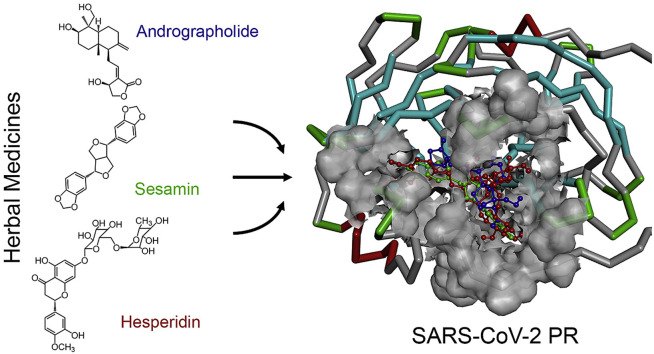
Abstract
The widespread problem of a 2019-novel coronavirus (SARS-CoV-2) strain outbreak in Wuhan, China has prompted a search for new drugs to protect against and treat this disease. It is necessary to immediately investigate this due to the mutation of the viral genome and there being no current protective vaccines or therapeutic drugs. Molecular modelling and molecular docking based on in silico screening strategies were employed to determine the potential activities of seven HIV protease (HIV-PR) inhibitors, two flu drugs, and eight natural compounds. The computational approach was carried out to discover the structural modes with a high binding affinity for these drugs on the homology structure of the Wuhan coronavirus protease (SARS-CoV-2 PR). From the theoretical calculations, all the drugs and natural compounds demonstrated various favorable binding affinities. An interesting finding was that the natural compounds tested had a higher potential binding activity with the pocket sites of SARS-CoV-2 PR compared to the groups of HIV-PR inhibitors. The binding modes of each complex illustrated between the drugs and compounds interacted with the functional group of amino acids in the binding pocket via hydrophilic, hydrophobic, and hydrogen bond interactions using the molecular dynamics simulation technique. This result supports the idea that existing protease inhibitors and natural compounds could be used to treat the new coronavirus. This report sought to provide fundamental knowledge as preliminary experimental data to propose an existing nutraceutical material against viral infection. Collectively, it is suggested that molecular modelling and molecular docking are suitable tools to search and screen for new drugs and natural compounds that can be used as future treatments for viral diseases.
Keywords: SARS-CoV-2, Protease inhibitors, Herbal medicines, Molecular docking, Molecular dynamics simulation
Graphical abstract
Highlights
-
•
We generated several docked complexes in order to evaluate the potential activities of novel coronavirus protease (SARS-CoV-2 PR) inhibitors.
-
•
The calculations of the binding affinity were interestingly found to predict the drug activities using the theoretical calculations.
-
•
The binding mode of each complex was found to observe at the potential site with slightly different amino acid binding poses in the SARS-CoV-2 PR.
-
•
Here, we are able to propose the first structure-activity relationships of alternative usage of the natural materials.
1. Introduction
The infection of 2019-novel coronavirus (SARS-CoV-2) occurred in Wuhan, Hubei China, in Early-December 2019. All initial cases were reported that linked to Hunan Seafood Wholesale Market [1], and has been designated a global health emergency by the World Health Organization since January 30, 2020 [2]. A positive-sense single-stranded RNA genome of this virus was elucidated on January 10th, 2020 from Wuhan, China, the original location of the primary outbreak [3]. As of February 9, 2020, 37,567 cases have been confirmed infections with 813 deaths and 2408 recoveries [4]. Two strains of coronavirus –– Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS) –– cause acute respiratory distress syndrome (ARDS), similar to the common cold and intestinal infections in animals and humans [5]. The genome of SARS-CoV-2 is distinct from SARS-CoV (∼79% identity), and MERS-CoV (∼50% identity), and several bat CoV (more than 85% identity) [6]. At the beginning of the SARS outbreak, it was found that almost all early index patients had animal exposure before developing the disease. However, human-to-human transmission has been confirmed for the SARS-CoV-2, according to Zhong Nanshan, the head of the health commission team investigating the outbreak [7].
Coronavirus has an RNA positive-strand as genetic material, which could be ready to synthesise RNA polymerase leading to the synthesis of the viral genome template using the biochemical tools inside the host cells [8,9]. One of these proteins contains protease (PR) activity, which has become a target for therapeutic agents against viral proliferation. The University of Hong Kong School of Medicine found that PR inhibitors and other drugs can effectively treat respiratory diseases such as SARS and other coronaviruses. In Thailand, doctors from Rajavithi Hospital, Bangkok have found a successful treatment for the new coronavirus by combining a drug cocktail of HIV-PR inhibitors, lopinavir, and ritonavir, and the flu drug oseltamivir. This combination has been reported to work successfully with several patients, although there is as yet no experimental evidence to prove this. Until now, Chinese researchers have reported that the antiviral drug remdesivir has been shown to be effective in clinical trials to treat patients with pneumonia who are infected with the novel coronavirus [10].
Thai herbal medicines have been used as medical treatment, with one type of dietary supplements. The natural products derived from plants are being evaluated for their therapeutic benefits based on the popularity of local medicinal plants. For examples, an andrographolide from the stem and leaves of Andrographis paniculata [11,12], sesamin from Sesamum indicum seeds [13,14], hesperidin from citrus fruits including sweet orange, mandarin, lemon, and lime [15], and curcumin from the rhizome of Curcuma longa [16] are found to present a variety of biological effects, therapeutic benefits, and are used in the treatment of many types of diseases, including anti-dengue virus activity [11], antiviral, antibacterial [12], anti-inflammation [13,14], antioxidant [14], and anticancer activities [15,16]. This discovery officially confirms the potential activity for the novel coronavirus treatments.
In this presentation, the researchers propose an alternative use for phytochemical materials from natural substances against the new strain of the novel coronavirus infection. Seven HIV-PR inhibitors, two flu drugs, and eight hebal medicines have been introduced to use against the SARS-CoV-2 PR as target protein using the theoretical approaches. The molecular structures, including the binding affinity of the SARS-CoV-2–drug complexes were determined. The docking experiment demonstrates and suggests results before use as treatment and for further molecular dynamics (MD) simulation study.
2. Methods
2.1. Protein-ligand preparation
The three-dimensional (3D) protease (PR) model of the SARS-CoV-2 was obtained from the Protein Data Bank (PDB code: 6LU7) as a monomer structure. To prepare the target protein structure for docking, charges and polar hydrogen atoms were added using the prepare_receptor4.py script from MGLTools 1.5.6 [17].
Eight of phytochemicals, seven of HIV-PR inhibitors, and two of neuraminidase inhibitors were chosen (Figure S1 and Figure S2, Supplementary materials). The 3D structure of each compound was drawn and followed by short minimization with a gradient tolerance of 0.0100 kcal mol−1 Å of root mean squared (RMS) using the Amber 18 program [18]. Individual PDB files were prepared for docking using the prepare_ligand4.py script from MGLTools [17], using only the largest non-bonded fragment present.
2.2. Docking parameters
The software package of AutoDock Vina [19] was performed for all molecular docking simulations to anchor the drugs into the potential binding site of the target protein. In general, the docking parameters were kept to their default values. The total size of the cubic docking box was set to be 60 × 60 × 60 Å along each dimension (x, y, and z) by the grid point spacing of 0.375 Å.
The dimension of the grid box was set to cover residues of the active site of the target protein. Exhaustiveness parameter corresponding to the amount of sampling effort was set to 200 with the energy range of 10 kcal mol−1, and the maximum amount of poses to report was set to 20 using the built-in clustering analysis with a 2.0 Å cut-off.
2.3. Molecular dynamics simulation
2.3.1. Molecular dynamics simulations and binding free energy calculation
All molecular dynamics (MD) simulations were performed by PMEMD.CUDA [20,21] from AMBER 18 suite of programs [18] on NVIDIA Geforce GTX-1070 Ti for speeding up the simulation times. The general AMBER force field (GAFF) parameters were carried out to generate the atomic parameters of each ligand and Gasteiger charge was used to assign the charge parameter for all ligands in MD simulations. Each complex structure under periodic boundary conditions was solvated in a cubic box of TIP3P water molecules extending to 10 Å along each direction from the complex model, and Na+ ions were added as neutralizing counterions. The cutoff distance was kept to 12 Å in order to compute the non-bonded interactions. All experimental simulations were performed under periodic boundary conditions [22]. The AMBER ff14SB force field parameters were used to apply the description of the complex characterization. The long-range electrostatic were treated using the particle mesh Ewald (PME) method [23,24]. The SHAKE algorithm and Langevin dynamics were applied to constrain the bonds that involved hydrogen atoms and to control the temperature. The time step of 2 fs was set and the trajectory was recorded every 0.2 ps. The temperature was gradually increased from 0 to 310.15 K over a period of 100 ps of NVT dynamics, and followed by 10 ns of NPT equilibration at 310.15 K and 1 atm pressure. Finally, a total 50 ns of the production phase MD simulations was performed for properties collection.
Trajectory analyses (root mean square deviation and fluctuation, dynamic cross-correlation, hydrogen bond) were carried out using CPPTRAJ module from Amber 18 program [25]. Binding free energy calculation of each simulation complex was performed based on selected MD snapshots using Amber molecular mechanics Poisson–Boltzmann surface area (MM-PBSA) method [26]. The 250 numbers of structural frequencies were used to extract the structures from all 50 ns trajectories data. The 1000 snapshots were collected from the trajectory data to calculate the binding free energy by the MM-PBSA method. The grid size from the PB calculations in MM-PBSA was 0.5 Å. The structural images were presented using DS software.
2.3.2. Dynamic cross-correlation matrix analysis
Dynamic movement and correlations between the Cα–atoms positions of SARS-CoV-2 PR protein over the simulation period were calculated by dynamic cross-correlation matrix (DCCM) analysis. DCCM was performed using CPPTRAJ module of the AMBER 18 suite. DCCM diagrams are displayed as a color coded matrix of Pearson correlation coefficients. Where there is a highly-correlated motion, the movement towards the same direction between the residue pairs shows a positive values in the color range from light green to deep red (+1); while in anti-correlated motions, the movement shows a negative values in the color range from grey to royal blue (−1) [27]. The diagonal square relates to the correlation of a residue with itself, i.e., only a region remarked to have highly-positive values (red). The potential amino acids in binding site of SARS-CoV-2 PR protein were clarified at residue number of (i) 21–50, and (ii) 141–190.
3. Results and discussion
Although many modelling properties of targeting coronavirus have been reported [28,29], the researchers opted to start afresh to circumvent any potential biases and focus on the protein class of the viral 2019-CoV protease. Fig. 1 compares the protease sequence based on multiple sequences alignments of the SARS-CoV-2, Wuhan-Hu-1 strain (protein ID: QHD43415.1) [30] with the others of SARS coronavirus proteases from the Protein Data Bank (PDB). The sequence alignment of the SARS-CoV-2 PR shows high similarity to the known crystal structure of the SARS coronavirus protease (1WOF, 2DUC, 2GTB, 2ZU4, 3SZN, 3TIU, 5B6O, 5C5O, 5N5O, and 6LU7).
Fig. 1.
Sequence alignment between SARS-CoV-2 (Wuhan-Hu-1 strain) protease (Protein ID: QHD43415.1) and various crystal structure of SARS coronavirus protease from Protein Data Bank using ClustalX2 [31].
Extensive studies on the crystal structures of SARS-CoV PR have shown that the PR families have similar overall 3D structures at their active binding sites. According to X-ray data, the crystal structures of SARS coronavirus main peptidase inhibited by an aza-peptide epoxide, such as from PDB entry 1WOF, 2A5I, 2HOB, 2ZU4, 3SZN, 5C5O, and 5N5O. By mapping the known crystal structure of SAR CoV-PR with the X-ray structure of SARS-CoV-2 PR (PDB code 6LU7), this observed agrees well with the reported data from Christian C. Gruber and Georg Steinkellner [32].
3.1. Molecular docking study
In this present work, molecular docking of the SARS-CoV-2 PR complexed with phytochemicals were reported to predict the bioactive binding modes and the binding affinity for the first time. As calculated results, all the selected drugs could be bound in the active pocket of the SARS-CoV-2 PR at the immediate vicinity of the binding sites, implying plausible inhibition on the protease activity (Table 1 ). Each inhibitor showed various favorable binding affinities (kcal mol−1) obtained from docking study in an energy range of −8.6 ∼ −5.2 kcal mol−1. More negative estimated binding affinity indicated a stronger favorable protein-ligand complex. Comparing the neutral drugs with the PR inhibitors, the bioflavonoid of hesperidin is predicted to lie on the pocket site of the SARS-CoV-2 PR with the highest binding affinity (−8.6 kcal mol−1). Residues within 5 Å of the ligand were identified (Table 1) to play a key role in the potential activity for PR inhibition. Sesamin is predicted to have comparable binding affinity as hesperidin. This result implies that natural compounds are a potential treatment option for viral infections. The binding mode of the other herbal drugs was observed to bind at the same site with slightly different amino acid binding poses by the energy difference 0.1–1.9 kcal mol−1.
Table 1.
Binding affinity (kcal mol−1) and common amino acid binding pocket residue in each system by AutoDock Vina.
| Compounds | Binding affinity | Common amino acid binding pocket residues within 5 Å |
|---|---|---|
| Herbal medicines | ||
| Andrographolide | −6.3 | His41, Leu141, Asn142, Gly143, Cys154, Met165, Glu166, Gln189 |
| Anthocyanin-β-D-glucoside | −7.3 | Leu141, Asn142, His163, Met165, Glu166, Leu167, Pro168, Thr190, Gln192 |
| Capsaicin | −5.5 | Thr25, His41, Asn142, Glu166, Met 165, Leu167, Pro168, Gln189, Thr190, Gln192, |
| Curcumin | −6.0 | Thr25, Thr45, Ser46, Cys145, Met165, Glu166, Pro168, Arg188, Gln189, Thr190 |
| Cyanidin | −6.7 | Leu141, Asn142, Cys145, Met165, Glu166, Leu167, Arg188, Gln189, Thr192, Gln192 |
| Cyanidin-3-O-glucoside | −7.4 | His41, Leu141, Asn142, Met165, Glu166, Leu167, Arg188, Gln189, Thr190, Gln192 |
| Sesamin | −7.7 | Thr25, Cys44, Met49, Asn142, Met165, Glu166, Leu167, Pro168, Gln189 |
| Hesperidin | −8.6 | Cys44, Thr45, Asn142, Cys145, Met165, Glu166, Leu167, Gln189, Thr190 |
| HIV-PR inhibitors | ||
| Amprenavir | −6.6 | Ser46, Met49, Leu141, His163, Met165, Glu166, Leu167, Pro168, Arg188, Gln189 |
| Atazanavir | −5.8 | Ser46, Met49, Leu141, Asn142, Gly143, Cys145, Met165, Glu166, Leu167, Pro168 |
| Darunavir | −6.4 | Thr26, Thr45, Ser46, Met49, Asn142, Gly143, Cys145, Met165, Glu166, Gln189 |
| Lopinavir | −7.4 | Thr26, Leu27, Ser46, Leu141, Asn142, Gly143, Cys145, Met 165, Glu166, Leu167, |
| Remdesivir | −6.5 | Thr45, Met49, Ser46, Asn142, Gly143, Cys145, Met165, Glu166, Leu167, Thr190, Gln192 |
| Ritonavir | −6.2 | Thr26, Asn119, Phe140, Leu141, Asn142, Cys145, Met165, Glu166, Leu167, Pro168, Gln189 |
| Saquinavir | −6.6 | Thr25, His41, Met49, Leu141, Asn142, Cys145, Pro168, Glu166, Leu167, Gln189, Gln192 |
| Neuraminidase inhibitors | ||
| Oseltamivir | −5.2 | Thr26, Leu27, Cys44, Ser46, Met49, Gly143, Met165, Gln189 |
| Zanamivir | −5.5 | Leu141, Asn142, Met165, Glu166, Leu167, Pro168, Arg188, Gln189, Gln192 |
For HIV-PR inhibitors, a marginally significant result was observed with an energy difference of around 0.4–1.8 kcal mol−1, indicating a probable finding of an inhibitor that binds to the protease of the Wuhan coronavirus as well. As per the results, lopinavir shows high potential as a better binder to the protease binding site compared to the other PR inhibitors. On the other hand, remdesivir that targets the RNA-dependent RNA polymerase presents weaker affinity to bind the protease of SARS-CoV-2 than that of lopinavir. Similarly, the interaction of neuraminidase inhibitors weakens potentially due to aiming for different target activity of the antiviral treatment.
3.2. Molecular dynamics simulations
To enhance the configuration space for sampling accessibility to the molecular geometries, 50 ns long-time simulations of the SARS-CoV-2 PR protein with and without the addition of the inhibitor were performed. The thus-obtained docked complexes with the lowest binding energy of the herbal medicines (hesperidin and sesamin) and HIV-PR inhibitors (lopinavir and remdesivir) have been choosen as representative model for the MD simulation study.
The structural stability of the proteins as well as the position of the ligands in the binding site cleft were monitored using the root mean square deviations (RMSD) with respect to their minimized structure (Figure S3 and Figure S4, Supplementary materials) [33]. Steady oscillation and small fluctuation of RMSD were observed in each complexe model compared to the bare SARS-CoV-2 PR protein, indicating that the complex models was more stable and endured lesser conformational changes during MD simulations.
3.2.1. Binding free energy calculation
To evaluate the binding affinity of the protein-ligand interactions, the relative binding free energy values obtained from MM-PBSA protocol were calculated as listed in Table 2 . The results revealed that the herbal medicines hesperidin (ΔGbinding = −11.96 kcal mol−1) binds to the SARS-CoV-2 PR protein better than sesamin (ΔGbinding = −9.93 kcal mol−1) as well as HIV-PR inhibitors lopinavir (ΔGbinding = −7.56 kcal mol−1) and remdesivir (ΔGbinding = −7.85 kcal mol−1). The calculated results are in a good agreement with the same tendency energy values from the docking calculation.
Table 2.
Energy component of the protein-ligand complex estimated by MM-PBSA method.
| Complex | Hesperidin | Sesamin | Lopinavir | Remdesivir |
|---|---|---|---|---|
| ΔGbinding | −11.96 ± 2.85 | −9.93 ± 2.02 | −7.56 ± 2.85 | −7.85 ± 3.16 |
| ΔS | −24.05 ± 1.38 | −16.89 ± 1.59 | −26.56 ± 2.00 | −21.90 ± 2.71 |
| vdW | −67.72 ± 3.04 | −35.63 ± 2.37 | −46.99 ± 2.78 | −54.15 ± 3.28 |
| EEL | −24.85 ± 6.66 | −8.99 ± 1.64 | −4.64 ± 2.37 | −13.78 ± 4.58 |
| EPB | 61.94 ± 6.50 | 20.85 ± 1.64 | 23.73 ± 2.36 | 43.41 ± 6.42 |
| ENPOLAR | −5.38 ± 0.11 | −3.03 ± 0.15 | −5.91 ± 0.29 | −5.23 ± 0.14 |
Note: The EEL and vdW represent the electrostatic and van der Waals contributions from MM, respectively. EPB stands for PB electrostatic contribution to the polar solvation energy, and ENPOLAR is the non-polar contribution to the solvation free energy. ΔS (kcal mol−1, at 298.15 K) is an entropically unfavorable protein-ligand complex energy calculated by normal mode analysis. ΔGbinding (kcal mol−1) is the final estimated binding free energy calculated from the terms above (ΔGbinding = ΔEMM + ΔEsolvation – TΔS) [33].
A recent literature revealed that there is no statistically significant difference was found between combining drug lopinavir/ritonavir treatment and a controlled trial to clinical improvement and a viral RNA detectability in patients with serious SARS-CoV-2 infection [34]. This observed data confirmed the high binding free energy of lopinavir to bind with the porket site of the SARS-CoV-2 PR.
The low binding affinity of remdesivir was observed to interact on the protease protein, which might be due to aiming for different target activity of remdesivir on the RNA-dependent RNA polymerase related with the low affinity energy value by docking calculation (Table 1). The free energy of the van der Waals (vdW) interactions and non-polar parts (ENPOLAR) of the solvation free energy contributes favorably to the binding where there is a slight energetic difference for each binding complex, as opposed to the unfavorable contributions of electrostatic energy (EEL + EPB). Each energy term (vdW, ENPOLAR, EEL + EPB) of hesperidin was found to be at least two times more favorable interaction than that of the other inhibitors. It can be noted that the bioactive natural products may allow better application of herbal drugs for future treatment of viral infected diseases.
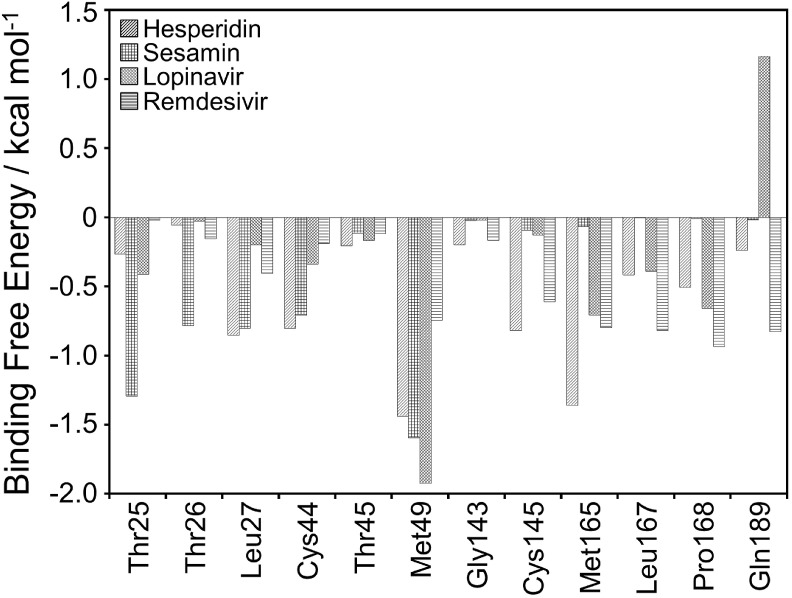
To define the essential amino acid residues that contribute to the binding affinity, per-residue free energy decomposition was calculated. Fig. 2 shows the decomposed per-residue energy upon the binding of each ligand-bound complex. The negative and positive values represent favorable and unfavorable contributions, respectively.
Fig. 2.
Per-residue free energy decomposition of the amino acid residues at the binding regions of the SARS-CoV-2 PR in each ligand-bound complex. All values were given in kcal mol−1.
As seen in Fig. 2, the amino acid residues of the SARS-CoV-2 PR involved in the binding regions and possessed favorable interaction energy including Thr25, Thr26, Leu27, Cys44, Thr45, Met49, Gly143, Cys145, Met165, Leu167, Pro168, and Gln189. Leu27, Cys44, Cys145, and Met165 contributed the most to the binding with strong affinity for hesperidin-bound system, while Thr25 and Thr26 showed more favorable interaction for sesamin-bound system. HIV-PR inhibitor lopinavir has more favorable contact with the residues Met49, while Gln189 has more favorable contact with lopinavir through the vdW decomposition energy (Figure S5, Supplementary materials). The high decomposed binding energies of Leu167, Pro168, and Gln189 residues have been justly obtained in the remdesivir-bound system.
3.2.2. Hydrogen bond analysis
Analysis of hydrogen bond formation of each complex system was conducted and hydrogen bond occupancy cutoff 3.5 Å from acceptor to donor of heavy atoms were selected as listed in Table 3 .
Table 3.
Hydrogen bond occupancy for the residue paris of each ligand-bound system during the production phase MD simulations.
| Complex | Acceptor | Donor | %Occupied | Average distanceA–D/Å |
|---|---|---|---|---|
| Hesperidin | Val186@O | HED@O | 27.10 | 2.95 |
| (HED) |
HED@O | Thr25@N | 26.33 | 3.36 |
| HED@O |
Glu166@N |
21.30 |
3.15 |
|
| Sesamin | SEM@O | Thr26@N | 50.32 | 3.19 |
| (SEM) |
SEM@O |
Asn142@N |
2.76 |
3.16 |
| Lopinavir | LOP@O | Gly143@N | 45.41 | 3.09 |
| (LOP) |
LOP@O |
Asn142@N |
5.42 |
3.13 |
| Remdesivir | REM@O | Thr26@N | 51.65 | 3.13 |
| (REM) | Asp187@O | REM@N | 13.60 | 3.26 |
Average distanceA–D = average distance between acceptor and donor of heavy atoms.
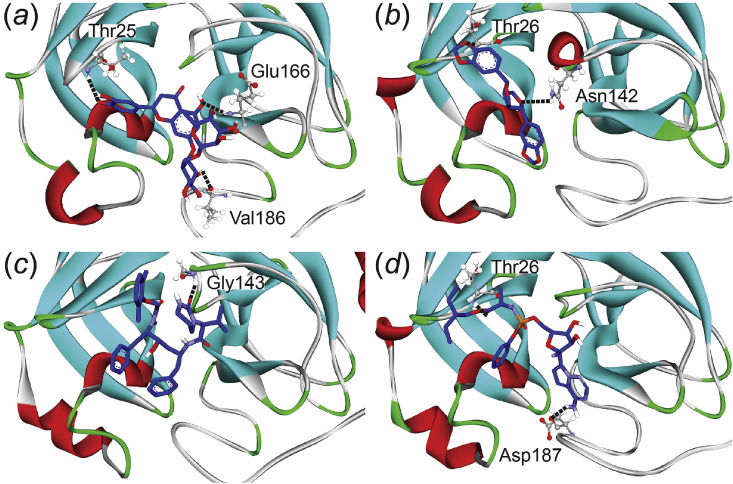
The hesperidin forms more hydrogen bonds compared to the other models when it interacts with the SARS-CoV-2 PR protein. Hesperidin is oriented in the potential site through favorable hydrogen bond interactions with proton-accepting amino acid of the SARS-CoV-2 PR including Thr25, Glu166, and Val186 with hydrogen bond length at around 2.95–3.17 Å. Thr25 of the PR protein that was conducted the hydrogen bond formation with more than 10% occupancy proportions, showed favorable binding interactions in the decomposition analysis through the vdW decomposition energy (Figure S5, Supplementary materials). On the others hands, Gly143 was found to bind to lopinavir with high occupancy proportions, while Thr26 and Asp187 was promoted a high occupancy of hydrogen bonding to interact with sesamin and remdesivir corresponding to the decomposed binding energy calculation.
Using the number of hydrogen bonds formation between the inhibitors and the potential residues in the SARS-CoV-2 PR protein alone is able to explain the reason why hesperidin binds with the protein exclusively from the other compounds. Fig. 3 shows the final conformations of the potential binding modes in each ligand-bound complexe at 50 ns from the production phase MD simulations with hydrogen bond-forming residues shown in stick representation.
Fig. 3.
Final conformations of (a) hesperidin, (b) sesamin, (c) lopinavir, and (d) remdesivir-bound systems in the binding pocket of the SARS-CoV-2 PR. The hydrogen bonds are shown in black stick line.
3.2.3. Dynamic cross-correlation matrix analysis
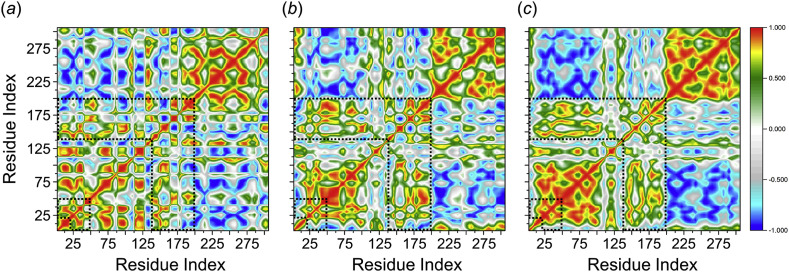
To further investigate the conformational dynamics that occurs on the SARS-CoV-2 PR protein by the effect of the drug ligands between hesperidin and lopinavir, dynamic cross-correlation matrix (DCCM) was calculated. The phenomenon of the dynamic motions during the simulations of the backbone Cα–atoms of SARS-CoV-2 PR protein in each bound system are shown in Fig. 4 .
Fig. 4.
Dynamic cross-correlation diagrams of the fluctuations of Cα–atoms of SARS-CoV-2 PR protein in the complex of (a) hesperidin-, (b) lopinavir-bound systems, and (c) bare SARS-CoV-2 PR protein. Positive and negative values are represented in range of color red to royal blue, respectively. The diagonal square relates to the correlation of a residue with itself, i.e., only a region remarked to have highly-positive values (red). The binding regions of the SARS-CoV-2 PR protein (at residues number of 21–50 and 141–190) are demarcated with dashed lines. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The DCCM map results showed that all inhibitors effect on the structural remodelling of the SARS-CoV-2 PR as illustrated by the change in the dynamic motions in comparison with the bare PR protein. As seen in dashed lines in Fig. 4, the hesperidin-bound system showed an overall anti-correlated motion at potential residues 141–190 highest than that of the lopinavir-bound complex. This may explains the lower decomposed binding free energy for the binding site observed by the MM-PBSA calculations. This calculation agrees well with the findings of previous studies that the anti-correlated motions of the protein residues may arise by means of an external perturbation effect, such as small ligand binding [35,36]. Moreover, a more anti-correlation motion was observed at residue number of 40–50 for the hesperidin-bound complex relative to low decomposed binding free energy in the decomposition analysis.
For lopinavir HIV-PR inhibitor, the biggest differences have been observed from the DCCM map at the key residues of 141–190 triggered a correlation motion on the binding regions as opposed to the hesperidin-bound system. The binding of lopinavir showed an anti-corelation motion at residues around 45–50 relative to more favorable interaction in the decomposition analysis. Using the correlation matrix, it seems that a high overall correlated motion was observed in the binding regions of the bare PR system, thus confirming conformational shifts by the effect of the ligand binding.
4. Conclusions
The researchers report molecular docking and molecular binding modes of potential activity for an alternative herbal supplement that binds to the protease binding pocket of the Wuhan coronavirus. The existing HIV-PR inhibitor, now in use as an anti-coronavirus drug, has side effects on liver and kidney function under continued use. The evidence presented in the present study proposes complementary and alternative neutraceuticals from natural resources which have fewer side effects, including andrographolide, sesamin, and hesperidin. These were found through a theoretical calculation using the molecular docking approach. According to the complete genomes of coronavirus from Wuhan, the protease becomes the first target for therapeutic agents against viral proliferation. Computer molecular modelling and molecular docking are therefore the most suitable tools to quickly screen and confirm existing and new drugs as well as nutraceuticals.
As our results, the molecular docking and molecular dynamics simulation techniques were used to successfully investigate the potential protease inhibitor approved for viral treatment. The natural compounds used as nutraceuticals could be used as co-protection and treatment with these existing drugs. Although the available drugs have not been considered in any pharmacological assay or biological experiment to have potential activity on the SARS-CoV-2, all the tested natural compounds were found to share high pharmacological benefits with alternative antiviral activity [11,12]. This report provides fundamental knowledge as a preliminary experiment for the consideration of further measurements and to propose an existing material as a nutraceutical. Consequently, subtle differences in the theoretical calculations corresponding with the SARS coronavirus experimental assay should prompt further investigation.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by grants from Thailand Excellence Center for Tissue Engineering and Stem Cells (PK and KK), Department of Biochemistry, Faculty of Medicine, Chiang Mai University, Chiang Mai, Thailand. The authors would like to thanks Dr. Christian C. Gruber and Dr. Georg Steinkellner, Innophore GmbH, Graz, Austria, for their information data of the Wuhan coronavirus (SARS-CoV-2) protease; and the Center of Excellence in Clinical Virology, Faculty of Medicine, Chulalongkorn University and the Research Chair Grant from the National Science and Technology Development Agency (P-15-50004) to support YP. The authors would like to appreciate Dr. Chanisorn Ngaojampa, Department of Chemistry, Faculty of Science, Chiang Mai University, for his valuable comments and suggestions.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmgm.2020.107717.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020 doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization (WHO) 20 January 2020. Statement on the second meeting of the international health regulations (2005) emergency committee regarding the outbreak of novel coronavirus (2019-nCoV) [Google Scholar]
- 3.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel Coronavirus I., Research T. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Report of BNO News . 2 February 2020. Tracking coronavirus: Map, data and timeline. Report of BNO News, Tracking coronavirus: Map, data and timeline. [Google Scholar]
- 5.Perlman S. Another decade, another coronavirus. N. Engl. J. Med. 2020 doi: 10.1056/NEJMe2001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet. 2020 doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Report of CBC News hina confirms human-to-human transmission of new coronavirus. 20 January 2020.
- 8.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Report of The Star News . 6 February 2020. China starts using antiviral drug Remdesivir in clinical trials against Wuhan virus. [Google Scholar]
- 11.Paemanee A., Hitakarun A., Wintachai P., Roytrakul S., Smith D.R. A proteomic analysis of the anti-dengue virus activity of andrographolide. Biomed. Pharmacother. 2019;109:322–332. doi: 10.1016/j.biopha.2018.10.054. [DOI] [PubMed] [Google Scholar]
- 12.Wen L., Xia N., Chen X., Li Y., Hong Y., Liu Y., Wang Z., Liu Y. Activity of antibacterial, antiviral, anti-inflammatory in compounds andrographolide salt. Eur. J. Pharmacol. 2014;740:421–427. doi: 10.1016/j.ejphar.2014.06.053. [DOI] [PubMed] [Google Scholar]
- 13.Fanhchaksai K., Kodchakorn K., Pothacharoen P., Kongtawelert P. Effect of sesamin against cytokine production from influenza type A H1N1-induced peripheral blood mononuclear cells: computational and experimental studies. In Vitro Cell Dev. Biol. Anim. 2016;52(1):107–119. doi: 10.1007/s11626-015-9950-7. [DOI] [PubMed] [Google Scholar]
- 14.Udomruk S., Kaewmool C., Pothacharoen P., Phitak T., Kongtawelert P. Sesamin suppresses LPS-induced microglial activation via regulation of TLR4 expression. J. Funct. Foods. 2018;49:32–43. [Google Scholar]
- 15.Byun E.B., Kim H.M., Song H.Y., Kim W.S. Hesperidin structurally modified by gamma irradiation induces apoptosis in murine melanoma B16BL6 cells and inhibits both subcutaneous tumor growth and metastasis in C57BL/6 mice. Food Chem. Toxicol. 2019;127:19–30. doi: 10.1016/j.fct.2019.02.042. [DOI] [PubMed] [Google Scholar]
- 16.Punfa W., Yodkeeree S., Pitchakarn P., Ampasavate C., Limtrakul P. Enhancement of cellular uptake and cytotoxicity of curcumin-loaded PLGA nanoparticles by conjugation with anti-P-glycoprotein in drug resistance cancer cells. Acta Pharmacol. Sin. 2012;33(6):823–831. doi: 10.1038/aps.2012.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanner M.F. Python: a programming language for software integration and development. J. Mol. Graph. Model. 1999;17:57–61. [PubMed] [Google Scholar]
- 18.Case D.A., Ben-Shalom I.Y., Brozell S.R., Cerutti D.S., Cheatham T.E., III., Cruzeiro V.W.D., Darden T.A., Duke R.E., Ghoreishi D., Gilson M.K., Gohlke H., Goetz A.W., Greene D., Harris R., Homeyer N., Huang Y., Izadi S., Kovalenko A., Kurtzman T., Lee T.S., LeGrand S., Li P., Lin C., Liu J., Luchko T., Luo R., Mermelstein D.J., Merz K.M., Miao Y., Monard G., Nguyen C., Nguyen H., Omelyan I., Onufriev A., Pan F., Qi R., Roe D.R., Roitberg A., Sagui C., Schott-Verdugo S., Shen J., Simmerling C.L., Smith J., Salomon-Ferrer R., Swails J., Walker R.C., Wang J., Wei H., Wolf R.M., Wu X., Xiao L., York D.M., Kollman P.A. University of California; San Francisco: 2018. AMBER 2018. [Google Scholar]
- 19.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Götz A.W., Williamson M.J., Xu D., Poole D., Le Grand S., Walker R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized born. J. Chem. Theor. Comput. 2012;8(5):1542–1555. doi: 10.1021/ct200909j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salomon-Ferrer R., Götz A.W., Poole D., Le Grand S., Walker R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J. Chem. Theor. Comput. 2013;9(9):3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 22.Weber W., Hünenberger P.H., McCammon J.A. Molecular dynamics simulations of a polyalanine octapeptide under Ewald boundary Conditions: influence of artificial periodicity on peptide conformation. J. Phys. Chem. B. 2000;104(15):3668–3675. [Google Scholar]
- 23.Darden T., York D.M., Pedersen L.G. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 24.Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103(19):8577–8593. [Google Scholar]
- 25.Roe D.R., Cheatham T.E. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theor. Comput. 2013;9(7):3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 26.Kollman P.A., Massova I., Reyes C., Kuhn B., Huo S., Chong L., Lee M., Lee T., Duan Y., Wang W., Donini O., Cieplak P., Srinivasan J., Case D.A., Cheatham T.E. Calculating structures and free energies of complex Molecules: combining molecular mechanics and continuum models. Acc. Chem. Res. 2000;33(12):889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 27.Hünenberger P.H., Mark A.E., van Gunsteren W.F. Fluctuation and cross-correlation analysis of protein motions observed in nanosecond molecular dynamics simulations. J. Mol. Biol. 1995;252(4):492–503. doi: 10.1006/jmbi.1995.0514. [DOI] [PubMed] [Google Scholar]
- 28.Dong N., Yang X., Ye L., Chen K., Chan E.W.-C., Yang M., Chen S. Genomic and protein structure modelling analysis depicts the origin and infectivity of 2019-nCoV, a new coronavirus which caused a pneumonia outbreak in Wuhan, China. bioRxiv. 2020 [Google Scholar]
- 29.Xue X., Yang H., Shen W., Zhao Q., Li J., Yang K., Chen C., Jin Y., Bartlam M., Rao Z. Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction. J. Mol. Biol. 2007;366(3):965–975. doi: 10.1016/j.jmb.2006.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. Shanghai Public Health Clinical Center & School of Public Health, Fudan University; Shanghai, China: 2020. A Novel Coronavirus Associated with a Respiratory Disease in Wuhan of Hubei Province, China: Wuhan Seafood Market Pneumonia Virus Isolate Wuhan-Hu-1, Complete Genome (GenBank: MN908947.3) [Google Scholar]
- 31.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D., Gibson T.J., Higgins D.G. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 32.Gruber C.C., Steinkellner G. 23 January 2020. Wuhan coronavirus 2019-nCoV - what we can find out on a structural bioinformatics level, Innophore, GmbH, Austria. [Google Scholar]
- 33.Wang E., Sun H., Wang J., Wang Z., Liu H., Zhang J.Z.H., Hou T. End-point binding free energy calculation with MM/PBSA and MM/GBSA: strategies and applications in drug design. Chem. Rev. 2019;119(16):9478–9508. doi: 10.1021/acs.chemrev.9b00055. [DOI] [PubMed] [Google Scholar]
- 34.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M., Li X., Xia J., Chen N., Xiang J., Yu T., Bai T., Xie X., Zhang L., Li C., Yuan Y., Chen H., Li H., Huang H., Tu S., Gong F., Liu Y., Wei Y., Dong C., Zhou F., Gu X., Xu J., Liu Z., Zhang Y., Li H., Shang L., Wang K., Li K., Zhou X., Dong X., Qu Z., Lu S., Hu X., Ruan S., Luo S., Wu J., Peng L., Cheng F., Pan L., Zou J., Jia C., Wang J., Liu X., Wang S., Wu X., Ge Q., He J., Zhan H., Qiu F., Guo L., Huang C., Jaki T., Hayden F.G., Horby P.W., Zhang D., Wang C. A trial of lopinavir-ritonavir in adults hospitalized with Severe Covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agoni C., Ramharack P., Soliman M.E.S. Allosteric inhibition induces an open WPD-loop: a new avenue towards glioblastoma therapy. RSC Adv. 2018;8(70):40187–40197. doi: 10.1039/c8ra08427k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kasahara K., Fukuda I., Nakamura H. A novel approach of dynamic cross correlation analysis on molecular dynamics simulations and its application to Ets1 dimer-DNA complex. PLoS One. 2014;9(11):112419–122432. doi: 10.1371/journal.pone.0112419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.