Abstract
De novo assembly of short DNA reads remains an essential technology, especially for large-scale projects and high-resolution variant analyses in epidemiology. However, the existing tools often lack sufficient accuracy required to compare closely related strains. To facilitate such studies on bacterial genomes, we developed Platanus_B, a de novo assembler that employs iterations of multiple error-removal algorithms. The benchmarks demonstrated the superior accuracy and high contiguity of Platanus_B, in addition to its ability to enhance the hybrid assembly of both short and nanopore long reads. Although the hybrid strategies for short and long reads were effective in achieving near full-length genomes, we found that short-read-only assemblies generated with Platanus_B were sufficient to obtain ≥90% of exact coding sequences in most cases. In addition, while nanopore long-read-only assemblies lacked fine-scale accuracies, inclusion of short reads was effective in improving the accuracies. Platanus_B can, therefore, be used for comprehensive genomic surveillances of bacterial pathogens and high-resolution phylogenomic analyses of a wide range of bacteria.
Keywords: Bacterial genome, de novo assembly, high-resolution phylogenomics, large-scale genomic surveillance
1. Introduction
Whole-genome sequencing (WGS) has become a crucial technique for investigation of the outbreaks and genomic evolution of pathogens.1–3 Technological advances have enabled large-scale studies that involve the surveillance of hundreds to thousands of isolated microbes.2,4–6 In addition, high-resolution variant analyses have been used to distinguish isolates whose genomes differ only by a few bases.1,7–10 The resulting phylogenomic information is, therefore, expected to help trace the infection route of pathogens.
Short-read sequencing is a fundamental technology for large-scale WGS analyses owing to its advantages of low error rates (<1%), high throughput, and low cost per isolate.11 Technically, some variant-calling tools, based on read mapping, were reported to output several false results, 12 and de novo assembly was found to be an effective method to improve such analyses for complex genomes.13,14 Even for bacterial genomes, read mapping or reference-guided assembly methods were reported to be influenced by the use of reference genomes15; de novo assembly can reduce biases that stem from the differences of richness of reference genomes among lineages.
Nanopore long-read sequencing is a powerful technology for assembling circular genomes16 and has been applied for the surveillance of outbreaks owing to its portability.17,18 However, the fine-scale (base-level) accuracy of long read-only assemblies could be inferior to that of hybrid assemblers that utilize short reads.11,19Single-molecule real-time (SMRT) sequencing by Pacific Biosciences (PacBio) is also considered effective in assembling gap-free genomes. Nonetheless, there are small and cheap units (flow cells) such as Flongle (Oxford Nanopore Technologies; throughput, ∼2 Gbp) for the nanopore sequencers, and they can be run without outsourcing the procedures. Because of these features, the nanopore sequencers may be cost- and time-efficient when targeting microbes with small genomes compared with the SMRT sequencers.
From the end of 2019 to February 2020, early investigations of the novel corona virus associated with the global outbreak employed the short read and de novo assembly.20,21 Nanopore long reads were also applied, but combined with short reads to improve the accuracy of the assembled sequences in certain studies.22 In other words, the short-read genomic sequencing is still a practical method in the emergent circumstances at present. Although the targets of these investigations are viral genomes of ∼30 kb in length, de novo assembly may be employed to capture variable genomic regions; similar situations will occur in bacterial genomes containing variable genomic regions derived from prophages and other types of mobile genetic elements that often encode drug-resistance genes and virulence-related genes.
Thus, to satisfy the stringent requirement of accuracy, especially the fine-scale accuracy, for high-resolution bacterial WGS analyses, short-read-based de novo assemblers require further improvement. Therefore, we developed an accurate assembler named Platanus_B.
2. Materials and methods
2.1. Overview of Platanus_B
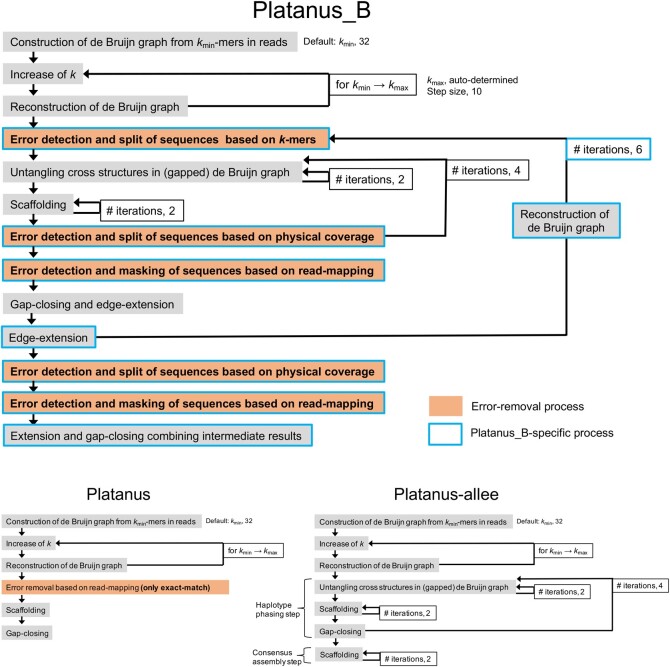
In this section and the following ones, the overview and specific functions of Platanus_B are described. The other miscellaneous procedures are described in the Supplementary Methods. The overview of Platanus_B and the differences between Platanus_B and related assemblers, Platanus23 and Platanus-allee,24 are illustrated in Fig. 1. These two related assemblers target diploid genomes, which do not principally exist in bacterial genomes, and try to merge (Platanus) or separate (Platanus-allee) heterozygous genomic regions. Although some elementary functions for de novo assembly, such as contig-assembly and scaffolding, are derived from these tools, the overall procedure is different and many specific functions are implemented (Fig. 1).
Figure 1.
Workflow of Platanus_B and the related assemblers. Orange- and blue-framed boxes correspond to error-correction and Platanus_B-specific processes, respectively.
The key features of Platanus_B are as follows: first, it employs multiple types of error-removal processes, and is thus expected to effectively handle a wide range of errors. Second, several functions can be iterated, which may be impractical for large eukaryotic genomes when considering the calculation cost.
Platanus_B first assembles short reads into contigs using de Bruijn graphs. This function is derived from Platanus but skips the bubble-removal step that is required for diploid genome assembly. Platanus_B primarily utilizes the paired ends of short reads and iterates the following steps six times:
Error detection and split of sequences based on k-mers
Untangling the cross-structures in a (gapped) de Bruijn graph
Scaffolding
Error detection and split of sequences based on physical coverage
Error detection and masking of sequences based on read mapping
Gap closing and edge extension.
When the process returns to Step (1) from (6), scaffolds and local contigs are then merged through the de Bruijn graph. Internally, Steps (2)–(4) are iterated further (Fig. 1). After these iterations of (1)–(6), the error-removal Steps (4) and (5) are applied again, and extension and gap closing are performed using the intermediate iteration results. Platanus_B can also use long reads in Steps (2)–(4) assisted by the Minimap2 aligner25; however, this function is optional.
2.2. Error detection and split of sequences based on k-mers
This function can remove small-scale misassemblies avoiding the issues derived from ambiguities in read mapping. The input comprises short reads and contigs assembled through the de Bruijn graph. k corresponds to the maximum value used in the previous contig-assembly step, and the numbers of k-mers in short reads are counted. A contig is split at a position where the k-mer coverage (number of occurrences in short reads) is <1/10th of the median value for the contig.
2.3. Untangling the cross-structures in a (gapped) de Bruijn graph
To simplify a de Bruijn graph and extend contigs, Platanus_B untangles the cross-structures that are generated from repetitive sequences in a genome in a de Bruijin graph. This untangling function is also applied to a gapped de Bruijn graph, which is a de Bruijn graph that allows gaps between contigs. This function is derived from Platanus-allee.24 However, Platanus_B uses the mean coverage depth instead of the depth of heterozygous regions to determine whether a cross-structure should be solved. This function is iterated twice within the outer loops (Fig. 1).
2.4. Scaffolding
To determine the arrangements of contigs and obtain long sequences (scaffolds) allowing gaps, Platanus_B performs a scaffolding step. This function is derived from Platanus and Platanus-allee, and can handle long reads and mate pairs in addition to paired-end reads. As a modification, sub-functions related with heterozygous regions, such as removal of bubbles in a scaffold graph, are omitted. This function is iterated twice within the outer loops (Fig. 1).
2.5. Error detection and split of sequences based on physical coverage
This function can remove relatively large-scale misassemblies. Physical coverage (the number of paired ends that span a position) is calculated by mapping the paired ends and/or other types of libraries to input scaffolds. In addition, for each position, the number of paired ends that are linked to a neighbouring position and another scaffold are counted and stored as the ‘diff-coverage’ value. Positions that indicate low physical coverage and high diff-coverage are then detected as misassembly candidates and the scaffolds are split at these positions. Specifically, a split will be performed if all the following conditions are satisfied:
diff-coverage/physical coverage > 2
physical coverage < median physical coverage/2
median physical coverage > 2
diff-coverage > 2.
2.6. Error detection and masking of sequences based on read mapping
This function can remove base-level errors. The reads are mapped to the input scaffolds using ungapped alignments (seed size, 32 bp), and the highest sequence identity and a corresponding position is calculated for each read, discarding results with multiple highest identities. Mapping results are categorized according to the threshold of identity (97%) and coverages, both are calculated for each category. If low-identity coverage exceeds high-identity coverage, the corresponding position is masked (the base is changed to ‘N’).
2.7. Gap closing and edge extension
Paired ends are mapped to the scaffolds. Reads corresponding to the gaps (‘N’s) and edges of scaffolds are collected and locally assembled. If the resultant contigs overlap the adjacent regions, the gaps are closed or the edges are extended. This function is derived from Platanus with the following modifications:
The k values of de Bruijn graphs for local assemblies are 20 and 80 rather than 24 and 72. In this step, contigs are generated from the two de Bruijn graphs of the two k values, and the two contig sets are merged.
A k-mer coverage cut-off of 2 instead of 3 is used for local assemblies.
If a gap is partially closed, it is filled and the gap size is reduced.
Edges of sequences are extended.
2.8. Reconstruction of a de Bruijn graph when starting the next iteration
To reduce redundancies and improve the lengths of contigs, scaffolds, and local contigs are merged through a de Bruijn graph. Local contigs represent the contigs that are assembled in the gap-closing step but are not used to close gaps. k is increased according to the progress of iterations, where k = mean read length × [1 + (current number of iterations − 1)/6]. Short branches are excluded from the graph in a similar manner as performed in the contig assembly step; sequences with errors can be excluded.
2.9. Benchmarks using actual bacterial genome data
Platanus_B and the existing assemblers were benchmarked with sequence data from five strains of four bacterial species26–30 (Table 1), using seven preparation kits for paired-end reads of MiSeq (Illumina, CA, USA; Supplementary Table S1). MinION (Oxford Nanopore Technologies, Oxford, UK) with a flowcell of R9.4.1 chemistry was used for long reads of two strains (Supplementary Table S1). The read length of the MiSeq sequencing was 300 bp, and the nominal insert sizes ranged from 300 to 600 bp. Although MiSeq has been one of the most popular sequencers so far, newer short-read sequencers such as NovaSeq are generally run with read length of 150 bp. To simulate the reads from the new sequencers, we additionally trimmed the reads of MiSeq and generated 150-bp reads. For details of sample preparation procedures, including culturing of Escherichia coli strains, sequencing, public data collection, pre-processes of reads, refer to Supplementary Methods. The complete genome sequences of all these strains were available and used for evaluation. As an indicator of the repetitiveness (i.e. difficulty) of the genomes, the occurrences of 100-mers in the reference genomes were counted. The rate of repetitive (occurrences ≥2) 100-mers is shown in Table 1. Escherichia coli O157 Sakai and Porphyromonas gingivalis ATCC 33277 exhibited a high repetitive 100-mer rate, which may reflect the presence of highly similar prophages27 and transposable elements,28 respectively.
Table 1.
Strains used in the benchmarks
| Strain | Genome size (bp) | Repetitive 100-mer rate (%) | No. of CDSs |
|---|---|---|---|
| E. coli O157 Sakai | 5,594,605 | 5.31 | 5,291 |
| E. coli K-12 MG1655 | 4,641,652 | 1.91 | 4,357 |
| B. bronchiseptica S798 | 5,191,712 | 0.95 | 4,824 |
| S. marcescens SM39 | 5,326,023 | 1.13 | 4,972 |
| P. gingivalis ATCC 33277 | 2,354,886 | 7.63 | 2,051 |
The reference genomic data were downloaded from the RefSeq database. Repetitive 100-mer rate indicates the rate of 100-mers that occur more than 1 time in a reference genome. The RefSeq assembly accessions of the reference genomes were GCF_000008865.2,27 GCF_000005845.2,26 GCF_000829175.1,30 GCF_000828775.1,29 and GCF_000010505.1.28
The popular assemblers, MaSuRCA,31 SPAdes,32 Unicycler,33 Canu,34 Flye,35 Wtdbg2,36 and miniasm37 were compared with Platanus_B. The first three assemblers accept either a single short-read library or a mixed (short and long reads) library. The last four assemblers accept long-read library. As miniasm does not have a function to construct consensus sequences, polishing with long reads was performed for its results by Racon38 three times. Additionally, we benchmarked DISCOVAR de novo,14 which is designed for paired ends whose read lengths ≥250 bp, for the 300 bp reads. A polishing tool, Pilon,39 was also tested for all assemblies. For long-read-based assemblers (Canu, Flye, Wtdbg2, and miniasm+Racon), Pilon was executed three times for each result.
Indicators to benchmark accuracy and contiguity of assemblies were measured by QUAST40 using reference genomes. Variable read coverage was tested using random sampling of reads. For each case, 10 replicated inputs were generated and indicators were averaged. Details of samples, preparation, and analysis are provided in the Supplementary Methods.
3. Results and discussion
3.1. Benchmark using short- and paired-end reads
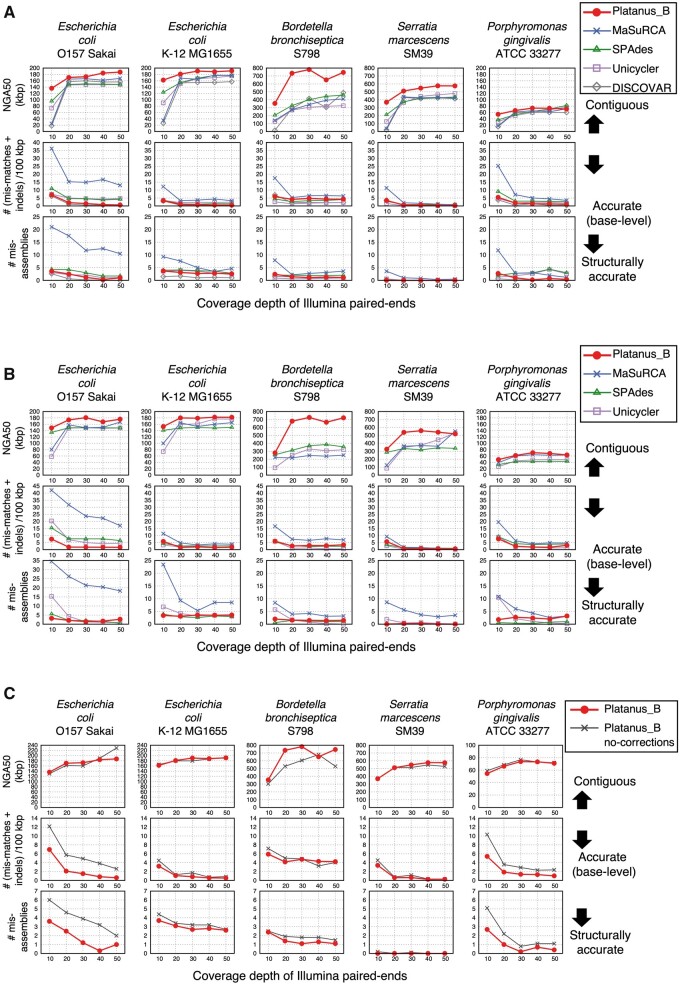
As a representative of the application of Platanus_B, Fig. 2A and Supplementary Table S2 show results for paired-end reads obtained from the TruSeq PCR-free kit. The nominal insert size was 600 bp. The coverage bias of this kit was similar to that of the relatively new kits.41 In most of the cases of 300 bp reads (24/25), among all assemblers tested, Platanus_B obtained the largest NGA50 values which are indicators of contiguity corrected for misassemblies. DISCOVAR provided the best lowest base error rate [(mismatches + indels)/100 kbp] and number of misassemblies in majority of the cases (16/25 and 17/25, respectively), which might reflect the effect of the special function for read length ≥250 bp. Among the versatile assemblers (Platanus_B, MaSuRCA, SPAdes, and Unicycler), Platanus_B provided the lowest values for these indicators in majority of the cases (13/25 and 15/25, respectively). For a repeat-rich strain, E. coli O157 Sakai, the base error rate advantage of Platanus_B was even more notable. The same trend was observed when polishing with Pilon (Supplementary Fig. S6 and Supplementary Table S3) or using 150-bp reads (Fig. 2B andSupplementary Table S4). To validate the effect of the error-correction functions of Platanus_B, the version in which these functions were deactivated were also benchmarked (Fig. 2C andSupplementary Table S5). NGA50 values of Platanus_B were comparable to the deactivated version for all strains, therefore rejecting the possibility of a poor effect on sequence contiguity. Base error rates and the number of misassemblies were approximately halved or lesser than those of the deactivated version for the repeat-rich strains, E. coli O157 Sakai and P. gingivalis. Consequently, the effectiveness of error corrections was validated especially for repeat-rich samples. With respect to the run time, in most cases, Platanus_B consumed less CPU and real time when compared with Unicycler [Table 2(A and B); 25/25 and 19/25, respectively]. Peak memory usages are shown in Table 2(C). Memory usage of Platanus_B depends on a value specified as an option (-m), and Platanus_B could assemble all the samples with memory usage <3 GB (-m 1). Therefore, Platanus_B shows good practicability in terms of both accuracy and time. As Platanus_B was designed to improve assemblies by introducing more steps than there are in Platanus and Platanus-allee (Fig. 1), it was expected to consume several-fold more real time than these; however, all real times of Platanus_B were <22 min (Table 2; the number of threads, 4), further confirming its practicability.
Figure 2.
Contiguity and accuracy of the short-read-based assemblers, and effects of the error-correction function in Platanus_B. (A and B) Platanus_B and other existing assemblers. Read lengths are 300 and 150 bp, respectively. (C) Deactivation test of the error-correction functions of Platanus_B. ‘Platanus_B no-corrections’ corresponds to the modified version where all correction functions (based on k-mers, physical coverage, and read mapping) are deactivated. Read length is 300 bp.
Table 2.
Run time and memory usage of benchmarks using the TruSeq PCR-free kit for multiple species
| Strain | Assembler | PE × 10 | PE × 20 | PE × 30 | PE × 40 | PE × 50 |
|---|---|---|---|---|---|---|
| E. coli O157 Sakai | Platanus_B | 796 | 919 | 1,080 | 1,125 | 1,284 |
| Platanus | 58 | 63 | 82 | 668 | 899 | |
| Platanus-allee | 278 | 289 | 353 | 420 | 481 | |
| MaSuRCA | 498 | 517 | 697 | 867 | 1,006 | |
| SPAdes | 621 | 927 | 1,283 | 1,626 | 2,023 | |
| Unicycler | 3,054 | 5,369 | 8,805 | 9,281 | 11,759 | |
| DISCOVAR | 575 | 276 | 330 | 411 | 471 | |
| E. coli K-12 MG1655 | Platanus_B | 672 | 741 | 856 | 853 | 996 |
| Platanus | 54 | 62 | 84 | 571 | 740 | |
| Platanus-allee | 235 | 241 | 283 | 308 | 351 | |
| MaSuRCA | 417 | 423 | 552 | 641 | 792 | |
| SPAdes | 516 | 803 | 1,109 | 1,446 | 1,709 | |
| Unicycler | 1,994 | 3,397 | 5,076 | 6,889 | 8,188 | |
| DISCOVAR | 484 | 204 | 225 | 279 | 326 | |
| B. bronchiseptica S798 | Platanus_B | 713 | 774 | 888 | 902 | 1,004 |
| Platanus | 62 | 63 | 80 | 513 | 706 | |
| Platanus-allee | 264 | 263 | 320 | 377 | 448 | |
| MaSuRCA | 445 | 487 | 641 | 791 | 945 | |
| SPAdes | 603 | 3,036 | 5,240 | 6,751 | 8,987 | |
| Unicycler | 1,856 | 5,237 | 8,389 | 10,602 | 13,830 | |
| DISCOVAR | 819 | 315 | 275 | 334 | 394 | |
| S. marcescens SM39 | Platanus_B | 744 | 801 | 970 | 993 | 1,128 |
| Platanus | 61 | 67 | 88 | 536 | 793 | |
| Platanus-allee | 263 | 250 | 300 | 340 | 374 | |
| MaSuRCA | 476 | 470 | 620 | 776 | 862 | |
| SPAdes | 543 | 887 | 1,146 | 1,458 | 1,840 | |
| Unicycler | 1,695 | 2,854 | 3,688 | 4,926 | 6,096 | |
| DISCOVAR | 657 | 262 | 274 | 339 | 384 | |
| P. gingivalis ATCC 33277 | Platanus_B | 419 | 476 | 529 | 559 | 666 |
| Platanus | 28 | 32 | 39 | 363 | 427 | |
| Platanus-allee | 158 | 167 | 190 | 211 | 246 | |
| MaSuRCA | 220 | 236 | 296 | 354 | 397 | |
| SPAdes | 273 | 380 | 505 | 666 | 813 | |
| Unicycler | 1,633 | 2,217 | 2,741 | 3,091 | 3,922 | |
| DISCOVAR | 236 | 128 | 154 | 189 | 225 | |
| (B) Real time (s) | ||||||
| E. coli O157 Sakai | Platanus_B | 977 | 1,056 | 1,116 | 1,116 | 1,274 |
| Platanus | 47 | 54 | 59 | 517 | 629 | |
| Platanus-allee | 292 | 308 | 337 | 365 | 393 | |
| MaSuRCA | 250 | 230 | 317 | 359 | 397 | |
| SPAdes | 227 | 295 | 401 | 505 | 624 | |
| Unicycler | 1,054 | 1,625 | 2,572 | 2,728 | 3,427 | |
| DISCOVAR | 177 | 89 | 113 | 134 | 166 | |
| E. coli K-12 MG1655 | Platanus_B | 869 | 921 | 963 | 951 | 1,040 |
| Platanus | 42 | 47 | 53 | 503 | 569 | |
| Platanus-allee | 269 | 281 | 297 | 311 | 340 | |
| MaSuRCA | 210 | 190 | 239 | 263 | 315 | |
| SPAdes | 180 | 256 | 348 | 449 | 526 | |
| Unicycler | 632 | 980 | 1,433 | 1,909 | 2,252 | |
| DISCOVAR | 139 | 66 | 80 | 95 | 104 | |
| B. bronchiseptica S798 | Platanus_B | 879 | 915 | 957 | 946 | 987 |
| Platanus | 43 | 49 | 56 | 410 | 533 | |
| Platanus-allee | 274 | 284 | 307 | 319 | 355 | |
| MaSuRCA | 217 | 220 | 273 | 322 | 371 | |
| SPAdes | 333 | 1,711 | 2,231 | 2,288 | 3,025 | |
| Unicycler | 610 | 2,164 | 2,737 | 3,027 | 4,216 | |
| DISCOVAR | 225 | 99 | 102 | 116 | 136 | |
| S. marcescens SM39 | Platanus_B | 919 | 942 | 1,017 | 1,005 | 1,084 |
| Platanus | 43 | 50 | 57 | 427 | 572 | |
| Platanus-allee | 268 | 268 | 293 | 302 | 313 | |
| MaSuRCA | 236 | 204 | 267 | 325 | 342 | |
| SPAdes | 188 | 303 | 359 | 497 | 646 | |
| Unicycler | 620 | 899 | 1,137 | 1,425 | 1,738 | |
| DISCOVAR | 182 | 79 | 94 | 108 | 127 | |
| P. gingivalis ATCC 33277 | Platanus_B | 637 | 678 | 695 | 697 | 808 |
| Platanus | 29 | 31 | 33 | 401 | 412 | |
| Platanus-allee | 217 | 220 | 229 | 235 | 252 | |
| MaSuRCA | 121 | 121 | 144 | 161 | 172 | |
| SPAdes | 110 | 126 | 174 | 230 | 263 | |
| Unicycler | 550 | 642 | 783 | 882 | 1,108 | |
| DISCOVAR | 74 | 47 | 52 | 60 | 69 | |
| E. coli O157 Sakai | Platanus_B (-m 16) | 14.98 | 15.02 | 15.08 | 15.00 | 14.95 |
| Platanus_B (-m 1) | 1.83 | 1.85 | 2.41 | 2.41 | 2.41 | |
| Platanus | 8.40 | 8.40 | 8.40 | 14.94 | 14.94 | |
| Platanus-allee | 14.95 | 14.96 | 14.96 | 14.95 | 14.95 | |
| MaSuRCA | 15.64 | 15.63 | 15.63 | 15.63 | 15.63 | |
| SPAdes | 1.37 | 2.61 | 2.62 | 2.62 | 2.62 | |
| Unicycler | 1.37 | 2.61 | 2.62 | 2.62 | 2.62 | |
| DISCOVAR | 2.36 | 4.21 | 6.02 | 7.97 | 9.88 | |
| (C) Peak memory usage (GB) | ||||||
| E. coli K-12 MG1655 | Platanus_B (-m 16) | 14.93 | 14.95 | 14.99 | 14.94 | 14.91 |
| Platanus_B (-m 1) | 1.63 | 1.64 | 1.64 | 2.40 | 2.40 | |
| Platanus | 8.40 | 8.40 | 8.40 | 14.90 | 14.90 | |
| Platanus-allee | 14.91 | 14.92 | 14.92 | 14.91 | 14.91 | |
| MaSuRCA | 15.64 | 15.63 | 12.92 | 7.51 | 12.92 | |
| SPAdes | 1.14 | 2.27 | 2.62 | 2.62 | 2.62 | |
| Unicycler | 1.14 | 2.27 | 2.62 | 2.62 | 2.62 | |
| DISCOVAR | 1.84 | 3.56 | 5.10 | 6.70 | 8.27 | |
| B. bronchiseptica S798 | Platanus_B (-m 16) | 14.95 | 14.98 | 15.02 | 14.95 | 14.95 |
| Platanus_B (-m 1) | 1.61 | 1.62 | 1.62 | 2.15 | 2.15 | |
| Platanus | 8.40 | 8.40 | 8.40 | 14.93 | 14.93 | |
| Platanus-allee | 14.94 | 14.95 | 14.95 | 14.94 | 14.94 | |
| MaSuRCA | 15.64 | 15.63 | 15.63 | 15.63 | 15.63 | |
| SPAdes | 1.17 | 2.34 | 2.62 | 2.62 | 2.62 | |
| Unicycler | 1.17 | 2.33 | 2.62 | 2.62 | 2.62 | |
| DISCOVAR | 2.03 | 3.93 | 5.67 | 7.45 | 9.21 | |
| S. marcescens SM39 | Platanus_B (-m 16) | 14.96 | 14.99 | 15.03 | 14.96 | 14.95 |
| Platanus_B (-m 1) | 1.80 | 1.81 | 1.81 | 2.15 | 2.15 | |
| Platanus | 8.40 | 8.40 | 8.40 | 14.93 | 14.93 | |
| Platanus-allee | 14.94 | 14.95 | 14.96 | 14.95 | 14.95 | |
| MaSuRCA | 15.64 | 15.63 | 11.56 | 11.57 | 6.16 | |
| SPAdes | 1.26 | 2.53 | 2.62 | 2.62 | 2.62 | |
| Unicycler | 1.26 | 2.52 | 2.62 | 2.62 | 2.62 | |
| DISCOVAR | 2.13 | 4.01 | 5.82 | 7.63 | 9.45 | |
| P. gingivalis ATCC 33277 | Platanus_B (-m 16) | 14.82 | 14.83 | 14.85 | 14.83 | 14.80 |
| Platanus_B (-m 1) | 1.07 | 1.08 | 1.09 | 1.20 | 1.20 | |
| Platanus | 8.40 | 8.40 | 8.40 | 14.79 | 14.79 | |
| Platanus-allee | 14.80 | 14.80 | 14.80 | 14.80 | 14.80 | |
| MaSuRCA | 15.63 | 15.63 | 14.27 | 10.19 | 7.47 | |
| SPAdes | 0.58 | 1.16 | 1.75 | 2.32 | 2.61 | |
| Unicycler | 0.60 | 1.16 | 1.74 | 2.32 | 2.61 | |
| DISCOVAR | 0.98 | 1.84 | 2.68 | 3.63 | 4.23 | |
As a machine environment, the number of CPUs were 24, the model name of CPU was Intel(R) Xeon(R) CPU E5-2687W v4, the clock rate of CPU was 3.00 GHz, and the amount of RAM was 256GB. Each tool was executed with the setting of 4 threads and the times (real and CPU time) were measured using GNU time (version 1.7). (A) CPU time (s), (B) Real time (s), and (C) Peak memory usage (GB). For Platanus_B, two values (16 and 1) are specified to an option of available memory amount (-m).
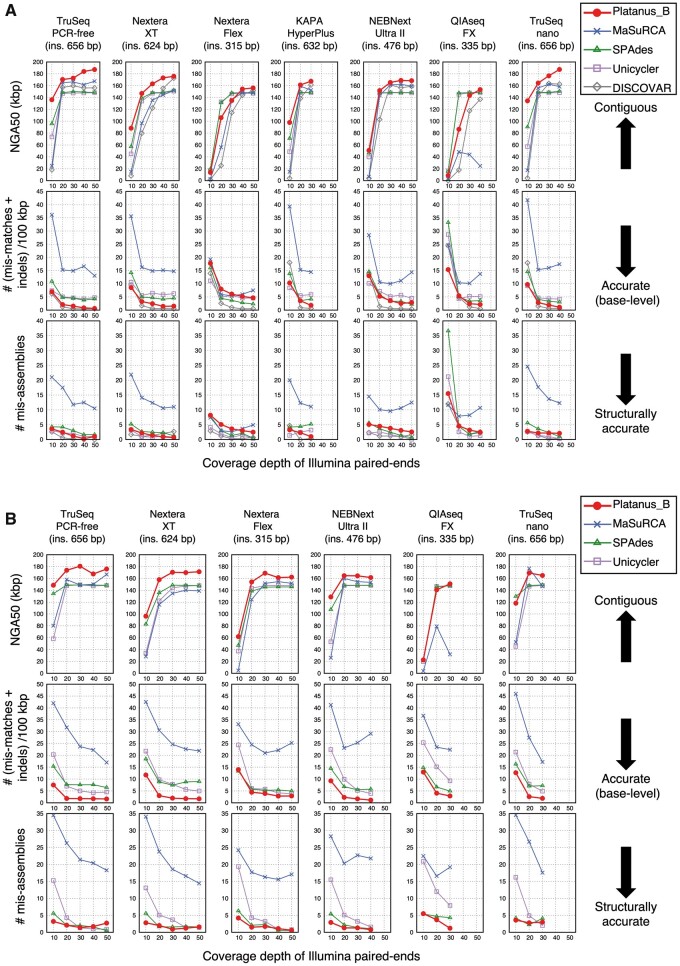
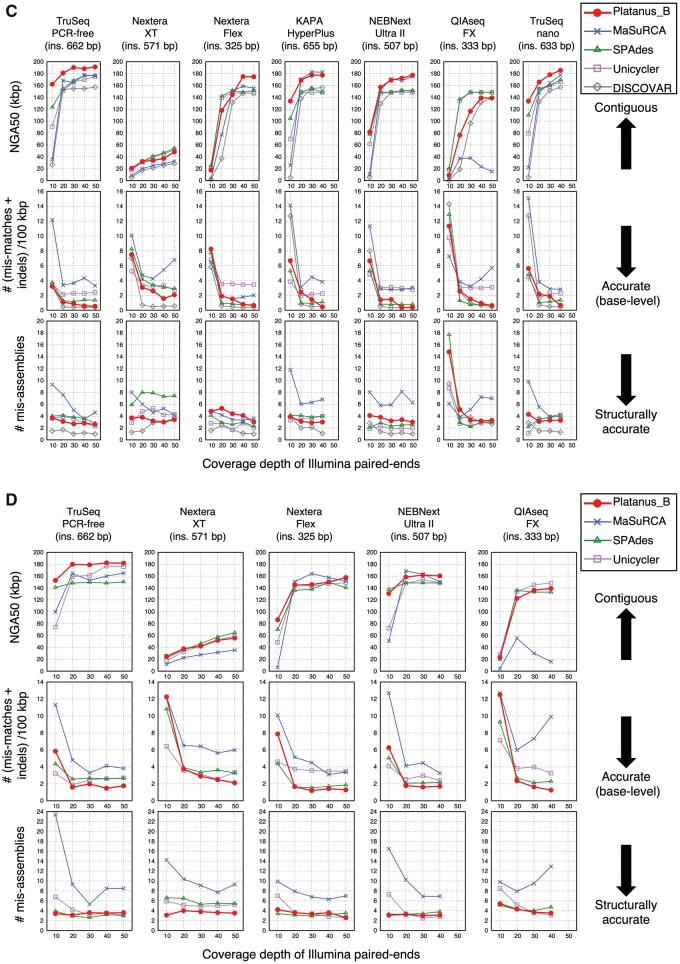
A similar advantage of Platanus_B was observed from the data obtained from libraries prepared by the Nextera-XT kit (Supplementary Figs S3 and S4 and Supplementary Tables S6 and S7). This kit was reported to have biased read coverage41; however, the results indicate that Platanus_B is robust to such bias. Similar results were obtained for other sequence library preparation kits (Fig. 3; Supplementary Figs S6 and S7 and Supplementary Tables S9–S13). Among the versatile assemblers, the benchmarks for all cases of 300-bp reads, Platanus_B provided the best NGA50 values and base error rates in 99 and 84 cases, respectively, out of 158 cases. The trend was found to be similar to that for 150-bp reads (Fig. 3B and D, Supplementary Fig. S5, and Supplementary Tables S8 and S11), which supports the versatility of Platanus_B.
Figure 3.
Benchmark using multiple preparation kits for short reads. (A and B) Escherichia coli O157 Sakai. Read lengths are 300 and 150 bp, respectively. (C and D) Escherichia coli K-12 MG1655. Read lengths are 300 and 150 bp, respectively.
Figure 3.
(Continued)
3.2. Benchmark using nanopore long-read sequencing
For benchmark of nanopore (R9.4.1) long-read inputs (Supplementary Figs S8–S11 and Supplementary Tables S15–S18), the long read-only assemblers (Canu, Flye, Wtdbg2, and miniasm+Racon) showed high base error rates and never achieved the best NGA50 values, even when Pilon was applied, confirming the importance of short reads for base-level accuracy. Although Platanus_B showed the best base error rate in many cases (49/120), the other hybrid assemblers were superior with respect to the other indicators (NGA50 and the number of misassemblies).
To test the feasibility of Platanus_B as a module in a hybrid assembler, we combined the results of Platanus_B into MaSuRCA and Unicycler using the integrated function of Platanus_B (‘combine’ command). This combination improved the base error rate in majority of the cases, and the Platanus_B + MaSuRCA combination resulted in the best NGA50 values in 65 out of 120 cases (Supplementary Figs S12–S15 and Supplementary Tables S20–S23). Therefore, Platanus_B is also expected to be used as a module for short reads in a hybrid assembler or a pipeline (e.g. SPAdes in Unicycler) to enhance the final sequence lengths and accuracy.
Additionally, we tested to assemble reads from MinION with new chemistry, R10.3. The targeted strain was E. coli K-12 MG1655. The reads are public data (SRA accession, ERR3890216) and the base-called reads (tool, Guppy v3.4.5) were downloaded from https://figshare.com/articles/Ecoli_K12_MG1655_R10_3_HAC/11823087. As a result (Supplementary Table S19), although base error rates of the long-read-based assemblers were lower than those of the R9.4.1 chemistry, the rates were still much higher than those of the short-read-based assemblers.
3.3. Comparison between short- and long-read-based assemblies using coding sequences
To verify the practicability of short-read-based assemblers and compare them with long-read-based assemblers, we calculated the success rate of constructions of protein-coding sequences (CDSs) for each assembly result. The target strains were E. coli O157 Sakai and E. coli K-12 MG1655 (the number of CDSs, 5291 and 4357, respectively), and we counted the number of the reference CDSs that were exactly matched to the assembled sequences to calculate the success rate (exact-match rate).
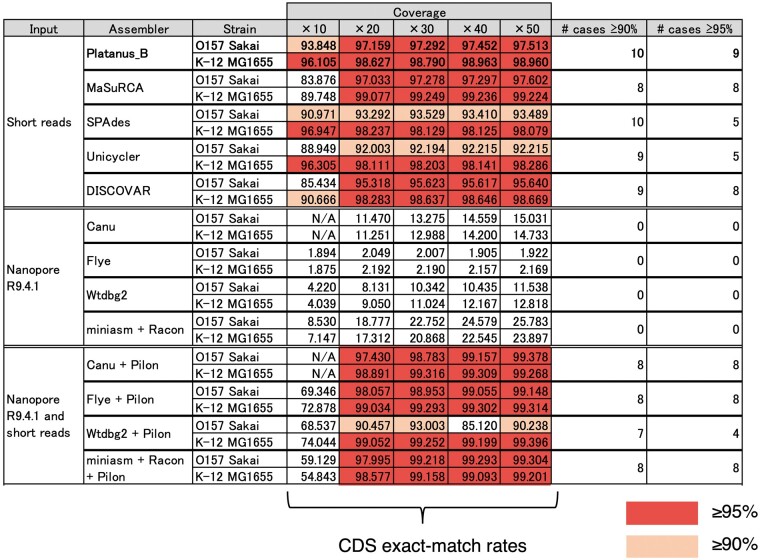
Platanus_B achieved the exact-match rates of over 90% in 10 case and over 95% in 9 other cases (out of 10 cases; 2 strains; coverage, 10–50; Fig. 4). Although the other short-read-based assemblers (MaSuRCA, SPAdes, Unicycler, and DISCOVAR) also had high exact-match rates, the number of high exact-match rates (≥90%, ≥95%) of Platanus_B was higher. Note that some tools are not designed to handle low-coverage data and we tested the data to investigate the limits of the tools. Although MaSuRCA exhibited the highest exact-match rates for K-12 MG1655 when coverage depths were ≥20, it indicated several-times higher base-level error rates and more misassemblies for many cases (Figs 2 and 3), suggesting that it produced many false sequences. We defined Platanus_B-specific CDS as a CDS of which the mean exact-match rate from Platanus_B was ≥80% while the values from the other assemblers were <50% for 10 random replicates. Among the versatile short-read-based assemblers for the E. coli O157 Sakai strain, there were 107 Platanus_B-specific CDSs, 55 of which contained the word ‘phage’ in their description. Considering that this strain contains multiple prophages similar to each other and these prophages encode genes associated with toxicity,27 the exact-match rates of Platanus_B were noteworthy.
Figure 4.
Coding sequence exact-match rates of short- and long-read-based assemblies for E. coli strains. With mixed input of long and short reads followed by polishing with Pilon a coverage depth corresponding to the column names, ×10–50, is obtained for each library. For example, the total coverage depth is 20 (long reads, 10; short reads, 10) if the coverage depth is denoted as ‘×10’. Pilon was executed three times for each long-read-based assembly (Canu, Flye, Wtdbg2, and miniasm+Racon).
The long-read-based assemblers (Canu, Flye, Wtdbg2, and miniasm+Racon) recorded low exact-match rates (<20%) for all cases when only nanopore long reads were analysed. When short-read-based polishing (Pilon) was applied to these results, the exact-match rates increased and often exceeded the values of short-read-only assemblies, inferring the high contiguity and low fine-scale accuracy of the long-read-based assemblers. In other words, the long-read-based methods require additional short-read sequencing and the short-read-based methods are cost-effective when collecting exact CDSs. To summarize, short-read-based assemblers exhibited good practicability and comprehensiveness in terms of CDS-construction, and it is recommended to combine long-read-based assemblers with short-read-based polishing.
3.4. Conclusion
From the benchmark results above, Platanus_B’s advantages of contiguities and accuracies for short-read inputs will be useful to proceed large-scale projects, which target hundreds of isolates and focus on a few variant sites between genomes. Short-read sequencing is still being utilized for many studies. Although it is difficult to estimate exact sequencing costs due to variations in market channels, there is remarkable improvement in the cost-performances of short-read sequencers. The sequencing costs of NovaSeq (Illumina) and MGISEQ-2000 (current name, DNBSEQ-G400; MGI Tech, Shenzhen, China) were reported to be 12–18 USD/Gbp and 10 USD/Gbp, respectively.42 Although read lengths of these sequencers are generally 150 bp, we confirmed the high performance of Platanus_B for the 150-bp reads (Figs 2B, 3B, and D, Supplementary Fig. S5, and Supplementary Tables S4, S8, S11, and S14), and the tool is expected to work for the reads from the new sequencers. We demonstrate that the combination of Platanus_B and short-read sequencers can be used for comprehensive scans of bacterial genomes. If sufficient data such as 95% of CDSs and/or core-genomic regions for a project are obtained using this tool, costs are reduced. For example, long-read sequencing is performed only if targeted regions or an appropriate fraction of a genome (judged using core-genes) are not assembled by the short-read-based method. Additionally, the fine-scale accuracy of Platanus_B can be utilized for high-resolution phylogenomic analyses that have not been discerned.
Supplementary Material
Acknowledgements
We thank Dr Yasuhiko Horiguchi and Dr Mariko Naito for providing the genomic DNA of B. bronchiseptica S798 and P. gingivalis ATCC 33277, respectively.
Supplementary data
Supplementary data are available at DNARES online.
Accession numbers
DDBJ/ENA/GenBank BioProject PRJDB9013.
Funding
This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI grant numbers 15H05979 and 18K19286.
Conflict of interest
None declared.
Data availability
Platanus_B is implemented in C++ and Perl and is freely available at http://platanus.bio.titech.ac.jp/platanus-b/ or GitHub (https://github.com/rkajitani/Platanus_B) under GNU General Public License (GNU GPL) version 3. The sequencing data generated in this study are available under DDBJ/ENA/GenBank BioProject PRJDB9013.
References
- 1. Harris S.R., Feil E.J., Holden M.T.G., et al. 2010, Evolution of MRSA during hospital transmission and intercontinental spread, Science, 327, 469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Coll F., Harrison E.M., Toleman M.S., et al. 2017, Longitudinal genomic surveillance of MRSA in the UK reveals transmission patterns in hospitals and the community, Sci. Transl. Med., 9, eaak9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baker S., Thomson N., Weill F.X., Holt K.E.. 2018, Genomic insights into the emergence and spread of antimicrobial-resistant bacterial pathogens, Science, 360, 733–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reuter S., Estee Torok M., Holden M.T.G., et al. 2016, Building a genomic framework for prospective MRSA surveillance in the United Kingdom and the Republic of Ireland, Genome Res., 26, 263–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wu L., Mccluskey K., Desmeth P., et al. 2018, The global catalogue of microorganisms 10K type strain sequencing project: closing the genomic gaps for the validly published prokaryotic and fungi species, Gigascience, 7, giy026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arimizu Y., Kirino Y., Sato M.P., et al. 2019, Large-scale genome analysis of bovine commensal Escherichia coli reveals that bovine-adapted E. coli lineages are serving as evolutionary sources of the emergence of human intestinal pathogenic strains, Genome Res., 29, 1495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Harris S.R., Cartwright E.J.P., Török M.E., et al. 2013, Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study, Lancet Infect. Dis., 13, 130–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Walker T.M., Ip C.L.C., Harrell R.H., et al. 2013, Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study, Lancet Infect. Dis., 13, 137–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pightling A.W., Pettengill J.B., Luo Y., Baugher J.D., Rand H., Strain E.. 2018, Interpreting whole-genome sequence analyses of foodborne bacteria for regulatory applications and outbreak investigations, Front. Microbiol., 9, 1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gotoh Y., Taniguchi T., Yoshimura D., et al. 2019, Multi-step genomic dissection of a suspected intra-hospital Helicobacter cinaedi outbreak, Microb. Genom., 5, e000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. De Maio N., Shaw L.P., Hubbard A., et al. 2019, Comparison of long-read sequencing technologies in the hybrid assembly of complex bacterial genomes, Microb. Genom., 5, e000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yoshimura D., Kajitani R., Gotoh Y., et al. 2019, Evaluation of SNP calling methods for closely related bacterial isolates and a novel high-accuracy pipeline: BactSNP, Microb. Genom., 5, e000261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Iqbal Z., Caccamo M., Turner I., Flicek P., McVean G.. 2012, De novo assembly and genotyping of variants using colored de Bruijn graphs, Nat. Genet., 44, 226–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Weisenfeld N.I., Yin S., Sharpe T., et al. 2014, Comprehensive variation discovery in single human genomes, Nat. Genet., 46, 1350–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pightling A.W., Petronella N., Pagotto F.. 2014, Choice of reference sequence and assembler for alignment of Listeria monocytogenes short-read sequence data greatly influences rates of error in SNP analyses, PLoS One, 9, e104579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wick R.R., Judd L.M., Gorrie C.L., Holt K.E.. 2017, Completing bacterial genome assemblies with multiplex MinION sequencing, Microb. Genom., 3, e000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Quick J., Loman N.J., Duraffour S., et al. 2016, Real-time, portable genome sequencing for Ebola surveillance, Nature, 530, 228–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gardy J.L., Loman N.J.. 2018, Towards a genomics-informed, real-time, global pathogen surveillance system, Nat. Rev. Genet., 19, 9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Giordano F., Aigrain L., Quail M.A., et al. 2017, De novo yeast genome assemblies from MinION, PacBio and MiSeq platforms, Sci. Rep., 7, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu F., Zhao S., Yu B., et al. 2020, A new coronavirus associated with human respiratory disease in China, Nature., 579, 265–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou P., Yang X.-L.L., Wang X.-G.G., et al. 2020, A pneumonia outbreak associated with a new coronavirus of probable bat origin, Nature., 579, 270–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhu N., Zhang D., Wang W., et al. ; China Novel Coronavirus Investigating and Research Team 2020, A novel coronavirus from patients with pneumonia in China, 2019, N Engl. J. Med., 382, 727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kajitani R., Toshimoto K., Noguchi H., et al. 2014, Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads, Genome Res., 24, 1384–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kajitani R., Yoshimura D., Okuno M., et al. 2019, Platanus-allee is a de novo haplotype assembler enabling a comprehensive access to divergent heterozygous regions, Nat. Commun., 10, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li H. 2018, Minimap2: pairwise alignment for nucleotide sequences, Bioinformatics, 34, 3094–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Blattner F.R., Plunkett G., Bloch C.A., et al. 1997, The complete genome sequence of Escherichia coli K-12, Science., 277, 1453–62. [DOI] [PubMed] [Google Scholar]
- 27. Hayashi T., Makino K., Ohnishi M., et al. 2001, Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12, DNA Res., 8, 11–22. [DOI] [PubMed] [Google Scholar]
- 28. Naito M., Hirakawa H., Yamashita A., et al. 2008, Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P. gingivalis, DNA Res., 15, 215–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Iguchi A., Nagaya Y., Pradel E., et al. 2014, Genome evolution and plasticity of Serratia marcescens, an important multidrug-resistant nosocomial pathogen, Genome Biol. Evol., 6, 2096–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Okada K., Ogura Y., Hayashi T., et al. 2014, Complete genome sequence of Bordetella bronchiseptica S798, an isolate from a pig with atrophic rhinitis, Genome Announc., 2, e00436–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zimin A.V., Marçais G., Puiu D., Roberts M., Salzberg S.L., Yorke J.A.. 2013, The MaSuRCA genome assembler, Bioinformatics., 29, 2669–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bankevich A., Nurk S., Antipov D., et al. 2012, SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing, J. Comput. Biol., 19, 455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wick R.R., Judd L.M., Gorrie C.L., Holt K.E.. 2017, Unicycler: resolving bacterial genome assemblies from short and long sequencing reads, PLoS Comput. Biol., 13, e1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Koren S., Walenz B.P., Berlin K., Miller J.R., Bergman N.H., Phillippy A.M.. 2017, Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation, Genome Res., 27, 722–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kolmogorov M., Yuan J., Lin Y., Pevzner P.A.. 2019, Assembly of long, error-prone reads using repeat graphs, Nat. Biotechnol., 37, 540–6. [DOI] [PubMed] [Google Scholar]
- 36. Ruan J., Li H.. 2020, Fast and accurate long-read assembly with wtdbg2, Nat. Methods, 17, 155–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li H. 2016, Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences, Bioinformatics, 32, 2103–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vaser R., Sović I., Nagarajan N., Šikić M.. 2017, Fast and accurate de novo genome assembly from long uncorrected reads, Genome Res., 27, 737–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Walker B.J., Abeel T., Shea T., et al. 2014, Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement, PLoS One., 9, e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gurevich A., Saveliev V., Vyahhi N., Tesler G.. 2013, QUAST: quality assessment tool for genome assemblies, Bioinformatics, 29, 1072–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sato M.P., Ogura Y., Nakamura K., et al. 2019, Comparison of the sequencing bias of currently available library preparation kits for Illumina sequencing of bacterial genomes and metagenomes, DNA Res., 26, 391–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jeon S.A., Park J.L., Kim J.H., et al. 2019, Comparison of the MGISEQ-2000 and Illumina HiSeq 4000 sequencing platforms for RNA sequencing, Genomics Inform., 17, e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Platanus_B is implemented in C++ and Perl and is freely available at http://platanus.bio.titech.ac.jp/platanus-b/ or GitHub (https://github.com/rkajitani/Platanus_B) under GNU General Public License (GNU GPL) version 3. The sequencing data generated in this study are available under DDBJ/ENA/GenBank BioProject PRJDB9013.