Abstract
Background
The development of the face occurs during the early days of intrauterine life by the formation of facial processes from the first Pharyngeal arch. Derangement in these well‐organized fusion events results in Orofacial clefts (OFC). Van der Woude syndrome (VWS) is one of the most common causes of syndromic cleft lip and/or palate accounting for 2% of all cases. Mutations in the IRF6 gene account for 70% of cases with the majority of these mutations located in the DNA‐binding (exon 3, 4) or protein‐binding domains (exon 7–9). The current study was designed to update the list of IRF6 variants reported for VWS by compiling all the published mutations from 2013 to date as well as including the previously unreported VWS cases from Africa and Puerto Rico.
Methods
We used PubMed with the search terms; "Van der Woude syndrome," “Popliteal pterygium syndrome,” "IRF6," and "Orofacial cleft" to identify eligible studies. We compiled the CADD score for all the mutations to determine the percentage of deleterious variants.
Results
Twenty‐one new mutations were identified from nine papers. The majority of these mutations were in exon 4. Mutations in exon 3 and 4 had CADD scores between 20 and 30 and mutations in exon 7–9 had CADD scores between 30 and 40. The presence of higher CADD scores in the protein‐binding domain (exon 7–9) further confirms the crucial role played by this domain in the function of IRF6. In the new cases, we identified five IRF6 mutations, three novel missense mutations (p.Phe36Tyr, p.Lys109Thr, and p.Gln438Leu), and two previously reported nonsense mutations (p.Ser424*and p.Arg250*).
Conclusion
Mutations in the protein and DNA‐binding domains of IRF6 ranked among the top 0.1% and 1% most deleterious genetic mutations, respectively. Overall, these findings expand the range of VWS mutations and are important for diagnostic and counseling purposes.
Keywords: Combined Annotation Dependent Depletion score, interferon regulatory factor 6, orofacial cleft, Popliteal pterygium syndrome, Van der Woude syndrome
The study reported novel variants in IRF6 from patients with VWS. Updated the list of all IRF6 variants reported from 2013 to date and provide an insight into on how to use the Combined Annotation Dependent Depletion score for the prioritization of IRF6 variants.
1. INTRODUCTION
Facial development occurs during the early days of intrauterine life by the formation of facial processes from the first pharyngeal arch and their subsequent fusion (Afshar, Brugmann, & Helms, 2012). Derangement in this process results in orofacial clefts. Orofacial clefts are a group of congenital defects that are present with the discontinuity of the affected part of the face, which can either occur in isolation (non‐syndromic), or together with other defects (syndromic), with a preponderance of the former (Gatta et al., 2004). Van der Woude syndrome (VWS) (OMIM: 119300 and 606713), an autosomal dominant disorder with variable expression and a high level of penetrance (Lam, David, Townsend, & Anderson, 2010), is the most common syndromic cleft accounting for 2% of all cases (Murray et al., 1997) It classically presents with lip pits and either cleft lip, cleft lip/palate, or cleft palate only. (Kondo et al., 2002; Malik, Wilcox, & Naz, 2013; Van Der Woude, 1954).
Mutations in the Interferon Regulatory Factor Six (IRF6) (OMIM: 607199) (Kondo et al., 2002; Ural, Bilgen, Çakmakli, & Bekerecioğlu, 2019), and Grainy Head Like Three (GRHL3) (OMIM: 608317) (Peyrard‐Janvid et al., 2014) genes have been shown to cause VWS and Popliteal pterygium syndrome (PPS) (OMIM: 119500). IRF6 is the only member of the IRF gene family involved in the craniofacial development (Starink et al., 2017). It is a protein‐coding gene with a highly conserved N terminal DNA‐binding domain and a less conserved C terminal protein‐binding domain (Ben, Jabs, & Chong, 2005; Kondo et al., 2002). Non‐ random mutations in the coding regions of IRF6 account for 70% of cases of VWS with the majority of these mutations located at the DNA‐binding domain (exon 3, 4) and the protein‐binding domain (exon 7–9) (Ferreira de Lima et al., 2009).
There are several bioinformatics tools used to predict the pathogenicity of genetic mutations (Adzhubei et al., 2010; Kumar, Henikoff, & Ng, 2009; Venselaar, Te Beek, Kuipers, Hekkelman, & Vriend, 2010) including SIFT, Polyphen and HOPE. However, the conflicting results from existing bioinformatics tools (Hicks, Wheeler, Plon, & Kimmel, 2011), the paucity of the functional data on reported variants, as well as the need for a more robust tool to accurately predict the clinical implications of these variants led to the development of the Combined Annotation Dependent Depletion score (CADD score, Kircher et al. 2014).
CADD is a comprehensive prediction tool developed by Kircher, Jain, O’Roak, Cooper, and Shendure (2014). It merges the algorithms of over 60 other previously developed tools to produce a single result. It thus compensates for the loopholes of the other individual tools and allowing for a more accurate prediction of the pathogenicity of these mutations. This is achieved via a scoring system linked to existing genomic databases using a score of 20 for the top 1% and 30 for the top 0.1% of the most deleterious genetic mutations in the human genome (Kircher et al., 2014).
The current study reviewed the literature to update the list of IRF6 variants reported for VWS, predicted their clinical significance using the CADD score, and reported novel IRF6 variants identified in Africans and Puerto Ricans with VWS.
2. METHODOLOGY
2.1. Literature search
We used PubMed with the search terms: "Van der Woude syndrome and Popliteal pterygium syndrome," "IRF6," and "Orofacial cleft" to identify studies that reported novel mutation(s) not found in the Leslie et al. (2013) study. We also compiled the CADD score for all the reported mutations using the CADD single nucleotide variant (SNV) look‐up feature (GRCh37‐v1.4 version).
2.2. New study population and sample information
2.2.1. Ethical compliance
The new study populations include Nigerians, Ghanaians, Ethiopians, and Puerto‐Ricans. Ethical approval for this project was received from the Institutional Review Boards (IRBs) at the University of Iowa (Iowa approvals numbers: 201101720), Lagos University Teaching Hospital Idi‐Araba, Lagos (IRB approval number: ADM/DCST/HREC/VOL.XV/321), Obafemi Awolowo University Teaching Hospital Ile‐Ife (IRB approval number: ERC/2011/12/01), Kwame Nkrumah University of Science and Technology (IRB approval number: CHRPE/RC/018/13), the Addis Ababa University (IRB approval number: 003/10/surg), the University of Puerto‐Rico (IRB approval number: 0640111), and Kwazulu‐Natal University (IRB approval number: BE309/18).
2.3. DNA sequencing
Primer sequences that were used to amplify exons 1 to 9 of IRF6 (RefSeq NM_006147.4) have been previously published (Ferreira Butali et al., 2014; de Lima et al., 2009) and available on request. All DNA processing protocols, PCR conditions, and electrophoretic procedures are available at the Murray laboratory website (http://genetics.uiowa.edu/protocols.php). The amplified DNA products were sent to Functional Biosciences in Madison, Wisconsin for sequencing using an ABI 3730XL DNA Sequencer. The sequence data and variants were viewed with CONSED. Each variant and genomic location that was revealed by CONSED were confirmed using the “Blat” function of THE UCSC Genome Browser. The functional consequences of variants were predicted using CADD, SIFT, and Polyphen‐2 to determine pathogenic and benign variants. HOPE was used to create a simulation of the mutant protein structure. The effect of a variant on mRNA splicing was ascertained using Human Splicing Finder 3.0 (http://www.umd.be/HSF3/). Furthermore, the effect of a variant on a regulatory region was predicted, using RegulomeDB (http://regulomedb.org/).
The Minor Allele Frequencies or the novelty of a variant was ascertained by comparing it to variants in the 1000 Genomes (http://browser.1000genomes.org/index.html), Exome Variant Server (http://evs.gs.washington.edu/EVS/), dbSNP (www.ncbi.nlm.nih.gov/SNP/) ExAC Browser (http://exac.broadinstitute.org/) and gnomAD (https://gnomad.broadinstitute.org/). If a variant has never been reported before in the literature or these databases, it was categorized as “novel.” When a variant of interest was found in a proband, the samples from their parents were sequenced. Identified variants were confirmed by sequencing in the reverse direction. The presence or absence of the mutation in the parent's DNA was used to determine if such variation was “de novo” or segregated in the family.
The data that support the findings of this study are openly available in [repository name e.g., “figshare”] at http://doi.org/[doi], reference number [reference number].
3. RESULTS
Following our literature search for variants reported after the Leslie et al. (2013) study, we identified 21 new mutations from nine papers and compiled a CADD score for them (Table 1, Table S1). The majority (69) of these mutations were in exon 4 with 194 mutations having CADD scores in the 20s and 30s (Table 1, Figure 1). There were 13 nonsense mutations with scores of 40+. A nonsense mutation in exon 8 was discovered in a patient with PPS and was predicted to be a dominant‐negative mutation with a CADD score of 54 (Table 1). These high CADD scores rank IRF6 mutations among the top 0.1 to 0.0001% most deleterious mutations in the human genome.
TABLE 1.
Showing the reported IRF6 mutations from 2013 till date
| Exon | cDNAPos | Mutation | Nucleotide change | Mutation type | CADD score | Population | References |
|---|---|---|---|---|---|---|---|
| 3 | 77 | p. Leu26Pro | T > C | missense | 32 | 1 | Sunny et al. (2019) |
| 7 | 720 | c.720del c | c.720del c | frameshift | 16.53 | 1 | Ural et al. (2019) |
| 4 | 294 | p. Asp98Glu | T > G | missense | 22.9 | 1 | Wang et al. (2019) |
| 8 | 1138 | p.Pro380Ser | C > T | missense | 29.6 | 1 | Wang et al. (2019) |
| chr1:209.872.038–210.246.107 | chr1:209.872.038–210.246.107 | Microdeletion | N/A | 1 | Mbuyi‐Musanzayi, (2019) | ||
| 4 | 254 | C85F | C > T | Missense | 25.8 | 1 | Leslie et al. (2016) |
| 7 | 1060+1 | Splice site | N/A | 1 | |||
| 4 | c.165delC | p.Ile56phefs*7 | Indels | N/A | 1 | ||
| 4 | c.379delG | p.Gly127valfs*43 | indels | N/A | 1 | ||
| 4 | 175‐2 | A > C | Splice site | 33 | 1 | Gowans, 2017 | |
| 4 | 194 | p.Gly65Val | G > T | Missense | 26.2 | 1 | |
| 4 | 205 | p.Glu69Lys | G > A | Missense | 32 | 2 | |
| 4 | 379+1 | G > T | Splice site | 34 | 1 | ||
| 6 | 554 | p.Asp185Thr | A > C | Missense | 21.7 | 1 | |
| 7 | 960 | p.Lys320Asp | G > C | Missense | 25.7 | 1 | |
| 3 | 113 | p.Iso38Thr | T > C | Missense | 27.8 | 1 | Tan et al, 2008 |
| 4 | 196 | p.Lys66X | A > T | Nonsense | 38 | 1 | Butali et al., 2014 |
| 4 | 551 | p.Pro126Pro | T > A | Splice site | N/A | 2 | |
| 7 | 690 | p.PheF230Leu | T > G | Missense | 23.8 | 5 | |
| 8 | 1139 | Pro380Gln | C > A | Missense | 28.5 | 1 | Khandelwal et al, (2017) |
| 5 | 673 | Asp225His | G > C | Missense | 32 | 1 | |
| 3 | 107 | p.Phe36Tyr | T > A | Missense | 29.3 | 1 | This study |
| 4 | 326 | p.Lys109Thr | A > C | Missense | 23.6 | 1 | This study |
| 9 | 1313 | p.Gln438Leu | A > T | Missense | 25.5 | 1 | This study |
[Correction added on 17 July 2020, after first online publication: in Table 1, the last 2 mutation details under ‘Mutation’ column have been changed from ‘p.Pro109Thr’ to ‘p.Lys109Thr’ and ‘p.Glu438Leu’ to ‘p.GLn109Thr’, respectively.]
FIGURE 1.
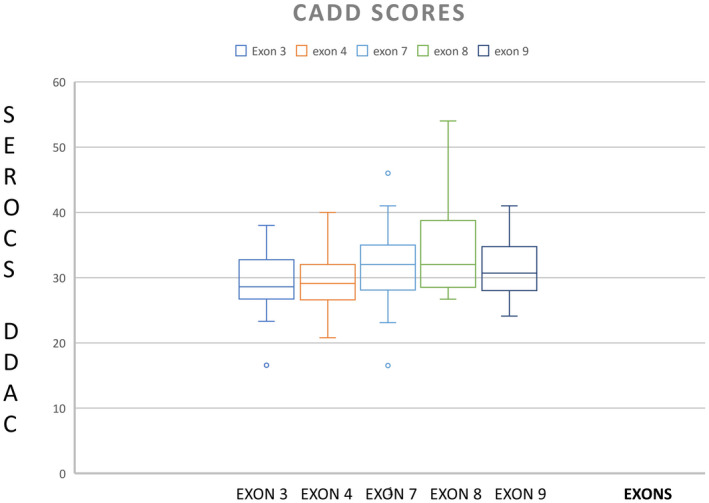
Box plot showing higher Combined Annotation Dependent Depletion score in the protein binding domain (exon 3 and 4) compared to the DNA binding domain (exon 7–9) of IRF6
In the current study, we identified five mutations, three novel missense mutations: p.Phe36Tyr, p.Lys109Thr, p.Gln438Leu, and two previously reported nonsense mutations: p.Ser424*and p.Arg250*. The three missense mutations were predicted to be probably damaging/ damaging by Polyphen‐2 and Sift prediction tools. The p.Phe36Tyr mutation located in exon 3 was identified in family A where both mother and child had shallow lip pits characteristic of VWS with an unaffected father (Figure 2). The p.Lys109Thr mutation located in exon 4 was identified in family B where samples for both parents were not available but were absent in the maternal grandmother. Thus, we were unable to determine the mode of inheritance. The proband with p.Gln438Leu mutation in exon 9, identified in family C, had incomplete cleft lip without lip pits and both parents appear clinically unaffected. The known nonsense mutations were in exon 9 and 7, respectively. The missense mutations (p.Phe36Tyr, p.Lys109Thr and p.Gln438Leu) had CADD scores 29, 23.6, and 25.5, while the nonsense mutations (p. Ser424* and p. Arg250*) had CADD scores of 40 and 43, respectively, as shown in (Table 2). The data that support the findings of this study are openly available in ClinVar at https://submit.ncbi.nlm.nih.gov/clinvar/, submission ID [SUB7353596].
FIGURE 2.
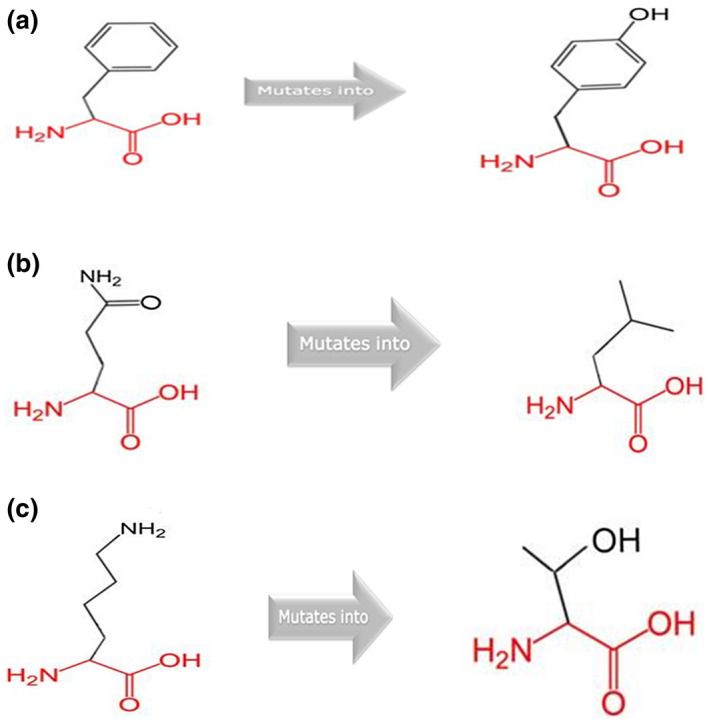
Showing the structural changes for the amino acid change p. Phe36Tyr (a), p. Gln438Leu (b) and p. Lys109thr (c) [Correction added on 22 July 2020, after first online publication: in Figure 2 caption, the order of the legends (b) and (c) has been interchanged so it reads ‘p. Gln438Leu (b) and p. Lys109thr (c)’.]
TABLE 2.
Showing the distribution of CADD score per exon
| CADD score | Exon 3 | Exon 4 | Exon 5 | Exon 6 | Exon 7 | EXON 8 | Exon 9 |
|---|---|---|---|---|---|---|---|
| 10–20 | 1 | 1 | |||||
| 20–30 | 23 | 43 | 1 | 4 | 17 | 7 | 10 |
| 30–40 | 20 | 24 | 1 | 4 | 23 | 7 | 7 |
| 40–50 | 2 | 1 | 2 | 6 | 3 | 3 | |
| >50 | 1 |
4. DISCUSSION
Non‐random mutations in the IRF6 highly conserved DNA‐binding domain (exon 3 and 4) and the less conserved protein‐binding domain (exon 7–9) have been implicated in the etiology of VWS and PPS (Ferreira de Lima et al., 2009). In this study, we found five IRF6 mutations after sequencing VWS families from Africa and Puerto Rico. These include p.Phe36Tyr, p.Lys109Thr, p.Ser424*, p.Arg250*, and p.Gln438Leu. The missense mutation (p.Phe36Tyr), identified in a Puerto Rican family segregates in the family (proband and mother) and confirms a maternal mode of inheritance. Based on the HOPE analyses, we observed that the mutant is in a domain that is important for the binding of other protein molecules. The mutant is large and will not fit into the core of the IRF6 protein where the wild type resides. Also, the wild‐type residue is more hydrophobic and thus the mutant might disturb the interaction between domains and thereby affect the function of the protein. The p.Lys109thr mutant is smaller compared to the wild type leading to an empty space in the core of the protein. Furthermore, the wild type is more hydrophobic and positively charge allowing it to form a salt bridge with aspartic acid at position 98, which is required for proper protein folding. Replacing it with a neutral and less hydrophobic mutant residue will distort the protein folding and also affect its ability to interact with other molecules—an important function of the domain. The mutant residue for the p.Gln438Leu is smaller and less hydrophobic, thus, affecting the interaction with other residues within the domain as well as the binding capacity of the domain. Unlike the previous two mutations where the wild type residue is highly conserved at their respective positions, the wild type is not conserved at this amino acid position (438). However, the mutant residue was also not among the residue types that have been observed at this position. Thus, why it was predicted as damaging to the protein by Sift and Polyphen‐2 would require further study, perhaps using zebrafish or a cell‐based assay as previously reported for other variants (Li et al., 2017).
When we reviewed the family data for the proband with the p.Phe36Tyr mutation, we observed that VWS was present in the mother. A family history of different types of clefts, the presence of lower lip pits, and types of cancer such as breast and colon cancer were found on the maternal side of the family. IRF6 has shown tumor‐suppressor activities in different cancer types (Botti et al., 2011) suggesting a possible connection. Other families did not provide access to the parental data so we were unable to ascertain the pattern of inheritance or the possibility of a de novo mutation.
A recent study functionally tested some IRF6 variants in zebrafish embryo and did not find any correlation between the Polyphen‐2 and SIFT prediction and functional consequences in zebrafish embryos (Li et al., 2017). However, when we reviewed the CADD score for the variants with functional consequences in zebrafish from the Li et al. (2017) study, we observed that over 95% of the variants had CADD scores >25 . This observation further buttresses the relationship between high CADD score and the likelihood of pathogenicity (van der Velde et al., 2017) . However, caution should be exercised so as not to use CADD score alone to assign pathogenicity and efforts should be made to follow the guidelines set by the American College of Medical Genetics and Genomics (ACMGG) for assigning pathogenicity to sequence variants (Richards et al., 2015). The previous work by De Lima et al. (2009) and Leslie et al. (2013) showed the distribution of mutations in IRF6 to be non‐random, segregating primarily in the DNA‐binding (exon 3 and 4), and protein‐binding domains (exon 7–9). Since most of these mutations were in exon 4, thus they proposed a major function for the DNA‐binding domain in the activities of IRF6. However, when 30 missense mutations (16 in the DNA‐binding domain and 14 in the protein‐binding domain) of IRF6 were isolated in patients with VWS and PPS were functionally tested in zebrafish embryos, 50% (15) were shown to be pathogenic. Most (11) of the pathogenic variants in zebrafish were in the protein‐binding domain (exons 7–9). Furthermore, we observed a higher rate of nonsense mutations with very high CADD scores in the protein‐coding domain which are pathogenic. Finally, we also observed that the mutation that resulted in G/C in the DNA‐binding domain and A/T in the protein‐binding domain tends to be more common among the variants that induced rupture (pathogenic) in the Li et al. (2017) study.
5. CONCLUSION
In conclusion, findings from the current study provided additional information on VWS mutations which is important for diagnostic and counseling purposes especially for families with a previous history.
CONFLICT OF INTERESTS
None declared.
AUTHOR CONTRIBUTIONS
Alade A, Carmen B, Ada M, and Carolina B wrote the initial draft, Alade, A, Tamara B, Ada M, and Carolina B performed the sequencing and analysis of the results. Peter D, Fareed A, Alexander, and Lord JJ coordinated patients’ recruitment with sample collection in Ghana and also contributed to the revised manuscript. James O, Modupe O, and Wasiu A coordinated patient recruitments with sample collection in Nigeria and also contributed to the revised manuscript. Thirona N, Collen A coordinated the sample collection in South Africa and contributed to the revised manuscript. Mairim S, Marilyn S, Ricardo L, Myrellis M, Jose F, Lydia M, Maria S, and Natalio D coordinated patient recruitment with sample collection in peurto Rico and also contributed to the revised manuscript. Peter M, Solomon O, oluwole A, John P, Waheed A, Chinyere A, Mary L, Joy O, Oluwole A, Olubukola O, Sara M, Martin D, Adebowale A, Mary M, Jeff M, Mekonem E, Aline P, fekir A, Taye H, Ibrahim M, Paul G, Millard D, and Mualalem G. All contributed to the revised manuscript. Azeez B supervised the analyses and draft of the manuscript.
Supporting information
Table S1
ACKNOWLEDGMENT
We are grateful to the families who voluntarily participated in this study in Puerto Rico, Nigeria, and Ghana. We are also grateful to all the administrative and research staff, students, Nurses, and Resident doctors who assisted with participant recruitment, consenting, and data collection. This research is supported by the National Institute of Health/National Institute of Dental and Craniofacial Research (R00 DE022378; A.B., R00 DE024571; C.B., R01 DE28300), National Institute on Minority Health and Health Disparities (S21 MD001830; C.B., U54 MD007587; C.B.).
Alade A, Buxo‐Martinez CJ, Mossey PA, et al. Non‐random distribution of deleterious mutations in the DNA and protein‐binding domains of IRF6 are associated with Van Der Woude syndrome. Mol Genet Genomic Med. 2020;8:e1355 10.1002/mgg3.1355
REFERENCES
- Adzhubei, I. A. , Schmidt, S. , Peshkin, L. , Ramensky, V. E. , Gerasimova, A. , Bork, P. , … Sunyaev, S. R. (2010). A method and server for predicting damaging missense mutations. Nature Methods, 7, 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben, J. , Jabs, E. W. , & Chong, S. S. (2005). Genomic, cDNA and embryonic expression analysis of zebrafish IRF6, the gene mutated in the human oral clefting disorders Van der Woude and popliteal pterygium syndromes. Gene Expression Patterns, 5(5), 629–638. 10.1016/j.modgep.2005.03.002 [DOI] [PubMed] [Google Scholar]
- Afshar, M. , Brugmann, S. A. , & Helms, J. A. (2012). Embryology of the craniofacial complex In Neligan P. (Ed.), Plastic surgery (pp. 503–516). Seattle, WA: Elsevier. [Google Scholar]
- Botti, E. , Spallone, G. , Moretti, F. , Marinari, B. , Pinetti, V. , Galanti, S. , … Costanzo, A. (2011). Developmental factor IRF6 exhibits tumor suppressor activity in squamous cell carcinomas. Proceedings of the National Academy of Sciences of the United States of America, 108, 13710–13715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butali, A. , Mossey, P. A. , Adeyemo, W. L. , Eshete, M. A. , Gaines, L. A. , Even, D. , … Murray, J. C. (2014). Novel IRF6 mutations in families with Van Der Woude syndrome and popliteal pterygium syndrome from sub‐Saharan Africa. Molecular Genetics & Genomic Medicine, 2, 254–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lima, R. L. , Hoper, S. A. , Ghassibe, M. , Cooper, M. E. , Rorick, N. K. , Kondo, S. , … Schutte, B. C. (2009). Prevalence and non‐random distribution of exonic mutations in Interferon Regulatory Factor 6 (IRF6) in 307 families with Van der Woude syndrome and 37 families with popliteal pterygium syndrome. Genetics in Medicine, 11, 241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatta, V. , Scarciolla, O. , Cupaioli, M. , Palka, C. , Chiesa, P. L. , & Stuppia, L. (2004). A novel mutation of the IRF6 gene in an Italian family with Van der Woude syndrome. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis, 547, 49–53. [DOI] [PubMed] [Google Scholar]
- Gowans, L. J. , Busch, T. D. , Mossey, P. A. , Eshete, M. A. , Adeyemo, W. L. , Aregbesola, B. … Butali, A. (2017). The prevalence, penetrance, and expressivity of etiologic IRF6 variants in orofacial clefts patients from sub‐Saharan Africa. Molecular Genetics & Genomic Medicine, 5(2), 164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks, S. , Wheeler, D. A. , Plon, S. E. , & Kimmel, M. (2011). Prediction of missense mutation functionality depends on both the algorithm and sequence alignment employed. Human Mutation, 32, 661–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandelwal, K. D. , Ishorst, N. , Zhou, H. , Ludwig, K. U. , Venselaar, H. , Gilissen, C. … Carels, C. E. (2017). Novel IRF6 mutations detected in orofacial cleft patients by targeted massively parallel sequencing. Journal of Dental Research, 96(2), 179–185. [DOI] [PubMed] [Google Scholar]
- Kircher, W. D. M. , Jain, P. , O’Roak, B. J. , Cooper, G. M. , & Shendure, J. (2014). A general framework for estimating the relative pathogenicity of human genetic variants. Nature Genetics, 46, 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher, M. , Witten, D. M. , Jain, P. , O'Roak, B. J. , Cooper, G. M. , & Shendure, J. (2014). A general framework for estimating the relative pathogenicity of human genetic variants. Nature Genetics, 6(3), 310–315. 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo, S. , Schutte, B. C. , Richardson, R. J. , Bjork, B. C. , Knight, A. S. , Watanabe, Y. , … Murray, J. C. (2002). Mutations in IRF 6 cause Van der Woude and popliteal pterygium syndromes. Nature Genetics, 32(2), 285–289. 10.1038/ng985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, P. , Henikoff, S. , & Ng, P. C. (2009). Predicting the effects of coding non‐synonymous variants on protein function using the SIFT algorithm. Nature Protocols, 4, 1073–1081. [DOI] [PubMed] [Google Scholar]
- Lam, A. K. , David, D. J. , Townsend, G. C. , & Anderson, P. J. (2010). Van der Woude syndrome: Dentofacial features and implications for clinical practice. Australian Dental Journal, 55, 51–58. [DOI] [PubMed] [Google Scholar]
- Leslie, E. J. , Liu, H. , Carlson, J. C. , Shaffer, J. R. , Feingold, E. , Wehby, G. , … Marazita, M. L. (2016). A Genome‐wide Association Study of Nonsyndromic Cleft Palate Identifies an Etiologic Missense Variant in GRHL3. American Journal of Human Genetics, 98, 744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie, E. J. , Standley, J. , Compton, J. , Bale, S. , Schutte, B. C. , & Murray, J. C. (2013). Comparative analysis of IRF6 mutationsin families with Van der Woude syndrome and popliteal pterygium syndrome using public whole‐exome databases. Genetics in Medicine, 15, 338–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, E. B. , Truong, D. , Hallett, S. A. , Mukherjee, K. , Schutte, B. C. , & Liao, E. C. (2017). Rapid functional analysis of computationally complex rare human IRF6 gene variants using a novel zebrafish model. PLoS Genetics, 13, e1007009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, S. , Wilcox, E. R. , & Naz, S. (2013). Novel lip pit phenotypes and mutations of IRF6 in Van der Woude syndrome patients from Pakistan. Clinical Genetics, 85, 487–491. [DOI] [PubMed] [Google Scholar]
- Mbuyi‐Musanzayi, S. , Kasamba, E. I. , Revencu, N. , Lukusa, P. T. , Kalenga, P. M. , Tshilombo, F. K. , … Devriendt, K. (2020). Microdeletion of the entire IRF6 gene in a Subsaharian African's family with Van Der Woude syndrome. Clinical Dysmorphology, 29(1), 24–27. [DOI] [PubMed] [Google Scholar]
- Murray, J. C. , Daack‐Hirsch, S. , Buetow, K. H. , Munger, R. , Espina, L. , Paglinawan, N. , … Magee, W. (1997). Clinical and epidemiologic studies of cleft lip and palate in the Philippines. Cleft Palate‐Craniofacial Journal, 34, 7–10. [DOI] [PubMed] [Google Scholar]
- Peyrard‐Janvid, M. , Leslie, E. J. , Kousa, Y. A. , Smith, T. L. , Dunnwald, M. , Magnusson, M. et al (2014). Dominant mutations in GRHL3 cause Van der Woude Syndrome and disrupt oral periderm development. American Journal of Human Genetics, 94, 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, S. , Aziz, N. , Bale, S. , Bick, D. , Das, S. , Gastier‐Foster, J. , … Rehm, H. L. (2015). Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in Medicine, 17, 405–423. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starink, E. , Hokken‐Koelega, A. C. S. , Visser, T. J. , Baan, J. , Peeters, R. P. , & de Graaff, L. C. G. (2017). Genetic analysis of IRF6, a gene involved in craniofacial midline formation, in relation to pituitary and facial morphology of patients with idiopathic growth hormone deficiency. Pituitary, 20, 499–508. 10.1007/s11102-017-0808-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunny, A. P. , Arunachal, G. , & Danda, S. (2019). Van Der Woude syndrome: IRF6 mutations. Indian Journal of Pediatrics, 86(11), 1070–1071. [DOI] [PubMed] [Google Scholar]
- Tan, E. C. , Lim, E. C. , Yap, S. H. , Lee, S. T. , Cheng, J. , Por, Y. C. , & Yeow, V. (2008). Identification of IRF6 gene variants in three families with Van der Woude syndrome. International Journal of Molecular Medicine, 21(6), 747–751. [PubMed] [Google Scholar]
- Ural, A. , Bilgen, F. , Çakmakli, S. , & Bekerecioğlu, M. (2019). Van der Woude syndrome with a novel mutation in the IRF6 Gene. Journal of Craniofacial Surgery, 30(5), e465–e467. 10.1097/SCS.0000000000005552 [DOI] [PubMed] [Google Scholar]
- van der Velde, K. J. , de Boer, E. N. , van Diemen, C. C. , Sikkema‐Raddatz, B. , Abbott, K. M. , Knopperts, A. , … Swertz, M. A. (2017). GAVIN: Gene‐Aware Variant INterpretation for medical sequencing. Genome Biology, 18, 6 10.1186/s13059-016-1141-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Woude, A. (1954). Fistula labii inferioris congenita and its association with cleft lip and palate. American Journal of Human Genetics, 2, 244–256. [PMC free article] [PubMed] [Google Scholar]
- Venselaar, H. , Te Beek, T. A. , Kuipers, R. K. , Hekkelman, M. L. , & Vriend, G. (2010). Protein structure analysis of mutations causing inheritable diseases. An e‐Science approach with life scientist friendly interfaces. BMC Bioinformatics, 11, 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, T.‐J. , Hsieh, K.‐S. , Lai, J.‐P. , Tsai, M.‐H. , Liang, Y.‐C. , & Chang, Y.‐H. (2019). Novel mutations of IRF6 gene in Taiwanese Van Der Woude syndrome patients. Pediatr Neonatol., 60(2), 218–220. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1


