Abstract
In social animals, affiliative behaviours bring many benefits, but also costs such as disease risk. The ways in which affiliation may affect the risk of infectious agent transmission remain unclear. Moreover, studies linking variation in affiliative interactions to infectious agent incidence/diversity have speculated that disease transmission may have occurred, rather than revealing that transmission did occur. We address these gaps using the phylogenetics of commensal gut Escherichia coli to determine whether affiliative grooming and huddling social networks mediated microbial transmission among rhesus macaques. We collected behavioural and microbial data from adult macaques across a 12-week period that was split into two 6-week phases to better detect dyadic transmission. We reconstructed undirected social networks from affiliative interactions and reconstructed microbial transmission networks from the pairwise phylogenetic similarity of E. coli pulsotypes from macaques within and across adjacent sampling events. Macaque E. coli pulsotypes were more phylogenetically similar to each other than to environmental isolates, which established a premise for socially mediated transmission. Dyadic grooming and huddling frequencies strongly influenced the likelihood of E. coli transmission during the second data collection phase, but not the first. Macaques that were more central/well connected in both their grooming and huddling networks were also more central in the E. coli transmission networks. Our results confirmed that affiliative grooming and huddling behaviours mediate the transmission of gut microbes among rhesus macaques, particularly among females and high-ranking individuals. The detectability of socially mediated E. coli transmission maybe partially masked by environmental acquisition in males, or by high frequencies of interactions in captivity. Predicting the potential transmission pathways of gastrointestinal parasites and pathogens, our findings add to current knowledge of the coevolutionary relationships between affiliative behaviour and health and may be used to identify ‘superspreader’ individuals as potential targets for disease control strategies.
Keywords: affiliative behavior, commensal E. coli, infectious disease, rhesus macaque, social network, superspreader, transmission
In group-living animals, affiliative social behaviours are important for establishment and maintenance of group cohesion and social structure (Clutton-Brock, 2016; Hinde, 1976; Kappeler & Van Schaik, 2002). Unsurprisingly, social affiliation is widespread in the animal kingdom, and varies broadly in distribution, form and function (Hinde, 1976; Kappeler & Van Schaik, 2002). From an evolutionary perspective, affiliation brings many health- and fitness-related benefits, such as (1) lowered stress levels (Cobb, 1976; Cohen, Janicki-Deverts, Turner, & Doyle, 2015; Young, Majolo, Heistermann, Schulke, & Ostner, 2014), (2) enhanced immune function (Kaplan et al., 1991; Segerstrom & Miller, 2004) and (3) the establishment and maintenance of long-term social bonds (Silk, 2014; Silk et al., 2003, 2010). On the other hand, a major cost of affiliation may be infectious disease risk (Alexander, 1974; Altizer et al., 2003; Kappeler, Cremer, & Nunn, 2015; Nunn, Thrall, Leendertz, & Boesch, 2011). Critical gaps remain in our understanding of whether and how affiliative social behaviours may mediate the acquisition and transmission of infectious agents (Kappeler et al., 2015). For example, empirical studies linking animal social behaviour and either infectious agent incidence or the sharing of genotypically similar gut microbes have only been able to speculate that socially mediated microbial transmission may have occurred, but fall short of establishing that transmission did occur. While behaviourally mediated sharing may be inferred based on genotypic similarity of microbes at single time points, the detection of socially mediated transmission requires comparing host attributes and social behavioural contact patterns with the genotypic similarity of microbes at longitudinal, overlapping time intervals (VanderWaal & Ezenwa, 2016). Here we exploit the phylogenetics of commensally living gut bacterium Escherichia coli and use comparisons of animals’ social and microbial transmission networks to assess whether affiliative social grooming and huddling behaviours may mediate longitudinal infectious agent transmission among rhesus macaques, Macaca mulatta.
Understanding the coevolutionary underpinnings of social life and infectious disease risk is a major challenge for researchers. For instance, affiliative behaviours may have unexpected opposing effects on health outcomes such as infectious disease risk. Although higher rates of affiliation may socially buffer individuals and lower their susceptibility to acquiring infectious agents by providing physiological or health-related benefits (Balasubramaniam, Beisner, Vandeleest, Atwill, & McCowan, 2016; Bartolomucci, 2007; Cohen, 2004; Cohen, Janicki-Deverts, & Miller, 2007; Klein, 2000a,b), some forms of affiliation often involve prolonged physical contact with conspecifics, which may increase the likelihood of acquiring infectious agents from previously infected individuals (Freeland, 1976; Kappeler et al., 2015; MacIntosh & Frias, 2017; Nunn et al., 2011). A typical behaviour that exhibits these trade-offs is social or allogrooming, i.e. cleaning or manipulating the fur or skin of another individual, which is a common form of affiliation in animal societies (e.g. social insects: Moore, Angel, Cheeseman, Robinson, & Fahrbach, 1995; bovids: Hart & Hart, 1992; bats: Carter & Leffer, 2015; nonhuman primates: Henzi & Barrett, 1999). In nonhuman primates, for instance, grooming may lower infectious disease risk by reducing ectoparasite load in recipients (Duboscq, Romano, Sueur, & MacIntosh, 2016; Tanaka & Takefushi, 1993) and by mitigating stress levels to socially buffer both givers and receivers from acquiring infectious agents (e.g. Balasubramaniam et al., 2016; Smutt, MacLarnon, Heistermann, & Semple, 2007; Young et al., 2014). Yet the prolonged periods of direct host-to-host contact during grooming may also increase the risk of acquiring and transmitting gastrointestinal parasites via faecale–oral contact routes (e.g. MacIntosh et al., 2012; Rimbach et al., 2015). Such contrasting mechanisms make unravelling the links between affiliative interactions and infectious agent risk highly complex.
Social network analysis is well suited for modelling such heterogeneous patterns in animals’ space use overlap and social interaction due to the consideration of both direct interactions and indirect secondary links among group members (Brent, Lehmann, & Ramos-Fernandez, 2011; Lusseau, Whitehead, & Gero, 2008; McCowan, Anderson, Heagarty, & Cameron, 2008; Sade, 1972a,b; Sueur, Jacobs, Amblard, Petit, & King, 2011). For this reason, networks have proven highly useful tools for modelling infectious agent acquisition and transmission (Craft, 2015; Craft & Caillaud, 2011; Drewe & Perkins, 2015; Godfrey, 2013; VanderWaal & Ezenwa, 2016). Indeed, recent efforts have revealed that, across a wide range of animal taxa, individuals that are well connected or highly central in their social networks are also more likely to be infected by gastrointestinal parasites and pathogens such as (1) protozoans (e.g. Crithidia sp. in bumblebees, Bombus impatiens: Otterstatter & Thomson, 2007; Entamoeba sp. in brown spider monkeys, Ateles hybridus: Rimbach et al., 2015; Cryptosporidium sp. in Belding’s ground squirrels, Urocitellus beldingi: VanderWaal, Atwill, Hooper, Buckle, & McCowan, 2013), (2) nematodes (e.g. Strongyloides sp. in Japanese macaques, Macaca fuscata: MacIntosh et al., 2012) and (3) pathogenic bacteria (e.g. Salmonella sp. and E. coli O157:H7 in African ungulates: VanderWaal, Atwill, Isbell, & McCowan, 2014; Shigella sp. in rhesus macaques: Balasubramaniam et al., 2016).
One aspect that remains unaddressed by the above studies is related to the detectability of socially mediated microbial transmission; these studies have been useful to speculate whether transmission may have occurred but cannot conclude that transmission did occur. One way to address this would be by assessing and comparing the phylogenetic relationships of commensal gut microbes to model the socially mediated transmission of microbes at high resolutions (reviewed in VanderWaal & Ezenwa, 2016). Commensal E. coli are ideal microbes for these purposes because two individuals with genotypically similar or identical E. coli likely acquired the strain either via faecale–oral contact-mediated transmission or from a common environmental source (Chiyo et al., 2014; Springer, Mellmann, Fichtel, & Kappeler, 2016; VanderWaal, Atwill, Isbell, & McCowan, 2013; VanderWaal & Ezenwa, 2016). Escherichia coli are facultative anaerobic bacteria that, by virtue of being highly prevalent in vertebrates, are potentially isolatable from the gastro-intestinal tracts of every individual in a group (Goldberg, Gillespie, & Singer, 2006; Sears, Janes, Saloum, Brownlee, & Lamoreaux, 1950, 1956; Tenaillon, Skurnik, Picard, & Denamur, 2010). They exhibit a clonal population structure that remains relatively unaffected by phenomena like horizontal gene transfer, mutation events within epidemiologically short time frames and/or host immune function (Goldberg et al., 2006; Tenaillon et al., 2010). In addition, although E. coli share close evolutionary histories and microecological niche space with gastrointestinal pathogens, commensal or nonpathogenic strains generally do not alter the behaviour of the host (Goldberg et al., 2006; VanderWaal & Ezenwa, 2016), nor is their acquisition likely to be influenced by stress-induced social buffering (Balasubramaniam et al., 2018).
A second gap in our understanding of the behavioural bases of disease risk is related to distinguishing between microbial acquisition from social contact (i.e. social transmission) versus from shared space use overlap (i.e. environmental acquisition). In reticulated giraffes, Giraffa camelopardalis, E. coli sharing was more strongly predicted by networks of social contact associations than by networks of shared space use overlap (VanderWaal, Atwill, Isbell et al., 2013). In contrast, shared habitat use and cohort membership, but not social contact interactions, seemed to influence E. coli genotypic diversity among African elephants, Loxodonta Africana (Chiyo et al., 2014). In brush-tailed possums, Trichosurus vulpecula, E. coli sharing seemed to depend more on social contact than space use, but specifically on more cryptic forms of contact rather than on long-term relationships (Blyton, Banks, Peakall, Lindenmayer, & Gordon, 2014). In Verreaux’s sifakas, Propithecus verreauxi, individuals’ group membership and rates of intergroup encounters both predicted the genotypic sharing of E. coli, suggesting indirect evidence for socially mediated bacterial transmission (Springer et al., 2016). In summary, these studies provide mixed evidence for whether animals’ shared use of space or their social contact behaviours is more likely to influence the genotypic diversity and sharing of E. coli.
Here we address the aforementioned gaps by examining whether affiliative social behaviours may facilitate the longitudinal transmission of gut E. coli among captive rhesus macaques. Rhesus macaques are a good study species for examining socially mediated microbial transmission. They show a high degree of heterogeneity in social interactions that arise from a nepotistic, despotic social structure, in which dominance rank, sex differences and kinship ties may all strongly influence affiliative interactions like social grooming (Balasubramaniam et al., 2012; Berman, 2011; Berman & Kapsalis, 2009; Berman & Thierry, 2010; Thierry, 2007). Moreover, the observed heterogeneity in affiliation is likely to influence socially mediated faecale–oral transmission of gut E. coli because our previous work has shown that genotypic diversity of E. coli is strongly linked to social network community membership across multiple rhesus groups (Balasubramaniam et al., 2018). Here we extend this work, and the work of others reviewed above, by adopting a longitudinal sampling approach that caters to detecting microbial transmission (see Methods).
Aim 1: Premise for Contact-mediated Microbial Transmission
Within a single group of captive rhesus macaques, we first establish a premise to expect socially mediated transmission of E. coli. Gastrointestinal microbes may be acquired by social contact or directly from contaminated substrates in the environment (Fenner, Godfrey, & Bull, 2011; Godfrey, Moore, Nelson, & Bull, 2010; Huffman et al., 2013; Hussain, Ram, Kumar, Shivaji, & Umapathy, 2013; VanderWaal, Atwill et al., 2014). As a premise for expecting social transmission despite potential environmental acquisition, we examined whether macaques’ gut E. coli were more phylogenetically similar to each other than to E. coli isolated from the environment. Furthermore, since dispersing males from female philopatric societies are often more exploratory in their environmental space use (Cords, 2013; Kappeler, 2000; Pusey & Packer, 1987; VanderWaal, Wang, McCowan, Fushing, & Isbell, 2014), we also examined whether E. coli genotypic similarity was stronger for females compared to males.
Aim 2: Social Contact Frequencies and the Dyadic Transmission of E. coli
Next, we asked whether macaques’ contact affiliation mediated the transmission of gut E. coli between animals, resulting in higher levels of phylogenetic similarity between their E. coli strains. We predicted that the frequency of dyadic social grooming and/or huddling interactions would be positively related to the likelihood of detecting high phylogenetic similarity (labelled here as a link) in their corresponding E. coli transmission network.
Aim 3: Social Network Connectedness and Superspreading of E. coli
Finally, we examined whether some macaques, due to being central both in the affiliation and microbial transmission networks, may also function as potential ‘superspreaders’ of infectious agents (Lloyd-Smith, Schreiber, Kopp, & Getz, 2005). We predicted that the most well-connected individuals in the grooming and/or huddling networks would also be central in the E. coli transmission network. Given that patterns of macaque affiliation are influenced by sex and dominance rank, we also examined whether these attributes interacted with individuals’ affiliation network positions to influence E. coli transmission.
METHODS
Study Location and Subjects
The study was conducted at the California National Primate Research Center (CNPRC) and the School of Veterinary Medicine (SVM), University of California at Davis, U.S.A. We collected behavioural and microbial data on a single group of rhesus macaques housed in a 0.2 ha outdoor enclosure. Subjects were 97 macaques >3 years of age (31 males, 66 females). Animals were fed a standard diet of monkey chow twice per day at approximately 0700 hours and between 1430 hours and 1530 hours. This diet was supplemented by fresh fruit or vegetables once a week, with seed mixture being provided daily. Water was available ad libitum from water spigots as well as sporadically from natural puddles. The protocols used for this research were approved by the University of California (UC) Davis Institutional Animal Care and Use Committee (protocol no. 18525) and were in accordance with the legal requirements of the jurisdictions in which the research was conducted.
Behavioural Data Collection
Three observers collected behavioural data for a total duration of 3 months of a single dry season (May–July), for 6 h per day (0900–1200 hours and 1300–1600 hours) and 4 days per week. The data collection period was split into two 6-week phases; this duration has been previously shown to yield adequate behavioural data to construct reliable social networks in this population (Balasubramaniam et al., 2018). Observers used an instantaneous scan sampling approach to record affiliative grooming and huddling interactions (Altmann, 1974). Social grooming or allogrooming was defined as an instance where an individual was observed cleaning or manipulating the fur of another individual. Huddling was recorded when an individual was observed in any kind of direct body contact, including (but not restricted to) ventral contact or an embrace, with another individual that did not involve an alternative social interaction such as grooming or contact aggression. Observers conducted scans once every 20 min during a 6 h duration of sampling per day, generating a total of 432 scans per 6-week phase, or 864 scans across the 3-month period. In addition, they used an event sampling approach to record dyadic aggressive interactions (Altmann, 1974), in order to determine dominance ranks of individuals using the Percolation-and-Conductance method available in the ‘Perc’ R package (Fujii et al., 2016; Fushing, McAssey, Beisner, & McCowan, 2011; for descriptions, see ; Balasubramaniam et al., 2016; Vandeleest et al., 2016).
Microbial Sampling, Isolation and DNA Fingerprinting
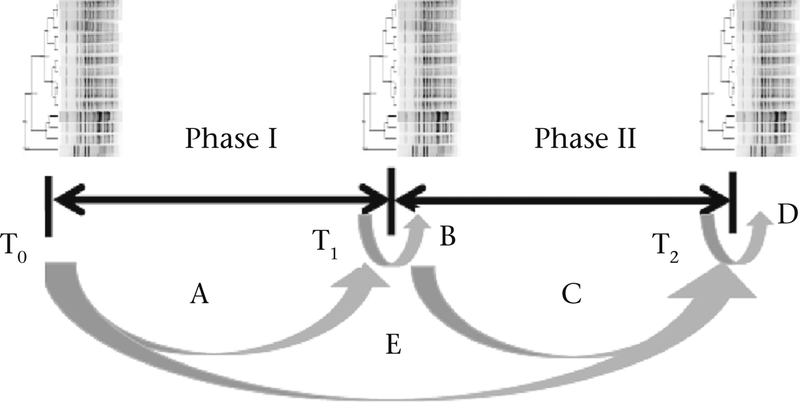
We used a longitudinal sampling approach to collect fresh faecal swabs from all individuals in the group during each of three sampling events. The first was just prior to the commencement of behavioural data collection (Fig. 1: sampling event T0). The second at the end of phase I, or after the first 6 weeks of behavioural data collection (Fig. 1: T1). The third was at the end of phase II, or after the second 6-week behavioural data collection period (Fig. 1: T2). This approach was aimed at reconstructing bacterial transmission networks from phylogenetic similarity of E. coli inferred both from within the same sampling event that followed a behavioural sampling period and from across adjacent sampling events that flanked a behavioural sampling period (Fig. 1; see details below regarding the construction of E. coli transmission networks). Since commensal E. coli typically survive in the mammalian gut for 8–25 weeks (Habteselassie et al., 2008; Sinton, Hall, & Braithwaite, 2007; Van Elsas, Semenov, Costa, & Trevors, 2011), we believed that collecting and processing microbial samples at the end of 6 weeks would be useful to detect at least some (if not all) dyadic transmission that may have occurred during this period. So we expected that our sampling approach would facilitate the establishment of dyadic transmission within shorter (6-week) time frames, and that ‘superspreader’ macaques from a more complete or well-connected transmission network could be inferred from across the entire duration of data collection (12 weeks).
Figure 1.
Behavioural and microbial data collection paradigm. T0, T1 and T2 represent microbial sampling events. Arrows (labelled A to E) connect sampling events used to construct E. coli transmission networks whereby the degree of similarity of E. coli pulsotypes between pairs of macaques either between sampling events or within the same sampling event that immediately followed a behavioural data collection phase was ≥90% similarity to indicate E. coli sharing or transmission (Table 2). Phase I and phase II represent two 6-week periods of behavioural data collection.
On the day of a sampling event, we immobilized each animal and collected fresh faecal swabs using previously published methods (Good, May, & Kawatomari, 1969). To rule out E. coli acquisition from environmental faeces and substrates, we also collected 18 samples by swabbing faecal-contaminated environmental features of the enclosure, three samples from within each of six equally divided sections of the enclosure. We immersed the swabs into prelabelled tubes (macaque samples) or small bags (environmental samples) containing Tryptic Soy Broth (TSB; BD Franklin Lakes, NJ, U.S.A.). We then isolated a single strain and biochemically confirmed this to be commensal E. coli from these TSB tubes in the majority of individuals, using previously standardized epidemiological procedures in the Atwill-McCowan laboratory in the UC Davis School of Veterinary Medicine (see Balasubramaniam et al., 2016, 2018, for more detailed explanations). The minority of individuals that tested negative during a sampling event were deemed ‘untypable’ but were included in the analyses if they tested positive for E. coli in at least one of the three sampling events. Confirmed E. coli isolates were banked and frozen within a −80 °C freezer. From these, we generated bacterial DNA fingerprint profiles, or pulsotypes, from each E. coli isolate using PulseNet Pulsed Field Gel Electrophoresis (PFGE: using the CDC protocol). PFGE is a well-established method for typing bacteria and has been shown to perform well for comparing large numbers of isolates of commensal gut E. coli (Cesaris et al., 2007; Kilonzo, Atwill, Mandrell, Garrick, & Villanueva, 2011; Kondo, Hoar, & Mandrell, 2010; Ribot et al., 2006; VanderWaal, Atwill et al., 2014). While we acknowledge that more novel techniques like whole genome sequencing (WGS) and metagenomic processing would add additional discriminatory power to our analyses, the prohibitory increase in cost would significantly reduce sample size and therefore prevent a network analysis. So we used PGFE given that this method (1) struck a balance between adequate sample size and molecular accuracy, (2) has a long history of success in past studies relying on molecular inference, (3) is sufficient to assess genus-typical bacterial diversity and (4) is adequate for our focus on a specific inhabitant of the gut microbiome (enteric E. coli). Upon generating E. coli genotypic profiles from the banked isolates, we determined the similarity of each genotype to all others, by comparing their standardized densitometric curves generated from the pulsotype images using the Bionumerics software (version 6.6, Applied Maths, Inc., Austin, TX, U.S.A.). These curves reveal both the locations of DNA fragments, as indicated peaks in the curve that correspond to the presence of bands on the pulsotype image, and the quantity of DNA in each fragment, as indicated by the peak dimensions that correspond to band intensity. Two genotypes were considered ‘identical’ if their densitometric curves ≥90% similar (cophenetic correlation coefficient). Reproducibility analyses conducted in a previous study revealed that this cutoff criterion minimizes both type I errors in matching to 1%, and limits type II error rates to <5% (VanderWaal, Atwill, Isbell et al., 2013).
Social and Microbial Transmission Networks
From the scan data of contact affiliative interactions, we reconstructed contact grooming and huddling social networks. All networks had the same number of individuals (N = 97) and were represented as square matrices. Networks were undirected but weighted based on the total number of interactions recorded between each pair of individuals. For each behaviour, we constructed three such networks, one each from the interactions recorded in phase I and phase II, respectively, and a third ‘overall’ network that combined behavioural interactions from both phases. From the latter, we computed grooming and huddling degree and betweenness for each individual. While being epidemiologically relevant to detecting microbial transmission, degree and betweenness capture slightly different aspects of individuals’ social positions. Weighted degree is an individuals’ total number of connections or partners, times the number of interactions with each partner (Opsahl, Agneessens, & Skvoretz, 2010). Being a measure of an individuals’ direct connections, degree has been useful in previous studies that have linked contact behavioural patterns to the likelihood of infectious agent acquisition (Balasubramaniam et al., 2016; Drewe, 2010; MacIntosh et al., 2012). In contrast, betweenness considers both direct interactions and indirect pathways of connections; it was calculated as the relative number of shortest paths that pass through a focal individual that connects other individuals in the network (Freeman, 1977; Newman, 2005; Wasserman & Faust, 1994). Its usefulness to identify ‘hubs’ that link key components of networks has led to its identification as being a key measure for the detection of potential ‘superspreaders’ of microbes (Drewe & Perkins, 2015; Friant, Ziegler, & Goldberg, 2016; Rushmore, Bisanzio, & Gillespie, 2017; VanderWaal, Atwill, Isbell et al., 2013; VanderWaal & Ezenwa, 2016).
From the data on pairwise similarity in E. coli, we constructed three microbial transmission networks using criteria that made them temporally overlap with the three social behavioural networks. These were undirected and unweighted, with links assigned based on whether pairs of macaques shared identical pulsotypes of E. coli (≥90% similarity), either across adjacent sampling events that flanked behavioural data collection phases, or within the same sampling event at the end of a behavioural data collection phase. Specifically, the transmission network of phase I was constructed by assigning links of E. coli similarity both (1) across sampling events To and T1 (arrow A in Fig. 1) and (2) within T1 (arrow B in Fig. 1). Likewise, shared E. coli similarity between sampling events T1 and T2 and within T2 were considered to construct the transmission network of phase II (arrows C and D in Fig. 1). As with the behavioural networks, we also constructed an ‘overall’ transmission network that included links based on E. coli similarity within and across all sampling events (arrows A to E in Fig. 1). As with the overall grooming and huddling networks, we estimated each individual’s degree and betweenness centrality in their overall E. coli transmission network.
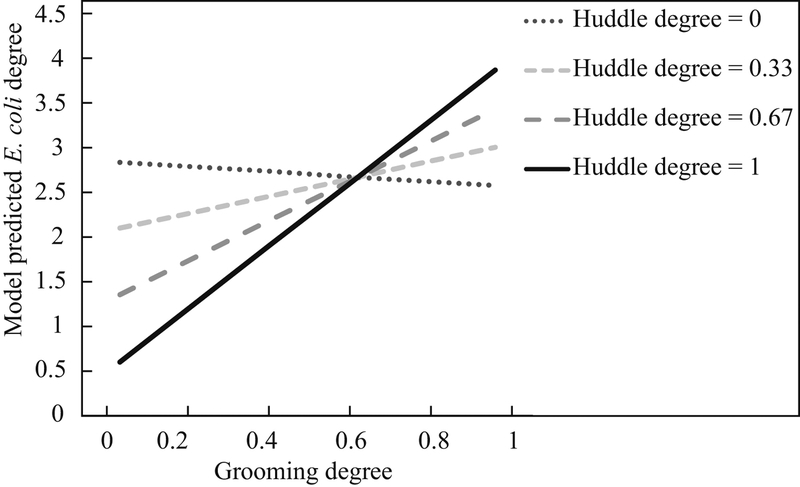
Figure 2.
Plot showing the predicted value of E. coli transmission degree at various values of social huddling and grooming degree (model A7) (Aim 3).
Statistical Analyses
To establish a premise for expecting social contact-mediated bacterial transmission (Aim 1), we used a series of Wilcoxon signed-rank tests. Within a given sampling event, these compared the mean dyadic macaque–macaque E. coli similarity coefficients with macaque–environmental E. coli similarity coefficients. We favoured a nonparametric test given (1) the interdependency of dyadic similarity coefficients and (2) the error distribution for dyadic macaque–macaque E. coli similarity coefficients deviating significantly from normality (e.g. at sampling event T0: Shapiro–Wilk test: W = 0.95, P < 0.01). To examine whether there were sex-based differences in this premise, we also ran separate Wilcoxon signed-rank tests for male–male and female–female dyads to compare the mean E. coli similarity of dyads to each other and to male–environmental or female–environmental dyads within each sampling event.
To examine whether the frequencies of dyadic behaviours in the social networks predicted the likelihood of E. coli sharing in the transmission networks (Aim 2), we ran logistic multiple regression quadratic assignment procedure (or MR-QAP) tests (Dekker, Krackhardt, & Snijders, 2007; Hubert, 1987; Krackhardt, 1987) using the ‘netlogit’ function in the ‘SNA’ R package (Butts, 2008). MR-QAP accounts for the interdependency of dyadic data sets by implementing a double Dekker semipartialling approach (Dekker et al., 2007; Hanneman & Riddle, 2005). In each model, the outcome variable was an E. coli transmission network (from phase I or phase II), represented as a binary matrix with the presence (1) or absence (0) of a shared link between each pair of macaques. Since grooming and huddling matrices were strongly correlated to each other (e.g. in phase I: r = 0.70, P < 0.01), we only ran univariate models, three for each phase. In the first two, the predictors were social grooming and huddling networks, respectively, with cells in the symmetrized matrices containing the frequencies of interactions between each dyad. To examine any potential effects of vertical microbial transmission from parents to offspring (Ley, Lozupone, Hamady, Knight, & Gordon, 2008; McCord et al., 2014), we ran a third model to examine the influence of kinship. This was a binary matrix of ‘close-kin dyads’ assigned based on whether (1) or not (0) they belonged to the same matriline. Kinship strongly influenced grooming and huddling frequencies, with related dyads affiliating with each other more than unrelated dyads (e.g. phase I: huddling among kin versus nonkin: Wilcoxon signed-rank test: Z = 19.4, P < 0.01; phase II: grooming among kin versus nonkin: Z = 12.2, P < 0.01). Because of this, we again refrained from running multivariate models. Given the limitations of MR-QAP with regards to providing meaningful R2 values, or accurate goodness-of-fit statistics (Ferrin, Dirks, & Shah, 2006; Gibbons, 2004; Zagenczyk, Gibney, Few, & Purvis, 2013), we interpreted each model based on the P values from the permutation tests.
To examine whether macaques that were well connected or central in their social networks also functioned as superspreaders of E. coli in their transmission network, we ran a series of GLMs using the ‘lme4’ package in R (Bates et al., 2016). We ran two model sets of seven models each, or 14 models in total. In the first model set, the outcome variable was a count of each individual’s degree in the E. coli transmission network. We therefore used a Poisson link function. We first ran two univariate models, one each for grooming and huddling degree centrality as predictors. We then ran four multivariate models in which we tested the interaction of each social network measure first with sex (two models) and then with dominance rank (two models). Finally, we ran one model that examined the interaction of grooming and huddling degree on the outcome of E. coli degree. To determine model fit, we calculated corrected Akaike information criteria (or AICc) scores and interpreted only those models that showed a significantly better fit (i.e. had a dAICc < 8) than a null or intercept-only model (Burnham, Anderson, & Huyvaert, 2011; Richards, Whittingham, & Stephens, 2011). In case of models with interaction terms, we interpreted these in the place of a simpler model with just a main effect if the interaction model was a significantly better fit (dAICc < 8) than the simpler model. In the second model set, the outcome variable was individuals’ betweenness centrality in the E. coli transmission network. This followed a negative binomial distribution, so we used the ‘glm.nb’ function with a log link function in the ‘lme4’ R package (Bates et al., 2016). As with the models for degree measures, we examined the effects of grooming and huddling betweenness, their interactions with sex and rank, and their interaction with each other. However, given interdependency concerns over the betweenness measure, we determined P values for each predictor in each model by using a post-network randomization, or a ‘node-swapping’ permutation approach, based on 1000 permutations of the outcome variable (Farine, 2017; Farine & Whitehead, 2015). Rather than using AIC, we interpreted all models in which predictors showed a significant P value from these randomization tests. All statistical analyses were performed using R (v.3.1.3), and the value of α was set as 0.05.
RESULTS
From 97 individual macaques and across the three sampling events, we collected and DNA fingerprinted a total of 254 bacterial isolates, 86 in the first sampling event (T0), 79 in the second sampling event (T1) and 89 in the third sampling event (T2). Across the three sampling events, we isolated and DNA fingerprinted one to three E. coli isolates for all 97 individuals. Individual consistency in E. coli isolate type was low – most of the 97 macaques (91 individuals, or 94%) had a different E. coli isolate at each of their sampling points. However, we found many cases where different individuals had identical isolates (>90% similarity) both across time points (T0 → T1; T1 → T2) and within the same time point that followed a behavioural data collection period (T1, T2). Out of the 54 environmental isolates that we processed (18 in each of three sampling events), 45 tested positive and were genotyped for E. coli, 17 in the first sampling event, 16 in the second event and 12 in the third sampling event.
Aim 1: Premise for Contact-mediated Microbial Transmission
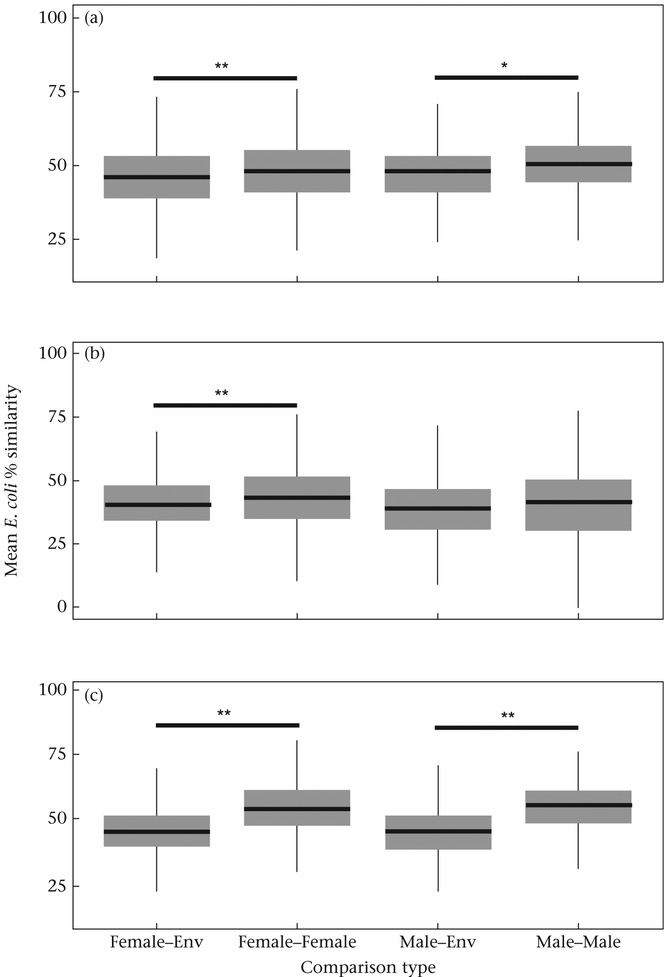
Our comparisons of macaque and environmental isolates established a strong premise for expecting social contact-based microbial transmission (Aim 1). Wilcoxon signed-rank tests revealed that, at all three sampling events, E. coli isolates from macaques were more similar to each other than they were to environmental isolates (Table 1). Similar analyses for different sexes revealed that this finding was more strongly sustained and consistent across sampling events for female macaques compared to males (Table 1, Fig. A1). At sampling event T1, the DNA similarity of E. coli isolates among males (1) did not differ significantly from E. coli from males compared to environmental isolates (Table 1, Fig. A1). Furthermore, (2) isolates among males were consistently less similar to each other than were isolates among females (Wilcoxon signed-rank test: Z = 3.8, P < 0.01).
Table 1.
Estimated differences in the degrees of DNA similarity between macaque–macaque and macaque–environmental E. coli isolates at each of the three sampling events (Aim 1)
| Sampling event | Wilcoxon Z (similarity of macaque–macaque isolates > macaque–environmental isolates) | ||
|---|---|---|---|
| Overall | Only females | Only males | |
| T0 | 6.8** | 5.34** | 3.03* |
| T1 | 4.45** | 4.46** | 0.58 |
| T2 | 26.28** | 19.8** | 12.97** |
Values indicate Z scores from Wilcoxon signed-rank tests.
P < 0.05;
P < 0.01.
Aim 2: Social Contact Frequencies and the Dyadic Transmission of E. coli
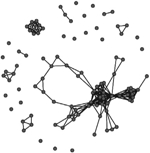
A comparison of the characteristics of the two phase-specific E. coli transmission networks revealed more microbial transmission links in phase II (between sampling periods T1 and T2) compared to phase I (between periods T0 and T1) (Table 2). The transmission network of phase I consisted of 244 transmission links yielding 48 network fragments, and 56 of 97 macaques shared E. coli isolates with at least one other group member (links assigned based on ≥90% DNA similarity). Mean E. coli degree was 6.8 links per individual. In comparison, the transmission network of phase II was more well connected. It contained more transmission links (348) yielding fewer fragments (27), and 78 of 97 macaques shared a strain with at least one other group member. Furthermore, the mean degree (11.2) was significantly greater for the phase II network than the phase I network (Wilcoxon signed-rank test: W = 3363, P < 0.01). Our criteria also yielded a fully connected overall transmission network across the three sampling events (Table 2).
Table 2.
Characteristics of the three E. coli transmission networks constructed using the bacterial genotypic profiles
| Phase I (T0 to T1,
arrows A and B in Fig. 1) |
Phase II (T1 to T2,
arrows C and D in Fig. 1) |
Overall across both
phases (T0 to T1 to T2, all arrows in Fig. 1) |
|
|---|---|---|---|
| Connected individuals (nodes with at least one edge) | 56 | 78 | 97 |
| Transmission links (edges) | 244 | 342 | 586 |
| Mean ± SE degree | 6.8 ± 5.6 | 11.7 ± 7.8 | 12.3 ± 8.7 |
| Graph | 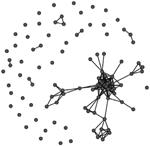 |
 |
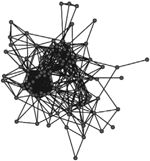 |
Unweighted links were assigned between any two macaques that shared an identical strain of E. coli (>90% similarity in pulsotypes) across adjacent sampling events, or within the same sampling event at the end of the corresponding behavioural data collection phase(s). Phases represent the two periods of behavioural data collection. T0, T1 and T2 represent sampling events that flanked behavioural data collection.
At the dyadic level, we found strong but somewhat inconsistent evidence for the frequencies of contact affiliative interactions to predict the likelihood of E. coli transmission (Aim 2) (Table 3). At phase II, the MR-QAP tests revealed a significant positive relationship between dyadic social grooming and huddling frequencies in the social networks and the likelihood of sharing a link in the E. coli transmission network (Table 3). In contrast, similar tests at phase I revealed no significant associations between dyadic grooming and huddling frequencies and the likelihood of detecting a link in the transmission network. However, the coefficients were positive in both univariate models, suggesting a possible tendency for dyadic contact-based transmission. We found no association between dyadic kinship (or matrilineal relatedness) and the likelihood of a link in the E. coli transmission network in either phase, in spite of kin dyads grooming and huddling more than nonkin dyads.
Table 3.
Univariate logistic MR-QAP regression models quantifying the association between grooming and huddling frequency and the likelihood of E. coli transmission (Aim 2)
| MR-QAP model | Coefficient (log odds) | |
|---|---|---|
| Phase I | Phase II | |
| E. coli links ~ Grooming frequency | 1.01 | 2.57* |
| E. coli links ~ Huddling frequency | 0.67 | 1.79* |
| E. coli links ~ Kinship | −0.13 | 0.09 |
P < 0.05.
Aim 3: Social Network Connectedness and Superspreading of E. coli
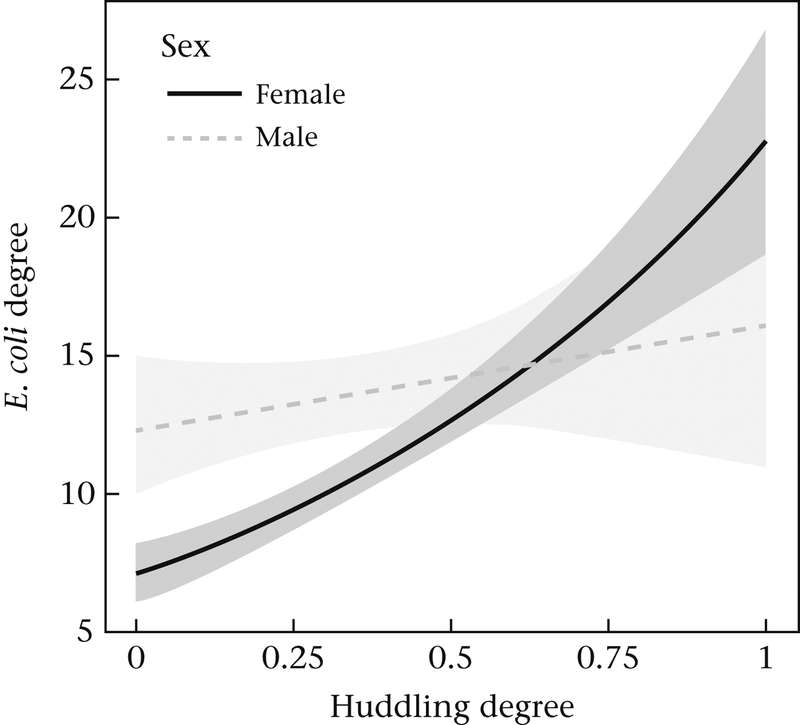
We found strong evidence to suggest that central or more well-connected macaques in their social network may also function as superspreaders of E. coli in their transmission networks (Aim 3). When we considered just direct connections or degree, the best-fit model revealed that E. coli degree in the overall transmission network was most strongly predicted by the interaction between grooming degree and huddling degree (Table 4, model A7). This model was also a significantly better fit than simpler models with just the main effects of grooming degree (model A1) and huddling degree (model A2). Grooming degree was positively associated with E. coli degree only at higher values of huddling degree (Fig. 2). Furthermore, two other models revealed that E. coli transmission degree was also impacted by interactions between individuals’ degree and both sex and dominance rank. There was a significant interaction between huddling degree and sex (Table 4, model A4), which revealed that females had a stronger positive relationship between huddling degree and E. coli degree than males (Fig. 3). There was also a significant interaction between grooming degree and dominance rank (Table 4, model A5), which suggested that grooming degree was more positively correlated with E. coli degree conditionally among high-ranking individuals.
Table 4.
Results of Poisson GLMs examining the effect of individuals’ degree centrality in their social grooming and huddling networks, and their interactions with sex and dominance rank, on the outcome of E. coli degree in their transmission networks (Aim 3)
| Model | Outcome variable | Predictors | β | SE | P | AICc | df |
|---|---|---|---|---|---|---|---|
| Null | E. coli degree | (Intercept) | 2.51 | 0.03 | <0.01** | 988.2 | 96 |
| Al | E. coli degree | (Intercept) | 2.61 | 0.06 | <0.01** | 987.05 | 95 |
| Groom degree | −0.27 | 0.15 | 0.07 | ||||
| A2 | E. coli degree | (Intercept) | 2.16 | 0.06 | <0.01** | 942.6 | 95 |
| Huddle degree | 0.88 | 0.13 | <0.01** | ||||
| A3 | E. coli degree | (Intercept) | 2.61 | 0.08 | <0.01 | 982.67 | 93 |
| Groom degree | −0.43 | 0.20 | 0.03* | ||||
| Sex | 0.00 | 0.12 | 1.00 | ||||
| Groom degree:Sex | 0.44 | 0.30 | 0.14 | ||||
| A4 | E. coli degree | (Intercept) | 1.96 | 0.08 | <0.01** | 926.39 | 93 |
| Huddle degree | 1.17 | 0.15 | <0.01** | ||||
| Sex | 0.56 | 0.13 | <0.01** | ||||
| Huddle degree:Sex | −0.90 | 0.29 | <0.01** | ||||
| A5 | E. coli degree | (Intercept) | 2.93 | 0.11 | <0.01** | 974.13 | 93 |
| Groom degree | 1.57 | 0.35 | <0.01** | ||||
| Rank | −0.56 | 0.22 | 0.01** | ||||
| Groom degree:Rank | 2.04 | 0.52 | <0.01** | ||||
| A6 | E. coli degree | (Intercept) | 2.20 | 0.11 | <0.01** | 942.28 | 93 |
| Huddle degree | 1.05 | 0.25 | <0.01** | ||||
| Rank | −0.17 | 0.22 | 0.43 | ||||
| Huddle degree:Rank | −0.13 | 0.42 | 0.76 | ||||
| A7 | E. coli degree | (Intercept) | 2.85 | 0.12 | <0.01** | 898.28 | 93 |
| Groom degree | −0.26 | 0.29 | 0.36 | ||||
| Huddle degree | 2.25 | 033 | <0.01** | ||||
| Groom degree:Huddle degree | 3.55 | 0.65 | <0.01** |
Models in bold indicate those that were interpreted as being significantly better-fit models (dAICc < 8) than the null, and (in the case of models with interaction terms) a better fit than a simpler model with just main effect(s).
P < 0.05;
P < 0.01.
Figure 3.
Plot showing the relationship between E. coli transmission degree and huddling degree for both males and females (model A4) (Aim 3). Shaded areas indicate the 95% confidence intervals around the two regression lines.
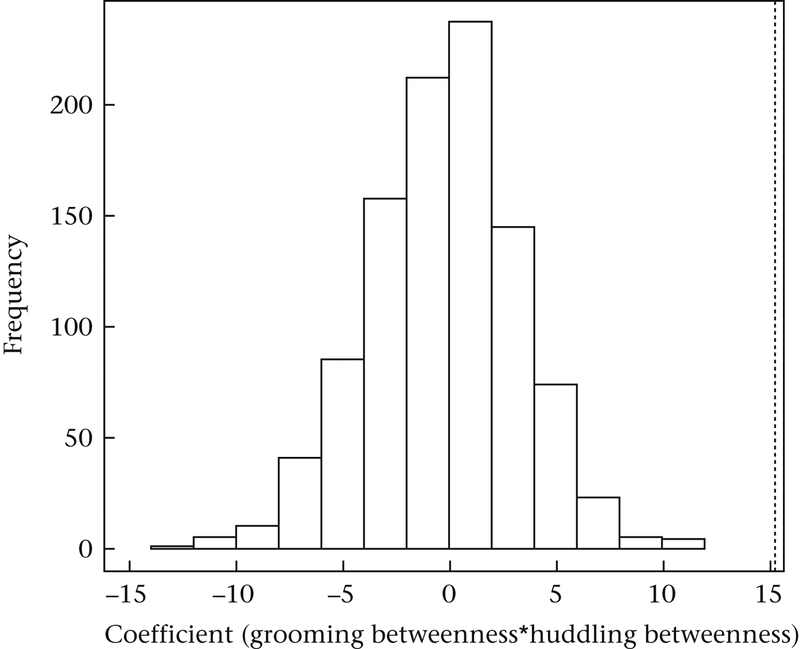
The inferences from our best-fit model were similar when we examined betweenness measures instead of degree (Table 5). Escherichia coli transmission betweenness was also significantly influenced by interactions between grooming and huddling betweenness (Table 5, model B7; Fig. 4), as indicated by the significant P values from the node-swapping permutation tests for these interaction terms. However, results from the other models were not consistent; we found no significant effects of interactions with sex (Table 5, model B4) and dominance rank (Table 5, model B5) when we examined behavioural and E. coli betweenness in place of degree.
Table 5.
Results of negative binomial GLMMs examining the effect of individuals’ betweenness centrality in their social grooming and huddling networks, and their interactions with sex and dominance rank, on the outcome of E. coli betweenness in their transmission networks (Aim 3)
| Model | Outcome variable | Predictors | β | SE | P | Ppermuted | df |
|---|---|---|---|---|---|---|---|
| Null | E. coli betweenness | (Intercept) | 4.89 | 0.16 | <0.01** | 96 | |
| B1 | E. coli betweenness | (Intercept) | 4.72 | 0.24 | <0.01** | 95 | |
| Groom betweenness | 0.78 | 0.93 | 0.40 | 0.27 | |||
| B2 | E. coli betweenness | (Intercept) | 4.79 | 0.25 | <0.01** | 95 | |
| Huddle betweenness | 0.40 | 0.82 | 0.63 | 0.49 | |||
| B3 | E. coli betweenness | (Intercept) | 4.67 | 0.31 | <0.01** | 93 | |
| Groom betweenness | 0.77 | 1.18 | 0.51 | 0.39 | |||
| Sex | 0.15 | 0.51 | 0.77 | 0.70 | |||
| Groom betweenness:Sex | 0.03 | 1.92 | 0.99 | 0.97 | |||
| B4 | E. coli betweenness | (Intercept) | 4.57 | 0.34 | <0.01** | 93 | |
| Huddle betweenness | 1.10 | 1.20 | 0.36 | 0.23 | |||
| Sex | 0.46 | 0.53 | 0.39 | 0.31 | |||
| Huddle betweenness:Sex | −1.27 | 1.65 | 0.44 | 0.43 | |||
| B5 | E. coli betweenness | (Intercept) | 4.79 | 0.47 | <0.01** | 93 | |
| Groom betweenness | 0.97 | 2.36 | 0.68 | 0.62 | |||
| Rank | −0.18 | 0.95 | 0.85 | 0.79 | |||
| Groom betweenness:Rank | −0.22 | 3.85 | 0.96 | 0.89 | |||
| B6 | E. coli betweenness | (Intercept) | 4.88 | 0.46 | <0.01** | 93 | |
| Huddle betweenness | 0.14 | 1.70 | 0.93 | 0.83 | |||
| Rank | −0.21 | 0.90 | 0.82 | 0.76 | |||
| Huddle Betweenness:Rank | 0.58 | 2.98 | 0.85 | 0.86 | |||
| B7 | E. coli betweenness | (Intercept) | 5.30 | 0.40 | <0.01** | 93 | |
| Groom betweenness | −3.56 | 1.91 | 0.06 | 0.06 | |||
| Huddle betweenness | −2.66 | 1.58 | 0.09 | 0.10 | |||
| Groom betweenness: Huddle betweenness | 15.26 | 6.43 | 0.02* | 0.01* |
Models in bold indicate those where the permuted P values, obtained by comparing the observed β coefficient with a distribution of 1000 coefficients generated by randomly swapping the nodes of the transmission network, reached significance (Ppermuted < 0.05).
P < 0.05;
P < 0.01.
Figure 4.
A node-swapping permutation test indicating the significant relationship between E. coli transmission betweenness centrality and the interaction between social grooming and huddling betweenness (Model B7) (Aim 3). The dotted line indicates the value of the coefficient from the original data against a null distribution of coefficients generated from 1000 permutations of node IDs.
DISCUSSION
In this study, we used the phylogenetic relationships of an enteric gut microbe to examine whether contact affiliative behaviours may mediate infectious agent transmission among captive rhesus macaques. We found that gut E. coli isolated directly from macaques were more genotypically similar to each other than to environmental isolates, which established a premise for contact transmission. Through longitudinal sampling and comparison, we then revealed that the frequencies of dyadic social grooming and huddling interactions among macaques positively predicted the likelihood of E. coli transmission, within one of two sampling periods. Finally, we found strong evidence for the ‘superspreader’ hypotheses: macaques that were the most well connected or central in both their grooming and their huddling networks were also the most well connected or central in their E. coli transmission network. We discuss these findings below and their relevance for understanding the links between social life and disease risk from evolutionary and conservation perspectives.
Gastrointestinal infectious agents may be transmitted through faecal–oral contact routes, by direct contact with a conspecific (e.g. Hamede, Bashford, McCallum, & Jones, 2009; MacIntosh et al., 2012; Otterstatter & Thomson, 2007), or indirectly in the environment (e.g. Fenner et al., 2011; Huffman et al., 2013; VanderWaal, Atwill, Isbell et al., 2014). Our comparisons of E. coli genotypic profiles revealed that, at all three sampling events, bacteria from rhesus macaque faecal swabs were more genotypically similar to each other than they were to bacteria from environmental faeces. Moreover, the pattern of E. coli genotypic diversity within and across sampling events revealed that there was a high turnover in macaque E. coli strains. In the majority of macaques, the same individual showed different E. coli strains across temporally adjacent sampling events. We also detected identical E. coli strains among different individuals either across sampling events or within the same sampling event that followed a period of behavioural observations. Linking such identical strains yielded a fully connected E. coli transmission network across the 12-week data collection period. This suggested that the overall genotypic diversity of E. coli appeared to be conserved within the group, with the same strains likely being transmitted among group members. Our previous work that showed ‘group membership’ had a strong effect on the population genetic structure of E. coli across three rhesus macaque groups also supports this argument (Balasubramaniam et al., 2018). Together, these lines of evidence established a premise to expect horizontal socially mediated acquisition and transmission of E. coli within our study group.
In one of the two study periods, we found that dyads with frequent affiliative interactions were the most likely to show genotypically identical E. coli. This is consistent with a previous study on free-living giraffes which, to our knowledge, is the only other study to reveal evidence for such contact-mediated microbial transmission among dyads (VanderWaal, Atwill, Isbell et al., 2013). In comparison to giraffes, rhesus macaques engage in more frequent contact affiliative interactions such as grooming and huddling, and with a wider range of conspecifics (Cords, 2013; Fooden, 2000; Sade, 1972a, b; Thierry, 2007); these may be especially more frequent in captivity where individuals come into closer proximity with each other (Judge & de Waal, 1997). Under such conditions, it is conceivable that the sharing of gut microbes is more readily discernible at higher organizational scales, such as among groups or communities of closely interacting individuals, rather than among dyads (Balasubramaniam et al., 2018). Thus, our ability to detect dyadic socially mediated transmission under these conditions may be somewhat unique and was likely facilitated by our longitudinal sampling approach that compared the phylogenetic similarity of E. coli across sampling time points. In free-living giraffes, VanderWaal, Atwill, Isbell et al. (2013) revealed that social networks were also strongly correlated to a network of shared space use, prompting speculation that E. coli transmission was more likely an outcome of spatiotemporal synchrony in animals’ environmental sources like watering-hole use patterns, rather than infrequently occurring tactile or other physical contact events. Our captive macaque group was exposed to consistent, similarly hot and dry environmental conditions, which are generally deemed unfavourable for the survival of E. coli (Habteselassie et al., 2008; Sinton et al., 2007; Van Elsas et al., 2011). Furthermore, artificial water sources within the enclosure tested negative for E. coli (Balasubramaniam et al., 2018). For these reasons, dyadic E. coli sharing in the macaques was more likely due to affiliative social contact rather than shared space use.
We found no effect of matriline membership on the likelihood of dyadic transmission, which suggests that our results were likely unaffected by potential vertical transmission of E. coli from mothers to their offspring. This is consistent with findings on baboons (Papio cynocephalus) that evidenced socially mediated sharing of gut microbiota independent of kinship (Tung et al., 2015), but contrasts other work on African elephants that reveals a strong effect of genetic relatedness on E. coli transmission (Chiyo et al., 2014). We speculate that in frequently interacting species like cercopithecine primates, horizontal contact transmission might mask the detectability of vertical transmission. Yet, since these primates, and in particular rhesus macaques, show strong matrilineal kin bias in affiliative contact patterns (Berman, 2011; Berman & Kapsalis, 2009), matriline membership might it-self affect the horizontal, social contact-mediated transmission of E. coli. So, our finding that kinship did not influence E. coli transmission is somewhat surprising but might be explained by the composition of the study group. Our group was composed of many small matrilines (26 matrilines; size range 1–8 individuals; see also Balasubramaniam et al., 2016), resulting in a disproportionately high percentage of nonkin dyads (4533 out of 4656, or 97.4%) compared to close-kin dyads (123 out of 4656, or 2.6%). We speculate that such small matriline sizes and the limited availability of close kin may increase individuals’ tendencies to seek out nonkin affiliation partners. In general, primates like macaques affiliate with nonkin for many reasons, such as social bond investment (Silk, 2014; Silk et al., 2003, 2010) and exchanging grooming for other social benefits (reviewed in Henzi & Barrett, 1999). Such interactions with nonkin might be even more frequent in spatially constrained environments like captive housing, where strong ties of affiliation might mitigate or offset the costs of heightened aggression (Judge & de Waal, 1997). So the lack of effect of kinship on the horizontal transmission of E. coli might be specific to group composition and living condition. Future work to confirm this should focus on animal groups with higher relationship coefficients.
Unlike in phase II, we detected no evidence for behaviourally mediated dyadic transmission in phase I. This may be due to the smaller number of total transmission links in the phase I network compared to the phase II network. It is also possible that greater frequency and diversity in macaque affiliation patterns may have masked the detectability of dyadic transmission in phase I, since the macaques engaged in more contact affiliative interactions with a wider range of partners in phase I compared to phase II (e.g. grooming: Wilcoxon signed-rank test: Z = 7.49, P < 0.01). This may have generated an even more frequent turnover of E. coli among the macaques in phase I, requiring longitudinal sampling within a shorter time frame to better detect transmission. More generally, our findings highlight the importance of possessing a priori knowledge of both microbe-specific characteristics (e.g. transmissibility, turnover rate) and host behaviours (e.g. the nature and frequency of affiliative contact) in order to formulate system-specific sampling paradigms for detecting dyadic transmission (see also below).
Across the 12-week data collection period, we found that macaques’ weighted degree and betweenness in their social networks strongly predicted their degree and betweenness in the E. coli transmission network. Whereas previous studies of other animal populations report a single type of host behaviour that may mediate E. coli transmission (e.g. home range use among African elephants: Chiyo et al., 2014; intergroup encounters in Verreaux’s sifakas: Springer et al., 2016; tactile contact in giraffes: VanderWaal, Atwill, Isbell et al., 2013), we found that E. coli transmission in rhesus macaques may occur through two inter-acting types of affiliative contact: grooming and huddling. In primates, increased centrality or connectedness in grooming networks has been previously linked to the incidence or diversity of gastro-intestinal (GIT) endoparasites (e.g. Japanese macaques: MacIntosh et al., 2012; brown spider monkeys: Rimbach et al., 2015). Contact huddling has been associated with the increased likelihood of infection from a pathogenic gut bacterium in this macaque group (Balasubramaniam et al., 2016). These studies have been able to speculate whether transmission may have occurred, but have stopped short of establishing that transmission did occur. Here we extend such findings by implementing longitudinal behavioural sampling and genotypic comparisons to establish the socially mediated transmission of commensal gut E. coli, which is considered a classic model microbe for detecting the potential transmission route(s) for a suite of gastrointestinal pathogens (e.g. enteric bacteria) and parasites (e.g. protozoans and nematodes) (Chiyo et al., 2014; VanderWaal, Atwill, Isbell et al., 2014; VanderWaal & Ezenwa, 2016).
Our results also indicate that macaques’ sex and dominance rank may influence E. coli transmission patterns. With respect to sex differences, huddling degree had a stronger positive correlation with E. coli degree among females compared to males. This may be due to features of rhesus macaque social organization. Female rhesus remain in their natal groups their entire lives (Cords, 2013; Pusey & Packer, 1987; Sade, 1972a, b) and typically have more social partners and spend more time in affiliative interactions with these partners than do males. In contrast to females, male macaques tend to disperse from their natal groups (Pusey & Packer, 1987). Although captivity prevents natural dispersal, young males may still exhibit greater exploratory tendencies than females in captive conditions in anticipation of dispersal opportunities (Judge & de Waal, 1997). Thus, male dispersal and associated tendencies for greater environmental exploration may explain our finding that males were more likely to acquire E. coli from environmental faeces than were females. Environmental acquisition may have masked or counteracted the detectability of socially mediated transmission among males. In general, males in many taxa may function as microbial transmitters between rather than within groups, due to their tendencies to disperse between groups (e.g. African elephants: Chiyo et al., 2014; grey-cheeked mangabeys, Lophocebus albigena: Arlet et al., 2015) or between communities (e.g. juvenile Belding’s ground squirrels: VanderWaal, Atwill, Hooper et al., 2013), or participate in intergroup encounters (e.g. Verreaux’s si-fakas: Springer et al., 2016).
With respect to rank-related differences in E. coli transmission, grooming degree was more strongly related to E. coli degree among high-ranking individuals. In many primate societies, grooming is preferentially directed towards higher-ranking individuals (Schino, 2001; Schino & Aureli, 2008; Seyfarth, 1977), and this could increase their exposure to infectious agents via social contact (MacIntosh et al., 2012). However, infectious agent risk is further complicated by the complex relationship between rank and social stress-induced susceptibility to infection. Although it is well established that social and psychological stress increase disease risk among mammals (Bartolomucci, 2007; Cohen, 2004; Cohen et al., 2007; Klein, 2000a, b), rank can impact stress in nonlinear ways (Sapolsky, 2005; Vandeleest et al., 2016). So, although previous studies have shown that dominance rank influences parasite risk in primates (Hausfater & Watson, 1976; MacIntosh et al., 2012; Muehlenbein, 2006; Muller-Graf, Collins, & Woolhouse, 1996), they have largely been unable to disentangle the relative effects of contact-mediated transmission from stress-induced acquisition (but see MacIntosh et al., 2012). Here our focus is on a commensal rather than a pathogenic microbe, possibly ruling out stress-induced susceptibility, which if true suggests that grooming promoted direct social contact-mediated microbial transmission more so among high-ranking compared to low-ranking macaques.
Our conclusions are unlikely to have been affected by our microbial sampling approach. One concern is related to the time window for microbial transmission to occur. We divided our 12-week study period into two 6-week phases. We chose a 6-week window to both yield enough behavioural data to build reliable grooming and huddling networks and to detect longitudinal E. coli transmission (see Methods). Our detection of sparsely connected transmission networks at 6 weeks suggested that this was not an optimal window to detect all transmission events. Nevertheless, our finding that social behaviour strongly predicted what may have been a fraction of the transmission links detected within this 6-week window is significant. A second concern is related to our definition and criteria for links in the E. coli transmission network. Our approach of considering dyadic E. coli similarity within and across longitudinally adjacent time points that flanked behavioural data collection is an advancement over previous studies that implemented more opportunistic microbial sampling and comparisons (e.g. Springer et al., 2016; VanderWaal, Atwill, Isbell et al., 2013). A more conservative approach to define the precise pathways of transmission may be to consider just a subset of the links we considered in Fig. 1, which involved two macaques sharing an identical strain at a follow-up time point (e.g. T1) that was also present only in one of the two individuals at the preceding time point (T0). However, this was impossible with our data given the small percentage of individuals for whom we detected identical strains at adjacent time points (6 out of 97, or 6%). Although more frequent microbial sampling at shorter intervals, or sampling multiple E. coli strains from the same macaque at each time point, would likely solve the above concerns, both were budgetarily beyond the scope of this study. These approaches may have only strengthened rather than changed our results, by generating more well-connected transmission networks within shorter time frames. Finally, although our approach highlights the importance of longitudinal sampling in general, specific choices related to sampling time windows and frequencies should remain flexible across studies, since they are affected both by feasibility and by animals’ physiology, socioecology and behaviour (VanderWaal & Ezenwa, 2016).
Our findings expand the current understanding of how social life impacts disease risk, by adding to a growing body of research demonstrating the complex effects of social behaviour on health (Kappeler et al., 2015). The fact that animals’ betweenness centrality in both their grooming and huddling networks strongly influenced their betweenness centrality in the E. coli transmission network supports this claim. Betweenness centrality identifies hubs or bridges that connect otherwise fragmented parts of a social network (Freeman, 1977; Newman, 2005; Wasserman & Faust, 1994), and social hubs are key for the maintenance of group cohesion and social stability and for the dissemination of socially acquired or learnt behaviours (Brent, 2015; Kanngiesser, Sueur, Riedl, Grossmann, & Call, 2011). Yet these same individuals, functioning as conduits of infectious agents, may be ‘superspreaders’ of microbial pathogens when they are infected and actively shedding the agent (Drewe & Perkins, 2015; Lloyd-Smith et al., 2005; VanderWaal & Ezenwa, 2016). So, from a conservation perspective, our findings indicate that pre-emptive control of the flow of infectious diseases may be possible if such superspreaders in a transmission network are targeted for disease control strategies like vaccination or antibiotic treatment (VanderWaal & Ezenwa, 2016).
Acknowledgments
We thank our dedicated team from the McCowan Animal Behavior Laboratory, including A. Nathman, A. Barnard, T. Boussina, A. Vitale, E. Cano, J. Greco, N. Sharpe and S. Seil, who participated in the behavioural data collection. We are grateful to members of the Atwill, McCowan and WIFSS (Western Institute for Food Safety and Security) laboratories, particularly C. Bonilla, J. Carabez, A. Maness, R. Pisano, I. Wong and C. Xiao, for playing key roles in the processing of faecal samples for bacterial isolation, characterization and fingerprinting at the School of Veterinary Medicine, University of California Davis. We also thank the Journal editor and two anonymous referees whose comments helped greatly improve the quality of the manuscript. The research was supported by NICHD of the National Institutes of Health under award number R01HD068335.
Appendix
Figure A1.
Box plots of comparisons between macaque–macaque and macaque–environmental E. coli isolates by sex, at sampling time points (a) T0, (b) T1 and (c) T2. Boxes indicate the 25th and 75th percentiles. The heavy line indicates the median. Whiskers represent 1.5 times the interquartile range of the data. P values are from Wilcoxon two-sample tests.
*P < 0.05; **P < 0.01.
Footnotes
Supplementary Material
Supplementary material associated with this article can be found, in the online version, at: https://doi.org/10.1016/j.anbehav.2019.03.009
References
- Alexander RD (1974). The evolution of social behaviour. Annual Review of Ecology and Systematics, 5, 325–383. [Google Scholar]
- Altizer S, Nunn C, Thrall P, Gittleman J, Antonovics J, Cunningham A, et al. (2003). Social organization and parasite risk in mammals: Integrating theory and empirical studies. Annual Review of Ecology, Evolution, and Systematics, 34, 517–547. [Google Scholar]
- Altmann J (1974). Observational study of behavior: Sampling methods. Behaviour, 49, 227–266. [DOI] [PubMed] [Google Scholar]
- Arlet ME, Chapman CA, Isbell LA, Mollerman F, Mand R, Horak P, et al. (2015). Social and ecological correlates of parasitic infections in adult male gray-cheeked mangabeys (Lophocebus albigena). International Journal of Primatology, 36, 967–986. [Google Scholar]
- Balasubramaniam KN, Beisner BA, Guan J, Vandeleest J, Fushing H, Atwill ER, et al. (2018). Social network community structure is associated with the sharing of commensal E. coli among captive rhesus macaques (Macaca mulatta). PeerJ, 6 e4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramaniam KN, Beisner BA, Vandeleest J, Atwill ER, & McCowan B (2016). Social buffering and contact transmission: Network connections have beneficial and detrimental effects on Shigella infection risk among captive rhesus macaques. PeerJ, 4 e2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramaniam KN, Dittmar K, Berman CM, Butovskaya M, Cooper MA, Majolo B, et al. (2012). Hierarchical steepness and phylogenetic models: Phylogenetic signals in Macaca. Animal Behaviour, 83, 1207–1218. [Google Scholar]
- Bartolomucci A (2007). Social stress, immune functions and disease in rodents. Frontiers in Neuroendocrinology, 28, 28–49. [DOI] [PubMed] [Google Scholar]
- Bates D, Maechler M, Bolker B, Walker S, Christensen RHB, Singmann H, et al. (2016). Linear mixed-effects models using ‘eigen’ and S4 (R package Version 1.1–21). https://CRAN.R-project.org/package = lme4. [Google Scholar]
- Berman CM (2011). Kinship: Family ties and social behavior In Campbell CJ, Fuentes A, MacKinnon KC, Panger M, & Bearder SK (Eds.), Primates in perspective (2nd ed, pp. 576–587). New York, NY: Oxford University Press. [Google Scholar]
- Berman CM, & Kapsalis E (2009). Variation over time in grooming kin bias among female rhesus macaques on Cayo Santiago supports the time constraints hypothesis. American Journal of Physical Anthropology, 48, 89–90.’ [Google Scholar]
- Berman CM, & Thierry B (2010). Variation in kin bias: Species differences and time constraints in macaques. Behaviour, 147(13), 1863–1887. [Google Scholar]
- Blyton MDJ, Banks SC, Peakall R, Lindenmayer DB, & Gordon DM (2014). Not all types of host contacts are equal when it comes to E. coli transmission. Ecology Letters, 17, 917–978. [DOI] [PubMed] [Google Scholar]
- Brent LJN (2015). Friends of friends: Are indirect connections in social networks important to animal behaviour? Animal Behaviour, 103, 211–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent LJN, Lehmann J, & Ramos-Fernandez G (2011). Social network analysis in the study of nonhuman primates: A historical perspective. American Journal of Primatology, 73(8), 720–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham KP, Anderson DR, & Huyvaert KP (2011). AIC model selection and multimodel inference in behavioral ecology: Some background, observations, and comparisons. Behavioral Ecology and Sociobiology, 65(1), 23–35. [Google Scholar]
- Butts CT (2008). Social network analysis with sna. Journal of Statistical Software, 24(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter G, & Leffer L (2015). Social grooming in bats: Are vampire bats exceptional? PLoS One, 10 e0138430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesaris L, Gillespie BE, Srinivasan V, Almeida RA, Zecconi A, & Oliver SP (2007). Discriminating between strains of Escherichia coli using pulse-field gel electrophoresis and BOX-PCR. Foodborne Pathogens and Disease, 4, 473–480. [DOI] [PubMed] [Google Scholar]
- Chiyo PI, Grieneisen LE, Wittemyer G, Moss CJ, Lee PC, Douglas-Hamilton I, et al. (2014). The influence of social structure, habitat, and host traits on the transmission of Escherichia coli in wild elephants. PLoS One, 9 e93408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clutton-Brock T (2016). Mammal societies. Chichester, U.K.: Wiley Blackwell. [Google Scholar]
- Cobb S (1976). Social support as a moderator of life stress. Psychosomatic Medicine, 38, 300–314. [DOI] [PubMed] [Google Scholar]
- Cohen S (2004). Social relationships and health. American Psychologist, 59, 676–684. [DOI] [PubMed] [Google Scholar]
- Cohen S, Janicki-Deverts D, & Miller GE (2007). Psychological stress and disease. Journal of the American Medical Association, 298, 1685–1687. [DOI] [PubMed] [Google Scholar]
- Cohen S, Janicki-Deverts D, Turner RB, & Doyle WJ (2015). Does hugging provide stress-buffering social support? A study of susceptibility to upper respiratory infection and illness. Psychological Science, 26, 135–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cords M (2013). The behavior, ecology, and social evolution of Cercopithecine monkeys In Mitani JC, Call J, Kappeler PM, Palombit RA, & Silk JB (Eds.), The evolution of primate societies (pp. 91–112). Chicago, IL: University of Chicago Press. [Google Scholar]
- Craft ME (2015). Infectious disease transmission and contact networks in wildlife and livestock. Philosophical Transactions of the Royal Society B: Biological Sciences, 370, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craft ME, & Caillaud D (2011). Network models: An underutilized tool in wildlife epidemiology. Interdisciplinary Perspectives on Infectious Diseases, 2011, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker D, Krackhardt D, & Snijders TAB (2007). Sensitivity of MR-QAP tests to collinearity and autocorrelation conditions. Psychometrika, 72, 563–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewe JA (2010). Who infects whom? Social networks and tuberculosis transmission in wild meerkats. Proceedings of the Royal Society B: Biological Sciences, 277, 633–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewe JA, & Perkins SE (2015). Disease transmission in animal social networks In Krause J, James R, Franks DW, & Croft DP (Eds.), Animal social networks (pp. 95–110). Oxford, U.K.: Oxford University Press. [Google Scholar]
- Duboscq J, Romano V, Sueur C, & MacIntosh AJJ (2016). Network centrality and seasonality interact to predict lice load in a social primate. Scientific Reports, 6, 22095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farine DR (2017). A guide to null models for animal social network analysis. Methods in Ecology and Evolution, 8, 1309–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farine DR, & Whitehead H (2015). Constructing, conducting and interpreting animal social network analysis. Journal of Animal Ecology, 84, 1144–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenner AL, Godfrey SS, & Bull MC (2011). Using social networks to deduce whether residents or dispersers spread parasites in a lizard population. Journal of Animal Ecology, 80, 835–843. [DOI] [PubMed] [Google Scholar]
- Ferrin DL, Dirks KT, & Shah PP (2006). Direct and indirect effects of third-party relationships on interpersonal trust. Journal of Applied Psychology, 91, 870–883. [DOI] [PubMed] [Google Scholar]
- Fooden J (2000). Systematic review of the rhesus macaque, Macaca mulatta (Zimmermann, 1780). Chicago, IL: Field Museum of Natural History. [Google Scholar]
- Freeland WJ (1976). Pathogens and the evolution of primate sociality. Biotropica, 8, 12–24. [Google Scholar]
- Freeman LC (1977). A set of measures of centrality based on betweenness. Sociometry, 40, 35–41. [Google Scholar]
- Friant S, Ziegler TE, & Goldberg TL (2016). Primate reinfection with gastrointestinal parasites: Behavioural and physiological predictors of parasite acquisition. Animal Behaviour, 117, 105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii K, Jin J, Shev A, Beisner B, McCowan B, & Fushing H (2016). Perc: Using percolation and conductance to find information flow certainty in a direct network (R package version 0.1.2). https://CRAN.R-project.org/package = Perc. [Google Scholar]
- Fushing H, McAssey MP, Beisner B, & McCowan B (2011). Ranking network of a captive rhesus macaque society: A sophisticated corporative kingdom. PLoS One, 6, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons DE (2004). Friendship and advice networks in the context of changing professional values. Administrative Science Quarterly, 49, 238–262. [Google Scholar]
- Godfrey SS (2013). Networks and the ecology of parasite transmission: A framework for wildlife parasitology. Parasites and Wildlife, 2, 235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey SS, Moore JA, Nelson NJ, & Bull CM (2010). Social network structure and parasite infection patterns in a territorial reptile, the tuatara (Sphenodon punctatus). International Journal for Parasitology, 40, 1575–1585. [DOI] [PubMed] [Google Scholar]
- Goldberg T, Gillespie TR, & Singer RS (2006). Optimization of analytical parameters for inferring relationships among Escherichia coli isolates from repetitive-element PCR by maximizing correspondence with multilocus sequence typing data. Applied and Environmental Microbiology, 72, 6049–6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good RC, May BD, & Kawatomari T (1969). Enteric pathogens in monkeys. Journal of Bacteriology, 97, 1048–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habteselassie M, Bischoff M, Blume E, Applegate B, Reuhs B, Brouder S, et al. (2008). Environmental controls of the fate of Escherichia coli in soil. Water, Air, & Soil Pollution, 190, 143–155. [Google Scholar]
- Hamede RK, Bashford J, McCallum H, & Jones M (2009). Contact networks in a wild Tasmanian devil (Sarcophilus harrisii) population: Using social network analysis to reveal seasonal variability in social behaviour and its implications for transmission of devil facial tumour disease. Ecology Letters, 12(11), 1147–1157. [DOI] [PubMed] [Google Scholar]
- Hanneman RA, & Riddle M (2005). Introduction to social network methods. Riverside, CA: University of California Press. [Google Scholar]
- Hart BL, & Hart L (1992). Reciprocal allogrooming in impala. Animal Behaviour, 44, 1073–1083. [Google Scholar]
- Hausfater G, & Watson DF (1976). Social and reproductive correlates of parasite ova emissions by baboons. Nature, 262, 688–689. [DOI] [PubMed] [Google Scholar]
- Henzi SP, & Barrett L (1999). The value of grooming to female primates. Primates, 40(1), 47–59. [DOI] [PubMed] [Google Scholar]
- Hinde RA (1976). Interactions, relationships and social structure. Man, 11, 1–17. [Google Scholar]
- Hubert LJ (1987). Assignment methods in combinatorial data analysis. New York, NY: Marcel Dekker. [Google Scholar]
- Huffman MA, Nahallage CAD, Hasegawa H, Ekanayake S, De Silva LDGG, & Athauda IRK (2013). Preliminary survey of the distribution of four potentially zoonotic parasite species among primates in Sri Lanka. Journal of the National Foundation of Science of Sri Lanka, 41, 319–326. [Google Scholar]
- Hussain S, Ram MS, Kumar A, Shivaji S, & Umapathy G (2013). Human presence increases parasitic load in endangered lion-tailed macaques (Macaca silenus) in its fragmented rainforest habitats in southern India. PLoS One, 8 e63685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judge PG, & de Waal FBM (1997). Rhesus monkey behaviour under diverse population densities: Coping with long-term crowding. Animal Behaviour, 54, 643–662. [DOI] [PubMed] [Google Scholar]
- Kanngiesser P, Sueur C, Riedl K, Grossmann J, & Call J (2011). Grooming network cohesion and the role of individuals in a captive chimpanzee group. American Journal of Primatology, 73, 758–767. [DOI] [PubMed] [Google Scholar]
- Kaplan JR, Heise ER, Manuck SB, Shively CA, Cohen S, Rabin BS, et al. (1991). The relationship of agonistic and affiliative behavior patterns to cellular immune function among cynomolgus monkeys Macaca fascicularis living in unstable social groups. American Journal of Primatology, 25(3), 157–174. [DOI] [PubMed] [Google Scholar]
- Kappeler PM (2000). Primate males: Causes and consequences of variation in group composition. Cambridge, U.K.: Cambridge University Press. [Google Scholar]
- Kappeler PM, Cremer S, & Nunn CL (2015). Sociality and health: Impacts of sociality on disease susceptibility and transmission in animal and human societies. Philosophical Transactions of the Royal Society B: Biological Sciences, 370, 20140116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappeler PM, & Van Schaik CP (2002). Evolution of primate social systems. International Journal of Primatology, 23, 707–740. [Google Scholar]
- Kilonzo C, Atwill ER, Mandrell R, Garrick M, & Villanueva V (2011). Prevalence and molecular characterization of Escherichia coli O157:H7 by multiple-locus variable-number tandem repeat analysis and pulsed-field gel electrophoresis in three sheep farming operations in California. Journal of Food Protection, 74, 1413–1421. [DOI] [PubMed] [Google Scholar]
- Klein SL (2000a). The effects of hormones on sex differences in infection: From genes to behavior. Neuroscience & Biobehavioral Reviews, 24, 627–638. [DOI] [PubMed] [Google Scholar]
- Klein SL (2000b). Hormones and mating system affect sex and species differences in immune function among vertebrates. Behavioural Processes, 51, 149–166. [DOI] [PubMed] [Google Scholar]
- Kondo S, Hoar BR, Mandrell R, & AE R (2010). Longitudinal prevalence and molecular typing of Escherichia coli O157:H7 using multiple-locus variable-number tandem-repeats analysis and pulsed field gel electrophoresis in a range cattle herd in California. American Journal of Veterinary Research, 71, 1339–1347. [DOI] [PubMed] [Google Scholar]
- Krackhardt D (1987). QAP partialling as a test of spuriousness. Social Networks, 9, 171–186. [Google Scholar]
- Ley RE, Lozupone CA, Hamady M, Knight R, & Gordon JI (2008). Worlds within worlds: Evolution of the vertebrate gut microbiota. Nature Reviews Microbiology, 6(10), 776–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Smith JO, Schreiber SJ, Kopp PE, & Getz WM (2005). Superspreading and the effect of individual variation on disease emergence. Nature, 438(7066), 355–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusseau D, Whitehead H, & Gero S (2008). Incorporating uncertainty into the study of animal social networks. Animal Behaviour, 75, 1809–1815. [Google Scholar]
- MacIntosh AJJ, & Frias L (2017). Coevolution of hosts and parasites In Bezanson M, MacKinnon KC, Riley E, Campbell CJ, Nekaris KAI, Estrada A, et al. (Eds.), The international encyclopedia of primatology (pp. 188–196). Chichester, U.K.: John Wiley. [Google Scholar]
- MacIntosh AJJ, Jacobs A, Garcia C, Shimizu K, Mouri K, Huffman MA, et al. (2012). Monkeys in the middle: Parasite transmission through a social network of a wild primate. PLoS One, 7, e51144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCord AI, Chapman CA, Weny G, Tumukunde A, Hyeroba D, Klotz K, et al. (2014). Fecal microbiomes of non-human primates in Western Uganda reveal species-specific communities largely resistant to habitat perturbation. American Journal of Primatology, 76(4), 347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCowan B, Anderson K, Heagarty A, & Cameron A (2008). Utility of social network analysis for primate behavioral management and well-being. Applied Animal Behaviour Science, 109, 396–405. [Google Scholar]
- Moore D, Angel JE, Cheeseman IM, Robinson GE, & Fahrbach SE (1995). A highly specialized social grooming honey bee (Hymenoptera: Apidae). Journal of Insect Behavior, 8, 855–861. [Google Scholar]
- Muehlenbein MP (2006). Intestinal parasite infections and fecal steroid levels in wild chimpanzees. American Journal of Physical Anthropology, 130, 546–550. [DOI] [PubMed] [Google Scholar]
- Muller-Graf CDM, Collins DA, & Woolhouse MEJ (1996). Intestinal parasite burden in five troops of olive baboons (Papio cynocephalus anubis) in Gombe Stream National Park, Tanzania. Parasitology, 112, 489–497. [DOI] [PubMed] [Google Scholar]
- Newman MEJ (2005). A measure of betweenness centrality based on random walks. Social Networks, 27, 39–54. [Google Scholar]
- Nunn CL, Thrall PH, Leendertz FH, & Boesch C (2011). The spread of fecally transmitted parasites in socially structured populations. PLoS One, 6 e21677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opsahl T, Agneessens F, & Skvoretz J (2010). Node centrality in weighted networks: Generalizing degree and shortest paths. Social Networks, 32(3), 245–251. [Google Scholar]
- Otterstatter MC, & Thomson JD (2007). Contact networks and transmission of an intestinal pathogen in bumble bee (Bombus impatiens) colonies. Oecologica, 154, 411–421. [DOI] [PubMed] [Google Scholar]
- Pusey AE, & Packer C (1987). Dispersal and philopatry In Smuts BB, Cheney DL, Seyfarth RM, Wrangham RW, & Struhsaker TT (Eds.), Primate societies (pp. 250–266). Chicago, IL: Chicago University Press. [Google Scholar]
- Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, et al. (2006). Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathogens and Disease, 3(1), 59–67. [DOI] [PubMed] [Google Scholar]
- Richards SA, Whittingham MJ, & Stephens PA (2011). Model selection and model averaging in behavioural ecology: The utility of the IT-AIC framework. Behavioral Ecology and Sociobiology, 65(1), 77–89. [Google Scholar]
- Rimbach R, Bisanzio D, Galvis N, Link A, Di Fiore A, & Gillespie TR (2015). Brown spider monkeys (Ateles hybridus): A model for differentiating the role of social networks and physical contact on parasite transmission dynamics. Philosophical Transactions of the Royal Society B: Biological Sciences, 370, 20140110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushmore J, Bisanzio D, & Gillespie TR (2017). Making new connections: Insights from primateeparasite networks. Trends in Parasitology, 33, 547–560. [DOI] [PubMed] [Google Scholar]
- Sade D (1972a). Sociometrics of Macaca mulatta I. Linkages and cliques in grooming matrices. Folia Primatologica, 18, 196–223. [DOI] [PubMed] [Google Scholar]
- Sade DS (1972b). A longitudinal study of social behavior of rhesus monkeys In Tuttle R (Ed.), The functional and evolutionary biology of primates (pp. 378–398). Chicago, IL: Aldine Atherton. [Google Scholar]
- Sapolsky RM (2005). The influence of social hierarchy on primate health. Science, 308, 648–652. [DOI] [PubMed] [Google Scholar]
- Schino G (2001). Grooming, competition and social rank among female primates: A meta-analysis. Animal Behaviour, 62, 265–271. [Google Scholar]
- Schino G, & Aureli F (2008). Tradeoffs in primate grooming reciprocation: Testing behavioral flexibility and correlated evolution. Biological Journal of the Linnean Society, 95, 439–446. [Google Scholar]
- Sears HJ, Brownlee I, & Uchiyama JK (1950). Persistence of individual strains of Escherichia coli in the intestinal tract of man. Journal of Bacteriology, 59, 293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears HJ, Janes H, Saloum R, Brownlee I, & Lamoreaux LF (1956). Persistence of individual strains of Escherichia coli in man and dog under varying conditions. Journal of Bacteriology, 71, 370–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segerstrom SC, & Miller GE (2004). Psychological stress and the human immune system: A meta-analytic study of 30 years of inquiry. Psychological Bulletin, 130, 601–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfarth RM (1977). A model of social grooming among adult female monkeys. Journal of Theoretical Biology, 65, 671–698. [DOI] [PubMed] [Google Scholar]
- Silk JB (2014). Evolutionary perspectives on the links between close social bonds, health, and fitness In Weinstein M, & Lane MA (Eds.), Sociality, hierarchy, health: Comparative biodemography: A collection of papers (pp. 120–143). Washington, D.C.: National Academies Press. [PubMed] [Google Scholar]
- Silk JB, Alberts SC, & Altmann J (2003). Social bonds of female baboons enhance infant survival. Science, 302, 1231–1234. [DOI] [PubMed] [Google Scholar]
- Silk JB, Beehner JC, Bergman C, Crockford AL, Engh LR, Moscovice RM, et al. (2010). Strong and consistent social bonds enhance the longevity of female baboons. Current Biology, 20, 1359–1361. [DOI] [PubMed] [Google Scholar]
- Sinton L, Hall C, & Braithwaite R (2007). Sunlight inactivation of Campylobacter jejuni and Salmonella enterica, compared with Escherichia coli, in seawater and river water. Journal of Water and Health, 5, 357–365. [DOI] [PubMed] [Google Scholar]
- Smutt K, MacLarnon A, Heistermann M, & Semple S (2007). Grooming in Barbary macaques: Better to give than to receive? Biology Letters, 3(3), 231–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer A, Mellmann A, Fichtel C, & Kappeler PM (2016). Social structure and Escherichia coli sharing in a group-living wild primate, Verreaux’s sifaka. BMC Ecology, 16, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueur C, Jacobs A, Amblard F, Petit O, & King AJ (2011). How can social network analysis improve the study of primate behavior? American Journal of Primatology, 73, 703–719. [DOI] [PubMed] [Google Scholar]
- Tanaka I, & Takefushi H (1993). Elimination of external parasites (lice) is the primary function of grooming in free-ranging Japanese macaques. Anthropological Science, 101, 187–193. [Google Scholar]
- Tenaillon O, Skurnik D, Picard B, & Denamur E (2010). The population genetics of commensal Escherichia coli. Nature Reviews Microbiology, 8, 207–217. [DOI] [PubMed] [Google Scholar]
- Thierry B (2007). Unity in diversity: Lessons from macaque societies. Evolutionary Anthropology, 16, 224–238. [Google Scholar]
- Tung J, Barreiro LB, Burns MB, Grenier J, Lynch J, Grieneisen LE, et al. (2015). Social networks predict gut microbiome composition in wild baboons. eLife, 4 e05224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Elsas JD, Semenov AV, Costa R, & Trevors JT (2011). Survival of Escherichia coli in the environment: Fundamental and public health aspects. IMSE Journal, 5, 173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeleest JV, Beisner BA, Hannibal D, Nathman A, Capitanio J, Fushing H, et al. (2016). Rank and dominance certainty influence multiple health outcomes in rhesus macaques (Macaca fuscata) (Manuscript submitted for publication). [Google Scholar]
- VanderWaal KL, Atwill ER, Hooper S, Buckle K, & McCowan B (2013a). Network structure and prevalence of Cryptosporidium in Belding’s ground squirrels. Behavioral Ecology and Sociobiology, 67(12), 1951–1959. [Google Scholar]
- VanderWaal KL, Atwill ER, Isbell LA, & McCowan B (2013b). Linking social and pathogen transmission networks using microbial genetics in giraffe (Giraffa camelopardalis). Journal of Animal Ecology, 83, 406–414. [DOI] [PubMed] [Google Scholar]
- VanderWaal KL, Atwill ER, Isbell LA, & McCowan B (2014a). Quantifying microbe transmission networks for wild and domestic ungulates in Kenya. Biological Conservation, 169, 136–146. [Google Scholar]
- VanderWaal KL, & Ezenwa VO (2016). Heterogeneity in pathogen transmission: Mechanisms and methodology. Functional Ecology, 30, 1606–1622. [Google Scholar]
- VanderWaal KL, Wang H, McCowan B, Fushing H, & Isbell L (2014b). Multilevel social organization and space use in reticulated giraffe (Giraffa camelopardis). Behavioral Ecology, 25, 17–26. [Google Scholar]
- Wasserman S, & Faust K (1994). Social network analysis: Methods and applications. Cambridge, U.K.: Cambridge University Press. [Google Scholar]
- Young C, Majolo B, Heistermann M, Schulke O, & Ostner J (2014). Responses to social and environmental stress are attenuated by strong male bonds in wild macaques. Proceedings of the National Academy of Sciences of the United States of America, 111, 18195–18200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagenczyk TJ, Gibney R, Few WT, & Purvis RL (2013). The ties that influence: A social network analysis of prototypical employees’ effects on job attitudes among coworkers. Journal of Management Policy and Practice, 14, 27–42. [Google Scholar]