Abstract
Wastewater-based monitoring of the spread of the new SARS-CoV-2 virus, also referred to as wastewater-based epidemiology (WBE), has been suggested as a tool to support epidemiology. An extensive sampling campaign, including nine municipal wastewater treatment plants, has been conducted in different cities of the Federal State of North Rhine-Westphalia (Germany) on the same day in April 2020, close to the first peak of the corona crisis. Samples were processed and analysed for a set of SARS-CoV-2-specific genes, as well as pan-genotypic gene sequences also covering other coronavirus types, using reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Additionally, a comprehensive set of chemical reference parameters and bioindicators was analysed to characterize the wastewater quality and composition. Results of the RT-qPCR based gene analysis indicate the presence of SARS-CoV-2 genetic traces in different raw wastewaters. Furthermore, selected samples have been sequenced using Sanger technology to confirm the specificity of the RT-qPCR and the origin of the coronavirus. A comparison of the particle-bound and the dissolved portion of SARS-CoV-2 virus genes shows that quantifications must not neglect the solid-phase reservoir. The infectivity of the raw wastewater has also been assessed by viral outgrowth assay with a potential SARS-CoV-2 host cell line in vitro, which were not infected when exposed to the samples. This first evidence suggests that wastewater might be no major route for transmission to humans. Our findings draw attention to the need for further methodological and molecular assay validation for enveloped viruses in wastewater.
Keywords: COVID-19, SARS-CoV-2, SARS-CoV-2 replication in vitro, Wastewater-based epidemiology (WBE), Wastewater treatment
Graphical abstract
1. Introduction
The current SARS-CoV-2 pandemic has far-reaching global consequences on public health, economic activities, and societies as a whole, which are unprecedented in many respects and cannot be fully assessed yet (WHO, 2020). The number and proportion of persons infected with COVID-19 are mainly determined based on individual testing and laboratory-based bio-molecular diagnostics using, e.g., reverse transcription-quantitative polymerase chain reaction (RT-qPCR). COVID-19 cases are reported by regional health authorities and aggregated on the level of, e.g., the Federal States in Germany, as well as on a national level (RKI, 2020). It is expected that, based on the individual testing that is often triggered by symptoms of test candidates or their respective risk profile, the actual state of infection in a specific region can only be very roughly estimated (Wurtzer et al., 2020). A study from China has also indicated that non-detected cases can make a significant contribution to the development of the infection as a whole (Li et al., 2020). Various researchers have shown that SARS-CoV-2 RNA is detectable in the feces of infected persons (e.g., Amirian, 2020). Furthermore, SARS-CoV-2 RNA can be found in stool samples weeks after the infection is no longer detectable in oral swab samples (Y. Wu et al., 2020). Therefore, it must be systematically assessed whether the virus might, in addition to respiratory droplets, also be transmitted via feces in wastewater (Nemudryi et al., 2020; Tang et al., 2020; Wang et al., 2020; Y. Wu et al., 2020; F. Wu et al., 2020). It could be shown that the duration of viral shedding differed among patients between 14 and 21 days past the onset of the infection (Wu et al. 2020, Xu et al., 2020). Furthermore, the magnitude of shedding varied from 102 to 108 RNA copies per gram feces (Lescure et al., 2020; Pan et al., 2020). Wastewater-based epidemiology (WBE) has been suggested as a potentially useful complementary tool to gain insights into the degree of infection spread in a population (Wu et al. 2020, Choi et al., 2018, Rodriguez-Manzano et al., 2010). Recently, various studies detected SARS-CoV-2 RNA in wastewater worldwide (cf. Table 1 ) like the Netherlands (Medema et al., 2020), France (Wurtzer et al., 2020), USA (Wu et al. 2020), Australia (Ahmed et al., 2020), and Italy (La Rosa et al., 2020). Wurtzer et al. (2020) reported the analysis of SARS-CoV-2 genes in the Greater Paris (France) area and were able to correlate trends in gene occurrence in the wastewater of different wastewater treatment plants (WWTP) with the number of infected individuals. Medema et al. (2020) have shown a good correlation between the number of COVID-19 cases and the gene concentration in the wastewater of different Dutch cities.
Table 1.
Overview of selected SARS-CoV2 studies in wastewater conducted in 2020.
| Study region | Genetic traces/genes analysed | Results | Reference |
|---|---|---|---|
| Paris (France) | RdRp | Samples in three WWTPs positive between 5th March and 23th April | Wurtzer et al., 2020 |
| Milan and Rome (Italy) | ORF1ab, S | 6 out of 12 samples positive in raw wastewater | La Rosa et al., 2020 |
| Netherlands | N1, N2, N3, E | 4 out of 7 wastewater samples positive at the beginning of March. 9 out 9 wastewater samples positive in the middle of March | Medema et al., 2020 |
| Milan (Italy) (WWTP and river) | ORF1ab, N, E | First sampling: 3 out of 4 raw wastewater samples positive 2 out of 2 effluent samples negative 2 out of 2 river samples positive |
Rimoldi et al., 2020 |
| Brisbane (Australia) | N and confirmation via Sanger and MiSeq Illumina sequencing | 2 out of 9 samples positive in raw wastewater | Ahmed et al., 2020 |
| Massachusetts (USA) | N1, N2, N3, | 10 out of 10 raw wastewater samples positive | F. Wu et al., 2020 |
| Israel (different cities and facilities) | E | 2 out of 15 samples positive in March 2020 9 out of 11 samples positive in April 2020 |
Bar Or et al., 2020 |
| Bozeman, Montana (USA) | N1, N2 | 7 out of 7 positive in March/April 2020 | Nemudryi et al., 2020 |
| Istanbul (Turkey) | RdRp | 9 out of 9 sludge samples positive | Kocamemi et al., 2020 |
| Valencia (Spain) | N1, N2, N3 | 35 out of 42 influent samples positive 2 out of 18 secondary treated samples positive 0 out of 12 tertiary effluent samples positive |
Randazzo et al., 2020 |
| Yamanashi Prefecture, Japan | N1, N2 | None of 5 influent samples positive 1 out of 5 secondary effluent samples positive None out of 3 river water samples positive |
Haramoto et al., 2020 |
| Ahmedabad, Gujara, India | ORF1ab, N and S | 2 out of 2 influent samples positive 2 out 2 effluent samples negative |
Kumar et al., 2020 |
| Louisiana, USA | N1 and N2 | 2 out of 15 raw wastewater samples positive Effluent samples all negative |
Sherchan et al., 2020 |
| Quito, Ecuador | N1 and N2 | 3 out of 3 river water samples positive | Guerrero-Latorre et al., 2020 |
However, the studies differed in, e.g., the type of samples, processing procedure, and targeted genes (like N1, N2, N3, and ORF1ab) in RT-qPCR analysis. Only a few studies were complemented by infectivity tests of the genes to determine whether the genetic material was present in intact virus particles or as free nucleic acids. Furthermore, only a few studies comprised phylogenetic analyses to better identify the genetic profile of the obtained material. An overview of recent studies conducted and reported in 2020 is given in Table 1.
Carducci et al. (2020) reviewed the current knowledge about the fate of coronaviruses in different environmental compartments, including wastewater. They revealed that only 22 studies were done since 1978 and that more in-depth studies are needed to gain a better understanding of the possible circulation of SARS-CoV-2 in water systems.
In particular, there are still open methodological questions on how to derive correlations between wastewater-based analysis and acute infection cases in the sampled region, as well as concerning the infectivity of the wastewater-borne genetic material of viral origin (Lodder and de Roda Husman, 2020). Additionally, little is known about the potential distribution of the virus in the aquatic environment and the survival of SARS-CoV-2 in water (Naddeo and Liu, 2020). On the one hand, it could be shown that SARS-CoV-2 from treated wastewater was potentially still infectious (Wurtzer et al., 2020). On the other hand, in the study of Rimoldi et al. (2020), the test of viability showed that pathogenicity of the virus in wastewater was negligible. Therefore, they assumed that the risk for public health was not significant.
The present study encompasses a comprehensive analysis of a set of samples from a coordinated campaign from nine wastewater treatment plants (WWTP) in the Federal State of North Rhine-Westphalia, Germany. The liquid and solid phase of the inflow samples were analysed, and a broad spectrum of reference parameters (e.g., bioindicators, nutrients, sum parameters, pharmaceuticals) was measured. Moreover, several SARS-CoV-2 specific genes were compared for their sensitivity and selectivity. A confirmation was conducted via Sanger sequencing. Replication tests were conducted to check the viability of the viral genetic material.
2. Material and methods
2.1. Sewage sampling
Nine municipal WWTP operated by six different water boards were selected for analysis throughout North-Rhine Westphalia (Germany). The plants differed in their design capacity, treatment processes, and connected catchment area characteristics (Table 2 ). Using installed autosampler devices, the operators of the WWTP collected 24 h flow-dependent composite samples on April 8th, 2020, during dry-weather conditions, either midnight to midnight or between the morning of April 8th, and the morning of April 9th. In addition to raw wastewater (sewage) inflow samples collected after the sand trap, treated sewage was sampled at selected locations (Table 2). All samples were transported to the laboratory in glass flasks on melting ice on the day of sampling.
Table 2.
Key properties of WWTPs sampled.
| Acronym | WWTP | Treatment process | Design capacitya PEdesign [–] |
Nominal number of connected residentsb PEnom [–] |
Annual wastewater flow in 2019a [m3/a] | Actual wastewater flow on April 8th, 2020b Qactual [m3/day] |
Sample ID and sampling location on April 8th, 2020 | |
|---|---|---|---|---|---|---|---|---|
| KLEM | Kläranlage Emscher-mündungc | AS | 2,400,000 | 2,228,933 | 347,602,053 | 1,159,488 | P1 | Inflow |
| P2 | Effluent | |||||||
| DU | Duisburg-Kaßlerfeld | AS | 450,000 | 257,262 | 33,028,604 | 82,957 | P3 | Inflow |
| E | Essen-Süd | AS | 135,000 | 119,226 | 12,543,902 | 29,695 | P4 | Inflow |
| MO | Moers-Gerdt | AS | 250,000 | 118,018 | 8,626,863 | 25,299 | P5 | Inflow |
| MG | Mönchenglad-bach-Neuwerk | AS | 632,500 | 494,077 | 33,382,628 | 85,131 | P6 | Inflow |
| EUS | Euskirchen-Kessenich | AS | 132,000 | 69,496 | 8,213,543 | 20,285 | P7 | Inflow |
| ERFT | Bergheim-Kenten | AS | 120,000 | 98,898 | 7,331,398 | 13,432 | P8 | Inflow |
| DN | Düren | AS | 310,000 | 134,723 | 20,104,329 | 54,253 | P9 | Inflow |
| P10 | Effluent | |||||||
| AC | Aachen-Soers | AS followed by full-scale ozonation and filtration | 458,000 | 205,000 | 25,057,198 | 66,052 | P11 | Inflow |
| P12 | After tertiary treatment | |||||||
| P13 | Effluent after ozonation and filtration | |||||||
AS: activated sludge treatment, PE: population equivalent.
Data from ELWAS (2020).
Data provided by operator.
WWTP KLEM treats the complete flow of river Emscher. PEnom refers to the complete upstream catchment area, including PEnom of all upstream WWTPs discharging into river Emscher.
For controls, sewage samples collected in earlier dry-weather sampling campaigns prior to the assumed onset of the COVID-19 pandemics in Germany that were kept frozen at −18 °C until analysis were used. Controls were sampled at WWTP Moers-Gerdt (C1-inflow sampled on July 18th, 2019), and Aachen-Soers (C2-effluent sampled in November 2018, C3-effluent in September 2019, and C4-inflow in January early 2017).
2.2. Sample processing for isolation and quantification of viral RNA
Frozen wastewater samples were thawed at 4 °C, and a total of 45 mL were further processed. First, wastewater was centrifuged at 4700 ×g for 30 min without break, and the clear supernatant was harvested. Purified wastewater was then concentrated using centrifugal ultrafiltration units (Amicon® Ultra-15 Centrifugal Filter Unit, Sigma). Therefore, 15 mL of wastewater was added to the filter unit and centrifuged for 15 min at 3500 ×g, and the concentrated supernatant was harvested. This was repeated twice until 45 mL of wastewater were completely concentrated (final volume of concentrated supernatant approximately 450 μL). For the solid phase of the wastewater sample, the pellet of the 45 mL sample was first washed with deionized water to remove aqueous remains from the sample and centrifuged at 4700 ×g for 5 min before being resuspended in 150 μL deionized water and centrifuged at 4700 ×g for 5 min again. A volume of 150 μL of the supernatant was then harvested for further RNA extraction, and the mass was determined (approx. 150 mg in influent and 5–10 mg in effluent). Contaminated equipment (e.g., reaction tubes, tips, filter units) were collected and autoclaved according to the daily cleaning program in the lab.
2.3. RNA extraction
RNA was isolated using the NucleoSpin RNA Virus kit (Macherey Nagel) following the manufacturer's instructions. Briefly, 150 μL supernatant was mixed with lysis buffer supplemented with carrier RNA. After binding on silica membranes, samples were washed several times and eluted in 50 μL RNase-free water. Isolated RNA was stored at −80 °C.
2.4. SARS-CoV-2 specific quantitative RT-qPCR
RNA was analysed by OneStep RT-qPCR using Luna Universal Probe One-Step RT-qPCR Kit (New England Biolabs) or LightCycler® Multiplex RNA Virus Master (Roche) and the CFX96 Real-Time System, with a C1000 Touch Thermal Cycler (Bio-Rad). Primer pairs for E-, N- and M-gene (Toptan et al., 2020) specific PCRs were used in equimolar concentrations (0.4 μM each per reaction), while RdRP-primer pairs were used according to Corman et al. (2020) with 0.6 μM and 0.8 μM for forward and reverse primers, respectively. The sequences of primers and probes are found in Table 3 . For probe-based Luna Universal One-Step RT-qPCR kits, 2 μL of RNA were subjected to reverse transcription performed at 55 °C for 10 min. Initial denaturation was performed for 1 min at 95 °C followed by 45 cycles of denaturation for 10 s and combined annealing and extension for 30 s at 60 °C. For the LightCycler® Multiplex RNA Virus Master kit, 5 μL of template RNA were used. Reverse transcription was performed at 55 °C for 10 min. Initial denaturation was allowed for 30 s at 95 °C followed by 45 cycles of denaturation for 5 s, extension for 30 s at 60 °C and final cool-down to 40 °C for 30 s. The PCR runs were analysed with Bio-Rad CFX Manager software version 3.1 (Bio-Rad Laboratories). For quantifications, standard curves using plasmid DNA (RdRP) or in vitro transcribed RNA (M-gene) were used as described previously (Toptan et al., 2020). Depending on the RT-qPCR for M-gene or RdRP the detection limit was 200 copies per reaction. If not indicated otherwise, a reaction was considered positive if the cycle threshold (CT) was below 40 cycles. To calculate from gene equivalents per reaction back to copies per mL, a PCR correction factor (cf) was determined with Luna Universal Probe One-Step RT-qPCR Kit (cf = 0.0037) and LightCycler® Multiplex RNA Virus Master (cf = 0.0015), respectively. To control PCR, viral RNA of SARS-CoV-2 (isolate SARS-CoV-2-FFM1) was used as a positive control (Hoehl et al., 2020; Toptan et al., 2020) and water as a negative control. Furthermore, to verify specificity and sensitivity, RNA of SARS-CoV (isolate SARS-CoV-FFM1) (Drosten et al., 2003) and HCoV-229E from cell culture supernatant was isolated and used in PCR.
Table 3.
Sequences of primers and probe established and verified for the detection of SARS-CoV-2 (in patient material) by RT-qPCR.
| Oligo name | Oligonucleotide sequence (5′-3′) | Gene | Reference |
|---|---|---|---|
| E_Sarbeco_F1 | ACAGGTACGTTAATAGTTAATAGCGT | E-gene | Corman et al., 2020 |
| E_Sarbeco_R2 | ATATTGCAGCAGTACGCACACA | ||
| E_Sarbeco_P1 | 6-Fam ACACTAGCCATCCTTACTGCGCTTCG BBQ1 | ||
| RdRP_SARSr-F2 | GTGARATGGTCATGTGTGGCGG | RdRP | Corman et al., 2020 |
| RdRP_SARSr-R1 | CARATGTTAAASACACTATTAGCATA | ||
| RdRP_SARSr-P2 | 6-Fam CAGGTGGAACCTCATCAGGAGATGC BBQ1 | ||
| M-475-F | TGTGACATCAAGGACCTGCC | M-gene | Toptan et al., 2020 |
| M-574-R | CTGAGTCACCTGCTACACGC | ||
| M-507-P | 6-Fam TGTTGCTACATCACGAACGC BHQ1 | ||
| SARS-CoV-2 N-gene F | TGGCCGCAAATTGCACAATT | N-gene | Toptan et al., 2020 |
| SARS-CoV-2 N-gene R | TGTAGGTCAACCACGTTCCC | ||
| SARS-CoV-2 Probe-N | 6-Fam CGCATTGGCATGGAAGTCAC BHQ1 |
2.5. Sanger sequencing analysis
RdRP-PCR products (wastewater sample P12, WTTP Aachen Soers after tertiary treatment; April 8th, 2020, effluent C2 sampled in November 2018 and inflow C4 sampled in January early 2017) were purified using the GenElute™ PCR Clean-Up Kit according to manufacturer's instructions and sequenced from both ends on the Applied Biosystems 3730 DNA Analyzer platform at the BiK-F Laboratory Centre, Frankfurt, Germany. Sequences were identified in terms of the closest homology sequence using BLAST, https://blast.ncbi.nlm.nih.gov/Blast.cgi (database Nucleotide collection (nr/nt) using Blastn (optimized for somewhat similar sequences) or Megablast (optimized for highly similar sequences)). Additional alignments were performed using Clustal Omega program for multiple sequence alignment (EMBL-EBI; https://www.ebi.ac.uk/Tools/msa/clustalo/). Results of the BLAST search and alignments can be found in Supplementary Fig. S1.
2.6. Replication studies with SARS-CoV-2 positive wastewater
Purified wastewater samples that tested positive for SARS-CoV-2 (P2, P5, P11, P12) were investigated for replication-competent viruses following a recently published procedure (Hoehl et al., 2020). Potential infectious work was performed under biosafety level 3 (BSL-3) conditions in a BSL-3 facility according to the Committee on Biological Agents (ABAS) and the Central Committee for Biological Safety (ZKBS). Briefly, wastewater samples were processed as described in Section 2.2. A human epithelial cell line from colon adenocarcinoma (Caco-2) was grown in cell culture tubes and infected with a 1:10 dilution of purified wastewater in Minimum Essential Media (MEM) supplemented with 1% fetal calf serum, 2% l-glutamine and 1% penicillin/streptomycin. As a control, cells were infected with SARS-CoV-2-containing cell culture supernatant. Cells were cultured for up to 10 days post-infection. Infection with replication-competent virus results in the development of a cytopathic effect, which was measured visually by light microscopy.
2.7. Analysis of chemical and physicochemical reference parameters
All physicochemical and chemical parameters were analysed immediately after sampling following DIN, EN, or ISO standard protocols. If this was not possible, the samples were chemically stabilized and then measured within the approved timeline of the standard protocols. The pH and conductivity were measured using a sensION MM374 Hach Lange instrument (EN ISO 10523 (2012-04), EN 27888 (1993-11)). For the content of the dry residues, a defined volume of wastewater sample was dried at 105 °C for 24 h (DIN 38409-1 (1987-01)), and the residue was weighed by a Sartorius A 120 S balance. Ready-to-use-cell test kits were used to analyse the chemical oxygen demand (COD) with a Macherey & Nagel PF12 and Vario 4 (test kits 985022, 985033 and 98029, ISO 15705 (2003-01)). Total organic carbon (TOC) and total bounded nitrogen (TNb) were analysed via combustion using a Shimadzu carbon/nitrogen analyser (EN 1484 (1997-08), EN 12260 (2003-12)). For the analysis of ammonia, nitrate, nitrite, total organic nitrogen, ortho-phosphate, and other ions such as chloride and sulfate, a Gallery discrete analyser with photometric and turbidity methods were used after filtration through a 0.45-μm filter (Thermo Fisher Scientific, ISO 15923-1 (2014-07)).
Two different types of population size markers (exogenous and endogenous) were measured by liquid chromatography coupled with mass spectrometric detection (LC-MS) and photometric detection with a discrete analyser (Choi et al., 2018). LC-MS analysis for the exogenous population size indicators was carried out by using an HP Series 1100 LC coupled to an MS/MS-System (Thermo Finnigan TSQ Quantum, Thermo Fisher Scientific) in positive electrospray ionization mode. Before measurement, 100-mL wastewater samples were spiked with internal standards (Supplementary Table S1) and then enriched and cleaned on an Oasis HLB cartridge, diluted with methanol, dried under nitrogen flow, and re-dissolved in 1 mL methanol/water (50/50 v/v) for injection. For separation, an RP C18-column with polar endcapping was used (Hypersil GOLD™ aQ C18, Thermo Fisher Scientific). The method was based on a German standard DIN 38407-47: 2017-07. The mobile phase was a gradient containing Millipore water and methanol. 2 mM ammonium acetate and 0.1% acetic acid were added to the methanol and water. The separation was run after injection of 10 μL sample with a linear gradient starting with 20% up to 90% and ending again with 20% methanol. All data were collected in SRM mode by using mass-labeled internal standards (Supplementary Table S1) and Xcalibur 2.0.7. SP1 software for data acquisition and quantification. The LOQs are in the range of 10 to 30 ng/L.
As endogenous parameters, creatinine and urea were quantified after filtration via a 0.45 μm filter by using a Gallery discrete analyser (Thermo Fisher Scientific). Creatinine was measured by photometry at 540 nm as quinonimine-chromogen after enzymatic reactions of creatine with creatinase, sarcosine oxidase, and ascorbate oxidase with a modified clinical method using Thermo Fisher Scientific reagent kit (enzyme. colortest PAP 981896) (Dörner, 2003). The Urea-method is based on a standardized bathwater method. In the first step, the contents of urea and ammonium are detected simultaneously. After enzymatic reaction of urea with urease to ammonium, ammonium reacts with nicotinamide adenine dinucleotide phosphate (NADPH) in a buffered solution at pH 8. The loss of NADPH is directly proportional to the ammonium content (Thermo Fisher Scientific test kit 981820). The urea content is calculated by subtraction of the ammonium content (ISO 15923-1 (2014-07)) from the total content of urea + ammonium.
2.8. Prevalence of reported COVID-19 cases in WWTP catchment area
In Germany, local health authorities report the number of COVID-19 cases confirmed by laboratory diagnosis after aggregation from the community to district level to state and federal authorities. Whenever available, the cumulative prevalence and the cumulative number of COVID-19 patients recovered and deceased were obtained from community-level reports published on the homepages of the responsible local health authorities on April 9th, 2020. If community-level reports were not available, district-level data published by the state health ministry of North Rhine-Westphalia (MAGS) on April 9th, 2020, was used. Since the catchment areas of WWTPs rarely correspond to administrative boundaries, the cases were estimated by calculating weighted sums relative to the residents of each community connected to that sewer network. Acute prevalence was calculated by subtracting reported recovered and deceased patients (Table 4 ). Nominal incidences (Inom) were calculated by dividing the estimated number of cases (Nnom) in a catchment area by the nominal number of connected residents to the sewer (PEnom). We note that these are estimates with considerable uncertainty since cases might be unevenly distributed between neighbouring districts, and recovered cases are often not based on laboratory diagnosis but the end of ordered quarantine. In addition, the actual number of persons staying in the catchment area on the day of sampling (PEactual) may also differ from the nominal number of connected residents (PEnom). Treating the complete flow of the river Emscher, WWTP KLEM is partly treating water that was treated upstreaming by other treatment plants. Based on the assumption of poor removal of SARS-CoV-2 in conventional WWTP, reported cases for KLEM in Table 4 are upper estimates covering the complete upstream catchment.
Table 4.
Estimated nominal cumulative and acute prevalence of COVID-19 cases on April 8th, 2020, in the catchment areas of the WWTP studied.
| WWTP catchment area | Cumulative prevalence of COVID-19 |
Acute prevalence of COVID-19 |
||
|---|---|---|---|---|
| Number of cases (Nnom) | Nominal incidence per 100,000 (Inom = Nnom / PEnom) | Number of cases (Nnom) | Nominal incidence per 100,000 (Inom = Nnom/PEnom) | |
| KLEM | 1924 | 86 | 1037 | 47 |
| DU | 271 | 105 | 144 | 56 |
| E | 117 | 98 | 62 | 52 |
| MO | 85 | 72 | 36 | 30 |
| MG | 554 | 112 | 299 | 60 |
| EUS | 110 | 158 | 74 | 107 |
| ERFT | 218 | 220 | 172 | 174 |
| DN | 158 | 117 | 82 | 61 |
| AC | 292 | 142 | 128 | 62 |
3. Results
3.1. SARS-CoV-2 detection in wastewater: aqueous phase
Wastewater samples from 9 distinct municipal WWTP were collected in April 2020 during the pandemic outbreak of SARS-CoV-2 in North Rhine Westphalia to investigate the presence of virus-specific RNA in untreated and treated wastewater samples by quantitative RT-qPCR. Using well-established quantitative RT-qPCRs for viral M (Fig. 1A) and RdRP genes (Fig. 1B), PCRs provided evidence of SARS-CoV-2-specific RNA traces in the aqueous phase of the tested sewage samples. SARS-CoV-2 RNA copies were found in a range from 3.0 and 20 gene equivalents per mL in the inflow, and 2.7 to 37 gene equivalents in the WWTP effluent (Fig. 1), which is in line with other reports in the Netherlands and Australia (Medema et al., 2020; Ahmed et al., 2020). CT values for the tested wastewater samples ranged between 32 and 35 for M-gene, and 33 and 37 for RdRP. Interestingly, all wastewater samples showed detectable virus RNA. The higher number of SARS-CoV-2 RNA copies and the higher standard deviation found for the viral RdRP analyses might possibly be attributed to variability between single PCR measurements given by RNA isolation and storage conditions for RNA extraction compared to samples analysed for M-genes. However, the tendencies obtained with both PCR targets were consistent.
Fig. 1.
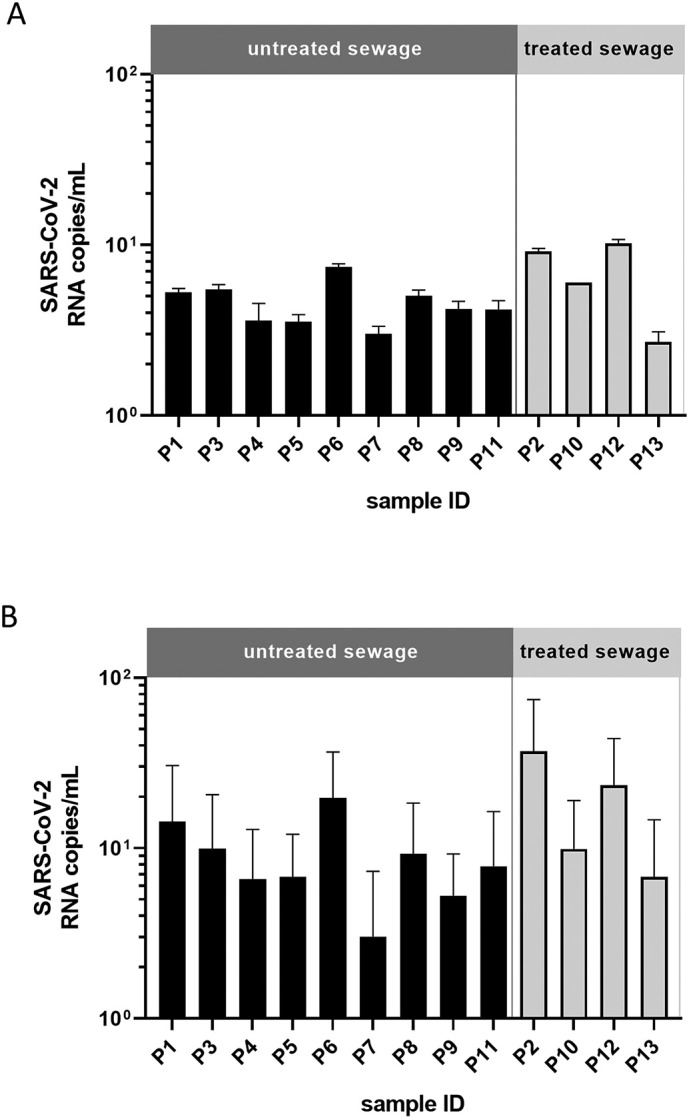
SARS-CoV-2 specific RNA fragment detected by (A) M-gene and (B) RdRP RT-qPCR in the aqueous phase of untreated and treated municipal sewage. Error bars indicate one standard deviation of duplicate samples. Results of a single PCR measurement are shown for M-gene. For M-gene RT-qPCR, CT values for the standard ranged between 8.6 (standard 1 = 108) and 38.4 (standard 6 = 102). Values of tested wastewater above CT 39 were considered negative for SARS-CoV-2. Standard curve was calculated using Bio-Rad CFX Manager software with E = 61.4%, R2 = 0.952 and slope = −4.813. Results of two independent PCR measurements of the same samples are shown for RdRp. For RdRP RT-qPCR CT values for standard range between 18 (standard 1 = 108) and 37 (standard 6 = 102). Values of tested wastewater above CT 38 were considered negative for SARS-CoV-2. Standard curve was calculated using Bio-Rad CFX Manager software with E = 96.5%, R2 = 0.990 and slope = −3.408.
3.2. Specificity and sensitivity of different SARS-CoV-2 target genes in RT-qPCR
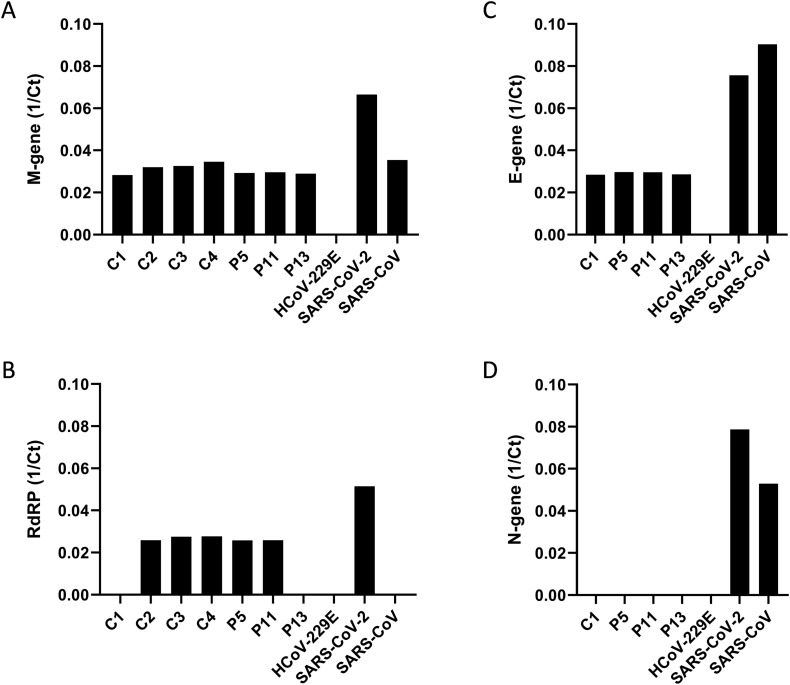
Given the complex nature of municipal sewage, disturbing factors accumulated during sample processing such as nucleic acids of other human coronaviruses circulating in the human population and potentially concentrated in the wastewater samples could influence analysis results. To verify the specificity and sensitivity of the PCR, we compared different viral genes (N-gene, E-gene, M-gene, and RdRP) and performed RT-qPCR with three different samples of sewage from April 2020, one control sample (C1) and RNA of three different coronavirus isolates (SARS-CoV-2, SARS-CoV, and HCoV-229E).
High specificity for SARS-CoV-2 with good sensitivity was found for the viral RdRP gene compared to the other tested viral genes (Fig. 2 ). Although no signal was detected for HCoV-229E with the RdRP gene PCRs, unspecific binding of primer and probe to other human coronaviruses like HCoV-NL63, HCoV-OC43, or HCoV-HKU1 cannot be excluded. Previous evaluation of the primers and probe within a different test setting analysing patient material or cell culture-derived SARS-CoV-2 showed minor cross-reactivity with other human coronaviruses (data not shown). Using a more sensitive and specific PCR for RdRP and M-gene may explain the detection of SARS-CoV-2 in all wastewater samples compared to other studies where PCR against N-gene was used to detect the presence of viral RNA in the wastewater. Comparing M-gene and RdRP PCR results to N- and E-gene PCR, specificity and sensitivity was clearly increased for RdRP and M-gene. However, taking into account that the M-gene and RdRP PCR may detect other endemic human coronaviruses, wastewater samples collected before the SARS-CoV-2 outbreak in Germany were investigated due to the detection of a positive PCR signal. Therefore, samples from WWTP Aachen-Soers collected in January 2017 (C4), November 2018 (C2), and September 2019 (C3) were processed comparably to the test samples, and RdRP-specific PCR was performed. Indeed, we could detect a minor signal in the PCR with CT values (2018: CT 38; 2019: CT 36 and 2020: CT 36) close to the detection limit of the PCR (CT 38) (data not shown). Comparing the amplification curve of SARS-CoV-2 positive control and retrained water controls showed comparable shape, but with 15 CT difference. A moderate correlation (R2 = 0.8733) of the inverse CT values of RdRP and M-gene PCR was detectable. Although there were positive results also for retained controls (C2 to C4), these samples grouped apart from the wastewater samples collected in April 2020 (Fig. 3 ).
Fig. 2.
Specificity and sensitivity of viral (A) M-gene, (B) RdRP-gene, (C) E-gene and (D) N-gene RT-qPCR with respect to control C1 sampled in July 2019 before the known onset of the pandemics in Germany, untreated sewage P5, P11, P13 sampled during the pandemics, SARS-CoV-2 control, and two other human coronaviruses HCoV-229E and SARS-CoV. Results are from wastewater samples measured in duplicates in a single PCR. Standard curve for M-gene PCR was calculated using Bio-Rad CFX Manager software with E = 90.8%, R2 = 0.996 and slope = −3.565. Efficiencies of other primer pairs were not calculated by serial dilution.
Fig. 3.
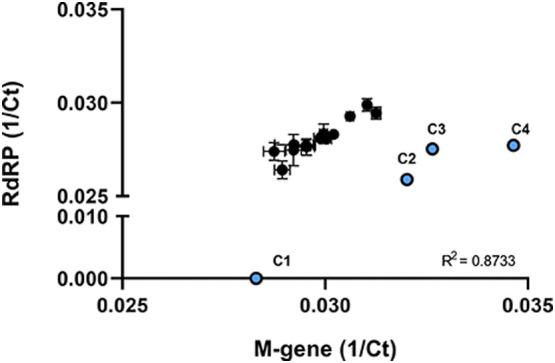
Correlation of reciprocal CT values between M-gene and RdRP-gene detection.
3.3. Sanger sequencing of PCR product
To further analyse retained wastewater samples showing a positive PCR result for M- and RdRP- gene, sequencing of the RdRP-PCR product of C2, C4 and P12 should evaluate if a specific signal was measured (Supplementary Fig. S1). While the sequence of the P12 PCR product showed significant similarity to SARS-CoV-2 sequence (isolate SARS-CoV-2/human/USA/CA-CZB-1525/2020; Accession: MT628199.1) confirming the positive identification of human SARS-CoV2 by means of RT-qPCR (Supplementary Fig. S1A), no match to SARS-CoV-2 was detectable for C2 (Supplementary Fig. S1B). In addition, a positive BLAST result for C4 with SARS-CoV-2 synthetic construct (accession: MT458696.1) (E value 0.002) (Supplementary Fig. S1C) was found. Aligning all three sequenced amplicons (P12, C2, and C4) with the SARS-CoV-2 specific sequence, only the amplicon of P12 showed a sequence identity above 90%. The similarity of C2 and C4 amplicons to the SARS-CoV-2 specific sequence was 50% and 47.3%, respectively (Fig. 4 ). Therefore, we conclude that the retained samples were negative for SARS-CoV-2.
Fig. 4.
Alignment of sequenced amplicons of P12, C2, and C4. Sequences of wastewater sample P12 and retrained samples C2 and C4 were analysed using Geneious - Bioinformatics Software for Sequence Data Analysis and compared to the SARS-CoV-2 synthetic construct sequence (GenBank: MT458696.1). In green primers binding sites are indicated. Dots in the sequence represent sequence mismatches to the SARS-CoV-2 synthetic construct sequence. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
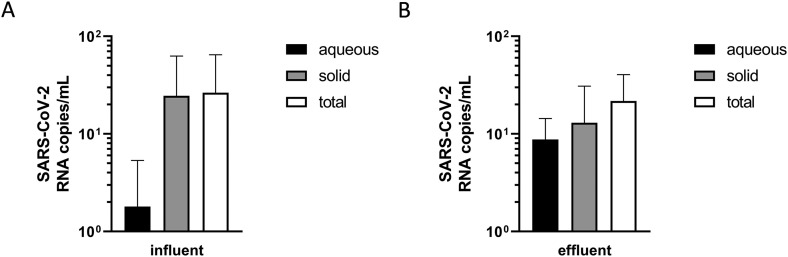
3.4. SARS-CoV-2 removal in wastewater treatment: role of the solid-phase reservoir
In wastewater treatment, solid residues are largely removed. We thus compared the amount of viral RNA in the aqueous and solid-phase of both inflow and effluent samples. During centrifugation in the processing of the wastewater samples, solid and aqueous phases are separated. To our understanding and as shown in other studies (Kocamemi et al., 2020), viral RNA of solid and liquid phase needs to be considered in wastewater surveillance. When comparing the aqueous and solid phase of the samples, we found a one log unit higher SARS-CoV-2 RNA copy number per mL in the solid phase (25 copies/mL) compared to the aqueous phase of the inflow sample (1.8 copies/mL), respectively (Fig. 5A). The difference between the aqueous and solid phase was less evident in the effluent sample, with 8.8 and 13 gene equivalents per mL, respectively (Fig. 5B). When comparing the aqueous phase only, the effluent exhibited a higher gene equivalent concentrations than the influent, which we attribute to repartitioning of gene material from the solid- to the liquid phase during wastewater treatment. In contrast, a difference of total viral copy numbers per mL between untreated and treated wastewater was not detectable (Fig. 5A and B). This might be explained by the sample conditions. While the residence time of the wastewater is about one day at the WWTP, sludge is continuously exchanged with an average sludge age of 10 to 14 days. We assume that solid-phase concentrations are lower-estimates since we did not test whether further extraction steps could mobilize additional SARS-CoV-2 RNA from the solid phase.
Fig. 5.
SARS-CoV-2 removal detected by RdRP RT-qPCR measured by the sum of aqueous and solid-phase of (A) untreated (AC-Inflow) and (B) treated sewage after ozonation. Results are shown as mean + SD of two independent PCR measurements from same sample material. RdRP RT-qPCR CT values for standard range between 18 (standard 1 = 108) and 37 (standard 6 = 102). Values of tested wastewater above CT 38 were considered negative for SARS-CoV-2. Standard curve was calculated using Bio-Rad CFX Manager software with E = 96.5%, R2 = 0.990 and slope = −3.408.
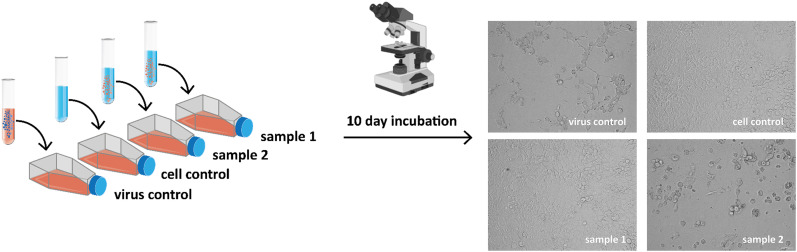
3.5. Infectious potential of sewage samples
Although the wastewater in the investigated plants was treated by different processes, viral RNA is not eliminated, as shown above. To test the infectious potential of untreated and treated wastewater, a cell culture model was used to analyse that impact (Fig. 6 ). Infection of differentiated Caco-2 cells with cell culture grown, replication-competent SARS-CoV-2 (MOI 0.01) led to the induction of a cytopathic effect (CPE after 48–72 h). The CPE is characterized by round and shrinking cells that detach from the culture surface. Observation of CPE after two to three days at a low MOI indicated rapid to moderate replication of the virus. Inoculation of differentiated Caco-2 cells for ten days with purified and concentrated wastewater (P2, P5, P11, and P12) did not result in the production of infectious SARS-CoV-2 particles (data not shown), which suggests that treated sewage appears to be non-infectious even though viral RNA fragments can be detected. Due to the lack of CPE, quantification and verification by RT-qPCR was not performed.
Fig. 6.
Viral outgrowth assay setup. Caco-2 cells were inoculated with either wastewater samples or with SASR-CoV-2 containing cell culture supernatant (positive control) or culture medium only (negative control). Cells were investigated daily by light microscopy for the appearance of a cytopathic effect caused by replication-competent SARS-CoV-2 infection for up to 10 days. The experiment was performed in a single replicate for four different wastewater samples (P2, P5, P11, P12) from April 8th 2020.
3.6. Reference parameters
The wastewater samples analysed showed typical wastewater characteristics for the different types of WWTPs studied (Brückner et al., 2018; Metcalf and Eddy, 2012; Supplementary Table S2). The measured concentrations of nutrients and physico-chemical parameters are in the same range as found in the last years in the official monitoring by the environmental protection agency of North Rhine-Westphalia (ELWAS, 2020). The samples P2, P10, P12, and P13 are taken in the effluent of the WWTP or the effluent of the clarification. As expected, the substance concentrations in these samples are lower than in the corresponding influent samples P1, P9, and P11. Only the effluent of the WWTP KLEM had a higher content of dry residues in the effluent than in the inflow, which is important for evaluating the partitioning between the liquid and solid phase.
Different bioindicators were analysed in the wastewater to evaluate whether they could serve to estimate the actual number of persons staying in the catchment area at the day of sampling (PEactual) more accurately than the nominal number of connected residents (PEnom) or the design capacity (PEdesign). Results are provided in Supplementary Table S3. The load of the bioindicators was found to correlate with the nominal number of connected residents (Fig. 7 ; Supplementary Table S5).
Fig. 7.
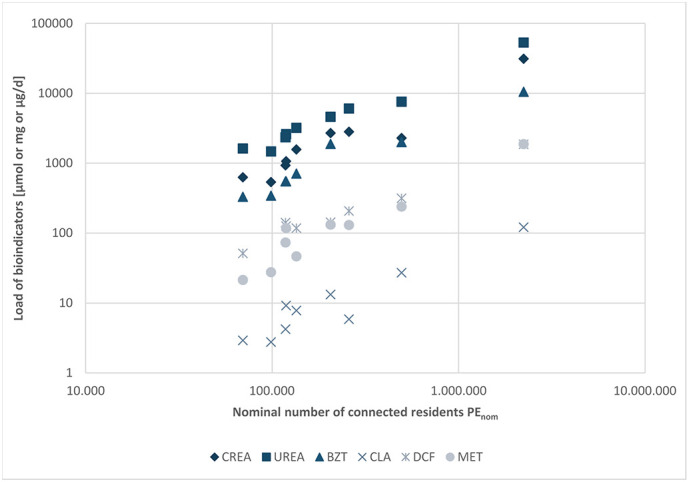
Bioindicator load for each WWTP: Creatinine [CREA μmol/day], urea [Urea mg/day], benzotriazole [BZT μg/day], clarithromycin [CLA μg/day], diclofenac [DCF μg/day], metoprolol [MET μg/day]. For regression lines and coefficients of determination, see Supplementary Table S5.
4. Discussion
4.1. Method validation: comparison to published papers and SOPs
During the SARS-CoV-2 pandemic, testing different sample material (e.g., throat swabs) was a crucial step in the detection of hotspots and limiting transmission. The first SARS-CoV-2 PCR assays were developed against the E-gene and the N-gene to detect the virus in patient samples, and numerous studies analysing wastewater for SARS-CoV-2 surveillance used E-gene or N-gene specific PCRs (Medema et al., 2020; Ahmed et al., 2020; Y. Wu et al., 2020; F. Wu et al., 2020). To a minor proportion, also PCR targeting SARS-CoV-2 RdRP was used (Wurtzer et al., 2020). In this study, wastewater samples were analysed with two different PCRs targeting SARS-CoV-2 RdRP and M-gene. In an initial analysis, a control sample (retained control) and wastewater test samples from April 2020 were tested for the presence of four different SARS-CoV-2 genes (N-, E-, M-gene, and RdRP), and specificity and sensitivity were evaluated based on other coronaviruses, including the closest relative SARS-CoV. RdRP-PCR was highly specific for SARS-CoV-2 and M-gene PCR being the most sensitive for SARS-CoV-2. These results are in line with recently published data by Toptan et al. (2020), where the efficiency of RT-qPCR primer and probes pairs to detect SARS-CoV-2 genes was compared. At least in this study, N-gene primers were less specific and were excluded due to limited linearity compared to other viral genes. Although other viral genes for SARS-CoV-2 detection in wastewater were used in this study, total gene equivalents per mL are comparable to other published studies (Ahmed et al., 2020; Y. Wu et al., 2020; F. Wu et al., 2020). To further control the specificity and sensitivity of our PCR, retained wastewater samples from 2017 to 2019, which should be negative for SARS-CoV-2, were analysed. Interestingly, a positive SARS-CoV-2 signal in PCR of retained samples from 2017 and 2018 was observed. One possible explanation could be a cross-contamination during the purification or RNA isolation step. To monitor the isolation process and identify possible cross-contamination, additional controls could be introduced such as isolation from a clean water control. So far unpublished data, analysing retained wastewater samples from Barcelona, Spain, taken in March 2019 showed positive RT-qPCR results for SARS-CoV-2 (Chavarria-Miró et al., 2020). The authors conclude that the virus was introduced to the Barcelona population due to global travel and remained undetected.
When sequencing the different PCR amplicons obtained using wastewater samples taken during the SARS-CoV-2 pandemic and long before the outbreak (retained samples), distinct results for the test sample (P12) which fits SARS-CoV-2/human/USA/CA-CZB-1525/2020 isolate from NCBI database was found, but not for the retained samples suggesting an unspecific signal in RT-qPCR, respectively. Although RNA samples used for PCR are highly purified, there might have been environmental contaminants affecting PCR. PCR is a highly sensitive method to detect nucleic acid and is strongly affected by pH, salt concentration, and contamination with extraneous nucleic acids that lead to false-negative or false-positive results, respectively (Schrader et al., 2012; Paul et al., 1991). BLASTn search showed minor sequence similarity (E values > 0.44) to Pithovirus, a giant DNA virus that infects amoebas (Legendre et al., 2014), bacteria like Streptococcus or animals (e.g., Xenopus laevis, Sparus aurata). To avoid false-positive results, optimization of PCR conditions can be taken in consideration. Primer pairs used in this study were optimized for RT-qPCR of patient material and cell culture-derived samples. The matrix of wastewater strongly differs from this defined sample material. Furthermore, primers adapted to RT-qPCR results in amplicons of approximately 100 bp, which are complicated to sequence. Establishing qualitative nested PCR could be an option to improve sequencing results. Furthermore, extraneous nucleic acids can be accumulated during the concentration process of the wastewater sample. While single studies may avoid the accumulation of extraneous nucleic acids using filter units with 10 kDa cut-off and therefore also could limit detection of SARS-CoV-2, most studies used filter units with 100 kDa cut-off or other methods for concentrating wastewater samples (Ahmed et al., 2020; Medema et al., 2020; Nemudryi et al., 2020; La Rosa et al., 2020). Although no false-positive results are described in these analyses, these studies did not use retained wastewater samples before the SARS-CoV-2 pandemic as control. Nevertheless, recently published data by La Rosa et al. (2020) or Chavarria-Miró et al. (2020) could detect SARS-CoV-2 RNA in retained wastewater samples taken before the first outbreak. Especially, wastewater analysis in Barcelona showed SARS-CoV-2 detection already in early 2019. However, results were not verified by an independent method like verifying PCR product size using gel electrophoresis, or sequencing (Chavarria-Miró et al., 2020). Taken together, it is important to consider environmental contaminants that might interfere with PCR and may lead to false-positive results. Appropriate controls, such as sequencing, are thus recommended to confirm PCR results.
4.2. Suitability for wastewater surveillance of COVID-19
To establish SARS-CoV-2 wastewater-based epidemiology as a reliable tool for early-warning and surveillance of the current or future COVID-19 outbreaks, the method's detection limit, precision, accuracy, and reliability must meet certain criteria to be useful for public surveillance. For example, early-warning requires a low detection limit to detect the very first cases, while in later phases of the epidemics, high precision and accuracy are needed to survey whether certain thresholds are exceeded, for example, the threshold currently set in Germany of 50 acute cases per 100,000 residents.
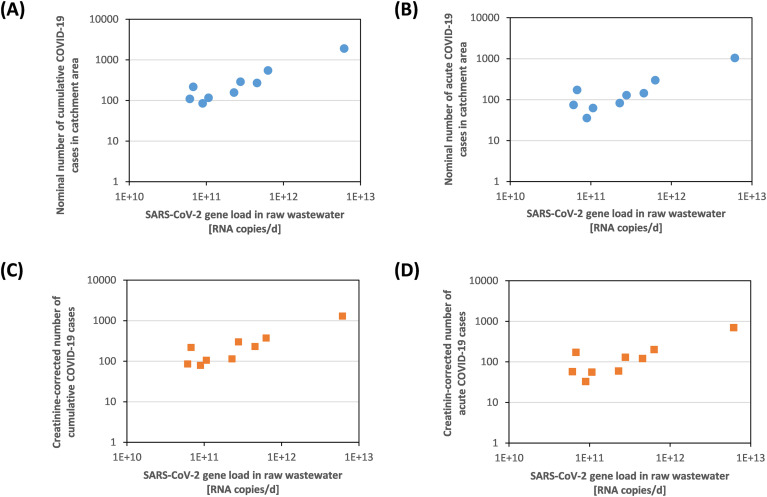
On the day of our sampling campaign, we estimate the nominal acute incidence of COVID-19 to range between 30 and 174 cases per 100,000 residents in the nine catchment areas studied (Table 4). Based on surveillance results (Fig. 4), our findings indicate that the RT-qPCR employed is capable of resolving acute incidences of 50 cases per 100,000 residents in dry-weather conditions, although the method has not yet been optimized in terms of sensitivity and precision to detect even lower incidence.
The nine catchment areas studied differ considerably in size, wastewater flow (Table 2), and nominal numbers of COVID-19 cases at the day of sampling ranging from 36 to 1037 acute cases and from 85 to 1924 cumulative cases (Table 4). Inter-comparing these nine catchment areas, we plotted the estimated cumulative and the acute prevalence against the measured SARS-CoV-2 load (Fig. 8 ), the latter calculated from RT-qPCR measured M-gene copy concentration (Fig. 4) and the actual wastewater flow Qactual on the day of sampling (Table 2). Clearly, the estimated nominal number of COVID-19 cases increases with increasing measured SARS-CoV-2 load, resulting in correlations both for cumulative and acute prevalence (Fig. 8A and B). Scatter in the correlations might be attributed to various reasons, including variations in virus RNA recovery and uncertainty in the estimated prevalence data. In contrast, plotting the incidence against SARS-CoV-2 concentration did not yield a conclusive correlation (not shown), likely because the precision of the qPCR employed was not sufficient to discriminate relatively minor differences in the incidence prevailing in the studied catchment areas at the time of sampling, ranging from 30 to 174 cases per 100,000 residents (less than an order of magnitude, Table 4).
Fig. 8.
Correlation between the measured SARS-CoV-2 M-gene copy loads and the nominal numbers of (A) cumulative and (B) acute COVID-19 cases on April 8th, 2020, in the catchment areas of the 9 WWTP studied. In comparison, creatinine-corrected numbers of (C) cumulative and (D) acute COVID-19 cases are displayed. For regressions and coefficients of determination, see Supplementary Table S5.
While the number of unreported cases is not known, also the number of persons staying in the catchment area (PEactual) may differ from the nominal number of residents connected to the sewer (PEnom). We evaluated the utilisation of various bioindicators to calculate the incidence in the catchment area by correcting PEnom to PEactual by multiplying PEnom with a bioindicator factor (BIF, Supplementary Table S4). Here we show results for creatinine that humans excrete in the urine (Fig. 8C and D). Although it cannot be evaluated based on the available data, if this correction is superior, it illustrates the potential of bioindicators to account for differences in wastewater composition.
In contrast to our approach, Medema et al. (2020) reported catchment-specific correlations plotting the increase in concentration (not load) of the N gene assays (and cycles of the E gene assays) against the increase in cumulative incidence from 0.1 to 100 cases per 100,000 residents (3 log10 units) over a 7-weeks outbreak of the pandemics. While catchment-specific temporal correlations may be less susceptible to interference with other factors, we consider RNA load-based evaluations more reliable to cope with variations in wastewater flow, for example, stormwater events in combined sewer systems or variations in industrial wastewater production during a lockdown of industry and businesses.
4.3. Potential transmission risks
Our results suggest that detected RNA fragments appear to be non-infectious based on small-volume laboratory studies testing about 1 to 10 gene copies in one reaction. However, given the large capacity and flow rate of WWTPs (Table 2), we calculate that each of the studied WWTP emits 6 ∗ 1010 to 6 ∗ 1012 SARS-CoV-2 gene equivalents per day to the receiving water bodies, some of which serve as crucial water resources for drinking water production, cooling water abstraction, public swimming, irrigation, recreation, and natural habitats. This viral load can be very roughly compared to the potential viral shedding by infected persons in the catchment area, although many uncertainties come with such an appraisal: Rose et al. (2015) report a mean amount of feces excreted of about 129 g/cap/day. Viral gene copies detected in human excreta can vary broadly, probably depending on factors such as the stage of infection, and are given in the range of 102–108 gene copies per g feces (Kitajima et al., 2020). Wölfel et al. (2020) showed the development of gene copy findings on the time course of the infection. With, e.g., 100 infected persons in a sampled catchment, these assumptions would yield a viral load in a range as broad as 106–1012 gene copies per d in the wastewater. The findings, as shown in Fig. 8, are within but on the upper end of this broad range. Few studies have evaluated the fate of coronaviruses and other enveloped viruses in wastewater treatment and surface water (Gundy et al., 2009; Ye et al., 2016). In a recent review by Kitajima et al. (2020), it is stated that currently no applicable dose-response models exist for SARS-CoV-2, which would allow for a better risk assessment. In a recent review by Foladori et al. (2020), the current knowledge about the fate of SARS-CoV-2 in wastewater treatment systems was summarized, and the few studies undertaken indicate that there is a significant reduction in viral load through the treatment process, but still, gene fragments are detectable in effluents. Although it cannot be excluded with the current testing system of RT-qPCR targeting SARS-CoV-2 genes that virus emitted in large quantities is still infectious, studies analysing SARS-CoV-2 environmental stability support the suggestion that infectivity and replication-competence of released viral particles are very unlikely (Liu et al., 2020; van Doremalen et al., 2020).
5. Conclusions
Based on our findings, we conclude the following:
-
•
SARS-CoV-2 RNA was detected in the inflow of all 9 studied WWTP at concentrations similar to those reported in other studies. Our screening of different target genes points to shortcomings in the selectivity of gene primers used in studies previously published by other authors that might also detect genes non-specific to SARS-CoV-2 viruses.
-
•
Using Sanger sequencing, we confirmed human SARS-CoV-2 for the investigated sample taken during the SARS-CoV-2 pandemic outbreak. In contrast, positive signals in RT-qPCR were not confirmed by sequencing for the retained samples from 2017 and 2018. These results are suggesting an unspecific signal in RT-qPCR, stressing again the urgent need for a confirmation of positive RT-qPCR results by sequencing or other appropriate techniques in order to avoid false-positive results.
-
•
Quantification of SARS-CoV-2 gene concentrations and loads in wastewater needs to consider both aqueous and solid-phase SARS-CoV-2 detection methods and establish robust standard protocols for clean-up and RT-qPCR measurements.
-
•
We observed poor removal of SARS-CoV-2 in all three of the studied conventional activated-sludge WWTP. Full-scale ozonation at one plant seemed to reduce SARS-CoV-2 fragments in the effluent. Membrane-based WWTP planned to be included in future studies.
-
•
We found that the total load of gene equivalents in wastewater correlated with the cumulative and the acute number of COVID-19 cases reported in the respective catchment areas. We consider load-based correlations superior to gene copy concentration-based approaches. Analysis of suitable bioindicators in the wastewater may improve the assessment of SARS-CoV-2 loads data in catchment areas.
-
•
While our results indicate that RNA fragments are not infectious to Caco-2 cells in cell culture-based assays, further studies are needed to evaluate the risk SARS-CoV-2 may pose in the water cycle.
It is recommended to further develop and implement the concept of wastewater-based epidemiology as a complementary measure to survey the outbreak of the SARS-CoV-2 pandemic and impose catchment-specific measures if necessary.
CRediT authorship contribution statement
Sandra Westhaus: Conceptualization, Methodology, Investigation, Writing - original draft, Data curation, Formal analysis. Frank-Andreas Weber: Conceptualization, Methodology, Resources, Writing - original draft, Data curation, Formal analysis. Sabrina Schiwy: Writing - original draft, Data curation, Visualization. Volker Linnemann: Conceptualization, Methodology, Resources, Writing - original draft, Data curation, Formal analysis. Markus Brinkmann: Writing - review & editing, Validation. Marek Widera: Methodology, Writing - review & editing. Carola Greve: Investigation, Writing - review & editing, Data curation. Axel Janke: Resources, Writing - review & editing. Henner Hollert: Conceptualization, Resources, Formal analysis, Supervision, Writing - review & editing. Thomas Wintgens: Conceptualization, Resources, Formal analysis, Supervision, Writing - review & editing. Sandra Ciesek: Resources, Supervision, Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank the water boards Emschergenossenschaft und Lippeverband (EGLV), Erftverband, Linksniederrheinische Entwässerungs-Genossenschaft (LINEG), Niersverband, Ruhrverband, and Wasserverband Eifel-Rur (WVER) for their participation in the sampling campaign on short notice and for ongoing support to FiW e.V. in difficult times. We acknowledge the work of all water-sector and health-care personnel for their continued service to society throughout the pandemics. M.B. was supported through the Global Water Futures (GWF) program that is funded through the Canada First Research Excellence Fund (CFREF).
Editor: Damia Barcelo
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2020.141750.
Appendix A. Supplementary data
Supplementary material
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amirian E.S. Potential fecal transmission of SARS-CoV-2: current evidence and implications for public health. Int. J. Infect. Dis. 2020;95:363–370. doi: 10.1016/j.ijid.2020.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar Or I., Yaniv K., Shagan M., Ozer E., Erster O., Mendelson E., Mannasse B., Shirazi R., Kramarsky-Winter E., Nir O., Abu-Ali H., Ronen Z., Rinott E., Lewis Y.E., Friedler E., Paitan Y., Bitkover E., Berchenko Y., Kushmaro A. Regressing SARS-CoV-2 sewage measurements onto COVID-19 burden in the population: a proof-of-concept for quantitative environmental surveillance. medRxiv. 2020 doi: 10.1101/2020.04.26.20073569. (2020.04.26.20073569) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brückner I., Kirchner K., Müller Y., Schiwy S., Klaer K., Dolny R., Wendt L., Könemann S., Pinnekamp J., Hollert H. Status quo report on wastewater treatment plant, receiving water’s biocoenosis and quality as basis for evaluation of large-scale ozonation process. Water Sci. Technol. 2018;77(2):337–345. doi: 10.2166/wst.2017.548. [DOI] [PubMed] [Google Scholar]
- Carducci A., Federigi I., Liu D., Thompson J.R., Verani M. Making Waves: coronavirus detection, presence and persistence in the water environment: state of the art and knowledge needs for public health. Water Res. 2020;179:115907. doi: 10.1016/j.watres.2020.115907. 15 July 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavarria-Miró G., Anfruns-Estrada E., Guix S., Paraira M., Galofré B., Sánchez G., Pintó R.M., Bosch A. Sentinel surveillance of SARS-CoV-2 in wastewater anticipates the occurence of COVID-19 cases. medRxiv. 2020 doi: 10.1101/2020.06.13.20129627. (Preprint, 2020.06.13.20129627) [DOI] [Google Scholar]
- Choi P.M., Tscharke B.J., Donner E., O’Brien J.W., Grant S.C., Kaserzon S.L., Mackie R., O’Malley E., Crosbie N.D., Thomas K.V., Mueller J.F. Wastewater-based epidemiology biomarkers: past, present and future. Trends Anal. Chem. 2018;105:453–469. doi: 10.1016/j.trac.2018.06.004. [DOI] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G.J.C., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J., Ellis J., Zambonm M., Peiris M., Goossens H., Reusken C., Koopmans M.P.G., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. (pii=2000045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörner K. ninth ed. Thieme; Oldenburg: 2003. Klinische Chemie und Hämatologie. [Google Scholar]
- Drosten C., Günther S., Preiser W., van der Werf S., Brodt H., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A.M., Berger A., Burguière A., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J., Müller S., Rickerts V., Stürmer M., Vieth S., Klenk H., Osterhaus A.D.M.E., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- ELWAS . Ministerium für Umwelt, Landwirtschaft, Natur- und Verbraucherschutz NRW; Düsseldorf: 2020. Elektronisches wasserwirtschaftliches Verbundsystem für die Wasserwirtschaftsverwaltung in NRW.https://www.elwasweb.nrw.de (Accessed 9 April 2020) [Google Scholar]
- Foladori P., Cutrupi F., Segata N., Manara S., Pinto F., Malpei F., Bruni L., La Rosa G. SARS-CoV-2 from faeces to wastewater treatment: what do we know? A review. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Latorre L., Ballesteros I., Villacrés-Granda I., Genoveva Grandam M., Freire-Paspuel B., Ríos-Touma B. SARS-CoV-2 in river water: implications in low sanitation countries. Sci. Total Environ. 2020;743:140832. doi: 10.1016/j.scitotenv.2020.140832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundy P.M., Gerba C.P., Pepper I.L. Survival of coronaviruses in water and wastewater. Food Environ. Virol. 2009;1(1):10. doi: 10.1007/s12560-008-9001-6. [DOI] [Google Scholar]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020;737 doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehl S., Rabenau H., Berger A., Kortenbusch M., Cinati J., Bojkova D., Behrens P., Böddinghaus B., Götsch U., Naujoks F., Neumann P., Schork J., Tiarks-Jungk P., Walczok A., Eickmann M., Vehresschild M.J.G.T., Kann G., Wolf T., Gottschalk R., Ciesek S. Evidence of SARS-CoV-2 infection in returning travelers from Wuhan, China. N. Engl. J. Med. 2020;382:1278–1280. doi: 10.1056/NEJMc2001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020;739:139076. doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocamemi B.A., Kurt H., Sait A., Sarac F., Saatci A.M., Pakdemirli B. SARS-CoV-2 detection in Istanbul wastewater treatment plant sludges. medRxiv. 2020 doi: 10.1101/2020.05.12.20099358. (Preprint) [DOI] [Google Scholar]
- Kumar M., Kumar Patel A., Shah A.V., Raval J., Rajpara N., Joshi M., Joshi C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;746 doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre M., Bartoli J., Shmakova L., Jeudy S., Labadie K., Adrait A., Lescot M., Poirot O., Bertaux L., Bruley C., Couté Y., Rivkina E., Abergel C., Claverie J. Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. PNAS. 2014;111(11):4274–4279. doi: 10.1073/pnas.1320670111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescure F., Boudma L., Nguyen D., Parisey M., Wicky P., Behillil S., Gaymard A., Bouscambert-Duchamp M., Donati F., Le Hingrat Q., Enouf V., Houhou-Fidouh N., Valette M., Mailles A., Lucet J., Mentre F., Duval X., Descamps D., Malvy D., Timsit J., Lina B., Van-der-Werf S. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect. Dis. 2020;20(6):697–706. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wony J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Li T., Deng Y., Liu S., Zhang D., Li H., Wang X., Jia L., Han J., Bei Z., Zhou Y., Li L., Li J. Stability of SARS-CoV-2 on environmental surfaces and in human excreta. medRxiv. 2020 doi: 10.1101/2020.05.07.20094805. (Preprint, 2020.05.07.20094805) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W., de Roda Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020;5(6):533–534. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020 doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Metcalf & Eddy . 4th edition. Publisher: TMH; 2012. Wastewater Engineering: Treatment and Reuse. ISBN: 0070495394/9780070495395. [Google Scholar]
- Naddeo V., Liu H. Editorial perspectives: 2019 novel coronavirus (SARS-CoV-2): what is its fate in urban water cycle and how can the water research community respond? Environ. Sci. Water Res. Technol. 2020;6:1213–1216. doi: 10.1039/D0EW90015J. [DOI] [Google Scholar]
- National Institutes of Health BLAST. 2020. https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch&BLAST_SPEC=Betacoronavirus (Accessed 14 July 2020)
- Nemudryi A., Nemudraiam A., Surya K., Wiegand T., Buyukyoruk M., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. medRxiv. 2020 doi: 10.1101/2020.04.15.20066746. (Preprint not yet peer reviewed. Preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Chen D., Xia Y., Wu X., Li T., Ou X., Zhou L., Liu J. Asymptomatic cases in a family cluster with SARS CoV-2 infection. Lancet Infect. Dis. 2020;20(4):410–411. doi: 10.1016/S1473-3099(20)30114-6. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J.H., Jiang S.C., Rose J.B. Concentration of viruses and dissolved DNA from aquatic environments by vortex flow filtration. Appl. Environ. Microbiol. 1991;57(8):2197–2204. doi: 10.1128/aem.57.8.2197-2204.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas Ferrando E., Simon P., Allende A., Sanchez G. SARS-CoV-2 RNA titers in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D., Maresca M., Longobardi C., Mancon A., Francesca R., Pagani C., Moja L., Gismondo M.R., Salerno F. Presence and vitality of SARS-CoV-2 virus in wastewaters and rivers. medRxiv. 2020 doi: 10.1101/2020.05.01.20086009. (Preprint 2020.05.01.20086009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- RKI COVID-19: Fallzahlen in Deutschland und weltweit. 2020. https://www.rki.de/DE/Content/InfAZ/N/Neuartiges_Coronavirus/Fallzahlen.html (Accessed 13 July 2020)
- Rodriguez-Manzano J., Miagostovich M., Hundesa A., Clemente-Casares P., Carratala A., Buti M., Jardi R., Girones R. Analysis of the evolution in the circulation of HAV and HEV in eastern Spain by testing urban sewage samples. J. Water Health. 2010;8(2):346–354. doi: 10.2166/wh.2009.042. [DOI] [PubMed] [Google Scholar]
- Rose C., Parker A., Jefferson B., Cartmell E. The characterization of feces and urine: a review of the literature to inform advanced treatment technology. Crit. Rev. Environ. Sci. Technol. 2015;45(17):1827–1879. doi: 10.1080/10643389.2014.1000761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader C., Schielke A., Ellerbroek L., Johne R. PCR inhibitors-occurrence, properties and removal. J. Appl. Microbiol. 2012;113(5) doi: 10.1111/j.1365-2672.2012.05384.x. [DOI] [PubMed] [Google Scholar]
- Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., Ahmed W., Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Wu C., Li X., Song Y., Yao X., Wu X., Duan Y., Zhang H., Wang Y., Qian Z., Cui J., Lu J. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020;7(6):1012–1023. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toptan T., Hoehl S., Westhaus S., et al. Optimized RT-qPCR approach for the detection of intra- and extra-cellular SARS-CoV-2 RNAs. Int. J. Mol. Sci. 2020;21(12):E4396. doi: 10.3390/ijms21124396. (Jun 20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., Tamin A., Harcourt J.L., Thornburg N.J., Gerber S.I., Lloyd-Smith J.O., de Wit E., Munster V.J. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020;382(16) doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q., Hu Y., Luo G., Wang K., Lu Y., Li H., Wang S., Ruan S., Yang C., Mei C., Wang Y., Ding D., Wu F., Tang X., Ye X., Ye Y., Liu B., Yamg J., Yin W., Wang A., Fan G., Zhou F., Liu Z., Gu X., Xu J., Shang L., Zhang Y., Cao L., Guo T., Wan Y., Qin H., Jiang Y., Jaki T., Hayden F.G., Horby P.W., Cao B., Wang C. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395(10236):1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Water, sanitation, hygiene, and waste management for the COVID-19 virus. Interim guidance March 19th 2020. 2020. https://www.who.int/publications-detail/water-sanitation-hygiene-and-waste-management-for-covid-19
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones Kelly T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten K., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(2020):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., Yin H., Xiao Q., Tang Y., Qu X., Kuang L., Fang X., Mishra N., Lu J., Shan H., Jiang G., Huang X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020;5:434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Gu X., Lee W.L., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Moniz K., Erickson T., Chai P., Thompson J., Alm E. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. medRxiv. 2020 doi: 10.1101/2020.04.05.20051540. preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J., Maday Y., Teyssou R., Richard E., Almayrac J.L., Moulin L. Evaluation o flockdown impact on SARS-CoV-2 dynamics through viral genome quantification in Paris wastewaters. medRxiv. 2020 doi: 10.1101/2020.04.12.20062679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Wu X., Jiang X., Xu K., Ying L., Ma C., Li S., Wang H., Zhang S., Gao H., Sheng J., C H., Qiu Y., Li L. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: retrospective case series. BMJ. 2020;368:m606. doi: 10.1136/bmj.m606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50(10):5077–5085. doi: 10.1021/acs.est.6b00876. 2016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material