Abstract
An abundance of information about lung development in animal models exists; however, comparatively little is known about lung development in humans. Recent advances using primary human lung tissue combined with the use of human in vitro model systems, such as human pluripotent stem cell-derived tissue, have led to a growing understanding of the mechanisms governing human lung development. They have illuminated key differences between animal models and humans, underscoring the need for continued advancements in modeling human lung development and utilizing human tissue. This review discusses the use of human tissue and the use of human in vitro model systems that have been leveraged to better understand key regulators of human lung development and that have identified uniquely human features of development. This review also examines the implementation and challenges of human model systems and discusses how they can be applied to address knowledge gaps.
Keywords: alveolosphere, branching morphogenesis, bronchopulmonary dysplasia, human development, lung, organoid, signaling
1. Introduction
The respiratory system is comprised of the trachea and airways of the lung, the branched network of epithelial tubes forming the bronchi and bronchioles, and the alveoli, where gas exchanges with the vascular system. Each of these structures is made of multiple specialized epithelial cell types that help carry out the lung’s unique functions of air intake and gas exchange, epithelial barrier function, protection from microbes and pathogens, and the maintenance of fluid and electrolyte homeostasis.[1] The diverse repertoire of respiratory epithelial cells that comprise the trachea, airways, and alveoli are derived from a common population of progenitor cells that are specified in the endodermal germ layer early during development.[2–4] In addition to the endoderm-derived epithelium, both the developing and mature respiratory systems contain cells derived from the mesoderm (e.g., smooth muscle) and ectoderm (e.g., neurons) germ layers, and the complex interactions between cells from all three germ layers are absolutely critical for proper respiratory system development and function.[5–8]
Development of the respiratory system is broadly divided into five stages, each representing major morphological changes that take place[9] (Figure 1a–c). The embryonic stage is defined by respiratory specification, the establishment of the nascent tracheal domain, and the emergence of two primary lung buds from the ventral anterior foregut endoderm (Figure 1a). Following these events, the lung enters the pseudoglandular stage where lung buds undergo repeated rounds of bifurcations during a process called branching morphogenesis, which establishes the arborized network of bronchi and bronchioles[10] (Figure 1b). The alveoli form across several stages, with alveolar cell-type specification beginning during branching morphogenesis and finalizing their differentiation in the terminal stages of lung development,[11] which includes the canalicular stage where alveolar ducts form at terminal bronchioles, the saccular stage where alveolar cells functionally mature and alveolar sacs form, and the alveolar stage where alveoli continue to mature and increase their surface area through septation (Figure 1c). The lung is one of the few organs that continues to develop in post-natal life as the alveoli continue to grow in size and complexity for seven years after birth in humans and one month after birth in mice.
Figure 1.
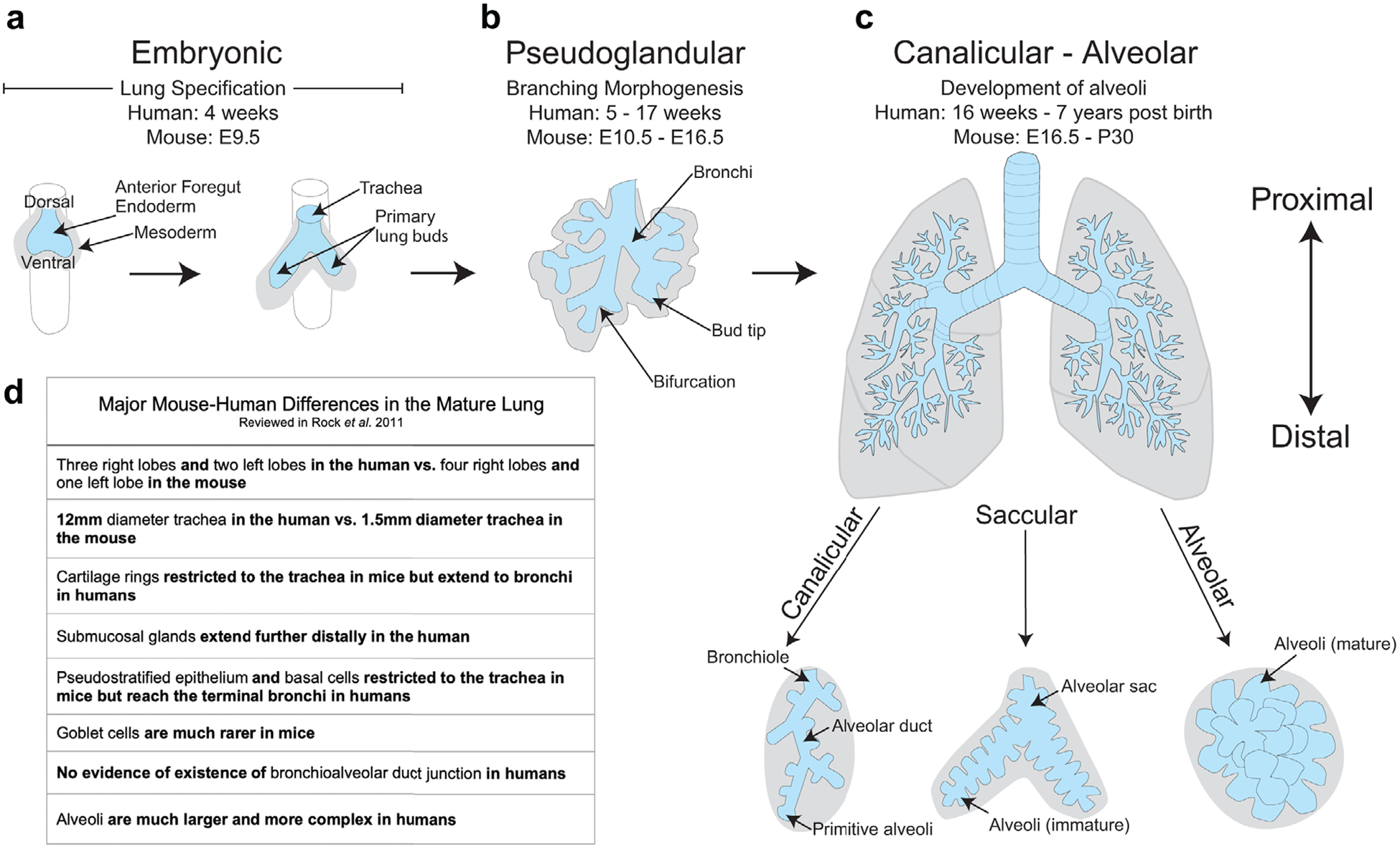
Morphology of the respiratory system during the five stages of respiratory development. a) During the embryonic stage, the lung arises ventrally from the anterior foregut endoderm, giving rise to two primary lung buds that branch off from the trachea into the surrounding mesoderm. b) The pseudoglandular stage is characterized by the processes of branching morphogenesis, whereby distal bud tips undergo repeated rounds of bifurcations to create the arborized network of airways. c) The alveoli, the air sacs that allow for gas exchange, are formed during the canalicular, saccular, and alveolar stages. This occurs as alveolar ducts form at the most distal airways, the bronchioles, which then form terminal sacs that will give rise to functional alveoli. d) The adult mouse and adult human lungs contain many morphological differences.
The structure and function of the adult mouse and adult human respiratory systems have multiple differences, including anatomical differences such as the number of airway branches, the identity and localization of adult stem cells, and the morphology of alveoli[12] (Figure 1d). These physiological differences likely contribute to the failure of most human clinical trials using lung therapeutics developed in mouse models.[13] Until recently, it has been difficult to assess the mechanistic differences that emerge during respiratory development that lead to differences in the mature lungs of mice and humans. However, contemporary research has addressed this issue by using primary human tissue and by developing in vitro model systems that mimic human respiratory development. Coupled with technological advances such as single-cell RNA sequencing (scRNAseq),[14–17] these studies have shed light on many of the similarities and differences between mouse and human respiratory development. In this review, we will discuss our understanding of human lung development during each stage of respiratory development, focusing on signaling and transcriptional networks that regulate the developing human respiratory system. We will also discuss the current state of human model systems to accurately model human respiratory development and disease and highlight the challenges that remain.
2. The Embryonic Stage of Respiratory Development
2.1. Contribution of Cellular Signaling Pathways to Respiratory Endoderm Specification
The respiratory system is specified at E9.5 in mice and at 4 weeks of gestation in humans as the trachea and primary lung buds separate ventrally from the esophagus in the anterior foregut endoderm.[18–20] The respiratory system is first marked by the transcription factor NKX2.1,[21,22] which is also necessary for lung specification.[23] Respiratory specification in mouse models has been reviewed extensively.[24–28] These studies have identified many of the signaling pathways that are essential during respiratory specification and have been used as a framework to differentiate human pluripotent stem cells (hPSCs) into respiratory lineages in vitro. This strategy, known as “directed differentiation,” is an attempt to recapitulate a series of developmental events in a stepwise manner by modifying the growth factor signaling environment in the tissue culture dish. This approach has allowed us to gain an appreciation of the signaling and transcriptional regulators that are necessary for respiratory specification in a human-specific context. The major developmental milestones for lung specification using directed differentiation include definitive endoderm differentiation,[29,30] followed by anterior-posterior patterning into anterior foregut endoderm,[31] at which point NKX2.1+ respiratory progenitor cells can be specified.[22,32–38]
Studies using directed differentiation from hPSCs as well as studies in animal models have stressed the importance of WNT signaling for initiating the expression of NKX2.1 from anterior foregut endoderm[37,39–41] (Figure 2a,b). However, activation of WNT signaling that induces NKX2.1 expression requires cooperation from multiple other signaling pathways (Figure 2a,b). The complex signaling network that induces the respiratory fate is dependent on retinoic acid (RA) signaling, which is required prior to respiratory specification and renders the ventral foregut endoderm competent to respond to cues that induce the respiratory lineage, although the mechanisms through which RA signaling acts are unknown.[33,35,37,42] It has been shown in mice that Sonic Hedgehog (SHH) ligands emanating from the endoderm induce the expression of WNT ligands in the mesoderm, which signal back to the endoderm to activate NKX2.1 expression.[37,43,44] NKX2.1+ cells have been induced from hPSCs without the addition of SHH signaling components to the media; however, since SHH signaling acts upstream of WNT in mice, it is possible that directed differentiation strategies using hPSCs bypass the need for SHH components through the addition of exogenous WNT ligands. In mice and humans, BMP signaling represses SOX2 in the endoderm, which is required for the endoderm to properly respond to WNT ligands and express NKX2.1.[37,45] Genetic gain- and loss-of-function studies in mice have also established a role for FGF signaling during respiratory specification[46–49]; however, like SHH, FGF has not played a prominent role in differentiation of hPSC into NKX2.1+ respiratory progenitor cells, and its role in human respiratory specification remains unknown.
Figure 2.
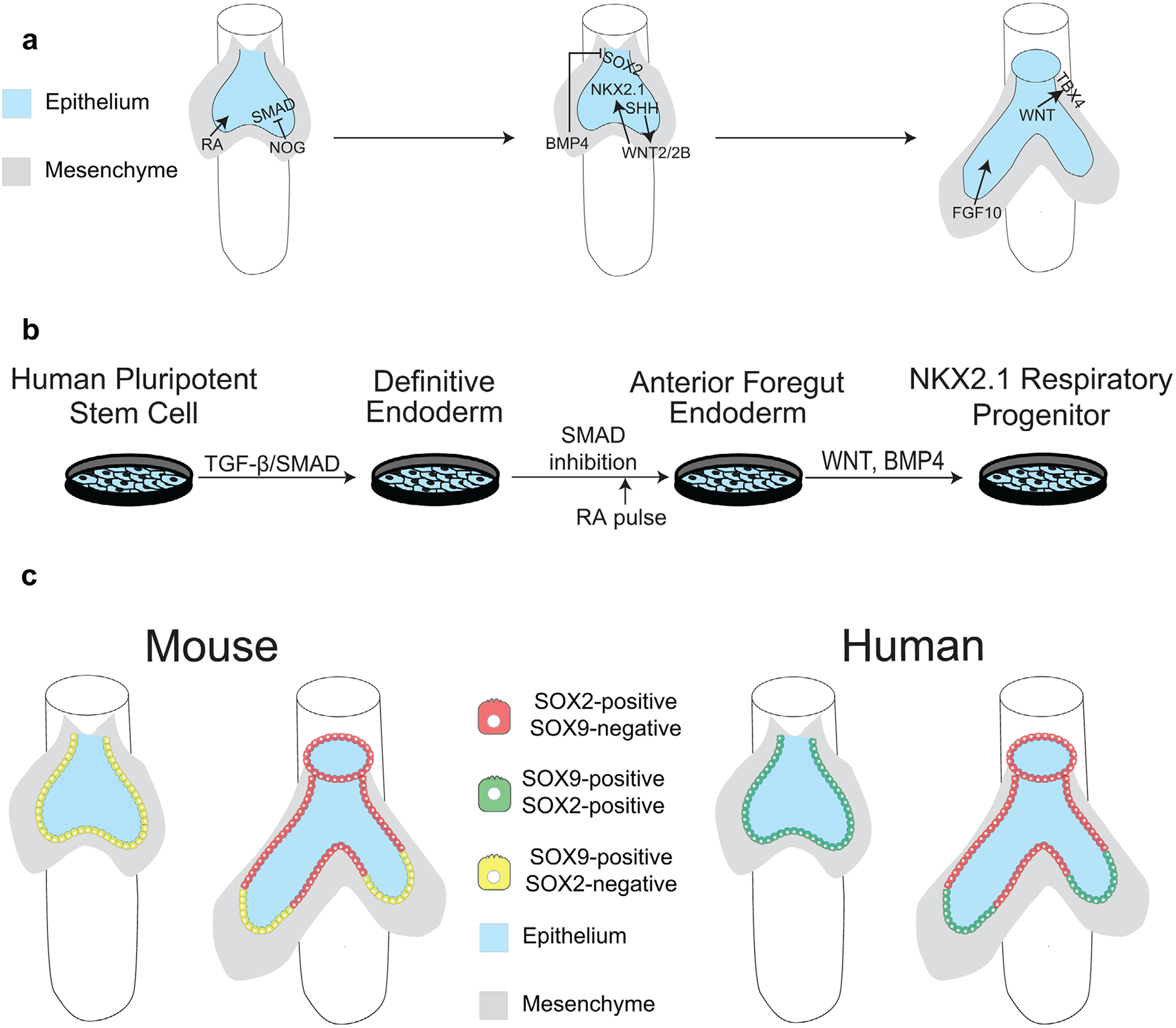
Signaling mechanisms required for respiratory specification a) in vivo and b) in vitro. TGF-β/SMAD signaling drives definitive endoderm specification, and SMAD inhibition through Noggin drives anteriorization of definitive endoderm. BMP4 from the mesoderm inhibits SOX2 expression in the mesoderm while SHH from the endoderm activates WNT ligands in the mesoderm that turn on NKX2.1 expression. RA is required for this process. WNT, BMP4, and SHH (humans only) from the endoderm specify the tracheal mesoderm, which is marked by TBX4. FGF10 is required for lung bud outgrowth. c) The mouse respiratory epithelium is initially made of SOX9+ bud tip progenitor cells, which become restricted to the budded tips of the lung as the primary lung buds grow out from the trachea. The bud tip progenitors that are left behind proximally become SOX2+. d) The human respiratory epithelium is initially made of SOX2+/SOX9+ bud tip progenitor cells, which become restricted to the budded tips of the lung as the primary lung buds grow out from the trachea. The bud tip progenitors that are left behind proximally lose SOX9 expression but remain SOX2+.
2.2. Signaling Involved in Self-Organization of 3D Lung Models
Many directed differentiation protocols that induce anterior foregut endoderm lineages from hPSCs use 2D cultures; however, it is also possible to generate 3D anterior foregut endoderm structures, called spheroids, using directed differentiation techniques.[36,50] Spheroids are immature multicellular tissue structures that arise during directed differentiation through unknown mechanisms and which mimic a primitive gut tube-like structure. Spheroids provide an opportunity to direct the differentiation of hPSCs into lung cells with the correct cellular organization. The cues that are needed to pattern hPSCs into 3D lung spheroids seem to require a different set of signals compared to cells grown in 2D. For example, Dye et al. have shown it is possible to derive 3D ventral anterior foregut structures that can give rise to mature lung lineages by simultaneously inhibiting SMAD, which is required for anterior foregut patterning, and by activating FGF4, WNT, and SHH, which are required for both inducing 3D spheroid formation and robust NKX2.1 expression.[36,51,52] The necessity of FGF4, WNT, and SHH for the formation of 3D structures suggest that these signaling pathways may be responsible for cell migration and patterning during respiratory fate specification in humans.
2.3. Different Signaling Pathways Contribute to Mouse and Human Respiratory Mesoderm Specification
In the mouse, respiratory mesoderm is Nkx2.1− but is marked by Tbx4 and Tbx5, both of which are necessary for respiratory mesoderm development and specification of the lung and trachea.[53] A recent study from Kishimoto et al. showed that WNT signaling originating from the mouse endoderm induces Tbx4 expression in the primitive lung mesoderm independent of Nkx2.1 expression (Figure 2a). Using mouse pluripotent stem cells and hPSCs, they showed that tracheal mesoderm (chondrocytes and proximal smooth muscle cells) could be specified from lateral plate mesoderm by BMP4 and WNT signaling in the mouse and SHH, BMP4, and WNT signaling in the human.[54] This demonstrates that the primary molecular mechanisms responsible for tracheal mesoderm specification are different between the mouse and human as mouse tracheal mesoderm specification does not require SHH. In another study, Wnt2+/Gli1+/Isl1+ mesodermal cells that arise prior to respiratory specification were shown to give rise to some lung and cardiac mesodermal lineages.[55] SHH signaling regulates specification of these “cardiopulmonary progenitors” into smooth muscle lineages in the lung[55]; however, the mechanisms regulating cardiopulmonary progenitor specification into other distal mesenchymal cell types are currently unknown.[56]
3. The Pseudoglandular Stage of Respiratory Development
The pseudoglandular stage occurs between E10.5 to E16.5 in mice and 5 to 17 weeks of gestation in humans. This stage is defined by branching morphogenesis, where progenitor-rich lung bud tips begin to undergo repeated bifurcations to create the complex arborized network of the airways[2,8,10,12,25,57–59] (Figure 1b). Humans undergo extended rounds of branching relative to mice (17–21 in humans, 7–17 in mice),[10,60] raising the possibility of regulatory divergence in human branching morphogenesis. Complex reciprocal signaling between the epithelium and mesenchyme during this stage creates a unique hurdle in characterizing the signaling pathways important for branching. Other changes in the lung during pseudoglandular development include the emergence of smooth muscle and vasculature, which both contribute to the environment that influences branching morphogenesis. Here, we discuss the emergence of lung cell types during branching morphogenesis, their role in establishing the lung microenvironment, and how these environments dictate local signaling.
3.1. Cellular Differentiation during Branching Morphogenesis
A significant event during the pseudoglandular stage is the specification of airway cell types in the lung epithelium. As branching tips of the epithelium continue to grow and bifurcate, bud tip progenitors leave progeny behind, which differentiate into airway cell types including basal, ciliated, secretory, and neuroepithelial cells (Figure 3b). Lineage tracing in mice suggest there is a specific developmental window where bud tip progenitors preferentially give rise to airway cell types.[61,62] Until recently, there was limited knowledge about how these processes differ in humans. Several groups performed scRNAseq on human fetal lung samples,[3,4,63] and these studies established important in vivo benchmarks of cellular transcriptional states that can be directly compared with in vitro-derived cells, providing a roadmap for developing directed differentiation approaches to generate specific airway cell types. For example, methods to direct the differentiation of bud tip progenitors to TP63+ basal cells have been developed by manipulating SMAD signaling[63,64] (Figure 3b). Alternatively, inhibition of NOTCH signaling directs hPSC-derived lung epithelium to differentiate into ciliated and neuroendocrine cells[65] (Figure 3b). There remains debate over the role of WNT on bud tip progenitor fate.[66,67] Some groups conclude that high WNT signaling supports a proximal airway cell fate and other groups conclude it supports alveolar cell types. As organoid models continue to improve, coupled with single cell studies, it is likely that more questions can be answered about cell lineage specification in the human airway.
Figure 3.
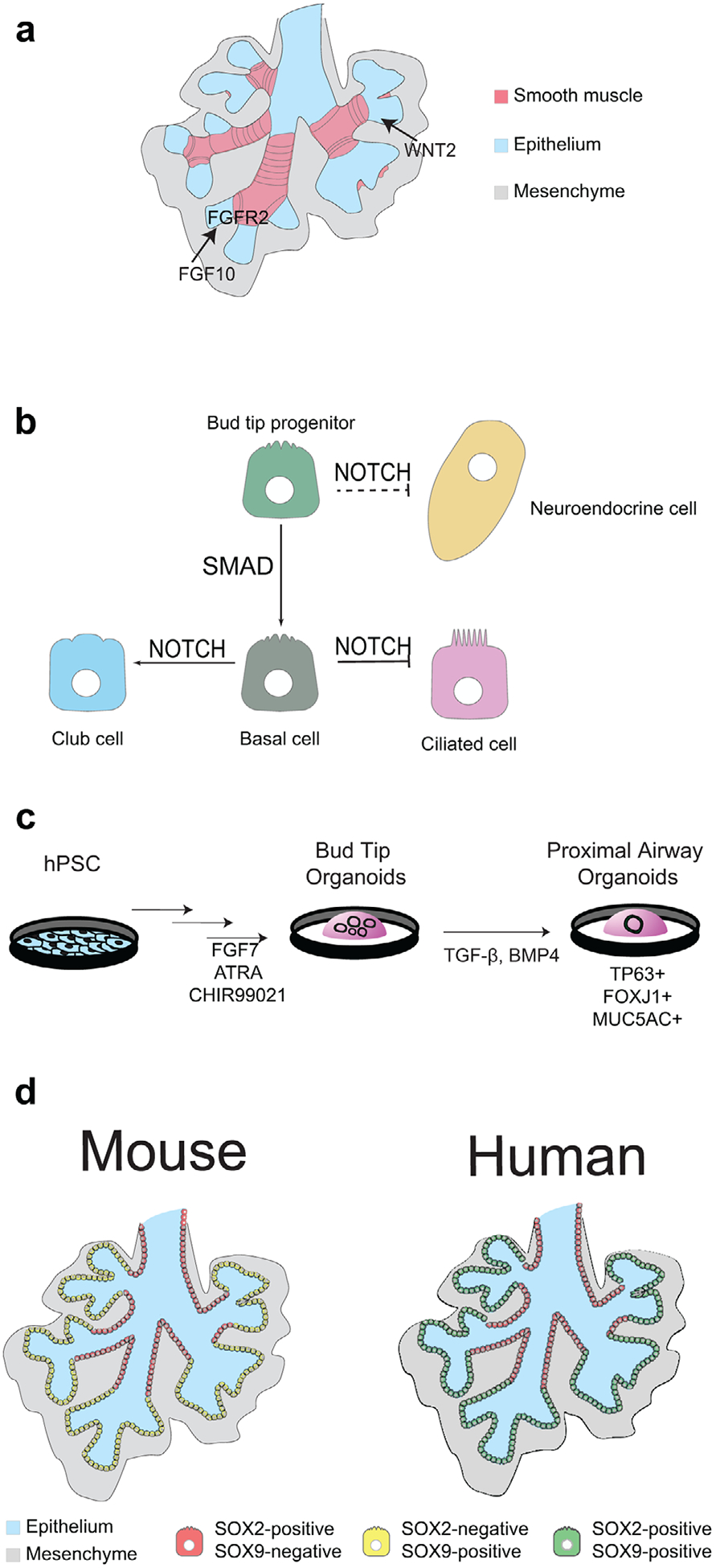
Signaling and cell types of branching morphogenesis. a) FGF10 signaling from the mesenchyme interacts with FGFR2 receptors on epithelium. WNT signaling from the mesenchyme also supports branching of the epithelium. An important physical cue for branching is the smooth muscle (pink). b) The signaling pathways important for airway cell differentiation include SMAD signaling from bud tip progenitors to TP63+ basal cells. Terminal differentiation into club/secretory cells is facilitated by active NOTCH signaling, and inhibition of NOTCH gives rise to multiciliated cells. Neuroendocrine cells also form from an epithelial progenitor through NOTCH inhibition, although it is less clear if they are specified directly from bud tip progenitors. c) In vitro directed differentiation approaches have enabled expansion of bud tip progenitors as well as their differentiation into airway cell types using mechanisms that mimic in vivo signaling. d) Organization of SOX2+ cells and SOX9+ cells vary between mice (left) and humans (right) where SOX2+ cells are limited to proximal airway cells, but bud tip progenitors are SOX9+ in mice and SOX2+/SOX9+ in humans.
3.2. Regulation of Bud Tips during Branching Morphogenesis
Branching morphogenesis is a complex morphological process that relies on highly proliferative progenitor-rich distal bud tips of the developing lung. RNAseq data on bud tips found differences in both gene and protein expression between human and mouse.[3,51,52,61] In mice, bud tip progenitors express N-myc, Id2, and Sox9 but are Sox2 negative.[61,68–70] This contrasts with humans where bud tip progenitors express SOX9 in addition to SOX2[3,4,71] (Figure 2c,3d). Loss of SOX2 does not occur in human bud tips until the canalicular stage. In cultured human lung explants where RAC1 inhibition causes decreased SOX9+/SOX2+ bud tip progenitors, there is also decreased epithelial proliferation and impaired branching.[71] As Sox2 has been shown to be essential for airway cell fates in mice,[72] longer perdurance of SOX2 expression in human bud tip progenitors may suggest that human bud tips retain the potential to differentiate into airway cell fates much later into development than in mice.
Studies of branching morphogenesis in the mouse have elucidated important mechanisms that regulate this process, which are reviewed extensively elsewhere.[26,73–75] Here, we focus on comparing the signaling regulation of murine and human bud tips during branching. A thoroughly investigated signaling pathway in branching is FGF10, which is expressed in the mesenchyme near the most distal bud tips and is critical for branching and proximal-distal patterning in mice[76,77] (Figure 3a). Fgf10−/− mice do not undergo branching and conditional knock-outs of Fgf10 or Fgfr2 also disrupt lobe growth and have fewer branches.[47,48,78] In the developing human lung, FGF10 is expressed from 10–21 weeks[76,77,79] diffusely throughout the lung parenchyma.[76,80] Murine lung explants cultured with FGF10 show increased branching, while in contrast, human lung explants cultured with FGF10 exhibit enlarged buds and fewer branches.[76,77] Human lung organoid models suggest that FGF10 is not required for bud tip maintenance,[3,4] though long term culture in FGF10-rich media results in differentiation of airway cell types.[36] Another important signaling pathway during branching is WNT (Figure 3a). In mice, loss of both Wnt2 and Wnt2b results in complete lung agenesis,[39] and conditional epithelial knock-out of β-catenin results in mal-formed distal airways with aberrant proximal airways.[81] Similarly, RAC1-mediated WNT inhibition in human lung explants decreases branching and results in loss of bud tip progenitors, although the molecular mechanisms remain to be investigated.[70,71] It was recently discovered that humans with mutations in the WNT activator R-spondin 2 (RSPO2) exhibit lung agenesis,[41] which is a surprising contrast to murine lung, where Rspo2 mutants have mild branching defects.[82] The continued use and advancement of human in vitro models are required to fully appreciate the molecular mechanisms of FGF and WNT signaling in human lung branching morphogenesis.
3.3. Molecular and Mechanical Cues from the Mesenchyme during Branching Morphogenesis
The mesenchyme undergoes significant morphological changes as the branching epithelium continues to bifurcate and alter the landscape of the lung. We are only beginning to understand the diversity of mesenchymal cell types and changes they undergo during human lung development.[83] Therefore, in vitro human models of lung mesenchyme are less developed compared to epithelial models. Engineering approaches using microfluidic chambers with mouse lung explants as well as in silico modeling have begun to examine the changes that occur during branching morphogenesis and show promising innovation for human models.[84,85] In mice, it has been shown that both Fgf10+ mesenchymal cells and Pdgfra+ mesenchymal cells give rise to airway smooth muscle,[86] the latter through WNT2 and WNT7b signaling.[87] Blocking smooth muscle differentiation also prevents epithelial buds from bifurcating,[88] and it was recently shown that smooth muscle differentiation defines specific domains along the airways that propagate branches in mice.[89] In humans, α-SMA+ smooth muscle cells support the proximal fate of the human airway and branching,[71] although the signaling mechanisms involved are unknown (Figure 3a). More in vitro models using organoid and explant-like cultures will be required for understanding the signaling changes in the mesenchyme during human lung development. Single cell analysis will continue to help shed light on the complexity of the mesenchyme and identify key signaling factors involved in the morphing landscape of the lung.
4. The Canalicular Stage of Respiratory Development
During the canalicular stage of respiratory development, the lung transitions from generating airway (bronchi, bronchioles) to generating the gas-exchange units of the lung, the alveoli. This is characterized by the continued differentiation of bud tip progenitor cells towards alveolar fates[3,4,61,90–94] and by the formation of the bronchoalveolar duct junction (BADJ) in mice,[95,96] which demarcates airway-fated epithelial cells from alveolar-fated epithelial cells and can be identified in mice by the terminal border of Sox2 expression.[14,97,98] The existence of a BADJ in humans has not been demonstrated, but both human and mouse bud tips lack expression of the airway cell fate marker SOX2 prior to generating alveolar cells[3,4,71] (Figure 4a).
Figure 4.
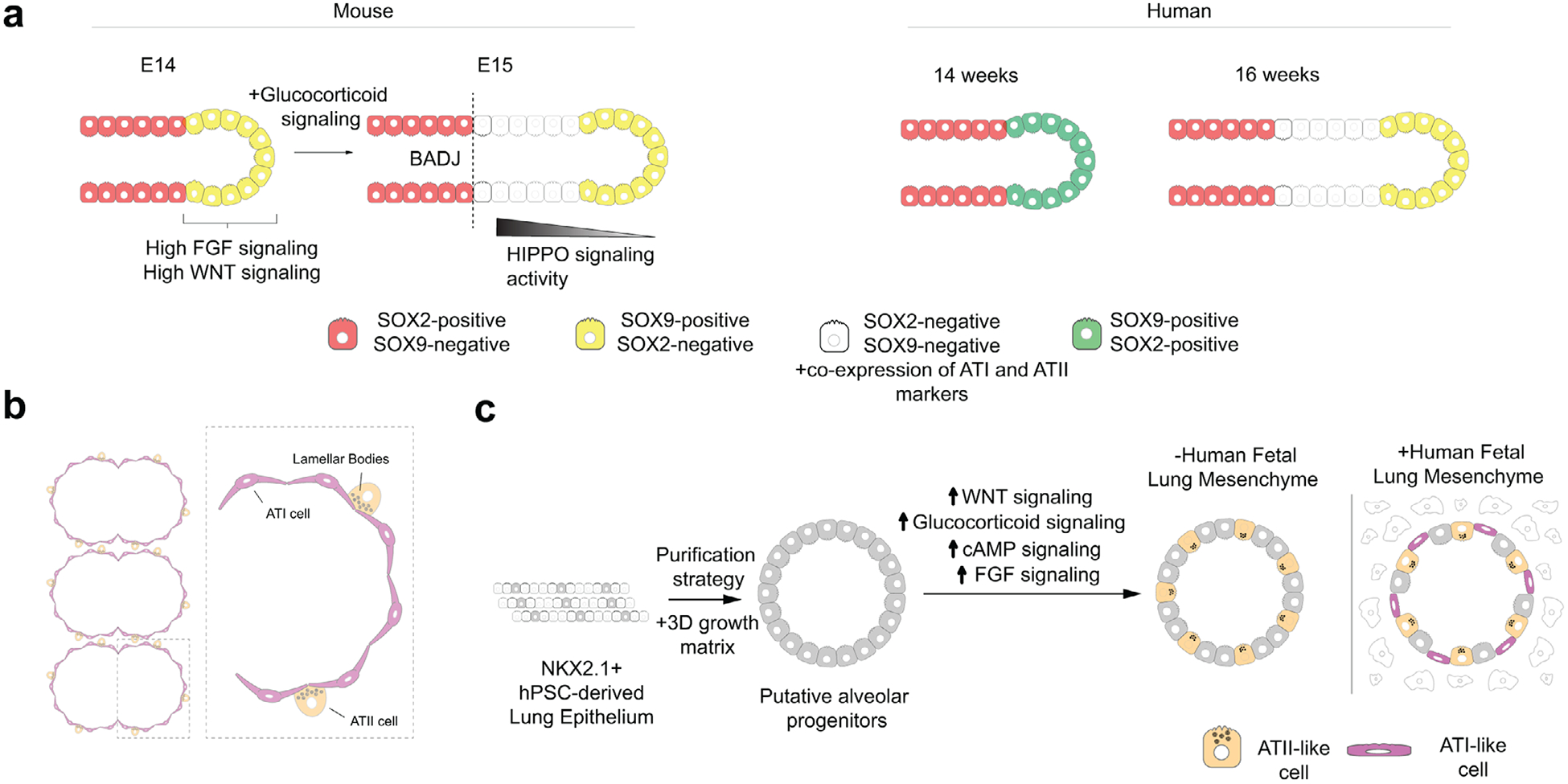
Alveolar cell fate specification. a) In mice, glucocorticoid signaling establishes the bronchoalveolar duct junction (BADJ), which demarcates the airway from the future site of alveoli formation. Bud tip progenitor identity is maintained by high levels of WNT and FGF signaling. High levels of HIPPO signaling in bud tip daughter cells born after BADJ formation leads to differentiation into alveolar progenitors, which co-express markers of ATI and ATII cells. In humans, whether BADJ formation occurs is unknown. Human bud tip progenitors downregulate SOX2 by week 16 of development, suggesting a change in the developmental potential of human bud tips occurs at 16 weeks. b) Morphology of alveoli. ATI cells are thin to facilitate gas exchange. ATII cells contain lamellar bodies, a surfactant producing organelle. c) In vitro models of alveolar cell fate specification. Putative alveolar progenitors are purified from hPSC-derived NXK2.1+ lung epithelium. Conditions of high WNT, glucocorticoid, cAMP, and FGF signaling lead to the formation of ATII-like cells containing lamellar bodies. Incorporation of human fetal lung mesenchyme leads to the generation of ATI-like cells.
4.1. Alveolar Cell Specification during the Canalicular Stage
Classic models of alveolar development proposed that alveolar cell types are specified in a sequential manner, with bud tip progenitors giving rise to alveolar progenitors, which give rise to alveolar type II (ATII) cells, which give rise to alveolar type I (ATI) cells.[99] More recent studies of alveolar cell specification at the single-cell level have proposed an alternative model that ATI and ATII cells are specified from a bipotent alveolar progenitor,[14,92] whose existence in mice was recently proved with lineage tracing strategies.[94] However, this latter study suggested that bipotent progenitors are rare and most likely remnant undifferentiated cells that remain at the end of branching morphogenesis.[94] It was further demonstrated that commitment to alveolar fates occurs much earlier than previously appreciated, taking place concurrently with branching morphogenesis, rather than afterwards. The majority of mature alveolar cells are the progeny of unipotent alveolar progenitor cells fated towards either an ATI or ATII cell early in development, with ATII cells being specified first at the most distal tip of the lung and ATI cells being specified just after ATII cells.[94] Interestingly, in humans, markers of ATI and ATII cell fate are not detected before 16 weeks of gestation (canalicular stage) and are not robust even at 20 weeks of gestation. It is likely that epithelial bud tip progenitors choose their eventual alveolar cell fate at the molecular level days before becoming morphologically and functionally distinct in mice, but alveolar specification may occur much later in humans. This data also poses a question about whether or not an alveolar progenitor cell state even exists; it is possible that ATI and ATII cells are directly specified from multipotent bud tip progenitor cells, obfuscating the timing of when it is appropriate to term a bud tip progenitor an alveolar progenitor.
In mice and humans, both paracrine signals from the mesenchyme and endocrine signals involving glucocorticoids appear to direct bud tip progenitor cells to give rise to alveolar cells.[90,100–104] Premature human infants are frequently given glucocorticoids in order to speed the maturation of ATII cells such that they begin producing surfactant to have functional lungs.[105–109] Although glucocorticoids are used to mature already specified alveolar cells in the human, studies using mice suggest that endocrine glucocorticoid signaling drives the formation of the BADJ. Interestingly, manipulation of glucocorticoid signal timing or strength alters the size of the future alveolar compartment of the lung without disrupting the appearance of mature alveolar cell types,[93,96] suggesting that glucocorticoid signaling acts to restrict the developmental potential of bud tip progenitor cells away from airway fates without being required for alveolar differentiation. Laresgoiti et al. showed that glucocorticoid signaling may interact with inflammatory pathways via STAT3 to initiate the switch from bud tip progenitors giving rise to airway cell types to alveolar cell types in the mouse.[93] Beyond this data, the signaling mechanisms that glucocorticoid signaling works through to propel alveolar formation and maturation are unknown, and given the clinical applications of glucocorticoid signaling in the developing human lung, understanding the precise role of glucocorticoid signaling in alveolar specification could have drastic impacts on preventing chronic respiratory disease in premature infants.
Signals originating locally from the mesenchyme are also involved in regulating the differentiation of bud tip progenitor cells into alveolar cells. Mesenchyme surrounding the bud tips in both humans and mice secrete FGF ligands.[46,76,110] In mice, it has been shown that mesenchyme-derived FGF acts on the epithelium through KRAS to maintain the progenitor state of bud tips.[91,96,111] Secretion of FGF from the mesenchyme is promoted by WNT ligands, which are thought to partially originate from the epithelium, creating a positive feedback loop.[39,112–115] HIPPO signaling terminates branching morphogenesis and promotes alveolar differentiation through degradation of β-catenin in the epithelium, disrupting the WNT-FGF feedback loop and directing bud tip progenitors to differentiate[115,116] (Figure 4a). Active FGF and WNT signaling are known to be important for maintaining bud tip progenitor identity in the human as well[3,4,71,76]; therefore, these pathways may perform analogous roles in maintaining the progenitor state of human canalicular stage bud tips. It is important to note that the specific FGF ligand(s) involved in human bud tip progenitor maintenance are likely different than those in mice.[76] A role for HIPPO signaling in the human lung has not been examined.
4.2. Mesenchyme Development during the Canalicular Stage
While the diversity of mesenchymal cell types in the developing lung is still being uncovered,[17] two distinct mesenchymal populations in mice have been defined to undergo significant changes during the canalicular stage: Fgf10+ mesenchymal cells and Pdgfra+ mesenchymal cells. Importantly, the appearance of lipofibroblasts in the human lung has not been confirmed.[117] However, in mice, Fgf10+ mesenchymal cells give rise to lipofibroblasts (LIFs), which are lipid droplet-containing mesenchymal cells that have a regulatory role during alveolar development.[79] Unlike during the pseudglandular stage when Fgf10+ mesenchymal cells give rise to myofibroblasts (MYFs), smooth muscle cells, and LIFs in the distal lung, the large majority of Fgf10+ cells give rise to LIFs (and other unknown mesenchymal cell types) but not MYFs during the canalicular and later stages of development.[118] Likewise, it was found that the majority of Pdgfra+ mesenchymal cells give rise to MYFs, which lay down much of the ECM important for alveolar formation and function,[119–121] during the canalicular and later stages of development.[121] TGF-β signaling negatively regulates FGF10 signaling in the mesenchyme to control the differentiation of mesenchymal progenitor cells to MYFs versus LIFs such that higher FGF10 signaling favors LIF identify and lower FGF10 signaling favors MYF identity.[79,122,123]
5. Saccular and Alveolar Stages of Respiratory Development
Sacculation and alveologenesis are the terminal stages of lung development, beginning late during development and completing sometime during the first decade of life. The saccular stage culminates in the formation of primitive alveoli called saccules. Saccules are further divided during alveologenesis through a process called septation that maximizes the area available for gasexchange. Similar to previous stages of lung development, formation of saccules (sacculation) and their maturation into alveoli are driven by changes occurring in both the epithelium and mesenchyme. In the epithelium, ATI cells transition from a cuboidal to a squamous morphology and then stretch to 10x their original size to form the majority of the surface area within the alveolar epithelium[124] (Figure 4b). ATII cells become highly proliferative and build specialized organelles dedicated to surfactant production called lamellar bodies (Figure 4b). Meanwhile, new cell types appear in the alveolar mesenchyme that secrete extracellular matrix (ECM) and further remodel it, thus contributing to development of saccules and setting the stage for further septation during alveologenesis. Importantly, defects in these late stages of lung development in model organisms mimic features of bronchopulmonary dysplasia,[125–129] a disease prevalent in premature births that leads to chronic respiratory difficulty throughout life. Thus, the mechanisms that ensure proper sacculation and alveologenesis are of great importance for developing interventions that will rescue lung function in the neonatal ward.
5.1. Sacculation and Alveologenesis—Signaling Active in the Epithelium
Sacculation occurs relatively late in human gestation (third trimester), making access to human lung tissue at this stage rare. To circumvent this limitation, several groups have developed methods to differentiate hPSCs into progenitors of the lung epithelium,[33,34,36,52,63,66] which can then give rise to alveolar cell types, partially recapitulating the development of the lung epithelium during sacculation and alveologenesis. Methods also exist to generate more purified populations of alveolar cells called alveolospheres,[22,34,101,102,130] which have already proven useful for modeling congenital disease of the alveoli[131,132] and alveolar injury.[133] These methods, although state-of-the-art, provide an incomplete picture of sacculation and alveolar development in humans because they either lack mesenchyme,[63,66,101] require exogenous mesenchyme for alveolar differentiation,[34,102] generate immature alveolar cells stochastically,[33,36,52,63,66] or give rise to ATII cells only.[101,102] Furthermore, although the methods mentioned above generate alveolar cells, it is unknown whether these cells pass through intermediate states that represent the true signaling, timing, and cell fate trajectories that occur in vivo. Nevertheless, establishment of these in vitro models has provided insights into cues necessary for alveolar cell specification and maturation in humans.
Methods to generate alveolospheres generally follow directed differentiation paradigms in order to induce lung progenitors from anterior foregut endoderm progenitors, followed by purification of putative alveolar progenitors. Alveolar progenitors are placed into various media types, but common to many protocols is the stimulation of cyclic AMP (cAMP) as well as the WNT, FGF, and glucocorticoid signaling pathways.[34,101,102] These studies suggest that WNT, FGF, glucocorticoid, and cAMP signaling pathways act to specify and/or mature alveolar cells in humans (Figure 4c).
A role for WNT in ATII cell specification and/or maturation is consistent with insights from animal models. During late sacculation and early alveologenesis, ATII cells exhibit an increase in WNT signaling activity that correlates with an expansion in ATII cell number,[11] and constitutive WNT increases ATII cell number while loss of β-catenin during sacculation reduces ATII cell number and leads to an increase in ATI cell number.[11] Together, this suggests that WNT signaling promotes the proliferation of ATII cells and may regulate the identity of alveolar progenitor progeny. The role of FGF and cAMP signaling in alveolar cell fate specification in animal models has yet to be elucidated.
Studies from animal models also suggest a key role for the HIPPO signaling pathway in promoting ATI cell fate. HIPPO signaling pathway mouse mutants exhibit defects in saccular architecture that phenocopy aspects of human emphysema and bronchopulmonary dysplasia.[127,128,134] Of note, mutations in HIPPO signaling pathway members leading to overactive TAZ activity generate lung epithelium with precocious and ectopic expression of markers of ATI cell identity,[135] suggesting that defects in the saccular architecture reflect a specific role for TAZ in promoting ATI cell fate. Intriguingly, physical association between the lung epithelial transcription factor NKX2.1 and TAZ has been demonstrated,[136] and more recently, NKX2.1 was demonstrated to perform a role in ATI cell specification distinct from its earlier role in specification of lung epithelium,[137] suggesting that NKX2.1 and TAZ may partner to drive development of ATI cells. Despite the important role of HIPPO signaling in lung epithelial progenitor specification (see Canalicular section) and the development of ATI cells in mice, the dynamics of HIPPO signaling in human models of lung development are not yet known.
5.2. Sacculation and Alveologenesis—Contributions from the Mesenchyme
Three major populations of alveolar fibroblasts have been defined that guide the development of alveolar epithelium through sacculation during the formation of mature alveoli. MYFs, expressing α-SMA, localize to developing septal tips where they remodel existing networks of elastin, which is necessary for proper formation of alveoli and provides elasticity for the lung during respiration.[119,129,138–140] Cues for remodeling the lung ECM may be primarily physical, as stretching induces the activity of elastase.[88] MYFs are also thought to play a key role in driving secondary septation during alveologenesis.[119,138,141] Similar to MYFs, matrix fibroblasts are intimately associated with the saccule during its development and are distinguished from MYFs by high levels of PDGF signaling activity and high levels of WNT5a production.[120,142] Matrix fibroblasts secret collagen and other ECM components[142] that are essential for sacculation and alveolar maturation.[143–147] In contrast to MYFs and matrix fibroblasts, which are thought to play more structural roles in sacculation and alveolar maturation, LIFs are thought to guide development and maturation of ATII cells through trafficking of lipids to ATII cells to assist in production of surfactant.[148] Interestingly, development of LIFs is dependent on signaling from ATII cells, which secrete PTHRP to antagonize Hedgehog and WNT signaling in LIF progenitors, which in turn leads to PPARγ mediated transcription of Leptin and ADRP,[149–151] molecules that induce surfactant production in ATII cells,[150,152] thus linking the co-maturation of ATII cells and LIFs.
hPSC-derived models of human lung development highlight the important contribution of mesenchyme to human sacculation and alveologenesis. For instance, hPSC-derived alveolospheres normally contain only ATII cells[101] but will give rise to cells with features of ATI cells when co-cultured with fetal lung fibroblasts (Figure 4c).[34,102] Likewise, fetal-derived lung bud tip progenitors cultured in vitro readily differentiate into airway cell types but require co-culture with fetal lung mesenchyme for alveolar cell fate specification to occur.[3] Together, these studies suggest that human fetal lung mesenchyme provides cues that induce alveolar cell fates in human lung epithelium. Mesenchyme-derived cues for alveolar cell fate specification are likely partially ECM-derived, as decellularized lung ECM supports the development of multiple alveolar cell types in hPSC-derived lung epithelium.[153,154] Notably, many hPSC-derived alveolospheres are grown in hydrogels that do not necessarily recapitulate the properties of the lung ECM during alveolar development. How ECM instructs alveolar differentiation is not known, but given the mechanosensitivity of the HIPPO signaling pathway, and the evidence for a central role of HIPPO signaling in ATI cell specification and maturation,[116,128,134,135,155] it is tempting to speculate that an ECM-to-HIPPO signaling axis guides the development of ATI cells in vivo. A greater understanding of the roles of mesenchyme during sacculation and alveologenesis will be essential to recapitulate cues that instruct hPSC-derived lung epithelium to specify alveolar cells.
6. Conclusions and Future Directions
We can never fully understand the unique aspects of human respiratory development without the use of in vitro model systems. In order to continue answering unknown questions in human respiratory development and properly model disease and genetic defects, several challenges must be overcome (Table 1). For example, most in vitro human lung model systems are still overly simplistic, where the epithelium is cultured alone and relies on the addition of signaling components to media, or where epithelium is co-cultured with poorly characterized mesenchymal cells that organize in an unclear way. Neither of these approaches meticulously recapitulate an in vivo environment, and it would be invaluable to develop model systems where the mesenchyme and epithelium are cultured together in the correct organization. It is also important to note that these systems often lack a functional vasculature and a nervous system, although efforts to improve complexity have been reported recently.[156,157] Access to developing human tissues as well as the advancement of technologies such as scRNAseq coupled with in situ hybridization and immunofluorescence have begun to provide temporal and spatial gene expression patterns and have laid a strong foundation for the description of cell types, cell type-associated gene expression signatures, transcription factors, and signaling pathway components.[14–16,63] Translation of genetic manipulation techniques such as CRISPR to in vitro human model systems is evolving and will be instrumental to the functional understanding of signaling pathways during human lung development.[158] In addition to molecular mechanisms and cellular functions guiding lung development, it is also appreciated that mechanical cues play important roles in lung development and function; thus, establishing complex in vitro human model systems that incorporate and/or mimic aspects cellular, signaling, and biomechanical cues important for human lung development remains a critical obstacle. Current challenges include incorporating mechanical forces that occur during lung development, such as local forces involved in branching morphogenesis, peristaltic contractions observed in the developing lung, blood sheer stress, transmural pressure, and surface tension.[84,85,88,89] It is likely that lung-on-chip[159] and microfluidic technologies[85] will serve as useful tools to understand the influence of mechanical forces on human lung development. As all of these technologies continue to be integrated into in vitro human model systems of respiratory development, we will better appreciate the mechanisms conserved among species as well as the uniqueness of human biology.
Table 1.
Summary of the major unknowns in human lung development.
| Respiratory specification | Branching morphogenesis |
|---|---|
| The role of SHH and FGF signaling in NKX2.1+ lung progenitor cell specification | The molecular mechanisms of FGF and WNT in branching morphogenesis |
| The signaling required for respiratory cell specification vs organization | Signaling mechanisms for mesenchymal cell maintenance and differentiation |
| Mechanisms involved in distal pulmonary mesenchymal cell specification during the embryonic stage | The role physical pressures (i.e., thoracic cavity) have on branching morphogenesis |
| Alveolar cell fate specification and maturation | |
| The timing of alveolar cell specification | |
| Signaling pathways regulating ATI vs ATII cell fate choice | |
| The role of glucocorticoid signaling in alveolar cell fate specificiation/maturation | |
| The role of mesenchyme-epithelial cross-talk in alveolar cell fate specification/maturation | |
In vitro models of human lung development will likely also play an important role in personalized medicine. With the ability to use cultured primary patient tissue or generate patient-derived induced pluripotent stem cells to generate human in vitro models, we have the capability to model human lung disease and perform large-scale screens for patient-specific reactions to toxins, new drugs, and therapies. This could serve as a powerful tool for diseases such as cystic fibrosis and COPD, where current therapies are often ineffective or can be extremely costly; personalized screens could save months of trial-and-error with various medications to determine the optimal drug regimen for a patient.[160,161] As chronic lung disease is a major cause of death worldwide,[160] the need for new therapies and better treatments is critical, and in vitro model systems of the human lung will provide a high-throughput opportunity to develop personalized treatments for lung diseases.
Acknowledgements
J.R.S. was supported by the National Heart, Lung, and Blood Institute (NHLBI - R01HL119215) and by the Cystic Fibrosis Foundation. This project has been made possible in part by grant number CZF2019-002440 from the Chan Zuckerberg Initiative DAF, an advised fund of Silicon Valley Community Foundation. R.F.C. was supported by the NIH Tissue Engineering, Regenerative Medicine Training Grant (NIH T-32-DE00007057-43) and by the University of Michigan Cell and Developmental Biology Patten Award. A.S.C. was funded by the T32 Michigan Medical Scientist Training Program (5T32GM007863-40).
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Renee F. Conway, Department of Cell and Developmental Biology, University of Michigan Medical School, Ann Arbor, MI 48104, USA
Tristan Frum, Department of Internal Medicine, Gastroenterology, University of Michigan Medical School, Ann Arbor, MI 48104, USA.
Ansley S. Conchola, Cell and Molecular Biology (CMB) Training Program, University of Michigan Medical School, Ann Arbor, MI 48104, USA
Jason R. Spence, Department of Cell and Developmental Biology, University of Michigan Medical School, Ann Arbor, MI 48104, USA Department of Internal Medicine, Gastroenterology, University of Michigan Medical School, Ann Arbor, MI 48104, USA; Cell and Molecular Biology (CMB) Training Program, University of Michigan Medical School, Ann Arbor, MI 48104, USA; Department of Biomedical Engineering, University of Michigan College of Engineering, Ann Arbor, MI 48104, USA.
References
- [1].Chakraborty M, Kotecha S, Breathe 2013, 9, 476. [Google Scholar]
- [2].Rawlins EL, Dev. Dyn 2011, 240, 463. [DOI] [PubMed] [Google Scholar]
- [3].Nikolić MZ, Caritg O, Jeng Q, Johnson J-A, Sun D, Howell KJ, Brady JL, Laresgoiti U, Allen G, Butler R, Zilbauer M, Giangreco A, Rawlins EL, eLife 2017, 6, e26575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Miller AJ, Hill DR, Nagy MS, Aoki Y, Dye BR, Chin AM, Huang S, Zhu F, White ES, Lama V, Spence JR, Stem Cell Rep. 2018, 10, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Alescio T, Cassini A, J. Exp. Zool 1962, 150, 83. [DOI] [PubMed] [Google Scholar]
- [6].Tollet J, Everett AW, Sparrow MP, Dev. Dyn 2001, 221, 48. [DOI] [PubMed] [Google Scholar]
- [7].Lazarus A, Del-Moral PM, Ilovich O, Mishani E, Warburton D, Keshet E, Development 2011, 138, 2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zepp JA, Morrisey EE, Nat. Rev. Mol. Cell Biol 2019, 20, 551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Schittny JC, Cell Tissue Res. 2017, 367, 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Metzger RJ, Klein OD, Martin GR, Krasnow MA, Nature 2008, 453, 745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Frank DB, Peng T, Zepp JA, Snitow M, Vincent TL, Penkala IJ, Cui Z, Herriges MJ, Morley MP, Zhou S, Lu MM, Morrisey EE, Cell Rep. 2016, 17, 2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Rock JR, Hogan BLM, Annu. Rev. Cell Dev. Biol 2011, 27, 493. [DOI] [PubMed] [Google Scholar]
- [13].Perrin S, Nature 2014, 507, 423. [DOI] [PubMed] [Google Scholar]
- [14].Treutlein B, Brownfield DG, Wu AR, Neff NF, Mantalas GL, Espinoza FH, Desai TJ, Krasnow MA, Quake SR, Nature 2014, 509, 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Brazovskaja A, Treutlein B, Camp JG, Curr. Opin. Biotechnol 2019, 55, 167. [DOI] [PubMed] [Google Scholar]
- [16].Travaglini KJ, Nabhan AN, Penland L, Sinha R, Gillich A, Sit RV, Chang S, Conley SD, Mori Y, Seita J, Berry GJ, Shrager JB, Metzger RJ, Kuo CS, Neff N, Weissman IL, Quake SR, Krasnow MA, bioRxiv 2019, 742320. [Google Scholar]
- [17].Guo M, Du Y, Gokey JJ, Ray S, Bell SM, Adam M, Sudha P, Perl AK, Deshmukh H, Potter SS, Whitsett JA, Xu Y, Nat. Commun 2019, 10, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zaw-Tun HA, Cells Tissues Organs 1982, 114, 1. [Google Scholar]
- [19].Perl A-KT, Wert SE, Nagy A, Lobe CG, Whitsett JA, Proc. Natl. Acad. Sci. USA 2002, 99, 10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Que J, Okubo T, Goldenring JR, Nam K-T, Kurotani R, Morrisey EE, Taranova O, Pevny LH, Hogan BLM, Development 2007, 134, 2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lazzaro D, Price M, de Felice M, Di Lauro R, Development 1991, 113, 1093. [DOI] [PubMed] [Google Scholar]
- [22].Hawkins F, Kramer P, Jacob A, Driver I, Thomas DC, McCauley KB, Skvir N, Crane AM, Kurmann AA, Hollenberg AN, Nguyen S, Wong BG, Khalil AS, Huang SXL, Guttentag S, Rock JR, Shannon JM, Davis BR, Kotton DN, J. Clin. Invest 2017, 127, 2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Minoo P, Hamdan H, Bu D, Warburton D, Stepanik P, deLemos R, Dev. Biol 1995, 172, 694. [DOI] [PubMed] [Google Scholar]
- [24].Warburton D, Bellusci S, De Langhe S, Del Moral P-M, Fleury V, Mailleux A, Tefft D, Unbekandt M, Wang K, Shi W, Pediatr. Res 2005, 57, 26R. [DOI] [PubMed] [Google Scholar]
- [25].V Cardoso W, Lü J, Xue Y, Hogan BLM, Development 2006, 133, 1611. [DOI] [PubMed] [Google Scholar]
- [26].Morrisey EE, Hogan BLM, Dev. Cell 2010, 18, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Rankin SA, Zorn AM, J. Cell. Biochem 2014, 115, 1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Swarr DT, Morrisey EE, Annu. Rev. Cell Dev. Biol 2015, 31, 553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].D’Amour KA, Bang AG, Eliazer S, Kelly OG, Agulnick AD, Smart NG, Moorman MA, Kroon E, Carpenter MK, Baetge EE, Nat. Biotechnol 2006, 24, 1392. [DOI] [PubMed] [Google Scholar]
- [30].Loh KM, Ang LT, Zhang J, Kumar V, Ang J, Auyeong JQ, Lee KL, Choo SH, Lim CYY, Nichane M, Tan J, Noghabi MS, Azzola L, Ng ES, Durruthy-Durruthy J, Sebastiano V, Poellinger L, Elefanty AG, Stanley EG, Chen Q, Prabhakar S, Weissman IL, Lim B, Cell Stem Cell 2014, 14, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Green MD, Chen A, Nostro M-C, D’Souza SL, Schaniel C, Lemischka IR, Gouon-Evans V, Keller G, Snoeck H-W, Nat. Biotechnol 2011, 29, 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wong AP, Bear CE, Chin S, Pasceri P, Thompson TO, Huan L-J, Ratjen F, Ellis J, Rossant J, Nat. Biotechnol 2012, 30, 876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Huang SXL, Islam MN, O’Neill J, Hu Z, Yang Y-G, Chen Y-W, Mumau M, Green MD, Vunjak-Novakovic G, Bhattacharya J, Snoeck H-W, Nat. Biotechnol 2014, 32, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gotoh S, Ito I, Nagasaki T, Yamamoto Y, Konishi S, Korogi Y, Matsumoto H, Muro S, Hirai T, Funato M, Mae S-I, Toyoda T, Sato-Otsubo A, Ogawa S, Osafune K, Mishima M, Stem Cell Rep. 2014, 3, 394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Huang SXL, Green MD, de Carvalho AT, Mumau M, Chen Y-W, D’Souza SL, Snoeck H-W, Nat. Protoc 2015, 10, 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Dye BR, Hill DR, Ferguson MA, Tsai Y-H, Nagy MS, Dyal R, Wells JM, Mayhew CN, Nattiv R, Klein OD, White ES, Deutsch GH, Spence JR, eLife 2015, 4, e05098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Rankin SA, Han L, McCracken KW, Kenny AP, Anglin CT, Grigg EA, Crawford CM, Wells JM, Shannon JM, Zorn AM, Cell Rep. 2016, 16, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Serra M, Alysandratos K-D, Hawkins F, McCauley KB, Jacob A, Choi J, Caballero IS, Vedaie M, Kurmann AA, Ikonomou L, Hollenberg AN, Shannon JM, Kotton DN, Development 2017, 144, 3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Goss AM, Tian Y, Tsukiyama T, Cohen ED, Zhou D, Lu MM, Yamaguchi TP, Morrisey EE, Dev. Cell 2009, 17, 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Harris-Johnson KS, Domyan ET, Vezina CM, Sun X, Proc. Natl. Acad. Sci. USA 2009, 106, 16287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Szenker-Ravi E, Altunoglu U, Leushacke M, Bosso-Lefèvre C, Khatoo M, Thi Tran H, Naert T, Noelanders R, Hajamohideen A, Beneteau C, de Sousa SB, Karaman B, Latypova X, Basaran S, Yücel EB, Tan TT, Vlaeminck L, Nayak SS, Shukla A, Girisha KM, Le Caignec C, Soshnikova N, Uyguner ZO, Vleminckx K, Barker N, Kayserili H, Reversade B, Nature 2018, 557, 564. [DOI] [PubMed] [Google Scholar]
- [42].Rankin SA, McCracken KW, Luedeke DM, Han L, Wells JM, Shannon JM, Zorn AM, Dev. Biol 2018, 434, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Litingtung Y, Lei L, Westphal H, Chiang C, Nat. Genet 1998, 20, 58. [DOI] [PubMed] [Google Scholar]
- [44].Motoyama J, Liu J, Mo R, Ding Q, Post M, Hui C, Nat. Genet 1998, 20, 54. [DOI] [PubMed] [Google Scholar]
- [45].Domyan ET, Ferretti E, Throckmorton K, Mishina Y, Nicolis SK, Sun X, Development 2011, 138, 971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Bellusci S, Grindley J, Emoto H, Itoh N, Hogan BL, Development 1997, 124, 4867. [DOI] [PubMed] [Google Scholar]
- [47].Min H, Danilenko DM, Scully SA, Bolon B, Ring BD, Tarpley JE, DeRose M, Simonet WS, Genes Dev. 1998, 12, 3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, Sato T, Yagishita N, Matsui D, Koga Y, Itoh N, Kato S, Nat. Genet 1999, 21, 138. [DOI] [PubMed] [Google Scholar]
- [49].Serls AE, Doherty S, Parvatiyar P, Wells JM, Deutsch GH, Development 2005, 132, 35. [DOI] [PubMed] [Google Scholar]
- [50].McCracken KW, Catá EM, Crawford CM, Sinagoga KL, Schumacher M, Rockich BE, Tsai Y-H, Mayhew CN, Spence JR, Zavros Y, Wells JM, Nature 2014, 516, 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Dye BR, Dedhia PH, Miller AJ, Nagy MS, White ES, Shea LD, Spence JR, eLife 2016, 5, e19732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Miller AJ, Dye BR, Ferrer-Torres D, Hill DR, Overeem AW, Shea LD, Spence JR, Nat. Protoc 2019, 14, 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Arora R, Metzger RJ, Papaioannou VE, PLoS Genet. 2012, 8, e1002866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kishimoto K, Furukawa KT, LuzMadrigal A, Yamaoka A, Matsuoka C, Habu M, Alev C, Zorn AM, Morimoto M, bioRxiv 2019, 758235. [Google Scholar]
- [55].Peng T, Tian Y, Boogerd CJ, Lu MM, Kadzik RS, Stewart KM, Evans SM, Morrisey EE, Nature 2013, 500, 589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Han L, Koike H, Chaturvedi P, Kishimoto K, Iwasawa K, Giesbrecht K, Witcher PC, Eicher A, Nasr T, Haines L, Shannon JM, Morimoto M, Wells JM, Takebe T, Zorn AM, bioRxiv 2019, 756825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Hogan BLM, Yinalina JM, Curr. Opin. Genet. Dev 1998, 8, 481. [DOI] [PubMed] [Google Scholar]
- [58].Dye BR, Miller AJ, Spence JR, Curr. Pathobiol. Rep 2016, 4, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Miller AJ, Spence JR, Physiology 2017, 32, 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Irvin CG, Bates JHT, Respir. Res 2003, 4, 10.1186/rr199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Rawlins EL, Clark CP, Xue Y, Hogan BLM, Development 2009, 136, 3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Yang Y, Riccio P, Schotsaert M, Mori M, Lu J, Lee D-K, García-Sastre A, Xu J, Cardoso WV, Dev. Cell 2018, 44, 752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Miller AJ, Yu Q, Czerwinski M, Tsai Y-H, Conway RF, Wu A, Holloway EM, Walker T, Glass IA, Treutlein B, Camp JG, Spence JR, Dev. Cell 2020, in press, pii: S1534–5807(20)30065–4. 10.1016/j.devcel.2020.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Mou H, Vinarsky V, Tata PR, Brazauskas K, Choi SH, Crooke AK, Zhang B, Solomon GM, Turner B, Bihler H, Harrington J, Lapey A, Channick C, Keyes C, Freund A, Artandi S, Mense M, Rowe S, Engelhardt JF, Hsu Y-C, Rajagopal J, Cell Stem Cell 2016, 19, 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Konishi S, Gotoh S, Tateishi K, Yamamoto Y, Korogi Y, Nagasaki T, Matsumoto H, Muro S, Hirai T, Ito I, Tsukita S, Mishima M, Stem Cell Rep. 2016, 6, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Chen Y-W, Huang SX, de Carvalho ALRT, Ho S-H, Islam MN, Volpi S, Notarangelo LD, Ciancanelli M, Casanova J-L, Bhattacharya J, Liang AF, Palermo LM, Porotto M, Moscona A, Snoeck H-W, Nat. Cell Biol 2017, 19, 542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].McCauley KB, Hawkins F, Serra M, Thomas DC, Jacob A, Kotton DN, Cell Stem Cell 2017, 20, 844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Okubo T, Development 2005, 132, 1363. [DOI] [PubMed] [Google Scholar]
- [69].Rockich BE, Hrycaj SM, Shih HP, Nagy MS, Ferguson MAH, Kopp JL, Sander M, Wellik DM, Spence JR, Proc. Natl. Acad. Sci. USA 2013, 110, E4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Danopoulos S, Krainock M, Toubat O, Thornton M, Grubbs B, Al Alam D, Am. J. Physiol. Lung Cell Mol. Physiol 2016, 311, L1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Danopoulos S, Alonso I, Thornton ME, Grubbs BH, Bellusci S, Warburton D, Al Alam D, Am. J. Physiol. Lung Cell Mol. Physiol 2018, 314, L144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Gontan C, de Munck A, Vermeij M, Grosveld F, Tibboel D, Rottier R, Dev. Biol 2008, 317, 296. [DOI] [PubMed] [Google Scholar]
- [73].Prince LS, Front. Genet 2018, 9, 10.3389/fgene.2018.00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Hines EA, Sun X, J. Cell. Biochem 2014, 115, 1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].McCulley D, Wienhold M, Sun X, Curr. Opin. Genet. Dev 2015, 32, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Danopoulos S, Thornton ME, Grubbs BH, Frey MR, Warburton D, Bellusci S, Al Alam D, J. Pathol 2018, 247, 5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Danopoulos S, Shiosaki J, Al Alam D, Front. Genet 2019, 10, 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Abler LL, Mansour SL, Sun X, Dev. Dyn 2009, 238, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Al Alam D, El Agha E, Sakurai R, Kheirollahi V, Moiseenko A, Danopoulos S, Shrestha A, Schmoldt C, Quantius J, Herold S, Chao C-M, Tiozzo C, De Langhe S, V Plikus M, Thornton M, Grubbs B, Minoo P, Rehan VK, Bellusci S, Development 2015, 142, 4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Jones MR, Dilai S, Lingampally A, Chao C-M, Danopoulos S, Carraro G, Mukhametshina R, Wilhelm J, Baumgart-Vogt E, Al Alam D, Chen C, Minoo P, Zhang JS, Bellusci S, Front. Genet 2019, 9, 746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Mucenski ML, Wert SE, Nation JM, Loudy DE, Huelsken J, Birchmeier W, Morrisey EE, J. Biol. Chem 2003, 278, 40231. [DOI] [PubMed] [Google Scholar]
- [82].Bell SM, Schreiner CM, Wert SE, Mucenski ML, Scott WJ, Whitsett JA, Development 2008, 135, 1049. [DOI] [PubMed] [Google Scholar]
- [83].Danopoulos S, Bhattacharya S, Mariani TJ, Al Alam D, Eur. Respir. J 2019, 1900746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Varner VD, Gleghorn JP, Miller E, Radisky DC, Nelson CM, Proc. Natl. Acad. Sci. USA 2015, 112, 9230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Nelson CM, Gleghorn JP, Pang MF, Jaslove JM, Goodwin K, Varner VD, Miller E, Radisky DC, Stone HA, Development 2017, 144, 4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Mailleux AA, Kelly R, Veltmaat JM, De Langhe SP, Zaffran S, Thiery JP, Bellusci S, Development 2005, 132, 2157. [DOI] [PubMed] [Google Scholar]
- [87].Miller MF, Cohen ED, Baggs JE, Lu MM, Hogenesch JB, Morrisey EE, Proc. Natl. Acad. Sci. USA 2012, 109, 15348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Kim HY, Pang M-F, Varner VD, Kojima L, Miller E, Radisky DC, Nelson CM, Dev. Cell 2015, 34, 719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Goodwin K, Mao S, Guyomar T, Miller E, Radisky DC, Košmrlj A, Nelson CM, Development 2019, 146, dev181172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Shannon JM, Gebb SA, Nielsen LD, Development 1999, 126, 1675. [DOI] [PubMed] [Google Scholar]
- [91].Chang DR, Martinez Alanis D, Miller RK, Ji H, Akiyama H, McCrea PD, Chen J, Proc. Natl. Acad. Sci. USA 2013, 110, 18042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Desai TJ, Brownfield DG, Krasnow MA, Nature 2014, 507, 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Laresgoiti U, Nikolić MZ, Rao C, Brady JL, Richardson RV, Batchen EJ, Chapman KE, Rawlins EL, Development 2016, 143, 3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Frank DB, Penkala IJ, Zepp JA, Sivakumar A, Linares-Saldana R, Zacharias WJ, Stolz KG, Pankin J, Lu M, Wang Q, Babu A, Li L, Zhou S, Morley MP, Jain R, Morrisey EE, Proc. Natl. Acad. Sci. USA 2019, 116, 4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Bal HS, Ghoshal NG, Lab. Anim 1988, 22, 76. [DOI] [PubMed] [Google Scholar]
- [96].Alanis DM, Chang DR, Akiyama H, Krasnow MA, Chen J, Nat. Commun 2014, 5, 3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Cole TJ, Solomon NM, Van Driel R, Monk JA, Bird D, Richardson SJ, Dilley RJ, Hooper SB, Am. J. Respir. Cell Mol. Biol 2004, 30, 613. [DOI] [PubMed] [Google Scholar]
- [98].Manwani N, Gagnon S, Post M, Joza S, Muglia L, Cornejo S, Kaplan F, Sweezey NB, Am. J. Respir. Cell Mol. Biol 2010, 43, 599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Adamson IY, Bowden DH, Lab. Invest 1975, 32, 736. [PubMed] [Google Scholar]
- [100].Deterding RR, Shimizu H, Fisher JH, Shannon JM, Am. J. Respir. Cell Mol. Biol 1994, 10, 30. [DOI] [PubMed] [Google Scholar]
- [101].Jacob A, Morley M, Hawkins F, McCauley KB, Jean JC, Heins H, Na C-L, Weaver TE, Vedaie M, Hurley K, Hinds A, Russo SJ, Kook S, Zacharias W, Ochs M, Traber K, Quinton LJ, Crane A, Davis BR, White FV, Wambach J, Whitsett JA, Cole FS, Morrisey EE, Guttentag SH, Beers MF, Kotton DN, Cell Stem Cell 2017, 21, 472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Yamamoto Y, Gotoh S, Korogi Y, Seki M, Konishi S, Ikeo S, Sone N, Nagasaki T, Matsumoto H, Muro S, Ito I, Hirai T, Kohno T, Suzuki Y, Mishima M, Nat. Methods 2017, 14, 1097. [DOI] [PubMed] [Google Scholar]
- [103].Sucre JMS, Jetter CS, Loomans H, Williams J, Plosa EJ, Benjamin JT, Young LR, Kropski JA, Calvi CL, Kook S, Wang P, Gleaves L, Eskaros A, Goetzl L, Blackwell TS, Guttentag SH, Zijlstra A, Am. J. Respir. Cell Mol. Biol 2018, 59, 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].de Carvalho ALRT, Strikoudis A, Liu H-Y, Chen Y-W, Dantas TJ, Vallee RB, Correia-Pinto J, Snoeck H-W, Development 2019, 146, dev171652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Liggins GC, Howie RN, Pediatrics 1972, 50, 515. [PubMed] [Google Scholar]
- [106].Dluholucký S, Babic J, Taufer I, Arch. Dis. Child 1976, 51, 420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Morrison JC, Whybrew WD, Bucovaz ET, Schneider JM, Am. J. Obstet. Gynecol 1978, 131, 358. [DOI] [PubMed] [Google Scholar]
- [108].Baud O, Maury L, Lebail F, Ramful D, El Moussawi F, Nicaise C, Zupan-Simunek V, Coursol A, Beuchée A, Bolot P, Andrini P, Mohamed D, Alberti C, Lancet 2016, 387, 1827. [DOI] [PubMed] [Google Scholar]
- [109].Roberts D, Brown J, Medley N, Dalziel SR, Cochrane database Syst. Rev 2017, 3, CD004454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Hirashima T, Iwasa Y, Morishita Y, Dev. Dyn 2009, 238, 2813. [DOI] [PubMed] [Google Scholar]
- [111].Volckaert T, De Langhe SP, Dev. Dyn 2015, 244, 342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Chen F, Cao Y, Qian J, Shao F, Niederreither K, Cardoso WV, J. Clin. Invest 2010, 120, 2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Goss AM, Tian Y, Cheng L, Yang J, Zhou D, Cohen ED, Morrisey EE, Dev. Biol 2011, 356, 541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Volckaert T, Campbell A, Dill E, Li C, Minoo P, De Langhe S, Development 2013, 140, 3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Volckaert T, Yuan T, Chao C-M, Bell H, Sitaula A, Szimmtenings L, El Agha E, Chanda D, Majka S, Bellusci S, Thannickal VJ, Fässler R, De Langhe SP, Dev. Cell 2017, 43, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Mahoney JE, Mori M, Szymaniak AD, Varelas X, V Cardoso W, Dev. Cell 2014, 30, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Ahlbrecht K, McGowan SE, American J Physiol. Lung Cell Mol. Physiol 2014, 307, L605. [DOI] [PubMed] [Google Scholar]
- [118].El Agha E, Herold S, Al Alam D, Quantius J, MacKenzie B, Carraro G, Moiseenko A, Chao C-M, Minoo P, Seeger W, Bellusci S, Development 2014, 141, 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Lindahl P, Karlsson L, Hellström M, Gebre-Medhin S, Willetts K, Heath JK, Betsholtz C, Development 1997, 124, 3943. [DOI] [PubMed] [Google Scholar]
- [120].Green J, Endale M, Auer H, Perl A-KT, Am. J. Respir. Cell Mol. Biol 2016, 54, 532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Li R, Bernau K, Sandbo N, Gu J, Preissl S, Sun X, eLife 2018, 7, e36865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].McQualter JL, McCarty RC, Van der Velden J, O’Donoghue RJJ, Asselin-Labat M-L, Bozinovski S, Bertoncello I, Stem Cell Res. 2013, 11, 1222. [DOI] [PubMed] [Google Scholar]
- [123].Li A, Ma S, Smith SM, Lee MK, Fischer A, Borok Z, Bellusci S, Li C, Minoo P, BMC Biol. 2016, 14, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Yang J, Hernandez BJ, Martinez Alanis D, Narvaez del Pilar O, Vila-Ellis L, Akiyama H, Evans SE, Ostrin EJ, Chen J, Development 2016, 143, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Husain AN, Siddiqui NH, Stocker JT, Hum. Pathol 1998, 29, 710. [DOI] [PubMed] [Google Scholar]
- [126].Jobe AJ, Pediatr. Res 1999, 46, 641. [DOI] [PubMed] [Google Scholar]
- [127].Makita R, Uchijima Y, Nishiyama K, Amano T, Chen Q, Takeuchi T, Mitani A, Nagase T, Yatomi Y, Aburatani H, Nakagawa O, V Small E, Cobo-Stark P, Igarashi P, Murakami M, Tominaga J, Sato T, Asano T, Kurihara Y, Kurihara H, Am. J. Physiol. Ren. Physiol 2008, 294, F542. [DOI] [PubMed] [Google Scholar]
- [128].Mitani A, Nagase T, Fukuchi K, Aburatani H, Makita R, Kurihara H, Am J Respir. Crit. Care Med 2009, 180, 326. [DOI] [PubMed] [Google Scholar]
- [129].Branchfield K, Li R, Lungova V, Verheyden JM, McCulley D, Sun X, Dev. Biol 2016, 409, 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Tamò L, Hibaoui Y, Kallol S, Alves MP, Albrecht C, Hostettler KE, Feki A, Rougier J-S, Abriel H, Knudsen L, Gazdhar A, Geiser T, Physiol J. Lung Cell Mol. Physiol 2018, 315, L921. [DOI] [PubMed] [Google Scholar]
- [131].Korogi Y, Gotoh S, Ikeo S, Yamamoto Y, Sone N, Tamai K, Konishi S, Nagasaki T, Matsumoto H, Ito I, Chen-Yoshikawa TF, Date H, Hagiwara M, Asaka I, Hotta A, Mishima M, Hirai T, Stem Cell Rep. 2019, 12, 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Leibel SL, Winquist A, Tseu I, Wang J, Luo D, Shojaie S, Nathan N, Snyder E, Post M, Sci. Rep 2019, 9, 13415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Heo H-R, Kim J, Kim WJ, Yang S-R, Han S-S, Lee SJ, Hong Y, Hong S-H, Sci. Rep 2019, 9, 505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Isago H, Mitani A, Mikami Y, Horie M, Urushiyama H, Hamamoto R, Terasaki Y, Nagase T, Am. J. Respir. Cell Mol. Biol 2019, 62, 256. [DOI] [PubMed] [Google Scholar]
- [135].Nantie LB, Young RE, Paltzer WG, Zhang Y, Johnson RL, Verheyden JM, Sun X, Development 2018, 145, dev163105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Park K-S, Whitsett JA, Di Palma T, Hong J-H, Yaffe MB, Zannini M, J. Biol. Chem 2004, 279, 17384. [DOI] [PubMed] [Google Scholar]
- [137].Little DR, Gerner-Mauro KN, Flodby P, Crandall ED, Borok Z, Akiyama H, Kimura S, Ostrin EJ, Chen J, Proc. Natl. Acad. Sci. USA 2019, 116, 20545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Boström H, Willetts K, Pekny M, Levéen P, Lindahl P, Hedstrand H, Pekna M, Hellström M, Gebre-Medhin S, Schalling M, Nilsson M, Kurland S, Törnell J, Heath JK, Betsholtz C, Cell 1996, 85, 863. [DOI] [PubMed] [Google Scholar]
- [139].Hrycaj SM, Dye BR, Baker NC, Larsen BM, Burke AC, Spence JR, Wellik DM, Cell Rep. 2015, 12, 903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Luo Y, Li N, Chen H, Fernandez GE, Warburton D, Moats R, Mecham RP, Krenitsky D, Pryhuber GS, Shi W, Sci. Rep 2018, 8, 8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].McGowan SE, Grossmann RE, Kimani PW, Holmes AJ, Anat. Rec 2008, 291, 1649. [DOI] [PubMed] [Google Scholar]
- [142].Endale M, Ahlfeld S, Bao E, Chen X, Green J, Bess Z, Weirauch MT, Xu Y, Perl AK, Dev. Biol 2017, 425, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Kida K, Thurlbeck WM, Am. J. Pathol 1980, 101, 693. [PMC free article] [PubMed] [Google Scholar]
- [144].Willem M, Miosge N, Halfter W, Smyth N, Jannetti I, Burghart E, Timpl R, Mayer U, Development 2002, 129, 2711. [DOI] [PubMed] [Google Scholar]
- [145].Bader BL, Smyth N, Nedbal S, Miosge N, Baranowsky A, Mokkapati S, Murshed M, Nischt R, Mol. Cell. Biol 2005, 25, 6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Loscertales M, Nicolaou F, Jeanne M, Longoni M, Gould DB, Sun Y, Maalouf FI, Nagy N, Donahoe PK, BMC Biol. 2016, 14, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Fumoto K, Takigawa-Imamura H, Sumiyama K, Yoshimura SH, Maehara N, Kikuchi A, J. Cell Sci 2019, 132, jcs235556. [DOI] [PubMed] [Google Scholar]
- [148].McGowan SE, Torday JS, Annu. Rev. Physiol 1997, 59, 43. [DOI] [PubMed] [Google Scholar]
- [149].Rubin LP, Kovacs CA, Tsai S-W, Pinar H, Torday JS, Kronenberg HM, Pediatric Research 1997, 41, 266.9029649 [Google Scholar]
- [150].Torday JS, Sun H, Wang L, Torres E, Sunday ME, Rubin LP, Am. J. Physiol. Lung Cell Mol. Physiol 2002, 282, L405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Torday JS, Rehan VK, Pediatr. Res 2006, 60, 382. [DOI] [PubMed] [Google Scholar]
- [152].Rubin LP, Kovacs CS, De Paepe ME, Tsai S-W, Torday JS, Kronenberg HM, Dev. Dyn 2004, 230, 278. [DOI] [PubMed] [Google Scholar]
- [153].Ghaedi M, Calle EA, Mendez JJ, Gard AL, Balestrini J, Booth A, Bove PF, Gui L, White ES, Niklason LE, J. Clin. Invest 2013, 123, 4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Gilpin SE, Ren X, Okamoto T, Guyette JP, Mou H, Rajagopal J, Mathisen DJ, Vacanti JP, Ott HC, Ann. Thoracic Surge 2014, 98, 1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Volckaert T, Yuan T, Yuan J, Boateng E, Hopkins S, Zhang J-S, Thannickal VJ, Fässler R, De Langhe SP, Development 2019, 146, dev166454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Tan Q, Choi KM, Sicard D, Tschumperlin DJ, Biomaterials 2017, 113, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Holloway EM, Capeling MM, Spence JR, Development 2019, 146, dev166173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Howden SE, Vanslambrouck JM, Wilson SB, Tan KS, Little MH, EMBO Rep. 2019, 20, e47483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Stucki JD, Hobi N, Galimov A, Stucki AO, Schneider-Daum N, Lehr C-M, Huwer H, Frick M, Funke-Chambour M, Geiser T, Guenat OT, Sci. Rep 2018, 8, 14359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Ferkol T, Schraufnagel D, Ann. Am. Thorac. Soc 2014, 11, 404. [DOI] [PubMed] [Google Scholar]
- [161].Pittman JE, Ferkol TW, Chest 2015, 148, 533. [DOI] [PMC free article] [PubMed] [Google Scholar]


