Abstract
With the COVID-19 pandemic, the evolutionary fate of SARS-CoV-2 becomes a matter of utmost concern. Mutation D614G in the spike (S) protein has become dominant, and recent evidence suggests it yields a more stable phenotype with higher transmission efficacy. We carry out a structural analysis that provides mechanistic clues on the enhanced infectivity. The D614G substitution creates a sticky packing defect in subunit S1, promoting its association with subunit S2 as a means to stabilize the structure of S1 within the S1/S2 complex. The results raise the therapeutic possibility of immunologically targeting the epitope involved in stabilizing the G614 phenotype as a means of reducing the infection efficacy of SARS-CoV-2. This therapeutic modality would not a-priori interfere directly with current efforts toward the immunological targeting of the RBD epitope; hence, it could be exploited as a complementary treatment.
The virus SARS-CoV-2 arose in China and rapidly propagated leading to the COVID-19 pandemic of 2020. With the awareness of this public health disaster, the evolutionary fate of the virus becomes a matter of the utmost concern.1−4 Its evolutionary change is likely to impact key functionalities associated with transmission efficacy and severity, bearing directly on the course of the pandemic.
Korber et al.1 provided epidemiological evidence that the amino acid substitution D614G in SARS-CoV-2 spike protein is rapidly becoming dominant, suggesting that the G614 mutant may entail a significant fitness advantage. Since the spike (S) protein mediates the viral recognition of the host receptor (angiotensin-converting enzyme 2, hACE2) as well as the fusion of viral and cellular membrane, it becomes imperative to find out how this mutation affects viral transmission and infectivity. Korber et al.1 argued that the rapid spread of the G614 mutant is related to higher infectivity. To support this hypothesis, they showed that the G614 variant yields higher titers in pseudoviruses from in vitro experiments. These results appear to be corroborated by other groups.2,3 In infected individuals, the variant G614 is likely to cause higher upper respiratory tract viral loads, yet the severity of the disease does not appear to increase. These findings prompt a mechanistic assessment of the impact of the S-protein mutation on virus transmission as a guidance to steer the development of novel immunological cures.
Transmission Efficacy of SARS-CoV-2 G614 Variant
The two competing phenotypes SD614 and SG614 were recently characterized and compared by Zhang et al.4 Their analysis revealed that retroviruses pseudotyped with SG614 infected hACE2-expressing cells significantly more efficiently than those with SD614. To interpret the results, we first note that the S protein is organized as a trimer with each monomer containing two tethered and mutually interacting domains S1 and S2, the first involved in receptor binding (RB) through its RBD domain, while S2 mediates the fusion of viral and cellular membranes.1 The greater transmission efficiency of the G614 mutant was proven not to correlate with higher epitope affinity or binding efficacy but with less S1 shedding and greater incorporation of the S protein into the pseudovirion, pointing to a higher stability of the SG614 phenotype.4 Because S1 residue 614 is proximal to the S2 domain in the S-protein quaternary structure, Zhang et al.4 first compared the ratio between the S1 and S2 domains in the virion that might signal altered release or shedding of the S1 domain after cleavage at the S1/S2 junction. The resulting S1:S2 ratio is markedly greater in PVG614 compared with PVD614, implying that G614 stabilizes the interaction between the S1 and S2 domains, which limits S1 shedding.
Structural and Mechanistic Impact of the D614G Mutation
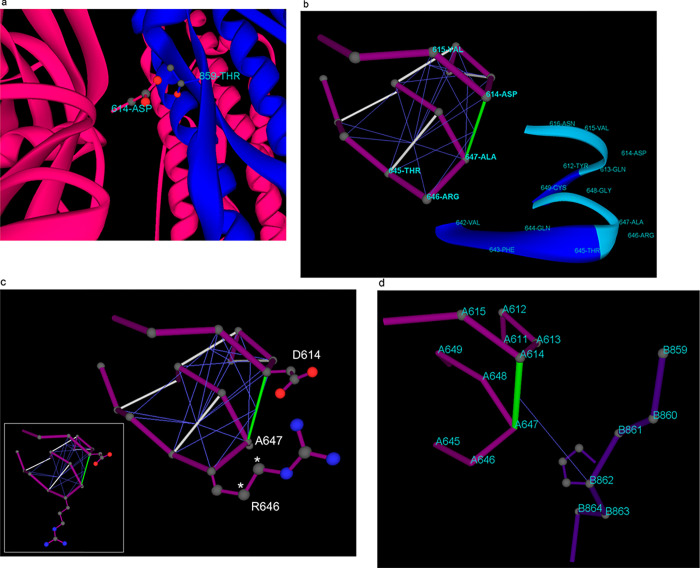
A biophysical/structural analysis of the impact of the D614G substitution on the S1/S2 interface supports this picture, yet as shown subsequently, the analysis needs to go beyond mere structural considerations (cf. ref (1)) and incorporate the relationship between structure and enveloping solvent, the so-called epistructure.5 It has been postulated1 that the carboxyl group in D614 forms a hydrogen bond with the hydroxyl group in T859 across the S1/S2 interface. The crystal structures of the complex do not support this picture, since the side-chain groups are not properly oriented for hydrogen bonding (Figure 1a). Furthermore, this conjecture is completely at odds with the experimental results of Zhang et al.4 because the substitution D614G would eliminate altogether the postulated hydrogen bond across the S1/S2 interface; hence, the SG614 phenotype would be characterized by more S1 shedding compared with SD614. The experimental evidence favors precisely the opposite conclusion.4
Figure 1.
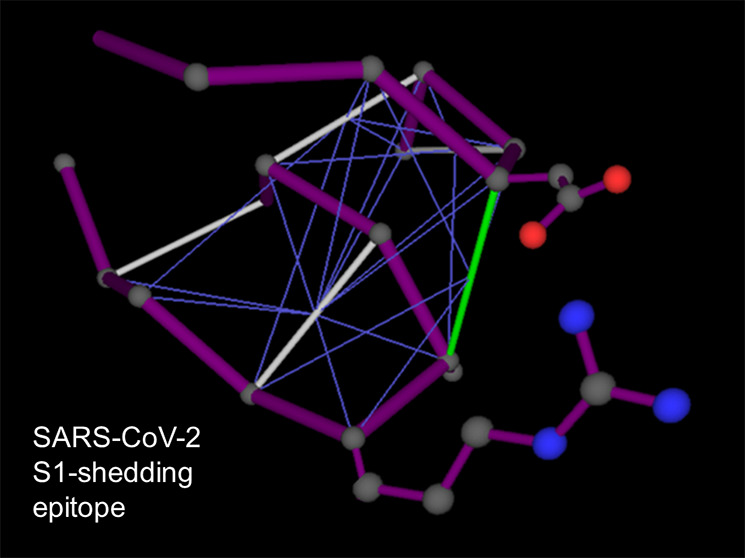
Structural and epistructural interactions at the S1/S2 interface in the spike protein of SARS-CoV-2. (a) Positioning of D614 in the S1 chain (magenta) relative to T859 in the S2 chain (blue) at the S1/S2 interface for the spike protein structure reported in PDB6VXX. The protein backbone is rendered in ribbon representation. (b) Intramolecular wrapping of backbone hydrogen bonds (BHBs) around dehydron D614-A647 for the S1 chain. The protein backbone is represented as a polygonal (magenta) with lines joining the α-carbons of consecutive residues. Well wrapped BHBs are shown as gray lines joining α-carbons of paired residues, while the dehydron is shown in green. The wrapping of each BHB is indicated by thin blue lines from the center of the bond to the α-carbon of the residue that contributes side-chain nonpolar groups to the desolvation domain of the BHB. The ribbon rendering is an aid to the eye. (c) Improved intramolecular wrapping of dehydron D614-A647 achieved by forming the D614-R646 salt bridge. In this configuration, R646 contributes two extra side-chain methylene groups (asterisks) to the wrapping of the D614-A647 BHB when compared with the fully hydrated R646 side chain that occurs when S1 becomes part of the S1/S2 complex (inset). (d) Intermolecular wrapping of dehydron D614-A647 by P862 across the S1/S2 interface. The chains are labeled A and B, corresponding respectively to S1 and S2, as reported in PDB 6VXX.
A more thorough analysis of the impact of the D614G substitution takes into account the epistructure of the interacting S1 and S2 domains. The preformed D614-A647 backbone hydrogen bond (BHB) is partially exposed to the solvent (Figure 1b), and hence it constitutes a special kind of packing defect known as dehydron(5−7) in the S1 domain. From a thermodynamic standpoint, the dehydron is an adhesive spot promoting removal of surrounding water7,8 for two mutually reinforcing reasons:5−7 (a) the preformed BHB gets stabilized when a protein association (i.e., S1/S2 interaction) causes removal of surrounding water thereby hindering the structurally disruptive hydration of amide and carbonyl (destabilizing the unbound state is tantamount to stabilize the bound state), and (b) the removal of backbone-solvating water molecules at the dehydron site is favorable because the partial confinement of such water molecules curtails their hydrogen bonding coordination possibilities; therefore, their transference to the bulk becomes thermodynamically supported.8 The dehydron adhesiveness can also be justified from an energetic standpoint, as the screening of partial charges paired by the BHB is removed concurrently with the removal of surrounding water molecules, thus strengthening the BHB.7
In general, dehydrons compromise the integrity of the protein structure, exposing it to disruptive backbone hydration, and promote protein associations as a means to exclude surrounding water.5−7 In this way, dehydrons become determinants for protein association because, by decreasing charge screening, exogenous removal of water from the dehydron microenvironment strengthens and stabilizes the electrostatic interaction that underlies the BHB. Bioinformatics evidence on the distribution of dehydrons at interfaces of protein complexes supports this picture, pointing to dehydrons as important factors driving complex formation.7
To identify dehydrons in the protein structure, we have introduced a descriptor named wrapping that determines the reliance on binding partnerships to maintain local structural integrity.6,7 A dehydron is then an underwrapped BHB which possesses an insufficient number of side-chain nonpolar groups clustered around the BHB, so that the BHB is exposed to structure-disruptive hydration. Dehydrons and wrapping may be computed directly from structural coordinates.7 The extent of hydrogen-bond wrapping gives the number of side chain nonpolar groups contained within a ‘“desolvation domain”’ (two intersecting balls centered at the α-carbons of the paired residues) that defines the BHB microenvironment in a reported structure. Thus, dehydrons are located in the tail of the distribution of wrapping values across BHBs in a structural database (Methods).7
In our case, the D614-A647 BHB in domain S1 is a dehydron wrapped intramolecularly by residues D614, A647, V615, T645, and R646 (Figure 1b). It achieves maximum wrapping from R646 when this residue forms a salt bridge with D614 (Figure 1c). In this way, the salt bridge D614-R646 contributes to stabilize BHB D614-A647 in the uncomplexed S1, while also blocking the S1/S2 association. As S1 gets associated with S2, the salt bridge gets dismantled (it is not present, as expected, in PDB structure 6VXX) and the dehydron D614-A647 completes its wrapping intermolecularly with a contribuition from P862 from the S2 domain (Figure 1d). The substitution D614G has a major impact on the epistructure of S1, vis-à-vis the previous considerations. The effect results from two contributions: (a) Mutation D614G eliminates the salt bridge that hampers the S1/S2 association as it improves the wrapping of the dehydron pairing residues at locations 614 and 647. (b) By reducing the dehydron-wrapping contributions from side chains at positions 614 and 646, the mutation destabilizes the uncomplexed S1 domain, making the dehydron G614-A647 a better promoter of the S1/S2 association. In the G614 mutant, the 614-647 BHB becomes more reliant on the contribution from P862 that occurs upon S1/S2 association (Figure 1d) to maintain its structural integrity.
In other words, the D614G mutation promotes the S1/S2 association because (a) it destabilizes the free (uncomplexed) S1 structure through the enhanced exposure of the BHB pairing residues G614 and A647 and (b) it stabilizes the S1/S2 interface as the D614G substitution decreases the intramolecular wrapping of the G614-A647 dehydron, thereby further promoting its intermolecular wrapping via the contribution from S2 residue P862 (Figure 1d). A conservative estimate drawn from experimental data on the cost of unwrapping the BHB (ref (8) Figure 3) gives 3 × 8 kJ/mol = 5.73 kcal/mol as the thermodynamic cost of destabilizing the S1 structure resulting from depriving the BHB 614–647 of three wrapping carbonaceous groups due to the D614G substitution. The lost wrapping contributions include one methylene group from the D → G substitution proper and two methylene groups from R646 that no longer can form the salt bridge with the glycine at position 614 (cf. Figure 1c).
Thus, the net gain in stability (loss in free energy) for the S1/S2 complex resulting from the D614G mutation is significant and may be conservatively estimated at ΔΔG = −5.73kcal/mol. We emphasize that the stabilization of the S1/S2 complex arises from the destabilization of the free (uncomplexed) S1 structure.
Conclusions and Discussion
The structural, or rather epistructural, impact of the mutation D614G is consistent with established phenotypic differences between SG614 and SD614 in the sense that SG614 has a greater stability resulting from less S1 shedding and greater incorporation of the intact S protein into the pseudovirion.4 In this way, the epistructural analysis sheds light on the mechanistic underpinnings for the superior transmission efficiency of the G614 strain of SARS-CoV-2.
The epistructural analysis presented raises the enticing therapeutic possibility of immunologically targeting the epitope involved in reducing S1-shedding as a means of impairing the infection efficacy of the G614 strain. Thus, a monoclonal antibody targeting the epitope associated with the dehydron 614-647 (Figure 1c) could significantly promote S1 shedding and destabilize the SG614 phenotype to levels representing severe impairment of the viral infectivity. This therapeutic modality would not a-priori interfere directly with current efforts toward an immunological targeting of the RBD epitope, hence it could serve as a complementary treatment.
Methods
Dehydrons were identified from PDB-reported structural coordinates of proteins5−8 using a plugin to the PyMol platform (https://pymolwiki.org/index.php/Dehydron). To that end, we define the extent of hydrogen-bond wrapping, w, giving the number of side chain nonpolar groups (CHn, n = 1, 2, 3) contained within a ‘“desolvation domain”’ around the BHB. The desolvation domain is defined as two intersecting balls of fixed radius commensurate with three water layers centered at the α-carbons of the residues paired by the BHB. A wrapping analysis of nonredundant 7476 high quality X-ray structures deposited in PDB was performed adopting the desolvation radius 6.5 Å (code publicly available at https://raw.githubusercontent.com/Pymol-Scripts/Pymol-script-repo/master/plugins/dehydron.py). The results revealed that backbone hydrogen bonds are wrapped by w = 27.41 ± 8.07 nonpolar groups. Being marginally wrapped, dehydrons are located in the tail of the distribution and hence identified by the relation w ≤ 19.
Glossary
Abbreviations
- ACE2
angiotensin-converting enzyme 2
- COVID-19
Coronavirus Disease 2019
- RBD
receptor-biding domain
- SARS-CoV
Severe Acute Respiratory Syndrome Coronavirus
- S protein
spike glycoprotein
Views expressed in this editorial are those of the author and not necessarily the views of the ACS.
The author declares no competing financial interest.
References
- Korber B.; Fischer W. M.; Gnanakaran S.; Yoon H.; Theiler J.; Abfalterer W.; Hengartner N.; Giorgi E. E.; Bhattacharya T.; Foley B.; Hastie K. M.; Parker M. D.; Partridge D. G.; Evans C. M.; Freeman T. M.; de Silva T. I.; McDanal C.; Perez L. G.; Tang H.; Moon-Walker A.; Whelan S. P.; LaBranche C. C.; Saphire E. O.; Montefiori D. C. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 2020, 182, 1–16. on behalf of the Sheffield COVID-19 Genomics Group 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J.; He C.-L.; Gao Q.-Z.; Zhang G.-J.; Cao X.-X.; Long Q.-X.; Deng H.-J.; Huang L.-Y.; Chen J.; Wang K.; Tang N.; Huang A.-L.. The D614G mutation of SARS-CoV-2 spike protein enhances viral infectivity and decreases neutralization sensitivity to individual convalescent sera. boRxiv 2020, 06.20.161323. 10.1101/2020.06.20.161323. [DOI] [Google Scholar]
- Ozono S.; Zhang Y.; Ode H.; Seng T. T.; Imai K.; Miyoshi K.; Kishigami S.; Ueno T.; Iwatani Y.; Suzuki T.; Tokunaga K.. Naturally mutated spike proteins of SARS-CoV-2 variants show differential levels of cell entry. boRxiv 2020, 06.15.151779. 10.1101/2020.06.15.151779. [DOI] [Google Scholar]
- Zhang L.; Jackson C. B.; Mou H.; Ojha A.; Rangarajan E. S.; Izard T.; Farzan M.; Choe H.. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. boRxiv 2020, 06.12.148726. 10.1101/2020.06.12.148726. [DOI] [Google Scholar]
- Fernández A.Physics at the Bomolecular Interface: Fundamentals for Molecular Targeted Therapy; Springer International Publishing: Switzerland, 2016; Chapters 1–3. [Google Scholar]
- Fernández A.; Crespo A. Protein wrapping: a marker for association, aggregation and molecular targeted therapy. Chem. Soc. Rev. 2008, 37, 2373–2382. 10.1039/b804150b. [DOI] [PubMed] [Google Scholar]
- Fernández A.; Scheraga H. A. Insufficiently dehydrated hydrogen bonds as determinants for protein interactions. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 113–118. 10.1073/pnas.0136888100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández A. Stickiness of the hydrogen bond. Ann. Phys. (Berlin, Ger.) 2018, 530, 1800162. 10.1002/andp.201800162. [DOI] [Google Scholar]