Abstract
BACKGROUND:
Accurate tissue diagnosis during ovarian cancer surgery is critical to maximize cancer excision and define treatment options. Yet, current methods for intraoperative tissue evaluation can be time intensive and subjective. We have developed a handheld and biocompatible device coupled to a mass spectrometer, the MasSpec Pen, which uses a discrete water droplet for molecular extraction and rapid tissue diagnosis. Here we evaluated the performance of this technology for ovarian cancer diagnosis across different sample sets, tissue types, and mass spectrometry systems.
METHODS:
MasSpec Pen analyses were performed on 192 ovarian, fallopian tube, and peritoneum tissue samples. Samples were evaluated by expert pathologists to confirm diagnosis. Performance using an Orbitrap and a linear ion trap mass spectrometer was tested. Statistical models were generated using machine learning and evaluated using validation and test sets.
RESULTS:
High performance for high-grade serous carcinoma (n = 131; clinical sensitivity, 96.7%; specificity, 95.7%) and overall cancer (n = 138; clinical sensitivity, 94.0%; specificity, 94.4%) diagnoses was achieved using Orbitrap data. Variations in the mass spectra from normal tissue, low-grade, and high-grade serous ovarian cancers were observed. Discrimination between cancer and fallopian tube or peritoneum tissues was also achieved with accuracies of 92.6% and 87.9%, respectively, and 100% clinical specificity for both. Using ion trap data, excellent results for high-grade serous cancer vs normal ovarian differentiation (n = 40; clinical sensitivity, 100%; specificity, 100%) were obtained.
CONCLUSIONS:
The MasSpec Pen, together with machine learning, provides robust molecular models for ovarian serous cancer prediction and thus has potential for clinical use for rapid and accurate ovarian cancer diagnosis.
Ovarian cancer is a highly lethal disease and the fifth leading cause of all cancer-related deaths in women (1, 2). Accurate diagnosis and stratification of ovarian cancer is important to develop personalized treatment approaches (3). High-grade serous carcinomas (HGSC)6 and low-grade serous carcinomas (LGSC) are common subtypes of ovarian cancers, with the latter accounting for just a small proportion of cases. HGSCs show aggressive features including rapid growth and invasive behavior, whereas LGSCs follow a more indolent course (4). Cytoreductive surgery in combination with chemotherapy is the primary course of treatment for HGSC and essential to maximize patient survival. However, the timing of cytoreductive surgery is of debate (5, 6). Patients likely to undergo complete resection commonly undergo surgery followed by chemotherapy, whereas patients likely to undergo incomplete resection are directed to neoadjuvant chemotherapy before surgery. In both scenarios, differentiation of tumor from normal tissue is critical to maximize cancer excision, although intraoperative identification through gross inspection can be difficult. For example, differentiation of tumor from scarring or fibrotic tissues resulting from neoadjuvant chemotherapy is challenging from metastatic sites, often requiring un-necessary resection of healthy tissue (7). Histopathologic analysis of tissue sections is commonly employed to diagnose surgical specimens, either intraoperatively through frozen section analysis, or postoperatively through permanent tissue sections. Despite its relatively rapid turnaround (approximately 30 min), intraoperative frozen section evaluation results can be limited owing to freezing artifacts affecting tissue histology. Further, frozen section evaluation of multiple tissue specimens can be impractical, because numerous areas of concern are often identified during surgery. Postoperative tissue analysis is time-consuming (approximately 1 week) and places the patient at additional health risks, discomfort, and anxiety (8). Thus, new technologies that allow rapid and accurate intraoperative tissue evaluation could improve diagnosis during cytoreductive surgery and the overall management of ovarian cancer patients (9, 10).
Mass spectrometry (MS) technologies have shown potential for clinical use and cancer diagnosis (11-23). Ambient ionization MS techniques are suitable for rapid and in situ analysis of biological tissues because of their operational simplicity at atmospheric pressure and minimal sample preparation requirements (24). Several ambient ionization techniques have been suggested and tested for intraoperative cancer diagnosis and surgical margin evaluation either through ex vivo tissue sections and tissue smears analyses (12-18), or in vivo (19-23). Desorption electrospray ionization-MS imaging, for example, has been applied for ex vivo analysis of metabolic profiles of borderline ovarian tumor, HGSC, and normal ovarian tissue sections, allowing cancer diagnosis and subtyping (25, 26). The rapid evaporative ionization MS technique was recently used for in vivo ovarian cancer analysis (27). Our team has previously described the development of a handheld device, named the MasSpec Pen, for ex vivo and in vivo nondestructive molecular analysis and diagnosis of tissues (21). We tested the MasSpec Pen for molecular evaluation and diagnosis of 253 human tissue samples (normal and cancer ovarian, lung, thyroid, and breast). For ovarian cancer (n = 29 normal and n = 28 HGSC), a clinical sensitivity of 100%, specificity of 89.7%, and overall accuracy of 94.7% were achieved using leave-one-out cross-validation analysis.
Biocompatibility and the nondestructive nature of the MasSpec Pen analyses are clinically attractive and could facilitate translation into clinical use. Nevertheless, testing of the statistical classifiers using different sample cohorts is critical to evaluate model overfitting and validate its performance. Analysis of other histological subtypes, such as LGSC, is also desirable to evaluate potential for ovarian cancer diagnosis. Moreover, distinguishing ovarian cancer from surrounding healthy peritoneum tissue where ovarian cancer often spreads could improve surgical resection (28). Because fallopian tubes (FTs) have been proposed as the most likely site of HGSC origin, evaluation of FT tissue is also relevant (5, 29). Finally, performance assessment using different mass spectrometers would demonstrate broad applicability of the technology. In particular, smaller, lower-cost, and lower-performance mass spectrometers, such as a linear ion trap (LIT), could ease technology translation into the clinical space (30-33). Yet, LIT exhibits lower mass resolving powers than Orbitrap systems, which could potentially prevent accurate tissue diagnosis. To address these questions, here we evaluated the MasSpec Pen for ovarian cancer diagnosis across different sample sets, tissue types, and MS systems. Training, validation, and test sample sets were used to evaluate the predictive performance and molecular information obtained. LGSC, FT, and peritoneum samples were analyzed to explore the MasSpec Pen capabilities for cancer subtyping and differentiation from other healthy tissues in the abdominal cavity. Lastly, performance using a LIT mass analyzer was investigated to test method versatility across systems.
Materials and Methods
HUMAN TISSUE SAMPLES
In total, 160 deidentified ovarian frozen tissue samples were obtained from the Cooperative Human Tissue Network (CHTN; Table 1). Samples were requested by the research team and collected from different tissue banks within the CHTN network in 3 different batches, the first in the fall of 2015, the second in the fall of 2017, and the third in the spring of 2018. Samples were selected following criteria based on tissue diagnosis (HGSC or LGSC) by gross anatomy and pathological evaluation, and specimen size (>200 mg). FT and peritoneum tissues (32 samples, 29 patients) were received as deidentified frozen specimens from the MD Anderson and CHTN Tissue Banks, or prospectively collected as fresh specimens during endometriosis surgery performed by MB (under approved IRB protocol#: 2017–08-0087) at the Seton Medical Center (Austin, TX; see Table S1 in the Data Supplement that accompanies the online version of this article at http://www.clinchem.org/content/vol65/issue5. Patients undergoing endometriosis surgery were consented. Information on disease severity (other than grade for ovarian cancer) and alternative diagnoses were not considered in our study. Samples were analyzed at room temperature in random order. The analyzed area of the tissue was demarcated and registered through optical images. The same demarcated tissue or a parallel piece was frozen and sectioned at 10–16 μm using a CryoStar NX50 cryostat (Thermo Fisher Scientific), stained by standard hematoxylin and eosin procedure, and evaluated by expert pathologists (not blinded from previous clinical information) to confirm diagnosis of the analyzed area (authors J.L. and S.L.). Of note, the final diagnoses were performed after MasSpec Pen analyses (Fig. 1). Only samples with clear diagnosis (n = 164) were used for statistical analysis (see Fig. S1 in the online Data Supplement).
Table 1.
Patient demographic information.a
| Parameters | Normal | HGSC | LGSC |
|---|---|---|---|
| Sample set 1 | |||
| Number of patients, n | 29 | 28 | – |
| Median age, y | 50 | 62 | – |
| Age Range, y | (31, 80) | (30, 83) | – |
| Number of patients by race (white, black, Asian, Hispanic, unknown) | (22, 7, 0, 0, 0) | (25, 2, 0, 0, 1) | – |
| Sample set 2 | |||
| Number of patients, n | 14 | 11 | 14 |
| Median age, y | 57 | 61 | 57.5 |
| Age range, y | (34, 82) | (47, 76) | (18, 82) |
| Number of patients by race (white, black, Asian, Hispanic, unknown) | (10, 3, 1, 0, 0) | (6, 1, 0, 0, 4) | (14, 0, 0, 0, 0) |
| Sample set 3 | |||
| Number of patients, n | 35 | 29 | – |
| Median age, y | 53 | 65 | – |
| Age range, y | (23, 86) | (44, 85) | – |
| Number of patients by race (white, black, Asian, Hispanic, unknown) | (22, 11, 0, 1, 1) | (24, 2, 1, 0, 2) | – |
Ovarian tissue samples were obtained from the CHTN Tissue Bank under approved institutional review board protocol. Samples were acquired as 3 different sample sets [set 1 (n = 57), set 2 (n = 39), set 3 (n = 64)].
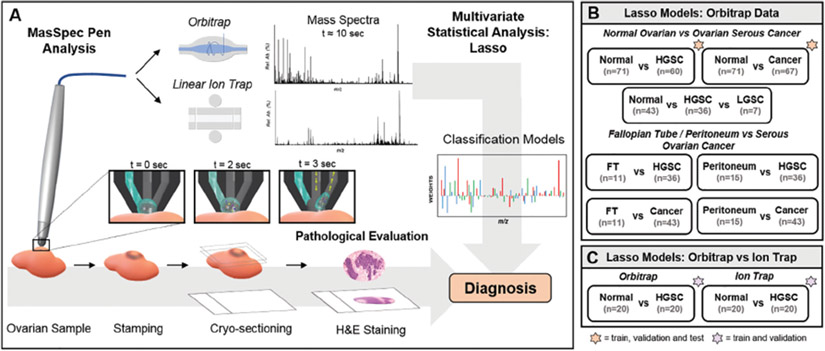
Fig. 1. Tissue sample analysis with the MasSpec Pen using a discrete water droplet (A).
For pathological evaluation, tissue samples were stamped post-analysis, frozen, sectioned, and stained. Mass spectra were obtained using (B) an Orbitrap and (C) a LIT mass analyzer and used to build molecular models for ovarian cancer classification (orange star = train, test, and validation; purple star = train and validation; no star = train). H&E, hematoxylin and eosin.
MASSPEC PEN ANALYSIS
The MasSpec Pen with a 2.7-mm pen tip diameter was used for analysis, using the same procedures and system previously described (see Methods in the online Data Supplement) (21). The MasSpec Pen uses a water droplet to extract molecules from tissues upon contact, which are then analyzed by an Orbitrap mass spectrometer and statistical classifiers, resulting in a total analysis time of approximately 10 s. A new MasSpec Pen device was used for each of the tissue analyses described in this study to prevent any potential carryover between samples. Experiments were performed on a Q Exactive Orbitrap and an LTQ mass spectrometer (Thermo Fisher Scientific). Orbitrap analyses were performed from m/z 120–1800 at a resolving power of 140000 at m/z 200. Ion trap analyses were performed from m/z 120–1000. Tandem MS of selected ions was performed during MasSpec Pen analyses using high-energy collisional dissociation in the Q Exactive. Of note, the MasSpec Pen Orbitrap analysis of sample set 1 (n = 57) and the resulting data were previously described (21), whereas all the remaining samples and analyses were new to this study.
STATISTICAL ANALYSIS
Orbitrap and ion trap data were binned, normalized, and filtered before being subjected to lasso (34) (see Methods in the online Data Supplement). Data from sample sets 1 and 2 were randomly split between training and validation sets. Classification performance was measured by sensitivity, specificity, and accuracy. The “pROC” R package was used to select an optimal threshold from the receiver operating characteristic curve (35). Cosine similarity analysis was performed in the CRAN R language using average mass spectra for each tissue type analyzed in each sample set.
Results
MASSPEC PEN ANALYSIS OF OVARIAN TISSUE SAMPLES USING AN ORBITRAP MASS ANALYZER
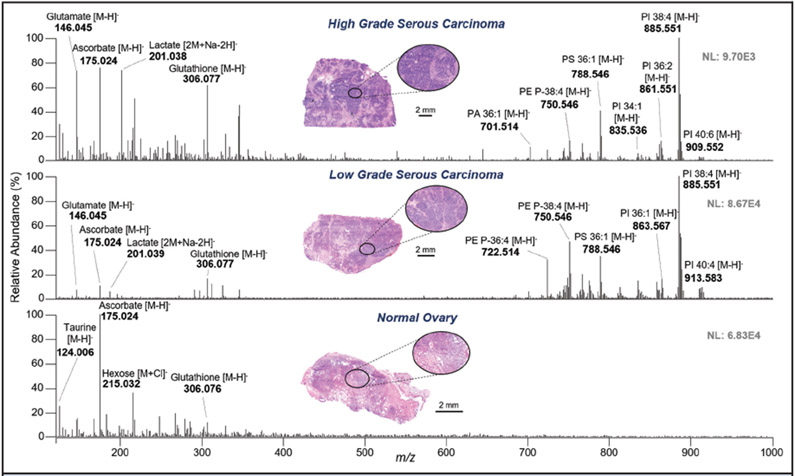
In our previous study, we analyzed a sample set including normal ovarian and HGSC tissue using the MasSpec Pen coupled to an Orbitrap mass spectrometer (21). Here, 2 additional sample sets were obtained and analyzed using the same approach (Table 1). Fig. 2 shows representative MasSpec Pen mass spectra from the 3 different tissue types from the second sample set. A variety of small metabolites, such as glutamate (m/z 146.045), ascorbate (m/z 175.025), and glutathione (m/z 306.077), as well as complex glyocerophospholipid (GP) species, such as glycerophosphoethanolamine (PE) P-18:0_20:4 (m/z 750.546), glycerophosphoserine (PS)_18:0 18:1 (m/z 788.546), and glycerophosphoinositol (PI) 18:0_20:4 (m/z 885.551), were detected. Identification and corresponding mass errors are provided in Table S2 in the online Data Supplement for species tentatively assigned using high mass accuracy and tandem MS/MS measurements. Representative MasSpec Pen MS/MS spectra of ovarian tissue samples are shown in Fig. S2 in the online Data Supplement. The molecular profiles obtained from normal and HGSC from the second and the third sample sets presented similar trends to those previously observed from tissues analyzed in the first sample set (see Fig. S3 in the online Data Supplement). In particular, mass spectra from normal ovarian tissue were characterized by high relative abundance of ascorbate and other small metabolites, whereas GP species appeared at higher relative abundances in the mass spectra from HGSC tissues (Fig. 2). The mass spectra obtained from LGSC also presented a rich variety of GP species at higher relative abundances than in the mass spectra from normal tissues. Further, when comparing LGSC to HGSC, qualitative changes in the relative abundances were observed between the mass spectra of the 2 cancer subtypes (Fig. 2). For example, higher relative abundances of plasmalogen PE species, such as m/z 722.514 and m/z 750.546, were observed in the mass spectra from LGSC, whereas glutamate and glycerophosphatidic acid (PA) 18:0 18:1 (m/z 701.514) were at higher relative abundances in HGSC tissue. Cosine analysis was performed to evaluate the similarity in the mass spectra between the 3 sample sets for the same tissue class. Note that cosine analysis provided similarity values ranging from 0 (dissimilar) to 1 (identical). Similarity values of 0.76 ± 0.06 and 0.80 ± 0.06 were achieved from HGSC and normal data, respectively, showing data comparability between the 3 sample sets for the same tissue type.
Fig. 2. Representative MasSpec Pen mass spectra of high-grade serous carcinoma (top), low-grade serous carcinoma (middle, background subtracted), and normal ovarian tissue (bottom) obtained with an Orbitrap mass analyzer.
Optical image of the stained parallel tissue section analyzed is provided. Tentative identifications are shown for selected ions. X:Y represents number of carbon atoms double bonds in the FA chains.
STATISTICAL PREDICTION OF OVARIAN CANCER USING TRAINING, VALIDATION, AND TEST SAMPLE SETS
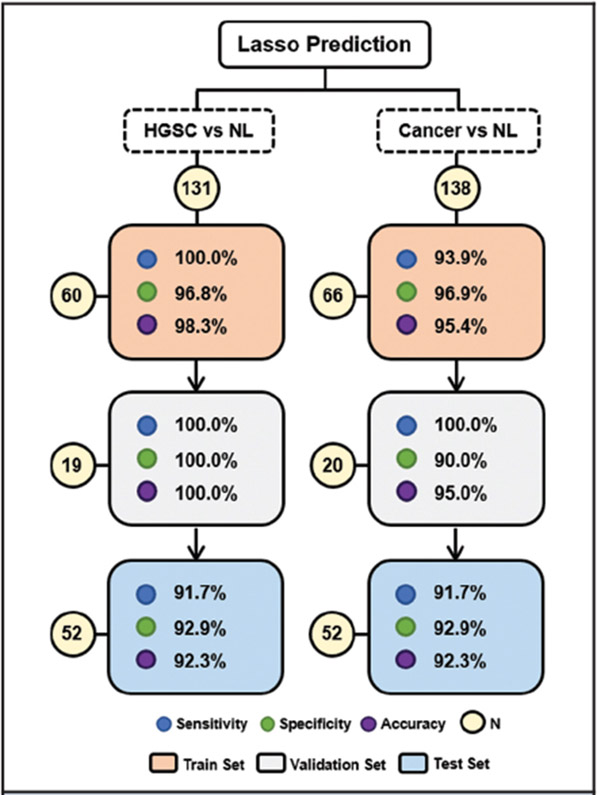
Next, we evaluated if the molecular information obtained from MasSpec Pen analysis using the Orbitrap mass spectrometer was predictive of normal, LGSC, and HGSC across different sample sets. In our previous study (21), a classifier for HGSC diagnosis was built from a sample set 1, a single sample batch, and evaluated using leave-one-out cross-validation only (21). Here, the histologically validated mass spectra obtained from the second set of samples were combined with the first sample set data to improve and validate the statistical classifiers (n = 43 normal, n = 36 HGSC). First, the combined data set was randomly split into a training set (n = 32 normal, n = 28 HGSC) to build the statistical model, and a validation set (n = 11 normal, n = 8 HGSC) to evaluate performance and test for possible overfitting. Using the training set, 100% clinical sensitivity, 96.8% clinical specificity, and 98.3% accuracy was achieved, which are an improvement over previously published results of 100% clinical sensitivity, 89.7% specificity, and 94.7% overall accuracy. In the validation set, 100% clinical sensitivity, specificity, and accuracy were obtained (Fig. 3). Using our previous model (built on sample set 1 alone) to predict on sample set 2, we achieved 100% clinical sensitivity, 75.0% specificity, and 90.9% overall accuracy (see Table S3 in the online Data Supplement), which was an overall lower performance than what we achieved by including samples from different batches in the training sample set. Note that tissue samples were grouped based on tissue type and, as such, the effect of sample source (or tissue bank) on method performance was not evaluated.
Fig. 3. Lasso classification performance for ovarian cancer prediction based on MasSpec Pen data for the training, validation, and test sets.
Results are shown in terms of sensitivity, specificity, and overall accuracy. Confusion matrices with per patient prediction are shown in Table 1 in the online Data Supplement. HG, high-grade serous carcinoma, NL, normal, N, sample number.
Next, a statistical classifier to discriminate between normal and serous cancer (SC) samples was also built by combining the HGSC and LGSC samples into a single class (n = 43 normal, n = 43 cancer). Similarly, the model exhibited high performance in both training (93.9% clinical sensitivity, 96.9% specificity, and 95.4% accuracy) and validation sample sets (100% clinical sensitivity, 90% specificity, and 95.0% accuracy). To further test the performance of the statistical models (HGSC vs normal and SC vs normal), prediction was performed on the third sample set (Fig. 3). Results are reported for samples of clear diagnosis by pathology (n = 28 normal, n = 24 HGSC), corresponding to 91.7% clinical sensitivity, 92.9% specificity, and 92.3% overall accuracy, with 4 out of 52 samples misclassified.
A 3-class statistical model was also created to differentiate between normal ovarian, HGSC, and LGSC tissues. Because of the limited LGSC sample size (n = 7), the model was built using cross-validation on the entire data set. Despite the introduction of the new LGSC subtype, all HGSC and normal ovarian samples were correctly classified (n = 79, 100% accuracy). For LGSC classification, an accuracy of 71.4% was achieved, with 2 out of the 7 samples misclassified as HGSC. Collectively, the 3-class model showed strong overall performance for cancer diagnosis, resulting in an overall accuracy of 97.7%. A summary of all results, including confusion matrices, classification performance, and number of metabolic species included in the models is provided in Table S4 in the online Data Supplement.
Among the species selected to generate the statistical models, ascorbate (m/z 175.024) and taurine (m/z 124.006) were selected as important for the discrimination of normal tissue, and lactate (m/z 201.038), glutathione (m/z 306.077), and the PI 18:0_20:4 (m/z 885.551) were characteristic of HGSC and the combined cancer class. For the 3-class model, PE P-18:0_20:4 at m/z 750.546 was given a positive weight for the characterization of LGSC, in agreement with the trends in relative abundances observed in the representative mass spectra shown in Fig. 2. Gluconate (m/z 195.051) and glutamate (m/z 146.046) were also selected as important for discrimination of LGSC and HGSC samples.
DISTINGUISHING OVARIAN CANCER FROM HEALTHY FT AND PERITONEUM TISSUE
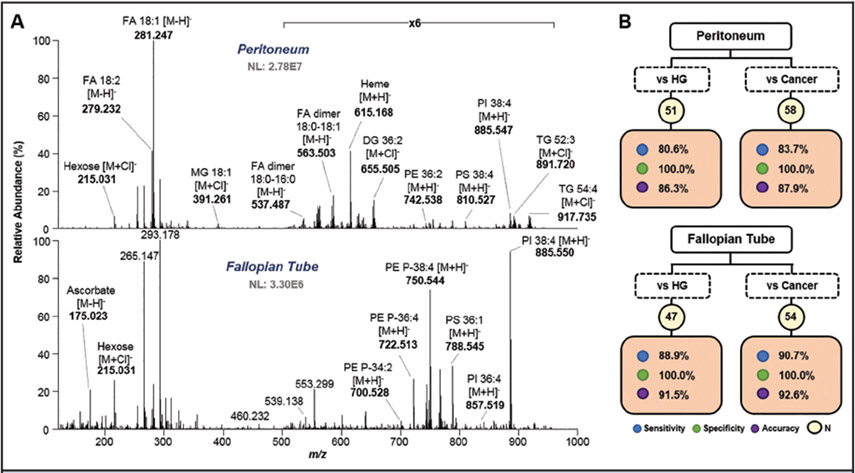
FT (n = 15) and peritoneum (n = 17) tissue samples were also analyzed with the MasSpec Pen and compared to ovarian cancer tissues. Mass spectra obtained from peritoneum tissue were commonly characterized by fatty acid (FA) species, detected both as monomers, such as FA 18:1 (m/z 281.247), and dimers, such as FA 18:0–16:0 (m/z 537.487). Chlorinated triacylglyceride (TG) species were also detected at higher mass range, such as TG 52:3 (m/z 891.720), reflecting on the fat content characteristic of peritoneal and omental tissues. High abundance of complex lipids, such as PI, PE, and PS species, were observed from the lipid and metabolite profiles obtained from FT samples (Fig. 4A). A list of tentatively identified species with mass errors is provided in Table S5 in the online Data Supplement.
Fig. 4. Differentiating ovarian cancer tissue from healthy peritoneum and FT tissue samples.
Representative mass spectra profiles obtained from MasSpec Pen analysis of peritoneum (top) and FT (bottom) tissue (A). Tentative identifications are shown for selective ions. Lasso results for peritoneum (top) and FT (bottom) vs HG and cancer tissue differentiation (B).
Molecular models to discriminate peritoneum and FT tissue from ovarian cancer samples from batch 1 and 2 were built by leave-one-out cross-validation analysis (Fig. 4B). A detailed summary of all classification results, including the number of metabolic species included in the models, is provided in Table S6 in the online Data Supplement. All peritoneum (n = 15) and FT (n = 11) samples were correctly classified when compared to HGSC and ovarian SC samples (100% clinical specificity). Clinical sensitivity values of 88.9% and 90.7% were achieved for HGSC (n = 36) and SC overall (n = 43) vs FT samples, respectively. When compared to peritoneum samples, lower clinical sensitivities were achieved for HG (80.6%) and SC (83.7%). Interestingly, many of the features selected by the models to characterize HGSC and ovarian SC tissue from FT and peritoneum tissue were similar to those used for discrimination against normal ovarian tissue, such as lactate, glutamate, or PI 38:4. Yet, ascorbate was given a positive weight for ovarian cancer classification. Hexose (m/z 215.031) was selected as predictive of both FT and peritoneum samples, whereas a species at m/z 267.073, tentatively identified as inosine, a purine nucleoside, was only associated to FT tissue.
VALIDATION OF THE MASPEC PEN USING AN ION TRAP MASS ANALYZER
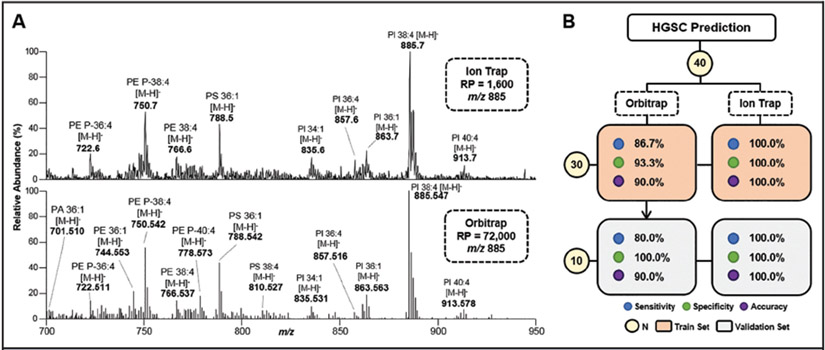
The MasSpec Pen was coupled to a LIT and used to evaluate a subset of HGSC and normal ovarian samples (n = 20 normal, 20 HGSC). Note that the LIT provides lower performance for mass analysis with limited resolving power (1600 at m/z 885.7) and mass accuracy when compared to the resolving power (72 000 at m/z 885.547) and mass accuracy (<5 ppm) achieved with Orbitrap mass analyzers. Fig. 5A shows that the mass spectra obtained for the same HGSC sample analyzed by both systems are highly comparable, although a higher number of resolved lipid species were detected by Orbitrap analysis, as anticipated. Distinctive metabolic profiles for HGSC and normal ovarian samples were obtained by MasSpec Pen and LIT analysis (see Fig. S4 in the online Data Supplement). Similar to the Orbitrap data, HGSC mass spectra were characterized by the high relative abundance of GP species, whereas normal samples displayed considerably lower lipid abundance and higher relative abundances of small metabolites.
Fig. 5. Representative mass spectra profile obtained from the same HGSC sample by ion trap (top, profile mode) and Orbitrap (bottom, centroid mode) analysis (A).
Tentative identifications are shown for selected ions. Lasso results for HGSC prediction comparing performance between the models built from Orbitrap and ion trap data acquired from the same sample set (n = 40) (B). RP, mass resolving power; N, sample number.
Statistical classifiers were built to evaluate the predictive performance for HGSC diagnosis based on LIT data. The data was randomly split into a training (n = 30) and validation sets (n = 10), yielding excellent classification performance, with 100% clinical sensitivity, specificity, and accuracy in both sets (Fig. 5B). For adequate comparison, a new model was built using Orbitrap data from the same set of samples analyzed using the LIT (n = 40). High specificity was achieved in both the training (93.3%) and validation sets (100%) by the new Orbitrap model, whereas lower clinical sensitivity was observed for the training (86.7%) and validation (80.0%) sets, respectively, with 3 HGSC samples misclassified as normal. Note that these 3 samples had been correctly classified by the model built using Orbitrap data acquired from the larger training set (n = 60). A detailed comparison of the results obtained for the Orbitrap and ion trap models is provided in Table S7 in the online Data Supplement.
Discussion
Validation of the diagnostic capabilities of the MasSpec Pen is paramount for potential clinical translation. Utilizing a cohort of 138 histologically validated ovarian cancer and normal tissue samples, we demonstrate here the predictive power of the statistical models built from MasSpec Pen analyses. Machine learning and classification algorithms are not parametrically constrained and thus are more susceptible to overfitting than other more traditional methods (36). Overfitting occurs when a model uses spurious correlations within a single data set rather than relationships within the population. Here, statistical validation and evaluation of model overfitting were performed using training (to fit the model), validation (to provide an unbiased evaluation while tuning parameters such as data normalization methods), and test (to provide an unbiased evaluation of a final model) sets of samples. Improved performance was achieved for HGSC diagnosis using a new training set that combines samples from different batches, compared to what we have previously reported using a single sample batch (21). These results show that including samples from different batches in the training set helps improve the predictive power of the classifiers. High prediction accuracies were also observed in both the validation and test sets, indicating that the model was not overfitting to the training set. Similar results were obtained for the normal vs SC classifier, demonstrating that the MasSpec Pen and machine learning provide robust predictive models for ovarian cancer diagnosis.
Discrimination between LGSC and HGSC tissue was also explored. Owing to the lower clinical occurrence of LGSC, sample availability was limited. Nevertheless, trends in lipid and metabolite composition between the 2 cancer types were and reflected by the moderate classification accuracy achieved. Collection of additional LGSC tissues is ongoing to continue MasSpec Pen validation for ovarian cancer subtyping. Four molecular models were also built to discriminate HGSC and SC from healthy FT and peritoneum samples, yielding 100% clinical specificity and sensitivity from 80.6% to 90.7% for cancer diagnosis. Assessment of the mass spectra from the cancer samples misclassified as FT revealed unusually high abundance of m/z 215.031, weighted toward FT classification, and an overall low lipid abundance in samples misclassified as peritoneum. Interestingly, 3 out of the 7 cancer tissues misclassified as peritoneum were obtained from ovarian cancer metastasis to the omentum. Segregation of primary or metastatic samples will be evaluated to investigate differences in molecular information that may be correlated to misclassification, as well as refinement of the statistical models. Nonetheless, the overall accuracies (>86%) achieved demonstrate the MasSpec Pen potential to differentiate SC from surrounding normal ovarian and abdominal healthy tissues.
The features chosen by the lasso models as predictive for ovarian cancer diagnosis and subtyping were identified as biologically relevant species. For example, ascorbic acid, which has an important role in normal ovarian functions (37), was selected as important for normal ovarian tissue characterization, whereas GP lipids, such as PI 18:0_20:4, were at high relative abundance in cancer tissue and selected as predictive markers. Gluconate and glutamate were selected as important for the discrimination of LGSC and HGSC samples. Similar trends and species have also been reported using desorption electrospray ionization MS (26). Moreover, increased abundance of glutamate has been reported previously in invasive ovarian carcinomas compared to borderline tumors by gas chromatography and MS analyses (38). Another interesting feature selected was inosine at m/z 267.073, which was associated with FT tissue. Inosine is rapidly metabolized from adenosine, a nucleoside known to modulate neurotransmission and contractile responses in the FTs (39), and has been previously detected in oviduct cells (40).
Smaller and lower-cost equipment could facilitate translation of MS technologies to the clinic (30). LIT mass spectrometers are attractive for clinical use due to their scalability, tolerance for higher operating pressure, and lower cost (31). However, LIT analyzers provide limited mass resolution, which could potentially compromise performance. To demonstrate that the MasSpec Pen can be compatible with a mass spectrometer platform other than an Orbitrap, and to further investigate how classification performance is affected by collecting data with lower performance mass analyzers, we coupled the MasSpec Pen to a LIT system. Using a smaller sample set than what collectively explored for Orbitrap analysis, we achieved 100% accuracy for HGSC classification in training and validation. The higher performance achieved compared to the Orbitrap data could be related to the absence of batch effects in the LIT data and will be further investigated. Overall, similarities between the detected metabolic profiles and statistical models observed between Orbitrap and LIT data support the validity and robustness of the predictive molecular species and further support that the MasSpec Pen provides consistent molecular analysis across MS systems. More testing of the LIT and other mass spectrometers is needed across multiple sample sets and cancer subtypes.
In conclusion, our study demonstrates high performance and versatility of the MasSpec Pen technology and statistical models across different sample sets and MS platforms, as well as feasibility for cancer subtyping. The MasSpec Pen leverages on its nondestructive nature for direct and gentle analysis of tissues and the sensitivity, specificity, and speed provided by MS for untargeted and accurate molecular diagnosis. Further, its simple design, ease of operation, and biocompatibility are attractive for clinical use. Thus, we envision the MasSpec Pen to be employed in the operating room (OR) for in vivo and/or ex vivo use in conjunction with standard surgical tools. Yet, this study represents a first step toward validating the performance of the MasSpec Pen for ovarian cancer diagnosis. Several challenges still need to be addressed and further validation pursued to demonstrate the usefulness of the MasSpec Pen in its envisioned application in the OR. Although our experiments performed on ex vivo tissues obtained from tissue banks yield diagnostic molecular results, in vivo and freshly excised tissues in the OR may present slightly different molecular profiles. Moreover, extensive analyses of benign ovarian disease and tissues with varied histologic composition are needed to further demonstrate its value in more complex surgical pathology cases. As such, in vivo OR experiments utilizing the MasSpec Pen for ovarian cancer diagnosis are currently planned to further evaluate its diagnostic capabilities. Performance evaluation across different institutions, instrumentation, and users are also envisioned to further validate our results and evaluate its usefulness in patient care.
Supplementary Material
Acknowledgments:
We thank Dr. Suzanne Ledet, Alena Bensussan, Rachel J. DeHoog, Abigail N. Gatmaitan, Mary King, Anna C. Krieger, and Ana I. Sariol for assistance with experiments, samples, and materials.
Research Funding: A.K. Sood, MTrap; L.S. Eberlin, the National Institutes of Health (Grant R00CA190783), the Cancer Prevention Research Institute of Texas (CPRIT, Grant RP180381), the Marion Milligan Mason Award for Women in the Chemical Sciences. Tissue samples were provided by the Cooperative Human Tissue Network (supported by the NCI), the MD Anderson Tissue Bank, and Seton Medical Center.
6 Nonstandard abbreviations:
- HGSC
high-grade serous carcinomas
- LGSC
low-grade serous carcinomas
- MS
mass spectrometry
- FT
fallopian tube
- LIT
linear ion trap
- CHTN
Cooperative Human Tissue Network
- GP
glyocerophospholipid
- PE
glycerophosphoethanolamine
- PS
glycerophosphoserine
- PI
glycerophosphoinositol
- PA
glycerophosphatidic acid
- SC
serous cancer
- FA
fatty acid
- TG
triacylglyceride
- OR
operating room
Footnotes
Authors’ Disclosures or Potential Conflicts of Interest: Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/or potential conflicts of interest:
Employment or Leadership: None declared.
Consultant or Advisory Role: A.K. Sood, Kiyatec.
Stock Ownership: A.K. Sood, Biopath.
Honoraria: L.S. Eberlin, UT Health San Antonio.
Expert Testimony: None declared.
Patents: J. Zhang, J.Q. Lin, and L.S. Eberlin are inventors in patent applications submitted by The University of Texas at Austin related to the MasSpec Pen technology. J. Zhang, US 15/692,167, US 62/591,179, US 62/640,385; J.Q. Lin, US Patent App. 15/692,167; N. Giese, US 62/591,179, US 62/640,385; A.K. Sood, SiRNA delivery (patents on DOPC, rHDL); L.S. Eberlin, US 15/692,167, US 62/591,179, US 62/640,385.
Role of Sponsor: The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, preparation of manuscript, or final approval of manuscript.
Data and materials availability: The data for this study have been deposited in Dataverse, which can be found at https://dataverse.harvard.edu/dataset.xhtml?persistentId=doi:10.7910/DVN/XD8LQ0.
M. Sans, statistical analysis; J.Q. Lin, statistical analysis; M. Breen, provision of study material or patients; K. Sebastian, administrative support, provision of study material or patients; J. Liu, provision of study material or patients; A.K. Sood, provision of study material or patients; L.S. Eberlin, financial support, statistical analysis, administrative support, provision of study material or patients.
References
- 1.Torre LA, Trabert B, DeSantis CE, Miller KD, Samimi G, Runowicz CD, et al. Ovarian cancer statistics, 2018. CA Cancer J Clin 2018;68:284–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL,Miller KD, Jemal A. Cancer statistics, 2018.CA Cancer J Clin 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- 3.Leong HS, Galletta L, Etemadmoghadam D, George J, Kobel M, Ramus SJ, et al. Efficient molecular subtype classification of high-grade serous ovarian cancer. J Pathol 2015;236:272–7. [DOI] [PubMed] [Google Scholar]
- 4.Vang R, Shih IM, Kurman RJ. Ovarian low-grade and high-grade serous carcinoma pathogenesis, clinicopathologic and molecular biologic features, and diagnostic problems.Adv Anat Pathol 2009;16:267–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coleman RL, Monk BJ, Sood AK, Herzog TJ. Latest research and treatment of advanced-stage epithelial ovarian cancer. Nat Rev Clin Oncol 2013;10:211–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams TI, Toups KL, Saggese DA, Kalli KR, Cliby WA, Muddiman DC. Epithelial ovarian cancer: disease etiology, treatment, detection, and investigational gene, metabolite, and protein biomarkers. J Proteome Res 2007;6:2936–62. [DOI] [PubMed] [Google Scholar]
- 7.Nick AM, Coleman RL, Ramirez PT, Sood AK. A frame-work for a personalized surgical approach to ovarian cancer. Nat Rev Clin Oncol 2015;12:239–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Macario A What does one minute of operating room time cost? J Clin Anesth 2010;22:233–6. [DOI] [PubMed] [Google Scholar]
- 9.Jones S,Anagnostou V, Lytle K, Parpart-Li S, Nesselbush M, Riley DR, et al. Personalized genomic analyses for cancer mutation discovery and interpretation. Sci Transl Med 2015;7:283ra53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mino-Kenudson M, Chirieac LR, Law K, Hornick JL, Lindeman N, Mark EJ, et al. A novel, highly sensitive antibody allows for the routine detection of ALK-rearranged lung adenocarcinomas by standard immunohisto-chemistry. Clin Cancer Res 2010;16:1561–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ifa DR, Eberlin LS.Ambient ionization mass spectrometry for cancer diagnosis and surgical margin evaluation. Clin Chem 2016;62:111–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarmusch AK, Pirro V, Baird Z, Hattab EM, Cohen-Gadol AA, Cooks RG. Lipid and metabolite profiles of human brain tumors by desorption electrospray ionization-MS. Proc Natl Acad Sci U S A 2016;113:1486–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eberlin LS, Tibshirani RJ, Zhang J, Longacre TA, Berry GJ, Bingham DB, et al. Molecular assessment of surgical-resection margins of gastric cancer by mass-spectrometric imaging. Proc Natl Acad Sci U S A 2014;111:2436–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calligaris D, Caragacianu D, Liu X, Norton I, Thompson CJ,Richardson AL, et al. Application of desorption electrospray ionization mass spectrometry imaging in breast cancer margin analysis. Proc Natl Acad Sci U S A 2014;111:15184–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eberlin LS, Norton I, Dill AL, Golby AJ, Ligon KL, Santagata S, et al. Classifying human brain tumors by lipid imaging with mass spectrometry. Cancer Res 2012;72:645–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guenther S, Muirhead LJ,Speller AV, Golf O, Strittmatter N, Ramakrishnan R, et al. Spatially resolved metabolic phenotyping of breast cancer by desorption electrospray ionization mass spectrometry. Cancer Res 2015;75:1828–37. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, Yu W, Ryu SW, Lin J, Buentello G, Tibshirani R, et al. Cardiolipins are biomarkers of mitochondria-rich thyroid oncocytic tumors. Cancer Res 2016;76:6588–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dill AL, Eberlin LS, Costa AB,Zheng C, Ifa DR, Cheng LA, et al. Multivariate statistical identification of human bladder carcinomas using ambient ionization imaging mass spectrometry. Chemistry 2011;17:2897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schafer KC, Denes J, Albrecht K, Szaniszlo T, Balog J, Skoumal R, et al. In vivo, in situ tissue analysis using rapid evaporative ionization mass spectrometry. Angew Chem Int Ed Engl 2009;48:8240–2. [DOI] [PubMed] [Google Scholar]
- 20.Fatou B, Saudemont P, Leblanc E, Vinatier D, Mesdag V, Wisztorski M, et al. In vivo real-time mass spectrometry for guided surgery application.Sci Rep 2016;6:25919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang JL,Rector J,Lin JQ,Young JH,Sans M,Katta N,et al. Nondestructive tissue analysis for ex vivo and in vivo cancer diagnosis using a handheld mass spectrometry system. Sci Transl Med 2017;9:eaan3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schafer KC, Szaniszlo T, Gunther S, Balog J, Denes J, Keseru M, et al. In situ, real-time identification of biological tissues by ultraviolet and infrared laser desorption ionization mass spectrometry. Anal Chem 2011;83:1632–40. [DOI] [PubMed] [Google Scholar]
- 23.Saudemont P, Quanico J, Robin YM, Baud A, Balog J, Fatou B, et al. Real-time molecular diagnosis of tumors using water-assisted laser desorption/ionization mass spectrometry technology. Cancer Cell 2018;34:840–51.e4. [DOI] [PubMed] [Google Scholar]
- 24.Laskin J, Lanekoff I. Ambient mass spectrometry imaging using direct liquid extraction techniques. Anal Chem 2016;88:52–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doria ML, McKenzie JS, Mroz A, Phelps DL, Speller A, Rosini F, et al. Epithelial ovarian carcinoma diagnosis by desorption electrospray ionization mass spectrometry imaging. Sci Rep 2016;6:39219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sans M, Gharpure K, Tibshirani R, Zhang J, Liang L, Liu J, et al. Metabolic markers and statistical prediction of serous ovarian cancer aggressiveness by ambient ionization mass spectrometry imaging. Cancer Res 2017; 77:2903–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phelps DL, Balog J, Gildea LF, Bodai Z, Savage A, El-Bahrawy MA, et al. The surgical intelligent knife distinguishes normal, borderline and malignant gynaecological tissues using rapid evaporative ionisation mass spectrometry (REIMS). Br J Cancer 2018;118:1349–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lengyel E Ovarian cancer development and metastasis.Am J Pathol 2010;177:1053–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurman RJ,Shih Ie M. Molecular pathogenesis and extraovarian origin of epithelial ovarian cancer-shifting the paradigm. Hum Pathol 2011;42:918–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang JL, Yu WD, Suliburk J, Eberlin LS. Will ambient ionization mass spectrometry become an integral technology in the operating room of the future? Clin Chem 2016;62:1172–4. [DOI] [PubMed] [Google Scholar]
- 31.Peng Y, Austin DE. New approaches to miniaturizing ion trap mass analyzers. Trends Analyt Chem 2011;30:1560–7. [Google Scholar]
- 32.Douglas DJ, Frank AJ, Mao DM. Linear ion trapsin mass spectrometry. Mass Spectrom Rev 2005;24:1–29. [DOI] [PubMed] [Google Scholar]
- 33.Marshall AG, Hendrickson CL. High-resolution mass spectrometers.Annu Rev Anal Chem 2008;1:579–99. [DOI] [PubMed] [Google Scholar]
- 34.Tibshirani R Regression shrinkage and selection via the lasso.J Roy Stat Soc B Met 1996;58:267–88. [Google Scholar]
- 35.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC,Muller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011;12:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.James G, Witten D, Hastie T, Tibshirani R. An introduction to statistical learning: with applications in R. New York: Springer; 2013426 pp. [Google Scholar]
- 37.Devine PJ, Perreault SD, Luderer U. Roles of reactive oxygen species and antioxidants in ovarian toxicity. Biol Reprod 2012;86:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Denkert C, Budczies J, Kind T, Weichert W, Tablack P, Sehouli J, et al. Mass spectrometry-based metabolic profiling reveals different metabolite patterns in invasive ovarian carcinomas and ovarian borderline tumors. Cancer Res 2006;66:10795–804. [DOI] [PubMed] [Google Scholar]
- 39.Wiklund NP, Samuelson UE, Brundin J. Adenosine modulation of adrenergic neurotransmission in the human fallopian tube.Eur J Pharmacol 1986;123:11–8. [DOI] [PubMed] [Google Scholar]
- 40.Cometti B, Dubey RK, Imthurn B, Jackson EK, Rosselli M.Oviduct cells express the cyclic amp-adenosine pathway. Biol Reprod 2003;69:868–75. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.