Introduction
Modern pig breeding schemes evaluate a large number of traits on specialized purebred breeding lines to improve the genetic makeup of crossbred animals used for pork production, but also for the next generation of purebred breeding animals. Much progress has been made regarding breeding for production and reproduction traits through the use of genomic information. However, pure line breeding is done under high health conditions for sanitary and trade reasons and the genetic variation in the group of traits around resilience is not fully exploited.
Resilience in the narrow sense of reactions upon infectious diseases is defined as the ability to maintain performance, regardless of pathogen burden (Mulder and Rashidi, 2017). This includes the ability of an animal to maintain performance under infection or to rapidly return to prior performance levels after infection. This can be due to resistance or tolerance or a combination of both. Resilience in a broader sense can also include this reaction in performance upon environmental challenges such as changes in diet, social grouping, or management procedures (Colditz and Hine, 2016). Urruty et al. (2016) defines resilience as the ability to absorb change and to anticipate future perturbations through adaptive capacity.
In this paper, resilience is defined as “minimal changes in the overall performance of an animal in spite of diseases.” Several studies attempt to define descriptors to quantify health challenge in pig production (Nakov et al., 2018; Guy et al., 2019). However, for practical breeding, easy and inexpensive descriptors are needed. These descriptors should be collected on a large number of animals, preferably from different production environments around the world. We describe several of our own efforts to 1) identify new phenotypes useful for breeding for resilience, including phenotypes collected under more challenging commercial conditions, and 2) genes and gene variants related to specific diseases. These (new) phenotypes include survival at different stages of life, as well as general and specific disease resistance. Resilience might also be approached by production performance of animals at commercial farms. Finally, carcass remarks collected at the slaughter line have been investigated. Figure 1 summarizes the different time points throughout the life of a finishing pig at which data were collected from different studies on our breeding or production animals to describe resilience.
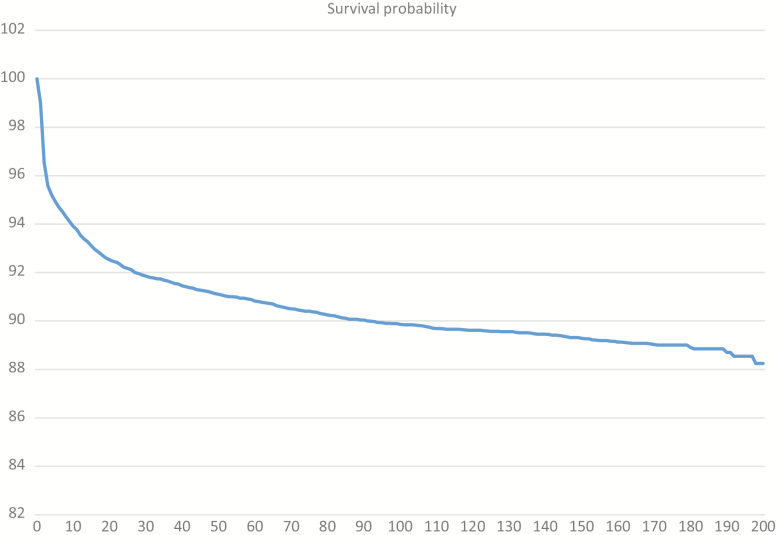
Figure 1.
Schematic overview of different phenotypes of commercial crossbred animals describing parts of resilience along the lifetime of an animal discussed in this article. Survival curve of a mean finisher with an average birth weight estimated from 9,506 crossbreds with a Cox proportional hazard model (Kleinbaum and Klein, 2011). 1a, Survival Through Life; 1b, Specific and General Disease Infection Trials; 1c, Performance Data From Commercial Farms; 1d, Meat Inspection Data. Data from experimental trials in italics. 2, Genomic Tools and Monogenic Effects. Embryonic recessive lethals estimated from 50K SNP genotype data.
a. Survival Through Life
This is the backbone of breeding for resilient pigs. Survival at birth and pre-weaning survival, each have two main genetic components, apart from other management factors: 1) genes of the piglet and 2) genes of the dam or foster dam. Survival during the finishing period is mainly governed by the genes of the pig itself. From these data, survival curves can be modeled along the lifetime of an animal and considerable genetic variation can be observed. The genetic standard deviation for farrowing and lactation survival, as a trait of the piglet or the sow, is between 3% and 4%. For finisher survival, we estimate 2.5%. The genetic standard variation for herd retention of sows at first parity is 6% and highly correlated to survival until parity 5. Therefore, ample genetic variation for survival traits can be observed from the field data of commercial crossbred animals.
b. Specific and General Disease Infection Trials
Breeders often have to decide if the breeding goal should include resilience to a specific disease, or if efforts should be diverted toward breeding for general disease resilience. Both types of approaches have been reported. Breeding for resilience to specific diseases, such as porcine reproductive and respiratory syndrome (PRRS), may be desirable, as the disease causes large economic losses and vaccination is not completely effective. Infection trials with PRRS virus (PRRSV) showed that selection for enhanced natural resilience to PRRSV infection is possible (Boddicker et al., 2014). Results of a recent challenge trial involving experimental infection of 1,400+ crossbred finishing pigs with a highly pathogenic PRRSV strain (and natural infection with multiple secondary pathogens) showed considerable genetic variation in mortality after infection. Genetic parameter estimates obtained from this trial indicate the possibility of reducing the incidence of mortality post-challenge by 5.7% after a single generation of single-trait selection (Dunkelberger et al., 2019).
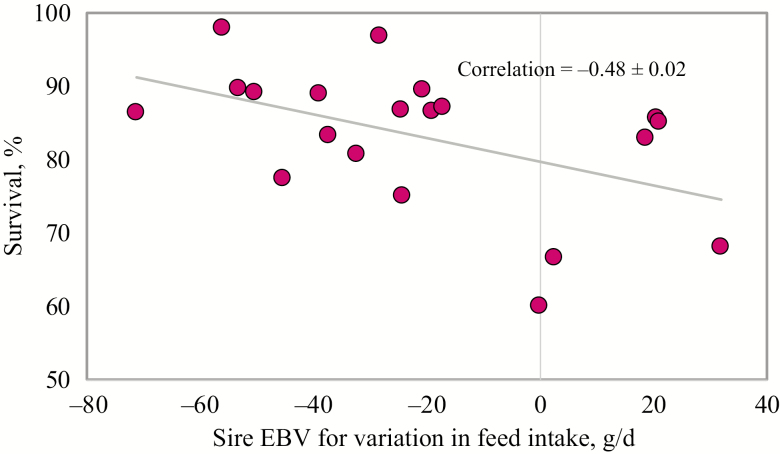
Given the number of pathogens that the pigs were exposed to in this trial (PRRSV plus numerous secondary pathogens), results obtained from this trial provide insight regarding genetic variation in overall resilience to disease. Another example of a study with the aim of investigating genetic variation in overall disease resilience is a series of natural challenge trials underway in Canada. For this project, pigs are placed in barns meant to mimic commercial conditions, including exposure to a mixture of field pathogens. Initial results from this project showed that variation in feed intake was correlated to mortality and number of treatments which, therefore, can be used as a novel phenotype for resilience (Putz et al., 2018). For this study, individual daily feed intake and mortality were recorded under the same conditions of disease infection. However, in practice, individual daily feed intake is recorded on nucleus pigs under high health conditions, due to cost and practical issues. Therefore, individual daily feed intake data collected on 5,726 purebred pigs from a nucleus farm were used to estimate breeding values for variation in feed intake. Of these 5,726 animals, 20 animals were used to sire 1,356 crossbred progeny that were subjected to an experimental PRRSV challenge, followed by natural challenge with numerous secondary pathogens. As shown in Figure 2, the estimated breeding value for variation in feed intake of the sires was negatively correlated (−0.48) with survival. These results suggest that genetic selection for reduced variation in feed intake could be used to improve finisher survival following a multifactorial PRRSV challenge.
Figure 2.
Relationship between estimated breeding value (EBV) for sires (n = 20) for variation in feed intake with progeny survival (n = 1,356) following a multifactorial porcine reproductive and respiratory syndrome virus challenge.
c. Resilience Through the Use of Performance Data From Commercial Farms
It is not always possible to conduct designed trials, such as those described above, to collect data on pathogen burden and mortality to estimate breeding values for resilience. In the absence of such data, field data, such as data collected during a disease outbreak, may prove useful for studying resilience to disease. For instance, reproduction data routinely collected at multiplication farms producing crossbred animals can be used to detect periods of PRRS outbreaks (Rashidi et al., 2014) and estimate resilience to the disease. In practice, it is often hard to clearly distinguish between the diseased and healthy phases. Therefore, it is important to consider the level of disease challenge, rather than the mere presence or absence of disease. Mathur et al. (2014a) suggested a method to estimate challenge load due to disease and other stressors using reproduction records. This method was tested to identify outbreaks of PRRS based on historical reproduction records from different countries in Europe, North America, and Brazil. It has been shown that the periods of outbreaks detected with the method matched with the clinical records from the farms (Mathur et al., 2014b). In addition, this method was used to estimate genetic parameters and breeding values across challenged environments (Herrero-Medrano et al., 2015). Results from this study suggest that as the level of disease challenge increases, so does the magnitude of genetic variation in response to challenge. This is particularly evident for traits such as number of stillborn and mummified pigs, since an increased level of challenge allows for increased expression of resilience. Selection for resilience using the above approach, with corresponding performance records, was shown to improve resistance as well as tolerance by Mulder and Rashidi (2017). Genomic selection can further enhance the genetic gains using such performance data (Mulder, 2016).
d. Meat Inspection Data
For the majority of slaughter plants, carcasses are inspected by trained meat inspectors and remarks are made with respect to different diseases, injuries, and other abnormalities. These remarks provide a summary of the disease challenges that the animal experienced throughout its life. Carcass remarks collected on 140,375 finisher pigs were obtained through a close cooperation between Topigs Norsvin and slaughter houses in Germany. Heritability estimates for pneumonia, pleuritis, pericarditis, liver lesions, and joint disorders were 0.10, 0.09, 0.14, 0.24, and 0.17, respectively, on the liability scale (Table 1) indicating substantial genetic variation for genetic improvement (Mathur et al., 2018). Consequently, these traits have been combined in a selection index for simultaneous genetic improvement in welfare, as well as production traits. Collection of such slaughter plant data is possible in Germany and most European countries, as the pigs are individually identified with RFID tags that are read and recorded through an automated system for food safety reasons. However, individual identification of slaughter pigs is still not a common practice in North America or several other parts of the world. This remains one of the major bottlenecks in using observations from a very large number of pigs slaughtered every day. In spite of this, there is a growing interest in genetic improvement of welfare traits in North America and in most other countries. Therefore, several large integrators are seriously considering adapting this system. This would be a desirable change, since use of this system would enable selection for enhanced resilience using multiple resilience traits, simultaneously.
Table 1.
Variance component and heritability estimates (SE) for slaughter remarks
| Component | Pneumonia | Pleuritis | Pericarditis | Liver lesions | Joint disorders |
|---|---|---|---|---|---|
| Genetic | 0.368 (0.033) | 0.336 (0.057) | 0.562 (0.098) | 0.939 (0.134) | 0.626 (0.046) |
| Residual | 3.290 (0.000) | 3.290 (0.000) | 3.290 (0.000) | 3.290 (0.000) | 3.290 (0.000) |
| Phenotypic | 3.609 (0.019) | 3.665 (0.036) | 3.885 (0.061) | 3.991 (0.072) | 3.638 (0.024) |
| Heritability | 0.10 (0.01) | 0.09 (0.02) | 0.14 (0.02) | 0.24 (0.03) | 0.17 (0.01) |
2. Genomic Tools and Monogenic Effects
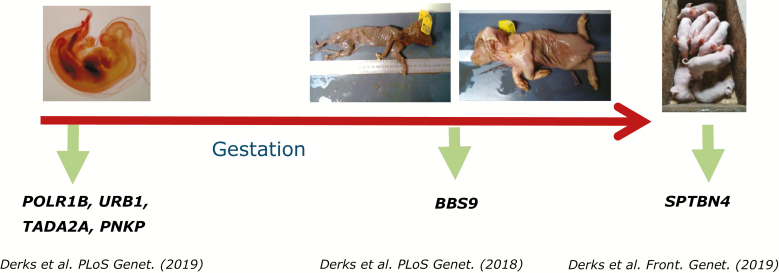
As genomic tools continue to develop and improve, so does the ability to identify deleterious genetic variants within specific genes. Using female fertility phenotypes registered at birth (total number born, number born alive, mummies, and stillborn) and lactation survival, several regions in the genome, as well as underlying recessive variants causing embryonic and fetal death, stillbirths or pre-weaning mortality, have recently been identified (Derks et al., 2017, 2018, 2019). Searching for missing haplotypes from single nucleotide polymorphisms (SNP) genotyping data and comparing fertility data from carrier × carrier and carrier × homozygous wild type matings of variants in specific genes located in these haplotypes could prove their deleteriousness. Overall, about one to four lethal variants per line have been identified with an allele frequency between 2% and 12% (Figure 3). Until now, deleterious recessive variants have mainly been shown to be line-specific and are, therefore, not expected to cause an increase in mortality in crossbred finishing pigs.
Figure 3.
Recessive lethal variants detected by missing homozygous haplotypes in different breeding lines. Genes for which the causative variants have been identified in italics (Derks et al., 2018, 2019).
Using 50K SNP genotype data as a template, the entire genome can be scanned to identify genomic regions with larger effects (quantitative trait loci [QTL]) on polygenic traits using a genome-wide association analysis (GWAS). For a large PRRSV-infection trial, including various genetic sources, results from a GWAS revealed a SNP (WUR10000125) that was associated with both viremia and weight gain following infection. The effect of this SNP was validated under alternative PRRSV-infection scenarios, including vaccination with a modified live PRRSV vaccine and co-infection with porcine circovirus type 2b (Dunkelberger et al., 2017). Results obtained by Boddicker et al. (2012) showed that this marker explained more than 11% of the total genetic variation in weight gain and 15.7% of the total genetic variation in viral load following infection. The WUR10000125 SNP is nearly in complete linkage disequilibrium with the functional mutation in the guanylate binding protein 5 (GBP5) gene (Koltes et al., 2015). At the molecular level, the favorable allele rescues the function of the GBP5 gene, thereby improving immune defenses of heterozygous animals by decreasing the efficiency of viral entry into host cells and subsequent viral replication (Schroyen et al., 2016). In an infection trial where pigs were vaccinated with a heterologous PRRSV strain, it was observed that pigs with the AB genotype had higher average daily gain and lower vaccine viral load compared to pigs with the AA genotype (Dunkelberger et al., 2017). These results suggest that certain genotypes could be more responsive to vaccination and, therefore, that genetic approaches could be used to enhance response to vaccination.
Another important disease in pigs is postweaning multisystemic wasting syndrome caused by porcine circovirus type 2 (PCV2) infection. Natural polygenic variation has also been described for PCV2 host susceptibility with two major resistance loci (Walker et al., 2018). For one QTL region, a missense mutation in the synaptogyrin-2 gene was associated with reduced viral load. This is an additional example of a case where part of the polygenic variation in resilience to infection could be explained at the molecular level. Going forward, such tools can be used to improve monogenic resilience to specific pathogens or specific or general resilience to infection.
Using genome editing, additional variation can even be introduced artificially. Recently, pigs exhibiting complete resistance to PRRSV infection were produced using gene editing to delete domain 5 of the CD163 gene (Burkard et al., 2018). This domain is responsible for attachment of PRRSV to host cells and deletion of this region was shown to prevent entry of PRRSV into host cells.
It remains to be seen to what extent specific resistance alleles or variants improving resilience can contribute to the continuous overall genetic improvement of resilience against the entire load of pathogens changing over time and environment. Selecting pigs to be more responsive to a specific disease can have serious drawbacks for their health or reduce their ability to defend other infective agents (Nakov et al., 2018). Therefore, overall negative genetic correlations with other traits need to be monitored.
Conclusions
Considerable natural genetic variation has been identified for a number of new resilience traits. Results from several studies show that the extent of genetic variation in resilience is most visible at the commercial production level where the level of disease challenge is greatest. Performance data, such as variation in feed intake and reproduction records, can contribute to the genetic evaluation of resilience. Availability of genomic information at lower costs, in addition to the availability of new genetic selection tools, has increased opportunities for breeding for enhanced resilience and monitoring lethal or deleterious variants. While genetics can contribute to increase resilience of our animals, disease surveillance, biosecurity, and vaccination remain important. Integrated approaches by geneticists, immunologists, virologists, veterinarians, and other disciplines are necessary for effective disease prevention, control, and eradication measures.
Glossary
Abbreviations
- GBP5
guanylate binding protein 5
- GWAS
genome-wide association analysis
- PCV2
porcine circovirus type 2
- PRRS
porcine reproductive and respiratory syndrome
- PRRSV
PRRS virus
- QTL
quantitative trait loci
Conflict of interest statement
The authors declare no potential or actual conflict of interest.
Literature Cited
- Boddicker N. J., Bjorkquist A., Rowland R. R., Lunney J. K., Reecy J. M., and Dekkers J. C.. 2014. Genome-wide association and genomic prediction for host response to porcine reproductive and respiratory syndrome virus infection. Genet. Sel. Evol. 46:18. doi: 10.1186/1297-9686-46-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddicker N., Waide E. H., Rowland R. R., Lunney J. K., Garrick D. J., Reecy J. M., and Dekkers J. C.. 2012. Evidence for a major QTL associated with host response to porcine reproductive and respiratory syndrome virus challenge. J. Anim. Sci. 90:1733–1746. doi: 10.2527/jas2011-4464 [DOI] [PubMed] [Google Scholar]
- Burkard C., Opriessnig T., Mileham A. J., Stadejek T., Ait-Ali T., Lillico S. G., Whitelaw C. B. A., Archibald A. L.. 2018. Pigs lacking the scavenger receptor cysteine-rich domain 5 of CD163 are resistant to porcine reproductive and respiratory syndrome virus 1 infection J. Virol. 92:e00415–e00418. doi: 10.1128/JVI.00415-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colditz I. G., and Hine B. C.. 2016. Resilience in farm animals: biology, management, breeding and implications for animal welfare. Anim. Prod. Sci. 56:1961. doi: 10.1071/AN15297 [DOI] [Google Scholar]
- Derks M. F. L., Gjuvsland A. B., Bosse M., Lopes M. S., van Son M., Harlizius B., Tan B. F., Hamland H., Grindflek E., Groenen M. A. M., et al. 2019. Loss of function mutations in essential genes cause embryonic lethality in pigs. PLoS Genet. 15:e1008055. doi: 10.1371/journal.pgen.1008055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derks M. F. L., Harlizius B., Lopes M. S., Greijdanus-van der Putten S. W. M., Dibbits B., Laport K., Megens H. J., and Groenen M. A. M.. 2019. Detection of a frameshift deletion in the SPTBN4 gene leads to prevention of severe myopathy and postnatal mortality in pigs. Front. Genet. 10:1226. doi: 10.3389/fgene.2019.01226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derks M. F. L., Lopes M. S., Bosse M., Madsen O., Dibbits B., Harlizius B., Groenen M. A. M., and Megens H. J.. 2018. Balancing selection on a recessive lethal deletion with pleiotropic effects on two neighboring genes in the porcine genome. PLoS Genet. 14:e1007661. doi: 10.1371/journal.pgen.1007661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derks M. F. L., Megens H. J., Bosse M., Lopes M. S., Harlizius B., and Groenen M. A. M.. 2017. A systematic survey to identify lethal recessive variation in highly managed pig populations. BMC Genomics 18:858. doi: 10.1186/s12864-017-4278-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkelberger J. R., Mathur P., Little E., Hanson D., Eggert J., Dee S., and Knol E. F.. 2019. Empirical evidence for genetic variation in survival following experimental infection with a highly pathogenic PRRSV strain. Leman Swine Conference; September 15, 2019; St. Paul (MN).
- Dunkelberger J. R., Mathur P. K., Lopes M. S., Knol E. F., and Dekkers J. C. M.. 2017. A major gene for host response to porcine reproductive and respiratory syndrome is not unfavorably associated with overall performance under nonchallenging conditions in commercial pig lines. J. Anim. Sci. 95:2838–2847. doi: 10.2527/jas.2017.1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy S. Z. Y., Li L., Thomson P. C., and Hermesch S.. 2019. Quantifying the health challenges in an Australian piggery using medication records for the definition of disease resilience1. J. Anim. Sci. 97:1076–1089. doi: 10.1093/jas/skz025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero-Medrano J. M., Mathur P. K., ten Napel J., Rashidi H., Alexandri P., Knol E. F., and Mulder H. A.. 2015. Estimation of genetic parameters and breeding values across challenged environments to select for robust pigs. J. Anim. Sci. 93:1494–1502. doi: 10.2527/jas.2014-8583 [DOI] [PubMed] [Google Scholar]
- Kleinbaum D. G. D., and Klein M.. 2011. Chapter 3 in Survival analysis: a self-learning text, third edition (statistics for biology and health). New York (NY): Springer Publishing. [Google Scholar]
- Koltes J. E., Fritz-Waters E., Eisley C. J., Choi I., Bao H., Kommadath A., Serão N. V., Boddicker N. J., Abrams S. M., Schroyen M., et al. 2015. Identification of a putative quantitative trait nucleotide in guanylate binding protein 5 for host response to PRRS virus infection. BMC Genomics 16:412. doi: 10.1186/s12864-015-1635-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur P. K., Herrero-Medrano J. M., Alexandri P., Knol E. F., ten Napel J., Rashidi H., and Mulder H. A.. 2014a. Estimating challenge load due to disease outbreaks and other challenges using reproduction records of sows. J. Anim. Sci. 92:5374–5381. doi: 10.2527/jas.2014-8059 [DOI] [PubMed] [Google Scholar]
- Mathur P., Herrero-Medrano J., Alexandri P., Knol E., Rashidi H., Mulder H., and ten Napel J.. 2014b. Genetic evaluation for disease resistance and tolerance in pigs using reproduction records. 10th World Congress on Genetics Applied to Livestock Production; August 18, 2014; Vancouver (Canada).
- Mathur P. K., Vogelzang R., Mulder H. A., and Knol E. F.. 2018. Genetic selection to enhance animal welfare using meat inspection data from slaughter plants. Animals 8:16. doi: 10.3390/ani8020016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder H. A. 2016. Genomic selection improves response to selection in resilience by exploiting genotype by environment interactions. Front. Genet. 7:178. doi: 10.3389/fgene.2016.00178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder H. A., and Rashidi H.. 2017. Selection on resilience improves disease resistance and tolerance to infections. J. Anim. Sci. 95:3346–3358. doi: 10.2527/jas.2017.1479 [DOI] [PubMed] [Google Scholar]
- Nakov D., Hristov S., Stankovic B., Pol F., Dimitrov I., Ilieski V., Mormede P., Hervé J., Terenina E., Lieubeau B., et al. 2018. Methodologies for assessing disease tolerance in pigs. Front. Vet. Sci. 5:329. doi: 10.3389/fvets.2018.00329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putz A. M., Harding J. C. S., Dyck M. K., Fortin F., Plastow G. S., and Dekkers J. C. M.; PigGen Canada 2018. Novel resilience phenotypes using feed intake data from a natural disease challenge model in wean-to-finish pigs. Front. Genet. 9:660. doi: 10.3389/fgene.2018.00660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashidi H., Mulder H. A., Mathur P., van Arendonk J. A., and Knol E. F.. 2014. Variation among sows in response to porcine reproductive and respiratory syndrome. J. Anim. Sci. 92:95–105. doi: 10.2527/jas.2013-6889 [DOI] [PubMed] [Google Scholar]
- Schroyen M., Eisley C., Koltes J. E., Fritz-Waters E., Choi I., Plastow G. S., Guan L., Stothard P., Bao H., Kommadath A., et al. 2016. Bioinformatic analyses in early host response to Porcine Reproductive and Respiratory Syndrome virus (PRRSV) reveals pathway differences between pigs with alternate genotypes for a major host response QTL. BMC Genomics 17:196. doi: 10.1186/s12864-016-2547-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urruty N., Tailliez-Lefebvre D., and Huyghe C.. 2016. Stability, robustness, vulnerability and resilience of agricultural systems. A review. Agron. Sustain. Dev. 36:15. doi: 10.1007/s13593-015-0347-5 [DOI] [Google Scholar]
- Walker L. R., Engle T. B., Vu H., Tosky E. R., Nonneman D. J., Smith T. P. L., Borza T., Burkey T. E., Plastow G. S., Kachman S. D., Ciobanu D. C.. 2018. Synaptogyrin-2 influences replication of Porcine circovirus 2. PLoS Genet. 14:e1007750. doi: 10.1371/journal.pgen.1007750 [DOI] [PMC free article] [PubMed] [Google Scholar]