Abstract
The microbiome and pregnancy are known to alter drug disposition, yet the interplay of the two physiologic factors on the expression and/or activity of drug metabolizing enzymes and transporters (DMETs) is unknown. This study investigated the effects of microbiome on host hepatic DMETs in mice during pregnancy by comparing four groups of conventional (CV) and germ-free (GF) female mice and pregnancy status, namely, CV nonpregnant, GF non-pregnant, CV pregnant, and GF pregnant mice. Transcriptomic and targeted proteomics of hepatic DMETs were profiled by using multiomics. Plasma bile acid and steroid hormone levels were quantified by liquid chromatography tandem mass spectrometry. CYP3A activities were measured by mouse liver microsome incubations. The trend of pregnancy-induced changes in the expression or activity of hepatic DMETs in CV and GF mice was similar; however, the magnitude of change was noticeably different. For certain DMETs, pregnancy status had paradoxical effects on mRNA and protein expression in both CV and GF mice. For instance, the mRNA levels of Cyp3a11, the murine homolog of human CYP3A4, were decreased by 1.7-fold and 3.3-fold by pregnancy in CV and GF mice, respectively; however, the protein levels of CYP3A11 were increased similarly ∼twofold by pregnancy in both CV and GF mice. Microsome incubations revealed a marked induction of CYP3A activity by pregnancy that was 10-fold greater in CV mice than that in GF mice. This is the first study to show that the microbiome can alter the expression and/or activity of hepatic DMETs in pregnancy.
SIGNIFICANCE STATEMENT
We demonstrated for the first time that microbiome and pregnancy can interplay to alter the expression and/or activity of hepatic drug metabolizing enzymes and transporters. Though the trend of pregnancy-induced changes in the expression or activity of hepatic drug metabolizing enzymes and transporters in conventional and germ-free mice was similar, the magnitude of change was noticeably different.
Introduction
Drug safety in pregnant women is a critical issue for drug development and regulatory agencies because of observed changes in efficacy and toxicity of medications during pregnancy (Beigi et al., 2016; Food and Drug Administration, 2018). Understanding how drug disposition is changed in pregnant women is therefore of utmost importance (see below). However, there remain gaps in knowledge of other factors that can affect drug disposition in this vulnerable population. The impact of the microbiome on drug disposition could be one such example (Sharma et al., 2019).
Pregnancy changes the maternal body in numerous ways to accommodate the developing fetus. Increased total body volume, decreased plasma albumin, increased cardiac output and renal glomerular filtration rate, and altered expression or activity of drug metabolizing enzymes and transporters (DMETs) profoundly alter drug disposition in pregnant women versus nonpregnant women or men (Helldén and Madadi, 2013; Isoherranen and Thummel, 2013; Pariente et al., 2016; Tasnif et al., 2016). Previous studies have shown increased activity of CYP3A4, CYP2D6, and CYP2C9 and decreased activity of CYP2C19 and CYP1A2 by using respective probe substrates prescribed to women during pregnancy (Feghali et al., 2015). Drug transporters also could have dynamic activity or mRNA expression changes during pregnancy. For example, the activity of P-glycoprotein and organic cation transporter 2 in the kidney appears to be induced during human pregnancy, whereas mRNA expression of multidrug resistance protein 3 (Mrp3/Abcc3) in the mouse liver is downregulated by pregnancy (Isoherranen and Thummel, 2013; Shuster et al., 2013). Pregnancy-related hormones such as progesterone, estradiol, and cortisol, which are ligands of nuclear receptors such as glucocorticoid receptor and pregnane X receptor (PXR), have been proposed to mediate induction of DMETs during pregnancy (Kliewer et al., 1998; Dussault et al., 2003; Papacleovoulou et al., 2011). Therefore, a mechanistic understanding of the changes in expression or activity of DMETs during pregnancy will help optimize the dosing regimens of drugs for improved efficacy and safety for both the mother and her fetus.
The microbiome is often referred to as the second human genome because of the abundance and diversity of bacteria. The microbiome can change drug disposition and toxicity profiles by directly metabolizing and inactivating compounds (Spanogiannopoulos et al., 2016). Indirectly, gut bacteria can alter drug disposition by changing the expression or activity of host DMETs (Spanogiannopoulos et al., 2016). It has been proposed that the predominant mechanism by which bacteria modulate DMET expression is by the modification of secondary bile acids (Claus et al., 2011; Klaassen and Cui, 2015). Primary bile acids are synthesized in the liver from cholesterol and subsequently transported into the bile duct by efflux transporters, such as the bile salt export pump. The bile duct leads primary bile acids into the duodenum where intestinal bacteria deconjugate, dehydroxylate, and epimerize them into secondary bile acids. Intestinal bacteria such as Clostridia from Firmicutes phylum express the enzymes (e.g., hydroxysteroid dehydrogenase and 7-dehydratase) necessary for these biotransformation reactions (Ridlon et al., 2006). The primary bile acids cholic acid (CA) and chenodeoxycholic acid (CDCA) as well as secondary bile acid deoxycholic acid (DCA), and their taurine conjugated forms have been shown to activate farnesoid X receptor (FXR), which regulates bile acid production (Wang et al., 1999; Kong et al., 2012). Lithocholic acid (LCA) has been shown to activate PXR (Staudinger et al., 2001), which in turn upregulates the expression of certain cytochrome P450s (P450). In mice, it has been demonstrated that the primary muricholic acids are antagonists of FXR, and thus the lack of transformation of primary bile acids into secondary bile acids in GF mice would result in accumulation of FXR antagonists (primary bile acids) and reduction of PXR agonists (secondary bile acids); ultimately, this decreases FXR and PXR signaling in the intestine and the liver (Klaassen and Cui, 2015; Wahlström et al., 2017). Recently, it was shown that the maternal gut microbiome shifts as pregnancy progresses in humans, with an increase in the abundance of Actinobacteria and Proteobacteria, which could alter bile acid profiles and DMET expression and/or activity (Nuriel-Ohayon et al., 2016).
Although both pregnancy and the microbiome individually have a profound impact on drug disposition, no studies to date have investigated their combined effects. The objective of this study was to explore the combined effects of pregnancy and the microbiome on the expression and/or activity of hepatic DMETs using CV and GF pregnant mice. We hypothesize that the magnitude and direction of changes in expression and/or activity reflect the combined effects of pregnancy and the microbiome.
Materials and Methods
Chemicals.
Liquid chromatography–mass spectrometry (LC-MS) grade methanol, water, and acetonitrile were purchased from Fisher Scientific (Fair Lawn, NJ). Steroid hormone standards and deuterated internal standards for bile acids (lithocholic acid, deoxycholic acid, cholic acid, glycochenodeoxycholic acid, and glycocholic acid) were purchased from Cerilliant Corporation (Round Rock, TX) and Steraloids Inc. (Newport, RI). All other chemicals were obtained from Thermo Fisher Scientific (Rockford, IL) unless stated otherwise.
Animal Studies.
Conventional C57BL/6J mice (JAX stock 000664) from The Jackson Laboratory (Bar Habor, ME) were habituated for 1 week at our own animal facility prior to experiments. Germ-free (GF) C57BL/6 colony were originated from mice purchased from the National Gnotobiotic Rodent Resource Center of the University of North Carolina at Chapel Hill. The conventional (CV) and GF mice have the same genetic background. All animals were cared for in accordance with the Guide for the Care and Use of Laboratory Animals published by the National Research Council. The animal protocol was approved by the Institutional Animal Care and Use Committee of the University of Washington (protocol 4035-04). GF mice were housed at the University of Washington Gnotobiotic Animal facility in isolators.
Both CV and GF mice were maintained on 12-hour light/dark cycles, and the same autoclaved Breeder Chow 5021 (LabDiet, St. Louis, MO), autoclaved nonacidified water, and autoclaved Enrich-N’Pure bedding (The Andersons Inc., Maumee, OH) were provided ad libitum. CV female mice at 8 weeks of age were mated with CV male mice overnight, and male mice were promptly separated from female mice in the morning. The day with the overnight housing was defined as gestation day (gd) 0. Because of difficulties in achieving pregnancies in GF mice, GF female mice of 11–15 weeks of age were mated with male mice for 72 hours prior to separation from GF male mice. Day 2 after introduction of male mice to the cage was assumed to be gd 0. For the purpose of tissue collection, all pregnant CV or GF mice were kept individually in separate cages during the period of gestation after mating. All pregnant mouse tissues were collected on gd 15, which was previously shown to be the optimal time point to observe peak levels of gene expression changes (Shuster et al., 2013). Nonpregnant female mice of similar ages (8–15 weeks) were housed exactly the same as pregnant mice as described above. On the day of tissue collection, pregnant or nonpregnant female mice were anesthetized with isoflurane, and blood was collected via cardiac puncture and immediately centrifuged (1500g for 10 minutes at 4°C) to isolate plasma. Tissues (e.g., liver) were removed, washed in cold saline, and immediately snap-frozen in liquid N2. In total, plasma samples and tissues from six CV nonpregnant (CVNP), five CV pregnant (CVP), six GF nonpregnant (GFNP), and five GF pregnant (GFP) mice were collected. All samples were then stored at −80°C until analysis.
RNA-Sequencing Analysis.
Total RNA was extracted from liver tissues by using Qiagen RNeasy Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. The concentration of total RNA was determined by using a Synergy HTX Multi-mode Plate Reader (BioTek, Winooski, VT) at 260 nm. Paired-end RNA sequencing (RNA-seq) (2 × 150 bp) was performed by Novogene Bioinformatics Technology Co., Ltd. (Sacramento, CA), using the Illumina NovaSEq. 6000 with an average 20 million reads. The library was prepared by using NEBNext Ultra RNA Library Prep Kit from Illumina. We aligned reads to the mouse GRCm38.p6 transcriptome (Gencode release M19) using the Salmon aligner (v0.11.3) (Patro et al., 2015), and then the transcript-level counts were imported into R and summarized at the gene level by using the Bioconductor tximport package (v1.10.1) (Soneson et al., 2015). Data were subsequently filtered to remove genes that had consistently low expression levels to improve the signal-to-noise ratio by using filterByExpr function in the edgeR package (v3.24.3) implemented in R (v3.5.1). After filtering, 18,849 genes remained. Differential gene expression analysis between groups was performed by using the edgeR package with a negative binomial generalized linear model and quasilikelihood F tests for a given contrast (Robinson et al., 2010; Chen et al., 2016). A false discovery rate (FDR) of 10% was selected to limit the number of false positives (Benjamini and Hochberg, 1995). RNA-seq data presented in this study were deposited in the National Center for Biotechnology Information Gene Expression Omnibus data repository under accession number GSE143391.
Quantitative Real-Time Polymerase Chain Reaction.
Quantitative real-time reverse-transcription polymerase chain reaction (qRT-PCR) was performed to determine the mRNA levels of selected genes, as previously described, to validate RNA sequencing data (Shuster et al., 2013). Aliquots from the same total RNA samples used for RNA-seq analysis were transcribed to cDNAs by using random primers from High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). We selected four enzyme genes for coding (Cyp2b13, Cyp2c50, Cyp3a16, and Sult4a1) and two transporter genes (Abcc3 and Slco1a4) for validation. The genes were selected based on the large magnitude of change in mRNA abundance observed from RNA-seq analysis. The cDNAs were amplified by using SsoAdvanced Universal SYBR Green Supermix and the Bio-Rad CFX384 Real-Time PCR Detection System (Hercules, CA). PCR primers were synthesized by Integrated DNA Technologies (Coralville, IA). Glyceraldehyde-3-phosphate dehydrogenase was used as a housekeeping gene for normalization of the gene expression data.
Targeted Proteomic Analysis of Hepatic DMETs by LC-MS/MS.
Quantification of relative protein abundance of major DMETs in the liver tissues of CV and GF mice was done by quantitative LC-MS/MS proteomic analysis using methods previously described (Bhatt and Prasad, 2018; Liao et al., 2018; Prasad et al., 2018). For P450 enzymes, changes in protein levels of CYP2C37, CYP2C50, CYP2C54, CYP2D22, CYP2D40, CYP3A11, CYP3A16, and CYP3A41 were quantified. For transporters, changes in protein levels of ABCB11, ABCB1A, ABCB1B, ABCC3, ABCG2, SLC22A2, SLC22A7, SLCO1A1, and SLCO1A4 were determined. We used the same procedures for protein isolation and digestion as previously published (Prasad et al., 2018). Briefly, approximately 50 mg of frozen liver tissue per mouse was homogenized in 2 ml of permeabilization buffer (the permeabilization and protease inhibitor solution mix) using dounce hand homogenizer. All steps were carried out on ice to minimize protein degradation. The resulting liver homogenate was shaken for 30 minutes at 4°C and then centrifuged at 16,000g for 15 minutes. The resulting supernatant was aliquoted for protein quantification via BCA analysis and diluted to 2 mg/ml prior to trypsin digestion. To 80 μl of post-treatment supernatant, 30 μl of ammonium bicarbonate buffer (100 mM, pH 7.8), 10 μl of human serum albumin, 20 μl of bovine serum albumin (BSA), and 10 μl of dithiothreitol were added and incubated at 95°C for 10 minutes. After cooling, 20 μl of iodoacetamide (500 mM) was added to incubate at room temperature for 30 minutes in the dark. Then, ice-cold methanol (0.5 ml), chloroform (0.1 ml), and water (0.4 ml) were added to desalt the samples. After centrifugation at 16,000g for 5 minutes at 4°C, the pellet was washed with ice-cold methanol (0.5 ml) and centrifuged at 8000g for 5 minutes at 4°C. Trypsin [20 μl, 1:10 trypsin: protein ratio (w/w)] was added and incubated for 16 hours at 37°C while shaking at 300 rpm. The reaction was quenched with dry ice, and internal standards were added. The relative levels of protein abundance of P450 enzymes and transporter were quantified using surrogate peptides for respective P450 enzymes or transporters as standards (Supplemental Table 1). Chromatographic and mass spectrometric conditions were the same as previously described (Prasad et al., 2018). Previous studies (Bhatt and Prasad, 2018) found that it was necessary to perform various normalization steps to minimize batch-to-batch and sample-to-sample variation. Therefore, BSA and a stable-isotope–labeled heavy peptide cocktail were added as normalization controls. The quality of peptide signal was verified by plotting the correlation between two or more peptide fragments or transitions derived from different samples with the same total protein concentration. Ion suppression was assessed with multiple peptides from the same protein. This approach measures whether the variability in peak responses reflects the biologic variability. In addition, by adding BSA, an exogenous protein, into the homogenized sample before protein denaturation, the sample loss during subsequent processing steps was addressed. Moreover, the use of heavy isotope–labeled peptides as internal standard provided adjustment for possible matrix effects and sample concentration changes because of evaporation and nonspecific binding to the vials. Relative protein abundance of each enzyme or transporter was calculated by taking the average signal for all peptides that passed linearity test for each enzyme or transporter in each individual liver sample and dividing it by the average signal of the corresponding heavy isotope–labeled peptide. The ratio was subsequently normalized to BSA and the group mean (pool of all samples) of each surrogate peptide. The resulting ratio-of-ratio estimates reflect relative protein abundance of each individual enzyme or transporter. The samples used for proteomic analysis were essentially the same as those used for RNA-seq analysis or activity assay (see below) except that the number of samples used for proteomic analysis in the CVP and GFNP groups was four instead of five or six because of a lack of sufficient tissue homogenates.
Liver Microsome Isolation and Activity Assay.
Liver microsomes were prepared from individual mouse livers as previously described (Paine et al., 1997; Shuster et al., 2014). All steps were carried out at 4°C to minimize protein degradation. Approximately 750 mg of frozen mouse liver tissue was homogenized in 2 ml homogenization buffer (50 mM KPi buffer containing 0.25 M sucrose and 1 mM EDTA) using Omni Bead Ruptor Homogenizer (Omni International, Kennesaw, GA). The homogenate was then centrifuged for 30 minutes at 15,000g, and the resulting supernatant was centrifuged for 70 minutes at 120,000g. The pellet containing microsomes was washed once with the buffer (10 mM KPi, 0.1 mM KCl, 1 mM EDTA, pH 7.4) before a final centrifugation for 70 minutes at 120,000g. The final pellet was resuspended in 1 ml of the storage buffer (50 mM KPi, 0.25 M sucrose, 10 mM EDTA, pH 7.4) and stored in 200 μl aliquots at −80°C until analysis. Microsomal protein concentration was determined by using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific).
CYP3A activities in individual liver microsomal preparations were determined by using the Promega P450-Glo Screening Kit (Promega Corporation, Madison, WI) according to the manufacturer’s instruction. Briefly, liver microsomes (7 μg per reaction) from CV and GF mice were preincubated with 3 μM luminogenic P450-Glo Luciferin-IPA for 10 minutes at 37°C. In the case of the inhibition assay, ketoconazole was added to the reaction mixture to a final concentration of 5 μM as previously described (Perloff et al., 1999). Then, NADPH regeneration system was added to the mixture to initiate reaction. Total reaction volume was 75 μl. After incubation for 10 minutes at 37°C, reaction was stopped by the addition of equal volume of the luciferin detection reagent at room temperature, and the mixture sat for 20 minutes. Cyp3a-mediated reaction results in generation of a luciferin product, which was measured by a Glomax 96 Microplate Luminometer (Promega Corporation). Light signal detected reflects the magnitude of Cyp3a activity. Both the substrate concentration and time of incubation were optimized to fall within the linear range of human CYP3A activity, as reported by Promega and described in its instruction. Control incubations using recombinant CYP3A4 microsomes were also performed in parallel as positive control. Incubations were done in triplicates and repeated once. The same assay kit has previously been used to measure mouse Cyp3a activity (Lee et al., 2013; Selwyn et al., 2015; Li et al., 2017).
Quantification of Plasma Bile Acids and Steroid Hormones.
Bile acids were extracted from mouse plasma as previously described (Alnouti et al., 2008). Briefly, 50 μl of plasma sample was mixed with 10 μl of internal standard solution (20 μg/ml d4-G-CDCA, 10 μg/ml d4-G-CDCA in 50% MeOH) and vortexed well. Protein was precipitated by adding 500 μl of ice-cold methanol. The mixture was centrifuged at 12,000g for 10 minutes at 4°C. The resulting supernatant was kept in a new tube, and the pellet was put through the same steps. The resulting supernatant was combined with the previous one, dried under vacuum (30°C), and reconstitute in 100 μl of 1:1 methanol-water (v/v). The suspension was centrifuged again at 12,000g for 10 minutes at 4°C, and 50 μl was subjected to ultraperformance LC-MS/MS for analysis. Chromatographic conditions and instrument settings were the same as previously described (Alnouti et al., 2008) with modifications. Samples were eluted using mobile phases consisting of 20% acetonitrile and 10 mmol/L ammonium acetate in aqueous solutions (A) and 80% acetonitrile and 10 mmol/L ammonium acetate in aqueous solutions (B) at a flow rate of 0.400 ml/min. Five microliters of each sample was injected on column for analysis using negative ionization mode.
Primary bile acids CA, CDCA, ursodeoxycholic acid (UDCA), α/β-murine cholic acid (α/β-MCA), and their respective taurine conjugates (TCA, T-CDCA, TUDCA, T-α/β-MCA) as well as secondary bile acids DCA, hyodeoxycholic acid (HDCA), LCA, murideoxycholic acid (MDCA), ω-muricholic acid (ω-MCA), and their respective taurine conjugates (T-ω-MCA, T-DCA, T-HDCA, and T-LCA) were quantified. Calibrator and different quality control samples were prepared by adding the appropriate amount of the different standard stock solutions and were extracted by using the similar sample preparation procedure described above.
Steroid hormones were extracted from mouse plasma as previously described (Basit et al., 2018). Briefly, 200 μl of methanol containing an internal standard cocktail (10 μg/ml estrone-d4, 10 μg/ml cortisol-d4, 10 μg/ml 11-deoxycortisol-d5, 10 μg/ml Dehydroepiandrosterone-d5 (DHEA-d5), 10 μg/ml progesterone-d9, 10 μg/ml 17OH-progesterone-d8, and 10 μg/ml 17OH-pregnenolone-d3) was added to 50 μl of plasma samples, and the mixture was mixed vigorously for 1 minute followed by a centrifugation at 3500g for 10 minutes at 4°C. The supernatant was moved to a new microcentrifuge tube and dried down under N2. Samples were reconstituted in 50% methanol and analyzed by LC-MS/MS, using the same instrument and conditions as previously described (Basit et al., 2018).
Statistical Analyses for Plasma Metabolite Quantification and Microsome Activity.
Data are presented as means ± S.E. (or S.D.) of independent samples from four to six different mice. Statistical significance of the difference between two groups was determined by the Wilcoxon t test, with P < 0.05 to be considered as significant. Data analyses were performed using R (v3.5.1 and v3.6.1) and GraphPad Prism (GraphPad Prism 5.01, La Jolla, CA).
Results
Validation of RNA-Seq Data by qRT-PCR.
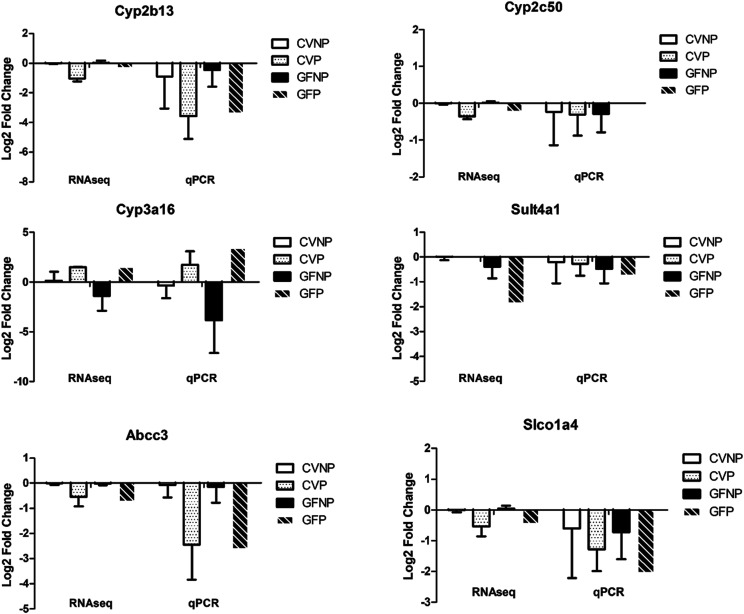
A direct comparison of the two approaches (RNA-seq and qRT-PCR) is shown in Fig. 1 for six selected genes (Cyp3a16, Cyp2b13, Cyp2c50, Sult4a1, Abcc3, and Slco1a4). Overall, gene expression levels determined by the two approaches showed the same pattern of pregnancy-induced changes in both CV and GF mice (upregulation or downregulation). More specifically, Cyp2b13, Cyp2c50, Sult4a1, Abcc3, and Slco1a4 demonstrated downregulation by both RNA-seq and qRT-PCR analyses. As for Cyp3a16, downregulation in GFNP group and upregulation in CVNP, CVP, and GFP groups were observed. The consistency of these results established the accuracy and reliability of the more comprehensive RNA-seq analysis.
Fig. 1.
Comparison of expression of selected genes determined by RNA-seq and qRT-PCR analysis. Log twofold change is relative to the CVNP group for both RNA-seq and qRT-PCR data. Shown are means ± S.D. of gene expression data from five to six different mouse liver tissues.
RNA-Seq Analysis of Hepatic DMETs.
To determine whether pregnancy and the microbiome interact to affect the expression of host hepatic DMETs, we performed RNA-seq analysis of liver tissues from CVNP (n = 6), CVP (n = 5), GFNP (n = 6), and GFP (n = 5) C57BL/6 mice. Specifically, we examined the mRNA expression profiles of genes important for xenobiotic, bile acid, and steroid hormone disposition, including phase I enzymes (P450 enzymes), phase II enzymes [UDP-glucuronosyltransferases (Ugt), sulfotransferases (Sult), and glutathione S-transferases (Gst)], and transporters [ATP-binding cassette (Abc) transporters, solute carrier (Slc) transporters, and solute carrier organic anion (Slco) transporters]. Differential changes in the expression of key DMETs by pregnancy in CV and GF mice are shown in Table 1. A complete list of DMETs as determined by RNA-seq analysis can be found in Supplemental Table 2.
TABLE 1.
Pregnancy and the microbiome alter mRNA expression of key DMETs in female C57BL/6 livers
This list of genes was generated using the following filtration criteria: fold-change >1.5 or <0.65 and FDR <0.1, on at least one comparison group between CVP and CVNP, GFP and GFNP, GFNP and CVNP, or GFP and CVP. Statistically significant differences with FDR values of <0.1 are highlighted in bold.
| Gene Symbol | CVP vs. CVNP | GFP vs. GFNP | GFNP vs. CVNP | GFP vs. CVP | ||||
|---|---|---|---|---|---|---|---|---|
| Fold Change | FDR | Fold Change | FDR | Fold Change | FDR | Fold Change | FDR | |
| Cyp17a1 | 1.7 | 0.28 | 2.9 | 0.01 | 0.9 | 1 | 1.6 | 0.704 |
| Cyp26a1 | 3.2 | 0.076 | 4.2 | 0.02 | 0.9 | 1 | 1.1 | 0.996 |
| Cyp26b1 | <0.1 | <0.001 | 0.1 | <0.001 | 1.9 | 0.982 | 5 | 0.289 |
| Cyp2b13 | 0.1 | <0.001 | 0.3 | <0.001 | 1.2 | 1 | 5.3 | <0.001 |
| Cyp2c37 | 0.3 | 0.002 | 0.6 | 0.152 | 1 | 1 | 1.8 | 0.401 |
| Cyp2c38 | 0.4 | 0.007 | 1 | 0.965 | 1.3 | 1 | 3 | 0.013 |
| Cyp2c39 | 1 | 0.983 | 2.4 | 0.022 | 1.1 | 1 | 2.7 | 0.154 |
| Cyp2c50 | 0.2 | <0.001 | 0.5 | 0.001 | 1 | 1 | 2 | 0.066 |
| Cyp2c54 | 0.4 | 0.001 | 0.8 | 0.561 | 1 | 1 | 2.2 | 0.074 |
| Cyp2c55 | 0.5 | 0.216 | 0.1 | <0.001 | 2.8 | 0.319 | 0.8 | 0.974 |
| Cyp2c67 | 0.4 | 0.012 | 0.6 | 0.279 | 1.4 | 1 | 2.3 | 0.238 |
| Cyp2c69 | 1 | 0.998 | 2.6 | 0.02 | 0.6 | 0.982 | 1.6 | 0.708 |
| Cyp2d40 | 3.9 | <0.001 | 5.4 | <0.001 | 0.7 | 1 | 1 | 1 |
| Cyp2d9 | 0.3 | 0.025 | 0.2 | 0.002 | 1.8 | 1 | 1.1 | 1 |
| Cyp2g1 | 2.8 | 0.047 | 3.6 | 0.01 | 0.6 | 1 | 0.8 | 0.964 |
| Cyp39a1 | 0.4 | 0.031 | 0.3 | 0.017 | 1 | 1 | 0.9 | 0.999 |
| Cyp3a11 | 0.6 | 0.29 | 0.3 | 0.005 | 0.6 | 0.953 | 0.3 | 0.131 |
| Cyp3a16 | 20.6 | 0.001 | 128 | <0.001 | 0.2 | 0.578 | 1.4 | 0.953 |
| Cyp3a41a | 4.8 | 0.047 | 10.8 | 0.002 | 0.6 | 1 | 1.4 | 0.941 |
| Cyp3a41b | 74.2 | <0.001 | 181.7 | <0.001 | 0.5 | 1 | 1.3 | 0.989 |
| Cyp3a44 | 14.3 | <0.001 | 30.5 | <0.001 | 0.8 | 1 | 1.7 | 0.816 |
| Cyp3a63-ps | 5.1 | 0.047 | 8.5 | 0.007 | 0.9 | 1 | 1.5 | 0.926 |
| Cyp4a14 | 0.4 | 0.021 | 0.3 | 0.002 | 1.2 | 1 | 0.9 | 0.958 |
| Cyp4a31 | 4.6 | <0.001 | 5.3 | <0.001 | 1.1 | 1 | 1.3 | 0.909 |
| Cyp4f13 | 0.6 | 0.042 | 0.8 | 0.329 | 0.8 | 1 | 1.1 | 0.987 |
| Cyp4f15 | 0.5 | 0.033 | 0.5 | 0.052 | 1 | 1 | 1.1 | 0.976 |
| Ugt1a10 | 0.9 | 0.977 | 9.6 | 0.004 | 0.2 | 0.439 | 2.4 | 0.689 |
| Ugt1a5 | 2.6 | 0.062 | 3.3 | 0.016 | 0.5 | 0.949 | 0.7 | 0.833 |
| Ugt1a6b | 0.3 | 0.042 | 0.3 | 0.023 | 1 | 1 | 0.9 | 0.999 |
| Ugt2b38 | 0.2 | 0.139 | <0.1 | 0.013 | 0.5 | 1 | 0.1 | 0.408 |
| Sult2a7 | 0.2 | 0.061 | 0.2 | 0.031 | 0.9 | 1 | 0.7 | 0.964 |
| Sult3a1 | 3.6 | 0.056 | 12.9 | <0.001 | 0.3 | 0.359 | 1 | 1 |
| Sult3a2 | 53.9 | <0.001 | 226.7 | <0.001 | 0.2 | 0.605 | 0.9 | 1 |
| Sult4a1 | 287.7 | <0.001 | 3.5 | 0.333 | 5.2 | 1 | 0.1 | 0.129 |
| Gsta2 | 0.6 | 0.215 | 0.4 | 0.033 | 2 | 0.605 | 1.5 | 0.782 |
| Gstk1 | 0.5 | 0.005 | 0.5 | 0.018 | 1.1 | 1 | 1.2 | 0.901 |
| Gstm2 | 1.8 | 0.041 | 1.4 | 0.248 | 1.4 | 1 | 1.1 | 0.984 |
| Gstm3 | 3.1 | 0.044 | 0.6 | 0.456 | 2.5 | 0.564 | 0.5 | 0.625 |
| Gstp1 | 3.4 | 0.002 | 2.2 | 0.039 | 0.9 | 1 | 0.6 | 0.635 |
| Gstp2 | 5.9 | 0.016 | 1.8 | 0.505 | 0.9 | 1 | 0.3 | 0.386 |
| Gstt1 | 0.4 | 0.005 | 0.4 | 0.017 | 0.8 | 1 | 0.9 | 0.976 |
| Gstt3 | 1.5 | 0.177 | 2 | 0.005 | 0.8 | 1 | 1.1 | 0.951 |
| Abca17 | 103.3 | <0.001 | 1779 | <0.001 | 0.1 | 0.757 | 1.2 | 0.989 |
| Abca5 | 2 | 0.025 | 3.3 | <0.001 | 0.8 | 1 | 1.3 | 0.768 |
| Abca7 | 0.9 | 0.851 | 0.5 | 0.006 | 1.1 | 1 | 0.6 | 0.298 |
| Abcb6 | 0.6 | 0.019 | 0.7 | 0.129 | 1 | 1 | 1.2 | 0.83 |
| Abcc3 | 0.3 | 0.025 | 0.1 | <0.001 | 1 | 1 | 0.5 | 0.613 |
| Abcg5 | 0.4 | 0.003 | 0.4 | 0.009 | 1.3 | 1 | 1.4 | 0.696 |
| Abcg8 | 0.5 | 0.008 | 0.6 | 0.061 | 1.2 | 1 | 1.4 | 0.613 |
| Slc10a1 | 0.4 | 0.02 | 0.5 | 0.045 | 0.8 | 1 | 1 | 1 |
| Slc16a1 | 2 | 0.018 | 1.6 | 0.142 | 0.9 | 1 | 0.7 | 0.625 |
| Slc16a6 | 16.2 | <0.001 | 14.4 | <0.001 | 1.1 | 1 | 1 | 1 |
| Slc17a2 | 0.3 | 0.057 | 0.2 | 0.009 | 0.9 | 1 | 0.6 | 0.816 |
| Slc22a15 | 0.5 | 0.008 | 0.5 | 0.028 | 1.1 | 1 | 1.2 | 0.869 |
| Slc22a2 | 260.5 | <0.001 | 561.5 | <0.001 | 0.4 | 1 | 0.8 | 0.984 |
| Slc22a7 | 1.3 | 0.713 | 3.1 | 0.004 | 0.5 | 0.437 | 1.1 | 0.975 |
| Slc24a3 | 9.5 | <0.001 | 6.6 | <0.001 | 1.3 | 1 | 0.9 | 0.969 |
| Slc25a19 | 0.8 | 0.291 | 0.6 | 0.015 | 1.1 | 1 | 0.9 | 0.857 |
| Slc25a28 | 0.6 | 0.007 | 0.7 | 0.024 | 0.9 | 1 | 0.9 | 0.953 |
| Slc25a30 | 1.7 | 0.128 | 2.2 | 0.01 | 0.5 | 0.315 | 0.7 | 0.639 |
| Slc25a37 | 0.5 | 0.012 | 0.6 | 0.027 | 0.9 | 1 | 1 | 1 |
| Slc25a51 | 0.6 | 0.026 | 0.3 | <0.001 | 1.8 | 0.181 | 1.1 | 0.951 |
| Slc26a10 | 0.4 | 0.079 | 0.3 | 0.006 | 1.3 | 1 | 0.9 | 0.988 |
| Slc26a4 | 0.1 | 0.009 | 0.2 | 0.016 | 1.2 | 1 | 1.6 | 0.947 |
| Slc27a1 | 0.7 | 0.21 | 0.6 | 0.018 | 1.3 | 1 | 1 | 1 |
| Slc35b1 | 1.8 | 0.006 | 2.5 | <0.001 | 1 | 1 | 1.3 | 0.617 |
| Slc35c1 | 1.3 | 0.254 | 1.6 | 0.029 | 0.9 | 1 | 1.1 | 0.953 |
| Slc35c2 | 1.7 | 0.003 | 2 | <0.001 | 0.9 | 1 | 1 | 1 |
| Slc36a1 | 2.3 | 0.001 | 2.7 | <0.001 | 0.9 | 1 | 1 | 1 |
| Slc37a1 | 11.6 | <0.001 | 6.2 | <0.001 | 1.5 | 1 | 0.8 | 0.946 |
| Slc38a4 | 0.3 | <0.001 | 0.3 | <0.001 | 1.1 | 1 | 1.2 | 0.951 |
| Slc39a11 | 1.4 | 0.18 | 2 | <0.001 | 0.9 | 1 | 1.3 | 0.626 |
| Slc39a14 | 2.3 | 0.013 | 2.1 | 0.022 | 1.5 | 0.978 | 1.4 | 0.775 |
| Slc41a2 | 30.2 | <0.001 | 28.1 | <0.001 | 1.1 | 1 | 1 | 1 |
| Slc41a3 | 4.3 | 0.001 | 3.5 | 0.002 | 1.9 | 0.781 | 1.5 | 0.753 |
| Slc43a1 | 2.6 | 0.037 | 3.1 | 0.01 | 1.1 | 1 | 1.3 | 0.934 |
| Slc45a3 | 3 | 0.001 | 2.1 | 0.027 | 1.2 | 1 | 0.9 | 0.948 |
| Slc4a1 | 8 | 0.002 | 24 | <0.001 | 0.9 | 1 | 2.6 | 0.415 |
| Slc4a9 | 0.1 | 0.025 | 0.1 | 0.001 | 3.9 | 0.359 | 1.9 | 0.914 |
| Slc6a9 | 6.1 | <0.001 | 9 | <0.001 | 0.7 | 1 | 1.1 | 1 |
| Slc7a15 | 5 | 0.102 | 19.1 | 0.01 | 0.1 | 0.554 | 0.4 | 0.786 |
| Slc7a2 | 0.4 | 0.013 | 0.5 | 0.058 | 1.3 | 1 | 1.6 | 0.625 |
| Slc7a7 | 5 | <0.001 | 2.9 | 0.003 | 1.1 | 1 | 0.6 | 0.601 |
| Slc8b1 | 0.5 | 0.023 | 0.6 | 0.055 | 0.8 | 1 | 0.9 | 0.958 |
| Slc9a3 | 0.4 | 0.766 | 74.3 | 0.005 | 0.1 | 1 | 23.5 | 0.238 |
| Slco1a4 | 0.4 | 0.045 | 0.4 | 0.063 | 1.1 | 1 | 1.2 | 0.945 |
| Slco4c1 | 4.8 | 0.519 | 17.8 | 0.035 | 2.6 | 1 | 9.6 | 0.376 |
There were no significant differences in the expression of DMETs between CVNP and GFNP mice, with either no change or slight trend of downregulation for most of the DMETs listed in Table 1 in GFNP mice compared with CVNP mice. Comparing pregnant mice, we found the mRNA levels of two genes, Cyp2b13 and Cyp2c38, were higher, 5.3-fold (FDR < 0.001) and threefold (FDR = 0.013), respectively, in GFP mice compared with CVP mice (Table 1). The expression of other DMET genes was not significantly different between CVP and GFP mice (Table 1).
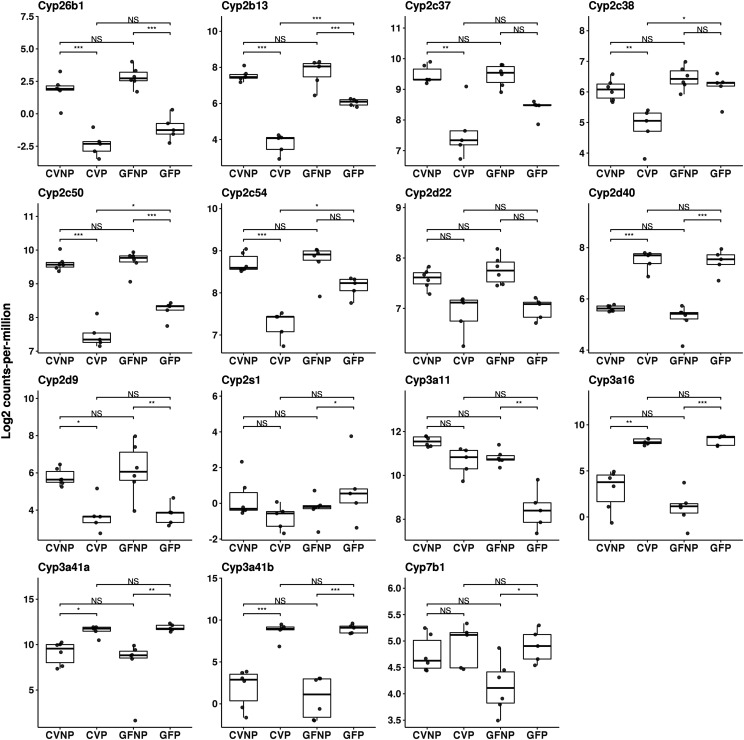
In both CV and GF mice, pregnancy induced significant changes in the expression of many DMETs in phase I (Fig. 2) and phase II (Fig. 3) enzymes as well as transporters (Fig. 4) relative to nonpregnant mice. Pregnancy altered the expression of DMETs with an overall similar trend but with different magnitudes of changes in CV and GF mice. For example, pregnancy decreased the mRNA expression of Cyp2c37, Cyp2c38, Cyp2c50, and Cyp2c54 by 70% (FDR < 0.01), 60% (FDR < 0.01), 80% (FDR < 0.001), and 60% (FDR < 0.01), respectively, in CV mice, whereas in GF mice, no significant or much smaller changes by pregnancy were observed for the same genes (Table 1). Similarly, Cyp2d40 was induced 3.9-fold (FDR < 0.001) and 5.4-fold (FDR < 0.001) by pregnancy, respectively, in CV and GF mice (Fig. 2; Table 1). Among all the P450 isoforms analyzed, CV and GF mice showed the most differences in the gene expressions of Cyp3a isoforms in response to pregnancy (Fig. 2; Table 1). We observed a 70% downregulation of Cyp3a11 by pregnancy in GF mice (FDR = 0.005) but a nonsignificant downregulation in CV mice by pregnancy. The mRNA levels of Cyp3a16 were induced 20.6-fold (FDR < 0.001) by pregnancy in CV mice versus 128-fold (FDR < 0.001) in GF mice. Likewise, Cyp3a41a and Cyp3a41b were induced by pregnancy 4.8-fold and 74.2-fold (FDR < 0.05 and FDR < 0.001, respectively) in CV mice compared with 10.8-fold and 181.7-fold (FDR = 0.002 and FDR < 0.001, respectively) in GF mice. Cyp3a44 was upregulated by 14.3-fold (FDR < 0.001) by pregnancy in CV mice versus 30.5-fold (FDR < 0.001) in GF mice.
Fig. 2.
Effect of pregnancy and microbiome on mRNA expression of hepatic phase I enzymes. Shown are boxplots with individual scatters of RNA-seq analysis data of hepatic phase I enzymes from female C57BL/6 mice. Data illustrates individual log2 counts per million for each DMET. All pregnant mice used were on gestation day 15. *FDR < 0.1; **FDR < 0.01; ***FDR < 0.001.
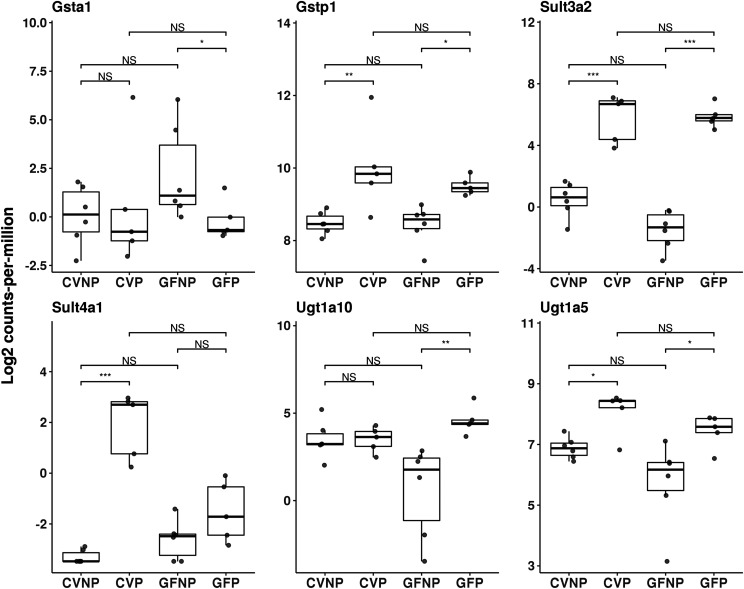
Fig. 3.
Effect of pregnancy and microbiome on mRNA expression of hepatic phase II enzymes. Shown are boxplots with individual scatters of RNA-seq analysis data of hepatic phase II enzymes from female C57BL/6 mice. Data illustrates individual log2 counts per million for each DMET. All pregnant mice used were on gestation day 15. *FDR < 0.1; **FDR < 0.01; ***FDR < 0.001.
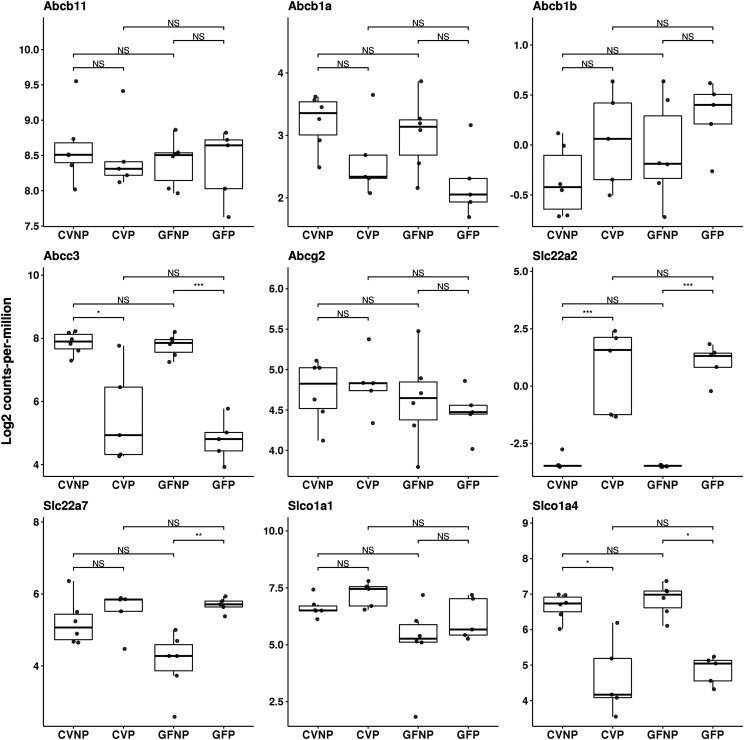
Fig. 4.
Effect of pregnancy and microbiome on mRNA expression of hepatic transporters. Shown are boxplots with individual scatters of RNA-seq analysis data of hepatic transporters from female C57BL/6 mice. Data illustrates individual log2 counts per million for each DMET. All pregnant mice used were on gestation day 15. *FDR < 0.1; **FDR < 0.01; ***FDR < 0.001.
Of all phase II enzymes that were differentially expressed during pregnancy, we noted that the most significant changes induced by pregnancy were associated with Sult3a2 and Sult4a1 in both CV and GF mice. We observed a 53.9-fold (FDR < 0.001) induction by pregnancy in CV mice versus a 226.7-fold (FDR < 0.001) induction in GF mice for Sult3a2 and a 287.7-fold (FDR < 0.001) induction in CV mice versus only a nonsignificant induction in GF mice for Sult4a1 (Fig. 3; Table 1).
For transporters, Abcc3 and Slco1a4 were downregulated by pregnancy to a similar extent in CV and GF mice (Fig. 4; Table 1). Abcc3 was downregulated by pregnancy by 70% (FDR < 0.05) and 90% (FDR < 0.001), respectively, in CV and GF mice. Slco1a4 was decreased 60% by pregnancy in CV mice (FDR < 0.05) but nonsignificantly in GF mice. Pregnancy induced Slc22a2 and Slc22a7 by 561.5-fold (FDR < 0.001) and 3.1-fold (FDR = 0.004), respectively, in GF mice versus 260.5-fold (FDR < 0.001) induction of Slc22a2 and a nonsignificant induction of Slc22a7 in CV mice (Fig. 4; Table 1).
Quantification of Relative Protein Abundance of Selected Hepatic DMETs by Targeted Proteomics.
To investigate and compare how protein levels of hepatic DMETs were changed by pregnancy in CV and GF mice, we performed quantitative proteomics of selected hepatic DMETs in liver tissues. Because of the limited amount of isolated liver membrane proteins available, we performed targeted proteomics only on a set of selected DMETs. Selection of DMETs was based on a priori knowledge of their importance in overall xenobiotic disposition (Nelson et al., 2004). Relative protein abundance of these DMETs was quantified using LC-MS/MS–based proteomics, and results are shown in Figs. 5 and 6.
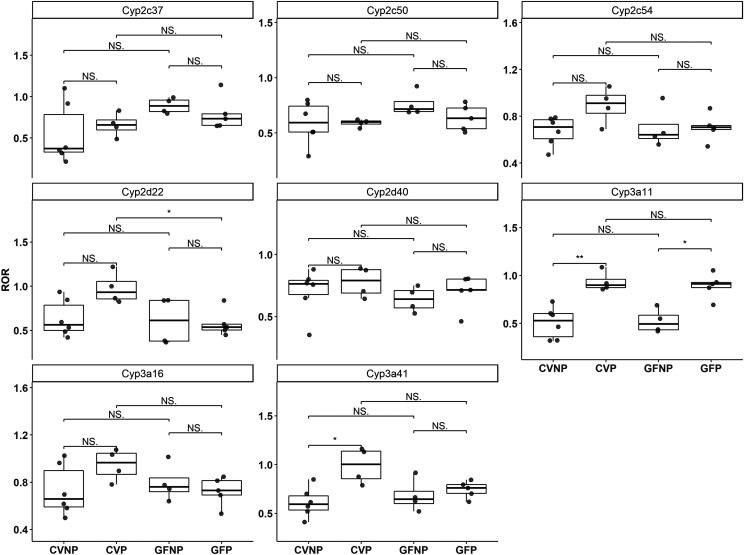
Fig. 5.
Effect of pregnancy and microbiome on protein expression of hepatic P450 enzymes. Shown are boxplots with individual data points of relative protein levels of hepatic P450 enzymes in C57BL/6 female mice. Shown are means ± S.E. for selected enzymes or transporters in the liver from four to six mice. Data illustrates ratio-of-ratio estimates for each DMET. All pregnant mice used were on gestation day 15. *P < 0.05; **P < 0.01 by Wilcoxon signed-rank test.
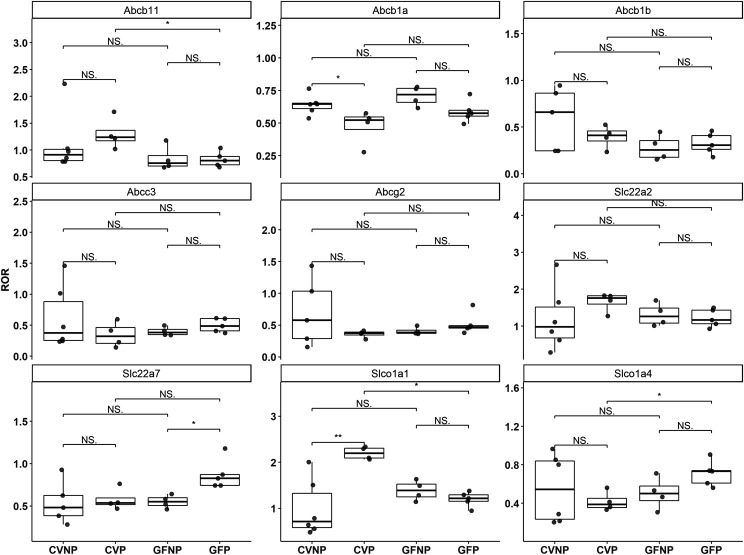
Fig. 6.
Effect of pregnancy and microbiome on protein expression of hepatic transporters. Shown are boxplots with individual data points of relative protein levels of hepatic transporters in C57BL/6 female mice. Shown are means ± S.E. for selected enzymes or transporters in the liver from four to six mice. Data illustrates ratio-of-ratio estimates for each DMET. All pregnant mice used were on gestation day 15. *P < 0.05; **P < 0.01 by Wilcoxon signed-rank test.
Of the phase I enzymes, we opted to quantify mouse isoforms homologous to human CYP2C19 (CYP2C37, CYP2C50, and CYP2C54), human CYP2D6 (CYP2D22 and CYP2D40), and human CYP3A4 (CYP3A11, CYP3A16, CYP3A41A, and CYP3A41B). Because there are no distinct methods to separate CYP3A41A and CYP3A41B, the results for these two enzymes were combined as CYP3A41. Overall, the effects of pregnancy on protein expression profiles of these P450 enzymes in CV and GF mice were notably different from the effects of pregnancy on respective mRNA expression profiles. Specifically, we found that pregnancy significantly increased the protein levels of CYP3A11 approximately twofold in both CV and GF mice (Fig. 5), whereas the mRNA levels of Cyp3a11 were downregulated by pregnancy in CV and GF mice (Fig. 2). There were no significant differences in the protein levels of CYP3A16 between CVNP and CVP or between GFNP and GFP; however, the protein level of CYP3A16 in CVP mice was significantly higher (∼25%) than in GFP mice (0.95 ± 0.04 vs. 0.72 ± 0.03, P < 0.05) (Fig. 5). The effect of pregnancy on protein expression of CYP3A41 was similar to its effect on mRNA expression with a trend of higher abundance in pregnancy for both CV and GF mice; however, a statistically significant increase by pregnancy was observed only in CV mice (Fig. 5). No significant differences in the protein levels of CYP2C and CYP2D isoforms between CVNP and CVP or between GFNP and GFP were observed (Fig. 5). Like CYP3A16, the protein levels of CYP2D22 in CVP mice were significantly higher by ∼40% than those in GFP mice (0.98 ± 0.03 vs. 0.58 ± 0.02, P < 0.05) (Fig. 5).
We also quantified various transporters because of their importance in drug disposition, including ABCB11 (bile salt export pump), ABCB1A/B (P-glycoprotein), ABCC3 (Mrp3), ABCG2 (breast cancer resistance protein), SLC22A2 (organic cation transporter 2), SLC22A7 [organic anion transporter (Oat) 2], SLCO1A1 (Oatp1a1), and SLCO1A4 (Oatp1a4). Similar to CYP3A16 and CYP2D22, there were no significant differences in the protein levels of ABCB11 between CVNP and CVP or between GFNP and GFP; however, the protein levels of ABCB11 in CVP mice were significantly greater (∼37%) than those in GFP mice (1.3 ± 0.07 vs. 0.82 ± 0.03, P < 0.05) (Fig. 6). We also observed significantly decreased protein abundance of SLCO1A1 in GFP mice compared with CVP mice (1.20 ± 0.03 vs. 2.20 ± 0.03, P < 0.001). The protein levels of SLCO1A4 in GFP mice were significantly higher than those in CVP mice (0.71 ± 0.03 vs. 0.42 ± 0.03, P < 0.05), but there were no significant differences between GFNP and CVNP mice (Fig. 6). Pregnancy significantly increased the protein levels of SLC22A7 ∼60% in GF mice (0.87 ± 0.04 vs. 0.55 ± 0.02, P < 0.05) but not in CV mice. No significant changes were observed in the abundance of ABCB1A/B, ABCC3, and ABCG2 proteins because of pregnancy or the microbiome.
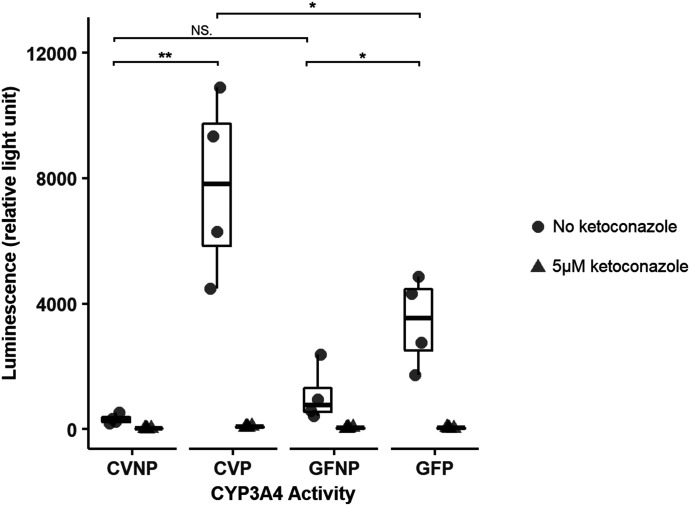
Activity of Hepatic Cyp3a Enzymes Determined by Liver Microsomal Incubations.
To confirm pregnancy-induced changes in protein expression of Cyp3a enzymes, microsomal incubations were performed to determine Cyp3a activity. We used a luciferin CYP3A probe to measure the amount of metabolite formed at a single incubation time point, which all fell within the linear range of product formation. Liver microsomes from CVP mice exhibited ∼30-fold higher CYP3A activities (P < 0.05) than those from CVNP mice, whereas liver microsomes from GFP mice showed only a ∼threefold induction (P < 0.05) of CYP3A activity compared with GFNP mice (Fig. 7). Furthermore, the addition of ketoconazole (a potent known CYP3A inhibitor) at 5 μM completely abolished metabolite formation with microsomes from both CV and GF mice (Fig. 7), suggesting that the enzymatic activity was due to CYP3A enzymes. Blank control and human recombinant CYP3A4 controls were also assayed in parallel to verify assay functionality (data not shown).
Fig. 7.
Effect of pregnancy and microbiome on hepatic Cyp3a activity. Shown are boxplots of individual data points of hepatic Cyp3a activity, which is presented as relative light unit. Circles represent incubations without an inhibitor, and triangles indicate incubations in the presence of the inhibitor ketoconazole at 5 μM. Data shown are means ± S.E. of hepatic Cyp3a activity in liver microsomes isolated from four to six mice from one representative experiment. The experiment was done in triplicate and repeated, and similar results were obtained. All pregnant mice used were on gestation day 15. *P < 0.05; **P < 0.01 by Wilcoxon signed-rank test.
Quantification of Plasma Bile Acids and Steroid Hormones.
Because secondary bile acids generated by the gut microbiome may play an important role in regulating the expression of certain DMETs including Cyp3a enzymes (Nelson et al., 2004; Chu et al., 2013), we quantified plasma concentrations of primary and secondary bile acids in all mice by LC-MS/MS.
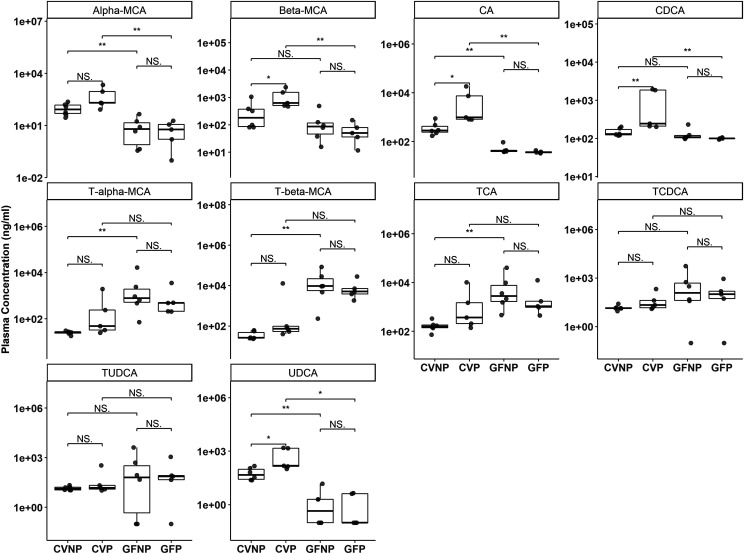
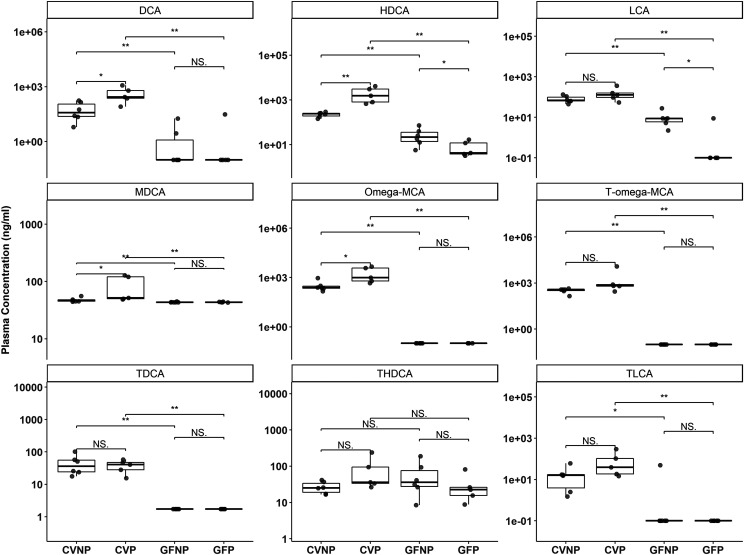
The plasma concentrations of the primary bile acids, α-MCA and β-MCA, in GFNP and GFP mice were lower than those in CVNP and CVP mice (Fig. 8). Pregnancy significantly increased the plasma concentrations of β-MCA in CV mice but had no effect in GF mice. The microbiome had the opposite effect on taurine conjugates, with higher plasma concentrations of T-α-MCA and T-β-MCA in GF mice than in CV mice, regardless of pregnancy. The plasma concentrations of CA were increased ∼14-fold by pregnancy in CV mice with no significant changes between GFP and GFNP mice. Moreover, the plasma concentrations of CA in GF mice were significantly lower than those in CV mice regardless of pregnancy (Fig. 8). The pattern of plasma concentrations of UDCA in all mice was similar to CA. Neither pregnancy nor the microbiome significantly affected the plasma concentrations of taurine conjugates of CA, CDCA, and UDCA, with the exception that the plasma concentrations of T-CA in GFNP mice were 55-times greater than those in CVNP mice (Fig. 8).
Fig. 8.
Effect of pregnancy and microbiome on plasma concentrations of primary bile acids. Shown are boxplots with individual data points of plasma concentrations of primary bile acids in C57BL/6 female mice. Concentrations were determined using targeted LC-MS/MS quantification and reported as nanograms per milliliter. Shown are means ± S.E. for selected bile acids or steroid hormones in the plasma from five to six mice. All pregnant mice used were on gestation day 15. *P < 0.05; **P < 0.01 by Wilcoxon signed-rank test.
As expected, the plasma concentrations of most of the secondary bile acids were lower in GF mice versus CV mice, likely because of the lack of gut bacteria to synthesize them from primary bile acids (Fig. 9; Supplemental Table 3). The plasma concentrations of DCA, HDCA, LCA, ω-MCA, T-ω-MCA, T-DCA, and T-LCA in GF mice were all significantly decreased versus those in CV mice, regardless of pregnancy (Fig. 9). Interestingly, in addition to a decrease in plasma concentrations of LCA in GFNP mice compared with CVNP mice (∼11-fold decrease, P < 0.05), there was a further decrease of ∼fivefold by pregnancy from GFNP to GFP mice (P < 0.05). Pregnancy also seemed to affect the production of some secondary bile acids. For example, pregnancy increased the plasma concentration of DCA ∼sevenfold (P < 0.05) in CV mice but had no effect in GF mice. Pregnancy also significantly increased the plasma concentrations of HDCA in CV mice (ninefold, P < 0.05) but caused a fourfold reduction (P < 0.05) in GF mice. Neither pregnancy nor the microbiome significantly affected the plasma concentrations of T-HDCA (Fig. 9). With respect to MDCA, pregnancy significantly increased its plasma concentrations only in CV mice but not in GF mice (Fig. 9). As expected, the plasma concentrations of MDCA were decreased in GF mice versus CV mice regardless of pregnancy, but the changes seemed to be small (Fig. 9; Supplemental Table 3).
Fig. 9.
Effect of pregnancy and microbiome on plasma concentrations of secondary bile acids. Shown are boxplots with individual data points of plasma concentrations of secondary bile acids in C57BL/6 female mice. Concentrations were determined using targeted LC-MS/MS quantification and reported as nanograms per milliliter. Shown are means ± S.E. for selected bile acids or steroid hormones in the plasma from five to six mice. All pregnant mice used were on gestation day 15. *P < 0.05; **P < 0.01; by Wilcoxon signed-rank test.
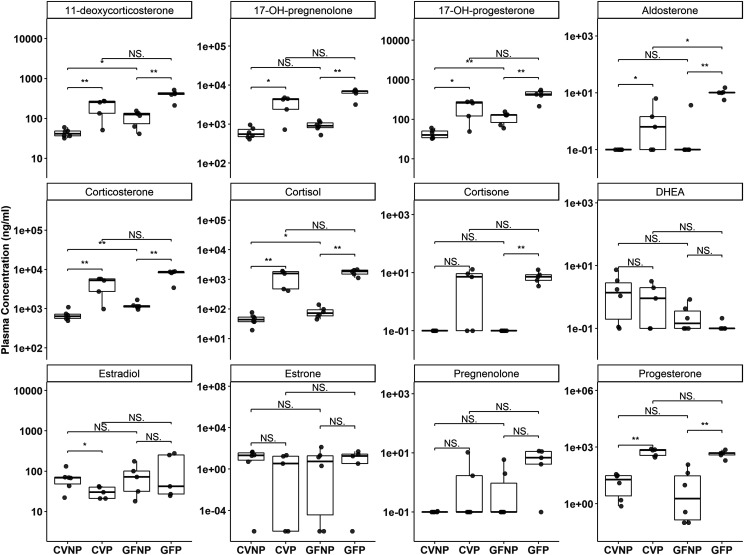
Pregnancy-related steroid hormones have been shown to regulate the expression of DMETs. Therefore, we also determined plasma concentrations of steroid hormones in all mice. As expected, pregnancy significantly increased the plasma concentrations of various steroid hormones, including 11-deoxycorticosterone, 17-OH-pregnenolone, 17-OH-progesterone, corticosterone, cortisol, and progesterone, 38-fold to 125-fold in both CV and GF mice (Fig. 10; Supplemental Table 4). The lack of the microbiome did not seem to alter pregnancy-induced levels of these hormones. The plasma concentrations of aldosterone and cortisone were significantly elevated by pregnancy in GF mice but not in CV mice (Fig. 10). The plasma concentrations of estradiol were slightly but significantly decreased by pregnancy in CV mice but not in GF mice. The lack of the microbiome did not affect the plasma concentrations of estradiol in both pregnant and nonpregnant mice (Fig. 10). On the other hand, we did not observe significant effects of pregnancy or the microbiome on pregnenolone, estrone, and Dehydroepiandrosterone (DHEA).
Fig. 10.
Effect of pregnancy and microbiome on plasma concentrations of steroid hormones. Shown are boxplots with individual data points of plasma concentrations of steroid hormones in C57BL/6 female mice. Concentrations were determined using targeted LC-MS/MS quantification and reported as nanograms per milliliter. Shown are means ± S.E. for selected bile acids or steroid hormones in the plasma from five to six mice. All pregnant mice used were on gestation day 15. *P < 0.05; **P < 0.01 by Wilcoxon signed-rank test.
Discussion
In this study, we investigated the combined effects of pregnancy and microbiome on hepatic DMETs. This study used the same C57BL/6 strain as previous studies that investigated the effects of microbiome on hepatic DMETs in male mice (Selwyn et al., 2015, 2016). Both this and the previous studies showed a downward trend in mRNA of Cyp3a isoforms in GF versus CV mice; however, the magnitude of downregulation observed in this study was much less pronounced. This could be due to sex difference. The previous studies also reported a significant induction of Cyp4a genes by the microbiome knockout, whereas we did not. Again, this could be due to Cyp4a genes, which are sex-divergent (Renaud et al., 2011). Regulation of Cyp4a14 was strongly influenced by androgen (Zhang and Klaassen, 2013), which is expectedly low in our female mice. Other factors that are known to control the gut microbiome ecosystems (e.g., age of mice, diet, and housing conditions) and might be different between this and previous studies could also potentially contribute to the discrepancy.
We previously showed that pregnancy alters hepatic DMETs in conventional FVB mice (Shuster et al., 2013). Overall, our results are in good agreement with our previous studies. For example, we reported that Cyp3a11 mRNA was decreased, whereas mRNA of Cyp3a16 and Cyp3a41 was increased during pregnancy. Consistently, this study also showed downregulation and induction of Cyp3a11 and Cyp3a16/Cyp3a41, respectively, by pregnancy in CV mice. We previously reported significant downregulation of Cyp2c genes and Mrp3/Abcc3 by pregnancy similar to this study in CV mice (Shuster et al., 2013). There are some inconsistencies in the magnitude of pregnancy-induced change for some DMETs between this and our previous studies. For example, this study showed a 20.6-fold induction of Cyp3a16 by pregnancy, whereas our previous studies reported an induction of only 60% on the same gd 15 (Shuster et al., 2013). Our previous studies used FVB mice. The two different mouse strains are known to possess significantly different basal microbiota (Ahn et al., 2018). Thus, the difference in mouse strain may contribute to the different magnitude of pregnancy-induced change for some DMETs between this and our previous studies.
The Cyp3a isoforms (Cyp3a11, Cyp3a16, Cyp3a41, and Cyp3a44) were of particular interest because of the prominent role of their human CYP3A4 homolog in drug metabolism. We therefore systematically examined their mRNA and protein expression along with catalytic activity. We found that Cyp3a11 mRNA was significantly decreased by pregnancy in both CV and GF mice but with a much greater magnitude of downregulation in GF versus CV mice. Downregulation of Cyp3a11 mRNA by pregnancy has been reported in our previous studies in CV mice, but the mechanistic reasoning is unknown (Zhang et al., 2008; Shuster et al., 2013). The greater magnitude of down-regulation of Cyp3a11 mRNA by pregnancy in GF mice could be due to the combined effects of the lack of secondary bile acids, which would otherwise compensate for the decreased Cyp3a11 gene expression during pregnancy in CV mice. Yet, targeted proteomics revealed an opposite effect of pregnancy on the protein levels of CYP3A11, with a comparable twofold induction in both CV and GF mice. In contrast to Cyp3a11, the mRNA levels of all other Cyp3a isoforms were induced by pregnancy in both CV and GF mice. Although the mRNA levels of these Cyp3a isoforms in CVP and GFP mice were comparable, the fold-induction in GF mice was generally much greater than that in CV mice (e.g., 128-fold in GF mice vs. 20.6-fold in CV mice for Cyp3a16) (Table 1). This was due to the lower baseline levels of these Cyp3a genes in GFNP mice compared with CVNP mice, which were likely caused by the lack of secondary bile acids. Surprisingly, pregnancy induced the protein levels of CYP3A16 and CYP3A41 in CV mice but had limited effect in GF mice. Although circadian patterns of P450 mRNA transcripts have been reported (Zhang et al., 2009), all mice were sacrificed around the same time of day, and thus the contribution of time of tissue collection is unlikely to be the source of changes observed. Next, we examined the effects of pregnancy and microbiome on the overall CYP3A activity. At present, it is not possible to determine activities of individual CYP3A isoforms because of the lack of specific probes or selective inhibitors for individual mouse CYP3A isoforms. Thus, the CYP3A activities we measured reflected the overall combined activities of all CYP3A isoforms. The CYP3A activities were significantly induced by pregnancy in both CV and GF mice but with a much greater induction in CV mice versus GF mice. The activity data agreed with the overall trend of pregnancy-induced increase in CYP3A protein and are also consistent with our previous studies showing that the mouse CYP3A activities for testosterone and glyburide were significantly induced by pregnancy (Zhang et al., 2008; Shuster et al., 2014). However, for the first time, we showed that the microbiome can significantly alter the magnitude of pregnancy induction of CYP3A activity. The lower pregnancy induction of CYP3A activity in GF mice could reflect only the activities of the CYP3A isoforms other than CYP3A11 that were not induced at the protein level (Fig. 5). Future studies are needed to explore the mechanisms of microbiome-induced changes in CYP3A activity.
There was a disconnection between the mRNA and protein expression data for many DMETs in addition to Cyp3a11 as described above. For example, pregnancy drastically decreased the mRNA levels of Cyp2c50 and Cyp2c54 but had no significant impact on their protein levels in both CV and GF mice. Likewise, the mRNA levels of Abcc3 were significantly decreased by pregnancy in CV and GF mice, but its protein levels were not affected by pregnancy at all. These inconsistencies could be due to the relatively small sample size and large variations in analysis (e.g., proteomics) for certain DMETs, yet the data for CYP3A isoforms seem to be reliable because both the mRNA and/or activity data for the CYP3A isoforms are fully consistent with previous studies. The discrepancy between mRNA and protein expression is not uncommon and could be due to post-transcriptional or epigenomic regulation of gene expression, as shown in numerous studies (Martínez-Jiménez et al., 2007; Takagi et al., 2008; Smutny et al., 2013; Wang et al., 2019). Recent studies showed that the gut bacteria Akkermansia muciniphila can affect the concentration of N6-methyladenosine in the intestines and the liver, and N6-methyladenosine methylation can greatly alter RNA transcription processes, leading to epitranscriptomic changes that may explain the gap between mRNA and protein data (Zhao et al., 2017; Miro-Blanch and Yanes, 2019). MicroRNA (miRNA) modifiers can also alter the transcript-protein relationship. A recent study reported bacteria phylum such as Firmicutes, Bacteroidetes, and Proteobacteria have significant effects on miRNA expression in humans (Yuan et al., 2018). Changes in miRNA expression because of bacteria shift or the microbiome knockout could also lead to changes in downstream gene regulation and result in variations in gene translation. Future studies are needed to explore the mechanisms of microbiome-induced disconnection between mRNA and protein expression of DMETs.
Cortisol has been shown to induce the expression of CYP3A through activation of glucocorticoid receptor, which in turn induces PXR, and then PXR induces transcription of CYP3A expression (Sachar et al., 2019). This could be the potential mechanism by which hepatic CYP3A is induced during pregnancy. Supporting this hypothesis, the plasma concentrations of cortisol were markedly increased by pregnancy in both CV and GF mice, and the mRNA levels of all Cyp3a isoforms but Cyp3a11 were induced by pregnancy. Secondary bile acids have been shown to be PXR ligands that can induce CYP3A (Dempsey et al., 2019). This explains our findings that the baseline mRNA levels of all Cyp3a isoforms in GFNP mice were generally lower than those in CVNP mice. However, the absence of secondary bile acids did not affect the induction of Cyp3a16 and Cyp3a41 mRNA by pregnancy, which is consistent with the finding that pregnancy-induced hormone production was generally not affected by the microbiome knockout. This suggests that the lack of microbiome does not interfere with pregnancy-mediated induction of Cyp3a gene transcription, which is mediated by increased production of steroid hormones during pregnancy. However, the microbiome can alter the magnitude of induction by changing the baseline expression of Cyp3a genes (Fig. 2) or influencing pregnancy-induced Cyp3a protein expression (Fig. 5) and, hence, activity (Fig. 7).
In summary, we have shown, for the first time, that the microbiome can affect hepatic DMETs in pregnant mice by altering the magnitude of pregnancy-induced fold-change in the expression (mRNA or protein) and/or the activity of hepatic DMETs. Caution should be taken to translate the data to drug disposition in vivo because of disconnection between mRNA and protein, relatively small sample size, and large variation in analysis of certain DMETs. Nevertheless, these results provide the basis for further mechanistic investigation of microbiome-mediated changes in hepatic DMETs and drug disposition during pregnancy. Future studies should also examine the impact of microbiome on DMETs in other organs of pregnant mice important for drug disposition, including the small intestine.
Acknowledgments
We would like to thank Dr. Adeline Hajjar for her assistance and guidance in the maintenance and breeding of germ-free mice.
Abbreviations
- Abc
ATP-binding cassette transporter family
- BSA
bovine serum albumin
- CA
cholic acid
- CDCA
chenodeoxycholic acid
- CV
conventional
- CVNP
conventional nonpregnant
- CVP
conventional pregnant
- DCA
deoxycholic acid
- DMET
drug metabolizing enzyme and transporter
- FDR
false discovery rate
- FXR
farnesoid X receptor
- gd
gestation day
- GF
germ-free
- GFNP
germ-free nonpregnant
- GFP
germ-free pregnant
- Gst
glutathione S-transferases
- HDCA
hyodeoxycholic acid
- LCA
lithocholic acid
- LC-MS/MS
liquid chromatography tandem mass spectrometry
- α/β-MCA
α/β-murine cholic acid
- ω-MCA
ω-muricholic acid
- MDCA
murideoxycholic acid
- miRNA
microRNA
- Mrp3/Abcc3
multidrug resistance–associated protein 3
- Oat2
organic anion transporter
- P450
cytochrome P450
- PXR
pregnane X receptor
- qRT-PCR
quantitative reverse-transcription polymerase chain reaction
- RNA-seq
RNA sequencing
- Slc
solute carrier family
- Slco
solute carrier organic anion transporter family
- Sult
sulfotransferases
- UDCA
ursodeoxycholic acid
- Ugt
UDP-glucuronosyltransferases
Author Contributions
Participated in research design: Han, Wang, Shi, Bammler, Cui, Mao.
Conducted experiments: Han, Pershutkina.
Contributed new reagents or analytic tools: Han, Wang, Shi, Dempsey, Dutta.
Performed data analysis: Han, Wang, Shi, Dempsey, Dutta.
Wrote or contributed to the writing of the manuscript: Han, Wang, Shi, Dempsey, Dutta, Bammler, Cui, Mao.
Footnotes
This work was supported by The Drug Metabolism, Transport and Pharmacogenomic Research Program at the University of Washington and in part by National Institutes of Health National Center for Advancing Translational Science [Grant TL1TR000422], National Institute of Environmental Health Sciences [Grant P30ES007033], National Institute on Drug Abuse [Grant P01DA032507], and National Institute of General Medical Sciences [Grant R01GM111381].
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Ahn I-S, Lang J, Olsen C, Ying Z, Zhang G, Byun HR, Zhao Y, Kurt Z, Lusis AJ, Hsaio E, et al. (2018) Host genetic background and gut microbiota contribute to differential metabolic responses to high fructose consumption in mice. Diabetes 67 (1), 10.1101/439786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alnouti Y, Csanaky IL, Klaassen CD. (2008) Quantitative-profiling of bile acids and their conjugates in mouse liver, bile, plasma, and urine using LC-MS/MS. J Chromatogr B Analyt Technol Biomed Life Sci 873:209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basit A, Amory JK, Prasad B. (2018) Effect of dose and 5α-reductase inhibition on the circulating testosterone metabolite profile of men administered oral testosterone. Clin Transl Sci 11:513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigi RH, Noguchi L, Brown G, Piper J, Watts DH. (2016) Performing drug safety research during pregnancy and lactation: biomedical HIV prevention research as a template. J Womens Health (Larchmt) 25:761–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57:289–300. [Google Scholar]
- Bhatt DK, Prasad B. (2018) Critical issues and optimized practices in quantification of protein abundance level to determine interindividual variability in DMET proteins by LC-MS/MS proteomics. Clin Pharmacol Ther 103:619–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Lun ATL, Smyth GK. (2016) From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000 Res 5:1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu X, Bleasby K, Evers R. (2013) Species differences in drug transporters and implications for translating preclinical findings to humans. Expert Opin Drug Metab Toxicol 9:237–252. [DOI] [PubMed] [Google Scholar]
- Claus SP, Ellero SL, Berger B, Krause L, Bruttin A, Molina J, Paris A, Want EJ, de Waziers I, Cloarec O, et al. (2011) Colonization-induced host-gut microbial metabolic interaction. MBio 2:e00271-e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey JL, Wang D, Siginir G, Fei Q, Raftery D, Gu H, Yue Cui J. (2019) Pharmacological activation of PXR and CAR downregulates distinct bile acid-metabolizing intestinal bacteria and alters bile acid homeostasis. Toxicol Sci 168:40–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussault I, Yoo HD, Lin M, Wang E, Fan M, Batta AK, Salen G, Erickson SK, Forman BM. (2003) Identification of an endogenous ligand that activates pregnane X receptor-mediated sterol clearance. Proc Natl Acad Sci USA 100:833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Food and Drug Administration (2018) Drug safety priorities 2018. https://www.fda.gov/media/124681/download
- Feghali M, Venkataramanan R, Caritis S. (2015) Pharmacokinetics of drugs in pregnancy. Semin Perinatol 39:512–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helldén A, Madadi P. (2013) Pregnancy and pharmacogenomics in the context of drug metabolism and response. Pharmacogenomics 14:1779–1791. [DOI] [PubMed] [Google Scholar]
- Isoherranen N, Thummel KE. (2013) Drug metabolism and transport during pregnancy: how does drug disposition change during pregnancy and what are the mechanisms that cause such changes? Drug Metab Dispos 41:256–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaassen CD, Cui JY. (2015) Review: mechanisms of how the intestinal microbiota alters the effects of drugs and bile acids. Drug Metab Dispos 43:1505–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, McKee DD, Oliver BB, Willson TM, Zetterström RH, et al. (1998) An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell 92:73–82. [DOI] [PubMed] [Google Scholar]
- Kong B, Wang L, Chiang JYL, Zhang Y, Klaassen CD, Guo GL. (2012) Mechanism of tissue-specific farnesoid X receptor in suppressing the expression of genes in bile-acid synthesis in mice. Hepatology 56:1034–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Ding X, Riddick DS. (2013) Downregulation of mouse hepatic CYP3A protein by 3-methylcholanthrene does not require cytochrome P450-dependent metabolism. Drug Metab Dispos 41:1782–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CY, Lee S, Cade S, Kuo L-J, Schultz IR, Bhatt DK, Prasad B, Bammler TK, Cui JY. (2017) Novel interactions between gut microbiome and host drug-processing genes modify the hepatic metabolism of the environmental chemicals polybrominated diphenyl ethers. Drug Metab Dispos 45:1197–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao MZ, Gao C, Bhatt DK, Prasad B, Mao Q. (2018) Quantitative proteomics reveals changes in transporter protein abundance in liver, kidney and brain of mice by pregnancy. Drug Metab Lett 12:145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Jiménez CP, Jover R, Donato MT, Castell JV, Gómez-Lechón MJ. (2007) Transcriptional regulation and expression of CYP3A4 in hepatocytes. Curr Drug Metab 8:185–194. [DOI] [PubMed] [Google Scholar]
- Miro-Blanch J, Yanes O. (2019) Epigenetic regulation at the interplay between gut microbiota and host metabolism. Front Genet 10:638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DR, Zeldin DC, Hoffman SMG, Maltais LJ, Wain HM, Nebert DW. (2004) Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics 14:1–18. [DOI] [PubMed] [Google Scholar]
- Nuriel-Ohayon M, Neuman H, Koren O. (2016) Microbial changes during pregnancy, birth, and infancy. Front Microbiol 7:1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paine MF, Khalighi M, Fisher JM, Shen DD, Kunze KL, Marsh CL, Perkins JD, Thummel KE. (1997) Characterization of interintestinal and intraintestinal variations in human CYP3A-dependent metabolism. J Pharmacol Exp Ther 283:1552–1562. [PubMed] [Google Scholar]
- Papacleovoulou G, Abu-Hayyeh S, Williamson C. (2011) Nuclear receptor-driven alterations in bile acid and lipid metabolic pathways during gestation. Biochim Biophys Acta 1812:879–887. [DOI] [PubMed] [Google Scholar]
- Pariente G, Leibson T, Carls A, Adams-Webber T, Ito S, Koren G. (2016) Pregnancy-associated changes in pharmacokinetics: a systematic review. PLoS Med 13:e1002160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love M, Irizarry R, Kingsford C. (2015) Salmon provides accurate, fast, and bias-aware transcript expression estimates using dual-phase inference. bioRxiv Available from: 10.1101/021592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perloff MD, von Moltke LL, Cotreau MM, Greenblatt DJ. (1999) Unchanged cytochrome P450 3A (CYP3A) expression and metabolism of midazolam, triazolam, and dexamethasone in mdr(-/-) mouse liver microsomes. Biochem Pharmacol 57:1227–1232. [DOI] [PubMed] [Google Scholar]
- Prasad B, Bhatt DK, Johnson K, Chapa R, Chu X, Salphati L, Xiao G, Lee C, Hop CECA, Mathias A, et al. (2018) Abundance of phase 1 and 2 drug-metabolizing enzymes in alcoholic and hepatitis C cirrhotic livers: a quantitative targeted proteomics study. Drug Metab Dispos 46:943–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud HJ, Cui JY, Khan M, Klaassen CD. (2011) Tissue distribution and gender-divergent expression of 78 cytochrome P450 mRNAs in mice. Toxicol Sci 124:261–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon JM, Kang DJ, Hylemon PB. (2006) Bile salt biotransformations by human intestinal bacteria. J Lipid Res 47:241–259. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachar M, Kelly EJ, Unadkat JD. (2019) Mechanisms of CYP3A induction during pregnancy: studies in HepaRG cells. AAPS J 21:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selwyn FP, Cheng SL, Bammler TK, Prasad B, Vrana M, Klaassen C, Cui JY. (2015) Developmental regulation of drug-processing genes in livers of germ-free mice. Toxicol Sci 147:84–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selwyn FP, Cheng SL, Klaassen CD, Cui JY. (2016) Regulation of hepatic drug-metabolizing enzymes in germ-free mice by conventionalization and probiotics. Drug Metab Dispos 44:262–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Buschmann MM, Gilbert JA. (2019) Pharmacomicrobiomics: the holy grail to variability in drug response? Clin Pharmacol Ther 106:317–328. [DOI] [PubMed] [Google Scholar]
- Shuster DL, Bammler TK, Beyer RP, Macdonald JW, Tsai JM, Farin FM, Hebert MF, Thummel KE, Mao Q. (2013) Gestational age-dependent changes in gene expression of metabolic enzymes and transporters in pregnant mice. Drug Metab Dispos 41:332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster DL, Risler LJ, Liang CKJ, Rice KM, Shen DD, Hebert MF, Thummel KE, Mao Q. (2014) Maternal-fetal disposition of glyburide in pregnant mice is dependent on gestational age. J Pharmacol Exp Ther 350:425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smutny T, Mani S, Pavek P. (2013) Post-translational and post-transcriptional modifications of pregnane X receptor (PXR) in regulation of the cytochrome P450 superfamily. Curr Drug Metab 14:1059–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C, Love MI, Robinson MD. (2015) Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000 Res 4:1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanogiannopoulos P, Bess EN, Carmody RN, Turnbaugh PJ. (2016) The microbial pharmacists within us: a metagenomic view of xenobiotic metabolism. Nat Rev Microbiol 14:273–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, LaTour A, Liu Y, Klaassen CD, Brown KK, Reinhard J, et al. (2001) The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci USA 98:3369–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi S, Nakajima M, Mohri T, Yokoi T. (2008) Post-transcriptional regulation of human pregnane X receptor by micro-RNA affects the expression of cytochrome P450 3A4. J Biol Chem 283:9674–9680. [DOI] [PubMed] [Google Scholar]
- Tasnif Y, Morado J, Hebert MF. (2016) Pregnancy-related pharmacokinetic changes. Clin Pharmacol Ther 100:53–62. [DOI] [PubMed] [Google Scholar]
- Wahlström A, Kovatcheva-Datchary P, Ståhlman M, Bäckhed F, Marschall H-U. (2017) Crosstalk between bile acids and gut microbiota and its impact on farnesoid X receptor signalling. Dig Dis 35:246–250. [DOI] [PubMed] [Google Scholar]
- Wang H, Chen J, Hollister K, Sowers LC, Forman BM. (1999) Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol Cell 3:543–553. [DOI] [PubMed] [Google Scholar]
- Wang S, Chen L, Wang Q, He Z, Chen S, Zhang H, Li H, Guo P, Li Q, Zhang R, et al. (2019) Strain differences between CD-1 and C57BL/6 mice in expression of metabolic enzymes and DNA methylation modifications of the primary hepatocytes. Toxicology 412:19–28. [DOI] [PubMed] [Google Scholar]
- Yuan C, Burns MB, Subramanian S, Blekhman R. (2018) Interaction between host MicroRNAs and the gut microbiota in colorectal cancer. mSystems 3:e00205–e00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wu X, Wang H, Mikheev AM, Mao Q, Unadkat JD. (2008) Effect of pregnancy on cytochrome P450 3a and P-glycoprotein expression and activity in the mouse: mechanisms, tissue specificity, and time course. Mol Pharmacol 74:714–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Klaassen CD. (2013) Hormonal regulation of Cyp4a isoforms in mouse liver and kidney. Xenobiotica 43:1055–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YKJ, Yeager RL, Klaassen CD. (2009) Circadian expression profiles of drug-processing genes and transcription factors in mouse liver. Drug Metab Dispos 37:106–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao BS, Roundtree IA, He C. (2017) Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol 18:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]