Reagents and conditions. (a) DIPEA, DMF, 90 °C; (b) TFA, DCM, rt; (c) i. carbonyldiimidazole, TEA, DCM, rt; ii. 18, DIPEA, rt; (d) i. 1,1’-thiocarbonyldiimidazole, TEA, DCM, rt; ii. 18, DIPEA, rt.
The syntheses of CRBN-based PROTACs with a C-N linkage are illustrated in Scheme 7. Since the pomalidomide moiety is unstable under the treatment of inorganic bases, they were synthesized either by nucleophilic substitution of mesylates with precursor 18/19 in the presence of DIPEA or by reductive amination of aldehydes with 18/19. Briefly, coupling of 38 with 49a- e/52a-d afforded 50a-e and 53a-d, respectively. Swern oxidation of 50a-e produced aldehyde 51a-e. Other oxidation methods, including PCC, PDC, Dess-Martin periodinane, and IBX, afforded unsatisfactory results. Subsequent reductive amination of the aldehydes with the corresponding precursor 18 or 19 yielded compounds 9a-c and 16a-e, respectively. Compounds 17a-c were obtained by using the same strategy. Compounds 12a-c were synthesized by using a different method. Mesylates 54a-c, which are more stable than the corresponding aldehyde intermediates, were prepared from 53a-c. Nucleophilic substitution of 54a-c with 18 in the presence of DIPEA and NaI afforded 12a-c. Similarly, XZ739-NC, the negative control compound of XZ739, was synthesized starting from compound 56.
4. Conclusions
In this study, we reported the design, synthesis, and evaluation of a series of PROTAC BCL-XL degraders. Through tethering BCL-2/BCL-XL dual inhibitor ABT-263 and a VHL ligand or a CRBN ligand, and varying the linker unit, we obtained a number of BCL-XL degraders that showed improved cytotoxicity against BCL-XL dependent MOLT-4 cells when compared with ABT-263. Protein degradation assay confirmed that these BCL-XL PROTACs were able to degrade BCL-XL but not BCL-2 despite that the warhead ABT-263 binds to both proteins. In addition, human platelets were more tolerant to the treatment of these degraders whereas the parent BCL-XL inhibitor ABT-263 showed no selectivity for MOLT-4 cells over platelets. Both CRBN and VHL-based PROTACs were able to achieve >100-fold selectivity in vitro for MOLT- 4 cells over human platelets. XZ739, a CRBN-based BCL-XL PROTAC, was the most potent analog against various cancer cell lines. The mouse plasma stability assay and preliminary pharmacokinetic study revealed that XZ739 is bioavailable (Figure S4). Overall, we have developed a novel strategy to reduce drug on-target toxicity by conversion of a conventional inhibitor into a protein degrader.
Despite the high molecular weight of PROTACs that are derived from PPI inhibitors, a number of PPI-targeting degraders have been successfully developed, including MCL-1 [38], BCL-2/MCL-1 [39], MDM2 [49], BCL-6 [50], and STAT3 [51,52]. Due to their catalytic mode of action, common ‘rules’ such as ‘rule-of-five’ may not be suitable to predict the drug-like properties of PROTACs. The size and molecular weight of our BCL-XL specific degraders are among the largest PROTACs, which further highlighted the broad utility of this therapeutic modality.
5. Experimental
5.1. Chemistry
THF, DCM, toluene, and acetonitrile were obtained via a solvent purification system by filtering through two columns packed with activated alumina and 4 Å molecular sieve, respectively. All other chemicals obtained from commercial sources were used without further purification. Flash chromatography was performed using silica gel (230–400 mesh) as the stationary phase. Reaction progress was monitored by thin-layer chromatography (silica-coated glass plates) and visualized by UV light, and/or by LC-MS. NMR spectra were recorded in CDCl3, CD3OD, or acetone-d6 at 400 or 600 MHz for 1H NMR. Chemical shifts δ are given in using tetramethylsilane as an internal standard. Multiplicities of NMR signals are designated as singlet (s), broad singlet (br s), doublet (d), doublet of doublets (dd), triplet (t), quartet (q), and multiplet (m). All final compounds for biological testing were of ≥95.0% purity as analyzed by LC-MS, performed on an Advion AVANT LC system with the expression CMS using two different columns: a Thermo Accucore™ Vanquish™ C18+ UHPLC Column (1.5 μm, 50 × 2.1 mm) at 40 °C and a Thermo Scientific™ BetaSil™ C18 Column (3.0 μm, 150 × 4.6 mm) at 25 °C. Gradient elution was used for UHPLC with a mobile phase of acetonitrile and water containing 0.1% formic acid.
5.1.1. General methods
5.1.1.1. General Method A.
A mixture of acid (1.0 equiv.), amine (1.0 equiv.), HATU (1.05 equiv.) and TEA (5.0 equiv.) in DCM was stirred at room temperature for 1 h. The mixture was poured into water and extracted with DCM. The combined organic layers were washed with NH4Cl (aq.) × 1, brine × 1, dried over anhydrous Na2SO4, filtered and concentrated under vacuum. The residue crude product was purified by flash column chromatography to afford the desired compound.
5.1.1.2. General Method B.
To a solution of ester (1.0 equiv.) in MeOH was added LiOH (5.0 equiv.). The mixture was stirred at room temperature for 2 h and the solvent was removed under reduced pressure. The residue crude product was purified by column chromatography to afford the corresponding carboxylic acid.
5.1.1.3. General Method C.
A mixture of amine (1.0 equiv.), an appropriate bromide or tosylate (5.0 equiv.), K2CO3 (2.0 equiv.), and NaI (0.2 equiv.) in DMSO was stirred at 80 °C overnight. The mixture was poured into water and extracted with DCM. The combined organic layers were washed with NH4Cl (aq.) × 1, brine × 1, dried over anhydrous Na2SO4, filtered, and concentrated under vacuum. The crude product was purified by column chromatography to afford the corresponding ester intermediate.
5.1.1.4. General Method D.
2-(2,6-Dioxopiperidin-3-yl)-4-fluoroisoindoline-1,3-dione (1.0 equiv.), amine (1.2 equiv.), and DIPEA (2.0 equiv.) in DMF were stirred at 90 °C overnight. The reaction was cooled to room temperature and poured into water. The resulting mixture was extracted with ethyl acetate and the combined organic layers were washed with water × 1, brine × 1, dried over anhydrous Na2SO4, filtered, and evaporated to dryness under vacuum. The residue crude product was purified by column chromatography to afford the desired compound.
5.1.1.5. General Method E.
To a solution of tert-butyl ester (1.0 equiv.) in DCM was added TFA (20–30 equiv.). The mixture was stirred at room temperature overnight and the solvent was evaporated under reduced pressure. The residue crude product was washed with Et2O to afford the desired product.
5.1.1.6. General Method F.
To a mixture of azide (1.0 equiv.), alkyne (1.2 equiv.) in tBuOH-THF (1:1, v/v) under argon was added CuSO4·5H2O (0.2 equiv.) and sodium ascorbate (0.2 equiv.) in water. The mixture was stirred at 50 °C for 3 h and cooled to room temperature before poured into water. The resulting mixture was extracted with DCM. The combined organic layers were washed with brine × 1, dried over anhydrous Na2SO4, filtered, and evaporated to dryness. The residue crude product was purified via flash column chromatography using DCM and MeOH as eluents to afford the desired product.
5.1.1.7. General Method G.
DMSO (3.0 equiv.) in DCM was cooled to −78 °C and (COCl)2 (1.5 equiv.) was added dropwise. The mixture was stirred for 10 min and alcohol (1.0 equiv.) in DCM was added dropwise into the solution. TEA (6.0 equiv.) was added after 10 min and the resulting mixture was kept at −78 °C for 30 min and then warmed to room temperature. The mixture was poured into water and extracted with DCM. The combined organic layers were washed with water × 1, brine × 1, dried over anhydrous Na2SO4, filtered, and evaporated to dryness. The residue crude product was purified by flash column chromatography using DCM and MeOH as eluents to afford the corresponding aldehyde, which was used directly in the next step.
5.1.1.8. General Method H.
A mixture of amine (1.0 equiv.) and aldehyde (1.5 equiv.) in DCM was treated with triethylamine (4.0 equiv.) and NaBH3CN (2.0 equiv.). The mixture was stirred at room temperature overnight. The solution was poured into water and extracted with DCM. The combined organic layers were washed with water × 1, brine × 1, dried over anhydrous Na2SO4, filtered, and evaporated to dryness. The residue crude product was purified by flash column chromatography using DCM and MeOH as eluents to afford the desired product.
5.1.1.9. General Method I.
To a mixture of alcohol (1.0 equiv.), TEA (4.0 equiv.) in DCM was added MsCl (1.2 equiv.). The mixture was stirred at room temperature for 3 h and poured into water. The resulting mixture was extracted with ethyl acetate and the combined organic layers were washed with water × 1, brine × 1, dried over anhydrous Na2SO4, filtered, and evaporated to dryness. The residue crude product was purified by flash column chromatography using DCM and MeOH as eluents.
5.1.1.10. General Method J.
A mixture of amine (1.0 equiv.), an appropriate mesylate (3.0 equiv.), DIPEA (10 equiv.), and NaI (0.2 equiv.) in 1,4-dioxane was stirred at 90 °C overnight. The mixture was poured into water and extracted with ethyl acetate. The combined organic phases were washed with water × 1, NH4Cl (aq.) × 1, brine × 1, dried over anhydrous Na2SO4, filtered, and evaporated to dryness. The residue crude product was purified by flash column chromatography using DCM and MeOH as eluents to afford the desired product.
(R)-tert-Butyl (4-(methylamino)-1-(phenylthio)butan-2-yl)carbamate (21).
A mixture of compound 20 (450 mg, 1.53 mmol), MeNH2 (2.0 M in MeOH; 7.5 mL, 15.0 mmol) and MgSO4 (3.0 g, 24.9 mmol) in THF (50 mL) was stirred at room temperature overnight. NaBH3CN (143 mg, 2.27 mmol) was then added. The resulting mixture was stirred at room temperature for 30 min. The solution was diluted with water and extracted with DCM. The combined organic phases were washed with brine × 1, dried over anhydrous Na2SO4, filtered and evaporated to dryness. The crude product was purified via column chromatography using DCM and MeOH as eluents to afford the title compound (280 mg, 56% yield). 1H NMR (400 MHz, CDCl3) δ 7.45–7.28 (m, 4H), 7.27–7.23 (m, 1H), 5.09 (d, J = 8.1 Hz, 1H), 4.52–4.31 (m, 1H), 3.86–3.66 (m, 1H), 3.27–2.81 (m, 3H), 2.67 (s, 3H), 2.30–2.17 (m, 1H), 1.88–1.72 (m, 1H), 1.44 (s, 9H). LC-MS (ESI): m/z 311.1 [M+H] +.
(R)-2,2,2-Trichloroethyl (3-((tert-butoxycarbonyl)amino)-4-(phenylthio)butyl)(methyl)carbamate (22).
A mixture of compound 21 (280 mg, 0.90 mmol), Troc-Cl (137 μL, 1.00 mmol) and TEA (250 μL, 1.80 mmol) in DCM (10 mL) was stirred at room temperature for 3 h. The mixture was poured into water and extracted with DCM. The combined organic phases were washed with brine × 1, dried over anhydrous Na2SO4, filtered, and evaporated to dryness. The residue crude product was purified via flash column chromatography using ethyl acetate and hexanes as eluents to afford the title compound. 1H NMR (400 MHz, CDCl3) δ 7.44–7.16 (m, 5H), 4.87–4.59 (m, 3H), 3.89–3.71 (m, 1H), 3.59–3.40 (m, 1H), 3.33–3.01 (m, 3H), 3.00–2.91 (m, 3H), 2.11–1.88 (m, 1H), 1.86–1.68 (m, 1H), 1.46– 1.37 (m, 9H). LC-MS (ESI): m/z 484.9 [M+H] +.
(R)-2,2,2-Trichloroethyl (3-amino-4-(phenylthio)butyl)(methyl)carbamate trifluoroacetate (23).
To a solution of compound 22 (from above) in DCM (10 mL) was added TFA (700 μL, 9.15 mmol). The mixture was stirred at room temperature for 3 h and the solvent was removed under reduced pressure. The residue solid was washed with Et2O to afford the title compound, which was used directly in the next step.
(R)-2,2,2-Trichloroethyl methyl(4-(phenylthio)-3-((4-sulfamoyl-2-((trifluoromethyl)sulfonyl)phenyl)amino)butyl)carbamate (25).
A mixture of compound 23 (from above), 4-fluoro-3-((trifluoromethyl)sulfonyl)benzenesulfonamide 24 (280 mg, 0.91 mmol) and TEA (750 μL, 5.39 mmol) in acetonitrile (30 mL) was refluxed overnight. The reaction was cooled to room temperature and the solvent was evaporated under reduced pressure. The residue was purified via flash column chromatography using ethyl acetate and hexanes as eluents to afford the title compound (220 mg, 36% yield in 3 steps). 1H NMR (400 MHz, CDCl3) δ 8.26 (s, 1H), 7.81 (dd, J = 9.1, 2.3 Hz, 1H), 7.45–7.29 (m, 5H), 7.03–6.94 (m, 1H), 6.51–6.37 (m, 1H), 4.92 (s, 2H), 4.83–4.55 (m, 2H), 3.81–3.61 (m, 1H), 3.56–3.24 (m, 2H), 3.19–2.90 (m, 5H), 2.46–2.19 (m, 1H), 1.93–1.71 (m, 1H). LC-MS (ESI): m/z 672.0 [M+H] +.
(R)-2,2,2-Trichloroethyl (3-((4-(N-(4-(4-((4’-chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzoyl)sulfamoyl)-2-((trifluoromethyl)sulfonyl)phenyl)amino)-4-(phenylthio)butyl)(methyl)carbamate (27).
A mixture of compound 25 (220 mg, 0.33 mmol), acid 26 (174 mg, 0.40 mmol), EDCI (151 mg, 0.79 mmol) and DMAP (96 mg, 0.79 mmol) in DCM (15 mL) was stirred at room temperature overnight. The mixture was poured into water and extracted with DCM. The combined organic layers were washed with 1N HCl (aq.) × 1, brine × 1, dried over anhydrous Na2SO4, filtered, and evaporated to dryness. The crude product was purified via flash column chromatography using DCM and MeOH as eluents to afford the title compound. 1H NMR (400 MHz, CDCl3) δ 8.38 (d, J = 2.2 Hz, 1H), 8.13 (dd, J = 9.2, 2.3 Hz, 1H), 7.66 (d, J = 8.7 Hz, 2H), 7.45–7.28 (m, 6H), 7.08–6.95 (m, 3H), 6.77 (d, J = 8.7 Hz, 2H), 6.48–6.38 (m, 1H), 4.83–4.50 (m, 2H), 3.82–3.61 (m, 1H), 3.48–3.26 (m, 6H), 3.22–2.88 (m, 7H), 2.60–2.19 (m, 7H), 2.12–2.06 (m, 2H), 1.93– 1.74 (m, 1H), 1.54–1.41 (m, 2H), 0.99 (s, 6H). LC-MS (ESI): m/z 1091.9 [M+H] +.
(R)-4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)-N-((4-((4-(methylamino)-1-(phenylthio)butan-2-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)benzamide (19).
Zinc powder (960 mg, 14.8 mmol) was added to a mixture of compound 27 and AcOH (600 μL, 10.5 mmol) in THF (20 mL). The reaction was stirred at room temperature overnight. Solid was removed by filtration and the filtrate was poured into water and extracted with ethyl acetate. The organic phases were washed brine × 1, dried over anhydrous Na2SO4, filtered and evaporated to dryness. The crude product was purified via column chromatography using DCM, MeOH and TEA as eluents to afford the title compound (180 mg, 60% yield in 2 steps). 1H NMR (600 MHz, CD3OD) δ 8.29 (d, J = 2.3 Hz, 1H), 8.09 (dd, J = 9.2, 2.3 Hz, 1H), 7.85–7.79 (m, 2H), 7.43–7.34 (m, 4H), 7.27– 7.20 (m, 2H), 7.19–7.13 (m, 3H), 6.96–6.91 (m, 2H), 6.88 (d, J = 9.3 Hz, 1H), 4.14–4.01 (m, 1H), 3.53–3.34 (m, 7H), 3.24–3.21 (m, 1H), 3.13–3.02 (m, 2H), 3.02–2.82 (m, 3H), 2.70 (s, 3H), 2.42–2.35 (m, 2H), 2.29–2.19 (m, 1H), 2.12–1.97 (m, 4H), 1.57 (t, J = 6.5 Hz, 2H), 1.07 (s, 6H). LC-MS (ESI): m/z 918.1 [M+H] +.
3-(((S)-1-((2S,4R)-4-Hydroxy-2-(((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)carbamoyl)pyrrolidin-1-yl)-3,3-dimethyl-1-oxobutan-2-yl)amino)-3- oxopropanoic acid (30a).
Following general methods A and B, compound 30a was obtained from 29a and 28 (27 mg, 88% yield). 1H NMR (400 MHz, CDCl3) δ 9.26 (s, 1H), 8.65 (s, 1H), 7.90 (s, 1H), 7.43–7.29 (m, 4H), 5.13–4.98 (m, 1H), 4.83–4.67 (m, 1H), 4.55–4.36 (m, 2H), 4.15 (d, J = 11.4 Hz, 1H), 3.64–3.49 (m, 1H), 3.27–3.09 (m, 2H), 2.50 (s, 3H), 2.43–2.23 (m, 1H), 2.21–2.06 (m, 1H), 1.45 (d, J = 7.0 Hz, 3H), 1.06 (s, 9H). LC-MS (ESI): m/z 531.1 [M+H] +.
4-(((S)-1-((2S,4R)-4-Hydroxy-2-(((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)carbamoyl)pyrrolidin-1-yl)-3,3-dimethyl-1-oxobutan-2-yl)amino)-4-oxobutanoic acid (30b).
Following general methods A and B, compound 30b was obtained from 29b and 28 (84 mg, 99% yield) 1H NMR (400 MHz, CDCl3) δ 8.67 (s, 1H), 7.94 (d, J = 8.2 Hz, 1H), 7.79 (d, J = 7.7 Hz, 1H), 7.44–7.34 (m, 4H), 5.13–5.03 (m, 1H), 4.81–4.73 (m, 1H), 4.51– 4.38 (m, 2H), 4.15 (d, J = 11.4 Hz, 1H), 3.54 (dd, J = 11.4, 3.5 Hz, 1H), 2.64–2.37 (m, 8H), 2.16–2.06 (m, 1H), 1.47 (d, J = 6.9 Hz, 3H), 1.05 (s, 9H). LC-MS (ESI): m/z 545.1 [M+H] +.
5-(((S)-1-((2S,4R)-4-Hydroxy-2-(((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)carbamoyl)pyrrolidin-1-yl)-3,3-dimethyl-1-oxobutan-2-yl)amino)-5- oxopentanoic acid (30c).
Following general methods A and B, compound 30c was obtained from 29c and 28. (10 mg, 31% yield). 1H NMR (400 MHz, CDCl3) δ 8.67 (s, 1H), 7.66 (d, J = 7.8 Hz, 1H), 7.45–7.32 (m, 4H), 7.19 (s, 1H), 5.15–5.02 (m, 1H), 4.80–4.69 (m, 1H), 4.57 (d, J = 8.4 Hz, 1H), 4.46 (s, 1H), 4.16–4.03 (m, 1H), 3.60 (dd, J = 11.1, 3.8 Hz, 1H), 2.52 (s, 3H), 2.47– 1.84 (m, 8H), 1.47 (d, J = 6.9 Hz, 3H), 1.05 (s, 9H). LC-MS (ESI): m/z 559.2 [M+H] +.
6-(((S)-1-((2S,4R)-4-Hydroxy-2-(((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)carbamoyl)pyrrolidin-1-yl)-3,3-dimethyl-1-oxobutan-2-yl)amino)-6-oxohexanoic acid (30d).
Following general methods A and B, compound 30d was obtained from 29d and 28 (33 mg, 59% yield). 1H NMR (400 MHz, CDCl3) δ 8.67 (s, 1H), 7.67 (d, J = 7.7 Hz, 1H), 7.43–7.32 (m, 4H), 6.85 (d, J = 8.9 Hz, 1H), 5.09 (t, J = 7.2 Hz, 1H), 4.72 (t, J = 8.1 Hz, 1H), 4.63 (d, J = 9.0 Hz, 1H), 4.46 (s, 1H), 4.06 (d, J = 11.3 Hz, 1H), 3.61 (dd, J = 11.2, 3.6 Hz, 1H), 2.52 (s, 3H), 2.41–2.05 (m, 6H), 1.73–1.52 (m, 4H), 1.47 (d, J = 6.9 Hz, 3H), 1.03 (s, 9H). LC-MS (ESI): m/z 573.2 [M+H] +.
7-(((S)-1-((2S,4R)-4-Hydroxy-2-(((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)carbamoyl)pyrrolidin-1-yl)-3,3-dimethyl-1-oxobutan-2-yl)amino)-7- oxoheptanoic acid (30e).
Following general methods A and B, compound 30e was obtained from 29e and 28 (100 mg, 88% yield). 1H NMR (400 MHz, CDCl3 and CD3OD) δ 8.72 (s, 1H), 8.05–7.89 (m, 1H), 7.43–7.33 (m, 4H), 7.24–7.08 (m, 1H), 5.14–4.95 (m, 1H), 4.73–4.40 (m, 3H), 4.00–3.93 (m, 1H), 3.76–3.59 (m, 1H), 2.52 (s, 3H), 2.38–2.05 (m, 6H), 1.71–1.49 (m, 9H), 1.04 (s, 9H). LC-MS (ESI): m/z 587.2 [M+H] +.
8-(((S)-1-((2S,4R)-4-Hydroxy-2-(((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)carbamoyl)pyrrolidin-1-yl)-3,3-dimethyl-1-oxobutan-2-yl)amino)-8-oxooctanoic acid (30f).
Following general methods A and B, compound 30f was obtained from 29f and 28 (90 mg, 97% yield). 1H NMR (400 MHz, CDCl3) δ 8.72 (s, 1H), 7.59 (d, J = 7.8 Hz, 1H), 7.40–7.33 (m, 4H), 6.92 (d, J = 8.7 Hz, 1H), 5.15–4.98 (m, 1H), 4.76–4.67 (m, 1H), 4.62 (d, J = 8.9 Hz, 1H), 4.52 (s, 1H), 4.04 (d, J = 11.2 Hz, 1H), 3.74–3.59 (m, 1H), 2.51 (s, 3H), 2.39– 2.10 (m, 6H), 1.66–1.45 (m, 7H), 1.35–1.27 (m, 4H), 1.03 (s, 9H). LC-MS (ESI): m/z 601.2 [M+H] +.
3-(3-(((S)-1-((2S,4R)-4-Hydroxy-2-(((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)carbamoyl)pyrrolidin-1-yl)-3,3-dimethyl-1-oxobutan-2-yl)amino)-3-oxopropoxy)propanoic acid (30g).
Following general method A, compound 30g was obtained from 29g and 28 (21.2 mg, 62% yield). 1H NMR (400 MHz, CDCl3) δ 8.66 (s, 1H), 7.60 (d, J = 7.8 Hz, 1H), 7.48–7.32 (m, 4H), 7.10 (d, J = 8.3 Hz, 1H), 5.12–4.98 (m, 1H), 4.76 (t, J = 8.0 Hz, 1H), 4.55 (d, J = 8.4 Hz, 1H), 4.49–4.41 (m, 1H), 4.08 (d, J = 11.4 Hz, 1H), 3.79–3.63 (m, 4H), 3.57 (dd, J = 11.3, 3.7 Hz, 1H), 2.53–2.42 (m, 8H), 2.15–2.05 (m, 1H), 1.47 (d, J = 6.9 Hz, 3H), 1.03 (s, 9H). LC-MS (ESI): m/z 589.2 [M+H] +.
2-(2-(2-(((S)-1-((2S,4R)-4-Hydroxy-2-(((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)carbamoyl)pyrrolidin-1-yl)-3,3-dimethyl-1-oxobutan-2-yl)amino)-2-oxoethoxy)ethoxy)acetic acid (30h).
Following general method A, compound 30h was obtained from 29h and 28 (22 mg, 65% yield). 1H NMR (400 MHz, CDCl3) δ 8.66 (s, 1H), 7.64–7.31 (m, 6H), 5.13–4.98 (m, 1H), 4.75 (t, J = 8.1 Hz, 1H), 4.57 (d, J = 8.8 Hz, 1H), 4.48–4.40 (m, 1H), 4.15–3.85 (m, 5H), 3.80–3.63 (m, 4H), 3.58 (dd, J = 11.3, 3.5 Hz, 1H), 2.55–2.37 (m, 4H), 2.16– 2.02 (m, 1H), 1.47 (d, J = 6.8 Hz, 3H), 1.05 (s, 9H). Yield: 11.3 mg, 31%. LC-MS (ESI): m/z 605.3 [M+H] +.
2-(3-(2-(((S)-1-((2S,4R)-4-Hydroxy-2-(((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)carbamoyl)pyrrolidin-1-yl)-3,3-dimethyl-1-oxobutan-2-yl)amino)-2-oxoethoxy)propoxy)acetic acid (30i).
Following general method A, compound 30i was obtained from 29i and 28 (32 mg, 89% yield). 1H NMR (400 MHz, CDCl3) δ 8.66 (s, 1H), 7.69 (d, J = 7.7 Hz, 1H), 7.40–7.26 (m, 5H), 5.11–5.02 (m, 1H), 4.71 (t, J = 7.9 Hz, 1H), 4.57 (d, J = 9.0 Hz, 1H), 4.49–4.43 (m, 1H), 4.03–3.85 (m, 5H), 3.69–3.57 (m, 5H), 2.51 (s, 3H), 2.45–2.34 (m, 1H), 2.12–2.03 (m, 1H), 1.96–1.87 (m, 2H), 1.47 (d, J = 7.0 Hz, 3H), 1.03 (s, 9H). LC-MS (ESI): m/z 619.4 [M+H] +.
(2S,4R)-1-((S)-2-(3-(4-((R)-3-((4-(N-(4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzoyl)sulfamoyl)-2-((trifluoromethyl)sulfonyl)phenyl)amino)-4-(phenylthio)butyl)piperazin-1-yl)-3-oxopropanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (1a).
Following general method A, compound 1a was obtained from 30a and 18 (13.1 mg, 57% yield). 1H NMR (400 MHz, CDCl3) δ 8.74–8.54 (m, 2H), 8.35 (d, J = 2.2 Hz, 1H), 8.10 (dd, J = 9.3, 2.3 Hz, 1H), 7.79–7.58 (m, 3H), 7.41–7.27 (m, 10H), 7.26–7.19 (m, 1H), 7.06 (d, J = 8.5 Hz, 1H), 7.01–6.94 (m, 2H), 6.75 (d, J = 8.7 Hz, 2H), 6.64 (d, J = 9.4 Hz, 1H), 5.13–5.01 (m, 1H), 4.80–4.71 (m, 1H), 4.54–4.44 (m, 2H), 4.19– 4.04 (m, 1H), 3.97–3.84 (m, 1H), 3.72–2.88 (m, 15H), 2.59–2.02 (m, 20H), 1.66 (d, J = 13.1 Hz, 1H), 1.45 (d, J = 6.9 Hz, 5H), 1.08 (s, 9H), 0.98 (s, 6H). LC-MS (ESI): m/z 1485.6 [M+H] +; HPLC: >95% purity.
(2S,4R)-1-((S)-2-(4-(4-((R)-3-((4-(N-(4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzoyl)sulfamoyl)-2-((trifluoromethyl)sulfonyl)phenyl)amino)-4-(phenylthio)butyl)piperazin-1-yl)-4-oxobutanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (1b).
Following general method A, compound 1b was obtained from 30b and 18 (14.2 mg, 66% yield). 1H NMR (400 MHz, CDCl3) δ 8.67 (s, 1H), 8.37 (d, J = 2.2 Hz, 1H), 8.09 (dd, J = 9.2, 2.3 Hz, 1H), 7.68 (d, J = 8.6 Hz, 2H), 7.60 (d, J = 7.8 Hz, 1H), 7.43–7.28 (m, 10H), 7.26–7.21 (m, 1H), 7.11–6.89 (m, 4H), 6.76 (d, J = 8.8 Hz, 2H), 6.62 (d, J = 9.4 Hz, 1H), 5.14–5.01 (m, 1H), 4.81–4.74 (m, 1H), 4.53 (d, J = 8.5 Hz, 1H), 4.47 (s, 1H), 4.06 (d, J = 11.5 Hz, 1H), 3.97–3.83 (m, 1H), 3.63–2.84 (m, 13H), 2.62–1.99 (m, 24H), 1.74–1.61 (m, 1H), 1.51–1.43 (m, 5H), 1.06 (s, 9H), 0.98 (s, 6H). LC-MS (ESI): m/z 1499.7 [M+H] +; HPLC: >95% purity.
(2S,4R)-1-((S)-2-(5-(4-((R)-3-((4-(N-(4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzoyl)sulfamoyl)-2-((trifluoromethyl)sulfonyl)phenyl)amino)-4-(phenylthio)butyl)piperazin-1-yl)-5-oxopentanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (1c).
Following general method A, compound 1c was obtained from 30c and 18. (19.9 mg, 64% yield). 1H NMR (400 MHz, CDCl3) δ 8.67 (s, 1H), 8.34 (d, J = 2.2 Hz, 1H), 8.09 (dd, J = 9.3, 2.3 Hz, 1H), 7.71 (d, J = 8.7 Hz, 2H), 7.52 (d, J = 7.8 Hz, 1H), 7.41–7.27 (m, 9H), 7.26–7.21 (m, 2H), 7.12–6.95 (m, 3H), 6.89 (d, J = 8.3 Hz, 1H), 6.76 (d, J = 8.8 Hz, 2H), 6.61 (d, J = 9.5 Hz, 1H), 5.14–5.02 (m, 1H), 4.79–4.68 (m, 1H), 4.58–4.44 (m, 2H), 4.12 (d, J = 11.3 Hz, 1H), 3.96–3.81 (m, 1H), 3.68–2.85 (m, 13H), 2.54–2.00 (m, 24H), 1.87 (m, J = 7.2 Hz, 2H), 1.78–1.60 (m, 1H), 1.46 (dd, J = 6.7, 3.6 Hz, 5H), 1.06 (s, 9H), 0.97 (s, 6H). LC-MS (ESI): m/z 1513.6 [M+H] +; HPLC: >95% purity.
(2S,4R)-1-((S)-2-(6-(4-((R)-3-((4-(N-(4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzoyl)sulfamoyl)-2-((trifluoromethyl)sulfonyl)phenyl)amino)-4-(phenylthio)butyl)piperazin-1-yl)-6-oxohexanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (1d).
Following general method A, compound 1d was obtained from 30d and 18 (19.4 mg, 88% yield). 1H NMR (400 MHz, CDCl3) δ 8.67 (s, 1H), 8.34 (d, J = 2.3 Hz, 1H), 8.09 (dd, J = 9.3, 2.3 Hz, 1H), 7.70 (d, J = 8.6 Hz, 2H), 7.45–7.27 (m, 10H), 7.26–7.22 (m, 2H), 7.12–6.94 (m, 3H), 6.76 (d, J = 8.7 Hz, 2H), 6.62 (d, J = 9.4 Hz, 1H), 6.56 (d, J = 8.7 Hz, 1H), 5.14–5.01 (m, 1H), 4.80–4.69 (m, 1H), 4.61 (d, J = 8.8 Hz, 1H), 4.49 (s, 1H), 4.10 (d, J = 11.4 Hz, 1H), 3.98–3.84 (m, 1H), 3.70–2.85 (m, 13H), 2.54–2.00 (m, 24H), 1.45 (d, J = 6.7 Hz, 10H), 1.05 (s, 9H), 0.97 (s, 6H). LC-MS (ESI): m/z 1527.7 [M+H] +; HPLC: >95% purity.
(2S,4R)-1-((S)-2-(7-(4-((R)-3-((4-(N-(4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzoyl)sulfamoyl)-2-((trifluoromethyl)sulfonyl)phenyl)amino)-4-(phenylthio)butyl)piperazin-1-yl)-7-oxoheptanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (1e).
Following general method A, compound 1e was obtained from 30e and 18 (12.7 mg, 40% yield). 1H NMR (400 MHz, CDCl3) δ 8.67 (s, 1H), 8.34 (d, J = 2.3 Hz, 1H), 8.08 (d, J = 9.1 Hz, 1H), 7.69 (d, J = 8.6 Hz, 2H), 7.46–7.27 (m, 12H), 7.14–6.93 (m, 3H), 6.76 (d, J = 8.6 Hz, 2H), 6.61 (d, J = 9.4 Hz, 1H), 6.34 (d, J = 8.7 Hz, 1H), 5.13–5.00 (m, 1H), 4.79–4.69 (m, 1H), 4.60 (d, J = 8.7 Hz, 1H), 4.49 (s, 1H), 4.10 (d, J = 11.4 Hz, 1H), 3.96–3.83 (m, 1H), 3.72–2.81 (m, 13H), 2.55–2.00 (m, 24H), 1.77–1.30 (m, 12H), 1.03 (s, 9H), 0.97 (s, 6H). LC-MS (ESI): m/z 1541.7 [M+H] +; HPLC: >95% purity.
(2S,4R)-1-((S)-2-(8-(4-((R)-3-((4-(N-(4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzoyl)sulfamoyl)-2-((trifluoromethyl)sulfonyl)phenyl)amino)-4-(phenylthio)butyl)piperazin-1-yl)-8-oxooctanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (1f).
Following general method A, compound 1f was obtained from 30f and 18 (21.7 mg, 57% yield). 1H NMR (400 MHz, CDCl3) δ 8.67 (s, 1H), 8.34 (d, J = 2.3 Hz, 1H), 8.14–8.05 (m, 1H), 7.71 (d, J = 8.5 Hz, 2H), 7.49–7.27 (m, 11H), 7.25– 7.18 (m, 1H), 7.13–6.94 (m, 3H), 6.75 (d, J = 8.6 Hz, 2H), 6.61 (d, J = 9.4 Hz, 1H), 6.32 (d, J = 8.7 Hz, 1H), 5.14–5.02 (m, 1H), 4.77–4.67 (m, 1H), 4.60 (d, J = 8.8 Hz, 1H), 4.49 (s, 1H), 4.10 (d, J = 11.4 Hz, 1H), 3.97–3.84 (m, 1H), 3.74–2.85 (m, 13H), 2.54–1.99 (m, 24H), 1.76–1.23 (m, 14H), 1.04 (s, 9H), 0.97 (s, 6H). LC-MS (ESI): m/z 1555.6 [M+H] +; HPLC: >95% purity.
(2S,4R)-1-((S)-2-(3-(3-(4-((R)-3-((4-(N-(4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzoyl)sulfamoyl)-2-((trifluoromethyl)sulfonyl)phenyl)amino)-4-(phenylthio)butyl)piperazin-1-yl)-3-oxopropoxy)propanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (3a).
Following general method A, compound 3a was obtained from 30g and 18 (25.0 mg, 95% yield). 1H NMR (400 MHz, CDCl3) δ 8.66 (s, 1H), 8.34 (d, J = 2.2 Hz, 1H), 8.00 (dd, J = 9.2, 2.2 Hz, 1H), 7.82 (d, J = 8.9 Hz, 2H), 7.44–7.32 (m, 7H), 7.31–7.26 (m, 3H), 7.26–7.21 (m, 2H), 7.01–6.91 (m, 4H), 6.76 (d, J = 9.0 Hz, 2H), 6.53 (d, J = 9.4 Hz, 1H), 5.12–4.99 (m, 1H), 4.73 (t, J = 8.0 Hz, 1H), 4.54 (d, J = 8.4 Hz, 1H), 4.50–4.42 (m, 1H), 4.09 (d, J = 11.4 Hz, 1H), 3.89–3.62 (m, 6H), 3.55 (dd, J = 11.4, 3.6 Hz, 1H), 3.48–3.30 (m, 3H), 3.27–3.20 (m, 4H), 3.09 (dd, J = 13.8, 4.8 Hz, 1H), 2.94 (dd, J = 13.8, 7.6 Hz, 1H), 2.82 (s, 2H), 2.59–2.00 (m, 24H), 1.69–1.60 (m, 1H), 1.48–1.41 (m, 5H), 1.03 (s, 9H), 0.97 (s, 6H). LC-MS (ESI): m/z 1543.7 [M+H] +; HPLC: >95% purity.
(2S,4R)-1-((S)-2-(2-(2-(2-(4-((R)-3-((4-(N-(4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzoyl)sulfamoyl)-2-((trifluoromethyl)sulfonyl)phenyl)amino)-4-(phenylthio)butyl)piperazin-1-yl)-2-oxoethoxy)ethoxy)acetamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (3b).
Following general method A, compound 3b was obtained from 30h and 18 (18.0 mg, 65% yield). 1H NMR (400 MHz, CDCl3) δ 8.67 (s, 1H), 8.35 (d, J = 2.3 Hz, 1H), 8.00 (dd, J = 9.2, 2.3 Hz, 1H), 7.78 (d, J = 8.8 Hz, 2H), 7.43–7.33 (m, 8H), 7.32–7.27 (m, 3H), 7.26–7.21 (m, 2H), 7.04–6.93 (m, 3H), 6.80– 6.72 (m, 2H), 6.57 (d, J = 9.4 Hz, 1H), 5.11–5.00 (m, 1H), 4.71 (t, J = 8.0 Hz, 1H), 4.58 (d, J = 8.8 Hz, 1H), 4.51–4.42 (m, 1H), 4.25–3.94 (m, 5H), 3.90–3.81 (m, 1H), 3.72–3.37 (m, 9H), 3.28–3.22 (m, 4H), 3.11–3.06 (m, 1H), 2.96 (dd, J = 13.8, 7.4 Hz, 1H), 2.82 (s, 2H), 2.51–2.00 (m, 20H), 1.71–1.59 (m, 1H), 1.49–1.42 (m, 5H), 1.05 (s, 9H), 0.97 (s, 6H). LC-MS (ESI): m/z 1559.6 [M+H] +; HPLC: >95% purity.
(2S,4R)-1-((S)-2-(2-(3-(2-(4-((R)-3-((4-(N-(4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzoyl)sulfamoyl)-2-((trifluoromethyl)sulfonyl)phenyl)amino)-4-(phenylthio)butyl)piperazin-1-yl)-2-oxoethoxy)propoxy)acetamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (3c).
Following general method A, compound 3c was obtained from 30i and 18 (15.9 mg, 63% yield). 1H NMR (400 MHz, CDCl3) δ 8.67 (s, 1H), 8.36 (d, J = 2.2 Hz, 1H), 8.03 (dd, J = 9.2, 2.3 Hz, 1H), 7.78–7.71 (m, 2H), 7.46–7.33 (m, 7H), 7.32–7.27 (m, 3H), 7.26–7.22 (m, 3H), 7.06 (d, J = 8.6 Hz, 1H), 7.02– 6.95 (m, 2H), 6.80–6.68 (m, 2H), 6.60 (d, J = 9.4 Hz, 1H), 5.12–5.02 (m, 1H), 4.74 (t, J = 7.9 Hz, 1H), 4.58 (d, J = 8.8 Hz, 1H), 4.53–4.46 (m, 1H), 4.19–3.81 (m, 6H), 3.72–3.54 (m, 6H), 3.52–3.33 (m, 3H), 3.32–3.22 (m, 4H), 3.12–3.07 (m, 1H), 2.98 (dd, J = 13.8, 7.3 Hz, 1H), 2.84 (s, 2H), 2.50–1.88 (m, 22H), 1.69–1.62 (m, 1H), 1.50–1.42 (m, 5H), 1.05 (s, 9H), 0.97 (s, 6H). LC-MS (ESI): m/z 1573.6 [M+H] +; HPLC: >95% purity.
N1-((R)-3-((4-(N-(4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzoyl)sulfamoyl)-2-((trifluoromethyl)sulfonyl)phenyl)amino)-4-(phenylthio)butyl)-N7-((S)-1-((2S,4R)-4-hydroxy-2-(((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)carbamoyl)pyrrolidin-1-yl)-3,3-dimethyl-1-oxobutan-2-yl)-N1-methylheptanediamide (5).
Following general method A, compound 5 was obtained from 30e and 19 (6.6 mg, 29% yield). 1H NMR (400 MHz, CDCl3 and CD3OD) δ 8.69 (s, 1H), 8.44–8.30 (m, 1H), 8.10–7.96 (m, 1H), 7.92–7.57 (m, 3H), 7.43–7.34 (m, 6H), 7.31–7.18 (m, 4H), 7.08– 6.73 (m, 6H), 6.56–6.26 (m, 1H), 5.12–4.94 (m, 1H), 4.72–4.40 (m, 3H), 4.12–3.88 (m, 1H), 3.81–2.77 (m, 15H), 2.51 (s, 3H), 2.45–1.93 (m, 15H), 1.83–1.16 (m, 12H), 1.08–0.94 (m, 15H). LC-MS (ESI): m/z 1486.6 [M+H] +; HPLC: >95% purity.
(2S,4R)-1-((S)-2-(5-(4-((R)-3-((4-(N-(4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzoyl)sulfamoyl)-2-((trifluoromethyl)sulfonyl)phenyl)amino)-4-(phenylthio)butyl)piperazin-1-yl)pentanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (2a).
Following general methods C, B and A, compound 2a was obtained from 31a, 18 and 28 (7.3 mg, 27% yield). 1H NMR (400 MHz, CDCl3 and CD3OD) δ 8.64 (s, 1H), 8.26 (d, J = 2.2 Hz, 1H), 8.05–7.94 (m, 1H), 7.78 (d, J = 8.6 Hz, 2H), 7.56 (d, J = 7.7 Hz, 1H), 7.38–7.29 (m, 6H), 7.25–7.15 (m, 5H), 7.00–6.83 (m, 4H), 6.73 (d, J = 8.6 Hz, 2H), 6.54 (d, J = 9.3 Hz, 1H), 5.08–4.95 (m, 1H), 4.59 (t, J = 8.2 Hz, 1H), 4.52 (d, J = 9.0 Hz, 1H), 4.42 (s, 1H), 3.95 (d, J = 11.3 Hz, 1H), 3.84–3.69 (m, 1H), 3.56 (dd, J = 11.3, 3.4 Hz, 1H), 3.23–3.15 (m, 4H), 3.05 (dd, J = 13.7, 5.0 Hz, 1H), 2.95 (dd, J = 13.8, 7.0 Hz, 1H), 2.78 (s, 2H), 2.67–1.92 (m, 28H), 1.69–1.40 (m, 10H), 0.99 (s, 9H), 0.94 (s, 6H). LC-MS (ESI): m/z 1499.8 [M+H] +; HPLC: >95% purity.
(2S,4R)-1-((S)-2-(6-(4-((R)-3-((4-(N-(4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzoyl)sulfamoyl)-2-((trifluoromethyl)sulfonyl)phenyl)amino)-4-(phenylthio)butyl)piperazin-1-yl)hexanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (2b).
Following general methods C, B and A, compound 2b was obtained from 31b, 18 and 28 (8.4 mg, 30% yield). 1H NMR (400 MHz, CDCl3 and CD3OD) δ 8.67 (s, 1H), 8.30 (d, J = 2.2 Hz, 1H), 8.01 (dd, J = 9.1, 2.2 Hz, 1H), 7.83 (d, J = 8.6 Hz, 2H), 7.57 (d, J = 7.7 Hz, 1H), 7.42–7.32 (m, 6H), 7.29–7.19 (m, 5H), 7.03–6.90 (m, 3H), 6.77 (t, J = 9.1 Hz, 3H), 6.54 (d, J = 9.3 Hz, 1H), 5.06 (p, J = 7.0 Hz, 1H), 4.66 (t, J = 8.2 Hz, 1H), 4.57 (d, J = 9.1 Hz, 1H), 4.45 (s, 1H), 4.01 (d, J = 11.4 Hz, 1H), 3.79 (s, 1H), 3.58 (dd, J = 11.2, 3.3 Hz, 1H), 3.22 (t, J = 4.9 Hz, 4H), 3.08 (dd, J = 13.7, 4.8 Hz, 1H), 3.01–2.92 (m, 1H), 2.82 (s, 2H), 2.79–1.94 (m, 28H), 1.75–1.18 (m, 12H), 1.02 (s, 9H), 0.97 (d, J = 1.8 Hz, 6H). LC-MS (ESI): m/z 1513.7 [M+H] +; HPLC: >95% purity.
(2S,4R)-1-((S)-2-(7-(4-((R)-3-((4-(N-(4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzoyl)sulfamoyl)-2-((trifluoromethyl)sulfonyl)phenyl)amino)-4-(phenylthio)butyl)piperazin-1-yl)heptanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (2c).
Following general methods C, B and A, compound 2c was obtained from 31c, 18 and 28 (22.6 mg, 81% yield). 1H NMR (400 MHz, CDCl3 and CD3OD) δ 8.75 (s, 1H), 8.33 (d, J = 2.2 Hz, 1H), 8.25 (d, J = 7.6 Hz, 1H), 8.07 (dd, J = 9.2, 2.3 Hz, 1H), 7.81 (d, J = 8.7 Hz, 2H), 7.56 (s, 1H), 7.41–7.17 (m, 11H), 7.13–7.00 (m, 3H), 6.84 (d, J = 9.0 Hz, 2H), 6.69 (d, J = 9.4 Hz, 1H), 5.09–4.98 (m, 1H), 4.65–4.46 (m, 3H), 4.00–3.82 (m, 2H), 3.77–3.65 (m, 1H), 3.39–2.92 (m, 12H), 2.75–1.95 (m, 24H), 1.86–1.21 (m, 14H), 1.04 (s, 9H), 1.02 (s, 6H). LC-MS (ESI): m/z 1527.7 [M+H] +; HPLC: >95% purity.
(2S,4R)-1-((S)-2-(8-(4-((R)-3-((4-(N-(4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzoyl)sulfamoyl)-2-((trifluoromethyl)sulfonyl)phenyl)amino)-4-(phenylthio)butyl)piperazin-1-yl)octanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (2d).
Following general methods C, B and A, compound 2d was obtained from 31d, 18 and 28 (18.3 mg, 65% yield). 1H NMR (400 MHz, CDCl3 and CD3OD) δ 8.75 (s, 1H), 8.34 (d, J = 2.2 Hz, 1H), 8.20 (d, J = 7.6 Hz, 1H), 8.08 (dd, J = 9.2, 2.3 Hz, 1H), 7.78 (d, J = 8.7 Hz, 2H), 7.56 (s, 1H), 7.46–7.19 (m, 11H), 7.13 (d, J = 8.4 Hz, 1H), 7.07–6.99 (m, 2H), 6.88–6.80 (m, 2H), 6.68 (d, J = 9.4 Hz, 1H), 5.11–4.98 (m, 1H), 4.61–4.41 (m, 3H), 3.99–3.82 (m, 2H), 3.78–3.67 (m, 1H), 3.37–2.91 (m, 12H), 2.65–1.99 (m, 24H), 1.85–1.30 (m, 16H), 1.04 (s, 9H), 1.01 (s, 6H). LC-MS (ESI): m/z 1541.7 [M+H] +; HPLC: >95% purity.
(2S,4R)-1-((S)-2-(5-(((R)-3-((4-(N-(4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzoyl)sulfamoyl)-2-((trifluoromethyl)sulfonyl)phenyl)amino)-4-(phenylthio)butyl)(methyl)amino)pentanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (6a).
Following general methods C, B and A, compound 6a was obtained from 31a, 19 and 28 (11.1 mg, 47% yield). 1H NMR (600 MHz, CDCl3) δ 8.69 (s, 1H), 8.43–8.34 (m, 1H), 7.93 (d, J = 9.0 Hz, 1H), 7.83 (d, J = 8.5 Hz, 2H), 7.48–7.29 (m, 8H), 7.28–7.18 (m, 4H), 7.05–6.92 (m, 3H), 6.84–6.74 (m, 3H), 6.34 (br s, 1H), 5.15–5.07 (m, 1H), 4.87–4.77 (m, 1H), 4.60 (d, J = 8.8 Hz, 1H), 4.53–4.46 (m, 1H), 4.19–4.02 (m, 2H), 3.66–3.58 (m, 1H), 3.30–3.20 (m, 4H), 3.13–3.03 (m, 2H), 2.86–2.81 (m, 2H), 2.74–2.49 (m, 7H), 2.40–2.02 (m, 16H), 1.85–1.73 (m, 1H), 1.49–1.36 (m, 9H), 1.05 (s, 9H), 1.00 (s, 6H). LC-MS (ESI): m/z 1444.5 [M+H] +; HPLC: >95% purity.
(2S,4R)-1-((S)-2-(6-(((R)-3-((4-(N-(4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzoyl)sulfamoyl)-2-((trifluoromethyl)sulfonyl)phenyl)amino)-4-(phenylthio)butyl)(methyl)amino)hexanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (6b).
Following general methods C, B and A, compound 6b was obtained from 31b, 19 and 28 (5.9 mg, 19% yield). 1H NMR (600 MHz, CDCl3) δ 8.68 (s, 1H), 8.32 (d, J = 2.2 Hz, 1H), 8.00–7.91 (m, 1H), 7.85 (d, J = 8.5 Hz, 2H), 7.48 (d, J = 7.8 Hz, 1H), 7.42–7.31 (m, 6H), 7.30–7.29 (m, 1H), 7.28–7.19 (m, 4H), 7.05–6.89 (m, 3H), 6.82–6.65 (m, 3H), 6.43 (d, J = 8.6 Hz, 1H), 5.12–5.03 (m, 1H), 4.78 (t, J = 8.2 Hz, 1H), 4.60 (d, J = 8.7 Hz, 1H), 4.50–4.42 (m, 1H), 4.09 (d, J = 11.3 Hz, 1H), 4.02–3.92 (m, 1H), 3.63–3.55 (m, 1H), 3.29–3.19 (m, 4H), 3.10 (dd, J = 13.9, 4.7 Hz, 1H), 3.03 (dd, J = 13.8, 6.4 Hz, 1H), 2.87–2.71 (m, 3H), 2.70–2.41 (m, 9H), 2.40–2.23 (m, 7H), 2.19–1.99 (m, 6H), 1.89– 1.80 (m, 1H), 1.48–1.38 (m, 7H), 1.35 (t, J = 7.3 Hz, 2H), 1.20–1.13 (m, 2H), 1.05 (s, 9H), 1.00 (s, 6H). LC-MS (ESI): m/z 1458.6 [M+H] +; HPLC: >95% purity.
(2S,4R)-1-((S)-2-(7-(((R)-3-((4-(N-(4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzoyl)sulfamoyl)-2-((trifluoromethyl)sulfonyl)phenyl)amino)-4-(phenylthio)butyl)(methyl)amino)heptanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (6c).
Following general methods C, B and A, compound 6c was obtained from 31c, 19 and 28 (7.3 mg, 23% yield). 1H NMR (600 MHz, CDCl3) δ 8.68 (s, 1H), 8.34 (d, J = 2.2 Hz, 1H), 7.93 (d, J = 9.0 Hz, 1H), 7.88 (d, J = 8.6 Hz, 2H), 7.45–7.29 (m, 8H), 7.28–7.21 (m, 4H), 7.03–6.94 (m, 3H), 6.78 (d, J = 9.0 Hz, 2H), 6.70–6.62 (m, 1H), 6.46–6.38 (m, 1H), 5.11–5.02 (m, 1H), 4.77 (t, J = 8.2 Hz, 1H), 4.62 (d, J = 8.8 Hz, 1H), 4.51–4.45 (m, 1H), 4.16–4.10 (m, 1H), 4.03–3.94 (m, 1H), 3.62–3.56 (m, 1H), 3.27–3.19 (m, 4H), 3.10 (dd, J = 13.8, 4.8 Hz, 1H), 3.01 (dd, J = 13.8, 6.7 Hz, 1H), 2.82–2.49 (m, 9H), 2.44–2.25 (m, 10H), 2.19–2.01 (m, 6H), 1.85–1.75 (m, 1H), 1.46–1.33 (m, 9H), 1.19–1.10 (m, 4H), 1.07 (s, 9H), 1.00 (s, 6H). LC-MS (ESI): m/z 1472.6 [M+H] +; HPLC: >95% purity.
(2S,4R)-1-((S)-2-(8-(((R)-3-((4-(N-(4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzoyl)sulfamoyl)-2-((trifluoromethyl)sulfonyl)phenyl)amino)-4-(phenylthio)butyl)(methyl)amino)octanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (6d).
Following general methods C, B and A, compound 6d was obtained from 31d, 19 and 28 (5.1 mg, 21% yield). 1H NMR (600 MHz, CDCl3) δ 8.69 (s, 1H), 8.51–8.39 (m, 1H), 8.10–8.02 (m, 1H), 7.91–7.79 (m, 2H), 7.44–7.29 (m, 8H), 7.28–7.19 (m, 4H), 7.06–6.97 (m, 3H), 6.85–6.76 (m, 3H), 6.46–6.36 (m, 1H), 5.06–4.93 (m, 1H), 4.80 (t, J = 8.5 Hz, 1H), 4.71–4.62 (m, 1H), 4.56–4.49 (m, 1H), 4.22–4.16 (m, 1H), 4.16–4.07 (m, 1H), 3.71–3.58 (m, 1H), 3.35–3.23 (m, 4H), 3.22–3.14 (m, 2H), 2.89–2.51 (m, 9H), 2.47–2.01 (m, 16H), 1.95–1.85 (m, 1H), 1.48 (t, J = 6.5 Hz, 9H), 1.27–1.16 (m, 6H), 1.06 (s, 9H), 1.01 (s, 6H). LC-MS (ESI): m/z 1486.6 [M+H] +; HPLC: >95% purity.
(2S,4R)-1-((S)-2-(9-(((R)-3-((4-(N-(4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzoyl)sulfamoyl)-2-((trifluoromethyl)sulfonyl)phenyl)amino)-4-(phenylthio)butyl)(methyl)amino)nonanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (6e).
Following general methods C, B and A, compound 6e was obtained from 31e, 19 and 28 (9.1 mg, 40% yield). 1H NMR (600 MHz, CDCl3) δ 8.69 (s, 1H), 8.36 (d, J = 2.1 Hz, 1H), 7.99 (d, J = 9.1 Hz, 1H), 7.84 (d, J = 8.6 Hz, 2H), 7.41–7.29 (m, 9H), 7.28–7.21 (m, 3H), 7.07–6.96 (m, 3H), 6.83–6.67 (m, 3H), 6.44 (d, J = 8.9 Hz, 1H), 5.10–5.01 (m, 1H), 4.76 (t, J = 8.1 Hz, 1H), 4.65 (d, J = 8.9 Hz, 1H), 4.54–4.49 (m, 1H), 4.18–4.00 (m, 2H), 3.65–3.58 (m, 1H), 3.32–3.24 (m, 4H), 3.13–3.03 (m, 2H), 2.88–2.46 (m, 11H), 2.44–2.01 (m, 14H), 1.95–1.85 (m, 1H), 1.54–1.43 (m, 9H), 1.22– 1.10 (m, 8H), 1.06 (s, 9H), 1.00 (s, 6H). LC-MS (ESI): m/z 1500.7 [M+H] +; HPLC: >95% purity.
(2S,4R)-1-((S)-2-(10-(((R)-3-((4-(N-(4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzoyl)sulfamoyl)-2-((trifluoromethyl)sulfonyl)phenyl)amino)-4-(phenylthio)butyl)(methyl)amino)decanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (6f).
Following general methods C, B and A, compound 6f was obtained from 31f, 19 and 28 (7.8 mg, 43% yield). 1H NMR (600 MHz, CDCl3) δ 8.69 (s, 1H), 8.33 (s, 1H), 8.04–7.94 (m, 1H), 7.90–7.76 (m, 2H), 7.46–7.32 (m, 7H), 7.30–7.29 (m, 1H), 7.28–7.19 (m, 4H), 7.07–6.95 (m, 3H), 6.84–6.57 (m, 3H), 6.46–6.29 (m, 1H), 5.15–4.99 (m, 1H), 4.76 (t, J = 8.0 Hz, 1H), 4.63 (d, J = 8.6 Hz, 1H), 4.57–4.47 (m, 1H), 4.26–3.97 (m, 2H), 3.65–3.54 (m, 1H), 3.32–3.19 (m, 4H), 3.15–3.03 (m, 2H), 2.89–2.52 (m, 8H), 2.51–2.02 (m, 17H), 1.87–1.84 (m, 1H), 1.53–1.45 (m, 9H), 1.22–1.09 (m, 10H), 1.04 (s, 9H), 1.00 (s, 6H). LC-MS (ESI): m/z 1514.7 [M+H] +; HPLC: >95% purity.
(2S,4R)-1-((S)-2-(2-(2-(4-((R)-3-((4-(N-(4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzoyl)sulfamoyl)-2-((trifluoromethyl)sulfonyl)phenyl)amino)-4-(phenylthio)butyl)piperazin-1-yl)ethoxy)acetamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (4a).
Following general methods C, B and A, compound 4a was obtained from 34a, 18 and 28 (7.4 mg, 49% yield). 1H NMR (400 MHz, CDCl3) δ 8.66 (s, 1H), 8.37 (d, J = 2.1 Hz, 1H), 8.09 (d, J = 8.9 Hz, 1H), 7.91–7.79 (m, 1H), 7.71 (d, J = 8.4 Hz, 2H), 7.46–7.28 (m, 11H), 7.25–7.16 (m, 1H), 6.99 (d, J = 8.1 Hz, 2H), 6.77 (d, J = 8.6 Hz, 2H), 6.70–6.57 (m, 1H), 5.20–5.00 (m, 1H), 4.91–4.77 (m, 1H), 4.67 (d, J = 9.3 Hz, 1H), 4.47 (s, 1H), 4.04–2.65 (m, 21H), 2.60–1.93 (m, 20H), 1.85–1.43 (m, 6H), 1.05 (s, 9H), 1.00 (s, 6H). LC-MS (ESI): m/z 1501.6 [M+H] +; HPLC: >95% purity.
(2S,4R)-1-((S)-2-(2-(2-(2-(4-((R)-3-((4-(N-(4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzoyl)sulfamoyl)-2-((trifluoromethyl)sulfonyl)phenyl)amino)-4-(phenylthio)butyl)piperazin-1-yl)ethoxy)ethoxy)acetamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (4b).
Following general methods C, B and A, compound 4b was obtained from 34b, 18 and 28 (5.4 mg, 54% yield). 1H NMR (400 MHz, CDCl3) δ 8.67 (s, 1H), 8.40 (d, J = 2.2 Hz, 1H), 8.01 (d, J = 9.3 Hz, 1H), 7.70 (d, J = 8.6 Hz, 2H), 7.60 (d, J = 7.4 Hz, 1H), 7.43–7.27 (m, 12H), 6.99 (d, J = 8.5 Hz, 2H), 6.78 (d, J = 8.7 Hz, 2H), 6.70–6.61 (m, 1H), 5.15–5.03 (m, 1H), 4.83–4.73 (m, 1H), 4.67 (d, J = 9.3 Hz, 1H), 4.45 (s, 1H), 4.18–2.65 (m, 25H), 2.61–1.93 (m, 20H), 1.85–1.44 (m, 6H), 1.05 (s, 9H), 0.99 (s, 6H). LC-MS (ESI): m/z 1545.7 [M+H] +; HPLC: >95% purity.
(2S,4R)-1-((2S,15R)-2-(tert-Butyl)-15-((4-(N-(4-(4-((4’-chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzoyl)sulfamoyl)-2-((trifluoromethyl)sulfonyl)phenyl)amino)-12-methyl-4-oxo-16-(phenylthio)-6,9-dioxa-3,12-diazahexadecanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (7a).
Following general methods C, B and A, compound 7a was obtained from 34b, 19 and 28 (7.5 mg, 48% yield). 1H NMR (400 MHz, CDCl3) δ 8.65 (s, 1H), 8.37 (d, J = 2.2 Hz, 1H), 8.07–7.99 (m, 1H), 7.73 (d, J = 8.6 Hz, 2H), 7.45 (d, J = 7.8 Hz, 1H), 7.39–7.27 (m, 8H), 7.25–7.10 (m, 4H), 7.01–6.95 (m, 2H), 6.79–6.70 (m, 3H), 5.13–5.03 (m, 1H), 4.84 (t, J = 8.1 Hz, 1H), 4.60 (d, J = 8.9 Hz, 1H), 4.53–4.46 (m, 1H), 4.11–3.98 (m, 2H), 3.87 (d, J = 15.4 Hz, 1H), 3.66–3.58 (m, 2H), 3.52–3.33 (m, 6H), 3.27–3.20 (m, 4H), 3.13–2.99 (m, 2H), 2.83– 2.78 (m, 2H), 2.75–2.49 (m, 6H), 2.47–1.98 (m, 15H), 1.79–1.68 (m, 1H), 1.51–1.43 (m, 5H), 1.04 (s, 9H), 0.98 (s, 6H). LC-MS (ESI): m/z 1490.7 [M+H] +; HPLC: >95% purity.
(2S,4R)-1-((2S,18R)-2-(tert-Butyl)-18-((4-(N-(4-(4-((4’-chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzoyl)sulfamoyl)-2-((trifluoromethyl)sulfonyl)phenyl)amino)-15-methyl-4-oxo-19-(phenylthio)-6,9,12-trioxa-3,15-diazanonadecanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (7b).
Following general methods C, B and A, compound 7b was obtained from 34c, 19 and 28 (7.8 mg, 60% yield). 1H NMR (400 MHz, CDCl3) δ 8.66 (s, 1H), 8.35 (d, J = 2.0 Hz, 1H), 8.01 (d, J = 8.6 Hz, 1H), 7.76 (d, J = 8.5 Hz, 2H), 7.49 (d, J = 7.8 Hz, 1H), 7.39–7.32 (m, 6H), 7.29–7.27 (m, 1H), 7.26–7.10 (m, 5H), 7.03– 6.95 (m, 2H), 6.82–6.68 (m, 3H), 5.13–5.01 (m, 1H), 4.82 (t, J = 8.1 Hz, 1H), 4.61 (d, J = 8.9 Hz, 1H), 4.48 (s, 1H), 4.12–4.00 (m, 2H), 3.91–3.48 (m, 13H), 3.30–3.19 (m, 4H), 3.13–3.06 (m, 2H), 2.84–2.45 (m, 10H), 2.39–1.98 (m, 13H), 1.93–1.81 (m, 1H), 1.52–1.43 (m, 5H), 1.04 (s, 9H), 0.98 (s, 6H). LC-MS (ESI): m/z 1534.7 [M+H] +; HPLC: >95% purity.
(2S,4R)-1-((2S,21R)-2-(tert-Butyl)-21-((4-(N-(4-(4-((4’-chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzoyl)sulfamoyl)-2-((trifluoromethyl)sulfonyl)phenyl)amino)-18-methyl-4-oxo-22-(phenylthio)-6,9,12,15-tetraoxa-3,18-diazadocosanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (7c).
Following general methods C, B and A, compound 7c was obtained from 34d, 19 and 28 (8.5 mg, 58% yield). 1H NMR (400 MHz, CDCl3) δ 8.66 (s, 1H), 8.40 (s, 1H), 8.03 (d, J = 9.1 Hz, 1H), 7.76 (d, J = 8.4 Hz, 2H), 7.48–7.32 (m, 8H), 7.31–7.28 (m, 2H), 7.25–7.12 (m, 3H), 7.02–6.96 (m, 2H), 6.92–6.67 (m, 3H), 5.17– 4.99 (m, 1H), 4.83 (t, J = 8.1 Hz, 1H), 4.61 (d, J = 8.9 Hz, 1H), 4.55–4.44 (m, 1H), 4.24–4.03 (m, 2H), 3.92–3.43 (m, 17H), 3.36–3.07 (m, 6H), 2.91–2.49 (m, 8H), 2.46–1.99 (m, 15H), 1.95– 1.85 (m, 1H), 1.52–1.44 (m, 5H), 1.05–0.96 (m, 15H). LC-MS (ESI): m/z 1578.7 [M+H] +; HPLC: >95% purity.
4-((2-(2,6-Dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)butanoic acid (39a).
Following general method D, compound 39a was obtained from 37a and 38 (34 mg, 26% yield). 1H NMR (400 MHz, CDCl3) δ 8.27 (s, 1H), 7.50 (dd, J = 8.5, 7.1 Hz, 1H), 7.11 (dd, J = 7.1, 0.6 Hz, 1H), 6.93 (d, J = 8.5 Hz, 1H), 6.36–6.25 (m, 1H), 4.92 (dd, J = 12.1, 5.3 Hz, 1H), 3.43–3.29 (m, 2H), 2.97–2.67 (m, 3H), 2.49 (t, J = 7.0 Hz, 2H), 2.16–2.08 (m, 1H), 2.03–1.96 (m, 2H). LC- MS (ESI): m/z 360.1 [M+H] +.
5-((2-(2,6-Dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)pentanoic acid (39b).
Following general method D, compound 39b was obtained from 37b and 38 (30 mg, 21% yield). 1H NMR (400 MHz, CDCl3) δ 8.28 (s, 1H), 7.49 (dd, J = 8.5, 7.1 Hz, 1H), 7.10 (dd, J = 7.1, 0.6 Hz, 1H), 6.93 (d, J = 8.5 Hz, 1H), 6.36–6.25 (m, 1H), 4.92 (dd, J = 12.1, 5.3 Hz, 1H), 3.30–3.20 (m, 2H), 2.97–2.67 (m, 3H), 2.45–2.35 (m, 2H), 2.16–2.05 (m, 1H), 1.80–1.65 (m, 4H). LC-MS (ESI): m/z 374.1 [M+H] +.
6-((2-(2,6-Dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)hexanoic acid (39c)
Following general method D, compound 39c was obtained from 37c and 38 (45 mg, 16% yield). 1H NMR (400 MHz, CDCl3) δ 8.29 (s, 1H), 7.54–7.44 (m, 1H), 7.09 (dd, J = 7.2, 0.6 Hz, 1H), 6.86 (d, J = 8.5 Hz, 1H), 6.27–6.10 (m, 1H), 4.91 (dd, J = 12.2, 5.3 Hz, 1H), 3.36–3.16 (m, 2H), 2.99–2.63 (m, 3H), 2.39 (t, J = 7.4 Hz, 2H), 2.18–2.06 (m, 1H), 1.82–1.62 (m, 4H), 1.52–1.39 (m, 2H). LC-MS (ESI): m/z 388.2 [M+H] +.
7-((2-(2,6-Dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)heptanoic acid (39d).
Following general method D, compound 39d was obtained from 37d and 38 (26 mg, 36% yield). 1H NMR (400 MHz, CDCl3) δ 8.39 (s, 1H), 7.48 (dd, J = 8.5, 7.1 Hz, 1H), 7.07 (dd, J = 7.1, 0.6 Hz, 1H), 6.87 (d, J = 8.5 Hz, 1H), 6.31–6.10 (m, 1H), 4.91 (dd, J = 12.0, 5.4 Hz, 1H), 3.32–3.18 (m, 2H), 2.97–2.66 (m, 3H), 2.35 (t, J = 7.4 Hz, 2H), 2.17–2.09 (m, 1H), 1.73–1.59 (m, 4H), 1.51–1.34 (m, 4H). LC-MS (ESI): m/z 402.1 [M+H] +.
8-((2-(2,6-Dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)octanoic acid (39e).
Following general method D, compound 39e was obtained from 37e and 38 (28 mg, 19% yield). 1H NMR (400 MHz, CDCl3) δ 8.37 (s, 1H), 7.49 (dd, J = 8.5, 7.1 Hz, 1H), 7.08 (d, J = 7.1 Hz, 1H), 6.88 (d, J = 8.5 Hz, 1H), 6.32–6.16 (m, 1H), 4.92 (dd, J = 12.1, 5.4 Hz, 1H), 3.31–3.20 (m, 2H), 3.02–2.65 (m, 3H), 2.35 (t, J = 7.4 Hz, 2H), 2.19–2.08 (m, 1H), 1.73–1.57 (m, 4H), 1.50– 1.32 (m, 6H). LC-MS (ESI): m/z 416.0 [M+H] +.
4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)-N-((4-(((2R)-4-(4-(4-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)butanoyl)piperazin-1-yl)-1-(phenylthio)butan-2-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)benzamide (8a).
Following general method A, compound 8a was obtained from 39a and 18 (3.1 mg, 14% yield). 1H NMR (400 MHz, CDCl3) δ 8.60–8.30 (m, 2H), 8.10–8.00 (m, 1H), 7.74–7.64 (m, 2H), 7.53–7.40 (m, 1H), 7.39–7.27 (m, 5H), 7.26–7.22 (m, 1H), 7.12–7.02 (m, 2H), 7.01–6.91 (m, 3H), 6.78–6.67 (m, 2H), 6.65–6.58 (m, 1H), 6.33–6.25 (m, 1H), 5.00–4.82 (m, 1H), 3.98–3.84 (m, 1H), 3.82–3.62 (m, 1H), 3.48– 3.21 (m, 9H), 3.13–2.96 (m, 2H), 2.91–2.66 (m, 5H), 2.47–1.93 (m, 20H), 1.73–1.58 (m, 1H), 1.46 (t, J = 6.5 Hz, 2H), 0.97 (s, 6H). LC-MS (ESI): m/z 1314.4 [M+H] +; HPLC: >95% purity.
4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)-N-((4-(((2R)-4-(4-(5-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)pentanoyl)piperazin-1-yl)-1-(phenylthio)butan-2-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)benzamide (8b).
Following general method A, compound 8b was obtained from 39b and 18 (11.9 mg, 63% yield). 1H NMR (400 MHz, CDCl3) δ 8.57–8.26 (m, 2H), 8.13–8.04 (m, 1H), 7.68 (d, J = 8.9 Hz, 2H), 7.50–7.44 (m, 1H), 7.39–7.26 (m, 5H), 7.26–7.23 (m, 1H), 7.11–6.95 (m, 4H), 6.87 (d, J = 8.5 Hz, 1H), 6.80–6.71 (m, 2H), 6.59 (dd, J = 9.6, 2.6 Hz, 1H), 6.24 (t, J = 5.6 Hz, 1H), 4.97–4.80 (m, 1H), 3.94–3.84 (m, 1H), 3.73–3.68 (m, 1H), 3.50–3.22 (m, 9H), 3.10–2.96 (m, 2H), 2.92–2.66 (m, 5H), 2.44–2.00 (m, 18H), 1.74–1.49 (m, 7H), 0.98 (s, 6H). LC-MS (ESI): m/z 1328.5 [M+H] +; HPLC: >95% purity.
4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)-N-((4-(((2R)-4-(4-(6-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)hexanoyl)piperazin-1-yl)-1-(phenylthio)butan-2-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)benzamide (8c).
Following general method A, compound 8c was obtained from 39c and 18 (15.4 mg, 61% yield). 1H NMR (400 MHz, CDCl3) δ 8.60–8.26 (m, 2H), 8.09 (t, J = 8.6 Hz, 1H), 7.71–7.59 (m, 2H), 7.51–7.45 (m, 1H), 7.42–7.27 (m, 5H), 7.26–7.25 (m, 1H), 7.16–6.94 (m, 4H), 6.87 (d, J = 8.5 Hz, 1H), 6.80–6.54 (m, 3H), 6.31–6.15 (m, 1H), 4.97–4.82 (m, 1H), 4.01–3.61 (m, 2H), 3.51–3.19 (m, 9H), 3.14–2.96 (m, 2H), 2.92–2.69 (m, 5H), 2.45–1.98 (m, 18H), 1.67–1.46 (m, 9H), 0.99 (s, 6H). LC-MS (ESI): m/z 1342.5 [M+H] +; HPLC: >95% purity.
4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)-N-((4-(((2R)-4-(4-(7-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)heptanoyl)piperazin-1-yl)-1-(phenylthio)butan-2-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)benzamide (8d).
Following general method A, compound 8d was obtained from 39d and 18 (17.6 mg, 79% yield). 1H NMR (400 MHz, CDCl3) δ 8.54 (d, J = 13.2 Hz, 1H), 8.36 (d, J = 2.1 Hz, 1H), 8.17–8.04 (m, 1H), 7.72–7.64 (m, 2H), 7.51–7.43 (m, 1H), 7.40–7.27 (m, 5H), 7.26–7.23 (m, 1H), 7.14–6.95 (m, 4H), 6.86 (d, J = 8.5 Hz, 1H), 6.74 (d, J = 8.7 Hz, 2H), 6.59 (dd, J = 9.5, 4.1 Hz, 1H), 6.21 (t, J = 5.6 Hz, 1H), 4.96– 4.85 (m, 1H), 3.94–3.62 (m, 2H), 3.50–3.21 (m, 9H), 3.13–2.97 (m, 2H), 2.92–2.65 (m, 5H), 2.52–1.98 (m, 18H), 1.72–1.39 (m, 11H), 0.98 (s, 6H). LC-MS (ESI): m/z 1356.5 [M+H] +; HPLC: >95% purity.
4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)-N-((4-(((2R)-4-(4-(8-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)octanoyl)piperazin-1-yl)-1-(phenylthio)butan-2-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)benzamide (8e).
Following general method A, compound 8e was obtained from 39e and 18 (2.3 mg, 13% yield). 1H NMR (400 MHz, CDCl3) δ 8.53 (s, 1H), 8.37 (s, 1H), 8.10 (d, J = 9.2 Hz, 1H), 7.63 (d, J = 8.5 Hz, 2H), 7.48 (t, J = 7.8 Hz, 1H), 7.42–7.27 (m, 6H), 7.13–6.95 (m, 4H), 6.87 (d, J = 8.6 Hz, 1H), 6.74 (d, J = 8.6 Hz, 2H), 6.61 (d, J = 9.3 Hz, 1H), 6.21 (t, J = 5.4 Hz, 1H), 4.98–4.84 (m, 1H), 3.95–3.57 (m, 2H), 3.55– 3.20 (m, 9H), 3.14–2.63 (m, 7H), 2.61–1.97 (m, 18H), 1.71–1.33 (m, 13H), 0.98 (s, 6H). LC-MS (ESI): m/z 1370.5 [M+H] +; HPLC: >95% purity.
2-(2-(2-((2-(2,6-Dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethoxy)acetic acid (41a).
Following general methods D and E, compound 41a was obtained from 40a and 38 (67 mg, 19% yield). 1H NMR (600 MHz, CDCl3) δ 9.05 (s, 1H), 7.51–7.43 (m, 1H), 7.08 (d, J = 7.1 Hz, 1H), 6.90 (d, J = 8.6 Hz, 1H), 5.01–4.87 (m, 1H), 4.18 (s, 2H), 3.80–3.65 (m, 6H), 3.53– 3.41 (m, 2H), 2.91–2.82 (m, 1H), 2.81–2.69 (m, 2H), 2.16–2.07 (m, 1H). LC-MS (ESI): m/z 420.2 [M+H] +.
2-(2-(2-(2-((2-(2,6-Dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethoxy)ethoxy)acetic acid (41b).
Following general methods D and E, compound 41b was obtained from 40b and 38 (19 mg, 37% yield). 1H NMR (600 MHz, CDCl3) δ 8.90 (s, 1H), 7.47 (dd, J = 8.5, 7.2 Hz, 1H), 7.08 (d, J = 7.0 Hz, 1H), 6.91 (d, J = 8.5 Hz, 1H), 4.97–4.89 (m, 1H), 4.14 (s, 2H), 3.77–3.65 (m, 10H), 3.49–3.43 (m, 2H), 2.91–2.81 (m, 1H), 2.80–2.69 (m, 2H), 2.15–2.06 (m, 1H). LC-MS (ESI): m/z 464.2 [M+H] +.
14-((2-(2,6-Dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)-3,6,9,12-tetraoxatetradecanoic acid (41c).
Following general methods D and E, compound 41c was obtained from 40c and 38 (14 mg, 15% yield). 1H NMR (600 MHz, CDCl3) δ 8.84 (s, 1H), 7.48 (dd, J = 8.4, 7.2 Hz, 1H), 7.08 (d, J = 7.1 Hz, 1H), 6.91 (d, J = 8.5 Hz, 1H), 4.97–4.89 (m, 1H), 4.13 (s, 2H), 3.74–3.69 (m, 4H), 3.69–3.62 (m, 10H), 3.48–3.44 (m, 2H), 2.91–2.81 (m, 1H), 2.81–2.68 (m, 2H), 2.15–2.04 (m, 1H). LC-MS (ESI): m/z 508.3 [M+H] +.
4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)-N-((4-(((2R)-4-(4-(2-(2-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethoxy)acetyl)piperazin-1-yl)-1-(phenylthio)butan-2-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)benzamide (10a).
Following general method A, compound 10a was obtained from 41a and 18 (6.6 mg, 47% yield). 1H NMR (400 MHz, CDCl3 and CD3OD) δ 9.11 (br s, 1H), 8.36 (s, 1H), 8.08 (d, J = 9.1 Hz, 1H), 7.66 (d, J = 8.2 Hz, 2H), 7.46 (t, J = 7.8 Hz, 1H), 7.39–7.27 (m, 6H), 7.14–7.02 (m, 2H), 6.98 (d, J = 8.3 Hz, 2H), 6.88 (d, J = 8.5 Hz, 1H), 6.75 (d, J = 8.6 Hz, 2H), 6.58 (d, J = 5.8 Hz, 1H), 6.52–6.43 (br s, 1H), 4.91– 4.81 (m, 1H), 4.26–4.15 (m, 2H), 3.94–3.81 (m, 1H), 3.70–3.33 (m, 12H), 3.31–3.22 (m, 4H), 3.12–2.93 (m, 2H), 2.89–2.55 (m, 5H), 2.49–2.00 (m, 16H), 1.74–1.70 (m, 1H), 1.51–1.45 (m, 2H), 0.98 (s, 6H). LC-MS (ESI): m/z 1374.5 [M+H] +; HPLC: >95% purity.
4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)-N-((4-(((2R)-4-(4-(2-(2-(2-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethoxy)ethoxy)acetyl)piperazin-1-yl)-1-(phenylthio)butan-2-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)benzamide (10b).
Following general method A, compound 10b was obtained from 41b and 18 (6.2 mg, 44% yield). 1H NMR (400 MHz, CDCl3 and CD3OD) δ 8.33–8.28 (m, 1H), 8.07 (d, J = 9.0 Hz, 1H), 7.78 (d, J = 8.8 Hz, 2H), 7.54–7.45 (m, 1H), 7.41–7.34 (m, 2H), 7.33–7.20 (m, 5H), 7.09 (d, J = 7.1 Hz, 1H), 7.06–6.98 (m, 3H), 6.94 (d, J = 8.6 Hz, 1H), 6.79 (d, J = 8.9 Hz, 2H), 6.61 (d, J = 9.4 Hz, 1H), 5.00–4.86 (m, 1H), 4.21 (s, 2H), 3.93–3.84 (m, 1H), 3.70–3.39 (m, 16H), 3.32–3.25 (m, 4H), 3.12–3.00 (m, 2H), 2.93–2.71 (m, 5H), 2.46–2.24 (m, 12H), 2.09–2.00 (m, 4H), 1.75–1.63 (m, 1H), 1.47 (t, J = 6.3 Hz, 2H), 0.99 (s, 6H). LC-MS (ESI): m/z 1418.5 [M+H] +; HPLC: >95% purity.
4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)-N-((4-(((2R)-4-(4-(14-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)-3,6,9,12-tetraoxatetradecanoyl)piperazin-1-yl)-1-(phenylthio)butan-2-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)benzamide (10c).
Following general method A, compound 10c was obtained from 41c and 18 (13.6 mg, 76% yield). 1H NMR (400 MHz, CDCl3 and CD3OD) δ 8.81 (br s, 1H), 8.33 (s, 1H), 8.13–7.99 (m, 1H), 7.79–7.60 (m, 2H), 7.45 (t, J = 7.8 Hz, 1H), 7.38–7.26 (m, 5H), 7.24–7.17 (m, 1H), 7.12–6.94 (m, 4H), 6.88 (d, J = 8.6 Hz, 1H), 6.73 (d, J = 8.8 Hz, 2H), 6.61–6.50 (m, 1H), 6.46 (br s, 1H), 4.95–4.84 (m, 1H), 4.16 (s, 2H), 3.88–3.80 (m, 1H), 3.72–3.39 (m, 20H), 3.32–3.25 (m, 4H), 3.12–3.01 (m, 2H), 2.93–2.71 (m, 5H), 2.46–2.24 (m, 12H), 2.09–2.00 (m, 4H), 1.75–1.63 (m, 1H), 1.47 (t, J = 6.3 Hz, 2H), 0.96 (s, 6H). LC-MS (ESI): m/z 1462.5 [M+H] +; HPLC: >95% purity.
(R)-N-((4-((4-(4-Azido-N-methylbutanamido)-1-(phenylthio)butan-2-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)-4-(4-((4’-chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzamide (43a).
Following general method A, compound 43a was obtained from 42a and 19 (24 mg, 65% yield). 1H NMR (400 MHz, CDCl3 and CD3OD) δ 8.49–8.35 (m, 1H), 8.23–8.12 (m, 1H), 7.75 (d, J = 8.5 Hz, 2H), 7.48–7.33 (m, 5H), 7.31–7.25 (m, 1H), 7.08–6.75 (m, 5H), 6.59–6.33 (m, 1H), 3.85–2.82 (m, 16H), 2.56–1.74 (m, 14H), 1.57–1.46 (m, 2H), 1.03 (s, 6H). LC-MS (ESI): m/z 1029.3 [M+H] +.
(R)-N-((4-((4-(7-Azido-N-methylheptanamido)-1-(phenylthio)butan-2-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)-4-(4-((4’-chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzamide (43b).
Following general method A, compound 43b was obtained from 42b and 19 (30 mg, 78% yield). 1H NMR (400 MHz, CDCl3 and CD3OD) δ 8.40–8.29 (m, 1H), 8.11 (dd, J = 9.2, 2.3 Hz, 1H), 7.68 (d, J = 8.7 Hz, 2H), 7.49–7.27 (m, 5H), 7.24–7.18 (m, 1H), 7.09–6.92 (m, 3H), 6.81–6.71 (m, 2H), 6.56– 6.25 (m, 1H), 3.77–2.85 (m, 16H), 2.52–1.15 (m, 22H), 0.96 (s, 6H). LC-MS (ESI): m/z 1071.1 [M+H] +.
4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)-N-((4-(((2R)-4-(4-(4-((2-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethoxy)methyl)-1H-1,2,3-triazol-1-yl)-N-methylbutanamido)-1-(phenylthio)butan-2-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)benzamide (13a).
Following general method F, compound 13a was obtained from 43a and 44a (14.1 mg, 84% yield). 1H NMR (400 MHz, CDCl3) δ 9.26 (s, 1H), 8.35 (s, 1H), 8.08 (d, J = 9.0 Hz, 1H), 7.82–7.27 (m, 7H), 7.26–7.15 (m, 3H), 7.11–6.81 (m, 5H), 6.78–6.36 (m, 4H), 5.00–4.86 (m, 1H), 4.73–4.56 (m, 2H), 4.39–4.17 (m, 2H), 3.85–2.64 (m, 25H), 2.47–1.95 (m, 14H), 1.83–1.66 (m, 1H), 1.51–1.40 (m, 2H), 0.97 (s, 6H). LC-MS (ESI): m/z 1428.5 [M+H] +; HPLC: >95% purity.
4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)-N-((4-(((2R)-4-(4-(4-((2-(2-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethoxy)ethoxy)methyl)-1H-1,2,3-triazol-1-yl)-N-methylbutanamido)-1-(phenylthio)butan-2-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)benzamide (13b).
Following general method F, compound 13b was obtained from 43a and 44b (14.3 mg, 83% yield). 1H NMR (400 MHz, CDCl3) δ 9.19 (s, 1H), 8.47–8.29 (m, 1H), 8.16–7.99 (m, 1H), 7.82–7.27 (m, 8H), 7.26–7.17 (m, 2H), 7.12–6.36 (m, 9H), 5.00–4.81 (m, 1H), 4.70–4.52 (m, 2H), 4.35–4.13 (m, 2H), 3.88–2.67 (m, 29H), 2.46–1.94 (m, 14H), 1.81–1.66 (m, 1H), 1.50–1.41 (m, 2H), 0.97 (s, 6H). LC-MS (ESI): m/z 1472.5 [M+H] +; HPLC: >95% purity.
4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)-N-((4-(((2R)-4-(7-(4-((2-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethoxy)methyl)-1H-1,2,3-triazol-1-yl)-N-methylheptanamido)-1-(phenylthio)butan-2-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)benzamide (14a).
Following general method F, compound 14a was obtained from 43b and 44a (16.8 mg, 82% yield). 1H NMR (400 MHz, CDCl3) δ 8.78 (s, 1H), 8.40–8.32 (m, 1H), 8.15–8.00 (m, 1H), 7.71 (d, J = 8.5 Hz, 2H), 7.60–7.52 (m, 1H), 7.50–7.42 (m, 1H), 7.39–7.27 (m, 4H), 7.26–7.18 (m, 2H), 7.10–7.04 (m, 1H), 7.03–6.86 (m, 4H), 6.80–6.65 (m, 2H), 6.56–6.40 (m, 2H), 4.97– 4.85 (m, 1H), 4.76–4.63 (m, 2H), 4.33–4.14 (m, 2H), 3.78–2.68 (m, 25H), 2.41–1.98 (m, 12H), 1.85–1.67 (m, 3H), 1.53–1.42 (m, 4H), 1.33–1.21 (m, 4H), 0.98 (s, 6H). LC-MS (ESI): m/z 1470.5 [M+H] +.
4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)-N-((4-(((2R)-4-(7-(4-((2-(2-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethoxy)ethoxy)methyl)-1H-1,2,3-triazol-1-yl)-N-methylheptanamido)-1-(phenylthio)butan-2-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)benzamide (14b).
Following general method F, compound 14b was obtained from 43b and 44b (16.4 mg, 77% yield). 1H NMR (400 MHz, CDCl3) δ 9.00–8.85 (m, 1H), 8.41–8.35 (m, 1H), 8.16–8.02 (m, 1H), 7.71 (d, J = 8.6 Hz, 2H), 7.57–7.27 (m, 6H), 7.26–7.19 (m, 2H), 7.11–6.85 (m, 5H), 6.78– 6.68 (m, 2H), 6.54–6.28 (m, 2H), 4.99–4.86 (m, 1H), 4.73–4.59 (m, 2H), 4.34–4.10 (m, 2H), 3.81–2.67 (m, 29H), 2.42–1.97 (m, 12H), 1.87–1.68 (m, 3H), 1.55–1.41 (m, 4H), 1.34–1.10 (m, 4H), 0.98 (s, 6H). LC-MS (ESI): m/z 1514.6 [M+H] +; HPLC: >95% purity.
(R)-N-((4-((4-((7-Azidoheptyl)(methyl)amino)-1-(phenylthio)butan-2-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)-4-(4-((4’-chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)benzamide (46).
Following general method C, compound 46 was obtained from 45 and 19 (11 mg, 47% yield). 1H NMR (400 MHz, CDCl3) δ 8.28 (d, J = 2.2 Hz, 1H), 7.93–7.78 (m, 3H), 7.37–7.27 (m, 3H), 7.26–7.18 (m, 3H), 7.05–6.88 (m, 3H), 6.75 (d, J = 8.6 Hz, 2H), 6.56 (d, J = 9.3 Hz, 1H), 3.96–3.79 (m, 1H), 3.29– 3.15 (m, 6H), 3.11–2.95 (m, 2H), 2.87–2.67 (m, 6H), 2.55 (s, 3H), 2.45–2.31 (m, 4H), 2.30–2.08 (m, 3H), 2.05–1.84 (m, 3H), 1.64–1.39 (m, 6H), 1.36–1.18 (m, 6H), 0.97 (s, 6H). LC-MS (ESI): m/z 1057.5 [M+H] +; HPLC: >95% purity.
4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)-N-((4-(((2R)-4-((7-(4-((2-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethoxy)methyl)-1H-1,2,3-triazol-1-yl)heptyl)(methyl)amino)-1-(phenylthio)butan-2-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)benzamide (15a).
Following general method F, compound 15a was obtained from 46 and 44a (12.4 mg, 90% yield). 1H NMR (400 MHz, CDCl3) δ 8.30 (s, 1H), 7.94 (d, J = 9.0 Hz, 1H), 7.84 (d, J = 8.4 Hz, 2H), 7.44 (t, J = 7.8 Hz, 1H), 7.38–7.30 (m, 2H), 7.28–7.26 (m, 1H), 7.25–7.16 (m, 3H), 7.05 (d, J = 7.1 Hz, 1H), 7.03–6.96 (m, 3H), 6.88 (d, J = 8.6 Hz, 1H), 6.71 (d, J = 8.5 Hz, 2H), 6.58 (d, J = 9.3 Hz, 1H), 6.47 (t, J = 5.7 Hz, 1H), 4.96–4.81 (m, 1H), 4.66 (s, 2H), 4.24 (t, J = 7.2 Hz, 2H), 4.02–3.84 (m, 1H), 3.79–3.60 (m, 6H), 3.46–3.38 (m, 2H), 3.27–3.14 (m, 4H), 3.11– 2.96 (m, 2H), 2.81–1.99 (m, 22H), 1.88–1.70 (m, 3H), 1.51–1.38 (m, 4H), 1.24–1.08 (m, 6H), 0.97 (s, 6H). LC-MS (ESI): m/z 1456.6 [M+H] +; HPLC: >95% purity.
4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)-N-((4-(((2R)-4-((7-(4-((2-(2-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethoxy)ethoxy)methyl)-1H-1,2,3-triazol-1-yl)heptyl)(methyl)amino)-1-(phenylthio)butan-2-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)benzamide (15b).
Following general method F, compound 15b was obtained from 46 and 44b (17.4 mg, 62% yield). 1H NMR (400 MHz, Acetone-d6) δ 10.00 (s, 1H), 8.31 (s, 1H), 8.12–7.97 (m, 2H), 7.93–7.80 (m, 2H), 7.61–7.52 (m, 1H), 7.46–7.31 (m, 4H), 7.31–7.23 (m, 2H), 7.22–6.99 (m, 6H), 6.92–6.81 (m, 2H), 6.66–6.58 (m, 1H), 5.16–5.00 (m, 1H), 4.75–4.51 (m, 2H), 4.46–4.19 (m, 3H), 3.82–3.48 (m, 12H), 3.45–3.22 (m, 6H), 3.11–2.65 (m, 13H), 2.55–2.13 (m, 9H), 1.94– 1.55 (m, 5H), 1.52–1.44 (m, 2H), 1.39–1.15 (m, 6H), 1.00 (s, 6H). LC-MS (ESI): m/z 1500.6 [M+H] +; HPLC: >95% purity.
4-((2-(2-(2-Aminoethoxy)ethoxy)ethyl)amino)-2-(2,6-dioxopiperidin-3-yl)isoindoline-1,3-dione trifluoroacetate (48).
Following general methods D and E, compound 48 was obtained from 47 and 38 (164 mg, 44% yield). 1H NMR (400 MHz, CDCl3) δ 9.63 (br s, 1H), 7.82 (br s, 2H), 7.48 (dd, J = 8.4, 7.2 Hz, 1H), 7.04 (d, J = 7.0 Hz, 1H), 6.87 (d, J = 8.5 Hz, 1H), 5.13–4.82 (m, 1H), 3.85–3.60 (m, 8H), 3.55–3.37 (m, 2H), 3.28–3.12 (m, 2H), 2.81–2.58 (m, 3H), 2.09– 1.88 (m, 1H). LC-MS (ESI): m/z 405.1 [M+H] +.
4-((R)-3-(4-(N-(4-(4-((2-(4-Chlorophenyl)-5,5-dimethylcyclohex-1-enyl)methyl)piperazin-1-yl)benzoyl)sulfamoyl)-2-(trifluoromethylsulfonyl)phenylamino)-4-(phenylthio)butyl)-N-(2-(2-(2-(2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-ylamino)ethoxy)ethoxy)ethyl)piperazine-1-carboxamide (11a).
A mixture of compound 48 (20 mg, 0.039 mmol), CDI (10 mg, 0.062 mmol) and TEA (7.0 μL, 0.050 mmol) in 3 mL DCM was stirred at room temperature for 2 h. Then compound 18 (15 mg, 0.015 mmol) and DIPEA (50 μL, 0.303 mmol) were added into the above solution. The reaction was stirred overnight and quenched by the addition of NH4Cl (aq.). Subsequently, it was poured into water and extracted with DCM. The organic phase was washed with water × 1, brine × 1, dried over anhydrous Na2SO4, filtered and evaporated to dryness. The crude product was purified by column chromatography using DCM and MeOH as eluents to afford the title compound (6.7 mg, 32% yield). 1H NMR (400 MHz, CDCl3) δ 9.24 (br s, 1H), 8.35 (s, 1H), 8.14–7.98 (m, 1H), 7.81–7.62 (m, 2H), 7.52–7.40 (m, 1H), 7.39–7.27 (m, 4H), 7.24–7.15 (m, 1H), 7.12–6.94 (m, 4H), 6.87 (d, J = 8.6 Hz, 1H), 6.73 (d, J = 7.2 Hz, 2H), 6.66–6.57 (m, 1H), 6.56–6.46 (m, 1H), 5.20–5.02 (br s, 1H), 5.00–4.83 (m, 1H), 3.95–3.81 (m, 1H), 3.75–3.69 (m, 2H), 3.67–3.61 (m, 4H), 3.61–3.53 (m, 2H), 3.49–3.38 (m, 4H), 3.38–3.18 (m, 8H), 3.12–2.95 (m, 2H), 2.88–2.66 (m, 5H), 2.47– 2.18 (m, 12H), 2.17–1.98 (m, 4H), 1.69–1.57 (m, 1H), 1.46 (t, J = 6.3 Hz, 2H), 0.97 (s, 6H). LC- MS (ESI): m/z 1403.5 [M+H] +; HPLC: >95% purity.
4-(4-((2-(4-Chlorophenyl)-5,5-dimethylcyclohex-1-enyl)methyl)piperazin-1-yl)-N-(4-((2R)-4-(4-(2-(2-(2-(2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-ylamino)ethoxy)ethoxy)ethylcarbamothioyl)piperazin-1-yl)-1-(phenylthio)butan-2-ylamino)-3-(trifluoromethylsulfonyl)phenylsulfonyl)benzamide (11b).
A mixture of compound 48 (12 mg, 0.023 mmol), 1,1’-thiocarbonyldiimidazole (6.0 mg, 0.034 mmol) and TEA (4.2 μL, 0.030 mmol) in 2 mL DCM was stirred at room temperature for 1 h. Then compound 18 (6.5 mg, 0.0067 mmol) and DIPEA (0.05 mL) were added into the above solution. The reaction was stirred overnight and quenched by the addition of NH4Cl (aq.). Subsequently, it was poured into water and extracted with DCM. The organic phase was washed with water x1, brine x1, dried over anhydrous Na2SO4, filtered and evaporated to dryness. The crude product was purified by column chromatography using DCM and MeOH as eluents to afford the title compound (6.4 mg, 68% yield). 1H NMR (400 MHz, CDCl3) δ 8.90 (br s, 1H), 8.37 (d, J = 2.1 Hz, 1H), 8.15–8.00 (m, 1H), 7.65 (d, J = 7.6 Hz, 2H), 7.51–7.42 (m, 1H), 7.38–7.27 (m, 5H), 7.12–7.05 (m, 2H), 6.99 (d, J = 8.4 Hz, 2H), 6.87 (d, J = 8.5 Hz, 1H), 6.75 (dd, J = 9.0, 3.8 Hz, 2H), 6.62 (dd, J = 12.9, 9.5 Hz, 1H), 6.51 (t, J = 5.2 Hz, 1H), 6.22 (br s, 1H), 4.95–4.80 (m, 1H), 3.90–3.61 (m, 15H), 3.48–3.41 (m, 2H), 3.30–3.20 (m, 4H), 3.13–2.96 (m, 2H), 2.88–2.71 (m, 5H), 2.47–2.22 (m, 12H), 2.13–2.00 (m, 4H), 1.75–1.70 (m, 1H), 1.46 (t, J = 6.4 Hz, 2H), 0.99 (s, 6H). LC-MS (ESI): m/z 1419.4 [M+H] +; HPLC: >95% purity.
2-(2,6-Dioxopiperidin-3-yl)-4-((3-hydroxypropyl)amino)isoindoline-1,3-dione (50a).
Following general method D, compound 50a was obtained from 49a and 38 (19 mg, 11% yield). 1H NMR (600 MHz, CDCl3) δ 8.18 (s, 1H), 7.52 (dd, J = 8.5, 7.1 Hz, 1H), 7.12 (d, J = 7.0 Hz, 1H), 6.96 (d, J = 8.5 Hz, 1H), 6.52–6.45 (m, 1H), 4.94 (dd, J = 12.4, 5.3 Hz, 1H), 3.84 (t, J = 5.8 Hz, 2H), 3.46 (q, J = 6.4 Hz, 2H), 2.95–2.69 (m, 3H), 2.18–2.12 (m, 1H), 1.94 (p, J = 6.3 Hz, 2H). LC-MS (ESI): m/z 332.1 [M+H] +.
2-(2,6-Dioxopiperidin-3-yl)-4-((4-hydroxybutyl)amino)isoindoline-1,3-dione (50b).
Following general method D, compound 50b was obtained from 49b and 38 (59 mg, 24% yield). 1H NMR (600 MHz, CDCl3) δ 7.97 (s, 1H), 7.52 (dd, J = 8.5, 7.2 Hz, 1H), 7.12 (d, J = 7.0 Hz, 1H), 6.92 (d, J = 8.5 Hz, 1H), 6.36–6.29 (m, 1H), 4.94 (dd, J = 12.4, 5.4 Hz, 1H), 3.75 (q, J = 5.8 Hz, 2H), 3.35 (q, J = 6.8 Hz, 2H), 2.97–2.72 (m, 3H), 2.19–2.13 (m, 1H), 1.84–1.78 (m, 2H), 1.75–1.69 (m, 2H), 1.37 (t, J = 5.1 Hz, 1H). LC-MS (ESI): m/z 346.1 [M+H] +.
2-(2,6-Dioxopiperidin-3-yl)-4-((5-hydroxypentyl)amino)isoindoline-1,3-dione (50c).
Following general method D, compound 50c was obtained from 49c and 38 (34 mg, 17% yield). 1H NMR (600 MHz, CDCl3) δ 7.96 (s, 1H), 7.52 (dd, J = 8.5, 7.1 Hz, 1H), 7.12 (d, J = 7.1 Hz, 1H), 6.91 (d, J = 8.5 Hz, 1H), 6.31–6.22 (m, 1H), 4.94 (dd, J = 12.4, 5.4 Hz, 1H), 3.76–3.67 (m, 2H), 3.37–3.28 (m, 2H), 2.96–2.71 (m, 3H), 2.20–2.12 (m, 1H), 1.78–1.70 (m, 2H), 1.69–1.62 (m, 2H), 1.56–1.51 (m, 2H). LC-MS (ESI): m/z 360.1 [M+H] +.
2-(2,6-Dioxopiperidin-3-yl)-4-((6-hydroxyhexyl)amino)isoindoline-1,3-dione (50d).
Following general method D, compound 50d was obtained from 49d and 38 (56 mg, 21% yield). 1H NMR (600 MHz, CDCl3) δ 8.01 (s, 1H), 7.52 (dd, J = 8.6, 7.1 Hz, 1H), 7.11 (d, J = 7.1 Hz, 1H), 6.91 (d, J = 8.5 Hz, 1H), 6.29–6.22 (m, 1H), 4.94 (dd, J = 12.4, 5.3 Hz, 1H), 3.72–3.65 (m, 2H), 3.33–3.27 (m, 2H), 2.92–2.72 (m, 3H), 2.18–2.12 (m, 1H), 1.76–1.68 (m, 2H), 1.65–1.60 (m, 2H), 1.52–1.42 (m, 4H). LC-MS (ESI): m/z 374.1 [M+H] +.
2-(2,6-Dioxopiperidin-3-yl)-4-((8-hydroxyoctyl)amino)isoindoline-1,3-dione (50e).
Following general method D, compound 50e was obtained from 49e and 38 (79 mg, 27% yield). 1H NMR (600 MHz, CDCl3) δ 8.06 (s, 1H), 7.57–7.48 (m, 1H), 7.14–7.09 (m, 1H), 6.90 (d, J = 8.5 Hz, 1H), 6.30–6.21 (m, 1H), 4.93 (dd, J = 12.4, 5.3 Hz, 1H), 3.67 (t, J = 6.6 Hz, 2H), 3.32– 3.24 (m, 2H), 2.94–2.71 (m, 3H), 2.19–2.13 (m, 1H), 1.72–1.66 (m, 2H), 1.59–1.36 (m, 10H). LC-MS (ESI): m/z 402.1 [M+H] +.
4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)-N-((4-(((2R)-4-(4-(3-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)propyl)piperazin-1-yl)-1-(phenylthio)butan-2-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)benzamide (9a).
Following general methods G and H, compound 9a was obtained from 50a and 18 (17.2 mg, 65% yield). 1H NMR (600 MHz, CDCl3) δ 8.72 (br s, 1H), 8.35 (s, 1H), 8.09–7.97 (m, 1H), 7.80 (d, J = 8.4 Hz, 2H), 7.51–7.43 (m, 1H), 7.40–7.34 (m, 2H), 7.33–7.29 (m, 2H), 7.28–7.19 (m, 2H), 7.09 (d, J = 7.1 Hz, 1H), 7.01 (d, J = 8.1 Hz, 2H), 6.96–6.85 (m, 2H), 6.78–6.68 (m, 2H), 6.60 (d, J = 9.2 Hz, 1H), 6.48– 6.35 (m, 1H), 4.98–4.90 (m, 1H), 3.88 (s, 1H), 3.34–3.18 (m, 6H), 3.13–3.06 (m, 1H), 3.00 (dd, J = 14.0, 6.9 Hz, 1H), 2.93–2.23 (m, 23H), 2.16–2.08 (m, 2H), 2.04–1.99 (m, 2H), 1.92–1.81 (m, 2H), 1.77–1.68 (m, 1H), 1.47 (t, J = 6.5 Hz, 2H), 0.98 (d, J = 4.1 Hz, 6H). LC-MS (ESI): m/z 1286.5 [M+H] +; HPLC: >95% purity.
4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)-N-((4-(((2R)-4-(4-(4-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)butyl)piperazin-1-yl)-1-(phenylthio)butan-2-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)benzamide (9b).
Following general methods G and H, compound 9b was obtained from 50b and 18 (3.5 mg, 19% yield). 1H NMR (600 MHz, CDCl3) δ 8.55 (br s, 1H), 8.37 (dd, J = 7.6, 2.2 Hz, 1H), 8.04 (t, J = 7.7 Hz, 1H), 7.77 (d, J = 8.4 Hz, 2H), 7.54–7.46 (m, 1H), 7.43–7.36 (m, 2H), 7.34–7.29 (m, 3H), 7.28–7.24 (m, 1H), 7.10 (d, J = 7.6 Hz, 1H), 7.05–6.97 (m, 3H), 6.89 (d, J = 8.5 Hz, 1H), 6.82–6.72 (m, 2H), 6.59 (t, J = 7.8 Hz, 1H), 6.29–6.20 (m, 1H), 4.97–4.90 (m, 1H), 3.94–3.83 (m, 1H), 3.34–3.21 (m, 6H), 3.10 (dd, J = 13.8, 5.0 Hz, 1H), 3.02–2.96 (m, 1H), 2.91–2.25 (m, 21H), 2.19–2.00 (m, 6H), 1.79–1.66 (m, 5H), 1.47 (t, J = 6.5 Hz, 2H), 0.99 (d, J = 5.0 Hz, 6H). LC-MS (ESI): m/z 1300.5 [M+H] +; HPLC: >95% purity.
4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)-N-((4-(((2R)-4-(4-(5-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)pentyl)piperazin-1-yl)-1-(phenylthio)butan-2-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)benzamide (9c).
Following general methods G and H, compound 9c was obtained from 50c and 18 (2.9 mg, 11% yield). 1H NMR (600 MHz, CDCl3) δ 8.77 (br s, 1H), 8.37–8.33 (m, 1H), 8.02 (d, J = 9.2 Hz, 1H), 7.83 (d, J = 8.6 Hz, 2H), 7.51–7.45 (m, 1H), 7.40–7.36 (m, 2H), 7.34–7.29 (m, 3H), 7.28–7.24 (m, 1H), 7.09 (d, J = 7.0 Hz, 1H), 7.03–6.94 (m, 3H), 6.86 (d, J = 8.6 Hz, 1H), 6.77 (d, J = 8.8 Hz, 2H), 6.60–6.45 (m, 1H), 6.20 (t, J = 5.6 Hz, 1H), 4.97–4.87 (m, 1H), 3.84–3.76 (m, 1H), 3.30–3.20 (m, 6H), 3.09 (dd, J = 13.8, 4.7 Hz, 1H), 2.99–2.93 (m, 1H), 2.91–2.66 (m, 9H), 2.58–2.00 (m, 18H), 1.74– 1.59 (m, 5H), 1.49–1.36 (m, 4H), 0.98 (d, J = 5.9 Hz, 6H). LC-MS (ESI): m/z 1314.5 [M+H] +; HPLC: >95% purity.
4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)-N-((4-(((2R)-4-((3-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)propyl)(methyl)amino)-1-(phenylthio)butan-2-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)benzamide (16a).
Following general methods G and H, compound 16a was obtained from 50a and 19 (9.0 mg, 33% yield). 1H NMR (600 MHz, CDCl3) δ 8.73–8.33 (m, 2H), 8.28–8.04 (m, 1H), 7.70–7.56 (m, 2H), 7.43–7.29 (m, 6H), 7.26–7.20 (m, 1H), 7.15–6.85 (m, 4H), 6.77–5.55 (m, 5H), 5.02–4.84 (m, 1H), 4.09–3.91 (m, 1H), 3.30–3.07 (m, 6H), 3.06–2.69 (m, 7H), 2.60–2.10 (m, 15H), 2.05–2.00 (m, 2H), 1.77–1.54 (m, 3H), 1.50–1.47 (m, 2H), 1.01 (d, J = 4.9 Hz, 6H). LC-MS (ESI): m/z 1231.4 [M+H] +; HPLC: >95% purity.
4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)-N-((4-(((2R)-4-((4-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)butyl)(methyl)amino)-1-(phenylthio)butan-2-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)benzamide (16b).
Following general methods G and H, compound 16b was obtained from 50b and 19 (3.0 mg, 16% yield). 1H NMR (600 MHz, CDCl3) δ 8.53–8.35 (m, 1H), 8.11 (s, 1H), 7.72–7.60 (m, 2H), 7.44–7.29 (m, 7H), 7.27–7.12 (m, 2H), 7.06–6.98 (m, 3H), 6.81–6.52 (m, 4H), 5.92–5.73 (m, 1H), 5.03–4.95 (m, 1H), 4.06–3.87 (m, 1H), 3.24–2.71 (m, 13H), 2.61–2.12 (m, 15H), 2.05–2.01 (m, 2H), 1.73–1.43 (m, 7H), 1.01 (d, J = 2.3 Hz, 6H). LC-MS (ESI): m/z 1245.5 [M+H] +; HPLC: >95% purity.
4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)-N-((4-(((2R)-4-((5-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)pentyl)(methyl)amino)-1-(phenylthio)butan-2-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)benzamide (16c).
Following general methods G and H, compound 16c was obtained from 50c and 19 (3.0 mg, 11% yield). 1H NMR (600 MHz, CDCl3) δ 8.38 (s, 1H), 8.18–8.01 (m, 1H), 7.71–7.62 (m, 2H), 7.47–7.29 (m, 7H), 7.27–7.19 (m, 2H), 7.07–6.96 (m, 3H), 6.82–5.60 (m, 5H), 4.98–4.90 (m, 1H), 4.06–3.90 (m, 1H), 3.25–2.69 (m, 13H), 2.59–2.11 (m, 15H), 2.03 (s, 2H), 1.61–1.47 (m, 9H), 1.01 (d, J = 3.7 Hz, 6H). LC-MS (ESI): m/z 1259.5 [M+H] +; HPLC: >95% purity.
4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)-N-((4-(((2R)-4-((6-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)hexyl)(methyl)amino)-1-(phenylthio)butan-2-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)benzamide (16d).
Following general methods G and H, compound 16d was obtained from 50d and 19 (5.9 mg, 21% yield). 1H NMR (600 MHz, CDCl3) δ 8.58–8.39 (m, 1H), 8.36–8.33 (m, 1H), 8.02–7.91 (m, 1H), 7.81–7.70 (m, 2H), 7.48–7.36 (m, 3H), 7.33–7.29 (m, 3H), 7.27–7.21 (m, 1H), 7.18–6.97 (m, 4H), 6.88–6.51 (m, 4H), 6.21–5.94 (m, 1H), 4.98–4.88 (m, 1H), 4.00–3.86 (m, 1H), 3.27–2.98 (m, 8H), 2.90–2.70 (m, 5H), 2.57–2.11 (m, 15H), 2.06–2.01 (m, 2H), 1.88–1.78 (m, 1H), 1.52–1.26 (m, 10H), 1.00 (s, 6H). LC-MS (ESI): m/z 1273.5 [M+H] +; HPLC: >95% purity.
4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)-N-((4-(((2R)-4-((8-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)octyl)(methyl)amino)-1-(phenylthio)butan-2-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)benzamide (16e).
Following general methods G and H, compound 16e was obtained from 50e and 19 (8.5 mg, 32% yield). 1H NMR (600 MHz, CDCl3) δ 8.31 (s, 1H), 8.01–7.88 (m, 1H), 7.79 (d, J = 8.3 Hz, 2H), 7.52–7.43 (m, 1H), 7.42–7.34 (m, 2H), 7.33–7.29 (m, 2H), 7.28–7.23 (m, 3H), 7.08 (d, J = 7.1 Hz, 2H), 7.04–6.98 (m, 2H), 6.90–6.82 (m, 1H), 6.78–6.70 (m, 2H), 6.58 (d, J = 9.2 Hz, 1H), 6.25–6.14 (m, 1H), 4.97– 4.88 (m, 1H), 3.98–3.85 (m, 1H), 3.23–2.68 (m, 13H), 2.62–2.13 (m, 15H), 2.03 (s, 2H), 1.89– 1.86 (m, 1H), 1.62–1.57 (m, 2H), 1.48 (t, J = 6.5 Hz, 2H), 1.37–1.24 (m, 10H), 1.00 (s, 6H). LC- MS (ESI): m/z 1301.5 [M+H] +; HPLC: >95% purity.
2-(2,6-Dioxopiperidin-3-yl)-4-((2-(2-hydroxyethoxy)ethyl)amino)isoindoline-1,3-dione (53a).
Following general methods D, compound 53a was obtained from 52a and 38 (83 mg, 40% yield). 1H NMR (400 MHz, CDCl3) δ 8.25 (br s, 1H), 7.58–7.46 (m, 1H), 7.11 (d, J = 7.1 Hz, 1H), 6.91 (d, J = 8.5 Hz, 1H), 6.60–6.52 (m, 1H), 4.92 (dd, J = 12.2, 5.3 Hz, 1H), 3.81–3.71 (m, 4H), 3.66–3.61 (m, 2H), 3.48 (dd, J = 10.7, 5.3 Hz, 2H), 2.92–2.67 (m, 3H), 2.32 (br s, 1H), 2.18–2.07 (m, 1H). LC-MS (ESI): m/z 362.1 [M+H] +.
2-(2,6-Dioxopiperidin-3-yl)-4-((2-(2-(2-hydroxyethoxy)ethoxy)ethyl)amino)isoindoline- 1,3-dione (53b).
Following general methods D, compound 53b was obtained from 52b and 38 (230 mg, 55% yield). 1H NMR (400 MHz, CDCl3) δ 8.19 (br s, 1H), 7.55–7.44 (m, 1H), 7.10 (d, J = 7.1 Hz, 1H), 6.91 (d, J = 8.5 Hz, 1H), 6.62–6.54 (m, 1H), 4.91 (dd, J = 12.0, 5.4 Hz, 1H), 3.85–3.65 (m, 8H), 3.64–3.59 (m, 2H), 3.51–3.43 (m, 2H), 2.92–2.68 (m, 3H), 2.57 (br s, 1H), 2.18–2.07 (m, 1H). LC-MS (ESI): m/z 406.0 [M+H] +.
2-(2,6-Dioxopiperidin-3-yl)-4-((2-(2-(2-(2-hydroxyethoxy)ethoxy)ethoxy)ethyl)amino)isoindoline-1,3-dione (53c).
Following general methods D, compound 53c was obtained from 52c and 38 (84 mg, 40% yield). 1H NMR (400 MHz, CDCl3) δ 8.23 (br s, 1H), 7.58–7.40 (m, 1H), 7.10 (d, J = 7.1 Hz, 1H), 6.92 (d, J = 8.6 Hz, 1H), 6.56–6.48 (m, 1H), 4.92 (dd, J = 12.0, 5.4 Hz, 1H), 3.77–3.65 (m, 12H), 3.63–3.58 (m, 2H), 3.52–3.44 (m, 2H), 3.00–2.59 (m, 4H), 2.24–2.04 (m, 1H). LC-MS (ESI): m/z 450.1 [M+H] +.
2-(2,6-Dioxopiperidin-3-yl)-4-((14-hydroxy-3,6,9,12-tetraoxatetradecyl)amino)isoindoline-1,3-dione (53d).
Following general methods D, compound 53d was obtained from 52d and 38 (92 mg, 19% yield). 1H NMR (400 MHz, CDCl3) δ 8.53 (br s, 1H), 7.49 (dd, J = 8.5, 7.1 Hz, 1H), 7.11 (d, J = 6.8 Hz, 1H), 6.93 (d, J = 8.5 Hz, 1H), 6.55–6.47 (m, 1H), 4.90 (dd, J = 12.0, 5.4 Hz, 1H), 3.76–3.58 (m, 18H), 3.50–3.43 (m, 2H), 2.92–2.74 (m, 3H), 2.18–2.07 (m, 1H). LC-MS (ESI): m/z 494.3 [M+H] +.
2-(2-((2-(2,6-Dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethyl methanesulfonate (54a).
Following general methods I, compound 54a was obtained from 53a (49 mg, 83% yield). 1H NMR (400 MHz, CDCl3) δ 8.10 (br s, 1H), 7.63–7.44 (m, 1H), 7.12 (d, J = 7.1 Hz, 1H), 6.93 (d, J = 8.5 Hz, 1H), 6.54–6.45 (m, 1H), 4.91 (dd, J = 12.1, 5.3 Hz, 1H), 4.48–4.35 (m, 2H), 3.86–3.66 (m, 4H), 3.58–3.41 (m, 2H), 3.13–2.69 (m, 6H), 2.23–2.05 (m, 1H). LC-MS (ESI): m/z 440.2 [M+H] +.
2-(2-(2-((2-(2,6-Dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethoxy)ethyl methanesulfonate (54b).
Following general methods I, compound 54b was obtained from 53b (20 mg, 83% yield). 1H NMR (400 MHz, CDCl3) δ 8.14 (br s, 1H), 7.65–7.45 (m, 1H), 7.12 (d, J = 7.1 Hz, 1H), 6.91 (d, J = 8.5 Hz, 1H), 6.56–6.48 (m, 1H), 4.94 (dd, J = 12.0, 5.3 Hz, 1H), 4.39 (dd, J = 5.3, 3.7 Hz, 2H), 4.00–3.66 (m, 8H), 3.52–3.44 (m, 2H), 3.05 (s, 3H), 2.93–2.62 (m, 3H), 2.28–2.06 (m, 1H). LC-MS (ESI): m/z 484.2 [M+H] +.
2-(2-(2-(2-((2-(2,6-Dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethoxy)ethoxy)ethyl methanesulfonate (54c).
Following general methods I, compound 54c was obtained from 53c (64 mg, 91% yield). 1H NMR (400 MHz, CDCl3) δ 8.20 (br s, 1H), 7.60–7.41 (m, 1H), 7.10 (d, J = 7.1 Hz, 1H), 6.92 (d, J = 8.5 Hz, 1H), 6.52–6.44 (m, 1H), 4.92 (dd, J = 11.8, 5.4 Hz, 1H), 4.36 (dd, J = 5.3, 3.7 Hz, 2H), 3.82–3.60 (m, 12H), 3.54– 3.40 (m, 2H), 3.07 (s, 3H), 2.98–2.65 (m, 3H), 2.24–2.06 (m, 1H). LC-MS (ESI): m/z 528.2 [M+H] +.
4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)-N-((4-(((2R)-4-(4-(2-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethyl)piperazin-1-yl)-1-(phenylthio)butan-2-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)benzamide (12a).
Following general method J, compound 12a was obtained from 54a and 18 (8.8 mg, 26% yield). 1H NMR (400 MHz, CDCl3) δ 8.35 (s, 1H), 8.09–7.98 (m, 1H), 7.72 (d, J = 8.7 Hz, 2H), 7.48 (t, J = 7.9 Hz, 1H), 7.40–7.29 (m, 5H), 7.25–7.20 (m, 1H), 7.10 (d, J = 7.1 Hz, 1H), 7.06–6.95 (m, 3H), 6.89 (d, J = 8.4 Hz, 1H), 6.73 (d, J = 9.1 Hz, 2H), 6.66–6.55 (m, 1H), 6.53–6.42 (m, 1H), 4.98–4.82 (m, 1H), 3.93– 3.80 (m, 1H), 3.76–3.40 (m, 6H), 3.32–2.64 (m, 17H), 2.43–1.97 (m, 16H), 1.70–1.66 (m, 1H), 1.52–1.41 (m, 2H), 1.01–0.95 (m, 6H). LC-MS (ESI): m/z 1316.5 [M+H] +; HPLC: >95% purity.
4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)-N-((4-(((2R)-4-(4-(2-(2-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethoxy)ethyl)piperazin-1-yl)-1-(phenylthio)butan-2-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)benzamide (12b).
Following general method J, compound 12b was obtained from 54b and 18 (5.8 mg, 42% yield). 1H NMR (400 MHz, CDCl3) δ 8.33 (s, 1H), 8.02 (t, J = 8.9 Hz, 1H), 7.76 (d, J = 7.0 Hz, 2H), 7.52–7.46 (m, 1H), 7.41–7.33 (m, 2H), 7.32–7.27 (m, 3H), 7.25–7.21 (m, 1H), 7.10 (dd, J = 7.1, 2.3 Hz, 1H), 7.04–6.92 (m, 3H), 6.88 (d, J = 8.6 Hz, 1H), 6.75 (d, J = 8.4 Hz, 2H), 6.56–6.40 (m, 2H), 4.96–4.73 (m, 1H), 3.86–3.40 (m, 11H), 3.33–2.51 (m, 17H), 2.50–1.79 (m, 16H), 1.74–1.60 (m, 1H), 1.48–1.37 (m, 2H), 0.95 (d, J = 5.4 Hz, 6H). LC-MS (ESI): m/z 1360.5 [M+H] +; HPLC: >95% purity.
4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)-N-((4-(((2R)-4-(4-(2-(2-(2-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethoxy)ethoxy)ethyl)piperazin-1-yl)-1-(phenylthio)butan-2-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)benzamide (12c).
Following general method J, compound 12c was obtained from 54c and 18 (16.4 mg, 25% yield). 1H NMR (400 MHz, CDCl3) δ 8.31 (s, 1H), 8.01 (d, J = 8.9 Hz, 1H), 7.81 (d, J = 8.5 Hz, 2H), 7.47 (t, J = 7.8 Hz, 1H), 7.38– 7.28 (m, 5H), 7.25–7.21 (m, 1H), 7.08 (d, J = 7.1 Hz, 1H), 6.99 (d, J = 8.3 Hz, 2H), 6.96–6.85 (m, 2H), 6.75 (d, J = 8.7 Hz, 2H), 6.54–6.43 (m, 2H), 4.96–4.83 (m, 1H), 3.90–3.39 (m, 15H), 3.28–2.68 (m, 17H), 2.51–1.95 (m, 16H), 1.61–1.57 (m, 1H), 1.47–1.41 (m, 2H), 0.97–0.93 (m, 6H). LC-MS (ESI): m/z 1404.5 [M+H] +; HPLC: >95% purity.
4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)-N-((4-(((2R)-4-((2-(2-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethoxy)ethyl)(methyl)amino)-1-(phenylthio)butan-2-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)benzamide (17a).
Following general methods G and H, compound 17a was obtained from 53b and 19 (16.2 mg, 60% yield). 1H NMR (400 MHz, CDCl3) δ 8.31 (s, 1H), 8.07–7.93 (m, 1H), 7.77 (d, J = 8.5 Hz, 2H), 7.48–7.39 (m, 1H), 7.33–7.27 (m, 3H), 7.25–7.14 (m, 3H), 7.09–6.95 (m, 4H), 6.84 (dd, J = 8.6, 3.2 Hz, 1H), 6.74–6.56 (m, 3H), 6.46–6.36 (m, 1H), 4.98–4.87 (m, 1H), 3.99–3.88 (m, 1H), 3.73–3.52 (m, 8H), 3.41– 3.33 (m, 2H), 3.29–3.18 (m, 4H), 3.10–2.69 (m, 11H), 2.60–1.99 (m, 13H), 1.95–1.85 (m, 1H), 1.45 (t, J = 6.5 Hz, 2H), 0.97 (s, 6H). LC-MS (ESI): m/z 1305.5 [M+H] +; HPLC: >95% purity.
4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)-N-((4-(((15R)-1-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)-12-methyl-16-(phenylthio)-3,6,9-trioxa-12-azahexadecan-15-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)benzamide (17b).
Following general methods G and H, compound 17b was obtained from 53c and 19 (14.6 mg, 53% yield). 1H NMR (400 MHz, CDCl3) δ 8.37–8.26 (m, 1H), 8.06–7.95 (m, 1H), 7.77 (d, J = 8.7 Hz, 2H), 7.50–7.39 (m, 1H), 7.38–7.31 (m, 2H), 7.29–7.27 (m, 1H), 7.26–7.17 (m, 3H), 7.16–7.04 (m, 2H), 7.02–6.95 (m, 2H), 6.87 (d, J = 8.5 Hz, 1H), 6.72 (d, J = 8.9 Hz, 2H), 6.61 (d, J = 9.4 Hz, 1H), 6.48–6.40 (m, 1H), 4.94–4.82 (m, 1H), 3.94–3.82 (m, 1H), 3.75–3.37 (m, 14H), 3.29–3.18 (m, 4H), 3.10–2.97 (m, 2H), 2.92–2.60 (m, 9H), 2.44–2.20 (m, 9H), 2.11–1.97 (m, 4H), 1.88–1.78 (m, 1H), 1.45 (t, J = 6.5 Hz, 2H), 0.97 (s, 6H). LC-MS (ESI): m/z 1349.5 [M+H] +; HPLC: >95% purity.
4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)-N-((4-(((18R)-1-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)-15-methyl-19-(phenylthio)-3,6,9,12-tetraoxa-15-azanonadecan-18-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)benzamide (17c).
Following general methods G and H, compound 17c was obtained from 53d and 19 (12.1 mg, 42% yield). 1H NMR (400 MHz, CDCl3) δ 8.40–8.33 (m, 1H), 8.12–8.04 (m, 1H), 7.76–7.68 (m, 2H), 7.51–7.42 (m, 1H), 7.39–7.33 (m, 2H), 7.31–7.27 (m, 2H), 7.26–7.20 (m, 3H), 7.12–7.06 (m, 1H), 7.03–6.94 (m, 2H), 6.87 (d, J = 8.6 Hz, 1H), 6.78–6.67 (m, 3H), 6.45 (t, J = 5.6 Hz, 1H), 4.96–4.80 (m, 1H), 4.02– 3.92 (m, 1H), 3.72–3.40 (m, 18H), 3.31–3.19 (m, 4H), 3.14–3.02 (m, 2H), 2.91–2.17 (m, 18H), 2.13–1.99 (m, 4H), 1.80–1.69 (m, 1H), 1.46 (t, J = 6.5 Hz, 2H), 0.98 (s, 6H). LC-MS (ESI): m/z 1393.5 [M+H] +; HPLC: >95% purity.
4-((2-(2-(2-(2-Hydroxyethoxy)ethoxy)ethoxy)ethyl)amino)-2-(1-methyl-2,6-dioxopiperidin-3-yl)isoindoline-1,3-dione (57).
Following general method D, compound 57 was obtained from 56 and 52c (80 mg, 34% yield). 1H NMR (600 MHz, CDCl3) δ 7.51 (dd, J = 8.5, 7.1 Hz, 1H), 7.12 (d, J = 7.1 Hz, 1H), 6.95 (d, J = 8.5 Hz, 1H), 6.55–6.49 (m, 1H), 4.97– 4.89 (m, 1H), 3.76–3.69 (m, 12H), 3.64–3.62 (m, 2H), 3.51 (q, J = 5.6 Hz, 2H), 3.23 (s, 3H), 3.05–2.95 (m, 1H), 2.84–2.72 (m, 2H), 2.17–2.05 (m, 1H). LC-MS (ESI): m/z 464.2 [M+H] +.
2-(2-(2-(2-((2-(1-Methyl-2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethoxy)ethoxy)ethyl methanesulfonate (58).
Following general method I, compound 58 was obtained from 57 (75.3 mg, 81% yield). 1H NMR (600 MHz, CDCl3) δ 7.52 (dd, J = 8.5, 7.1 Hz, 1H), 7.13 (d, J = 7.1 Hz, 1H), 6.95 (d, J = 8.5 Hz, 1H), 6.55–6.45 (m, 1H), 4.98–4.89 (m, 1H), 4.42–4.34 (m, 2H), 3.81–3.77 (m, 2H), 3.74 (t, J = 5.4 Hz, 2H), 3.72–3.66 (m, 8H), 3.50 (q, J = 5.4 Hz, 2H), 3.23 (s, 3H), 3.08 (s, 3H), 3.03–2.95 (m, 1H), 2.85–2.72 (m, 2H), 2.17–2.09 (m, 1H). LC-MS (ESI): m/z 542.1 [M+H] +.
4-(4-((4’-Chloro-4,4-dimethyl-3,4,5,6-tetrahydro-[1,1’-biphenyl]-2-yl)methyl)piperazin-1-yl)-N-((4-(((15R)-12-methyl-1-((2-(1-methyl-2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)-16-(phenylthio)-3,6,9-trioxa-12-azahexadecan-15-yl)amino)-3-((trifluoromethyl)sulfonyl)phenyl)sulfonyl)benzamide (XZ739-NC).
Following general method J, compound XZ739-NC was obtained from 58 and 19 (4.9 mg, 11% yield). 1H NMR (600 MHz, CDCl3) δ 8.37 (d, J = 2.3 Hz, 1H), 8.12–8.05 (m, 1H), 7.70 (d, J = 8.7 Hz, 2H), 7.51–7.45 (m, 1H), 7.42–7.29 (m, 6H), 7.27–7.22 (m, 1H), 7.09 (d, J = 7.0 Hz, 1H), 7.03–6.97 (m, 2H), 6.91 (d, J = 8.5 Hz, 1H), 6.77 (d, J = 8.8 Hz, 2H), 6.68 (dd, J = 9.4, 3.9 Hz, 1H), 6.49–6.42 (m, 1H), 4.97–4.88 (m, 1H), 4.02–3.92 (m, 1H), 3.74–3.43 (m, 14H), 3.31–3.24 (m, 4H), 3.21 (s, 3H), 3.13–3.05 (m, 2H), 3.00–2.47 (m, 9H), 2.39–2.26 (m, 9H), 2.12–2.02 (m, 4H), 1.81–1.76 (m, 1H), 1.48 (t, J = 6.5 Hz, 2H), 1.00 (s, 6H). LC-MS (ESI): m/z 1363.5 [M+H] +; HPLC: >95% purity.
5.2. Biology
5.2.1. Cell lines and culture.
T-ALL MOLT-4 (Cat# CRL-1582), B-ALL RS4;11 (RS4, Cat# CRL-1873), and SCLC NCI-H146 (H146, Cat# HTB-173) cell lines were purchased from American Type Culture Collection (ATCC, Manassas, VA). MOLT-4, RS4;11 and H146 cells were cultured in RPMI 1640 media (Life Technologies, Carlsbad, CA, USA) supplemented with 10% FBS (Atlanta Biologicals, Flowery Branch, GA, USA) and 1% penicillin-streptomycin solution (Thermo Fisher Scientific, Waltham, MA, USA).
5.2.2. Cell viability assay.
Cell viability was measured by Tetrazolium-based MTS assay (Promega, Madison, WI, USA). 5×104 to 1×105 suspension cells were seeded and treated in 96- well plates for 48 h or 72 h. The EC50 values of individual agents were calculated with GraphPad Prism 7 software (GraphPad Software, La Jolla, CA, USA).
5.2.3. Immunoblotting.
Cells were collected and lysed in Lysis buffer (Boston Bio Products, Ashland, MA, USA) supplied with protease and phosphatase inhibitor cocktails (Sigma-Aldrich, St. Louis, MO, USA). The equal amount of protein lysates was separated on pre-casted 4–20% Tris-glycine gels (Bio-Rad, Hercules, CA, USA). Thereafter, the proteins were transferred to PVDF membranes (MilliporeSigma, Billerica, MA, USA). The membranes were blocked with 5% w/v nonfat dry milk in TBS + Tween-20 (0.1% v/v), and then probed with primary antibodies overnight at 4° C. Next day, the membranes were washed and incubated with appropriate HRP-conjugated secondary antibodies. The signal was detected using ECL substrate (MilliporeSigma) and captured on X-ray films or ChemiDoc MP Imaging System (Bio-Rad). The band intensities were calculated on ImageJ software and normalized to equal loading control β-actin.
5.2.4. Human platelet isolation and viability assays.
Platelet-rich plasma (PRP) was purchased from Life South Community Blood Center (Gainesville, FL, USA). Platelets were separated from PRP as previously described [34]. Briefly, PRP was transferred into a 50 mL tube containing 5 mL acid citrate buffer (Cat. No. sc-214744, Santa Cruz Biotechnology). To prevent clotting, prostaglandin E1 (PGE1, Cat. No. sc-201223A, Santa Cruz Biotechnology) and apyrase (Cat. No. A6237, Sigma-Aldrich) were added to final concentrations of 1 μM and 0.2 units/mL, respectively. After centrifugation, platelet number was adjusted to 2 × 108/mL in HEPES Tyrode’s buffer containing 10% FBS, 1 μM PGE1 and 0.2 units/mL apyrase, and treated with various compounds. After 48 h of treatment, platelet viability was measured using the MTS reagent (Cat. No. G1111, Promega, Madison, WI, USA). The data were analyzed by GraphPad Prism 7 software for the calculation of IC50 values.
5.2.5. Flow cytometry.
MOLT-4 cells were plated in 12 well plates at 4 × 105 cells/well and treated with Veh, ABT-263, or XZ739 for 48 h. Caspase activity was blocked using Q-VD-OPh (QVD, Cat. No. S7311, Selleckchem, Houston, TX, USA) pretreatment for 2 h. After treatment, cells were stained with Alexa Fluor 647-Annexin V (1: 50, Cat. No. 640912, BioLegend, San Diego, CA, USA) and PI (10 μg/mL, Cat. No. 421301, BioLegend, San Diego, CA, USA) at room temperature for 30 min. For apoptosis analysis, the stained cells were analyzed on an Aurora flow cytometer (Cytek Aurora, Fremont, CA, USA).
Supplementary Material
BCL-XL PROTACs have significantly improved cytotoxicity in MOLT-4 cells
XZ739 treatment leads to remarkable BCL-XL degradation with a DC50 at 2.5 nM
XZ739 exhibits >100-fold selectivity for MOLT-4 cells over human platelets, whereas ABT-263 has no selectivity
Acknowledgements
This study was supported in part by NIH grants R01CA211963, R01CA219836, and R21CA223371.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare the following competing financial interest(s): X. Zhang, P. Zhang, S. Khan, D. Zhou, and G. Zheng are co-inventors for BCL-XL PROTACs disclosed in this study. D. Zhou and G. Zheng are co-founders and shareholders of Dialectic Therapeutics, a company that is developing BCL-XL PROTACs to treat cancers.
Declaration of interests
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Appendix A. Supplementary data
Supplementary data related to this article can be found at xxxxxx. CRBN and VHL expression in human platelets and cancer cell lines; DC50 of representative PROTACs; BCL-XL and BCL-2 binding affinities of ABT-263, DT2216, and XZ739; pharmacokinetics (PK) profile of XZ739; 1H-NMR spectrum of XZ739; and protein levels of IZFK1 and IZFK3 in MOLT-4 cells treated with XZ739 or pomalidomide are available in SI (PDF).
References
- [1].Singh R; Letai A; Sarosiek K Regulation of apoptosis in health and disease: the balancing act of BCL-2 family proteins. Nat. Rev. Mol. Cell Biol 2019, 20, 175–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hanahan D; Weinberg RA Hallmarks of cancer: the next generation. Cell. 2011, 144, 646–674. [DOI] [PubMed] [Google Scholar]
- [3].Delbridge ARD, Grabow S, Strasser A, Vaux DL. (2016). Thirty years of BCL-2: Translating cell death discoveries into novel cancer therapies. Nat. Rev. Cancer. 2016, 16, 99–109. [DOI] [PubMed] [Google Scholar]
- [4].Yap JL; Chen L; Lanning ME; Fletcher S Expanding the cancer arsenal with targeted therapies: Disarmament of the antiapoptotic Bcl‑2 proteins by small molecules. J. Med. Chem 2017, 60, 821–838. [DOI] [PubMed] [Google Scholar]
- [5].Garner TP; Lopez A; Reyna DE; Spitz AZ; Gavathiotis E Progress in targeting the BCL-2 family of proteins. Curr. Opin. Chem. Biol 2017, 39, 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Oltersdorf T; Elmore SW; Shoemaker AR; Armstrong RC; Augeri DJ; Belli BA; Bruncko M; Deckwerth TL; Dinges J; Hajduk PJ; Joseph MK; Kitada S; Korsmeyer SJ; Kunzer AR; Letai A; Li C; Mitten MJ; Nettesheim DG; Ng S; Nimmer PM; O’Connor JM; Oleksijew A; Petros AM; Reed JC; Shen W; Tahir SK; Thompson CB; Tomaselli KJ; Wang B; Wendt MD; Zhang H; Fesik SW; Rosenberg SH An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005, 435, 677–681. [DOI] [PubMed] [Google Scholar]
- [7].Bruncko M; Oost TK; Belli BA; Ding H; Joseph MK; Kunzer A; Martineau D; McClellan WJ; Mitten M; Ng S-C; Nimmer PM; Oltersdorf T; Park C-M; Petros AM; Shoemaker AR; Song X; Wang B; Wendt MD; Zhang H; Fesik SW; Rosenberg SH; Elmore SW Studies leading to potent, dual inhibitors of Bcl-2 and Bcl-xL. J. Med. Chem 2007, 50, 641–662. [DOI] [PubMed] [Google Scholar]
- [8].Tse C; Shoemaker AR; Adickes J; Anderson MG; Chen J; Jin S; Johnson EF; Marsh KC; Mitten MJ; Nimmer P; Roberts L; Tahir SK; Xiao Y; Yang X; Zhang H; Fesik S; Rosenberg SH; Elmore SW ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Research 2008, 68, 3421–3428. [DOI] [PubMed] [Google Scholar]
- [9].Park C-M; Bruncko M; Adickes J; Bauch J; Ding H; Kunzer A; Marsh KC; Nimmer P; Shoemaker AR; Song X; Tahir SK; Tse C; Wang X; Wendt MD; Yang X; Zhang H; Fesik S; Rosenberg SH; Elmore SW Discovery of an orally bioavailable small molecule inhibitor of prosurvival B-cell lymphoma 2 proteins. J. Med. Chem 2008, 51, 6902–6915. [DOI] [PubMed] [Google Scholar]
- [10].Wilson WH; O’Connor OA; Czuczman MS; LaCasce AS; Gerecitano JF; Leonard JP; Tulpule A; Dunleavy K; Xiong H; Chiu YL; Cui Y; Busman T; Elmore SW; Rosenberg SH; Krivoshik SP; Enschede SH; Humerickhouse RA Navitoclax, a targeted high-affi nity inhibitor of BCL-2, in lymphoid malignancies: a phase 1 dose-escalation study of safety, pharmacokinetics, pharmacodynamics, and antitumour activity. Lancet Oncol 2010, 11, 1149–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Mason KD; Carpinelli MR; Fletcher JI; Collinge JE; Hilton AA; Ellis S; Kelly PN; Ekert PG; Metcalf D; Roberts AW; Huang DCS; Kile BT Programmed anuclear cell death delimits platelet life span. Cell. 2007, 128, 1173–1186. [DOI] [PubMed] [Google Scholar]
- [12].Zhang H; Nimmer PM; Tahir SK; Chen J; Fryer RM; Hahn KR; Iciek LA; Morgan SJ; Nasarre MC; Nelson R; Preusser LC; Reinhart GA; Smith ML; Rosenberg SH; Elmore SW; Tse C Bcl-2 family proteins are essential for platelet survival. Cell Death Differ 2007, 14, 943–951. [DOI] [PubMed] [Google Scholar]
- [13].Souers AJ; Leverson JD; Boghaert ER; Ackler SL; Catron ND; Chen J; Dayton BD; Ding H; Enschede SH; Fairbrother WJ; Huang DC; Hymowitz SG; Jin S; Khaw SL; Kovar PJ; Lam LT; Lee J; Maecker HL; Marsh KC; Mason KD; Mitten MJ; Nimmer PM; Oleksijew A; Park CH; Park CM; Phillips DC; Roberts AW; Sampath D; Seymour JF; Smith ML; Sullivan GM; Tahir SK; Tse C; Wendt MD; Xiao Y; Xue JC; Zhang H; Humerickhouse RA; Rosenberg SH; Elmore SW ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat. Med 2013, 19, 202–208. [DOI] [PubMed] [Google Scholar]
- [14].DiNardo CD; Pratz K; Pullarkat V; Jonas BA; Arellano M; Becker PS; Frankfurt O; Konopleva M; Wei AH; Kantarjian HM; Xu T; Hong WJ; Chyla B; Potluri J; Pollyea DA; Letai A Venetoclax combined with decitabine or azacitidine in treatment-naive, elderly patients with acute myeloid leukemia. Blood. 2018, 133, 7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Roberts AW; Davids MS; Pagel JM; Kahl BS; Puvvada SD; Gerecitano JF; Kipps TJ; Anderson MA; Brown JR; Gressick L; Wong S; Dunbar M; Zhu M; Desai MB; Cerri E; Enschede SH; Humerickhouse RA; Wierda WG; Seymour JF Targeting BCL2 with venetoclax in relapsed chronic lymphocytic leukemia. N. Engl. J. Med 2016, 374, 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Oppermann S; Ylanko J; Shi Y; Hariharan S; Oakes CC; Brauer PM; Zúñiga-Pflücker JC; Leber B; Spaner DE; Andrews DW (2016). From high-content screening identifies kinase inhibitors that overcome venetoclax resistance in activated CLL cells. Blood. 2016, 128, 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Mihalyova J; Jelinek T; Growkova K; Hrdinka M; Simicek M; Hajek R Venetoclax: A new wave in hematooncology. Exp. Hematol 2018, 61, 10–25. [DOI] [PubMed] [Google Scholar]
- [18].Perini GF; Ribeiro GN; Neto JVP; Campos LT; Hamerschlak N BCL-2 as therapeutic target for hematological malignancies. J. Hematol. Oncol 2018, 11, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Vogler M Targeting BCL2-proteins for the treatment of solid tumours. Adv. Med 2014, Article ID 943648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Amundson SA; Myers TG; Scudiero D; Kitada S; Reed JC; Fornace AJ An informatics approach identifying markers of chemosensitivity in human cancer cell lines. Cancer Res 2000, 60, 6101–6110. [PubMed] [Google Scholar]
- [21].Leverson JD; Phillips DC; Mitten MJ; Boghaert ER; Diaz D; Tahir SK; Belmont LD; Nimmer P; Xiao Y; Ma XM; Lowes KN; Kovar P; Chen J; Jin S; Smith M; Xue J; Zhang H; Oleksijew A; Magoc TJ; Vaidya KS; Albert DH; Tarrant JM; La N; Wang L; Tao ZF; Wendt MD; Sampath D; Rosenberg SH; Tse C; Huang DCS; Fairbrother WJ; Elmore SW; Souers AJ Exploiting selective BCL-2 family inhibitors to dissect cell survival dependencies and define improved strategies for cancer therapy. Sci. Transl. Med 2015, 7, 279ra40. [DOI] [PubMed] [Google Scholar]
- [22].Mullard A Pioneering apoptosis-targeted cancer drug poised for FDA approval. Nat. Rev. Drug Discov 2016, 15, 147–149. [DOI] [PubMed] [Google Scholar]
- [23].Chang J; Wang Y; Shao L; Laberge RM; Demaria M; Campisi J; Janakiraman K; Sharpless NE; Ding S; Feng W; Luo Y; Wang X; Aykin-Burns N; Krager K; Ponnappan U; Hauer-Jensen M; Meng A; Zhou D Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat. Med 2016, 22, 78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhu Y; Tchkonia T; Fuhrmann-Stroissnigg H; Dai HM; Ling YY; Stout MB; Pirtskhalava T; Giorgadze N; Johnson KO; Giles CB; Wren JD; Niedernhofer LJ; Robbins PD; Kirkland JL Identification of a novel senolytic agent, navitoclax, targeting the Bcl-2 family of anti-apoptotic factors. Aging Cell. 2016, 15, 428–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yosef R; Pilpel N; Tokarsky-Amiel R; Biran A; Ovadya Y; Cohen S; Vadai E; Dassa L; Shahar E; Condiotti R; Ben-Porath I; Krizhanovsky V Directed elimination of senescent cells by inhibition of BCL-W and BCL-XL. Nat. Comm 2016, 7, 11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhu Y; Doornebal EJ; Pirtskhalava T; Giorgadze N; Wentworth M; Fuhrmann- Stroissnigg H; Niedernhofer LJ; Robbins PD; Tchkonia T; Kirkland JL New agents that target senescent cells: the flavone, fisetin, and the BCL-XL inhibitors, A1331852 and A1155463. Aging. 2017, 9, 955–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Demaria M; O’Leary MN; Chang J; Shao L; Liu S; Alimirah F; Koenig K; Le C; Mitin N; Deal AM; Alston S; Academia EC; Kilmarx S; Valdovinos A; Wang B; de Bruin A; Kennedy BK; Melov S; Zhou D; Sharpless NE; Muss H; Campisi J Cellular senescence promotes adverse effects of chemotherapy and cancer relapse. Cancer Discov 2017, 7, 165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bai L; Chen J; Liu L; McEachern D; Aguilar A; Zhou H; Yang CY; Wang H; Wen J; Wang G; Zhai Y; Guo M; Yang D; Wang S BM-1252 (APG-1252): a potent dual specific Bcl-2/Bcl-xL inhibitor that achieves complete tumor regression with minimal platelet toxicity. Eur. J. Cancer. 2014, 50, 109–110. [Google Scholar]
- [29].Lakhani NJ; Rasco DW; Tolcher AW; Huang Y; Ji J; Wang H; Dong Q; Men L; O’Rourke TJ; Chandana SR; Amaya A; Cole Y; Kaiser B; Mays TA; Patnaik A; Papadopoulos KP; Yang D; Zhai Y A phase I study of novel dual Bcl- 2/Bcl-xL inhibitor APG-1252 in patients with advanced small cell lung cancer (SCLC) or other solid tumor. J. Clin. Oncol 2018, 36, suppl. abstr. 2594. [Google Scholar]
- [30]. https://clinicaltrials.gov/ct2/show/NCT03595059.
- [31].Lai AC; Crews CM Induced protein degradation: an emerging drug discovery paradigm. Nature Reviews Drug Discovery. Nat. Rev. Drug Discov 2017, 16, 101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Churcher I Protac-induced protein degradation in drug discovery: breaking the rules or just making new ones? J. Med. Chem 2018, 61, 444–452. [DOI] [PubMed] [Google Scholar]
- [33].Schapira M; Calabrese MF; Bullock AN; Crews CM Targeted protein degradation: expanding the toolbox. Nat. Rev. Drug Discov 2019, 19, DOI: 10.1038/s41573-019-0047-y [DOI] [PubMed] [Google Scholar]
- [34].Khan S; Zhang X; Lv D; Zhang Q; He Y; Zhang P; Liu X; Thummuri D; Yuan Y; Wiegand JS; Pei J; Zhang W; Sharma A; McCurdy CR; Kuruvilla VM; Baran N; Ferrando AA; Kim Y; Rogojina A; Houghton PJ; Huang G; Hromas R; Konopleva M; Zheng G; Zhou D A selective BCL-XL PROTAC degrader achieves safe and potent antitumor activity. Nat. Med 2019, DOI: 10.1038/s41591-019-0668-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bray PF; McKenzie SE; Edelstein LC; Nagalla S; Delgrosso K; Ertel A; Kupper J; Jing Y; Londin E; Loher P; Chen HW; Fortina P; Rigoutsos I The complex transcriptional landscape of the anucleate human platelet. BMC Genomics. 2013, 14, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kissopoulou A; Jonasson J; Lindahl TL; Osman A Next generation sequencing analysis of human platelet PolyA+ mRNAs and rRNA-depleted total RNA. PLoS One. 2013, 8, e81809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhang X; Thummuri D; He Y; Liu X; Zhang P; Zhou D; Zheng G Utilizing PROTAC technology to address the on-target platelet toxicity associated with inhibition of BCL-XL. Chem. Commun 2019, 55, 14765–14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Papatzimas JW; Gorobets E; Maity R; Muniyat MI; MacCallum JL; Neri P; Bahlis NJ; Derksen DJ From inhibition to degradation: targeting the antiapoptotic protein myeloid cell leukemia 1 (MCL1). J. Med. Chem 2019, 62, 5522–5540; [DOI] [PubMed] [Google Scholar]
- [39].Wang Z; He N; Guo Z; Niu C; Song T; Guo Y; Cao K; Wang A; Zhu J; Zhang X; Zhang Z Proteolysis targeting chimeras for the selective degradation of Mcl-1/Bcl-2 derived from nonselective target binding ligands. J. Med. Chem 2019, 62, 8152–8163. [DOI] [PubMed] [Google Scholar]
- [40].Lai AC; Toure M; Hellerschmied D; Salami J; JaimeFigueroa S; Ko E; Hines J; Crews CM Modular PROTAC Design for the Degradation of Oncogenic BCR-ABL. Angew. Chem. Int. Ed 2016, 55, 807–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Han X; Wang C; Qin C; Xiang W; Fernandez-Salas E; Yang C-Y; Wang M; Zhao L; Xu T; Chinnaswamy K; Delproposto J; Stuckey J; Wang S Discovery of ARD-69 as a highly potent proteolysis targeting chimera (PROTAC) degrader of androgen receptor (AR) for the treatment of prostate cancer. J. Med. Chem 2019, 62, 941–964. [DOI] [PubMed] [Google Scholar]
- [42].Hu J; Hu B; Wang M; Xu F; Miao B; Yang C-Y; Wang M; Liu Z; Hayes DF; Chinnaswamy K; Delproposto J; Stuckey J; Wang S Discovery of ERD-308 as a highly potent proteolysis targeting chimera (PROTAC) degrader of estrogen receptor (ER). J. Med. Chem 2019, 62, 1420–1442. [DOI] [PubMed] [Google Scholar]
- [43].Crew AP; Raina K; Dong H; Qian Y; Wang J; Vigil D; Serebrenik YV; Hamman BD; Morgan A; Ferraro C; Siu K Identification and characterization of Von Hippel-Lindau-recruiting proteolysis targeting chimeras (PROTACs) of TANK-binding kinase 1. J. Med. Chem 2018, 61, 583–598. [DOI] [PubMed] [Google Scholar]
- [44].Zoppi V; Hughes SJ; Maniaci C; Testa A; Gmaschitz T; Wieshofer C; Koegl M; Riching KM; Daniels DL; Spallarossa A; Ciulli A Iterative design and optimization of initially inactive proteolysis targeting chimeras (PROTACs) identify VZ185 as a potent, fast, and selective von Hippel–Lindau (VHL) based dual degrader probe of BRD9 and BRD7. J. Med. Chem 2018, 62, 699–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Wurz RP; Dellamaggiore K; Dou H; Javier N; Lo MC; McCarter JD; Mohl D; Sastri C; Lipford JR; Cee VJA “click chemistry platform” for the rapid synthesis of bispecific molecules for inducing protein degradation. J. Med. Chem 2018, 61, 453–461. [DOI] [PubMed] [Google Scholar]
- [46].Raina K; Lu J; Qian Y; Altieri M; Gordon D; Rossi AM; Wang J; Chen X; Dong H; Siu K; Winkler JD; Crew AP; Crews CM; Coleman KG PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc. Natl. Acad. Sci. USA. 2016, 113, 7124–7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Riching KM; Mahan S; Corona CR; McDougall M; Vasta JD; Robers MB; Urh M; Daniels DL Quantitative live-cell kinetic degradation and mechanistic profiling of PROTAC mode of action. ACS Chem. Biol 2018, 13, 2758–2770. [DOI] [PubMed] [Google Scholar]
- [48].Moreau K; Coen M; Zhang AX; Pachl F; Castaldi MP; Dahl G; Boyd H; Scott C; Newham P PROTAC (PROteolysis TArgeting Chimeras) in drug development: a safety perspective. Br. J. Pharmacol 2020, 10.1111/bph.15014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Li Y; Yang J; Aguilar A; et al. Discovery of MD-224 as a first-in-class, highly potent, and efficacious proteolysis targeting chimera murine double minute 2 degrader capable of achieving complete and durable tumor regression. J. Med. Chem 2018, 62, 448–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].McCoull W; Cheung T; Anderson E; Barton P; Burgess J;Byth K; Cao Q; Castaldi MP; Chen H; Chiarparin E; Carbajo RJ; Code E; Cowan S; Davey PR; Ferguson AD; Fillery S; Fuller NO; Gao N; Hargreaves D; Howard MR; Hu J; Kawatkar A; Kemmitt PD; Leo E; Molina DM; O’Connell N; Petteruti P; Rasmusson T; Raubo P; Rawlins PB; Ricchiuto P; Robb GR; Schenone M; Waring MJ; Zinda M; Fawell S; Wilson DM Development of a novel B-cell lymphoma 6 (BCL6) PROTAC to provide insight into small molecule targeting of BCL6. ACS Chem. Biol 2018, 13, 3131–3141. [DOI] [PubMed] [Google Scholar]
- [51].Bai L; Zhou H; Xu R; Zhao Y; Chinnaswamy K; McEachern D; Chen J; Yang C-Y; Liu Z; Wang M; Liu L; Jiang H; Wen B; Kumar P; Meagher JL; Sun D; Stuckey JA; Wang S A potent and selective small-molecule degrader of STAT3 achieves complete tumor regression in vivo. Cancer Cell, 2019, 36, 498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Zhou H; Bai L; Xu R; Zhao Y; Chen J; McEachern D; Chinnaswamy K; Wen B; Dai L; Kumar P; Yang C-Y; Liu Z; Wang M; Liu L; Meagher JL; Yi H; Sun D; Stuckey JA; Wang S Structure-based discovery of SD-36 as a potent, selective and efficacious PROTAC degrader of STAT3 protein. J. Med. Chem DOI: 10.1021/acs.jmedchem.9b01530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


