SUMMARY
Nonsense-mediated decay (NMD) is a pathway that degrades mRNAs containing premature termination codons. Here we describe a genome-wide screen for NMD factors that uncovers an unexpected mechanism that broadly governs 3′ untranslated region (UTR)-directed regulation. The screen reveals that NMD requires lysosomal acidification, which allows transferrin-mediated iron uptake, which, in turn, is necessary for iron-sulfur (Fe-S) cluster biogenesis. This pathway maintains the activity of the Fe-S cluster-containing ribosome recycling factor ABCE1, whose impaired function results in movement of ribosomes into 3′ UTRs, where they displace exon junction complexes, abrogating NMD. Importantly, these effects extend beyond NMD substrates, with ABCE1 activity required to maintain the accessibility of 3′ UTRs to diverse regulators, including microRNAs and RNA binding proteins. Because of the sensitivity of the Fe-S cluster of ABCE1 to iron availability and reactive oxygen species, these findings reveal an unanticipated vulnerability of 3′ UTR-directed regulation to lysosomal dysfunction, iron deficiency, and oxidative stress.
Graphical Abstract
In Brief
Zhu et al. find a critical role of the ribosome recycling factor ABCE1 in nonsense-mediated decay and other forms of 3′ UTR-mediated regulation. The iron-sulfur cluster of ABCE1 is highly sensitive to lysosomal dysfunction and reactive oxygen species, linking these pathologic stresses to defective post-transcriptional control.
INTRODUCTION
Nonsense-mediated decay (NMD) is a ubiquitous pathway in eukaryotic cells that recognizes and degrades messenger RNAs (mRNAs) containing premature termination codons (PTCs) (Kurosaki et al., 2019). Transcripts that harbor nonsense or frameshift mutations frequently undergo rapid degradation by NMD, limiting production of truncated proteins. As a result, the NMD pathway is a potent modifier of genetic disease phenotypes, protecting from deleterious activity of dominant-negative truncated proteins or exacerbating loss-of-function phenotypes by dampening production of truncated proteins with residual activity (Inoue et al., 2007; Kerr et al., 2001; Kugler et al., 1995; Miller and Pearce, 2014). Although first discovered because of this mRNA quality control function, it is now appreciated that a broad set of physiologic transcripts is targeted for accelerated decay by NMD, with approximately 10% of the mammalian transcriptome under control of this pathway (Colombo et al., 2017; Mendell et al., 2004; Tani et al., 2012). Accordingly, the NMD pathway is essential for embryogenesis in mammals, and its impairment is associated with neurodevelopmental disorders and cancer (Jaffrey and Wilkinson, 2018; Li et al., 2015; McIlwain et al., 2010; Medghalchi et al., 2001; Tarpey et al., 2007; Wang et al., 2011; Weischenfeldt et al., 2008).
The events that occur when a ribosome reaches a termination codon dictate whether the NMD pathway will be engaged. For the vast majority of mammalian transcripts, the termination codon is located in the final exon or close to the final exon-exon junction. During splicing, the position of exon-exon junctions is stably marked by deposition of exon junction complexes (EJCs), which remain associated with the mRNA as it is exported to the cytoplasm (Le Hir et al., 2000). As a translating ribosome traverses the mRNA, it displaces EJCs, normally leading to their complete removal by the time it reaches the termination codon at the 3′ end of the open reading frame (ORF). The release factor eRF1 then enters the empty A site of the ribosome and, together with eRF3, promotes peptide release (Hellen, 2018). Finally, ABCE1, an iron-sulfur (Fe-S) cluster-containing protein, is recruited to the terminating ribosome by eRF1 and facilitates splitting of the 40S and 60S subunits, a process known as ribosome recycling (Barthelme et al., 2011; Pisarev et al., 2010; Young et al., 2015). In the absence of ABCE1 or its yeast homolog Rli1, unrecycled ribosomes move into the 3′ untranslated region (UTR), where they may aberrantly re-initiate translation (Mills et al., 2016; Sudmant et al., 2018; Young et al., 2015).
When a stop codon is present at least 50 nucleotides (nt) upstream of the final exon-exon junction, translation termination occurs in the context of a downstream EJC remaining bound to the mRNA. This configuration represents the classic trigger for activation of NMD in mammalian cells (Bhuvanagiri et al., 2010; He and Jacobson, 2015; Kurosaki et al., 2019). In this scenario, the core NMD factors UPF1 and SMG1 are recruited to the terminating ribosome by eRF1 and eRF3 and interact with UPF2 and UPF3B, which are anchored on the downstream EJC. The formation of this complex stimulates phosphorylation of UPF1 by SMG1, leading to recruitment of SMG5, SMG6, and SMG7. SMG6, an endonuclease, cleaves the mRNA, whereas SMG5 and SMG7 recruit the deadenylase and decapping machinery to promote degradation of the targeted nonsense transcript. NMD may also be triggered when a message has an unusually long 3′ UTR (Bühler et al., 2006; Hogg and Goff, 2010; Kebaara and Atkin, 2009), a particularly important feature of the pathway in budding yeast, where most genes lack introns. The mechanisms that lead to recruitment of NMD factors and subsequent activation of the pathway in the absence of a downstream EJC, however, remain incompletely understood.
Evidence suggests that ribosome recycling occurs through distinct mechanisms at normal termination codons versus PTCs. Binding of UPF1 and SMG1 to eRF1/3 at PTCs likely precludes recruitment of ABCE1 (Celik et al., 2015). Moreover, it has been reported that the ATPase activity of yeast Upf1 is required for translation termination and ribosome recycling at PTCs (Serdar et al., 2016), and human UPF3B has been shown to exhibit ribosome recycling activity in an in vitro translation assay (Neu-Yilik et al., 2017). These data suggest that the UPF complex performs the ribosome recycling function at nonsense codons, although a role of ABCE1 in post-termination events at PTCs has not yet been directly examined.
Genetic screens have played a key role in elucidating the mechanism of NMD, with the UPF and SMG proteins having been identified through screens performed in yeast and nematodes, respectively (Hodgkin et al., 1989; Leeds et al., 1991). More recently, advances in CRISPR-Cas9 and RNA interference (RNAi) have enabled extension of this strategy to mammalian cells. Alexandrov et al. (2017) utilized a lentiviral CRISPR library coupled with fluorescence-activated cell sorting (FACS) of a reporter cell line to identify NMD regulators in HeLa cells. Although new potential NMD factors were identified in this study, the use of multiple rounds of sorting likely reduced the genome-wide coverage of this screen. A recent small interfering RNA (siRNA) screen that uncovered NMD factors in HEK293 cells likely avoided this caveat (Baird et al., 2018), but the short duration of siRNA-mediated knockdown may have precluded identification of long-lived NMD regulators or pathways that act upstream of core NMD factors. Thus, additional NMD factors may remain to be discovered in mammalian cells.
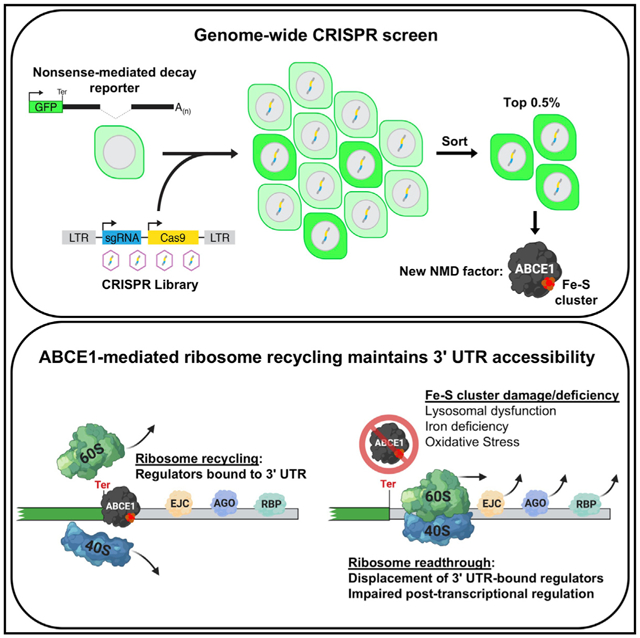
We recently developed a highly effective genome-wide CRISPR screening approach that utilizes fluorescent reporters coupled with a single round of high-stringency sorting that allows recovery of essential genes as hits (Golden et al., 2017; Manjunath et al., 2019). Here we describe application of this strategy to identify components of the NMD pathway in mammalian cells. Surprisingly, these experiments revealed that NMD requires activity of the vacuolar ATPase (V-ATPase), which is responsible for lysosomal acidification, as well as Fe-S cluster biogenesis and the Fe-S cluster-containing ribosome recycling factor ABCE1. We demonstrate that inhibition of lysosomal acidification compromises transferrin-mediated iron uptake, which, in turn, impairs Fe-S cluster biogenesis and abrogates ABCE1 activity. Loss of ABCE1 leads to accumulation of ribosomes in the 3′ UTRs of NMD targets, which displace downstream EJCs and reduce recruitment of UPF1 to nonsense transcripts. Remarkably, these effects are not limited to NMD targets, with ABCE1 deficiency leading to broad impairment of 3′ UTR-mediated post-transcriptional control, including mRNA turnover induced by microRNAs (miRNAs), AU-rich elements (AREs), N6-methyladenosine (m6A), and various RNA binding proteins (RBPs). Thus, ABCE1-mediated ribosome recycling is critical for preventing displacement of diverse 3′ UTR-bound messenger ribonucleoprotein (mRNP) complexes that confer post-transcriptional gene regulation. Importantly, the sensitivity of Fe-S clusters to intracellular iron levels and reactive oxygen species (ROS) results in vulnerability of 3′ UTR-directed regulation to lysosomal dysfunction, iron deficiency, and oxidative stress.
RESULTS
A Genome-wide CRISPR Screen in Human Cells Identifies Regulators of NMD
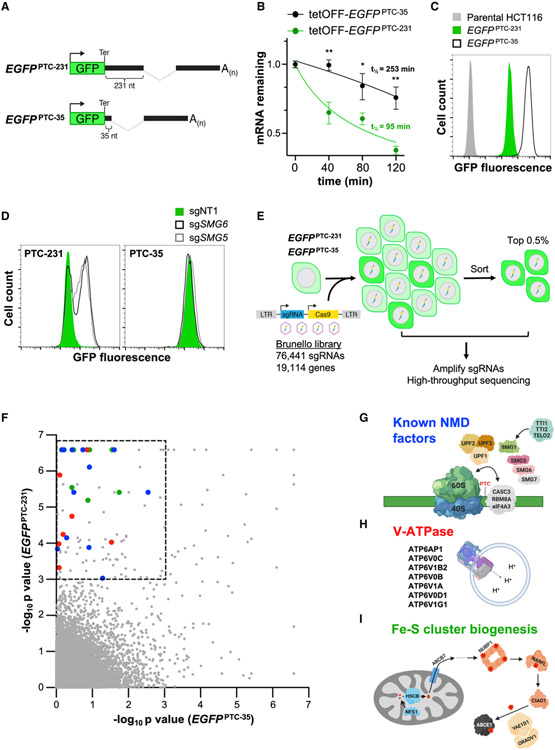
To carry out a genome-scale CRISPR-Cas9 screen to identify new components or regulators of the NMD pathway in human cells, we expressed an enhanced green fluorescent protein (EGFP) reporter transcript that is efficiently targeted for degradation by NMD. Exons 2 and 3 as well as the intervening intron of the human β-globin (HBB) gene were cloned downstream of the EGFP ORF, positioning the exon-exon junction 231 nt downstream of the termination codon, creating an NMD substrate termed EGFPPTC-231 (Figure 1A). A control transcript with the HBB intron placed 35 nt downstream of the EGFP stop codon, too close to trigger NMD (Cheng et al., 1994; Cheng et al., 1990), was also constructed (EGFPPTC-35; Figure 1A). Half-life measurements of these transcripts expressed from a doxycycline-repressible promoter confirmed that EGFPPTC-231 is degraded much more rapidly than EGFPPTC-35 (Figure 1B), consistent with elimination by the NMD pathway.
Figure 1. A Genome-wide CRISPR-Cas9 Screen Identifies NMD Regulators.
(A) Schematic of EGFPPTC-231 and EGFPPTC-35 reporter constructs. Ter, termination codon.
(B) Analysis of EGFPPTC-231 mRNA and EGFPPTC-35 mRNA stability in respective doxycycline-repressible (tetOFF)-EGFP cell lines. Cells were treated with 1 μg/mL doxycycline, and the EGFP mRNA level relative to GAPDH abundance at the indicated time points was determined by qRT-PCR.
(C) Flow cytometry analysis of EGFP fluorescence in the indicated cell lines.
(D) Flow cytometry analysis of EGFPPTC-231 cells and EGFPPTC-35 cells after lentiviral delivery of Cas9 and non-target (NT) or SMG5- or SMG6-targeting sgRNAs.
(E) Schematic of the CRISPR-Cas9 screening strategy.
(F) MAGeCK analysis of screening data. Hits within the dashed box represent those that met the significance threshold of p < 0.001 in EGFPPTC-231 cells and p > 0.001 in EGFPPTC-35 cells. Blue dots, known NMD factors; red dots, V-ATPase components; green dots, Fe-S biogenesis factors.
(G–I) Known NMD factors (G), V-ATPase subunits (H), and genes required for Fe-S cluster biogenesis (I) that were recovered as significant hits in the screen. Data are represented as mean ± SD (n = 3 biological replicates for all qRT-PCR experiments). *p < 0.05, **p < 0.01; calculated by Student’s t test. See also Figure S1 and Table S1.
The EGFPPTC-231 and EGFPPTC-35 constructs were then placed under the control of a strong constitutive promoter and integrated into the AAVS1 locus, a region of open chromatin that allows high-level transgene expression (Hockemeyer et al., 2009; Lamartina et al., 2000), in HCT116 cells, a stably diploid human cell line amenable to pooled CRISPR screening (Golden et al., 2017; Manjunath et al., 2019). As expected, EGFPPTC-231 cells exhibited approximately 10-fold lower GFP fluorescence compared with EGFPPTC-35 cells (Figure 1C). To verify that decreased expression of EGFPPTC-231 was a consequence of downregulation by NMD, reporter cells were infected with lentiviruses expressing Cas9 and single guide RNAs (sgRNAs) targeting the established NMD factors SMG5 and SMG6 (Figure 1D). Deficiency of either gene robustly increased EGFP expression in EGFPPTC-231 but not EGFPPTC-35 cells, demonstrating that expression of the EGFPPTC-231 reporter provided a reliable readout of NMD activity.
EGFPPTC-231 and EGFPPTC-35 cells were then infected with a genome-wide lentiviral CRISPR library targeting approximately 19,000 human genes (Doench et al., 2016; Figure 1E). Ten days after transduction, the brightest 0.5% of cells, representing those with impaired repression of the EGFP reporter, were collected by FACS. sgRNA representation in sorted and unsorted cells was determined by high-throughput sequencing, and model-based analysis of genome-wide CRISPR-Cas9 knockout (MAGeCK) (Li et al., 2014) was used to identify genes targeted by sgRNAs that were enriched in sorted populations (Table S1). Genes that scored as significant hits in EGFPPTC-231 cells (p < 0.001), but not in EGFPPTC-35 cells, represented potential NMD factors and were selected for further analysis (Figure 1F). Based on this cutoff, the majority of established NMD factors were recovered as significant hits (Figures 1G and S1A), confirming the ability of this screening approach to identify components and regulators of the NMD pathway.
The screen identified numerous potential NMD regulators that have not been associated previously with the pathway (Table S1). Gene Ontology (GO) analysis revealed that, in addition to factors linked to mRNA processing, transport, and metabolism, genes associated with lysosomal acidification and Fe-S cluster assembly were unexpectedly enriched among the hits (Figure S1B; Table S2). Further examination of these classes of genes uncovered several components of the V-ATPase, which is responsible for acidification of the endosome/lysosome (Mindell, 2012; Figure 1H), and several factors necessary for Fe-S cluster biogenesis as well as the Fe-S cluster-containing protein ABCE1 (Figure 1I) as significant hits. Because of the unknown mechanisms through which these factors influence NMD, we elected to further study their role in the pathway.
V-ATPase Activity Is Required for NMD
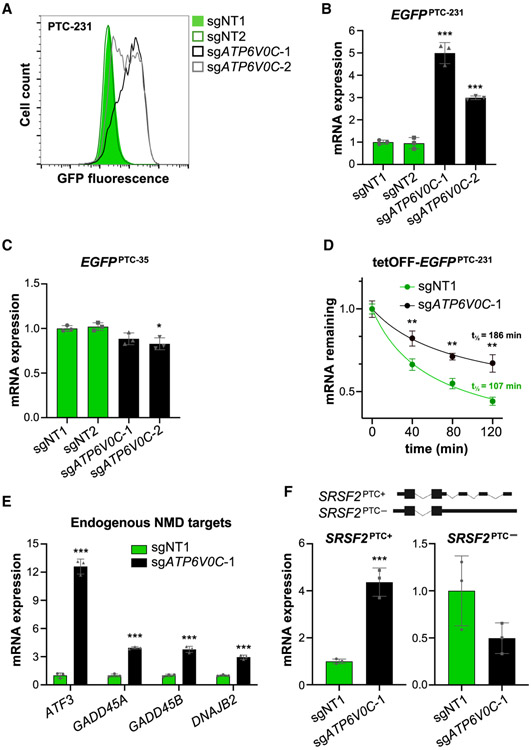
To validate the requirement for V-ATPase function in NMD, EGFPPTC-231 and EGFPPTC-35 cells were infected with lentiviral vectors encoding Cas9 and sgRNAs targeting 4 subunits of the complex (ATP6V0C, ATP6V0B, ATP6AP1, or ATP6V1B2) (Figures 2A, S2A, and S2B). Knockout of each of these genes robustly increased the GFP signal in EGFPPTC-231 cells while only minimally affecting expression of the EGFPPTC-35 reporter. Western blotting confirmed a strong increase in EGFP protein levels in ATP6V0C knockout EGFPPTC-231 cells (Figure S2C). In agreement with these results, treatment with bafilomycin, a V-ATPase inhibitor, significantly increased GFP fluorescence in EGFPPTC-231 cells, with minor effects on expression of EGFPPTC-35 (Figure S2D).
Figure 2. The V-ATPase Is Required for NMD.
(A) Flow cytometry analysis of EGFPPTC-231 cells after lentiviral delivery of Cas9 and NT or ATP6V0C-targeting sgRNAs.
(B and C) qRT-PCR analysis of EGFPPTC-231 (B) or EGFPPTC-35 mRNA (C) in reporter cell lines after lentiviral delivery of Cas9 and the indicated sgRNAs. All mRNA expression levels shown in this figure were first normalized to GAPDH expression and then normalized to expression level in sgNT1-infected cells.
(D) Analysis of EGFPPTC-231 mRNA stability in tet-OFF-EGFPPTC-231 cells after lentiviral delivery of Cas9 and the indicated sgRNAs. Cells were treated with 1 μg/mL doxycycline, and the EGFP mRNA level relative to GAPDH abundance at the indicated time points was determined by qRT-PCR.
(E and F) qRT-PCR analysis of endogenous NMD targets (E) or PTC+ and PTC− isoforms of SRSF2 (F) in HCT116 cells after lentiviral delivery of Cas9 and the indicated sgRNAs.
Data are represented as mean ± SD (n = 3 biological replicates for all qRT-PCR experiments). *p < 0.05, **p < 0.01, ***p < 0.001; calculated by Student’s t test. See also Figure S2.
Because V-ATPase-mediated lysosomal acidification is essential for protein hydrolysis within the lysosome, we initially postulated that the protein encoded by the EGFPPTC-231 mRNA might be targeted for lysosome-mediated degradation by the NMD machinery. Nevertheless, surprisingly, we observed that the increase in EGFP expression in ATP6V0C knockout EGFPPTC-231 cells was due to an increase in reporter mRNA expression (Figure 2B). Conversely, no increase in EGFPPTC-35 mRNA abundance was detected upon knockout of ATP6V0C (Figure 2C). Similar results were observed in bafilomycin-treated cells (Figures S2E and S2F). These results suggested an unexpected role of V-ATPase in decay of NMD target transcripts. Further corroborating this conclusion, half-life measurements documented that EGFPPTC-231 mRNA was stabilized by ATP6V0C knockout or bafilomycin treatment (Figures 2D and S2G).
To verify that these effects were not limited to the EGFPPTC-231 reporter, we assessed the expression of several established endogenous NMD targets, including ATF3, GADD45A, GADD45B, DNAJB2, and an alternatively spliced isoform of SRSF2 with multiple introns in its 3′ UTR (Hauer et al., 2016; Mendell et al., 2004; Tani et al., 2012). All tested NMD targets were significantly upregulated in ATP6V0C knockout and bafilomycin-treated cells (Figures 2E, 2F, and S2H). Last, we demonstrated that the requirement for V-ATPase activity in NMD was not limited to HCT116 cells because treatment of Huh7, HeLa, and HEK293T cells with bafilomycin similarly increased expression of endogenous NMD target genes (Figure S2I). Taken together, these data document that accelerated transcript degradation through the NMD pathway requires activity of the V-ATPase complex.
Iron Depletion in V-ATPase-Deficient Cells Inhibits NMD
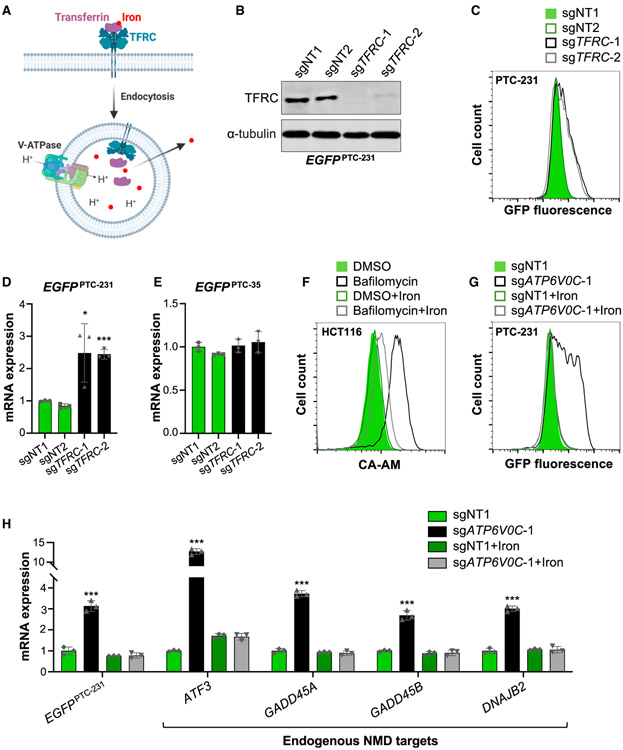
Transferrin-mediated iron uptake, the major mechanism of iron absorption in mammalian cells, requires V-ATPase activity. Extracellular iron in complex with transferrin is bound by the transferrin receptor (TFRC) and taken up by receptor-mediated endocytosis (Crielaard et al., 2017; Figure 3A). V-ATPase-mediated acidification of the endosomal/lysosomal compartment triggers release of iron, which is transported to the cytoplasm for use in cellular metabolism, including incorporation into Fe-S clusters. Given the preponderance of genes required for Fe-S cluster biogenesis among the top hits in the screen (Figure 1I), we hypothesized that the requirement for V-ATPase activity in NMD may be due to its role in iron absorption.
Figure 3. Iron Deficiency Compromises NMD in V-ATPase-Deficient cells.
(A) Schematic representation of transferrin (TF)-mediated iron uptake. After receptor-mediated endocytosis via the TF receptor (TFRC), acidification of the endosome/lysosome by V-ATPase promotes iron release and transport to the cytoplasm.
(B) Western blot analysis of the TFRC in EGFPPTC-231 cells after lentiviral delivery of Cas9 and NT or TFPC-targeting sgRNAs. α-Tubulin served as a loading control.
(C) Flow Cytometry analysis of EGFPPTC-231 cells after lentiviral delivery of Cas9 and the indicated sgRNAs.
(D and E) qRT-PCR analysis of EGFPPTC-231 (D) or EGFPPTC-35 mRNA(E) in reporter cell lines after lentiviral delivery of Cas9 and the indicated sgRNAs. All mRNA expression levels shown in this figure were first normalized to GAPDH expression and then normalized to expression level in sgNT1-infected cells.
(F) Flow cytometry assay for intracellular iron content, based on calcein-AM (CA-AM) fluorescence, in control (DMSO)- or bafilomycin (5 nM)-treated HCT116 cells with or without 0.5 mM iron citrate supplementation.
(G) Flow cytometry analysis of EGFPPTC-231 cells after lentiviral delivery of Cas9 and the indicated sgRNAs with or without 0.5 mM iron citrate supplementation.
(H) qRT-PCR analysis of NMD targets in EGFPPTC-231 cells after lentiviral delivery of Cas9 and the indicated sgRNAs with or without 0.5 mM iron citrate supplementation.
Data are represented as mean ± SD (n = 3 biological replicates for all qRT-PCR experiments). *p < 0.05, ***p < 0.001; calculated by Student’s t test. See also Figure S3.
Although TFRC did not initially score among the top hits, the gene was highly ranked in the EGFPPTC-231 screen (rank 283, top 1.5%; Table S1), and transferrin transport was highlighted as a significantly enriched GO term among the hits (Figure S1B). Accordingly, TFRC knockout increased reporter fluorescence and mRNA abundance in EGFPPTC-231 but not EGFPPTC-35 cells (Figures 3B-3E, S3A, and S3B). Moreover, treatment with the iron chelator desferrioxamine (DFO) resulted in similar effects on reporter expression (Figures S3C and S3D) and stabilized the EGFPPTC-231 transcript (Figure S3E). These results indicated that TFRC-mediated iron uptake is required for NMD.
confirmed that inhibition of lysosomal acidification with bafilomycin reduced intracellular iron levels, as measured by an established flow cytometry assay based on iron-mediated quenching of calcein-AM (CA-AM) fluorescence (Prus and Fibach, 2008; Figure 3F). Treatment with iron citrate, which is taken up through an endocytosis-independent mechanism (Sturrock et al., 1990), replenished intracellular iron. Furthermore, iron citrate supplementation fully reversed the increase in reporter fluorescence in ATP6V0C knockout and bafilomycin-treated EGFPPTC-231 cells (Figures 3G and S3F). Additionally, EGFPPTC-231 mRNA and endogenous NMD target abundance were restored to control levels under these conditions (Figures 3H and S3G). Thus, impairment of NMD in V-ATPase-deficient cells is attributable to intracellular iron deficiency.
Deficiency of Fe-S Cluster Biogenesis or the Fe-S Cluster-Containing Protein ABCE1 Impairs NMD
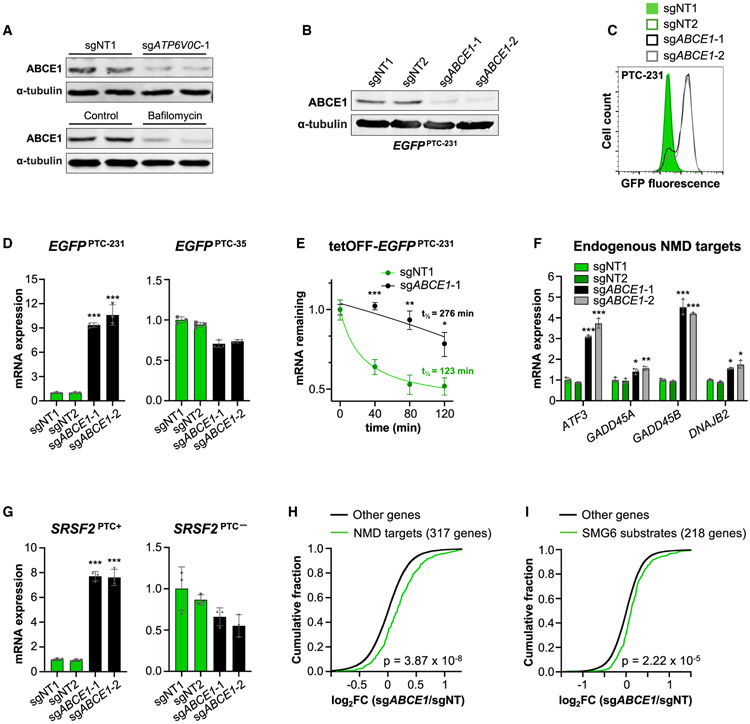
Sufficient intracellular iron is necessary for biogenesis of Fe-S clusters, which are assembled in mitochondria and then exported to the cytoplasm for incorporation into target proteins (Rouault and Tong, 2005; Figure 1I). Several proteins in this pathway were recovered as significant hits in the screen, including the mitochondrial assembly factors NFS1 and HSCB, the ABCB7 transporter, and the cytosolic Fe-S chaperones NUBP1, NARFL, CIAO1, YAE1D1, and ORAOV1. Interestingly, the only Fe-S cluster recipient protein among the top hits was the ribosome recycling factor ABCE1, strongly implicating an essential role of ABCE1 and its associated Fe-S cluster in the NMD pathway. Notably, we observed that ATP6V0C knockout or bafilomycin treatment potently reduced ABCE1 protein abundance (Figure 4A). This finding is consistent with the requirement for adequate intracellular iron for Fe-S cluster biogenesis, which, in turn, is necessary for ABCE1 protein stability (Sudmant et al., 2018).
Figure 4. ABCE1 Is Required for NMD.
(A) Western blot analysis of ABCE1 in EGFPPTC-231 cells after lentiviral delivery of Cas9 and NT or ATP6V0C-targeting sgRNAs (top) or treatment with bafilomycin (5 nM) or a control (DMSO) (bottom). α-Tubulin served as a loading control.
(B) Western blot analysis of ABCE1 in EGFPPTC-231 cells after lentiviral delivery of Cas9 and NT or ABCE1-targeting sgRNAs.
(C) Flow cytometry analysis of EGFPPTC-231 cells after lentiviral delivery of Cas9 and the indicated sgRNAs.
(D) qRT-PCR analysis of EGFPPTC-231 or EGFPPTC-35 mRNA in reporter cell lines after lentiviral delivery of Cas9 and the indicated sgRNAs. All mRNA expression levels shown in this figure were first normalized to GAPDH expression and then normalized to expression level in sgNT1-infected cells.
(E) Analysis of EGFPPTC-231 mRNA stability in tetOFF-EGFPPTC-231 cells after lentiviral delivery of Cas9 and the indicated sgRNAs. Cells were treated with 1 μg/mL doxycycline, and the EGFP mRNA level relative to GAPDH abundance at the indicated time points was determined by qRT-PCR.
(F and G) qRT-PCR analysis of endogenous NMD targets (F) or PTC+ and PTC− isoforms of SRSF2 (G) in HCT116 cells after lentiviral delivery of Cas9 and the indicated sgRNAs.
(H and I) Cumulative distribution function (CDF) plot demonstrating the effect of ABCE1 deficiency on expression of NMD target genes (Colombo et al., 2017; El-Bchiri et al., 2008; H) or SMG6 substrates (Schmidt et al., 2015; I). FC, fold change.
Data are represented as mean ± SD (n = 3 biological replicates for all qRT-PCR experiments). *p < 0.05, **p < 0.01, ***p < 0.001; calculated by Student’s t test; p values for CDF plots were calculated by one-sided Kolmogorov-Smirnov test. See also Figure S4.
In keeping with the screening results, targeting of HSCB, NARFL, YAE1D1, and ORAOV1 with lentivirally delivered CRISPR components increased GFP fluorescence in EGFPPTC-231 but not EGFPPTC-35 cells (Figure S4A). Similarly, ABCE1 knockout strongly increased the fluorescence and mRNA abundance of the EGFPPTC-231 but not the EGFPPTC-35 reporter (Figures 4B-4D, S4B, and S4C) and stabilized the EGFPPTC-231 transcript (Figure 4E). Endogenous NMD target transcripts were similarly increased in multiple cell lines after ABCE1 depletion (Figures 4F, 4G, S4D, and S4E). These data support a requirement for Fe-S cluster biogenesis and ABCE1 in NMD.
It has been reported that depletion of ABCE1 globally reduces translation (Toompuu et al., 2016). Because of the dependency of NMD on active translation, this could potentially account for impaired decay of nonsense transcripts in ABCE1-deficient cells. Nevertheless, under the conditions utilized in these experiments, we observed only a minor reduction in overall translation upon ABCE1 loss of function, as assessed by incorporation of fluorescent O-propargyl-puromycin (OPP) into newly translated proteins (Figure S4F). Further arguing against an indirect effect on general translation as the cause of NMD inhibition, we observed that knockout of the Fe-S cluster biogenesis factor ORAOV1 or ABCE1 resulted in a robust increase in EGFP protein in EGFPPTC-231 cells (Figure S4G). Thus, NMD targets were still actively translated under these conditions. Additionally, the lack of translation factors among the significant hits in the CRISPR screen suggested that globally reduced translation does not result in an increase in EGFPPTC-231 reporter fluorescence.
To investigate the broader effects of ABCE1 deficiency on NMD-regulated transcripts, RNA sequencing (RNA-seq) was performed on ABCE1-depleted HCT116 cells. Examination of a high-stringency set of NMD target genes, representing the overlap of two previous gene expression studies of cells with UPF1, SMG6, and/or SMG7 deficiency (Colombo et al., 2017; El-Bchiri et al., 2008), documented that NMD targets were globally upregulated in ABCE1 knockout cells (Figure 4H). Transcripts that are targeted for cleavage by SMG6 (Schmidt et al., 2015) were also upregulated (Figure 4I). NMD of the EGFPPTC-231 reporter is initiated by an EJC downstream of the termination codon. Upstream ORFs (uORFs) and long 3′ UTRs have also been shown to induce NMD, albeit with lower potency (Kurosaki et al., 2019). Expression of transcripts harboring these features was also globally increased upon ABCE1 depletion (Figures S4H and S4I). Taken together, these data establish that NMD induced by a variety of triggers requires the activity of ABCE1.
Unrecycled Ribosomes Downstream of Termination Codons Displace EJCs on NMD Targets in ABCE1-Deficient Cells
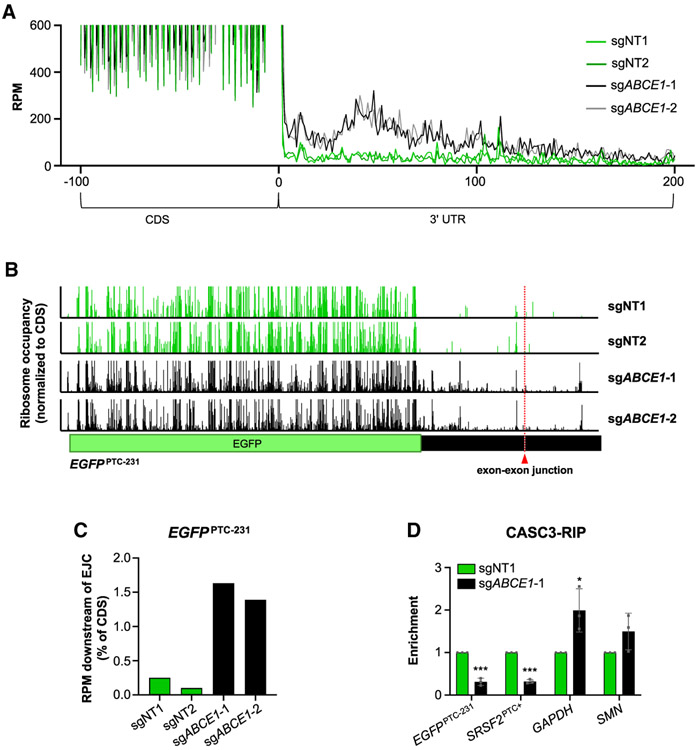
Although it has been suggested that the UPF complex carries out the ribosome recycling function at PTCs (Celik et al., 2015; Neu-Yilik et al., 2017; Serdar et al., 2016), our data demonstrating a requirement for ABCE1 in NMD suggested an essential role of canonical ribosome recycling in decay of nonsense transcripts. To examine this directly, we performed ribosome profiling in ABCEI-deficient cells, which allowed determination of the positions of ribosomes on transcripts undergoing NMD. As expected, ribosome-protected fragments exhibited 3-nt periodicity within ORFs, consistent with the stepwise translation of each codon (Figure S5A). In accordance with data reported previously (Mills et al., 2016; Sudmant et al., 2018; Young et al., 2015), loss of ABCE1 resulted in queuing of ribosomes at termination codons and an increase in ribosomes within 3′ UTRs (Figures 5A, S5A, and S5B).
Figure 5. Unrecycled Ribosomes Downstream of Ters in ABCE1-Depleted Cells Displace EJCs.
(A) Metagene analysis demonstrating ribosome occupancy in 3′ UTRs. A window spanning 100 nt upstream of termination codon (position 0) to 200 nt downstream of termination codon is shown. RPM, ribosome-protected fragments per million mapped reads.
(B) Ribosome occupancy on the EGFPPTC-231 transcript after lentiviral delivery of Cas9 and the indicated sgRNAs. The position of the exon-exon junction in the in 3′ UTR of the reporter transcript is indicated. Ribosome occupancy is represented as the RPM value at a given position, normalized to the total RPM value in the EGFP ORF.
(C) Total ribosome occupancy downstream of the exon-exon junction in the 3′ UTR of the EGFPPTC-231 transcript after lentiviral delivery of Cas9 and the indicated sgRNAs. For each condition, the total RPM downstream of the exon-exon junction was normalized to the total RPM value in the EGFP ORF.
(D) qRT-PCR analysis of NMD targets (EGFPPTC-231 and SRSF2PTC+) andcontrol transcripts (GAPDH and SMN) in CASC3 RIP samples from cells expressing Cas9 and the indicated sgRNAs. For each transcript, enrichment was first normalized to input and then normalized to a paired control sample.
Data are represented as mean ± SD (n = 3 biological replicates for all qRT-PCR experiments). *p < 0.05, ***p < 0.001; calculated by Student’s t test. See also Figure S5.
To determine whether ABCE1 is required for ribosome recycling on an NMD substrate transcript, we examined ribosome occupancy on the EGFPPTC-231 mRNA. Indeed, a large increase in ribosomes within the EGFPPTC-231 3′ UTR was observed in ABCE1-depleted cells (Figure 5B). Moreover, increased ribosome occupancy was clearly detectable downstream of the exon-exon junction (Figures 5B and 5C), strongly suggesting that unrecycled ribosomes are able to displace EJCs as they move through the 3′ UTR.
We next used RNA immunoprecipitation (RIP) to directly assess EJC occupancy downstream of termination codons on NMD sub-strates. The EGFPPTC-231 transcript has only a single exon-exon junction, located within the 3′ UTR (Figure 5B). The alternatively spliced variant of SRSF2 that is an NMD substrate has multiple exon-exon junctions in its 3′ UTR (Figure 2F), and the single EJC in the SRSF2 coding region will be efficiently removed during translation. Thus, steady-state EJC association with these transcripts largely or entirely represents binding of this complex downstream of the termination codon. CASC3 binding sites are highly specific markers of the location of EJCs (Hauer et al., 2016), and CASC3 knockout results in a global increase in NMD targets (Gerbracht et al., 2019). Indeed, CASC3 was the most highly ranked hit in our screen (Table S1) and was also identified in other published NMD screens (Alexandrov et al., 2017; Baird et al., 2018). Pull-down of CASC3 revealed a significant decrease in EJC occupancy within the 3′ UTRs of EGFPPTC-231 and NMD-sensitive SRSF2 transcripts in ABCE1-deficient cells (Figures 5D, S5C, and S5D). Finally, in keeping with the presence of unrecycled ribosomes in 3′ UTRs and displacement of EJCs, the association of UPF1 with EGFPPTC-231 and the NMD-sensitive isoform of SRSF2 was significantly decreased upon loss of ABCE1 (Figures S5E-S5G). Collectively, these ribosome profiling and RIP results demonstrate that deficiency of ABCE1 leads to accumulation of ribosomes distal to the termination codon on NMD target transcripts, resulting in dissociation of downstream EJCs and impaired recruitment of the NMD machinery.
Oxidative Stress or Lysosomal Dysfunction Impairs Ribosome Recycling and NMD
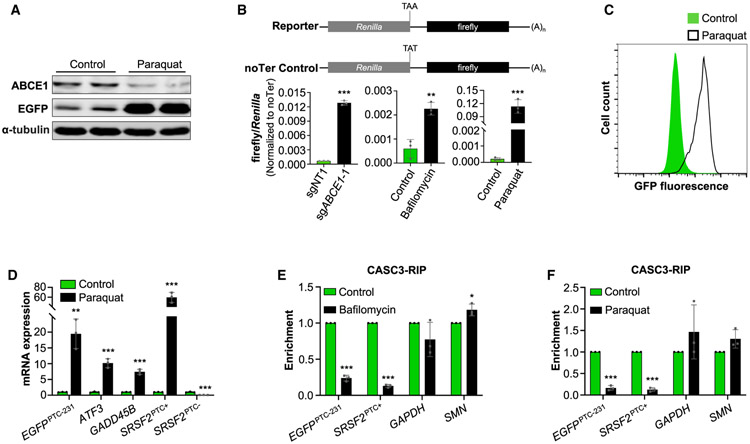
It has been reported that ROS damage Fe-S clusters, reducing ABCE1 protein stability and leading to defective ribosome recycling (Alhebshi et al., 2012; Gerashchenko et al., 2012; Sudmant et al., 2018). The effect of ROS on ABCE1 protein abundance was recapitulated in HCT116 cells by exposure to paraquat, a potent ROS-inducing compound (Figure 6A). As shown above, impaired lysosomal acidification has a similar effect on ABCE1 levels (Figure 4A). To confirm that these forms of cellular stress manifest in a ribosome recycling defect, we employed a dual-luciferase reporter system in which a Renilla luciferase ORF is followed by a firefly luciferase ORF, the latter of which will be translated when the intervening stop codon is bypassed by unrecycled ribosomes (Figure 6B). Indeed, we observed that loss of ABCE1, impairment of lysosomal acidification, or generation of ROS strongly induced translation of the downstream ORF. Moreover, paraquat treatment potently inhibited NMD, as evidenced by a robust increase in fluorescence, EGFP mRNA, and EGFP protein in EGFPPTC-231 reporter cells (Figures 6A, 6C, and 6D). Endogenous NMD target genes were markedly upregulated as well. Supporting the model that impaired ABCE1-mediated ribosome recycling causes displacement of EJCs downstream of termination codons under these conditions, CASC3 RIP demonstrated that paraquat or bafilomycin treatment reduced EJC occupancy in the 3′ UTRs of EGFPPTC-231 and NMD-sensitive SRSF2 transcripts (Figures 6E, 6F, and S6). These data demonstrate that the Fe-S cluster of ABCE1 links oxidative stress and lysosomal dysfunction to the NMD pathway.
Figure 6. Oxidative Stress Inhibits ABCE1 and NMD.
(A) Western blot analysis of ABCE1 and EGFP protein in paraquat-treated (200 μg/mL) EGFPPTC-231 cells.
(B) Activity of dual-luciferase reporters in HCT116 cells after lentiviral delivery of control or ABCE1-targeting sgRNAs, bafilomycin treatment (5 nM), or paraquat treatment (200 μg/mL). Firefly/Renilla activity in reporter-transfected cells was normalized to firefly/Renilla activity in noTer control-transfected cells.
(C) Flow cytometry analysis of paraquat-treated EGFPPTC-231 cells.
(D) qRT-PCR analysis of NMD targets or PTC+ and PTC− isoforms of SRSF2 in paraquat- or control-treated EGFPPTC-231 cells. mRNA expression was first normalized to GAPDH expression and then normalized to expression level in control-treated cells.
(E and F) qRT-PCR analysis of NMD targets (EGFPPTC-231 and SRSF2PTC+) and control transcripts (GAPDH and SMN) in CASC3 RIP samples from bafilomycin-treated (5 nM, E) or paraquat-treated (200 μg/mL, F) cells. For each transcript, enrichment was first normalized to input and then normalized to a paired control sample.
Data are represented as mean ± SD (n = 3 biological replicates for all qRT-PCR and luciferase experiments). *p < 0.05, **p < 0.01, ***p < 0.001; calculated by Student’s t test. See also Figure S6.
Loss of ABCE1-Mediated Ribosome Recycling Broadly Impairs 3′ UTR-Directed Post-transcriptional Regulation
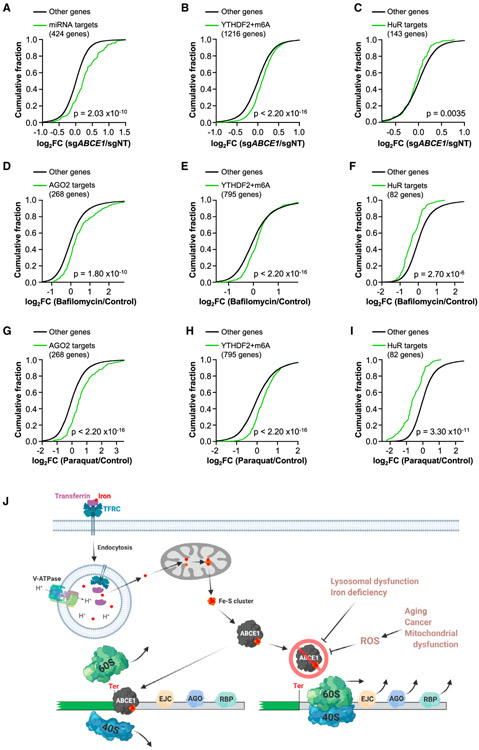
The binding of diverse RNP complexes to mRNA 3′ UTRs results in a broad array of post-transcriptional regulatory outcomes, including altered mRNA stability, translation, or localization (Mayr, 2017). Given our finding that loss of ABCE1-mediated ribosome recycling results in displacement of EJCs as ribosomes move beyond the termination codon on NMD substrates, we hypothesized that unrecycled ribosomes might similarly displace other regulatory factors that are encountered as 3′ UTRs are traversed. miRNAs represent a highly studied class of post-transcriptional repressors that recruit Argonaute (AGO) proteins to target sites predominantly within 3′ UTRs (Gebert and MacRae, 2019). Remarkably, we observed global upregulation of mRNAs that are regulated by AGO2 (Golden et al., 2017) in ABCE1-deficient HCT116 cells (Figure 7A). AREs represent another class of 3′ UTR cis elements that recruit proteins such as KHSPR, TIA1/TIAL1, AUF1, and TTP to accelerate mRNA decay (Shen and Malter, 2015). As observed for miRNA targets, ARE-containing transcripts (Bakheet et al., 2018) were broadly upregulated upon depletion of ABCE1 (Figure S7A). Similar effects were observed for transcripts modified by m6A (Wang et al., 2014), which recruits YTHDF2 to accelerate mRNA turnover (Shi et al., 2019; Wang et al., 2014; Figure 7B) as well as for transcripts bound by TIA1 or RC3H1 (Meyer et al., 2018; Murakawa et al., 2015; Figures S7B and S7C), RBPs that promote decay of their targets (Murakawa et al., 2015; Yamasaki et al., 2007). To confirm that upregulation of these classes of transcripts was not due to general impairment of mRNA turnover, we examined targets of the RBP HuR, which can stabilize mRNAs to which it binds. Indeed, downregulation of HuR targets (Lebedeva et al., 2011) was observed in ABCE1-deficient cells (Figure 7C).
Figure 7. Depletion of ABCE1 Broadly Impairs 3′ UTR-Directed Regulation.
(A–C) CDF plots showing the effects of ABCE1 deficiency on the expression of miRNA targets, defined as mRNAs with FC > 1.5 in AGO2−/− HCT116 cells compared with wild-type cells (Golden et al., 2017) (A), mRNAs with m6A modification that were bound by YTHDF2 (Wang et al., 2014) (B), or HuR targets defined as the 150 most downregulated HuR-bound mRNAs in HuR-deficient cells (Lebedeva et al., 2011) (143 were expressed in HCT116 cells; C).
(D–I) CDF plots showing the effect of bafilomycin treatment (5 nM, D–F) or paraquat treatment (200 μg/mL, G–I) on the targets of post-transcriptional regulators shown in (A–C). Targets in each plot were restricted to those that were increased (AGO2 and YTHDF2 targets) or decreased (HuR targets) in ABCE1 knockout cells to specifically examine transcripts whose levels are regulated by ribosome recycling.
(J) ABCE1-mediated ribosome recycling links lysosomal dysfunction, iron-deficiency, and oxidative stress to 3′ UTR-directed post-transcriptional regulation and NMD. Lysosomal acidification is necessary for TF-mediated iron uptake, which is required for biogenesis of the Fe-S cluster that serves as a critical co-factor for ABCE1. Impaired ABCE1 function, occurring in the setting of reduced iron availability or elevated ROS, leads to movement of unrecycled ribosomes into 3′ UTRs, where they displace EJCs and other regulatory RNPs, disabling NMD and other forms of post-transcriptional regulation.
The p values for CDF plots were calculated by one-sides Kolmogorov-Smirnov test. See also Figure S7.
We further assessed whether impaired lysosomal acidification or oxidative stress, which compromise ABCE1 function, similarly inhibit 3′ UTR-directed post-transcriptional regulation. The expression of transcripts that are regulated by ABCE1 and are targets of various post-transcriptional regulatory pathways were examined by RNA-seq after bafilomycin or paraquat treatment. Similar to the effects of ABCE1 depletion, bafilomycin (Figures 7D, 7E, and S7D-S7F) or paraquat (Figures 7G, 7H, and S7G-S7I) upregulated targets of AGO2, YTHDF2, TIA1, and RC3H1 as well as ARE-containing mRNAs, whereas HuR targets were downregulated under these conditions (Figures 7F and 7I). In addition, we documented that miRNA targets, m6A-modified mRNAs, and TIA1-bound transcripts that were highly upregulated in ABCE1 knockout cells were similarly upregulated upon iron chelation (Figure S7J). Altogether, these data demonstrate that efficient ABCE1-mediated ribosome recycling, which is impaired by lysosomal dysfunction, disruption of iron homeostasis, or oxidative stress, is essential for maintaining the activity of a broad set of post-transcriptional regulatory pathways.
DISCUSSION
The 3′ UTR is a privileged region of an mRNA that is critical for appropriate post-transcriptional regulation. Although the 5′ UTR and ORF must be traversed by ribosomal subunits or translating ribosomes, 3′ UTRs are free of this constraint and are instead bound by diverse mRNP complexes that confer a variety of regulatory outcomes on the transcript, including altered mRNA localization, stability, and translation (Mayr, 2017). NMD can be thought of as a specialized form of 3′ UTR-directed regulation where the presence of an EJC downstream of the termination codon or other features indicative of an atypical 3‱ UTR, such as excessive length, trigger accelerated mRNA decay (Celik et al., 2015). Here we describe a previously unrecognized mechanism that governs 3′ UTR-directed regulation. Specifically, we demonstrated that efficient removal of ribosomal subunits at termination codons by the ribosome recycling factor ABCE1 is essential to maintain the accessibility of 3′ UTRs to a broad array of regulatory assemblies, including EJCs, miRNA-AGO complexes, and RBPs. Furthermore, the Fe-S cluster that serves as a crucial co-factor for ABCE1 is a key vulnerability that connects lysosomal function, iron metabolism, and oxidative stress to 3′ UTR-mediated post-transcriptional control.
A Genetic Screen Reveals a V-ATPase/Fe-S Cluster/ABCE1 Pathway that Is Essential for NMD
This study reports the results of a comprehensive genome-wide CRISPR-Cas9 screen to identify components and regulators of the human NMD pathway. Because of the ability of our screening methodology to achieve prolonged gene knockout while retaining the sensitivity to recover essential genes as hits, these experiments uncovered many putative NMD factors that have not been associated previously with the pathway. We anticipate that this dataset will be a valuable resource for the field, complementing previous genetic screens performed in model organisms (Hodgkin et al., 1989; Leeds et al., 1991) and other human cell lines (Alexandrov et al., 2017; Baird et al., 2018).
We were particularly intrigued by two classes of genes that uniquely emerged from our screen: components of the V-ATPase and proteins required for Fe-S cluster biogenesis. Functional dissection of these pathways revealed that NMD requires lysosomal acidification to maintain transferrin-mediated iron uptake, which, in turn, is necessary for biogenesis of the Fe-S cluster that serves as an essential co-factor for ABCE1. ABCE1 promotes efficient dissociation of ribosomal subunits at termination codons, and its loss results in movement of ribosomes into 3′ UTRs. On NMD substrates, unrecycled 3′ UTR ribosomes displace EJCs, preventing recruitment of the NMD machinery and subsequent accelerated decay of the transcript (Figure 7J).
Ribosome Recycling, NMD, and 3′ UTR-Directed Regulation
ABCE1-mediated ribosome recycling is initiated when the eRF1-eRF3 complex completes the process of peptide release at a terminating ribosome. At this stage, eRF3 dissociates, enabling ABCE1 to bind to the site it previously occupied on eRF1 (Becker et al., 2012; Preis et al., 2014). However, at NMD-inducing termination codons, UPF1 and SMG1 are recruited to terminating ribosomes that are thought to be still occupied by eRF1 and eRF3 (Kashima et al., 2006). In this scenario, ABCE1 would be physically constrained from joining the termination complex. Furthermore, ATP hydrolysis by Upf1 is required to dissociate ribosomes at PTCs in yeast (Serdar et al., 2016), whereas human UPF3B can promote ribosome recycling in vitro (Neu-Yilik et al., 2017). These findings support the existence of distinct ribosome recycling mechanisms at NMD- and non-NMD-inducing termination codons (Celik et al., 2015).
How can a requirement for ABCE1 in efficient decay of NMD substrates be reconciled with the evidence suggesting an alternative mechanism of ribosome recycling at stop codons that initiate NMD? Recent results from single-molecule studies of the kinetics of NMD in living cells provide key insights into this question (Hoek et al., 2019). It has been proposed that NMD exclusively occurs during a pioneer round of translation (Ishigaki et al., 2001; Maquat et al., 2010; Sato et al., 2008), although evidence supporting activation of NMD during later rounds of translation has also been reported (Durand and Lykke-Andersen, 2013; Rufener and Mühlemann, 2013). The evidence provided by Hoek et al. (2019) indicates that each translating ribosome has an equal probability of inducing NMD, with approximately eight rounds of translation occurring before accelerated decay of a nonsense transcript occurs. This scenario provides an opportunity for ABCE1-dependent and ABCE1-independent ribosome recycling to operate on the same transcript. Because the probability of NMD activation by the first ribosome is relatively low, ABCE1 activity is necessary to prevent ribosome leakage into the 3′ UTR, which would displace EJCs and impair recruitment of the NMD machinery during later rounds of translation. It is worth noting that movement of only a single ribosome through the 3′ UTR is likely sufficient to clear bound EJCs, rendering a transcript resistant to NMD, which likely explains the potent inhibitory effect of ABCE1 deficiency on NMD despite the relatively low steady-state 3′ UTR ribosome occupancy observed under this condition (Figure 5A).
The removal of 3′ UTR-bound regulatory factors by unrecycled ribosomes is not limited to NMD-inducing complexes such as EJCs. In principle, any trans-effector bound to the 3′ UTR may be displaced as ribosomes aberrantly move downstream of the termination codon. Indeed, by examining the targets of diverse post-transcriptional regulators, we found evidence of widespread impairment of 3′ UTR-directed mRNA control in ABCE1-deficient cells. Observed defects included impaired miRNA-, ARE-, and m6A-mediated transcript decay as well as reduced regulation by diverse RBPs. Thus, efficient ribosome recycling by ABCE1 is needed to maintain the appropriate mRNP configuration downstream of the stop codon to enable activity of a broad set of post-transcriptional regulatory pathways (Figure 7J).
The Fe-S Cluster of ABCE1 Links Lysosomal Function, Iron Availability, and Oxidative Stress to 3′ UTR-Directed Regulation
Impaired lysosomal acidification is strongly associated with many diseases, particularly multiple forms of congenital or acquired neurodegeneration (Colacurcio and Nixon, 2016). In addition to disease-associated mutations in genes encoding V-ATPase subunits that directly disable lysosomal acidification, defects in enzymes that cause lysosomal storage diseases also impair acidification of this organelle (Fukuda et al., 2006; Jansen et al., 2016; Ramirez et al., 2006; Yambire et al., 2019; Yuan et al., 2014). Because an acidic pH is essential for hydrolysis of proteins and other cellular constituents within the lysosome, pathogenesis of diseases in this context has traditionally been ascribed to toxic accumulation of lysosomal material. It is now understood, however, that lysosomal dysfunction results in much broader effects on cellular signaling and metabolism (Lieand Nixon, 2019). For example, because of the dependence of transferrin-mediated iron uptake upon an acidic lysosomal pH, lysosomal defects cause a deficiency of intracellular iron. Iron depletion is a major toxic consequence of lysosomal dysfunction in yeast and mammalian cells (Hughes et al., 2020; Weber et al., 2020). Previous work largely attributed the deleterious effects of iron deficiency to mitochondrial dysfunction. The results presented here add an additional dimension to our understanding of the consequences of lysosomal dysfunction and iron deficiency on cellular physiology. The impairment of Fe-S cluster biogenesis that occurs under these conditions ultimately inactivates ABCE1, leading to widespread loss of 3′ UTR-mediated regulation and inactivation of the NMD pathway. Thus, the Fe-S cluster of ABCE1 broadly links lysosomal function and iron availability to post-transcriptional control. These effects represent an important additional mechanism that likely contributes to the deleterious phenotypic consequences resulting from lysosomal abnormalities.
The presence of oxidative stress is another condition that leads to inhibition of NMD. Previously, this effect was attributed to the ability of ROS to activate the integrated stress response (Karam et al., 2013; Liu et al., 2008), in which phosphorylation of eIF2a globally reduces bulk translation while selectively increasing translation of factors that facilitate stress adaptation. Because of the dependency of NMD on ongoing translation, this scenario would be expected to lead to global stabilization of nonsense transcripts. Importantly, the Fe-S cluster of ABCE1 is also known to be highly susceptible to damage by ROS (Alhebshi et al., 2012). Accordingly, ROS-inducing agents abrogate ABCE1-mediated ribosome recycling and cause ribosomal accumulation within 3′ UTRs (Sudmant et al., 2018). Our studies confirmed that ROS generation potently inhibits NMD and is associated with removal of EJCs downstream of termination codons. Thus, loss of ribosome recycling contributes to impairment of NMD in the presence of oxidative stress. Importantly, the widespread requirement for efficient ribosome recycling in 3′ UTR-directed regulation provides a mechanism through which oxidative stress may affect gene expression much more broadly. Given the prevalence of elevated ROS in many pathologic conditions, including mitochondrial dysfunction, aging, and cancer (Davalli et al., 2016; Sabharwal and Schumacker, 2014), the Fe-S cluster of ABCE1 is a key vulnerability that may connect diverse disease conditions to defective post-transcriptional control (Figure 7J).
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Requests for further information or reagents should be directed to the lead contact, Joshua T. Mendell (Joshua.Mendell@UTSouthwestern.edu).
Materials Availability
All reagents generated in this study are available upon request from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
All high-throughput sequencing data generated in this study (CRISPR screening, RNA-seq, and ribosome profiling) have been deposited in GEO (Accession number GEO: GSE144165). No custom code was generated in this study.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human cell lines were obtained from ATCC. HCT116 cells (male) were cultured in McCoy′s 5A media (Invitrogen) supplemented with 10% (v/v) fetal bovine serum (Sigma) and 1X Antibiotic-Antimycotic (Invitrogen). HEK293T (most likely female due to the presence of multiple X chromosomes and no detectable Y chromosome), Huh7 (male), and HeLa (female) were cultured in Dulbecco′s Modified Eagle′s Medium (DMEM) (Invitrogen) supplemented with 10% (v/v) fetal bovine serum (Sigma) and 1X Antibiotic-Antimycotic (Invitrogen). Cell lines were confirmed to be free of mycoplasma contamination.
METHOD DETAILS
Construction of NMD reporter plasmids
A DNA fragment containing HBB exon 2 (231bp), intron 2, and exon 3 was amplified from human genomic DNA by PCR with Phusion High-Fidelity DNA Polymerase (New England Biolabs) and cloned into pAAVS-EGFP-DONOR (Manjunath et al., 2019) XbaI and MfeI sites using NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs) to generate pAAVS-EGFPPTC-231. A sub-fragment of HBB, beginning 35 bp upstream of the of the 3′ end of exon 2, was amplified from pAAVS-EGFPPTC-231 and cloned into pAAVS-EGFP-DONOR XbaI and MfeI sites to generate pAAVS-EGFPPTC-35. Primer sequences are provided in Table S3.
Generation of NMD reporter cell lines
Plasmids expressing TALEN pairs targeting the human AAVS1/PPP1R12C locus (Addgene #35431 and # 35432) (Sanjana et al., 2012) and pAAVS-EGFPPTC-231 or pAAVS-EGFPPTC-35 were co-transfected into HCT116 cells using FuGENE HD transfection reagent (Promega) at a ratio of Left TALEN:Right TALEN:EGFPPTC-231 or pAAVS-EGFPPTC-35 = 1:1:5. Two days after transfection, selection with 200 μg/ml hygromycin (ThermoFisher Scientific) was initiated and maintained for ten days. Single cell clones were then derived and screened for reporter fluorescence.
Genome-wide CRISPR-Cas9 screening
Virus preparation
The Brunello library (Addgene #73179) (Doench et al., 2016) was amplified in Endura Electrocompetent cells (Lucigen) and sequenced to confirm maintenance of sgRNA representation. Virus was generated by transfecting HEK293T cells growing in 10 cm dishes with 5 μg Brunello library plasmid, 3 μg psPAX2 (Addgene #12260), and 2 μg pMD2.G (Addgene #12259) using 30 μL FuGENE HD transfection reagent. Supernatant was collected at 48 hours and 72 hours post-transfection and stored at −80°C.
Cell transduction and sorting
Screening was performed in two independent EGFPPTC-231 clones and two independent EGFPPTC-35 clones. For each clone, 1.35 × 108 cells were seeded into nine 15 cm dishes (1.5 × 107 cells/dish), followed immediately by addition of 8 μg/ml of polybrene (Millipore) and lentiviral library at a multiplicity of infection of ~0.3 in order to achieve ~500X coverage of the library. Two days post-transduction, cells were re-seeded into media containing 1 μg/ml puromycin (ThermoFisher Scientific) and selected for eight days, during which time cells were passaged every 2 or 3 days as needed. After selection, 6 × 107 cells were sorted per clone using a Mo-Flo cell sorter (Beckman Coulter), collecting the brightest 0.5% of cells. At least 2 × 105 sorted cells and 4 × 107 unsorted cells for each clone were used for genomic DNA isolation.
Genomic DNA isolation, sgRNA amplification, sequencing, and data analysis
Genomic DNA (gDNA) was isolated from sorted cells by phenol–chloroform extraction as described (Golden et al., 2017). gDNA was isolated from unsorted cells using the MasterPureTM Complete DNA Purification Kit (Lucigen). sgRNAs were amplified from gDNA using Herculase II Fusion DNA Polymerase (Agilent) using two sequential rounds of PCR. For unsorted cells, 6.6 μg of gDNA (repre-senting approximately 1 × 106 cells) were used in each 100 μL reaction for PCR 1, with 40 reactions used in total for each sample. For sorted cells, all recovered gDNA was split into two 100 μL reactions. After 18 cycles of amplification for PCR 1, reactions were pooled and 5 μL were used for a second round of PCR, consisting of 8-12 cycles, to append barcodes and adapters for Illumina sequencing. Primer sequences are provided in Table S3. Amplicons were purified using Agencourt AMPure XP beads (Beckman Coulter Life Sciences) and sequenced on an Illumina NextSeq500 with 75 bp single-end reads. Bowtie 2 (Langmead and Salzberg, 2012) was used to map reads obtained from the demultiplexed FASTQ files to the reference library, allowing no mismatches, and normalized read counts were calculated as described (Shalem et al., 2014). Approximately 2 × 107 reads were obtained for each sample. MAGeCK (Li et al., 2014) was used to identify genes targeted by sgRNAs that were enriched in sorted cells versus unsorted cells. DAVID 6.8 was used for GO analysis of EGFPPTC-231-specific hits (https://david.ncifcrf.gov/home.jsp) (Dennis et al., 2003).
CRISPR-Cas9 mediated gene knockout
sgRNAs targeting genes of interest, as well as non-target (NT) sgRNAs (sequences provided in Table S3), were cloned into the lenti-CRISPR v2 vector (Addgene #52961) as previously described (Sanjana et al., 2014). Knockout pools were generated as described previously (Manjunath et al., 2019) with minor modifications. To generate ABCE1-knockout pools, transduced cells were selected with 1 μg/ml puromycin for 4 days (HCT116, Huh7, and HeLa) or 5 days (HEK293T) before analysis. To produce knockout pools for all other genes, transduced cells were selected with 1 μg/ml puromycin for 6 days before harvesting.
Ribosome profiling
Ribosome profiling was performed as described previously (McGlincy and Ingolia, 2017) with the following modifications:
Ribosome footprints were size selected by gel electrophoresis as described previously (Ingolia et al., 2012), and fragments between 26-34nt were excised for library preparation.
Samples were not pooled after linker ligation, and the subsequent steps were performed individually for each sample.
3′ linker-ligated RNA fragments were purified using gel extraction as previously described (Ingolia et al., 2012).
Ribosomal RNA was depleted as described previously (Ingolia et al., 2012).
RNA-seq
RNA was isolated using the RNeasy Mini kit (QIAGEN) and gDNA contamination was removed using the TURBO DNA-Free kit (ThermoFisher Scientific). Sequencing libraries were generated with the TruSeq Stranded mRNA Library Prep Kit (Illumina) and sequenced on a NextSeq 500 (Illumina) with 75-bp single-end reads.
Measurement of EGFP reporter mRNA stability
The piggyBac donor plasmid XLone-GFP (Addgene #96930) (Randolph et al., 2017) was modified by replacing the GFP (between KpnI and SpeI sites) with a DNA fragment containing NheI, BmtI, NotI, PstI, BglII, and PmeI sites to generate XLone-MCS. The rtTA sequence was then replaced with tTA, producing XLone-tTA-MCS. The EGFPPTC-231 or EGFPPTC-35 sequences were excised from pAAVS-EGFPPTC-231 or pAAVS-EGFPPTC-35, respectively, using KpnI and HpaI and cloned into the KpnI and PmeI sites of XLone-tTA-MCS. The resulting tetOFF-EGFPPTC-231 or tetOFF-EGFPPTC-35 plasmids, together with pCMV-hyPBase plasmid (Yusa et al., 2011) which encodes the piggyBac transposase, were co-transfected into HCT116 cells and cells were selected with 5 μg/ml blasticidin (ThermoFisher Scientific) for 14 days to generate stable cell lines. For mRNA stability measurements, cells were treated with 1 μg/ml doxycycline (Sigma) for 0, 40, 80, or 120 minutes. qRT-PCR was used to assess reporter mRNA abundance relative to GAPDH mRNA at each time point.
Western blotting
Cells were lysed in RIPA buffer (Sigma). Proteins were separated by SDS-PAGE electrophoresis, transferred to nitrocellulose membranes, and detected using an infrared fluorescent antibody detection system (LI-COR). Antibodies are provided in the Key Resources Table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| ABCE1 antibody | Novus Biologicals | Cat# NB400-116; RRID:AB_10002055 |
| CASC3 Antibody | Bethyl | Cat# A302-472A; RRID:AB_1944210 |
| CASC3 Antibody | Bethyl | Cat# A302-471A; RRID:AB_1944212 |
| UPF1 antibody | Bethyl | Cat# A300-038A; RRID:AB_2288326 |
| UPF1 antibody | Santa Cruz | Cat# sc-393594 |
| TFRC antibody | Bethyl | Cat# A304-381A-T; RRID:AB_2781867 |
| α-tubulin antibody | Sigma | Cat# T6199; RRID:AB_477583 |
| GFP antibody | Cell Signaling Technology | Cat# 2956S; RRID:AB_1196615 |
| Bacterial and Virus Strains | ||
| Endura Electrocompetent cells | Lucigen | Cat# 60242-2 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| FuGENE HD Transfection Reagent | Promega Corporation | Cat# E2311 |
| Hygromycin B | ThermoFisher Scientific | Cat# 10687010 |
| Puromycin (Puromycin Dihydrochloride) | ThermoFisher Scientific | Cat# A11138-03 |
| Polybrene | Millipore | Cat# TR-1003-G |
| Blasticidin | ThermoFisher Scientific | Cat# A1113903 |
| Bafilomycin A1 | Fisher Scientific | Cat# AAJ61835MCR |
| Deferoxamine mesylate (DFO) | Sigma | Cat# D9533 |
| Iron(III) citrate | Sigma | Cat# F6129 |
| Paraquat dichloride hydrate | Sigma | Cat# 36541 |
| Protease Inhibitor Cocktail Set III, EDTA-Free | Millipore | Cat# 539134 |
| Doxycycline monohydrate | Sigma | Cat# D1822 |
| Dulbecco′s Modified Eagle′s Medium (DMEM) | Invitrogen | Cat# 11995-073 |
| McCoy’s 5A medium | Invitrogen | Cat# 12330031 |
| Fetal Bovine Serum (FBS) | Sigma | Cat# F2442 |
| Antibiotic-antimycotic | Invitrogen | Cat# 15240-112 |
| PhosSTOP Phosphatase inhibitor | Sigma | Cat# 4906837001 |
| RNase Inhibitor | New England Biolabs | Cat# M0314L |
| Laemmli buffer | Fisher Scientific | Cat# NC9165175 |
| TRIzol | Invitrogen | Cat# 15596026 |
| HBSS buffer | ThermoFisher Scientific | Cat# 14025092 |
| RIPA Buffer | Sigma | Cat# R0278 |
| Calcein acetoxymethyl ester (CA-AM) | Abcam | Cat# ab141420 |
| Critical Commercial Assays | ||
| Agencourt AMPure XP beads | Beckman Coulter Life Sciences | Cat# A63882 |
| Qubit dsDNA HS Assay Kit | ThermoFisher Scientific | Cat# Q32851 |
| Direct-zol RNA Miniprep Kit | Zymo Research | Cat# R2051 |
| SYBR Green PCR Master Mix | ThermoFisher Scientific | Cat# 4309155 |
| Phusion High-Fidelity DNA Polymerase | New England Biolabs | Cat# M0530S |
| NEBuilder HiFi DNA Assembly Master Mix | New England Biolabs | Cat# E2621S |
| TURBO DNA-free kit | ThermoFisher Scientific | Cat# AM1907 |
| PrimeScript RT Master Mix (Perfect Real Time) | Clontech | Cat# RR036A |
| RNeasy Mini Kit | QIAGEN | Cat# 74106 |
| Herculase II Fusion DNA Polymerase | Agilent | Cat# 600679 |
| Protein Synthesis Assay Kit | Cayman Chemical | Cat# 601100 |
| TruSeq Stranded mRNA Library Prep Kit | Illumina | Cat# RS-122-2101 |
| Dynabeads Protein G | Invitrogen | Cat# 10004D |
| Dynabeads® MyOne Streptavidin T1 | Invitrogen | Cat# 65601 |
| Dual-Luciferase® Reporter Assay System | Promega Corporation | Cat# E1980 |
| MasterPureTM Complete DNA Purification Kit | Lucigen | Cat# MC85200 |
| Deposited Data | ||
| CRISPR screening data | This paper | GEO: GSE144165 |
| Ribosome profiling data from ABCE1-knockout cells | This paper | GEO: GSE144165 |
| RNA-seq data from ABCE1-knockout cells | This paper | GEO: GSE144165 |
| RNA-seq data from Bafilomycin-treated cells | This paper | GEO: GSE144165 |
| RNA-seq data from Paraquat-treated cells | This paper | GEO: GSE144165 |
| Experimental Models: Cell Lines | ||
| pAAVS-EGFPPTC-231 cells | This paper | N/A |
| pAAVS-EGFPPTC-35 cells | This paper | N/A |
| tetOFF-EGFPPTC-231 cells | This paper | N/A |
| tetOFF-EGFPPTC-35 cells | This paper | N/A |
| HCT116 | ATCC | CCL-247 |
| HEK293T | ATCC | CRL-3216 |
| HeLa | ATCC | CCL-2 |
| Oligonucleotides | ||
| Sequences of oligonucleotides used in this study provided in Table S3 | This paper | N/A |
| Recombinant DNA | ||
| pAAVS-EGFPPTC-231 | This paper | N/A |
| pAAVS-EGFPPTC-35 | This paper | N/A |
| tetOFF-EGFPPTC-231 | This paper | N/A |
| tetOFF-EGFPPTC-35 | This paper | N/A |
| XLone-MCS | This paper | N/A |
| XLone-tTA-MCS | This paper | N/A |
| XLone-GFP | Addgene (Xiaojun Lian) | Cat# 96930 (Randolph et al., 2017) |
| pCMV-hyPBase | Nancy Craig | Yusa et al., 2011 |
| Human CRISPR Knockout Pooled Library (Brunello) | Addgene (David Root, John Doench) | Cat# 73179 (Doench et al., 2016) |
| lentiCRISPR v2 | Addgene (Feng Zhang) | Cat# 52961 (Sanjana et al., 2014) |
| psPAX2 | Addgene (Didier Trono) | Cat# 12260 |
| pMD2.G | Addgene (Didier Trono) | Cat# 12259 |
| hAAVS1 1L TALEN | Addgene (Feng Zhang) | Cat# 35431 (Sanjana et al., 2012) |
| hAAVS1 1R TALEN | Addgene (Feng Zhang) | Cat# 35432 (Sanjana et al., 2012) |
| pAAVS-EGFP-DONOR | Joshua Mendell | Manjunath et al., 2019 |
| pGL3-Renilla-Ter-Firefiy | This paper | N/A |
| pGL3-Renilla-noTer-Firefly | This paper | N/A |
| pGL3-Control | Promega Corporation | Cat# E1741 |
| Software and Algorithms | ||
| Bowtie2 | Langmead and Salzberg, 2012 | N/A |
| MAGeCK | Li et al., 2014 | N/A |
| STAR | Dobin et al., 2013 | N/A |
| featureCounts | Liao et al., 2014 | N/A |
| edgeR | Robinson et al., 2010 | N/A |
qRT-PCR
RNA was isolated using the RNeasy Mini kit (QIAGEN) using on-column DNase digestion. PrimeScript RT Master Mix (Clontech) was used to synthesize cDNA from 1 μg total RNA and qRT-PCR was performed with the SYBR Green Master Mix (ThermoFisher Scientific). mRNA expression was normalized to GAPDH expression in all experiments. Primer sequences are provided in Table S3.
Measurement of intracellular iron
Calcein acetoxymethyl ester (CA-AM) (Abcam) was used to measure the intracellular labile iron pool in HCT116 cells as previously described (Prus and Fibach, 2008) with the following modifications. Cells were trypsinized and washed twice with Hank’s Balanced Salt Solution (HBSS, ThermoFisher Scientific) followed by incubation with 0.05 mM CA-AM in HBSS for 15 minutes with gentle shaking. After staining, cells were washed twice with HBSS and analyzed by flow cytometry.
Bafilomycin, DFO, and paraquat treatment
Bafilomycin (Fisher Scientific) was dissolved in DMSO, while desferrioxamine (DFO) (Sigma) and paraquat (Sigma) were dissolved in distilled water. An equal volume of vehicle was used as a control for all experiments. 6 × 104 cells were seeded into each well of 12-well plate and drug treatments were initiated 24 hours later. For bafilomycin and DFO treatment, cells were treated with 5-10 nM bafilomycin or 8 mM DFO for 48 hours. For paraquat treatment, cells were treated with 200 μg/mL paraquat for a total of 96 hours. Media was changed 24 hours after the beginning of the paraquat treatment and cells were split 1:2 into a new 12-well plate 48 hours after drug addition.
Iron (III) citrate treatment
Iron (III) citrate (Sigma) was dissolved in distilled water. 24 hours after lentiviral delivery of Cas9 and non-target (NT) or ATP6V0C-targeting sgRNAs, 0.5 mM Iron (III) citrate was added to the medium. 48 hours after lentiviral transduction, fresh medium with 1 μg/ml puromycin and 0.5 mM Iron (III) citrate was added and cells were grown for an additional 6 days before harvesting. For bafilomycin/Iron (III) citrate co-treatment, 6 × 104 cells were seeded into each well of 12-well plate with 0.5 mM Iron (III) citrate. 24 hours later, 5 nM bafilomycin was added to culture medium and cells were incubated for an additional 48 hours.
Analysis of total protein synthesis with OPP
Protein synthesis was assessed using the Protein Synthesis Assay Kit (Cayman Chemical) as described by the manufacturer. Briefly, cells were incubated with OPP Working Solution at 37°C for 30 minutes. Cells were then washed, fixed, and incubated in 5FAM-Azide staining solution in the dark at room temperature for 30 minutes followed by analysis by flow cytometry.
RNA immunoprecipitation (RIP)
RIP was performed as previously described (Kurosaki et al., 2018). Briefly, 2 × 107 cells grown in a 15 cm dish were harvested by scraping in 1 mL cold PBS. Cells were then washed twice with cold PBS and lysed in 1 mL hypotonic lysis buffer [10 mM Tris–HCl (pH 7.4), 10 mM NaCl, 10 mM EDTA, 0.5% Triton X-100, 1x EDTA-free Protease inhibitor (Millipore), 1x PhosSTOP Phosphatase inhibitor (Sigma), 1x RNase inhibitor (New England Biolabs)]. Following centrifugation at 15,000 g for 10 min at 4°C, the supernatant was pre-cleared using 50 μL Dynabeads Protein G (Invitrogen). Precleared lysate was added to 50 μL Dynabeads Protein G coupled with 3 μg antibody (anti-CASC3, Bethyl, A302-471A; anti-UPF1, Bethyl, A300-038A) and incubated at 4°C for 2 hours with end-over-end rotation. Beads were washed 2 times each in ice-cold NET-2 buffer-1 [50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 0.1% NP-40], ice-cold NET-2 buffer-2 [50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 0.5% NP-40], and high salt wash buffer [20 mM Tris–HCl (pH 7.4), 325 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.1% SDS]. Washed beads were resuspended in 120 μL lysis buffer. 20 μL was added to Laemmli buffer (Fisher Scientific) and used for western blot analysis (anti-CASC3, Bethyl, 302-472A; anti-UPF1, Santa Cruz, sc-393594), while the remaining 100 μL was added to 900 μL Trizol (Invitrogen) and used for RNA isolation and qRT-PCR analysis.
Dual luciferase assay
A Renilla luciferase ORF with or without a TAA termination codon was inserted upstream of firefly luciferase in pGL3-Control (Promega) to generate pGL3-Renilla-Ter-firefly and pGL3-Renilla-noTer-firefly plasmids, respectively. 3 × 104 HCT116 cells were seeded into each well of a 12-well plate. 24 hours later, cells were transfected with 0.3 μg pGL3-Renilla-Ter-firefly or pGL3-Renilla-noTer-firefly plasmids using FuGENE HD transfection reagent (Promega). Beginning 12 hours post-transfection, cells were treated with 5 nM bafilomycin for 48 hours or 200 μg/mL paraquat for 72 hours. For analysis of ABCE1 knockout, cells were transduced with lentiCRISPR vectors and then, 72 hours later, transfected with luciferase reporter plasmids. Beginning 24 hours after transfection, cells were selected in puromycin for 48 hours prior to luciferase assays. Luciferase activity was determined using the Dual-Luciferase Reporter Assay System (Promega Corporation). firefly/Renilla activity in pGL3-Renilla-Ter-firefly transfected cells was normalized to firefly/Renilla activity in pGL3-Renilla-noTer-firefly plasmid cells.
QUANTIFICATION AND STATISTICAL ANALYSIS
Analysis of RNA-seq and ribosome profiling data
For RNA-seq, reads were mapped to hg19 using STAR (Dobin et al., 2013), read counts were generated using featureCounts (Liao et al., 2014), and edgeR (Robinson et al., 2010) was used to analyze differential gene expression. Only transcripts with CPM > 0.5 were used for CDF analyses. Ribosome profiling data were analyzed as previously described (Manjunath et al., 2019).
Statistical analysis
All experiments were repeated with a minimum of three biological replicates. The Student′s t test and Kolmogorov-Smirnov test were used to determine statistical significance. All values are reported as mean ± SD in each figure.
Supplementary Material
Highlights.
A genome-wide CRISPR screen identifies regulators and components of the NMD pathway
ABCE1 is required for NMD and other forms of 3′ UTR-mediated regulation
Upon ABCE1 loss, ribosomes move into 3′ UTRs and displace bound regulators
Iron deficiency and oxidative stress impair ABCE1 and post-transcriptional control
ACKNOWLEDGMENTS
We thank David Root, John Doench, Feng Zhang, Didier Trono, Xiaojun Lian, and Nancy L. Craig for plasmids; Vanessa Schmid, Rachel Bruce, and Caitlin Eaton in the McDermott Center Next Generation Sequencing Core for assistance with high-throughput sequencing; Angie Mobley and the UT Southwestern Flow Cytometry Core facility for FACS; and Michael Buszczak, Kathryn O’Donnell, and members of the Mendell laboratory for helpful comments on the manuscript. This work was supported by grantsfrom CPRIT (RP150596 to the UTSW Bioinformatics Core Facility), the NIH (R35CA197311, P30CA142543, and P50CA196516 to J.T.M.), and the Welch Foundation (I-1961-20180324 to J.T.M.). J.T.M. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.107895.
DECLARATION OF INTERESTS
J.T.M. is a member of the scientific advisory board of Ribometrix.
REFERENCES
- Alexandrov A, Shu MD, and Steitz JA (2017). Fluorescence Amplification Method for Forward Genetic Discovery of Factors in Human mRNA Degradation. Mol. Cell 65,191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhebshi A, Sideri TC, Holland SL, and Avery SV (2012). The essential iron-sulfur protein Rli1 is an important target accounting for inhibition of cell growth by reactive oxygen species. Mol. Biol. Cell 23, 3582–3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird TD, Cheng KC, Chen YC, Buehler E, Martin SE, Inglese J, and Hogg JR (2018). ICE1 promotes the link between splicing and nonsense-mediated mRNA decay. eLife 7, e33178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakheet T, Hitti E, and Khabar KSA (2018). ARED-Plus: an updated and expanded database of AU-rich element-containing mRNAs and pre-mRNAs. Nucleic Acids Res. 46 (D1), D218–D220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelme D, Dinkelaker S, Albers SV, Londei P, Ermler U, and Tampé R (2011). Ribosome recycling dependson a mechanistic link between the FeS clusterdomain and a conformational switch of the twin-ATPase ABCE1. Proc. Natl. Acad. Sci. USA 108, 3228–3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Franckenberg S, Wickles S, Shoemaker CJ, Anger AM, Armache JP, Sieber H, Ungewickell C, Berninghausen O, Daberkow I, et al. (2012). Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature 482, 501–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuvanagiri M, Schlitter AM, Hentze MW, and Kulozik AE (2010). NMD: RNA biology meets human genetic medicine. Biochem. J 430, 365–377. [DOI] [PubMed] [Google Scholar]
- Behler M, Steiner S, Mohn F, Paillusson A, and Mühlemann O (2006). EJC-independent degradation of nonsense immunoglobulin-mu mRNA depends on 3′ UTR length. Nat. Struct. Mol. Biol 73, 462–64. [DOI] [PubMed] [Google Scholar]
- Celik A, Kervestin S, and Jacobson A (2015). NMD: At the crossroads between translation termination and ribosome recycling. Biochimie 114, 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Fogel-Petrovic M, and Maquat LE (1990). Translation to near the distal end of the penultimate exon is required for normal levels of spliced triosephosphate isomerase mRNA. Mol. Cell. Biol 10, 5215–5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Belgrader P, Zhou X, and Maquat LE (1994). Introns are cis effectors of the nonsense-codon-mediated reduction in nuclear mRNA abundance. Mol. Cell. Biol 14, 6317–6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colacurcio DJ, and Nixon RA (2016). Disorders of lysosomal acidification- The emerging role of v-ATPase in aging and neurodegenerative disease. Ageing Res. Rev 32, 75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M, Karousis ED, Bourquin J, Bruggmann R, and Mühlemann O (2017). Transcriptome-wide identification of NMD-targeted human mRNAs reveals extensive redundancy between SMG6- and SMG7-mediated degradation pathways. RNA 23, 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crielaard BJ, Lammers T, and Rivella S (2017). Targeting iron metabolism in drug discovery and delivery. Nat. Rev. Drug Discov 76, 400–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalli P, Mitic T, Caporali A, Lauriola A, and D’Arca D (2016). ROS, Cell Senescence, and Novel Molecular Mechanisms in Aging and Age-Related Diseases. Oxid. Med. Cell. Longev 2016, 3565127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, and Lempicki RA (2003). DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, 3. [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, et al. (2016). Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol 34, 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand S, and Lykke-Andersen J (2013). Nonsense-mediated mRNA decay occurs during eIF4F-dependent translation in human cells. Nat. Struct. Mol. Biol 20, 702–709. [DOI] [PubMed] [Google Scholar]
- El-Bchiri J, Guilloux A, Dartigues P, Loire E, Mercier D, Buhard O, Sobhani I, de la Grange P, Auboeuf D, Praz F, et al. (2008). Nonsense-mediated mRNA decay impacts MSI-driven carcinogenesis and anti-tumor immunity in colorectal cancers. PLoS ONE 3, e2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T, Ewan L, Bauer M, Mattaliano RJ, Zaal K, Ralston E, Plotz PH, and Raben N (2006). Dysfunction of endocytic and autophagic pathways in a lysosomal storage disease. Ann. Neurol 59, 700–708. [DOI] [PubMed] [Google Scholar]
- Gebert LFR, and MacRae IJ (2019). Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol 20, 21–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerashchenko MV, Lobanov AV, and Gladyshev VN (2012). Genome-wide ribosome profiling reveals complex translational regulation in response to oxidative stress. Proc. Natl. Acad. Sci. USA 109, 17394–17399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbracht JV, Boehm V, and Britto-Borges T (2019). CASC3 promotes transcriptome-wide activation of nonsense-mediated decay by the exon junction complex. bioRxiv. 10.1101/811018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden RJ, Chen B, Li T, Braun J, Manjunath H, Chen X, Wu J, Schmid V, Chang TC, Kopp F, et al. (2017). An Argonaute phosphorylation cycle promotes microRNA-mediated silencing. Nature 542, 197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauer C, Sieber J, Schwarzl T, Hollerer I, Curk T, Alleaume AM, Hentze MW, and Kulozik AE (2016). Exon Junction Complexes Show a Distributional Biastoward Alternatively Spliced mRNAs and against mRNAs Coding for Ribosomal Proteins. Cell Rep. 76, 1588–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, and Jacobson A (2015). Nonsense-Mediated mRNA Decay: Degradation of Defective Transcripts Is Only Part of the Story. Annu. Rev. Genet 49, 339–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen CUT (2018). Translation Termination and Ribosome Recycling in Eukaryotes. Cold Spring Harb. Perspect. Biol 10, a032656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, et al. (2009). Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol 27, 851–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J, Papp A, Pulak R, Ambros V, and Anderson P (1989). A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics 723, 301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek TA, Khuperkar D, Lindeboom RGH, Sonneveld S, Verhagen BMP, Boersma S, Vermeulen M, and Tanenbaum ME (2019). Single-Molecule Imaging Uncovers Rules Governing Nonsense-Mediated mRNA Decay. Mol. Cell 75, 324–339.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg JR, and Goff SP (2010). Upf1 senses 3′UTR length to potentiate mRNA decay. Cell 143, 379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CE, Coody TK, Jeong MY, Berg JA, Winge DR, and Hughes AL (2020). Cysteine Toxicity Drives Age-Related Mitochondrial Decline by Altering Iron Homeostasis. Cell 180, 296–310.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, and Weissman JS (2012). The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc 7, 1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Ohyama T, Sakuragi Y, Yamamoto R, Inoue NA, Yu LH, Goto Y, Wegner M, and Lupski JR (2007). Translation of SOX10 3′ untranslated region causes a complex severe neurocristopathy by generation of a deleterious functional domain. Hum. Mol. Genet 16, 3037–3046. [DOI] [PubMed] [Google Scholar]
- Ishigaki Y, Li X, Serin G, and Maquat LE (2001). Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106, 607–617. [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, and Wilkinson MF (2018). Nonsense-mediated RNA decay in the brain: emerging modulator of neural development and disease. Nat. Rev. Neurosci 19, 715–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen EJ, Timal S, Ryan M, Ashikov A, van Scherpenzeel M, Graham LA, Mandel H, Hoischen A, lancu TC, Raymond K, et al. (2016). ATP6AP1 deficiency causes an immunodeficiency with hepatopathy, cognitive impairment and abnormal protein glycosylation. Nat. Commun. 7, 11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karam R, Wengrod J, Gardner LB, and Wilkinson MF (2013). Regulation of nonsense-mediated mRNA decay: implications for physiology and disease. Biochim. Biophys. Acta 1829, 624–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, and Ohno S (2006). Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complextriggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 20, 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebaara BW, and Atkin AL (2009). Long 3′-UTRs target wild-type mRNAs for nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Nucleic Acids Res. 37, 2771–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr TP, Sewry CA, Robb SA, and Roberts RG (2001). Long mutant dystrophins and variable phenotypes: evasion of nonsense-mediated decay? Hum. Genet. 109, 402–407. [DOI] [PubMed] [Google Scholar]
- Kugler W, Enssle J, Hentze MW, and Kulozik AE (1995). Nuclear degradation of nonsense mutated beta-globin mRNA: a post-transcriptional mechanism to protect heterozygotes from severe clinical manifestations of betathalassemia? Nucleic Acids Res. 23, 413–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T, Hoque M, and Maquat LE (2018). Identifying Cellular Nonsense-Mediated mRNA Decay (NMD) Targets: Immunoprecipitation of Phosphorylated UPF1 Followed by RNA Sequencing (p-UPF1 RIP-Seq). Methods Mol. Biol 1720, 175–186. [DOI] [PubMed] [Google Scholar]
- Kurosaki T, Popp MW, and Maquat LE (2019). Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat. Rev. Mol. Cell Biol 20, 406–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamartina S, Sporeno E, Fattori E, and Toniatti C (2000). Characteristics of the adeno-associated virus preintegration site in human chromosome 19: open chromatin conformation and transcription-competent environment. J. Virol 74, 7671–7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, and Moore MJ (2000). The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 19, 6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedeva S, Jens M, Theil K, Schwanhäusser B, Selbach M, Landthaler M, and Rajewsky N (2011). Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol. Cell 43, 340–352. [DOI] [PubMed] [Google Scholar]
- Leeds P, Peltz SW, Jacobson A, and Culbertson MR (1991). The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 5 (12A), 2303–2314. [DOI] [PubMed] [Google Scholar]
- Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, Irizarry RA, Liu JS, Brown M, and Liu XS (2014). MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Shi Y, Wang P, Guachalla LM, Sun B, Joerss T, Chen YS, Groth M, Krueger A, Platzer M, et al. (2015). Smg6/Est1 licenses embryonic stem cell differentiation via nonsense-mediated mRNA decay. EMBO J. 34, 1630–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Lie PPY, and Nixon RA (2019). Lysosome trafficking and signaling in health and neurodegenerative diseases. Neurobiol. Dis 122, 94–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Wise DR, Diehl JA, and Simon MC (2008). Hypoxic reactive oxygen species regulate the integrated stress response and cell survival. J. Biol. Chem 283,31153–31162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjunath H, Zhang H, Rehfeld F, Han J, Chang TC, and Mendell JT (2019). Suppression of Ribosomal Pausing by eIF5A Is Necessary to Maintain the Fidelity of Start Codon Selection. Cell Rep. 29, 3134–3146.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE, Hwang J, Sato H, and Tang Y (2010). CBP80-promoted mRNP rearrangements during the pioneer round of translation, nonsense-mediated mRNA decay, and thereafter. Cold Spring Harb. Symp. Quant. Biol 75, 127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C(2017). Regulation by 3′-Untranslated Regions. Annu. Rev.Genet 51, 171–194. [DOI] [PubMed] [Google Scholar]
- McGlincy NJ, and Ingolia NT (2017). Transcriptome-wide measurement of translation by ribosome profiling. Methods 126, 112–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlwain DR, Pan Q, Reilly PT, Elia AJ, McCracken S, Wakeham AC, Itie-Youten A, Blencowe BJ, and Mak TW (2010). Smg1 is required for embryogenesis and regulates diverse genes via alternative splicing coupled to nonsense-mediated mRNA decay. Proc. Natl. Acad. Sci. USA 107, 12186–12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, and Dietz HC (2001). Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum. Mol. Genet 10, 99–105. [DOI] [PubMed] [Google Scholar]
- Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, and Dietz HC (2004). Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet 36, 1073–1078. [DOI] [PubMed] [Google Scholar]
- Meyer C, Garzia A, Mazzola M, Gerstberger S, Molina H, and Tuschl T (2018). The TIA1 RNA-Binding Protein Family Regulates EIF2AK2-Mediated Stress Response and Cell Cycle Progression. Mol. Cell 69, 622–635.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JN, and Pearce DA (2014). Nonsense-mediated decay in genetic disease: friend or foe? Mutat. Res. Rev. Mutat. Res 762, 52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills EW, Wangen J, Green R, and Ingolia NT (2016). Dynamic Regulation of a Ribosome Rescue Pathway in Erythroid Cells and Platelets. Cell Rep. 17, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA (2012). Lysosomal acidification mechanisms. Annu. Rev. Physiol 74, 69–86. [DOI] [PubMed] [Google Scholar]
- Murakawa Y, Hinz M, Mothes J, Schuetz A, Uhl M, Wyler E, Yasuda T, Mastrobuoni G, Friedel CC, Dolken L, et al. (2015). RC3H1 post-transcriptionally regulates A20 mRNA and modulates the activity of the IKK/NF-κB pathway. Nat. Commun 6, 7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu-Yilik G, Raimondeau E, Eliseev B, Yeramala L, Amthor B, Deniaud A, Huard K, Kerschgens K, Hentze MW, Schaffitzel C, and Kulozik AE (2017). Dual function of UPF3B in early and late translation termination. EMBO J. 36, 2968–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CU, and Pestova TV (2010). The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol. Cell 37, 196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preis A, Heuer A, Barrio-Garcia C, Hauser A, Eyler DE, Berninghausen O, Green R, Becker T, and Beckmann R (2014). Cryoelectron microscopic structures of eukaryotic translation termination complexes containing eRF1-eRF3 or eRF1-ABCE1. Cell Rep. 8, 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prus E, and Fibach E (2008). Flow cytometry measurement of the labile iron pool in human hematopoietic cells. Cytometry A 73, 22–27. [DOI] [PubMed] [Google Scholar]
- Ramirez A, Heimbach A, Griindemann J, Stiller B, Hampshire D, Cid LP, Goebel I, Mubaidin AF, Wriekat AL, Roeper J, et al. (2006). Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat. Genet 38, 1184–1191. [DOI] [PubMed] [Google Scholar]
- Randolph LN, Bao X, Zhou C, and Lian X (2017). An all-in-one, Tet-On 3G inducible PiggyBac system for human pluripotent stem cells and derivatives. Sci. Rep 7, 1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault TA, and Tong WH (2005). Iron-sulphur cluster biogenesis and mitochondrial iron homeostasis. Nat. Rev. Mol. Cell Biol 6, 345–351. [DOI] [PubMed] [Google Scholar]
- Rufener SC, and MGhlemann O (2013). eIF4E-bound mRNPs are substrates for nonsense-mediated mRNA decay in mammalian cells. Nat. Struct. Mol. Biol 20,710–717. [DOI] [PubMed] [Google Scholar]
- Sabharwal SS, and Schumacker PT (2014). Mitochondrial ROS in cancer: initiators, amplifiers or an Achilles’ heel? Nat. Rev. Cancer 14, 709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, and Zhang F (2012). A transcription activator-like effector toolbox for genome engineering. Nat. Protoc 7, 171–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, and Zhang F (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Hosoda N, and Maquat LE (2008). Efficiency of the pioneer round of translation affects the cellular site of nonsense-mediated mRNA decay. Mol. Cell 29, 255–262. [DOI] [PubMed] [Google Scholar]
- Schmidt SA, Foley PL, Jeong DH, Rymarquis LA, Doyle F, Tenenbaum SA, Belasco JG, and Green PJ (2015). Identification of SMG6 cleavage sites and a preferred RNA cleavage motif by global analysis of endogenous NMD targets in human cells. Nucleic Acids Res. 43, 309–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serdar LD, Whiteside DL, and Baker KE (2016). ATP hydrolysis by UPF1 is required for efficient translation termination at premature stop codons. Nat. Commun 7, 14021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, and Zhang F (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen ZJ, and Malter JS (2015). Regulation of AU-Rich Element RNA Binding Proteins by Phosphorylation and the Prolyl Isomerase Pin1. Biomolecules 5,412–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Wei J, and He C (2019). Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol. Cell 74, 640–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturrock A, Alexander J, Lamb J, Craven CM, and Kaplan J (1990). Characterization of a transferrin-independent uptake system for iron in HeLa cells. J. Biol. Chem 265, 3139–3145. [PubMed] [Google Scholar]
- Sudmant PH, Lee H, Dominguez D, Heiman M, and Burge CB (2018). Widespread Accumulation of Ribosome-Associated Isolated 3′ UTRs in Neuronal Cell Populations of the Aging Brain. Cell Rep. 25, 2447–2456.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H, Imamachi N, Salam KA, Mizutani R, Ijiri K, Irie T, Yada T, Suzuki Y, and Akimitsu N (2012). Identification of hundreds of novel UPF1 target transcripts by direct determination of whole transcriptome stability. RNA Biol. 9, 1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarpey PS, Raymond FL, Nguyen LS, Rodriguez J, Hackett A, Vandeleur L, Smith R, Shoubridge C, Edkins S, Stevens C, et al. (2007). Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat. Genet 39, 1127–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toompuu M, Karblane K, Pata P, Truve E, and Sarmiento C (2016). ABCE1 is essential for S phase progression in human cells. Cell Cycle 15, 1234–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Zavadil J, Martin L, Parisi F, Friedman E, Levy D, Harding H, Ron D, and Gardner LB (2011). Inhibition of nonsense-mediated RNA decay by the tumor microenvironment promotes tumorigenesis. Mol. Cell. Biol 31, 3670–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber RA, Yen FS, Nicholson SPV, Alwaseem H, Bayraktar EC, Alam M, Timson RC, La K, Abu-Remaileh M, Molina H, and Birsoy K (2020). Maintaining Iron Homeostasis Is the Key Role of Lysosomal Acidity for Cell Proliferation. Mol. Cell 77, 645–655.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J, Damgaard I, Bryder D, Theilgaard-Mönch K, Thoren LA, Nielsen FC, Jacobsen SE, Nerlov C, and Porse BT (2008). NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev. 22, 1381–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki S, Stoecklin G, Kedersha N, Simarro M, and Anderson P (2007). T-cell intracellular antigen-1 (TIA-1)-induced translational silencing promotes the decay of selected mRNAs. J. Biol. Chem 282, 30070–30077. [DOI] [PubMed] [Google Scholar]
- Yambire KF, Rostosky C, Watanabe T, Pacheu-Grau D, Torres-Odio S, Sanchez-Guerrero A, Senderovich O, Meyron-Holtz EG, Milosevic I, Frahm J, et al. (2019). Impaired lysosomal acidification triggers iron deficiency and inflammation in vivo. eLife 8, e51031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DJ, Guydosh NR, Zhang F, Hinnebusch AG, and Green R (2015). Rli1/ABCE1 Recycles Terminating Ribosomes and Controls Translation Reinitiation in 3’UTRs In Vivo. Cell 162, 872–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Zhang J, Chang Q, Zeng J, Xin F, Wang J, Zhu Q, Wu J, Lu J, Guo W, et al. (2014). De novo mutation in ATP6V1B2 impairs lysosome acidification and causes dominant deafness-onychodystrophy syndrome. Cell Res. 24, 1370–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusa K, Zhou L, Li MA, Bradley A, and Craig NL (2011). A hyperactive piggyBac transposase for mammalian applications. Proc. Natl. Acad. Sci. USA 108, 1531–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All high-throughput sequencing data generated in this study (CRISPR screening, RNA-seq, and ribosome profiling) have been deposited in GEO (Accession number GEO: GSE144165). No custom code was generated in this study.