Abstract
DNA carries important genetic instructions and plays vital roles in regulating biological activities in living cells. Proteins such as transcription factors binds to DNA to regulate the biological functions of DNA, and similarly many drug molecules also bind to DNA to modulate its functions. Due to the importance of protein-DNA and drug-DNA binding, there has been intense effort in developing novel nanosensors in the same length scale as DNA, to effectively study these binding interactions in details. In addition, aptamers can be artificially selected to detect metal ions and pathogens such as bacteria and viruses, making nucleic acid nanosensors more versatile in detecting a large variety of analytes. In this minireview, we first explained the different types and binding modes of protein-DNA and drug-DNA interactions in the biological systems, as well as aptamer-target binding. This was followed by the review of five types of nucleic acid nanosensors based on optical or electrochemical detection. The five types of nucleic acid nanosensors utilizing colorimetric, dynamic light scattering (DLS), surface-enhanced Raman spectroscopy (SERS), fluorescence and electrochemical detections have been recently developed to tackle some of the challenges in high-throughput screening technology for large scale analysis, which is especially useful for drug development and mass screening for pandemic outbreak such as SARS or COVID-19.
Keywords: Nucleic acid nanosensor, Protein-DNA binding, Drug-DNA binding, Aptamer sensor, Nanostructures
Highlights
-
•
Novel nucleic acid based nanosensors are being developed to investigate protein-DNA and drug-DNA binding interactions.
-
•
Artificially selected aptamers expand the range of analytes from ions to small molecules to larger pathogenic cell.
-
•
Nanoparticles function effectively as the signal enhancer or transducer for improving nucleic acid nanosensor performance.
-
•
The advancement in nucleic acid nanosensors is useful for mass screening during pandemic outbreaks such as SARS or COVID-19.
1. Introduction
Deoxyribonucleic acid (DNA) is a polymeric chain comprising of repeating units called nucleotides, which carries important genetic instructions that play vital roles in regulating biological activities in living cells. Mutations in DNA have always been correlated with various diseases including cancer. DNA typically exists as two strands of oligonucleotides wrapped around each other to form a double helix [1,2]. The double helix structure of DNA is characterized by two grooves which allow binding with a large variety of proteins or drug molecules [3]. Proteins such as transcription factors, nucleases or enzymes bind to DNA and regulate the biological functions of DNA, usually gene expression. On the other hand, DNA is the main target of many drug molecules that interacts with the DNA in various modes to manipulate its function [4]. Today, DNA is strategically set as a target for ligands in treating many human diseases especially for cancer therapeutics [5,6]. Therefore, in-depth understanding of protein-DNA and drug-DNA binding interactions is the key to understand disease mechanisms which helps in the drug design in order to successfully tackle a range of diseases [[7], [8], [9]].
Besides performing critical cellular functions, there is an artificially selected group of short, single-stranded DNA/RNA called aptamers that can bind to a diverse range of analytes including metal ions, small molecules, proteins and even pathogens with high affinity and selectivity [[10], [11], [12]]. Although aptamers are frequently considered as the nucleic acid equivalent of antibodies, they offer many unique advantages due to their relatively small size, versatility in binding to any target of choice (especially small molecular analytes of <1700 kDa), higher target selectivity, better stability under non-physiological conditions, prolonged shelf life, ease of chemical modification and cost-effectiveness for large quantity synthesis, which are all beneficial in developing nucleic acid nanosensors [10,12]. Cai et al. have reviewed the recent advances in studying the interactions of aptamers with their binding targets, which offers great insights into the range of binding targets as well as the binding constants and binding forces between the aptamer and their targets [13]. These insights are especially useful in the design of aptamer based nanosensors for detecting these specific binding interactions.
The advancement of nanotechnology has contributed significantly in developing novel nucleic acid-based biosensors involving the use of different types of nanostructures. Plasmonic nanomaterials such as gold and silver nanoparticles, fluorescent nanomaterials like quantum dots and carbon dots, electrochemically active materials including carbon nanotubes and graphene are frequently used as the key components in these nanosensors [[14], [15], [16]]. The interaction of DNA with nanomaterials for biosensing applications has been reviewed by He et al. [17]. In this review, we focus on the design of nucleic acid nanosensors for detecting a broader range of biological analytes, which include ions, protein, bacteria, etc. According to the signal generation mechanism, nanosensors can be detected in different sensing modes such as optical or electrochemical detection [14]. The uniqueness of a nanosensor lies in the similar length scale of the nanostructures to the biological targets to facilitate better binding interactions mimicking natural interactions, which may further allow in vivo applications. In addition, large surface area-to-volume ratios provided by nanostructures enable the binding of a large number of bio-recognition elements to capture the target analyte, thus greatly improving the detection sensitivity.
Due to the importance of understanding protein-DNA and drug-DNA interactions and the prevalent applications of aptamers in various fields, it is not surprising to observe synergistic combination of nucleic acids as the recognition element and nanostructures as the signalling element for the advancement of nucleic acid nanosensors to detect sequence specific binding interactions. Many of these nucleic acid nanosensors incorporate natural DNA sequences or in vitro selected aptamers as the bio-recognition element to investigate a wide range of targets from metal ions, drug -molecules, proteinaceous targets such as antibodies or enzymes to the entire cells such as viruses or bacteria. Both optical and electrochemical sensing principles are widely employed in these nucleic acid nanosensors to study the binding interactions between nucleic acids and targets of interest.
2. Types of nucleic acid binding interactions in biological systems
As mentioned earlier, DNA is a polymer comprising of repeating units of four nucleotides [18]. The nucleotides typically constitute a five-carbon sugar called deoxyribose and a nitrogen containing base, which are connected through phosphodiester linkages within a strand. The bases include thymine (T), cytosine (C), guanine (G) and adenine (A) [19]. The secondary structure of DNA is a double helix with two antiparallel polynucleotide chains firmly held together by hydrogen bonds, where A forms two hydrogen bonds with T while C forms three hydrogen bonds with G. In the double helix, the sequences of the strands are complementary to each other. The interactions of biomolecules or drugs with DNA double helix are varied. Typically, proteins interact with DNA through specific base-pair recognition, multi-specific binding or non-specific binding independent of DNA sequences [20], whereas, drug molecules can interact in covalent or non-covalent interactions [19,21,22].
2.1. Protein-DNA binding
Proteins that possess DNA-binding domains can bind to DNA through non-specific, specific or multi-specific interactions [3]. The most extensively studied DNA-binding protein is called transcription factor (TF), which regulates gene transcription such as the famous tumour suppressor protein, p53 [23]. When the amino acid residues of a TF interact with DNA, they recognize a specific sequence stringently to form a stable complex, although there is still an underlying difference in the extent of stability depending on the particular family of TFs. On the other hand, non-specific DNA-binding proteins prefer to bind in the minor grooves of double helix, without a good discrimination between the bases. As for the multi-specific binding, this group of DNA-binding proteins recognize target sequences less stringently, even though some bases are frequently mutated [20,24].
For the sequence-specific protein-DNA binding interactions, TFs bind to a specific DNA sequence called the response elements (RE) through hydrogen bonding and van der Waals forces [3,25]. Generally, TFs interact with the major groove of DNA double helix as the bases are exposed for ease of identification and interaction [26]. There are approximately 3000 TFs with sequence-specific DNA binding characteristics [27,28], and they are classified based on their DNA-binding domains [29] which may predict their unique functions. Among the various binding domains existed in TFs (Fig. 1 ), the three main groups of TFs include 1) C2H2 zinc fingers, 2) homeodomain (HD) and 3) helix-loop-helix (HLH).
Fig. 1.
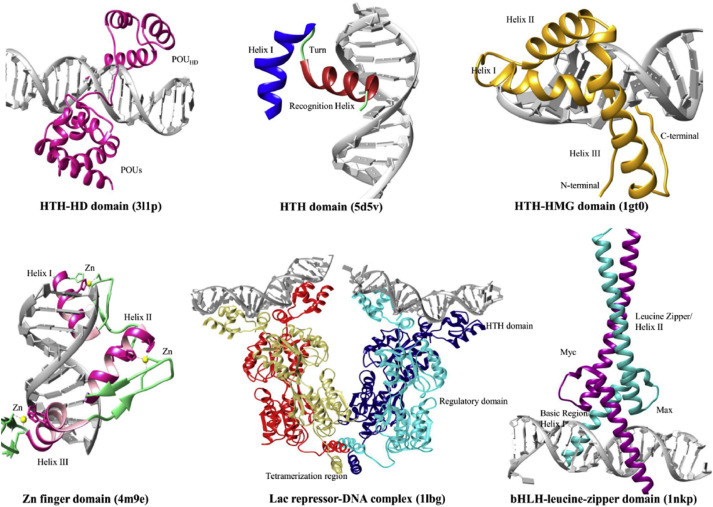
Representative binding domains of transcription factor (3l1p, 4m9e, 5d5v, 1lbg, 1gt0, and 1nkp). The crystal structures were obtained from the Protein Data Bank (PDB) and redrawn using chimera. The respective domains and important regions have been labelled. HTH stands for helix-turn-helix domain. bHLH stands for basic helix-loop-helix motif. HD and HMG stand for homeodomain and high-mobility group box domain, respectively. Adapted from Refs. [30].
Detailed investigations have discovered that TFs can bind their RE as homo or hetero dimers. Homodimeric TFs clamp the DNA on both strands. In addition, higher oligomerization has also been observed, for example, heat shock factors act as trimers and p53 proteins bind as tetramers. The tumor suppressor p53 protein plays a critical role in regulating a myriad of cellular processes such as replication, transcription, cell division and DNA repair. Many diseases related mutations take place at the DNA binding domain of p53 proteins, making them a potential target in clinical diagnosis and drug development. The DNA binding domain of p53 protein have four canonical binding sites (→← →←), and these sites made up the RE. In principle, full-length p53 proteins dimerize through C-terminal oligomerization domain and then the RE half-sites bind to the p53 core domain in a symmetric interface. Finally, the dimers tetramerize with the second half-site. This example illustrates how complicated TF-DNA binding can be, thus, smart and sensitive detection methods need to be developed to gain an in-depth understanding of varied protein-DNA binding interactions including binding affinity evaluation, the mode of binding, the effect of protein mutation, etc, which will be discussed in details in Section 3.
2.2. Drug-DNA binding
DNA, especially in its double-stranded form, is able to complex with a large number of natural or synthetic molecules, therefore DNA has become the pharmacological target of many drugs. For example, chemotherapeutic drugs bind to DNA and tend to alter the properties of DNA which then influence critical cellular processes [31]. Elucidating the detailed interactions between drug molecules and DNA has always been a hot research topic which aids drug design and drug screening.
In general, drug-DNA interactions can be classified as covalent or non-covalent [31]. The covalent interaction is an irreversible reaction which inhibits the functions of DNA and consequently causes cell apoptosis. A popular example is a platinum based anticancer drug, cisplatin, which is frequently used in cancer treatment due to their strong interactions with DNAs (Fig. 2 a). This drug typically forms bifunctional intra- and inter-strand DNA crosslinks through covalent bonds with purine nucleobases, which subsequently disrupts DNA replication, inhibits transcription and consequently kills the cancer cells. Unfortunately, the platinum-based drugs are limited by side effects and poor activity in certain types of cancer because their detailed mechanisms of action are still not clearly understood [32]. Another example is alkylator which interacts covalently with the nucleophilic group of DNA through a strong electrostatic interaction to form an irreversible adduct (Fig. 2b). Typical alkylators such as ethyleneimines and methane sulfonates can undergo substitution reactions with the N atoms of guanine or adenine.
Fig. 2.
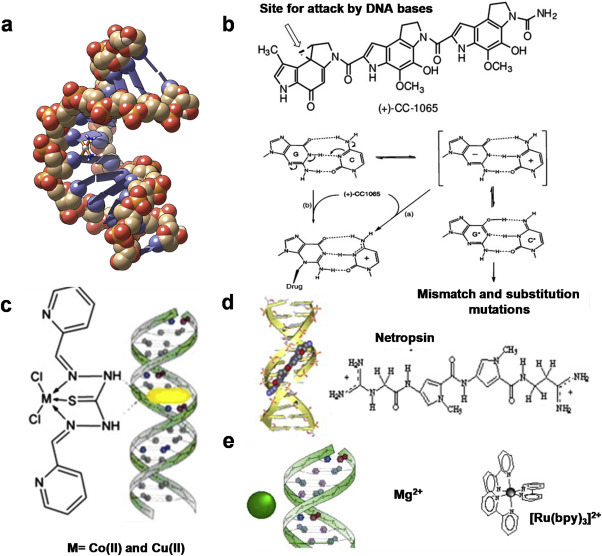
Different modes of drug-DNA interactions (a) Cisplatin is covalently bonded to DNA. Adapted with permission from Ref. [36] (b) Proposed mechanisms for the alkylation of N3 guanine by (+)-CC1065 (c) Intercalation of Co(II) and Cu(II) metal complexes of N1,N5-bis[pyridine-2-methylene]-thiocarbohydrazone (d) DNA complexed with netropsin, a minor groove binder (e) External association of the complex in the atmosphere of ions of the DNA polyelectrolyte. Adapted with permission from Ref. [19].
On the other hand, the three major modes of non-covalent drug-DNA interactions include (1) groove binding, (2) intercalative binding, (3) external binding [31]. Groove binders possess free rotating heterocyclic or aromatic hydrocarbon ring groups that allow them to fit into the major or minor groove of DNA double helix. For example, netropsin and distamycin interact with DNA minor groove through hydrogen bonding and hydrophobic interactions (Fig. 2d) [33]. Intercalators, such as flat aromatic or heteroaromatic molecules, slide in between adjacent base pairs of DNA, and form stable complexes via hydrophobic stacking interactions (Fig. 2c) [34,35]. Intercalation can result in DNA unwinding, which prevents transcription or replication. The external binding is typically electrostatic in nature. Some positively charged ligands such as metal complexes can bind electrostatically to the negatively charged phosphate-sugar backbone (Fig. 2e).
2.3. Aptamer-target binding
Aptamers are single-stranded DNA or RNA that can bind to their targets with high affinity and specificity similar to the antigen-antibody interactions. Aptamers, often 25–90 bases in length, are derived from an in vitro selection process called systematic evolution of ligands by exponential enrichment (SELEX) [37]. The SELEX process is used to screen oligonucleotides from large libraries of RNA or single-stranded DNA. This in vitro selection process is iteratively performed and subsequently the selected sequences are amplified by polymerase chain reaction (PCR). Aptamers bind to their targets via the complementary shape interactions and three-dimensional folding with high affinity. The most common secondary or tertiary structural motifs for aptamer include G-quadraplexes, stem-loops, pseudoknots, kissing complexes, three-way junctions, and hairpins [12]. Compared to the most commonly used biorecognition element-antibodies that mainly targets proteins, aptamers can be generated against a myriad of targets ranging from metal ions (e.g. Cu2+, Hg2+), small molecules (e.g. ATP, cocaine, aflatoxin B1) to proteins (e.g. thrombin, streptavidin) and even whole cells (e.g. viruses, bacteria, cancer cells) [12,13,[38], [39], [40], [41]]. This gives great versatility to design nucleic acid sensors to target any analyte of interest.
3. Nanosensors designed for detecting DNA binding interactions
To detect DNA binding interactions, such as protein-DNA binding, drug-DNA binding, aptamer-target binding, researchers have developed various sensing methods. Among which, the most promising one is the nanosensor design, which uses either the plasmonic nanostructures for direct optical sensing or employ various type of nanomaterials such as graphene, metal oxides, rare-earth metal, magnetic nanoparticles, etc. to achieve signal amplification or enhancement in sensing performance [42]. Particularly, optical and electrochemical detection principles have been widely applied in developing nanosensors for various analytes, which will be discussed in details with specific examples highlighted in the following subsections.
3.1. Metal nanoparticles-based colorimetric detection
Colorimetric sensors are simple, low cost due to their easy read out, which is even possible by naked eyes detection [43]. Noble metal nanoparticles such as gold, silver are suitable colorimetric indicators due to their excellent extinction coefficients and also unique distance-dependent optical properties [44]. For example, Zhang's group reported a sensitive approach to detect transcription factor NF-kB p50 by converting them into reporter oligonucleotides through protein-DNA interaction, exonuclease (III) digestion and isothermal exponential amplification [45]. If NF-kB is present, its binding to DNA protects the DNA from nuclease digestion, the DNAs are then amplified to cause aggregation of the DNA-modified gold nanoparticle (AuNP) probes leading to precipitation of AuNPs. In the absence of NF-kB, DNAs are digested and no sequences can be amplified, therefore the AuNP probes remain well suspended and the solution is still red in color. This sensing method is very sensitive, achieving a limit of detection of 3.8 pM. However, the procedure of this assay is quite time consuming and tedious, requiring many steps of heating and cooling to allow DNA digestion and signal amplification.
Our lab has been developing a series of easy-to-perform gold nanoparticles (AuNP)-enabled colorimetric sensors to quickly detect protein-DNA binding with various sensing strategies (Fig. 3 ) [[46], [47], [48], [49], [50], [51], [52]]. Tan et al. first studied the binding interactions between estrogen receptors (ER) to their corresponding DNA-binding element known as estrogen response element (ERE) using unmodified AuNPs. It was found that the protein-DNA complexes can better stabilize the citrate anion-capped AuNPs against salt-induced aggregation as compared to the protein or DNA alone (Fig. 3a). Moreover, this assay offers high sequence specificity which distinguishes single-base variation in the ERE [46,52]. A similar detection strategy is later applied to monitor the binding of single strand binding protein (SSB) to a single-stranded DNA (ssDNA) of various base sequences [48]. It was successfully revealed that the minimum DNA length required to form a stable SSB-ssDNA complex is more than 10-mer, and the SSB proteins binds to homopyrimidines with a higher affinity than the homopurines of similar length (Fig. 3b). In these two nanosensor designs, non-crosslinking AuNPs aggregation strategy has been employed successfully to detect not only the sequence-specific protein-DNA interactions without any tedious surface modification steps, but also provide useful insights into their intrinsic binding mechanisms in homogenous solution similar to that in the biological systems.
Fig. 3.
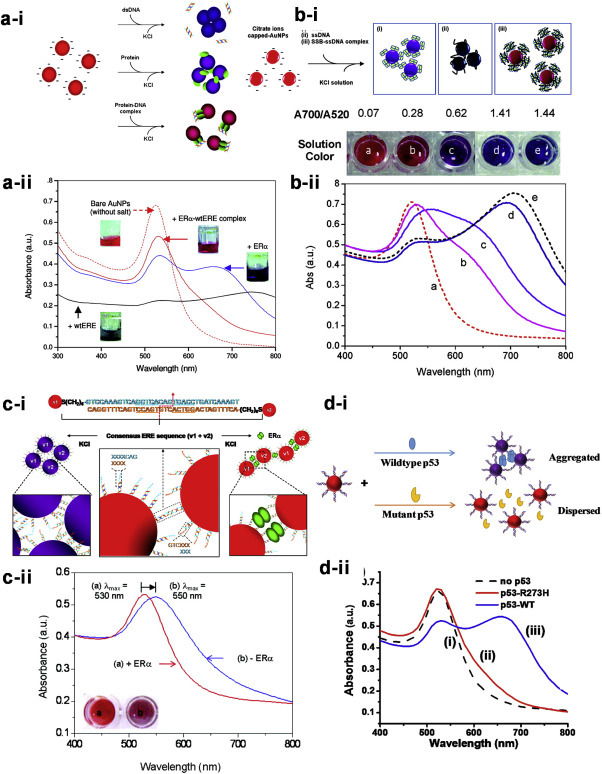
Development of gold nanoparticles (AuNPs)-based colorimetric biosensors to detect sequence-specific protein-DNA binding: (a) ER-ERE binding (b) SSB-ssDNA binding (d) segmented ER-ERE binding (d) p53-RE binding, using different biomolecular interactions induced particle aggregation strategies. Row (i) show the sensing principles of the colorimetric assay and row (ii) show the representative photos and UV–vis spectra for each nanoparticle biosensor. Adapted with permission from Ref. [[46], [47], [48],50]. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
As the citrate-capped AuNPs may be prone to aggregation induced by environmental factors, our group has further developed a new sensing strategy based on the use of segmented DNA-modified AuNPs and non-crosslinking mechanism to detect sequence-specific ER (protein)-ERE (DNA) interactions. Specifically, two sets of double-stranded DNA (dsDNA) modified AuNPs, each carrying a half site segment of the ERE was designed to contain a short complementary sticky end for base pairing to cause particle aggregation and change in solution colour from red to purple (Fig. 3c). In the presence of ER, the sequence-specific protein binding is able to stabilize the transient nanostructures formed by the two sets of segmented ERE-linked AuNPs upon mixing due to steric forces, thus the particle solution remains red [47]. In this work, silver nanoparticles (AgNPs) has been demonstrated to give a more sensitive detection limit with a different visual color change from yellow to brown. Tan and co-workers later carried out a systematic investigation on the decorating DNA design to gain better insight into the non-crosslinking AuNPs aggregation using DNA-conjugated AuNPs for more efficient assay development [49]. It was found that (1) the dsDNA spacer of 11-base pairs (bp) or longer are more effective than the ssDNA spacer and (2) the symmetrically spaced sticky ends could facilitate the dsDNA-AuNPs aggregation better than the asymmetrical combination. More recently, our lab developed a new “mix-and-measure” protein-DNA binding assay utilizing only one set of dsDNA-modified AuNPs, which contains the p53 response element (RE-AuNP) for rapid detection of wildtype p53 proteins from the complex cell lysates [50]. As shown in Fig. 3d, the wildtype p53 proteins bind to RE-AuNPs in tetrameric forms causing particle aggregations and thus solution color change from red to purple. As no tetramerization occurs in the presence of mutant p53 proteins, the RE-AuNPs are still dispersed and the solution remains red. The colorimetric difference in this assay is very distinct which can be easily observed by naked eyes and quantified by the UV–visible spectroscopy. This protein tetramerization-induced RE-AuNPs aggregation sensing principle has been further employed for small molecular drug screening and cancer diagnostics in the reported studies [50,53].
Other than utilizing natural DNA sequences in designing colorimetric nucleic acid nanosensors, aptamer represents a popular choice due to its wide-ranging targets. For example, Lu's group pioneered the development of a general sensor design involving aptamers and AuNPs for the detection of small molecules such as adenosine and cocaine. Two sets of DNA-modified AuNPs are first crosslinked via a third sequence containing a complementary sequence to both sets of DNAs on AuNPs with an aptamer sequence appended. This causes the AuNPs to aggregate and display purple color. Upon target addition, the aptamer binds to the target, which weakens the binding to the two sets of DNA-modified AuNPs, thus causing the aggregated particles to re-disperse and display red color. This general colorimetric sensing strategy based on the disassembly of aggregated nanoparticle linked by aptamers can be applied to a wide range of analytes, suggesting the versatility of aptamer-based nanosensors [10,54].
3.2. Nanoparticles-enabled dynamic light scattering (nano-DLS) detection
Dynamic light scattering (DLS) is usually applied to measure particle size by monitoring the scattered light intensity fluctuation of particles undergoing Brownian motion in solution. Due to the large scattering dimensions of gold nanoparticles (AuNPs), it can be coupled with DLS as a readout system to construct a series of nanoparticle-enabled DLS (nano-DLS) bioassays to monitor various binding interactions. Huo's group pioneered the application of nano-DLS assays to successfully study DNA-DNA binding and protein-protein interactions [15,55,56].
Our lab focuses on designing nano-DLS biosensors to monitor protein-DNA interactions and mRNA detection. Seow et al. developed a unique DNA-assembled AuNP dimers for DLS sensing of transcription factor-DNA binding. As shown in Fig. 4 a, estrogen response element (ERE) is incorporated into the DNA linkers that bridges the AuNP dimers. Upon ER binding, a complex peak at a larger diameter appears, which marks the first time that this dimeric nanostructure can be used to study protein-DNA interactions by hydrodynamic size changes accurately measured by DLS [57]. As a step further, Zheng et al. later designed an ultrasensitive nano-DLS biosensor for rapid therapeutic drug screening. In this work, dsDNA-conjugated dumbbell-shaped AuNPs can be used to directly detect sequence-specific p53 protein-RE binding in drug-treated cancer cells [53]. Specifically, binding of wildtype p53 protein to the RE-modified nano-dumbbell AuNP probes causing tetramerization to form large complex, leading to significant size changes in DLS detection, while the mutant p53 proteins do not cause any change in the hydrodynamic size (Rh) as illustrated in Fig. 4b. A competitive assay is developed to allow fast screening and ranking of the p53 binding affinity to different DNA sequences. Most critically, this nano-DLS biosensor enables the screening of p53 activating or reactivating drugs directly using complex cell lysates. If the drug molecule is effective, mutant p53 protein can be reactivated and restore its binding to the target DNA (i.e., p53 RE) functionalized on the nano-dumbbell probe, resulting in a significant increase in hydrodynamic sizes (measured by DLS) similar to the wildtype p53 protein. These works confirm that nano-DLS is a very promising technique in studying a wide spectrum of protein-DNA and even drug effect on protein-DNA interactions in cells. On the other hand, we have also developed the AuNPs-DLS tandem for rapid and quantitative detection of the let7 microRNA family which associates with many medical conditions [58]. In this work, we have achieved an ultralow detection limit of 100 fmol and a selectivity discriminating close members of the let7 family. Most importantly, this nano-DLS detection can be completed in 5 min, which is much faster than the conventional techniques such as quantitative reverse transcriptase polymerase chain reaction and microarray technologies for miRNAs detection.
Fig. 4.
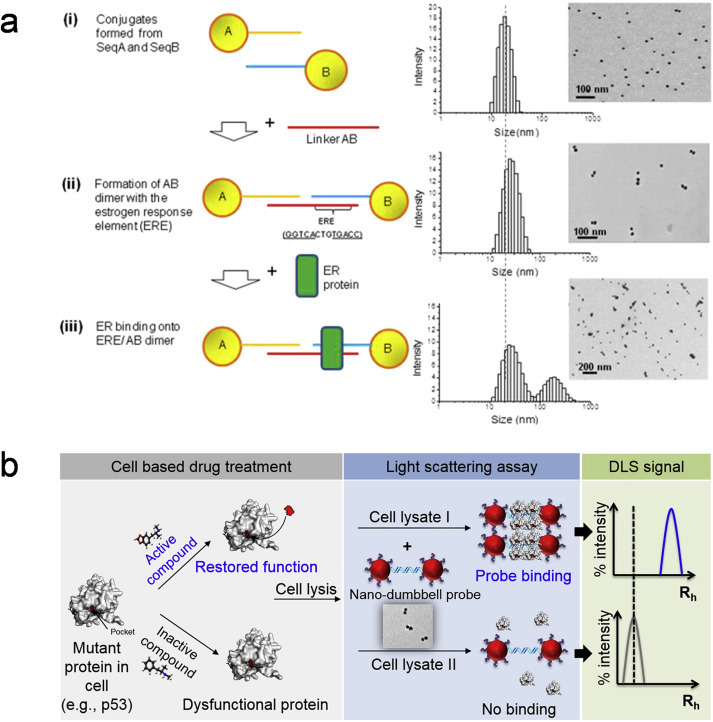
(a) Schematic illustration of the DLS sensing principle for transcription factor-DNA detection. DLS readout of (i) ssDNA-AuNPs conjugates formed from Seq A and Seq B respectively to AuNPs, exhibiting a single peak/population on DLS (ii) ERE-containing AuNP dimer formation, showing a single peak with a ∼10 nm right-shift in size compared to the ssDNA-AuNPs conjugates (iii) Addition of ERβ to the as prepared ERE-containing AuNP dimer sensing probes, showing a two-peak readout with an additional complex peak at around 200 nm. TEM images showed the AuNP size and distributions generally corresponded to the DLS hydrodynamic diameter readouts. Adapted with permission from Ref. [57] (b) (Left) Principle of nano-DLS biosensor for drug screening using dsDNA-modified nano-dumbbell AuNP probes (left). Detecting wildtype p53 or reactivation of mutant p53 proteins by drugs via tetramerization mechanism (middle) causing a significant increase in size (Rh: Hydrodynamic diameter) as measured by DLS (top right). Adapted with permission from Ref. [53].
3.3. Surface-enhanced Raman spectroscopy (SERS) detection
SERS is one of the most promising technique for molecular detection which merges the benefits of molecular fingerprint specificity with single molecule sensitivity [59]. Simulation studies have shown that metallic nanostructures of higher orders (e.g., trimer and tetramer) and different configurations can lead to better SERS enhancement effects [60,61]. For instance, SERS have been employed to study protein-DNA or drug-DNA interactions, achieving enhanced signal and therefore improved detection sensitivity [62]. Bonham et al. designed a short DNA duplex to have a core sequence to which the protein binds specifically and two short single-strand overhangs that hybridize with a pair of DNA-modified gold nanoparticles. DNA-bridged nanoparticle assemblies were formed and then silver plated to detect sequence-dependent protein-DNA interactions using confocal Raman microprobe [62]. In addition, the Raman intensity correlates with the protein concentration in a sequence specific manner leading to the estimation of the apparent dissociation constant (Fig. 5 a) [62].
Fig. 5.
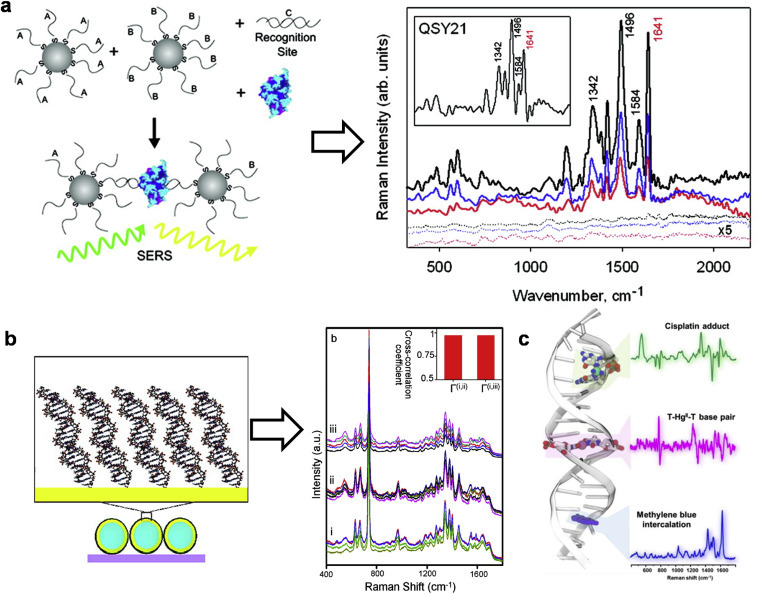
(a) Detection of protein binding to ABC assemblies of DNA-modified AuNPs via SERS (left); averaged spectra of labelled proteins: streptavidin (black), TBP (blue), and M.HhaI (red) with cognate-DNA (solid lines) and noncognate-DNA (x5 signal, dotted lines). Adapted with permission from Ref. [62] (b) SERS spectra of transplatin bound to 30 bp dsDNA SN5. (i) before, (ii) immediately following, and (iii) after overnight incubation with transplatin. Adapted with permission from Ref. [63] (c) Applying SERS to reveal covalent binding of cisplatin, intercalation of methylene blue and formation of T-Hg2-T base pairs with dsDNA. Adapted with permission from Ref. [64]. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
SERS is also an excellent technique to study drug-DNA interaction. Barhoumi et al. detected the drug induced conformational changes of DNA on gold nanoshell SERS substrate. The SERS spectra of DNA before and after interaction of cisplatin and transplatin were similar with only one new peak at 450 cm−1, which can be attributed to the platinum-amine stretching, thereby verifying covalent bonding of these two drugs to DNA (Fig. 5b) [63]. Matteo et al. then continued the effort to carefully investigate DNA interactions with exogenous agents using SERS enhanced by spermine-coated silver nanoparticles (AgNP@Sp) [64]. The complexation of cisplatin with DNA duplex to form covalent DNA adducts, the intercalation of methylene blue into DNA, and the formation of DNA-metal ion coordination by a toxic metal ion (HgII) were all revealed by their respective unique vibrational alterations in SERS spectra (Fig. 5c). With the use of appropriate nanostructures, SERS is an efficient analytical tool to detect the mode and quantify the extent of drug-DNA binding at ultra-sensitivity.
3.4. Quantum dots-based fluorescence detection
Fluorescence detection is one of the most used technique to study the interactions of drugs and proteins with DNA, as it offers high sensitivity, selectivity and wide linear range [65]. For drug-DNA interaction, the orientation of the fluorescent drug molecules and their proximity to DNA base pairs can be easily studied through Fluorescence resonance energy transfer (FRET) or fluorescence anisotropy. For example, Zhao et al. developed a quantum dot (QD) based FRET sensor to study the interaction between the common platinum drugs (cisplatin, oxaliplatin and carboplatin) with DNAs. As shown in Fig. 6 , the QD fluorescence was first quenched by the platinum drugs and can be recovered when the drugs covalently interact with the added DNAs [66].
Fig. 6.
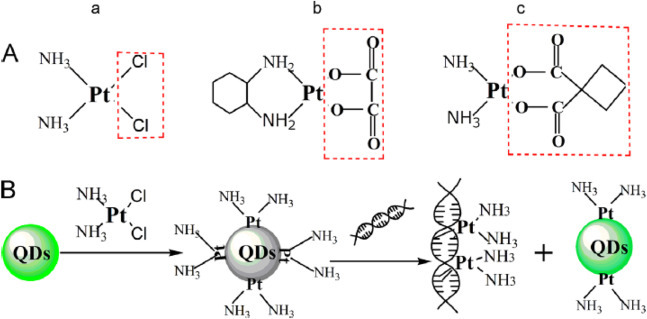
(A) Chemical structures of (a) cisplaitin, (b) oxaliplatin and (c) carboplatin The leaving groups are marked by dashed boxes (B) Schematic illustration of the signal transduction mechanism of QDs–cisplatin modulated by DNA. Adapted with permission from Ref. [66].
To study protein-DNA interactions, protein or oligonucleotides need to be fluorescently labelled. So far, most of the FRET assays for detecting DNA-binding proteins work in a “turn off” mode by monitoring fluorescence quenched upon binding, which tend to give false positive results due to non-specific quenching by environmental factors. Recently, a FRET assay utilizing cationic conjugated polymer (CCP) nanoparticles, hairpin DNA and an intercalative dye is developed to detect nuclear factor-kappa B (NF-kB) (Fig. 7 ) [67]. If NF-kB is present, it binds to the hairpin DNA probe and protect it from digestion by exonuclease III, thus resulting in efficient FRET between the intercalative dye and the CCP nanoparticles. As low as 1 pg/mL of NF-kB in HeLa nuclear extracts were successfully detected, which is 10,000-fold more sensitive than the previously reported methods. Fluorescent nanosensors are promising tools in studying both drug-DNA and protein-DNA interaction with high sensitivity. In addition to fluorescent dyes, semiconductor quantum dots, fluorescent conjugated polymers, novel fluorescent nanomaterials such as bioinspired carbon dots [[68], [69], [70], [71], [72]], metal nanoclusters [[73], [74], [75], [76], [77]] and aggregation induced emission dyes [78,79] can also be used in developing sensitive assays to monitoring biomolecular interactions including protein-DNA and drug-DNA binding.
Fig. 7.
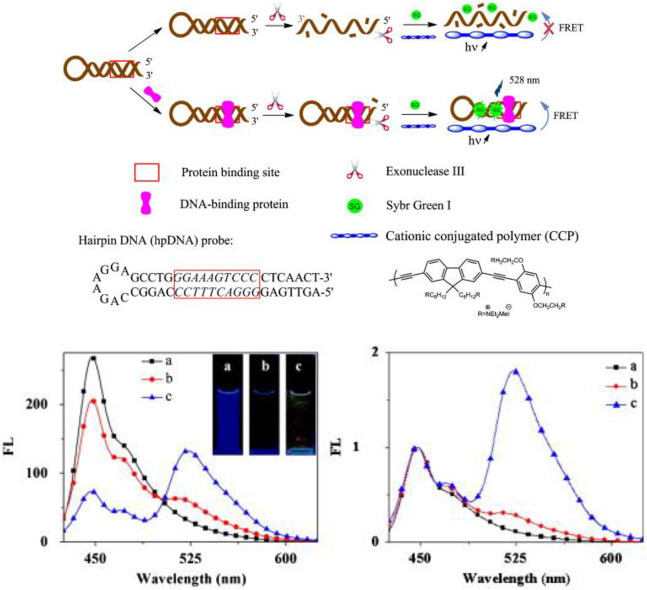
(a) Scheme for the detection of DNA-binding protein (transcription factor) using hairpin DNA (hpDNA), nano-sized cationic conjugated polymer (CCP) and an intercalative dye Sybr Green I (SG) (b) Fluorescence spectra and images of a: CCP; b: DNA/ExoIII/SG/CCP; c: DNA/NF-kB/ExoIII/SG/CCP (right) Normalized fluorescence spectra of the samples in (left). Adapted with permission from Ref. [67]. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
As more and more aptamer sequences are determined from the SELEX or cell-SELEX process, aptamers are becoming a very popular affinity reagent in designing fluorescent nanosensors. For example, Gao et al. engineered a fluorescence resonance energy transfer (FRET) based aptasensor for bacterial detection utilizing a fluorescent labelled DNA, a P. aeruginosa specific aptamer and graphene oxide quantum dot (GOQD) as the quencher. The fluorescent DNA can hybridize with the central portion of the aptamer, leaving the flanking region of aptamer as single strands. In the absence of P. aeruginosa, the DNA-aptamer complex is adsorbed on the GOQDs surface which leads to fluorescence quenching. With the addition of P. aeruginosa, the DNA-aptamer hybrids bind to the bacteria specifically, leading to the dissociation of fluorescently labelled DNA from GOQDs and subsequently fluorescence recovery. The detection limit of this aptasensor is 100 cfu/mL and can be applied to detect pathogens in complex samples such as drinking water, orange juice and popsicle [80]. Besides food-borne bacteria, aptamer based nanosensors have also been developed for food toxins. Zhang et al. successfully developed a fluorescent nanosensor which relied on the nanosized graphene oxide (GO nanosheet)-aptamer interaction to detect aflatoxin B1 (AFB1). The fluorescently labelled aptamer was first incubated with GO to form a complex, causing fluorescence quenching. This was followed by the addition of AFB1 and DNase I. If AFB1 is present, the aptamer will preferentially bind to the AFB1 and then get digested by DNase I, leaving the fluorescent molecules free in the solution, and thus fluorescence recovery is observed. Most intriguingly, the dynamic ranges of this nanosensor can be tuned by changing the size of the GO nanosheet [81]. Similar fluorescent sensors based on GO-aptamer interactions have been developed to detect other biologically important molecules such as ATP [82]. The use of aptamers has greatly expanded the range of targets that nucleic acid nanosensors can detect nowadays.
3.5. Nanostructure-enhanced electrochemical detection
Electrochemical investigations of specific DNA-target binding interactions render a good alternative to the optical detection methods as presented in sections 1, 2, 3 as they can also provide useful information for evaluating and predicting the binding process [83]. Electrochemical sensing of biomolecular-DNA/aptamer interactions usually involve three steps: (1) DNA (or aptamer) immobilization on electrode surface; (2) analyte addition to the electrode (3) output electrochemical signal measurement. The output signal which could be current or voltage response, is related to the protein-DNA, drug-DNA or aptamer-target binding interactions. The electrochemical signal potentially provides evidence for interaction mechanism, nature of the complex formed. With in-depth analysis, it is even possible to calculate the binding constants and also estimate the size of binding site.
Bonham et al. demonstrated a simple electrochemical sensor to detect the binding of transcription factor (TF) in crude nuclear extract to their target DNA [84]. This detection strategy is based on a structure-switching mechanism. The methylene blue (MB) modified DNA is first bound to the electrode surface to construct a sensor. Without TF binding, the DNA structure is relaxed and the redox reporter MB is positioned far away from the electrode surface, leading to lower amperometric signal. When TF binds, the DNA probe configuration switches and now MB is brought nearer to the electrode surface, leading to an increase in current. Zhang et al. then reported a similar strategy utilizing a different sensing principle which relies on repressed electrolyte diffusion associated with the protein-DNA binding, leading to a reduction in electrochemical signal [85]. Electrochemical sensors are usually very sensitive and fast in detecting protein-DNA binding. Gold nanoparticles (AuNPs) and AuNPs-enhanced silver deposition strategies have been used to further improve the electrochemical signal to achieve even lower detection limits [86].
In addition to act as the electrochemical signal enhancer, gold nanoparticles can be synthesized to exhibit peroxidase like activities. This enzyme mimic activity can be combined with the high affinity offered by aptamers to develop sensitive electrochemical nanosensor for bacteria, as demonstrated by Das et al. In their approach, P. aeruginosa specific aptamer (F23) can adsorb onto the surfaces of AuNPs to inhibit its inherent peroxidase like activity. In the presence of P. aeruginosa, the aptamer leaves the AuNP surface to bind strongly with the bacteria causing the restoration of the peroxidase activity. With the addition of an electrochemically active species, this approach enables rapid detection of P. aeruginosa with a detection limit of 60 cfu/mL [87].
Other than metal nanoparticles, chemically modified graphene is also excellent electroactive indicators. Hernandez et al. created an electrochemical nanosensor by non-covalently conjugating S. aureus specific aptamer to graphene via π- π interactions. With the addition of S. aureus, the aptamer prefers to bind to the bacteria and thus separate its negatively ionized phosphodiester groups from the graphene surface at pH 7.4. Since the graphene sheet act as asymmetric capacitors, this charge separation leads to a change of the recorded potential. This nanosensor is highly sensitive, achieving an ultralow detection limit of 1 cfu/mL [88]. Similarly, Shahrokhian et al. designed an electrochemical aptamer-based nanosensor for the detection of P. aeruginosa, by utilizing an engineered ZIF-8 layer to increase the aptamer immobilization density and ferocene-functionalized graphene oxide (Fc-GO) as the electroactive indicator. In the detection scheme as shown in Fig. 8 , aptamer conjugated ZIF-8 is first functionalized on the electrode surface, and Fc-GO is adsorbed via π- π stacking. However, when P. aeruginosa is added, the Fc-GO is displaced from the electrode surface causing electrochemical signal reduction due to their higher affinity binding with the aptamer modified ZIF-8. This particular nanosensor design also achieves an excellent detection limit of 12 cfu/mL and can be used to directly diagnose P. aeruginosa infection in urine [89]. We believe that these aptamer-based nanosensor designs offers a new angle to develop novel sensors to detect SARS-COV-2 in a rapid and sensitive manner.
Fig. 8.
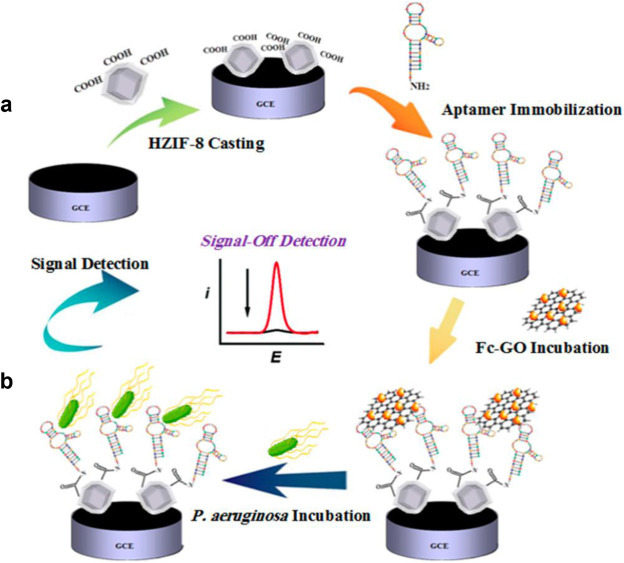
(a) Schematic illustration of aptasensor fabrication and (b)P. aeruginosa signal-off detection. Adapted with permission from Ref. [89].
Table 1 shows the comparison of the five types of nucleic nanosensors as reviewed herein for studying DNA-binding interactions based on different detection methods, including the colorimetric and DLS, SERS, fluorescence and electrochemical nanosensors. Different type of nanomaterials (e.g., metal nanoparticles, quantum dots, conducting polymers) are employed for the design of each type of nanosensor, having their own advantages and limitations.
Table 1.
Comparison of five types of nanosensors based on different detection method for studying sequence specific DNA-binding interactions.
| Detection method | Nanoparticle applied | Advantages | Limitations |
|---|---|---|---|
| Colorimetric | Noble metal nanoparticles |
|
|
| Dynamic light scattering (DLS) | Metal nanoparticle |
|
|
| Surface enhanced Raman spectroscopy (SERS) | Metal nanostructures |
|
|
| Fluorescence | Quantum dots; Carbon dots; Metal nanoclusters; Conjugated polymers; AIE dyes |
|
|
| Electrochemical | Metal nanoparticles; Carbon nanomaterials; Conducting polymers |
|
|
As we are in the midst of COVID-19 pandemic, a lot of effort has gone into developing fast and accurate diagnostic kits for mass screening of infections as well as to accelerate the development of effective anti-infective drugs or vaccines. In addition to the widely used nucleic acid amplification (e.g., PCR) and serological assays in detecting COVID-19 infections, the recent discovery of specific aptamer sequence that targets the SARS-CoV-2 spike protein [90] will allow the research community to quickly design aptamer based nanosensors for rapid and accurate detection of COVID-19 infections. Furthermore, nanoDLS assay also presents a feasible way to screen drug candidate targeting the virus with high-throughput. It is expected that these nucleic acid nanosensors would play very critical roles in both drug development and mass screening for pandemic outbreak such as SARS or COVID-19.
4. Conclusions
Understanding and detecting DNA binding interactions, specifically protein-DNA binding and drug-DNA binding are very crucial in revealing disease mechanisms and mode of actions for drugs which will then guide diagnosis and drug development. With the recent development of various nanosensors to detect various types of DNA binding interactions, we are expecting to see promising progresses with in-depth understanding of these interactions in various aspects, including binding kinetics, binding affinities, stoichoimetry, multiple component interactions and many more, which will certainly provide better guidance on the development of disease diagnosis and help to inform drug design. Furthermore, the advancement in aptamer design and synthesis has helped to greatly expand the range of analytes detectable by nucleic acid nanosensors. The development of many easy-to-use nanosensors that only requires one-step mix-and-measure protocol to operate are similarly important, which will help to greatly improve the throughput of screening platform for large-scale analysis and mass screening for infectious diseases containment applications.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to acknowledge the Institute of Materials Research and Engineering (IMRE), Agency for Science, Technology and Research (A∗STAR), Singapore for the Biomimetic and Biomedical Program exploratory fund IMRE/16–1P1401. The authors also acknowledge the support from Newcastle University (RSA/CCEAMD5010).
References
- 1.Misteli T. The cell biology of genomes: bringing the double helix to life. Cell. 2013;152:1209–1212. doi: 10.1016/j.cell.2013.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seeman N.C. DNA in a material world. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- 3.Jolma A., Yan J., Whitington T., Toivonen J., Nitta K.R., Rastas P., Morgunova E., Enge M., Taipale M., Wei G. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 4.Yu H., Lee H., Herrmann A., Buettner R., Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat. Rev. Canc. 2014;14:736–746. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 5.Kristensen L.S., Hansen L.L. PCR-based methods for detecting single-locus DNA methylation biomarkers in cancer diagnostics, prognostics, and response to treatment. Clin. Chem. 2009;55:1471–1483. doi: 10.1373/clinchem.2008.121962. [DOI] [PubMed] [Google Scholar]
- 6.Sozzi G., Conte D., Leon M., Cirincione R., Roz L., Ratcliffe C., Roz E., Cirenei N., Bellomi M., Pelosi G. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J. Clin. Oncol. 2003;21:3902–3908. doi: 10.1200/JCO.2003.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Kara H.E.S. Redox mechanism of anticancer drug idarubicin and in-situ evaluation of interaction with DNA using an electrochemical biosensor. Bioelectrochemistry. 2014;99:17–23. doi: 10.1016/j.bioelechem.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 8.Cheung-Ong K., Giaever G., Nislow C. DNA-damaging agents in cancer chemotherapy: serendipity and chemical biology. Chem. Biol. 2013;20:648–659. doi: 10.1016/j.chembiol.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 9.Gupta R.K., Sharma G., Pandey R., Kumar A., Koch B., Li P.-Z., Xu Q., Pandey D.S. DNA/protein binding, molecular docking, and in vitro anticancer activity of some thioether-dipyrrinato complexes. Inorg. Chem. 2013;52:13984–13996. doi: 10.1021/ic401662d. [DOI] [PubMed] [Google Scholar]
- 10.Liu J., Cao Z., Lu Y. Functional nucleic acid sensors. Chem. Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan W., Donovan M.J., Jiang J. Aptamers from cell-based selection for bioanalytical applications. Chem. Rev. 2013;113:2842–2862. doi: 10.1021/cr300468w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Y., Belmonte I., Sykes K.S., Xiao Y., White R.J. Perspective on the future role of aptamers in analytical chemistry. Anal. Chem. 2019;91:15335–15344. doi: 10.1021/acs.analchem.9b03853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai S., Yan J., Xiong H., Liu Y., Peng D., Liu Z. Investigations on the interface of nucleic acid aptamers and binding targets. Analyst. 2018;143:5317–5338. doi: 10.1039/c8an01467a. [DOI] [PubMed] [Google Scholar]
- 14.Chen G., Roy I., Yang C., Prasad P.N. Nanochemistry and nanomedicine for nanoparticle-based diagnostics and therapy. Chem. Rev. 2016;116:2826–2885. doi: 10.1021/acs.chemrev.5b00148. [DOI] [PubMed] [Google Scholar]
- 15.Jans H., Huo Q. Gold nanoparticle-enabled biological and chemical detection and analysis. Chem. Soc. Rev. 2012;41:2849–2866. doi: 10.1039/c1cs15280g. [DOI] [PubMed] [Google Scholar]
- 16.Xiao Z., Farokhzad O.C. Aptamer-functionalized nanoparticles for medical applications: challenges and opportunities. ACS Nano. 2012;6:3670–3676. doi: 10.1021/nn301869z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He Q., Wu Q., Feng X., Liao Z., Peng W., Liu Y., Peng D., Liu Z., Mo M. Interfacing DNA with nanoparticles: surface science and its applications in biosensing. Int. J. Biol. Macromol. 2020;151:757–780. doi: 10.1016/j.ijbiomac.2020.02.217. [DOI] [PubMed] [Google Scholar]
- 18.Watson J.D., Crick F.H. Cold Spring Harbor Laboratory Press; 1953. Cold Spring Harbor Symposia on Quantitative Biology; pp. 123–131. [DOI] [PubMed] [Google Scholar]
- 19.Sirajuddin M., Ali S., Badshah A. Drug–DNA interactions and their study by UV–Visible, fluorescence spectroscopies and cyclic voltametry. J. Photochem. Photobiol. B Biol. 2013;124:1–19. doi: 10.1016/j.jphotobiol.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 20.Luscombe N.M., Thornton J.M. Protein–DNA interactions: amino acid conservation and the effects of mutations on binding specificity. J. Mol. Biol. 2002;320:991–1009. doi: 10.1016/s0022-2836(02)00571-5. [DOI] [PubMed] [Google Scholar]
- 21.Chaires J.B. Drug—DNA interactions. Curr. Opin. Struct. Biol. 1998;8:314–320. doi: 10.1016/s0959-440x(98)80064-x. [DOI] [PubMed] [Google Scholar]
- 22.Aleksić M.M., Kapetanović V. An overview of the optical and electrochemical methods for detection of DNA - drug interactions. Acta Chim. Slov. 2014;61:555–573. [PubMed] [Google Scholar]
- 23.Muller P.A., Vousden K.H. p53 mutations in cancer. Nat. Cell Biol. 2013;15:2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 24.Afek A., Schipper J.L., Horton J., Gordân R., Lukatsky D.B. Protein−DNA binding in the absence of specific base-pair recognition. Proc. Natl. Acad. Sci. Unit. States Am. 2014;111:17140–17145. doi: 10.1073/pnas.1410569111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goh W.L., Assah E., Zheng X.T., Lane D.P., Ghadessy F.J., Tan Y.N. Transcription factors as detection and diagnostic biomarkers in cancer. In: Chandra P., Tan Y.N., Singh S.P., editors. Next Generation Point-of-care Biomedical Sensors Technologies for Cancer Diagnosis. Springer Singapore; Singapore: 2017. pp. 31–58. [Google Scholar]
- 26.Smith N.C., Matthews J.M. Mechanisms of DNA-binding specificity and functional gene regulation by transcription factors. Curr. Opin. Struct. Biol. 2016;38:68–74. doi: 10.1016/j.sbi.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 27.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 28.Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A. The sequence of the human genome. science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 29.Atchley W.R., Fitch W.M. A natural classification of the basic helix–loop–helix class of transcription factors. Proc. Natl. Acad. Sci. Unit. States Am. 1997;94:5172–5176. doi: 10.1073/pnas.94.10.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yesudhas D., Batool M., Anwar M.A., Panneerselvam S., Choi S. Proteins recognizing DNA: structural uniqueness and versatility of DNA-binding domains in stem cell transcription factors. Genes. 2017;8:192. doi: 10.3390/genes8080192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rehman S.U., Sarwar T., Husain M.A., Ishqi H.M., Tabish M. Studying non-covalent drug–DNA interactions. Arch. Biochem. Biophys. 2015;576:49–60. doi: 10.1016/j.abb.2015.03.024. [DOI] [PubMed] [Google Scholar]
- 32.Hato S.V., Khong A., de Vries I.J.M., Lesterhuis W.J. Molecular pathways: the immunogenic effects of platinum-based chemotherapeutics. Clin. Canc. Res. 2014;20:2831–2837. doi: 10.1158/1078-0432.CCR-13-3141. [DOI] [PubMed] [Google Scholar]
- 33.Barrett M.P., Gemmell C.G., Suckling C.J. Minor groove binders as anti-infective agents. Pharmacol. Ther. 2013;139:12–23. doi: 10.1016/j.pharmthera.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 34.Biebricher A.S., Heller I., Roijmans R.F., Hoekstra T.P., Peterman E.J., Wuite G.J. The impact of DNA intercalators on DNA and DNA-processing enzymes elucidated through force-dependent binding kinetics. Nat. Commun. 2015;6 doi: 10.1038/ncomms8304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rocha M., Cavalcante A., Silva R., Ramos E. On the effects of intercalators in dna condensation: a force spectroscopy and gel electrophoresis study. J. Phys. Chem. B. 2014;118:4832–4839. doi: 10.1021/jp501589d. [DOI] [PubMed] [Google Scholar]
- 36.Deo K.M., Pages B.J., Ang D.L., Gordon C.P., Aldrich-Wright J.R. Transition metal intercalators as anticancer agents-recent advances. Int. J. Mol. Sci. 2016;17 doi: 10.3390/ijms17111818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan J., Xiong H., Cai S., Wen N., He Q., Liu Y., Peng D., Liu Z. Advances in aptamer screening technologies. Talanta. 2019;200:124–144. doi: 10.1016/j.talanta.2019.03.015. [DOI] [PubMed] [Google Scholar]
- 38.Li F., Yu Z., Han X., Lai R.Y. Electrochemical aptamer-based sensors for food and water analysis: a review. Anal. Chim. Acta. 2019;1051:1–23. doi: 10.1016/j.aca.2018.10.058. [DOI] [PubMed] [Google Scholar]
- 39.Alkhamis O., Canoura J., Yu H., Liu Y., Xiao Y. Innovative engineering and sensing strategies for aptamer-based small-molecule detection. Trac. Trends Anal. Chem. 2019;121:115699. doi: 10.1016/j.trac.2019.115699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharifi S., Vahed S.Z., Ahmadian E., Dizaj S.M., Eftekhari A., Khalilov R., Ahmadi M., Hamidi-Asl E., Labib M. Detection of pathogenic bacteria via nanomaterials-modified aptasensors. Biosens. Bioelectron. 2020;150:111933. doi: 10.1016/j.bios.2019.111933. [DOI] [PubMed] [Google Scholar]
- 41.Kaur H., Shorie M. Nanomaterial based aptasensors for clinical and environmental diagnostic applications. Nanoscale Advances. 2019;1:2123–2138. doi: 10.1039/c9na00153k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chandra P., Tan Y.N., Singh S.P. vol. 10. Springer; 2017. (Next Generation Point-of-care Biomedical Sensors Technologies for Cancer Diagnosis). [Google Scholar]
- 43.Zhao V.X.T., Wong T.I., Zheng X.T., Tan Y.N., Zhou X. Colorimetric biosensors for point-of-care virus detections. Materials Science for Energy Technologies. 2020;3:237–249. doi: 10.1016/j.mset.2019.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan Y.N., Zheng X.T., Yu Y. Institution of Engineering and Technology; 2016. Nanobiosensors For Personalized And Onsite Biomedical Diagnosis; pp. 537–566. [Google Scholar]
- 45.Zhang Y., Hu J., Zhang C.-y. Sensitive detection of transcription factors by isothermal exponential amplification-based colorimetric assay. Anal. Chem. 2012;84:9544–9549. doi: 10.1021/ac3024087. [DOI] [PubMed] [Google Scholar]
- 46.Tan Y.N., Su X., Liu E.T., Thomsen J.S. Gold-nanoparticle-based assay for instantaneous detection of nuclear hormone Receptor−Response elements interactions. Anal. Chem. 2010;82:2759–2765. doi: 10.1021/ac9026498. [DOI] [PubMed] [Google Scholar]
- 47.Tan Y.N., Su X., Zhu Y., Lee J.Y. Sensing of transcription factor through controlled-assembly of metal nanoparticles modified with segmented DNA elements. ACS Nano. 2010;4:5101–5110. doi: 10.1021/nn100943d. [DOI] [PubMed] [Google Scholar]
- 48.Tan Y.N., Lee K.H., Su X. Study of single-stranded DNA binding protein–nucleic acids interactions using unmodified gold nanoparticles and its application for detection of single nucleotide polymorphisms. Anal. Chem. 2011;83:4251–4257. doi: 10.1021/ac200525a. [DOI] [PubMed] [Google Scholar]
- 49.Tan Y.N., Lee K.H., Su X. A study of DNA design dependency of segmented DNA-induced gold nanoparticle aggregation towards versatile bioassay development. RSC Adv. 2013;3:21604–21612. [Google Scholar]
- 50.Assah E., Goh W., Zheng X.T., Lim T.X., Li J., Lane D., Ghadessy F., Tan Y.N. Rapid colorimetric detection of p53 protein function using DNA-gold nanoconjugates with applications for drug discovery and cancer diagnostics. Colloids Surf. B Biointerfaces. 2018;169:214–221. doi: 10.1016/j.colsurfb.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 51.Tan Y.N., Lai A., Su X. Interrogating cooperative interactions of transcription factors with composite DNA elements using gold nanoparticles. Sci. Adv. Mater. 2014;6:1460–1466. [Google Scholar]
- 52.Aung K.M.M., Tan Y.N., Desai K.V., Su X. Interrogating oestrogen receptor–DNA interactions using metallic nanoparticles and surface plasmon resonance technique. Aust. J. Chem. 2011;64:1288–1294. [Google Scholar]
- 53.Zheng X.T., Goh W.L., Yeow P., Lane D.P., Ghadessy F.J., Tan Y.N. Ultrasensitive dynamic light scattering based nanobiosensor for rapid anticancer drug screening. Sensor. Actuator. B Chem. 2019;279:79–86. [Google Scholar]
- 54.Liu J., Lu Y. Fast colorimetric sensing of adenosine and cocaine based on a general sensor design involving aptamers and nanoparticles. Angew. Chem. Int. Ed. 2006;45:90–94. doi: 10.1002/anie.200502589. [DOI] [PubMed] [Google Scholar]
- 55.Pierre-Pierre N., Huo Q. Recent Progress in Colloid and Surface Chemistry with Biological Applications. American Chemical Society; 2015. Dynamic light scattering coupled with gold nanoparticle probes as a powerful sensing technique for chemical and biological target detection; pp. 157–179. [Google Scholar]
- 56.Zheng T., Bott S., Huo Q. Techniques for accurate sizing of gold nanoparticles using dynamic light scattering with particular application to chemical and biological sensing based on aggregate formation. ACS Appl. Mater. Interfaces. 2016;8:21585–21594. doi: 10.1021/acsami.6b06903. [DOI] [PubMed] [Google Scholar]
- 57.Seow N., Tan Y.N., Yung L.-Y.L., Su X. DNA-directed assembly of nanogold dimers: a unique dynamic light scattering sensing probe for transcription factor detection. Sci. Rep. 2015;5:18293. doi: 10.1038/srep18293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seow N., Tan Y.N., Yung L.-Y.L. Gold nanoparticle–dynamic light scattering tandem for the rapid and quantitative detection of the let7 MicroRNA family. Part. Part. Syst. Char. 2014;31:1260–1268. [Google Scholar]
- 59.Cialla D., März A., Böhme R., Theil F., Weber K., Schmitt M., Popp J. Surface-enhanced Raman spectroscopy (SERS): progress and trends. Anal. Bioanal. Chem. 2012;403:27–54. doi: 10.1007/s00216-011-5631-x. [DOI] [PubMed] [Google Scholar]
- 60.Tavakkoli Yaraki M., Daqiqeh Rezaei S., Tan Y.N. Simulation guided design of silver nanostructures for plasmon-enhanced fluorescence, singlet oxygen generation and SERS applications. Phys. Chem. Chem. Phys. 2020;22:5673–5687. doi: 10.1039/c9cp06029d. [DOI] [PubMed] [Google Scholar]
- 61.Tavakkoli Yaraki M., Daqiqeh Rezaei S., Middha E., Tan Y.N. Synthesis and simulation study of right silver bipyramids via seed-mediated growth cum selective oxidative etching approach. Part. Part. Syst. Char. 2020;37:2000027. [Google Scholar]
- 62.Bonham A.J., Braun G., Pavel I., Moskovits M., Reich N.O. Detection of sequence-specific protein-DNA interactions via surface enhanced resonance Raman scattering. J. Am. Chem. Soc. 2007;129:14572–14573. doi: 10.1021/ja0767837. [DOI] [PubMed] [Google Scholar]
- 63.Barhoumi A., Zhang D., Tam F., Halas N.J. Surface-enhanced Raman spectroscopy of DNA. J. Am. Chem. Soc. 2008;130:5523–5529. doi: 10.1021/ja800023j. [DOI] [PubMed] [Google Scholar]
- 64.Masetti M., Xie H.-n., Krpetić Ž., Recanatini M., Alvarez-Puebla R.A., Guerrini L. Revealing DNA interactions with exogenous agents by surface-enhanced Raman scattering. J. Am. Chem. Soc. 2015;137:469–476. doi: 10.1021/ja511398w. [DOI] [PubMed] [Google Scholar]
- 65.Yaseen Z., Banday A.R., Hussain M.A., Tabish M. Determination of the cationic amphiphilic drug–DNA binding mode and DNA-assisted fluorescence resonance energy transfer amplification. Spectrochim. Acta Mol. Biomol. Spectrosc. 2014;122:553–564. doi: 10.1016/j.saa.2013.11.030. [DOI] [PubMed] [Google Scholar]
- 66.Zhao D., Li J., Yang T., He Z. “Turn off–on” fluorescent sensor for platinum drugs-DNA interactions based on quantum dots. Biosens. Bioelectron. 2014;52:29–35. doi: 10.1016/j.bios.2013.08.031. [DOI] [PubMed] [Google Scholar]
- 67.Liu X., Ouyang L., Cai X., Huang Y., Feng X., Fan Q., Huang W. An ultrasensitive label-free biosensor for assaying of sequence-specific DNA-binding protein based on amplifying fluorescent conjugated polymer. Biosens. Bioelectron. 2013;41:218–224. doi: 10.1016/j.bios.2012.08.027. [DOI] [PubMed] [Google Scholar]
- 68.Xu H.V., Zheng X.T., Zhao Y., Tan Y.N. Uncovering the design principle of amino acid-derived photoluminescent biodots with tailor-made structure–properties and applications for cellular bioimaging. ACS Appl. Mater. Interfaces. 2018;10:19881–19888. doi: 10.1021/acsami.8b04864. [DOI] [PubMed] [Google Scholar]
- 69.Zheng X.T., Lai Y.C., Tan Y.N. Nucleotide-derived theranostic nanodots with intrinsic fluorescence and singlet oxygen generation for bioimaging and photodynamic therapy. Nanoscale Advances. 2019;1:2250–2257. doi: 10.1039/c9na00058e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng X.T., Tan Y.N. Development of blood-cell-selective fluorescent biodots for lysis-free leukocyte imaging and differential counting in whole blood. Small. 2019:1903328. doi: 10.1002/smll.201903328. 0. [DOI] [PubMed] [Google Scholar]
- 71.Choi Y., Zheng X.T., Tan Y.N. Bioinspired carbon dots (biodots): emerging fluorophores with tailored multiple functionalities for biomedical, agricultural and environmental applications. Molecular Systems Design & Engineering. 2020;5:67–90. [Google Scholar]
- 72.Zheng X.T., Choi Y., Phua D.G.G., Tan Y.N. Noncovalent fluorescent biodot–protein conjugates with well-preserved native functions for improved sweat glucose detection. Bioconjugate Chem. 2020;20:754–763. doi: 10.1021/acs.bioconjchem.9b00856. [DOI] [PubMed] [Google Scholar]
- 73.Yu Y., Luo Z., Teo C.S., Tan Y.N., Xie J. Tailoring the protein conformation to synthesize different-sized gold nanoclusters. Chem. Commun. 2013;49:9740–9742. doi: 10.1039/c3cc46005c. [DOI] [PubMed] [Google Scholar]
- 74.Yu Y., New S.Y., Xie J., Su X., Tan Y.N. Protein-based fluorescent metal nanoclusters for small molecular drug screening. Chem. Commun. 2014;50:13805–13808. doi: 10.1039/c4cc06914e. [DOI] [PubMed] [Google Scholar]
- 75.Yu Y., Mok B.Y.L., Loh X.J., Tan Y.N. Rational design of biomolecular templates for synthesizing multifunctional noble metal nanoclusters toward personalized theranostic applications. Advanced Healthcare Materials. 2016;5:1844–1859. doi: 10.1002/adhm.201600192. [DOI] [PubMed] [Google Scholar]
- 76.Yu Y., Zheng X.T., Yee B.W., Tan Y.N. Biomimicking synthesis of photoluminescent molecular lantern catalyzed by in-situ formation of nanogold catalysts. Mater. Sci. Eng. C. 2017;77:1111–1116. doi: 10.1016/j.msec.2017.04.029. [DOI] [PubMed] [Google Scholar]
- 77.Yu Y., Lee W.D., Tan Y.N. Protein-protected gold/silver alloy nanoclusters in metal-enhanced singlet oxygen generation and their correlation with photoluminescence. Mater. Sci. Eng. C. 2020;109:110525. doi: 10.1016/j.msec.2019.110525. [DOI] [PubMed] [Google Scholar]
- 78.Geng J., Goh W.L., Zhang C., Lane D.P., Liu B., Ghadessy F., Tan Y.N. A highly sensitive fluorescent light-up probe for real-time detection of the endogenous protein target and its antagonism in live cells. J. Mater. Chem. B. 2015;3:5933–5937. doi: 10.1039/c5tb00819k. [DOI] [PubMed] [Google Scholar]
- 79.Tavakkoli Yaraki M., Hu F., Daqiqeh Rezaei S., Liu B., Tan Y.N. Metal-enhancement study of dual functional photosensitizers with aggregation-induced emission and singlet oxygen generation. Nanoscale Advances. 2020;2:2859–2869. doi: 10.1039/d0na00182a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gao R., Zhong Z., Gao X., Jia L. Graphene oxide quantum dots assisted construction of fluorescent aptasensor for rapid detection of Pseudomonas aeruginosa in food samples. J. Agric. Food Chem. 2018;66:10898–10905. doi: 10.1021/acs.jafc.8b02164. [DOI] [PubMed] [Google Scholar]
- 81.Zhang J., Li Z., Zhao S., Lu Y. Size-dependent modulation of graphene oxide–aptamer interactions for an amplified fluorescence-based detection of aflatoxin B1 with a tunable dynamic range. Analyst. 2016;141:4029–4034. doi: 10.1039/c6an00368k. [DOI] [PubMed] [Google Scholar]
- 82.Liu Z., Liu B., Ding J., Liu J. Fluorescent sensors using DNA-functionalized graphene oxide. Anal. Bioanal. Chem. 2014;406:6885–6902. doi: 10.1007/s00216-014-7888-3. [DOI] [PubMed] [Google Scholar]
- 83.Rauf S., Gooding J., Akhtar K., Ghauri M., Rahman M., Anwar M., Khalid A. Electrochemical approach of anticancer drugs–DNA interaction. J. Pharmaceut. Biomed. Anal. 2005;37:205–217. doi: 10.1016/j.jpba.2004.10.037. [DOI] [PubMed] [Google Scholar]
- 84.Bonham A.J., Hsieh K., Ferguson B.S., Vallée-Bélisle A., Ricci F., Soh H.T., Plaxco K.W. Quantification of transcription factor binding in cell extracts using an electrochemical, structure-switching biosensor. J. Am. Chem. Soc. 2012;134:3346–3348. doi: 10.1021/ja2115663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang Y., Liu F., Nie J., Jiang F., Zhou C., Yang J., Fan J., Li J. An electrochemical sensing platform based on local repression of electrolyte diffusion for single-step, reagentless, sensitive detection of a sequence-specific DNA-binding protein. Analyst. 2014;139:2193–2198. doi: 10.1039/c4an00096j. [DOI] [PubMed] [Google Scholar]
- 86.Pan Q., Zhang R., Bai Y., He N., Lu Z. An electrochemical approach for detection of specific DNA-binding protein by gold nanoparticle-catalyzed silver enhancement. Anal. Biochem. 2008;375:179–186. doi: 10.1016/j.ab.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 87.Das R., Dhiman A., Kapil A., Bansal V., Sharma T.K. Aptamer-mediated colorimetric and electrochemical detection of Pseudomonas aeruginosa utilizing peroxidase-mimic activity of gold NanoZyme. Anal. Bioanal. Chem. 2019;411:1229–1238. doi: 10.1007/s00216-018-1555-z. [DOI] [PubMed] [Google Scholar]
- 88.Hernández R., Vallés C., Benito A.M., Maser W.K., Xavier Rius F., Riu J. Graphene-based potentiometric biosensor for the immediate detection of living bacteria. Biosens. Bioelectron. 2014;54:553–557. doi: 10.1016/j.bios.2013.11.053. [DOI] [PubMed] [Google Scholar]
- 89.Shahrokhian S., Ranjbar S. Development of a sensitive diagnostic device based on zeolitic imidazolate frameworks-8 using ferrocene–graphene oxide as electroactive indicator for Pseudomonas aeruginosa detection. ACS Sustain. Chem. Eng. 2019;7:12760–12769. [Google Scholar]
- 90.Song Y., Song J., Wei X., Huang M., Sun M., Zhu L., Lin B., Shen H., Zhu Z., Yang C. Discovery of aptamers targeting the receptor-binding domain of the SARS-CoV-2 spike glycoprotein. Anal. Chem. 2020;92:9895–9900. doi: 10.1021/acs.analchem.0c01394. [DOI] [PubMed] [Google Scholar]


