Abstract
The histological and molecular subtypes of breast cancer demand distinct therapeutic approaches. Invasive ductal carcinoma (IDC) is subtyped according to estrogen-receptor (ER), progesterone-receptor (PR), and HER2 status, among other markers. Desorption-electrospray-ionization-mass-spectrometry imaging (DESI-MSI) is an ambient-ionization MS technique that has been previously used to diagnose IDC. Aiming to investigate the robustness of ambient-ionization MS for IDC diagnosis and subtyping over diverse patient populations and interlaboratory use, we report a multicenter study using DESI-MSI to analyze samples from 103 patients independently analyzed in the United States and Brazil. The lipid profiles of IDC and normal breast tissues were consistent across different patient races and were unrelated to country of sample collection. Similar experimental parameters used in both laboratories yielded consistent mass-spectral data in mass-to-charge ratios (m/z) above 700, where complex lipids are observed. Statistical classifiers built using data acquired in the United States yielded 97.6% sensitivity, 96.7% specificity, and 97.6% accuracy for cancer diagnosis. Equivalent performance was observed for the intralaboratory validation set (99.2% accuracy) and, most remarkably, for the interlaboratory validation set independently acquired in Brazil (95.3% accuracy). Separate classification models built for ER and PR statuses as well as the status of their combined hormone receptor (HR) provided predictive accuracies (>89.0%), although low classification accuracies were achieved for HER2 status. Altogether, our multicenter study demonstrates that DESI-MSI is a robust and reproducible technology for rapid breast-cancer-tissue diagnosis and therefore is of value for clinical use.
Graphical Abstract
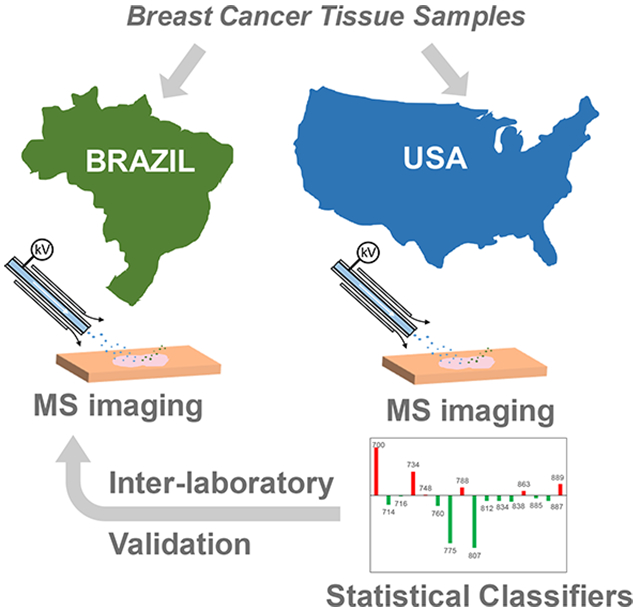
Breast cancer is a complex and heterogeneous disease, and the leading cause of cancer deaths among females worldwide.1,2 Breast cancer exhibits distinct gene-expression patterns depending on the molecular subtype, which is defined mostly by the estrogen-receptor (ER), progesterone-receptor (PR), and human-epidermal-growth-factor-receptor-2 (HER2) statuses. Precise diagnosis and subtyping of breast cancer at the molecular level is pivotal for managing cancer patients because each subtype presents distinct clinical outcomes and therefore requires targeted treatment regimens.3 Diagnosis and molecular subtyping of breast cancer is routinely performed in the clinic on the basis of histopathologic interpretation of hematoxylin and eosin (H&E) staining of tissue sections, immunohistochemistry (IHC) assays that are specific for ER and PR determination, and fluorescence in situ hybridization (FISH) for the evaluation of the amplification of the HER2 gene.4-7 Although they are relatively simple techniques, H&E and IHC are time-consuming assays that may present bias as a result of reaction conditions and subjectivity in data interpretation.8 FISH assays are time-consuming and require experienced personnel to perform and interpret results.9,10 Implementation of new clinical technologies that provide precise and rapid diagnosis and characterization of breast cancer are therefore desirable to guide treatment and improve patient care. Molecular technologies offer the opportunity to incorporate cancer-specific biomarkers into clinical decision making. Genetic mutational signatures associated with the molecular and clinical differences of breast cancers, for example, have been incorporated into clinical workflows.11,12 New technologies that allow rapid assessment of metabolic and protein alterations in breast cancer have also been increasingly explored and have shown great potential for clinical use to expedite diagnosis and treatment decisions.13
Mass-spectrometry-imaging (MSI) technologies offer a powerful tool for chemical and spatial characterization of biological tissues with high specificity and sensitivity and have been widely explored for human-cancer-tissue analysis,14-19 including that of breast cancer.20-26 Ambient-ionization MSI techniques, such as desorption electrospray ionization (DESI), allow analysis of tissue sections with high-throughput and minimal sample preprocessing,27 features that are attractive for clinical use in preoperative cancer diagnosis and intraoperative surgical-margin evaluation.28 A few studies have employed DESI-MSI to investigate metabolic information on human breast cancer in an effort to improve diagnosis and surgical-margin evaluation.21,29,30 Dill et al. used DESI to image altered glycerophospholipids in a small set of breast-cancer-tissue samples.29 Calligaris et al. used DESI-MSI to analyze 61 breast-tissue samples from 14 patients who had undergone mastectomies.30 Guenther et al. used DESI-MSI to characterize surgical biopsies from lesions (28 patients, 28 samples) and tumor beds (22 patients, 98 samples) and achieved an overall accuracy of 98.2% for breast-cancer diagnosis.21 The latter two studies also showed correlations with hormone-receptor (HR) status, which combines PR and ER statuses into a single HR category, although no correlations with lipid information and HER2 status were found. More recently, we have used DESI-MSI to detect and characterize metastatic breast cancer in lymph nodes.31 Whereas these studies strongly showcase the potential of MSI techniques to complement histological and histochemical characterization and diagnosis of breast cancer, these isolated investigations have not evaluated the validity of determining biomarker status for breast cancer or the analytical performance of the methods for breast-cancer diagnosis across different patient populations and laboratories. Racial, dietary, and environmental traits have been associated with molecular and prognostic differences within breast-cancer patients, which may result in molecular variability and thus failure to properly categorize tissue samples.32,33 Analytically, tissue preparation, tissue storage conditions, choice of instrumentation and experimental parameters have been associated with variabilities in imaging and mass-spectra quality by MSI,34 which could affect the method’s performance. Larger studies using diverse cohorts of samples are therefore needed to properly evaluate the robustness of molecular markers and workflows of DESI-MSI for clinical use in breast-cancer diagnosis and characterization. Herein, we report a multicenter study using DESI-MSI to investigate the lipid signatures of a diverse set of breast tissues and to validate the predictive performance of the method for breast-cancer diagnosis. Samples from 103 patients of various races were independently investigated in the United States and Brazil to validate predictive molecular signatures and evaluate the sensitivity, specificity, and accuracy of the method for breast-cancer diagnosis.
MATERIALS AND METHODS
Human Breast-Cancer Tissues.
For our study, 131 frozen human-breast-tissue samples were obtained. Demographic and clinicopathologic characteristics of the samples are provided in Table S1. Tissue procurement, handling, and shipment were performed under approved IRB protocols at the respective institutions. After DESI-MSI, the same tissue sections analyzed by DESI-MSI were subjected to standard H&E-staining protocol. For more information on tissue samples, histopathology, and light microscopy, please see the Supporting Information.
DESI-MSI.
A 2D Omni Spray DESI imaging platform (Prosolia Inc., Indianapolis, IN) coupled to a Q-Exactive (Thermo Fisher Scientific, San Jose, CA) in Brazil and an LTQ-Orbitrap Elite (Thermo Fisher Scientific, San Jose, CA) in the United States were used for tissue imaging. Lab-built sprayers were adapted to the commercial Omni Spray DESI imaging stages. DESI-MSI was performed using the histologically compatible solvent system dimethylformamide/acetonitrile (DMF/ACN) 1:1 (v/v) in negative-ion mode.35 Other experimental parameters for each center are described in Table 1. Ion images were assembled using Biomap and MSiReader software.36 For ion identification, high-mass-resolution and -accuracy measurements were conducted using CID and HCD methods, using the Orbitrap for analysis.
Table 1.
Summary of the Main Experimental Parameters Used in the United States and Brazil DESI-MSI Experiments
| United States | Brazil | |
|---|---|---|
| mass spectrometer | LTQ-Orbitrap Elite | Q-Exactive Hybrid Quadrupole-Orbitrap |
| S-lens RF value | 60 | 100 |
| resolving power (at m/z 400) | 60 000 | 70 000 |
| m/z range | 100–1500 | 100–1200 |
| DESI solvent | ACN/DMF (1:1) | ACN/DMF (1:1) |
| DESI gas pressure | 180 psi | 150 psi |
| DESI flow rate | 1.5 μL/mL | 0.7–1.2 μL/mL |
| spatial resolution | 250 μm | 200 μm |
Statistical Analysis.
MS data corresponding to the areas of interest were extracted from the ion images using MSiReader software.36 After data preprocessing, logistic regression was performed with Lasso regularization using the glmnet package in the R language. Regularization parameters were determined by 5-fold cross-validation (CV) analysis. PCA was performed by centering the preprocessed data to mean zero and computing principal components using the prcomp function in R. To quantify tissue similarity, the cosine similarity method was used from the lsa package in CRAN. For more information, please see the Supporting Information.
RESULTS
Molecular Imaging of Breast Tissues by DESI-MSI.
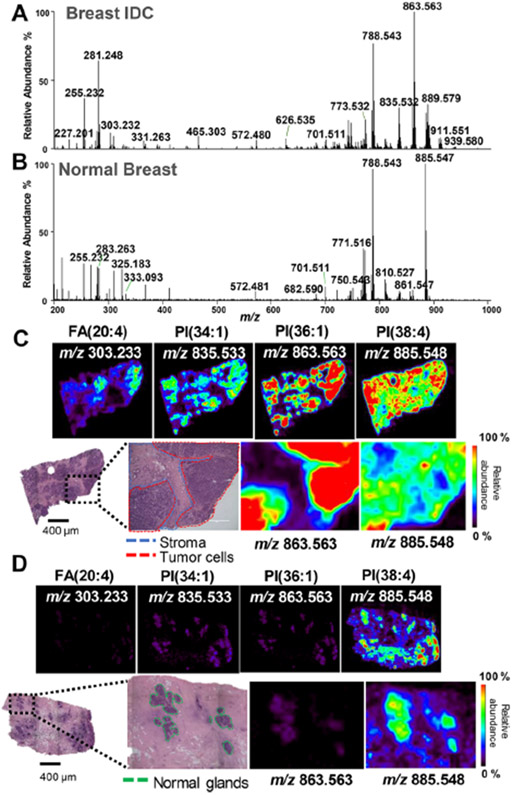
DESI-MSI in negative-ion mode was performed on a total of 131 human-breast-tissue samples including 86 invasive ductal carcinomas (IDC) and 45 normal-breast-tissue samples obtained from 103 breast-cancer patients. IDC is the most common histologic subtype of breast cancer, accounting for approximately 80% of all invasive breast tumors. Because of its high incidence and relevance, only IDC tumors were used for our study. IDC tissue sections typically present regions of predominantly tumor cells neighboring adjacent stroma and adipose tissues. Figure 1A,C show a representative mass spectrum extracted from the tumor region of an IDC sample, selected ion images, and optical images of the H&E-stained tissue section. The spatial resolution used for DESI-MSI (250 μm) enabled visualization of histologic features within the IDC tissues, allowing correlation between histology and molecular information. For example, the distribution of the ions corresponding to deprotonated PI(34:1), PI(36:1), and FA(20:4) spatially correlate with regions of IDC (outlined in red), whereas PI(38:4) was detected throughout the entire tissue section, including regions of adjacent normal stroma tissue (outlined in blue). A magnification of the optical image obtained from the H&E-stained tissue section shows an overlap between the spatial distribution of PI(36:1) and regions of IDC as well as a decrease in abundance of this ion within the surrounding normal stroma cells.
Figure 1.
Negative-ion-mode DESI-MSI of IDC and normal breast tissues. (A) Representative profile for the IDC-tissue region. (B) Representative profile for the normal-breast-tissue region. (C) Representative ion images for the IDC-tissue sample and optical images of the H&E stained tissue section. (D) Representative ion images for the normal-tissue sample and optical images of the H&E stained tissue section. Tumor areas are outlined in red on H&E slides. Areas of red intensity within the ion images represent the highest (100%) relative abundances, whereas black represents the lowest (0%). PI: glycerophosphoinositol, PS: glycerophosphoserine, FA: fatty acid. Lipid species are described by the numbers of fatty acid chain carbons and double bonds.
Normal breast tissues typically present stroma or adipose tissue surrounding focal regions of normal epithelial glands. Note that tissues classified as “normal” correspond to specimens deemed grossly normal at the time of specimen allocation and further confirmed by histopathologic evaluation. Normal samples were acquired from different breast regions depending on the patient, including contralateral breasts from bilateral mastectomies, tissues from breast-reduction surgeries for non-neoplastic purposes and from prophylactic mastectomies, and noncancerous regions adjacent to the tumor (for more information, please see the Supporting Information). Figure 1B,D shows a representative mass spectrum extracted from normal epithelial cells of a normal-tissue sample, selected ion images, and optical images of the H&E-stained tissue section. DESI-MSI allowed visualization of focal epithelial glands (outlined in green) within adjacent stroma, which spatially overlapped with regions of high relative abundances of PS(36:1) and PI(38:4), for example. Qualitatively, the mass spectra from normal epithelial glands and IDC tissues presented distinct molecular profiles. For example, lower relative abundances of the ion of m/z 863.563, attributed to PI(36:1), were observed in the mass spectra of normal epithelial glands when compared with those of IDC, whereas the relative abundances of the ion of m/z 771.516, PG(36:3), were higher in the mass spectra of normal epithelial glands. These results showcase the usefulness of DESI-MSI in spatially investigating the lipid profiles of IDC and normal-breast-cancer tissue sections, even within fine histological features.
DESI-MSI of Breast-Tissue Samples from Patients of Different Races.
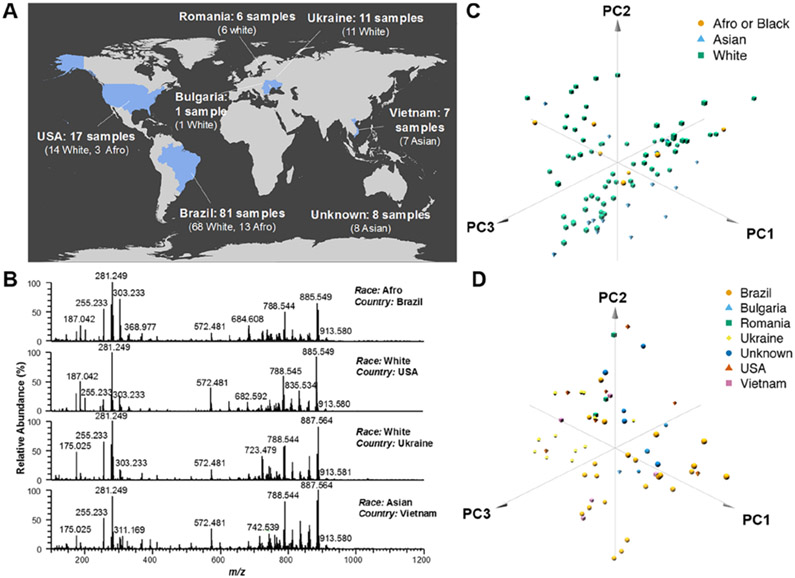
Next, we evaluated whether the metabolite and lipid profiles obtained from breast tissues by DESI-MSI were consistent across patients of different races or ethnicities. Normal- and cancerous-tissue samples from a diverse patient population were obtained from collection sites in different countries, including 81 tissue samples from 53 patients from Brazil, 17 tissue samples from 17 patients from the United States, 18 tissue samples from 18 patients from Europe, 7 tissue samples from 7 patients from Vietnam, as well as 8 tissue samples from Asia (specific country of origin unavailable, Figure 2A and Table S1). For racial classification, we grouped white Americans and white Hispanics into a single “White” group, and afro-Americans and afro-Hispanics into a single “Afro” group, resulting in three possible racial groups: White (99 tissue samples from 76 patients), Afro (16 tissue samples from 11 patients), and Asian (15 tissue samples from 15 patients).37 In the United States, all tissue samples were analyzed using standardized DESI-MSI experimental parameters. Figure 2B shows representative DESI mass spectra obtained from IDC-tissue samples from an Afro patient from Brazil, a white patient from the United States, a white patient from Ukraine, and an Asian patient from Vietnam. Overall, consistent trends in the relative abundances of the molecular species (deprotonated molecules) were observed across the mass spectra. For example, the ion of m/z 281.249, attributed to FA(18:1), was detected at the highest relative abundance in comparison with other FA species. In the higher m/z range where complex lipids are detected, high relative abundances of the ions of m/z 835.533, PI(34:1); m/z 861.550, PI(36:2); m/z 863.563, PI(36:1); m/z 885.548, PI(38:4); and m/z 887.564, PI(38:3), were seen across all mass spectra, although with some variations in their relative abundances. To evaluate the level of variance in the mass spectra from all the patients, we employed unsupervised PCA. No significant separation was observed in the score plots due to patient race based on the DESI mass spectra for cancerous (Figure 2C) and normal tissues (Figure S1A). Further, no significant separation was seen in the score plots due to country of collection based on the DESI mass spectra for cancerous (Figure 2D) and normal tissues (Figure S1B). These results suggest that the metabolite and lipid profiles obtained by DESI-MSI are characteristic of IDC and normal breast tissues across different patient races and are unrelated to sample-collection site.
Figure 2.
(A) DESI-MSI performed on a large cohort of human tissue samples collected from different countries and from patients of different races. (B) Similar negative-ion-mode DESI mass spectra were obtained for breast-IDC-tissue samples from patients of different races and collection sites. Projections of the 86 mass spectra from the breast-IDC-tissue samples onto the first three principal components (PC) do not separate patients by (C) race or (D) country of collection, as observed in the 3D PCA plots. The first three PCs explain 63.0% and 60.0% of the total variance of the full data set for race and country, respectively.
Interlaboratory Assessment of the Reproducibility of DESI-MSI.
Assessment of interlaboratory reproducibility is essential to demonstrate the robustness of an analytical technology for a targeted application. We evaluated the interlaboratory reproducibility of DESI-MS for breast-cancer-tissue imaging by independently analyzing serial tissue sections of the same tissue sample in the United States and in Brazil, using similar experimental parameters (Table 1 and Figure S2). The mass spectra obtained in the United States and Brazil from the same region of the tissue sections showed similar patterns, with variations in the low-mass range from m/z 100–700 observed more clearly for the normal-tissue samples. Cosine similarity analyses were conducted to quantitatively evaluate similarity between the mass spectra obtained in Brazil and in the United States. As Table S2 summarizes, a low cosine value of 0.51 was obtained when comparing the full mass spectra of the normal samples analyzed in Brazil with the normal samples analyzed in the United States. When the m/z range was restricted to m/z 700–1200, the cosine value increased to 0.85, reflecting higher similarity at the higher m/z range. Further evaluation of the interlaboratory data revealed inconsistencies in the relative and total abundances of metabolites, FA, and background ions detected at the low m/z range. As the DESI ion images and mass spectra of Figure S2A show, higher relative abundances of ions attributed to ascorbic acid and FA(20:4) were detected in the tissue sections analyzed in the United States compared with in the adjacent tissue sections analyzed in Brazil. An unidentified background ion of m/z 415.140 was only observed in the data acquired in Brazil. Higher consistency in the relative and total ion abundances and spatial distribution was observed for the complex lipid region of the mass spectra (m/z > 700), where glycerophospholipids are commonly seen. The inconsistency in the ions detected at the low m/z range was not as pronounced for the data obtained from cancer regions of IDC samples (Figure S2B), which was also reflected in the similar cosine values obtained for the full m/z range (0.91) and the restricted m/z range (0.92).
Predictive Models of IDC Based on Lipid DESI-MSI Data.
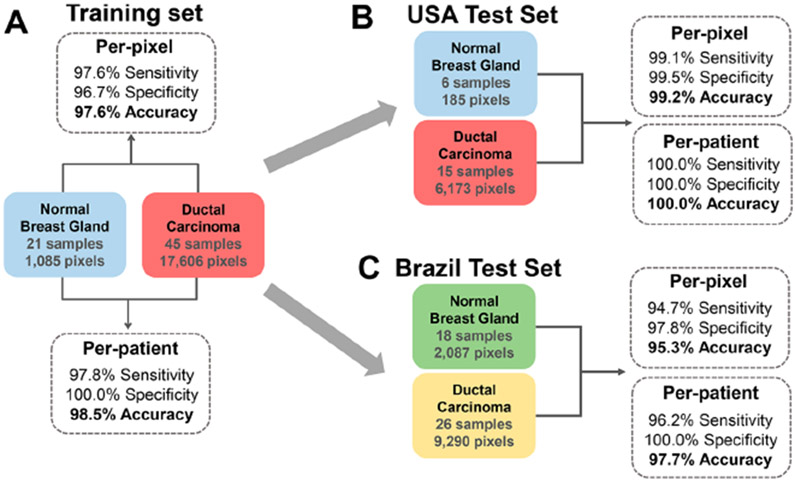
To evaluate if the metabolite and lipid information obtained by DESI-MSI are predictive of breast cancer, we used the Lasso method to build a classification model for IDC.38,39 DESI-mass-spectra data were extracted from areas within the ion images that presented predominantly IDC tumor or normal epithelial glands, yielding a total of 36 426 individual pixels per mass spectrum for all data acquired (United States and Brazil). The data was restricted to the high range of m/z 700–1200, which provided reproducible results as previously described. Next, 44 samples from 27 of the 53 Brazilian patients were randomly selected and excluded from the sample set with the goal of being used as an independent interlaboratory validation set in our study. The remaining 87 tissue samples were randomly divided into a training set and a validation set using a 75–25% split, respectively. The training set (45 IDC and 21 normal) yielded a total of 18 691 pixels (17 606 IDC and 1085 normal, all acquired in the United States), which were used to build the classification model. Note that many of the normal-breast-tissue samples analyzed were predominantly composed of fat; therefore, the total number of pixels extracted from focal regions presenting epithelial cells were limited. The remaining set of samples (15 IDC and 6 normal) were later used as an independent intralaboratory validation set. Prediction results are presented as sensitivity, specificity, and overall agreement on a per-pixel and per-patient basis (Figure 3 and Table S3). For the training set, 97.6% sensitivity, 96.7% specificity, and 97.6% accuracy were achieved on a per-pixel basis for IDC diagnosis using 5-fold cross-validation (Figure 3A). For the per-patient analysis, just a single cancer-tissue sample was misclassified as normal tissue. A total of 44 predictive m/z values were selected for the classification model with assigned mathematical weights related to their importance in distinguishing between IDC and normal tissues (Table 2). For an extended discussion on the features selected, please see the Supporting Information.
Figure 3.
Classification per-pixel and per-patient prediction results for normal and IDC samples including sensitivity, specificity, and overall accuracy in (A) cross-validation performed with data acquired in the United States, (B) test set with data acquired in the United States, and (C) test set with data acquired in Brazil.
Table 2.
Identified Species Selected by Lasso as Significant Contributors to the Molecular Model for IDC- and Normal-Breast-Tissue Classification with Attributed Statistical Weightsa
| measured m/z | lipid assignment | exact m/z | mass error (ppm) | proposed formula | Lasso weight |
|---|---|---|---|---|---|
| Characteristic of Normal Breast Tissue | |||||
| 742.539 | PE(36:2) | 742.539 | −0.4 | C41H77NO8P | −213.0 |
| 769.502 | PG(36:4) | 769.503 | −0.9 | C42H74O10P | −189.6 |
| 792.530 | PC(34:2) | −174.7 | |||
| 750.545 | PE(O-38:5) | 750.544 | 0.7 | C43H77NO7P | −169.4 |
| 786.529 | PS(36:2) | 786.529 | −0.4 | C42H77NO10P | −166.3 |
| 778.539 | PE(39:5) | 778.539 | −0.6 | C44H77NO8P | −149.5 |
| 748.528 | PE(O-38:6) | 748.529 | −0.8 | C43H75NO7P | −148.1 |
| 727.527 | PA(38:2) | 727.528 | −1.9 | C41H76O8P | −133.6 |
| 734.534 | PS(O-33:0) | 734.534 | −0.3 | C39H77NO9P | −105.0 |
| 891.722 | TG(52:3) | 891.721 | 0.3 | C55H100O6Cl | −104.6 |
| 770.534 | PS(P-36:2) or PS(O-36:3) | 770.534 | 0.1 | C42H77NO9P | −39.8 |
| 700.602 | Cer(t42:1) | 700.602 | 0.4 | C42H83NO4Cl | −38.8 |
| 816.576 | PS(38:1) | 816.576 | −0.6 | C44H83NO10P | −32.9 |
| 788.545 | PS(36:1) | 788.544 | 0.6 | C42H79NO10P | −25.3 |
| 752.553 | PE(O-38:4) | 752.560 | −9.0 | C43H79NO7P | −24.8 |
| 885.550 | PI(38:4) | 885.550 | −0.3 | C47H82O13P | −19.9 |
| 837.547 | PI(34:0) | 837.550 | −3.9 | C43H82O13P | −14.3 |
| 794.55 | PC(34:1) | −14.1 | |||
| 723.478 | CL(72:8) | 723.479 | −0.8 | C81H140O17P2 | −4.7 |
| 724.485 | CL(72:7) | 724.487 | −2.6 | C81H142O17P2 | −2.5 |
| Characteristic of IDC Tissue | |||||
| 846.657 | PS(O-41:0) | 846.659 | −3.3 | C47H93NO9P | +12.3 |
| 835.534 | PI(34:1) | 835.534 | −0.7 | C43H80O13P | +17.2 |
| 887.564 | PI(38:3) | 887.566 | −2.3 | C47H84O13P | +18.1 |
| 838.559 | PS(40:4) | 838.560 | −1.6 | C46H81NO10P | +34.3 |
| 773.533 | PG(36:2) | 773.534 | −0.8 | C42H78O10P | +56.2 |
| 760.513 | PS(34:1) | 760.513 | −0.3 | C40H75NO10P | +90.8 |
| 766.539 | PE(38:4) | 766.539 | 0.0 | C43H77NO8P | +116.4 |
| 810.525 | PS(38:4) | 810.529 | −4.9 | C44H77NO10P | +120.4 |
| 819.517 | PG(40:7) | 819.518 | −0.9 | C46H76O10P | +169.3 |
| 747.517 | PG(34:1) | 747.518 | −0.9 | C40H76O10P | +187.9 |
| 861.549 | PI(36:2) | 861.550 | −0.7 | C45H82O13P | +237.7 |
Chemical species were tentatively identified by high-mass-accuracy–high-mass-resolution and tandem MS analyses. Positive weights are given to m/z that are indicative of IDC, whereas negative weights are given to m/z that are indicative of normal breast tissues.
Intra- and Interlaboratory Validation of the Predictive Power of the Lasso Classification Model.
We have also evaluated the robustness and performance of our classification model in predicting breast-cancer diagnosis from the DESI data (m/z 700–1200) acquired from independent sample sets analyzed in the United States (intralaboratory) and in Brazil (interlaboratory). Our model was first tested using the excluded test set analyzed in the United States (15 IDC and 6 normal), which yielded a total of 6385 pixels (6173 IDC and 185 normal). Excellent performance was achieved: 99.1% sensitivity, 99.5% specificity, and 99.2% accuracy per-pixel, as well as 100.0% sensitivity, specificity, and accuracy per-patient (Figure 3B). The classification model was then used to predict data acquired in Brazil for the excluded validation set of 44 tissue samples from 27 Brazilian patients, which yielded a total of 11 377 pixels (9290 IDC and 2087 normal). Remarkably, excellent performance was seen on a per-pixel basis for IDC diagnosis (Figure 3C): 94.7% sensitivity, 97.8% specificity, and 95.3% accuracy. In the per-patient analysis, only a single cancer sample was misclassified as normal. These results suggest that a single Lasso model built from DESI-MSI data (m/z 700–1200) can be used to classify intra- and interlaboratory data acquired under optimized conditions from independent sample sets. Further, the predictive complex lipids selected by the Lasso model are robust for breast-cancer diagnosis.
Prediction of Breast-Cancer-Hormone-Receptor and HER2 Status.
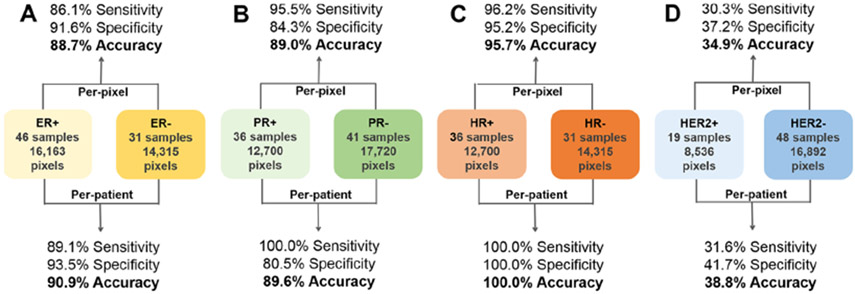
Next, we investigated if the molecular information obtained by DESI-MSI enabled prediction of positive or negative status for ER, PR, combined hormone receptor (HR), and HER2 by building classification models for each class using IHC or FISH results as the gold standard. For the 77 IDC samples with ER status, 46 samples were ER+ (16 163 pixels), and 31 samples were ER− (14 315 pixels). The classification model built to predict ER status achieved 86.1% sensitivity, 91.6% specificity, and 88.7% accuracy on a per-pixel basis (Figure 4). In the per-patient analysis, five ER+ tissue samples were misclassified as ER− tissues, and two ER− tissue samples were misclassified as ER+ tissues, resulting in 89.1% sensitivity, 93.5% specificity, and 90.9% accuracy. For PR status, the classification model built using 36 PR+ samples (12 700 pixels) and 41 PR− samples (17 720 pixels) yielded 95.5% sensitivity, 84.3% specificity, and 89.0% accuracy on a per-pixel basis. In the per-patient analysis, eight PR− tissue samples were misclassified as PR+ tissues. Samples were further combined into hormone-receptor (HR) positive (ER+ and PR+) and HR negative (ER− and PR−) groups on the basis of the combined ER and PR status. The classification model built using 36 HR− samples (14 315 pixels) and 31 HR+ samples (12 700 pixels) yielded 96.2% sensitivity, 95.2% specificity, and 95.7% accuracy on a per-pixel basis and 100.0% sensitivity, specificity, and accuracy on a per-patient basis. These accuracies are similar to the overall 96.0% accuracy achieved in the per-patient analysis (n = 27) for HR positive and negative statuses described by Guenther et al.21 and corroborate the PCA clustering observed for HR+ versus HR− samples (n = 14) by Calligaris et al.30 Next, we built a HER2 classification model using 19 HER2+ samples (8536 pixels) and 48 HER2− samples (16 892 pixels), but poor classification results were achieved (30.3% sensitivity, 37.2% specificity, and 34.9% accuracy on a per-pixel basis). Interestingly, no separation or discrimination achieved on the basis of the HER2 status was previously reported by Guenther et al. (n = 27) and Calligaris et al. (n = 14), which again corroborated our results.
Figure 4.
Per-pixel and per-patient sensitivity, specificity, and overall accuracy of the prediction results for positive or negative (A) ER, (B) PR, (C) HR, and (D) HER2 statuses. Prediction results were obtained using a 5-fold CV approach.
DISCUSSION
We have performed a multicenter study using DESI-MSI to investigate the metabolic signatures of a diverse set of 131 breast-tissue samples from 103 breast-cancer patients from various countries, including the United States, Brazil, Ukraine, Vietnam, and others. DESI-MSI enabled clear visualization of fine histologic features in breast tissues and thus a detailed investigation of metabolic profiles characteristic of breast cancer, normal breast glands, and adjacent stroma. Using tissue samples obtained from patients from various countries, we showed that DESI-MSI allows for the detection of lipid profiles that are characteristic of cancer tissue independent of patient race or ethnicity. These molecular profiles, when used to build classification models for cancer diagnosis, provided high sensitivity, specificity, and accuracy for both training and test sample sets using data acquired in the United States and revealed predictive lipid markers. Most notably, the molecular classifiers showed high performance for cancer diagnosis of an independent data set acquired in a laboratory in Brazil using standardized experimental conditions. Classifiers built to characterize PR and ER statuses in breast-cancer samples also showed high accuracy in determining hormone-receptor status. Altogether, our study provide strong evidence that DESI-MSI is a robust molecular technology able to provide rapid diagnosis and characterization of breast tissues, with potential use in the clinical setting across different institutions.
Alterations in the abundances of FA and glycerophospholipids were detected in our study, which reflect known abnormalities in cancer-cell metabolism40 and breast-cancer tissue.13,41,42 Fatty acid synthesis, for example, is highly relevant in breast-cancer-tumor biology because of the ability of these molecules to modulate the fluidity of lipid membranes and to affect cellular machinery.43 Several, single-center studies suggest that lipid MSI signatures are diagnostic of breast cancer.22,24,30 A MALDI-MSI study has reported higher relative abundances of monounsaturated FA, in comparison with those of polyunsaturated FA, in breast cancer than in adjacent normal tissues.22 Mao et al. also found FA(18:2) and FA(18:1) to be more abundant in cancer tissue when compared with in normal tissue analyzed by air-flow-assisted ionization MSI,24 whereas Calligaris et al. also reported FA(18:1) as a DESI-MSI discriminator for cancer detection and margin analysis.30 Similarly, we consistently noted high relative abundances of FA(18:1), at m/z 281.249, in breast-cancer tissues in our study and identified several glycerophospholipid ions as predictive markers of breast cancer, with increased relative abundance in malignant histologic regions of tissue sections. Also using MALDI-MSI, Toi et al. have previously reported altered relative abundances of PIs in malignant epithelial regions of breast-cancer tissues.23 In their study, PI(36:1), for example, was observed in high relative abundances in cancer tissues when compared with those in benign epithelial regions, whereas PI(38:3) was putatively associated with cancer-cell invasion. Similarly, high relative abundances of the PI(36:1) ion of m/z 863.563 as well as the PI(36:2) ion of m/z 861.549 were consistently observed in breast-cancer tissues in comparison with normal-tissue samples (Figure 1C); hence, PI(36:2) was selected and shown to be highly predictive of breast IDC tissue by the Lasso classification model. Lipid information acquired by DESI-MSI was also used by Guenther et al. to build classification models for breast-cancer diagnosis based on a subset of 19 predictive features, including various FA and glycerophospholipids, such as PE, PC, and PI.44 Within these, three glycerophospholipid species, PC(34:2), PC(34:1), and PI(38:3), were also selected as predictive markers by our classifier. Our study now confirms with comprehensive data that lipid profiles are robust diagnostic markers of breast cancer and that such markers are valid across different patient populations. Our PCA results also showed great similarity for the breast-IDC DESI-MS data regardless of race, further demonstrating the commonality of diagnostic lipid profiles. Future studies are needed to validate these findings on larger patient cohorts.
The main goal of our multicenter study was to evaluate the performance of the DESI-MSI workflow in producing reproducible data for breast-cancer diagnosis across institutions and operators. A few multicenter studies have been performed to evaluate the performance of proteomics analysis for breast-cancer diagnosis using traditional proteomics assays45 and MALDI-MSI.46 In the MALDI-MSI study, 40 breast-cancer tissues were analyzed in two centers in Europe (n = 12 in Munich, and n = 18 in Leiden) using independent sample sets, methods, and operators.46 Hierarchical clustering yielded 100 and 80.9% classification accuracies for discriminating extra-tumoral and intratumoral stromal profiles in the Munich and Leiden data sets, respectively. In our study, sample exchange among centers was implemented, and a standardized experimental workflow was adopted to ensure uniformity in the analytical approach of DESI-MSI employed in the United States and Brazil, despite differences in the mass-spectrometer platforms and minor operational parameters. Our interlaboratory analysis using the same tissue samples analyzed in Brazil and the United States revealed high mass-spectra similarity in the high m/z region (m/z 700–1200) of the data for both normal (cosine similarity = 0.86) and cancerous (cosine similarity = 0.93) tissues, and moderate similarity in the relative abundances of low m/z ions (m/z < 400) was observed for normal breast tissue (cosine similarity = 0.48), despite high similarity in the low m/z region for cancerous tissue (cosine similarity = 0.91). The DESI mass spectra in the low m/z region is mostly composed of ions identified as background solvent peaks, small metabolites, and fatty acids. Whereas variations in chemical noise and background ions were anticipated across different mass spectrometers, differences in the relative abundances of biologically relevant ions such as FA were unexpected. We suggest that these differences could be due to small variations in the DESI spray-geometry parameters or the mass-spectrometer S-lens ion-optics RF values, which may lead to more efficient desorption or transmission of ions. Further, these differences could be due to lipid degradation owing to possible variations in temperatures and freezing conditions during international sample shipment in dry ice, factors which have been associated with increases in FA abundance in DESI-mass-spectra profiles (for more information, please see the Supporting Information).47 As normal breast tissue provides poorer molecular profiles than IDC, higher variance was observed in the low m/z range for normal breast tissues. Nevertheless, high reproducibility in the higher m/z range, in which glycerophospholipids and other complex lipid markers are detected, was seen for normal and cancerous tissues analyzed in the United States and Brazil. Lasso classifiers built using 44 predictive features detected at the restrictive m/z range provided outstanding performance in cross-validation both per-pixel (97.6% accuracy, n = 18 691) and per-patient (98.5% accuracy, n = 66). The classification models provided high accuracy in cancer diagnosis using an independent data set acquired in the United States (99.2% per-pixel accuracy, n = 6358) and, most remarkably, acquired in Brazil (95.3% per-pixel accuracy, n = 11 377). These results validate the predictive power of the lipid-ion markers and classification models built from DESI-MSI data for breast-cancer diagnosis and provides strong evidence that this technology is robust for breast-cancer diagnosis across centers and populations.
Determining ER, PR, and HER2 status is essential for identifying breast-cancer molecular subtypes to help guide treatment for patients. Previous studies have also suggested a relationship between lipid profiles and breast-cancer molecular subtypes.42 In our study, we further evaluated the ability of using the lipid data acquired by DESI-MSI in the United States to build predictive models for positive or negative statuses of HER2, PR, and ER separately, as well as combined HR. Per-patient accuracy of 90.9% was achieved for predicting positive or negative ER status (n = 77), whereas 89.6% per-patient accuracy was achieved for predicting positive or negative PR status (n = 77). When evaluating the combined HR status, we achieved 100% per-patient accuracy. Although prediction of combined HR status has been previously reported by DESI-MSI (accuracy of 86.7%, n = 27), our results now indicate that the lipid information is also predictive of ER and PR status separately. When evaluating HER2 status, we found no relationship between lipid profiles detected by DESI-MSI and HER2 status (38.8% per-patient accuracy, n = 67), as was previously found using DESI-MSI. This confusion could be due to intratumor heterogeneity of HER2-positive breast cancers,48,49 which may lead to variations in lipid metabolism within the cancer-tissue regions from which the mass spectra were extracted for statistical analysis. The use of FISH to determine HER2 status was also only employed when IHC results were “undetermined”, potentially leading to higher incidence of inaccurate HER2 statuses.50 Further studies using IHC and FISH assays to investigate the spatial expression of HER2 in each tissue section correlated with lipid abundances will be performed to better investigate these observations. In addition, further studies using larger sample cohorts will be performed to investigate possible correlations between lipid profiles detected by DESI-MS imaging and other patient and clinical characteristics, including breast-cancer molecular subtypes, stage, and treatment choice.
In conclusion, our study provides strong evidence that the lipid information acquired by DESI-MSI is highly accurate in predicting breast cancer as well as ER and PR status. Most importantly, our multicenter study has demonstrated that DESI-MSI is a robust, highly reproducible technology for rapid breast-cancer-tissue section diagnosis and may be useful in the clinical setting.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Cancer Institute (NCI) of the National Institutes of Health under Award Number R00CA190783 to L.S.E., FAPESP grants 2013/25683-3 and 2015/18830-5 to L.O.S., and a grant from the Brazilian National Research Council - CNPq (project no. 470451/2014-9) to A.M.P. A sub-set of the tissue samples were provided by the Cooperative Human Tissue Network, which is funded by the NCI.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.8b01961.
Detailed materials and methods, predictive features selected by Lasso, demographic and clinicopathologic characteristics of samples, statistical results, DESI mass spectra, ion images, and PCA results (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Koboldt DC; Fulton RS; McLellan MD; Schmidt H; Kalicki-Veizer J; McMichael JF; Fulton LL; Dooling DJ; Ding L; Mardis ER; et al. Nature 2012, 490, 61–70.23000897 [Google Scholar]
- (2).Jemal A; Bray F; Center MM; Ferlay J; Ward E; Forman D Ca-Cancer J. Clin 2011, 61, 69–90. [DOI] [PubMed] [Google Scholar]
- (3).Esserman LJ; Berry DA; DeMichele A; Carey L; Davis SE; Buxton M; Hudis C; Gray JW; Perou C; Yau C; et al. J. Clin. Oncol 2012, 30, 3242–3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Kos Z; Dabbs DJ Histopathology 2016, 68, 70–85. [DOI] [PubMed] [Google Scholar]
- (5).Perou CM; Sorlie T; Eisen MB; van de Rijn M; Jeffrey SS; Rees CA; Pollack JR; Ross DT; Johnsen H; Akslen LA; et al. Nature 2000, 406, 747–752. [DOI] [PubMed] [Google Scholar]
- (6).Perou CM Oncologist 2010, 15, 39–48. [DOI] [PubMed] [Google Scholar]
- (7).Sorlie T; Perou CM; Tibshirani R; Aas T; Geisler S; Johnsen H; Hastie T; Eisen MB; van de Rijn M; Jeffrey SS; et al. Proc. Natl. Acad. Sci. U. S. A 2001, 98, 10869–10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Yaziji H; Barry T Adv. Anat. Pathol 2006, 13, 238–246. [DOI] [PubMed] [Google Scholar]
- (9).Hicks DG; Tubbs RR Hum. Pathol 2005, 36, 250–261. [DOI] [PubMed] [Google Scholar]
- (10).Furrer D; Sanschagrin F; Jacob S; Diorio C Am. J. Clin. Pathol 2015, 144, 686–703. [DOI] [PubMed] [Google Scholar]
- (11).Frampton GM; Fichtenholtz A; Otto GA; Wang K; Downing SR; He J; Schnall-Levin M; White J; Sanford EM; An P; et al. Nat. Biotechnol 2013, 31, 1023–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Alexandrov LB; Nik-Zainal S; Wedge DC; Aparicio S; Behjati S; Biankin AV; Bignell GR; Bolli N; Borg A; Borresen-Dale AL; et al. Nature 2013, 500, 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Budczies J; Brockmoller SF; Muller BM; Barupal DK; Richter-Ehrenstein C; Kleine-Tebbe A; Griffin JL; Oresic M; Dietel M; Denkert C; et al. J. Proteomics 2013, 94, 279–288. [DOI] [PubMed] [Google Scholar]
- (14).Heeren RMA Int. J. Mass Spectrom 2015, 377, 672–680. [Google Scholar]
- (15).Golf O; Strittmatter N; Karancsi T; Pringle SD; Speller AV; Mroz A; Kinross JM; Abbassi-Ghadi N; Jones EA; Takats Z Anal. Chem 2015, 87, 2527–2534. [DOI] [PubMed] [Google Scholar]
- (16).Laskin J; Heath BS; Roach PJ; Cazares L; Semmes OJ Anal. Chem 2012, 84, 141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Walsh CM; Reschke BR; Fortney J; Piktel D; Razunguzwa TT; Powell MJ; Gibson LF Cancer Res. 2012, 72, 4793. [Google Scholar]
- (18).Hsu C-C; Dorrestein PC Curr. Opin. Biotechnol 2015, 31, 24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Laskin J; Lanekoff I Anal. Chem 2016, 88, 52–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Bluestein BM; Morrish F; Graham DJ; Guenthoer J; Hockenbery D; Porter PL; Gamble LJ Analyst 2016, 141, 1947–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Guenther S; Muirhead LJ; Speller AVM; Golf O; Strittmatter N; Ramakrishnan R; Goldin RD; Jones E; Veselkov K; Nicholson J; et al. Cancer Res. 2015, 75, 1828–1837. [DOI] [PubMed] [Google Scholar]
- (22).Guo S; Wang YM; Zhou D; Li ZL Sci. Rep 2015, 4, 5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Kawashima M; Iwamoto N; Kawaguchi-Sakita N; Sugimoto M; Ueno T; Mikami Y; Terasawa K; Sato TA; Tanaka K; Shimizu K; et al. Cancer science 2013, 104, 1372–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Mao X; He J; Li T; Lu Z; Sun J; Meng Y; Abliz Z; Chen J Sci. Rep 2016, 6, 21043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Rauser S; Marquardt C; Balluff B; Deininger SO; Albers C; Belau E; Hartmer R; Suckau D; Specht K; Ebert MP; et al. J. Proteome Res 2010, 9, 1854–1863. [DOI] [PubMed] [Google Scholar]
- (26).Tata A; Woolman M; Ventura M; Bernards N; Ganguly M; Gribble A; Shrestha B; Bluemke E; Ginsberg HJ; Vitkin A; et al. Sci. Rep 2016, 6, 35374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Alberici RM; Simas RC; Sanvido GB; Romao W; Lalli PM; Benassi M; Cunha IBS; Eberlin MN Anal. Bioanal. Chem 2010, 398, 265–294. [DOI] [PubMed] [Google Scholar]
- (28).Ifa DR; Eberlin LS Clin. Chem 2016, 62, 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Dill AL; Ifa DR; Manicke NE; Ouyang Z; Cooks RG J. Chromatogr. B: Anal. Technol. Biomed. Life Sci 2009, 877, 2883–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Calligaris D; Caragacianu D; Liu X; Norton I; Thompson CJ; Richardson AL; Golshan M; Easterling ML; Santagata S; Dillon DA; et al. Proc. Natl. Acad. Sci. U. S. A 2014, 111, 15184–15189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Zhang J; Feider CL; Nagi C; Yu W; Carter SA; Suliburk J; Cao HS; Eberlin LS J. Am. Soc. Mass Spectrom 2017, 28, 1166–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Huo DZ; Hu H; Rhie SK; Gamazon ER; Cherniack AD; Liu JF; Yoshimatsu TF; Pitt JJ; Hoadley KA; Troester M; et al. Jama Oncology 2017, 3, 1654–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Michels KB; Mohllajee AR; Roset-Bahmanyar E; Beehler GP; Moysich KB Cancer 2007, 109, 2712–2749. [DOI] [PubMed] [Google Scholar]
- (34).Dill AL; Eberlin LS; Costa AB; Ifa DR; Cooks RG Anal. Bioanal. Chem 2011, 401, 1949–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Eberlin LS; Ferreira CR; Dill AL; Ifa DR; Cheng L; Cooks RG ChemBioChem 2011, 12, 2129–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Bokhart MT; Nazari M; Garrard KP; Muddiman DC J. Am. Soc. Mass Spectrom 2018, 29, 8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).O’Brien KM; Cole SR; Tse CK; Perou CM; Carey LA; Foulkes WD; Dressler LG; Geradts J; Millikan RC Clin. Cancer Res 2010, 16, 6100–6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Eberlin LS; Tibshirani RJ; Zhang J; Longacre TA; Berry GJ; Bingham DB; Norton JA; Zare RN ; Poultsides GA Proc. Natl. Acad. Sci. U. S. A 2014, 111, 2436–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Sans M; Gharpure K; Tibshirani R; Zhang J; Liang L; Liu J; Young JH; Dood RL; Sood AK; Eberlin LS Cancer Res. 2017, 77, 2903–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Cairns RA ; Harris IS; Mak TW Nat. Rev. Cancer 2011, 11, 85–95. [DOI] [PubMed] [Google Scholar]
- (41).Chughtai K; Jiang L; Greenwood TR; Glunde K; Heeren RM J. Lipid Res 2013, 54, 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Baumann J; Sevinsky C; Conklin DS Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2013, 1831, 1509–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Baenke F; Peck B; Miess H; Schulze A Dis. Models & Mech 2013, 6, 1353–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Guenther S; Muirhead LJ; Speller AV; Golf O; Strittmatter N; Ramakrishnan R; Goldin RD; Jones E; Veselkov K; Nicholson J; et al. Cancer Res. 2015, 75, 1828–1837. [DOI] [PubMed] [Google Scholar]
- (45).Mertens BJA J. Proteomics 2009, 72, 785–790. [DOI] [PubMed] [Google Scholar]
- (46).Dekker TJ; Balluff BD; Jones EA; Schone CD; Schmitt M; Aubele M; Kroep JR; Smit VT; Tollenaar RA ; Mesker WE; et al. J. Proteome Res 2014, 13, 4730–4738. [DOI] [PubMed] [Google Scholar]
- (47).Dill AL; Eberlin LS; Costa AB; Ifa DR; Cooks RG Anal. Bioanal. Chem 2011, 401, 1949–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Buckley NE; Forde C; McArt DG; Boyle DP; Mullan PB; James JA; Maxwell P; McQuaid S; Salto-Tellez M Sci. Rep 2016, 6, 23383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Burstein HJ N. Engl. J. Med 2005, 353, 1652–1654. [DOI] [PubMed] [Google Scholar]
- (50).Wesola M; Jelen M Advances in Clinical and Experimental Medicine 2015, 24, 899–903. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.